Abstract
Human AlkB homolog 6, ALKBH6, plays key roles in nucleic acid damage repair and tumor therapy. However, no precise structural and functional information are available for this protein. In this study, we determined atomic resolution crystal structures of human holo-ALKBH6 and its complex with ligands. AlkB members bind nucleic acids by NRLs (nucleotide recognition lids, also called Flips), which can recognize DNA/RNA and flip methylated lesions. We found that ALKBH6 has unusual Flip1 and Flip2 domains, distinct from other AlkB family members both in sequence and conformation. Moreover, we show that its unique Flip3 domain has multiple unreported functions, such as discriminating against double-stranded nucleic acids, blocking the active center, binding other proteins, and in suppressing tumor growth. Structural analyses and substrate screening reveal how ALKBH6 discriminates between different types of nucleic acids and may also function as a nucleic acid demethylase. Structure-based interacting partner screening not only uncovered an unidentified interaction of transcription repressor ZMYND11 and ALKBH6 in tumor suppression but also revealed cross talk between histone modification and nucleic acid modification in epigenetic regulation. Taken together, these results shed light on the molecular mechanism underlying ALKBH6-associated nucleic acid damage repair and tumor therapy.
Keywords: ALKBH6, AlkB homolog, crystal structure, protein–nucleic acid interaction, protein–protein interaction
Abbreviations: α-KG, α-ketoglutarate; ALKBH, AlkB homolog; DSBH, double-stranded β-helix; Flips (Flip1, Flip2 and Flip3), the components of nucleotide recognition lids which can recognize nucleic acids and flip the methylated lesion; MST, microscale thermophoresis; NRLs, nucleotide recognition lids; PHD, plant homeodomain; RRM, RNA recognition motif; TEV, tobacco etch virus
As α-ketoglutarate (α-KG) and Fe(II)-dependent dioxygenases, AlkB family members are widespread in the biological kingdom and play important roles in epigenetics. The AlkB gene was found in Escherichia coli, and its product was involved in repairing alkylated bases (1). Human genome encodes nine AlkB homologs named ALKBH1–8 and FTO (fat mass and obesity-associated) with low sequence identity (2, 3). All the nine AlkB homologs have different substrates and functions including DNA repair, RNA modification, actomyosin demethylation, and fatty acid metabolism (4, 5, 6). Most of them have essential roles in metabolism and diseases curbing such as cancers (7, 8, 9). Currently, more than 20 kinds of nucleic acids methylated adducts have been reported as AlkB members substrates (10). The AlkB family members have broad substrates selectivity: ALKBH1 can demethylate DNA 6mA modification enriched in human glioblastoma (7); it also can demethylate 3mC, 1mA, et al. (11, 12). ALKBH2 demethylates 1mA and 3mC both in ssDNA and dsDNA; ALKBH3 preferentially demethylates 1mA and 3mC in ssDNA (13). ALKBH4, without any structural information, can demethylate 6mA on dsDNA and might also on actin K84me1 (6). ALKBH5 and ALKBH8 can demethylate m6A RNA or mcm5U RNA, respectively (10). ALKBH7 is recently reported to be able to demethylate m22G and m1A of mt-RNA (14). FTO prefers to catalyze RNA substrates; it can suppress colorectal cancer with a better prognosis in an m6A-dependent manner, which highlights its clinical functions (9). Besides, we previously solved structures of human ALKBH5, ALKBH7, and ALKBH1 (15, 16, 17), providing precise structural information for function research.
Being a member of the AlkB family, ALKBH6 located in nuclei and cytoplasm is widely distributed with the highest expression in testis and pancreas (2, 18). Chromosomal gain of ALKBH6 is frequently detected in embryonal rhabdomyosarcoma but not in alveolar rhabdomyosarcoma, suggesting that ALKBH6 may play important roles in embryonal rhabdomyosarcoma (19). Besides, mice immunized with a mutant peptide of ALKBH6 can suppress tumor growth significantly, whereas the wildtype cannot (20). ALKBH6 has been identified as a “cancer gene” in endometrial tumors (21). Recently, researchers showed that ALKBH6 can complement AlkB-deficient E. coli strain resistance against alkylating agent methyl methane sulfonate (22). During exposure to the alkylating agent, ALKBH6 plays a crucial role in protecting pancreatic cancer from alkylating-induced DNA damage and promoting cell survival. Furthermore, the overexpression of ALKBH6 in patients with pancreatic cancer has been linked to better survival outcomes (22). The above information indicates that ALKBH6 may play key roles in DNA damage repair and tumor therapy. Huong et al. (23) reported that Arabidopsis ALKBH6, with 36% identity to human ALKBH6, could affect seed germination and survival under abiotic stress serving as a potential eraser protein of RNA methylation. Besides, when rice faced drought, cold, or ABA treatment, the expression level of ALKBH6 was decreased. These data suggest that plant ALKBH6 might be instrumental in abiotic stress responses (24). However, the structure, interacting partner, substrates, and activity of human ALKBH6 remain fantasy to date. Moreover, ALKBH6 has a low sequence identity (less than 18.5%) to all the released structures, hindering further research of its function.
In this article, we present two high-resolution structures of human ALKBH6, in complex with Tris or cofactor α-KG. These structures reveal a conventional β-strand jelly-roll fold with multiple unique features. With multiple kinds of nucleic acids screened, we demonstrated that ALKBH6 could bind nucleic acids and it preferred single-stranded nucleic acids. Structure-based interacting partner screening found that ALKBH6 could physically interact with both itself and ZMYND11, which offered us novel clues for functional research.
Results
Structure determination of ALKBH6
The human ALKBH6 protein expressed from E. coli failed to form crystal after extensive screening both fragments and crystallization conditions. By using the Pichia pastoris X-33 expression system, we successfully got the full-length ALKBH6 protein and performed crystal screening. Finally, high-resolution crystals of ALKBH6 in complex with Tris and cofactor α-KG were obtained after optimization. However, owing to the low sequence homology (less than 18.5%) to all structures in the Protein Data Bank (PDB), all molecular replacements failed when using single or multiple homologous structures as models. Then we tried different truncations of the predicted structure by Raptorx (25) as molecular replacement (MR) models, such as removing disordered loops or short β-sheets. After substantial trials of different resolution and space groups, the best solution was found by BALBES (26). After numerous cycles of manual rebuilding and refining, the final structure of the ALKBH6∙Ni2+∙Tris was refined to a resolution of 1.79 Å in the space group P212121, with an Rwork of 19.8% and an Rfree of 21.8% (Table 1). The structure of ALKBH6 complexed with cofactor α-KG and Mn2+ (holo-ALKBH6) was determined by molecular replacement using the ALKBH6∙Ni2+∙Tris structure as a model and was refined to a final Rwork of 17.2% and Rfree of 19.7% at 1.75 Å (Table 1). Each cell contains one ALKBH6 molecule (Fig. S1A). An overlay of holo-ALKBH6 and ALKBH6∙Ni2+∙Tris structures revealed high similarity in the overall conformation with a root-mean-square deviation (r.m.s.d.) of 0.2 Å (Fig. S1A).
Table 1.
Data collection and refinement statistics of ALKBH6 complexes
| Data | ALKBH6∙Ni2+∙Tris | ALKBH6∙Mn2+∙α-KG |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a,b,c (Å) | 46.4, 64.2, 89.0 | 46.1, 64.4, 88.4 |
| α,β,γ (˚) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 50.0–1.79 (1.84–1.79) | 50.0–1.75 (1.78–1.75) |
| Rmerge | 9.0% (79.0%) | 9.9% (46.9%) |
| I/σ | 31.5 (2.4) | 15.0 (2.1) |
| Completeness (%) | 99.8 (87.1) | 99.4(95.1) |
| Total no. of reflections | 180,599 | 122,192 |
| Unique reflections | 25,643 | 27,487 |
| Redundancy | 7.1 (3.3) | 4.5 (3.5) |
| Refinement | ||
| Resolution (Å) | 50.0–1.79 (1.84–1.79) | 50.0–1.75 (1.80–1.75) |
| No. of reflections | 24,111 | 25,773 |
| Rwork/Rfree (%) | 19.8/21.8 | 17.2/19.7 |
| No. of atoms | ||
| Protein | 1569 | 1706 |
| Ligand/ions | 9 | 11 |
| Water | 89 | 86 |
| Average B-factors (Åb) | ||
| Protein | 24.6 | 22.2 |
| Ligand/ion | 25.3 | 16.9 |
| Water | 29.1 | 23.6 |
| rms deviations | ||
| Bond lengths (Å) | 0.008 | 0.009 |
| Bond angles (º) | 1.467 | 1.495 |
| Ramachandran plot (%)b | 97.5/2.5/0 | 99.5/0.5/0 |
| Molprobity score | ||
| Overall | 1.26 (98th percentile) | 1.01 (100th percentile) |
| Clashcore | 1.91 (100th percentile) | 2.07 (99th percentile) |
Statistics for highest resolution shell.
Residues in favored, allowed, and outlier regions of the Ramachandran plot, respectively.
Structural characterization of ALKBH6
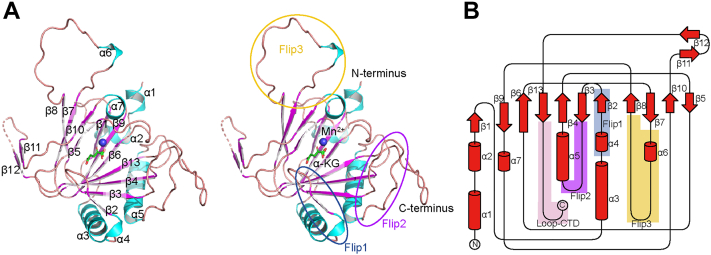
The overall structure of ALKBH6 shows a double-stranded β-helix (DSBH) fold at the catalytic core, which is conserved in the Fe(II)/α-KG-dependent dioxygenase superfamily (27). The overall similarity is evident in the central DSBH core. DALI search (http://www2.ebi.ac.uk/dali) reveals that S. Pombe OFD2 (PDB: 5YL6) is the most similar structural homolog of the ALKBH6 in the PDB database, with the Z-score of 19.2 and an r.m.s.d. of 2.4 Å over 164 Cα atoms. The ALKBH6 structure also has values comparable with other solved human AlkB family members, such as ALKBH8 (the Z-score of 19.0 and r.m.s.d. of 2.2 Å), ALKBH5 (the Z-score of 18.5 and r.m.s.d. of 2.5 Å), and ALKBH3 (the Z-score of 18.4 and r.m.s.d. of 2.1 Å). Significant differences occur in the secondary structures out of the DSBH fold, such as the protein-interacting regions and the substrate-binding regions near the active sites (Fig. S1B). In ALKBH6, 11 antiparallel β-strands constitute the DSBH domain, which is surrounded by seven α-helices. The DSBH contains a major sheet formed by seven β-strands (β1, β2, β3, β4, β6, β9, and β13), a minor sheet formed by four β-strands (β5, β7, β8, and β10), and sandwiches Mn2+ and α-KG at the active center (Fig. 1A). Two large α-helices (α3 and α5) and three small α-helices (α1, α2, and α4) buttress the major sheet. Consistent with other AlkB family members, ALKBH6 also has Flip1 (residues from A45 to R56) and Flip2 (residues from G62 to P76) (Fig. 1). Interesting, a unique loop (residues from E139 to P158) including a 310 helix (α6) inserts between β7 and β8 in the ALKBH6∙Mn2+∙α-KG structure, nearly overhanging vertical to DSBH, hereafter named Flip3 (Fig. 1). However, in the ALKBH6∙Ni2+∙Tris structure, most of the Flip3 (residues from P140 to P155) had no visible electron density and, therefore, could not be built (Fig. S1A). The reason may be that the cofactor α-KG from the symmetric unit induces the stabilization of the Flip3 conformation in the crystal packing. Meanwhile, residues from A195 to G210 between short β11 and β12 had no visible electron density in either ALKBH6∙Ni2+∙Tris or ALKBH6∙Mn2+∙α-KG structures and thus could not be built. At the same time, these two short β-sheets (β11 and β12) were far away from the potential active sites with high temperature factors (Fig. S1C) and they had an almost 90-degree bend conformation. These two β-sheets and the disorder region between them might mediate regulatory interactions with the DSBH minor sheet as in the ALKBH8.
Figure 1.
Overall structure of human ALKBH6 complexes.A, overall structure of human ALKBH6∙Mn2+∙α-KG in stereo mode. Mn2+ ion is shown as a blue sphere, α-KG and key secondary structures are labeled respectively. α-Helices, cyan; β-sheets, magenta; loops, salmon. Flip1, Flip2, and Flip3 are circled in dark blue, purple, and orange, respectively. B, topology of ALKBH6. Flip1, dark blue; Flip2, purple; Flip3, orange; Loop-CTD, pink.
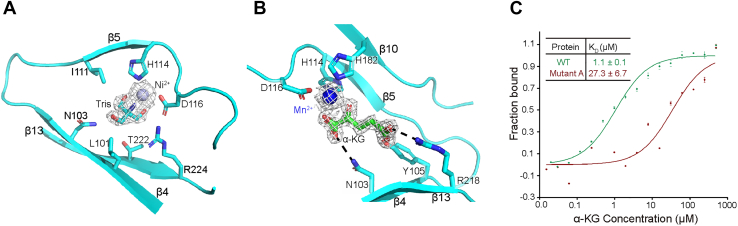
In a previous study (27), the H × D…H motif is conserved in the active center of the Fe(II)/α-KG-dependent dioxygenase superfamily. Here in ALKBH6, H114, D116, and H182 compose the motif of H × D…H and coordinate the metal ion stably. In the active center of the apo-ALKBH6 (ALKBH6∙Ni2+∙Tris) structure, an electron density of Tris, from the buffer, was found unexpectedly, which had not been reported in other AlkB family members (Fig. 2A). Surprising, Tris tridentately chelates Ni2+, which forms hexadentate coordination and forms two hydrogen bonds with N103 and D116 of ALKBH6. In the ALKBH6∙Mn2+∙α-KG complex, cofactor α-KG coordinates the Mn2+ bidentately in a standard conformation resembling that seen in other iron(II)/α-KG-based oxygenases and binds to the side chains of N103, Y105, R218, and S220 from β4 or β13 by hydrogen bonds or salt bridges (Fig. 2B). Thus, there are fewer interactions between Tris and ALKBH6 compared with those in the ALKBH6∙Mn2+∙α-KG complex. Besides, the key residues in the above interaction network have no evident conformational difference between the ALKBH6∙Ni2+∙Tris and ALKBH6∙Mn2+∙α-KG structures. Consistently, the defective mutant of three residues (mutant A, N103A/Y105A/R218A) can sharply reduce the binding of α-KG to ALKBH6 (Fig. 2C).
Figure 2.
ALKBH6 has an unusual active center.A, the interaction network around Tris and Ni2+ in the ALKBH6∙Ni2+∙Tris structure, Fo-Fc (3.0 σ, gray mesh). B, the interaction network around α-KG (Fo-Fc, 3.0 σ, gray mesh) and distribution of binding sites. C, binding analysis of ALKBH6 or mutant A with α-KG (mutant A: N103A/Y105A/R218A). The experiments were repeated three times.
The distinct structural features of ALKBH6 Flip1 and Flip2
Although ALKBH4 was reported to have protein substrates (6), all structure-solved human AlkB family members had nucleic acid oxygenase activity. ALKBH7 was recently reported to be able to demethylate m22G and m1A of mt-RNA (14). However, the substrate(s) of ALKBH6 remains a mystery for decades. Structural analyses of secondary structures out of the DSBH domain might give some clues.
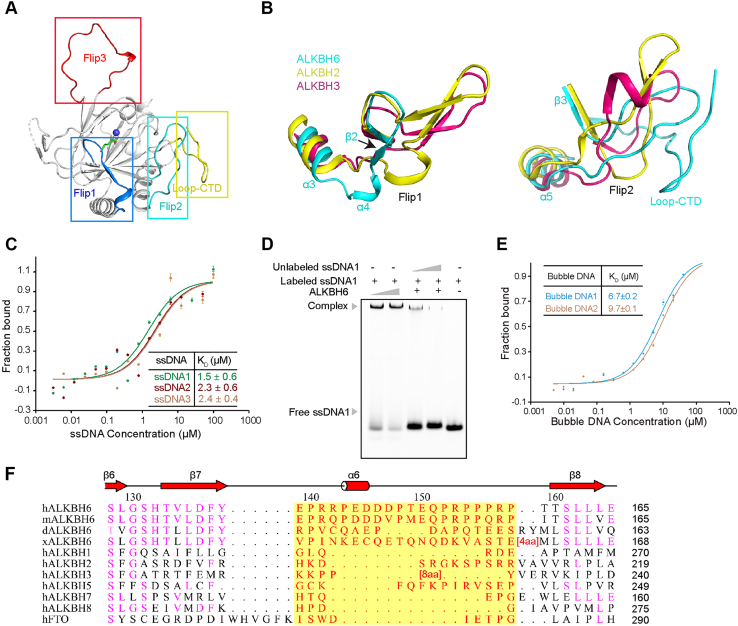
To characterize the difference between ALKBH6 and other AlkB proteins, we overlayed their structures. AlkB family proteins bind and immobilize substrates by nucleotide recognition lids (NRLs, also called Flips), containing several key loops around the catalytic domain (28). The NRLs contribute to substrates’ selectivity (28, 29) and are less conserved among AlkB members. The Flip1 and Flip2 of ALKBH6 (Fig. 3A) form the NRL domain and are notably different from those of other human AlkB family proteins (Figs. 3B and S1B).
Figure 3.
Key Flips have impacts on the binding of ALKBH6 to ssDNA.A, key loops of ALKBH6. B, overlay analysis of ALKBH6, ALKBH2, and ALKBH3 around the Flip domains; secondary structures are labeled based on ALKBH6 (ALKBH6, cyan; ALKBH2, yellow, PDB code 3S57; ALKBH3, warm pink, PDB code 2IUW). C, the binding of His-tag labeled ALKBH6 with different unlabeled ssDNAs, KD measured by MST. D, EMSA assay shows the binding of ALKBH6 with FAM-labeled ssDNA1. The EMSA assay was detected by the fluorescence of FAM. Unlabeled ssDNA1 is a competitor of the labeled ssDNA1 to rule out spurious interactions. E, ALKBH6 binding with different FAM-labeled bubble DNAs. The experiments were repeated three times. F, structure-based sequence alignment of ALKBH6 in different species or its family members within the Flip3 region. Secondary structural elements are represented according to the structure of ALKBH6. hALKBH6, human ALKBH6; mALKBH6, mouse ALKBH6 (Q8K2U2); dALKBH6, Danio rerio ALKBH6 (Q6IQE9); xALKBH6, Xenopus laevis (Q5PQ59). Cylinders and arrows represent α-helices and β-sheets, respectively. Invariable residues are highlighted. The length of sequences omitted for the clarity of the presentation are shown in brackets. Evidently, Flip3 in red font shaded yellow is a unique motif of human ALKBH6.
The Flip1 of ALKBH6 contains a short α-helix α4, a short β-sheet β2, and a loop (Fig. 3B). It is the shortest one among the family members, which may affect substrates’ binding. The β2 is the outermost β-sheet of the seven-stranded major sheet and is less conserved in the family. Owing to the high temperature factors of Flip1 (Fig. S1C), it may take a conformational change upon substrate binding. In the ALKBH2–dsDNA complex structure, F102, which locates in the Flip1 of ALKBH2, intercalates into the duplex stack and fills the DNA gap (30), whereas ALKBH6 lacks any aromatic residues in the corresponding position (Fig. S2A). The Flip2 of ALKBH6 is solved as a loop (Fig. 3A). Surprising, among the human AlkB family proteins, only the Flip2 of ALKBH6 does not have any aromatic residue (Fig. S2A). In the total 27 residues of Flip1 and Flip2, only 1 aromatic residue W50 exists. Of note, Flip1 and Flip2 of ALKBH6 have a higher basic amino acids ratio (7/27) compared with ALKBH2 (5/40), ALKBH3 (6/35), or ALKBH5 (6/45). Moreover, a prominent feature of the NRL domain is that Flip2 is restrained by the C-terminal loop (hereafter Loop-CTD, residues V226–K238). Flip2 has hydrophobic interaction network with Loop-CTD. Especially, the ALKBH6 Loop-CTD has the highest content of hydrophobic residues (7/13) among the family (Fig. S2A). Consistently, an extensive hydrophobic network is formed by P65, M70, V71, P72 from Flip2 and V226, V229, L230, L235 from Loop-CTD. Moreover, the main chain of residue M70 forms two hydrogen bonds with main chains of residues R228 and L230 from Loop-CTD. The side chain of R68 forms two hydrogen bonds with the carbonyl group of residue G237 (Fig. S2B). Thus, Loop-CTD greatly stabilizes the conformation of Flip2 and the average temperature factor of Flip2 is lower than that of Flip1 (Fig. S1C).
Substrates screen for ALKBH6
As the substrate for ALKBH6 is still unknown, we carefully analyze the structure and superpose with homologous members. The distribution of the positive charge of ALKBH6 is different from that of the other family proteins (Fig. S2C). ALKBH6 has fewer positively charged residues compared with ALKBH1 or ALKBH2 at the surface of the active site. But it has some similarities to ALKBH3 and ALKBH5. A positively charged groove traverses the potential active site between Flip1 and Flip2; however, it is very narrow. The positively charged groove may directly bind the substrates. Most of the structure-solved AlkB family members have dynamic flexible Flips to dock substrates into the active site. However, it is difficult to accurately figure out the natural substrates of ALKBH6 because of these characteristic flexible Flips.
As the residues in the DSBH core shift less when binding nucleotides, here we analyze some of these residues involved in nucleotides’ recognition. In the structure of the ALKBH2-dsDNA complex (30), the nucleobase ring of cytidine forms aromatic stacking interaction with the Fe2+-ligating residue H171, while H114 is in the same conserved conformation in ALKBH6. In structures of the human AlkB family members, the C terminus of Flip1 (except in ALKBH7) has a key positively charged residue that is involved in interacting with the nucleic acid (Fig. S2, A and D). R110 in ALKBH2 directly binds the phosphate backbone (30); ALKBH6 R56 and ALKBH8 R174 are in similar positions (31). In addition, the positions of I111 and R224 are invariant among ALKBH2, ALKBH6, and ALKBH8. Moreover, the ALKBH6 active center contains four residues (L58, N60, L101, and Y120) that are similar to those in ALKBH2 (Fig. S2D). Therefore, these similarities reveal that ALKBH6 might act as a DNA/RNA demethylase like other human AlkB family members. As reported, expression of ALKBH6 in AlkB-deficient E. coli strain increases survival dramatically after methyl methane sulfonate treatment (22).
Based on the structural clues, we proposed that ALKBH6 could interact with nucleic acids directly. Then we explored various substrates, such as dsDNA, ssDNA, RNA, and bubble or bulge DNA with different numbers of mismatched base pairs in the middle of dsDNA (17, 32). With extensive and multiple screening, we finally found that ALKBH6 could bind ssDNA (Fig. 3, C and D and Table S1), bubble DNA (Figs. 3E and S3A), bulge DNA (Fig. S3B), and RNA (Fig. S3, C and D) but not dsDNA (Fig. S3E). All the nucleic acids have no notable conservative motif or any characteristics such as GC rich or AT rich. To eliminate the nonspecific binding, an unlabeled ssDNA1 competitor was used to rule out spurious interaction. With the competitor concentration increasing, the FAM-labeled complex signal decreased (Fig. 3D).
In Arabidopsis, ALKBH6 binds to m6A and m5C RNA (23), but the enzymatic activity has not been reported. Human ALKBH6 has a 36% identity with its homology in Arabidopsis. It implies that human ALKBH6 might bind RNA. We used FAM-labeled 30-nt poly(A) as a potential substrate to incubate with ALKBH6. We got a similar binding affinity as ssDNA by microscale thermophoresis (MST) and a complex band in EMSA (Fig. S3C). In eukaryotes, the mRNA has a 3′-poly(A); the above results prompt us to think if ALKBH6 could bind to mRNA and this awaits further demonstration.
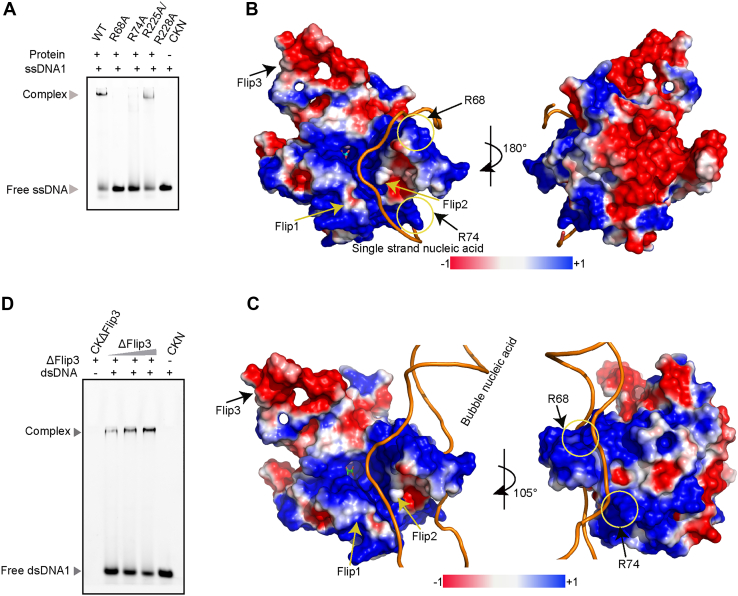
When the mutant ALKBH6ΔLoop-CTD is used to test the binding with bubble or bulge DNA, it shows a compromised binding affinity (Figs. S3A and 3B). Therefore, the Loop-CTD has a moderately positive effect on binding bubble or bulge DNA. To know the key residues in binding nucleic acids, we carefully chose several residues to make sure the essential position for binding. R68A or R74A mutants almost completely abolished the ssDNA binding activity, whereas the R225A/R228A mutant was comparable with the wildtype (Figs. 4A and S3F). As both R68 and R74 are located in Flip2, Flip2 plays key roles during substrates binding. According to the above analysis and experimental data, two possible binding models between ALKBH6 and single-stranded/bubble nucleic acid were proposed. One end of the single-stranded nucleic acid extends along the groove formed by Flip2 and Flip3, and the other end extends along the groove formed by Flip2 and Flip1 (Fig. 4B). To bind bubble nucleic acid, the other strand might interact with Loop-CTD, which has four positive residues and no negative residues (Fig. 4C). To a certain extent, the establishment of these models provides a theoretical basis for understanding the molecular mechanism of ALKBH6 binding to substrates and has an ongoing significance to uncover the recognition mechanism.
Figure 4.
ALKBH6 binds with different types of DNAs.A, ALKBH6 and mutants interact with FAM-ssDNA1. R68A and R74A mutants almost completely abolished the ssDNA binding ability, whereas the R225A/R228A mutant was comparable with the wildtype. Proposed model of ALKBH6 binding with single-stranded (B) or bubble nucleic acid (C), the key residues for binding are circled in yellow and labeled. D, ALKBH6ΔFlip3 binds with dsDNA. CKΔFlip3, only ALKBH6ΔFlip3 protein; CKN, only nucleic acid.
A unique Flip3 of ALKBH6 determines the preference of unpaired substrates
According to the phylogenetic position in the human AlkB family, ALKBH6 has the highest homology with ALKBH8, which can catalyze RNA hypermodification (2, 31). A DALI search also showed that ALKBH8 is the most similar structural homolog of ALKBH6 among human AlkB family members. We superposed ALKBH6 and ALKBH8; the unique Flip3 of ALKBH6 has a partial overlay with the RNA recognition motif (RRM) of ALKBH8 (Fig. S4A). To test whether Flip3 has a similar function as RRM, we constructed an ALKBH6ΔFlip3 mutant. Surprising, when Flip3 was deficient, the binding affinity of this mutant to ssDNA was higher than that of the wildtype (Fig. S4B). Thus, Flip3 may have a self-inhibitory effect on the binding of ALKBH6 to nucleic acids, contrary to the function of RRM. To explore the reason, we further compared the structures of this family.
In the human AlkB family, only ALKBH6, ALKBH5, and ALKBH2 have a long Flip3 (Figs. S1B and S2A). The ALKBH2 Flip3 binds to the complementary strand of the methylated strand with residues Gly204 and Lys205 to grasp dsDNA (30). The ALKBH5 Flip3 is fixed to the minor sheet of the DSBH fold by a disulfide bond between C230 and C267, which impedes the binding of the unmethylated strand and thus only selects single-stranded substrates (15). Of interest, ALKBH6 has the longest Flip3 among this family. There are eight proline residues in total 20 residues of Flip3. As we know, proline is the most rigid among the 20 usual residues. The high proline content rigidifies the Flip3 region significantly, making it hard to change orientation. When the AlkB–dsDNA or ALKBH2–dsDNA complex structures are superimposed with ALKBH6, the methylated strand is well accommodated by ALKBH6, whereas the unmethylated strand has a stereo clash with the Flip3 region (Fig. S4C). Moreover, six negative residues in Flip3 form a negatively charged surface and might electrostatically repulse the phosphate backbone of the neighbor unmethylated strand. The rich negative surface may explain its self-inhibition that the deletion of Flip3 can enhance the binding affinity to ssDNA (Fig. S4B). To further confirm the above analysis, we tested the binding ability of the ALKBH6ΔFlip3 to dsDNA. EMSA results showed that this mutant could bind dsDNA and the binding was concentration dependent (Figs. 4D and S3E). Taken together, Flip3 of ALKBH6 impedes the access of paired double-stranded nucleic acids to the active site and may confer ALKBH6 the ability to discriminate against double-stranded nucleic acids.
Structure-based interaction screens
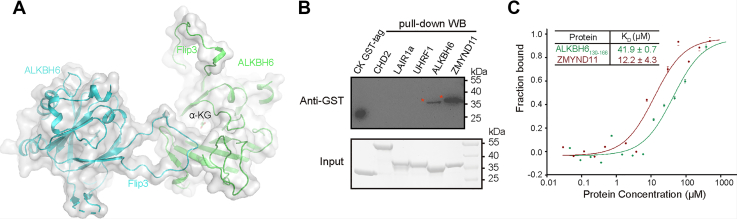
Immunization of mice with a mutant peptide of ALKBH6 Flip3, which binds the major histocompatibility complex and renders the overall peptide more rigid, can suppress tumor growth significantly (20). As the unique Flip3 has the highest temperature factor in the solved structure (Fig. S1C) and is far away from the potential catalytic core, we wonder if it is able to bind proteins. Based on the symmetry of the structure, we found that Flip3 could insert into the adjacent ALKBH6 active center (Figs. 5A and S5A). Thus, we suspected that ALKBH6 might interact with Flip3 from another ALKBH6. And then, we verified it by pull-down and Western blotting. We detected the signal band clearly (Figs. 5B and S5B). Wondering if there were any similar peptides in the human proteome that could interact with ALKBH6, we searched the human database based on the sequence (143PEDDDPTE150) to find if there are any similar peptides. Initially, we used the motif PEDDDPTE and only found ALKBH6 itself. Because no obvious interactions were observed for D145 and D147, we used PE×D×PT(E/D) (× means any residue) as searching motifs; ultimately, only three proteins were found (Table S2). Based on the structural information that positive charges are rich in the surface of the active center of ALKBH6 (Fig. S5A), the binding might be enhanced by introducing additional negative charges. Finally, we optimized the searching motifs by mutating proline to glutamic or aspartic acid, so two motifs PE×D×(E/D)T(E/D) and (E/D)E×D×(E/D)T(E/D) were used. A total of 5 and 19 candidates were found, respectively (Table S2). We chose several candidates to verify according to their sublocation and functions. In the pull-down experiments (Fig. 5B), LAIR1 (the corresponding sequence: PEKDRETD), with one proline replacement, did not bind ALKBH6. Fortunately, a peptide from V71 to K151 of ZMYND11 (the found motif: DEIDWETE), with two proline replacements, could interact with ALKBH6 by pull-down experiments (Fig. 5B), and the binding was concentration dependent (Fig. S5C). Moreover, the binding affinity of ZMYND11 to ALKBH6 was 3.5-fold higher than ALKBH6 itself (Fig. 5C), giving us a new insight into the ALKBH6 function regulated by ZMYND11. As the peptide of ZMYND11 has a zinc finger that belongs to the plant homeodomain (PHD) type, we then checked whether ALKBH6 can bind other PHD-type zinc fingers without a similar sequence. UHRF1, an E3 ubiquitin-protein ligase, also has a zinc finger that belongs to the PHD type from N310 to D366 whose sequence does not show any similarity to the Flip3 of ALKBH6. Then we made a construct from R296 to A367 of UHRF1 containing its PHD domain and tested the interaction between ALKBH6 and UHRF1 by pull-down. As the results shown in Figure 5B, ALKBH6 had no physical interaction with the UHRF1 PHD domain. Thus, the interaction between ALKBH6 and the ZMYND11 PHD domain has sequence selectivity.
Figure 5.
ALKBH6 interacts with proteins.A, the Flip3 of ALKBH6 interacts with a neighboring ALKBH6 molecule in crystal packing. B, ALKBH6 (MBP-ALKBH6) interacts with different GST-tag fusion proteins by MBP pull-down assay. CK GST-tag is a positive marker only for transferring membrane in the upper panel during Western blot of anti-GST tag antibody and a reference for input samples in the bottom panel. Size markers (kDa) are shown on the right. C, MST measured ALKBH6 interacting with ZMYND11 peptide or ALKBH6 itself. The experiments were repeated three times.
Discussion
Epigenetics has been of increasing importance in human development, diseases diagnosis, and treatment (33, 34). AlkB family members can participate in the modification of nucleic acids and have great impacts on human diseases especially carcinomas (8, 35, 36, 37, 38). Despite being a member of the AlkB family, ALKBH6 has been a mystery for decades. Here we report the structure and interacting partners of ALKBH6 and provide many clues for functional research.
In this study, we found that ALKBH6 could bind Tris in the active center (Fig. 2A), which has never been found in other members of this family. Tris, commonly used in experiments, occupies the position of α-KG and may thus interfere with the activity and should be avoided in the functional assay of ALKBH6. It is intriguing that Tris chelates the metal ion in the active center in a tridentate manner. To the best of our knowledge, this is the first report of a chemical compound that coordinates the metal ion tridentately in the AlkB family. Thus, it may be a special start point for the selective drug design against AlkB family proteins.
As reported, human AlkB homologs seem to work on ssDNA, dsDNA with a damaged base on an unstable base pair, or modified RNA (15, 39, 40, 41). In the human AlkB homologs, ALKBH1 prefers bubble or bulge DNA (17, 42); ALKBH2 binds dsDNA as substrate (30); ALKBH3 chooses ssDNA or RNA as substrates (37, 42); and ALKBH5, ALKBH8, and FTO function as RNA demethylases (43, 44, 45). In our study, we found that the unique Flip3 was key for the binding of ALKBH6 to unpaired nucleic acids rather than dsDNA. Moreover, ALKBH6 prefers ssDNA or ssRNA to other types of DNAs without notable sequence preference. Consistently, self-hydroxylation of L101 in ALKBH6 was observed (42), revealing that ALKBH6 has enzymatic activity. We did not observe obvious demethylase activity of ALKBH6 on several kinds of modified nucleic acids, such as m1A and m6A ssDNA/ssRNA. Structural alignments reveal that ALKBH6 has unique features. First, the Flip1 of ALKBH6 is the shortest one among the family members and lacks an aromatic residue corresponding to F102 in ALKBH2 to intercalate into the duplex stack and fill the DNA gap (30) (Fig. S2A). Second, in the Flip2 of AlkB members, Y122 and F124 from ALKBH2, Y143 in ALKBH3, Y139 and Y141 in ALKBH5 could interact with substrates by hydrogen bonds or van der Waals. Unfortunately, the ALKBH6 Flip2 loses any aromatic residues. Finally, ALKBH6 has the self-inhibitory ability as its Flip3 deletion has higher affinity than the wildtype and the active center is occupied by the negatively charged Flip3 intermolecularly. Structure-based sequence alignment shows that the ALKBH6 Flip3 in human is a unique loop compared with other family members or that in other nonmammal species (Figs. 3F and S2A). In short, a combination of Flip3, Flip1, and Flip2 results in ALKBH6 self-inhibition and weak binding to substrate; this may be the main reason why it is difficult to find the natural substrates of ALKBH6 under normal physiological situations.
In a previous study, Kernstock et al. (46) used yeast two-hybrid to screen and define the potential binding partner of ALKBH6; however, they failed at the confirmed progress. Here, from the crystal packing of the ALKBH6∙Mn2+∙α-KG structure, we found that the unique Flip3 binds the active center with a buried interface area of 309 Å2. The intermolecular interaction between Flip3 and ALKBH6 may be caused by the opposite charges distribution in Flip3 and the active center and is concentration dependent. Then we conducted the small-angle X-ray scattering (47) assay (Table S3), with the results showing that ALKBH6 was mainly monomeric in solution (Fig. S6). However, when the dimer component was added, the Minimal Ensemble Search fit was improved significantly, revealing the existence of dimer.
Furthermore, to find potential interacting partners of ALKBH6, we used structure-based sequence similarity or mutation enhancement strategies to search the human database and found many candidates. Our experiments revealed that ALKBH6 not only interacts with itself but also has a stronger binding affinity to ZMYND11, giving new insights for functional research. ZMYND11 can link histone H3.3K36 trimethylation (H3.3K36me3) to transcription elongation and tumor suppression (48). ZMYND11 has several “reader” domains especially for histone, including a PHD, a bromodomain, and a PWWP domain (named for a conserved Pro-Trp-Trp-Pro motif). The peptide from V71 to K151 covers the PHD domain (from Y102 to C145). When the PHD domain is deficient, the binding between ZMYND11 and H3.3K36me3 peptide can be greatly diminished (48). In the beginning, we found that ALKBH6 could bind nucleic acids; now our results showed that it also could interact with proteins, highlighting the importance of ALKBH6 in vivo. We speculated that the binding of ZMYND11 may affect the activity of ALKBH6 and thus influence the demethylation of modified nucleic acids, even though no experimental evidence has been reported. ZMYND11, as a reader protein for modified histones, interacts with ALKBH6 and thus may bidirectionally influence their functions in the cell.
Finally, analysis of the COSMIC database revealed that many cancer-associated ALKBH6 mutations occur in Flip1, Flip2, and Flip3 (Table S4). These mutations might be able to disrupt the enzymatic function, protein interaction, and immunity, thereby contributing to tumorigenesis. We made several cancer-associated mutations based on the cosmic data and validated the interaction between ALKBH6 and ZMYND11. The R68Q or R68P mutants located in Flip2 can diminish the interaction between ALKBH6 and ZMYND11, whereas P65H or P65S mutants have comparable ability to the wildtype ALKBH6 (Fig. S7). These observations might be caused by the loss of the positive charge of Flip2. Our results showed that the cancer-relevant mutations of the key positive residue could greatly diminish the interaction between ALKBH6 and ZMYND11, underscoring the vital role of ALKBH6 in tumor suppression.
Briefly, we successfully solved structures of ALKBH6, which broaden the structural knowledge of the AlkB family. Besides, based on the structural information we demonstrated that ALKBH6 can bind several different types of nucleic acids; also, we figured out the key Flips for its binding ability, providing new insights into its substrates screen and even for its enzymatic activity research. We also found that ALKBH6 could interact with ZMYND11 directly, which may act as a new point of view for ALKBH6 function illustration. In general, the above results provide the bases for characterizing ALKBH6 functions such as nucleic acid damage repair and tumor therapy.
Experimental procedures
Protein expression
The full-length ALKBH6 was amplified from cDNA. For eukaryotic expression, the fragment was inserted into EcoR I and Kpn I sites in a modified pPICZ-B vector with a tobacco etch virus (TEV) protease cleavage site, followed by a GFP-6×His tag. GFP was used as a reporter to show the expression of target gene, which can simplify the screening process. P. pastoris X-33 cells transformed with pPICZ-B-ALKBH6 were incubated in yeast extract peptone dextrose medium at 30 °C until the absorbance at 600 nm (A600) reached 4 to 5 and then induced at 28 °C with BMMH medium for an additional 72 h.
For prokaryotic expression, full-length ALKBH6, truncations, or mutants were amplified and cloned into a modified pET-28a vector with a TEV site. The fragments were inserted into the BamH I and Xho I sites. Final clones were verified by DNA sequencing and transformed into Rosetta competent cells. Cells were grown in LB medium at 37 °C until A600 reached 0.6 to 0.8. Target protein overexpression was induced overnight at 18 °C by the addition of 0.2 mM isopropyl-β-D-thiogalactoside. The MBP-fused proteins, constructed by vector V28E4 (49), were prepared by prokaryotic expression and used for pull-down and EMSA.
Protein purification
Both yeast and E. coli cultures were harvested by centrifugation at 3500g for 10 min and resuspended in buffer A (20 mM Tris pH 7.5, 1 M NaCl) and disrupted by high-pressure (1800 bar, P. pastoris) cell disruptor or sonication (E. coli). After centrifugation at 47,000g for 60 min to remove cells debris, the soluble fraction was applied to a Ni2+-chelating column (GE Healthcare).
After protein binding, the column was washed with buffer B (20 mM Tris pH 7.5, 1 M NaCl, 20 mM Imidazole) and eluted with buffer C (20 mM Tris pH 7.5, 150 mM NaCl, 100 mM Imidazole, 3 mM β-Me). TEV protease was added to the protein-containing eluent to remove the His-tag at a 1:5 weight ratio overnight at 4 °C. The digestion was reloaded onto a Ni2+ affinity column to remove His-tag and His-tagged TEV protease. The protein sample was then loaded onto an SP sepharose column (GE Healthcare) and eluted with a NaCl gradient from 0 to 1 M NaCl in 20 mM Tris pH 7.5. The eluent was further purified by gel filtration (Superdex increase 200 10/300, GE Healthcare) with buffer D (20 mM Tris pH 7.5, 150 mM NaCl). Peak fractions were collected and visualized by SDS-PAGE followed by Coomassie Bright Blue staining. The purified protein was concentrated by ultrafiltration (Amicon Ultra-15 centrifugal filter units, Millipore) to 10 to 13 mg/ml for crystallization. All the protein purification procedures were performed at 4 °C. The protein used for pull-down, EMSA, and MST was purified in Hepes (pH 7.5) buffer instead of Tris buffer.
Crystallization and data collection
Extensive crystallization screens were performed at both 18 and 4 °C with many commercial kits. Small crystals of ALKBH6 expressed from P. pastoris were initially obtained in reservoir solution using the sitting drop vapor diffusion method. To get high-quality crystals, we optimized the concentration of ALKBH6, different buffer pH, and precipitant concentration at both 18 and 4 °C. Finally, for apo-ALKBH6, purified ALKBH6 10 mg/ml was mixed with a well solution containing 10% PEG 8000, 0.1 M Tris pH 8.5 at 4 °C. For ALKBH6∙Mn2+∙α-KG complex, the purified protein was mixed with 2 mM MnCl2, 10 mM α-KG and incubated on ice for more than 5 h. Mn(II) was used to replace Fe(II). Crystallization trials were performed at 4 °C.
The crystals were transferred to a solution containing 30% (v/v) glycerol for cryoprotection and flash cooled in liquid nitrogen before data collection. The data were collected on beamlines BL17U1 and BL19U1 at the Shanghai Synchrotron Radiation Facility. Data were indexed, integrated, and scaled with HKL2000 suite of programs (50). Data collection statistics are summarized in Table 1.
Structure determination
ALKBH6 has a low sequence identity (less than 18.5%) to all the released structures in the PDB, so we failed to find a good molecular replacement result for the ALKBH6∙Ni2+∙Tris. But fortunately, a new idea came to our mind and we used a predicted result of ALKBH6 structure by Raptorx (25) as an MR model. We tried different truncations of the predicted structure as MR models, such as removing disordered loops or short β-sheets. After extensive trials of different resolution and space groups, the best solution was found by BALBES (26) with an MR score of 3.61 while the Rwork/Rfree were both above 0.50. After removing the inappropriate main and side chains in the model using the program COOT (51), the optimized structure was used to find the right position by BALBES again. The primitive density map has a relatively clear outline with recognizable β-sheets and α-helices with an MR score of 14.56 and an Rwork/Rfree of 0.410/0.488.
Then we iteratively refined and built the model using CNS (52), the CCP4 suite (53), and autobuild program in the Phenix (54). According to the density, the nonrational main and side chains were adjusted using the program COOT followed by REFMAC5 (55) refinement. After numerous cycles of manual rebuilding and refining, the best solution stopped at an Rwork/Rfree value of 0.198/0.218. The complex structure of ALKBH6∙Mn2+∙α-KG was determined by molecular replacement using the above structure as a model. Structure validations were carried out with Molprobity (56). The ALKBH6∙Ni2+∙Tris has a Molprobity score of 1.26 (98th percentile) and a clashscore of 1.91 (100th percentile). The ALKBH6∙Mn2+∙α-KG has a Molprobity score of 1.01 (100th percentile) with a clashscore of 2.07 (99th percentile) (Table 1).
Microscale thermophoresis
MST was used to measure the dissociation constant for the interaction of wildtype or mutated proteins with α-KG cofactor, proteins, and DNA substrates.
To measure the binding affinities between α-KG and ALKBH6, ALKBH6 wildtype and mutants were labeled with a fluorescent dye NT-647 according to the manufacturer’s manual. 10 μM of 10 μl labeled protein in buffer containing 20 mM Hepes pH 7.5, 80 mM NaCl, 0.4% NP40 was mixed with 10 μl α-KG at various concentrations. After incubation for 30 min at room temperature, the samples were loaded into capillaries. Thermophoresis was measured at 22 °C for 30 s with 40% infrared laser power. Datasets were combined and analyzed using the MO-Affinity analysis software.
To measure the binding affinities between DNA oligonucleotides and ALKBH6 using MST, the oligonucleotides were labeled with 5′-FAM or the protein was labeled by using His-tag labeling kit RED-tris-NTA. The MST buffer was 20 mM Hepes pH 7.5, 80 mM KCl, 200 μM α-KG, 5 μM MnCl2, 0.4% NP40. We illustrated two experimental setups by using labeled DNA with unlabeled protein or labeled protein with unlabeled DNA. The mixtures are incubated at room temperature for 15 min then loaded into capillaries and measured at 22 °C for 30 s with 40% or 60% infrared laser power.
His-tag labeled ALKBH6 was used to measure protein–protein interaction in MST buffer containing 20 mM Hepes pH 7.5, 80 mM KCl, 200 μM α-KG, 5 μM MnCl2, 0.05% NP40, 0.15% Tween 20, 0.025% PEG 8000, 0.05% Triton X-100, and 1 mM DTT with 60% infrared laser power.
Electrophoretic mobility shift assay
EMSA separates unbound nucleic acid from protein-bound nucleic acid through polyacrylamide. EMSA was performed to detect the nucleic acid binding ability of ALKBH6 or mutants fused with the MBP tag. Protein, 10 to 300 pmol, was incubated with 10 pmol FAM-labeled DNA (Table S1) in 20 μl reaction mixture containing 20 mM Hepes pH 7.5, 80 mM KCl, 200 μM α-KG, 5 μM MnCl2 for 30 min at 4 °C. After incubation, 5 μl 30% glycerol was added to each sample and 5 μl of the mixture was then separated on a 5% to 7% native polyacrylamide gel (acrylamide:bisacrylamide = 39:1 or 79:1) in 1×TB (45 mM Tris pH 8.3, 45 mM boric acid) at 170 V for 40 min. The gel was scanned with ChemiDoc MP Imaging System (Bio-Rad) at the wavelength of 488 nm to detect the fluorescent signal of FAM-DNA.
Pull-down assay
ALKBH6 was fused with MBP tag, and the interacting candidates were GST tag fusions. One hundred micrograms of MBP-ALKBH6-6×His was incubated with different GST-proteins for 2 h at 4 °C in 1 ml binding buffer (20 mM Hepes pH 7.5, 75 mM NaCl, 1 mM MnCl2, 10 mM α-KG, 5% Glycerol, 3 mM β-Me, 0.1% Tween-20, and 1 mM benzamidine hydrochloride) in the presence of amylose resin. Then the resin was rinsed with the same buffer three times to remove the unbound and nonspecifically bound proteins. Proteins left on the beads were separated by SDS-PAGE and analyzed by Western blotting of anti-GST tag antibody.
Small-angle X-ray scattering
Small-angle X-ray scattering measurements were performed on beamline BL19U2 at the Shanghai Synchrotron Radiation Facility. Proteins were purified by gel filtration in a buffer containing 20 mM Hepes pH 7.5, 150 mM NaCl, 2 mM β-Me. Various concentrations of protein were used from 1 to 5 mg/ml. Measurements were carried out at 10 °C. Individual data were processed by RAW (57). The control buffer was measured before and after each sample measurement.
Data availability
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession numbers 7VJS and 7VJV.
Supporting information
This article contains supporting information (58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank the staff of beamlines BL17U1, BL19U1, and BL19U2 at the Shanghai Synchrotron Radiation Facility for the excellent technical assistance. This work was supported by National Key Research and Development Program of China (2018YFE0113100), National Natural Science Foundation of China (31872713 and 32071210), and Extramural Scientists of State Key Laboratory of Agrobiotechnology Grant (2022SKLAB6-1). Funding for open access charge: National Key Research and Development Program of China (2018YFE0113100).
Author contributions
Z. C. methodology; H. L., Z. T., M. Y., J. M., G. S., Y. L., and G. W. software; Z. C. formal analysis; L. M. investigation; L. M., M. X., and Z. C. writing – original draft; W. W., Z. Z., and S. D. writing – review & editing.
Edited by Joseph Jez
Supporting information
References
- 1.Kataoka H., Yamamoto Y., Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 1983;153:1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurowski M.A., Bhagwat A.S., Papaj G., Bujnicki J.M. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A.P., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falnes P.O. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee D.H., Jin S.G., Cai S., Chen Y., Pfeifer G.P., O'Connor T.R. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 6.Li M.-M., Nilsen A., Shi Y., Fusser M., Ding Y.-H., Fu Y., Liu B., Niu Y., Wu Y.-S., Huang C.-M., Olofsson M., Jin K.-X., Lv Y., Xu X.-Z., He C., et al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nat. Commun. 2013;4:1832. doi: 10.1038/ncomms2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U., Zhang C., Dowiak A.V., Lin K., Zeng C., Sui Y., et al. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018;175:1228–1243.e20. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao C.L., Zhu S., He M., Chen D., Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F., Liang Z., Wang D.P., Bo X.C., Gu X.F., Wang K., et al. N(6)-Methyladenine DNA modification in the human genome. Mol. Cell. 2018;71:306–318.e7. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Ruan D.Y., Li T., Wang Y.N., Meng Q., Li Y., Yu K., Wang M., Lin J.F., Luo L.Z., Wang D.S., Lin J.Z., Bai L., Liu Z.X., Zhao Q., Wu X.Y., et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene. 2021;40:5168–5181. doi: 10.1038/s41388-021-01916-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry G.S., Das M., Woon E.C.Y. Inhibition of AlkB nucleic acid demethylases: Promising new epigenetic targets. J. Med. Chem. 2021;64:16974–17003. doi: 10.1021/acs.jmedchem.1c01694. [DOI] [PubMed] [Google Scholar]
- 11.Westbye M.P., Feyzi E., Aas P.A., Vagbo C.B., Talstad V.A., Kavli B., Hagen L., Sundheim O., Akbari M., Liabakk N.B., Slupphaug G., Otterlei M., Krokan H.E. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J. Biol. Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G., Ma H., Han D., Evans M., Klungland A., Pan T., et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:816–828.e16. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan T., Trewick S.C., Koivisto P., Bates P.A., Lindahl T., Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L.S., Xiong Q.P., Pena Perez S., Liu C., Wei J., Le C., Zhang L., Harada B.T., Dai Q., Feng X., Hao Z., Wang Y., Dong X., Hu L., Wang E.D., et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat. Cell Biol. 2021;23:684–691. doi: 10.1038/s41556-021-00709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng C., Liu Y., Wang G., Deng Z., Zhang Q., Wu W., Tong Y., Cheng C., Chen Z. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem. 2014;289:11571–11583. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G., He Q., Feng C., Liu Y., Deng Z., Qi X., Wu W., Mei P., Chen Z. The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism. J. Biol. Chem. 2014;289:27924–27936. doi: 10.1074/jbc.M114.590505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian L.F., Liu Y.P., Chen L., Tang Q., Wu W., Sun W., Chen Z., Yan X.X. Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res. 2020;30:272–275. doi: 10.1038/s41422-019-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsujikawa K., Koike K., Kitae K., Shinkawa A., Arima H., Suzuki T., Tsuchiya M., Makino Y., Furukawa T., Konishi N., Yamamoto H. Expression and sub-cellular localization of human ABH family molecules. J. Cell. Mol. Med. 2007;11:1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C., Li D., Hu J., Jiang J., Zhang W., Chen Y., Cui X., Qi Y., Zou H., Zhang W., Li F. Chromosomal and genetic imbalances in Chinese patients with rhabdomyosarcoma detected by high-resolution array comparative genomic hybridization. Int. J. Clin. Exp. Pathol. 2014;7:690–698. [PMC free article] [PubMed] [Google Scholar]
- 20.Duan F., Duitama J., Al Seesi S., Ayres C.M., Corcelli S.A., Pawashe A.P., Blanchard T., McMahon D., Sidney J., Sette A., Baker B.M., Mandoiu I.I., Srivastava P.K. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao S., Devega R., Francois A., Kidane D. Human ALKBH6 is required for maintenance of genomic stability and promoting cell survival during exposure of alkylating agents in pancreatic cancer. Front. Genet. 2021;12:635808. doi: 10.3389/fgene.2021.635808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huong T.T., Ngoc L.N.T., Kang H. Functional characterization of a putative RNA demethylase ALKBH6 in Arabidopsis growth and abiotic stress responses. Int. J. Mol. Sci. 2020;21:6707. doi: 10.3390/ijms21186707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J., Manduzio S., Kang H. Epitranscriptomic RNA methylation in plant development and abiotic stress responses. Front. Plant Sci. 2019;10:500. doi: 10.3389/fpls.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long F., Vagin A.A., Young P., Murshudov G.N. BALBES: A molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonough M.A., Loenarz C., Chowdhury R., Clifton I.J., Schofield C.J. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr. Opin. Struct. Biol. 2010;20:659–672. doi: 10.1016/j.sbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Xu B., Liu D., Wang Z., Tian R., Zuo Y. Multi-substrate selectivity based on key loops and non-homologous domains: New insight into ALKBH family. Cell. Mol. Life Sci. 2020;78:129–141. doi: 10.1007/s00018-020-03594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundheim O., Talstad V.A., Vågbø C.B., Slupphaug G., Krokan H.E. AlkB demethylases flip out in different ways. DNA Repair. 2008;7:1916–1923. doi: 10.1016/j.dnarep.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Yang C.G., Yi C., Duguid E.M., Sullivan C.T., Jian X., Rice P.A., He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastore C., Topalidou I., Forouhar F., Yan A.C., Levy M., Hunt J.F. Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J. Biol. Chem. 2012;287:2130–2143. doi: 10.1074/jbc.M111.286187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M., Yang S., Nelakanti R., Zhao W., Liu G., Li Z., Liu X., Wu T., Xiao A., Li H. Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA. Cell Res. 2020;30:197–210. doi: 10.1038/s41422-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herceg Z., Ghantous A., Wild C.P., Sklias A., Casati L., Duthie S.J., Fry R., Issa J.P., Kellermayer R., Koturbash I., Kondo Y., Lepeule J., Lima S.C.S., Marsit C.J., Rakyan V., et al. Roadmap for investigating epigenome deregulation and environmental origins of cancer. Int. J. Cancer. 2018;142:874–882. doi: 10.1002/ijc.31014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa Y., Kuribayashi K., Minami T., Ohmuraya M., Kijima T. Epigenetic alterations and biomarkers for immune checkpoint inhibitors-current standards and future perspectives in malignant pleural mesothelioma treatment. Front. Oncol. 2020;10:554–570. doi: 10.3389/fonc.2020.554570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Wang J., Gu Q., Ma Y., Yang Y., Zhu J., Zhang Q. The biological function of m6A demethylase ALKBH5 and its role in human disease. Cancer Cell Int. 2020;20:347. doi: 10.1186/s12935-020-01450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao M.-Z., Liu J.-M., Xian C.-L., Chen K.-Y., Liu Z.-Q., Cheng Y.-Y. Therapeutic potential of ALKB homologs for cardiovascular disease. Biomed. Pharmacother. 2020;131:110645. doi: 10.1016/j.biopha.2020.110645. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z., Qi M., Shen B., Luo G., Wu Y., Li J., Lu Z., Zheng Z., Dai Q., Wang H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–2545. doi: 10.1093/nar/gky1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilžys T., Marcinkowski M., Kukwa W., Garbicz D., Dylewska M., Ferenc K., Mieczkowski A., Kukwa A., Migacz E., Wołosz D., Mielecki D., Klungland A., Piwowarski J., Poznański J., Grzesiuk E. ALKBH overexpression in head and neck cancer: Potential target for novel anticancer therapy. Sci. Rep. 2019;9:13249. doi: 10.1038/s41598-019-49550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishina Y., Chen L.X., He C. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli. J. Am. Chem. Soc. 2004;126:16930–16936. doi: 10.1021/ja045066z. [DOI] [PubMed] [Google Scholar]
- 40.Mishina Y., Lee C.H., He C. Interaction of human and bacterial AlkB proteins with DNA as probed through chemical cross-linking studies. Nucleic Acids Res. 2004;32:1548–1554. doi: 10.1093/nar/gkh319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janin M., Coll-SanMartin L., Esteller M. Disruption of the RNA modifications that target the ribosome translation machinery in human cancer. Mol. Cancer. 2020;19:70. doi: 10.1186/s12943-020-01192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundheim O., Vågbø C.B., Bjørås M., Sousa M.M., Talstad V., Aas P.A., Drabløs F., Krokan H.E., Tainer J.A., Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H., Lu Z., Bosmans R.P.G., Dai Q., Hao Y.-J., Yang X., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu D., Brophy J.A., Chan C.T., Atmore K.A., Begley U., Paules R.S., Dedon P.C., Begley T.J., Samson L.D. Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Maelandsmo G.M., Egge-Jacobsen W., Falnes P.O. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 47.Tian Z., Li X., Li M., Wu W., Zhang M., Tang C., Li Z., Liu Y., Chen Z., Yang M., Ma L., Caba C., Tong Y., Lam H.M., Dai S., et al. Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms. Cell Discov. 2020;6:17. doi: 10.1038/s41421-020-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J., Li B., Barton M.C., Li W., Li H., Shi X. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin T., Chuenchor W., Jiang J., Cheng J., Li Y., Fang K., Huang M., Smith P., Xiao T.S. Design of an expression system to enhance MBP-mediated crystallization. Sci. Rep. 2017;7:40991. doi: 10.1038/srep40991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brunger A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 53.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen S.S., Toft K.N., Snakenborg D., Jeppesen M.G., Jacobsen J.K., Vestergaard B., Kutter J.P., Arleth L. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Crystallogr. 2009;42:959–964. [Google Scholar]
- 58.Mimaki S., Totsuka Y., Suzuki Y., Nakai C., Goto M., Kojima M., Arakawa H., Takemura S., Tanaka S., Marubashi S., Kinoshita M., Matsuda T., Shibata T., Nakagama H., Ochiai A., et al. Hypermutation and unique mutational signatures of occupational cholangiocarcinoma in printing workers exposed to haloalkanes. Carcinogenesis. 2016;37:817–826. doi: 10.1093/carcin/bgw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giannakis M., Mu X.J., Shukla S.A., Qian Z.R., Cohen O., Nishihara R., Bahl S., Cao Y., Amin-Mansour A., Yamauchi M., Sukawa Y., Stewart C., Rosenberg M., Mima K., Inamura K., et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonilla X., Parmentier L., King B., Bezrukov F., Kaya G., Zoete V., Seplyarskiy V.B., Sharpe H.J., McKee T., Letourneau A., Ribaux P.G., Popadin K., Basset-Seguin N., Ben Chaabene R., Santoni F.A., et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 61.Zou S., Li J., Zhou H., Frech C., Jiang X., Chu J.S., Zhao X., Li Y., Li Q., Wang H., Hu J., Kong G., Wu M., Ding C., Chen N., et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat. Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 62.Liu Z., Yang C., Li X., Luo W., Roy B., Xiong T., Zhang X., Yang H., Wang J., Ye Z., Chen Y., Song J., Ma S., Zhou Y., Yang M., et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget. 2018;9:27412–27422. doi: 10.18632/oncotarget.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pickering C.R., Zhou J.H., Lee J.J., Drummond J.A., Peng S.A., Saade R.E., Tsai K.Y., Curry J.L., Tetzlaff M.T., Lai S.Y., Yu J., Muzny D.M., Doddapaneni H., Shinbrot E., Covington K.R., et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. 2014;20:6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krauthammer M., Kong Y., Ha B.H., Evans P., Bacchiocchi A., McCusker J.P., Cheng E., Davis M.J., Goh G., Choi M., Ariyan S., Narayan D., Dutton-Regester K., Capatana A., Holman E.C., et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vinayanuwattikun C., Le Calvez-Kelm F., Abedi-Ardekani B., Zaridze D., Mukeria A., Voegele C., Vallee M., Purnomosari D., Forey N., Durand G., Byrnes G., McKay J., Brennan P., Scelo G. Elucidating genomic characteristics of lung cancer progression from in situ to invasive adenocarcinoma. Sci. Rep. 2016;6:31628. doi: 10.1038/srep31628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nayar U., Cohen O., Kapstad C., Cuoco M.S., Waks A.G., Wander S.A., Painter C., Freeman S., Persky N.S., Marini L., Helvie K., Oliver N., Rozenblatt-Rosen O., Ma C.X., Regev A., et al. Acquired HER2 mutations in ER(+) metastatic breast cancer confer resistance to estrogen receptor-directed therapies. Nat. Genet. 2019;51:207–216. doi: 10.1038/s41588-018-0287-5. [DOI] [PubMed] [Google Scholar]
- 67.Giannakis M., Hodis E., Jasmine Mu X., Yamauchi M., Rosenbluh J., Cibulskis K., Saksena G., Lawrence M.S., Qian Z.R., Nishihara R., Van Allen E.M., Hahn W.C., Gabriel S.B., Lander E.S., Getz G., et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hayward N.K., Wilmott J.S., Waddell N., Johansson P.A., Field M.A., Nones K., Patch A.M., Kakavand H., Alexandrov L.B., Burke H., Jakrot V., Kazakoff S., Holmes O., Leonard C., Sabarinathan R., et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 69.Rabbie R., Ferguson P., Wong K., Couturier D.L., Moran U., Turner C., Emanuel P., Haas K., Saunus J.M., Davidson M.R., Lakhani S.R., Shivalingam B., Long G.V., Parkinson C., Osman I., et al. The mutational landscape of melanoma brain metastases presenting as the first visceral site of recurrence. Br. J. Cancer. 2021;124:156–160. doi: 10.1038/s41416-020-01090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J.K., Wang J., Sa J.K., Ladewig E., Lee H.O., Lee I.H., Kang H.J., Rosenbloom D.S., Camara P.G., Liu Z., van Nieuwenhuizen P., Jung S.W., Choi S.W., Kim J., Chen A., et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 2017;49:594–599. doi: 10.1038/ng.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cha S., Lee J., Shin J.Y., Kim J.Y., Sim S.H., Keam B., Kim T.M., Kim D.W., Heo D.S., Lee S.H., Kim J.I. Clinical application of genomic profiling to find druggable targets for adolescent and young adult (AYA) cancer patients with metastasis. BMC Cancer. 2016;16:170. doi: 10.1186/s12885-016-2209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.South A.P., Purdie K.J., Watt S.A., Haldenby S., den Breems N., Dimon M., Arron S.T., Kluk M.J., Aster J.C., McHugh A., Xue D.J., Dayal J.H., Robinson K.S., Rizvi S.H., Proby C.M., et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J. Invest. Dermatol. 2014;134:2630–2638. doi: 10.1038/jid.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sossey-Alaoui K., Vaca-Paniagua F., Alvarez-Gomez R.M., Maldonado-Martínez H.A., Pérez-Plasencia C., Fragoso-Ontiveros V., Lasa-Gonsebatt F., Herrera L.A., Cantú D., Bargallo-Rocha E., Mohar A., Durand G., Forey N., Voegele C., Vallée M., et al. Revealing the molecular portrait of triple negative breast tumors in an understudied population through omics analysis of formalin-fixed and paraffin-embedded tissues. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durinck S., Ho C., Wang N.J., Liao W., Jakkula L.R., Collisson E.A., Pons J., Chan S.W., Lam E.T., Chu C., Park K., Hong S.W., Hur J.S., Huh N., Neuhaus I.M., et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMillan E.A., Ryu M.J., Diep C.H., Mendiratta S., Clemenceau J.R., Vaden R.M., Kim J.H., Motoyaji T., Covington K.R., Peyton M., Huffman K., Wu X., Girard L., Sung Y., Chen P.H., et al. Chemistry-first approach for nomination of personalized treatment in lung cancer. Cell. 2018;173:864–878.e29. doi: 10.1016/j.cell.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seidlitz T., Merker S.R., Rothe A., Zakrzewski F., von Neubeck C., Grützmann K., Sommer U., Schweitzer C., Schölch S., Uhlemann H., Gaebler A.M., Werner K., Krause M., Baretton G.B., Welsch T., et al. Human gastric cancer modelling using organoids. Gut. 2019;68:207–217. doi: 10.1136/gutjnl-2017-314549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession numbers 7VJS and 7VJV.