Abstract
Increasing evidences indicate that unlimited capacity for self-renewal and pluripotency, two unique properties of embryonic stem cells (ESCs), are intrinsically linked to cell cycle control. However, the precise mechanisms coordinating cell fate decisions and cell cycle regulation remain to be fully explored. Here, using CRISPR/Cas9-mediated genome editing, we show that in ESCs, deficiency of components of the cell cycle regulatory MuvB complex Lin54 or Lin52, but not Lin9 or Lin37, triggers G2/M arrest, loss of pluripotency, and spontaneous differentiation. Further dissection of these phenotypes demonstrated that this cell cycle arrest is accompanied by the gradual activation of mesoendodermal lineage-specifying genes. Strikingly, the abnormalities observed in Lin54-null ESCs were partially but significantly rescued by ectopic coexpression of genes encoding G2/M proteins Cyclin B1 and Cdk1. Thus, our study provides new insights into the mechanisms by which the MuvB complex determines cell fate through regulation of the cell cycle machinery.
Keywords: Lin54, Lin52, Lin37, Lin9, MuvB, embryonic stem cells, pluripotency, germ layer lineages, cell cycle, G2/M
Abbreviations: ChIP, chromatin immunoprecipitation; DREAM, Drosophila RBF, dE2F2, and dMyb-interacting proteins; ESC, embryonic stem cell; LIF, leukemia inhibitory factor; MEF, mouse embryonic fibroblast; MMB, Myb-MuvB; MuvB, multivulva class B; RT-qPCR, quantitative reverse transcriptase PCR; sgRNA, single-guide RNA; SMA, smooth muscle actin
Embryonic stem cells (ESCs) possess an indefinite proliferative and self-renewal potential, while retaining the capacity to differentiate into a wide array of cell types found in the body. It has been demonstrated that several transcription factors, including Oct4, Sox2, and Nanog along with other genetic and epigenetic factors, form the core of a transcription factor network that maintain ESC identity while simultaneously repressing the expression of genes that promote differentiation (1, 2). Pluripotency is also linked with core cell cycle machinery because studies in ESCs have suggested that their pluripotent status is connected with a specific cell cycle profile (3, 4). Therefore, dissecting the potential interactions coordinating cell fate choice and cell cycle progression will unravel the intricate mechanisms governing ESC pluripotency.
ESCs exhibit very unusual cell cycle features, characterized by a prolonged S phase and an abbreviated G1 phase, as compared with most other cell types, including adult stem cells, which are typically quiescent (5, 6). The conserved “multi-vulva class B” (MuvB) complex acts as a switchable pivotal cell cycle regulator, governing expression of cell cycle regulatory genes during the cell cycle and contributing to repression of cell cycle genes during G0 or quiescence (7, 8, 9, 10). The mammalian core MuvB complex, consisting of five subunits Lin9, Lin37, Lin52, Lin54, and Rbbp4, associates with the retinoblastoma protein (pRB)-related pocket proteins, p107 and p130, and with the transcription factor heterodimer, E2F4-DP1, to form DREAM (Drosophila RBF, dE2F2 and dMyb-interacting proteins), which globally represses cell cycle genes in G0/G1, maintaining the cell in a quiescent state (7, 11, 12). When cells enter the cell cycle, the MuvB core complex is released from the p130-E2F4-DP1-containing DREAM complex and independently partners with B-Myb transcription factor to form Myb-MuvB (MMB) complex that activates expression of a subset of late cell cycle genes required for progression through mitosis and cytokinesis (7, 8, 9, 10, 13). The importance of MMB in transcriptional activation of mitotic genes is emphasized by the observation that disruption of the MMB complex causes mitotic defects and G2/M arrest (14, 15). Among the core MuvB complex components, Lin54 is the only sequence-specific DNA-binding protein. It possesses dual tandem cysteine-rich (CXC) domains (with the consensus sequence CXCX4CX3YCXCX6CX3CXCX2C) separated by a conserved hinge domain. The CXC domains of Lin54 direct MuvB binding to the cell cycle genes homology region (CHR) of target genes (16, 17). Interestingly, the CHR element is located in the promoters of majority of the DREAM-bound G2/M genes (7, 17, 18).
Despite recent advances in our understanding of cell cycle progression, the precise role of the cell cycle machinery in maintaining ESC self-renewal and pluripotency remains largely uncharacterized. Here we examined the role of MuvB core complex members in the cell cycle and the maintenance of self-renewal and pluripotency in ESCs. Our results show that ablation of Lin54 or Lin52, but not Lin9 or Lin37, resulted in cell cycle arrest at G2/M transition accompanied by rapid exit of undifferentiated state in ESCs and the initiation of differentiation toward the mesoendoderm lineages. Mechanistically, we demonstrate that MuvB complex-mediated activation of Cyclin B1/Cdk1, key components in the control of cell cycle progression from G2 to M phase, exerts control over cell fate decisions and pluripotency.
Results
Ablation of the MuvB components Lin54 and Lin52 causes exit from pluripotency in ESCs
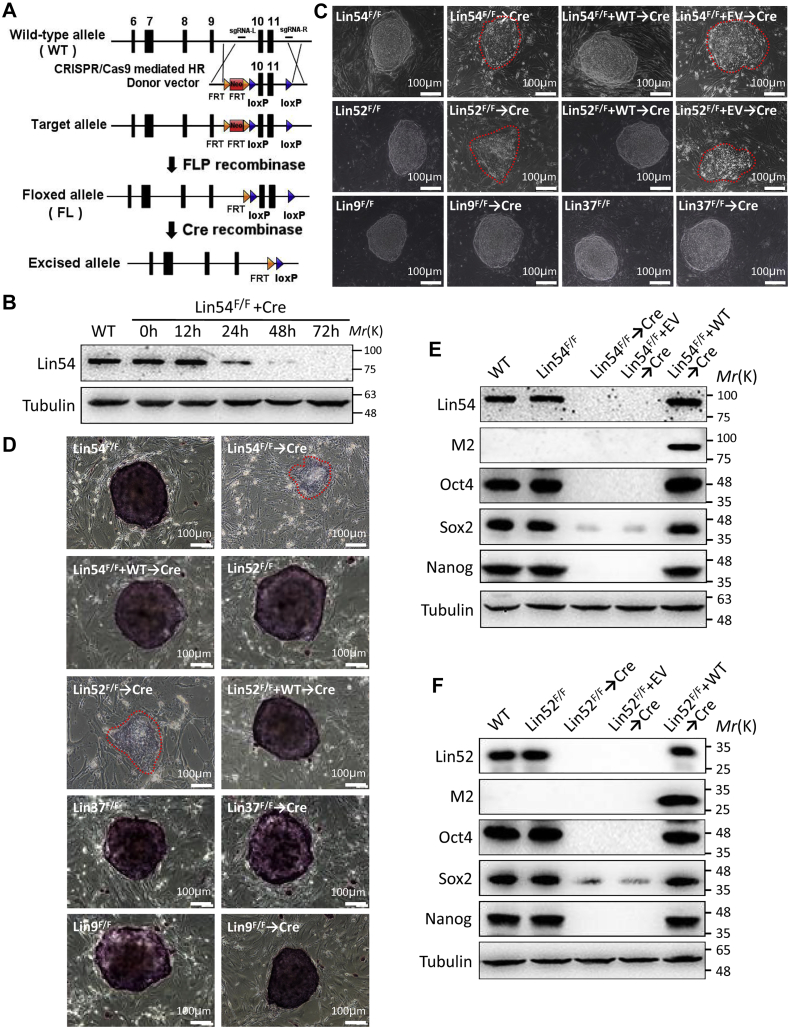
To systematically assess the role for MuvB core family in ESCs, we first determined the expression status of MuvB genes in ESCs. Each MuvB core member was expressed in ESCs assayed by Western blot analysis (Fig. S1A). To circumvent the potential lethality associated with complete loss of MuvB members in ESCs, we generated ESCs with conditional knockout of the MuvB genes, in which the critical exons of both alleles were flanked by parallel loxP sites, based on Cre-loxP system (Figs. 1A and S1–S4). As studies in Rbbp4 knockout ESCs have been reported elsewhere (19), we decided to further examine the potential role of the remaining members of MuvB complex in maintaining ESC identity in this study. In these conditional floxed ESC lines (Lin54F/F, Lin52F/F, Lin9F/F, and Lin37F/F), Cre-mediated ablation of the floxed genes was substantially achieved by 48 h after transfection with a plasmid encoding Cre recombinase, with their proteins being essentially absent by 72 h after transfection, as evident in Western blot (Figs. 1B, S2D, S3D and S4D). Therefore, unless otherwise stated, the cells were used for the experiments after 72 h of transfection throughout this study. These mutant cells were first monitored for their ability to form colonies from single cells after seeding on mitotically inactivated murine embryonic fibroblast (MEF) feeder layer (Fig. 1C). ESCs harboring the floxed alleles showed normal protein levels and formed tightly packed domed colonies that were morphologically indistinguishable from wild-type controls (Figs. 1, B and C, S2D, S3D and S4D). However, in contrast to their floxed cells, ablation of Lin54 (Lin54Δ/Δ) or Lin52 (Lin52Δ/Δ) resulted in flat and spreading colonies with irregular edges (Fig. 1C). To determine whether the observed growth defects coincided with exit from pluripotency and spontaneous differentiation, we performed alkaline phosphatase (AP) staining (Fig. 1D). The floxed as well as wild-type ESC colonies were stained uniform and bright, while Lin54Δ/Δ or Lin52Δ/Δ colonies appeared very weak or negative, suggesting loss of self-renewal ability and spontaneous differentiation (Fig. 1D). Consistent with the AP staining results, gene expression of pluripotency markers Oct4, Sox2, and Nanog was enormously reduced upon loss of Lin54 or Lin52 gene expression (Fig. 1, E and F). Importantly, lentivirus-mediated reintroduction of FLAG-tagged full-length Lin54 or Lin52 fully restored all these defective phenotypes observed in these mutants (Fig. 1, C–F). Unexpectedly, ESCs deficient for either Lin9 or Lin37 were viable and maintained a typical undifferentiated state as characterized by tightly packed morphology (Fig. 1C), AP staining (Fig. 1D), high levels of the core pluripotency-associated transcription factors Oct4, Sox2, and Nanog even after long-term culture (Figs. S3E and S4E). Pluripotency of Lin9Δ/Δ or Lin37Δ/Δ lines was confirmed by teratoma formation assay, which is often considered as the most stringent standard of pluripotency for ESCs. After 4 to 6 weeks of subcutaneous injection of Lin9Δ/Δ or Lin37Δ/Δ ESCs into immunodeficient mice, teratomas containing tissue or structures derived from all three embryonic germ layers were observed, indicating that these cells retained pluripotency (Fig. S5). Together, these results suggest that inactivation of MuvB core components Lin54 and Lin52 is sufficient to cause loss of self-renewal and pluripotency of ESCs.
Figure 1.
Ablation of the MuvB components Lin54 and Lin52 causes exit from pluripotency in ESCs.A, schematic overview of the generation for Lin54 conditional inactivation via CRISPR/Cas9-mediated gene targeting in ESCs. B, Western blot revealing the loss of Lin54 protein in ESCs after Cre recombinase expression. Tubulin was used as a loading control. C, morphological images of ESC colonies of indicated genotypes. ESC colonies were photographed at day 7 after seeding single-cell suspensions on MEF. Images were taken at 100× magnification. Red circles represent the borders of ESC colonies. D, shown were alkaline phosphatase staining images of indicated ESC colonies. Scale bar, 100 μm. E and F, Western blot demonstrating the expression levels of Lin54, Lin52, M2, Sox2, Oct4 and Nanog in ESCs of indicated genotypes. Tubulin served as a loading control. Results shown in (B–F) are representative of three independent experiments performed with three different clones of each genotype. ESC, embryonic stem cell; HR, homologous recombination; MuvB, multivulva class B; WT, wild-type.
Inactivation of MuvB genes Lin54 and Lin52 leads to G2/M arrest in cell cycle
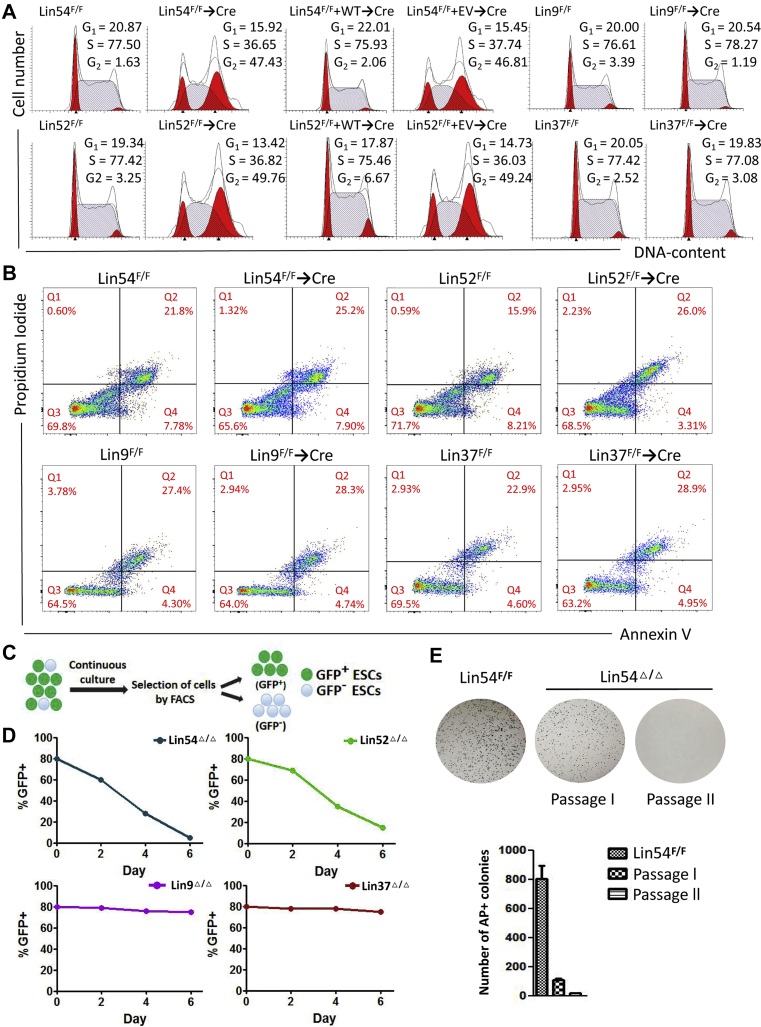
The severe growth defects of Lin54Δ/Δ or Lin52Δ/Δ ESCs might be due to cell cycle redistribution and/or an increase in apoptosis. To exploit these possibilities, we compared cell cycle profile and survival in Lin54Δ/Δ or Lin52Δ/Δ ESCs and their corresponding controls. Cell-cycle analysis indicated that Lin54Δ/Δ or Lin52Δ/Δ ESCs had a remarkably extended G2/M phase and a shortened S phase (Fig. 2A). The alteration of cell cycle profile in Lin54-and Lin52-deficient cells was almost completely rescued by ectopic expression of Lin54 and Lin52, respectively. In contrast, the loss of either Lin9 or Lin37 had no effect on cell cycle progression. Furthermore, Lin54Δ/Δ or Lin52Δ/Δ ESCs exhibited the same apoptotic rates as their floxed cells, as determined by annexin V staining (Fig. 2B), suggesting that the impaired growth of Lin54Δ/Δ or Lin52Δ/Δ cells was due to changed cell cycle progression. To get a further understanding of MuvB genes in mediating ESC self-renewal, we conducted a competition assay in which MuvB genes-deleted cells (GFP+) were mixed with their parental floxed ESCs (GFP−) at an 8:2 ratio, and the percentage of GFP(+) cells was determined over time by fluorescence-activated cell sorting (FACS) (Fig. 2C). Lin54Δ/Δ or Lin52Δ/Δ ESCs got rapidly eliminated, whereas cell survival of Lin9Δ/Δ or Lin37Δ/Δ ESCs was unaffected (Fig. 2D). Additionally, inactivation of Lin54 or Lin52 also almost abrogated secondary ESC colony formation (Fig. 2E). Together, these results indicate that inactivation of MuvB core components Lin54 and Lin52 is sufficient to trigger G2/M cell cycle arrest and loss of self-renewal of ESCs.
Figure 2.
Inactivation of MuvB genes Lin54 and Lin52 leads to G2/M arrest in cell cycle.A, cell cycle analysis of ESCs of indicated genotypes by flow cytometry. Representative histograms showing the cell cycle distribution in different phases of ESCs (G0/G1, bright red; S, light blue; G2/M, bright red). B, flow cytometric analysis of cell apoptosis with annexin V and propidium iodine (PI) staining. The percentages of four quadrants were labeled. C, schematic depicting the concept behind the GFP competition assay. ESCs deficient for MuvB genes (GFP+) were mixed with their parental Floxed ESCs (GFP−) at a 8:2 ratio. The mixed cells were cultured for 6 days and percentage of GFP in the population was analyzed by FACS analyzer. D, % GFP ratio measured at day 2, 4, and 6 by FACS. E, secondary ESC colony-forming assay. Bar chart represents the number of AP-positive ESC colonies. Three independent experiments with three different clones of each genotype were conducted with similar results to those shown in (A–E). ESC, embryonic stem cell; FACS, fluorescence-activated cell sorting; MuvB, multivulva class B.
Disruption of MuvB complex triggers activation of lineage-specific genes
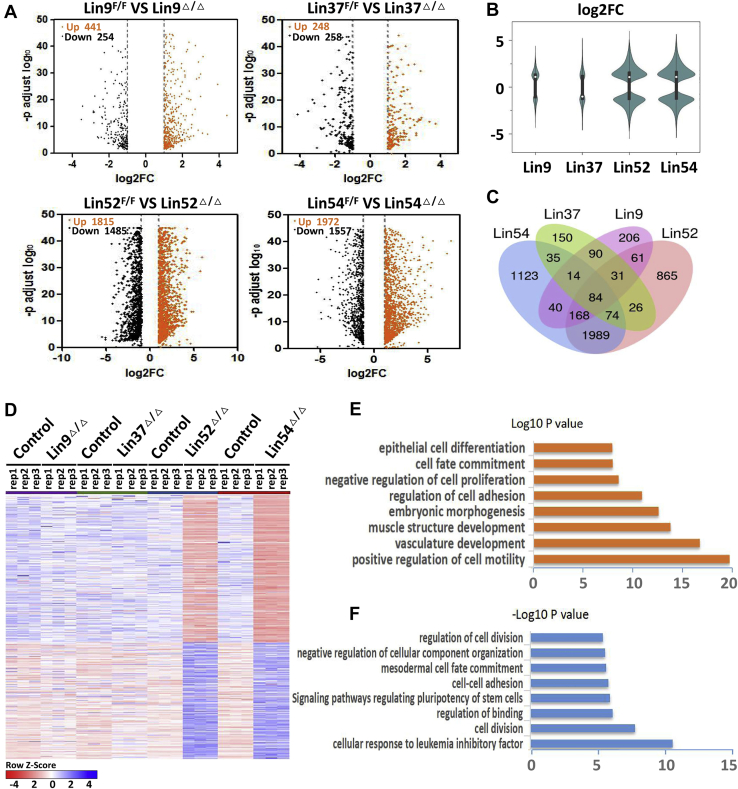
To gain insight into the molecular mechanisms underlying the observed phenotypic alterations associated with MuvB inactivation, we performed RNA-seq analysis in ESCs deficient in the MuvB components (Tables S1–S4). Complete loss of Lin54 and Lin52 in ESCs resulted in 3529 (1972 up and 1557 down) and 3300 (1815 up and 1485 down) differentially expressed genes, respectively, compared with their corresponding controls. In contrast, only a total of 695 (441 up and 254 down) and 506 (248 up and 258 down) genes were deregulated in Lin9 and Lin37 single mutants, respectively, further supporting a minor role of Lin9 and Lin37 in maintaining ESC identity (Fig. 3, A and B). As expected, a large number of the genes deregulated in Lin54Δ/Δ were also observed in ESCs deficient for Lin52 and vice versa, consistent with the similar phenotypes of the Lin54 and Lin52 null mutants (Fig. 3, C and D). As expected, a substantial number of the genes deregulated in Lin9Δ/Δ and Lin37Δ/Δ cells were also observed in ESCs deficient for Lin54 or Lin52, consistent with the notion that they exist as components of the same complex. Gene ontology (GO) analysis showed that among genes upregulated in Lin54Δ/Δ and Lin52Δ/Δ ESCs were genes primarily linked to epithelial cell differentiation, cell fate commitment, negative regulation of cell proliferation, and embryonic morphogenesis (Fig. 3E). In contrast, processes related to regulation of cell division, mesodermal cell fate commitment, and cell–cell adhesion were overrepresented among the genes downregulated in Lin54Δ/Δ and Lin52Δ/Δ cells (Fig. 3F).
Figure 3.
MuvB complex preserves transcriptional identity in ESCs.A, volcano plots showing the differentially expressed genes with 2-fold changes in ESCs of indicated genotypes. Orange and black points represent up-regulated and down-regulated genes respectively. RNA was harvested from Lin9F/F, Lin37F/F, Lin52F/F and Lin54F/F cells after transfecting with Cre recombinase for 72 h. B, a violin plot showing the comparison of differentially expressed genes in ESCs deficient for Lin9, Lin37, Lin52 or Lin54. C, Venn diagram showing the significant overlapping of differentially expressed genes in ESCs of indicated genotypes. D, heatmap showing the RNA expression of twofold changes in ESCs of indicated genotypes. E and F, gene ontology enrichment analysis of the overlapped differentially expressed genes in ESCs deficient for Lin54 or Lin52. Orange (E) and blue (F) histograms depict upregulated and downregulated genes respectively. ESC, embryonic stem cell; FC, fold change; MuvB, multivulva class B.
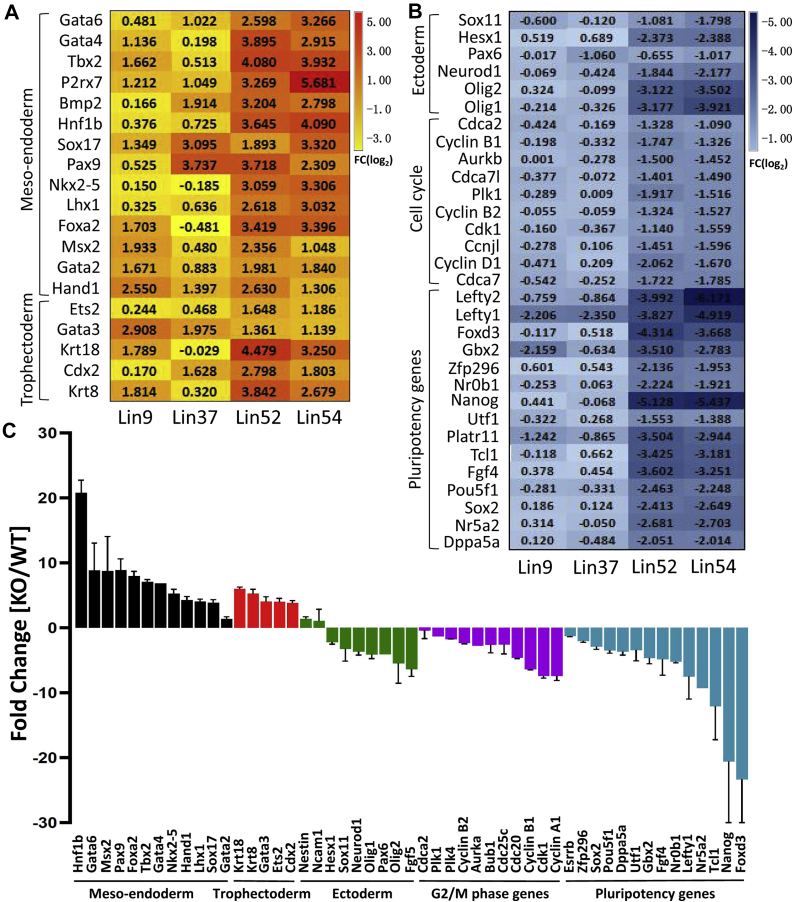
Importantly, among the genes downregulated in Lin54Δ/Δ and Lin52Δ/Δ cells, our analysis revealed the pluripotency genes, including Nanog, Oct4 (Pou5f1), Sox2, Nr5a2, and Dppa5a (Fig. 4, A and B), which have been demonstrated to have critical roles in maintaining pluripotency in ESCs. Additionally, RNA-Seq analysis demonstrated that loss of Lin54 or Lin52 led to the upregulation of mesoendoderm markers (Gata6, Gata4, Tbx2, Sox17, Foxa2, Msx2, and Hand1) and trophectoderm markers (Ets2, Gata3, and Krt18, Cdx2, and Krt8), whereas ectoderm markers (Sox11, Pax6, Neurod1, Olig1, and Olig2) were instead either unchanged or slightly reduced (Fig. 4, A and B). By performing reverse transcription followed by qPCR (RT-qPCR), we confirmed the differential expression of selected genes observed by RNA-seq analysis and observed that Lin54Δ/Δ ESCs displayed downregulation and upregulation in expression of core pluripotency factors and key mesoendodermal lineage identity genes, respectively (Fig. 4C). Consistent with the cell cycle analysis (Fig. 2A), we found a substantial number of genes involved in cell cycle control to be differentially expressed in both Lin54Δ/Δ and Lin52Δ/Δ cells. These include Cyclin B1 (CCNB1) and Cdk1, crucial regulators of the G2/M phase of the cell cycle (Fig. 4, B and C). Together, these results suggest that Lin54 and Lin52 normally function to maintain ESCs in an undifferentiated state by repressing differentiation toward mesoendoderm lineages.
Figure 4.
Disruption of MuvB complex triggers activation of lineage-specific genes.A and B, heatmap showing the expression levels of genes involved in germ layer specification, pluripotency maintenance and cell cycle progression in ESCs individually deficient for Lin9, Lin37, Lin52, or Lin54. C, RNA-Seq data were verified by RT-qPCR. RNA was obtained from Lin54F/F ESCs 3 days after Cre transfection. The RT-qPCR data of selected genes were normalized to β-actin. Error bars denote standard deviation of triplicate data, n = 3. ESC, embryonic stem cell; MuvB, multivulva class B.
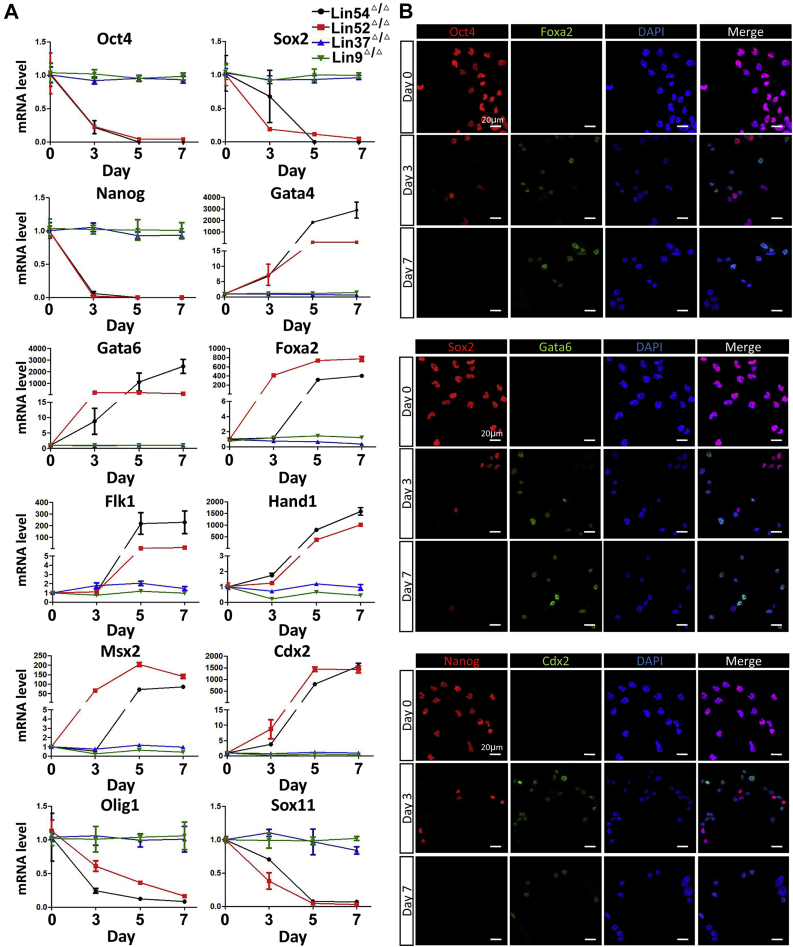
To rigorously evaluate the importance of transcriptional mediators responsible for loss of pluripotency and lineage commitment upon Lin54 or Lin52 deficiency, we assessed the temporal mRNA expression changes of pluripotency genes as well as three germ layer-specific genes in Lin54F/F or Lin52F/F ESCs after transfection with Cre recombinase plasmid. RT-qPCR analysis indicated that the mRNA levels of Oct4, Nanog, and Sox2 were significantly reduced within 3 days after Cre transfection and declined steadily thereafter to baseline levels on day 7 (Fig. 5A). The loss of expression of key pluripotency markers was accompanied by the aberrant activation of lineage specific genes. Markers for mesoendoderm (Gata4, Gata6, Foxa2, Hand1, Msx2, and Flk1) and trophectoderm (Cdx2) were upregulated. These findings were corroborated by immunofluorescence (IF) microscopy, which demonstrated that loss of either Lin54 or Lin52 led to precocious differentiation of ESCs and activation of a subset of lineage-specific genes (Figs. 5B and S6). In Lin54F/F and Lin52F/F ESCs, the expression of pluripotency markers (Oct4, Sox2, and Nanog) was high, whereas positivity for different lineage-associated markers (Foxa2, Gata6, and Cdx2) was rarely observed, indicating the undifferentiated state of the cells. After transient transfection of plasmid DNA expressing Cre, the expression of these lineage-associated markers gradually increased in a time-dependent manner, which was accompanied by the loss of expression of pluripotency markers Oct4, Sox2, and Nanog. In accordance with the Western blot results (Figs. S3E and S4E), our IF analysis also supports the idea that Lin9Δ/Δ and Lin37Δ/Δ cells retained their high expression of pluripotency factors without displaying any obvious differentiation toward three germ layer lineages (Fig. S6).
Figure 5.
Lin54 deficiency results in dynamic changes in gene expression characteristic of spontaneous differentiation.A, time-course analysis of selected pluripotency and lineage-specific transcripts in Lin54F/F ESCs after Cre recombinase expression. Error bars mean ± SD (n = 3). B, immunofluorescence showing the expression of pluripotency-associated genes (Oct4, Sox2, and Nanog) and lineage-specific markers (Foxa2, Gata6, and Cdx2) in Lin54F/F ESCs following Cre recombinase expression. Images were taken at 63× magnification using confocal microscopy. Results shown in (A) and (B) are representative of three independent experiments performed with three independent Lin54F/F clones. ESC, embryonic stem cell.
CXC domains are required for binding of Lin54 to the promoter of its target genes
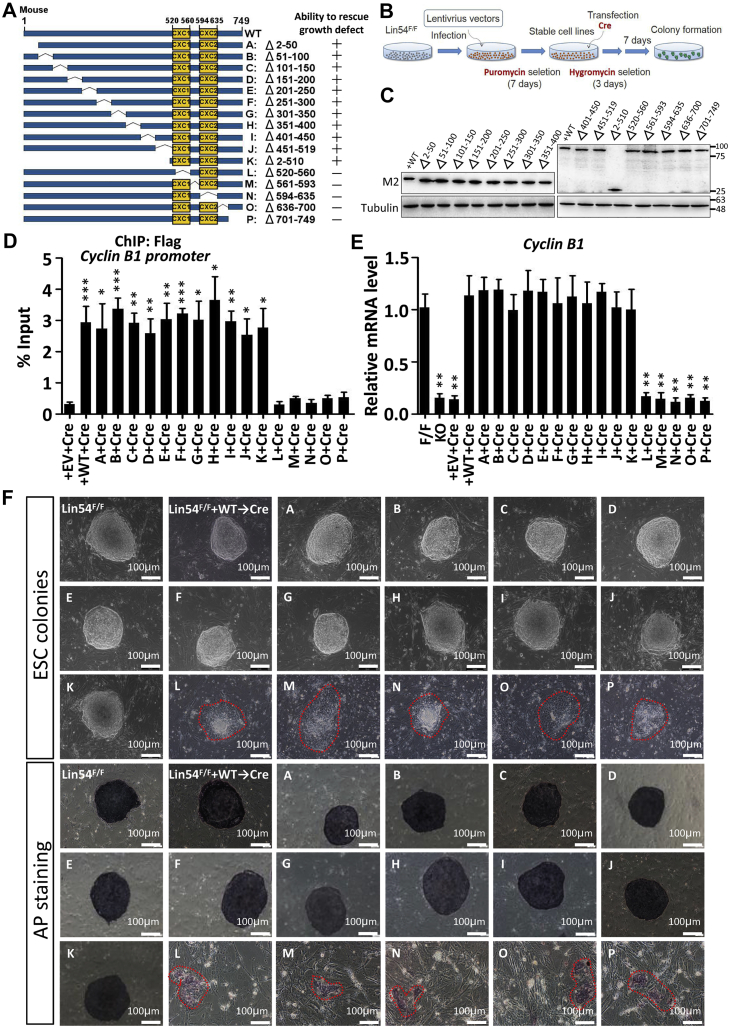
As mentioned above, all of the cellular phenotypes observed in Lin54Δ/Δ cells were fully rescued by reexpression of wild-type full-length Lin54 (Fig. 1, C–F), thus providing us with an excellent opportunity to study Lin54 structure and function in ESCs. Lin54 contains two tandem CXC domains, each consisting of nine cysteines that enable its DNA-binding function (16, 18, 20), separated by a short spacer called hinge. Next, we performed a structure–function analysis to map the regions within the Lin54 protein that are responsible for its chromatin tethering activity. The series of FLAG-tagged Lin54 deletion mutants shown in Figure 6A was constructed and introduced into Lin54F/F ESCs (Fig. 6B). Of note, all the mutants were overall expressed at similar levels as determined by Western blotting (Fig. 6C). To determine whether these mutants could keep the DNA-binding activities and to examine the capacity of these mutants to maintain the expression of cell cycle target genes in the absence of endogenous Lin54, the cells were transiently transfected with Cre recombinase and analyzed 12 days later. ChIP-qPCR analyses showed that the C-terminally deleted protein and the mutants with deletion of CXC domains (including the spacer connecting the two CXC domains) did not bind to the promoter of Cyclin B1 (Fig. 6D). Consistent with their failure to tightly associate with chromatin in ESCs, these mutants lost completely the ability to rescue target gene expression (Fig. 6E). In contrast, the N terminus of Lin54 was dispensable for DNA binding and thus retained these gene expressions as effectively as the wild-type. To gain insight into the capacity of these mutants to restore the self-renewal defect of Lin54-deficient ESCs, we tested their ability to form colonies after seeding single-cell suspensions onto inactivated MEF feeder layers. As shown in Figure 6F, a mutant lacking the first 519 amino acids completely rescued wild-type levels of self-renewal growth in ESCs, while Lin54 mutants in which the CXC domains or C-terminal regions are deleted abolished their ability to rescue the colony growth defect. Next, to examine whether the conserved cysteines in the two CXC domains were essential for binding to the Cyclin B1 promoter, the two conserved cysteine residues in the CXC domains (C525 and C613) were mutated to alanines (Fig. S7) (16, 20). We found that mutation of C525 and C613 abolished the binding of Lin54 to the Cyclin B1 promoter. Consistently, substitution of these two conserved residues also abolished the ability of Lin54 to rescue defects associated with Lin54 deficiency in ESCs (Fig. S7). Overall, these results suggest that both C-terminal and CXC domains are essential for the targeting of Lin54 to chromatin and show that these regions confer upon Lin54 the ability to maintain the identity of ESC.
Figure 6.
CXC domains are required for binding of Lin54 to the promoter of its target genes.A, a schematic diagram showing the structures of wild-type and deletion mutant Lin54 (left). Numbers represent the positions of amino acid residues of Lin54. The thin bent lines refer to deleted regions. A concise summary represents the ability of mutants to rescue growth defect upon Lin54 ablation (right). B, a schema of rescue assay. C, the expression levels of different Lin54 deletion mutants in Lin54Δ/Δ ESCs were examined by Western blot with anti-Flag antibody. Tubulin was used as a loading control. D, ChIP-qPCR analysis of Lin54 binding to Cyclin B1 promoter in wild-type and deletion mutants. Data were obtained from three independent experiments and analyzed using two-tailed Student’s t test by GraphPad Prime 5 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). E, RT-qPCR detecting Cyclin B1 expression level in ESCs of indicated genotypes. Data were normalized to β-actin. F, representative images of wild-type and Lin54 deletion mutant ESC colonies (top) and AP-staining (bottom) images of indicated ESC colonies. Scale bar is 100 μm. ESC, embryonic stem cell.
Ectopic expression of G2/M genes largely rescues the phenotypic defects caused by Lin54 deficiency
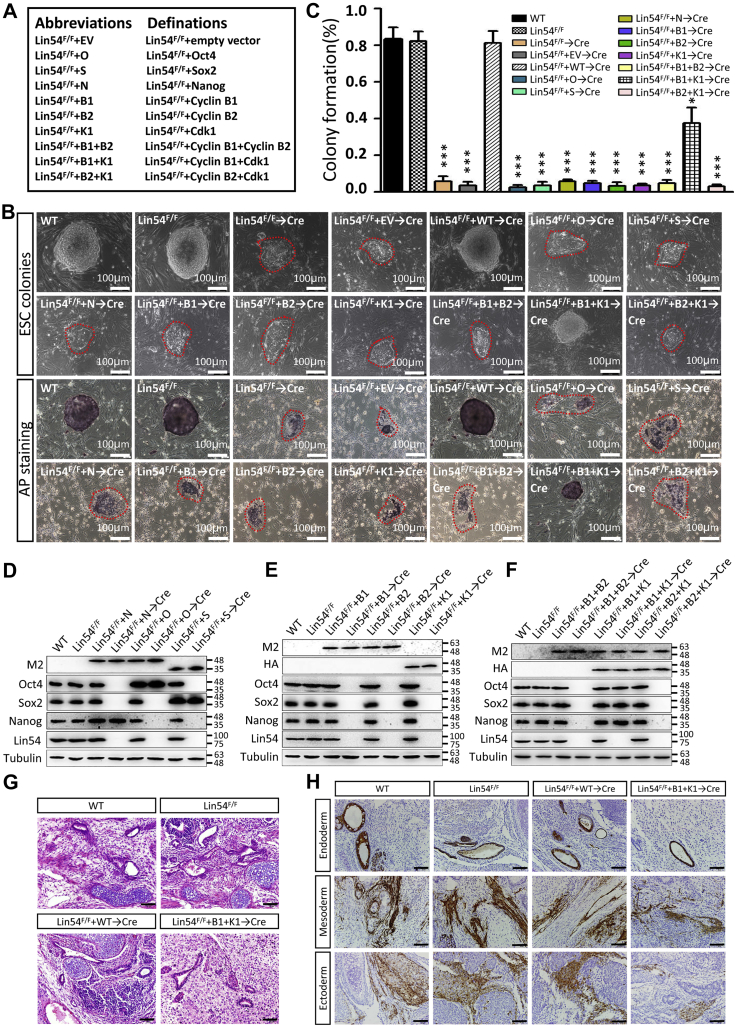
Because the inactivation of Lin54 in ESCs was accompanied by a coincidental decline in the mRNA and protein levels of Oct4, Sox2, and Nanog (Figs. 1E and 5, A and B and Table S1), we assessed whether Lin54 contributed to the maintenance of ESC property via regulation of these core pluripotency transcription factors. The Lin54-null rescue system utilized Lin54F/F ESCs, in which the floxed alleles were excised upon Cre-recombinase. Lentivirus-mediated expression of Oct4, Sox2, or Nanog did not restore the defects observed in Lin54Δ/Δ ESCs (Fig. 7, A–D), suggesting that keeping the requisite levels of Oct4, Sox2 and Nanog might not be responsible for Lin54-mediated ESC maintenance.
Figure 7.
Ectopic expression of G2/M genes largely rescues the phenotypic defects caused by Lin54 deficiency.A, a legend box containing reference cell lines. B, morphological images of ESC colonies of indicated genotypes without (top) and with AP staining (bottom). The scale bar represents 100 μm. C, bar graphs represent the percentages of isolated ESCs that grow into visible colonies. D, Lin54F/F ESCs were infected with lentiviruses encoding FLAG-tagged Oct4, Nanog, and Sox2, and anti-Flag M2 antibody was used to determine their protein levels. E and F, Lin54F/F ESCs were coinfected with lentiviruses encoding FLAG-tagged Cyclin B1 or Cyclin B2 and HA-tagged Cdk1 as indicated. The anti-Flag M2 and anti-HA antibodies were used to determine their protein levels. G, representative images of H&E staining of teratomas generated from indicated ESCs. The scale bar represents 50 μm. H, immunohistochemical analysis of three embryonic germ layer markers of teratomas. The scale bar represents 50 μm. ESC, embryonic stem cell.
Our above-noted data indicate that inactivation of Lin54 resulted in G2/M arrest and in transcriptional inhibition of G2/M genes. One prediction from these observations is that Lin54 preserves the undifferentiated state of ESCs through regulation of these G2/M genes.
To test this possibility, we attempted to rescue Lin54Δ/Δ ESCs by introducing the transgenes of Cyclin B1, Cyclin B2, or Cdk1. Introducing Cyclin B1, Cyclin B2, or Cdk1 did not restore the self-renewal defect observed in Lin54Δ/Δ ESCs. In contrast, ectopic expression of Cdk1 together with Cyclin B1, but not Cyclin B2, partially rescued the proliferation defect of Lin54-deficient ESCs (Fig. 7, A–C), indicating that Lin54 controls ESC self-renewal through a Cyclin B1/Cdk1-dependent mechanism. The rescued cells were able to form viable colonies on feeder cells and could be continuously propagated (for at least 30 passages) without any significant change in colony morphology. Although these colonies were much smaller than usual, they maintained an undifferentiated state as characterized by their morphology, staining for AP, expression of the pluripotency markers, Oct4, Sox2, and Nanog (Fig. 7, B–F). Consistent with these results, these rescued cells formed teratoma when injected subcutaneously into nude mice. In histologic analysis, Cyclin B1/Cdk1 teratomas featured a paucity of mature elements (Fig. 7G). Yet examples of differentiation into tissues from all three germ layers could be found. Immunohistochemistry of wild-type ESC-derived teratomas with antibody specific to α-smooth muscle actin (SMA; mesoderm marker), TROMA1 (endoderm), and GFAP (ectoderm) revealed large numbers of positive cells, whereas teratomas derived from Cyclin B1/Cdk1 ESC had decreases in all three germ layer derivatives (Fig. 7H). Therefore, our data suggest a critical role for Lin54 in the transcriptional regulation of G2/M genes that are essential for cell cycle progression and maintenance of pluripotency in ESCs.
Discussion
The nature of the relationship between the cell cycle machinery and pluripotency of ESCs has remained largely elusive, although it has been proposed that pluripotency and self-renewal are intimately linked to cell cycle control in ESCs (4, 21). Lin52, Lin54, Lin9, Lin37, and Rbbp4 are integral components of the mammalian MuvB complex, which is a master regulator of cell-cycle-dependent gene expression (10). Multiple studies have indicated that they have important roles in MuvB function (7, 8, 9, 10, 14). However, their precise roles in transcription of mitotic genes and ESC self-renewal and maintenance have not been clearly defined. Here, by using the CRISPR/Cas9 system for generating loss-of-function mutants in ESCs, we have systematically examined the role of MuvB components in cell cycle progression and cell fate decisions. We show that Lin52, Lin54, Lin9, Lin37 of MuvB were differentially required for ESC identity maintenance. Specifically, we find that ablation of Lin52 or Lin54 resulted in aberrant activation of genes encoding key lineage-specific regulators and triggered loss of self-renewal and spontaneous differentiation. In contrast, loss of Lin9 or Lin37 does not substantially perturb gene expression and self-renewal capacity. Our observation is generally consistent with previous biochemical data suggesting the critical roles of Lin54 and Lin52 in recognition of CHR DNA elements in cell-cycle-regulated promoters and facilitating proper assembly of DREAM and MMB complexes, respectively (13, 16, 18, 22). Of note, our previous study has indicated that Rbbp4 is also essential for ESC self-renewal and pluripotency, consistent with the finding that Rbbp4 is a histone-binding protein (19, 23). Interestingly, germline knockout mouse model with loss of Lin9 has been characterized, revealing an essential role of the MuvB core in early embryonic development (15). Furthermore, knockdown of Lin9 by RNAi in human fibroblasts or F9 embryonal carcinoma cells severely impairs proliferation and delays progression G2 phase to mitosis (14, 24), suggesting that MuvB-mediated cell cycle control appears to be executed in a cell-type-specific manner. Notably, consistent with our finding in ESCs, human cells deprived of Lin54 grew significantly slower than control cells, accumulating in the G2/M phase (16, 25).
The potential interplay between cell cycle control and cell fate decision has remained largely elusive due to the lack of appropriate tools for exploring cell cycle regulation in pluripotent cells. Of particular interest, it has been reported that human ESCs (hESCs) in early G1 phase can only initiate differentiation into endodermal lineage, whereas these cells in late G1 are only permissive for neuroectoderm differentiation (3). Further experiments reveal that the divergent ability of differentiation is governed by Cyclin D1–3/CDK4–6, which are expressed during the late G1 phase (3). Similarly, we have recently shown that deficiency of Rbbp4 in ESCs causes a prolonged G1 phase of the cell cycle, which is accompanied by spontaneous differentiation toward mesendodermal lineages (19). Intriguingly, the importance of the G2/M phase of the cell cycle in ESC self-renewal and pluripotency is by far less explored. Our study remedies this shortfall by uncovering that the G2/M transition could also have a pivotal function in the mechanisms governing ESC differentiation. We found that ablation of either Lin54 or Lin52, but not Lin9 or Lin37, is associated with a number of cell-cycle-relevant phenotypes including reduced proliferation, accumulation of cells at G2/M stage consistent with the decreased expression of G2/M genes in Lin54 and Lin52 null cells. Importantly, the resulting defects of Lin54 ablation were largely rescued by ectopic coexpression of G2/M genes Cyclin B1 and Cdk1. Ongoing efforts are focused on defining the mechanisms by which members of the MuvB complex cooperate to mediate transcriptional activation of their target genes, including the G2/M genes. Nevertheless, our new discoveries place the MuvB complex at the forefront of cell fate determination in ESCs, paving the way for more detailed mechanistic understanding of the coordination of the cell cycle with cell identity specification.
Overall, we have demonstrated that self-renewal and pluripotency in ESCs depend upon MuvB-mediated cell cycle progression. Our observations imply that simple manipulation of the cell cycle using small molecules is a promising strategy for directing differentiation of ESCs toward specific cell types of interest. Collectively, we provide strong evidence that the cell cycle progression and the maintenance of pluripotency are tightly connected to safeguard ESC from premature differentiation.
Experimental procedures
Cell culture
Mouse ESCs, 293FT cells, and MEFs were used in this study. ESCs were cultured in DMEM (Gibco) with 15% fetal calf serum (Gibco), 1:100 nonessential amino acids (Gibico), 1000 U/ml leukemia inhibitory factor (LIF), 2 mM L-glutamine (Invitrogen), 0.5 mM β-mercaptoethanol (Sigma), and penicillin/streptomycin (Thermo Fisher) on 0.1% gelatin-treated culture dishes at 37 °C and 5% CO2. MEFs and 293FT cells were cultured in DMEM with 10% fetal calf serum and penicillin/streptomycin as previously described (26, 27).
CRISPR/Cas9-mediated genomic deletion
The sgRNA sequences of interested genes were designed on http://crispor.tefor.net/. Subsequently, the fragments were cloned into BbsI-linearized pSpCas9 (BB)-2A-Puro vectors (Addgene plasmid #62988; http://n2t.net/) as per previous description (27, 28). To establish conditional knockout ESCs, SacII-digested targeting vectors with loxP sequence and Cas9-vectors with sgRNA sequence were transfected into ESCs using Lipofectamine 2000 (Invitrogen). ESCs were selected with puromycin (Sigma, finial concentration of 2 μg/ml) 24 h after transfection. Residual ESCs were seeded on mitomycin-irradiated MEFs 48 h after selection. Individual clones were picked after seeding ESCs on MEFs for 7 days. ESCs were genotyped after cultured for 5 days. The primer sequences were listed in Table S5.
Alkaline phosphatase (AP) staining
ESCs were washed with PBS twice and fixed with 4% paraformaldehyde for 2 min. Subsequently, fixed cells were washed with TBST twice and stained with Alkaline Phosphatase Stain Kit (Yeasen #40749ES60) according to the manufacturer’s instructions. The ESC colonies were photographed with microscope (Olympus IX73). Images were taken at 100 × magnification.
Cell cycle and apoptosis analysis
Cell cycle analysis was performed as described (27). Cell cycle analysis kit (Vazyme A411-01/02) was used in this study. Briefly, ESCs were trypsinized, washed twice with cold PBS, and fixed in ice-cold 70% ethanol at 4 °C overnight. Then, fixed cells were washed twice with cold PBS, incubated with ribonuclease A (RNase A) (20 mg/ml) for 30 min at 37 °C, and stained with propidium iodide (PI) for 30 min at 4 °C. Following analysis was performed with an LSRFortessa flow cytometer equipped with Cell Quest software (BD Biosciences). The data were analyzed using Modfit software. Cell apoptosis detection was performed according to the manufacturer’s instructions (Yeason #40302ES60). Briefly, 2 × 106 ESCs were digested with trypsin without EDTA and washed twice with cold PBS. Then, ESCs were incubated in 100 μl binding buffer containing 5 μl Annexin V-FITC and 10 μl PI for 10 min at 4 °C and detected using an LSRFortessa flow cytometer (BD Biosciences) as described (27). The data were analyzed using FlowJo software.
Plasmid construction
Full-length Lin54, Lin52, Lin37, and Lin9 cDNA were amplified from ESCs using Phanta Max Super-Fidelity DNA Polymerase (Vazyme #P505-d1) with primers carrying the Flag sequence (DYKDDDDK) and inserted into lentivirus vectors using ClonExpress II One Step Cloning Kit (Vazyme #C112-01). The sequences of Lin54 mutants were generated by PCR using Quick-change II site-directed mutagenesis kit (Aligent) and confirmed by DNA sequencing. All the verified plasmids were transfected into ESCs.
Lentiviral supernatant production and infection
Lentiviral supernatant was prepared as described (26, 27). Briefly, packaging cells (293FT) were seeded on 6 cm dish. For transfection, 3 μg insert plasmid along with 0.3 μg VSVG-plasmid, 0.15 μg gag/pol-plasmid, 0.15 μg tat-plasmid, and 0.15 μg rev-plasmid were mixed in 150 μl DMEM, and Lipofectamine 2000 was added into 150 μl DMEM for 5 min at room temperature. The transfection admixture was incubated for 15 min at room temperature and added to packaging cells. Lentiviral supernatant was harvested 48 h after transfection and concentrated by ultracentrifugation for 2 h at 4 °C. For infection, concentrated lentiviral supernatant was added to ESCs in the presence of polybrene (Sigma, final concentration of 8 μg/ml). Puromycin was used to select ESCs 24 h after infection.
Quantitative real-time PCR
To isolate total RNA (2 μg) from cells, the cells were harvested and resuspended in Trizol reagent (Invitrogen) as previous description (25). Reverse transcription was performed with 2 μg total RNA using HiScriptTM first Strand cDNA Synthesis Kit (Vazyme #R111-01). Quantitative PCR was performed using PowerUp TM SYBR Green Master Mix (Vazyme) on a StepOne TM Software v2.3 (Applied Biosystems) according to the manufacturer's protocol. The RT-qPCR primer sequences were listed in Table S6. The mRNA expression was normalized by β-actin.
Teratoma formation and analysis
Teratoma formation was performed as previous description (26, 27). Briefly, ESCs were trypsinized, washed twice by PBS, resuspended in PBS, and injected subcutaneously into the backside of 6-week-old immunodeficient mice. Teratomas were dissected from mice 3 weeks after injection. For histological analysis, teratomas were fixed in 4% paraformaldehyde for 6 h at 4 °C, dehydrated by ethanol, and embedded in paraffin. Paraffin-embedded tissues were sliced to 5 μm thickness slides on microslides. Subsequently, the paraffin sections were fixed at 37 °C overnight. Paraffin sections were used for H&E staining and immunohistochemistry. For immunohistochemistry analysis, the experiments were performed according to the manufacturer's instructions (UltraSensitive SP IHC Kit). The antibodies used in this study were listed in Table S7. All experimental procedures about animals were approved by Institutional Animal Care and Use Committee (IACUC) of the Model Animal Research Center of Nanjing University.
Immunofluorescence
Immunofluorescence was performed as previously described (27). Briefly, ESCs were seeded on gelatin coated coverslips, washed twice by PBS, and fixed by 4% paraformaldehyde for 15 min at room temperature. Then, ESCs were incubated in 0.5% Triton X-100 for 20 min at room temperature, washed twice by PBS, and permeabilized with 1% BSA for 30 min at room temperature. ESCs were incubated with primary antibodies in 1% BSA at 4 °C overnight. Subsequently, samples were rinsed twice with PBST and incubated with secondary antibodies for 1 h at room temperature. Then, coverslips were incubated with DAPI. The antibodies used were listed in Table S7. Finally, ESCs were mounted with glycerinum and imaged by Zeiss LSM880 laser scanning confocal microscope at 63 × magnification.
Western blot
Western blot was performed as described (26). Briefly, cells were collected, lysed in RIPA buffer [50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% TtitonX-100, 1 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, phenylmethylsulfonyl fluoride (10 mg/ml), 1 mM dithiothreitol, 100 mM NaF, and protease inhibitors] for 30 min on ice and centrifuged at 12,000 rpm, 4 °C for 20 min. Then, supernatant was mixed with loading buffer, boiled at 95 °C for 5 min, and subjected to Western blot analysis. The blots were incubated with primary antibodies at 4 °C overnight with rotation. Subsequently, blots were permeabilized with horseradish-peroxidase-marked secondary antibodies for 2 h at room temperature. The antibodies were listed in Table S7. Finally, enhanced ECL Chemiluminescence Test Kit (Vazyme #E412-01) was used to detect signal.
RNA-seq analysis
For RNA-Seq analysis, ESCs were harvested and suspended in Trizol (Invitrogen) according to the manufacturer’s instructions. DNase I (Takara) was used to remove genomic DNA. RNA isolation, purification, quality testing, and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology as per previous description (29). Samples were from three independent biological replicates. For RNA-Seq data analysis, genes with twofold changes and p value <0.05 were considered as differentially expressed.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as per previous description (26, 27). Briefly, ESCs were trypsinized and harvested. Cross-linking was performed with 37% formaldehyde (Sigma) to a working concentration of 1% for 10 min and stopped with 2.5 M glycine to the finial concentration of 125 mM for 5 min at room temperature by gentle shaking. ESCs were washed twice with precooling PBS and lysed in lysis buffer [1% SDS, 50 mM Tris (pH8.0), 10 mM EDTA, protease inhibitors] for 30 min on ice. Samples were interrupted by sonication using Bioruptor Sonication System (Diagenode). The average size of sheared DNA fragments is 250 ∼ 500 bp. After sonication, 100 μl sonicate lysate was removed to a new microtube and served as “input.” The sonicated chromatin samples were incubated with anti-Flag M2 antibody in dilution buffer [0.01% SDS, 16.7 mM Tris (pH 8.0), 1% Triton-X 100, 165 mM NaCl, 1.2 mM EDTA, protease inhibitors] at 4 °C overnight under rotation. Subsequently, immunoprecipitated samples were centrifuged for 10 min at 2000g, and the supernatant was removed. The immune complexes were washed with low-salt buffer (twice) [0.1% SDS, 20 mM Tris (pH 8.0), 1% Triton-X 100, 150 mM NaCl, 1 mM EDTA, protease inhibitors], high-salt buffer (twice) [0.1% SDS, 20 mM Tris (pH 8.0), 1% Triton-X 100, 500 mM NaCl, 1 mM EDTA, protease inhibitors], and TE buffer (twice) [0.25 mM EDTA, 10 mM Tris (pH 8.0), and protease inhibitors]. Subsequently, the immune complexes were eluted in elution buffer (twice) (1% SDS, 100 mM NaHCO3) at 65 °C. To reverse cross-linking, RNase (0.2 mg/ml) and 5 M NaCl (0.2 ∼ 0.3 M working concentration) were mixed with elution buffer. The immunoprecipitated DNA was purified with DNA gel extraction kit (Axygen). ChIP enrichment analysis was performed by qPCR and data were normalized to input. The primer sequences for ChIP-qPCR were listed on Table S6.
Data availability
RNA-Seq results are available on NCBI Gene Expression Omnibus under accession GSE186139. All original data pertaining to this study will be made available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are indebted to all members of the Qin laboratory and to Yikai Huang and Ting Su for experimental advice and helpful discussions.
Author contributions
J. Q. and C. W. conceptualization; C. W. data curation; J. Q. and C. W. formal analysis; C. W. and J. Q. investigation; C. W., J. W., L. Z., L. X., K. H., and L. D. methodology; C. W. and K. H. visualization; J. Q. and C. W. writing–original draft; J. Q., C. W., Y. X., and Q. J. writing– review and editing.
Funding and additional information
This work was supported by grants from the National Natural Science Foundation of China (31970810) to J. Q.
Edited by Qi Qun Tang
Supporting information
References
- 1.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Gifford D.K., Melton D.A., Jaenisch R., Young R.A. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronado D., Godet M., Bourillot P.Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R., Anastassiadis K., Savatier P. A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res. 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 5.White J., Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 6.Orford K.W., Scadden D.T. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 7.Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Pilkinton M., Sandoval R., Colamonici O.R. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene. 2007;26:7535–7543. doi: 10.1038/sj.onc.1210562. [DOI] [PubMed] [Google Scholar]
- 9.Sadasivam S., Duan S., DeCaprio J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walston H., Iness A.N., Litovchick L. DREAM on: Cell cycle control in development and disease. Annu. Rev. Genet. 2021;55:309–329. doi: 10.1146/annurev-genet-071819-103836. [DOI] [PubMed] [Google Scholar]
- 11.Lewis P.W., Beall E.L., Fleischer T.C., Georlette D., Link A.J., Botchan M.R. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korenjak M., Taylor-Harding B., Binné U.K., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 13.Litovchick L., Florens L.A., Swanson S.K., Washburn M.P., DeCaprio J.A. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–813. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterloh L., von Eyss B., Schmit F., Rein L., Hübner D., Samans B., Hauser S., Gaubatz S. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007;26:144–157. doi: 10.1038/sj.emboj.7601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reichert N., Wurster S., Ulrich T., Schmitt K., Hauser S., Probst L., Götz R., Ceteci F., Moll R., Rapp U., Gaubatz S. Lin9, a subunit of the mammalian DREAM complex, is essential for embryonic development, for survival of adult mice, and for tumor suppression. Mol. Cell. Biol. 2010;30:2896–2908. doi: 10.1128/MCB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmit F., Cremer S., Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276:5703–5716. doi: 10.1111/j.1742-4658.2009.07261.x. [DOI] [PubMed] [Google Scholar]
- 17.Müller G.A., Wintsche A., Stangner K., Prohaska S.J., Stadler P.F., Engeland K. The CHR site: Definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42:10331–10350. doi: 10.1093/nar/gku696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marceau A.H., Felthousen J.G., Goetsch P.D., Iness A.N., Lee H.W., Tripathi S.M., Strome S., Litovchick L., Rubin S.M. Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters. Nat. Commun. 2016;7:12301. doi: 10.1038/ncomms12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y., Su T., Wang C., Dong L., Liu S., Zhu Y., Hao K., Xia Y., Jiang Q., Qin J. Rbbp4 suppresses premature differentiation of embryonic stem cells. Stem Cell Rep. 2021;16:566–581. doi: 10.1016/j.stemcr.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuo T., Kuramoto H., Kumazaki T., Mitsui Y., Takahashi T. LIN54 harboring a mutation in CHC domain is localized to the cytoplasm and inhibits cell cycle progression. Cell Cycle. 2012;11:3227–3236. doi: 10.4161/cc.21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz S., Panopoulos A.D., Herrerías A., Bissig K.D., Lutz M., Berggren W.T., Verma I.M., Izpisua Belmonte J.C. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiley K.Z., Iness A.N., Saini S., Tripathi S., Lipsick J.S., Litovchick L., Rubin S.M. Structural mechanism of Myb-MuvB assembly. Proc. Natl. Acad. Sci. U. S. A. 2018;115:10016–10021. doi: 10.1073/pnas.1808136115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuyama T., Dalal Y., Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight A.S., Notaridou M., Watson R.J. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28:1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- 25.Kittler R., Pelletier L., Heninger A.K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., Wagner J., Surendranath V., Richter C., Bowen W., Jackson A.L., et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 26.Qin J., Whyte W.A., Anderssen E., Apostolou E., Chen H.H., Akbarian S., Bronson R.T., Hochedlinger K., Ramaswamy S., Young R.A., Hock H. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–332. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin J., Wang C., Zhu Y., Su T., Dong L., Huang Y., Hao K. Mga safeguards embryonic stem cells from acquiring extraembryonic endoderm fates. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y., Zhao W., Wang C., Zhu Y., Liu M., Tong H., Xia Y., Jiang Q., Qin J. Combinatorial control of recruitment of a variant PRC1.6 complex in embryonic stem cells. Cell Rep. 2018;22:3032–3043. doi: 10.1016/j.celrep.2018.02.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq results are available on NCBI Gene Expression Omnibus under accession GSE186139. All original data pertaining to this study will be made available upon request.