Abstract
N-demethylases have been reported to remove the methyl groups on primary or secondary amines, which could further affect the properties and functions of biomacromolecules or chemical compounds; however, the substrate scope and the robustness of N-demethylases have not been systematically investigated. Here we report the recreation of natural evolution in key microdomains of the Thermomicrobium roseum sarcosine oxidase (TrSOX), an N-demethylase with marked stability (melting temperature over 100 °C) and enantioselectivity, for enhanced substrate scope and catalytic efficiency on -C-N- bonds. We obtained the structure of TrSOX by crystallization and X-ray diffraction (XRD) for the initial framework. The natural evolution in the nonconserved residues of key microdomains—including the catalytic loop, coenzyme pocket, substrate pocket, and entrance site—was then identified using ancestral sequence reconstruction (ASR), and the substitutions that accrued during natural evolution were recreated by site-directed mutagenesis. The single and double substitution variants catalyzed the N-demethylation of N-methyl-L-amino acids up to 1800- and 6000-fold faster than the wild type, respectively. Additionally, these single substitution variants catalyzed the terminal N-demethylation of non-amino-acid compounds and the oxidation of the main chain -C-N- bond to a -C=N- bond in the nitrogen-containing heterocycle. Notably, these variants retained the enantioselectivity and stability of the initial framework. We conclude that the variants of TrSOX are of great potential use in N-methyl enantiomer resolution, main-chain Schiff base synthesis, and alkaloid modification or degradation.
Keywords: Thermomicrobium roseum sarcosine oxidase, key microdomains, natural evolutionary trend, terminal N-demethylation, main chain Schiff base
Abbreviations: ASR, ancestral sequence reconstruction; HRP, horseradish peroxidase; SOX, sarcosine oxidase; TrSOX, Thermomicrobium roseum sarcosine oxidase; XRD, X-ray diffraction
N-methylation/-demethylation is a vital chemical or biochemical modification (1) and plays a vital role in various bioprocesses, including epigenetics, chromosomal segregation, and cell cycle progression (2). In addition, N-methylation/-demethylation has been demonstrated to modulate the structural and functional properties of proteins (3, 4), peptides (5, 6, 7), and pharmaceutical compounds (8). Protocols for N-methylation have been well developed (9, 10), while N-demethylation has always been considered a challenge. The von Braun reaction (Fig. 1A) (11) or nonclassical Polonovski reaction (12), N-demethylation processes catalyzed by 1-chloroethyl chloroformate (Fig. 1B) (13), hard electrophiles (14), dialkyl azodicarboxylates (15), N-oxides (Fig. 1C) (16), or metals (Fig. 1D) (17), have been developed, and recently, organophotocatalytic (18) or electrochemical (19) protocols have also been reported. These chemical reactions usually require toxic reagents, strong oxidants, high energy input, relatively long reaction times; in addition, in some cases, when the N-methyl group was next to a chiral carbon atom, the chemical reaction did not show enantioselectivity (12). In addition, N-demethylation of N-dimethyl compounds could be catalyzed by horseradish peroxidase (HRP) in the presence of strong oxidants (Fig. 1E) (20).
Figure 1.
The reported N-demethylation reactions.A–D, traditional chemical protocols for N-demethylation; E, N-demethylation catalyzed by HRP.
Sarcosine oxidase (SOX, EC. 1.5.3.1) oxidizes the -C-N- bond of sarcosine (N-methyl-Gly) or L-abrine (N-methyl-L-Trp) into the -C=N- bond and finally exposes the primary amine (Fig. 2, 1→2a→2b and 3→4a→4b), using a covalently linked FAD or flavin mononucleotide (FMN) as the cofactor (21). Recently, most members of the SOX family were screened from fungi and bacteria and have been used in the detection of serum creatinine for clinical diagnosis of kidney diseases or in the monitoring of glycerol or organic acid in the food industry (22, 23). SOXs have not been well investigated in terms of other potential substrates except N-methyl-Gly or N-methyl-L-Trp. In addition, L-abrine is an acute toxic alkaloid (24), and it can be converted into L-Trp by SOXs without toxicity. Recently, because the alkaloid compounds have been widely used in the chemical and pharmaceutical industry (25, 26), they have become significant pollutants in food (27), pharmaceuticals, plant extracts, municipal and hospital wastewaters (24, 28), and they have also been considered a potential risk for bioterrorism (29). Alkaloids contain different types of terminal and main chain -C-N- bonds (30, 31), and the N-demethylation or break down of -C-N- bonds could help to produce the corresponding compounds or promote their degradation (16). Therefore, the N-demethylation activity of SOXs can be a potential method for detoxification treatment after alkaloid pollution.
Figure 2.
The reactions catalyzed by TrSOXs.
In our previous studies, the SOX enzyme (TrSOX, GenBank: ACM06094.1) was obtained from Thermomicrobium roseum surviving in the basic hot springs of the Yellowstone National Park, and it had outstanding resistance against temperature (Tm > 100 °C), pH, and organic solvents; however, the poor recombinant expression level was a challenge for the characterization and application of this enzyme; and through surface design, a single mutant-TrSOX (S320R) far from the catalytic center was built with an over 20-fold promotion in soluble expression (22, 23). Although the structures of several modern SOXs from Escherichia coli or Bacillus have been elucidated (32, 33, 34, 35), the structural details of TrSOX are still unclear because of the low sequence identity between TrSOX and these homologs (no more than 40%).
To optimize the structural and catalytic properties of enzymes, directed evolution and rational design are considered universal protocols. Directed evolution requires large-scale mutant libraries and high-throughput screening protocols. The rational design is more efficient for regulating the folding–unfolding equilibrium by destabilizing unfolding and stabilizing the folded conformation (36), localizing potential hotspots for rational design, or using semirational and de novo design (37) and the B-FIT-directed evolution method (38). However, significantly promoting the stability of a specific enzyme is always a challenge. Recently, starting from the sequences of well-studied modern enzymes, ancestral sequence reconstruction (ASR) has been applied to studying the evolutionary history of homologous families because ancestors in older nodes may possess specific properties produced in certain environments over billions of years of evolution, enhancing the thermostability of target enzymes (39, 40, 41). Properties of ancestors have previously been used as references to promote the stability of modern enzymes (42).
In most of the ASR-associated studies, some fractional features of ancestral enzymes were introduced into modern ones in order to promote the stability, but it did not always work. We supposed that the stability of an enzyme should be associated with the whole framework but not only several limited sites; therefore, in this work, a different strategy was employed. After the structure of TrSOX was elucidated using crystallization and XRD, the stable framework of TrSOX was used as the starting model. The natural evolutionary trends of nonconserved residues in the catalytic loop, substrate pocket, coenzyme pocket, and entrance site were identified by ASR, using approximately 100 homologous sequences, including modern and hypothetical SOXs. Furthermore, site-directed mutations were employed to recreate the substitutions occurring in these four regions during evolution, and the structural and catalytic properties were characterized.
Results
The structure of TrSOX and the natural evolutionary trends in key microdomains
The structure of TrSOX (S320R) was solved by molecular replacement at 1.42 Å resolution, using a homologous model (PDB ID: 2GF3) as reference, and the data were deposited in the RCSB Protein Data Bank (PDB ID: 7EXS) (Fig. 3A). In sequence alignment and ASR, almost all the homologous enzymes sharing over 50% sequence identities with TrSOX were uncharacterized ones (Fig. 3B). The low sequence identities of TrSOX with homologous enzymes, e.g., PDB ID: 2UZZ (35.10%), 2A89 (39.90%), 3BHF (40.18%), 2GF3 (40.00%), 3M13 (40.18%), and 1ZOV (39.67%), indicated that there were significant evolutionary distances. In our previous work, the model of TrSOX was simulated using the above structures, and the low-soluble expression level of TrSOX was attributed to the high hydrophobic area (over 40%) on the solvent-accessible surface (23); however, in the crystal structure of TrSOX, hydrophobic area on the surface was less than 30%—almost the same as those of the modern homologs. This difference indicated that the models with significant evolutionary distances could not help to understand the structure of an unsolved one. Therefore, in this work, the structural resolution of TrSOX helped fill the informational gaps in the phylogenetic tree of this family, and it could be considered as the reference for the research of other homologs with closer evolutionary distances (Fig. 3B).
Figure 3.
Structural resolution and ancestral sequence reconstruction.A, to resolve the crystal structure of TrSOX (S320R), the rod-shaped crystal was subjected to X-ray diffraction, and the structure was resolved at 1.42 Å by molecular replacement, using 2GF3 as reference. FAD was covalently linked with Cys312, α-helixes were labeled in red, β-sheets were labeled in light blue, β-turns were labeled in green. The structure of TrSOX (S320R) was overlapped with 3BHF (In white). B, molecular phylogenetic analysis was performed by Maximum Likelihood method, ▼ TrSOX, ● homologous sequences with 3D structures. C, the catalytic loop was labeled in yellow, the residuals in 4 Å from the coenzyme (FAD) were labeled in purple, the residuals in 3 Å from the substrate pocket (green mesh surface) were labeled in red, and S320 at the entrance site was labeled in green, and four Cys residues were labeled in blue. D, in ancestral sequence reconstruction, the natural evolutionary trends of the nonconserved residues in the catalytic loop, coenzyme pocket, substrate pocket, and entrance site were identified. TrSOX, Thermomicrobium roseum sarcosine oxidase.
In the structure of TrSOX, there were four Cys residues at sites 153, 158, 274, and 312, C312 was covalently linked with FAD, and there was no disulfide bridge (Fig. 3C). To maintain the stable framework of TrSOX, we hope to bring as few changes as possible to limited sites. Through ASR, the natural evolutionary process of nonconserved residuals in key microdomains, which should play vital roles in enzymatic reactions, including catalytic loop, coenzyme pocket, substrate pocket, and entrance site, were identified (Fig. 3, C and D).
Recreation of natural evolution in catalytic loop
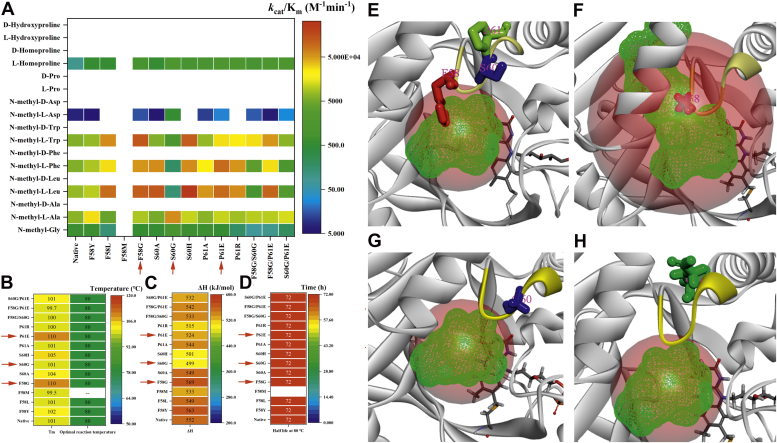
Ten of the 18 identified substitutions accruing in natural evolution in the catalytic loop (residues 57–62: -YFESPE-) (Fig. 3, C and D) were prepared and characterized (Table S1). In Figure 4, A–D, as for the native TrSOX, none of the mutants were active against N-methyl-D-amino acids; all these substitutions showed outstanding thermostability similar to the initial frame work, with Tm at ∼100 °C, ΔH above 500 kJ/mol, optimal reaction temperature at 80 °C, and the 80 °C active half-lives at ∼72 h. F58G and P61E could promote the catalytic efficiency against most of the initial substrates (Fig. 2, 3→4a, 5→6a, 7→8a, 9→10a, 11→12a and 17→18a). However, S60G led to an over 40-fold enhanced activity against N-methyl-L-Asp (Fig. 2, 9→10a), with a dramatic decrease in activity for substrates with hydrophobic side chains (Fig. 2, 3→4a, 7→8a, 11→12a). The benzene ring of F58 was close to the substrate pocket, while S60 and P61 were relatively distanced (Fig. 4E). The substitution of Phe by Gly at site-58 might reduce the steric hindrance outside of the substrate cavity and enlarge its volume (Fig. 4F) and then promote the accessibility of most of the initial substrates except N-methyl-Gly and N-methyl-L-Phe, as F58G showed increased Km values for these two substrates; furthermore, the expanded substrate cavity led to the enhanced catalytic ratio (Table S1). S60G and P61E were unable to affect the size of the substrate pocket (Fig. 4, G and H), and they may act as navigators of the substrate outside of the pocket, as described in our previous study (23). Phe and Pro at site-58 and 61 were hydrophobic and rigid residues (43), and they should play vital roles in the structure and function of the catalytic loop. Double substitutions containing one of these two residues, e.g., F58G/S60G and S60G/P61E, exhibited higher Km values and lower catalytic efficiency against substrates with hydrophobic side chains (Tables 1 and S1). Another double substitution, F58G/P61E was not as efficient as the two single substitutions; it should be attributed to the substitutions of these two rigid and hydrophobic residues by soft and hydrophilic ones, which might promote the flexibility and vibration of the catalytic loop.
Figure 4.
The substitutions in the catalytic loop.A, kcat/Km, (B) melting temperature (Tm), optimal reaction temperature, (C) denaturation enthalpy (ΔH), and (D) activity half-life of substitutions in the catalytic loop. The catalytic loop (57–62: -YFESPE-) was labeled in yellow. E, in native TrSOX, F58, S60, and P61 were labeled in red, blue, and green, respectively; and the volume of the substrate pocket was 112.875; (F) In F58G, G58 was labeled in red, and the volume of the substrate pocket was 199.250; (G) In S60G, G60 was labeled in blue, and the volume of the substrate pocket was 112.875; (H) In P61E, E61 was labeled in green, and the volume of the substrate pocket was 112.875. TrSOX, Thermomicrobium roseum sarcosine oxidase.
Table 1.
The specific catalytic efficiency of positive mutants in key microdomains
| Substrate | Specific catalytic efficiency (kcat/Km) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Native |
Catalytic loop |
Coenzyme pocket |
Substrate pocket |
Entrance sit |
||||||||||||||
| kcat/Km (M−1 × min−1) | Specific efficiency | F58G | S60G | P61E | F58G/S60G | S60G/P61E | F58G/P61E | S46T | I52V | V174A | L209V | F243Y | I244T | E251V | Y253A | S320R | S320K | |
| 1 | 1002.9 | 1 | 0.718 | 1.95 | 0.745 | 0.125 | 0.323 | 0.167 | 0.096 | 1.27 | 2.55 | 0.557 | 8.76 | 3.45 | 0.141 | 0.037 | 2.41 | 1.05 |
| 3 | 7231.0 | 1 | 33.3 | 0.344 | 5.17 | 5.33 | 1.52 | 8.94 | 6.86 | 6.72 | 0.175 | 0.261 | 0.27 | 0.304 | 0.497 | 0.203 | 32.45 | 25.74 |
| 5 | 10070.9 | 1 | 1.69 | 8.18 | 2.04 | 1.83 | 2.83 | 1.87 | 0.407 | 0.429 | 0.703 | 0.323 | 6.92 | 3.03 | 1.19 | 0.064 | 4.17 | 4.49 |
| 7 | 12461.5 | 1 | 16.9 | 0.02 | 13.8 | 0.114 | 0.299 | 14.8 | 3.98 | 4.16 | 0.596 | 5.19 | 17.7 | 346 | 31.1 | 1.65 | 6.63 | 282.23 |
| 9 | 12.6 | 1 | 2.03 | 43.3 | 2.9 | 2.19 | 3.04 | 0.794 | - | - | 3.26 | 66.9 | 750 | 188 | 11.4 | - | 13.57 | 1760.38 |
| 11 | 8413.2 | 1 | 6.13 | 0.036 | 18.7 | 0.517 | 0.3 | 4.57 | 8.76 | 2.78 | 3.39 | 0.468 | 26.5 | 282 | 29.2 | 2.85 | 33.24 | 55.41 |
| 13 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | - | - | - |
| 15 L-Pro | - | - | - | - | - | - | - | - | - | Novel | - | Novel | - | Novel | Novel | Novel | - | Novel |
| D-Pro | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | - | - | - | - |
| 17 | 176.8 | 1 | 8.94 | 11.6 | 9.99 | 0.013 | 0.007 | 0.009 | 32.8 | - | 5.93 | - | 60.4 | 5.2 | 1.06 | 13.3 | 13.05 | 65.84 |
| 19 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | Novel | - | - |
| 21 | - | - | - | - | - | - | - | - | - | - | Novel | - | - | - | Novel | Novel | - | - |
| 23 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | - | - | |
| 25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | Novel | - | - |
| 27 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | Novel | - | - |
| 29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Novel | Novel | - | - |
The kcat/Km of native TrSOX for substrate 1, 3, 5, 7, 9, 11, and 17 were set as 1.
The natural evolution in coenzyme pocket
Ten of the 15 substitutions accruing in natural evolution in the coenzyme pocket (Fig. 3, C and D) were prepared and characterized (Table S2). Of note, the enantioselectivity for substrates and thermostability were always retained (Fig. 5, A–D); S46T promoted the catalytic efficiency against substrates with large and hydrophobic side chains (Fig. 2, 3→4a, 7→8a, 11→12a); I52V and L209V revealed activity against L-Pro (Fig. 2, 15→16), L209V showed a ∼67-fold promotion against N-methyl-L-Asp (Fig. 2, 9→10a), while V174A revealed activity against (R)-2-methyl-pyrrolidine (Fig. 2, 21→22) (Table 1). For the nonconserved sites in coenzyme pocket, S46T (Fig. 5F), V174A (Fig. 5H), and L209V (Fig. 5I) were relatively far away and resulted in no significant differences in substrate pocket, and they might affect the whole structure of the enzyme by binding with the cofactor and regulate catalytic properties by affecting the electron transfer chain through FAD; the detailed mechanism should be investigated through crystallization and XRD in our further work; I52 (Fig. 5G) was located at the bottom of the substrate pocket, and it could enlarge the volume of substrate pocket. Double mutants including S46T/V174A, S46T/L209V, and V174A/L209V were designed; however, these could not be expressed in the soluble form as FAD was covalently linked with the enzyme, and it may play a vital role in the natural folding process, multiple mutants close to the coenzyme might have disrupted the entire structure of the enzyme.
Figure 5.
The substitutions in the coenzyme pocket. The (A) kcat/Km, (B) melting temperature (Tm) and optimal reaction temperature, (C) denaturation enthalpy (ΔH), and (D) activity half-life of TrSOXs. E, in native TrSOX, the S46, I52, V174, and L209 were labeled in red, yellow, blue, and green, respectively; the substrate pocket volume was 112.875. F, in S46T, T46 was labeled in red, and the volume of the substrate pocket was 112.875. G, in I52V, V52 was labeled in yellow, and the volume of the substrate pocket was 161.250. H, in V174A, A174 was labeled in blue, and the volume of the substrate pocket was 112.875. I, in L209V, V209 was labeled in green, and the volume of the substrate pocket was 112.875. TrSOX, Thermomicrobium roseum sarcosine oxidase.
Rebuilding the natural evolution in substrate pocket
Thirty one of the 41 substitutions accruing in natural evolution in substrate pocket (Fig. 3, C and D) were prepared and characterized (Table S3). Notably, the enantioselectivity for substrates and thermostability were also always maintained (Fig. 6, A–D); F243Y and I244T dramatically enhanced the catalytic efficiency against N-methyl-L-amino acids except for N-methyl-L-Trp (Fig. 2, 3→4a); F243Y brought a ∼750-fold enhanced activity for N-methyl-L-Asp (Fig. 2, 9→10a), while I244T showed activities against both of L- and D-Pro without enantioselectivity. Mutants at residues 245, 251, and 253 showed significantly decreased activity against N-methyl-L-Gly (Fig. 2, 1→2a); however, the substrate scope was enlarged, and especially for E251V, which could recognize non-amino-acid compounds or nitrogen-containing heterocycles, including N-Methyl-1-naphthalenemethylamine (Fig. 2, 13→14a), L-Pro (Fig. 2, 15→16a), L-homoproline (Fig. 2, 17→18a), L-Hydroxyproline (Fig. 2, 19→20), (R)-2-methyl-pyrrolidine (Fig. 2, 21→22), (3R)-(+)-3-(ethylamino)pyrrolidine (Fig. 2, 23→24), 1,2,3,4-tetrahydroisoquinoline (Fig. 2, 25→26,), 1-methyl-1,2,3,4-tetrahydroisoquinoline (Fig. 2, 27→28), and hexamethyleneimine (Fig. 2, 29→30) (Table 1).
Figure 6.
The substitutions in the substrate pocket. The (A) kcat/Km, (B) melting temperature (Tm) and optimal reaction temperature, (C) denaturation enthalpy (ΔH), and (D) activity half-life of TrSOXs. E, in native TrSOX, F243, I244, E251, and Y253 were labeled in red, yellow, blue, and green, respectively; the substrate pocket volume was 112.875. There was direct interaction between F243 and surrounding residues, including V92, G254, L240, and L299. F, in F243Y, Y243 was labeled in red, and the volume of the substrate pocket was 112.875; Y253 and V92 were closer to Y243. G, in I244T, T244 was labeled in yellow, and the volume of the substrate pocket was 197.125. H, in E251V, V251 was labeled in blue, and the volume of the substrate pocket was 107.000. I, in Y253A, A253 was labeled in green, and the volume was 234.750. TrSOX, Thermomicrobium roseum sarcosine oxidase.
E251 and Y253 were located in one beta-sheet next to the substrate pocket, while F243 and I244 were in another parallel sheet (Fig. 6E). F243Y was positioned at the bottom of the substrate pocket, and it resulted in no significant difference in the volume of the pocket; but Y243 was closer to V92 (carbon hydrogen bond) and Y253 (newly formed amide-Pi stacked), which might affect the arrangement of the two-parallel beta-sheets in substrate pocket (Fig. 6F). With the enlarged substrate pocket volume (Fig. 6G), I244T promoted the activity except for N-methyl-L-Trp (Fig. 2, 3→4a), and D or L-Pro could both be recognized. E251V was positioned at the top of the substrate pocket, and it led to the decreasing of the pocket volume and the hydrophilicity (Fig. 6H), which could promote the accessibility of substrates with hydrophobic side chains. Y253A was positioned at the bottom of the substrate pocket, which enlarged the volume of the pocket (Fig. 6I). As the above four sites were all located on two parallel beta-sheets (Fig. 6E), the proximity of the multiple substitutions might disrupt the corrected folding, the double mutants designed of these sites could not be expressed in soluble form. As reported (21), the carboxylic acid group might participate in the reaction catalyzed by SOXs; and in this work, TrSOX (E251V) catalyzed the terminal N-demethylation of N-Methyl-1-naphthalenemethylamine (Fig. 2, 13→14a→14b), which suggested that the carboxylic acid group in the substrate should not be necessary for this reaction.
Natural evolution at entrance site
To follow the natural evolutionary trend at S320, six of the eight mutants were prepared and characterized (Table S4). Figure 7A shows that the two mutants—S320R and S320K—could enhance the catalytic efficiency; with S320K showing the most effective promotion of the terminal N-demethylation of N-methyl-L-amino acids (Fig. 2, 1→2a (∼1.05-fold), 3→4a (∼25.7-fold), 5→6a (∼4.49-fold), 7→8a (∼282-fold), 9→10a (∼1760-fold), 11→12a (∼55.4-fold)) and the Schiff base formation of the L-Pro associated compounds (Fig. 2, 15→16 (novel), 17→18 (∼65.8-fold), 19→20 (novel)) (Table 1). Notably, the thermostability of the mutants at the entrance site remained similar to the native TrSOX (Fig. 7, B–D). S320 was located near the entrance of the substrate pocket (Fig. 7E), but far away from the catalytic center, while in S320K (Fig. 7F) and S320R (Fig. 7G), a hydrophilic lid was formed extending from the substrate pocket with the enlarged pocket volume, and the hydrophilic lids could contribute to navigate substrates with an acidic sidechain and to reject those without these groups.
Figure 7.
The entrance site and the multiple substitutions. The (A) kcat/Km, (B) melting temperature (Tm) and optimal reaction temperature, (C) denaturation enthalpy (ΔH), and (D) activity half-life of substitutions at entrance site. E, in native TrSOX, S320 were labeled in red, and the volume of the substrate pocket was 112.875; on the surface, S320 was labeled with pink mesh. F, in S320K, K320 was labeled in blue, and the volume of the substrate pocket was 192.750. On the surface, a hydrophilic lid was formed, and it was labeled with pink mesh. G, in S320R, RS320 was labeled in blue; the volume of the substrate pocket was 143.250; on the surface, it was labeled with pink mesh. The (H) kcat/Km, (I) melting temperature (Tm) and optimal reaction temperature, (J) denaturation enthalpy (ΔH), and (K) activity half-life of multiple substitutions. TrSOX, Thermomicrobium roseum sarcosine oxidase.
Multiple substitutions across the key microdomains
Multiple substitution variants across the key microdomains were prepared and characterized (Table S5). Regarding Figure 7H and Table 2, most S320K-containing substitutions promoted catalytic efficiency against N-methyl-L-amino acids, and S320K/F243Y was the most remarkable one; it significantly enhanced the N-demethylation of N-methyl-L-amino acids (Fig. 2, 1→2b (∼11.4-fold), 3→4b (∼1.58-fold), 5→6b (∼8.38-fold), 7→8b (∼478-fold), 9→10b (∼6230-fold), 11→12b (∼93.4-fold)) and the Schiff base formation of L-Pro associated compounds (Fig. 2, 15→16 (∼11.4-fold), 17→18 (∼128-fold)). In addition, E251V-containing substitutions revealed the novel activities against nitrogen-containing heterocycles; it suggested that the native residues inside the active pocket could recognize and catalyze these substrates; however, they were rejected by the hydrophilic Glu 251 residues on the top of the pocket, while the substitution by a hydrophobic and soft residues-Val could promote the entering of nitrogen-containing heterocycles. Furthermore, the combined triple substitutions, S320K/E251V/P61E and S320K/E251V/V174A, could enhance the catalytic efficiency against some nitrogen-containing heterocycles. In addition, multiple mutagenesis did not decrease the marked thermostability of TrSOX (Fig. 7, I–K). However, these novel activities against the main chain -C-N- bonds in nitrogen-containing heterocycles were relatively lower, and these substrates were seldom reported in the whole homologous family, which might be attributed to the natural evolution. In future studies, artificial evolution of the conserved residues in key microdomains will be carried out to enhance these novel activities.
Table 2.
The specific catalytic efficiency of positive multiple mutants
| Substrate | Specific catalytic efficiency (kcat/Km) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Native |
E251V |
Multiple mutants |
|||||||
| kcat/Km (M−1 × min−1) | Specific efficiency | kcat/Km (M−1 × min−1) | Specific efficiency | S320K/F243Y | S320K/P61E | S320K/E251V | E251V/P61E | S320K/E251V/P61E | |
| 1 | 1002.9 | 1 | 141.1 | 0.141 | 11.4 | 0.237 | 0.084 | 0.057 | 0.045 |
| 3 | 7231.0 | 1 | 3595.8 | 0.497 | 1.58 | 1.69 | 0.318 | 0.223 | 0.13 |
| 5 | 10070.9 | 1 | 11976.6 | 1.19 | 8.38 | 1.16 | 0.591 | 1.84 | 0.994 |
| 7 | 12461.5 | 1 | 387,237.3 | 31.1 | 478 | 12.1 | 8.69 | 12.8 | 29.6 |
| 9 | 12.6 | 1 | 144.0 | 11.4 | 6230 | 14 | 9.3 | 5.52 | 9.19 |
| 11 | 8413.2 | 1 | 245751.3 | 29.2 | 93.4 | 11 | 8.29 | 449 | 18.1 |
| 13 | - | - | 13.4 | 1 | - | - | 0.253 | - | 2.74 |
| 15 | - | - | 22.6 | 1 | 11.4 | 0.957 | 3.08 | 1.12 | 1.98 |
| 17 | 176.8 | 1 | 187.9 | 1.06 | 128 | 4.41 | 2.76 | 1.38 | 1.89 |
| 19 | - | - | 7.7 | 1 | - | - | 1.96 | 0.366 | 2.64 |
| 21 | - | - | 0.2 | 1 | - | - | 4.79 | - | - |
| 23 | - | - | 0.3 | 1 | - | - | 2.46 | 3.54 | 4.19 |
| 25 | - | - | 4.0 | 1 | - | - | 2.62 | 2.94 | 3.46 |
| 27 | - | - | 17.7 | 1 | - | - | 0.123 | 0.034 | 0.319 |
| 29 | - | - | 16.3 | 1 | - | - | 0.124 | 0.166 | 0.075 |
The kcat/Km of native TrSOX for substrate 1, 3, 5, 7, 9, 11, and 17 were set as 1; the kcat/Km of E251V for substrate 13, 15, 19, 21, 23, 25, 27 and 29 were set as 1.
Characterization of the products
Regarding the TLC, LC-MS, NMR, and optical rotation assays, except for N-methyl-Gly, TrSOX variants presented specific enantioselectivity for substrates and products, and there was no racemization in the reactions. N-methyl-L-amino acids were finally converted to L-amino acids (Fig. 2, 1→2b, 3→4b, 5→6b, 7→8b, 9→10b, and 11→12b) (Figs. S1, S8–S13; Table S6), and this indicated that TrSOXs should be of great potential for the enantio-separation of N-methyl- substrates. As reported, N-methyl-D-Asp is an essential molecule in the development of the brain and nervous system, mediating different signal pathways (44, 45), and has been widely used in neurological research and the synthesis of anticancer drugs or used as a functional food and feed additive (46). In this study, the S320K/F243Y variant significantly promoted N-demethylation activity against N-methyl-L-Asp, and it might be of great potential in the preparation of N-methyl-D-Asp by enantio-separation from N-methyl-DL-Asp. In addition, in previous studies (21), SOXs converted N-methyl-glycine to 2-(methyleneamino)acetic acid (Fig. 2, 1→2a) as an intermediate product. However, because of the fast nonenzymatic hydrolysis reaction, all the Schiff bases in the intermediate states of the N-methyl-L-amino acids (Fig. 2, 2a, 4a, 6a, 8a, 10a, 12a) could not be detected here. Similar to amino-acid-associated substrates, catalyzed by TrSOX (E251V), N-Methyl-1-naphthalenemethylamine was converted to naphthalene-1-ylmethanamine (Fig. 2, 13→14b) (Figs. S3 and S17), and resulting from the nonenzymatic hydrolysis process (Fig. 2, 14a→14b), the intermediate product, N-(naphthalen-1-ylmethyl)methanimine (Fig. 2, 14a) could not be detected. The terminal N-demethylation activity of TrSOXs with specific enantioselectivity could be employed in the detoxification treatment for alkaloid pollution, such as the N-demethylation of L-abrine (N-methyl-L-Trp) (24).
In the Pro-associated metabolism processes of the human body, L-Pro and L-homoproline could be converted to Δ1-pyrroline-5-carboxylate (P5C) and Δ1-piperideine-6-carboxylate (P6C) (Fig. 2, 15→16a and 17→18a) and were associated with the development of diseases; however, without efficient preparation protocol and commercial standards, they could only be detected by LC-MS in previous studies (47, 48). In this study, these two products were prepared, recovered, and confirmed by TLC, LC-MS, and NMR assays (Figs. S2, S14, and S15). Although the activity for L-hydroxyproline, a typical derivative of L-Pro, was relatively low, and the product was not enough for NMR analysis, LC-MS assays suggested that L-hydroxyproline was converted to (2S)-3-hydroxy-3,4-dihydro-2H-pyrrole-2-carboxylic acid (Fig. 2, 19→20) (Figs. S2 and S16). In addition, the main chain Schiff-base in P5C and P6C were more stable than the terminal ones, and there was no hydrolysis in enzymatic reaction and recovery. However, after incubation at 37 °C and pH 5.0 (adjusted by 0.1 M HCl) for 12 h, the hydrolyzed P5C and P6C were detected (Fig. 2, 16a→16b and 18a→18b), while the substrates were stable (Figs. S14 and S15). These results suggested that TrSOXs could promote the degradation of nitrogen-containing heterocycles.
Similar to L-Pro, the pyrrolidine ring was the key structure of (R)-2-methyl-pyrrolidine and 3(R)-(+)-3-(ethylamino)pyrrolidine, and they were converted to 2-methyl-1-pyrroline (Fig. 2, 21→22) (Figs. S4 and S18) and (R)-N-ethyl-3,4-dihydro-2H-pyrrol-4-amine (Fig. 2, 23→24) (Figs. S5 and S19), respectively. Notably, the -C=N- bonds of these two products were close to the side chain, opposite to those of P5C and P6C. Regarding the CDOCKER results, on the binding of L-Pro and L-homoproline with E251V (Fig. 8, A and B), the guanidine group of the conserved residues R54 could hold the position of both substrates. In contrast, for the binding of (R)-2-methyl-pyrrolidine and 3(R)-(+)-3-(ethylamino)pyrrolidine (Fig. 8, C and D), the hydrophobic side chains were rejected by R54 and were positioned on an opposing site. These results indicated that R54 should play a vital role in substrate binding and could also determine the sites of -C=N- bonds in nitrogen-containing heterocycles. In addition, there was much more direct interaction between L-homoproline and surrounding residues in the substrate pocket compared with those of L-Pro, (R)-2-methyl-pyrrolidine, and 3(R)-(+)-3-(ethylamino)pyrrolidine, which should be associated with the relatively higher activity.
Figure 8.
The binding of substrate pocket with L-Pro, L-homoproline, (R)-2-methyl-pyrrolidine, and (3R)-(+)-3-(ethylamino)pyrrolidine. After CDOCKER simulation, for L-Pro (A), R54 could interact with the carboxylic acid group; for L-homoproline (B), R54 could also directly interact with the carboxylic acid group; for (R)-2-methyl-pyrrolidine (C), there was no direct interaction between R54 and the side chain of the substrate; for (3R)-(+)-3-(ethylamino)pyrrolidine (D), R54 could directly interact with the secondary amine but not the side chain. FAD was labeled in yellow, and the distances between the substrates and surrounding residues were revealed.
For other nitrogen-containing heterocycles, 1,2,3,4-tetrahydroisoquinoline and 1-methyl-1,2,3,4-tetrahydroisoquinoline were converted to 3,4-dihydroisoquinoline and 1-methyl-3,4-dihydroisoquinoline, respectively (Fig. 2, 25→26 and 27→28) (Figs. S6, S7, S20 and S21), while hexamethyleneimine was converted to 3,4,5,6-tetrahydro-2H-azepine (Fig. 2, 29→30) (Figs. S8 and S22). These results indicated that this enzyme could recognize pyrrolidine and piperidine-associated substrates or even larger rings.
As reported, the -C=N- bonds in Schiff bases play a unique role in conferring broad-spectrum biological activities, and compounds containing Schiff bases have been widely used in chemical or pharmaceutical applications (49). Additionally, alkaloids are now widely used in the agricultural, chemical, and pharmaceutical industries, and the alkaloids residue in exhausted wastewater have been considered as an environmental risk (24, 25, 26, 27, 28). We believe that the biofunctions of TrSOXs should be of great value for these problems. Furthermore, in our recent work, using nicotine and atropinol, several specific mutants might work as morphinan N-demethylases to release the methyl group on tertiary amines (50). The coproduct (H2O2) has been detected using a colorimetric method in the reaction, and product extraction and identification are still in progress.
The enantioselective conversion of -C-N- bonds containing substrates
Regarding Table 3, S320K/F243Y could completely convert 100 mM of most of the N-methyl-L-amino acids to demethylated products in 2 h, but it was always inactive to D-form substrates. L-homoproline was completely converted into P6C in 2 h while L-Pro required 72 h, and this suggested that this variant was efficient in the enantio-separation of these kinds of racemic compounds.
Table 3.
Enzymatic conversion of substrates
| Enzyme | Substrate (100 mM) | Time (h) | Conversion ratio (%) |
|---|---|---|---|
| S320K/F243Y (10 μg/ml) | 1: N-methyl-Gly | 2 | >99 |
| 3: N-methyl-L-Trp | 2 | >99 | |
| N-methyl-D-Trp | 2 | - | |
| 5: N-methyl-L-Ala | 2 | >99 | |
| N-methyl-D-Ala | 2 | - | |
| 7: N-methyl-L-Leu | 2 | >99 | |
| N-methyl-D-Leu | 2 | - | |
| 9: N-methyl-L-Asp | 2 | >99 | |
| N-methyl-D-Asp | 2 | - | |
| 11: N-methyl-L-Phe | 2 | >99 | |
| N-methyl-D-Phe | 2 | - | |
| 15: L-Pro | 72 | >99 | |
| D-Pro | 72 | - | |
| 17: L-homoproline | 2 | >99 | |
| D-homoproline | 2 | - | |
| E251V (50 μg/ml) | 13: N-Methyl-1-naphthalenemethylamine | 72 | 90.3 |
| 19: L-Hydroxyproline | 72 | 32.5 | |
| D-Hydroxyproline | 72 | - | |
| 21: (R)-2-methyl-pyrrolidine | 72 | 25.3 | |
| (S)-2-Methyl-pyrrolidine | 72 | - | |
| 23: (3R)-(+)-3-(ethylamino)pyrrolidine | 72 | 37.6 | |
| (3S)-(-)-3-(ethylamino)pyrrolidine | 72 | - | |
| 25: 1,2,3,4-tetrahydroisoquinoline | 72 | 95.5 | |
| 27: 1-methyl-1,2,3,4-tetrahydroisoquinoline | 72 | 45.7 | |
| 29: Hexamethyleneimine | 72 | 90.6 |
In a 72-h reaction with E251V, the conversion ratio of N-methyl-1-naphthalenemethylamine, 1,2,3,4-tetrahydroisoquinoline, and hexamethyleneimine were over 90% (Fig. 2, 12, 25, and 29), while those of other substrate was not efficient. The conversion ratio of 1-methyl-1,2,3,4-tetrahydroisoquinoline (Fig. 2, 27) was ∼45%, which should be attributed to the enantioselectivity of this enzyme. These results indicated that E251V should be of great potential in the enantioselective Schiff base synthesis or the degradation of nitrogen-containing heterocycles. To promote these activities, the artificial evolution of conserved residues in key microdomains would be considered in further work.
Discussion
N-demethylation, especially the reaction with substrate enantioselectivity, has always been considered a challenge, and the chemical processes are carried out with toxic reagents or strong oxidants and usually without enantioselectivity (11, 12, 13, 14, 15, 16, 17, 18, 19). On the other hand, enzymatic N-demethylation reactions are seldom reported. Owing to their oxidation activity on terminal -C-N- bonds of N-methyl-Gly, we supposed that SOXs should be of great potential in the N-demethylation; however, the systematic investigation of SOXs on substrate scope has not been reported. Therefore, the aim of this work is to build robust enzymes for N-demethylation through protein engineering.
For protein engineering, direct evolution and design are the most widely used strategy. And the universal protocol is to start from an efficient enzyme and to promote its stability through different drive forces; however, it is a challenge to maintain the balance between the catalytic efficiency and stability, and the uncertainty of these kinds of work sometimes brings results out of design (36, 37, 38). Recently, ASR has been applied in enzymatic engineering; after the ancestors are simulated from modern enzymes, some residues would be introduced to modern enzyme to promote the stability (39, 40, 41, 42).
However, in this work, we supposed that the stability of a protein was associated with the whole framework but not only several limited sites; the structural resolution of TrSOX, a hypothetical SOX from the genome of T. roseum surviving in basic hot spring in Yellowstone, was in good accordance with this hypothesis, although this enzyme showed outstanding stability against temperature, pH, and organic solvents (21, 22, 23), it was overlapped with homologous models, without significant structural differences (Fig. 3A). Therefore, we hoped to start from a stable model, to expand the substrate scope, and to promote the catalytic efficiency. Additionally, in order not to disrupt the stable framework, as few mutant sites as possible should be considered. ASR was used to trace the natural evolutionary trends in key microdomains, which were necessary for the recognition and binding of substrates and for the reaction, including catalytic loop, coenzyme pocket, substrate pocket, and entrance site, and characterized or hypothetical homologues with close or far evolutionary distances were used as references. Of note, TrSOX was not an ancestral enzyme, but a homolog on a special branch.
Fortunately, this strategy did narrow the screening scale and shorten the experiment, and some variants with initial stability were built together with promoted catalytic efficiency or expanded substrate adaptivity. However, not all the sites predicted by ASR worked, some of the designed substitutions led to misfolding and partially or completely inactivation; this phenomenon might be attributed to the limited coverage ratio of homologs in ASR, because there should always be unknown ones in natural environment. In addition, recreating the natural evolutionary substitutions in key microdomains might not alter some specific properties in the homologous family, e.g., the enantioselectivity; and our further work will be focused on conserved residues in key microdomains to alter the enantioselectivity for substrates.
The TrSOX variants could catalyze the terminal N-demethylation and form main-chain Schiff bases in nitrogen-containing heterocycles with significant enantioselectivity. In addition, in these reactions, the electron was transferred to O2 through FAD; therefore, an additional coenzyme regeneration system was not required. These reactions required mild conditions, low energy consumption, without toxic reagents, and they should be of great potential in chemical synthesis, pharmaceutical design, and the synthesis, modification, or degradation of alkaloids.
The natural environment on earth provides different possibilities for structural or functional evolution. Enzymes of organisms from specific conditions might reveal associated traits, such as resistance to high or low temperature, high or low pressure, acidic or basic environments, which may play vital roles in their large-scale application. Introducing substitutions that occurred during natural evolution into such a specific homolog is a good protein engineering strategy to expand its substrate scope and increase its catalytic activity.
Experimental procedures
Materials
The TrSOX gene (GeneBank: ACM06094.1) was cloned in a pMA5-Pxyl-trsox plasmid and kept in our laboratory. The HisTrap HP Ni-column was purchased from GE Healthcare; AmiconUltra centrifugal filter tube (10 kDa) was from Merck Millipore; Thin-layer chromatography (TLC) plates (Merck, 105626, 105729, 105721 and 105554) were purchased from Merck Millipore; Q5 Site-Directed Mutagenesis Kit was from NEB Inc.; NeXtal Tubes JCSG Core Suites I-IV, NeXtal Tubes Classics Suite, NeXtal Tubes Classics II Suite, and NeXtal Tubes Cryos Suite were from Qiagen; and Pierce BCA Protein Assay Kit was from Thermo Scientific. N-methyl or alkaloid substrates were from MACKLIN and ALADDIN. Other reagents and chemicals were purchased from local companies and were of superior analytical grade.
Structural resolution of TrSOX
In our previous work, the expression of TrSOX was promoted with a single mutant-S320R (23). The crystallization of TrSOX (S320R) was screened using the hanging-drop vapor diffusion method with NeXtal Tubes JCSG Core Suites I-IV, NeXtal Tubes Classics Suite, NeXtal Tubes Classics II Suite, and NeXtal Tubes Cryos Suite (Qiagen). Diffraction datasets were collected at the Beamline 17U, Shanghai Synchrotron Radiation Facility (SSRF). The crystal structure of TrSOX (S320R) was resolved by molecular replacement and further refinement using the CCP4 Program Suite 6.1.3. The models of native TrSOX and the mutants were built and refined using the “Mutate Protein” module in the software Discovery Studio 2019, and the “Receptor-Ligand Interactions” module was employed to screen the substrate pocket and to investigate the interaction between FAD and the residues in the coenzyme pocket, and the “CDOCKER” module of Discovery Studio 2019 was employed to simulate the binding of substrates in the substrate pocket.
Ancestral sequence reconstruction analysis
The amino acid sequence of TrSOX was interrogated using BLAST (Basic Local Alignment Search Tool, nih.gov), and Blastp (protein-protein BLAST) was employed to search database nr. From the result of BLAST, approximately 100 homologous enzymes showing 25% to 99% sequence identities with TrSOX were applied for ancestral sequence reconstruction analysis using the Maximum Likelihood method in MEGA-X (51).
Site-directed mutagenesis
The TrSOX gene sequence was reconstructed in a pMA5-Pxyl-trsox plasmid in our previous work (23). The primers for each site were designed and synthesized (Tables S7–S10), and a Q5 Site-Directed Mutagenesis Kit (NEB) was used for the designed substitutions according to the manufacturer’s recommendations. The mutants were expressed in Bacillus subtilis W600 and prepared as described in our previous study (23).
Activity assay
As described in our previous study (22, 23), to detect N-demethylation-associated activities, the amount of coproduct (H2O2) was measured using a real-time colorimetric method at 37 °C and pH 8.0; to detect the catalytic constants, enzyme (5–50 μg) was added to a 1 ml reaction mixture containing substrates (1–200 mM). The produced H2O2 in the reaction was calculated with A500nm, and each experiment was repeated at least three times. Km and kcat were calculated through “Nonlinear Curve Fit” using the Michaelis–Menten equation in Origin software. The optimal reaction temperature and the half-lives of the activities were also investigated.
Nano-DSC assay
According to the Nano-DSC (TA Instruments) instructions, 300 μl of Tris-HCl (20 mM, pH 8) buffer was used as control, and then an equal volume of purified TrSOX (1 mg/ml) was injected into the sample bath for the experiment. The scanning was performed at −10 to 130 °C at 2 °C/min, and the response time was 5 s with a built-in pressurization perturbation of up to 6 atm. The melting temperature (Tm) and enthalpy of denaturation (ΔH) of the samples were then measured (22, 23, 52).
TLC analysis
After the enzymatic reaction with TrSOXs, the products of N-methyl-amino acids/non-amino-acid compounds and nitrogen-containing heterocycles were applied to TLC assays (Table 4), and most of the product dots containing -C=N- bonds revealed fluorescent signals under 365 nm. For preparation, the products were applied to classical silica TLC plates (20 × 20 cm) (Merck, 105721) or silica TLC plates F254 (20 × 20 cm) (Merck, 105554), using substrates as references, and the products were extracted using ddH2O, methanol or chloroform.
Table 4.
TLC protocols
| Substrate | TLC plate | Developer | Detection |
|---|---|---|---|
| 1 | Classical silica TLC plate (10 × 10 cm) (Merck, 105626) | 8:1:1 N-butanol: glacial acetic acid: water (v/v) | Sprayed with 1% ninhydrin in acetone (m/v) and heated at 80 °C for 5–10 min |
| 3 | |||
| 5 | |||
| 7 | |||
| 9 | |||
| 11 | |||
| 13 | Classical silica TLC plate (10 × 10 cm) (Merck, 105626) | 2.5:1 ethyl acetate: acetic acid (v/v), | Sprayed with 1% ninhydrin in acetone (m/v) and heated at 80 °C for 5–10 min |
| 15 | Silica TLC plates F254 (10 × 10 cm) (Merck, 105729) | 1:1 N-propanol: ammonia (v/v) | Sprayed with 1% ninhydrin in acetone (m/v) and heated at 80 °C for 5–10 min, or scanned under 365 nm |
| 17 | |||
| 19 | |||
| 21 | Silica TLC plates F254 (10 × 10 cm) (Merck, 105729) | 8:2:1:1 methanol: formic acid: acetic acid: water (v/v) | Sprayed with 1% ninhydrin in acetone (m/v) and heated at 80 °C for 5–10 min, or scanned under 254 and 365 nm |
| 23 | Classical silica TLC plate (10 × 10 cm) (Merck, 105626) | 2.5:1 N-propanol: ammonia (v/v) | Sprayed with 1% ninhydrin in acetone (m/v) and heated at 80 °C for 5–10 min, or scanned under 254 and 365 nm |
| 25 | Silica TLC plates F254 (10 × 10 cm) (Merck, 105729) | 10:1:1:2 ethyl acetate: formic acid: acetic acid: water (v/v) | Scanned under 254 and 365 nm |
| 27 | |||
| 29 | Classical silica TLC plate (10 × 10 cm) (Merck, 105626) | 10:1:1:1 methanol: formic acid: acetic acid: water (v/v) | Scanned under 365 nm |
LS-MS analysis
Using a Waters MALDI SYNAPT MS spectrometer, 5 μl of recovered products (∼0.5 mg/ml) was injected into a C18 column (Waters BEH 2.1 mm × 1 50 mm, 1.7 μm) at 0.3 ml/min and 45 °C. Monitored at 200 to 400 nm, the compounds were eluted in a 6 min linear gradient of A (CH3CN) 2% and B (0.1% HCOOH), ionized using a 3.5 kV capillary voltage, 30 V cone voltage, and 100 °C source block temperature, and further scanned using an m/z scanning range of 20 to 1000, 1 s scan time, 0.1 s interscan delay, and 50 l/h cone gas flow rate. The MS data were obtained and analyzed using Mass Lynx software (version 4.1).
NMR analysis
The 1H-NMR and 13C-NMR spectra of products recovered from TLC plates were analyzed using a Bruker Advance 400 MHz spectrometer (51, 53).
Optical rotation assay
The products of N-methyl-amino acids extracted from the TLC plate were applied to a Rudolph Autopol IV Polarimeter for optical rotation assay.
Enzymatic conversion
TrSOXs were employed for the bioconversion of N-methyl-amino acids and nitrogen-containing heterocycles, respectively. The enzyme (10–50 μg/ml) was added to a 20 ml reaction solution (100 mM substrate, 20 mM Tris-HCl, pH 8.0), and the mixtures were incubated at 37 °C for 2 to 72 h. The reactions were monitored by detecting the coproduced H2O2 and LC-MS.
Data availability
The structure presented in this article has been deposited in the Protein Data Bank (PDB) with the following code: 7EXS. All remaining data are contained within the article.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by National Key Research and Development Program of China (2021YFC2100303); National Natural Science Foundation of China (Grant No. 81970819); Natural Science Foundation of Jiangsu Province (BK20160053 and BE2018055); Top Talent Support Program for Young and Middle-aged People of Wuxi Health Committee (2020012); Priority Academic Program Development of Jiangsu Higher Education Institutions, 111 Project (No. 111-2-06). The authors are grateful to Shanghai Medicilon Inc for technological support. We thank the staff at the Beamline 17U, Shanghai Synchrotron Radiation Facility (SSRF).
Author contributions
Y. X., J. S., and L. Z. conceptualization; Y. X., J. S., and L. Z. funding acquisition; Y. X., C. S., M. W. T., Z. H. G., Z. T. G., and Y. S. investigation; Y. X., C. S., M. W. T., Z. H. G., Z. T. G., and Y. S. methodology; Y. X., J. S., and L. Z. supervision; Y. X., C. S., M. W. T., Z. H. G., Z. T. G., and Y. S. writing—original draft; Y. X., J. S., and L. Z. writing—review and editing.
Edited by Ruma Banerjee
Contributor Information
Yu Xin, Email: yuxin@jiangnan.edu.cn.
Jun Shao, Email: shaojun@njmu.educn.
Liang Zhang, Email: zhangl@jiangnan.edu.cn.
Supporting information
References
- 1.Chatterjee J., Rechenmacher F., Kessler H. N-methylation of peptides and proteins: An important element for modulating biological functions. Angew. Chem. Int. Ed. Engl. 2013;52:254–269. doi: 10.1002/anie.201205674. [DOI] [PubMed] [Google Scholar]
- 2.Ongpipattanakul C., Nair S.K. Molecular basis for autocatalytic backbone N-methylation in RiPP natural product biosynthesis. ACS Chem. Biol. 2018;13:2989–2999. doi: 10.1021/acschembio.8b00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBrayer D.N., Gantman B.K., Tal-Gan Y. N-Methylation of amino acids in gelatinase biosynthesis-activating pheromone identifies key site for stability enhancement with retention of the Enterococcus faecalis fsr quorum sensing circuit response. ACS Infect. Dis. 2019;5:1035–1041. doi: 10.1021/acsinfecdis.9b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori1 S., Pang A.H., Lundy T.A., Garzan A., Tsodikov O.V., Garneau-Tsodikova S. Structural basis for backbone N-methylation by an interrupted adenylation domain. Nat. Chem. Biol. 2018;14:428–430. doi: 10.1038/s41589-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 5.van der Velden N.S., Kälin N., Helf M.J., Piel J., Freeman M.F., Künzler M. Autocatalytic backbone N -methylationin a family of ribosomal peptide natural products. Nat. Chem. Biol. 2017;13:833–835. doi: 10.1038/nchembio.2393. [DOI] [PubMed] [Google Scholar]
- 6.Li W.J., Hu K., Zhang Q.Z., Wang D.Y., Ma Y., Hou Z.F., Yin F., Li Z.G. N terminal N-methylation modulates chiral centre induced helical (CIH) peptides' biophysical properties. Chem. Commun. 2018;54:1865–1868. doi: 10.1039/c7cc09201f. [DOI] [PubMed] [Google Scholar]
- 7.Räder A.F.B., Reichart F., Weinmüller M., Kessler H. Improving oral bioavailability of cyclic peptides by N-methylation. Bioorg. Med. Chem. 2018;26:2766–2773. doi: 10.1016/j.bmc.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Sciabola S., Goetz G.H., Bai G.Y., Rogers B.N., Gray D.L., Duplantier A., Fonseca K.R., Vanase-Frawley M.A., Kablaoui N.M. Systematic N-methylation of oxytocin: Impact on pharmacology and intramolecular hydrogen bonding network. Bioorg. Med. Chem. 2016;24:3513–3520. doi: 10.1016/j.bmc.2016.05.062. [DOI] [PubMed] [Google Scholar]
- 9.Dang T.T., Ramalingam B., Seayad A.M. Efficient ruthenium catalyzed N-methylation of amines using methanol. ACS Catal. 2015;5:4082–4088. [Google Scholar]
- 10.Bobbink F.D., Das S., Dyson P.J. N-formylation and N-methylation of amines using metal-free N-heterocyclic carbene catalysts and CO2 as carbon source. Nat. Protoc. 2017;12:417–428. doi: 10.1038/nprot.2016.175. [DOI] [PubMed] [Google Scholar]
- 11.von Braun J. Die Aufspaltung cyclischer Basen durch Bromcyan. Ber. Dtsch. Chem. Ges. 1909;42:2035–2057. [Google Scholar]
- 12.Rosenau T., Hofinger A., Potthast A., Kosma P. A general, selective, high-yield N-demethylation procedure for tertiary amines by solid reagents in a convenient column chromatography-like setup. Org. Lett. 2004;6:541–544. doi: 10.1021/ol036319g. [DOI] [PubMed] [Google Scholar]
- 13.Peat A.J., Buchwald S.L. Novel syntheses of tetrahydropyrroloquinolines: Applications to alkaloid synthesis. J. Am. Chem. Soc. 1996;118:1028–1030. [Google Scholar]
- 14.Selfridge B.R., Wang X., Zhang Y., Yin H., Grace P.M., Watkins L.R., Jacobson A.E., Rice K.C. Structure-activity relationships of (+)-naltrexone-inspired toll-like receptor 4 (TLR4) antagonists. J. Med. Chem. 2015;58:5038–5052. doi: 10.1021/acs.jmedchem.5b00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauterbach E.H., Dinkel T., Heller S., Bertogg A. 2009. Demethylation of 14-hydroxy substituted alkaloid derivatives. US 2009/0163717 A1. [Google Scholar]
- 16.Kok G.B., Pye C.C., Singer R.D., Scammells P.J. Two-step iron(o)-mediated N-demethylation of N-methyl alkaloids. J. Org. Chem. 2010;75:4806–4811. doi: 10.1021/jo1008492. [DOI] [PubMed] [Google Scholar]
- 17.Yi X.W., Lei S.Y., Liu W.S., Che F.R., Yu C.Z., Liu X.S., Wang Z.H., Zhou X., Zhang Y.X. Copper-catalyzed radical N-demethylation of amides using N-fluorobenzenesulfonimide as an oxidant. Org. Lett. 2020;22:4583–4587. doi: 10.1021/acs.orglett.0c00863. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y.S., Glotz G., Cantillo D., Kappe C.O. Organophotocatalytic N-demethylation of oxycodone using molecular oxygen. Chem. Eur. J. 2020;26:2973–2979. doi: 10.1002/chem.201905505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glotz G., Kappe C.O., Cantillo D. Electrochemical N-demethylation of 14-hydroxy morphinans: Sustainable access to opioid antagonists. Org. Lett. 2020;22:6891–6896. doi: 10.1021/acs.orglett.0c02424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedderis G.L., Hollenberg P.F. Characterization of the N-demethylation reactions catalyzed by horseradish peroxidase. J. Biol. Chem. 1983;258:8129–8138. [PubMed] [Google Scholar]
- 21.Abe Y., Shoji M., Nishiya Y., Aiba H., Kishimotod T., Kitaurae K. The reaction mechanism of sarcosine oxidase elucidated using FMO and QM/MM methods. Phys. Chem. Chem. Phys. 2017;19:9811–9822. doi: 10.1039/c6cp08172j. [DOI] [PubMed] [Google Scholar]
- 22.Xin Y., Hao M.Y., Fan G.M., Zhang Y., Zhang L. Soluble expression of Thermomicrobium roseum sarcosine oxidase and characterization of N-demethylation activity. Mol. Catal. 2019;464:48–56. [Google Scholar]
- 23.Gao Q.Y., Shao J., Tang M.W., Xin Y., Zhang L. Promote the expression and corrected folding of an extremely stable N-demethylase by promoter reconstruction, native environment simulation and surface design. Int. J. Biol. Macromol. 2021;178:434–443. doi: 10.1016/j.ijbiomac.2021.02.176. [DOI] [PubMed] [Google Scholar]
- 24.Johnson R.C., Zhou Y.T., Jain R., Lemire S.W., Fox S., Sabourin P., Barr J.R. Quantification of L-abrine in human and rat urine: A biomarker for the toxin abrin. J. Anal. Toxicol. 2009;33:77–84. doi: 10.1093/jat/33.2.77. [DOI] [PubMed] [Google Scholar]
- 25.Zhao S.Z., Sirasani G., Andrade R.B. Aspidosperma and strychnos alkaloids: Chemistry and biology. Alkaloids Chem. Biol. 2021;86:1–143. doi: 10.1016/bs.alkal.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Wicks C., Hudlicky T., Rinner U. Morphine alkaloids: History, biology, and synthesis. Alkaloids Chem. Biol. 2021;86:145–342. doi: 10.1016/bs.alkal.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Han H.L., Jiang C.L., Wang C., Liu X., Lu C.Y., Chen H.P. Pyrrolizidine alkaloids in tea: A review of analytical methods, contamination levels and health risk. Food Sci. 2021;42:255–266. [Google Scholar]
- 28.Negreira N., de Alda M.L., Barceló D. Cytostatic drugs and metabolites in municipal and hospital wastewaters in Spain: Filtration, occurrence, and environmental risk. Sci. Total Environ. 2014;497-498:68–77. doi: 10.1016/j.scitotenv.2014.07.101. [DOI] [PubMed] [Google Scholar]
- 29.Janik E., Ceremuga M., Saluk-Bijak J., Bijak M. Biological toxins as the potential tools for bioterrorism. Int. J. Mol. Sci. 2019;20:1181. doi: 10.3390/ijms20051181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin J., Zhang S.Y., Zhang Y.B., Chen L.F., Chen N.H., Wu Z.N., Luo D., Wang G.C., Li Y.L. Two new isoquinoline alkaloids from the seeds of Nandina domestica. Nat. Prod. Res. 2021;35:3254–3260. doi: 10.1080/14786419.2019.1696334. [DOI] [PubMed] [Google Scholar]
- 31.Vallejo-López M., Écija P., Vogt N., Demaison J., Lesarri A., Basterretxea F.J., Cocinero E.J. N-methyl inversion and accurate equilibrium structures in alkaloids: Pseudopelletierine. Chemistry. 2017;23:16491–16496. doi: 10.1002/chem.201702232. [DOI] [PubMed] [Google Scholar]
- 32.Ilari A., Bonamore A., Franceschini S., Fiorillo A., Boffi A., Colotti G. The X-ray structure of N-methyltryptophan oxidase reveals the structural determinants of substrate specificity. Proteins. 2008;71:2065–2075. doi: 10.1002/prot.21898. [DOI] [PubMed] [Google Scholar]
- 33.Nagata K., Sasaki H., Hua M., Okai M., Kubota K., Kamo M., Ito K., Ichikawa T., Koyama Y., Tanokura M. Crystal structure of monomeric sarcosine oxidase from Bacillus sp. NS-129 reveals multiple conformations at the active-site loop. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2005;81:220–224. [Google Scholar]
- 34.Jorns M.S., Chen Z.W., Mathews F.S. Structural characterization of mutations at the oxygen activation site in monomeric sarcosine oxidase. Biochemistry. 2010;49:3631–3639. doi: 10.1021/bi100160j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassan-Abdallah A., Zhao G., Chen Z.W., Mathews F.S., Jorns M.S. Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: A context-sensitive model for the electrostatic impact of arginine to lysine mutations. Biochemistry. 2008;47:2913–2922. doi: 10.1021/bi702351v. [DOI] [PubMed] [Google Scholar]
- 36.Kazlauskas R. Engineering more stable proteins. Chem. Soc. Rev. 2018;47:9026–9045. doi: 10.1039/c8cs00014j. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q., Xun G.H., Feng Y. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnol. Adv. 2019;37:530–537. doi: 10.1016/j.biotechadv.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Sun Z.T., Liu Q., Qu G., Feng Y., Reetz M.T. Utility of B-factors in protein science: Interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem. Rev. 2019;119:1626–1665. doi: 10.1021/acs.chemrev.8b00290. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen V., Wilson C., Hoemberger M., Stiller J.B., Agafonov R.V., Kutter S., English H., Theobald D.L., Kern D. Evolutionary drivers of thermoadaptation in enzyme catalysis. Science. 2017;355:289–294. doi: 10.1126/science.aah3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayuso-Fernández I., Rencoret J., Gutiérrez A., Ruiz-Dueñas F.J., Martínez A.T. Peroxidase evolution in white-rot fungi follows wood lignin evolution in plants. Proc. Natl. Acad. Sci. U. S. A. 2019;116:17900–17905. doi: 10.1073/pnas.1905040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clifton B.E., Kaczmarski J.A., Carr P.D., Gerth M.L., Tokuriki N., Jackson C.J. Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nat. Chem. Biol. 2018;14:542–547. doi: 10.1038/s41589-018-0043-2. [DOI] [PubMed] [Google Scholar]
- 42.Schriever K., Saenz-Mendez P., Rudraraju R.S., Hendrikse N.M., Hudson E.P., Biundo A., Schnell R., Syrén P. Engineering of ancestors as a tool to elucidate structure, mechanism, and specificity of extant terpene cyclase. J. Am. Chem. Soc. 2021;143:3794–3807. doi: 10.1021/jacs.0c10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H.R., Zhao Y., Guo C., Gan Y.R., Huang H. The role of proline substitutions within flexible regions on thermostability of luciferase. Biochim. Biophys. Acta. 2015;1854:65–72. doi: 10.1016/j.bbapap.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 44.Zamanian G., Shayan M., Rahimi N., Bahremand T., Shafaroodi H., Ejtemaei-Mehr S., Aghaei I., Dehpour A.R. Interaction of morphine tolerance with pentylenetetrazole-induced seizure threshold in mice: The role of NMDA-receptor/NO pathway. Epilepsy Behav. 2020;112:107343. doi: 10.1016/j.yebeh.2020.107343. [DOI] [PubMed] [Google Scholar]
- 45.Ghislandi A.B., Garcez M.L., Zambon G.M., Constantino L.C., Matos D.N., Pescador B.B., Tasca C.I., Boeck C.R. Adenosine and NMDA receptors modulate neuroprotection-induced NMDA preconditioning in mice. J. Mol. Neurosci. 2020;70:590–599. doi: 10.1007/s12031-019-01463-0. [DOI] [PubMed] [Google Scholar]
- 46.Farber N.B. NMDA antagonists for treatment-resistant depression. Handb. Exp. Pharmacol. 2019;250:287–305. doi: 10.1007/164_2018_165. [DOI] [PubMed] [Google Scholar]
- 47.Mathis D., Beese K., Rüegg C., Plecko B., Hersberger M. LC-MS/MS method for the differential diagnosis of treatable early onset inherited metabolic epilepsies. J. Inherit. Metab. Dis. 2020;43:1102–1111. doi: 10.1002/jimd.12244. [DOI] [PubMed] [Google Scholar]
- 48.Cappelletti P., Tallarita E., Rabattoni V., Campomenosi P., Sacchi S., Pollegioni L. Proline oxidase controls proline, glutamate, and glutamine cellular concentrations in a U87 glioblastoma cell line. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hameed A., al-Rashida M., Uroos M., Ali S.A., Khan K.M. Schiff bases in medicinal chemistry: A patent review (2010-2015) Expert Opin. Ther. Pat. 2017;27:63–79. doi: 10.1080/13543776.2017.1252752. [DOI] [PubMed] [Google Scholar]
- 50.Augustin M.M., Augustin J.M., Brock J.R., Kutchan T.M. Enzyme morphinan N-demethylase for more sustainable opiate processing. Nat. Sustain. 2019;2:465–474. [Google Scholar]
- 51.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin Y., Gao Q.Y., Gu Y., Hao M.Y., Fan G.M., Zhang L. Self-assembly of metal-cholesterol oxidase hybrid nanostructures and application in bioconversion of steroids derivatives. Front. Chem. Sci. Eng. 2021;15:615–629. [Google Scholar]
- 53.Hsu A., O'Brien P.A., Bhattacharya S., Rance M., Palmer A.G., 3rd Enhanced spectral density mapping through combined multiple-field deuterium 13CH2D methyl spin relaxation NMR spectroscopy. Methods. 2018;138:76–84. doi: 10.1016/j.ymeth.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure presented in this article has been deposited in the Protein Data Bank (PDB) with the following code: 7EXS. All remaining data are contained within the article.