Highlights
-
•
Sharing data from cooperative studies is essential to develop new biomarkers in MS.
-
•
Differences in MRI acquisition, analysis, storage represent a substantial constraint.
-
•
We review the state of the art and developments in the harmonization of MRI.
-
•
We provide recommendations to harmonize large MRI datasets in the MS field.
Keywords: MRI, Harmonization, Multiple sclerosis
Abstract
There is an increasing need of sharing harmonized data from large, cooperative studies as this is essential to develop new diagnostic and prognostic biomarkers. In the field of multiple sclerosis (MS), the issue has become of paramount importance due to the need to translate into the clinical setting some of the most recent MRI achievements. However, differences in MRI acquisition parameters, image analysis and data storage across sites, with their potential bias, represent a substantial constraint. This review focuses on the state of the art, recent technical advances, and desirable future developments of the harmonization of acquisition, analysis and storage of large-scale multicentre MRI data of MS cohorts. Huge efforts are currently being made to achieve all the requirements needed to provide harmonized MRI datasets in the MS field, as proper management of large imaging datasets is one of our greatest opportunities and challenges in the coming years. Recommendations based on these achievements will be provided here. Despite the advances that have been made, the complexity of these tasks requires further research by specialized academical centres, with dedicated technical and human resources. Such collective efforts involving different professional figures are of crucial importance to offer to MS patients a personalised management while minimizing consumption of resources.
1. Introduction
The increasing need of sharing data from large, cooperative studies is of paramount importance as this is essential to develop new diagnostic and prognostic biomarkers. In the field of multiple sclerosis (MS), the issue has become prominent due to the need to translate into the clinical setting some of the most important magnetic resonance imaging (MRI) achievements. However, the recent increase of large-scale MRI studies has highlighted the need to control the MR-related source of variability introduced by differences in scanners, acquisition protocols and image analysis when combining MRI data across multiple sites (Shinohara et al., 2017, Biberacher et al., 2016). Such unwanted variability can have unpredictable effects and potentially influence the detection of pathological findings, leading to inconsistent results and spurious relationships with clinical outcomes (Biberacher et al., 2016). Recent studies have tackled the challenges related to the lack of harmonization of MRI data from multiple sites (Dewey et al., 2019) showing that, with some new procedures, it can be possible to remove unwanted sources of scan variability while simultaneously increasing the power and reproducibility of statistical analyses. However, the issue of harmonization of MRI data is far from being solved, with open challenges involving image acquisition, analysis and data storage.
Issues with image acquisition include standardization across scanners of certain characteristics (e.g., field strength, vendor, sequence type, coil channels, gradient distortions) and subject-related factors (e.g., positioning, motion, etc.). The key concept is the reproducibility of the MR examination, which is difficult to obtain even from the same vendor (Shinohara et al., 2017). Practical solutions have been proposed by the 2021 MAGNIMS-CMSC-NAIMS guidelines, merging recommendations from the Magnetic Resonance Imaging in Multiple Sclerosis study group, Consortium of Multiple Sclerosis Centres, and North American Imaging in Multiple Sclerosis Cooperative, on the use of MRI in MS for clinical implementation in the diagnostic process and for establishing prognosis and monitoring patients (Wattjes et al., 2021a). While some serious attempts to standardize image acquisition protocol have been performed, this is much less the case for image analysis harmonization, which should include: i) quality assessment (QA) to homogeneously check the initial images, classifying or excluding suboptimal examinations, and fixing possible errors through a modified pipeline, and ii) choice of the most appropriate analysis method for a given task. Recommendations to improve imaging analysis of brain lesions and volume in longitudinal MS studies have been published by the MAGNIMS group (Vrenken et al., 2013), but many issues are presently unsolved and the harmonization of imaging analysis remains a concern. Finally, it is now clear that the availability of efficient data storage and sharing methods can have a huge impact to facilitate the management of large imaging datasets.
Against this background, this review focuses on the state of the art and desirable future developments in the harmonization of acquisition, analysis, and storage of large-scale multicenter MRI data of MS cohorts. We will refer here to strategies aiming at reducing, after merging brain MRI scans from different datasets, the technical sources of variability due to acquisition and analysis but preserving the variability due to the disease-related abnormalities. To do this, we will organize this review as follows: first, we will describe the impact on imaging biomarkers used in MS of the most common sources of variability related to MRI scan acquisition and analysis and the possible solutions to deal with them; second, we will describe the latest statistical strategies to merge multicentre MRI data and to store and retrieve these large datasets. Third, we will highlight recent experiences with large multicenter MRI datasets with a focus on how the issue of data harmonization has been faced. Finally, we will describe future perspectives and provide conclusive recommendations.
2. Methods
An international workshop was held in Siena, Italy in 2019 under the auspices of the MAGNIMS study group, an intellectually independent European network of academic centers with a common interest in the study of MS with MRI (www.magnims.eu). The panel convened to the workshop was composed of experts in the use of MRI for the diagnosis and management of MS, and included neurologists, neuroradiologists and physicists from MAGNIMS institutions and one invited non-MAGNIMS expert. The panel met to present and discuss data from MS multicenter research studies (published in English) leading to advances in knowledge in the harmonization of MRI data. The first draft of the manuscript was written by the first and last authors and was based on contributions from each panelist. The draft was then circulated to all members, who iteratively modified the document until a final consensus was reached by all panel members.
2.1. Harmonization of MRI biomarkers in MS
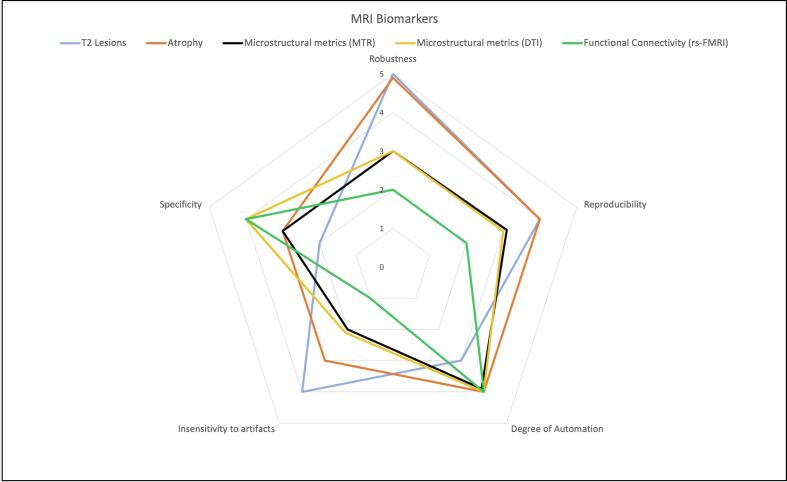
We will tackle here issues related to the lack of harmonization of the MRI data from multiple sites and provide tailored solutions to reduce sources of variability for MS imaging biomarkers that have been extensively used in multicenter studies. For each MR-related biomarker, solutions to harmonize image acquisition and analysis will be considered. Fig. 1 shows the suitability of each MRI-derived metric towards harmonization, and Table 1 summarizes the main sources of variability in MRI acquisition and analysis and provides recommendation for their harmonization.
Fig. 1.
Consensus Chart on the maturity of the main MRI biomarkers used in MS towards harmonization.
Table 1.
Sources of variability related to MRI scan acquisition and analysis and recommendation to reduce their impact for the most common imaging biomarkers used in MS.
| Biomarkers | Sources of variability in MRI acquisition | Sources of variability in MRI analysis | Recommendations for harmonization of MRI acquisition and analysis |
|---|---|---|---|
| WM lesions |
|
|
|
| Atrophy |
|
|
|
| MTR-derived metrics |
|
|
|
| DTI-derived metrics |
|
|
|
| Functional MRI-derived metrics |
|
|
|
Abbreviations: MR: Magnetic resonance; DL: Deep learning; QC: Quality Control; MTR: Magnetization Transfer Ratio; GM: Grey Matter; WM: White Matter; ROI: Region of Interest; TE: Echo Time; TR: Repetition Time; ICA: Independent Component Analysis; EPI: Echo planar imaging.
Using the radar chart, each biomarker was ranked on 5 features that make it suitable to be used in multi-centers studies. The values range from 0 (no suitability) to 5 (excellent suitability). The scores of the different items were decided via consensus after being proposed by the first author. All co-authors agreed with the proposed scores. This multivariate, qualitative representation allows to compare the most used MRI biomarkers in MS. The chart shows that while atrophy measures (orange) and T2-Lesions (light blue) are more reliable measures in multi-center studies than functional connectivity related metrics (green), they do lack in specificity.
2.2. White matter lesions
The presence of T2-weighted (T2-w) hyperintense brain white matter (WM) lesions as detected by MRI is the hallmark of MS and play a crucial role in diagnosis, prognosis and treatment response of MS patients (Wattjes et al., 2021a). The 2017 revision of the MRI criteria for the diagnosis of MS recommended a strict standardization of MRI acquisition to avoid misdiagnosis (Thompson et al., 2018). Recently, the 2021 MAGNIMS–CMSC–NAIMS consensus recommendations provided guidance on how and when to use MRI for the management of patients with MS and suggested changes in MRI acquisition protocols to improve diagnostic accuracy (Wattjes et al., 2021a).
To minimize between-site variability while maintaining maximum sensitivity in detecting brain WM lesions on clinical scanners, 3D T2w-fluid attenuated inversion recovery (FLAIR) images with spatial resolution of 1 mm3 at a minimum strength of 1.5 T are recommended, as they lead to both improved lesion detection and realignment of anatomic orientation to detect new lesions in serial MRI scans (Wattjes et al., 2021b). Magnetic field strength also may play a role, as the visibility of brain WM lesions is higher at 3 T than 1.5 T (Hagens et al., 2019).
For quantitative measurement of brain WM lesion volume (LV) in MS, early work using a semi-automated thresholding technique observed that, while LVs differed between fast-FLAIR, RARE (Rapid Acquisition with Relaxation Enhancement) and GRASE (GRadient And Spin Echo) sequences, repeatability was similar (Rovaris et al., 1999). For automated methods, while the performance on 2D FLAIR is limited, some methods on 3D FLAIR exhibit good performance (Egger et al., 2017). Direct comparisons between scanners are scarce but between-scanner variability of LV is not as large as the variability found between different types of analysis software. However, even with harmonized protocols and scanners from a single vendor, small systematic between-scanner LV differences remain (Shinohara et al., 2017).
Quantitative measurement of spinal LV is under development and so the detection of these lesions represents the main goal. New automatic frameworks for segmenting the spinal cord and intramedullary MS lesions have been recently developed (De Leener et al., 2017, Gros et al., 2019). The standardization of imaging protocols may potentially improve the performance of these tools and help the identification of very small lesions while reducing the number of false positives (Cohen-Adad et al., 2021a, Cohen-Adad et al., 2021b). Various studies showed higher sensitivity to lesions for axial compared to sagittal images, including lesions located in the lateral columns, lesions of smaller size and gadolinium (Gd) enhancing lesions (Breckwoldt et al., 2017). Studies also showed that proton density (PD) images are sensitive (Karavasilis et al., 2019), making dual-echo or similar multi-contrast sequences the preferred imaging approach, given the technical challenges still associated with cerebrospinal fluid suppression in spinal cord FLAIR scans. Ideally, pooling together 3D T2w and T2* weighted axial images would allow a better automated MS cord lesion segmentation (Gros et al., 2019).
Harmonization of the MRI acquisition
Lesion number and volume should be assessed on images acquired on scanners with the same magnetic field strength (Di Perri et al., 2009, Stankiewicz et al., 2011). The automated detection of lesions can be improved by correcting for B1 inhomogeneity (Sajja et al., 2006). To our knowledge, no studies have been performed to assess, using the same MR scanner and sequence, the impact of a change in TR or TE on the evaluation of brain and spinal MS lesions. By contrast, for voxel dimensions, such a comparison was performed and suggested that lesional measures should be obtained from images with the same voxel dimensions (Molyneux et al., 1998).
Harmonization of MRI analysis
Manual lesion segmentation has always been considered the gold standard of LV measurement. However, this approach is time-consuming and not feasible in large MR datasets. In addition, the growing use of 3D FLAIR images makes this task nearly unfeasible in a lab setting (Filippi et al., 2019). The first step to obtain comparable results in LV number and estimation is to perform a careful QA to exclude images with low quality and artefacts. LV can be biased also by the intrinsic variability in MRI intensity. In a recent paper (Shinohara et al., 2017), where a single MS patient was scanned-rescanned (with replacement) seven times at seven different centers all equipped with a 3 T scanner and performing the same MRI acquisition protocol, the center explained 80% of variability in manual lesion outlining. The same dataset was used to evaluate the improvement in manual lesion segmentation when MICA (multisite image harmonization by CDF), a non-linear intensity transformation, was first applied to each MR image (Wrobel et al., 2020). This preprocessing step reduced the inter-center variability by one third, leading to a substantial improvement in lesion estimation.
In last few years, an impressive number of new tools to automatically quantify lesions in MS have been based on artificial intelligence (AI) algorithms (Zeng et al., 2020). Deep learning (DL) based algorithms have shown good performance on independent datasets acquired with the same MR protocol. Studies showed, however, that their performance decreases when applied to images with features (i.e., signal to noise ratio [SNR], contrast to noise ratio [CNR]) different from those used in the training dataset (Mårtensson et al., 2020) or when simple procedures such as the facial features removal (FFR) (a step implemented before data sharing in order to protect the subject’s privacy) that could change the overall histogram intensity of the image are applied (de Sitter et al., 2020). Nonetheless, using transfer learning, specific features of data from a new scanner can be learned to refine an existing trained network, which can yield good performance even when re-training based only on a single subject (Weeda et al., 2019a, Valverde et al., 2019).
Recommendations
Against this background, 3D FLAIR images with same/similar acquisition parameters should be used to outline lesions, after inhomogeneity correction and intensity normalization. Isotropic 3D FLAIR images should be preferred to anisotropic 2D as they are more sensitive to lesion detection and less sensitive to artifacts. (Filippi et al., 2019). The criteria used to perform the QA should be provided. DL methods should be applied to images similar to those of the training dataset. These automated methods take advantage of and improve their performance, when trained on more than one MR modality. Thus, for example, isotropic 3D T1-W, 3D T2-W and 3D FLAIR images could be used together to increase the precision of the trained algorithms. The magnetic field strength should be taken into account when merging lesion outputs from scanners with different magnetic fields.
2.3. Atrophy
Atrophy measurements have been extensively used in group studies to chart the MS disease course and to test treatment effects (Sastre-Garriga et al., 2020). However, brain volume measurements, both cross-sectional and longitudinal, can differ systematically between images acquired i) from scanners of different vendors (with rather poor agreement) (Biberacher et al., 2016), ii) before and after scanner upgrade (Steenwijk et al., 2016), and iii) even from two different scanners of the same model in a single site (Takao et al., 2011). Moreover, changes of T1w sequence (i.e., from magnetization prepared-rapid gradient echo [MP-RAGE] to inversion recovery fast spoiled gradient recalled echo [IR-SPGR]) may have greater effects on brain volume measurements than intra-vendor scanner upgrades (Lee et al., 2019).
Finally, the effects are not the same for all types of measurements (i.e., whole brain versus smaller brain structures) or analysis software used, as some seem to be more robust than others (Steenwijk et al., 2017). Measurement of spinal cord volume in MS has mostly focused on the cervical segment, with issues similar to those of the measurement of brain volumes.
Harmonization of the MRI acquisition
Unenhanced 3D T1-weighted MRI is the reference sequence for brain volume quantification, although T2-FLAIR data have shown to provide robust and reproducible results (Goodkin et al., 2021). Specific strategies can be adopted to reduce the impact of B0- and B1-inhomogeneities. The former can be reduced by repositioning the patient at the magnet isocenter (Caramanos et al., 2010) and using distortion correction techniques commonly implemented by scanner vendors (Jovicich et al., 2006), including pre-computed displacement tables, phase mapping, and forward/reverse frequency-encoding gradients (Doran et al., 2005). The latter can be corrected after image acquisition by using several open-source online software tools (Song et al., 2017), often included in the analysis algorithms used for biomarker evaluation, e.g. the N3 and N4 methods (Sled et al., 1998, Tustison et al., 2010). In addition, the variability in MRI-based global and regional volume measurements can be reduced at the statistical analysis taking into account magnet vendor and magnetic field as confounding factors (Potvin et al., 2016, Battaglini et al., 2019). A standardized protocol should be always used in multicenter studies, as it has been shown that the variance of brain volume measures increases with greater deviation from the standardized protocol in terms of repetition time, echo time and spatial resolution (Potvin et al., 2019).
Atrophy of the upper cervical cord can be assessed through head scans rather than dedicated spinal cord scans (intra-class correlation coefficient [ICC] = 0.987) making reliable standardized acquisition more accessible (Liu et al., 2016), although correction for distortion due to gradient nonlinearity is needed (Papinutto et al., 2018). Systematic between-scanner differences in mean upper cervical cord area (MUCCA) have been reported for patients scanned on multiple scanners as well as for multi-site travelling volunteers (Weeda et al., 2019b, Lukas et al., 2021). Within-subject identical scanning protocol is mandatory to have reliable quantification of longitudinal spinal cord atrophy (Weeda et al., 2019b, Papinutto and Henry, 2019).
Harmonization of MRI analysis
The first preliminary step to obtain comparable results in brain volume assessment is to perform a careful QC to exclude images with a suboptimal quality. Strategies of voxel intensity normalization can reduce atrophy measurement error (Wrobel et al., 2020, Battaglini et al., 2018). DeepHarmony, a method of DL architecture aiming at creating intensity transformation maps to equalize images from two different MR scanners (Dewey et al., 2019); showed a significant decrease in the brain volume measurement error on a dataset of 12 subjects scanned at two different MR centers. To face the global intensity changes due to the pathological features such as the presence of T1-weighted hypointense lesions, a number of methods include the “filling” of WM lesions before image segmentation (Nakamura and Fisher, 2009, Sdika and Pelletier, 2009, Battaglini et al., 2012), with a great reduction of the measurement error. The defacing step showed a significant systematic increase in the variability of brain volume measurement with over- or underestimation depending on the software used (de Sitter et al., 2020).
Over the last decade, various studies assessing the variability in brain volume measurements (Sastre-Garriga et al., 2020) showed that different software tools have a weak ICC (Guizard et al., 2015, Popescu et al., 2016, Battaglini et al., 2009). Most of these tools showed increased variability when applied to MR images of the same subject that were acquired with different scans and/or vendors, in comparison with those acquired with the same MRI equipment (Storelli et al., 2018). Several studies have tried to optimize the software setting options in order to minimize the technical variability (Popescu et al., 2012, Lutkenhoff et al., 2014). More recently, several studies investigated the performance of DL architectures trained on binarized segmented GM regions obtained by specific software packages (Gabr et al., 2020, McKinley et al., 2021) and therefore referred to here as “silver standard”, as opposed to the “gold standard” that is represented by the binarised GM map manually segmented. Results showed comparable accuracy between the images used for training and those used for the evaluation, also when a small sample size (e.g., 10–20 subjects) was used for training the DL architecture (Narayana et al., 2020). Although promising, these results need confirmation when the training dataset is acquired with a different protocol compared to the evaluation dataset.
Semiautomated software packages have been developed to assess spinal cord atrophy both cross-sectionally (De Leener et al., 2017, Horsfield et al., 2010), measuring the spinal cord cross-sectional area (CSA), and longitudinally, measuring the generalized boundary shift integral (GBSI) (Prados et al., 2020).
Recommendations
In general, the method of choice for quantification of brain and spinal cord volume remains the consistent use of the most appropriate MR sequence for the type of software package that is going to be used (e.g., unenhanced 3D gradient-echo T1-weighted), with the identical scanning protocol and as little hardware or software change as possible. When dealing with 2D acquisitions, almost all methods reformat input scans to 1 mm isotropic resolution to improve measurement precision (Amann et al., 2015). Hence, a direct acquisition of isotropic 3D T1-weighted images is preferable as it is less distorted by blurring due to interpolation. Thus, we recommend that brain volume measurement should be assessed on lesion-filled 3D T1-weighted images of patients placed at magnet iso-center, bias-field corrected, and intensity normalized. The analysis pipeline should be clearly detailed as well as the criteria used to perform the QC.
DL methods are promising but need to be applied to images similar to those used for the training, where the output “to be learnt” has been obtained using the same software.
Spinal cord sequences should be acquired according to the selected software for the quantification of volume. As for brain atrophy, strictly harmonized protocols should be used.
2.4. Microstructural biomarkers
Using computational models of biophysical properties of tissues, it is possible to derive metrics sensitive to microstructural integrity and metabolic information of the underlying tissue (Granziera et al., 2021). Ultimately, the aim would be to provide patient-specific metrics able to predict outcomes and contribute to decision making on disease management (Granziera et al., 2021).
To improve the reliability of microstructural biomarkers (e.g., using magnetization transfer ratio [MTR], myelin water imaging [MWI]), inter- and intra- intensity image harmonization is required to increase the sensitivity to disease-related effects and to better assess longitudinal changes. For specificity to biophysical properties, which allows to understand mechanisms of disease progression and treatment response, harmonization is needed to evaluate the tissue integrity (e.g. neurite density index, bound pool fraction).
Despite their potential value, non-conventional MRI metrics are still dependent on acquisition parameters, which are heavily influenced by coil geometry, gradient systems and magnetic field strengths. This, together with the restricted availability of some non-conventional MRI sequences, has limited the use of some promising metrics (e.g., MWI) in multicenter studies and is important to consider in other, more widely used measures (e.g., MTR, diffusion metrics), which will be discussed here.
Microstructural damage assessment using MTR
Magnetization transfer (MT) images are obtained by the normalized difference between two images with and without MT pulse. They showed sensitivity to the inherent relaxation properties of heterogeneous tissues (Ropele et al., 2005). MTR is a semi-quantitative metric obtained by the two-pool model (restricted protons of macromolecules and free protons of bulk water), predominantly sensitive to myelin content, but also influenced by axonal loss and inflammation. MTR can be measured on multi-vendor MRI images using a standardized acquisition protocol (Barker et al., 2005) and has been used as outcome measure in clinical trials in relapsing-remitting (RR) and progressive MS (Romme Christensen et al., 2014, Miller et al., 2015). Advances of modelling of the MT effect led to the development of multi-modal acquisition protocols such as the multi-parametric mapping, which allows to quantify several parameters sensitive to tissue microstructural changes (MT saturation, apparent proton density, relaxation times). These parameters have been shown to be comparable, at least between scanners of the same vendor (Weiskopf et al., 2013).
Harmonization of the MRI acquisition
Due to dependencies on the MT weighting radiofrequency pulse, harmonizing this protocol is challenging, although recent developments in acquisition and scanner parameters (i.e. use of different resolutions and selective pulses for saturation) attempted to reduce confounding factors, thus improving MT feasibility (Zhang et al., 2019). Indeed, i) MT images from scanners with the same strength of magnet could be merged if they are equipped with the same transmission coil (Tofts et al., 2006); ii) impact of B1-inhomogeneity could be corrected by computing the linear regression between the B1 errors and MTR values (Ropele et al., 2005); iii) acquisition parameters should be kept constant, as variability in the MTR measurement can be up to 15% when subjects are acquired on the same scanner but with different MR protocols (Sormani et al., 2000).
Harmonization of MRI analysis
Features of MTR intensity histograms (Tortorella et al., 2000, Fernando et al., 2005) and averaged MTR values (De Stefano et al., 2006) of the whole brain, GM, WM and lesions are the most widely used measures obtained from the MT images. There are currently no open-source software tools able to automatically assess MTR values from specific ROIs. However, this should not represent a major limitation for MTR measurement, once registration and segmentation steps are fine-tuned on MR images of each center and visually checked.
Recommendations
MT images need to be acquired with the same transmission coil and corrected for B1 errors. The same acquisition protocol needs to be used. To obtain a precise assessment of average MTR values within the ROIs, registration needs to be carefully checked to avoid the inclusion of inappropriate voxels.
Microstructural damage assessment using Diffusion Tensor Imaging (DTI)
Information on microstructural integrity can also be assessed using diffusion weighted imaging (DWI) with a minimum of 6 gradient directions, through either model-free approaches or computational modelling of DWI signal (Cercignani and Gandini Wheeler‐Kingshott, 2019). The most simplistic model is diffusion tensor imaging (DTI), which reduces the complex motion of water molecules within a tissue to three eigenvectors and corresponding eigenvalues. DTI provides metrics such as fractional anisotropy (FA), mean diffusivity (MD) and axial and radial diffusivity that are rotationally invariant, within the subject’s frame of reference and heavily influenced by the underlying tissue microstructure that poses hindrance or restriction to free motion of water.
Recent studies reported that DTI can be highly reproducible and standardized at brain (Palacios et al., 2017) and spinal cord (Samson et al., 2016) levels. However, DTI metrics are non-specific since they are highly sensitive to microstructural alterations and higher order models are required to disentangle the source of signal anisotropy. Moreover, the time-dependence of the diffusion signal, that is how the signal changes by increasing the observational diffusion time, depends on the size of the underlying axons. This has been extensively studied in the brain (De Santis et al., 2016) and is particularly important for imaging the spinal cord, which has larger axons than brain and where, with a short diffusion time, a high number of large axons may not contribute to the signal (Grussu et al., 2019). To overcome these limitations, advanced DWI approaches have been developed (Assaf et al., 2004). However, although more specific to tissue microstructure, further studies are needed to prove the translation of these multiple-scale approaches into clinical practice. (Cercignani and Gandini Wheeler‐Kingshott, 2019).
Harmonization of MRI acquisition and data-preprocessing
When applying a standardized MRI protocol across different centers, averaged FA and MD measures from large homogeneous ROIs (e.g., the mid-sagittal section of the corpus callosum) can be merged. On the other hand, differences in vendors, magnetic field strength and number of diffusion gradients should be statistically taken into account when measures from smaller ROIs are obtained, in order to reduce the intra-scanner and intra-protocol sources of variability (Pagani et al., 2010). B0 susceptibility-induced distortions can be corrected using parallel imaging and acquiring MRI data with opposite phase-encoding directions. Susceptibility induced distortions can influence both intra- and inter-subject reproducibility (Ganzetti et al., 2016), but its effect can be reduced using the DTI-specific software (e.g., FSL topup (Andersson et al., 2003). Finally, specific DTI related sources of variability such as the eddy-current distortions, the Gibbs ringing artefacts (Perrone et al., 2015) and those related to table vibration during the acquisition (Gallichan et al., 2009) can be controlled by specific software tools (Maximov et al., 2019). Same voxel size, b = 0 (i.e., no -diffusion weighting) volumes, echo time and repetition time should be implemented across centers, to reduce the difference in SNR measure and harmonize the overall quality of the images (Laganà et al., 2010).
Harmonization of MRI analysis
Several software tools are able to derive FA and MD images from DTI data (Soares et al., 2013, Tournier et al., 2019). A comparison among four of them on the reconstruction of the main WM tracts showed poor between-software agreement for FA, axial and radial diffusivity (Christidi et al., 2016).
Recently, various studies in the DTI field have applied DL techniques in order to reduce the scan time (Tian et al., 2020), reliably reconstructing WM tracts (Li et al., 2020) and characterizing pathological conditions (Marzban et al., 2020). Although promising, such techniques need a further extensive validation. A few studies applied AI methods to DTI data, with the aim to characterize different stages of MS (Marzullo et al., 2019, Oladosu et al., 2021, Kontopodis et al., 2021).
Recommendations
Analysis from multicentre DTI can be more precise if images are acquired on MR scanners with the same magnetic field strength, number of gradients and opposite phase-encoding directions. Then, both pre-processing (i.e., correction for eddy currents, motion, B0- and B1-inhomogeneity) and post-processing software tools should be used with identical and well documented analysis algorithms.
2.5. Functional biomarkers
An important remark should be made on functional MRI (fMRI) and functional connectivity (FC). From a set of T2*-weighted EPI volumes it is possible to model the local tissue changes in the concentration of oxyhaemoglobin, which is diamagnetic, compared to deoxyhaemoglobin, which is paramagnetic. Such changes are also linked to the baseline neuronal activity at rest, thus leading to a remarkable increase in research studies of resting state fMRI (rs-fMRI) in MS. It is well known that low frequency signal fluctuations of regions belonging to networks supporting specific functions are synchronized, with altered FC strength occurring in pathologic conditions (Castellazzi et al., 2018). Harmonization of rs-fMRI acquisition should aim to strengthen the statistical inference of FC metrics and potentially propose an approach based on the single-subject comparison to healthy population. It is important that rs-fMRI acquisition protocols across scanners have the same sensitivity, which can be measured with parameters such as the temporal signal-to-fluctuation-noise-ratio (SFNR) (Glover et al., 2012).
Harmonization of the rs-fMRI acquisition
3 T scanners showed significantly higher reproducibility in blood-oxygenation level dependent (BOLD) signal than 1.5 T scanners (Zou et al., 2005, Badhwar et al., 2020). B0-susceptibility induced distortion can be reduced with the use of parallel imaging and the acquisition of data with opposite phase encoding directions (Andersson et al., 2003). Sophisticated strategies have been recently introduced to reduce the impact of B1-inhomogeneity through the creation of B1 maps by modelling the B1- receive and B1 + transmit fields (Glasser et al., 2013). Head motion is usually corrected by aligning EPI images (Graedel et al., 2017), and the impact of patient movements can be further reduced by applying independent component analysis (ICA)-based method (Pruim et al., 2015); eye movement artefacts can be reduced by asking the subject to keep the eyes closed during the acquisition or to fix on a point (Costumero et al., 2020); physiological noise can be reduced by recording electrocardiogram (ECG) signal and regressing it out in the statistical analysis (Glover et al., 2000); ICA-based methods can be a good alternative when ECG recording is not available (Behzadi et al., 2007). A short repetition time (e.g., 1,5 s or less) can provide a reliable rs-fMRI sequence to be potentially used in the clinical studies (Jahanian et al., 2019), by reducing the impact of eye motion and physiological noise compared with longer (i.e., >2 s) EPI sequences (Yang et al., 2007). However, this requires the availability of efficient simultaneous multi-slice acquisition techniques on the scanner. The echo time selection is important for distinguishing between intra- and extra-vascular BOLD contributions (Ragot and Chen, 2019). Finally, larger voxel sizes can potentially increase SNR (Bennett and Miller, 2010).
Harmonization of rs-fMRI analysis
In general, rs-fMRI analysis methods can be divided into two groups: model-dependent, such as seed-based methods (Andrews-Hanna et al., 2007), and model-free methods, including principal component analysis (PCA) (Friston et al., 1993), singular value decomposition (SVD) (Worsley et al., 2005) and ICA (Beckmann et al., 2005). In a recent study (Botvinik-Nezer et al., 2020), 108 datasets with the same task (cognitive)-fMRI were independently analyzed by 70 teams over the world, and no two of them chose identical workflows for data analysis. Different sources of variability were identified, from the smoothing level (the higher the spatial smoothing, the larger the activated region) to the head-motion correction procedures and the software package used. Overall, this study showed that “analytical choices have a major effect on reported results”.
Although AI methods have extensively been applied to extract significant features for disease classification (Wen et al., 2018), they need yet to be extensively validated on external datasets to prove reliability and robustness.
Recommendations
The most useful recommendation to date is that a harmonized dataset should be acquired on scanners with the same magnetic field strength, using opposite phase-encoding directions and the same MRI protocol. Both pre-processing (i.e., correction for head- cardiac- and eyes-related motion artefacts, correction for eddy currents, B0- and B1-inhomogeneity) and post-processing analysis software tools should use an identical and well documented pipeline.
2.6. Mega and Meta-analysis
Two distinct strategies can be followed to statistically analyze MRI data from multicenter studies, named mega- and meta-analysis. In the former, raw/individual data are pooled and analyzed centrally. In the latter, data of each center are locally analyzed and only the summary statistics from all centers are brought together, after weighting the role of each center-specific statistic. Both procedures must indicate the criteria followed to perform the QA of the images before the analysis, in order to avoid bias. This is particularly important when, as in the meta-analysis procedure, data can be collected retrospectively, with the risk of merging MR images with substantial differences in quality.
An example of mega-analysis is the creation of normative data for biomarkers such as atrophy rate (Battaglini et al., 2019) and volume of deep GM structures (Potvin et al., 2016), where a single regression is performed on the pooled MRI data. In these cases, statistical analysis acts as a harmonization procedure per se considering, for example, different vendors or magnetic field strength as confounding variables. However, statistical procedures, such as regressions, can be performed also in the context of data harmonization at voxel-level, for modelling and subsequently removing the local unwanted inter-site variability. A notable example is ComBat (Fortin et al., 2016, Fortin et al., 2018), a batch-effect correction tool used in genomics and recently implemented for multi-center MRI data analysis.
In performing meta-analysis, different criteria of gauging the role of each center-specific statistics can be employed. METAL (Willer et al., 2010), one of the most commonly used software platforms to perform meta-analysis, has implemented two strategies of weighting, which are based on the inverse variance and the sample size. Using METAL with a sample size approach, recently a genome-wide association study (GWAS) discovered four new loci associated with hippocampal volume (Hibar et al., 2017). These jointed analyses, combining genomic and imaging biomarkers, may have a great potential implication in the MS field.
Recommendations
Mega– and meta-analyses are able to reduce the impact of the heterogeneity in acquisition protocols but have both pros and cons (Kochunov et al., 2014). On the one hand, mega-analysis, unlike meta-analysis, allows considering the individual variability and reducing the inter-center variability, even at voxel level. On the other hand, meta-analysis can be used to easily analyze retrospective data and without image data sharing, thus managing large MRI datasets without facing issues related to data organization and storage. Although both statistical approaches may be useful for merging images acquired in different centers with different protocols, a clear indication on the type of images included in the analysis remains mandatory. For example, if datasets of both 3D and 2D-T1W are merged for atrophy studies a reliability analysis must be performed (Eshaghi et al., 2021).
3. Image and analysis pipeline transfer and storage
Over the past few years, we moved from small studies with hundreds of subjects acquired cross-sectionally, to large multi-modal studies with thousands of subjects and several time points. Collaboration has increased among centers and consequently the need to merge different studies and manage large MRI datasets. This new scenario highlighted the key role of efficient data storage, managing and sharing, which can positively impact research results, ease longitudinal studies, facilitate data transfer, allow multiple users to work on the same dataset, optimize computational resources and, ultimately, reduce costs. Recently, Dojat et. al presented a comprehensive work analyzing different software and hardware infrastructures for image transfer and storage (Dojat et al., 2017). All these features are essential in medical image-based research in the big data era where there is an increasing demand to keep track of the projects to avoid starting from scratch every time that new people start working on the data. In this context, tools such as XNAT (eXtensible Neuroimaging Archive Toolkit) (Marcus et al., 2007) are platforms that support medical image-based research data handling by efficiently managing clinical and non-clinical data for multi-center studies or data sharing.
Sometimes, however, it might not be possible to install the infrastructure to adopt a data management platform. Despite this, a reliable and robust data sharing between centres is still possible using a common folder and data structure. In that sense, BIDS (Brain Imaging Data Structure) (Gorgolewski et al., 2016) has become a standard for annotating and storing neuroimaging data. The majority of data management platforms support importing or exporting data in BIDS format and BIDS is becoming popular across the neuroimage community. Noteworthy, specific platforms such as XNAT do facilitate sharing of analysis pipelines where the optimized settings for each procedure used are “frozen” and therefore the analysis can be easily run using an identical setting from MR images of different centers.
Recommendations
The implementation of a state-of-the-art platform that efficiently stores, manages and shares a large amount of medical image-based research data that are being acquired nowadays is an investment that will have a major mid- and long-term impact in daily work. The common and centralized data access ensures to avoid repetitions (i.e., to compute several times brain lesion filling and tissue segmentation), promotes collaborations across research groups, keeps track of all the changes and data manipulation, avoids data loss and helps to keep a fluent and consistent workflow across people and along the projects. Finally, it is strongly recommended to share the analysis pipeline, as it is known that the same dataset can provide non-comparable results if analyzed with the same software but with different options (Botvinik-Nezer et al., 2020).
4. Current experience on harmonization of big data
4.1. Experience of ADNI and European initiatives on Alzheimer’s disease
The Alzheimer’s disease neuroimaging initiative (ADNI) is currently the most successful and valuable example of a freely accessible longitudinal neuroimaging resource available for the study of Alzheimer’s disease (AD), mild cognitive impairment (MCI), and healthy aging brain (Weiner et al., 2017). This public-private partnership, now at its fourth wave of funding, has the primary goal to test whether serial imaging data (MRI and positron emission tomography [PET]) can be combined with clinical, neuropsychological and biological markers to measure the progression from normal aging to AD (Weiner et al., 2017). The most important changes over time have been: (i) inclusion of new subjects especially cognitively normal controls and patients with MCI in addition to previously included subjects that are still “on study” (total expected number >1000), (ii) regular update of available datasets (last update: 10 February 2020), (iii) continuous effort to provide uniform neuroimaging protocols for reaching an improved consistency in data analysis. Assessments are performed 6-monthly with now many years of follow-up available for the original subjects.
From the release of ADNI dataset, a large amount of studies has enhanced our knowledge about AD so far, ranging from machine learning neuroimaging studies for a correct AD/MCI classification and prediction of AD conversion (Dimitriadis et al., 2018) to discoveries far beyond imaging, such as showing the importance of genetic and biological factors for AD heterogeneity (Ramanan et al., 2021).
In terms of infrastructure, the free access to raw DICOM images through the LONI website (http://adni.loni.usc.edu/) poses some challenges of use, leading to duplication of efforts and inconsistent data usage. However, ADNI is under constant improvement and progressively focusing on advanced MRI techniques and novel PET tracers. Indeed, in the last version (ADNI-3), the scan protocol moved to accelerated 1 mm3 3D T1-weighted sequences, from 2D T2-weighted to 3D T2-weighted FLAIR sequences, a wide use of arterial spin labelling (ASL) and improved DTI and rs-fMRI protocols (Jack et al., 2015).
Similar European initiatives include PharmaCog (www.alzheimer-europe.org/Research/PharmaCo), which has a smaller dataset of healthy controls and MCI patients with high quality MRI, with access only through the principal investigators. Using this dataset, it has been shown that fMRI and DTI metrics are quite reproducible across centers (Jovicich et al., 2016). Recently, MRI structural biomarkers have been identified from this cohort to improve subject selection and to be used as surrogate outcomes of disease progression. Among these, hippocampus has been confirmed to be the most sensitive region to disease progression in MCI with prodromal AD (Marizzoni et al., 2019).
More recently, the European prevention of Alzheimer's dementia (EPAD) program (www.ep-ad.org) aims to deliver a platform, adaptive, phase 2 proof of concept trial for the secondary prevention of AD, including a longitudinal cohort study (Ritchie et al., 2020).
EPAD currently involves 21 centers across Europe, and it has enrolled >2000 subjects with normal cognition or MCI with yearly follow-up assessment including biological and cognitive markers. The EPAD imaging protocol includes conventional MRI sequences for all subjects, advanced techniques such as rsFMRI, DTI, ASL and susceptibility weighted imaging (SWI), in >50% of the dataset with repeated measurements (ten Kate et al., 2018). A subset of subjects will also have amyloid-PET for a specific substudy called AMYPAD (www.amypad.eu). The EPAD and AMYPAD data are structured in XNAT and will be made publicly available in the future.
4.2. UK Biobank experience
UK Biobank (www.ukbiobank.ac.uk) is a large prospective cohort study that recruited 500,000 people aged between 40 and 69 years in 2006–2010 from across UK. Participants underwent a range of physical measures, provided their details, and gave blood, urine, and saliva samples for future analyses. They also agreed to be followed through linkage to their medical records. The proposal for an imaging enhancement in 100,000 participants was made in 2012. Meanwhile, three imaging centers have been operating (Stockport, Newcastle, Reading) and over 50,000 participants have been scanned. Access to the imaging datasets is provided (since March 2020) by previous examination of the researcher and its research proposal. The simultaneous imaging of brain, heart, arteries, abdomen, and bone in each individual and the combination of these data will provide a dataset that is uniquely able to address questions regarding the relationships between phenotypes across organs and to assess pathogenic mechanisms operating at a systemic level.
To meet the temporal, financial, and geographic constraints of scanning 100,000 participants, the measurements are made in different locations equipped with two scanners each (one 3 T MAGNETOM Skyra and one 1.5 T MAGNETOM Aera). Scanning sites were selected to optimize volunteer recruitment from UK Biobank participants across the UK. The comprehensive brain program runs for about 30 min exclusively on the 3 T systems while an integrated body and cardiac program is executed on the 1.5 T scanners for 10 and 20 min, respectively.
In general, MRI at UK Biobank is subject to the same considerations as other long-term investigations with respect to protocol design, data storage and QA. However, the intended number of participants requires specific operational decisions to provide a most consistent dataset: 18 participants are invited to each center every day. This implies that a delay or scan repeat must remain confined to the corresponding participant and lead to a shortening of the imaging program.
Along with the examination protocols, a QA procedure was developed with three main aspects to consider over time: status of scanners, actual image quality, and data completeness (Alfaro-Almagro et al., 2018). Specific to UK Biobank in comparison to many other large-scale imaging studies is the use of the same type of scanner and software version over the complete duration of the project. Identical procedures and protocol settings apply at all centers. This provides the potential to differentiate the effect of hardware variations with a great precision and to study individual variations in detail. In order to further separate technical from individual effects, phantom scans are executed daily and the time characteristics of field and noise are analyzed.
4.3. INNI experience
Given the paramount importance of MRI to diagnose and monitor MS, recent collaborations promoted the acquisition of relatively standard conventional MRI protocols. Good examples are the German Competence Network Multiple Sclerosis (https://www.kompetenznetz-multiplesklerose.de), the North American Registry for Care and Research in Multiple Sclerosis (NARCRMS, https://www.narcrms.org/), the Canadian Prospective Cohort Study to Understand Progression in MS (CanProCo) (Dimitriadis et al., 2018) and the MS PATHS (Mowry et al., 2020). With the exception of CanProCo, these initiatives do not include advanced MRI techniques, such as DTI and rs-fMRI, which are very useful to characterize microstructural damage and functional reorganization occurring in this condition. The Italian Neuroimaging Network Initiative (INNI) (www.inni-ms.org/) is one of the latest and promising data sharing initiatives in the field of MRI in MS. It started from four Italian research centers with recognized expertise in the study of neuroimaging in MS (Filippi et al., 2017), with the support of the Italian MS Society, to promote the collection of conventional and advanced structural and functional (including DTI and rs-fMRI) MRI scans as well as clinical and neuropsychological data in a centralized repository. MRI data are added from each centre both retrospectively and prospectively. All imaging biomarkers are analyzed using advanced procedures for harmonization.
The main goals of INNI are to identify and validate novel MRI biomarkers, which can be used as predictors and/or outcomes in future MS studies, and to support a standardized use of neuroimaging protocols in MS at national level. According to the first INNI survey in 2017, >1300 subjects were uploaded on the online platform (Filippi et al., 2017). This number has progressively increased over time reaching, to date, >2500 MS patients and healthy controls (with>4200 MRI scans, including longitudinal assessments). Although similar MR acquisition protocols were implemented by different centers and data have to fulfil minimum requirements to be included, a full standardization of the MRI acquisition protocols across sites was not requested, at least in the first project phase (Filippi et al., 2017). However, a systematic quantitative QA procedure, which is essential to ensure rigorous results, was recently implemented for all data uploaded to the INNI repository, as an initial pre-processing step (Storelli et al., 2019). Thus, when centers upload the MRI scans on the INNI platform, an image QA is performed within database content to rate image artefacts, distortions and inhomogeneities. Only scans with an adequate image quality and a certain degree of harmonization will be used for the INNI research projects.
Table 2 summarizes the main problems related to harmonization and if they have been addressed or not addressed yet by these large MRI datasets.
Table 2.
Examples of how currently available large MRI datasets face the issue of data harmonization from the acquisition to the storage point of view. The tick indicates that the issue is completely (green) or partially (yellow) faced; the red cross means that the issue is still open.
| MRI datasets | Steps towards harmonization |
||||||
|---|---|---|---|---|---|---|---|
| Standardization of acquisition parameters and implementation in clinical protocols | Standardization of procedures for quality control | Availability of objective criteria for inclusion/exclusion of images depending on the endpoint and software | Availability of pipeline of analyses for testing the reproducibility | Incorporation of analysis pipelines in software containers^ to be shared between centers | Automatic control of the conformity of imported data with a predefined study protocol | Availability of mechanisms to ensure privacy and security of the stored data | |
| ADNI |  |
||||||
| Uk Biobank | |||||||
| INNI | |||||||
*Possible loss of privacy for genetic material to identify relatives.
^ Software containers, or dockers, are application build and deployment tool. They are based on the idea that you can package your code with dependencies into a deployable unit called container.
5. Future perspectives and conclusions
Multicenter demographic, clinical and MRI data, ideally combined with blood, cerebrospinal fluid and genetic biomarkers are undoubtedly needed to create the required large datasets to generate models based on artificial intelligence, which should be able to predict disability status and treatment response/failure in MS patients and thus provide a personalized management of patients. However, one of the major challenges to use MRI measures with this approach is the lack of harmonization on acquisition parameters, post-processing analysis tools and storage of the huge amount of imaging data.
Homogeneous acquisition of MR images cross-sectionally and longitudinally across centers is still an unsolved issue, and this article analyses some solutions that could minimize this problem. These solutions should be, ideally, independent of MR vendor and field strength, and of software and hardware upgrades in order to be widely implemented. There is no doubt that the first requirement is to use a core standardized MRI protocol that should be obtained in a reasonable acquisition time in both clinical and research studies. Different scientific organizations in Europe (MAGNIMS) and in North America (CMSC, NAIMS) have actively worked to provide standardized protocols that can be easily implemented in clinical practice (Wattjes et al., 2021a). In this context, the use of synthetic images (e.g., Fingerprinting (Hsieh and Svalbe, 2020), Synthetic MR (Gonçalves et al., 2018) could be of extremely high value as they can provide harmonized, co-registered and simultaneous measures of different contrast-weighted images (Iglesias et al., 2021).
Another essential requirement is to harmonize and validate the different post-processing tools that will facilitate the use of MRI quantitative data for both clinical and research purposes. Ideally, these tools should be available online, open to QA, and harmonized among different MR vendors. Availability of post-processing validated pipeline standards and analysis methods for specific aspects is an unmet need, which clearly requires the cooperation between academic medical centers, biopharmaceutical and bioinformatic companies and MR vendors. This is undoubtedly a win-win strategy between all these partners, whose final goal is to use quantitative and robust data to incorporate in learning health system models intended to provide continual improvement in patient care, particularly regarding personalized selection of optimal treatment initiation and sequencing.
Finally, one recent challenge for researchers using medical imaging is storing, indexing, and sharing their data, which requires professionals with information technology skills, and experts in the complex legal regulations on data management transfer. The complexity of the above-mentioned tasks overwhelms academic researchers and requires technical and human resources that should be provided by the academical centers to support research efforts. Huge efforts are currently being undertaken in order to achieve all the requirements needed to harmonize the acquisition, analysis, storage and secure transfer of imaging data. Proper management of large imaging datasets is one of our greatest opportunities and challenges in the coming years. This is particularly important for offering MS patients an optimal individualized management while minimizing consumption of resources.
CRediT authorship contribution statement
Nicola De Stefano: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Marco Battaglini: Methodology, Writing – original draft, Writing – review & editing. Deborah Pareto: Methodology, Writing – original draft, Writing – review & editing. Rosa Cortese: Methodology, Writing – original draft, Writing – review & editing. Jian Zhang: Methodology, Writing – original draft, Writing – review & editing. Niels Oesingmann: Methodology, Writing – original draft, Writing – review & editing. Ferran Prados: Methodology, Writing – original draft, Writing – review & editing. Maria A. Rocca: Methodology, Writing – original draft, Writing – review & editing. Paola Valsasina: Methodology, Writing – original draft, Writing – review & editing. Hugo Vrenken: Methodology, Writing – original draft, Writing – review & editing. Claudia A.M. Gandini Wheeler-Kingshott: Methodology, Writing – original draft, Writing – review & editing. Massimo Filippi: Methodology, Writing – original draft, Writing – review & editing. Frederik Barkhof: Methodology, Writing – original draft, Writing – review & editing. Àlex Rovira: Methodology, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Shinohara R.T., Oh J., Nair G., Calabresi P.A., Davatzikos C., Doshi J., Henry R.G., Kim G., Linn K.A., Papinutto N., Pelletier D., Pham D.L., Reich D.S., Rooney W., Roy S., Stern W., Tummala S., Yousuf F., Zhu A., Sicotte N.L., Bakshi R. Volumetric Analysis from a Harmonized Multisite Brain MRI Study of a Single Subject with Multiple Sclerosis. AJNR Am J Neuroradiol. 2017;38(8):1501–1509. doi: 10.3174/ajnr.A5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biberacher V., Schmidt P., Keshavan A., Boucard C.C., Righart R., Sämann P., Preibisch C., Fröbel D., Aly L., Hemmer B., Zimmer C., Henry R.G., Mühlau M. Intra- and interscanner variability of magnetic resonance imaging based volumetry in multiple sclerosis. Neuroimage. 2016;142:188–197. doi: 10.1016/j.neuroimage.2016.07.035. [DOI] [PubMed] [Google Scholar]
- Dewey B.E., Zhao C., Reinhold J.C., Carass A., Fitzgerald K.C., Sotirchos E.S., Saidha S., Oh J., Pham D.L., Calabresi P.A., van Zijl P.C.M., Prince J.L. DeepHarmony: A deep learning approach to contrast harmonization across scanner changes. Magn Reson Imaging. 2019;64:160–170. doi: 10.1016/j.mri.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattjes M.P., Ciccarelli O., Reich D.S., Banwell B., de Stefano N., Enzinger C., Fazekas F., Filippi M., Frederiksen J., Gasperini C., Hacohen Y., Kappos L., Li D.K.B., Mankad K., Montalban X., Newsome S.D., Oh J., Palace J., Rocca M.A., Sastre-Garriga J., Tintoré M., Traboulsee A., Vrenken H., Yousry T., Barkhof F., Rovira À., Wattjes M.P., Ciccarelli O., de Stefano N., Enzinger C., Fazekas F., Filippi M., Frederiksen J., Gasperini C., Hacohen Y., Kappos L., Mankad K., Montalban X., Palace J., Rocca M.A., Sastre-Garriga J., Tintore M., Vrenken H., Yousry T., Barkhof F., Rovira A., Li D.K.B., Traboulsee A., Newsome S.D., Banwell B., Oh J., Reich D.S., Reich D.S., Oh J. 2021 MAGNIMS-CMSC-NAIMS consensus recommendations on the use of MRI in patients with multiple sclerosis. Lancet Neurol. 2021;20(8):653–670. doi: 10.1016/S1474-4422(21)00095-8. [DOI] [PubMed] [Google Scholar]
- Vrenken H., Jenkinson M., Horsfield M.A., Battaglini M., van Schijndel R.A., Rostrup E., Geurts J.J.G., Fisher E., Zijdenbos A., Ashburner J., Miller D.H., Filippi M., Fazekas F., Rovaris M., Rovira A., Barkhof F., de Stefano N. Recommendations to improve imaging and analysis of brain lesion load and atrophy in longitudinal studies of multiple sclerosis. J Neurol. 2013;260(10):2458–2471. doi: 10.1007/s00415-012-6762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G., Correale J., Fazekas F., Filippi M., Freedman M.S., Fujihara K., Galetta S.L., Hartung H.P., Kappos L., Lublin F.D., Marrie R.A., Miller A.E., Miller D.H., Montalban X., Mowry E.M., Sorensen P.S., Tintoré M., Traboulsee A.L., Trojano M., Uitdehaag B.M.J., Vukusic S., Waubant E., Weinshenker B.G., Reingold S.C., Cohen J.A. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17(2):162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Wattjes MP, Ciccarelli O, Reich DS, et al (2021) International 2020 MAGNIMS-CMSC-NAIMS consensus recommendations on the use of MRI in multiple sclerosis. The Lancet Neurology in press. [DOI] [PubMed]
- Hagens M.HJ., Burggraaff J., Kilsdonk I.D., Ruggieri S., Collorone S., Cortese R., Cawley N., Sbardella E., Andelova M., Amann M., Lieb J.M., Pantano P., Lissenberg-Witte B.I., Killestein J., Oreja-Guevara C., Wuerfel J., Ciccarelli O., Gasperini C., Lukas C., Rovira A., Barkhof F., Wattjes M.P. Impact of 3 Tesla MRI on interobserver agreement in clinically isolated syndrome: A MAGNIMS multicentre study. Mult Scler. 2019;25(3):352–360. doi: 10.1177/1352458517751647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovaris M., Rocca M.A., Yousry I., Yousry T.A., Colombo B., Comi G., Filippi M. Lesion load quantification on fast-FLAIR, rapid acquisition relaxation-enhanced, and gradient spin echo brain MRI scans from multiple sclerosis patients. Magn Reson Imaging. 1999;17(8):1105–1110. doi: 10.1016/s0730-725x(99)00068-5. [DOI] [PubMed] [Google Scholar]
- Egger C., Opfer R., Wang C., Kepp T., Sormani M.P., Spies L., Barnett M., Schippling S. MRI FLAIR lesion segmentation in multiple sclerosis: Does automated segmentation hold up with manual annotation? Neuroimage Clin. 2017;13:264–270. doi: 10.1016/j.nicl.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leener B., Lévy S., Dupont S.M., Fonov V.S., Stikov N., Louis Collins D., Callot V., Cohen-Adad J. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage. 2017;145:24–43. doi: 10.1016/j.neuroimage.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Gros C., De Leener B., Badji A., Maranzano J., Eden D., Dupont S.M., Talbott J., Zhuoquiong R., Liu Y., Granberg T., Ouellette R., Tachibana Y., Hori M., Kamiya K., Chougar L., Stawiarz L., Hillert J., Bannier E., Kerbrat A., Edan G., Labauge P., Callot V., Pelletier J., Audoin B., Rasoanandrianina H., Brisset J.-C., Valsasina P., Rocca M.A., Filippi M., Bakshi R., Tauhid S., Prados F., Yiannakas M., Kearney H., Ciccarelli O., Smith S., Treaba C.A., Mainero C., Lefeuvre J., Reich D.S., Nair G., Auclair V., McLaren D.G., Martin A.R., Fehlings M.G., Vahdat S., Khatibi A., Doyon J., Shepherd T., Charlson E., Narayanan S., Cohen-Adad J. Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. Neuroimage. 2019;184:901–915. doi: 10.1016/j.neuroimage.2018.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Adad J., Alonso-Ortiz E., Abramovic M., Arneitz C., Atcheson N., Barlow L., Barry R.L., Barth M., Battiston M., Büchel C., Budde M., Callot V., Combes A.J.E., De Leener B., Descoteaux M., de Sousa P.L., Dostál M., Doyon J., Dvorak A., Eippert F., Epperson K.R., Epperson K.S., Freund P., Finsterbusch J., Foias A., Fratini M., Fukunaga I., Wheeler-Kingshott C.A.M.G., Germani G., Gilbert G., Giove F., Gros C., Grussu F., Hagiwara A., Henry P.-G., Horák T., Hori M., Joers J., Kamiya K., Karbasforoushan H., Keřkovský M., Khatibi A., Kim J.-W., Kinany N., Kitzler H., Kolind S., Kong Y., Kudlička P., Kuntke P., Kurniawan N.D., Kusmia S., Labounek R., Laganà M.M., Laule C., Law C.S., Lenglet C., Leutritz T., Liu Y., Llufriu S., Mackey S., Martinez-Heras E., Mattera L., Nestrasil I., O’Grady K.P., Papinutto N., Papp D., Pareto D., Parrish T.B., Pichiecchio A., Prados F., Rovira À., Ruitenberg M.J., Samson R.S., Savini G., Seif M., Seifert A.C., Smith A.K., Smith S.A., Smith Z.A., Solana E., Suzuki Y., Tackley G., Tinnermann A., Valošek J., Van De Ville D., Yiannakas M.C., Weber K.A., Weiskopf N., Wise R.G., Wyss P.O., Xu J. Generic acquisition protocol for quantitative MRI of the spinal cord. Nat Protoc. 2021;16(10):4611–4632. doi: 10.1038/s41596-021-00588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Adad J., Alonso-Ortiz E., Abramovic M., Arneitz C., Atcheson N., Barlow L., Barry R.L., Barth M., Battiston M., Büchel C., Budde M., Callot V., Combes A.J.E., De Leener B., Descoteaux M., de Sousa P.L., Dostál M., Doyon J., Dvorak A., Eippert F., Epperson K.R., Epperson K.S., Freund P., Finsterbusch J., Foias A., Fratini M., Fukunaga I., Gandini Wheeler-Kingshott C.A.M., Germani G., Gilbert G., Giove F., Gros C., Grussu F., Hagiwara A., Henry P.-G., Horák T., Hori M., Joers J., Kamiya K., Karbasforoushan H., Keřkovský M., Khatibi A., Kim J.-W., Kinany N., Kitzler H.H., Kolind S., Kong Y., Kudlička P., Kuntke P., Kurniawan N.D., Kusmia S., Labounek R., Laganà M.M., Laule C., Law C.S., Lenglet C., Leutritz T., Liu Y., Llufriu S., Mackey S., Martinez-Heras E., Mattera L., Nestrasil I., O’Grady K.P., Papinutto N., Papp D., Pareto D., Parrish T.B., Pichiecchio A., Prados F., Rovira À., Ruitenberg M.J., Samson R.S., Savini G., Seif M., Seifert A.C., Smith A.K., Smith S.A., Smith Z.A., Solana E., Suzuki Y., Tackley G., Tinnermann A., Valošek J., Van De Ville D., Yiannakas M.C., Weber II K.A., Weiskopf N., Wise R.G., Wyss P.O., Xu J. Open-access quantitative MRI data of the spinal cord and reproducibility across participants, sites and manufacturers. Sci Data. 2021;8(1) doi: 10.1038/s41597-021-00941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckwoldt M.O., Gradl J., Hähnel S., Hielscher T., Wildemann B., Diem R., Platten M., Wick W., Heiland S., Bendszus M. Increasing the sensitivity of MRI for the detection of multiple sclerosis lesions by long axial coverage of the spinal cord: A prospective study in 119 patients. J Neurol. 2017;264(2):341–349. doi: 10.1007/s00415-016-8353-3. [DOI] [PubMed] [Google Scholar]
- Karavasilis E., Velonakis G., Argiropoulos G., Athanasakos A., Poulou L.S., Toulas P., Kelekis N.L., Efstathopoulos E.P. Proton Density Fat Suppressed MRI in 3T Increases the Sensitivity of Multiple Sclerosis Lesion Detection in the Cervical Spinal Cord. Clin Neuroradiol. 2019;29(1):45–50. doi: 10.1007/s00062-017-0626-4. [DOI] [PubMed] [Google Scholar]
- Di Perri C., Dwyer M.G., Wack D.S., et al. Signal abnormalities on 1.5 and 3 Tesla brain MRI in multiple sclerosis patients and healthy controls. A morphological and spatial quantitative comparison study. Neuroimage. 2009;47:1352–1362. doi: 10.1016/j.neuroimage.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Stankiewicz J.M., Glanz B.I., Healy B.C., Arora A., Neema M., Benedict R.H.B., Guss Z.D., Tauhid S., Buckle G.J., Houtchens M.K., Khoury S.J., Weiner H.L., Guttmann C.R.G., Bakshi R. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging. 2011;21(2):e50–e56. doi: 10.1111/j.1552-6569.2009.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajja B.R., Datta S., He R., Mehta M., Gupta R.K., Wolinsky J.S., Narayana P.A. Unified approach for multiple sclerosis lesion segmentation on brain MRI. Ann Biomed Eng. 2006;34(1):142–151. doi: 10.1007/s10439-005-9009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux P.D., Tubridy N., Parker G.J., et al. The effect of section thickness on MR lesion detection and quantification in multiple sclerosis. AJNR Am J Neuroradiol. 1998;19:1715–1720. [PMC free article] [PubMed] [Google Scholar]
- Filippi M., Preziosa P., Banwell B.L., Barkhof F., Ciccarelli O., De Stefano N., Geurts J.J.G., Paul F., Reich D.S., Toosy A.T., Traboulsee A., Wattjes M.P., Yousry T.A., Gass A., Lubetzki C., Weinshenker B.G., Rocca M.A. Assessment of lesions on magnetic resonance imaging in multiple sclerosis: Practical guidelines. Brain. 2019;142(7):1858–1875. doi: 10.1093/brain/awz144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel J., Martin M.L., Bakshi R., Calabresi P.A., Elliot M., Roalf D., Gur R.C., Gur R.E., Henry R.G., Nair G., Oh J., Papinutto N., Pelletier D., Reich D.S., Rooney W.D., Satterthwaite T.D., Stern W., Prabhakaran K., Sicotte N.L., Shinohara R.T., Goldsmith J. Intensity warping for multisite MRI harmonization. Neuroimage. 2020;223:117242. doi: 10.1016/j.neuroimage.2020.117242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Gu L., Liu Z., Zhao S. Review of Deep Learning Approaches for the Segmentation of Multiple Sclerosis Lesions on Brain MRI. Front Neuroinform. 2020;14 doi: 10.3389/fninf.2020.610967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårtensson G., Ferreira D., Granberg T., Cavallin L., Oppedal K., Padovani A., Rektorova I., Bonanni L., Pardini M., Kramberger M.G., Taylor J.-P., Hort J., Snædal J., Kulisevsky J., Blanc F., Antonini A., Mecocci P., Vellas B., Tsolaki M., Kłoszewska I., Soininen H., Lovestone S., Simmons A., Aarsland D., Westman E. The reliability of a deep learning model in clinical out-of-distribution MRI data: A multicohort study. Med Image Anal. 2020;66:101714. doi: 10.1016/j.media.2020.101714. [DOI] [PubMed] [Google Scholar]
- de Sitter A., Visser M., Brouwer I., Cover K.S., van Schijndel R.A., Eijgelaar R.S., Müller D.M.J., Ropele S., Kappos L., Rovira Á., Filippi M., Enzinger C., Frederiksen J., Ciccarelli O., Guttmann C.R.G., Wattjes M.P., Witte M.G., de Witt Hamer P.C., Barkhof F., Vrenken H. Facing privacy in neuroimaging: Removing facial features degrades performance of image analysis methods. Eur Radiol. 2020;30(2):1062–1074. doi: 10.1007/s00330-019-06459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda M.M., Brouwer I., de Vos M.L., de Vries M.S., Barkhof F., Pouwels P.J.W., Vrenken H. Comparing lesion segmentation methods in multiple sclerosis: Input from one manually delineated subject is sufficient for accurate lesion segmentation. NeuroImage: Clinical. 2019;24:102074. doi: 10.1016/j.nicl.2019.102074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde S., Salem M., Cabezas M., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira À., Salvi J., Oliver A., Lladó X. One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. Neuroimage Clin. 2019;21:101638. doi: 10.1016/j.nicl.2018.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre-Garriga J., Pareto D., Battaglini M., Rocca M.A., Ciccarelli O., Enzinger C., Wuerfel J., Sormani M.P., Barkhof F., Yousry T.A., De Stefano N., Tintoré M., Filippi M., Gasperini C., Kappos L., Río J., Frederiksen J., Palace J., Vrenken H., Montalban X., Rovira À. MAGNIMS consensus recommendations on the use of brain and spinal cord atrophy measures in clinical practice. Nature Reviews Neurology. 2020;16(3):171–182. doi: 10.1038/s41582-020-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwijk M.D., Vrenken H., Jonkman L.E., Daams M., Geurts J.JG., Barkhof F., Pouwels P.JW. High-resolution T1-relaxation time mapping displays subtle, clinically relevant, gray matter damage in long-standing multiple sclerosis. Mult Scler. 2016;22(10):1279–1288. doi: 10.1177/1352458515615953. [DOI] [PubMed] [Google Scholar]
- Takao H., Hayashi N., Ohtomo K. Effect of scanner in longitudinal studies of brain volume changes. J Magn Reson Imaging. 2011;34(2):438–444. doi: 10.1002/jmri.22636. [DOI] [PubMed] [Google Scholar]
- Lee H., Nakamura K., Narayanan S., Brown R.A., Arnold D.L. Estimating and accounting for the effect of MRI scanner changes on longitudinal whole-brain volume change measurements. NeuroImage. 2019;184:555–565. doi: 10.1016/j.neuroimage.2018.09.062. [DOI] [PubMed] [Google Scholar]
- Steenwijk M.D., Amiri H., Schoonheim M.M., de Sitter A., Barkhof F., Pouwels P.J.W., Vrenken H. Agreement of MSmetrix with established methods for measuring cross-sectional and longitudinal brain atrophy. NeuroImage: Clinical. 2017;15:843–853. doi: 10.1016/j.nicl.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodkin O., Prados F., Vos S.B., Pemberton H., Collorone S., Hagens M.H.J., Cardoso M.J., Yousry T.A., Thornton J.S., Sudre C.H., Barkhof F. FLAIR-only joint volumetric analysis of brain lesions and atrophy in clinically isolated syndrome (CIS) suggestive of multiple sclerosis. Neuroimage Clin. 2021;29:102542. doi: 10.1016/j.nicl.2020.102542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramanos Z., Fonov V.S., Francis S.J., Narayanan S., Pike G.B., Collins D.L., Arnold D.L. Gradient distortions in MRI: Characterizing and correcting for their effects on SIENA-generated measures of brain volume change. Neuroimage. 2010;49(2):1601–1611. doi: 10.1016/j.neuroimage.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Jovicich J., Czanner S., Greve D., Haley E., van der Kouwe A., Gollub R., Kennedy D., Schmitt F., Brown G., MacFall J., Fischl B., Dale A. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30(2):436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Doran S.J., Charles-Edwards L., Reinsberg S.A., Leach M.O. A complete distortion correction for MR images: I. Gradient warp correction. Phys Med Biol. 2005;50(7):1343–1361. doi: 10.1088/0031-9155/50/7/001. [DOI] [PubMed] [Google Scholar]
- Song S., Zheng Y., He Y. A review of Methods for Bias Correction in Medical Images. BMER. 2017;3(1) doi: 10.18103/bme10.18103/bme.V3.N110.18103/bme.v3i1.1550. [DOI] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Yuanjie Zheng, Egan A., Yushkevich P.A., Gee J.C. N4ITK: Improved N3 Bias Correction. IEEE Transactions on Medical Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin O., Mouiha A., Dieumegarde L., Duchesne S. Normative data for subcortical regional volumes over the lifetime of the adult human brain. NeuroImage. 2016;137:9–20. doi: 10.1016/j.neuroimage.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Battaglini M., Gentile G., Luchetti L., et al. Lifespan normative data on rates of brain volume changes. Neurobiology of Aging. 2019;81:30–37. doi: 10.1016/j.neurobiolaging.2019.05.0100. [DOI] [PubMed] [Google Scholar]
- Potvin O., Chouinard I., Dieumegarde L., Bartha R., Bellec P., Collins D.L., Descoteaux M., Hoge R., Ramirez J., Scott C.J.M., Smith E.E., Strother S.C., Black S.E., Duchesne S. The Canadian Dementia Imaging Protocol: Harmonization validity for morphometry measurements. Neuroimage Clin. 2019;24:101943. doi: 10.1016/j.nicl.2019.101943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lukas C., Steenwijk M.D., Daams M., Versteeg A., Duan Y., Li K., Weiler F., Hahn H.K., Wattjes M.P., Barkhof F., Vrenken H. Multicenter Validation of Mean Upper Cervical Cord Area Measurements from Head 3D T1-Weighted MR Imaging in Patients with Multiple Sclerosis. AJNR Am J Neuroradiol. 2016;37(4):749–754. doi: 10.3174/ajnr.A4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papinutto N., Bakshi R., Bischof A., Calabresi P.A., Caverzasi E., Constable R.T., Datta E., Kirkish G., Nair G., Oh J., Pelletier D., Pham D.L., Reich D.S., Rooney W., Roy S., Schwartz D., Shinohara R.T., Sicotte N.L., Stern W.A., Tagge I., Tauhid S., Tummala S., Henry R.G. Gradient nonlinearity effects on upper cervical spinal cord area measurement from 3D T1 -weighted brain MRI acquisitions. Magn Reson Med. 2018;79(3):1595–1601. doi: 10.1002/mrm.26776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda M.M., Middelkoop S.M., Steenwijk M.D., Daams M., Amiri H., Brouwer I., Killestein J., Uitdehaag B.M.J., Dekker I., Lukas C., Bellenberg B., Barkhof F., Pouwels P.J.W., Vrenken H. Validation of mean upper cervical cord area (MUCCA) measurement techniques in multiple sclerosis (MS): High reproducibility and robustness to lesions, but large software and scanner effects. Neuroimage Clin. 2019;24:101962. doi: 10.1016/j.nicl.2019.101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Bellenberg B., Prados F., et al. Quantification of cervical cord cross-sectional area: Which acquisition, vertebra level and analysis software? A multi-center repeatability study on a travelling healthy volunteer. Front Neurol. 2021;12 doi: 10.3389/fneur.2021.693333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papinutto N., Henry R.G. Evaluation of Intra- and Interscanner Reliability of MRI Protocols for Spinal Cord Gray Matter and Total Cross-Sectional Area Measurements. J Magn Reson Imaging. 2019;49(4):1078–1090. doi: 10.1002/jmri.26269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglini M., Jenkinson M., De Stefano N. SIENA-XL for improving the assessment of gray and white matter volume changes on brain MRI. Human Brain Mapping. 2018;39(3):1063–1077. doi: 10.1002/hbm.23828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Fisher E. Segmentation of brain magnetic resonance images for measurement of gray matter atrophy in multiple sclerosis patients. Neuroimage. 2009;44(3):769–776. doi: 10.1016/j.neuroimage.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdika M., Pelletier D. Nonrigid registration of multiple sclerosis brain images using lesion inpainting for morphometry or lesion mapping. Hum Brain Mapp. 2009;30(4):1060–1067. doi: 10.1002/hbm.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglini M., Jenkinson M., De Stefano N. Evaluating and reducing the impact of white matter lesions on brain volume measurements. Hum Brain Mapp. 2012;33(9):2062–2071. doi: 10.1002/hbm.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizard N., Fonov V.S., García-Lorenzo D., Nakamura K., Aubert-Broche B., Collins D.L., Muñoz-Barrutia A. Spatio-Temporal Regularization for Longitudinal Registration to Subject-Specific 3d Template. PLoS One. 2015;10(8):e0133352. doi: 10.1371/journal.pone.0133352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu V., Schoonheim M.M., Versteeg A., Chaturvedi N., Jonker M., Xavier de Menezes R., Gallindo Garre F., Uitdehaag B.M.J., Barkhof F., Vrenken H., Derfuss T. Grey Matter Atrophy in Multiple Sclerosis: Clinical Interpretation Depends on Choice of Analysis Method. PLoS ONE. 2016;11(1):e0143942. doi: 10.1371/journal.pone.0143942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglini M., Giorgio A., Stromillo M.L., Bartolozzi M.L., Guidi L., Federico A., De Stefano N. Voxel-wise assessment of progression of regional brain atrophy in relapsing-remitting multiple sclerosis. J Neurol Sci. 2009;282(1-2):55–60. doi: 10.1016/j.jns.2009.02.322. [DOI] [PubMed] [Google Scholar]
- Storelli L., Rocca M.A., Pagani E., Van Hecke W., Horsfield M.A., De Stefano N., Rovira A., Sastre-Garriga J., Palace J., Sima D., Smeets D., Filippi M. Measurement of Whole-Brain and Gray Matter Atrophy in Multiple Sclerosis: Assessment with MR Imaging. Radiology. 2018;288(2):554–564. doi: 10.1148/radiol.2018172468. [DOI] [PubMed] [Google Scholar]
- Popescu V., Battaglini M., Hoogstrate W.S., Verfaillie S.C.J., Sluimer I.C., van Schijndel R.A., van Dijk B.W., Cover K.S., Knol D.L., Jenkinson M., Barkhof F., de Stefano N., Vrenken H. Optimizing parameter choice for FSL-Brain Extraction Tool (BET) on 3D T1 images in multiple sclerosis. Neuroimage. 2012;61(4):1484–1494. doi: 10.1016/j.neuroimage.2012.03.074. [DOI] [PubMed] [Google Scholar]
- Lutkenhoff E.S., Rosenberg M., Chiang J., Zhang K., Pickard J.D., Owen A.M., Monti M.M., Wu X. Optimized brain extraction for pathological brains (optiBET) PLoS One. 2014;9(12):e115551. doi: 10.1371/journal.pone.0115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabr R.E., Coronado I., Robinson M., Sujit S.J., Datta S., Sun X., Allen W.J., Lublin F.D., Wolinsky J.S., Narayana P.A. Brain and lesion segmentation in multiple sclerosis using fully convolutional neural networks: A large-scale study. Mult Scler. 2020;26(10):1217–1226. doi: 10.1177/1352458519856843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley R., Wepfer R., Aschwanden F., Grunder L., Muri R., Rummel C., Verma R., Weisstanner C., Reyes M., Salmen A., Chan A., Wagner F., Wiest R. Simultaneous lesion and brain segmentation in multiple sclerosis using deep neural networks. Sci Rep. 2021;11(1) doi: 10.1038/s41598-020-79925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana P.A., Coronado I., Sujit S.J., Wolinsky J.S., Lublin F.D., Gabr R.E. Deep-Learning-Based Neural Tissue Segmentation of MRI in Multiple Sclerosis: Effect of Training Set Size. J Magn Reson Imaging. 2020;51(5):1487–1496. doi: 10.1002/jmri.26959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield M.A., Sala S., Neema M., Absinta M., Bakshi A., Sormani M.P., Rocca M.A., Bakshi R., Filippi M. Rapid semi-automatic segmentation of the spinal cord from magnetic resonance images: Application in multiple sclerosis. Neuroimage. 2010;50(2):446–455. doi: 10.1016/j.neuroimage.2009.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados F., Moccia M., Johnson A., Yiannakas M., Grussu F., Cardoso M.J., Ciccarelli O., Ourselin S., Barkhof F., Wheeler-Kingshott C. Generalised boundary shift integral for longitudinal assessment of spinal cord atrophy. Neuroimage. 2020;209:116489. doi: 10.1016/j.neuroimage.2019.116489. [DOI] [PubMed] [Google Scholar]
- Amann M., Andělová M., Pfister A., Mueller-Lenke N., Traud S., Reinhardt J., Magon S., Bendfeldt K., Kappos L., Radue E.-W., Stippich C., Sprenger T. Subcortical brain segmentation of two dimensional T1-weighted data sets with FMRIB’s Integrated Registration and Segmentation Tool (FIRST) Neuroimage Clin. 2015;7:43–52. doi: 10.1016/j.nicl.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granziera C., Wuerfel J., Barkhof F., Calabrese M., De Stefano N., Enzinger C., Evangelou N., Filippi M., Geurts J.J.G., Reich D.S., Rocca M.A., Ropele S., Rovira À., Sati P., Toosy A.T., Vrenken H., Gandini Wheeler-Kingshott C.A.M., Kappos L., Barkhof F., de Stefano N., Sastre-Garriga J., Ciccarelli O., Enzinger C., Filippi M., Gasperini C., Kappos L., Palace J., Vrenken H., Rovira À., Rocca M.A., Yousry T. Quantitative magnetic resonance imaging towards clinical application in multiple sclerosis. Brain. 2021;144(5):1296–1311. doi: 10.1093/brain/awab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropele S., Filippi M., Valsasina P., Korteweg T., Barkhof F., Tofts P.S., Samson R., Miller D.H., Fazekas F. Assessment and correction of B1-induced errors in magnetization transfer ratio measurements. Magn Reson Med. 2005;53(1):134–140. doi: 10.1002/mrm.20310. [DOI] [PubMed] [Google Scholar]
- Barker G.J., Schreiber W.G., Gass A., Ranjeva J.P., Campi A., Waesberghe J.H.T.M.V., Franconi J.-M., Watt H.C., Tofts P.S. A standardised method for measuring magnetisation transfer ratio on MR imagers from different manufacturers–the EuroMT sequence. MAGMA. 2005;18(2):76–80. doi: 10.1007/s10334-004-0095-z. [DOI] [PubMed] [Google Scholar]
- Romme Christensen J., Ratzer R., Bornsen L., Lyksborg M., Garde E., Dyrby T.B., Siebner H.R., Sorensen P.S., Sellebjerg F. Natalizumab in progressive MS: Results of an open-label, phase 2A, proof-of-concept trial. Neurology. 2014;82(17):1499–1507. doi: 10.1212/WNL.0000000000000361. [DOI] [PubMed] [Google Scholar]
- Miller D.H., Fox R.J., Phillips J.T., Hutchinson M., Havrdova E., Kita M., Wheeler-Kingshott C.A.M., Tozer D.J., MacManus D.G., Yousry T.A., Goodsell M., Yang M., Zhang R., Viglietta V., Dawson K.T. Effects of delayed-release dimethyl fumarate on MRI measures in the phase 3 CONFIRM study. Neurology. 2015;84(11):1145–1152. doi: 10.1212/WNL.0000000000001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf N., Suckling J., Williams G., Correia M.M., Inkster B., Tait R., Ooi C., Bullmore E.T., Lutti A. Quantitative multi-parameter mapping of R1, PD(*), MT, and R2(*) at 3T: A multi-center validation. Front Neurosci. 2013;7 doi: 10.3389/fnins.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen T., Tian H., Xue H., Ren H., Li L.i., Fan Q., Wen B., Ren Z. Reproducibility of inhomogeneous magnetization transfer (ihMT): A test-retest, multi-site study. Magn Reson Imaging. 2019;57:243–249. doi: 10.1016/j.mri.2018.11.010. [DOI] [PubMed] [Google Scholar]
- Tofts P.S., Steens S.C.A., Cercignani M., Admiraal-Behloul F., Hofman P.A.M., van Osch M.J.P., Teeuwisse W.M., Tozer D.J., van Waesberghe J.H.T.M., Yeung R., Barker G.J., van Buchem M.A. Sources of variation in multi-centre brain MTR histogram studies: Body-coil transmission eliminates inter-centre differences. MAGMA. 2006;19(4):209–222. doi: 10.1007/s10334-006-0049-8. [DOI] [PubMed] [Google Scholar]
- Sormani M.P., Iannucci G., Rocca M.A., et al. Reproducibility of magnetization transfer ratio histogram-derived measures of the brain in healthy volunteers. AJNR Am J Neuroradiol. 2000;21:133–136. [PMC free article] [PubMed] [Google Scholar]
- Tortorella C., Viti B., Bozzali M., Sormani M.P., Rizzo G., Gilardi M.F., Comi G., Filippi M. A magnetization transfer histogram study of normal-appearing brain tissue in MS. Neurology. 2000;54(1):186. doi: 10.1212/wnl.54.1.186. [DOI] [PubMed] [Google Scholar]
- Fernando K.T.M., Tozer D.J., Miszkiel K.A., Gordon R.M., Swanton J.K., Dalton C.M., Barker G.J., Plant G.T., Thompson A.J., Miller D.H. Magnetization transfer histograms in clinically isolated syndromes suggestive of multiple sclerosis. Brain. 2005;128(12):2911–2925. doi: 10.1093/brain/awh654. [DOI] [PubMed] [Google Scholar]
- De Stefano N., Battaglini M., Stromillo M.L., et al. Brain damage as detected by magnetization transfer imaging is less pronounced in benign than in early relapsing multiple sclerosis. Brain. 2006;129:2008–2016. doi: 10.1093/brain/awl152. [DOI] [PubMed] [Google Scholar]
- Cercignani M., Gandini Wheeler‐Kingshott C. From micro- to macro-structures in multiple sclerosis: what is the added value of diffusion imaging. NMR in biomedicine. 2019;32(4) doi: 10.1002/nbm.3888. [DOI] [PubMed] [Google Scholar]
- Palacios E.M., Martin A.J., Boss M.A., Ezekiel F., Chang Y.S., Yuh E.L., Vassar M.J., Schnyer D.M., MacDonald C.L., Crawford K.L., Irimia A., Toga A.W., Mukherjee P. Toward Precision and Reproducibility of Diffusion Tensor Imaging: A Multicenter Diffusion Phantom and Traveling Volunteer Study. AJNR Am J Neuroradiol. 2017;38(3):537–545. doi: 10.3174/ajnr.A5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.S., Lévy S., Schneider T., Smith A.K., Smith S.A., Cohen-Adad J., Gandini Wheeler-Kingshott C.A.M., Leemans A. ZOOM or Non-ZOOM? Assessing Spinal Cord Diffusion Tensor Imaging Protocols for Multi-Centre Studies. PLoS One. 2016;11(5):e0155557. doi: 10.1371/journal.pone.0155557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis S., Jones D.K., Roebroeck A. Including diffusion time dependence in the extra-axonal space improves in vivo estimates of axonal diameter and density in human white matter. Neuroimage. 2016;130:91–103. doi: 10.1016/j.neuroimage.2016.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grussu F., Ianuş A., Tur C., Prados F., Schneider T., Kaden E., Ourselin S., Drobnjak I., Zhang H., Alexander D.C., Gandini Wheeler‐Kingshott C.A.M. Relevance of time-dependence for clinically viable diffusion imaging of the spinal cord. Magn Reson Med. 2019;81(2):1247–1264. doi: 10.1002/mrm.27463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y., Freidlin R.Z., Rohde G.K., Basser P.J. New modeling and experimental framework to characterize hindered and restricted water diffusion in brain white matter. Magn Reson Med. 2004;52(5):965–978. doi: 10.1002/mrm.20274. [DOI] [PubMed] [Google Scholar]
- Pagani E., Hirsch J.G., Pouwels P.J.W., Horsfield M.A., Perego E., Gass A., Roosendaal S.D., Barkhof F., Agosta F., Rovaris M., Caputo D., Giorgio A., Palace J., Marino S., De Stefano N., Ropele S., Fazekas F., Filippi M. Intercenter differences in diffusion tensor MRI acquisition. J Magn Reson Imaging. 2010;31(6):1458–1468. doi: 10.1002/jmri.22186. [DOI] [PubMed] [Google Scholar]
- Ganzetti M., Wenderoth N., Mantini D. Intensity Inhomogeneity Correction of Structural MR Images: A Data-Driven Approach to Define Input Algorithm Parameters. Front Neuroinform. 2016;10:10. doi: 10.3389/fninf.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.L.R., Skare S., Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: Application to diffusion tensor imaging. Neuroimage. 2003;20(2):870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Perrone D., Aelterman J., Pižurica A., Jeurissen B., Philips W., Leemans A. The effect of Gibbs ringing artifacts on measures derived from diffusion MRI. NeuroImage. 2015;120:441–455. doi: 10.1016/j.neuroimage.2015.06.068. [DOI] [PubMed] [Google Scholar]
- Gallichan D., Scholz J., Bartsch A., Behrens T.E., Robson M.D., Miller K.L. Addressing a systematic vibration artifact in diffusion-weighted MRI. Hum Brain Mapp. 2009:NA–NA. doi: 10.1002/hbm.20856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov I.I., Alnæs D., Westlye L.T. Towards an optimised processing pipeline for diffusion magnetic resonance imaging data: Effects of artefact corrections on diffusion metrics and their age associations in UK Biobank. Hum Brain Mapp. 2019;40(14):4146–4162. doi: 10.1002/hbm.24691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganà M., Rovaris M., Ceccarelli A., Venturelli C., Marini S., Baselli G. DTI Parameter Optimisation for Acquisition at 1.5T: SNR Analysis and Clinical Application. Computational Intelligence and Neuroscience. 2010;2010:1–8. doi: 10.1155/2010/254032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J.M., Marques P., Alves V., Sousa N. A hitchhiker’s guide to diffusion tensor imaging. Front Neurosci. 2013;7:31. doi: 10.3389/fnins.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier J.-D., Smith R., Raffelt D., Tabbara R., Dhollander T., Pietsch M., Christiaens D., Jeurissen B., Yeh C.-H., Connelly A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage. 2019;202:116137. doi: 10.1016/j.neuroimage.2019.116137. [DOI] [PubMed] [Google Scholar]
- Christidi F., Karavasilis E., Samiotis K., Bisdas S., Papanikolaou N. Fiber tracking: A qualitative and quantitative comparison between four different software tools on the reconstruction of major white matter tracts. Eur J Radiol Open. 2016;3:153–161. doi: 10.1016/j.ejro.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Bilgic B., Fan Q., Liao C., Ngamsombat C., Hu Y., Witzel T., Setsompop K., Polimeni J.R., Huang S.Y. DeepDTI: High-fidelity six-direction diffusion tensor imaging using deep learning. Neuroimage. 2020;219:117017. doi: 10.1016/j.neuroimage.2020.117017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.o., de Groot M., Steketee R.M.E., Meijboom R., Smits M., Vernooij M.W., Ikram M.A., Liu J., Niessen W.J., Bron E.E. Neuro4Neuro: A neural network approach for neural tract segmentation using large-scale population-based diffusion imaging. Neuroimage. 2020;218:116993. doi: 10.1016/j.neuroimage.2020.116993. [DOI] [PubMed] [Google Scholar]
- Marzban E.N., Eldeib A.M., Yassine I.A., Kadah Y.M., Ginsberg S.D. Alzheimer’s disease diagnosis from diffusion tensor images using convolutional neural networks. PLoS One. 2020;15(3):e0230409. doi: 10.1371/journal.pone.0230409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzullo A., Kocevar G., Stamile C., Durand-Dubief F., Terracina G., Calimeri F., Sappey-Marinier D. Classification of Multiple Sclerosis Clinical Profiles via Graph Convolutional Neural Networks. Front Neurosci. 2019;13 doi: 10.3389/fnins.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladosu O., Liu W.-Q., Pike B.G., Koch M., Metz L.M., Zhang Y. Advanced Analysis of Diffusion Tensor Imaging Along With Machine Learning Provides New Sensitive Measures of Tissue Pathology and Intra-Lesion Activity in Multiple Sclerosis. Front Neurosci. 2021;15 doi: 10.3389/fnins.2021.634063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontopodis E., Papadaki E., Trivzakis E., Maris T., Simos P., Papadakis G., Tsatsakis A., Spandidos D., Karantanas A., Marias K. Emerging deep learning techniques using magnetic resonance imaging data applied in multiple sclerosis and clinical isolated syndrome patients (Review) Exp Ther Med. 2021;22(4) doi: 10.3892/etm.2021.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellazzi G., Debernard L., Melzer T.R., Dalrymple-Alford J.C., D'Angelo E., Miller D.H., Gandini Wheeler-Kingshott C.A.M., Mason D.F. Functional Connectivity Alterations Reveal Complex Mechanisms Based on Clinical and Radiological Status in Mild Relapsing Remitting Multiple Sclerosis. Front Neurol. 2018;9 doi: 10.3389/fneur.2018.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover G.H., Mueller B.A., Turner J.A., van Erp T.G.M., Liu T.T., Greve D.N., Voyvodic J.T., Rasmussen J., Brown G.G., Keator D.B., Calhoun V.D., Lee H.J., Ford J.M., Mathalon D.H., Diaz M., O'Leary D.S., Gadde S., Preda A., Lim K.O., Wible C.G., Stern H.S., Belger A., McCarthy G., Ozyurt B., Potkin S.G. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J Magn Reson Imaging. 2012;36(1):39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K.H., Greve D.N., Wang M., Pieper S.D., Warfield S.K., White N.S., Manandhar S., Brown G.G., Vangel M.G., Kikinis R., Wells W.M. Reproducibility of functional MR imaging: Preliminary results of prospective multi-institutional study performed by Biomedical Informatics Research Network. Radiology. 2005;237(3):781–789. doi: 10.1148/radiol.2373041630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badhwar AmanPreet, Collin-Verreault Y., Orban P., Urchs S., Chouinard I., Vogel J., Potvin O., Duchesne S., Bellec P. Multivariate consistency of resting-state fMRI connectivity maps acquired on a single individual over 2.5 years, 13 sites and 3 vendors. NeuroImage. 2020;205:116210. doi: 10.1016/j.neuroimage.2019.116210. [DOI] [PubMed] [Google Scholar]
- Glasser M.F., Sotiropoulos S.N., Wilson J.A., Coalson T.S., Fischl B., Andersson J.L., Xu J., Jbabdi S., Webster M., Polimeni J.R., Van Essen D.C., Jenkinson M. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graedel N.N., McNab J.A., Chiew M., Miller K.L. Motion correction for functional MRI with three-dimensional hybrid radial-Cartesian EPI. Magn Reson Med. 2017;78(2):527–540. doi: 10.1002/mrm.26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R.H.R., Mennes M., van Rooij D., Llera A., Buitelaar J.K., Beckmann C.F. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage. 2015;112:267–277. doi: 10.1016/j.neuroimage.2015.02.064. [DOI] [PubMed] [Google Scholar]
- Costumero V., Bueichekú E., Adrián-Ventura J., Ávila C. Opening or closing eyes at rest modulates the functional connectivity of V1 with default and salience networks. Sci Rep. 2020;10:9137. doi: 10.1038/s41598-020-66100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover G.H., Li T.Q., Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Behzadi Y., Restom K., Liau J., Liu T.T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37(1):90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanian H., Holdsworth S., Christen T., Wu H., Zhu K., Kerr A.B., Middione M.J., Dougherty R.F., Moseley M., Zaharchuk G. Advantages of short repetition time resting-state functional MRI enabled by simultaneous multi-slice imaging. J Neurosci Methods. 2019;311:122–132. doi: 10.1016/j.jneumeth.2018.09.033. [DOI] [PubMed] [Google Scholar]
- Yang H., Long X.-Y., Yang Y., Yan H., Zhu C.-Z., Zhou X.-P., Zang Y.-F., Gong Q.-Y. Amplitude of low frequency fluctuation within visual areas revealed by resting-state functional MRI. Neuroimage. 2007;36(1):144–152. doi: 10.1016/j.neuroimage.2007.01.054. [DOI] [PubMed] [Google Scholar]
- Ragot D.M., Chen J.J. Characterizing contrast origins and noise contribution in spin-echo EPI BOLD at 3 T. Magn Reson Imaging. 2019;57:328–336. doi: 10.1016/j.mri.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Bennett C.M., Miller M.B. How reliable are the results from functional magnetic resonance imaging? Ann N Y Acad Sci. 2010;1191:133–155. doi: 10.1111/j.1749-6632.2010.05446.x. [DOI] [PubMed] [Google Scholar]
- Andrews-Hanna J.R., Snyder A.Z., Vincent J.L., Lustig C., Head D., Raichle M., Buckner R.L. Disruption of large-scale brain systems in advanced aging. Neuron. 2007;56(5):924–935. doi: 10.1016/j.neuron.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J., Frith C.D., Liddle P.F., Frackowiak R.S.J. Functional connectivity: The principal-component analysis of large (PET) data sets. J Cereb Blood Flow Metab. 1993;13(1):5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- Worsley K.J., Chen J.-I., Lerch J., Evans A.C. Comparing functional connectivity via thresholding correlations and singular value decomposition. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):913–920. doi: 10.1098/rstb.2005.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C.F., DeLuca M., Devlin J.T., Smith S.M. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinik-Nezer R., Holzmeister F., Camerer C.F., Dreber A., Huber J., Johannesson M., Kirchler M., Iwanir R., Mumford J.A., Adcock R.A., Avesani P., Baczkowski B.M., Bajracharya A., Bakst L., Ball S., Barilari M., Bault N., Beaton D., Beitner J., Benoit R.G., Berkers R.M.W.J., Bhanji J.P., Biswal B.B., Bobadilla-Suarez S., Bortolini T., Bottenhorn K.L., Bowring A., Braem S., Brooks H.R., Brudner E.G., Calderon C.B., Camilleri J.A., Castrellon J.J., Cecchetti L., Cieslik E.C., Cole Z.J., Collignon O., Cox R.W., Cunningham W.A., Czoschke S., Dadi K., Davis C.P., Luca A.D., Delgado M.R., Demetriou L., Dennison J.B., Di X., Dickie E.W., Dobryakova E., Donnat C.L., Dukart J., Duncan N.W., Durnez J., Eed A., Eickhoff S.B., Erhart A., Fontanesi L., Fricke G.M., Fu S., Galván A., Gau R., Genon S., Glatard T., Glerean E., Goeman J.J., Golowin S.A.E., González-García C., Gorgolewski K.J., Grady C.L., Green M.A., Guassi Moreira J.F., Guest O., Hakimi S., Hamilton J.P., Hancock R., Handjaras G., Harry B.B., Hawco C., Herholz P., Herman G., Heunis S., Hoffstaedter F., Hogeveen J., Holmes S., Hu C.-P., Huettel S.A., Hughes M.E., Iacovella V., Iordan A.D., Isager P.M., Isik A.I., Jahn A., Johnson M.R., Johnstone T., Joseph M.J.E., Juliano A.C., Kable J.W., Kassinopoulos M., Koba C., Kong X.-Z., Koscik T.R., Kucukboyaci N.E., Kuhl B.A., Kupek S., Laird A.R., Lamm C., Langner R., Lauharatanahirun N., Lee H., Lee S., Leemans A., Leo A., Lesage E., Li F., Li M.Y.C., Lim P.C., Lintz E.N., Liphardt S.W., Losecaat Vermeer A.B., Love B.C., Mack M.L., Malpica N., Marins T., Maumet C., McDonald K., McGuire J.T., Melero H., Méndez Leal A.S., Meyer B., Meyer K.N., Mihai G., Mitsis G.D., Moll J., Nielson D.M., Nilsonne G., Notter M.P., Olivetti E., Onicas A.I., Papale P., Patil K.R., Peelle J.E., Pérez A., Pischedda D., Poline J.-B., Prystauka Y., Ray S., Reuter-Lorenz P.A., Reynolds R.C., Ricciardi E., Rieck J.R., Rodriguez-Thompson A.M., Romyn A., Salo T., Samanez-Larkin G.R., Sanz-Morales E., Schlichting M.L., Schultz D.H., Shen Q., Sheridan M.A., Silvers J.A., Skagerlund K., Smith A., Smith D.V., Sokol-Hessner P., Steinkamp S.R., Tashjian S.M., Thirion B., Thorp J.N., Tinghög G., Tisdall L., Tompson S.H., Toro-Serey C., Torre Tresols J.J., Tozzi L., Truong V., Turella L., van ‘t Veer A.E., Verguts T., Vettel J.M., Vijayarajah S., Vo K., Wall M.B., Weeda W.D., Weis S., White D.J., Wisniewski D., Xifra-Porxas A., Yearling E.A., Yoon S., Yuan R., Yuen K.S.L., Zhang L., Zhang X.u., Zosky J.E., Nichols T.E., Poldrack R.A., Schonberg T. Variability in the analysis of a single neuroimaging dataset by many teams. Nature. 2020;582(7810):84–88. doi: 10.1038/s41586-020-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D., Wei Z., Zhou Y., Li G., Zhang X.u., Han W. Deep Learning Methods to Process fMRI Data and Their Application in the Diagnosis of Cognitive Impairment: A Brief Overview and Our Opinion. Front Neuroinform. 2018;12 doi: 10.3389/fninf.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P., Sweeney E.M., Muschelli J., Crainiceanu C.M., Shinohara R.T. Removing inter-subject technical variability in magnetic resonance imaging studies. Neuroimage. 2016;132:198–212. doi: 10.1016/j.neuroimage.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P., Cullen N., Sheline Y.I., Taylor W.D., Aselcioglu I., Cook P.A., Adams P., Cooper C., Fava M., McGrath P.J., McInnis M., Phillips M.L., Trivedi M.H., Weissman M.M., Shinohara R.T. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage. 2018;167:104–120. doi: 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C.J., Li Y., Abecasis G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar D.P., Adams H.H.H., Jahanshad N., Chauhan G., Stein J.L., Hofer E., Renteria M.E., Bis J.C., Arias-Vasquez A., Ikram M.K., Desrivières S., Vernooij M.W., Abramovic L., Alhusaini S., Amin N., Andersson M., Arfanakis K., Aribisala B.S., Armstrong N.J., Athanasiu L., Axelsson T., Beecham A.H., Beiser A., Bernard M., Blanton S.H., Bohlken M.M., Boks M.P., Bralten J., Brickman A.M., Carmichael O., Chakravarty M.M., Chen Q., Ching C.R.K., Chouraki V., Cuellar-Partida G., Crivello F., Den Braber A., Doan N.T., Ehrlich S., Giddaluru S., Goldman A.L., Gottesman R.F., Grimm O., Griswold M.E., Guadalupe T., Gutman B.A., Hass J., Haukvik U.K., Hoehn D., Holmes A.J., Hoogman M., Janowitz D., Jia T., Jørgensen K.N., Karbalai N., Kasperaviciute D., Kim S., Klein M., Kraemer B., Lee P.H., Liewald D.C.M., Lopez L.M., Luciano M., Macare C., Marquand A.F., Matarin M., Mather K.A., Mattheisen M., McKay D.R., Milaneschi Y., Muñoz Maniega S., Nho K., Nugent A.C., Nyquist P., Loohuis L.M.O., Oosterlaan J., Papmeyer M., Pirpamer L., Pütz B., Ramasamy A., Richards J.S., Risacher S.L., Roiz-Santiañez R., Rommelse N., Ropele S., Rose E.J., Royle N.A., Rundek T., Sämann P.G., Saremi A., Satizabal C.L., Schmaal L., Schork A.J., Shen L.i., Shin J., Shumskaya E., Smith A.V., Sprooten E., Strike L.T., Teumer A., Tordesillas-Gutierrez D., Toro R., Trabzuni D., Trompet S., Vaidya D., Van der Grond J., Van der Lee S.J., Van der Meer D., Van Donkelaar M.M.J., Van Eijk K.R., Van Erp T.G.M., Van Rooij D., Walton E., Westlye L.T., Whelan C.D., Windham B.G., Winkler A.M., Wittfeld K., Woldehawariat G., Wolf C., Wolfers T., Yanek L.R., Yang J., Zijdenbos A., Zwiers M.P., Agartz I., Almasy L., Ames D., Amouyel P., Andreassen O.A., Arepalli S., Assareh A.A., Barral S., Bastin M.E., Becker D.M., Becker J.T., Bennett D.A., Blangero J., van Bokhoven H., Boomsma D.I., Brodaty H., Brouwer R.M., Brunner H.G., Buckner R.L., Buitelaar J.K., Bulayeva K.B., Cahn W., Calhoun V.D., Cannon D.M., Cavalleri G.L., Cheng C.-Y., Cichon S., Cookson M.R., Corvin A., Crespo-Facorro B., Curran J.E., Czisch M., Dale A.M., Davies G.E., De Craen A.J.M., De Geus E.J.C., De Jager P.L., De Zubicaray G.I., Deary I.J., Debette S., DeCarli C., Delanty N., Depondt C., DeStefano A., Dillman A., Djurovic S., Donohoe G., Drevets W.C., Duggirala R., Dyer T.D., Enzinger C., Erk S., Espeseth T., Fedko I.O., Fernández G., Ferrucci L., Fisher S.E., Fleischman D.A., Ford I., Fornage M., Foroud T.M., Fox P.T., Francks C., Fukunaga M., Gibbs J.R., Glahn D.C., Gollub R.L., Göring H.H.H., Green R.C., Gruber O., Gudnason V., Guelfi S., Håberg A.K., Hansell N.K., Hardy J., Hartman C.A., Hashimoto R., Hegenscheid K., Heinz A., Le Hellard S., Hernandez D.G., Heslenfeld D.J., Ho B.-C., Hoekstra P.J., Hoffmann W., Hofman A., Holsboer F., Homuth G., Hosten N., Hottenga J.-J., Huentelman M., Hulshoff Pol H.E., Ikeda M., Jack Jr C.R., Jenkinson M., Johnson R., Jönsson E.G., Jukema J.W., Kahn R.S., Kanai R., Kloszewska I., Knopman D.S., Kochunov P., Kwok J.B., Lawrie S.M., Lemaître H., Liu X., Longo D.L., Lopez O.L., Lovestone S., Martinez O., Martinot J.-L., Mattay V.S., McDonald C., McIntosh A.M., McMahon F.J., McMahon K.L., Mecocci P., Melle I., Meyer-Lindenberg A., Mohnke S., Montgomery G.W., Morris D.W., Mosley T.H., Mühleisen T.W., Müller-Myhsok B., Nalls M.A., Nauck M., Nichols T.E., Niessen W.J., Nöthen M.M., Nyberg L., Ohi K., Olvera R.L., Ophoff R.A., Pandolfo M., Paus T., Pausova Z., Penninx B.W.J.H., Pike G.B., Potkin S.G., Psaty B.M., Reppermund S., Rietschel M., Roffman J.L., Romanczuk-Seiferth N., Rotter J.I., Ryten M., Sacco R.L., Sachdev P.S., Saykin A.J., Schmidt R., Schmidt H., Schofield P.R., Sigursson S., Simmons A., Singleton A., Sisodiya S.M., Smith C., Smoller J.W., Soininen H., Steen V.M., Stott D.J., Sussmann J.E., Thalamuthu A., Toga A.W., Traynor B.J., Troncoso J., Tsolaki M., Tzourio C., Uitterlinden A.G., Hernández M.C.V., Van der Brug M., van der Lugt A., van der Wee N.J.A., Van Haren N.E.M., van ’t Ent D., Van Tol M.-J., Vardarajan B.N., Vellas B., Veltman D.J., Völzke H., Walter H., Wardlaw J.M., Wassink T.H., Weale M.E., Weinberger D.R., Weiner M.W., Wen W., Westman E., White T., Wong T.Y., Wright C.B., Zielke R.H., Zonderman A.B., Martin N.G., Van Duijn C.M., Wright M.J., Longstreth W.T., Schumann G., Grabe H.J., Franke B., Launer L.J., Medland S.E., Seshadri S., Thompson P.M., Ikram M.A. Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8(1) doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P., Jahanshad N., Sprooten E., Nichols T.E., Mandl R.C., Almasy L., Booth T., Brouwer R.M., Curran J.E., de Zubicaray G.I., Dimitrova R., Duggirala R., Fox P.T., Elliot Hong L., Landman B.A., Lemaitre H., Lopez L.M., Martin N.G., McMahon K.L., Mitchell B.D., Olvera R.L., Peterson C.P., Starr J.M., Sussmann J.E., Toga A.W., Wardlaw J.M., Wright M.J., Wright S.N., Bastin M.E., McIntosh A.M., Boomsma D.I., Kahn R.S., den Braber A., de Geus E.J.C., Deary I.J., Hulshoff Pol H.E., Williamson D.E., Blangero J., van 't Ent D., Thompson P.M., Glahn D.C. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: Comparing meta and megaanalytical approaches for data pooling. Neuroimage. 2014;95:136–150. doi: 10.1016/j.neuroimage.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A., Young A.L., Wijeratne P.A., Prados F., Arnold D.L., Narayanan S., Guttmann C.R.G., Barkhof F., Alexander D.C., Thompson A.J., Chard D., Ciccarelli O. Identifying multiple sclerosis subtypes using unsupervised machine learning and MRI data. Nature Communications. 2021;12(1) doi: 10.1038/s41467-021-22265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dojat M., Kennedy D.N., Niessen W. Editorial: MAPPING: MAnagement and Processing of Images for Population ImagiNG. Front ICT. 2017;4 doi: 10.3389/fict.2017.00018. [DOI] [Google Scholar]
- Marcus D.S., Olsen T.R., Ramaratnam M., Buckner R.L. The Extensible Neuroimaging Archive Toolkit: An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–33. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Gorgolewski K.J., Auer T., Calhoun V.D., Craddock R.C., Das S., Duff E.P., Flandin G., Ghosh S.S., Glatard T., Halchenko Y.O., Handwerker D.A., Hanke M., Keator D., Li X., Michael Z., Maumet C., Nichols B.N., Nichols T.E., Pellman J., Poline J.-B., Rokem A., Schaefer G., Sochat V., Triplett W., Turner J.A., Varoquaux G., Poldrack R.A. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data. 2016;3(1) doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Green R.C., Harvey D., Jack C.R., Jagust W., Morris J.C., Petersen R.C., Salazar J., Saykin A.J., Shaw L.M., Toga A.W., Trojanowski J.Q. The Alzheimer’s Disease Neuroimaging Initiative 3: Continued innovation for clinical trial improvement. Alzheimers Dement. 2017;13(5):561–571. doi: 10.1016/j.jalz.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis S.I., Liparas D., Alzheimer’s Disease Neuroimaging Initiative How random is the random forest? Random forest algorithm on the service of structural imaging biomarkers for Alzheimer’s disease: from Alzheimer’s disease neuroimaging initiative (ADNI) database. Neural Regen Res. 2018;13:962–970. doi: 10.4103/1673-5374.233433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V.K., Lesnick T.G., Przybelski S.A., Heckman M.G., Knopman D.S., Graff-Radford J., Lowe V.J., Machulda M.M., Mielke M.M., Jack C.R., Petersen R.C., Ross O.A., Vemuri P. Coping with brain amyloid: Genetic heterogeneity and cognitive resilience to Alzheimer’s pathophysiology. Acta Neuropathol Commun. 2021;9(1) doi: 10.1186/s40478-021-01154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Barnes J., Bernstein M.A., Borowski B.J., Brewer J., Clegg S., Dale A.M., Carmichael O., Ching C., DeCarli C., Desikan R.S., Fennema‐Notestine C., Fjell A.M., Fletcher E., Fox N.C., Gunter J., Gutman B.A., Holland D., Hua X., Insel P., Kantarci K., Killiany R.J., Krueger G., Leung K.K., Mackin S., Maillard P., Malone I.B., Mattsson N., McEvoy L., Modat M., Mueller S., Nosheny R., Ourselin S., Schuff N., Senjem M.L., Simonson A., Thompson P.M., Rettmann D., Vemuri P., Walhovd K., Zhao Y., Zuk S., Weiner M. Magnetic resonance imaging in Alzheimer’s Disease Neuroimaging Initiative 2. Alzheimers Dement. 2015;11(7):740–756. doi: 10.1016/j.jalz.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J., Minati L., Marizzoni M., Marchitelli R., Sala-Llonch R., Bartrés-Faz D., Arnold J., Benninghoff J., Fiedler U., Roccatagliata L., Picco A., Nobili F., Blin O., Bombois S., Lopes R., Bordet R., Sein J., Ranjeva J.-P., Didic M., Gros-Dagnac H., Payoux P., Zoccatelli G., Alessandrini F., Beltramello A., Bargalló N., Ferretti A., Caulo M., Aiello M., Cavaliere C., Soricelli A., Parnetti L., Tarducci R., Floridi P., Tsolaki M., Constantinidis M., Drevelegas A., Rossini P.M., Marra C., Schönknecht P., Hensch T., Hoffmann K.-T., Kuijer J.P., Visser P.J., Barkhof F., Frisoni G.B. Longitudinal reproducibility of default-mode network connectivity in healthy elderly participants: A multicentric resting-state fMRI study. Neuroimage. 2016;124:442–454. doi: 10.1016/j.neuroimage.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Marizzoni M., Ferrari C., Jovicich J., Albani D., Babiloni C., Cavaliere L., Didic M., Forloni G., Galluzzi S., Hoffmann K.-T., Molinuevo J.L., Nobili F., Parnetti L., Payoux P., Ribaldi F., Rossini P.M., Schönknecht P., Salvatore M., Soricelli A., Hensch T., Tsolaki M., Visser P.J., Wiltfang J., Richardson J.C., Bordet R., Blin O., Frisoni G.B., Perry G. Predicting and Tracking Short Term Disease Progression in Amnestic Mild Cognitive Impairment Patients with Prodromal Alzheimer’s Disease: Structural Brain Biomarkers. J Alzheimers Dis. 2019;69(1):3–14. doi: 10.3233/JAD-180152. [DOI] [PubMed] [Google Scholar]
- Ritchie C.W., Muniz-Terrera G., Kivipelto M., Solomon A., Tom B., Molinuevo J.L. The European Prevention of Alzheimer’s Dementia (EPAD) Longitudinal Cohort Study: Baseline Data Release V500.0. J Prev Alzheimers Dis. 2020:1–7. doi: 10.14283/jpad.2019.46. [DOI] [PubMed] [Google Scholar]
- ten Kate M., Ingala S., Schwarz A.J., Fox N.C., Chételat G., van Berckel B.N.M., Ewers M., Foley C., Gispert J.D., Hill D., Irizarry M.C., Lammertsma A.A., Molinuevo J.L., Ritchie C., Scheltens P., Schmidt M.E., Visser P.J., Waldman A., Wardlaw J., Haller S., Barkhof F. Secondary prevention of Alzheimer’s dementia: Neuroimaging contributions. Alzheimers Res Ther. 2018;10(1) doi: 10.1186/s13195-018-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Almagro F., Jenkinson M., Bangerter N.K., Andersson J.L.R., Griffanti L., Douaud G., Sotiropoulos S.N., Jbabdi S., Hernandez-Fernandez M., Vallee E., Vidaurre D., Webster M., McCarthy P., Rorden C., Daducci A., Alexander D.C., Zhang H., Dragonu I., Matthews P.M., Miller K.L., Smith S.M. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. NeuroImage. 2018;166:400–424. doi: 10.1016/j.neuroimage.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry E.M., Bermel R.A., Williams J.R., Benzinger T.L.S., de Moor C., Fisher E., Hersh C.M., Hyland M.H., Izbudak I., Jones S.E., Kieseier B.C., Kitzler H.H., Krupp L., Lui Y.W., Montalban X., Naismith R.T., Nicholas J.A., Pellegrini F., Rovira A., Schulze M., Tackenberg B., Tintore M., Tivarus M.E., Ziemssen T., Rudick R.A. Harnessing Real-World Data to Inform Decision-Making: Multiple Sclerosis Partners Advancing Technology and Health Solutions (MS PATHS) Front Neurol. 2020;11 doi: 10.3389/fneur.2020.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M., Tedeschi G., Pantano P., De Stefano N., Zaratin P., Rocca M.A. The Italian Neuroimaging Network Initiative (INNI): Enabling the use of advanced MRI techniques in patients with MS. Neurol Sci. 2017;38(6):1029–1038. doi: 10.1007/s10072-017-2903-z. [DOI] [PubMed] [Google Scholar]
- Storelli L., Rocca M.A., Pantano P., Pagani E., De Stefano N., Tedeschi G., Zaratin P., Filippi M. MRI quality control for the Italian Neuroimaging Network Initiative: Moving towards big data in multiple sclerosis. J Neurol. 2019;266(11):2848–2858. doi: 10.1007/s00415-019-09509-4. [DOI] [PubMed] [Google Scholar]
- Hsieh J.J.L., Svalbe I. Magnetic resonance fingerprinting: From evolution to clinical applications. J Med Radiat Sci. 2020;67(4):333–344. doi: 10.1002/jmrs.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves F.G., Serai S.D., Zuccoli G. Synthetic Brain MRI: Review of Current Concepts and Future Directions. Top Magn Reson Imaging. 2018;27:387–393. doi: 10.1097/RMR.0000000000000189. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Billot B., Balbastre Y., Tabari A., Conklin J., Gilberto González R., Alexander D.C., Golland P., Edlow B.L., Fischl B. Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast. Neuroimage. 2021;237:118206. doi: 10.1016/j.neuroimage.2021.118206. [DOI] [PMC free article] [PubMed] [Google Scholar]