Abstract
The increased use of antibacterial and antifungal agents in recent years has resulted in the development of resistance to these drugs. The significant clinical implication of resistance has led to heightened interest in the study of antimicrobial resistance from different angles. Areas addressed include mechanisms underlying this resistance, improved methods to detect resistance when it occurs, alternate options for the treatment of infections caused by resistant organisms, and strategies to prevent and control the emergence and spread of resistance. In this review, the mode of action of antifungals and their mechanisms of resistance are discussed. Additionally, an attempt is made to discuss the correlation between fungal and bacterial resistance. Antifungals can be grouped into three classes based on their site of action: azoles, which inhibit the synthesis of ergosterol (the main fungal sterol); polyenes, which interact with fungal membrane sterols physicochemically; and 5-fluorocytosine, which inhibits macromolecular synthesis. Many different types of mechanisms contribute to the development of resistance to antifungals. These mechanisms include alteration in drug target, alteration in sterol biosynthesis, reduction in the intercellular concentration of target enzyme, and overexpression of the antifungal drug target. Although the comparison between the mechanisms of resistance to antifungals and antibacterials is necessarily limited by several factors defined in the review, a correlation between the two exists. For example, modification of enzymes which serve as targets for antimicrobial action and the involvement of membrane pumps in the extrusion of drugs are well characterized in both the eukaryotic and prokaryotic cells.
The past decade has witnessed a significant increase in the prevalence of resistance to antibacterial and antifungal agents. Resistance to antimicrobial agents has important implications for morbidity, mortality and health care costs in U.S. hospitals, as well as in the community. Hence, substantial attention has been focused on developing a more detailed understanding of the mechanisms of antimicrobial resistance, improved methods to detect resistance when it occurs, new antimicrobial options for the treatment of infections caused by resistant organisms, and methods to prevent the emergence and spread of resistance in the first place. Most of this attention has been devoted to the study of antibiotic resistance in bacteria for several reasons: (i) bacterial infections are responsible for the bulk of community-acquired and nosocomial infections; (ii) the large and expanding number of antibacterial classes offers a more diverse range of resistance mechanisms to study; and (iii) the ability to move bacterial resistance determinants into standard well-characterized bacterial strains facilitates the detailed study of molecular mechanisms of resistance in bacterial species.
The study of resistance to antifungal agents has lagged behind that of antibacterial resistance for several reasons. Perhaps most importantly, fungal diseases were not recognized as important pathogens until relatively recently (2, 148). For example, the annual death rate due to candidiasis was steady between 1950 and about 1970. Since 1970, this rate increased significantly in association with several changes in medical practice, including more widespread use of therapies that depress the immune system, the frequent and often indiscriminate use of broad-spectrum antibacterial agents, the common use of indwelling intravenous devices, and the advent of chronic immunosuppressive viral infections such as AIDS. These developments and the associated increase in fungal infections (5) intensified the search for new, safer, and more efficacious agents to combat serious fungal infections.
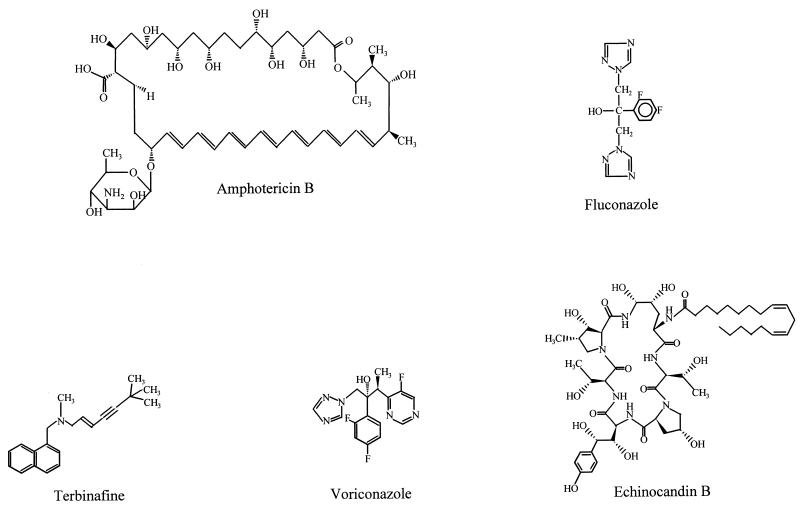
For nearly 30 years, amphotericin B (Fig. 1), which is known to cause significant nephrotoxicity, was the sole drug available to control serious fungal infections. The approval of the imidazoles and the triazoles in late 1980s and early 1990s were major advances in our ability to safely and effectively treat local and systemic fungal infections. The high safety profile of triazoles, in particular fluconazole (Fig. 1), has led to their extensive use. Fluconazole has been used to treat in excess of 16 million patients, including over 300,000 AIDS patients, in the United States alone since the launch of this drug (124a). Concomitant with this widespread use, there have been increasing reports of antifungal resistance (115). The clinical impact of antifungal resistance has been recently reviewed (115). Also, three excellent reviews concentrating on various aspects of antifungal resistance including clinical implications have been published recently (27, 86, 153). Therefore, the clinical impact of resistance is not covered in this review. Instead, our goal is to focus on the molecular mechanisms of antifungal resistance. Since mechanisms of antibacterial resistance are characterized in considerably more detail than those of antifungal resistance, we have chosen to use well-described mechanisms of bacterial resistance as a framework for understanding fungal mechanisms of resistance, insofar as such comparisons can be logically applied. In so doing, we hope to make an understanding of antifungal resistance mechanisms accessible to those who use these agents clinically, as well as those who may wish to study them in the future.
FIG. 1.
Structures of representative antifungal agents.
PROBLEMS WITH COMPARING ANTIFUNGAL AND ANTIBACTERIAL RESISTANCE
Although it is our premise that a comparison between mechanisms of resistance to antifungals and antibacterials is a useful way of developing a perspective on antimicrobial resistance in the two kingdoms, the comparison is necessarily limited by several factors. First, the structures of fungi and bacteria differ in very significant ways (such as the diploid nature of most fungi and the longer generation time of fungi compared to bacteria), and the available antibacterial and antifungal agents target structures and functions most relevant to the organisms to be inhibited. For example, many antibacterial agents inhibit steps important for the formation of peptidoglycan, the essential component of the bacterial cell wall. In contrast, most antifungal compounds target either the formation or the function of ergosterol, an important component of the fungal cell membrane. Nevertheless, there are important parallels between the mechanisms by which fungi develop resistance to ergosterol biosynthesis inhibitors and bacteria develop resistance to anti-cell wall agents. Regarding other types of bacterial resistance, comparisons are limited by the fact that antifungal analogues of many classes of antibacterial agents (protein synthesis inhibitors such as aminoglycosides, macrolides, and tetracyclines; topoisomerase inhibitors such as fluoroquinolones; and metabolic pathway inhibitors such as trimethoprim-sulfamethoxazole) do not exist. Conversely, antifungal nucleoside analogues such as 5-fluorocytosine (5FC) have no counterparts among clinically available antibacterial agents (although they are represented among the antiviral compounds). As such, the capacity for fungi to develop ribosomal resistance or topoisomerase mutations is unknown, as is the capacity for bacteria to develop resistance to nucleoside analogues. Interestingly, the antibacterial RNA polymerase inhibitor rifampin, which demonstrates no intrinsic activity against fungi, appears quite active against several fungal species when used in combination with amphotericin B (8). This synergistic activity has been attributed to increased uptake of the rifampin into the fungal cell resulting from the action of amphotericin B on the fungal membrane. Similar synergism has been demonstrated between amphotericin B and 5FC by Polak et al. (109), using murine models of candidiasis. The mechanism for this synergism has been postulated by some investigators to be improved uptake of the 5FC as a result of membrane disorganization due to amphotericin B-ergosterol interaction (87). This synergistic effect resembles the postulated mechanism of bactericidal synergism between cell wall-active agents and aminoglycosides against enterococci, in which increased intracellular concentrations of streptomycin are detectable when streptomycin is combined with penicillin in vitro against Enterococcus faecalis (91). In contrast to the notion that amphotericin B improves the uptake of 5FC, data obtained by Beggs et al. (7) suggest that these two agents act sequentially and not in combination against Candida albicans to affect synergy.
The second limitation to the comparison between antifungal and antibacterial resistance mechanisms is that some general classes of resistance mechanisms have not yet been identified in fungi. Resistance to antibacterial agents results from modification of the antibiotic, modification of the antimicrobial target, reduced access to the target, or some combination of these mechanisms. Antibiotic modification is arguably the most important mechanism of resistance to the β-lactam (β-lactamases) and aminoglycoside (aminoglycoside-modifying enzymes) classes of antibacterials. In contrast, although there has been a single, unconfirmed report of degradation of nystatin by dermatophytic fungi (13), there are no data to suggest that antibiotic modification is an important mechanism of antifungal resistance. On the other hand, accumulating evidence suggests that both target alterations and reduced access to targets (sometimes in combination) are important mechanisms of resistance to antifungal agents. These mechanisms have important parallels in antibacterial resistance.
The third limitation to the comparison is that our knowledge of genetic exchange mechanisms in bacteria is far more advanced than our knowledge of exchange mechanisms, if they exist, in fungi. Bacteria employ an extensive repertoire of plasmids, transposons, and bacteriophages to facilitate the exchange of resistance and virulence determinants among and between species. As a result, the opportunity for rapid emergence of high-level resistance and the potential for emergence and dissemination of resistance even in the absence of direct selection by specific antimicrobial pressure abound. Conversely, antifungal resistance described to date generally involves the emergence of naturally resistant species (as in the increasing importance of Candida krusei in areas of extensive use in certain medical centers) or the progressive, stepwise alterations of cellular structures or functions to avoid the activity of an antifungal agent to which there has been extensive exposure.
The final important limitation in comparing mechanisms of resistance to antifungals and antibacterials lies in the availability of standardized bacterial strains and plasmids for use in the study of antimicrobial resistance in bacteria. This availability allows the isolation of resistance determinants in well-characterized backgrounds, so that the specific contribution of different resistance mechanisms can be assessed. The availability of well-characterized strains and systems for DNA delivery allows a much more rigorous approach to the study of the genetics and physiology of bacterial resistance mechanisms, in comparison to fungal mechanisms of resistance. For example, the first step in analyzing plasmid-mediated β-lactamases in bacteria is the transfer of the plasmid to a well-characterized strain, generally Escherichia coli. In this way, complicating mechanisms such as membrane alteration can be controlled and reasonable comparisons of the level of resistance conferred by different β-lactamases can be made. The fact that similar standardized systems are not available in fungi means that the study of resistance almost always occurs in the clinical strains themselves, making an assessment of the precise contribution of individual resistance mechanisms to the phenotypic expression of resistance difficult and often impossible.
DEFINITION OF RESISTANCE
Since clinicians tend to depend, predominantly, on MIC breakpoints to determine the susceptibility of an isolate, it is important to briefly update the status of antifungal susceptibility testing. Unlike antibacterial agents, for which standardized susceptibility testing methods with interpretive breakpoints are well established, acceptable testing methods and tentative breakpoints for antifungal agents have only recently been suggested (116) (Table 1). Even these breakpoints are limited to yeast, particularly Candida, and fluconazole and itraconazole. Suggested breakpoints were based on the analysis of in vitro MIC (involving 883 isolates) and clinical data from 729 patients treated with these two antifungal agents. Based on currently available data, it appears that appropriately measured resistance in vitro (MICs of fluconazole and itraconazole of ≥64 and ≥1 μg/ml, respectively) often correlates with failure in the treatment of clinical infections (116).
TABLE 1.
Tentative interpretive guidelines for susceptibility testing in vitro of Candida speciesa
| Antifungal agent | MIC (μg/ml) for:
|
||
|---|---|---|---|
| Susceptible strains | Susceptible (dose-dependent) strains | Resistant strains | |
| Fluconazole | ≤8 | 16–32 | ≥64 |
| Itraconazole | ≤0.125 | 0.25–0.5 | ≥1 |
Conditions used are those recommended by the M27 method (112).
It is important to emphasize that the predictability of clinical success or failure in response to the administration of a specific antifungal (or antibacterial) agent depends on more than just its MIC for the infecting organism. Other factors (clinical status of the patient, presence of foreign material, location of infection) have a significant impact on the likelihood of therapeutic success. The role of these other factors in determining the clinical outcome of fungal infections is best illustrated by a study investigating the correlation of MIC and clinical outcome from patients with AIDS-associated cryptococcal meningitis (155). In this study, our group determined the fluconazole MIC against Cryptococcus neoformans isolates from 76 patients enrolled in two collaborative clinical trials (75, 89). When the MIC was correlated with the clinical response, several interesting observations were made. First, a statistically significant correlation between MIC data and clinical success or failure was apparent (P = 0.012). Further, upon multivariate analysis, it became clear that use of 5FC and the presence of a positive blood culture at the time of study entry also had a significant impact on response (155). When the patients were subdivided into these four possible groups (did/did not receive 5FC, did/did not have a positive blood culture), a distinct and strong correlation of MIC with outcome became apparent. For example, the predicted probability of treatment failure for a patient who did not receive 5FC, had a negative blood culture, and an isolate with a fluconazole MIC of 16 μg/ml was over 40%. This probability decreased to roughly 10% if the patient received 5FC. The need to consider other factors to predict clinical outcome, in addition to MICs, was also emphasized by the findings of others (115), who showed that host factors, such as the presence of an indwelling catheter, influenced clinical outcome.
MECHANISMS OF ACTION AND RESISTANCE
Understanding the mechanism(s) of action of different antimicrobial agents is an important prerequisite to understanding mechanisms of resistance. In fact, in many cases an elucidation of resistance mechanisms has allowed or enhanced our understanding of specific mechanisms of action. We therefore combine our discussions of mechanisms of action and resistance to individual antimicrobial classes, although the bulk of our attention is focused on mechanisms of resistance. Readers interested in a more in-depth discussion of the mechanisms of action of different antifungal agents are referred to thorough reviews (35, 36, 48, 52, 118, 124).
Antimicrobial Agents Affecting Fungal Sterols
The three major groups of antifungal agents in clinical use, azoles, polyenes, and allylamine/thiocarbamates, all owe their antifungal activities to inhibition of synthesis of or direct interaction with ergosterol. Ergosterol is the predominant component of the fungal cell membrane (104).
Azole-based antimycotic agents.
(i) Mechanism of action.
The first reports of the antifungal properties of N-substituted imidazoles were published in the late 1960s (55, 125). These original compounds, such as miconazole and econazole, and those that followed, such as ketoconazole, fluconazole, and itraconazole, proved to be important drugs for combating human fungal infections. The clinical efficacy and safety of fluconazole in particular has resulted in widespread use. The resultant emergence of resistance to azoles has intensified the search for new compounds that are active against resistant organisms (29, 47, 58, 76, 93, 94, 106, 129, 133, 136, 156). A review of the abstracts presented at the 1995 and 1996 Interscience Conference on Antimicrobial Agents and Chemotherapy revealed that 10 azole-related agents are currently under development for the treatment of fungal infections.
Ergosterol serves as a bioregulator of membrane fluidity and asymmetry and consequently of membrane integrity in fungal cells (100). Integrity of the cell membrane requires that inserted sterols lack C-4 methyl groups. Several lines of evidence suggest that the primary target of azoles is the heme protein, which cocatalyzes cytochrome P-450-dependent 14α-demethylation of lanosterol (51). Inhibition of 14α-demethylase leads to depletion of ergosterol and accumulation of sterol precursors, including 14α-methylated sterols (lanosterol, 4,14-dimethylzymosterol, and 24-methylenedihydrolanosterol), resulting in the formation of a plasma membrane with altered structure and function. The more recent triazole derivatives, such as fluconazole, itraconazole, and voriconazole (a triazole in development), owe their antifungal activity at least in part to inhibition of cytochrome P-450-dependent 14α-sterol demethylase (121). Compelling data in support of this mechanism of action comes from studies in which Geber et al. (34) cloned the structural genes encoding the 14α-methyl sterol demethylase (ERG11) and the Δ5,6 sterol desaturase (ERG3) from C. glabrata and used these cloned genes to create knockout mutants of each gene individually and both genes together. Phenotypic analysis revealed that the ERG3 deletion mutant remained susceptible to fluconazole and itraconazole. In contrast, the ERG11 deletion mutant and a double mutant in which both genes were deleted were resistant to 100, 16, and 2 μg of fluconazole, itraconazole, and amphotericin B per ml, respectively. These data suggest an inhibitory interaction between azoles and 14α-demethylase.
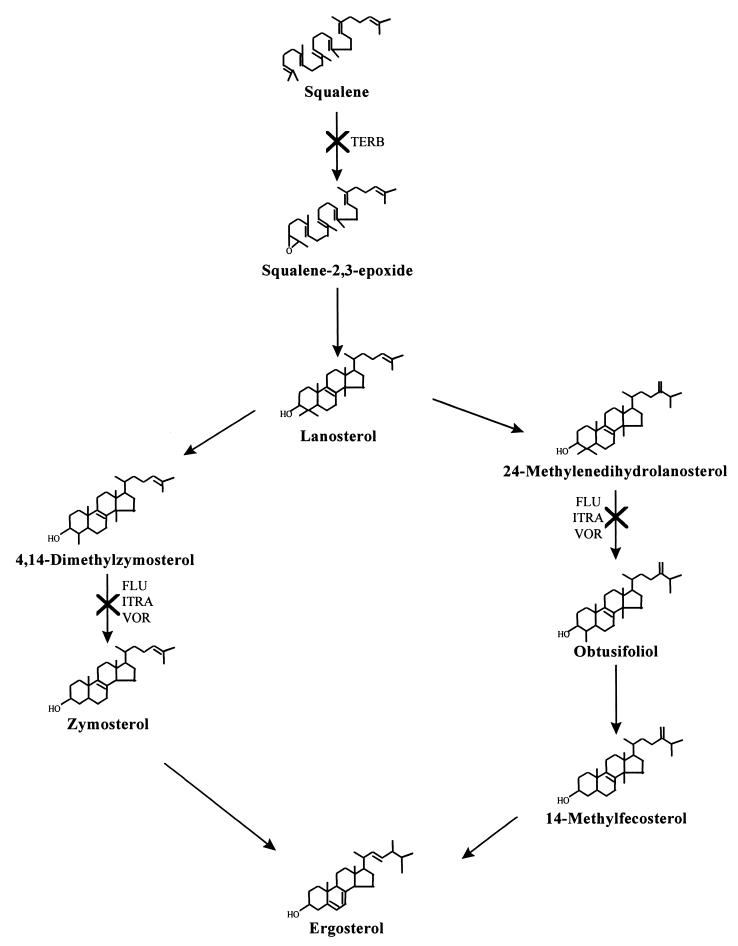
Although more recent azole antifungals are 14α-demethylase inhibitors, there exists a heterogeneity of action among these antifungals (6, 37, 131). The earlier imidazole derivatives (such as miconazole, econazole, and ketoconazole) have a complex mode of action, inhibiting several membrane-bound enzymes as well as membrane lipid biosynthesis (for a review, see Sheehan et al. [125] and Hitchcock and Whittle [52]). An accumulation of zymosterol and squalene synthesis was observed when C. albicans cells were treated with voriconazole (121). It is unclear whether the accumulation of these intermediates results from voriconazole interaction with various (non-14α-demethylase) enzymes involved in ergosterol synthesis or from secondary effects of 14α-demethylase inhibition. Azole activity may also vary with the genus tested. In addition to inhibiting the 14α-demethylase in Cryptococcus neoformans, fluconazole and itraconazole affect the reduction of obtusifolione to obtusifoliol, which results in the accumulation of methylated sterol precursors (39, 140). Mammalian cholesterol synthesis is also blocked by azoles at the stage of 14α-demethylation; however, the dose required to effect the same degree of inhibition is much higher than that required for fungi (51, 142, 143). For example, Hitchcock et al. (54) showed that voriconazole had a 50% inhibitory concentration of 7.4 μM against P-450-dependent 14α-sterol demethylase (P-450DM) of rat liver cholesterol. In contrast, the 50% inhibitory concentration of this antifungal agent against fungal P-450DM was as low as 0.03 μM (about 250-fold more active against the fungal enzyme than against the mammalian enzyme). The clinical effects of inhibition of human sterol biosynthesis are most prominently seen with ketoconazole. Figure 2 is a summary of the ergosterol biosynthetic pathway showing sites of action of antifungal agents.
FIG. 2.
Ergosterol biosynthetic pathway. Steps at which various antifungal agents exert their inhibitory activities are shown. TERB, terbinafine; FLU, fluconazole; ITRA, itraconazole; VOR, voriconazole.
(ii) Mechanisms of resistance to azoles.
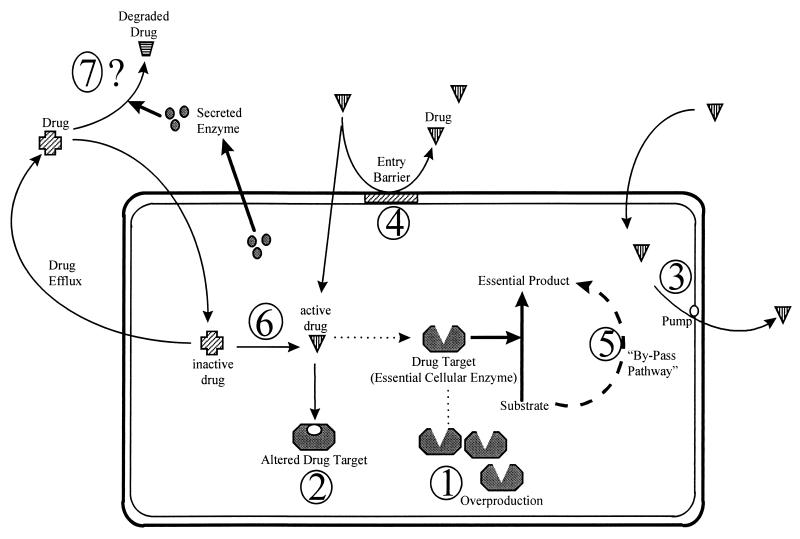
As noted above, there are as yet no reports of modification of azole antimicrobials as a mechanism of resistance. Resistant strains therefore either exhibit a modification in the quality or quantity of target enzyme, reduced access to the target, or some combination of these mechanisms. These mechanisms are discussed in detail below and are summarized in Fig. 3 and Table 2.
FIG. 3.
Mechanisms by which microbial cells might develop resistance. 1, The target enzyme is overproduced, so that the drug does not inhibit the biochemical reaction completely. 2, The drug target is altered so that the drug cannot bind to the target. 3, The drug is pumped out by an efflux pump. 4, The entry of the drug is prevented at the cell membrane/cell wall level. 5, The cell has a bypass pathway that compensates for the loss-of-function inhibition due to the drug activity. 6, Some fungal “enzymes” that convert an inactive drug to its active form are inhibited. 7, The cell secretes some enzymes to the extracellular medium, which degrade the drug.
TABLE 2.
Biochemical basis of azole resistance
| Mechanism | Caused by: | Comments |
|---|---|---|
| Alteration in drug target (14α-demethylase) | Mutations which alter drug binding but not binding of the endogenous substrate | Target is active (i.e., can catalyze demethylation) but has a reduced affinity towards azoles |
| Alteration in sterol biosynthesis | Lesions in the Δ5(6)-desaturase | Results in accumulation of 14α-methyl fecosterol instead of ergosterol |
| Reduction in the intercellular concentration of target enzyme | Change in membrane lipid and sterols; overexpression of specific drug efflux pumps (CDR1, PDR5, and BENr) | Poor penetration across the fungal membrane; active drug efflux |
| Overexpression of antifungal drug target | Increased copy number of the target enzyme | Results in increased ergosterol synthesis; contributes to cross-resistance between fluconazole and itraconazole |
(a) Modification of target. Several lines of evidence implicate a modification in the quantity or quality of 14α-demethylase in the expression of resistance to azole antifungal agents. A recent study examined the biochemical mechanisms for resistance to fluconazole by comparing sterol composition, fluconazole accumulation, and inhibition of 14α-demethylase by fluconazole in two clinical C. krusei strains (expressing intrinsic resistance to fluconazole) and a susceptible C. albicans isolate (101). No significant differences in the sterol content of C. krusei and C. albicans were detected (ergosterol was the major sterol in both species). Studies performed on cell extracts indicated that the concentration of fluconazole required to inhibit the synthesis of ergosterol by 50% was approximately 24- to 46-fold higher in C. krusei than in C. albicans, suggesting that affinity of the target enzyme is different in the two species (101). A comparison of fluconazole accumulation by C. albicans and C. krusei indicated that fluconazole accumulation in the first 60 min was similar in all study strains. However, analysis after 90 min of incubation revealed that C. krusei accumulated 60% less fluconazole than did C. albicans, implicating active efflux in the fluconazole resistance expressed by these C. krusei strains (see below). The potential coexistence of two resistance mechanisms precludes a precise calculation of the level of resistance contributed by the low-affinity 14α-demethylase.
Other studies have implicated altered 14α-demethylase in resistance to azoles. Reduced susceptibility of C. albicans B41628 (isolated from a patient with chronic mucocutaneous candidiasis who relapsed following an extended period of treatment with ketoconazole) to miconazole, ketoconazole, itraconazole, and fluconazole was attributed to differences in the microsomal cytochrome P-450 enzyme. Analysis of carbon monoxide (CO) difference spectra of microsomes from this strain revealed that it contained cytochrome P-450 with a Soret peak different from that characteristic of the cytochrome in azole-susceptible cells (127, 128). Additionally, the enzyme had a low binding affinity for azole antifungals (144). Whether the altered 14α-demethylase is solely responsible for the level of resistance observed in this strain is unclear, since C. albicans B41628 is a clinical isolate and the contribution of other resistance mechanisms to the reduced susceptibility of this isolate cannot be excluded (53).
Overexpression of 14α-demethylase has also been implicated as a mechanism of resistance to azole antifungals. Vanden Bossche et al. (141) characterized an azole-resistant C. glabrata strain and showed that its ergosterol content was increased compared with that of the pretreatment isolate. This increase was accompanied by a decrease in susceptibility to both azoles and amphotericin B. The increase in ergosterol synthesis was attributed to an elevated microsomal P-450 content in the resistant strain, suggesting an overexpression of the enzyme. Although the intracellular content of fluconazole in the resistant strain was 1.5- to 3-fold lower than that in the pretreatment isolate, suggesting active efflux of this antifungal, the amount of itraconazole retained by the resistant strain did not differ from that found in the pretreatment isolate (141). This finding suggests that the increased P-450 levels were responsible for the cross-resistance to these two triazoles. The scarcity of clinical isolates in which overproduction of 14α-demethylase has been observed, the fact that this phenomenon was observed in C. glabrata only, and the finding that other resistance mechanisms may be operative in the same strain suggest that overexpression of target enzyme plays only a limited role in clinical resistance to the azoles.
White (151) investigated the target enzyme (Erg11p) in the C. albicans series (which consists of 17 isolates obtained from the same patient over a 2-year period) described by Redding et al. (114) by using biochemical and molecular techniques. Testing the susceptibility of Erg11p to fluconazole in cell extracts revealed that a substantial decrease occurred in isolate 13, corresponding to resistance development. To determine whether the ERG11 gene acquired any alterations in response to drug pressure, this gene was sequenced. Sequence analysis identified a single point mutation that resulted in a single-amino-acid substitution (R467K) (152). This substitution resides between two residues known to be involved in interactions with the heme moiety in the active site of the enzyme. A similar point mutation (T315A) that alters the susceptibility of the target enzyme has been observed in close proximity to the active site of this enzyme in C. albicans (72). A second significant change observed in the ERG11 gene of the resistant isolate was reported by White (152), namely, loss of allelic variation in the ERG11 promoter and in the downstream THR1 gene (which encodes homoserine kinase, which is involved in threonine synthesis). Although these changes may account for resistance development, they are not the only factors involved (see below).
(1) Correlation with antibacterial resistance. Modification of enzymes that serve as targets for antibacterial action is a well-characterized mechanism of resistance to β-lactam antimicrobials. For example, the creation of mosaic penicillin-binding proteins (PBPs) through homologous recombination is the primary mechanism of resistance to penicillin in Streptococcus pneumoniae and is an important mechanism of resistance to penicillin in Neisseria gonorrhoeae (25). In these instances, PBPs are modified by splicing in segments of PBP genes from more resistant bacteria that are taken up by the pathogenic bacteria through the process of natural transformation. Point mutations in PBPs associated with decreased susceptibility to penicillin or its derivatives have also been described in several bacterial species, including Staphylococcus aureus and Enterococcus faecium (44, 158).
Resistance resulting from increased expression of the target enzyme has also been described in bacteria. It is well established that penicillin resistance expressed by Enterococcus hirae (and E. faecium) can be increased from roughly 4 to 64 μg/ml in association with increased expression of low-affinity PBP5 (28). Further increases in the MIC of penicillin for these strains appear to require additional mutations within the pbp5 gene itself (158). Overexpression of target enzyme has also been described as a primary mechanism of resistance to the β-lactam–β-lactamase inhibitor combinations (28). Overexpression of β-lactamase enzyme may overwhelm the amount of β-lactamase inhibitor entering the periplasmic space, resulting in increased levels of resistance. Mutations within the β-lactamases themselves, resulting in decreased affinity for the inhibitor molecule, have also been implicated in resistance to these agents (157).
(b) Active efflux. Considerable evidence has now been accumulated to suggest that active efflux is an important mechanism of resistance to azole antifungals. Recent studies indicate that fungi possess at least two efflux systems: (i) proteins belonging to the major facilitator superfamily (MFS) and (ii) ATP-binding cassette (ABC) superfamily of proteins. The MFS drug efflux proteins are associated with the transport of structurally diverse compounds and account for a range of resistance to toxic compounds in microorganisms (60). An example of MFS protein associated with drug resistance in Candida is BENr (CaMDR1), which is implicated in resistance to several drugs, including benomyl, methotrexate, and fluconazole. The ABC superfamily of proteins bind ATP, which is essential for substrate transport, through a highly conserved amino acid sequence (known as the binding cassette) (60). Four families of ABC transporters have been identified in Saccharomyces cerevisiae (MDR, CFTR, YEF, and PDR). These transporters have a common four-core domain structure (49) consisting of two integral membrane domains that span the membrane multiple times and two ATP-binding cytoplasmic domains that couple ATP hydrolysis to substrate transport (60). To date, eight genes for ABC transporters have been identified in Candida. An example of an ABC transporter found in both Candida and, more recently, in Cryptococcus neoformans is CDR1, which is involved in resistance to fluconazole and other azoles. The gene encoding this transporter was cloned by Prasad et al. (111) and appears to be similar in structure to human P-glycoprotein, which functions as a multidrug pump and is associated with resistance to a number of chemotherapeutic agents in neoplasms (40). Recently, Walsh et al. (146) provided evidence that C. albicans may possess one or more additional genes encoding ATP-binding cassette MDR-like proteins that are distinct from CDR1, which could participate in the development of azole resistance. In this regard, five CDR genes (CDR1 to CDR5) which belong to the PDR family have been identified in C. albicans (88, 117, 151). Additionally, one member each of the MDR, CFTR, and YEF families were identified (HST6, YCF1, and ELF1, respectively).
Evidence implicating drug efflux as a mechanism of resistance in Candida species has been forthcoming recently. Parkinson et al. (103) compared pretreatment (susceptible) and posttreatment (resistant) isolates of C. glabrata and showed that while no change in sterol biosynthesis between these two isolates was observed, the resistant isolate accumulated less fluconazole than the susceptible one did. The reduced ability of the resistant strain to accumulate fluconazole was a consequence of energy-dependent drug efflux (103). In an extension of these studies, Hitchcock and coworkers examined the mechanism of resistance to azoles in C. albicans, C. glabrata, and C. krusei by using the fluorescent dye rhodamine 123 (Rh123), which is known to be transported by a number of MDR (multidrug-resistant) organisms (18). Their results showed that resistant isolates accumulated less Rh123 than susceptible cells did. Furthermore, active efflux of Rh123 was observed in azole-resistant isolates of C. albicans and C. glabrata, consistent with the activity of an MDR transporter. The efflux mechanism associated with movement of Rh123 appears to play a role in azole resistance in C. glabrata but not in C. albicans, suggesting that azole resistance in C. albicans may be mediated by an alternative efflux pump (74).
Sanglard et al. (123) studied a set of 16 sequential C. albicans isolates obtained from five AIDS patients. The strains were selected on the basis of increasing fluconazole resistance following prolonged treatment. In some resistant strains, decreased accumulation of fluconazole was associated with up to a 10-fold increase in the mRNA levels of the CDR1 gene. Other resistant isolates overexpressed mRNA from the gene encoding BENr (CaMDR1) and had normal levels of CDR1 mRNA. Data from this study suggests that CDR1 is involved in the export of several azole derivatives (including fluconazole, itraconazole, and ketoconazole) while BENr confers resistance specifically to fluconazole.
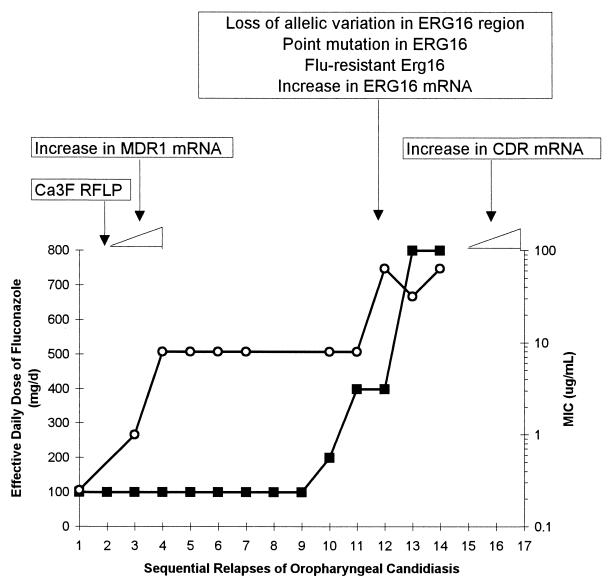
Redding et al. (114) studied a series of 17 C. albicans isolates cultured from a patient with recurrent episodes of oropharyngeal candidiasis who required progressively higher doses of fluconazole to control the infection. Over a 2-year period, the patient experienced 15 relapses, each of which was treated with fluconazole. Isolates from the early relapses had fluconazole MICs of <8 μg/ml, and the infection responded to fluconazole (100 mg/day). Fluconazole MICs for subsequent isolates rose steadily to 64 μg/ml, requiring progressively greater doses of fluconazole to produce a clinical response. Fluconazole was ineffective after the 14th relapse. This is shown graphically in Fig. 4, in which the minimum effective dose of fluconazole at each relapse is plotted against the MIC for the isolate from that episode. The approximate breakpoints suggested by these data correlate roughly with achievable levels of fluconazole in blood: 100 mg/day produces peak concentrations of approximately 6 μg/ml in serum, 400 mg/day produces peak concentrations of 20 to 30 μg/ml, and the linear pharmacokinetics of fluconazole would predict concentrations of 40 to 60 μg/ml in serum at 800 mg/day. Analysis of all isolates by contour-clamped homogeneous electric field electrophoresis confirmed the persistence of the same C. albicans strain throughout all infectious episodes (114).
FIG. 4.
Relationship between MIC, fluconazole dose, and emergence of resistance in oropharyngeal candidiasis. ○, MIC of fluconazole for the clinical isolate; ■, effective daily dose of fluconazole. MICs are represented on the secondary y axis, in logarithmic scale. Boxes above the graph represent genetic changes identified at each stage. Based on data from references 114 and 151.
The sterol content did not differ between susceptible and resistant isolates in this collection (37a), suggesting that the mechanism(s) of resistance does not involve alteration in sterol composition. White (151) recently examined the expression of several genes of interest in all 17 of these isolates, including ERG16 (encoding 14-demethylase), MDR1 (encoding a member of the MFS), and CDR1 (encoding an ABC transporter), as well as other genes potentially involved in resistance. A number of interesting findings were reported (Fig. 4): (i) MDR1 expression was increased early in the series, while the CDR1 mRNA level was increased only in the final isolates; (ii) ERG16 signal increased toward the end of the series; (iii) increases in mRNA levels of ERG16 and CDR1 correlated with increased resistance to ketoconazole and itraconazole but not to amphotericin B; and (iv) no changes in the mRNA signals for genes encoding members of the YEF and CFTR gene families (members of the ABC transporter family) were detected in the series, and no expression of ERG1 and ERG7 (genes involved in the ergosterol biosynthetic pathway) was detected (151). These data suggest that high-level azole resistance, at least in this series of isolates, results from the contributions of several mechanisms. They also suggest that prolonged exposure of a strain to one azole may lead to overexpression of genes, such as ERG16 and CDR1, that result in cross-resistance to other azoles.
The general availability of such a set is rare, and Redding and colleagues are to be commended for preserving these isolates and making them available to the scientific community for use as a tool in the investigation of resistance mechanisms as well as for preclinical evaluation of antifungals under development (9, 121).
(1) Correlation with antibacterial resistance. Membrane pumps involved in the efflux of antimicrobials are well described in many different species of bacteria. Among the best characterized of the systems involving multidrug membrane pumps is that regulated by the mar (multiple antibiotic resistance) gene locus found in Escherichia coli. Increased expression of mar-stimulated membrane pumps has been associated with resistance to chloramphenicol and tetracycline, as well as other compounds (20). Multidrug efflux pumps have also been implicated in resistance to β-lactams in Pseudomonas aeruginosa and fluoroquinolones in Staphylococcus aureus (79, 96). Similar to the situation in fungi, these pumps may combine with other resistance mechanisms (such as mutations in DNA gyrase genes) to yield higher levels of resistance than would be achievable with either mechanism alone.
(2) Alteration in membrane composition. Interactions between sterols and phospholipids in the cytoplasmic membrane affect membrane fluidity and asymmetry (104) and consequently influence the transport of materials across the membranes. A decrease in the amount of drug taken up by the fungal cell may result from changes in the sterol and/or the phospholipid composition of the fungal cell membrane. Using cerulenin as a lipid modulator, Mago and Khuller (83) demonstrated that altered phospholipids and fatty acid profiles affected C. albicans cell permeability and rendered the cells more resistant to miconazole. Hitchcock et al. (53) showed that an azole- and polyene-resistant C. albicans mutant had a larger lipid content and lower polar-lipid-to-neutral-lipid ratio than did strains susceptible to azoles. However, the most significant change in the lipid of the resistant strain was in the membrane sterol pattern, where ergosterol was replaced by methylated sterols, such as methylfecosterol. The authors hypothesized that an altered membrane sterol pattern is responsible for the doubly resistant phenotype observed in this strain (52).
Although alteration in the sterol pattern could explain the resistance mechanism in certain fungal strains (particularly in cases where ergosterol is replaced by fecosterol), we were unable to demonstrate a correlation between the sterol composition of C. albicans and resistance to azoles and polyenes. Two strain sets of C. albicans were analyzed for their sterol pattern: the first set was obtained from David Kerridge (University of Cambridge, Cambridge, England) and consisted of seven isolates that differ in their susceptibility to azoles and polyenes, while the second consisted of two clinical C. albicans isolates with different susceptibilities to fluconazole. Thin-layer and gas-liquid chromatography analyses showed that the major sterol present in all the strains tested was ergosterol (data not shown). Lower levels of lanosterol, obtusifoliol, 4,14-dimethylzymosterol, and squalene were also detected. Comparison of the sterol pattern between these resistant and susceptible strains revealed no correlation between sterol composition and susceptibility to antifungals (unpublished data). Therefore, resistance to azoles and/or polyenes in these strains is attributable to another mechanism(s) not related to the sterol pattern.
(1) Correlation with antibacterial resistance. Permeability barriers conferred by cytoplasmic membranes have been implicated in the natural resistance of anaerobic bacteria to the activity of aminoglycosides because aminoglycoside transport across the cytoplasmic membrane is an oxygen-dependent process. The intrinsic resistance of enterococci to aminoglycosides has also been hypothesized to be the result of the essentially anaerobic metabolism of these species (77).
The presence of an outer membrane in gram-negative bacteria has offered a much more varied array of opportunities for mutation to development of resistance to antibacterial compounds. Imipenem resistance in P. aeruginosa results from a combination of decreased expression of outer membrane protein D2 (a porin through which imipenem traverses the outer membrane) and increased expression of the chromosomal AmpC β-lactamase (81). Neither mutation by itself results in resistance to imipenem. Membrane changes in concert with β-lactamase production have also been implicated as mechanisms of resistance to cefepime and cefoxitin (80, 102). Vancomycin resistance in all aerobic gram-negative rods has also been attributed to the exclusion of the vancomycin molecule by the bacterial outer membrane. This exclusion is presumably based on the size of vancomycin rather than the absence of a specific porin.
Polyenes.
(i) Mechanism of action.
From the 1950s until the discovery of the azoles, polyene antifungal agents such as amphotericin B represented the standard of therapy for systemic fungal infections (132). There is an association between polyene susceptibility and the presence of sterols in the plasma membrane of the cells. All organisms susceptible to polyenes, e.g., yeasts, algae, and protozoa, contain sterols in their outer membrane, while resistant organisms do not (97). The importance of membrane sterols for polyene activity is also supported by earlier studies, where it was shown that fungi can be protected from the inhibitory action of certain polyenes by the addition of sterol to the growth medium (41, 73, 159). It was suggested that this effect is due to a physicochemical interaction between added sterols and the polyenes, which prevents the drug from binding with the cellular sterols. The interaction between the sterols and polyenes is further supported by direct spectrophotometric evidence that adding sterols to aqueous solutions of the polyene filipin or nystatin decreases the UV absorbance significantly (73), suggesting a direct interaction between the added sterol and the antifungal agent (69, 98).
For larger polyenes, such as amphotericin B, it has been proposed that the interaction of the antifungal with membrane sterol results in the production of aqueous pores consisting of an annulus of eight amphotericin B molecules linked hydrophobically to the membrane sterols (22, 56) (Fig. 5). This configuration gives rise to a pore in which the polyene hydroxyl residues face inward, leading to altered permeability, leakage of vital cytoplasmic components, and death of the organism (66, 67). The fatty acyl composition of the phospholipids has also been implicated in polyene susceptibility of yeast (57, 112, 113, 144). In addition, killing of C. albicans has been attributed to oxidative damage caused by polyenes (43, 137). The reader is referred to the review by Bolard and Milhaud (11) for a full discussion of the interaction of polyenes with lipids.
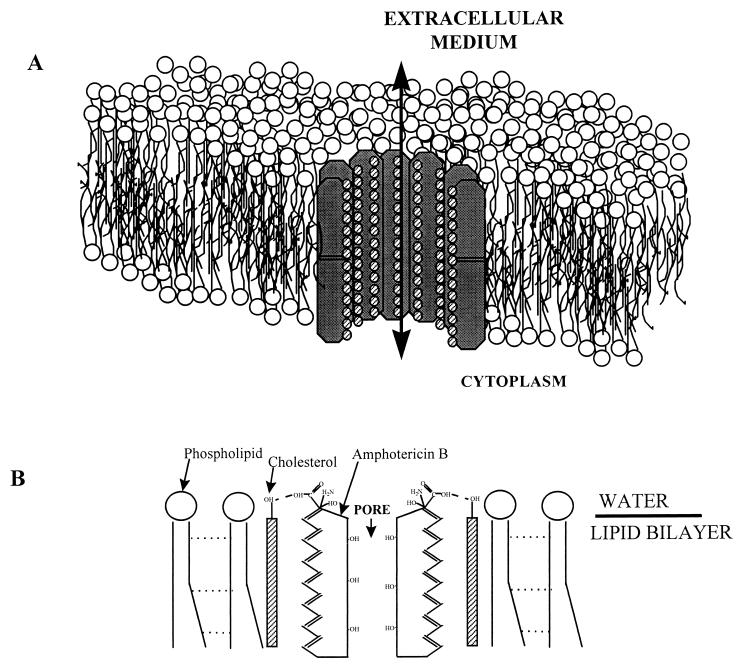
FIG. 5.
Schematic representation of the interaction between amphotericin B and cholesterol in a phospholipid bilayer. (A) The conducting pore is formed by the end-to-end union of two wells or half pores. Adapted from reference 21 with permission of the publisher. (B) Molecular orientation in an amphotericin B-cholesterol pore. The dotted lines between the hydrocarbon chains of phospholipids represent short-range London-van der Waals forces. The dashed lines represent hydrogen bonds formed between amphotericin B and cholesterol molecules.
Although amphotericin B is the most effective antifungal drug available, its narrow therapeutic index continues to limit its clinical utility (17, 33, 82, 154). To reduce untoward effects, amphotericin B has been formulated in liposomes to allow the transfer of higher doses of amphotericin B with less toxicity to mammalian cells (62, 105). Several amphotericin B liposomal preparations have been developed, including ABELCET, Amphoteck, and AmBisome. A liposomal preparation of nystatin, a polyene antifungal agent (Nyotran), is currently undergoing preclinical and clinical evaluation (145). It is hypothesized that once amphotericin B is incorporated into liposomes, it may participate in a selective transfer mechanism, which involves its transfer from the “donor” liposome to the ergosterol-containing “target” in the fungal cell membrane aided by the fungal and/or host phospholipases (62).
(ii) Mechanism of resistance to polyenes.
Despite more than 30 years of clinical use, resistance to polyene antibiotics, such as amphotericin B and nystatin, is rare, with resistant isolates being confined mostly to the less common species of Candida, such as C. lusitaniae, C. glabrata, and C. guilliermondii (84). Fryberg (31) suggested that development of resistance occurs by selection of naturally occurring resistant cells, present in small numbers in the population. These naturally resistant cells produce modified sterols that bind nystatin with lower affinity. The growth rate in the presence of nystatin is therefore dependent upon the normal growth rate (in the absence of nystatin) and the rate at which nystatin causes cell membrane damage. This latter rate is presumed to be a function of the affinity of nystatin for the membrane sterols: the greater the nystatin-sterol affinity, the greater the rate of membrane damage. As a corollary, one would expect each resistant strain to exhibit slower growth than its more susceptible parent. A difference between resistant and susceptible strains was in fact observed (31). The fact that resistance to polyenes is gradually lost after serial passage on media devoid of nystatin presumably represents repopulation of the culture by cells producing sterols with a higher affinity for nystatin. The molecular genetics underlying these shifts in sterol content are not well worked out. Athar and Winner (4), however, have suggested that resistance results from mutation rather than selection.
Most of our knowledge of the mechanisms of resistance to polyenes in fungal species has come from studies using mutants generated by (i) growing cells in the presence of increasing concentrations of antifungal agents (multistep mutants), (ii) exposing the cells to a gradient concentration (4), or (iii) creating mutants by one-step mutation with mutagenic agents (45). Hamilton-Miller (46) proposed a “biochemical” hypothesis that resistance arises due to changes, either quantitative or qualitative, in the sterol content of the cells. According to this hypothesis, resistant cells with altered sterol content should bind smaller amounts of polyene than do susceptible cells. This decreased binding of polyenes in C. albicans mutants could be attributed to (i) a decrease in the total ergosterol content of the cell, without concomitant changes in sterol composition; (ii) replacement of some or all of the polyene-binding sterols by ones which bind polyene less well, e.g., substitution of ergosterol, cholesterol, or stigmasterol by a 3-hydroxy or 3-oxo sterol (88); or (iii) reorientation, or masking, of existing ergosterol, so that binding with polyenes is sterically or thermodynamically less favored.
Different investigators have furnished evidence in support of all of these possibilities. Capek et al. (14) demonstrated that development of inducible resistance (induced by adaptation mechanism) in a strain of C. albicans was accompanied by a decrease in the ergosterol content of the cells. This decrease in ergosterol content was due not to enzymatic degradation of preformed ergosterol but to inhibition of its synthesis. Similarly, Dick et al. (24) studied 27 polyene-resistant C. albicans isolates obtained from neutropenic patients and showed that these strains had a 74 to 85% decrease in their ergosterol content. Thus, decreased ergosterol content may lead to decreased susceptibility to polyenes.
Fryberg (31) tested a number of resistant Candida strains and showed that incrementally more resistant isolates possessed principal sterols arising from blockage of the biosynthetic pathway (leading to ergosterol) at successively earlier stages. They reported that cultures possessing Δ8-sterols are more resistant to polyenes than those possessing Δ7-sterols, which, in turn, are more resistant than those possessing Δ5,7-sterols. Kelly et al. (65) compared the susceptibility and sterol pattern of two Cryptococcus neoformans (pre- and posttreatment) isolates from an AIDS patient who failed antifungal therapy. These authors observed a correlation between resistance to amphotericin B and sterol pattern. The resistant, posttreatment isolate had a defect in Δ8,7-sterol isomerase, leading to accumulation of ergosta-5,8,22-dienol, ergosta-8,22-dienol, fecosterol, and ergosta-8-enol, with a concomitant depletion of ergosterol, the major sterol in the susceptible pretreatment isolate. In a recent study, Mbongo et al. (85) provided further evidence that the mechanism of amphotericin B resistance in Leishmania donovani involves the substitution of another sterol for ergosterol in the cell membrane. This substitution is associated with a change in membrane fluidity and a lower affinity of amphotericin B for such modified membranes.
The role played by cell wall components in affecting the interaction of polyenes with their primary site of action, the cytoplasmic membrane, was studied extensively by Kerridge and coworkers (32, 68). These authors compared the polyene susceptibility of exponential- and stationary-phase candidal cells and showed that stationary-phase cells were more resistant than exponential-phase ones. This observation was attributed to the fact that in the exponential-phase cells, breakdown and resynthesis of cell wall constituents occurs at a high rate, resulting in improved polyene access to the cell membrane. In contrast, stationary-phase cells would be expected to break down and synthesize cell wall at a much lower rate (68).
In the early 1970s, Capek and Simek (13) reported on the degradation of nystatin by an induced enzyme system elicited by dermatophytic fungi. No other study has confirmed this finding. It is therefore considered unlikely that drug modification represents a prominent mechanism of resistance to polyene antimicrobial agents. Furthermore, since polyenes do not require entrance into the cell, efflux mechanisms are unlikely to be involved in resistance development.
Limited numbers of studies have addressed the genetic basis of polyene resistance and have focused mainly on Saccharomyces cerevisiae. Molzahn and Woods (92) reported the isolation and characterization of S. cerevisiae mutants (n = 103) which were resistant to polyenes including nystatin, filipin, and pemaricin. The mutants were allocated to four unlinked genes, pol1, pol2, pol3, and pol5. These authors found a correlation between the polyene used for mutant isolation and (i) the extent of cross-resistance and (ii) the selection of mutants with mutations at particular pol genes. Analysis of sterols found in the parent and mutants revealed that ergosterol and 24,(28)-dehydroergosterol were predominant in the wild type. In contrast, the latter sterol was not detected in any of the mutants, while ergosterol was lacking in the pol2 mutant and present at only very low levels in the pol3 mutant. Although the interaction between the pol genes is unknown, derived data obtained by using UV absorption spectra suggested that these mutants have an epistatic relationship, i.e., that they act in series rather than parallel (92).
(a) Correlation with antibacterial resistance. Since little is known about the mechanisms by which fungi alter their ergosterol content in association with polyene resistance, it is difficult to draw parallels with antimicrobial resistance mechanisms. Insofar as the mechanism of polyene action involves direct interaction with a structural cellular component (rather than an enzyme or a part of the protein synthesis machinery like a ribosome), it resembles the action of the glycopeptide antibiotics vancomycin and teicoplanin. Glycopeptide antibiotics act by binding to the terminal d-alanyl-d-alanine of the pentapeptide peptidoglycan precursors. This binding inhibits the cleavage of the terminal d-alanine that provides the energy for formation of the bond creating the cross-bridge between different peptide side changes, as well as sterically inhibiting the transglycosylation necessary for peptidoglycan biosynthesis. In the most common form of vancomycin resistance found in gram-positive bacteria, an acquired set of genes sets in motion a process that results in the formation of pentapeptide precursors terminating in d-lactate, to which glycopeptides bind with roughly 1,000-fold lower affinity than to those terminating in d-alanine (3).
Since the above-described mechanism of resistance to glycopeptide antibiotics results from the acquisition of a resistance operon, it is not clear how relevant it is for comparison to polyene resistance in fungi. Perhaps more relevant is the recently described resistance to glycopeptides in Staphylococcus haemolyticus (10). This resistance, similar to polyene resistance, occurs as a result of serial passage on antimicrobial-containing plates and presumably as a result of repeated exposure to vancomycin in patients undergoing peritoneal dialysis for renal failure. Although the exact mechanism of this type of resistance is not clear, levels of resistance appears to correlate with substitutions in the bridge linking the peptide side chains. These alterations in bridge composition may inhibit cooperative binding of glycopeptides to the target, resulting in increased MICs of these antimicrobial agents.
Allylamines.
(i) Mechanism of action.
Allylamines, such as terbinafine and naftifine, have been developed as a new class of ergosterol biosynthetic inhibitors that are functionally as well as chemically distinct from the other major classes of ergosterol-inhibiting antifungal agents (118, 119). Terbinafine (Fig. 1) is highly effective against dermatophytes in vivo and in vitro. A recent study of terbinafine by the National Committee for Clinical Laboratory Standards M27 method showed that its geometric mean MIC against 179 clinical isolates of C. albicans was 1.2 μg/ml (61, 118). Furthermore, preliminary evidence from our group and from Ryder and coworkers indicates that terbinafine has good activity against at least some azole-resistant C. albicans strains (61, 118). By using the same assay system, terbinafine appears highly active against Cryptococcus neoformans (118). Studies investigating the efficacy of this agent against disseminated candidiasis in an animal model are under way.
Allylamines act by inhibiting early steps of ergosterol biosynthesis (Fig. 2). This inhibition coincides with accumulation of the sterol precursor squalene and the absence of any other sterol intermediate (66), suggesting that allylamine inhibition of sterol synthesis occurs at the point of squalene epoxidation, a reaction catalyzed by squalene epoxidase. Studies with isolated squalene epoxidase indicate that it is the target for allylamine activity (118). Fungal cell death is related primarily to the accumulation of squalene rather than to ergosterol deficiency (118). High levels of squalene may increase membrane permeability (74), leading to disruption of cellular organization.
(ii) Mechanism of resistance to allylamines.
Although clinical failure has been observed in patients treated with terbinafine, allylamine resistance in association with clinical use of terbinafine and naftifine has not been found in human pathogenic fungi. However, with the increased use of this agent, resistance may be expected, since Vanden Bossche et al. (141) have reported a C. glabrata strain that became resistant to fluconazole and expressed cross-resistance to terbinafine. Other investigators report that CDR1 can use terbinafine as a substrate (122). The machinery to develop resistance to allylamines is therefore already present in some fungal species.
(a) Correlation with antibacterial resistance. Since allylamine-resistant fungi are as yet not well described, comparisons of resistance mechanisms are moot. It is worth noting, however, that the different sites of action of the azoles, polyenes, and allylamine resemble the sequential actions on cell wall synthesis exhibited by different antibacterial agents, including phosphomycin (a phosphoenolpyruvate analogue that acts at an early step in peptidoglycan synthesis) (64), penicillin (which acts at an intermediate step), and vancomycin (which acts at the final step in cross-linking). As in the study of cell wall synthesis in bacteria, some of the mechanisms of action of antifungal agents have been elucidated by analyzing the accumulation of specific precursors after exposure to the antibiotic. Since all of the antibiotics act at different steps of the same process, it is perhaps not surprising that specific mutations will result in cross-resistance to several of the compounds.
Compounds Active against Fungal Cell Walls
The fungal cell wall contains compounds, such as mannan, chitin, and α- and β-glucans, that are unique to the fungal kingdom. Since these components are not found elsewhere in nature, they have been identified as possible targets that provide selective toxicity advantages (48). Our knowledge of the cell wall composition of medically important fungi comes mainly from studies conducted with C. albicans. The cell wall of this yeast is a multilayered structure composed of chitin, β-glucan and mannoprotein, with the last two constituents making up to 80% of the wall mass (16, 110, 134). The outer layers are composed of mannan, mannoprotein, and β-(1,6)-glucan, while the inner layers are predominantly β-(1,3)-glucan and chitin with some mannoprotein (135).
A number of compounds that have the ability to affect the cell walls of fungi have been discovered and described over the past 30 years (48). We will concentrate in this review on glucan synthesis inhibitors only, since at least one antifungal agent that belongs to this class of compounds is being evaluated in clinical trials (MK-0991, being developed by Merck & Co.). Chitin synthesis inhibitors, such as nikkomycins, have been extensively investigated, but no product has been commercially developed.
Inhibitors of glucan synthesis.
Of the three groups of compounds (aculeacins, echinocandins, and papulacandins) that are specific inhibitors of fungal 3β-glucan synthase, only echinocandins (Fig. 1) are being actively pursued in clinical trials to evaluate their safety, tolerability, and efficacy against candidiasis. Echinocandins, which are lipopeptides, have fungicidal activity both in vitro and in vivo against Candida and Aspergillus species (15, 138, 147).
(i) Mechanism of action.
β-Glucan inhibitors act as specific noncompetitive inhibitors of β-(1,3)-glucan synthetase, a large (210-kDa) integral membrane heterodimeric protein (48). Treatment of fungi with these compounds inhibits the synthesis of the structural glucan component without affecting nucleic acid or mannan synthesis (90, 137). Inhibitors of glucan synthesis also have secondary effects on other components of intact cells including a reduction in the ergosterol and lanosterol content and an increase in the chitin content of the cell wall (107). Inhibition of β-(1,3)-glucan synthetase results in cytological and ultrastructural changes in fungi characterized by growth as pseudohyphae, thickened cell wall, and buds failing to separate from mother cells. Cells also become osmotically sensitive (15, 139), with lysis being restricted largely to the growing tips of budding cells (12).
(ii) Mechanism of resistance to glucan synthesis inhibitors.
Since clinical use of glucan synthesis inhibitors has not occurred, resistant mutants resulting from clinical therapy are not available. Therefore, knowledge of mechanisms of glucan synthesis inhibitors resistance is based entirely on analysis of laboratory-derived mutants. The following discussion is based on the results of laboratory mutation experiments reported by Kurtz and coworkers (70, 71), who analyzed resistant mutants of S. cerevisiae. The target of lipopeptides, including echinocandins, is glucan synthase (a heterodimeric enzyme), which in S. cerevisiae is encoded by FKS1 and RHO1. S. cerevisiae also contains another gene, FKS2, which is highly homologous to FKS1. Mutations in the FKS1 gene confer high-level in vitro resistance to lipopeptides. Low-level resistance (<10-fold) is associated with mutations in another cell wall synthesis gene, GNS1, that encodes an enzyme involved in fatty acid elongation. Mutations in FKS2 gene do not confer resistance. Additionally, activation of MDR-like genes or selection of pathway bypass mutations does not seem to be important as resistance mechanisms to the lipopeptides. Finally, since lipopeptides do not traverse the cell membrane, entry mechanisms may not play a role in their action and thus probably cannot play a role in the response of the fungal cell to them. These findings, taken together with the low rate of mutation (10−8) per generation of fungal cells, suggest that at least in vitro, S. cerevisiae develops resistance to lipopeptide antimycotic agents via mutations that alter the protein encoded by FKS1, which is the main target of the inhibitor and is presumed to be the catalytic component of the fungal cell wall glucan synthase.
Further experimentation with C. albicans mutants indicated that resistance to pneumocandin in S. cerevisiae and C. albicans is very much alike, once the fact that Candida is diploid is taken into account. Of clinical significance is the finding that the C. albicans mutants recovered by Kurtz and coworkers were unchanged in their susceptibility to other clinically used antifungals such as fluconazole, itraconazole, 5FC, and amphotericin B when tested in vitro and in animal models of candidiasis (70, 71).
Compounds Inhibiting Nucleic Acids
5-Fluorocytosine.
(i) Mechanism of action.
5FC is a fluorinated pyrimidine with inhibitory activity against many yeasts, including Candida and Cryptococcus neoformans. The initial promise of this agent has been diminished by the high prevalence of primary resistance in many fungal species. Two surveys of C. albicans conducted by Stiller et al. (130) and Defever et al. (21) provided estimates of resistance frequencies. The majority of the candidal isolates studied were susceptible (60 and 57%), but significant percentages were partially resistant (36 and 37%) or highly resistant (4 and 6%). Today, 5FC is used in combination with other antifungals, such as amphotericin B and fluconazole, but only rarely as a single agent.
5FC enters fungal cells aided by a permease enzyme. Once inside, it is converted to 5-fluorouracil (5FU) by the enzyme cytosine deaminase. Subsequently, 5FU is converted by UMP pyrophosphorylase into 5-fluorouridylic acid (FUMP), which is phosphorylated further and incorporated into RNA, resulting in disruption of protein synthesis (108). 5FU also is converted to 5-fluorodeoxyuridine monophosphate, a potent inhibitor of thymidylate synthase, an enzyme involved in DNA synthesis and nuclear division (23). Thus, 5FC acts by interfering with pyrimidine metabolism, as well as RNA, DNA, and protein synthesis in the fungal cell.
(ii) Mechanism of resistance to 5-fluorocytosine.
5FC resistance mechanisms have been fully investigated and reviewed in depth (59, 149). In principle, resistance to 5FC may result from decreased uptake (loss of permease activity) or loss of enzymatic activity responsible for conversion to FUMP. Although resistance due to decreased 5FC uptake has been found in S. cerevisiae and C. glabrata, this mechanism does not seem to be important in C. albicans or Cryptococcus neoformans (63, 149).
Available data suggest that blocking the formation of FUMP by loss of cytosine deaminase activity or by loss of uracil phosphoribosyltransfnerase (UPRTase) activity is sufficient to confer 5FC resistance. Cytosine deaminase and UPRTase constitute the pyrimidine salvage pathway and are not essential for growth under normal circumstances in which pyrimidines are synthesized de novo. Resistance in the large majority of both clinical and laboratory strains of 5FC-resistant C. albicans and Cryptococcus neoformans is attributable to mutational loss of one of the pyrimidine salvage enzymes (99, 108, 150). Decreased UPRTase activity was associated with resistance in a gene dosage-dependent manner in C. albicans (150). FCY/FCY homozygotes possessed high UPRTase specific activity (approximately 3 U), whereas FCY/fcy heterozygotes possessed less activity (approximately 1.5 U) and fcy/fcy homozygotes possessed barely detectable activity.
(a) Correlation with antibacterial resistance.
Although there are no nucleoside analogues among antibacterial compounds, there are agents that require chemical modification for activity inside the bacterial cell. Among these is metronidazole, a 5-nitroimidazole molecule whose activation depends on reduction of the nitro group in the absence of oxygen. Resistance to metronidazole is rare (perhaps because of its relatively infrequent use in comparison to other agents) and is believed to be due to decreased uptake or reduced rate of reduction (26). A more relevant comparison involves resistance of herpes simplex virus to the antiviral compound acyclovir. This agent is phosphorylated intracellularly by virus-encoded thymidine kinase. Acyclovir monophosphate then becomes converted to acyclovir triphosphate by cellular enzymes, at which point the molecule becomes integrated into replicating viral DNA, where it acts as a chain terminator. The most common mechanism of resistance to acyclovir is either a deficiency or altered substrate specificity of the viral thymidine kinase (50). These alterations result in the failure to convert acyclovir to its active form inside the cell.
IS THERE A RELATIONSHIP BETWEEN RESISTANCE DEVELOPMENT AND VIRULENCE?
It is generally accepted that a difference in the pathogenicity and resistance pattern of various candidal species exists. For example, C. albicans is both more pathogenic and more susceptible to antifungals than C. krusei (115). This raises the question whether there is a correlation between the ability of an organism to cause infection and its resistance to antifungals.
A number of investigators have attempted to answer this question. Anaissie et al. (1) evaluated the pathogenicity of C. krusei in normal and immunocompromised mice and compared its virulence to C. albicans. Unlike C. albicans, a combination of a large inoculum and immunosuppression was needed to establish severe infection. A high inoculum was also required to establish hematogenously disseminated candidal infection in a neutropenic guinea pig model (38). These data underline the low pathogenicity of C. krusei. In another study, Graybill et al. (43) demonstrated decreased virulence of serial C. albicans isolates with increasing fluconazole MICs in a mouse model of systemic candidiasis. In another study by the same group (42), a murine model of systemic candidiasis was used to assess the virulence of serial C. albicans strains (obtained from five patients with 17 episodes of oropharyngeal candidiasis) for which the fluconazole MICs were increasing. The fluconazole MICs for these isolates exhibited at least an eightfold progressive increase from susceptible (MIC < 8 μg/ml) to resistant (MIC > 16 μg/ml). When the virulence of these isolates was tested in the animal model, a fivefold progressive decrease in the dose, accounting for a 50% mortality rate, was noted. Consistent with a reduction in virulence of the serial isolates was the finding that a decreased fungal burden in the kidneys occurred in mice challenged with two of three resistant strains. Therefore, there is a suggestion from animal studies with species that are innately resistant, such as C. krusei strains and C. albicans isolates that have attained resistance, that the presence of resistance is correlated with diminished virulence. However, establishing a direct cause-and-effect relationship requires more investigation.
There is evidence that virulence is an intrinsic trait related to species-specific genetic determinants. Studies show that unlike C. albicans, C. krusei lacks virulence determinants. Therefore, virulence is not associated with resistance or susceptibility of an organism per se. Data from our group and others show that C. krusei adheres poorly to host cells (epithelial and endothelial) as well as to nonbiological surfaces (30, 120). Moreover, C. krusei-mediated endothelial-cell damage requires a longer incubation period and higher initial inoculum compared to C. albicans (1 × 106 and 2 × 105 cells, respectively) (30). Additionally, C. krusei showed less invasiveness of dorsal tongue mucosal cultured cells than did C. albicans or C. tropicalis (120). These characteristics seem to be maintained across different C. krusei strains, suggesting that virulence attenuation is a general characteristic of C. krusei and is not related to whether the strain is susceptible or resistant to antifungal agents. Studies comparing the virulence of a number of C. krusei strains that differ in their susceptibility to antifungals are necessary to prove this hypothesis.
Examples abound of bacterial species in which resistant variants are no less virulent than their more susceptible counterparts. Methicillin-resistant staphylococci are just as virulent as their susceptible counterparts, as are penicillin-resistant pneumococci and ampicillin-resistant E. coli strains. It is conceivable that some resistance traits, such as those mediated by the decreased expression of outer membrane proteins, may confer a selective disadvantage in the absence of antibiotic selective pressure, since these channels presumably perform other functions important for the survival of the cell. Conversely, in the setting of widespread antimicrobial use, resistance determinants may indeed behave as virulence determinants by favoring colonization, which predisposes to infection. Much more work is required to define mechanisms of bacterial virulence before a precise correlation between virulence and resistance can be made.
PREVENTION AND CONTROL OF ANTIFUNGAL RESISTANCE
Strategies to avoid and suppress the emergence of antifungal resistance have not been defined. However, approaches analogous to those recommended for antibacterials (19, 78, 126) could be suggested. These measures include (i) prudent use of antifungals, (ii) appropriate dosing with special emphasis on avoiding treatment with low antifungal dosage, (iii) therapy with combinations of existing agents, (iv) treatment with the appropriate antifungal (in cases where the etiological agent is known), and (v) use of surveillance studies to determine the true frequency of antifungal resistance. It should be emphasized that data supporting the use of the suggested measures is largely lacking, and ongoing studies may provide some specific guidelines in the near future. Additionally, advances in rapid diagnosis of fungi may be helpful in reducing the use of inappropriate antifungals to treat organisms that are resistant to a particular agent. Unfortunately, progress in developing diagnostic methods specific to fungi has been slow. The recent approval of a reference method for the antifungal susceptibility testing of yeast (95) is encouraging and provides a means for performing surveillance studies.
CONCLUSION
The expression of resistance to antimicrobial agents is the logical and inevitable consequence of using these agents to treat human infections. The availability of molecular genetic tools has led to a rapid expansion in our understanding of the mechanisms by which antibacterial resistance emerges and spreads and promises to greatly inform efforts to develop novel and effective compounds for future use. With increased use and availability of different classes of antifungal agents, it is anticipated that we will see an increasing number and variety of fungal species resistant to these agents. Continued efforts to study the mechanisms of antifungal resistance and the development of experimental systems (similar to those available in bacteria) in which individual resistance mechanisms can be studied will be important components of a strategy to limit the emergence of resistance to these agents and to develop safer and more potent compounds for the future.
ACKNOWLEDGMENTS
We thank Pranab Mukherjee for drawing the figures and for assistance in putting the manuscript together. Also, we thank Kathleen Smith for assisting in typing the manuscript and Mike Petit for performing the literature search.
REFERENCES
- 1.Anaissie E, Hachem R, Tin-U C K-, Stephens L C, Bodey G P. Experimental hematogenous candidiasis caused by Candida krusei and Candida albicans: species differences in pathogenicity. Infect Immun. 1993;61:1268–1271. doi: 10.1128/iai.61.4.1268-1271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anaissie E J, Bodey G P. Nosocomial fungal infections—old problems and new challenges. Infect Dis Clin North Am. 1989;3:867–882. [PubMed] [Google Scholar]
- 3.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 4.Ather M A, Winner H I. Development of resistance by Candida species to polyene antibiotics in vitro. J Med Microbiol. 1971;4:505–517. doi: 10.1099/00222615-4-4-505. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Sagué C M, Jarvis W R National Nosocomial Infections Surveillance System. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 6.Beggs W H. Comparison of miconazole- and ketoconazole-induced release of K+ from Candida species. J Antimicrob Chemother. 1983;11:381–383. doi: 10.1093/jac/11.4.381. [DOI] [PubMed] [Google Scholar]
- 7.Beggs W H, Andrews F A, Sarosi G A. Combined action of amphotericin B and 5-fluorocytosine on pathogenic yeasts susceptible to either drug alone. Chemotherapy. 1981;27:247–251. doi: 10.1159/000237987. [DOI] [PubMed] [Google Scholar]
- 8.Beggs W H, Sarosi A, Walker M I. Synergistic action of amphotericin B and rifampin against Candida species. J Infect Dis. 1976;133:206–209. doi: 10.1093/infdis/133.2.206. [DOI] [PubMed] [Google Scholar]
- 9.Belanger P, Nast C, Fratti R, Sanati H, Ghannoum M. Voriconazole (UK-109, 496) inhibits the growth and alters the morphology of fluconazole-susceptible and -resistant Candida species. Antimicrob Agents Chemother. 1997;41:1840–1842. doi: 10.1128/aac.41.8.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billot-Klein D, Gutmann L, Bryant D, Bell D, van Heijenoort J, Grewal J, Shlaes D M. Peptidoglycan synthesis and structure in Staphylococcus haemolyticus expressing increasing levels of resistance to glycopeptide antibiotics. J Bacteriol. 1996;178:4696–4703. doi: 10.1128/jb.178.15.4696-4703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolard J, Milhaud J. Interaction of the anti-Candida amphotericin B (and other polyene antibiotics) with lipids. In: Prasad R, Ghannoum M, editors. Lipids of pathogenic fungi. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 253–274. [Google Scholar]
- 12.Bozzola J J, Mehta R, Nisbet L, Valenta J. The effect of aculeacin A and papulacandin B on morphology and cell wall ultrastructure in Candida albicans. Can J Microbiol. 1984;30:857–863. doi: 10.1139/m84-133. [DOI] [PubMed] [Google Scholar]
- 13.Capek A, Simek A. Antimicrobial agents. XII. Relationship between biochemical resistance and microbial degradation of antimycotics. Folia Microbiol. 1971;16:472–475. doi: 10.1007/BF02872721. [DOI] [PubMed] [Google Scholar]
- 14.Capek A, Simek A, Bruna L, Svab A, Budesinsk Z. Antimicrobial agents. XXI. Dependence of antifungal activity on the structure of the side chain in the pyrimidine group. Folia Microbiol. 1974;19:169–171. doi: 10.1007/BF02872853. [DOI] [PubMed] [Google Scholar]
- 15.Cassone A, Mason R, Kerridge D. Lysis of growing yeast-form cells of Candida albicans by echinocandin: a cytological study. Sabouraudia. 1981;19:97–110. [PubMed] [Google Scholar]
- 16.Cassone A, Mason R E, Kerridge D. Lysis of growing yeast-form cells of Candida albicans following cessation of growth and their possible relationship to the development of polyene resistance. J Gen Microbiol. 1979;110:339–349. doi: 10.1099/00221287-110-2-339. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J T, Witty R T, Robinson R R, Yarger W E. Amphotericin B nephrotoxicity: increased renal resistance and tubule permeability. Kidney Int. 1982;22:626–633. doi: 10.1038/ki.1982.221. [DOI] [PubMed] [Google Scholar]
- 18.Clark F S, Parkinson T, Hitchcock C A, Gow N A R. Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species: possible role for drug efflux in drug resistance. Antimicrob Agents Chemother. 1996;40:429–425. doi: 10.1128/aac.40.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen M L. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 20.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Defever K, Whelan W, Rogers A, Beneke E, Veselenak J, Soll D. Candida albicans resistance to 5-fluorocytosine: frequency of partially resistant strains among clinical isolates. Antimicrob Agents Chemother. 1982;22:810–815. doi: 10.1128/aac.22.5.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Kruijff B, Demel R A. Polyene antibiotic-sterol interactions in membranes of Acholeplasma laidlawaii cells and lecithin liposomes. III. Molecular structure of the polyene antibiotic-cholesterol complexes. Biochim Biophys Acta. 1974;339:57–70. doi: 10.1016/0005-2736(74)90332-0. [DOI] [PubMed] [Google Scholar]
- 23.Diasio R, Bennett J, Myers C. Mode of action of 5-fluorocytosine. Biochem Pharmacol. 1978;27:703–707. doi: 10.1016/0006-2952(78)90507-5. [DOI] [PubMed] [Google Scholar]
- 24.Dick J D, Merz W G, Saral R. Incidence of polyene-resistant yeasts recovered from clinical specimens. Antimicrob Agents Chemother. 1980;18:158–163. doi: 10.1128/aac.18.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowson C, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 26.Edwards D I. Nitroimidazole drugs—action and resistance mechanisms. II. Mechanisms of resistance. J Antimicrob Chemother. 1993;31:201–210. doi: 10.1093/jac/31.2.201. [DOI] [PubMed] [Google Scholar]
- 27.Espinel-Ingroff A. Clinical relevance of antifungal resistance. Antimicrob Resist. 1997;11:929–944. doi: 10.1016/s0891-5520(05)70398-6. [DOI] [PubMed] [Google Scholar]
- 28.Fontana R, Aldegheri M, Ligozzi M, Lopez H, Sucari A, Satta G. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1994;38:1980–1983. doi: 10.1128/aac.38.9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fothergill W A, Sutton D A, Rinaldi M G. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemother. Washington, D.C.: American Society for Microbiology; 1996. An in vitro head-to-head comparison of Schering 5692, amphotericin B, fluconazole, and itraconazole against a spectrum of filamentous fungi, abstr. F89; p. 115. [Google Scholar]
- 30.Fratti R A, Belanger P H, Sanati H, Ibrahim A S, Ghannoum M A. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C.: American Society for Microbiology; 1996. The effect of the new triazole, UK-109,496 on the interactions between Candida albicans and endothelial cells in vitro, abstr. F-76; p. 155. [Google Scholar]
- 31.Fryberg M. Sterol biosynthesis in antibiotic-resistant yeast: nystatin. Arch Biochem Biophys. 1974;160:83–89. doi: 10.1016/s0003-9861(74)80011-1. [DOI] [PubMed] [Google Scholar]
- 32.Gale E F, Johnson A M, Kerridge D, Koh T Y. Factors affecting the changes in amphotericin B sensitivity of Candida albicans during growth. J Gen Microbiol. 1975;87:20–36. doi: 10.1099/00221287-87-1-20. [DOI] [PubMed] [Google Scholar]
- 33.Gallis H A, Drew R H, Pickard W W. Amphotericin B: 30 years of clinical experience. Rev Infect Dis. 1990;12:308–329. doi: 10.1093/clinids/12.2.308. [DOI] [PubMed] [Google Scholar]
- 34.Geber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung K J, Bennett J. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgopapadakou N H, Walsh T J. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 36.Georgopapadakou N H, Walsh T J. Antifungal agents: chemotherapeutic targets and immunologic strategies. Antimicrob Agents Chemother. 1996;40:279–291. doi: 10.1128/aac.40.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghannoum M, Witt M, Espinel-Ingroff A, Ritchie J, Larsen R, Edwards J E., Jr . Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C.: American Society for Microbiology; 1994. Correlation of in vitro susceptibility testing of Cryptococcus neoformans with in vivo outcome: a comparison of two microdilution methods, abstr. F-103; p. 606. [Google Scholar]
- 37a.Ghannoum, M. Unpublished data.
- 38.Ghannoum M, Okagbule-Wonodi I, Bhat N, Sanati H. Antifungal activity of voriconazole (UK-109,496), fluconazole and amphotericin B against hematogenous Candida krusei infection in neutropenic guinea pig model. J Chemother. 1999;11:34–39. doi: 10.1179/joc.1999.11.1.34. [DOI] [PubMed] [Google Scholar]
- 39.Ghannoum M, Spellberg B J, Ibrahim A, Ritchie J A, Currie B, Spitzer E, Edwards J E, Jr, Casadevall A. Sterol composition of Cryptococcus neoformans in the presence and absence of fluconazole. Antimicrob Agents Chemother. 1994;38:2029–2033. doi: 10.1128/aac.38.9.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottesman M M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 41.Gottleib D H, Carter H E, Sloneker J H, Ammann A. Protection of fungi against polyene antibiotics by sterols. Science. 1958;128:361–365. doi: 10.1126/science.128.3320.361. [DOI] [PubMed] [Google Scholar]
- 42.Graybill J R, Montalbo E, Kirkpatrick W R, Luther M F, Revankar S G, Patterson T F. Fluconazole versus Candida albicans: a complex relationship. Antimicrob Agents Chemother. 1998;42:2938–2942. doi: 10.1128/aac.42.11.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graybill J R, Montalbo E, Kirkpatrick W R, Revankar S, Rinaldi M, Patterson T. 13th International Conference for Human and Animal Mycology. 1997. Candida albicans: less predictable than we may think, abstr. O49. [Google Scholar]
- 44.Hackbarth C J, Kocagoz T, Kocagoz S, Chambers H F. Point mutations in Staphylococcus aureus PBP 2 gene affect penicillin-binding kinetics and are associated with resistance. Antimicrob Agents Chemother. 1995;39:103–106. doi: 10.1128/aac.39.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton-Miller J M T. Physiological properties of mutagen induced variants of Candida albicans resistant to polyene antibiotics. J Med Microbiol. 1972;5:425–440. doi: 10.1099/00222615-5-4-425. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton-Miller J M T. Chemistry and biology of the polyene macrolide antibiotics. Bacteriol Rev. 1973;37:166–196. [PMC free article] [PubMed] [Google Scholar]
- 47.Hata K, Ueno J, Miki H, et al. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1995. ER-30346, a novel antifungal triazole. III. In vitro activity and the mode of action, abstr. F92. [Google Scholar]
- 48.Hector R F. Compounds active against cell walls of medically important fungi. Clin Microbiol Rev. 1993;6:1–21. doi: 10.1128/cmr.6.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 50.Hill E L, Hunter G A, Ellis M N. In vitro and in vivo characterization of herpes simplex virus clinical isolates recovered from patients infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1991;35:2322–2328. doi: 10.1128/aac.35.11.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hitchcock C, Dickinson K, Brown S B, Evans E G, Adams D J. Interaction of azole antifungal antibiotics with cytochrome P450-dependent 14 α-sterol demethylase purified from Candida albicans. J Biochem. 1990;266:475–480. doi: 10.1042/bj2660475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hitchcock C, Whittle P T. Chemistry and mode of action of fluconazole. In: Rippon J W, Fromtling R A, editors. Cutaneous antifungal agents: selected compounds in clinical practice and development. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 183–197. [Google Scholar]
- 53.Hitchcock C A, Barrett-Bee K A, Russell N J. The lipid composition and permeability to azole of an azole- and polyene-resistant mutant of Candida albicans. J Med Vet Mycol. 1987;25:29–37. doi: 10.1080/02681218780000041. [DOI] [PubMed] [Google Scholar]
- 54.Hitchcock C A, Pye G W, Oliver G P, Troke P F. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1995. UK-109,496: a novel, wide-spectrum triazole derivative for the treatment of fungal infections: antifungal activity and selectivity in vitro, abstr. 2739. [Google Scholar]
- 55.Holt R J. The imidazoles. In: Speller D C E, editor. Antifungal chemotherapy. Chichester, England: John Wiley & Sons, Ltd.; 1980. pp. 107–148. [Google Scholar]
- 56.Holz R W. The effects of the polyene antibiotics nystatin and amphotericin B on thin lipid membranes. Ann N Y Acad Sci. 1974;235:469–479. doi: 10.1111/j.1749-6632.1974.tb43284.x. [DOI] [PubMed] [Google Scholar]
- 57.HsuChen C C, Feingold D S. Polyene antibiotic action on lecithin liposomes: effect of cholesterol and fatty acyl chains. Biochem Biophys Res Commun. 1973;51:972–978. doi: 10.1016/0006-291x(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 58.Itoh K, Okonagi K, Tasaka A, Hayashi R, Tamura N, Tsuchimori N, Kitazaki T, Matsushita Y, Obita J. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1996. TAK 187, a new antifungal triazole: Synthesis and antifungal activity, abstr. F74; p. 112. [Google Scholar]
- 59.Iwata K. Drug resistance in human pathogenic fungi. Eur J Epidemiol. 1992;8:407–421. doi: 10.1007/BF00158576. [DOI] [PubMed] [Google Scholar]
- 60.Jenkinson H F. Ins and outs of antimicrobial resistance: era of the drug pumps. J Dent Res. 1996;75:736–742. doi: 10.1177/00220345960750020201. [DOI] [PubMed] [Google Scholar]
- 61.Jessup C J, Ryder N S, Ghannoum M A. Program and Abstracts of the 36th Annual Meeting of the Infectious Diseases Society of America. 1998. Evaluation of antifungal activity of terbinafine against a broad range of pathogenic fungi, abstr. 246 Sa; p. 122. [Google Scholar]
- 62.Juliano R, Grant C W M, Barber K R, Kalp M A. Mechanism of the selective toxicity of amphotericin B incorporated into liposomes. Mol Pharmacol. 1987;31:1–11. [PubMed] [Google Scholar]
- 63.Jund R, Lacroute F. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J Bacteriol. 1970;102:607–615. doi: 10.1128/jb.102.3.607-615.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kahan F M, Kahan J S, Cassidy P J, Kropp H. The mechanism of action of fosfomycin (phosphomycin) Ann N Y Acad Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 65.Kelly S L, Lamb D C, Taylor M, Corran A J, Baldwin B C, Powderly W G. Resistance to amphotericin B associated with defective sterol Δ8,7 isomerase in a Cryptococcus neoformans strain from an AIDS patients. FEMS Microbiol Lett. 1994;122:39–42. doi: 10.1111/j.1574-6968.1994.tb07140.x. [DOI] [PubMed] [Google Scholar]
- 66.Kerridge D. The plasma membrane of Candida albicans and its role in the action of antifungal drugs. In: Gooday G W, Lloyd D, Trinci A P J, editors. The eukaryotic microbial cell. Cambridge, England: Cambridge University Press; 1980. p. 103. [Google Scholar]
- 67.Kerridge D. The protoplast membrane and antifungal drugs. In: Peberdy J F, Ferenczy L, editors. Fungal protoplasts: applications in biochemistry and genetics. New York, N.Y: Marcel Dekker Inc.; 1985. p. 135. [Google Scholar]
- 68.Kerridge D, Koh T Y, Marriott M S, Gale E F. Microbiology and plant protoplasts. In: Peberdy J F, Rose A H, Rodger H J, Cocking E C, editors. Microbiology and plant protoplasts. London, England: Churchill Livingston; 1976. pp. 23–38. [Google Scholar]
- 69.Kleinschmidt M G, Chough K S, Mudd J B. Effect of filipin on liposomes prepared with different types of steroids. Plant Physiol. 1972;49:852. doi: 10.1104/pp.49.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kurtz M B. New antifungal drug targets: a vision for the future. ASM News. 1997;64:31–39. [Google Scholar]
- 71.Kurtz M B, Douglas C M. Lipopeptide inhibitors of fungal glucan synthase. J Med Vet Mycol. 1997;35:79–86. doi: 10.1080/02681219780000961. [DOI] [PubMed] [Google Scholar]
- 72.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14 alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 73.Lampen J O, Arnow P M, Safferman R S. Mechanism of protection by sterol against polyene antibiotics. J Bacteriol. 1960;80:200–206. doi: 10.1128/jb.80.2.200-206.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lanyi J, Plachy W Z, Kates M. Lipid interactions in membranes of extremely halophilic bacteria. II. Modification of the bilayer structure by squalene. Biochemistry. 1974;13:4914–4920. doi: 10.1021/bi00721a006. [DOI] [PubMed] [Google Scholar]
- 75.Larsen R A, Bozzette S A, Jones B E. Fluconazole combined with flucytosine for cryptococcal meningitis in persons with AIDS. Clin Infect Dis. 1994;19:741–745. doi: 10.1093/clinids/19.4.741. [DOI] [PubMed] [Google Scholar]
- 76.Law D, Denning D W. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1996. In vitro activity of Schering 56592, compared with fluconazole and itraconazole against Candida spp., abstr. F88; p. 115. [Google Scholar]
- 77.Leclerq R, Dutka-Malen S, Brisson-Noel A, Molinas C, Derlot E, Arthur M, Duval J, Courvalin P. Resistance of enterococci to aminoglycosides and glycopeptides. Clin Infect Dis. 1992;15:495–501. doi: 10.1093/clind/15.3.495. [DOI] [PubMed] [Google Scholar]
- 78.Levy S B. Starting life resistance-free. N Engl J Med. 1990;323:335–337. doi: 10.1056/NEJM199008023230509. [DOI] [PubMed] [Google Scholar]
- 79.Li X-Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Limaye A P, Gautom R K, Black D, Fritsche T R. Rapid emergence of resistance to cefepime during treatment. Clin Infect Dis. 1997;25:339–340. doi: 10.1086/516917. [DOI] [PubMed] [Google Scholar]
- 81.Livermore D M. Interplay of impermeability and chromosomal β-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2046–2048. doi: 10.1128/aac.36.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lopez-Berestein G, Bodey G, Frankel L S, Mehta K. Treatment of hepatosplenic candidiasis with liposomal amphotericin. Br Clin Oncol. 1987;5:310–317. doi: 10.1200/JCO.1987.5.2.310. [DOI] [PubMed] [Google Scholar]
- 83.Mago N, Khuller G K. Influence of lipid composition on the sensitivity of Candida albicans to antifungal agents. Indian J Biochem Biophys. 1989;26:30–33. [PubMed] [Google Scholar]
- 84.Martins M D, Rex J H. Resistance to antifungal agents in the critical care setting: problems and perspectives. New Horizons. 1996;4:338–344. [PubMed] [Google Scholar]
- 85.Mbongo N, Loiseau P M, Billion M A, Robert-Gero M. Mechanism of amphotericin B resistance in Leishmania donovani promastigotes. Antimicrob Agents Chemother. 1998;42:352–357. doi: 10.1128/aac.42.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGinnis M, Rinaldi M G. Antifungal drugs: mechanisms of action, drug resistance, susceptibility testing, and assays of activity in biological fluids. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 176–211. [Google Scholar]
- 87.Medoff G, Comfort M, Kobayashi G S. Synergistic action of amphotericin B and 5-fluorocytosine against yeast-like organisms. Proc Soc Exp Biol Med. 1971;138:571–574. doi: 10.3181/00379727-138-35943. [DOI] [PubMed] [Google Scholar]
- 88.Michaelis S, Berkower C. Sequence comparison of yeast ATP binding cassette (ABC) proteins. Cold Spring Harbor Symp Quant Biol. 1995;60:291–309. doi: 10.1101/sqb.1995.060.01.034. [DOI] [PubMed] [Google Scholar]
- 89.Milefchik E, Leal M A, Haubrich R, See R, Bozzette S A, Larsen R. Programs and Abstracts of the 9th International Conference on AIDS/4th STD World Congress. 1993. High dose fluconazole with and without flucytosine for AIDS associated cryptococcal meningitis, abstr. WS-B5-12. [Google Scholar]
- 90.Mizoguchi J, Saito T, Mizuno K, Hayano K. On the mode of action of a new antifungal antibiotic, aculeacin A: inhibition of cell wall synthesis of Saccharomyces cerevisiae. J Antibiot. 1977;30:308–313. doi: 10.7164/antibiotics.30.308. [DOI] [PubMed] [Google Scholar]
- 91.Moellering R C, Weinberg A N. Studies on antibiotic synergism against enterococci. II. Effect of various antibiotics on the uptake of 14C-labelled streptomycin by enterococci. J Clin Investig. 1971;50:2580–2584. doi: 10.1172/JCI106758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Molzahn S W, Woods R A. Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J Gen Microbiol. 1972;72:339–348. doi: 10.1099/00221287-72-2-339. [DOI] [PubMed] [Google Scholar]
- 93.Munayyer H, Shaw K J, Hare R S, Salisbury B, Heimark L, Pramanik B, Greene J R. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1996. SCH 56592 is a potent inhibitor of sterol C-14 demethylation in fungi, abstr. F92; p. 115. [Google Scholar]
- 94.Nakamura T, Sakai R, Sonada J, Katoh T, Kaneko T, Horie T. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1995. ER-30346, a novel antifungal triazole. IV. Pharmacokinetic and toxicological studies in mice, rats, and dogs, abstr. F94; p. 129. [Google Scholar]
- 95.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 96.Neyfakh A A, Borsch C M, Kaatz G W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Norman A W, Demel R A, de Kruijff B, Guerts-van Kessel W S M, Van Deenen L L M. Studies on the biological properties of polyene antibiotics: comparison of other polyenes with filipin in their ability to interact specifically with sterol. Biochim Biophys Acta. 1972;290:1–14. doi: 10.1016/0005-2736(72)90046-6. [DOI] [PubMed] [Google Scholar]
- 98.Norman A W, Demel R A, de Kruijff B, Van Deenen L L M. Studies on the biological properties of polyene antibiotics. Evidence for the direct interaction of filipin with cholesterol. J Biol Chem. 1972;247:1918–1929. [PubMed] [Google Scholar]
- 99.Normark S, Schonebeck J. In vitro studies of 5-fluorocytosine resistance in Candida albicans and Torulopsis glabrata. Antimicrob Agents Chemother. 1972;2:114–121. doi: 10.1128/aac.2.3.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nozawa Y, Morita T. Molecular mechanisms of antifungal agents associated with membrane ergosterol. Dysfunction of membrane ergosterol and inhibition of ergosterol biosynthesis. In: Iwata K, Vanden Bossche H, editors. In vitro and in vivo evaluation of antifungal agents. Amsterdam, The Netherlands: Elsevier Science Publishers, B. V.; 1986. p. 111. [Google Scholar]
- 101.Orozco A, Higginbotham L, Hitchcock C, Parkinson T, Falconer D, Ibrahim A, Ghannoum M, Filler S G. Mechanism of fluconazole resistance in Candida krusei. Antimicrob Agents Chemother. 1998;42:2645–2649. doi: 10.1128/aac.42.10.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pangon B, Bizet C, Bure A, Pichon F, Phillipon A, Regnier B, Gutmann L. In vivo selection of a cephamycin-resistant, porin-deficient mutant of Klebsiella pneumoniae producing a TEM-3 β-lactamase. J Infect Dis. 1989;159:1005–1006. doi: 10.1093/infdis/159.5.1005. [DOI] [PubMed] [Google Scholar]
- 103.Parkinson T, Falconer D J, Hitchcock C. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob Agents Chemother. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parks L W, Casey W M. Fungal sterols. In: Prasad R, Ghannoum M, editors. Lipids of pathogenic fungi. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 63–82. [Google Scholar]
- 105.Payne N I, Cosgrove R F, Green A P, Liu L. In vivo studies of amphotericin B liposomes derived from proliposomes; effect of formulation on toxicity and tissue disposition of the drug in mice. J Pharm Pharmacol. 1987;39:24–28. doi: 10.1111/j.2042-7158.1987.tb07156.x. [DOI] [PubMed] [Google Scholar]
- 106.Perfect J R, Schell W A. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1995. In vitro efficacy of the azole, SCH 56592, compared to amphotericin B, fluconazole, and itraconazole versus Cryptococcus neoformans, abstr. F64; p. 124. [Google Scholar]
- 107.Pfaller M, Riley J, Koerner T. Effect of cilofungin ( LY121019) on carbohydrate and sterol composition of Candida albicans. Eur J Clin Microbiol Infect Dis. 1989;8:1067–1070. doi: 10.1007/BF01975172. [DOI] [PubMed] [Google Scholar]
- 108.Polak A, Scholer H. Mode of action of 5-fluorocytosine and mechanisms of resistance. Chemotherapia. 1975;21:113–130. doi: 10.1159/000221854. [DOI] [PubMed] [Google Scholar]
- 109.Polak A, Scholer H J, Wall M. Combination therapy of experimental candidiasis, cryptococcosis and aspergillosis in mice. Exp Chemother. 1982;28:461–479. doi: 10.1159/000238138. [DOI] [PubMed] [Google Scholar]
- 110.Poulain D, Tronchin Dubremetz J F, Biguet J. Ultrastructure of the cell wall of Candida albicans blastospores: study of the constitutive layers by the use of a cytochemical technique revealing polysaccharides. Ann Microbiol. 1978;129A:141–153. [PubMed] [Google Scholar]
- 111.Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 112.Rao T V G, Das S, Prasad R. Effect of phospholipid enrichment on nystatin action: differences in antibiotic sensitivity between in vivo and in vitro conditions. Microbios. 1985;42:145–153. [PubMed] [Google Scholar]
- 113.Rao T V G, Trivedi A, Prasad R. Phospholipid enrichment of Saccharomyces cerevisiae and its effect on polyene sensitivity. Can J Microbiol. 1985;31:322–326. doi: 10.1139/m85-061. [DOI] [PubMed] [Google Scholar]
- 114.Redding S J, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhine-Chalberg J, Pfaller M. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin Infect Dis. 1994;18:240–242. doi: 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 115.Rex J H, Rinaldi M, Pfaller M. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rex J H, Pfaller M A, Galgiani J N, Bartlett M S, Espinel-Ingroff A, Ghannoum M A, Lancaster M, Odds F C, Rinaldi M G, Walsh T J, Barry A L. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin Infect Dis. 1996;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 117.Roessner C A, Min C, Hardin S H, Harris H L, McColum J C, Scott A I. Sequence of the Candida albicans erg7 gene. Gene. 1993;127:149–150. doi: 10.1016/0378-1119(93)90631-c. [DOI] [PubMed] [Google Scholar]
- 118.Ryder N, Favre B. Antifungal activity and mechanism of action of terbinafine. Rev Contemp Pharmacother. 1997;8:275–287. [Google Scholar]
- 119.Ryder N S, Seidl G, Troke P. Effect of the antimycotic drug naftifine on growth of and sterol biosynthesis in Candida albicans. Antimicrob Agents Chemother. 1984;25:483–487. doi: 10.1128/aac.25.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Samaranayake Y H, Samaranayake L P. Candida krusei: biology, epidemiology, pathogenicity and clinical manifestations of an emerging pathogen. Med Microbiol. 1994;41:295–310. doi: 10.1099/00222615-41-5-295. [DOI] [PubMed] [Google Scholar]
- 121.Sanati H, Belanger P, Fratti R, Ghannoum M. A new triazole, voriconazole (UK-109,496), blocks sterol biosynthesis in Candida albicans and Candida krusei. Antimicrob Agents Chemother. 1997;41:2492–2496. doi: 10.1128/aac.41.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sanglard D, Ischer F, Monod M, Bille J. Susceptibility of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schulman J A, et al. Fatal disseminated cryptococcosis following intraocular involvement. Br J Ophthalmol. 1988;72:171–175. doi: 10.1136/bjo.72.3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124a.Sheehan, D. J. (Pfizer Pharmaceuticals Group). Personal communication.
- 125.Sheehan D J, Hitchcock C A, Sibley C M. Current and emerging azole antifungal agents. Clin Microbiol Rev. 1999;12:40–79. doi: 10.1128/cmr.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shlaes D M, Gerding D N, John J F, Jr, Craig W A, Bornstein D L, Duncan R A, et al. Society for Healthcare Epidemiology of America and Infectious Diseases Society of American Joint Committee on the Prevention of Antimicrobial Resistance: guidelines for the prevention of antimicrobial resistance in hospitals. Clin Infect Dis. 1997;25:584–599. doi: 10.1086/513766. [DOI] [PubMed] [Google Scholar]
- 127.Smith K J, Warnock D W, Kennedy C T C, Johnson E M, Hopwood V, Van Cutsem J, Vanden Bossche H. Azole resistance in Candida albicans. J Med Vet Mycology. 1986;24:133–144. [PubMed] [Google Scholar]
- 128.Sokol-Anderson M L, Brajtburg J, Medoff G. Sensitivity of Candida albicans to amphotericin B administered as single or fractionated doses. Antimicrob Agents Chemother. 1986;29:701. doi: 10.1128/aac.29.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stevens D A, Devine M J, Martinez M, Aristizabal B. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1995. In vitro antifungal activity of novel azole derivatives with a morpholine ring, abstr. F85; p. 127. [Google Scholar]
- 130.Stiller R, Bennett J, Scholer J, Wall M, Polak A, Stevens D A. Susceptibility to 5-fluorocytosine and prevalence of serotype in 402 Candida albicans isolates from the United States. Antimicrob Agents Chemother. 1982;22:482–487. doi: 10.1128/aac.22.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sud I J, Feingold D S. Mechanisms of action of the antimycotic imidazoles. J Investig Dermatol. 1981;76:428–441. doi: 10.1111/1523-1747.ep12521036. [DOI] [PubMed] [Google Scholar]
- 132.Sugar A M. The polyene macrolide antifungal drugs. In: Peterson P K, Verhoef J, editors. Antimicrobial agents. Vol. 1. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1986. pp. 229–244. [Google Scholar]
- 133.Sugar A M, Liu X P. Program and Abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1995. In vitro activity of SCH against Blastomyces dermatitidis (Bd), abstr. F65; p. 124. [Google Scholar]
- 134.Sullivan P A, Chiew Y Y, Molloy C, Templeton M D, Shepard M G. An analysis of the metabolism and cell wall composition of Candida albicans during germ tube formation. Can J Microbiol. 1983;29:1514–1525. doi: 10.1139/m83-233. [DOI] [PubMed] [Google Scholar]
- 135.Surarit R, Gopal P K, Shepard M G. Evidence for a glycosidic linkage between chitin and glucan in the cell wall of Candida albicans. J Gen Microbiol. 1988;134:1723–1730. doi: 10.1099/00221287-134-6-1723. [DOI] [PubMed] [Google Scholar]
- 136.Tatsumi Y, Yokoo M, Aarika T, Ogura K, Nagal K, Naito T. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1996. Therapeutic efficacy of KP-103, a novel topical antifungal triazole, on experimental superficial mycosis, abstr. F80; p. 113. [Google Scholar]
- 137.Titsworth E, Grunberg E. Chemotherapeutic activity of 5-fluorocytosine and amphotericin B against Candida albicans in mice. Antimicrob Agents Chemother. 1973;4:306–308. doi: 10.1128/aac.4.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tkacz J S. Glucan biosynthesis in fungi and its inhibition. In: Sutcliffe J, Georgopapadakou N H, editors. Emerging targets in antibacterial and antifungal chemotherapy. New York, N.Y: Chapman & Hall; 1996. pp. 495–523. [Google Scholar]
- 139.Traxler P, Gruner J, Auden J. Papulacandins, a new family of antibiotics with antifungal activity. II. Isolation, chemical and biological characterization of papulacandins A, B, C, D, and E. J Antibiot. 1977;30:289–296. doi: 10.7164/antibiotics.30.289. [DOI] [PubMed] [Google Scholar]
- 140.Vanden Bossche H, Marichal H P, Le Jeune L, Coene M, Gorrens J, Cools W. Effects of itraconazole on cytochrome P-450-dependent 14 α-demethylation and reduction of 3-ketosteroids in Cryptococcus neoformans. Antimicrob Agents Chemother. 1993;37:2101–2105. doi: 10.1128/aac.37.10.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vanden Bossche H, Marichal H P, Odds F, Le Jeune L, Coene M-C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vanden Bossche H, Willemsens G. Effect of the antimycotics, miconazole and ketoconazole on cytochrome P450 in yeast microsomes and rat liver microsomes. Arch Int Physiol Biochem. 1982;90:B218–B219. [Google Scholar]
- 143.Vanden Bossche H, Willemsens G, Cools W, Lauwers W F J, Le Jeune L. Biochemical effect of miconazole on fungi. II. Inhibition of ergosterol biosynthesis in Candida albicans. Chem Biol Interact. 1978;21:59–78. doi: 10.1016/0009-2797(78)90068-6. [DOI] [PubMed] [Google Scholar]
- 144.Vanden Bossche H, Willemsens G, Cools W, Marichal H P, Lauwers W F J. Hypothesis on the molecular basis of the antifungal activity of N-substituted imidazoles and triazoles. Biochem Soc Trans. 1983;11:665–667. doi: 10.1042/bst0110665. [DOI] [PubMed] [Google Scholar]
- 145.Wallace T L, Paeznick V, Cassum P A, Anaissie E. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1996. Nyotran (Liposomal nystatin) against disseminated Aspergillus fumigatus in neutropenic mice, abstr. B53; p. 31. [Google Scholar]
- 146.Walsh T J, Kasai M, Francesconi A, Landsman D, Chanock S J. New evidence that Candida albicans possesses additional ATP-binding cassette MDR-like genes: implications for antifungal azole resistance. J Med Vet Mycol. 1996;35:133–137. [PubMed] [Google Scholar]
- 147.Walsh T J, Lee J W, Kelley P, Bacher J, Lecciones J, Thomas V, Lyman C, Coleman D, Gordee R S. The antifungal effects of the nonlinear pharmacokinetics of cilofungin, a 1,3 β-glucan synthetase inhibitor, during continuous and intermittent intravenous infusions of cilofungin in treatment of experimental disseminated candidiasis. Antimicrob Agents Chemother. 1991;35:1321–1328. doi: 10.1128/aac.35.7.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wey S B, Mori M, Pfaller M, Woolson R F, Wenzel R P. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch Intern Med. 1988;148:2642–2645. doi: 10.1001/archinte.148.12.2642. [DOI] [PubMed] [Google Scholar]
- 149.Whelan W L. The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans. Crit Rev Microbiol. 1987;15:45–56. doi: 10.3109/10408418709104447. [DOI] [PubMed] [Google Scholar]
- 150.Whelan W L, Kerridge D. Decreased activity of UMP pyrophosphorylase associated with resistance to 5-fluorocytosine in Candida albicans. Antimicrob Agents Chemother. 1984;26:570–574. doi: 10.1128/aac.26.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14 α-demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.White T C, Marr K A, Bowden R A. Clinical, cellular and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wiebe V J, DeGregorio M W. Liposome-encapsulated amphotericin B: a promising new treatment for disseminated fungal infections. Rev Infect Dis. 1988;10:1097–1101. doi: 10.1093/clinids/10.6.1097. [DOI] [PubMed] [Google Scholar]
- 155.Witt M, Larsen R, Milefchik E, Leal M A, Haubrich R, Ritchie J, Edwards J E, Jr, Ghannoum M. Identification of patients with acute AIDS-associated cryptococcal meningitis who can be effectively treated with fluconazole therapy: the role of antifungal testing. Clin Infect Dis. 1996;22:322–328. doi: 10.1093/clinids/22.2.322. [DOI] [PubMed] [Google Scholar]
- 156.Yokoyama K, Wang L, Miyaji M. Program and Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1996. SSY726, a new triazole antifungal agent: in vitro and in vivo evaluation, abstr. F75; p. 113. [Google Scholar]
- 157.Zhou X Y, Bordon F, Sirot D, Kitzis M D, Gutmann L. Emergence of clinical isolates of Escherichia coli producing TEM-1 derivatives or an OXA-1 β-lactamase conferring resistance to β-lactamase inhibitors. Antimicrob Agents Chemother. 1994;38:1085–1089. doi: 10.1128/aac.38.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zorzi W, Zhou X Y, Dardenne O, Lamotte J, Raze D, Pierre J, Gutmann L, Coyette J. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J Bacteriol. 1996;178:4948–4957. doi: 10.1128/jb.178.16.4948-4957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zygmunt W A, Tavormina P A. Steroid interference with antifungal activity of polyene antibiotics. Appl Microbiol. 1966;14:865–869. doi: 10.1128/am.14.6.865-869.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]