Significance
Parkinson’s disease (PD) is the second-most common neurodegenerative disorder. Mutations leading to overactivation of LRRK2 are a leading cause of familial PD, and this protein is therefore considered as an appealing target for drug design. Here, we describe the discovery and characterization of a diverse set of LRRK2-targeting nanobodies. A subset of these nanobodies inhibit LRRK2 via a mechanism that differs from the commonly used LRRK2 kinase inhibitors. Importantly, some of these nanobodies selectively inhibit certain LRRK2 activities (Rab phosphorylation) while leaving other activities (autophosphorylation) unaffected. We anticipate that these nanobodies will find multiple applications as research tools and will open up opportunities for the development of new PD diagnostics and therapeutics in parallel to other currently pursued strategies.
Keywords: Parkinson’s disease, LRRK2, drug design, allosteric inhibitor, nanobody
Abstract
Mutations in the gene coding for leucine-rich repeat kinase 2 (LRRK2) are a leading cause of the inherited form of Parkinson’s disease (PD), while LRRK2 overactivation is also associated with the more common idiopathic form of PD. LRRK2 is a large multidomain protein, including a GTPase as well as a Ser/Thr protein kinase domain. Common, disease-causing mutations increase LRRK2 kinase activity, presenting LRRK2 as an attractive target for drug discovery. Currently, drug development has mainly focused on ATP-competitive kinase inhibitors. Here, we report the identification and characterization of a variety of nanobodies that bind to different LRRK2 domains and inhibit or activate LRRK2 in cells and in in vitro. Importantly, nanobodies were identified that inhibit LRRK2 kinase activity while binding to a site that is topographically distinct from the active site and thus act through an allosteric inhibitory mechanism that does not involve binding to the ATP pocket or even to the kinase domain. Moreover, while certain nanobodies completely inhibit the LRRK2 kinase activity, we also identified nanobodies that specifically inhibit the phosphorylation of Rab protein substrates. Finally, in contrast to current type I kinase inhibitors, the studied kinase-inhibitory nanobodies did not induce LRRK2 microtubule association. These comprehensively characterized nanobodies represent versatile tools to study the LRRK2 function and mechanism and can pave the way toward novel diagnostic and therapeutic strategies for PD.
Parkinson’s disease (PD) is a common and devastating neurodegenerative movement disorder affecting around 2% of the global population (1). The disease is characterized by degeneration of dopaminergic neurons, which leads to the typical symptoms, including resting tremor, bradykinesia, and postural instability. Although treatments to alleviate these symptoms have been available for a long time, there is still no cure. A very promising strategy that is currently being intensively pursued is the targeting of the protein leucine-rich repeat kinase 2 (LRRK2). Mutations in LRRK2 are among the most common causes of familial PD (2), while an increased LRRK2 activity has also been associated with the more frequent idiopathic form of PD (3, 4). Moreover, LRRK2 mutations and/or overexpression have also been linked to a number of chronic inflammatory conditions, including Crohn’s disease (5, 6).
LRRK2 is a large multidomain protein belonging to the ROCO family (Fig. 1A) that bears a rather unique combination of two catalytic activities: GTPase activity mediated by the Roc domain and Ser/Thr protein kinase activity (7). Recently, several Rab GTPases were identified as the physiological substrates of LRRK2 kinase activity (8, 9), while LRRK2 is also known to autophosphorylate (10). Although the details of the regulatory mechanism of LRRK2 are not yet completely understood, we previously showed, using a more simple LRRK2 homolog from the bacterium Chlorobium tepidum (CtRoco), that the RocCOR supradomain undergoes a dimer–monomer cycle concomitant with GTP binding and hydrolysis (11). This is in line with findings for LRRK2 in cells, which show that the protein predominantly occurs as a monomer with low-kinase activity in the cytosol and as a dimer with high-kinase activity at the membrane (12, 13). These results point toward a complex interplay between the GTPase and kinase domains of LRRK2, regulated by large-scale, conformational changes.
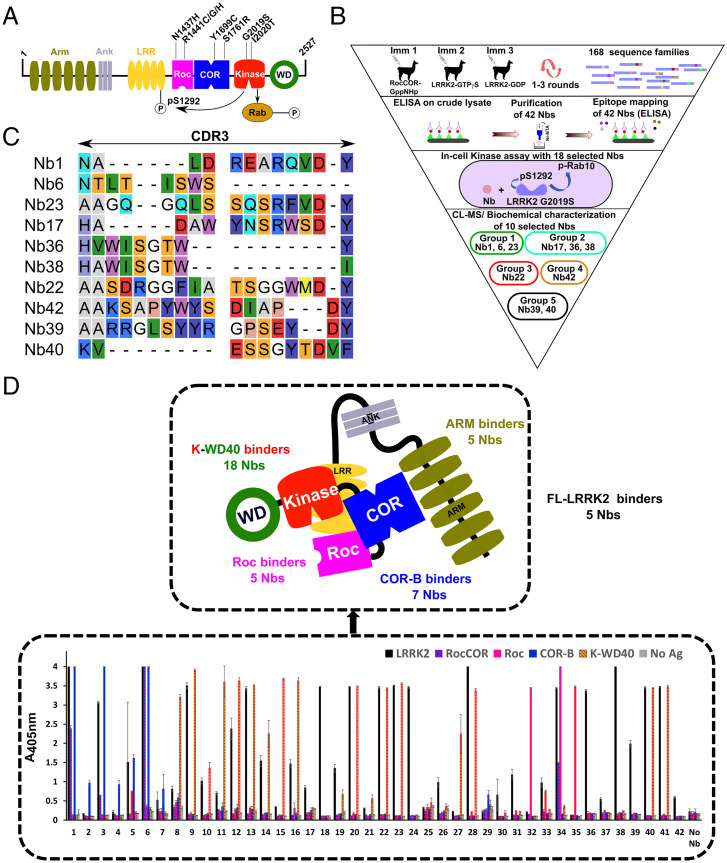
Fig. 1.
Identification of LRRK2-targeting Nbs. (A) Domain arrangement of LRRK2, with important PD mutations indicated. Two LRRK2 kinase activities relevant to this study are also indicated: phosphorylation of Rab proteins and autophosphorylation at position S1292. (B) Funnel approach used in this study to identify and characterize LRRK2-binding and modulating Nbs. The 10 Nbs that are characterized in detail are categorized into five functional groups: group1, inhibit all LRRK2 kinase activities (dark green); group 2, specifically inhibit LRRK2 Rab phosphorylation (light green); group 3, activate LRRK2 kinase (red); group 4, inhibit LRRK2 activity in cells (orange); and group 5, no effect on LRRK2 activity (black). (C) Sequences of the CDR3 regions of the 10 Nbs that were analyzed in detail. Nb36 and Nb38 belong to the same CDR3 sequence family. (D, Lower) Domain mapping of the purified Nbs using ELISA on either FL-LRRK2, the RocCOR, Roc, COR-B, or K-WD40 constructs. The Nbs that only show binding to FL-LRRK2 were additionally tested for binding on the RCKW and ARM domain constructs (SI Appendix, Fig. S3). Each ELISA signal is the average of three experiments. (Upper) The results of both of these domain-mapping experiments are schematically shown.
The most prevalent PD mutations in LRRK2 are clustered within the catalytic RocCOR and kinase domains (Fig. 1A), and several PD mutations lead to a decrease in GTPase and/or an increase in kinase activity (14, 15). Most notably, autophosphorylation of serine-1292 (16) and Rab protein phosphorylation (8) are increased by pathogenic LRRK2 variants and particularly by the most common G2019S mutation. These findings support the idea that LRRK2 mutations cause PD through a gain‐of‐function mechanism, and inhibition of LRRK2 kinase activity is therefore considered to be a particularly promising strategy for the treatment of PD (17, 18). However, preclinical studies have indicated that long‐term inhibition of LRRK2 with ATP-competitive inhibitors might potentially be associated with toxic side effects, including kidney abnormalities in rodents and an accumulation of lamellar bodies in type II pneumocytes in the nonhuman primate lung (19–21). Targeting the multiple enzymatic functions and regulatory mechanisms of LRRK2 in an alternative way, using compounds that bind outside the ATP-binding pocket, could form a very attractive strategy complementing the currently explored approaches (22). One way to modulate the dynamics, regulation, and activity of proteins is via the use of nanobodies (Nbs), the small and stable single-domain fragments derived from camelid heavy chain–only antibodies (23). Accordingly, we recently reported the identification of Nbs that allosterically activate the GTPase activity of a bacterial LRRK2 homolog by interfering with the protein’s monomer–dimer cycle (24).
Here, we report the identification and in vitro and in-cell characterization of a wide range of Nbs acting as modulators of human LRRK2 kinase activity. Some of these Nbs bind to sites that are topographically distinct from the active site and thus act via a completely different mode-of-action compared to the currently available, ATP-competitive kinase inhibitors.
Results
Identification of a Wide Variety of LRRK2-Binding Nbs.
To identify Nbs that bind LRRK2 and to maximize chances of finding LRRK2 activity–modulating Nbs, three immunization strategies were followed, each time using a different llama (Fig. 1B). In a first immunization strategy, we immunized a llama with the LRRK2 RocCOR construct (SI Appendix, Fig. S1), and after immunization, Nbs were selected via a phage display–panning approach using full-length LRRK2 (FL-LRRK2) as the bait protein. In immunization strategies 2 and 3, we immunized llamas with LRRK2 either in the presence of a large excess of GTPγS (a nonhydrolysable GTP analog) or in the presence of a large excess of GDP, and phage display selections were performed using the corresponding proteins. Additionally, to enrich for Nbs binding to the Roc domain, phage display selections were performed using either the guanosine-5′-(γ-thio)-triphosphate– (GTPγS) or guanosine-5′-diphosphate (GDP)–bound Roc domain protein. These different strategies finally resulted in 168 Nb families (each Nb family displaying a unique complimentary determining region 3 [CDR3] sequence). Based on an initial screening of crude Escherichia coli cell extracts, expressing these Nbs, in an enzyme-linked immunosorbent assay (ELISA) and in LRRK2 activity assays, 42 Nbs belonging to 41 different CDR3 sequence families were selected, expressed, and purified to homogeneity (SI Appendix, Fig. S2 and Table S1).
Subsequently, binding to FL-LRRK2 was confirmed for this set of purified Nbs using ELISA, while in parallel also, a first mapping of the binding regions of these Nbs was performed by assessing the binding to purified RocCOR, Roc, C-terminal subdomain of COR (COR-B) and kinase-WD40 (K-WD40) domains of LRRK2 (Fig. 1D). Nearly all tested Nbs show binding to FL-LRRK2 and/or at least one of the domain constructs, except for two Nbs (Nb25 and Nb29). All seven purified Nbs that resulted from the immunization with the RocCOR domain construct (Immunization 1) specifically interacted with the COR-B. Among the Nbs obtained from the immunizations and selections using FL-LRRK2, the majority bound within the K-WD40 domain of the protein (18 Nbs). Another subset of 10 Nbs showed robust binding to LRRK2, while no binding was observed to any of the individual LRRK2 domain constructs. We therefore assume that these Nbs either bind exclusively to the N-terminal region of LRRK2 (armadillo-ankyrin-LRR domains) that was not covered by individual domains in the ELISA and/or bind on an epitope on the interface of two or more domains and thus require the FL-LRRK2 protein for binding. To further distinguish between these two scenarios, we performed an additional ELISA experiment for those 10 Nbs using FL-LRRK2 and its N-terminal armadillo (ARM) and C-terminal Roc-COR-Kinase-WD40 (RCKW) domain constructs (SI Appendix, Fig. S3). This experiment showed that while none of the Nbs bound exclusively to the C-terminal RCKW part of LRRK2, five Nbs bound to the ARM domain and five Nbs required the full-length protein for binding. Finally, a last set of Nbs were specifically selected by phage display panning on the LRRK2 Roc domain, resulting in five Roc domain–binding Nbs (Nb32, Nb33, Nb34, Nb35, and Nb42; note that Nb42 did not show clear binding to Roc in ELISA, but the binding was confirmed by size-exclusion chromatography; SI Appendix, Fig. S4). The results of this initial domain mapping are schematically summarized in Fig. 1D.
Nbs Bind LRRK2 and Modulate Its Activity in Cells.
After confirming the binding of 40 purified Nbs to LRRK2 in vitro, we next wanted to test whether these Nbs bound to LRRK2 in human embryonic kidney 293 (HEK293) cells overexpressing LRRK2. Hereto, we selected a subset of 18 Nbs, considering Nbs originating from the three immunization strategies and targeting different LRRK2 domains. The Nbs were expressed as GFP-fusions from a pEGFP vector in LRRK2 (wild type)-overexpressing HEK293 cells, and a pull-down experiment was performed using magnetic GFP-nanotrap beads (SI Appendix, Fig. S5). This experiment showed that all tested Nbs were able to pull-down LRRK2 under these conditions. This thus indicates that these 18 Nbs are functional as intrabodies in the context of the cytoplasm of human cells and have sufficiently high affinity to pull-down their target protein.
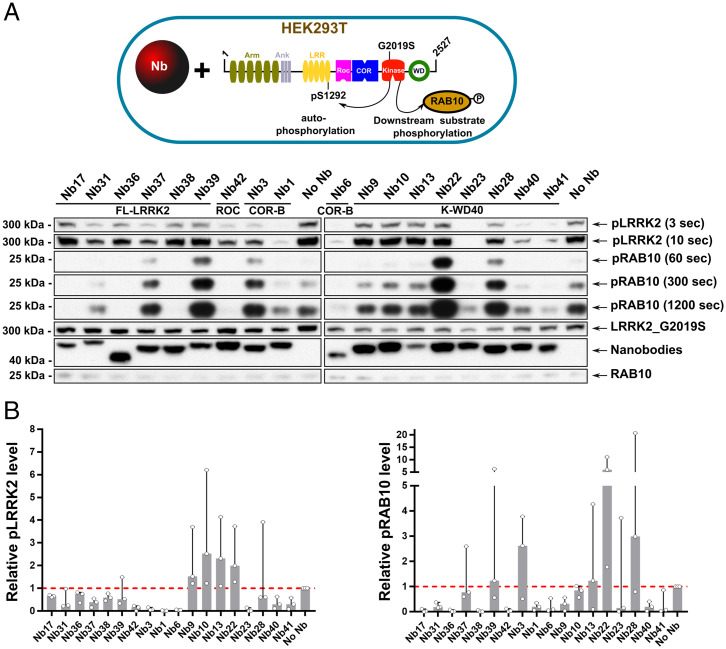
Subsequently, to assess the influence of these 18 Nbs on in-cell LRRK2 kinase activity, we monitored two activities of LRRK2: phosphorylation of the endogenous substrate Rab10 at position T73 and LRRK2 autophosphorylation at position S1292 (8, 9, 16, 25). Both activities have previously been shown to be increased in the most relevant LRRK2 PD mutants, including the common G2019S mutant. In this case, we thus coexpressed the LRRK2 (G2019S) mutant together with the different Nb-GFP fusions in HEK293T cells (Fig. 2 and SI Appendix, Fig. S6). Among the 18 selected Nbs, four Nbs strongly and consistently decreased both LRRK2 autophosphorylation and Rab10 phosphorylation in cells compared to the negative control: Nb1, Nb6, Nb23, and Nb42. Another three inhibitory Nbs only decrease the level of Rab10 phosphorylation, while they largely leave autophosphorylation unaffected: Nb17, Nb36, and Nb38. Finally, two Nbs increase the levels of LRRK2-mediated Rab10 phosphorylation: Nb22 and Nb28. The other Nbs have either no effect or a somewhat less outspoken or more variable effect on the kinase activities. Based on their domain specificity and their effect on LRRK2 kinase activity (representatives of inhibiting, activating, and neutral Nbs), we selected the following 10 Nbs for further in-depth characterization: Nb1, Nb6, Nb17, Nb22, Nb23, Nb36, Nb38, Nb39, Nb40, and Nb42 (Fig. 1C).
Fig. 2.
Nbs modulate LRRK2 activity in cells. (A) Influence of Nbs on the kinase activity of the LRRK2(G2019S) variant in HEK293T cells. LRRK2 (G2019S) and its effector Rab29 were overexpressed together with GFP-tagged Nbs in HEK293T cells. A negative control, in which no Nb is overexpressed (“No Nb”), is also included. In rows labeled “pLRRK2,” LRRK2 pS1292 levels are determined by Western blot using a site-specific anti-pLRRK2 (pS1292) antibody (Abcam, ab203181) (shown at different times of development). In the rows labeled “pRAB10,” endogenous pT73-Rab10 levels are determined by Western blot using the MJFF (Michael J. Fox Foundation)/Abcam antibody MJF-R21 (Abcam, ab230261) (shown at different times of development). The three lower rows contain controls of LRRK2, GFP-Nb, and Rab10 expression levels, determined on a different Western blot than pLRRK2 and pRab10. Blot is representative of n = 3 (SI Appendix, Fig. S6). (B) Quantification of three different replicates (A and SI Appendix, Fig. S6) based on the densitometric analysis of the Western blots relative to the control (no Nb expressed). (Left) LRRK2 autophosphorylation at S1292. (Right) LRRK2-mediated Rab10 phosphorylation at T73. Median values (bars) and SDs (error bars) are shown. Individual values representing the three replicates are shown as individual datapoints.
Epitope Mapping and Affinity of LRRK2-Modulating Nbs.
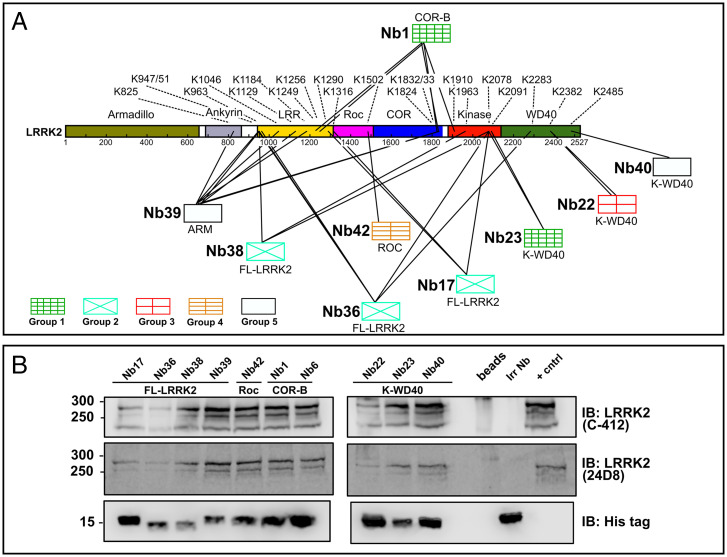
In addition to the ELISA experiment described in the paragraph "Identification of a Wide Variety of LRRK2-binding Nbs," we used a cross-linking mass spectrometry (CL-MS) approach to obtain more detailed insight in the binding epitopes of the 10 selected Nbs. The CL-MS data revealed that the different Nbs cross-link to lysines within LRRK2 predominantly via a conserved lysine residue in the framework 3 region of the Nb. This lysine residue is present in all selected Nbs except for Nb6, for which correspondingly no cross-linking data could be obtained. Overall, the CL-MS data are in very good agreement with the results of the domain mapping using ELISA (Fig. 3A) (26). Based on the ELISA experiments, Nb17, Nb36, and Nb38 were found to bind either on the N-terminal ANK or LRR domains of LRRK2 or to bind on the interface of several domains (“FL-LRRK2 binders”). Correspondingly, CL-MS reveals that all these Nbs indeed make multiple contacts to the N-terminal LRR domain as well as C-terminal parts of LRRK2, including the kinase domain (Nb17 and Nb38) and both the kinase and WD40 domain (Nb36). While the domain mapping suggested that Nb39 binds to the N-terminal ARM domain, the CL-MS experiment shows multiple cross-links with N-terminal domains and the COR domain. This could reflect a high motility of the ARM domain with respect to the other domains, as also shown in the recent cryogenic electron microscopy (cryo-EM) structure (27), bringing the Nb (transiently) in proximity to these other domains. For Nb42, only one cross-link with a lysine (K1502) within the Roc domain is identified, in good agreement with the domain mapping in ELISA. Nb22, Nb23, and Nb40 were all identified as K-WD40 binders in ELISA, and correspondingly, the cross-linking data reveals interactions with the WD40 domain (Nb22 and Nb40) or the kinase domain (Nb23). Interestingly, Nb1, which was unequivocally identified as a COR-B binder in ELISA, correspondingly makes multiple cross-links with the COR-B domain (K1824, K1832, and K1833), but also, cross-links are found with residues within other LRRK2 domains, including the kinase domain and the leucine-rich repeats. This finding is in good agreement with a very central localization of the COR-B domain within recent structural models of LRRK2 (27–30).
Fig. 3.
Nbs bind LRRK2 via interactions with different domains. (A) Mapping of the Nb-binding epitopes using CL-MS. The Nbs are divided into five groups according to their effect on LRRK2 kinase activity, as defined in Fig. 1B. The observed cross-links between the Nbs and LRRK2 are indicated by lines, with the corresponding lysine residues on LRRK2 indicated by their residue number. The domain specificity of the Nbs, as determined in ELISA, is given below the respective Nbs. (B) Nbs immunoprecipitate endogenous (mouse) LRRK2. Lysates derived from RAW264.7 cells were incubated with 1.5 µM purified His-tagged Nbs or an irrelevant control Nb (Irr Nb), and pull-downs were performed using magnetic Dynabeads. As a positive control (+ cntrl) LRRK2 was pulled-down using a LRRK2-specific Nb. LRRK2 was detected via immunoblotting (IB) using two different antibodies (C-412 and 24D8). The blot is representative of n = 3.
To determine the binding affinity, KD, (dissociation constant) of the 10 Nbs two methods were used in parallel: microscale thermophoresis (MST) and biolayer interferometry (BLI). For the MST experiment, we site specifically labeled the 10 Nbs with an m-TAMRA fluorophore at its C terminus using sortase-mediated coupling (24) and then titrated increasing amounts of FL-LRRK2 to these Nbs (SI Appendix, Fig. S7). For all 10 Nbs, an MST signal was observed, except for Nb23, which did not generate a change in thermophoresis behavior upon binding to LRRK2. For the other Nbs, KD values are found in the range of 25 to 150 nM (Table 1 and SI Appendix, Fig. S7). For the BLI experiment, LRRK2 was first trapped on a streptavidin-coated biosensor using biotinylated Nb40 as trapping agent (except to assess binding of Nb40, in which Nb42 was used as trapping agent), after which binding of all Nbs to LRRK2 was determined. A clear binding signal was obtained for all Nbs (including Nb23), with some Nbs showing an indication of a second slow phase in the binding and dissociation curves, which could either reflect conformational changes upon Nb binding or the presence of preexisting conformational heterogeneity in LRRK2. Fitting of the sensorgrams was performed initially using a 1:1 binding model, and the resulting equilibrium response (Req) values were subsequently used to calculate KD values from the corresponding dose–response curves fitted on a Langmuir model. This yields KD values ranging from 2 to 60 nM (Table 1 and SI Appendix, Fig. S8). Overall, a (nearly 5- to 10-fold) higher affinity is obtained with BLI compared to MST. These differences are probably due to the experimental setup, with MST using LRRK2 in solution and BLI using LRRK2 trapped on a surface by means of a second Nb, which might also result in different LRRK2 conformations. Additionally, the KD values obtained via the MST titration will scale with the concentration of properly folded LRRK2 in the sample during the measurement, and denaturation of a part of the LRRK2 protein could hence lead to an overestimation of the KD values (note that in MST LRRK2 is used at varying concentrations, while in BLI the Nbs are used at varying concentrations). Subsequently, a BLI experiment was performed using the biotinylated RCKW construct of LRRK2 coupled to the streptavidine-coated biosensors and titration with increasing amount of Nbs (SI Appendix, Fig. S9). In agreement with the ELISA and CL-MS experiments, no binding to the RCKW construct is observed for Nb17, Nb36, Nb38, and Nb39. Comparison of the KD values obtained with BLI of Nb1, Nb6, Nb22, Nb23, and Nb40 for FL-LRRK2 and the RCKW protein (Table 1) shows that these values are in relatively good agreement, indicating that the N-terminal domains of LRRK2 only play a minor role in the equilibrium-binding affinities of those Nbs.
Table 1.
Equilibrium KD (KD in nanomolar, nM) for binding of the set of 10 Nbs (belonging to five functional groups based on their effect on kinase activity) to either FL-LRRK2 or its RCKW domain construct, as assessed by two methods in parallel: MST and BLI
| Nb | Binding epitope (ELISA) | Nb group | FL-LRRK2 KD (nM) | RCKW KD (nM) | |
| MST | BLI | BLI | |||
| Nb1 | COR-B | Group 1 | 91 ± 28 | 55 ± 12 | 39 ± 4 |
| Nb6 | COR-B | Group 1 | 83 ± 24 | 8 ± 1 | 5 ± 1 |
| Nb23 | K-WD40 | Group 1 | NBS* | 20 ± 5 | 17 ± 7 |
| Nb17 | FL-LRRK2‡ | Group 2 | 67 ± 24 | 2.0 ± 0.3 | NB† |
| Nb36 | FL-LRRK2‡ | Group 2 | 78 ± 25 | 16 ± 8 | NB† |
| Nb38 | FL-LRRK2‡ | Group 2 | 48 ± 11 | 37 ± 4 | NB† |
| Nb22 | K-WD40 | Group 3 | 145 ± 55 | 10 ± 1 | 5 ± 1 |
| Nb42 | Roc | Group 4 | 94 ± 30§ | 58 ± 18 | 18 ± 2 |
| Nb39 | FL-LRRK2‡ | Group 5 | 79 ± 19 | 12 ± 3 | NB† |
| Nb40 | K-WD40 | Group 5 | 26 ± 10 | 2.5 ± 0.3 | 2.6 ± 0.3 |
*NBS: no binding signal observed in MST.
†NB: no binding on RCKW.
‡FL-LRRK2: full-length LRRK2.
§Value determined in presence of GTPγS instead of GDP, which was used in all the other measurements.
Considering their high binding affinity, we next tested whether those Nbs would also be able to pull-down LRRK2 at endogenous/physiological expression levels. Therefore, we turned to lysates of mouse RAW264.7 cells, which express LRRK2 at relatively high levels (31). Interestingly, we find that all 10 Nbs efficiently pull-down LRRK2 when added to these lysates (Fig. 3B), showing that 1) the Nbs are cross-reactive toward mouse LRRK2 and 2) their affinity is sufficiently high to pull-down endogenous levels of LRRK2 from cell lysates.
Nbs Inhibit LRRK2 Peptide and/or Rab Phosphorylation In Vitro.
To test if the 10 Nbs directly influence LRRK2 (wild type) kinase activity, we next performed in vitro assays using purified proteins. In a first approach, we measured LRRK2 kinase activity vis-à-vis an optimized LRRK2 peptide substrate (AQT0615) using the commercially available PhosphoSens Protein Kinase Assay (AssayQuant Technologies Inc.) (Fig. 4A), while in a second approach we also measured the influence of the Nbs on LRRK2-mediated Rab8a phosphorylation using a Western blot–based method (Fig. 4B). The influence of all 10 Nbs on LRRK2 activity was screened at a fixed Nb concentration of 25 µM and compared to a no-Nb negative control and a positive control in which the ATP-competitive LRRK2 inhibitor MLi-2 was added (32–34). These experiments were performed with LRRK2 either in the presence of a large excess (500 µM) of GDP or GTPγS, but no significant influence of the nucleotide on the inhibition profile was observed (SI Appendix, Fig. S10). Consistent with the data in cells, the WD40-binding Nb22 activates the kinase activity of LRRK2, both toward the peptide and Rab8a substrate, by about 50% compared to the controls. The group of Nbs, which showed an influence on Rab phosphorylation in cells while leaving autophosphorylation unaffected (Nb17, Nb36, and Nb38; Fig. 2), very consistently only show a very small or no effect on LRRK2 peptide phosphorylation, while they have a prominent effect on Rab8a phosphorylation in vitro. Combined, our cellular and in vitro assays thus suggest that these three Nbs do not directly affect LRRK2 kinase activity per se but rather specifically influence Rab phosphorylation. In contrast, Nb1, Nb6, and (to a lesser extent) Nb23 have a clear inhibitory effect on LRRK2 kinase activity toward both peptide and Rab substrates in vitro. This is again in very good agreement with our data in cells (Fig. 2) which showed that these Nbs inhibit both LRRK2 autophosphorylation and Rab phosphorylation, thus suggesting that these Nbs act as true inhibitors of total LRRK2 kinase activity. One exception seems to be Nb42, which inhibited both LRRK2 autophosphorylation and Rab phosphorylation in cells, while no inhibitory effect could be observed in vitro.
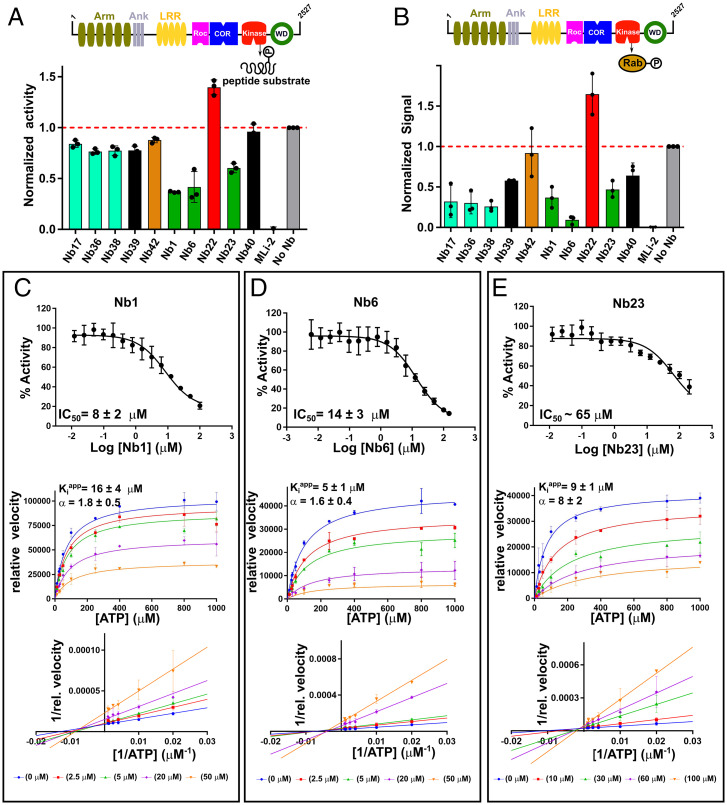
Fig. 4.
Modulation of in vitro kinase activity by LRRK2-targeting Nbs. (A) Effect of Nbs on LRRK2 kinase activity measured using the LRRK2-optimized AQT0615 peptide as substrate. (B) Effect of Nbs on LRRK2-mediated phosphorylation of Rab8a determined via a Western blot assay (SI Appendix, Fig. S10). In both A and B, the influence of the Nbs (25 µM) on the relative kinase activity compared to the “No-Nb” control is plotted, and a positive MLi-2 control is included. Each bar reflects the average (±SD) of three independent measurements. (C–E, Upper) Dose–response curves for the inhibition of the in vitro LRRK2 kinase activity by the group1 Nbs: Nb1 (C), Nb6 (D), and Nb23 (E), using a serial dilution of the Nb and a fixed concentration of AQT0615 peptide (10 µM) and ATP (1 mM). (Middle) The Michaelis–Menten curves obtained for LRRK2 at varying concentrations of ATP and a fixed (subsaturating) concentration of peptide substrate (AQT0615) and at varying concentrations of the respective Nbs. (Lower) The corresponding linearizations according to the Lineweaver–Burk method (double-reciprocal plot). The Nb concentrations used are indicated below the plots. Each datapoint reflects the average (±SE) of three independent measurements. The IC50 (±SD) values resulting from fitting on a three-parameter logistic equation and the Kiapp and α values (±SD) resulting from global fitting on a mixed-type inhibition mechanism are indicated on the graphs.
Since we found that Nb1, Nb6, and Nb23 inhibit LRRK2 kinase activity with respect to peptides and Rab proteins in vitro, we continued to perform a dose–response analysis with these three Nbs using the same model peptide substrate and at a fixed concentration of 1 mM ATP (Fig. 4 C–E). Fitting of these dose–response curves yielded half-maximum inhibitory concentration (IC50) values of 8 ± 2 µM and 14 ± 3 µM for Nb1 and Nb6, respectively, and an approximate IC50 value of 65 µM for Nb23. The difference between the observed KD and IC50 values of these Nbs could be indicative of a complex mechanism of inhibition, involving (slow) conformational changes in LRRK2 upon Nb binding, thereby differentially affecting equilibrium (KD) and kinetic (IC50) values. Such differences in KD and IC50 values have been previously observed for Nbs (24, 35). Similarly, a dose–response was observed for Nb17, Nb36, and Nb38 with respect to LRRK2 Rab8a phosphorylation (SI Appendix, Fig. S10), but saturation of the blot at high-Nb concentrations prevented a more qualitative treatment of these curves.
LRRK2-Inhibiting Nbs Act via a Non–ATP-Competitive Mechanism.
Three Nbs (Nb1, Nb6, and Nb23) were identified that robustly inhibit LRRK2 autophosphorylation and Rab phosphorylation activity in cells, as well as LRRK2 kinase activity toward peptide and Rab substrates in vitro. Combined, these experiments suggest that these Nbs target the LRRK2 kinase activity per se. Interestingly, while the ELISA and CL-MS experiments suggest that Nb23 binds to the kinase domain, Nb1 and Nb6 bind to the COR-B domain. This strongly suggests that at least the latter two Nbs act via binding to a pocket that is different from the ATP-binding pocket. To further confirm this, we set out to determine the mechanism of inhibition (competitive versus uncompetitive versus mixed/noncompetitive) of these three Nbs with regard to ATP. Full Michaelis–Menten curves were obtained at a fixed (probably subsaturating) concentration of peptide substrate (10 µM) and varying concentrations of ATP (Fig. 4 C–E). For Nb1 and Nb6, linearization of the curves using the Lineweaver–Burk plot clearly shows intersecting lines left of the y-axis, indicative of a mixed-type inhibition. This confirms that these Nbs are not competing with ATP for binding and that they inhibit the reaction by binding on a topographically different site, in agreement with the ELISA and CL-MS domain/epitope-mapping data. For Nb1, a global fit of the kinetic data using a mixed inhibition model accordingly gives an apparent inhibition constant (Kiapp) of 16 ± 4 µM with an α-value of 1.8 ± 0.5, corresponding to a Kicapp (competitive Kiapp - equal to apparent Ki for apo-LRRK2) of 16 µM and a Kiuapp (uncompetitive Kiapp - equal to apparent Ki for ATP-bound LRRK2) of 30 µM. For Nb6, fitting on the same model yields a Kiapp of 5 ± 1 µM with an α-value of 1.6 ± 0.4, corresponding a Kicapp of 5 µM and a Kiuapp of 8 µM. For the kinase domain–binding Nb23, the linearized curves intersect closer to the y-axis, indicating a mechanism which is more ATP-competitive like. Yet the lines do not exactly intersect on the y-axis and a systematic decrease in the apparent maximal velocity (Vmaxapp) and increase (or no effect) in the apparent Michaelis constant (KMapp) with increasing Nb concentration is observed, which also here indicates a mixed-inhibition mechanism (SI Appendix, Fig. S11). Fitting on a mixed-inhibition model for Nb23 gives a Kiapp of 9 ± 1 µM with an α-value of 8 ± 2, corresponding a Kicapp of 9 µM and a Kiuapp of 72 µM.
To confirm that Nb1, Nb6, and Nb23 bind outside the kinase ATP-binding pocket and thus target a different site than the “classical” ATP-competitive inhibitors, such as MLi-2 (32–34), we performed a competition ELISA titration experiment (SI Appendix, Fig. S12). In this setup, the ELISA experiment was performed using a fixed concentration of LRRK2 coated on the bottom of the ELISA plate and using a dilution series (from 450 to 0.2 nM) of either of the three Nbs, resulting in a dose–response titration curve reflecting an apparent affinity of the Nbs. Subsequently, a repetition of the setup in the presence of a large excess of MLi-2 (1 µM), or of the corresponding nontagged Nb (at 9 µM) as a positive control, was performed. While a very prominent rightward shift in the titration curves is observed when adding the corresponding untagged Nb as a direct orthosteric competitor, no rightward shift is observed for any of the three Nbs in presence of MLi-2. This thus further proves that neither of these three Nbs bind in the same pocket as the type I ATP-competitive inhibitor MLi-2 (32–34, 36), thus confirming that these Nbs act via a non–ATP-competitive inhibitory mechanism. To further assess whether Nb1, Nb6, and Nb23 compete with either peptide or Rab substrate binding, a similar competition ELISA experiment was performed in absence or presence of Rab8a (at 10 µM) or excess peptide substrate (LRRKtide at 100 µM) (SI Appendix, Fig. S12). At the concentrations of LRRKtide and Rab8a used, no shift in the titration curves can be observed, indicating that the Nbs do not compete with, or at least do not completely block, the binding of those substrates.
Since Nb17, Nb36, and Nb38 specifically inhibit Rab phosphorylation, a similar experiment was also performed with these Nbs in presence of Rab8a (SI Appendix, Fig. S13). ELISA signals for the FL-LRRK2–binding Nbs are generally low, probably due to the solid-phase coating of LRRK2 making their binding epitopes less accessible. Therefore, both the concentrations of the Nbs (900 to 0.4 nM) and Rab8a (20 µM) were increased in this experiment. This higher-Rab8a concentration led to a high background because of nonspecific binding of the primary antibodies used in the ELISA, complicating the analysis of these experiments. Nevertheless, the experiments suggest that the binding of Nb17 is hardly affected by the presence of Rab8a (approximate EC50 values of 3 versus 5 nM in absence or presence of Rab8a, respectively). For Nb36 and Nb38, an exact analysis is complicated by the high-Rab8a background, and, while still a clear binding of Nb36 is observed in presence of Rab8a, the curves suggest that, at least in the case of Nb38, Rab8a might interfere with binding of the Nb.
Together, our data show that Nb1, Nb6, and Nb23, which inhibit LRRK2 peptide and Rab phosphorylation, clearly bind outside the active site pocket, since they do not compete with either MLi-2, peptide, or Rab8a binding and correspondingly act as noncompetitive inhibitors. For Nb1 and Nb6, this is in very good agreement with their binding to the COR-B domain. Since Nb17, Nb36, and Nb38 do not affect peptide phosphorylation, it is clear that they do not block the binding of ATP or peptide substrates, while our data does not rule out that they might interfere to a certain extent with Rab8a binding (especially in case of Nb36 and Nb38).
Expression of LRRK2-Targeting Nbs Did Not Result in LRRK2 Relocalization to Microtubules.
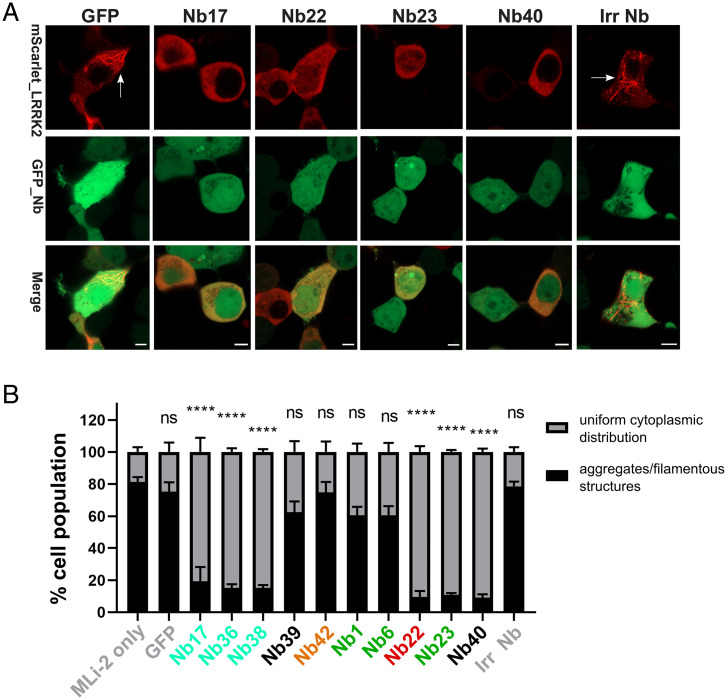
LRRK2 pharmacological kinase inhibitors of different structural classes induce cellular recruitment of overexpressed LRRK2 to microtubules, similar to four out of the five major PD-causing mutations (37, 38). The binding of LRRK2 to microtubules subsequently reduces the kinesin- and dynein-mediated transport along microtubules (29). In order to investigate whether our 10 identified LRRK2 kinase-modulating Nbs result in a similar phenotype, HEK293 cells were cotransfected with constructs coding for mScarlet-LRRK2 and GFP-Nbs. For all 10 Nbs analyzed, confocal microscopy analysis shows that, at the expression levels obtained in our experiments, no relocalization to microtubules is observed 48 h after cotransfection with the Nbs, in contrast to MLi-2 treatment in which filamentous, skein-like structures of LRRK2 are observed, indicating that the Nbs trap LRRK2 in a different conformation compared to classical inhibitors (SI Appendix, Fig. S14) (39). Next, we wanted to investigate whether the LRRK2-targeting Nbs have the ability to alter the recruitment of LRRK2 to microtubules induced by inhibition of LRRK2 kinase activity by the specific, ATP-competitive inhibitor MLi-2 (32–34, 36). To do that, HEK293 cells cotransfected with mScarlet-LRRK2 and GFP-Nb constructs were treated with 1 µM MLi-2 (Fig. 5 and SI Appendix, Fig. S15). The confocal images were analyzed, and cells that showed coexpression of both LRRK2 and Nbs were scored as the following: 1) show LRRK2 aggregates and/or filamentous structures or 2) display uniform cytoplasmic distribution of LRRK2 (SI Appendix, Fig. S16). Interestingly, cells cotransfected with a subset of the tested Nbs showed significantly reduced, MLi-2–induced LRRK2 recruitment to microtubules, and LRRK2 cytoplasmic distribution was maintained in contrast to controls with MLi-2–treated cells cotransfected with GFP or an irrelevant Nb. At the expression levels obtained, which is varying among different Nbs, this rescuing effect was observed for Nb17, Nb22, Nb23, Nb36, Nb38, and Nb40.
Fig. 5.
Nbs can rescue MLi-2–induced LRRK2 relocalization to microtubules. (A) Effect of Nbs on MLi-2–induced microtubule relocalization of LRRK2 (results for a selected set of Nbs are shown, see SI Appendix, Fig. S15 for all data). HEK293 cells were cotransfected with GFP or the GFP-tagged Nbs and mScarlet-LRRK2 for 24 h and then treated with 1 µM MLi-2 for 90 min. Cotransfection of mScarlet-LRRK2 with only GFP or an irrelevant control Nb (Irr Nb) show the MLi-2–induced, filamentous, skein-like structures of LRRK2 indicated by white arrows. Cotransfection with a subset of GFP-Nbs inhibited the MLi-2–induced LRRK2 relocalization. (Scale Bar, 5 µM.) (B) Quantification of results shown in A and SI Appendix, Fig. S15. A minimum of 200 transfected cells were analyzed. Only cells coexpressing both LRRK2 and Nbs were considered and grouped as either showing LRRK2 aggregates and/or filamentous structures or possessing a uniform cytoplasmic distribution of LRRK2 (as illustrated in SI Appendix, Fig. S16). For each Nb, the average percentage of cells showing a uniform cytoplasmic LRRK2 distribution (gray) versus aggregates and/or filamentous structures (black), together with the SEMs for three biological replicates, are shown with P values: one-way ANOVA and Dunnett’s multiple comparisons test (MLi-2 only as a control), ****P ≤ 0.0001 and ns: not significantly different (P > 0.05).
Discussion
LRRK2 is an intensively pursued target for drug development in the fight against PD and might also hold promise to treat certain inflammatory diseases such as Crohn’s disease (6, 17, 18, 32). However, certain concerns for potential adverse side effects associated with the use of small-molecule kinase inhibitors directed toward the LRRK2 ATP-binding pocket remain (19, 20, 40). Here, we report the identification of a large and diverse repertoire of Nbs directed toward different domains of LRRK2 (Fig. 1). After a preselection based on their effect on LRRK2 kinase activity in cells, we chose to characterize 10 of these Nbs in detail. All 10 Nbs were able to efficiently pull-down LRRK2 from cell lysates, both upon LRRK2 overexpression and at endogenous LRRK2 expression levels. This reflects a high-affinity binding, which was subsequently confirmed using a combination of two biophysical methods which yielded KD values ranging from 2 to 150 nM.
We analyzed the influence of this set of Nbs on LRRK2 autophosphorylation and Rab10 phosphorylation in cells, as well as their effect on in vitro LRRK2 kinase activity toward a peptide and a Rab-GTPase substrate. This allows us to classify these Nbs in five functional groups (Fig. 1). Nbs belonging to functional group1 (containing Nb1, Nb6, and Nb23) inhibit all tested LRRK2 kinase activities, including auto- and Rab phosphorylation in cells and peptide and Rab phosphorylation in vitro. This indicates that these Nbs probably block LRRK2 kinase activity per se. Group2 Nbs (containing Nb17, Nb36, and Nb38) only inhibited Rab phosphorylation in cells and in in vitro, while leaving autophosphorylation and peptide phosphorylation unaffected. Group3 Nbs (containing Nb22) activated LRRK2 kinase activity both in cells and in in vitro. The group4 Nb, Nb42, had a strong inhibitory effect on LRRK2 autophosphorylation and Rab phosphorylation in cells, while leaving LRRK2 activity unaffected in vitro. At this moment, we can only speculate that this Nb specifically influences the in-cell behavior of LRRK2, potentially by influencing important protein–protein interactions or by preventing Rab29-mediated recruitment to a relevant subcellular compartment, such as the Golgi. Finally, binding of the fifth group of Nbs (Nb39 and potentially Nb40) does not have a large influence on LRRK2 kinase activity. In general, an effector molecule (either inhibitor or activator) is defined as being allosteric according to the following three criteria: 1) the effector is not chemically identical to the substrate, 2) the effector elicits a change in the activity or functional properties of the protein, and 3) the effector binds at a site of the protein that is topographically distinct from the active site (41). While kinase-activating Nbs belonging to group3 would automatically classify as allosteric, this definition also clearly shows that the inhibitory group1 Nbs can be considered as allosteric modulators of LRRK2 activity. Indeed, the group1 Nbs (Nb1, Nb6, and Nb23) act as mixed-type inhibitors of LRRK2 activity with respect to ATP and cannot be outcompeted by MLi-2 or peptide and Rab substrates, showing that they do not bind in the active site. Nb1 and Nb6 moreover bind to an entirely different domain (COR-B), further illustrating their allosteric mode of inhibition. Since the group2 Nbs (Nb17, Nb36, and Nb38) only affect Rab phosphorylation, they most likely also do not overlap with the ATP- or peptide substrate–binding pockets, and they probably either sterically interfere with the binding of the Rab substrates or stabilize LRRK2 in a conformation that precludes Rab binding or that renders LRRK2 specifically unable to phosphorylate Rab substrates. The competition ELISA experiments (SI Appendix, Fig. S13) show that at least Nb17 still binds in presence of Rab8a, allowing us to speculate that this Nb might trap LRRK2 in a conformation which only allows the Rab proteins to bind in a nonfunctional conformation that does not allow entry of the Rab into the active site pocket.
The CL-MS experiments in combination with the recent cryo-EM structures of FL-LRRK2 and its C-terminal RCKW construct (27, 29) also allow us to map the approximate binding positions of the Nbs and to speculate more on the mechanism of inhibition or activation of the group1, group2, and group3 Nbs (Fig. 6 and SI Appendix, Fig. S17). The kinase inhibitory group1 Nbs fall in two categories based on their binding domains. According to ELISA-based domain mapping, Nb1 and Nb6 are able to bind exclusively to the COR-B, and the CL-MS data for Nb1 suggest close contacts with residues of the COR-B, LRR, and kinase domains (Figs. 3A and 6). An overall view on this cross-linking pattern mapped on the FL-LRRK2 structure (SI Appendix, Fig. S17A) suggests that Nb1 binds via COR-B in a very central cavity in the middle of the interfaces formed by the LRR, COR-B, Roc, and kinase domains. As such, we could hypothesize that the inhibition observed for Nb1 and Nb6 could be due to either sterically “pushing” LRRK2 toward an inactive more open conformation or “pulling” it together toward a closed, inhibited conformation. The structure of FL-LRRK2 that is currently available indeed presumably shows LRRK2 in an inactive conformation in which the LRR domain wraps around the kinase domain, thereby keeping the kinase in an inactive conformation (27). Nb1 and Nb6 could act by stabilizing this inactive conformation. Another intriguing observation is that Nb1 also forms a cross-link with K1910 on the N-terminal lobe of the kinase domain. This latter residue is located between K1906 and E1920, two of the conserved regulatory triad residues formed by K1906-E1920-D2017. In the inactive state, K1906 and E1920 interact with Y2018, thereby preventing a direct salt bridge between K1906 and E1920 and thus keeping the kinase in an inactive state (27). It has indeed been shown that via this mechanism Y2018 acts as a break on the kinase activity (42). Potentially, Nb1 can contribute to the stabilization of this inactive conformation by contacting the peptide region connecting K1906 and E1920 (SI Appendix, Fig. S17A). On the other hand, the other group1 Nb, Nb23, binds to the K-WD40 domain in ELISA and cross-links exclusively with K2078 and K2091 in the C-terminal lobe of the kinase domain (Fig. 6 and SI Appendix, Fig. S17B). The latter residues are located in relatively close proximity to each other but are located quite far from the ATP-binding pocket. Interestingly, both cross-linking sites lay adjacent to residue N2081, of which the N2081D mutation has been identified as a major susceptibility factor for Crohn’s disease (6). N2081 makes a hydrogen bond with N1269 within the LRR domain, which might contribute to the stabilization of the LRR domain in an inactivating conformation, and correspondingly, the N2081D mutation has been shown to increase Rab10 phosphorylation. We could hence speculate that Nb23 inhibits LRRK2 kinase activity by further locking LRRK2 in this inactive conformation. The fact that none of the group1 Nbs directly bind in the kinase ATP-binding pocket also corresponds with the observation that none of these Nbs induce microtubule relocalization of LRRK2 in contrast to ATP-competitive type I inhibitors.
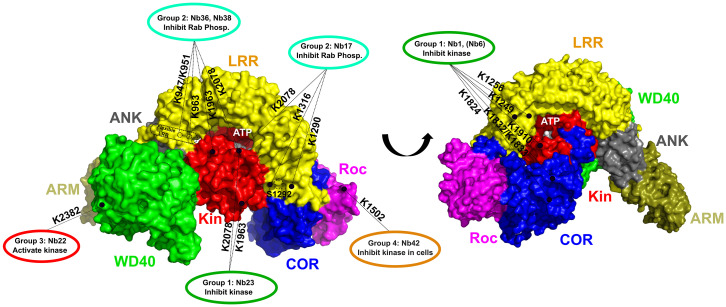
Fig. 6.
Schematic representation of the relation between the activity and binding epitopes of the different, functional Nb groups. A surface representation of the cryo-EM structure of FL-LRRK2 is shown with the domains colored as indicated (Protein Data Bank identification code 7LHW) (27). The ATP-binding pocket and the S1292 autophosphorylation site are indicated. The binding epitopes, determined by combining the results from ELISA and CL-MS experiments, of the Nbs belonging to different functional groups are indicated with dotted lines, with the LRRK2 lysine residues that form cross-links with the respective Nbs indicated adjacent to the lines.
All group2 Nbs belong to what we called “FL-LRRK2 binders” that bind on the interface of N-terminal and C-terminal domains. The CL-MS data show that Nb17 binds close to K1290 and K1316 on the LRR domain and to K2078 in the C-terminal lobe of the kinase domain (Fig. 6 and SI Appendix, Fig. S17C). These three residues are in very close proximity to each other in the FL-LRRK2 structure, again in proximity to the “Crohn’s disease residue” N2081. The binding epitopes of Nb17 and Nb23 thus seem to partially overlap, although Nb23 is not interacting directly with the LRR domain (Fig. 6 and SI Appendix, Fig. S17C). Nb17 could hence inhibit the phosphorylation of bulky substrates, like Rab proteins, by keeping the LRR domain in the closed inhibitory conformation, while still a certain degree of flexibility or conformational change would be possible to allow the phosphorylation of smaller peptide substrates and autophosphorylation.
Nb36 and Nb38, which belong to the same Nb family, cross-link with residues on the C-terminal lobe of the kinase domain (K1963 and K2078) and the N terminus of the LRR domain (K947, K951, and K963). Although the latter three residues are not resolved in the cryo-EM structure of FL-LRRK2, this again suggests a mechanism in which the LRR domain is blocked in an inhibitory conformation (Fig. 6). Together, these observations suggest a common mechanism for the group2 Nbs, which bind on the interface of several LRRK2 domains, thereby keeping LRRK2 in a “tight” conformation with the LRR domain wrapped as a lid around the kinase domains and preventing either the binding or the entry of bulky Rab substrates into the active site pocket to allow their phosphorylation. In this respect, it is also worth noting that “unleashing” of the N-terminal domains from the catalytic RCKW domains, as induced by type I kinase inhibitors and multiple PD mutations, has been linked to the association of LRRK2 to microtubules. Correspondingly, we found that group2 Nbs inhibit MLi-2–induced LRRK2 microtubule association (Fig. 5).
Finally, the kinase-activating Nb22 cross-links with residue K2382 on the WD40 domain. This binding epitope is very close to the interface of the WD40 domain with the ankyrin domain and the so-called “hinge helix” of the LRR domain (amino acid 832 to 854). This hinge helix bridges the ARM, ANK, and WD40 domains and thereby inhibits LRRK2 oligomerization via the WD40 domains, which in turn is associated with filament formation (27) (Fig. 6 and SI Appendix, Fig. S17D). K2382 is located very close to G2385. The G2385R mutation is a PD-associated variant that is common in the Chinese population (43) and was shown to disrupt WD40 dimer formation and the formation of pathogenic filaments in cells (30, 44). In very good agreement, we also find that Nb22 very efficiently inhibits MLi-2–induced LRRK2 filament formation in our experiments (Fig. 5). Moreover, probably via disrupting the interaction between the WD40 and the LRR hinge helix, the G2385R mutation leads to an activation of Rab10 phosphorylation (44), and we can thus anticipate that Nb22 increases the kinase activity via a similar mechanism.
The discovery of Nbs with a wide variety of LRRK2-modulating activities and with different mechanisms of inhibition forms a treasure chest for the further development of research tools to study the cellular role of LRRK2 and the associated disease mechanisms, as well as for future development of new diagnostic and therapeutic strategies. In particular, the finding that a subset of Nbs inhibits total LRRK2 kinase activity (group1), while others only inhibit Rab phosphorylation (group2), can be exploited to disentangle the role of the different LRRK2 activities and substrates in pathology. Owing to their ease of recombinant production, small size, and stability, Nbs are also being intensively exploited for the development of diagnostics and therapeutics (23). This presents the Nbs described here as a potentially promising route for the development of new PD therapeutics next to other currently explored strategies, including but not restricted to small-molecule kinase inhibitors (36, 45), inhibitors of GTP binding (46), and LRRK2-targeting antisense oligonucleotides (47, 48). However, therapeutic targeting of LRRK2 has so far mainly focused on the development of small-molecule, ATP-competitive inhibitors, and recently, two molecules from Denali (DNL151 and DNL201) entered clinical trials (49, 50). But despite these successes, some concerns regarding the observed adverse side effect upon prolonged treatment with high concentrations of ATP-competitive inhibitors remain (19, 20, 40). Recent studies identified oligomerization of overexpressed LRRK2 on microtubules, with concomitant blocking of microtubule-associated motor proteins, as a potential underlying cause of LRRK2 pathology. Similar to four out of the five common PD mutations, also treatment with type I ATP-competitive inhibitors increases such microtubule association. In contrast, the LRRK2-inhibiting Nbs act via completely novel, non–ATP-competitive, allosteric mechanisms. Correspondingly, none of these Nbs induce LRRK2 association with microtubules, while some even revert MLi-2–induced LRRK2 relocalization. These observations will set the stage for further development of those Nbs into LRRK2 inhibitors, with a completely different mode of action and cellular profile to the currently existing, small-molecule inhibitors. While Nbs have already been tested as therapeutics at the (pre)clinical stage for different diseases, therapeutic targeting of LRRK2 with Nbs presents a number of challenges, including the need for obtaining sufficient levels of Nb within cells in the brain for a sustained period. One possible approach is to deliver the genes encoding the Nbs using a viral, vector-based system (51). The field of gene therapy for the nervous system has undergone explosive growth in the last 5 y (52, 53), and gene therapy strategies have also been developed for PD (54–57). Different approaches for the development of the Nbs into next-generation therapeutics to target LRRK2 in PD are currently being explored.
Materials and Methods
Protein Expression and Purification.
Full-length human LRRK2 was expressed and purified based on previously developed protocols (28, 58), with minor adaptations to obtain purified LRRK2 either bound to GTPγS or GDP, as described in SI Appendix, SI Materials and Methods. Also, the expression and purification of Rab8a and the LRRK2 four-domain RCKW construct; the two-domain K-WD40 construct; and the ARM, Roc-COR, Roc, and COR-B domains are described in detail in SI Appendix, SI Materials and Methods.
Immunizations and Nb Selection.
In total, three immunizations using different llamas were performed with the following: 1) with the RocCOR domain construct of human LRRK2; 2) with FL-LRRK2 in the presence of an excess of GTPγS; and 3) with FL-LRRK2 in the presence of an excess of GDP. Additionally, in case of immunization 2 and 3, a mild cross-linking was performed on the protein after purification and prior to immunization in order to “trap” at least part of the injected proteins in their respective, nucleotide-specific conformation during immunization (note that during all phage display selection steps non-crosslinked LRRK2 was used). A 6-wk protocol with weekly immunizations in presence of GERBU adjuvant was used, and blood was collected 4 d after the last injection. All animal vaccinations were performed in strict accordance with good practices and the European Union animal welfare legislation. Next, construction of immune libraries and Nb selection via phage display were performed using previously described protocols (59), as described in detail in SI Appendix, SI Materials and Methods.
Nb Expression, Purification, and Binding Analysis via ELISA and Pull-down Experiments.
The expression and purification of Nbs, as well as their analysis via ELISA and pull-down experiments, were performed according to standard and previously published protocols (24) and are described in detail in SI Appendix, SI Materials and Methods.
Phospho-Rab Assay in Cells.
HEK293T cells were transfected with the individual Nb-GFP expression constructs, SF-tagged LRRK2(G2019S), and FLAG-HA Rab29. After 48 h, cells were lysed, and lysates were cleared by centrifugation and adjusted to a protein concentration of 1 µg/µL in 1× Laemmli Buffer. LRRK2 pS1292 and Rab10 T73 phosphorylation levels were determined by Western blot analysis, as detailed in SI Appendix, SI Materials and Methods.
Chemical CL-MS.
For chemical cross-linking, the LRRK2 concentration was adjusted to 3 µM, and each Nb was added at a 2:1 molar ratio and incubated for 1 h at 4 °C. The cross-linking reaction was performed using the N-hydroxysuccinimide (NHS)-ester–based and collision-induced dissociation (CID)-cleavable reagent disuccinimidyl sulfoxide (Thermo Fisher Scientific) (60) at a molar excess of 60:1 (referred to the Nbs) and carried out for 30 min at room temperature. Proteins were precipitated by chloroform/methanol and subjected to tryptic proteolysis (61). The tryptic peptide solutions were cleaned up by StageTips and subjected to size exclusion chromatography (SEC) separation to enrich for cross-linked peptides (28). Vacuum-dried fractions were analyzed on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) using the MS2_MS3 fragmentation method (version 3.0), and the Thermo Raw files were analyzed with the MS2_MS3 workflow provided by in Proteome Discoverer 2.5 (build 2.5.0.400) using XlinkX (version 2.5) (62), as described in SI Appendix, SI Materials and Methods.
MST and BLI Measurements.
For the MST experiments, the Nbs were first expressed and purified from a pHEN29 vector with a C-terminal LPETGG-His6-EPEA tag, and the latter tag was exchanged with an m-TAMRA–labeled GGGYK peptide (GenicBio) using Sortase (63). MST measurements were performed using a Monolith NT.115 instrument (Nanotemper technologies) by titrating a fixed concentration of m-TAMRA–labeled Nb (50 to 100 nM) with varying concentrations of LRRK2 (16 points, 3:1 dilution series) in 50 mM Hepes pH 8.0, 150 mM NaCl, 10 mM MgCl2, 5% glycerol, 0.1% bovine serum albumin (BSA), 0.05% Tween, and 500 µM GDP (GTPγS was used for Nb42). After incubation, measurements were performed (in triplicate) at 25 °C using 50 to 70% LED (light-emitting diode) power and 80% laser intensity. Data were initially processed using the MO.affinity software, and final KD values were obtained by fitting the MST signal (at 5 to 15 s on-time) versus LRRK2 curves to a quadratic binding isotherm in GraphPad Prism 7.
BLI measurements were performed using an Octet Red96 (FortéBio, Inc.) system in the same buffer at 25 °C. Binding of the Nbs to RCKW was measured using the biotinylated RCKW protein loaded onto streptavidin-coated (SA) biosensors at a concentration of 10 µg/mL. Binding of Nbs to FL-LRRK2 was measured by trapping LRRK2 on SA biosensors using an LRRK2-specific, biotinylated Nb (either Nb40 or Nb42). This sensor was then used to monitor association and dissociation of the whole set of Nbs (in triplicate). The association/dissociation traces were fitted with a 1:1 binding model using either the local, partial, or global (full) options (implemented in the FortéBio Analysis Software). The resulting Req values were subsequently plotted against the Nb concentration and used to derive the KD values from the corresponding dose–response curves fitted on a Langmuir model. Final figures were generated using GraphPad Prism7.
In Vitro Peptide and Rab8a Phosphorylation Assay.
The LRRK2 kinase activity toward a peptide substrate was determined using the PhosphoSens Protein Kinase Assay (AssayQuant Technologies Inc.) using the optimized AQT0615 peptide as substrate, according to the manufacturers’ instructions and as described in the SI Appendix, SI Materials and Methods.
To determine LRRK2 kinase activity toward Rab8a, 100 nM LRRK2 and 25 µM of the respective Nbs were incubated for 30 min at 4 °C in kinase assay buffer (50 mM Hepes pH 7.5, 50 mM NaCl, 10 mM MgCl2, 500 µM GDP or GTPγS, 1 mM ATP, and 0.1 mg/mL BSA). A positive control, containing no Nb, and two negative controls, containing either 25 µM MLi-2 or no LRRK2, were treated the same way. The protein kinase reaction was initiated by adding 2.5 µM His6-Rab8a and performed for 3 min at 30 °C. For Western blotting (WB), samples were transferred after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) to 0.45 µm nitrocellulose (GE Healthcare), and membranes were blocked. Monoclonal mouse-anti–His6 (1:1,000, GE Healthcare) and monoclonal rabbit-anti–pT72-Rab8a (1:1,000, Abcam) were used as primary antibodies. For LRRK2 detection, a monoclonal mouse-anti–Flag antibody (1:1,000, SIGMA) was used as primary antibody. IRDye680RD goat-anti–mouse and IRDye800CW donkey-anti–rabbit (both 1:15,000, Li-COR) were used as secondary antibodies. Quantification of the Western blots was performed using an Odyssey FC imaging system and Image Studio Lite software version 5.2 (both LiCOR). The ratio of the pT72-Rab8a signal to the according total His6-Rab8a signal was used for quantification. Additionally, the signal ratios were normalized by using the positive control without Nb as a reference. To determine (semiquantitative) dose–response curves, Nbs in concentrations ranging from 4 nM to 40 µM were incubated with 2.5 µM His6-Rab8a, and Rab8a phosphorylation was quantified via WB analysis (upon loading of 250 ng Rab8a on SDS gel). Relative values were quantified with Image Studio Lite software version 5.2 (LiCOR), and dose–response curves plotted against the concentration of the Nbs were calculated using GraphPad Prism 6.
Confocal Microscopy and Microtubule Localization.
A HEK293 cell line was cultured in complete media (high-glucose Dulbecco’s modified Eagle’s medium, 10% fetal bovine serum, and penicillin–streptomycin–glutamine [Gibco]). Cells were seeded on 8-well µ-slide (Ibidi) and transfected at a confluency of 50 to 70% with GFP-Nb and mScarlet-LRRK2 constructs, using JetPEI reagent (Polyplus transfection). After 24 h, cells were treated with either DMSO or 1 µM LRRK2 kinase inhibitor (MLi-2, catalog No. 5756, TOCRIS) for 90 min and then examined for localization. Data acquisition was done with a ×100 oil-immersion objective with a Zeiss LSM800 confocal laser-scanning microscope. Image analysis of z-scan was done using the Zeiss microscope software ZEN.
Supplementary Material
Acknowledgments
This work was supported by the Michael J. Fox Foundation for Parkinson’s Research (Grant No. 14527 to A.K., C.J.G., and W.V.; 14527.01 to F.W.H., A.K., C.J.G., and W.V.; and 002977 to D.C. and S.K.), the Fonds voor Wetenschappelijk Onderzoek (FWO; G005219N to W.V.), the DFG (Grant No. HE 1818/11-1 to D.C. and S.K.), the ASAP program (to D.C. and S.K.), and a Strategic Research Program Financing from the Vrije Universiteit Brussel (SRP50 to W.V.). L.V.R. was supported by a VUB OZR-bridging grant. We acknowledge Instruct-ERIC and the FWO for their support to the Nb discovery. We thank the staff of the Core Facility for Medical Bioanalytics at the University of Tubingen for technical assistance and Bernd Gilsbach and Michaela Hansch for help in protein purification. We thank Sebastian Mathea for fruitful scientific discussions.
Footnotes
Competing interest statement: R.K.S., A.K., C.J.G, and W.V. are inventors on a filed patent covering findings described in this manuscript (publication number: WO/2021/170540). All other authors declare no competing interests.
This article is a PNAS Direct Submission. D.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112712119/-/DCSupplemental.
Data Availability
Source datasets (26, 39) are published as supporting information on the journal website along with the manuscript or deposited to dedicated online repositories (the ProteomeXchange Consortium via the Proteomics Identification [PRIDE] Database with dataset identifier PXD030063 for the CL-MS dataset, and to Zenodo [https://doi.org/10.5281/zenodo.6028265] for the microscopy data). The full amino acid sequences of the Nbs that were thoroughly biochemically/biophysically characterized in this study are provided in Dataset S1 linked to this manuscript and available via the journal webpage, and the materials can also be obtained from the Versées Laboratory by contacting mta.requests@vib.be. All other study data are included in the article and/or supporting information.
References
- 1.Elbaz A., Carcaillon L., Kab S., Moisan F., Epidemiology of Parkinson’s disease. Rev. Neurol. (Paris) 172, 14–26 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Monfrini E., Di Fonzo A., “Leucine-rich repeat kinase (LRRK2) genetics and Parkinson’s disease” in Leucine-Rich Repeat Kinase 2 (LRRK2). Advances in Neurobiology, H. Rideout, Ed. (2017), pp. 3–30. [DOI] [PubMed] [Google Scholar]
- 3.Di Maio R., et al. , LRRK2 activation in idiopathic Parkinson’s disease. Sci. Transl. Med. 10, eaar5429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilks W. P., et al. , A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet 365, 415–416 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Barrett J. C., et al. , NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium, Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40, 955–962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui K. Y., et al. , Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci. Transl. Med. 10, eaai7795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wauters L., Versées W., Kortholt A., Roco proteins: GTPases with a baroque structure and mechanism. Int. J. Mol. Sci. 20, 147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steger M., et al. , Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z., et al. , LRRK2 phosphorylates membrane-bound Rabs and is activated by GTP-bound Rab7L1 to promote recruitment to the trans-Golgi network. Hum. Mol. Genet. 27, 385–395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchand A., Drouyer M., Sarchione A., Chartier-Harlin M.-C., Taymans J.-M., LRRK2 phosphorylation, more than an epiphenomenon. Front. Neurosci. 14, 527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deyaert E., et al. , A homologue of the Parkinson’s disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover. Nat. Commun. 8, 1008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen S., Webber P. J., West A. B., Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J. Biol. Chem. 284, 36346–36356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger Z., Smith K. A., Lavoie M. J., Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry 49, 5511–5523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis P. A., et al. , The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 357, 668–671 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West A. B., et al. , Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U.S.A. 102, 16842–16847 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng Z., et al. , Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med. 4, 164ra161 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Taymans J.-M., Greggio E., LRRK2 kinase inhibition as a therapeutic strategy for Parkinson’s disease, where do we stand? Curr. Neuropharmacol. 14, 214–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domingos S., Duarte T., Saraiva L., Guedes R. C., Moreira R., Targeting leucine-rich repeat kinase 2 (LRRK2) for the treatment of Parkinson’s disease. Future Med. Chem. 11, 1953–1977 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Fuji R. N., et al. , Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci. Transl. Med. 7, 273ra15 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Baptista M. A. S., et al. , Loss of leucine-rich repeat kinase 2 (LRRK2) in rats leads to progressive abnormal phenotypes in peripheral organs. PLoS One 8, e80705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryce D., et al. , Characterization of the onset, progression, and reversibility of morphological changes in mouse lung following pharmacological inhibition of LRRK2 kinase activity. J. Pharmacol. Exp. Ther., 10.1124/jpet.120.000217 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Lu X., Smaill J. B., Ding K., New promise and opportunities for allosteric kinase inhibitors. Angew. Chem. Int. Ed. Engl. 59, 13764–13776 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Muyldermans S., Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Leemans M., et al. , Allosteric modulation of the GTPase activity of a bacterial LRRK2 homolog by conformation-specific nanobodies. Biochem. J. 477, 1203–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kluss J. H., et al. , Detection of endogenous S1292 LRRK2 autophosphorylation in mouse tissue as a readout for kinase activity. NPJ Parkinsons Dis. 4, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. J. Gloeckner, Mapping of nanobody binding sites on human LRRK2 by CX-MS. ProteomeXchange Consortium via the PRIDE partner repository. https://www.ebi.ac.uk/pride/archive/projects/PXD030063. Deposited 30 November 2021. [Google Scholar]
- 27.Myasnikov A., et al. , Structural analysis of the full-length human LRRK2. Cell 184, 3519–3527.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guaitoli G., et al. , Structural model of the dimeric Parkinson’s protein LRRK2 reveals a compact architecture involving distant interdomain contacts. Proc. Natl. Acad. Sci. U.S.A. 113, E4357–E4366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deniston C. K., et al. , Structure of LRRK2 in Parkinson’s disease and model for microtubule interaction. Nature 588, 344–349 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe R., et al. , The in situ structure of Parkinson’s disease-linked LRRK2. Cell 182, 1508–1518.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardet A., et al. , LRRK2 is involved in the IFN-γ response and host response to pathogens. J. Immunol. 185, 5577–5585 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott J. D., et al. , Discovery of a 3-(4-Pyrimidinyl) indazole (MLi-2), an orally available and selective leucine-rich repeat kinase 2 (LRRK2) inhibitor that reduces brain kinase activity. J. Med. Chem. 60, 2983–2992 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Fell M. J., et al. , MLi-2, a potent, selective, and centrally active compound for exploring the therapeutic potential and safety of LRRK2 kinase inhibition. J. Pharmacol. Exp. Ther. 355, 397–409 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Williamson D. S., et al. , Design of leucine-rich repeat kinase 2 (LRRK2) inhibitors using a crystallographic surrogate derived from checkpoint kinase 1 (CHK1). J. Med. Chem. 60, 8945–8962 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Demeestere D., et al. , Development and validation of a small single-domain antibody that effectively inhibits matrix metalloproteinase 8. Mol. Ther. 24, 890–902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding X., Ren F., Leucine-rich repeat kinase 2 inhibitors: A patent review (2014-present). Expert Opin. Ther. Pat. 30, 275–286 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Dzamko N., et al. , Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem. J. 430, 405–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kett L. R., et al. , LRRK2 Parkinson disease mutations enhance its microtubule association. Hum. Mol. Genet. 21, 890–899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. A. Soliman, R. K. Singh, W. Versées, A. Kortholt, Nanobodies as allosteric modulators of Parkinson's disease-associated LRRK2. Zenodo. https://zenodo.org/record/6028265#.Yhaa9OjMLcs. Deposited 2 October 2021. [Google Scholar]
- 40.Andersen M. A., et al. , PFE-360-induced LRRK2 inhibition induces reversible, non-adverse renal changes in rats. Toxicology 395, 15–22 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Fenton A. W., Allostery: An illustrated definition for the ‘second secret of life’. Trends Biochem. Sci. 33, 420–425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt S. H., et al. , The dynamic switch mechanism that leads to activation of LRRK2 is embedded in the DFGψ motif in the kinase domain. Proc. Natl. Acad. Sci. U.S.A. 116, 14979–14988 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mata I. F., et al. , Lrrk2 pathogenic substitutions in Parkinson’s disease. Neurogenetics 6, 171–177 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Zhang P., et al. , Crystal structure of the WD40 domain dimer of LRRK2. Proc. Natl. Acad. Sci. U.S.A. 116, 1579–1584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baptista M. A. S., et al. , LRRK2 inhibitors induce reversible changes in nonhuman primate lungs without measurable pulmonary deficits. Sci. Transl. Med. 12, eaav0820 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Li T., et al. , Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson’s disease cell and mouse models. Hum. Mol. Genet. 23, 6212–6222 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Korecka J. A., et al. , Splice-switching antisense oligonucleotides reduce LRRK2 kinase activity in human LRRK2 transgenic mice. Mol. Ther. Nucleic Acids 21, 623–635 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao H. T., et al. , LRRK2 antisense oligonucleotides ameliorate α-synuclein inclusion formation in a Parkinson’s disease mouse model. Mol. Ther. Nucleic Acids 8, 508–519 (2017). Correction in: Mol. Ther. Nucleic Acids 24, 1051–1053 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Study to Evaluate DNL201 in Subjects With Parkinson’s Disease, https://clinicaltrials.gov/ct2/show/NCT03710707. Accessed 18 January 2022.
- 50.Study to Evaluate DNL151 in Subjects With Parkinson’s Disease, https://clinicaltrials.gov/ct2/show/NCT04056689. Accessed 18 January 2022.
- 51.Messer A., Butler D. C., Optimizing intracellular antibodies (intrabodies/nanobodies) to treat neurodegenerative disorders. Neurobiol. Dis. 134, 104619 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Hudry E., Vandenberghe L. H., Therapeutic AAV gene transfer to the nervous system: A clinical reality. Neuron 101, 839–862 (2019). Correction in: Neuron 102, 263 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Kimura S., Harashima H., Current status and challenges associated with CNS-targeted gene delivery across the BBB. Pharmaceutics 12, 1216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merola A., et al. , Gene therapy in movement disorders: A systematic review of ongoing and completed clinical trials. Front. Neurol. 12, 648532 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buttery P. C., Barker R. A., Gene and cell-based therapies for Parkinson’s disease: Where are we? Neurotherapeutics 17, 1539–1562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hitti F. L., Yang A. I., Gonzalez-Alegre P., Baltuch G. H., Human gene therapy approaches for the treatment of Parkinson’s disease: An overview of current and completed clinical trials. Parkinsonism Relat. Disord. 66, 16–24 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N., Adeno-associated virus-based gene therapy for CNS diseases. Hum. Gene Ther. 27, 478–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gloeckner C. J., et al. , Phosphopeptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. J. Proteome Res. 9, 1738–1745 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Pardon E., et al. , A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kandur W. V., Kao A., Vellucci D., Huang L., Rychnovsky S. D., Design of CID-cleavable protein cross-linkers: Identical mass modifications for simpler sequence analysis. Org. Biomol. Chem. 13, 9793–9807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gloeckner C. J., Boldt K., Ueffing M., Strep/FLAG tandem affinity purification (SF-TAP) to study protein interactions. Curr. Protoc. Protein Sci. 57, 19.20.1–19.20.19 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Liu F., Rijkers D. T. S., Post H., Heck A. J. R., Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Massa S., et al. , Sortase A-mediated site-specific labeling of camelid single-domain antibody-fragments: A versatile strategy for multiple molecular imaging modalities. Contrast Media Mol. Imaging 11, 328–339 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source datasets (26, 39) are published as supporting information on the journal website along with the manuscript or deposited to dedicated online repositories (the ProteomeXchange Consortium via the Proteomics Identification [PRIDE] Database with dataset identifier PXD030063 for the CL-MS dataset, and to Zenodo [https://doi.org/10.5281/zenodo.6028265] for the microscopy data). The full amino acid sequences of the Nbs that were thoroughly biochemically/biophysically characterized in this study are provided in Dataset S1 linked to this manuscript and available via the journal webpage, and the materials can also be obtained from the Versées Laboratory by contacting mta.requests@vib.be. All other study data are included in the article and/or supporting information.