Significance
This proof-of-concept modeling study offers quantitative insights into the potential epidemiological benefits of gene-editing technologies and how these may be most effectively implemented to control one of the most harmful pig diseases to date. In the future, the epidemiological benefits will need to be complemented by systematic assessment of economic and technological feasibility to enable balancing these against ethical and societal concerns.
Keywords: gene editing, PRRS, CRISPR/Cas9, mathematical model, infectious disease
Abstract
Recent breakthroughs in gene-editing technologies that can render individual animals fully resistant to infections may offer unprecedented opportunities for controlling future epidemics in farm animals. Yet, their potential for reducing disease spread is poorly understood as the necessary theoretical framework for estimating epidemiological effects arising from gene-editing applications is currently lacking. Here, we develop semistochastic modeling approaches to investigate how the adoption of gene editing may affect infectious disease prevalence in farmed animal populations and the prospects and time scale for disease elimination. We apply our models to the porcine reproductive and respiratory syndrome (PRRS), one of the most persistent global livestock diseases to date. Whereas extensive control efforts have shown limited success, recent production of gene-edited pigs that are fully resistant to the PRRS virus have raised expectations for eliminating this deadly disease. Our models predict that disease elimination on a national scale would be difficult to achieve if gene editing was used as the only disease control. However, from a purely epidemiological perspective, disease elimination may be achievable within 3 to 6 y, if gene editing were complemented with widespread and sufficiently effective vaccination. Besides strategic distribution of genetically resistant animals, several other key determinants underpinning the epidemiological impact of gene editing were identified.
Novel genomic technologies such as gene editing offer promising opportunities to tackle some of the most pressing global challenges humanity faces today. They provide new prospects to solving emerging threats such as the global COVID-19 pandemic (1) as well as to long-standing global health issues such as the HIV/AIDS crisis (2) or malnutrition (3, 4), with minimal side effects. Besides the medical field, food production stands to gain most from widespread use of genome editing technologies. Currently 11% of the human population suffers malnourishment (5), and this is expected to increase with the projected growth of the human population to 10.9 billion by 2100 (42%) (6). Meeting the 60% increase of agricultural production needed to provide sustainable and nutritious diets will likely require transformative innovations to existing production methods (7). While genome-editing technologies have been applied widely in plant breeding to simultaneously improve production and resilience to diverse stressors (see ref. 8 for examples), their application in the livestock sector is still in its infancy, primarily due to technical limitations associated with the gene-editing process itself and the safe and fast dissemination of edits, as well as ethical and societal concerns (9). Nevertheless, breakthroughs in genetic modification of farm animals through genome editing start to emerge with drastic improvements in efficiency traits (10, 11), animal welfare (12), and disease resistance (13, 14). Improving disease resistance in livestock seems particularly pertinent, as infectious diseases affect the entire food production chain and its economic viability (15).
The recent scientific breakthroughs in genome editing raise expectations for radical shifts in infectious disease control in livestock (14). Although many countries currently lack specific regulations covering the application of genome-edited animals in the food chain, this technology currently falls under genetically modified organism legislation in countries that have such processes. Reflecting this, we are seeing the rapid development of gene-editing regulations worldwide [see the Global Gene Editing Regulation Tracker (16) for an up-to-date status of gene-editing regulations per country]. Specifically, some countries have identified that some genome-editing strategies are exempt from regulatory approval. This is reflected in the recent announcement in Japan that a genome-edited seabream does not need to be regulated as no gene has been introduced into the genome (17, 18). These developments make it realistic that application of gene editing to help control infectious disease is likely in the near future. This prospect evokes pressing questions concerning the theoretical and practical feasibility of tackling diseases for which conventional control methods have failed. It is currently not known how to best implement gene-editing-induced disease resistance to achieve noticeable reduction in disease prevalence and possibly even eliminate the disease on a national scale, and on what time scale such ambitious goals could be achieved.
These questions are impossible to address in an entirely hypothetical context since epidemiological characteristics affecting the spread of the disease in question and the dynamics of the dispersal of resistant animals within the population play important roles in the success of the scheme. In this study, we focus on a particular disease, porcine reproductive and respiratory syndrome (PRRS), for the development of a mathematical modeling framework to investigate the feasibility of the application of gene editing to achieve disease elimination. PRRS represents one of the most important infectious disease problems for the pig industry worldwide, with economic losses estimated at $2.5 billion per annum in the United States and Europe alone (19, 20). Despite extensive global control efforts, the disease continues to persist in national commercial pig populations, largely due to high genetic heterogeneity of the PRRS virus, PRRSV (21), and the associated limited effectiveness of all PRRS vaccines (22, 23) and limited reliability of diagnostic tests (24, 25). There is considerable natural genetic variation in pigs’ responses to PRRSV infection, but evidence to date suggests that no pig strain is naturally fully resistant to it (26). However, recent advances in gene editing of porcine macrophages, in which a simple disruption of the CD163 gene confers complete resistance to infection with PRRSV, may revolutionize future PRRS control (27–29).
To exploit the full potential of gene editing for PRRS control, we here develop a theoretical proof-of-concept model to address a number of crucial research questions. To what extent can gene editing help reduce PRRS prevalence in national commercial pig populations? Is it possible to eliminate this disease through gene editing by creating a disease-resistant subpopulation adequately dispersed within the national susceptible population? If so, what proportion of pigs would have to be PRRSV-resistant and how would these animals need to be distributed across herds?
It is unlikely that gene editing will fully replace existing control methods, such as widespread vaccination. Hence, we also use our model to investigate the epidemiological effects of gene editing and vaccination combined. Finally, we investigate how fast the required proportion of resistant animals could be introduced in a national commercial pig population, if gene editing was strictly limited to breeding programs and resistance alleles propagate to commercial pigs using current industry practices with their diverse technical limitations. This last question becomes particularly important for an RNA virus with a high evolutionary rate such as PRRSV, since escape mutants of the virus might limit the shelf-life of gene editing and vaccines in terms of effectiveness (14, 30).
We address these questions with two linked simulation models: 1) an epidemiological model to simulate the effects of different disease control schemes on PRRS prevalence in a national commercial pig population and 2) a gene flow model to simulate the propagation of PRRSV resistance alleles from breeding programs that routinely carry out gene editing for PRRS resistance into the commercial population. The epidemiological model provides insight into the numbers and distribution of genetically resistant pigs required to eliminate PRRS under a range of realistic scenarios. The allele propagation model subsequently provides estimates for the time required to realistically produce this required number of genetically resistant pigs.
Our proof-of-concept model provides quantitative estimates for how gene editing may reduce infectious disease prevalence in farm animals and the required time frame and criteria for eliminating a disease on a national level.
Results
Impact of Gene Editing on Disease Prevalence and Chance of Disease Elimination.
Gene editing as the only disease control.
We first investigated how gene editing of pigs may affect PRRS prevalence at a national level. We assessed whether disease elimination through gene editing alone is possible and what proportion of a population would have to be genetically resistant to achieve this goal. Epidemiological theory for herd immunity stipulates that disease elimination is possible provided that individual subpopulations or herds contain sufficient proportions of resistant individuals (31). The required proportion of resistant individuals (Pe*) in a population depends largely on the disease transmission potential, otherwise known as the reproductive ratio R, which is defined as the expected number of secondary cases caused by a primary case over its infectious period (32).
To predict the potential effects of gene editing on PRRS prevalence at a national level, we simulated national commercial pig populations consisting of herds that varied in size, PRRS virus exposure, and the baseline transmission potential R0 in the absence of genetically resistant or vaccinated animals (see Methods). We then simulated four different distribution scenarios according to which given numbers of available genetically resistant pigs are distributed across the herds. These scenarios mimic different degrees of regulations concerning the distribution of these pigs, ranging from a centrally regulated scheme that may be based on either little or accurate information about the baseline transmission potential R0 to an entirely voluntary uptake by the farmers (see Methods and Table 1). Following epidemiological theory (32), the presence of genetically resistant pigs in a herd reduces the herd-specific R0 value to the effective reproductive rate
where Pe denotes the fraction of genetically resistant in the herd (see Methods for the more generic model also including vaccination effects). PRRS prevalence on a national level was then defined as the proportion of herds with R above 1.
Table 1.
Overview of the scenarios for the distribution of genetically resistant pigs across herds in the epidemiological model
| Distribution scenario | Optimum | Comprehensive | Concentrated | Unregulated |
| Baseline risk R0 known? | Yes, for each herd | No | Not necessarily, though estimates for average R0 may exist | No |
| Proportion Pe of resistant pigs per herd | Optimal proportion to achieve herd-specific R < 1 | Equal proportion | Equal proportion in herds that adopt gene-editing technology* | Arbitrary variable proportion in herds that adopt gene-editing technology* |
| Herds that receive genetically resistant pigs | Only herds with R0 >1 | All herds | Only herds that adopt gene-editing technology* | Only herds that adopt gene-editing technology* |
| Interpretation | Fully informed and regulated. Optimal distribution for elimination depending on demand; only (theoretically) possible if R0 was known for each herd | Supply of resistant pigs is uniform across all herds; supply is managed by breeding companies or national control programs | Voluntary adoption of gene-editing; all adopting herds are supplied with a fixed proportion of resistant pigs†; supply is managed by breeding companies | Voluntary adoption of gene editing, with farmers deciding how many resistant pigs they receive |
*See The Epidemiological Simulation Model for information how these herds were chosen.
†This fixed proportion may or may not be informed by estimates of the baseline disease risk R0; see The Epidemiological Simulation Model for further information.
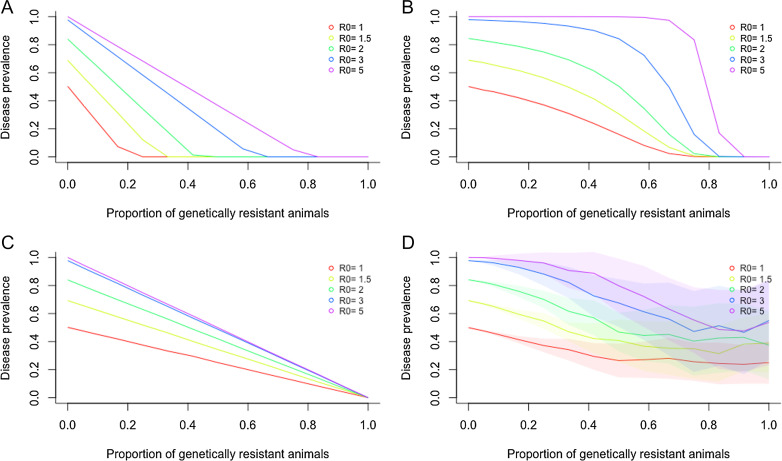
Fig. 1 demonstrates that gene editing can contribute to considerable reduction of disease prevalence and even lead to full elimination under optimal conditions. However, the rate at which disease prevalence reduces with increasing proportions of genetically resistant individuals depends strongly on both R0 and how resistant pigs are distributed across herds. In particular, the latter plays a significant role in whether or not a strategy achieves full disease elimination (Fig. 1).
Fig. 1.
Predicted reduction in PRRS prevalence achieved by using genetically PRRSV-resistant pigs, depending on the average baseline PRRSV transmission potential R0, the available proportion of resistant individuals, and their distribution across herds. PRRS prevalence is defined as the proportion of herds with effective disease transmission potential R above 1. The four graphs show four different distribution scenarios of resistant animals into herds (see Table 1 and main text for details). (A) Optimum distribution, (B) comprehensive distribution, (C) concentrated distribution, (D) unregulated distribution. Shaded areas correspond to confidence intervals comprising 95% of the predicted values from 100 simulated replicates (note that in A–C these are too narrow to be visible). Note that in the unregulated distribution scenario (D) the actual proportion of genetically resistant animals across all herds may be lower than the available proportion (presented on the x axis), explaining why elimination is not possible even if there is unlimited supply of genetically resistant pigs.
Specifically, under optimal conditions where the herd-specific R0 is known or accurately estimated, resistant animals, if sufficiently available, can be distributed according to demand to reduce the herd-specific R to below 1 (see above equation, or Eq. 1 in Methods). This optimum distribution leads to a significant reduction in disease prevalence even under higher average R0 (Fig. 1A) and could achieve disease elimination when less than half of the national pig population is genetically resistant for relatively high average R0 (i.e., average R0 <3). For a moderate average R0 of 1.5, as estimated for PRRS (33–35), the required proportion of genetically resistant pigs drops to 30% (Fig. 1A). While this ideal situation would provide the best environment for PRRS elimination using genetically resistant animals, it is unlikely to occur in a real pig production system, where the herd-specific R0 is unknown and farmers can be expected to differ in their willingness and capability to invest in adopting the new technology. A perhaps more realistic distribution scenario, hereafter called comprehensive distribution, assumes that all herds are supplied with an equal proportion of available genetically resistant animals, and the sourcing of resistant pigs is managed by the supplying breeding companies rather than the farmer (Fig. 1B). Under these circumstances, disease prevalence only decreases considerably when the available proportion of genetically resistant individuals in the population is high. In particular, disease elimination is only possible if the majority of individuals are genetically resistant (e.g., 74% for average R0 of 1.5; Fig. 1A). The third alternative model scenario, hereafter called concentrated distribution, considers that not all farmers may embrace gene-editing and splits farmers into “adopters” and “nonadopters.” Randomly chosen adopters are supplied with an equal fixed proportion of genetically resistant animals (where the proportion may or may not be based on national or regional estimates for R0), whereas nonadopters opt out of this supply entirely. In contrast to the other scenarios, this concentrated distribution leads to a linear reduction in disease prevalence with increasing proportion of genetically resistant animals. Disease elimination is, however, unachievable unless the supply is based on reasonably accurate estimates of R0. For moderate average R0 of 1.5 this implies that most herds (>98%) would need to contain a large proportion (∼75%) of genetically resistant animals (Figs. 1C and 2A). In contrast to all regulated scenarios, the fourth scenario simulated an entirely unregulated distribution of genetically resistant animals, where adoption of these animals was assumed to be entirely optional to the farmer. Thus, from a modeling perspective, arbitrary herds are supplied with arbitrary proportions of resistant animals independent of herd size or herd-specific R0. This scenario leads to a relatively small reduction in disease prevalence with high uncertainty, as represented by the wide confidence intervals in the simulations (Fig. 1D). Disease elimination through gene editing alone is out of reach for this unregulated distribution scenario.
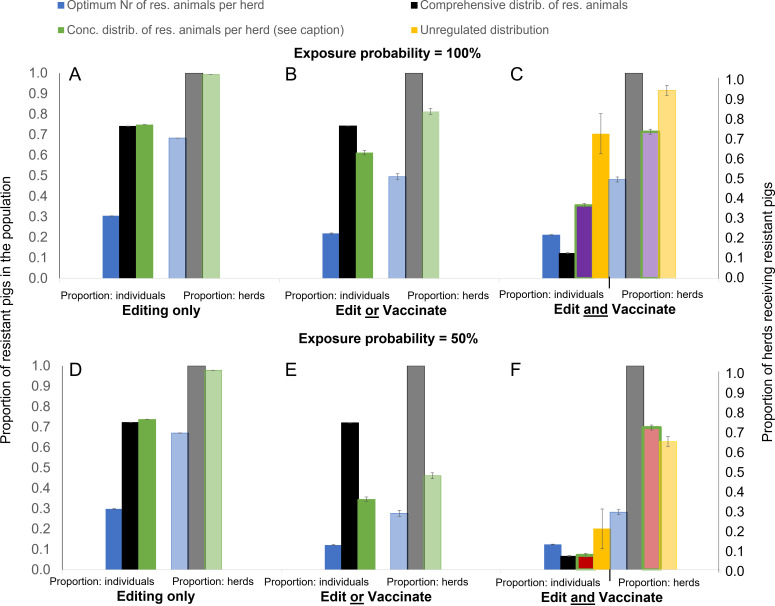
Fig. 2.
Minimum required proportion of genetically resistant animals (solid bars) and corresponding herds adopting gene editing (transparent bars) for achieving disease elimination through gene editing alone or with vaccination combined, depending on how edited animals are distributed across the herds. Results are shown for average R0 value of 1.5 and exposure probability of either 100% (A–C) and 50% (D–F) and vaccine effectiveness of 70%. Different colors refer to different distribution scenarios (see Table 1) with blue = optimum, black = comprehensive, green = concentrated and yellow = unregulated. The proportion of edited animals in the concentrated scenarios is chosen at the smallest possible proportion for elimination under each scenario, resulting in a Pe of 0.75 for scenarios A, B, D, and E (green bars), a Pe of 0.5 for scenario C (green bars, purple fill), and a Pe of 0.1 for scenario F (green bars, red fill). For further explanation of editing and vaccination strategies and the different distribution of edited individuals across herds see the main text.
The above model scenarios assume a pessimistic situation where all herds are exposed to PRRSV-infected pigs. Reducing the exposure probability of each herd to 50% had, however, little effect on the overall model predictions: Unless the baseline transmission potential R0 is known and the distribution of genetically resistant pigs is regulated accordingly, PRRS elimination through gene editing alone is only achievable if almost all herds (>95% for R0 = 1.5) consist primarily (>70% for R0 = 1.5) of genetically resistant animals (see Fig. 2 D–F for moderate R0 = 1.5 and SI Appendix, Fig. S1 for high R0 = 5).
Gene editing and vaccination as combined disease control.
Controversial technologies such as gene editing are unlikely to fully replace existing measures of disease control soon. The second question we therefore sought to answer is how gene editing can effectively complement existing disease control measures. Mass vaccination of pigs against PRRS is already widespread in many countries but has limited effectiveness (22, 36) and subsequently cannot serve as a singular elimination tool. To investigate the combined impact of gene editing and vaccination on the feasibility of eliminating PRRS, we calculated the PRRSV transmission potential (see Methods) for scenarios where either vaccination or gene editing is applied as the sole disease control strategy or applied either as complementary alternatives (hereafter referred to as the Edit or Vaccinate scenario; see Methods) or jointly (hereafter referred to as the Edit and Vaccinate scenario; see Methods).
In line with existing estimates, our model (with R values calculated using Eq. 1 in Methods) predicts that PRRS elimination cannot be achieved through mass vaccination alone when vaccine effectiveness is 70% or less and the average R0 is 1.5 and exposure probability is 50% or higher (37, 38). Elimination could, however, be achievable if vaccination and gene editing are deployed together (Fig. 2). Compared to the requirements for eliminating PRRS through gene editing alone, the required amount of genetically resistant animals and herds adopting such animals reduces considerably if gene editing is complemented by mass vaccination (Fig. 2). The biggest gains occur if vaccination is applied to all susceptible animals (Edit and Vaccinate scenario, Fig. 2 C and F) rather than just in herds that deploy vaccination as an alternative disease control to gene editing (Edit or Vaccinate scenario, Fig. 2 B and E). For example, when the average R0 is 1.5 and all herds are exposed to PRRSV infection, the required proportion of genetically resistant pigs drops by 83% from 74 to as little as 12% resistant pigs for the centrally regulated comprehensive distribution scenario when gene editing is complemented by vaccination of all susceptible animals with a vaccine of 70% effectiveness (Fig. 2C).
Perhaps most importantly, the model predicts that PRRS elimination becomes possible even when the adoption of genetically resistant animals is unregulated if mass vaccination is simultaneously applied, although it would still require most herds in a population to purchase genetically resistant animals (Fig. 2 C and F). The exact percentage of herds and genetically resistant animals required depends strongly on the baseline transmission potential (see Fig. 2 and SI Appendix, Fig. S1) and the exposure probability. Whereas the voluntary scheme would require 70% of pigs to be genetically resistant in over 91% of herds when the average R0 is 1.5 and PRRSV exposure is 100%, only 20% of genetically resistant pigs distributed across 63% of herds would suffice if the exposure probability dropped to 50% (Fig. 2 C and F).
As would be expected, the required number of resistant pigs increases when the transmission potential of PRRS is higher. However, even in a severe scenario corresponding to average R0 of 5 and 100% exposure, the model predicts that disease can be eliminated when all herds are supplied with a set proportion of 53% genetically resistant animals and all susceptible pigs are vaccinated (see SI Appendix, Fig. S1).
Impact of vaccine effectiveness on disease elimination.
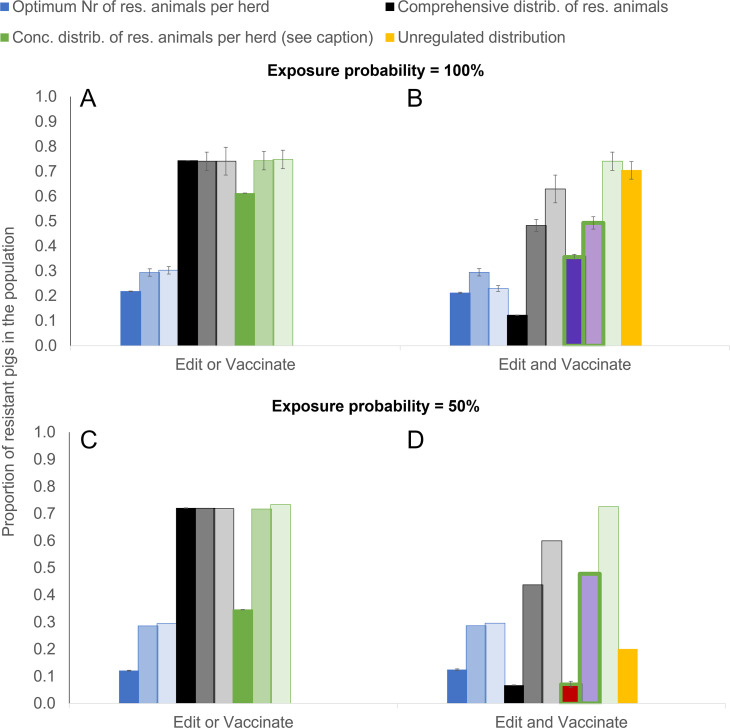
Whereas gene editing and vaccination with vaccines of relatively high effectiveness (≥70%) emerges as a highly effective PRRS elimination strategy in our models, vaccination with poorly effective vaccines is predicted to contribute relatively little to disease elimination. This is illustrated in Fig. 3 (and SI Appendix, Fig. S2 for higher R0), which also shows that for a voluntary distribution scheme disease elimination is no longer possible when vaccine effectiveness is 50% or less.
Fig. 3.
Minimum required proportion of genetically resistant animals for achieving disease elimination through gene editing and vaccination combined, depending on vaccine effectiveness εV and exposure probability. Dark bars: εV = 0.7, medium bars: εV = 0.5; light bars: εV = 0.3. Different colors refer to different distribution scenarios (see Table 1) with blue = optimum, black = comprehensive, green = concentrated and yellow = unregulated. The proportion of edited animals in the concentrated scenarios is chosen at the smallest possible proportion for elimination under each scenario, resulting in a Pe of 0.75 for scenarios A and C (green bars), a Pe of 0.5 for scenario B (green bars, purple fill), and a Pe of 0.1 for scenario D (green bars, red fill). An average transmission potential of R0 = 1.5 was assumed.
Time scale for achieving disease elimination.
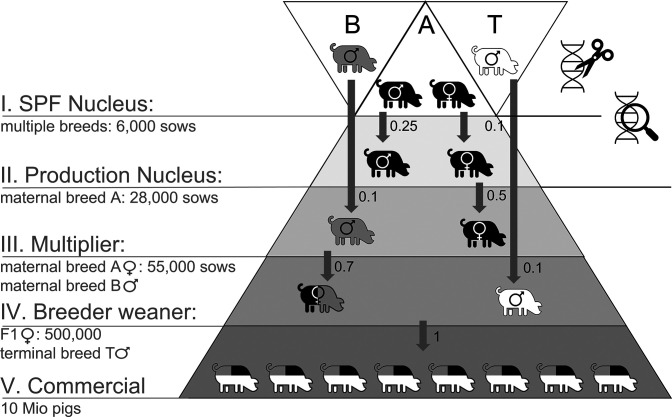
With the required proportions of genetically resistant animals under different strategies defined, the third question surrounding the feasibility of gene editing can be addressed: How long does it take to produce the required numbers of genetically resistant animals using current breeding techniques within existing technical constraints? Given the potentially limited shelf life of gene editing caused by the emergence of escape mutants, fast dissemination of genetically resistant pigs into the commercial level is crucial. This could be hampered by the fact that gene-editing technology will be limited to the top tier of the multitier pig production pyramid (Fig. 4) and that the PRRS resistance allele is recessive (14). Genotyping of pigs to trace resistance genotypes could help to identify both resistant and heterozygous carrier selection candidates and propagate the resistance allele efficiently through the production pyramid. However, genotyping is costly and not usually applied in the lower tiers. Despite these and various other technical limitations, which were considered in our gene flow simulation model (see Methods for details), we found that gene-edited resistance alleles can be efficiently disseminated through the tiers of the population without continuous genotyping of selection candidates in lower population tiers. Through selective mating of both homozygous resistant and heterozygous carrier animals in the top two tiers where genotyping is conventionally carried out, the resistance allele effectively propagates through the whole production pyramid, eventually resulting in genetically resistant animals carrying two copies of the resistance allele in the commercial tier (Fig. 4).
Fig. 4.
Schematic diagram of a typical five-tier pig production structure implemented into the gene-flow model. Two maternal breeds, A (black, e.g., Yorkshire) and B (gray, e.g., Landrace), are crossed to create hybrid females. Hybrid sows are mated to males from a terminal breed T (white, e.g., Duroc) to produce commercial animals. The color composition in individual animals represents the relative breed contribution. Numbers next to the arrows denote selection proportions transferred into subsequent tiers. Gene editing is performed in all three breeds but limited to tier I only; genotyping of selection candidates is carried out in tiers I and II (see Methods: The Epidemiological Simulation Model for more details).
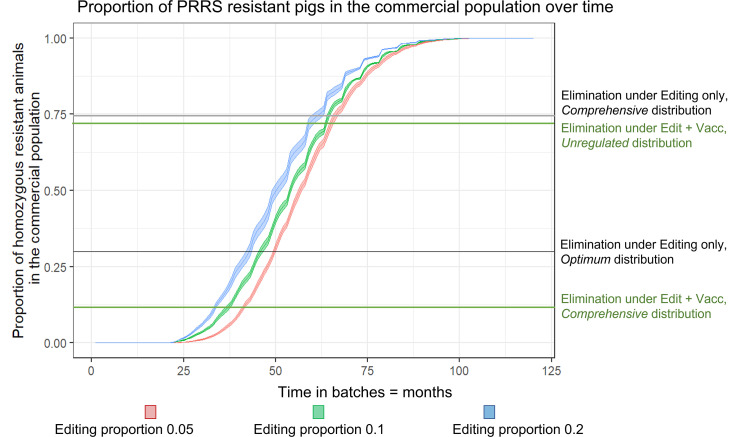
Our natural gene flow model predicts that the required proportion of resistant animals in the commercial population to achieve disease elimination under the different distribution and vaccination scenarios above can be reached within less than 6 y (see Fig. 5). In the best-case scenario, where genetically resistant animals are distributed optimally across herds and this is augmented by mass vaccination with a vaccine of at least 70% effectiveness (either only in herds that do not receive resistant animals or of susceptible animals in general), this can be achieved within less than 3 y (green lines, Fig. 5; for details on timepoints see SI Appendix, Table S1). Gene-editing a higher percentage of selection candidates in the top tier of the production pyramid does not result in a proportional reduction of the time needed to produce the required proportion of resistant animals (e.g., in the example above, increasing the editing proportion from 5 to 20% only reduces the time before required numbers are reached by 20%).
Fig. 5.
Time to reach proportions of resistant pigs in the population needed for PRRS elimination under different gene-editing scenarios. The indicated threshold levels refer to required numbers of genetically resistant pigs for achieving elimination under different distribution scenarios of pigs in the commercial tier (average R0 = 1.5 and exposure probability = 100%). For visibility, not all scenarios are depicted.
Discussion
The results of our modeling study suggest that gene editing could drastically reduce PRRS prevalence and may succeed in eliminating PRRS within 3 to 6 y of selective breeding. If gene editing was the only disease elimination tool, this would, however, require a highly regulated distribution scheme that supplies the majority of herds with a disproportionally large percentage of genetically resistant pigs. Given that adoption by farmers remains one of the biggest barriers to implementation of biotechnology (39), this blanket distribution of a novel genomic technology seems unlikely under current conditions. Nonetheless, we found PRRS elimination still to be feasible for a more realistic scenario where gene editing and mass vaccination are used conjunctively, allowing individual farmers to choose their management tool. Effective application of both control strategies simultaneously drastically reduces the required number of genetically resistant pigs and herds needed to adopt these and can achieve elimination even without stringent regulations concerning their distribution. Since PRRS has proven difficult to combat with conventional disease control (22, 40), this finding is encouraging, as it illustrates that effective combination of existing control tools with novel genomic technologies may achieve the so-far-impossible outcome of much desired disease elimination.
Our model, despite its simplicity, provides important insights into the key determinants and their interactions that underpin the success of gene editing in controlling livestock epidemics.
Determinant 1: The Baseline Transmission Potential R0.
As expected, the higher the baseline transmission potential R0, the more stringent control measures (e.g., more genetically resistant pigs) are needed to achieve a desired outcome (compare Fig. 2 [mean R0 = 1.5] and SI Appendix, Fig. S1 [mean R0 = 5]). In practice, the implementation of effective disease control is hampered by the fact that R0 typically varies across subpopulations and that precise estimates of R0 are rarely available (41, 42). Our model accommodates for heterogeneities in R0 implicitly by drawing herd-specific R0 values from normal distributions. The results highlight the importance for obtaining precise subpopulation-specific estimates of R0, as such estimates allow for more effective targeted disease control with minimum wastage of valuable resources, such as genetically resistant pigs. The optimal distribution scenario in our model, which assumes that herd-specific R0 values are known, required up to 60% fewer genetically resistant pigs for disease elimination compared to other distribution scenarios with less precise or no knowledge of R0. However, given the high uncertainty in herd-specific R0 values in practice (42), we incorporated different degrees of knowledge about R0 in the modeled distribution scenarios, ranging from full knowledge of herd-specific R0 represented by the optimum distribution scenario to partial knowledge (e.g., national or regional average R0 estimates) accommodated within the concentrated scenario to potentially zero knowledge represented by the other scenarios. Based on our model predictions, PRRS elimination through gene editing was only possible if R0 was at least partially known or complemented by mass vaccination of all susceptible individuals with a sufficiently effective vaccine.
Determinant 2: Distribution of Genetically Resistant Animals across Herds.
Our model results show that reduction in disease prevalence and the prospect of disease elimination depend strongly on how available genetically resistant animals are distributed across herds. Whereas the modeled optimum distribution was able to eliminate PRRS from a national commercial pig population without complementary vaccination with only as little as 30% of pigs carrying the PRRS resistance genotype, the unregulated distribution could only achieve elimination if 70% of all pigs were genetically resistant and the remaining pigs were vaccinated with a sufficiently effective vaccine. Feasibility issues with regard to the appropriate dissemination of genetically resistant individuals in commercial populations warranted modeling a variety of potential scenarios.
The optimum distribution scenario provides valuable insights into the potential scope of gene editing for controlling epidemics in a hypothetical world, where the full-scale benefits of gene editing for disease control can be realized. However, it is unlikely to be met in practice as it not only assumes that herd-specific R0 values are known but also that PRRSV-resistant pigs are identifiable, and that no obstacles for providing each herd with the required number of genetically resistant pigs exist. Identifying PRRSV-resistant pigs would require either tracing the parentage across the production pyramid or genotyping all commercial pigs, neither of which is current industry practice. Unless adoption of genetically resistant pigs was made compulsory (comprehensive scenario), only a fraction of herds is therefore likely to contain these pigs in practice. Furthermore, the proportion of genetically resistant pigs that each of these herds receive could be either controlled by the supplier (concentrated scenario) or by the farmer (see unregulated scenario). Either of them could base their decisions on estimates of R0, which are realistically only available on a national or regional level. Our choice of distribution scenarios aimed to capture this wide spectrum of potential scenarios and to provide useful quantitative estimates of the associated impact. To accommodate the common lack of herd-specific R0 estimates, all distribution scenarios except for the optimum scenario assumed that the proportion of genetically resistant pigs per herd is independent of the herd-specific R0. It should be noted that predictions for all alternative scenarios to the optimum scenario also apply if the resistance genotype of pigs was not exactly known, as long as the overall proportion of genetically resistant pigs in the population was known by the suppliers and pigs were distributed randomly to the receiving herds. Our model provides quantitative estimates of how each distribution scenario may affect PRRS prevalence and importantly reveals that gene editing can substantially reduce the prevalence even if adopted in restricted, suboptimal capacity. However, PRRS elimination would realistically require a widespread uptake of genetically resistant pigs and a regulated distribution of these across a significant proportion of herds (i.e., over 50% for average R0 = 1.5), coupled with a disease surveillance and vaccination program.
Determinant 3: Combination of Alternative Control Measures with Different Effectiveness.
There is general acceptance that no single silver bullet can eliminate persistent diseases such as PRRS, but that this would require a combination of effective control measures (43–45). Correspondingly, our epidemiological model predicts that PRRS elimination cannot be realistically achieved through the sole application of gene editing or vaccination but becomes feasible if both measures are effectively used in conjunction. Importantly, our results suggest that the likely presence of staunch nonadopters, e.g., farmers that cannot be incentivized to participate in an elimination scheme on the basis of gene editing, may not necessarily stand in the way of realizing the full potential of gene editing since not all herds have to receive genetically resistant animals if simultaneous vaccination is applied.
Our model results also demonstrate that the success of combined control strategies hinges on their relative effectiveness. Whereas evidence to date suggests that pigs carrying two copies of the PRRS resistance alleles are fully resistant to PRRSV infection (i.e., effectiveness of gene editing = 1) (27, 28, 46), the effectiveness of existing PRRS vaccines is severely compromised among other factors by the limited cross-protectivity of a given vaccine to different PRRSV strains, resulting in vaccine efficacies below 50% (47, 48), suboptimal vaccine administration (37, 49), or host heterogeneity in vaccine responsiveness (50). In our model, elimination of PRRS falls out of reach for the less stringent unregulated and concentrated distribution scenarios if vaccine effectiveness drops below 50%. Published field-study estimates of vaccine effectiveness for PRRS are rare; however, a recent PRRS modeling study calibrated with weekly PRRSV outbreak data from over 2,100 US pig farms estimated that a 50% vaccine effectiveness as defined in Eq. 1 could be achieved with vaccines with 12% efficacy, whereas efficacies above 50% would be required to pass the 70% effectiveness threshold (38). These predictions clearly demonstrate the need for continued support of vaccine development even when new and perhaps at first sight more promising technologies such as gene editing appear on the horizon.
Similar to gene editing, the impact of vaccination also depends strongly on vaccine coverage (37). Here we deliberately made the strong assumption that mass vaccination is applied either in all herds that do not adopt gene editing or in all herds altogether. Although PRRS vaccination is widespread in practice, these assumptions are obviously unlikely to be met in reality. Incomplete vaccine coverage would prevent disease elimination when the adoption of genetically resistant pigs is sparse and exposure risk is high, as indicated by the high proportion of resistant pigs needed when vaccines with lower effectiveness are being used (see SI Appendix, Fig. S2). This highlights the need to consider additional determinants that may underpin the success of gene editing for disease control in future studies, such as natural genetic variation in pigs’ PRRS resistance. Indeed, genetic selection of pigs for increased natural PRRS resistance has been advocated as a viable complement to existing PRRS control (51, 52). Combined application of these complementary genetic disease control strategies may effectively eliminate PRRS even under restricted vaccine usage.
Determinant 4: Exposure Risk.
It is unlikely that all herds are simultaneously and equally exposed to the PRRS virus. Heterogeneity in exposure was included in our model through a random uniform exposure probability distribution. While reduction of the average exposure risk from 100 to 50% had little influence on the model results associated with gene editing as sole disease control strategy, it drastically reduced the requirements for genetically resistant animals when gene editing and vaccination were used in conjunction. In reality, exposure risk will likely depend on PRRS prevalence in herds that are in close spatial proximity or linked through, e.g., transport or trading (53, 54). While spatial factors were not explicitly considered in our model presented here, exploration of these is an important avenue for future modeling studies as they would allow more strategic and targeted distribution of genetically resistant animals in epidemic hotspots. Furthermore, some countries or regions contain high frequency of smallholder farms with small herd size, which are unlikely to adopt gene-editing technologies or even vaccination. The impact of these farms on the overall exposure risk and subsequent prospects for elimination warrants further investigation.
Timeliness and Other Considerations for Practical Applications.
PRRSV has been estimated to have the highest evolutionary rate (on the order of 10−2 per site per year) of all known RNA viruses (with rates ranging from 10−3 to 10−5 per site per year) (55). This alarming evolutionary rate, together with observations that the virus evolves toward increased virulence with ability to evade vaccine-induced immunity (36), raises concerns about how long the current gene-editing process confers complete resistance to this virus. Hence, ambitious goals such as disease elimination would need to be achievable within a short time frame. Coupling the epidemiological model with a gene flow model suggests that PRRS can be potentially eliminated through use of gene editing within 3 to 6 y. Although it is impossible to predict whether this is sufficiently fast to win the race against virus evolution, this time scale fits well within the anticipated time scale of current national or regional elimination programs for PRRS and other livestock diseases (45, 56).
A number of simplifying assumptions in our gene-flow model warrant further investigations with regard to their impact on the predicted time scales. Our model describes the national pig industry by a five-tier breeding pyramid originating from three pure breeds. Although this structure is common for modeling pig breeding schemes (57, 58), it does not take into account the multitude of different breeding companies and different breeds that often form part of the cross-breeding schemes behind hybrid pig production. Furthermore, we assumed that all selection candidates for selection in the top pyramid tier are also candidates for gene editing, thus ignoring the possibility that some breeding companies may not apply the technology to all selection candidates, or not apply it at all if this meets best their costumers’ demand. Our model could easily accommodate this increased complexity by increasing the proportion of gene edits carried out to a subset of selection candidates in the top tier. In the current model gene editing of 20% of animals in the top tier was sufficient to satisfy the demands for genetically resistant animals in the lower tiers. Increasing this proportion in a subset of breeds composing the top production tier would generate the required number of genetically resistant animals in the commercial population in a similar time frame. An additional limitation of the current model is the absence of a strategy for the management of inbreeding, which could be incorporated alongside the implementation of separate breed-specific populations.
Our gene-flow model assumes gene-editing technologies to be incorporated into traditional breeding schemes based on natural mating or artificial insemination of selection candidates. However, a number of more efficient methods for fast propagation of genetically resistant to the commercial tier have been recently proposed, such as, e.g., the use of surrogate sire technology (59) or gene-drives (60) for the faster propagation of the resistance allele, or the use of, e.g., self-terminating “daisy chain” gene-drives that disappear from the population after a few generations (61). These may not only accelerate the rate at which genetically resistant animals can be produced but may also help to limit potential contamination effects of gene editing on the wider population (62), e.g. organic producers that need to ensure that their animals do not carry any artificially altered genetic material.
Finally, it is important to remind readers that this study focused purely on the epidemiological impact of gene editing. Implementation of this controversial technology into practical disease control will also largely depend on economic and societal aspects. Estimated annual economic losses due to PRRSV range between $24 million and $664 million in European countries and the United States alone (63, 64). Future studies are therefore required to assess the economic feasibility of the approaches presented here and to weigh the associated economic costs against the considerable potential economic benefits of eliminating one of the costliest livestock diseases to date. A thorough cost–benefit analysis is beyond the scope of this study. However, one major cost factor flagged by our models concerns the investment into routine genotyping of commercial pigs, which would allow identification and targeted distribution of genetically resistant pigs, thus increasing the chance of disease elimination. In addition, economic assessments should consider potential trade-offs arising from selection for gene-edited pigs with selection for other important livestock traits in multitrait improvement programs. Preferential selection of animals carrying the resistance allele likely results in a loss in genetic gain for other traits in the breeding goal, as do selection decisions due to inbreeding avoidance. While these weighted selection decisions are expected to have a limited impact on the time needed to reach sufficient numbers of genetically PRRSV-resistant individuals for PRRS elimination, the scale of these trade-offs will greatly influence the willingness of livestock breeders and farmers to produce and adopt genetically resistant animals. This willingness may drop considerably when PRRS prevalence has reduced to low levels, or if elimination has been achieved. As such, including scale of adoption over time in a cost–benefit analysis framework would inform a feasible level of investment in gene editing for PRRS resistance. In the context of adoption, an important aspect to consider, with epidemiological and economic consequences, is the likelihood that reducing the number of genetically resistant pigs in the national population increases the risk of reintroduction of PRRSV through international trading of domestic pigs, and possibly also through natural reservoirs such as wild boars infected with PRRSV (65, 66).
Conclusions
In summary, our proof-of-concept study highlights hitherto unprecedented opportunities for eliminating infectious diseases in livestock by complementing existing control methods with novel gene-editing technologies. The model provides some first quantitative estimates of how many edited individuals may be required, and how these would need to be distributed depending on the overall transmission potential of the disease and the quality and application of available vaccines. It particularly highlights the continued need to develop vaccines with high effectiveness and to consider additional control options such as genomic selection for natural (yet incomplete) PRRS resistance. Effective combination of these alternatives increases the chance for disease elimination and reduces the requirements for stringent regulations concerning the application of each of these measures. Finally, our study provides some first estimates of resource requirements to balance epidemiological benefits against economic trade-offs and stresses the urgent need to carefully investigate and weigh epidemiological and economic benefits against ethical and other societal concerns.
Methods
The Epidemiological Simulation Model.
We simulated a commercial pig population representative for many countries in Europe or pig-producing regions in North America or China (67), which consisted of 12 million pigs distributed into 5,000 herds. Herd size was assumed normally distributed around a mean of 2,400 with an SD of 1,000 pigs. Note that this excludes countries or regions in which pigs are predominantly reared in smallholder or backyard farms. Furthermore, we assumed that each herd is exposed to PRRSV infection with a given exposure probability pexp. This value was originally set to 1 to model the worst-case scenario and then reduced to 0.5 to mimic the more realistic situation of heterogenous exposure risk.
Once exposed, epidemiological theory stipulates that an infectious disease cannot invade a herd if its transmission potential, i.e., the so-called reproductive ratio R, is below 1, whereas invasion is possible when R > 1 (32). The R value is usually not precisely known and is expected to differ between individual herds, depending on the circulating pathogen strain, the pig breed, individual variation in resistance to the infection, environmental factors, and herd management and biosecurity characteristics (37, 68). Detailed epidemiological modeling of PRRSV transmission dynamics that considers these demographic characteristics as well as within- and between-herd contact structures affecting disease transmission will be an important avenue for future predictive modeling, but as a first step we here sought to gain initial qualitative and quantitative understanding about the potential impact of gene editing on PRRS control. To achieve this, we simply assumed that in the absence of gene editing or vaccination the baseline PRRSV transmission potential R0 for the different herds follows a normal distribution ∼N(μR0, σR0), which is independent of the herd size, i.e., PRRS transmission was assumed to be density-dependent (69).
Following epidemiological theory (32) and assuming no interactive effects between genetic resistance and vaccination, the presence of genetically resistant and/or vaccinated pigs in a herd reduces the herd-specific R0 value to the effective reproductive rate
| [1] |
where the parameters and denote the effectiveness (i.e., proportional reduction in PRRSV infection) of gene editing and vaccination, respectively, and Pe and Pv denote the fraction of genetically resistant or vaccinated animals in the current herd, respectively, with . For scenarios representing heterogeneous exposure, herds (chosen at random with probability ) that are not exposed to PRRSV infection are assigned a value . Input parameters with the assumed ranges for the epidemiological simulation model are listed in SI Appendix, Table S2.
In this study we define PRRS prevalence as the proportion of herds for which the effective reproductive rate as per Eq. 1. PRRSV is considered to be eliminated from the population if in over 99% of herds.
Eq. 1 allows calculation of the required proportion of genetically resistant and vaccinated individuals to achieve herd immunity, i.e., . In particular, in a nonvaccinated herd ) and assuming gene-editing efficacy =1, the required minimum proportion of edited pigs per herd for preventing disease invasion (i.e., achieving ) is
| [2] |
Expression 2 implies that PRRS can in principle be eliminated from a national pig population if the herd-specific R0 values were known or could be reliably estimated and each herd contains the critical number of genetically resistant individuals .
According to Eqs. 1 and 2, disease prevalence and elimination on a national scale depend not only on the proportion of genetically resistant and vaccinated animals in a population, and on the effectiveness of the corresponding control measure, but also on how these animals are distributed across the herds. The proportions of genetically resistant pigs in each herd were specified by the corresponding distribution and vaccination scenarios. Specifically, for the optimal, concentrated, and unregulated distribution scenarios (Table 1), herds were selected at random to receive the required proportion (optimal), or a given fixed proportion (concentrated) or arbitrary proportion (unregulated) of genetically resistant animals, respectively, until the available stock of genetically resistant animals was either depleted or the demand was satisfied, whichever was achieved first. In the comprehensive distribution strategy, the available stock of genetically resistant animals was distributed uniformly across all herds, thus resulting in an average equal fraction of edited animals in each herd.
For simulations that also included vaccination, the distribution of genetically resistant animals across herds was carried out first, and vaccination was subsequently assumed to be applied to either all animals in herds that contained no genetically resistant animals (Edit or Vaccinate strategy) or to all remaining susceptible animals across all herds (Edit and Vaccinate strategy). Thus, for the Edit or Vaccinate strategy the proportion of vaccinated individual per herd is either 0 or 1, depending on whether the farmer adopts genetically resistant animals or applies mass vaccination to control PRRS. For the Edit or Vaccinate scenario, where all nonresistant animals (and possibly also resistant animals if their resistance status is unknown) are vaccinated, was set to .
For each model scenario, 100 replicates were produced, and the means and SEs over the replicates were calculated. The minimum number of herds and genetically resistant animals required to achieve disease elimination for each simulated scenario was calculated using the Newton–Raphson optimization method (70).
Gene-Flow Simulation Model.
We developed a stochastic gene-flow simulation model to track the propagation of PRRS resistance alleles through a typical five-tier pig production pyramid into the commercial pig population (57), where gene editing can realistically only be carried out on a subset of pigs at the top pyramid tier. This specific pathogen-free (SPF) nucleus tier typically consists of purebred animals (here from three distinct breeds) that are selectively bred at high health and management level and of which a proportion are then sold or provide semen to farms in the lower tiers of the pyramid, as outlined in Fig. 4. Pigs in each tier are produced through mating (or artificial insemination) a fixed proportion of males and females from the same or upper pyramid tiers that have been selected to act as parents for the next generation (see SI Appendix, Table S3 for selected proportions and mating ratios), thus propagating their genetic material to offspring in the same or subsequent tier.
To assign a time scale to the natural propagation of the resistance alleles through the production pyramid, offspring in each tier are produced in the simulations in discrete monthly batches to represent a management system that is aligned with the natural reproductive and life cycle of pigs (see assumed parameter values in SI Appendix, Table S4).
PRRS resistance was assumed to be controlled by a single gene in our model and to follow Mendelian inheritance patterns. Since PRRS resistance is expected to be just one of multiple genetic traits on which selection decisions are based, each animal was also assigned a single value representing its total genetic merit that it passes on to its offspring. This value represents a combination of genetically correlated and uncorrelated traits controlled by many genes with standard polygenic inheritance patterns (71) and allows for the calculation of mean genetic merit of the entire population.
In the beginning of the simulation, a stable starting population was generated for each tier of the production pyramid in the absence of gene editing. This was achieved by first creating founder populations for each of the three breeds represented in the top pyramid tier, where each animal was assigned a genetic merit drawn from a random normal distribution. Specified proportions of individuals were then selected for mating within the nucleus based on their genetic merit (for proportions, see SI Appendix, Table S3). Once a stable base population was obtained within the nucleus (after about 18 mo), individuals (or semen) were selected for transfer to subsequent tiers as shown in Fig. 4. The burn-in phase was then run for an additional 33 mo to create base populations in all pyramid tiers. The maximum numbers of sows in each tier were back-calculated based on the number of commercial piglets produced annually (12 million), the selection proportions, and the underlying pig life-cycle parameters (SI Appendix, Table S4). The burn-in period resulted in a homogenously susceptible population that contained no animals carrying the PRRS resistance allele.
PRRS resistance was introduced into the population by selecting a fixed proportion (5%, 10%, or 20%, respectively) of animals with the highest genetic merit from each breed in the top tier of the production pyramid, the SPF nucleus, to undergo the gene-editing process. Gene editing was limited to tier I to test the feasibility of reaching sufficient numbers of resistant animals in the commercial tier without applying repeated gene editing throughout the breeding pyramid. Editing success using CRISPR/Cas9 and embryo survival rates were assumed to be 0.81 and 0.61, respectively (72). Animals in the top pyramid tier were then preferentially selected based on their PRRS resistance genotype as well as (if there were not enough animals carrying at least one PRRS resistance allele) their genetic merit value, thus allowing the resistance alleles to be naturally propagated to the subsequent tiers following Mendelian inheritance patterns.
As selection candidates in tier II, the production nucleus tier, were assumed to be genotyped to determine their resistance genotype, preferential selection for the successful gene edit occurred here as well. Since only high-merit selection candidates are selected in the gene-editing process inside the SPF nucleus, selection in the absence of genotyping in the lower tiers of the pyramid is expected to also be skewed toward animals carrying the PRRS resistance allele. However, genotyping in the top two tiers accelerates the flow of resistant individuals from the top of the breeding pyramid into the lower tiers while reflecting current industry practices.
In tiers III and IV, animals were selected based on their genetic merit alone. The gene-flow simulation model generated estimates for the number of animals carrying one or two copies of the resistance allele in each pyramid tier, and in particularly for the number of PRRSV resistant animals in the commercial population, over time.
Supplementary Material
Acknowledgments
This study was funded through the UK Research and Innovation Industry Strategic Challenge Fund Transforming Food Production Seeding Award. A.D.-W.’s contribution was funded by the Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant BBS/E/D/20002172-4 (ISP2). We thank Prof. Bruce Whitelaw from the Roslin Institute, United Kingdom, for his advice regarding the technological and regulatory aspects of gene editing for disease resistance in livestock included in this study and for providing feedback to the draft manuscripts.
Footnotes
Competing interest statement: Although the University of Edinburgh’s Roslin Institute and the animal genetics company Genus plc have signed an agreement to produce pigs that are resistant to a respiratory disease affecting livestock worldwide (https://www.ed.ac.uk/roslin/news-events/latest-news/agreement-targets-disease-resistant-gene-edited-pi), this study, carried out prior to the agreement, builds solely on published findings and rigorous scientific methods for model development and assessment. As such the results are entirely objective, and neither the results nor their interpretation are in any way influenced by this agreement or by personal beliefs or self-interest.
This article is a PNAS Direct Submission. E.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107224119/-/DCSupplemental.
Data Availability
Code and simulated data have been deposited in GitHub (https://github.com/awilso17/GeneEdit-sim).
References
- 1.Dhama K., et al. , Coronavirus disease 2019-COVID-19. Clin. Microbiol. Rev. 33, 1–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L., et al. , CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol. Ther. 25, 1782–1789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang S., Shen Q. W., Principles of gene editing techniques and applications in animal husbandry. 3 Biotech. 9, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubock A., Golden rice: Instructions for use. Agric. Food Secur. 6, 60 (2017). [Google Scholar]
- 5.World Food Programme, 2019 - Hunger map. https://www.wfp.org/publications/2019-hunger-map. Accessed 13 August 2019.
- 6.United Nations, World Population Prospects 2019: Highlights (United Nations, New York, 2019). [Google Scholar]
- 7.Thomson K., World agriculture: Towards 2015/2030: An FAO perspective. Land Use Policy 20, 375 (2003). [Google Scholar]
- 8.Bahadur B., Rajam M. V., Sahijram L., Krishnamurthy K. V., Eds., Plant Biology and Biotechnology (Springer India, 2015). [Google Scholar]
- 9.de Graeff N., Jongsma K. R., Johnston J., Hartley S., Bredenoord A. L., The ethics of genome editing in non-human animals: A systematic review of reasons reported in the academic literature. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., et al. , Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 6, 32271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., et al. , Disruption of FGF5 in cashmere goats using CRISPR/Cas9 results in more secondary hair follicles and longer fibers. PLoS One 11, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson D. F., et al. , Production of hornless dairy cattle from genome-edited cell lines. Nat. Biotechnol. 34, 479–481 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Proudfoot C., Burkard C., Genome editing for disease resistance in livestock. Emerg. Top. Life Sci. 1, 209–219 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Tait-Burkard C., et al. , Livestock 2.0—Genome editing for fitter, healthier, and more productive farmed animals. Genome Biol. 19, 204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rushton J., Ed. The Economics of Animal Health and Production (CABI, 2008). [Google Scholar]
- 16.Genetic Literacy Project, Human and agriculture gene editing: Regulations and index. https://crispr-gene-editing-regs-tracker.geneticliteracyproject.org/. Accessed 18 October 2021.
- 17.Jinguji M., Segawa S., Fleshier sea bream due to genome editing hits the market. Asahi Shimbum, Accessed 30 September 2021. https://www.asahi.com/ajw/articles/14445610.
- 18.Kishimoto K., et al. , Production of a breed of red sea bream Pagrus major with an increase of skeletal muscle muss and reduced body length by genome editing with CRISPR/Cas9. Aquaculture 495, 415–427 (2018). [Google Scholar]
- 19.Holtkamp D. J., et al. , “Economic impact of porcine reproductive and respiratory syndrome virus on U.S. pork producers” (Iowa State University Animal Industry Report AS 658, Ames, IA, 2012).
- 20.Nathues H., et al. , Cost of porcine reproductive and respiratory syndrome virus at individual farm level - An economic disease model. Prev. Vet. Med. 142, 16–29 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Guo Z., Chen X. X., Li R., Qiao S., Zhang G., The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: A molecular epidemiological perspective. Virol. J. 15, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowland R. R. R., Morrison R. B., Challenges and opportunities for the control and elimination of porcine reproductive and respiratory syndrome virus. Transbound. Emerg. Dis. 59 (suppl. 1), 55–59 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Chae C., Commercial PRRS modified-live virus vaccines. Vaccines (Basel) 9, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimman T. G., Cornelissen L. A., Moormann R. J., Rebel J. M. J., Stockhofe-Zurwieden N., Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27, 3704–3718 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Trevisan G., et al. , Macroepidemiological aspects of porcine reproductive and respiratory syndrome virus detection by major United States veterinary diagnostic laboratories over time, age group, and specimen. PLoS One 14, e0223544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boddicker N. J., et al. , Genome-wide association and genomic prediction for host response to porcine reproductive and respiratory syndrome virus infection. Genet. Sel. Evol. 46, 18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkard C., et al. , Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. J. Virol. 92, e00415-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitworth K. M., et al. , Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Chen J., et al. , Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163. Int. J. Biol. Sci. 15, 481–492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Marthaler D., Rovira A., Rossow S., Murtaugh M. P., Emergence of a virulent porcine reproductive and respiratory syndrome virus in vaccinated herds in the United States. Virus Res. 210, 34–41 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Anderson R. M., The concept of herd immunity and the design of community-based immunization programmes. Vaccine 10, 928–935 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Anderson R. M., May R. M., Infectious Diseases of Humans: Dynamics and Control (Oxford University Press, 1992). [Google Scholar]
- 33.Arruda A. G., Alkhamis M. A., VanderWaal K., Morrison R. B., Perez A. M., Estimation of time-dependent reproduction numbers for porcine reproductive and respiratory syndrome across different regions and production systems of the US. Front. Vet. Sci. 4, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nodelijk G., et al. , Introduction, persistence and fade-out of porcine reproductive and respiratory syndrome virus in a Dutch breeding herd: A mathematical analysis. Epidemiol. Infect. 124, 173–182 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pileri E., et al. , Vaccination with a genotype 1 modified live vaccine against porcine reproductive and respiratory syndrome virus significantly reduces viremia, viral shedding and transmission of the virus in a quasi-natural experimental model. Vet. Microbiol. 175, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Nan Y., et al. , Improved vaccine against PRRSV: Current progress and future perspective. Front. Microbiol. 8, 1635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bitsouni V., Lycett S., Opriessnig T., Doeschl-Wilson A., Predicting vaccine effectiveness in livestock populations: A theoretical framework applied to PRRS virus infections in pigs. PLoS One 14, e0220738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galvis J. A., Corzo C. A., Prada J. M., Machado G., Modelling the transmission and vaccination strategy for porcine reproductive and respiratory syndrome virus. Transbound. Emerg. Dis. 2021, 1–16 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Ezezika O. C., et al. , Factors influencing agbiotech adoption and development in sub-Saharan Africa. Nat. Biotechnol. 30, 38–40 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Amadori M., Razzuoli E., Immune control of PRRS: Lessons to be learned and possible ways forward. Front. Vet. Sci. 1, 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keeling M. J., Grenfell B. T., Individual-based perspectives on R(0). J. Theor. Biol. 203, 51–61 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Britton T., Epidemics in heterogeneous communities: Estimation of R0 and secure vaccination coverage. J. R. Stat. Soc. Series B Stat. Methodol. 63, 705–715 (2001). [Google Scholar]
- 43.Corzo C. A., et al. , Control and elimination of porcine reproductive and respiratory syndrome virus. Virus Res. 154, 185–192 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Rowland R. R. R., Lunney J., Dekkers J., Control of porcine reproductive and respiratory syndrome (PRRS) through genetic improvements in disease resistance and tolerance. Front. Genet. 3, 260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen A. R., Skuce R. A., Byrne A. W., Bovine tuberculosis in Britain and Ireland—A perfect storm? The confluence of potential ecological and epidemiological impediments to controlling a chronic infectious disease. Front. Vet. Sci. 5, 109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkard C., et al. , Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 13, e1006206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng X. J., Heterogeneity of porcine reproductive and respiratory syndrome virus: Implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 74, 309–329 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mateu E., Diaz I., The challenge of PRRS immunology. Vet. J. 177, 345–351 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phoo-ngurn P., Kiataramkul C., Chamchod F., Modeling the spread of porcine reproductive and respiratory syndrome virus (PRRSV) in a swine population: Transmission dynamics, immunity information, and optimal control strategies. Adv. Differ. Equ. 2019, 432 (2019). [Google Scholar]
- 50.Chase-Topping M., et al. , New insights about vaccine effectiveness: Impact of attenuated PRRS-strain vaccination on heterologous strain transmission. Vaccine 38, 3050–3061 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Reiner G., Genetic resistance—An alternative for controlling PRRS? Porcine Health Manag. 2, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunkelberger J. R., Mathur P. K., Lopes M. S., Knol E. F., Dekkers J. C. M., “Pigs can be selected for increased natural resistance to PRRS without affecting overall economic value in the absence of PRRS” (Iowa State University Animal Industry Report AS 663, Ames, IA, 2017).
- 53.Pileri E., Mateu E., Review on the transmission porcine reproductive and respiratory syndrome virus between pigs and farms and impact on vaccination. Vet. Res. (Faisalabad) 47, 108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fahrion A. S., Beilage Eg., Nathues H., Dürr S., Doherr M. G., Evaluating perspectives for PRRS virus elimination from pig dense areas with a risk factor based herd index. Prev. Vet. Med. 114, 247–258 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Hanada K., Suzuki Y., Nakane T., Hirose O., Gojobori T., The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol. Biol. Evol. 22, 1024–1031 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rathkjen P. H., Dall J., Control and eradication of porcine reproductive and respiratory syndrome virus type 2 using a modified-live type 2 vaccine in combination with a load, close, homogenise model: An area elimination study. Acta Vet. Scand. 59, 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visscher P., Pong-Wong R., Whittemore C., Haley C., Impact of biotechnology on (cross)breeding programmes in pigs. Livest. Prod. Sci. 65, 57–70 (2000). [Google Scholar]
- 58.Dekkers J. C. M., Mathur P. M., Huff-Lonergan E. J., “Genetic improvement of the pig” in The Genetics of the Pig, Rothschild M., Ruvinsky A., Eds. (CABI, ed. 2, 2011), pp. 390–425. [Google Scholar]
- 59.Gottardo P., et al. , A strategy to exploit surrogate sire technology in livestock breeding programs. G3 Genes, Genomes. G3 (Bethesda) 9, 203–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonen S., et al. , Potential of gene drives with genome editing to increase genetic gain in livestock breeding programs. Genet. Sel. Evol. 49, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noble C., et al. , Daisy-chain gene drives for the alteration of local populations. Proc. Natl. Acad. Sci. U.S.A. 116, 8275–8282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Norris A. L., et al. , Author Correction: Template plasmid integration in germline genome-edited cattle. Nat. Biotechnol. 38, 503 (2020). [DOI] [PubMed] [Google Scholar]
- 63.de Paz X., PRRS cost for the European swine industry. Pig333. https://www.pig333.com/articles/prrs-cost-for-the-european-swine-industry_10069/. Accessed 18 October 2021.
- 64.Holtkamp D. J., et al. , Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 21, 72–84 (2013). [Google Scholar]
- 65.Reiner G., Fresen C., Bronnert S., Willems H., Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) infection in wild boars. Vet. Microbiol. 136, 250–258 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Stankevicius A., et al. , Detection and molecular characterization of porcine reproductive and respiratory syndrome virus in Lithuanian wild boar populations. Acta Vet. Scand. 58, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pig333, Professional pig community, pig production data - Annual pig census. Pig333. https://www.ed.ac.uk/news/2021/agreement-targets-disease-resistant-geneedited-pigs. Accessed 18 October 2021.
- 68.Hall W., Neumann E., Fresh pork and porcine reproductive and respiratory syndrome virus: Factors related to the risk of disease transmission. Transbound. Emerg. Dis. 62, 350–366 (2015). [DOI] [PubMed] [Google Scholar]
- 69.McCallum H., Barlow N., Hone J., How should pathogen transmission be modelled? Trends Ecol. Evol. 16, 295–300 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Polyak B. T., Newton’s method and its use in optimization. Eur. J. Oper. Res. 181, 1086–1096 (2007). [Google Scholar]
- 71.Oldenbroek K., van der Waaij L., Textbook Animal Breeding: Animal Breeding and Genetics for BSc Students (Centre for Genetic Resources and Animal Breeding and Genomic Centre, Wageningen University and Research Centre, 2015). [Google Scholar]
- 72.Hai T., Teng F., Guo R., Li W., Zhou Q., One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24, 372–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and simulated data have been deposited in GitHub (https://github.com/awilso17/GeneEdit-sim).