Abstract
There are genetic risk factors that influence the outcome of COVID-19 [COVID-19 Host Genetics Initiative, Nature 600, 472–477 (2021)]. The major genetic risk factor for severe COIVD-19 resides on chromosome 3 and is inherited from Neandertals [H. Zeberg, S. Pääbo, Nature 587, 610–612 (2020)]. The risk-associated DNA segment modulates the expression of several chemokine receptors, among them CCR5, a coreceptor for HIV which is down-regulated in carriers of the COVID-19 risk haplotype. Here I show that carriers of the risk variant have an ∼27% lower risk of HIV infection.
Keywords: COVID-19, HIV, genetics
The major genetic risk factor for severe COVID-19 was introduced into modern human populations via gene flow from Neandertals 50,000 to 70,000 y ago. Although there is no direct evidence for positive selection on the risk haplotype, it has increased in frequency since the Last Glacial Maximum (1) and is unusually common today, reaching carrier frequencies of 16% and 50% in Europe and South Asia, respectively (2). Given the prevalence of this genetic variant, it is of interest to consider whether it may offer protection against some pathogen other than severe acute respiratory syndrome coronavirus 2, either today or in the past.
The major genetic risk factor for COVID-19 severity is located on chromosome 3 in a genomic region encompassing a gene cluster encoding chemokine receptors (see Data Availability for an interactive diagram of the association in this genomic region). The chemokine genes CCR1, CCR2, CCR3, CCR5, CCR9, XCR1, and CXCR6 are all located within 0.55 megabases of the genetic variant (3) likely to confer risk for severe COVID-19 (rs17713054, chr3:45859651:G/A, hg19). Using expression data from whole blood of ∼30,000 individuals (4), I find that all of the chemokine receptor genes, with the exception of XCR1, are differentially expressed in carriers of the risk variant (Table 1; P < 1e-6), with reduced expression for all genes except for CCR9. All associations pass genome-wide correction for multiple comparisons (pFDR < 0.01) and are also seen in a smaller dataset containing 1,331 individuals (Table 1).
Table 1.
Differential expression of chemokine receptors for carriers of the major genetic risk factor for COVID-19
| eQTLgen | eQTL cat. | NES (eQTL cat.) | P value (eQTL cat.) | P value (eQTLgen) | |
| Whole blood | |||||
| CCR1 | ↓ | ↓ | −0.10 | 1.8e-2 | 8.7e-14 |
| CCR2 | ↓ | ↓ | −0.05 | 3.4e-2 | 5.0e-7 |
| CCR3 | ↓ | ↓ | −0.31 | 8.3e-7 | 8.8e-47 |
| CCR5 | ↓ | ↓ | −0.05 | 1.7e-1 | 1.6e-7 |
| CCR9 | ↑ | ↑ | +0.13 | 2.6e-2 | 2.7e-7 |
| CXCR6 | ↓ | ↓ | −0.16 | 1.2e-3 | 7.9e-44 |
| CCR5 | eQTL cat. | NES (eQTL cat.) | P value | Study ref. | |
| Macrophages | ↓ | −1.15 | 7.1e-5 | 19 | |
| Monocytes | ↓ | −0.59 | 1.7e-3 | 20 | |
| Memory T-follicular helper cells | ↓ | −1.09 | 5.4e-3 | 21 | |
| Memory T helper 2 cells | ↓ | −0.76 | 8.7e-3 | 21 | |
| Memory T helper 1 cells | ↓ | −0.34 | 2.6e-2 | 21 | |
| Monocytes | ↓ | −0.31 | 3.1e-2 | 22 | |
| CD8+ T cells | ↓ | −0.69 | 5.3e-2 | 21 | |
| Macrophages | ↓ | −0.24 | 5.5e-2 | 23 | |
| Memory T helper 1/17 cells | ↓ | −0.24 | 1.5e-1 | 21 | |
| NK cells | ↓ | −0.42 | 1.6e-1 | 21 | |
| Monocytes | ↓ | −0.20 | 2.1e-1 | 21 | |
| CD16+ Monocytes | ↓ | −0.45 | 3.4e-1 | 21 | |
| T cells | ↓ | −0.16 | 4.0e-1 | 20 | |
| Memory T regulatory cells | ↓ | −0.07 | 6.2e-1 | 21 | |
| T cells | ↓ | −0.07 | 6.6e-1 | 24 | |
| T regulatory cells | ↓ | −0.10 | 7.4e-1 | 21 | |
| Memory T helper 17 cells | ↓ | −0.004 | 9.8e-1 | 21 | |
Downward arrow and negative normalized effect sizes (NES) represent reduced expression for carriers of the COVID-19 risk allele at rs17713054. NES available from the eQTL catalog. The risk allele for COVID-19 is associated with reduced expression of all receptors except CCR9 in whole blood. Expression data from whole blood aggregated by the eQTLGen Consortium includes 26,000 to 31,569 samples, whereas the eQTL catalog comprises 1,331 whole blood samples (meta-analyzed using inverse-variance weighting of the contributing studies). CCR5 is down-regulated in all investigated immunological cell types (19–24). For CCR5 in macrophages, the NES translates to an allelic fold change of ∼3.7 (log2) based on the transcript levels (19).
One of the most well-studied genetic variants modulating infectious disease risk is a 32-base pair deletion that introduces a premature stop codon in CCR5, resulting in a nonfunctional receptor. This mutation, CCR5-Δ32, which confers protection against HIV-1 infection and likely also against smallpox, has been positively selected (5). HIV-1 relies on CD4 for viral entry and commonly requires also the chemokine receptors CCR5 and/or CXCR4 as coreceptors, even if it can also utilize CCR3 (6) and CXCR6 (7).
Macrophages, dendritic cells, and memory T cells patrolling the mucosa are the first cells infected by HIV-1 (8). Indeed, a major route for HIV-1 infection among men is urethral macrophages (9), and HIV-1 can be present in macrophages in the vagina (10). Strikingly, the down-regulation of CCR5 in carriers of the COVID-19 risk variant on chromosome 3 is primarily seen in macrophages, monocytes (which are related to dendritic cells), and memory T cells (Table 1).
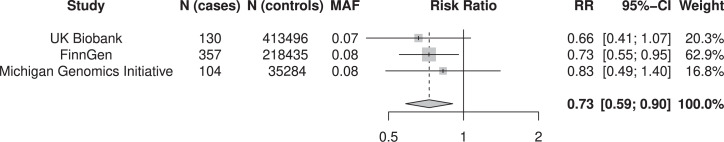
Since individuals carrying the major genetic risk factor for COVID-19 have lower CCR5 as well as CCR3 and CXCR6, I hypothesized that they might have lower prevalence of HIV infection. A limiting factor for such an analysis is the low prevalence of HIV in European cohorts available for analysis. To increase statistical power, I conducted a meta-analysis of three of the largest currently available biobanks, the UK Biobank (11), the Michigan Genomics Initiative, and FinnGen. Together these cohorts contain 591 European individuals with HIV infection and 667,215 controls. In all three cohorts, carriers of the risk allele for COVID-19 have a risk ratio for HIV infection between 0.66 and 0.83 (Fig. 1). Meta-analysis of the three cohorts results in an overall risk ratio of 0.73 (95% CI: 0.59 to 0.90, P = 4.1e-3). Thus, carriers of the chromosome 3 COVID-19 risk allele has a 27% reduction in risk for HIV infection (95% CI: 9 to 40%). There was no detectable heterogeneity across the cohorts (I2 = 0%, P = 0.82).
Fig. 1.
Risk ratios for HIV disease for carriers of the major genetic risk factor for COVID-19. A meta-analysis by inverse-variance weighting of 591 individuals with HIV disease and 667,215 controls. Risk ratios (OR) are calculated for a linear model with effect estimates per copy of the Neandertal-derived allele at rs17713054. The MAF column shows the minor allele frequency in the cohorts. Meta-analysis was performed in R (version 4.1.2) using the package meta (version 5.1).
Sequence diversity in the envelope spike of HIV-1 is known to influence coreceptor use (12). Moreover, selective pressures arising from host genetic variation affect HIV-1 sequence variation (13). I therefore investigated whether variation in the HIV-1 envelope gene Env is associated with carrier status of the COVID-19 risk haplotype. There are 34 genetic variants that cosegregate (r2 > 0.5) with rs17713054 among individuals in the 1000 Genomes Project (14). In a published dataset (13), four of these variants (rs17764980, rs17714101, rs17714228, and rs71325092) are associated (P < 7e-4) with the amino acid replacement M115L in the HIV-1 envelope protein Gp41 encoded by Env. Specifically, carriers of the risk variant for COVID-19 were more likely to be infected with a virus variant carrying a leucine residue at position 115 in Gp41 (odds ratio, OR = 1.3), a viral variant less sensitive to the fusion inhibitor enfuvirtide (15). That the association between the host genome and the viral genome involves the coreceptor and the envelope spike suggests that the protective effect of the haplotype is mediated by reduced viral entry.
The major genetic risk factor for COVID-19 rose in frequency between 20,000 to 10,000 y ago (1); since this significantly predates the HIV pandemic, it is unlikely that this increase in frequency resulted from positive selection driven by HIV. I can only speculate about the pathogen that exerted the genuine selective pressure on this allele. Variola virus emerged more than 10,000 y ago (16), making smallpox a likely candidate, whereas as Yersina pestis emerged later, ∼7,000 y ago (17). I also note that the highest allele frequencies today (2) coincide with regions where cholera is endemic.
The association described here highlights that gene flow from Neandertals was a double-edged sword. Whereas this genetic variant has had tragic consequences during the last 2 y in the COVID-19 pandemic, it appears to have offered considerable protection against HIV during the last 40 y. Its role in past and future pandemics remains to be seen.
Acknowledgments
I thank Megan Michel, Benjamin Vernot, Janet Kelso, and Svante Pääbo for careful reading of the manuscript and for helpful comments. I want to acknowledge the participants and investigators of the FinnGen study, the UK Biobank, the Michigan Genomics Initiative, and the COVID-19 Host Genetics Initiative. This research was supported by Jeanssons Stiftelser, Magnus Bergsvalls Stiftelse, and the Swedish Research Council (2021-03050). These funding agencies had no role in the design, implementation, or interpretation of this study.
Footnotes
The author declares no competing interest.
Data Availability
Summary statistics for HIV infection among people of primarily European descent were obtained from FinnGen (freeze 5, https://r5.finngen.fi), Michigan Genomics Initiative (freeze 2, https://pheweb.org/MGI-freeze2), and UK Biobank (https://pan.ukbb.broadinstitute.org/). Covariates included in the analysis are age, sex, and the first four principal components (Michigan Genomics Initiative) or the first 10 principal components (UK Biobank and FinnGen). In addition, Michigan Genomics Initiative and UK Biobank controlled for genetic relatedness, and FinnGen and Michigan Genomics Initiative included genotyping batch and chip version, respectively. The HIV-summary statistics provided by these resources are on the OR scale but were here directly translated to risk ratios under the rare disease assumption. Summary statistics for expression differences were obtained from the eQTLGen Consortium (https://www.eqtlgen.org) and the eQTL Catalogue (https://www.ebi.ac.uk/eqtl/). Data were meta-analyzed using inverse-variance weighting. Linkage disequilibrium was calculated using data from the 1000 Genomes Project (15). The data on the interaction between host and viral genetic sequence variation were taken from the supplementary material of a previous study (14) which had been deposited online (https://dx.doi.org/10.5281/zenodo.7139). An interactive diagram of the association for the major risk factor for COVID-19 (COVID-19 Host Genetics Initiative, release 6) has been deposited on LocusZoom (https://my.locuszoom.org/gwas/815606/) (18).
References
- 1.Zeberg H., Pääbo S., A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc. Natl. Acad. Sci. U.S.A. 118, e2026309118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeberg H., Pääbo S., The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 587, 610–612 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Downes D. J., et al. ; COvid-19 Multi-omics Blood ATlas (COMBAT) Consortium, Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat. Genet. 53, 1606–1615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Võsa U., et al. , Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv [Preprint] (2018). 10.1101/447367 (Accessed 3 September 2021). [DOI]
- 5.Galvani A. P., Slatkin M., Evaluating plague and smallpox as historical selective pressures for the CCR5-Delta 32 HIV-resistance allele. Proc. Natl. Acad. Sci. U.S.A. 100, 15276–15279 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J., et al. , CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature 385, 645–649 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Deng H. K., Unutmaz D., KewalRamani V. N., Littman D. R., Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388, 296–300 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Koppensteiner H., Brack-Werner R., Schindler M., Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology 9, 82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganor Y., et al. , The adult penile urethra is a novel entry site for HIV-1 that preferentially targets resident urethral macrophages. Mucosal Immunol. 6, 776–786 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Shen R., et al. , Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J. Virol. 83, 3258–3267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bycroft C., et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor B. M., et al. , An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J. Virol. 82, 5460–5471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartha I., et al. , A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. eLife 2, e01123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton A., et al. ; 1000 Genomes Project Consortium, A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su C., et al. , The relationship between susceptibility to enfuvirtide of baseline viral recombinants and polymorphisms in the env region of R5-tropic HIV-1. Antivir. Ther. 8, S59 (2003). [Google Scholar]
- 16.Li Y., et al. , On the origin of smallpox: Correlating variola phylogenics with historical smallpox records. Proc. Natl. Acad. Sci. U.S.A. 104, 15787–15792 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Susat J., et al. , A 5,000-year-old hunter-gatherer already plagued by Yersinia pestis. Cell Rep. 35, 109278 (2021). [DOI] [PubMed] [Google Scholar]
- 18.H. Zeberg, Chr3 COVID HGI B2 20210607 (all including 23andMe). LocusZoom. https://my.locuszoom.org/gwas/815606/. Deposited 5 November 2021. [Google Scholar]
- 19.Alasoo K., et al. ; HIPSCI Consortium, Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50, 424–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L., et al. , Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmiedel B. J., et al. , Impact of genetic polymorphisms on human immune cell gene expression. Cell 175, 1701–1715.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quach H., et al. , Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167, 643–656.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nédélec Y., et al. , Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167, 657–669.e21 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez-Arcelus M., et al. , Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2, e00523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Summary statistics for HIV infection among people of primarily European descent were obtained from FinnGen (freeze 5, https://r5.finngen.fi), Michigan Genomics Initiative (freeze 2, https://pheweb.org/MGI-freeze2), and UK Biobank (https://pan.ukbb.broadinstitute.org/). Covariates included in the analysis are age, sex, and the first four principal components (Michigan Genomics Initiative) or the first 10 principal components (UK Biobank and FinnGen). In addition, Michigan Genomics Initiative and UK Biobank controlled for genetic relatedness, and FinnGen and Michigan Genomics Initiative included genotyping batch and chip version, respectively. The HIV-summary statistics provided by these resources are on the OR scale but were here directly translated to risk ratios under the rare disease assumption. Summary statistics for expression differences were obtained from the eQTLGen Consortium (https://www.eqtlgen.org) and the eQTL Catalogue (https://www.ebi.ac.uk/eqtl/). Data were meta-analyzed using inverse-variance weighting. Linkage disequilibrium was calculated using data from the 1000 Genomes Project (15). The data on the interaction between host and viral genetic sequence variation were taken from the supplementary material of a previous study (14) which had been deposited online (https://dx.doi.org/10.5281/zenodo.7139). An interactive diagram of the association for the major risk factor for COVID-19 (COVID-19 Host Genetics Initiative, release 6) has been deposited on LocusZoom (https://my.locuszoom.org/gwas/815606/) (18).