Significance
Seed vigor is important to farmers, breeders, and industries, with critical implications for seed quality and germplasm preservation. We included RNA sequencing data, searching the differentially expressed genes, separately, in seeds of two rice varieties with huge differences in seed vigor. We then compared the transcriptome profiles between them. We compared the metabolic profiling of these two seeds. We uncovered the genetic architecture of two rice varieties with distinct seed vigor. We identified PER1A that acts as a positive regulator and bZIP23 that functions upstream and as a transcription activator of PER1A and is also a positive regulator of seed vigor. Finally, we demonstrated that the bZIP23-PER1A regulatory module for rice seed vigor operates via the abscisic acid signaling pathway.
Keywords: seed vigor, ABA signaling, reactive oxygen species
Abstract
Seed vigor in crops is important in terms of improving grain quality and germplasm conservation; however, little is known about its regulatory mechanisms through the encoded proteome and gene network. Comparative analyses of transcriptome (RNA sequencing [RNA-seq]) and broadly targeted metabolic profiling of two subspecific rice cultivars with distinct seed vigor during accelerated aging revealed various biological pathways and metabolic processes as key influences explaining trait differences. RNA-seq coexpression regulatory network analyses identified several transcription factors, including bZIP23 and bZIP42, that act as nodes in the gene network. Importantly, transgenic seeds of overexpression of bZIP23 enhanced seed vigor, whereas its gene knockout reduced seed vigor, suggesting that the protein it encodes functions as a positive regulator. Similarly, overexpression and knockout of PER1A that encodes a key player in the detoxification pathway enhanced and decreased seed vigor, respectively. We further demonstrated a direct interaction of the PER1A promoter with bZIP23 in seeds, which activates the expression of PER1A, and the genetic evidence suggested that bZIP23 most likely functions in a common pathway with and acts upstream of PER1A to modulate seed vigor. In addition, the control of seed vigor by the bZIP23-PER1A module was connected with that of the abscisic acid signaling pathway. Collectively, we revealed the genetic architecture of variation in seed vigor and uncovered the bZIP23-PER1A–mediated detoxification pathway that enhances the trait in rice.
Seeds gradually lose their viability during storage (i.e., seed aging), the rate of which is heavily affected by environmental conditions—mainly of storage temperature and moisture content (1–3). Seed vigor can be synonymous with seed germination vigor under favorable or adverse environmental conditions (4, 5). Seed vigor has important implications for seed banking and biodiversity conservation, and it is an important aspect of seed quality (4–6). Seed-accelerated aging is regularly used for the rapid assessment of seed vigor (7, 8).
Reactive oxygen species (ROS) are often described as a common and key factor in the regulation of seed vigor, which must be under a strict control because they are usually considered to be highly toxic to proteins, DNA, and lipids (9–12). The balance between the generation and removal of ROS could be disturbed by various adverse biotic and abiotic stress factors such as high temperature and high light (13, 14). Importantly, plants have evolved sophisticated strategies of ROS detoxification to avoid oxidative damage such as nonenzymatic antioxidants of ascorbate, reduced glutathione (GSH), tocopherol (vitamin E), flavonoids, alkaloids, and carotenoids as well as enzymatic scavenging mechanisms of SUPEROXIDE DISMUTASE (SOD), ASCORDASE PEROXIDASE (APX), GLUTATHIONE PEROXIDASE (GPX), and CATALASE (CAT) (10, 11). For example, mutations of the Arabidopsis thaliana tocopherol biosynthesis genes VTE1 and VTE2 whose protein products are involved in tocopherol biosynthesis significantly reduced seed vigor (15). Notably, 1-CYS PEROXIREDOXIN (also called PER1) is a seed-specific antioxidant in many plants that uses cysteine residues to scavenge ROS (16); interestingly, ectopic expression of its Nymphaea tetragona (sacred lotus) homolog NnPER1 in A. thaliana enhanced seed vigor and longevity (16). However, the biological functions and mechanisms of PER1 and ROS-related regulation of seed vigor in crops have remained largely unknown.
The plant hormone abscisic acid (ABA) assumes an indispensable role in the regulation of seed maturation and vigor (5, 17–21). The extreme alleles of abscisic acid insensitive 3 (abi3), the protein product of which is a crucial downstream component of ABA signaling, has severely compromised seed longevity and vigor, and the ABA biosynthesis mutant abscisic acid deficient 1 also showed reduced traits (17, 22). In addition, the bZIP transcription factor (TF) encoded by ABSCISIC ACID INSENSITIVE 5 (ABI5) is a prominent regulator of seed maturation and vigor in legumes (23). Interestingly, the Arabidopsis AtPER1 transcript level is strongly reduced in the abi3 mutation, and ABI5 likely binds to the ABA-responsive element of the AtPER1 promoter (24). DELAY OF GERMINATION 1 (DOG1) is critical for the induction of dormancy, and the abi3/dog1 double mutant is highly insensitive to ABA treatment, very similar to the phenotype of severe abi3 mutants, suggesting a genetic interaction between these two factors; besides, the Arabidopsis dog1 mutants show reduced seed vigor in which the ABI5 expression was significantly down-regulated (18, 25).
These progresses have greatly improved our understanding of the molecular mechanism of seed vigor regulation; however, whether and/or how the control of seed vigor by ABA was connected with the antioxidant signaling pathway remains unclear. In this study, we integrated transcriptomic and metabolomic techniques to dissect the genetic architecture of two Oryza sativa (rice) varieties with distinct seed vigor. The comparative analyses of these omic data allow us to identify several biological pathways likely contributing to the altered seed vigor. We constructed a coexpression regulatory network and identified the TFs bZIP23 and bZIP42 that regulate seed vigor positively. We further showed that PER1A functioned as a positive regulator of seed vigor and was a direct target of bZIP23. In addition, we also showed that the ABA signaling pathway was most likely involved in the bZIP23-PER1A–mediated regulation of seed vigor in rice.
Results
Dissecting the Transcriptome Profile for Different Seed Vigor.
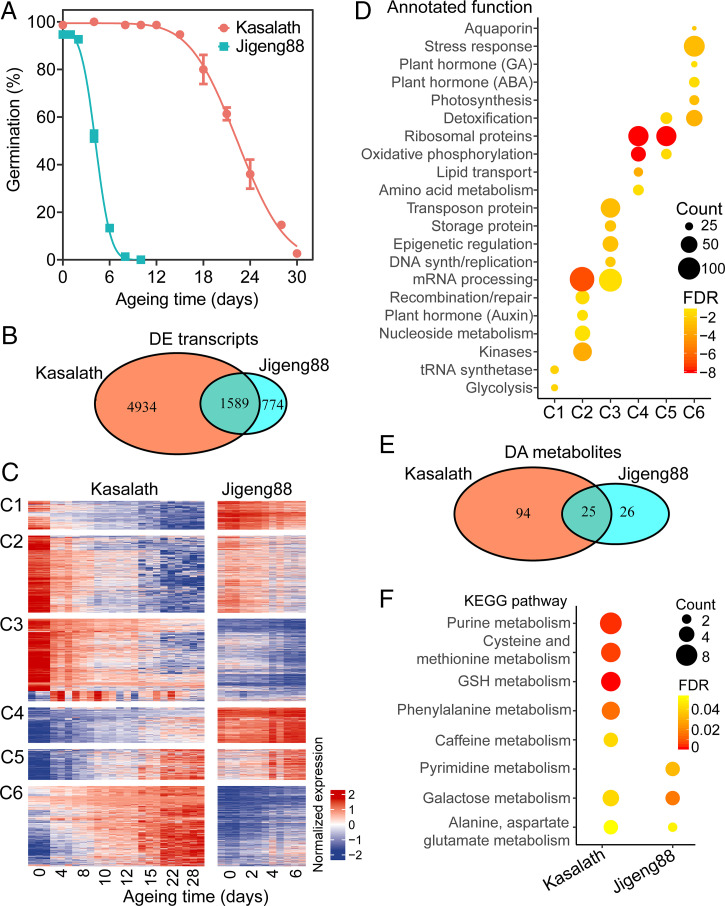
We sought to identify the determinants of seed vigor in cultivated rice (O. sativa L.). We used an accelerated aging experiment to assess seed vigor. The dynamic feature of germination during aging showed that, at day 12 of treatment, the seeds of the indica cultivar Kasalath maintained a final cumulative germination of 98.7%, whereas those of Jigeng88, a cultivar growing widely in the northern region of China, have completely lost the capability to germinate (Fig. 1A).
Fig. 1.
Global comparison of transcriptomic and metabolomic change during Kasalath and Jigeng88 seed aging. (A) The change in Kasalath and Jigeng88 seed germination during accelerated aging. The seeds were aged under 45 °C and 80% RH. The error bars indicate ± SE (n = 3). (B) A Venn diagram of the comparison of DE transcripts during Kasalath and Jigeng88 seed aging. (C) The expression profile of transcripts DE during Kasalath seed aging. The expression of transcripts DE during Kasalath seed aging was grouped into six clusters in contrast to their corresponding expression during Jigeng88 seed aging. At each aging time point, the data were shown in three biological replicates. (D) The enrichment of biological processes in different expression clusters. The biological processes were then manually annotated. P values were determined from the hypergeometric test with FDR calibration. (E) A Venn diagram of comparison of DA metabolites during Kasalath and Jigeng88 seed aging. (F) KEGG enrichment analysis of DA metabolites. KEGG pathways that were significantly enriched (FDR < 0.05) are shown.
We reasoned that the huge difference in seed vigor between Kasalath and Jigeng88 originated from differential gene expression. Our strategy was to identify the differentially expressed (DE) genes, separately, in Kasalath and Jigeng88 seeds during accelerated aging and then compare their transcriptome profiles. The embryos of the seeds of Kasalath aged for 0, 4, 8, 10, 12, 15, 22, and 28 d, and those of Jigeng88 aged for 0, 2, 4, and 6 d were sampled in triplicates for RNA sequencing (RNA-seq). Ultimately, on average, 24 million paired-end reads per replicate of the samples were generated from the RNA-seq clean data, and more than 92% of which could be aligned to the Nipponbare reference genome (Dataset S1). Considering that many genes have splice variants and different promoters that require measuring expression changes of individual transcripts (26), we applied RNA-seq analysis of differential expression at transcript resolution to capture the transcriptome dynamics.
We observed highly positive correlations between the replicates of each sample (R > 0.95; SI Appendix, Fig. S1). A principal component analysis (PCA) showed that the first principal component apparently separated the gene expression of Kasalath from those of Jigeng88, suggesting that genotype plays a major role (SI Appendix, Fig. S2A). We used the edgeR and DESeq2 for differential expression analysis and picked up the overlapped differential expression of the two algorithms under a stringent criteria of false discovery rate (FDR) < 0.01. We compared the gene expression between the unaged seeds of Kasalath and Jigeng88 and identified 7,674 DE transcripts (derived from 7,594 genes; Dataset S2), reflecting the great difference of the genotypes. We next made every effort to identify the DE genes in response to seed aging. The resultant altered 6,523 transcripts (derived from 6,379 genes), and 2,363 transcripts (2,361 genes) were identified, respectively, from the Kasalath and Jigeng88 samples. Surprisingly, 67.2% (1,589) of the DE transcripts of Jigeng88 were shared with 24.4% of those of Kasalath (Fig. 1B and Dataset S3). Nevertheless, we found that the rice PROTEIN L-ISOASPARTYL (isoAsp) O-METHYLTRANSFERASE 1 (OsPIMT1) and OsPIMT2 that encode the enzymes restricting deleterious isoAsp and ROS accumulation and function in the regulation of seed vigor and longevity (27) have significantly altered transcript abundance in the samples of Kasalath (SI Appendix, Fig. S3). In addition, the homologs of AtTIP3-1, AtTIP3-2, AtLIG4, AtLIG6, AtPLDα1, and AtHB25 that involved in seed longevity regulation in Arabidopsis (28–31) have also changed transcript accumulation in our RNA-seq data (SI Appendix, Fig. S3).
To discover biological pathways that confer to the seeds of Kasalath the ability to live longer, we performed a cluster analysis of the DE genes. We determined the optimal number of clusters to be 10 and then manually classified them into six groups: the DE transcripts were generally categorized into two groups, the down-regulated and up-regulated ones of Kasalath, and each of them was further divided into three clusters (C1 to C3 versus C4 to C6) by comparing the abundance of transcripts between Kasalath and Jigeng88 (Fig. 1C and SI Appendix, Fig. S4). We manually annotated the DE genes as documented previously (32). Ultimately, up to 83.6% (5,331) of the Kasalath DE genes were assigned to a specific biological process (Dataset S3). We then evaluated the significance of enrichment for each biological process (annotated function) in the clusters C1 to C6 (FDR < 0.05). These different clusters have enriched distinct annotated functions (Fig. 1D). In particular, the DE genes of cluster C6 in Kasalath were up-regulated and accumulated much higher abundance than those in Jigeng88 (Fig. 1C). Interestingly, several biological processes, including the ABA, gibberellin (GA), stress response, and aquaporin (water transport) pathways that were reported to be involved in seed vigor (21–23, 30, 31), were specially enriched in this cluster (Fig. 1D). In addition, DE genes involved in the detoxification pathway were enriched in both clusters C5 and C6 (Fig. 1D).
Associating Seed Metabolome Variations with Altered Seed Vigor.
We further compared the variation at the metabolite levels of the Kasalath versus Jigeng88 seeds during accelerated aging. For this purpose, we collected the seeds of Kasalath under the aging treatment of 0, 4, 12, and 22 d, and those of Jigeng88 treated for 0, 2, and 4 d, respectively, as starting samples for high-throughput metabolite analysis. We quantified a total of 557 and 553 metabolites, respectively, in the samples of Kasalath and Jigeng88 (Dataset S4). PCA revealed that genotypes contribute to the major variation of the metabolite accumulation (SI Appendix, Fig. S2B). We distinguished the differentially accumulated (DA) metabolites under the criteria of the variable importance (VIP) scores ≥ 1 and absolute fold change ≥ 2 and identified 119 and 51 metabolites to be DA in the Kasalath and Jigeng88 samples, respectively (Fig. 1E and Dataset S5). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment revealed that many metabolic pathways were significantly altered in Kasalath, including the purine and GSH metabolism (Fig. 1F). To further uncover a possible relationship between these metabolites, we constructed a metabolic network using the MetaboAnalyst website services. Interestingly, adenosine monophosphate, glutamine, adenine, GSH, methionine, and ornithine represented the hub nodes that associated with a huge number of other metabolites (SI Appendix, Fig. S5), suggesting that they might play an important role in the functional regulation of seed vigor.
The TFs bZIP23 and bZIP42 Positively Control Seed Vigor in Rice.
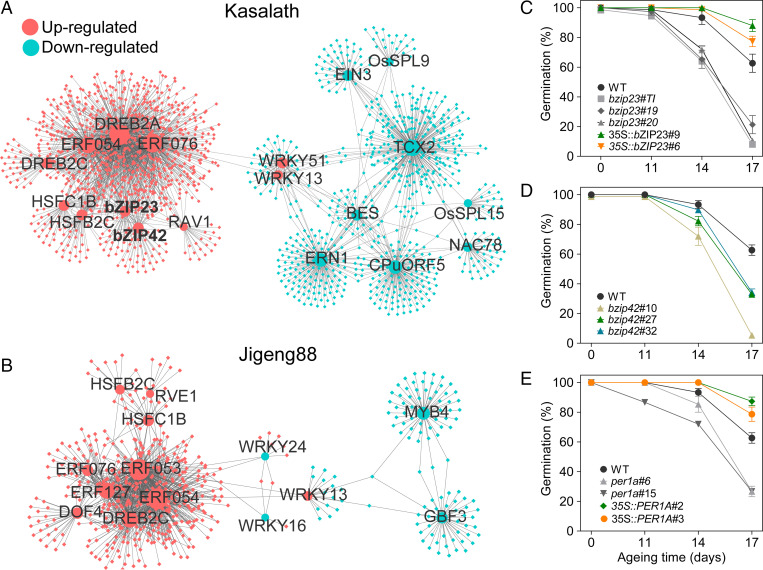
To identify potentially key TFs responsible for the gene differential expression, we searched the plant regulatory data and analysis platform PlantRegMap using the correspondingly DE genes of Kasalath or Jigeng88 as a query. The resultant 21 and 16 TFs that harbored significantly overrepresented targets were identified, respectively, in Kasalath and Jigeng88 seeds (Fig. 2 and Dataset S6). We then constructed a coexpression regulatory network of these TFs and their corresponding targets with a threshold of Pearson correlation > 0.7 or < −0.7. Interestingly, several of the candidate TFs that significantly up-regulated and involved in ABA signaling, including bZIP23 and bZIP42, were exclusively present in the gene networks of Kasalath (Fig. 2 A and B).
Fig. 2.
The identification of gene-regulated seed vigor from the coexpression network of TFs and targeted genes. (A and B) The coexpression network of TFs and targeted genes both DE during Kasalath and Jigeng88 seed aging, respectively. Only TF–gene targets showing Pearson correlation > 0.7 or < −0.7 in expression were projected onto the network. TFs were plotted in different sizes according to the count of their targeted genes. Note that WRKY family TFs were shown as negative regulators for targeted gene expression. The genes in the figure are detailed in SI Appendix, Dataset S6. (C) The change of seed germination of WT, bZIP23 knockout (bzip23#TI, bzip23#19, and bzip23#20), and overexpression transgenic (35S:bZIP23#6 and 35S:bZIP23#9) plants during seed aging. bZIP23 mutations were mediated by both T-DNA insertion (bzip23#TI) and CRISPR/Cas9 (bzip23#19 and bzip23#20). (D) The change of seed germination of WT and bZIP42 CRISPR knockout (bzip42#10, bzip42#27, and bzip42#32) plants during aging. (E) The change of seed germination in WT, PER1A knockout (per1a#6 and per1a#9), and overexpressed (35S:PER1A#2 and 35S:PER1A#3) plants during aging. The seeds were aged under 42 °C and 80% RH for 0, 11, 14, and 17 d and then germinated at 30 °C in darkness. Error bars in C and D indicate ± SE (n = 3).
The bZIP23/42 genes have been characterized as an important genetic factor for conferring ABA-dependent tolerance to abiotic stresses, such as salinity and drought tolerance in rice (33, 34). It was noteworthy that the bZIP family gene ABI5 was reported to be involved in the regulation of seed maturation and longevity in legumes (23). However, so far, whether bZIP23/42 could regulate seed vigor has remained unclear. To verify the biological function of bZIP23, we produced transgenic rice seeds with an altered bZIP23 expression (ZH11 was a transgenic recipient and otherwise will be noted; SI Appendix, Fig. S6A). We observed that, upon accelerated aging, knockouts of the bZIP23 gene (bzip23 T1, #19, and #20; SI Appendix, Fig. S6B) dramatically reduced the completion of seed germination (when aged for 17 d, less than 25% for bzip23 versus 62% for wild type [WT]). By contrast, its overexpression substantially improved seed vigor (Fig. 2C). Similarly, we knocked out bZIP42 using the CRISPR-Cas9 method (SI Appendix, Fig. S7) and observed that the seeds of three independent bZIP42 knockout lines exhibited reduced seed vigor compared to those of WT (Fig. 2D). In addition, we generated the bZIP42 overexpressing plants; however, the seed vigor of these lines did not differ significantly from that of WT (SI Appendix, Fig. S8). We also produced transgenic plants with bZIP23 overexpression in Jigeng88. However, although the corresponding transgenic plants have substantially enhanced expression levels of bZIP23, they can hardly set seeds because of severe sterility (SI Appendix, Fig. S9), indicating that Agrobacterium-mediated transformation might exert differently undesirable side effects on different rice genotypes.
We next asked whether the expression variation of bZIP23/42 positively correlates with seed vigor. To solve this challenge, we investigated the correlation between relative expression levels of bZIP23 with seed vigor among six japonica and five indica cultivars that exhibited great differences of the trait (SI Appendix, Fig. S10A). In particular, the germinations of these cultivars upon aging treatment of 8 d showed obvious differences (SI Appendix, Fig. S10B). Meanwhile, consistent with earlier observation, we observed that the expression of the bZIP23 alleles was up-regulated upon aging treatment with distinct variations among these cultivars (SI Appendix, Fig. S10C). Strikingly, the seed vigor of these cultivars was highly correlated with their relative expression of bZIP23 (R2 = 0.63, P < 0.0001; SI Appendix, Fig. S10D). We also observed that the seed vigor of these cultivars was relatively mild but significantly correlated with their relative expression of bZIP42 (R2 = 0.48, P < 0.0001; SI Appendix, Fig. S10 E and F).
The Peroxiredoxin PER1A Is Also a Positive Regulator of Rice Seed Vigor.
RNA-seq data showed that detoxification was specifically enriched in the C6 cluster of DE transcripts (Fig. 1D) in which the transcripts accumulated abundantly and increased markedly during Kasalath seed aging compared to Jigeng88 (Fig. 1C). Consistently, the metabolome data revealed a similar enrichment of a dozen metabolites involved in cysteine and methionine and GSH metabolism (Fig. 1F). Given that GSH, a major antioxidant in detoxifying ROS (and other toxic substances) was proposed to play a central role in seed germination and longevity (10, 35), our results suggested that a detoxification pathway could play an important role in regulating seed vigor.
We next integrated the transcriptional with the metabolic profile related to GSH biosynthesis and found that core metabolites decreased in their contents, and the majority of those genes encoding the synthetases in the pathway were down-regulated (SI Appendix, Fig. S11A). It was noteworthy that, at the onset of the seed aging treatment, the GSH content of Kasalath was 50% higher than that of Jigeng88 (SI Appendix, Fig. S11A). Thus, the higher GSH stockpile in the Kasalath seeds might endow it with enhanced detoxification ability. We observed that most of genes involved in GSH-dependent and GSH-independent detoxification pathways were significantly up-regulated in Kasalath (SI Appendix, Fig. S11 B–D). Furthermore, we examined the generation of reactive oxygen intermediates (H2O2) to assess the overall oxidative stress. We note that, upon accelerated aging treatment of seeds, the final germination percentage of Kasalath aged for 4 d and of Jigeng88 aged for 2 d did not differ from those of unaged controls (Fig. 1A). However, we observed that the seeds of Jigeng88 had been stained more heavily than those of Kasalath with both the 2′,7′-dichlorofluorescin diacetate (DCFH-DA) (SI Appendix, Fig. S11E) and 3,3′-diaminobenzidine,tetrahydrochloride (DAB) treatment (SI Appendix, Fig. S11F). Accordingly, the measurement of H2O2 concentration found that, relative to those of Kasalath, the embryos of Jigeng88 accumulated higher levels of H2O2 content (SI Appendix, Fig. S11G).
Interestingly, a recent report revealed that the Arabidopsis seeds of ectopic expressions of NnPER1 had improved tolerance to accelerated aging (16). These observations prompted us to pay particular attention to the PEROXIREDOXIN (PRX) family—five of which were DE genes (SI Appendix, Figs. S11D and S12). The rice genome contains 10 members of the PRX genes with diversified spatiotemporal expression patterns, and one of which, PER1A (also called Os1-CysPrxA), was preferentially expressed in the seeds (36). We also observed that PER1A was expressed far more pronounced than the other four identified DE members, with its allelic transcript level of Kasalath stronger than that of Jigeng88 (SI Appendix, Fig. S12). Furthermore, a phylogenetic tree analysis indicated that PER1A was very different in protein sequence from other members (SI Appendix, Fig. S13).
To know the function of PER1A in rice, we produced the transgenic rice overexpressing PER1A under the control of the 35S promoter (SI Appendix, Fig. S14A) and found that overexpressed PER1A seeds had a slower rate of decrease of final percentage germination than that of WT during aging. Accordingly, PER1A knockout through the CRISPR/Cas9 approach (SI Appendix, Fig. S14B) increased the rate of loss of seed viability during aging compared to WT (Fig. 2E). Notably, the seed vigor of the heterozygous per1a mutants did not differ significantly from their homozygous mutants (SI Appendix, Fig. S15), revealing the dominant effect of per1a mutation on seed vigor.
PER1A Is Directly Regulated by the TFs bZIP23 and bZIP42.
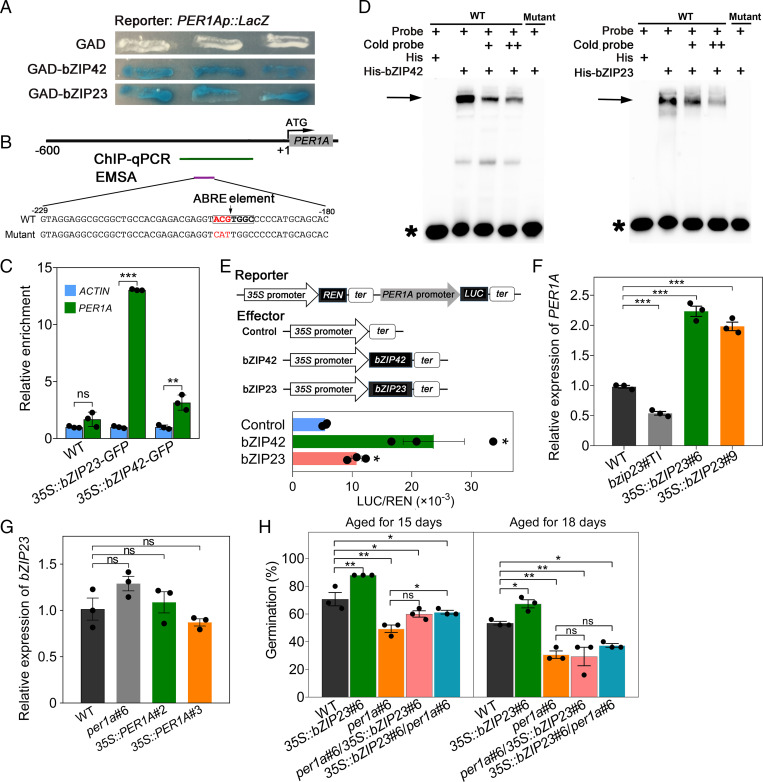
All of bZIP23, bZIP42, and PER1A act as positive regulators of seed vigor. The functional similarity prompted us to assume that PER1A could be under the control of these bZIP TFs. To test this idea, we employed a yeast one-hybrid (Y1H) assay to test whether these TFs could bind to the PER1A promoter. The GAL4 transcriptional activation domain (GAD) fused with the protein bZIP42 (GAD-bZIP42) or GAD-bZIP23 activated the LacZ reporter genes driven by the PER1A promoter but that fused with GAD alone did not (Fig. 3A). We constructed both the native promoter and 35S promoter-driven fusion bZIP23-GFP/Myc and bZIP42-GFP constructs, which were then individually transformed into rice protoplasts as indicated. The following chromatin immunoprecipitation coupled with quantitative polymerase chain reaction (ChIP-qPCR) using an anti-GFP antibody suggested that both bZIP23 and bZIP42 could bind to the PER1A promoter (SI Appendix, Fig. S16). The binding was further validated by examining the binding of bZIP23 or bZIP42 on PER1A fragment in relative to a control of ACTIN fragments in the transgenic 35S::bZIP23-GFP and 35S::bZIP42-GFP rice seeds (Fig. 3C). Consistent with these results, an electrophoretic mobility shift assay (EMSA) revealed that recombinant His-bZIP42 or His-bZIP23 fusion protein, but not His alone, caused an upshift of the biotin-labeled WT PER1A probe but not of the mutant probe (mutating the predicted core sequence “ACGTGGC” of the ABRE element; Fig. 3B); furthermore, the nonlabeled (cold) probe addition could significantly reduce the level of the shift bands (Fig. 3D). Thus, the identified ABRE element confers specific and direct binding for both bZIP42 and bZIP23.
Fig. 3.
The genetic regulation of seed vigor through transactivation of PER1A by bZIP23. (A) The Y1H analysis of the binding of bZIP23 and bZIP42 to the PER1 promoter. The PER1A promoter (−3 to −1,545 bp from the start codon ATG) was inserted upstream of the LacZ reporter gene. bZIP23 and bZIP42 were infused with GAD. Vectors containing GAD along were used as the control. (B) A schematic diagram indicating fragments in the PER1A promoter for ChIP-qPCR and EMSA analysis. The green and purple lines indicate the targeted fragments for ChIP-qPCR and EMSA analyses, respectively. The indicated fragments contain an ABRE element binding by the bZIP family protein (boxed sequence) and were mutated (the red colored sequence) for EMSA assay. (C) The ChIP-qPCR analysis of relative binding of bZIP23 and bZIP42 to PER1A promoter. The anti-GFP IgG affinity bead was used for DNA immunoprecipitation from embryos. The ACTIN fragment was used as an internal control. The relative enrichment was calculated as Input% of the PER1A fragment/Input% of ACTIN fragment. (D) The EMSA identification of bZIP42- and bZIP23-binding element in the PER1 promoter. The arrow indicates the shift bands, and the asterisk indicates the free probe. (E) The transient DLR assay of bZIP23 and bZIP42 binding to PER1A promoter. The constructs of pGreenII 0800-LUC containing the PER1A promoter and of pGreenII 62-SK with or without bZIP23 or bZIP42 were transiently transformed into the rice protoplasts. The LUC/REN ratio represents the relative activity of transactivation. (F) The relative expression of PER1A in seeds of WT, bZIP23 T-DNA insertion (bzip23), and overexpression transgenic (35S:bZIP23#6 and 35S:bZIP23#9) plants. (G) The relative expression of bZIP23 in seeds of WT, CRISPR knockout (per1a), and overexpressed (35S::PER1A#2 and 35S::PER1A#3) plants. (H) The germination of WT, 35S::bZIP23, per1a, 35S::bZIP23/per1a, and per1a/35S::bZIP23 seeds after being aged for different days. The seeds were aged under 42 °C and 80% RH for 15 and 18 d. ns, no significant difference. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test). Error bars indicate ± SE (n = 3).
To further examine whether bZIP42 and/or bZIP23 has an intrinsic transcriptional regulatory activity, we performed a dual-luciferase reporter (DLR) assay; the transient expression in the rice protoplasts of the firefly (Photinus pyralis) LUCIFERASE (LUC) gene driven by the PER1A promoter as a reporter construct and an effector construct containing the bZIP42 or bZIP23 gene driven by the CaMV35S promoter suggested that bZIP42 and bZIP23 could directly bind to the promoter and significantly activate the expression of PER1A (Fig. 3E).
Furthermore, real-time qPCR (qRT-PCR analysis showed that, compared to that in WT, PER1A expression was down-regulated in the seeds of bzip23 and was significantly up-regulated in the bZIP23-overexpressed seeds, whereas in the PER1A-knockout line, bZIP23 expression exhibited no alteration (Fig. 3 F and G and SI Appendix, Fig. S17). However, we further found that, in the seeds of bZIP42 knockout mutants, PER1A expression exhibited no change, and in those of the per1a mutant, the expression of bZIP42 also did not alter (SI Appendix, Fig. S18). Furthermore, to examine the genetic interaction between bZIP23 and PER1A, we crossed the loss-of-function PER1A mutant per1a #6 (male parent) and the bZIP23 overexpressing line (35S::bZIP23 #6; female parent) to obtain 35S::bZIP23 #6/per1a #6 and also crossed 35S::bZIP23 #6 (male parent) and per1a #6 (female parent) to obtain per1a #6/35S::bZIP23 #6. As expected, we observed that the seed vigor of both 35S::bZIP23 #6/per1a #6 and per1a #6/35S::bZIP23 #6 that are hemizygous for the 35S::bZIP23 #6 transgene and heterozygous for the per1a allele were very similar to those of per1a #6 (Fig. 3H and SI Appendix, Fig. S19). Thus, we concluded that PER1A functions in a common genetic pathway with and acts downstream of bZIP23, yet the regulatory relationship between bZIP42 and PER1A is currently complicated.
To know whether bZIP23 and PER1A function in detoxifying H2O2, we examined H2O2 contents in transgenic seeds. We observed that, during accelerated aging, the H2O2 contents in seeds of both the per1a and bzip23 lines were significantly higher than that of WT (SI Appendix, Fig. S20A). Consistently, both the embryos of the per1a and bzip23 seeds were stained by DAB more heavily than that of WT (SI Appendix, Fig. S20B).
The Regulation of Seed Vigor by the bZIP23-PER1A Regulatory Module via the ABA Signaling Pathway.
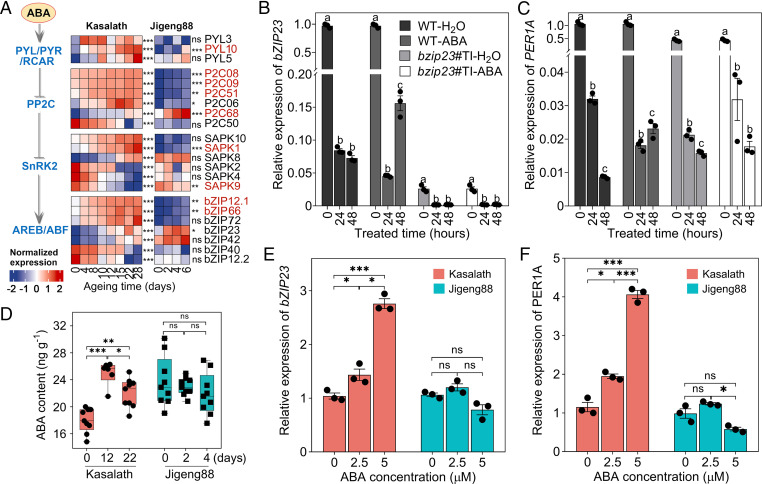
The bZIP23 gene product mediated the sensitivity to exogenous ABA treatment in rice (33). We thus assumed that the ABA signaling pathway might play an important role in controlling seed vigor in rice. Consistent with this assumption, like PER1A, many other genes encoding core players in the ABA pathway, including PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PYL/RCAR), PROTEIN PHOSPHATASE 2C (PP2C), SUCROSE NONFERMENTING-RELATED PROTEIN KINASE 2, and ABA-REPONSIVE ELEMENT-BINDING FACTOR (AREB/ABF), were significantly up-regulated in response to accelerated aging treatment (Fig. 4A). Consistently, our qRT-PCR analysis revealed that a transiently up-regulated expression of bZIP23 was observed 48 h after the treatment of the WT seeds with exogenous ABA; however, the induced bZIP23 expression was not evident in the knockout mutant bzip23 seeds (Fig. 4B); as anticipated, we observed a similar expression pattern of PER1A to that of bZIP23 in response to ABA treatment (Fig. 4C). These evidences supported the conclusion that an ABA-induced signaling pathway is most likely involved in the bZIP23-PER1A–mediated regulation of seed vigor.
Fig. 4.
The involvement of ABA signaling in rice seed longevity. (A) A schematic overview of the ABA signaling pathway alongside DE gene expression during seed aging. ns, no significant difference. *FDR < 0.05; **FDR < 0.01; ***FDR < 0.001 (likelihood ratio test by DEseq2 program). The transcripts showing significant difference in abundance at FDR < 0.01 were considered to be DE. The transcripts DE in both varieties were colored in red. Expression was scaled across all the samples of Kasalath and Jigeng88 (z-score). SnRK2, SUCROSE NONFERMENTING-RELATED PROTEIN KINASE 2. (B and C) The effect of ABA treatment on the expression of bZIP23 and PER1A, respectively, in WT and bzip23 mutant. The seeds were imbibed in water and 5 μM ABA for 0, 24, and 48 h, and the embryos were excised for qRT-PCR analysis of bZIP23 and PER1A expression. Different letters above the boxes indicate a significant difference (P < 0.05, Welch’s ANOVA) among different time points of a given treatment. (D) The change of ABA content during seed aging. The box plots show medians with interquartile (n = 9). ns, no significant difference. *P < 0.05; **P < 0.01; ***P < 0.001 (Welch’s ANOVA test). (E and F) The effect of ABA treatment on the expression of bZIP23 and PER1A, respectively, in Kasalath and Jigeng88 seeds. The seeds were treated with 0, 2.5, and 5 μM ABA for 24 h, and then the embryo was excised for RNA extraction and qPCR analysis. Significant differences were determined as in B. The error bars in B, C, E, and F indicate ± SE (n = 3).
Furthermore, we noticed that the expression of several genes encoding the crucial enzymes in the ABA biosynthesis pathway, including ALDEHYDE OXIDASE and MOLYBDENUM COFACTOR SULFURASE that catalyze the final formation of ABA from abscisic aldehyde, ABSCISIC ACID 8′-HYDROXYLASE for ABA degradation, and BETA-GLUCOSIDASE for de-conjugating ABA–glucose ester, have significantly altered during aging (SI Appendix, Fig. S21). Nevertheless, we observed that endogenous ABA content was substantially elevated at day 12 and then moderately reduced at day 22 during aging in Kasalath seeds but kept relatively constant in Jigeng88 seeds, although the absolute ABA level in Jigeng88 was generally higher than that in Kasalath seeds (Fig. 4D). We thus propose that the dynamic concentration alteration but not the absolute level of ABA acts as a signal to prime the bZIP23-PER1A–mediated signaling pathway that confers rice seeds to survive high temperature and humidity. The results also hint that the sensitivities of the Kasalath and Jigeng88 seeds to ABA treatment might differ. To test this idea, we investigated expression of bZIP23 and PER1A in imbibed seeds of Kasalath versus Jigeng88 treated with a rising concentration of ABA and observed that bZIP23 was substantially up-regulated in Kasalath but not in Jigeng88 (Fig. 4E). Similarly, PER1A was also markedly induced by the rising concentration of ABA treatment (Fig. 4F). Consistent with the results, we observed that the repressive effect on germination by the increased concentration of ABA in Kasalath seeds was much more severe than those of Jigene88 (SI Appendix, Fig. S22). Hence, the seeds of Kasalath were much more sensitive to ABA treatment than those of Jigeng88. Taken together, we conclude that, in response to the aging treatment, the seeds of Kasalath have a sudden surge of ABA level, which activates the bZIP23-PER1A pathway because of sensitivity, yet those of Jigeng88 did not.
Discussion
In the present study, we observed that the seed vigor of indica rice cultivar Kasalath was superior to that of japonica Jigeng88 (Fig. 1A). The following comparative analysis of the transcriptome and metabolome of the two cultivars during seed-accelerated aging revealed that the expression of a number of genes, the products from which function in various biological pathways and metabolism processes, such as detoxification, GA and ABA biosynthesis and signaling pathways, and GSH metabolism, have significantly altered (Fig. 1 and Dataset S3). Ultimately, we identified the PRX family gene PER1A, which encodes a protein involved in detoxification of ROS (24, 37) as a positive regulator of rice seed vigor. Although as a homolog of PER1A, NnPER1 has been characterized to regulate seed vigor in Arabidopsis (16), its regulatory mechanisms remained largely unknown. We here revealed that the AREB factor bZIP23 functions upstream of PER1A and is also a positive regulator of the seed vigor trait (Figs. 2 and 3). In addition, our results also indicated that the bZIP23-PER1A–mediated regulation of seed vigor presumably occurs through the ABA signaling pathway (Fig. 4). We proposed that, in Kasalath seeds (higher vigor), endogenous ABA is initially increased in response to accelerated aging treatment, thereby promoting the AREB factor bZIP23 expression, the protein from which activates the expression of its downstream PER1A target gene and enhances seed vigor. However, in Jigeng88 seeds (lower vigor), the endogenous ABA is unaltered during accelerated aging, and the bZIP23-PER1A–mediated signaling pathway presumably could not act. Nevertheless, we identified the bZIP23-PER1A–mediated detoxification pathway for rice seed vigor regulation and provided potentially useful targets for improving crop quality.
Our results have revealed that the control of seed vigor by ABA in hydrated rice seeds might therefore be connected with the maintenance of an antioxidant scavenging pathway. This cross-talk has often associated with various plant stress responses (38–40). It was worth noting that a series of genes coding for key players in the ABA signaling pathway were up-regulated (Fig. 4A); consistent with this result, we identified several TFs such as DREB2A, DREB2C, bZIP23, and bZIP42 that were involved in ABA signaling, acting as nodes in the transcription regulatory network in Kasalath seeds in response to accelerated aging (Fig. 2A). Coincidentally, the transcriptional expression levels of many genes of both the GSH-dependent and -independent detoxification pathways were significantly altered during accelerated aging (SI Appendix, Fig. S11). More interestingly, we found that ABRE elements are present in the promoters of many DE genes related to detoxification (SI Appendix, Fig. S23 and Dataset S7). We investigated the expression of eight of these genes (SODC1, SODM, GPX4B, GRX10, GRXS9, TRX9, TRX15, and PRX2C) in transgenic rice seeds of bZIP23 and PER1A without aging (0 d) and aged for 14 d. We found that the expressions of most (five in eight) of these ROS-related genes were significantly up-regulated in WT during seed aging (SI Appendix, Figs. S24 and S25). As expected, nearly all (seven in eight) of these genes exhibited higher expressions in the seeds of 35S::bZIP23 than those of WT (SI Appendix, Fig. S24). In contrast, the expressions of these genes did not differ in the seeds of 35S::PER1A from those of WT under without aging condition (SI Appendix, Fig. S25). These results suggest that these detoxification pathways most likely are also under the control of the ABA signaling pathway.
Apart from ABA, GA was also reported to be involved in the regulation of Arabidopsis seed vigor. Overexpression of the zinc finger TF ARABIDOPSIS THALIANA HOMEOBOX 25 increases the transcriptional expression of GIBBERELLIC ACID3-OXIDASE 2 coding for a GA biosynthesis enzyme and hence increases the concentrations of active GA1 and GA4. Plants treated with GA produced seeds with enhanced vigor, and mutations causing the activation (quintuple DELLA mutant) or repression (pre1 mutant) of GA signaling had seeds with increased or decreased seed vigor, respectively (30). Similarly, we observed that several genes coding for crucial enzymes in the GA metabolic pathway such as GIBBERELLIN 20 OXIDASE and GIBBERELLIN 2-BETA-DIOXYGENASE and GIBBERELLIN RECEPTOR 1 were up-regulated in our RNA-seq data. In concurrence with this, the GA1 concentration of the seeds of Kasalath was elevated, whereas that of Jigeng88 was decreased during accelerated aging treatment (SI Appendix, Fig. S26A). We also observed that, upon accelerated aging treatment, only a small number of auxin signaling genes such as IAA1 and IAA3 were up-regulated, whereas most of them, including TIR1, were down-regulated (SI Appendix, Fig. S26B). In agreement with these observations, several lines of evidence have suggested that auxin signaling might also play a role in seed vigor (41–44). The Arabidopsis seed vigor gene ABI3 was induced by auxin and deregulated in auxin biosynthesis mutants (43). In addition, the Helianthus annuus (sunflower) Aux/IAA protein HaIAA27 interacts with the heat shock TF HaHSFA9 and represses the transcriptional activation imposed by the latter. Accordingly, the overexpression of an auxin-resistant form of HaIAA27 resulted in reduced seed vigor comparable to ectopic down-regulation of HaHSFA9 in seeds (41). Moreover, we found that several genes functioning in the brassinosteroid (BR) signaling pathway such as BRI1, BAK1, GSK1, and LIC were significantly down-regulated (SI Appendix, Fig. S26C). Coincidently, prior transcriptomic analysis revealed that genes related to BR biosynthesis and signaling were highly expressed in primed seeds with shorter longevity (45).
However, whether the metabolic and signaling pathways of other plant hormones function to assist in the determination of seed vigor currently remains an interesting open question. Unexpectedly, we observed that the transcription levels of several genes functioning in the ethylene signaling pathway were significantly altered, including ETR4, EBF1, EIL3, and various ethylene responsive factors (SI Appendix, Fig. S26D). In particular, we identified several TFs such as ERF053, ERF054, ERF076, and ERF127 of the ethylene signaling pathway as nodes of transcription regulatory network in Jigeng88 seeds (Fig. 2B). We also observed that an abundance of transcripts encoding various proteins involved in the metabolic and signaling pathways of cytokinin, jasmonic acid, and salicylic acid and contents of these hormones were altered upon seed aging (SI Appendix, Fig. S26 E–G). These results have provided interesting clues for possible associations of these plant hormones with seed vigor, which deserves to test in future experiments.
Detoxification has long been known to associate with seed vigor (11, 12, 19, 35). In this study, we identified the bZIP23-PER1A–mediated detoxification pathway for seed vigor regulation in rice. In addition to PER1A, we identified PER1B in rice, which is very similar in sequence to PER1A; however, upon accelerated aging, we observed that the expression of PER1A, but not that of PER1B, was significantly altered (SI Appendix, Fig. S12). bZIP23 belongs to the class E subclass of the bZIP family (46). To date, eight out of the total of 11 members of the subfamily, including bZIP66/TRAB1 (47), bZIP62 (48), bZIP46 (49), bZIP72 (50), bZIP12/OsABF1 (51), ABI5 (52), bZIP42 (34), and bZIP23 (33), have been characterized to be involved in abiotic stress regulation. Seven of these eight genes, except ABI5, were also reported to regulate the vegetative drought response (33, 34, 47–51). However, their roles in seed vigor have remained elusive. Furthermore, we observed that the cis-acting element ABRE to which bZIP proteins presumably bind was present in the promoters of all the DE PRX genes (SI Appendix, Fig. S23 and Dataset S7). Interestingly, we observed that the expression of several bZIP genes, including bZIP23, bZIP42, bZIP40, bZIP66, and bZIP12, were significantly altered in our RNA-seq data (Fig. 4A). Thus, we cannot exclude the possibility that, in addition to bZIP23-PER1A, other bZIP-PRX combinations might also contribute to the regulation of seed vigor through the PRX pathway, which deserves future investigation.
Materials and Methods
Plant Materials.
Rice varieties Kasalath (O. sativa L. var. indica) and Jigeng88 (O. sativa L. var. japonica) as well as five other indica and six japonica rice varieties (SI Appendix, Table S1) were cultivated in paddy fields of the Institute of Botany in Beijing. The seeds were harvested 45 d after flowering and dried in the sunlight for 1 wk to approximately 9% moisture content on a fresh weight basis before being stored in hermetically sealed plastic bags at −20 °C. Transgenic plants were grown in greenhouse (30 °C with a photoperiod of 14-h light/10-h dark).
Transgenic plants were produced in the background of Zhonghua11. The open reading frame (ORF) complementary DNA (cDNA) segments of PER1A, bZIP23, and bZIP42 were amplified and inserted into binary vectors PHB, pSUPER1300, and pCAMBIA2300, respectively, to construct transgenic overexpression plants. To construct the per1a, bzip23, and bzip42 mutant, single guide RNAs (sgRNAs) corresponding to the second exon of PER1A, the first exon of bZIP23 and the nucleotide fragment that encodes the bZIP domain of bZIP42, respectively, were designed, and the sgRNA expression cassettes were cloned into the pCAMBIA1300 vector. The bzip23 transfer DNA (T-DNA) insertion mutant was kindly provided by L.-Z. Xiong (Huazhong Agricultural University, Wuhan, China). The primers used in this study are listed in SI Appendix, Table S2.
The per1a#6/35S::bZIP23#6 and 35S::bZIP23#6/per1a#6 plants were generated through crossing per1a and 35S::bZIP23#6 with each other. Overexpression of bZIP23 and knockout of per1a were identified for the hybrid plants and then used for a test of seed vigor.
Seed-Accelerated Aging Test.
Seed-accelerated aging was performed as described before (7). The seeds were suspended over 2.5 L saturated NaCl solution (80% relative humidity [RH]) and aged at 45 °C (for seeds of different rice cultivars) or 42 °C (for transgenic seeds) in a sealed desiccator after equilibrating at 20 °C for 3 d. For germination, 25 seeds were imbibed on moistened filter paper at 30 °C in darkness. The completion of germination was scored at day 7 and was defined as when the radical protrudes out of the hull. The embryos of seeds of different rice cultivars were excised immediately after being aged for different times and rapidly frozen in liquid nitrogen and stored at −80 °C until used.
RNA-Seq and Analysis.
A total of 50 mature, desiccated embryos were ground into powder in liquid nitrogen, and total RNA was extracted using a kit (Magen Biotech Co. Ltd.). RNA was evaluated on an Impen NanoPhotometer and Agilent 2100 Bioanalyzer. RNA samples passing a cutoff of RNA integrity number (RIN) ≥ 5 were used for RNA-seq library construction. Paired-end libraries were constructed using a kit (Illumina) and sequenced using an Illumina HiSeq X Ten platform. Reads were trimmed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and mapped to the Nipponbare reference using the Hierarchical Indexing for Spliced Alignment of Transcripts program. The featureCounts was employed to summarize the reads. Differential expression of transcript was analyzed between the unaged seeds of Kasalath and Jigeng88 rice and across all the aging times for each cultivar using the likelihood ratio test in the DESeq2 or quasi-likelihood method in the EdgeR. The significance level (P value) was adjusted using the Benjamini–Hochberg method (53). Transcripts were considered to be DE when the FDR was examined to be less than 0.01 by both EdgeR and DEseq2. PCA and heatmap plotting were performed using R software. Clustering of DE transcripts was performed using the K-means clustering algorithm with manual adjustment.
Functional Enrichment Analysis.
KEGG enrichment was performed using KEGG Orthology Based Annotation System 3.0 (http://kobas.cbi.pku.edu.cn/kobas3/?t=1). The threshold significance of P value uses FDR calibration at 0.05. We manually assigned a gene to a function category according to Bevan et al. (32) and also to gene annotations in the gene ontology or KEGG databases, protein homolog annotation in the Swiss-Prot protein database, or TF annotation in the plantTFDB database (http://planttfdb.cbi.pku.edu.cn/). A hypergeometric test with FDR calibration computing with all the DE genes as background was used for enrichment analysis.
Coexpression Regulatory Network.
The PlantRegMap online tool (http://plantregmap.gao-lab.org/tf_enrichment.php) was employed to predict TFs (FDR < 0.05). Pearson’s correlation coefficient between TF and targeted gene of >0.7 (positive regulation) or <−0.7 (negative regulation) were used as a threshold and visualized using Cytoscape3.6. The PlantCARE web service (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to identify the cis element in the gene promoter.
Metabolite Quantification.
Metabolite quantification was conducted according to Chen et al. (54). A total of 1 g rice seeds was ground into powder with a mixer mill (MM 400, Retsch). A total of 100 mg powder was mixed with 1.2 mL 70% aqueous methanol containing 1 ppm an internal standard of 3,4-Dichloro-phenylalanine and incubated overnight at 4 °C with periodical stirring three times. The extracts were centrifuged at 10,000 × g for 10 min, and the supernatant was filtered using a microporous membrane (SCAA-104, 0.22 μm, ANPEL) before liquid chromatography mass spectrometry analysis.
The sample extracts were analyzed using an ultra-performance liquid chromatography (UPLC) (Shim-pack UFLC SHIMADZU CBM30A) and an electrospray ionization mass spectrum (ESI-MS/MS) system (Applied Biosystems 6500 Q TRAP) as described before (54). The peak area for each metabolite was obtained using the MultiQuant software (v 3.0.3) and normalized and log2 transformed for statistical analysis. VIP scores were extracted from orthogonal partial least squares discriminant analysis using R package MetaboAnalystR (55). Metabolites with VIP ≥ 1 and absolute fold change ≥ 2 between two aging time points were considered as DA. PCA was performed using the prcomp function in R. KEGG pathway analyses were carried out on MBROLE2.0 online services (http://csbg.cnb.csic.es/mbrole2/index.php). An FDR at 0.05 was used to determine significance. Metabolite–metabolite interaction network was predicted using MetaboAnalyst web service (https://www.metaboanalyst.ca) and visualized using Cytoscape 3.6.
Phytohormone Quantification.
A total of 120 mg seed powder was prepared as described earlier for phytohormone extraction and added to 1.2 mL 80% methanol and incubated overnight at 4 °C with periodical stirring three times. The supernatant after centrifuge was evaporated to dryness under a nitrogen gas stream and resuspended in 30% methanol. Phytohormones were quantified using the UPLC-coupled ESI-MS/MS platform and analytical conditions as described earlier. Absolute hormone content was calculated using the standard curves of each standard hormone quantified as nanogram per gram of fresh weight of seeds.
qRT-PCR.
Total RNA was extracted from 50 embryos and reverse transcription performed using a kit (TransGen Biotech). The transcripts were quantified by qRT-PCR in a Bio-Rad CFX96 system using a kit (Kapa Biosystems). The relative expression of messenger RNA was calculated according to Livak and Schmittgen (56) using NADPH (LOC_Os04g40950) as the reference gene. Three biological replications were carried out for each treatment. The primer list can be found in SI Appendix, Table S3.
H2O2 Histochemical Staining and Quantification.
Dehulled seeds were transected, rinsed with distilled water, and imbibed in 2 µM DCFH-DA in 0.1 M phosphate buffered saline buffer (pH7.0) for 30 min or in 0.1% DAB.4HCl (adjusted to pH 6.0 with Na2HPO4) for 3 h at 25 °C in darkness. After being rinsed three times with distilled water, the staining was imaged under a laser-scanning confocal microscope (ZEISS LSM 510 META, excitation 488 nm, emission 500 to 550 nm) and an optical microscope (Nikon DXM1200C). H2O2 content was determined according to Mukherjee and Choudhuri (57). The embryos were ground in 2 mL precooled acetone. Homogenate was centrifuged for 10 min at 15,000 × g. A total of 1 ml supernatant, 0.1 mL titanium reagent, and 0.2 mL ammonium solution was mixed to precipitate the titanium–hydroperoxide complex. After centrifugation at 5,000 × g for 10 min, the precipitate was dissolved in 1 mL 2M H2SO4. H2O2 was measured using an ultraviolet (UV)-visible spectrophotometer (SHIMADZU UV-2450), and the absolute amount was calculated according to a standard curve.
Y1H.
For the Y1H assay, the PER1A promoter sequence and ORF cDNA of putative TFs were amplified using primers listed in SI Appendix, Table S4. The TF segment was cloned into pB42AD, and the promoter segment was cloned into the pLacZi vector.
ChIP-qPCR Assay.
The full-length of bZIP23 and bZIP42 cDNA and promoter sequences were amplified using primers listed in SI Appendix, Table S5 and cloned into an Myc-fused pSUPER1300 or a GFP-fused pCAMBIA2300 vector, respectively. The resultant vectors were transformed into the rice protoplasts extracted from 5-d-old shoots. The protoplasts were suspended in 20 mL ChIP buffer containing 0.4 M sucrose, 10 mM Tris HCl (pH 8.0), 10 mM MgCl2, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), and 1 tablet/50 mL protease inhibitor mixture, cross-linked with 1% formaldehyde under a vacuum, and quenched with 0.125 M glycine. Thereafter, the nuclei were purified through sucrose gradient centrifugation at 14,000 g for 60 min at 4 °C and suspended in the buffer component of 50 mm Tris HCl (pH 8.0), 10 mm ethylene diamine tetraacetic acid, 1% sodium dodecyl sulfate, 1 mm PMSF, 1 mm DTT, and 1 tablet/50 mL protease inhibitor mixture. After sonication, Anti-c-Myc agarose or anti-GFP affinity beads were used for DNA immunoprecipitation for bZIP23 and bZIP42 constructs or empty vector. For the ChIP assay in the seeds, three replicates of 500 embryos of the transgenic 35S::bZIP23-GFP, 35S::bZIP42-GFP or WT seeds were excised and ground into powder in liquid nitrogen. The powder was suspended in the ChIP buffer and treated as the protoplast in the following except for an additional filter of embryo tissues. Monoclonal anti-GFP IgG affinity beads were used for DNA immunoprecipitation. The DNA sample was analyzed by qRT-PCR using primers listed in SI Appendix, Table S5 with a kit (Kapa Biosystems or Foreverstar Biotech). For the ChIP assay in seeds, the ACTIN fragment was used as an internal control. The relative enrichment was calculated as the ratio between Input% of PER1A and ACTIN fragments.
EMSA.
The bZIP23 or bZIP42 ORF cDNA segments were amplified using primers listed in SI Appendix, Table S6 and inserted into the vector pET32a+ to produce a fusion protein with an amino-terminal hexahistidyl tag. The protein was expressed in Escherichia coli BL21 (DE3) cells and purified with HisPur nickel nitrilotriaceate Resin (Thermo Fisher). EMSA was performed twice using the LightShift Chemiluminescent EMSA kit (Thermo). The EMSA bands were visualized using a Tannon-5200 image system.
DLR Assay.
The bZIP23 and bZIP42 ORF cDNA segments were amplified using primers in SI Appendix, Table S7 and inserted into pGreenII 62-SK as an effector, and the PER1A promoter segment was fused into pGreenII 0800-LUC as a reporter; these constructs were cotransformed into rice protoplasts. The activities of firefly LUC and Renilla reniformis LUCIFERASE (REN) used as a normalization control were measured using the DLR Assay System Reagents of Promega Glomax20/20. The binding activity of bZIP23 and bZIP42 to the PER1A promoter was calculated as LUC to REN ratio.
ABA Treatment Assays.
Kasalath and Jigeng88 seeds were imbibed in 8 mL different concentrations of ABA (0, 1, 2.5, and 5 μM) and incubated at 30 °C in darkness. Seed germination was monitored at 12 or 24 h internal. The embryos of the seeds imbibed in 0, 2.5, and 5 μM ABA for 24 h were excised immediately and rapidly frozen in liquid nitrogen and stored at −80 °C until used. WT or the bzip23 mutant was imbibed in 0 and 5 μM ABA at 30 °C in darkness for 0, 24, and 48 h, and the embryos were excised immediately and rapidly frozen in liquid nitrogen and stored at −80 °C until used. Three repeats were performed for each experiment.
Supplementary Material
Acknowledgments
We thank Prof. Y.-H.L. (Institute of Genetic and Developmental Biology, Chinese Academy of Sciences [CAS]), Prof. Q.-Y.B. (Northeast Institute of Geography and Agroecology, CAS), and J.-Z.Y. (China National Rice Research Institute) for sharing the indica and japonica rice materials and Prof. L.-Z.X. (Huazhong Agricultural University) for sharing the bzip23 T-DNA insertion rice, Dr. Y.W. for the RNA-seq data analysis, and Dr. J.-Q.L. for the use of the laser-scanning confocal microscope. This work was supported by grants from the CAS (XDA24010101-2), the National Key Research and Development Program of China (2016YFD0100402), and the National Natural Science Foundation of China (91735302, 91435113, 31471466, and 31770357). W.-Q.W. is supported by a grant from the Youth Innovation Promotion Association, CAS.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026355119/-/DCSupplemental.
Data Availability
Raw RNA-seq data were deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject identification PRJNA637155. All other study data are included in the article and/or supporting information.
References
- 1.Harrington J. F., Drying, storing, and packaging seed to maintain germination and vigor. Seedsmen’s Dig. 11, 16 (1960). [Google Scholar]
- 2.Roberts E. H., “Loss of viability and crop yields” in Viability of Seeds, Roberts E. H., Ed. (Chapman and Hall, London, 1972), pp. 307–359. [Google Scholar]
- 3.Ellis R. H., Hong T. D., Jackson M. T., Seed production environment, time of harvest, and the potential longevity of seeds of three cultivars of rice (Oryza sativa L.). Ann. Bot. 72, 583–590 (1993). [Google Scholar]
- 4.Finch-Savage W. E., Bassel G. W., Seed vigour and crop establishment: Extending performance beyond adaptation. J. Exp. Bot. 67, 567–591 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Leprince O., Pellizzaro A., Berriri S., Buitink J., Late seed maturation: Drying without dying. J. Exp. Bot. 68, 827–841 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Lee J. S., et al. , Variation in seed longevity among diverse Indica rice varieties. Ann. Bot. 124, 447–460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delouche J. C., Baskin C. C., Accelerated aging techniques for predicting the relative storability of seed lots. Seed Sci. Technol. 1, 427–452 (1973). [Google Scholar]
- 8.Hampton J. G., TeKrony D. M., Handbook of Vigour Test Methods (The International Seed Testing Association, Zurich, ed. 3, 1995). [Google Scholar]
- 9.Hendry G. A. F., Oxygen, free radical processes and seed longevity. Seed Sci. Res. 3, 141–153 (1993). [Google Scholar]
- 10.Apel K., Hirt H., Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Bailly C., Active oxygen species and antioxidants in seed biology. Seed Sci. Res. 14, 93–107 (2004). [Google Scholar]
- 12.Kranner I., Minibayeva F. V., Beckett R. P., Seal C. E., What is stress? Concepts, definitions and applications in seed science. New Phytol. 188, 655–673 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Apostol I., Heinstein P. F., Low P. S., Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: Role in defense and signal transduction. Plant Physiol. 90, 109–116 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittler R., ROS are good. Trends Plant Sci. 22, 11–19 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Sattler S. E., Gilliland L. U., Magallanes-Lundback M., Pollard M., DellaPenna D., Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16, 1419–1432 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H. H., et al. , Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J. 88, 608–619 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Clerkx E. J. M., Blankestijn-De Vries H., Ruys G. J., Groot S. P. C., Koornneef M., Genetic differences in seed longevity of various Arabidopsis mutants. Physiol. Plant. 121, 448–461 (2004). [Google Scholar]
- 18.Dekkers B. J. W., et al. , The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 85, 451–465 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Sano N., et al. , Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol. 57, 660–674 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Zhou W., et al. , A matter of life and death: Molecular, physiological, and environmental regulation of seed longevity. Plant Cell Environ. 43, 293–302 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Yoong F.-Y., O’Neill C. M., Penfield S., Temperature during seed maturation controls seed vigour through ABA breakdown in the endosperm and causes a passive effect on DOG1 mRNA levels during entry into quiescence. New Phytol. 232, 1311–1322 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Ooms J., Leon-Kloosterziel K. M., Bartels D., Koornneef M., Karssen C. M., Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (A comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol. 102, 1185–1191 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zinsmeister J., et al. , ABI5 is a regulator of seed maturation and longevity in legumes. Plant Cell 28, 2735–2754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haslekås C., et al. , ABI3 mediates expression of the peroxiredoxin antioxidant AtPER1 gene and induction by oxidative stress. Plant Mol. Biol. 53, 313–326 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Bentsink L., Jowett J., Hanhart C. J., Koornneef M., Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 17042–17047 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C., et al. , Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petla B. P., et al. , Rice PROTEIN l-ISOASPARTYL METHYLTRANSFERASE isoforms differentially accumulate during seed maturation to restrict deleterious isoAsp and reactive oxygen species accumulation and are implicated in seed vigor and longevity. New Phytol. 211, 627–645 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Devaiah S. P., et al. , Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J. 50, 950–957 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Waterworth W. M., et al. , A plant DNA ligase is an important determinant of seed longevity. Plant J. 63, 848–860 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Bueso E., et al. , ARABIDOPSIS THALIANA HOMEOBOX25 uncovers a role for Gibberellins in seed longevity. Plant Physiol. 164, 999–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Z., Sun W., Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of ABSCISIC ACID INSENSITIVE 3. J. Exp. Bot. 66, 4781–4794 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevan M., et al. , Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Xiang Y., Tang N., Du H., Ye H., Xiong L., Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joo J., Lee Y. H., Song S. I., OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 249, 1521–1533 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Roach T., Nagel M., Borner A., Eberle C., Kranner I., Changes in tocochromanols and glutathione reveal differences in the mechanisms of seed ageing under seedbank conditions and controlled deterioration in barley. Environ. Exp. Bot. 156, 8–15 (2018). [Google Scholar]
- 36.Gho Y. S., et al. , Comparative expression analysis of rice and Arabidopsis peroxiredoxin genes suggests conserved or diversified roles between the two species and leads to the identification of tandemly duplicated rice peroxiredoxin genes differentially expressed in seeds. Rice (N. Y.) 10, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K. O., et al. , Rice 1Cys-peroxiredoxin over-expressed in transgenic tobacco does not maintain dormancy but enhances antioxidant activity. FEBS Lett. 486, 103–106 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Xing Y., Jia W., Zhang J., AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Zhang H., et al. , The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice. J. Integr. Plant Biol. 54, 500–510 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Zhang H., et al. , A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 65, 5795–5809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carranco R., Espinosa J. M., Prieto-Dapena P., Almoguera C., Jordano J., Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc. Natl. Acad. Sci. U.S.A. 107, 21908–21913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Q., et al. , ZmDREB2A regulates ZmGH3.2 and ZmRAFS, shifting metabolism towards seed aging tolerance over seedling growth. Plant J. 104, 268–282 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Pellizzaro A., et al. , A role for auxin signaling in the acquisition of longevity during seed maturation. New Phytol. 225, 284–296 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Yuan Z., et al. , OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid and protective substances. Plant Physiol. 186, 469–482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sano N., et al. , RNA-seq using bulked recombinant inbred line populations uncovers the importance of brassinosteroid for seed longevity after priming treatments. Sci. Rep. 7, 8095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nijhawan A., Jain M., Tyagi A. K., Khurana J. P., Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333–350 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobo T., Kowyama Y., Hattori T., A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. U.S.A. 96, 15348–15353 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang S., et al. , A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 19, 260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang N., Zhang H., Li X., Xiao J., Xiong L., Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu G., Gao C., Zheng X., Han B., Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229, 605–615 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Zhang C., et al. , The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 68, 4695–4707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou M., Guan Y., Ren H., Zhang F., Chen F., A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66, 675–683 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 54.Chen W., et al. , Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46, 714–721 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Chong J., Xia J., MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34, 4313–4314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Mukherjee S. P., Choudhuri M. A., Implications of water stress-induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plant. 58, 166–170 (1983). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq data were deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject identification PRJNA637155. All other study data are included in the article and/or supporting information.