Significance
The molecular basis of how the BEN domain–containing gene family regulates chromatin function and transcription remains to be elucidated. We report that BEND3 is highly expressed in pluripotent cells and binds to promoters of genes involved in differentiation. BEND3 regulates the expression of differentiation-associated genes by modulating the chromatin architecture at promoters. We propose that transcription repression mediated by BEND3 is essential for normal development and maintenance of pluripotency.
Keywords: BEND3, differentiation, p21, promoter, transcription repression
Abstract
BEN domain–containing proteins are emerging rapidly as an important class of factors involved in modulating gene expression, yet the molecular basis of how they regulate chromatin function and transcription remains to be established. BEND3 is a quadruple BEN domain–containing protein that associates with heterochromatin and functions as a transcriptional repressor. We find that BEND3 is highly expressed in pluripotent cells, and the induction of differentiation results in the down-regulation of BEND3. The removal of BEND3 from pluripotent cells results in cells exhibiting upregulation of the differentiation-inducing gene expression signature. We find that BEND3 binds to the promoters of differentiation-associated factors and key cell cycle regulators, including CDKN1A, encoding the cell cycle inhibitor p21, and represses the expression of differentiation-associated genes by enhancing H3K27me3 decoration at these promoters. Our results support a model in which transcription repression mediated by BEND3 is essential for normal development and to prevent differentiation.
The function of a protein can be predicted through its amino acid sequence and three-dimensional structure. Both of these attributes mediate the function of a protein by virtue of enabling the binding to specific partners, including other protein factors or nucleic acids. BEN domain–containing proteins, presented widely across metazoans, constitute an important class of proteins involved in gene regulation (1). It is generally believed that BEN proteins act as cellular transcription factors that have been co-opted by viruses. Recent studies have pointed out that several BEN domain–containing proteins function as transcription repressors. These include Drosophila Mod(mdg4) (2), Insensitive (3), mammalian BANP (4–6), NAC1 (7, 8), and BEND3 (9–11). The mammalian genome encodes several BEN domain proteins, but we know little about the functionality of these proteins. Previous work from the Lai laboratory has predicted that >100 annotated BEN proteins may function as transcription factors (12). Yet the molecular basis of how the BEN domain–containing gene family regulates chromatin function and transcription remains to be elucidated.
We previously identified BEN domain–containing protein BEND3, a highly conserved protein among vertebrates that associates with heterochromatic structures and functions as a transcriptional repressor (11). More recently, a proteomic screen identified BEND3 as a factor bound exclusively to gene promoters, indicative of its role in transcriptional regulation (13). We have also previously shown that BEND3 mediates ribosomal RNA (rRNA) gene repression by stabilizing the NoRC, a member of mammalian ISWI-containing chromatin remodeling machines (9). The association of BEND3 with NoRC, NuRD (nuclear remodeling and deacetylase complex), and PRC2 (polycomb repressive complex) complexes in a context-dependent manner has been linked to ribosomal DNA (rDNA) silencing and mediating the switching from constitutive to facultative heterochromatin (14). In addition, BEND3 has also been reported to interact with PICH, a multidomain DNA translocase required for the maintenance of chromosome stability in human cells (15). BEND3 and NoRC play a concerted role in not just rRNA gene repression but also global chromatin organization. However, the physiological context of BEND3-mediated transcription repression remains to be determined.
NTERA2 is a clonally derived, pluripotent human embryonic carcinoma cell line that shares a number of characteristics with human embryonic stem cells and exhibits biochemical and developmental characteristics similar to the cells of the early embryo (16, 17). We find that BEND3 is expressed at relatively high levels in the dividing NTERA2 cells. BEND3 levels decline dramatically upon differentiation of NTERA2 as well as mouse embryonic stem cells. Furthermore, loss of BEND3 in pluripotent NTERA2 resulted in cells exhibiting a prodifferentiation gene expression signature. Genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) analysis revealed that BEND3 occupies gene promoters and represses transcription of differentiation-associated genes. Our results support that BEND3-mediated repression of differentiation-inducing genes including the cell cycle inhibitor CDKN1A is critical to safeguard pluripotency.
Results
Loss of BEND3 Causes Cells to Enter a “Differentiation State.”
Recent work has demonstrated that a BEN domain–containing protein, BEND6, contributes to neural stem cell fate decisions and influences neuronal migration (18). Furthermore, BEND6 functions as a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. This motivated us to evaluate if other BEN domain proteins might influence stem cell fate decisions. We found that BEND3 is expressed at relatively high levels in the NTERA2 embryonal carcinoma cell line (SI Appendix, Fig. S1A). Upon treatment with retinoic acid (RA), NTERA2 cells differentiate into the postmitotic neuron-like lineage (16). We observed that BEND3 levels decline dramatically upon RA treatment (Fig. 1A). Upon differentiation, a reduction in pluripotency markers Oct4 and NANOG (protein and RNA levels) (19) was observed (Fig. 1 A and B). Similar results were observed in mouse embryonic stem (ES) cells in which the BEND3 expression was highest in pluripotent cells and declined upon differentiation (SI Appendix, Fig. S1B). To gain functional insights into the role of BEND3, we depleted BEND3 using multiple independent short hairpin RNAs (shRNAs). The reduction of BEND3 resulted in a decrease in Oct4 and NANOG RNA levels and NANOG protein levels (Fig. 1 C and D). We found that the loss of BEND3 resulted in a prominent reduction in S-phase cells consistent with the loss of cycling cells (Fig. 1E).
Fig. 1.
Loss of BEND3 leads to differentiation state. (A) Western blot showing a reduction of BEND3 after RA-mediated differentiation of NTERA2. Note the reduction in Oct4 and NANOG, the pluripotency markers upon differentiation. β” U2snRNP staining was used as a loading control. The asterisk denotes a cross-reacting band. (B) RNA levels of Oct4 and NANOG upon differentiation. (C) Depletion of BEND3 with different shRNAs (sh60, 61, and 62). Immunoblot of BEND3, Oct4, and NANOG is shown. Tubulin Western blot is shown as a loading control. (D) RNA levels of BEND3, Oct4, and NANOG upon BEND3 KD. (E) Propidium iodide (PI) flow cytometry analysis of BEND3 shRNA-treated cells. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by unpaired Student’s two-tailed t test.
To examine how the loss of BEND3 affects expression of genes involved in cellular differentiation, we performed RNA sequencing (RNA-seq) of wild-type and BEND3-depleted NTERA2 cells (in duplicates, Sh60, Fig. 2A). The heatmap showed the differential expression of 4,495 genes (2,517 up-regulated and 1,978 down-regulated; fold change > 1 and false discovery rate (FDR) < 0.05) in BEND3-depleted cells (Fig. 2A, SI Appendix, Fig. S2A, and Dataset S1). It was striking to note that the down-regulated genes were those involved in DNA replication, chromosome condensation, and cell cycle progression, while the up-regulated genes were those involved in cell–cell interactions (Fig. 2B and Dataset S2). Gene Ontology (GO) analysis within these 4,495 genes in biological processes showed that there were significant changes in differentiation pathways (Fig. 2C, SI Appendix, Fig. S2 B and C, and Datasets S3 and S4). Strikingly, gene set enrichment analyses revealed down-regulation of genes involved in embryonic organ development and up-regulation of genes involved in tissue development (Fig. 2 D and E, SI Appendix, Fig. S2 D and E, and Dataset S4). We observed that the expression of several genes involved in the neuronal differentiation pathway (as noted in Datasets S3 and S4) was significantly altered in the absence of BEND3. Detailed analyses of the pathways that were specifically affected upon the loss of BEND3 showed changes in mitogen-activated protein kinase pathway genes and the p53 pathway, known to play an active role in promoting differentiation in human ES cells (20). These were further validated by qPCR analysis (SI Appendix, Fig. S2 F and G). For example, BDNF and TGFB2, two vital genes of the neuronal lineage, showed significant up-regulation after BEND3 knock down (KD) (SI Appendix, Fig. S2F).
Fig. 2.
Loss of BEND3 is correlated with changes in expression of genes involved in cellular differentiation. (A) Heatmap showing the expression of 4,495 DE genes from RNA-seq (sh60) with fold change > 1 and FDR < 0.05. (B) GO analysis of DE up-regulated and down-regulated genes between control and BEND3 knockdown cells. (C) Top 10 biological processes involved in cell differentiation associated with DE genes between control and BEND3 knockdown cells. (D and E) Gene set enrichment analysis (GSEA) and heatmap showing the expression of developmental genes present in 4,495 DE genes of the BEND3 RNA-seq data. When performing GSEA analysis, only significant pathways were selected and included in the figures. The P values, FDR values, and normalized enrichment score (NES) values can be found in Dataset S4. (F, a) Venn diagram showing overlap genes between 21 d post-RA NTERA2-treated cell (GSE125370) and BEND3 KD RNA-seq. (F, b) Detailed Venn diagram showing the gene expression directionality of the 3,200 overlap genes between 21 d post-RA NTERA2-treated cell (GSE125370) and BEND3 KD RNA-seq. (G) RNA levels of genes (that are up-regulated upon BEND3 KD) during differentiation. *P < 0.05 and **P < 0.01 by unpaired Student’s two-tailed t test.
It is noteworthy that BEND3 expression was high in embryonic stem cells and in induced pluripotent stem cells (iPSCs) (SI Appendix, Fig. S3A). We further compared the altered gene expression in our BEND3 shRNA-treated RNA-seq data (FDR < 0.05, 4,495 genes) with the RNA-seq data set from NT2 cells treated with RA (to induce differentiation; RA21_GSE125370). Of the 4,495 genes differentially expressed (DE) upon BEND3 KD, we found that 3,200 showed overlap with the NT2-RA DE genes (Fig. 2 F, a and Dataset S5). Of these, 2,344 genes showed changes in the same direction (1,362 up and 982 down; Fig. 2 F, b and Dataset S5). Of the common genes, ∼75% changed in the same direction, which was significantly higher than a randomly shuffled set of genes (∼50% of genes would have similar fold change direction) with a P < 0.0001 chi-squared test value. Common genes with positive fold change are involved in cell–cell interactions, and the common genes with negative fold change are involved in the cell cycle, consistent with these cells having activated a differentiation pathway (SI Appendix, Fig. S3 B and C and Dataset S5). GO analyses further corroborated these observations (SI Appendix, Fig. S3 D–G and Dataset S6). Consistent with the above observations, we found that the genes whose expression was induced in BEND3-depleted cells showed an up-regulation upon RA-mediated differentiation (Fig. 2G). Changes in gene expression upon BEND3 loss are attributed to the enhanced differentiation, supporting the model that BEND3 plays a vital role in modulating cellular differentiation.
BEND3 Associates with G-quadruplex–Rich Sequences at Gene Promoters.
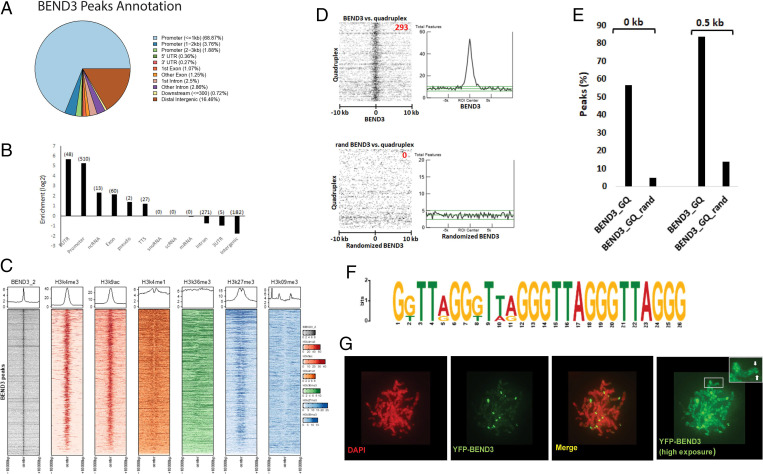
Even though we observed earlier that BEND3 functions as a transcription repressor, its endogenous targets remained to be identified. Toward this, we performed ChIP-seq of BEND3 in NTERA2 cells. ChIP-seq was performed in duplicate with two independent biological replicates. The distribution and chromosomal visualization of the mapped BEND3 peaks showed genome-wide distribution without specific enrichment on a particular chromosome (SI Appendix, Fig. S4 A and B). Strikingly, BEND3 was mostly found to be enriched at the promoters of genes but not so at the introns, exons, and intergenic regions (Fig. 3 A and B, SI Appendix, Fig. S4C, and Dataset S7). The heatmap revealed a strong colocalization of BEND3 occupancy with H3K4me3 and H3K9ac, consistent with BEND3 being at the promoters (Fig. 3C). Similar co-occupancy of histone marks and BEND3 was also obtained in the H1 ES dataset (SI Appendix, Fig. S4E). Similar to our studies, chromatin profiling using ChIP-mass spectrometry to identify proteins associated with genomic regions marked by histones posttranslational modifications at specific lysine residues (H3K27ac, H3K4me3, H3K79me2, H3K36me3, H3K9me3, and H4K20me3) in ES cells revealed enrichment of BEND3 at H3K27ac and H3K4me3, suggesting it is enriched at the promoters (13). BEND3-bound promoters were enriched for H3K27me3 in addition to the active marks (Fig. 3C). BEND3-bound genes in H1 human embryonic stem cells were also enriched with H3K27me3 (SI Appendix, Fig. S4F). We have looked at the distribution of a transcription corepressor Rb with the repressors REST, H3K4me3, and H3K27me3 and find that Rb is also enriched at promoters that have H3K4me3 marks (SI Appendix, Fig. S4D). Our data on BEND3 are consistent with the fact that repressors are found at promoters enriched for active histone marks. BEND3 localizes at regions occupied by active marks at the transcription start site (TSS), whereas it preferentially localizes with H3K27me3 marks on either side of the TSS.
Fig. 3.
BEND3 associates with GQ-rich sequences at promoters. (A) Percentage of BEND3 peaks at promoters and at nonpromoter regions. (B) Enrichment score of BEND3 peaks in multiple genome sites. Annotation enrichment analysis performed by the Homer tool compares the annotation distribution of the input peaks compared to a background set of peaks. The log2 ratios in the figures show the enrichment of different genomic locations between the input peak annotation and background peak set. Positive enrichment values mean that the input peak set was more enriched, while negative enrichment values mean that the background peak set was more enriched. (C) Heatmap showing the colocalization between BEND3 peaks, with different histone marks in NTERA2 cells. (D) Heatmap (Left) and graphs (Right) showing the colocalization between BEND3 peaks (Top) or randomized peaks (Bottom) with quadruplex regions in NTERA2 cells. The randomized genomic file was generated by a script to randomize genomic regions (41). (E) Graph depicting the percentage of colocalization between BEND3 peaks and quadruplex regions. A 0-kb gap and a 0.5-kb gap (details in Materials and Methods) were used for the analysis. (F) Consensus sequence for BEND3 binding identified using a subset of BEND3-bound regions. (G) Localization of YFP-BEND3 at rDNA and telomeres.
Our analyses found that BEND3 bound to the promoters of ∼800 genes. BEND3-ChIP peaks were also significantly enriched within the G-quadruplex (GQ) regions (Fig. 3 D and E). Detailed analyses identified a 26-nt G-rich sequence as the BEND3-interacting consensus sequence (Fig. 3F). Such sequences are reminiscent of those observed in human telomeric GQs, and many gene promoters are known to be enriched for GQ (21).
Consistent with our ChIP-seq experiments, we found that BEND3 also associates with telomeres, structures enriched for the TTAGGG sequence and GQs (Fig. 3G, note the higher exposure). More recently, an independent study confirmed the association of BEND3 in telomeres where it colocalized with telomeric repeat binding factor 2 (TRF2) (22). These results confirm that BEND3 associates with GQs enriched at telomeres and at gene promoters.
BEND3 Represses the Expression of Differentiation-Associated Genes.
Our ChIP-seq experiments revealed that BEND3 associated with the promoters of nearly 800 genes (Fig. 4A, SI Appendix, Fig. S4 G and H, and Dataset S7). We performed an intersection of the ChIP-seq data with the BEND3-depleted cells’ RNA-seq dataset (FDR < 0.05, 4,495 genes) to uncover the direct targets of BEND3 (Fig. 4A and Dataset S7). A total of 275 genes were bound by BEND3 whose expression was altered significantly in the absence of BEND3 (Fig. 4A and Dataset S8). Furthermore, we found that two-thirds of these BEND3-bound genes were associated with GQs. Analyses of these 275 genes revealed differentiation pathways as the most enriched biological process, with DNA binding and SMAD binding as the key molecular functions that were affected (SI Appendix, Fig. S5 A and B and Dataset S9). Out of these 275 genes, we have found that 117 were down-regulated and 158 were up-regulated. Furthermore, >65% of the BEND3-bound genes that are up-regulated upon the loss of BEND3 showed concordant up-regulation in the RA-induced differentiation dataset. Of the 158 up-regulated genes, 83 genes were involved in differentiation. We validated the differential expression of ∼20 of these genes by real-time qPCR (two shRNAs) (Fig. 4C). RT-qPCR validation of the expression of these genes in BEND3 KD cells demonstrated that genes like CDKN1A, TMEM130, CSF1, NRG1, RCAN1, KDM5B, GFPT1, and LRRN1 showed significant up-regulation upon the loss of BEND3 (Fig. 4C). BEND3 was also found to bind to the regulatory sequence of the abovementioned genes (SI Appendix, Fig. S5C). This was further confirmed by the enrichment of chromatin marks H3K4me3 and the transcription factor SP1 used as a promoter reference (SI Appendix, Fig. S5C). The up-regulation of these genes is positively correlated with cellular differentiation, consistent with our observations that BEND3 repressed the expression of genes required for differentiation. Furthermore, overexpression of BEND3 in NT2 cells caused down-regulation of a similar subset of differentiation-associated genes, including CDKN1A, TMEM130, CSF1, KDM5B, GFPT1, and LRRN1 (Fig. 3 D and E). On the other hand, overexpression of BEND3 did not alter the expression of genes involved in cell proliferation (SI Appendix, Fig. S5D). HA-ChIP in HA-BEND3–expressing cells further confirmed the binding of BEND3 to the differentiation-associated gene promoters (SI Appendix, Fig. S5E).
Fig. 4.
BEND3 associates with GQ within the promoter region to regulate gene expression. (A) Venn diagram showing overlap genes (orange) between BEND3 ChIP-seq (yellow) and BEND3 KD RNA-seq (FDR < 0.05, fold change > 1) (red). (B) Top 10 biological processes involved in cell differentiation of 275 overlap genes between BEND3 ChIP-seq and BEND3 KD RNA-seq (FDR < 0.05, fold change > 1). (C) qPCR validation of the expression of BEND3-bound differentiation-associated genes in BEND3 KD cells. (D) RNA level of BEND3 in cells overexpressing BEND3. (E) Validation of differentiation-associated genes by qPCR analysis in BEND3 overexpression (OE) experiment. (F) Luciferase reporter assay showing BEND3 repressive activity on CDKN1A, KDM5B, and GFPT1 promoter and relief of repression in the GQ promoter mutants. GQ mutations are marked with red color. (G) H3K27me3 ChIP-qPCR analysis at differentiation-associated gene promoter regions in cells overexpressing BEND3. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by unpaired Student’s two-tailed t test.
BEND3 bound to the promoters of differentiation-associated genes (shown in SI Appendix, Fig. S5F are CDKN1A, KDM5B, and GFPT1); each of these contain GQ sequences at their promoter, and BEND3 repressed the transcription of these genes in vivo (SI Appendix, Fig. S5F). We have used a luciferase assay to examine the BEND3-mediated repression of the reporter driven by the above promoters (wild-type promoter or the promoter harboring GQ mutation) (23) (Fig. 4F and SI Appendix, Fig. S5F). We observed that BEND3 repressed the luciferase expression driven by CDKN1A, KDM5B, and GFPT1 promoters. However, this repression was relieved when the luciferase expression was driven by a GQ mutant promoter.
To gain insights into the mechanism of BEND3 action at the promoters, we determined the occupancy of histone H3K27me3 and POL II at the promoters of the differentiation-associated genes ANXA5, CDKN1A, CSF1, and TDRD7 in cells overexpressing BEND3. We observed an increase in the H3K27me3 marks at the promoter of these genes upon BEND3 overexpression (Fig. 4G). We had previously reported similar data in which BEND3 promoted a repressive chromatin state at rDNA loci by modulating H3K27me3, thereby repressing rRNA transcription (9). Finally, the increase in the repressive histone mark at the promoter caused POL II to pause at the promoter (SI Appendix, Fig. S5G), suggesting that BEND3 inhibits transcription by inducing repressive chromatin changes at BEND3-bound gene promoters (Fig. 5).
Fig. 5.
Model representing the mechanism behind BEND3 transcriptional repressive activity at promoter regions. We show that BEND3 is required to fine-tune the expression of differentiation-associated genes by modulating the levels of H3K27me3 to BEND3-bound promoters to generate repressive chromatin.
Discussion
The bioinformatically recognized BEN domain (BANP, E5R, and NAC1) is found in proteins in metazoans as well as in viruses. Some of the BEN domain proteins have been implicated to regulate chromatin organization, whereas others act as transcription repressors. Several years ago, our laboratory identified BEND3 (KIAA1553), a protein with four BEN domains, as a heterochromatin-associated factor that regulates heterochromatin maintenance (11). BEN domain 4 was found to be necessary and sufficient for the localization of BEND3 to heterochromatic regions (11). We found that BEND3 could repress transcription and that its SUMOylation was required for its repressive ability. More recently, we demonstrated that BEND3 mediates rRNA gene repression by stabilizing NoRC (9).
A proteomic screen identified BEND3 as a factor binding to gene promoters and supported its role in transcriptional regulation (13). We observed that BEND3 levels are high in cycling cells and, upon differentiation, BEND3 levels decreased dramatically both at the protein and transcript levels. Consistent with these observations, we found that the loss of BEND3 caused the cells to exhibit a pro-differentiation gene expression signature. It is interesting to note that BEND3 binds to its own promoter and is likely to repress its transcription (SI Appendix, Fig. S5 E and H). This may be an important mode of regulation to ensure that BEND3 levels are carefully regulated in the cells. In support of this, an accompanying study confirmed that BEND3 is autoregulatory (24). Evaluating how the loss of BEND3 affects certain sets of genes, we found that, most prominently, the expression of genes involved in differentiation was significantly enhanced in the absence of BEND3. This included several genes involved in the neuronal differentiation pathway. Recent work has demonstrated that another BEN domain–containing protein, BEND6, contributes to neural stem cell fate decisions and influences neuronal migration (18). In that study, the authors demonstrated that BEND6 functions as a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Even though BEN domains are loosely related to one another, studies from the Lai laboratory demonstrated that the knowledge of the Insv, a single BEN domain–containing protein structure, could be used to deduce the importance of base-specific amino acids in other BEN domain proteins (18). Based on these predictions, mammalian BEND5 was found to function as a sequence-specific repressor that regulates neurogenesis, implying that in addition to BEND3, several of the BEN domain–containing proteins play vital roles in differentiation.
We found that BEND3 binds to ∼800 gene promoters, and a subset of these genes showed altered expression upon loss of BEND3, consistent with the notion that BEND3 impacts the expression of these genes directly. BEND3 binds to the promoters of differentiation-associated genes (RCAN1, TMEM130, NRG1, CDKN1A, MAP2, CSF1, CXADR, KDM5B, GFPT1, LRRN1, SLC1A1, and TDRD7). BEND3-depleted cells showed elevated expression of these genes, and the overexpression of BEND3 results in decreased expression of these genes, supporting BEND3’s role as a transcriptional repressor. Prominent among this list was the key cell cycle inhibitor p21 encoded by CDKN1A (25, 26). In the developing mouse embryo, CDKN1A expression is correlated with cell cycle arrest that precedes terminal differentiation in a variety of tissues (27). It is well established that the overexpression of p21 is able to induce cellular differentiation in a variety of normal and tumor cells, and this is mediated by the induction of cell cycle exit (28). P21 functions as a positive or negative regulator of differentiation in a context-dependent manner (29). The expression of p21 is reported to be essential for the survival of differentiating neuroblastoma cells (30). We find that BEND3 binds to the CDKN1A gene promoter and represses its expression, enabling the cells to be active in the cell cycle. Upon loss of BEND3, its repressive ability is abolished, and p21 levels increase in the cell. The binding of BEND3 to the promoters of several differentiation regulators and repressing their transcription demonstrates its pivotal role in safeguarding pluripotency.
We have previously shown that BEND3 is required for rDNA silencing, and it interacts with members of the NuRD complex (11). This complex plays an important role in transcriptional repression (31, 32). In addition to the rRNA genes, the NuRD complex can modulate fundamental physiological processes, including the maintenance of pluripotency (33–36), reprogramming of neural stem cells into iPSCs (37), S-phase progression, and pericentric heterochromatin formation (38). Furthermore, mass spectrometry analyses of BEND3-associated proteins revealed that BEND3 associates strongly with most of the NuRD complex members (14). The NuRD complex is also known to maintain genes in a “poised/bivalent” configuration in embryonic stem cells (ESCs) (39). The association of BEND3 to active (H3K4me3) as well as inactive (H3K27me3) histone marks in NTERA2 cells suggests that BEND3 might associate with “poised/bivalent” promoters in stem cells. We have previously demonstrated that the deregulation of BEND3 causes defects in the cell cycle as well as changes in the chromatin structure in human cells. Furthermore, we have reported that BEND3-overexpressed cells show an increase in repressive marks H3K27me3 and H4K20me3 at the rDNA loci, resulting in down-regulation of rDNA transcription. We find that overexpression of BEND3 results in increased H3K27me3 at the BEND3-bound gene promoters, and this causes pausing of RNA polymerase II at the promoter region (Fig. 5). BEND3 has been implicated in polycomb recruitment and H3K27me3 deposition to specific chromatin to generate repressive chromatin, especially in the absence of H3K9me3 and DNA methylation (14). Furthermore, Ezh2 was found to methylate Elongin A to restrict RNAPII elongation in mouse embryonic stem cells and control differentiation (40). We propose that BEND3 is required to fine-tune the expression of differentiation-associated genes by modulating the levels of H3K27me3, perhaps by recruiting more Ezh2 to restrict elongation and increase Pol II pausing. Future work will entail whether the BEND3/PRC2/NuRD axis governs the repression of genes, regulating cellular differentiation.
Materials and Methods
A detailed description of all the plasmids, primers, shRNA and experimental procedures can be found in SI Appendix, Materials and Methods. Experimental procedures include details of chromatin immunoprecipitation and the dual-luciferase reporter assay. Bioinformatics and statistical analyses of RNA-seq data, ChIP, ChIP-seq, and ChIP-seq analyses are included in SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank Drs. C. Fields, A. Hernandez, R. Hsu, M. Jadaliha, E. Lai, Y. Wang, and You Jin Song for providing reagents and suggestions. This work was supported by NIH (GM132458 and AG065748), a Cancer Center at Illinois (CCIL) Seed Grant and the Prairie Dragon Paddlers, NSF Early-Concept Grants for Exploratory Research (EAGER) (MCB1723008) awards to K.V.P., and NSF (1243372 and 1818286), NIH (GM125196), and CCIL awards to S.G.P.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107406119/-/DCSupplemental.
Data Availability
RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus database (accession nos. GSE171091 and GSE151235, respectively).
References
- 1.Abhiman S., Iyer L. M., Aravind L., BEN: A novel domain in chromatin factors and DNA viral proteins. Bioinformatics 24, 458–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai H. N., Levine M., The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16, 1732–1741 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan H., et al. , Insensitive is a corepressor for Suppressor of Hairless and regulates Notch signalling during neural development. EMBO J. 30, 3120–3133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavithra L., et al. , SMAR1 forms a ternary complex with p53-MDM2 and negatively regulates p53-mediated transcription. J. Mol. Biol. 388, 691–702 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Rampalli S., Pavithra L., Bhatt A., Kundu T. K., Chattopadhyay S., Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25, 8415–8429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sreenath K., et al. , Nuclear matrix protein SMAR1 represses HIV-1 LTR mediated transcription through chromatin remodeling. Virology 400, 76–85 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Korutla L., Degnan R., Wang P., Mackler S. A., NAC1, a cocaine-regulated POZ/BTB protein interacts with CoREST. J. Neurochem. 101, 611–618 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Korutla L., Wang P. J., Mackler S. A., The POZ/BTB protein NAC1 interacts with two different histone deacetylases in neuronal-like cultures. J. Neurochem. 94, 786–793 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Khan A., et al. , BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8338–8343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan A., Prasanth S. G., BEND3 mediates transcriptional repression and heterochromatin organization. Transcription 6, 102–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathyan K. M., Shen Z., Tripathi V., Prasanth K. V., Prasanth S. G., A BEN-domain-containing protein associates with heterochromatin and represses transcription. J. Cell Sci. 124, 3149–3163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Q., et al. , Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev. 29, 48–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji X., et al. , Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl. Acad. Sci. U.S.A. 112, 3841–3846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saksouk N., et al. , Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell 56, 580–594 (2014). Correction in: Mol. Cell 57, 202 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Pitchai G. P., et al. , A novel TPR-BEN domain interaction mediates PICH-BEND3 association. Nucleic Acids Res. 45, 11413–11424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews P. W., Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev. Biol. 103, 285–293 (1984). [DOI] [PubMed] [Google Scholar]
- 17.Pleasure S. J., Lee V. M., NTera 2 cells: A human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J. Neurosci. Res. 35, 585–602 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Dai Q., et al. , The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 27, 602–614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer L. A., et al. , Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain A. K., et al. , p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 10, e1001268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H., Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita T., et al. , Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci. Rep. 3, 3171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain T., et al. , Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci. Rep. 7, 11541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L. Zheng et al., Distinct structural bases for sequencespecific DNA binding by mammalian BEN domain proteins. Genes and Development, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J., The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993). [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Xiong Y., Beach D., Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol. Biol. Cell 4, 897–906 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker S. B., et al. , p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267, 1024–1027 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Warfel N. A., El-Deiry W. S., p21WAF1 and tumourigenesis: 20 years after. Curr. Opin. Oncol. 25, 52–58 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Rowland B. D., Peeper D. S., KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 6, 11–23 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Poluha W., et al. , The cyclin-dependent kinase inhibitor p21 (WAF1) is required for survival of differentiating neuroblastoma cells. Mol. Cell. Biol. 16, 1335–1341 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denslow S. A., Wade P. A., The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433–5438 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Xue Y., et al. , NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Kaji K., et al. , The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8, 285–292 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Zhu D., Fang J., Li Y., Zhang J., Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One 4, e7684 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Berg D. L., et al. , An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie W., et al. , The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. U.S.A. 109, 8161–8166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.dos Santos R. L., et al. , MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell 15, 102–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims J. K., Wade P. A., Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol. Biol. Cell 22, 3094–3102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harikumar A., Meshorer E., Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 16, 1609–1619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ardehali M. B., et al. , Polycomb repressive complex 2 methylates elongin A to regulate transcription. Mol. Cell 68, 872–884.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith O. K., et al. , Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin 9, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus database (accession nos. GSE171091 and GSE151235, respectively).