Significance
Duchenne muscular dystrophy (DMD) is a fatal disorder of progressive body-wide muscle weakness, considered the most common muscular dystrophy worldwide. Most patients have out-of-frame deletions in the DMD gene, leading to dystrophin absence in muscle. There is no cure for DMD, but exon skipping is emerging as a potential therapy that uses antisense oligonucleotides to convert out-of-frame to in-frame mutations, enabling the production of truncated, partially functional dystrophin. Currently approved exon skipping therapies, however, have limited applicability and efficacy. Here, we developed a more economical approach to skip DMD exons 45 to 55 (a strategy that could treat nearly half of all DMD patients) and identified DG9 peptide conjugation as a powerful way to improve exon skipping efficiencies in vivo.
Keywords: Duchenne muscular dystrophy, exons 45 to 55 skipping, exon 51 skipping, minimized multi-exon skipping cocktail, PMO
Abstract
Duchenne muscular dystrophy (DMD) is primarily caused by out-of-frame deletions in the dystrophin gene. Exon skipping using phosphorodiamidate morpholino oligomers (PMOs) converts out-of-frame to in-frame mutations, producing partially functional dystrophin. Four single-exon skipping PMOs are approved for DMD but treat only 8 to 14% of patients each, and some exhibit poor efficacy. Alternatively, exons 45 to 55 skipping could treat 40 to 47% of all patients and is associated with improved clinical outcomes. Here, we report the development of peptide-conjugated PMOs for exons 45 to 55 skipping. Experiments with immortalized patient myotubes revealed that exons 45 to 55 could be skipped by targeting as few as five exons. We also found that conjugating DG9, a cell-penetrating peptide, to PMOs improved single-exon 51 skipping, dystrophin restoration, and muscle function in hDMDdel52;mdx mice. Local administration of a minimized exons 45 to 55–skipping DG9-PMO mixture restored dystrophin production. This study provides proof of concept toward the development of a more economical and effective exons 45 to 55–skipping DMD therapy.
Duchenne muscular dystrophy (DMD) is a fatal, X-linked recessive disorder caused by mutations in the DMD gene that lead to absence of dystrophin in muscle. Dystrophin stabilizes the sarcolemma by bridging cytoskeletal actin to the extracellular matrix via forming a membrane-associated glycoprotein complex (1, 2). Dystrophin loss results in progressive body-wide muscle degeneration, loss of ambulation before the teens, and cardiorespiratory malfunction during the twenties that typically leads to death (3). DMD is present in 19.8 per 100,000 male births and is the most common inherited neuromuscular disorder worldwide (4).
Exon skipping is emerging as a promising therapy for DMD. Most patients (∼70%) have large out-of-frame deletions in DMD (5). Exon skipping is based on the observation that, at least ∼90% of the time, in-frame mutations in DMD give rise to milder phenotypes, as found in Becker muscular dystrophy (BMD) (5, 6). By excluding out-of-frame exons from the DMD transcript using antisense oligonucleotides, exon skipping converts out-of-frame into in-frame mutations, producing truncated but partially functional dystrophin. Four exon-skipping therapies have been approved by the US Food and Drug Administration: eteplirsen/Exondys 51 (Sarepta) (7), golodirsen/Vyondys 53 (Sarepta) (8), viltolarsen/Viltepso (NS Pharma) (9), and casimersen/Amondys 45 (Sarepta) (10). All are phosphorodiamidate morpholino oligomer (PMO) antisense oligonucleotides.
The applicability of single-exon skipping is, however, limited due to its mutation-specific nature. The therapies above could each treat at most only 8 to 13% of all patients (11). Multiexon skipping overcomes this issue, particularly skipping exons 45 to 55. Exons 45 to 55 is a mutation hotspot in the DMD gene, harboring 66% of large (≥1 exon) deletions and 15% of large duplications in patients (5). Exons 45 to 55 skipping could theoretically treat more than 40% of all DMD patients (12). An exons 45 to 55 deletion is also commonly associated with asymptomatic to mild phenotypes. In the Leiden DMD database, 90% of patients with the deletion have BMD; in other databases, including UMD-TREAT-NMD (Universal Mutation Database-Translational Research in Europe for the Assessment and Treatment of Neuromuscular Diseases), and clinical studies, the deletion is associated with BMD or asymptomatic individuals in all examined cases (12 to 15). Previous works from our group targeted each exon in exons 45 to 55 for skipping. Using in silico tools to minimize detrimental interactions between PMOs, we developed exons 45 to 55–skipping mixtures that restored dystrophin synthesis in the muscles of dystrophic mice (16). With similar approaches, we developed a PMO mixture that skipped human DMD exons 45 to 55 in immortalized patient myotubes and humanized DMD mice (13, 17). Average skipping efficacies of 27 to 61% and 15 to 22% were observed, respectively, and treatment produced up to 14% dystrophin of normal levels in vitro (13). However, this mixture used one PMO per exon (except exon 48, which required two) to skip exons 45 to 55. As all PMOs have to be present in the same nuclei to induce exon skipping, using fewer PMOs may improve efficacy and lower the risk of off-target effects. Moreover, using a large number of PMOs is not economically favorable given how expensive PMOs are. We aim to reduce the number of PMOs required to skip exons 45 to 55, an objective also inspired by previous observations, where skipping a target exon leads to the skipping of an adjacent nontarget exon; e.g., when skipping dystrophin exons 6 to 8, we tend to see exon 9 spontaneously skipped (18).
Another issue associated with exon-skipping therapies is their efficacy. In the case of PMOs, this is tied to their rapid clearance from the bloodstream, poor uptake into muscle, and inability to escape from endosomes (19, 20). Eteplirsen, for instance, only restored 0.93% dystrophin of normal levels in patients after 180 wk of treatment with a once-weekly 30 or 50 mg/kg dose (7). One solution is the conjugation of cell-penetrating peptides to PMOs (20). By enhancing PMO uptake in vivo, these peptides have been widely documented to improve exon skipping efficiencies and treatment outcomes in preclinical trials. Using a zebrafish reporter, we identified DG9 as a promising, novel PMO peptide conjugate that induced strong exon skipping in skeletal muscle and higher skipping in the heart (21). Here, we test the efficacy of DG9-conjugated exon skipping PMOs (DG9-PMOs) in dystrophic hDMDdel52;mdx mice, which have an integrated human DMD transgene in chromosome 5, with an out-of-frame partial deletion of exon 52 (22, 23). These mice also have the mdx mutation, a nonsense point mutation in exon 23 of the mouse Dmd gene (24), and so do not have detectable human or mouse dystrophin. This genetic configuration makes the model excellent for testing the efficacy of human sequence-specific exon-skipping PMOs in vivo. We first evaluate the efficacy of systemic DG9-PMO treatment in this model using a single-exon (exon 51)–skipping approach. We then apply the DG9 modification to the minimized exons 45 to 55–skipping mixture developed in this study, and determine its potential as a multiexon-skipping therapy for DMD.
Results
Minimizing the Exons 45 to 55–Skipping Mixture.
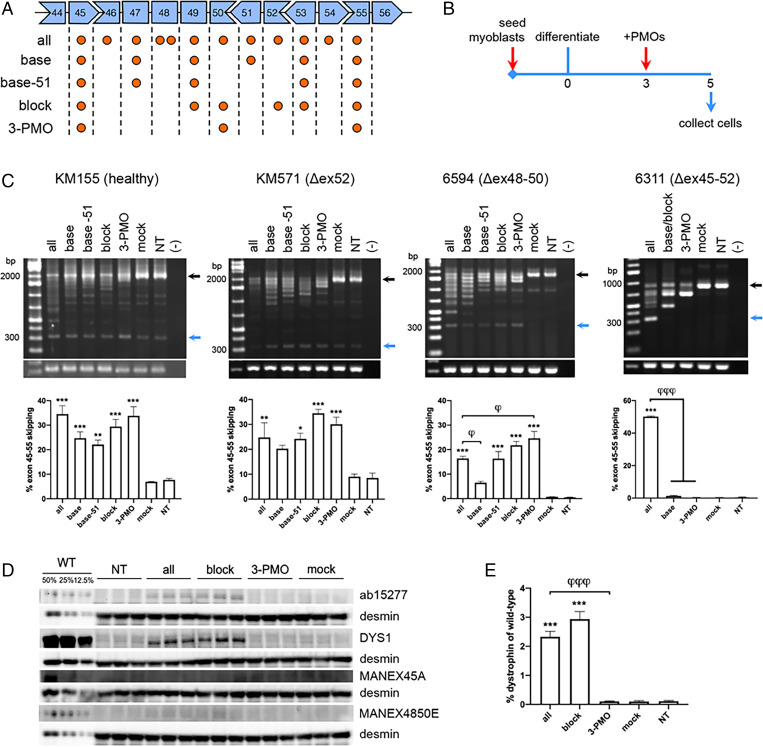
We minimized the number of PMOs to skip DMD exons 45 to 55 using two strategies. First, we took our previously developed exons 45 to 55–skipping mixture (13) (SI Appendix, Table S1) and prepared derivatives where we each removed a PMO (or PMOs) that targeted an exon in the region. We refer to the full mixture, with PMOs targeting all exons within exons 45 to 55, as the “all” mixture (Fig. 1A). Upon transfecting the “all” mixture and its derivatives into healthy immortalized patient-derived KM155 myotubes (Fig. 1B), only the “all” mixture skipped exons 45 to 55 significantly higher than the mock control (P < 0.005) (SI Appendix, Fig. S1A). PMOs whose absence led to a considerable drop in skipping were kept as part of the minimized mixture, i.e., those targeting exons 45, 47, and 53. We also decided to retain PMOs targeting exons 49, 51, and 55 based on their position, to presumably keep the mixture working at an appreciable efficiency. This minimized mixture was called the “base” mixture, and was subjected to another round of minimization in immortalized exon 52–deleted patient-derived KM571 myotubes. The “base” mixture showed significant exons 45 to 55 skipping (P < 0.05) compared to the mock, as well as its derivatives where exon 47 (P < 0.05) or 51 (P < 0.05) were not targeted (SI Appendix, Fig. S1A). We decided to move forward with the “base” and “base −51” mixtures since these showed the highest skipping efficiencies in this batch (Fig. 1A).
Fig. 1.
Testing minimized exons 45 to 55–skipping mixtures in immortalized patient myotubes. (A) The DMD exons targeted by the “all” mixture and its derivatives are indicated by orange circles. (B) Culture scheme for PMO transfection in immortalized patient myotubes. (C) RT-PCR exons 45 to 55–skipping efficiency results upon transfection of PMO mixtures in KM155, KM571, 6594, and 6311 myotubes. Black arrows indicate native/unskipped bands; blue arrows indicate exons 45 to 55-skipped bands. GAPDH is shown below as a control. Quantification is shown at the Bottom. (D) Western blot for dystrophin in PMO-treated and nontreated (NT) KM571 myotubes using various antibodies (ab15277, DYS1, MANEX45A, MANEX4850E). Protein extracts were loaded at 40 μg for treated and NT samples, and at indicated percentages of this for wild-type KM155 samples (WT). Desmin was detected as a loading control. (E) Quantification of DYS1 signals in (D), relative to the intensity of the 12.5% WT band (n = 3 for C–E). Error: SEM *P < 0.05, **P < 0.005, ***P < 0.001, one-way ANOVA with Dunnett’s test versus mock, φP < 0.05, φφφP < 0.001 one-way ANOVA with Dunnett’s test versus “all.”
For the second strategy, we prepared minimized mixtures based on the endogenous splicing of DMD exons 45 to 55. A model proposes that exons 45 to 49, 50 to 52, and 53 to 55 are spliced first, after which these groups are spliced together to complete the exons 45 to 55 region (25). We designed derivative mixtures from the “all” mixture with PMOs targeting the terminal exons of these three groups. Transfection into healthy KM155 myotubes revealed three derivative mixtures to skip exons 45 to 55 significantly higher than the mock (P < 0.05) (SI Appendix, Fig. S1B), with skipping efficiencies not significantly different from the “all” mixture. Based on this, we selected mixtures targeting exons 45, 49, 50, 52, 53, and 55 (“block” mixture) as well as targeting exons 45, 50, and 55 (“3-PMO” mixture) for further experiments (Fig. 1A).
Minimized Exon-Skipping Mixtures Effectively Skip Exons 45 to 55 in Various Patient Cells.
The “all,” “base,” “base -51,” “block,” and “3-PMO” exons 45 to 55–skipping mixtures were subsequently tested in the following immortalized patient-derived muscle cell lines: KM155 (healthy), KM571 (Δex52), 6594 (Δex48-50), and 6311 (Δex45-52) (Fig. 1A). Mutation-tailored versions of the mixtures were used for each cell line, with unnecessary PMOs removed. Upon transfection into myotubes (Fig. 1B) and RT-PCR analysis, the “all” mixture significantly skipped exons 45 to 55 in all lines compared to the mock (P < 0.005 or P < 0.001) (Fig. 1C). The “base -51,” “block,” and “3-PMO” mixtures induced significant exons 45 to 55 skipping in KM155, KM571, and 6594 myotubes compared to the mock (at least P < 0.05); the “base” mixture only showed significant skipping in KM155 cells (P < 0.001). In 6594 myotubes, the “3-PMO” mixture induced significantly higher skipping than the “all” mixture. Intriguingly, none of the minimized mixtures showed any exons 45 to 55 skipping in 6311 myotubes.
We then evaluated the dystrophin restoration capabilities of the “block” and “3-PMO” mixtures in KM571 myotubes, since these mixtures induced the highest levels of exons 45 to 55 skipping in this line. Western blot using antibodies against the rod (DYS1, exons 26 to 30) and C-terminal domains (ab15277) of dystrophin detected dystrophin upon treatment with the “all” and “block” mixtures, but not with the “3-PMO” mixture (Fig. 1D). Quantification of the DYS1 signal showed that “block” mixture treatment restored an average 2.94% dystrophin of wild-type levels, slightly higher than the average 2.33% dystrophin restored by the “all” mixture and elevated compared to the 0.10% dystrophin level in the mock (P < 0.001) (Fig. 1E). Western blot with the MANEX45A (corresponding to exons 45 to 46) and MANEX4850E (corresponding to exons 48 to 50) antibodies did not detect dystrophin in treated myotubes (Fig. 1D), suggesting that the dystrophin-induced posttreatment likely came from exons 45 to 55–skipped transcripts.
Single Intravenous Treatment with DG9-PMO Induces Higher Dystrophin Production than Unconjugated PMO in the Skeletal Muscles and the Heart.
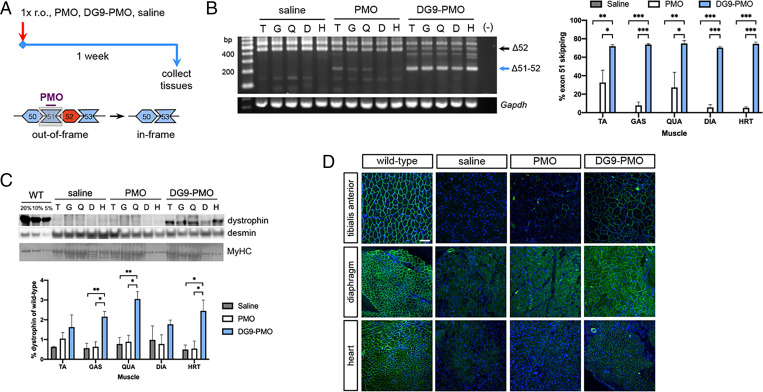
To evaluate DG9 as a conjugate for our minimized exons 45 to 55–skipping mixture, we decided to first test its efficacy in vivo as applied to single-exon skipping. DG9 was conjugated to Ex51_Ac0, an exon 51–skipping PMO from the “all” mixture that was up to 7 times more effective than eteplirsen in restoring dystrophin production in vitro (13, 26). hDMDdel52;mdx mice at 3 mo were given a single retro-orbital injection of either saline, 50 mg/kg unconjugated PMO, or 64 mg/kg DG9-PMO (equimolar to the PMO dose) and assessed a week later (Fig. 2A). DG9-PMO–treated mice had significantly higher levels of exon 51 skipping than the saline- or PMO-treated groups across various skeletal muscles (at least P < 0.05) and the heart (P < 0.001) (Fig. 2B). DG9-PMO induced 2.2- to 12.3-fold higher skipping in skeletal muscles and 14.4-fold higher skipping in the heart on average compared to unconjugated PMO. DG9-PMO treatment significantly restored dystrophin production compared to the saline (P < 0.005) and PMO (P < 0.05) treatments in the gastrocnemius and quadriceps, reaching up to 3% of wild-type levels (Fig. 2C). In the heart, DG9-PMO restored an average 2.5% dystrophin of wild-type levels, significantly higher than that in the saline or PMO groups (P < 0.05). Compared to PMO-treated mice, mice that received DG9-PMO had 1.5- to 3.4-fold and 4.5-fold higher dystrophin protein levels on average in the skeletal muscles and heart, respectively.
Fig. 2.
Single-dose exon 51 skipping treatment with DG9-PMO. (A) Male, 3-mo-old hDMDdel52;mdx mice were given a single retro-orbital injection (1× r.o.) of saline, 50 mg/kg PMO, or equimolar 64 mg/kg DG9-PMO for exon 51 skipping. Tissues were collected 1 wk later. (B) RT-PCR exon 51–skipping efficiency results posttreatment in various muscles, with quantification on the Right. Gapdh is shown as a control. (C) Western blot for dystrophin (DYS1), with wild-type (WT) shown for reference. Protein extracts were loaded at 40 μg for saline-injected and treated muscles, and at indicated percentages of this for WT tibialis anterior samples. Desmin and myosin heavy chain (MyHC) serve as loading controls. Quantification of dystrophin signals are shown relative to the intensity of the 5% WT band. (D) Representative immunofluorescence images for dystrophin (DYS1, green) and nuclei (DAPI, blue) in various muscles and treatment conditions. Scale bar: 100 μm (n = 3/group for B–D). Error: SEM *P < 0.05, **P < 0.005, ***P < 0.001, one-way ANOVA with Tukey’s test. TA/T, tibialis anterior; GAS/G, gastrocnemius; QUA/Q, quadriceps; DIA/D, diaphragm; HRT/H, heart.
Widespread dystrophin-positive fibers were observed in the tibialis anterior, diaphragm, and heart of DG9-PMO–treated mice, and very few to none in saline- or PMO-treated mice (Fig. 2D). However, histological analysis revealed no reductions in the percentage of centrally nucleated fibers (CNFs, a marker of cumulative muscle regeneration) in the tibialis anterior and diaphragm (SI Appendix, Fig. S2 A and B), or any improvements in muscle fiber size (SI Appendix, Fig. S2 A, C, and D) after PMO or DG9-PMO treatment.
Repeated Intravenous Treatment with DG9-PMO Improves Dystrophin Production, Muscle Function, and Fiber Size in Dystrophic Mice.
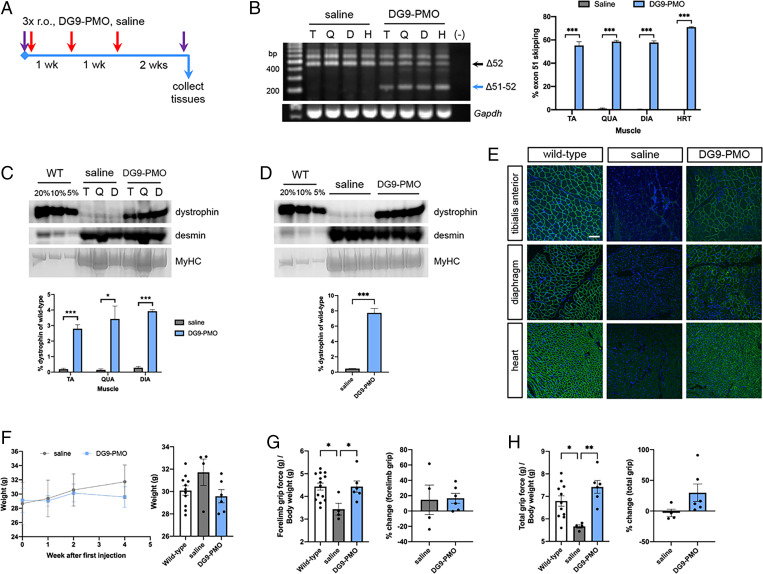
A repeated-dose treatment study was performed to provide more insight into the efficacy of DG9-PMO exon-skipping therapy with regard to ameliorating dystrophic symptoms. hDMDdel52;mdx mice at 2 mo were retro-orbitally injected with saline or 30 mg/kg of DG9-PMO, once weekly for 3 wk. Functional assessments were done at baseline and at 2 wk following the last injection, after which tissues were collected (Fig. 3A). Once again, DG9-PMO treatment significantly induced exon 51 skipping at high levels across skeletal muscles and the heart (55 to 71% on average) compared to the saline control (P < 0.001) (Fig. 3B). This resulted in significant dystrophin production in various skeletal muscles ranging an average of 2.8 to 3.9% compared to 0.1 to 0.3% in saline-treated mice (at least P < 0.05) (Fig. 3C), as well as in the heart at an average of 7.7% versus 0.5% in the saline group (P < 0.001) (Fig. 3D). Immunofluorescence confirmed the presence of widespread dystrophin-positive fibers in the tibialis anterior, diaphragm, and heart (Fig. 3E).
Fig. 3.
Repeated-dose exon 51 skipping treatment with DG9-PMO. (A) Male, 2-mo-old hDMDdel52;mdx mice were given three once-weekly retro-orbital injections (3× r.o.) of saline or 30 mg/kg DG9-PMO for exon 51 skipping. Purple arrows indicate functional testing. Tissues were collected 2 wk later. (B) RT-PCR exon 51–skipping efficiency results posttreatment in various muscles, with quantification on the Right. Gapdh is shown as a control. (C) Western blot for dystrophin (DYS1) in various skeletal muscles or (D) the heart, with corresponding wild-type (WT) samples used for reference. Protein extracts were loaded at 40 μg for saline-injected and treated muscles, and at indicated percentages of this for WT. Desmin and myosin heavy chain (MyHC) serve as loading controls. Quantification of dystrophin signals are shown relative to the intensity of the 5% WT band (n = 3/group for B–D). Error: SEM *P < 0.05, ***P < 0.001, unpaired two-tailed t test for (B–D). (E) Representative immunofluorescence images for dystrophin (DYS1, green) and nuclei (DAPI, blue) in various muscles and conditions. Scale bar: 100 μm (n = 3/group). (F) Body weights of saline- and DG9-PMO–treated mice throughout the experiment. (G) Forelimb grip strength for saline- and DG9-PMO–treated mice, normalized to body weight. The percent change from baseline is on the right. (H) Similar to (G), but for total limb grip strength. (n = 11 to 14, WT; n = 4, saline; n = 6, DG9-PMO for F–H) Error: SEM *P < 0.05, **P < 0.005, one-way ANOVA with Tukey’s test for (G and H). TA/T, tibialis anterior; QUA/Q, quadriceps; DIA/D, diaphragm; HRT/H, heart.
Body weights between groups did not significantly differ over the course of treatment (Fig. 3F). Most impressively, repeated DG9-PMO treatment significantly improved forelimb (P < 0.05) (Fig. 3G) and total limb (P < 0.005) (Fig. 3H) grip strength in hDMDdel52;mdx mice, such that they were not significantly different from wild-type controls. Nearly all mice had improved forelimb/total limb grip strength from baseline, except one which showed a −0.3% difference in forelimb grip strength posttreatment. This is in contrast to the saline controls, which showed no direction of change (Fig. 3G) or no change from baseline (Fig. 3H). Rotarod and treadmill tests showed similar improvements; in particular, the treadmill distance traveled significantly improved from baseline in treated versus saline control mice (SI Appendix, Fig. S3 A and B).
Histological analysis still did not show any significant reductions in CNFs in the tibialis anterior and diaphragm (SI Appendix, Fig. S3 C and D). However, we did observe a significant increase in fiber size with DG9-PMO treatment (P < 0.001) (SI Appendix, Fig. S3 E and F), with most fibers having a minimum Feret’s diameter of 45 to 50 μm in the tibialis anterior and 25 to 30 μm in the diaphragm. This is in contrast to most fibers having diameters of 30 to 35 μm and 20 to 25 μm in saline control muscles, respectively. We also performed a qualitative histological analysis of the liver and kidney in single- and repeated-dose treatment mice, but found no evidence of toxicity due to PMO or DG9-PMO treatment (SI Appendix, Fig. S4 A and B).
Local Treatment with the DG9-Conjugated Minimized Exons 45 to 55–Skipping Mixture Induces Successful Skipping and Dystrophin Production.
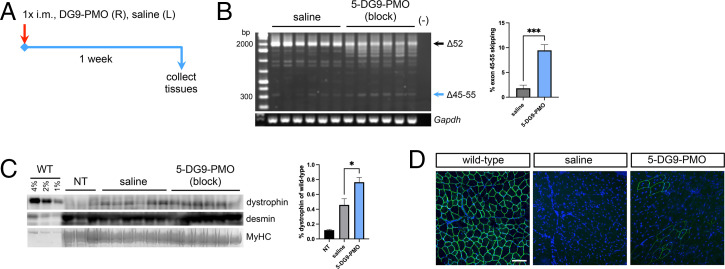
Finally, we conjugated DG9 to each PMO of the minimized “block” exons 45 to 55–skipping mixture for in vivo testing. The “block” mixture was chosen as it induced high levels of exons 45 to 55 skipping and dystrophin protein restoration in KM571 myotubes (Fig. 1 C to E). Thus, 5- to 6-mo-old hDMDdel52;mdx mice were intramuscularly injected in the tibialis anterior with either saline or the mutation-tailored DG9-conjugated version of the “block” mixture (five DG9-PMOs, 5 μg/DG9-PMO, 25 μg total dose) and assessed a week later (Fig. 4A). Significant exons 45 to 55–skipping was observed compared to the saline control (P < 0.001) (Fig. 4B). Western blot revealed dystrophin restoration in DG9-PMO–treated muscles at 0.76% of wild-type levels on average, significantly higher than that in saline-treated muscles at 0.46% (P < 0.05) (Fig. 4C). As the same mouse received DG9-PMO in one leg and saline in the contralateral leg, we included protein from a nontreated mouse to account for possible leakage between legs. Nontreated tibialis anterior muscles had 0.12% dystrophin of wild-type levels, which was lower than the level in the saline control. A few scattered dystrophin-positive fibers in DG9-PMO–treated muscles were observed, and very few to none in the saline controls, confirming dystrophin restoration (Fig. 4D).
Fig. 4.
Local treatment with the minimized “block” DG9-PMO exons 45 to 55–skipping mixture. (A) Male 5- to 6-mo-old hDMDdel52;mdx mice were injected with the DG9-conjugated “block” mixture at 5 μg/DG9-PMO (25 μg total dose) in the right tibialis anterior (R), and saline in the left (L). Tissues were collected 1 wk later. (B) RT-PCR exons 45 to 55–skipping efficiency results posttreatment, with quantification on the Right. Gapdh is shown as a control. (C) Western blot for dystrophin (DYS1), with wild-type (WT) and nontreated (NT) tibialis anterior samples used for reference. Protein extracts were loaded at 40 μg for NT, saline-treated, and DG9-PMO–treated muscles, and at indicated percentages of this for WT. Desmin and myosin heavy chain (MyHC) serve as loading controls. Quantification of dystrophin signals are shown relative to the intensity of the 1% WT band. (D) Representative immunofluorescence images for dystrophin (DYS1, green) and nuclei (DAPI, blue). Scale bar: 100 μm (n = 3, wild-type; n = 6, saline; n = 6, DG9-PMO). Error: SEM *P < 0.05, ***P < 0.001, unpaired two-tailed t test. i.m., intramuscular injection.
Discussion
We have successfully developed a minimized exons 45 to 55–skipping mixture that induces dystrophin restoration in immortalized patient cells and dystrophic hDMDdel52;mdx mice. For exon 52–deleted DMD transcripts, the number of PMOs used for exons 45 to 55 skipping was reduced from 11 in the “all” to 5 in the “block” mixture (Fig. 1A), a more than 50% decrease in PMO content. While most minimized mixtures significantly skipped exons 45 to 55 in KM571 (Δex52) and 6594 (Δex48-50) myotubes, apparently all remaining exons have to be targeted in 6311 myotubes (Δex45-52). Since we only have an idea of endogenous exons 45 to 55 splicing in normal DMD transcripts (25), studies on how this occurs in mutant transcripts are warranted. The “block” mixture was identified as a promising candidate for in vivo study, which restored dystrophin production to ∼3% of wild-type levels, near the amount seen with the “all” mixture (Fig. 1E). Intriguingly, despite showing strong exons 45 to 55 skipping, treatment with the “3-PMO” mixture did not restore dystrophin in KM571 myotubes (Fig. 1 D and E). We do not know why, but it is possible that the exons 45 to 55–skipped transcript produced by the “3-PMO” mixture either was unstable or may not have been the predominant skipping product. In the latter scenario, splicing in the region may have been perturbed such that adjacent exons outside of exons 45 to 55 were skipped out as well. Further experiments will be done to elucidate the reasons behind this result.
We were likewise successful in demonstrating the efficacy of conjugating DG9 to PMOs in single- and multiexon skipping applications. DG9 is a cell-penetrating peptide based on the protein transduction domain of the human Hph-1 transcription factor. Fusion proteins containing this Hph-1 domain have improved delivery in a wide variety of tissues, including the heart (27); DG9 is comprised of two such Hph-1 domains. Treatment with DG9-PMO resulted in higher single-exon skipping and dystrophin restoration levels in vivo compared to unconjugated PMO (Fig. 2 B and D), similar to what has been extensively observed for other cell-penetrating peptides (20, 28). Repeated treatment resulted in greater dystrophin restoration in all examined tissues, especially the heart, from 2.5 to 7.7% of wild-type levels (Figs. 2C and 3D).
The advantage of DG9 is in its potentially better toxicity profile compared to other peptides. Peptide-conjugated PMOs have induced dose-dependent toxic effects in preclinical studies, including lethargy, weight loss, and kidney damage (20). This is linked to the membrane-disruptive properties of cell-penetrating peptides, which are largely influenced by their amino acid compositions (20, 29). Certain L-arginine residues in DG9 were converted to D-arginine, as this improves the viability of peptide-conjugated PMO-treated cells in vitro (30). DG9 also does not contain any 6-aminohexanoic acid residues, which have been associated with increased toxicity (30). Compared to DG9, previous peptides mostly used L-arginine and contained multiple 6-aminohexanoic acid residues, but were of similar length and overall arginine content (20). Even though the above modifications decrease the antisense activity of peptide-conjugated PMOs, it comes with the benefit of increased safety. A balance must be struck between efficacy and safety for peptide-conjugated PMOs, as too frequent or too high doses increase toxicity (20, 29). We did not find any evidence of toxicity caused by DG9-PMO on the liver and kidney (SI Appendix, Fig. S4). However, we plan to undertake a more extensive study of the pharmacokinetics and safety of DG9 versus other published peptide conjugates in the future.
On the topic of efficacy, even though high exon 51 skipping was induced by our DG9-PMO, the dystrophin restoration observed was generally lower than that achieved by other peptide-conjugated PMOs (20, 29). This may be due to a difference in strategy, as the majority of peptide-conjugated PMO studies performed exon 23 skipping (20). Similar discrepancies between exon skipping and dystrophin restoration levels in vivo were observed by Aoki et al. (2010), who skipped exon 51 with PMOs (31), and Aupy et al. (2020), who also skipped exon 51 but with adeno-associated virus-delivered U7 small nuclear RNAs (32). Exon 51–skipped transcripts or proteins may be less stable than their exon 23–skipped counterparts, leading to the reduced dystrophin levels observed. An in vitro study using various internally truncated dystrophin proteins revealed that exon 51–skipped dystrophins were mostly as stable as full-length dystrophin (33), favoring a hypothesis of decreased transcript stability. While previous work has shown that DMD transcript stability has an impact on dystrophin protein production (34, 35), it remains to be determined if this explains the differences in the case of exon 23– and exon 51–skipped transcripts. Another factor would be the animal model used for testing. As humanized dystrophic mice such as hDMDdel52;mdx have only recently been developed, it would be important to characterize the stabilities of the human DMD transcript and protein in these models, to determine if this would have an impact on the efficacy of tested exon skipping therapies.
Despite its relatively reduced activity, we encouragingly observed functional improvement in mice given repeated doses of DG9-PMO (Fig. 3 G and H; SI Appendix, Fig. S3 A and B). Treated hDMDdel52;mdx mice had average dystrophin restoration levels at 2.8 to 3.9% of wild-type in the skeletal muscles (Fig. 3C). This was accompanied by improvements in fiber size, as seen in the tibialis anterior and diaphragm (SI Appendix, Fig. S3 C, E, and F). Our finding supports the notion that not much dystrophin may be needed to achieve functional benefit in vivo. Indeed, previous studies in mdx mice with nonrandom X-chromosome inactivation (mdx-XistΔhs) have shown that as little as 3 to 14% dystrophin of normal levels was sufficient to improve performance in hanging wire and grip strength tests to wild-type levels (36). Future work will focus on evaluating the effects of repeated DG9-PMO therapy on cardiac function, given the importance of this phenotype in determining the survival of DMD patients.
In terms of multiexon skipping, it was promising that the DG9-conjugated version of our minimized “block” mixture successfully skipped exons 45 to 55 at 9.5% efficiency and restored dystrophin production in hDMDdel52;mdx mice at nearly 0.8% of wild-type levels, on average (Fig. 4 B and C). The “all” mixture was previously shown to induce 15% exons 45 to 55–skipping efficiency upon intramuscular treatment of a different humanized DMD mouse model, at a dose of 1.67 μg/PMO (13). Unlike what we saw in vitro, a higher dose of the “block” mixture is apparently needed to induce comparable skipping levels to the “all” mixture in vivo. Because the human and mouse transcript target sequences for the DG9-PMOs in the “block” mixture are 70 to 93% identical, the mouse Dmd transcript expressed in hDMDdel52;mdx mice may have sequestered some of the administered DG9-PMOs. Crossing the hDMDdel52 transgene over to a mouse Dmd-null background (37) would eliminate this possibility, and may yield more representative results of efficacy. Furthermore, understanding how DG9 affects PMO interactions in a multiexon-skipping mixture may be necessary, similar to the in silico work we did to minimize self- and cross-annealing between PMOs in the “all” mixture (13). Aside from conducting this in silico analysis, we will investigate how the efficacy of our minimized multiexon skipping mixture could be increased by varying treatment doses and regimens. The possibility of systemic treatment would also be explored. In summary, we have shown proof of concept that a minimized PMO mixture for skipping human DMD exons 45 to 55 could be developed. We have also identified DG9 to be a promising, effective cell-penetrating peptide for PMO conjugation for both single-exon and multiexon skipping approaches.
Materials and Methods
Experiments with immortalized patient-derived cells were approved by the University of Alberta Health Research Ethics Board (Pr00079871). Mice were housed at the University of Alberta Health Sciences Laboratory Animal Services. All experiments were approved by the Animal Care and Use Committee at the University of Alberta Research Ethics Office (AUP00000365). Detailed information on cell culture, transfections, animal testing, molecular work, histology, and statistical analysis is in the SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Dr. Annemieke Aartsma-Rus (Leiden University Medical Center, Leiden, The Nethderlands) for providing the hDMDdel52;mdx mice used to start our colony for the study. The MANEX45A monoclonal antibody developed by G.E. Morris was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA. Funding was provided by the Canadian Institutes of Health Research (FDN 143251, PS 169193), Jesse’s Journey, Heart and Stroke Foundation (G-20-0029382), Women and Children’s Health Research Institute (IG 2115), The Friends of Garrett Cumming Research & Muscular Dystrophy Canada Endowed Research Chair, Henri M. Toupin Chair in Neurological Science, and Alberta Innovates Graduate Studentship (201810476).
Footnotes
Competing interest statement: T.Y. and R.M. are cofounders of OligomicsTx. The authors plan to file a patent application relevant to the technology described in this article.
This article is a PNAS Direct Submission. S.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112546119/-/DCSupplemental.
Data Availability
All other data are included in the manuscript and/or SI Appendix.
References
- 1.Hoffman E. P., R. H. Brown, Jr, Kunkel L. M., Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Petrof B. J., Shrager J. B., Stedman H. H., Kelly A. M., Sweeney H. L., Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl. Acad. Sci. U.S.A. 90, 3710–3714 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manzur A. Y., Kinali M., Muntoni F., Update on the management of Duchenne muscular dystrophy. Arch. Dis. Child. 93, 986–990 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Crisafulli S., et al. , Global epidemiology of Duchenne muscular dystrophy: An updated systematic review and meta-analysis. Orphanet J. Rare Dis. 15, 141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bladen C. L., et al. , The TREAT-NMD DMD Global Database: Analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 36, 395–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monaco A. P., Bertelson C. J., Liechti-Gallati S., Moser H., Kunkel L. M., An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2, 90–95 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Lim K. R. Q., Maruyama R., Yokota T., Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 11, 533–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anwar S., Yokota T., Golodirsen for Duchenne muscular dystrophy. Drugs Today (Barc) 56, 491–504 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Roshmi R. R., Yokota T., Viltolarsen for the treatment of Duchenne muscular dystrophy. Drugs Today (Barc) 55, 627–639 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Shirley M., Casimersen: First approval. Drugs 81, 875–879 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Aartsma-Rus A., et al. , Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 30, 293–299 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Béroud C., et al. , Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum. Mutat. 28, 196–202 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Echigoya Y., et al. , Exons 45-55 skipping using mutation-tailored cocktails of antisense morpholinos in the DMD Gene. Mol. Ther. 27, 2005–2017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim K. R. Q., Nguyen Q., Yokota T., Genotype-phenotype correlations in Duchenne and Becker muscular dystrophy patients from the Canadian Neuromuscular Disease Registry. J. Pers. Med. 10, 241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura A., et al. , Comparison of the phenotypes of patients harboring in-frame deletions starting at exon 45 in the Duchenne muscular dystrophy gene indicates potential for the development of exon skipping therapy. J. Hum. Genet. 62, 459–463 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Aoki Y., et al. , Bodywide skipping of exons 45-55 in dystrophic mdx52 mice by systemic antisense delivery. Proc. Natl. Acad. Sci. U.S.A. 109, 13763–13768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echigoya Y., Mouly V., Garcia L., Yokota T., Duddy W., In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS One 10, e0120058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim K. R. Q., et al. , Efficacy of multi-exon skipping treatment in Duchenne muscular dystrophy dog model neonates. Mol. Ther. 27, 76–86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehto T., et al. , Cellular trafficking determines the exon skipping activity of Pip6a-PMO in MDX skeletal and cardiac muscle cells. Nucleic Acids Res. 42, 3207–3217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsoumpra M. K., et al. , Peptide-conjugate antisense based splice-correction for Duchenne muscular dystrophy and other neuromuscular diseases. EBioMedicine 45, 630–645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Clark K., Barton C., Tanguay R., Moulton H., “A novel zebrafish model for assessing in vivo delivery of morpholino oligomers” in Exon Skipping and Inclusion Therapies (Humana Press, New York, NY, 2018), pp. 293–306. [DOI] [PubMed] [Google Scholar]
- 22.Veltrop M., et al. , A dystrophic Duchenne mouse model for testing human antisense oligonucleotides. PLoS One 13, e0193289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yavas A., et al. , Detailed genetic and functional analysis of the hDMDdel52/mdx mouse model. PLoS One 15, e0244215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sicinski P., et al. , The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science 244, 1578–1580 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Suzuki H., et al. , Endogenous multiple exon skipping and back-splicing at the DMD mutation hotspot. Int. J. Mol. Sci. 17, 1722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Echigoya Y., et al. , Quantitative antisense screening and optimization for Exon 51 skipping in Duchenne Muscular Dystrophy. Mol. Ther. 25, 2561–2572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi J.-M., et al. , Intranasal delivery of the cytoplasmic domain of CTLA-4 using a novel protein transduction domain prevents allergic inflammation. Nat. Med. 12, 574–579 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Lehto T., Ezzat K., Wood M. J. A., El Andaloussi S., Peptides for nucleic acid delivery. Adv. Drug. Deliv. Rev. 106, 172–182 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Moulton H. M., Moulton J. D., Morpholinos and their peptide conjugates: Therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta 1798, 2296–2303 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Wu R. P., et al. , Cell-penetrating peptides as transporters for morpholino oligomers: Effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 35, 5182–5191 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki Y., et al. , In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol. Ther. 18, 1995–2005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aupy P., et al. , Long-term efficacy of AAV9-U7snRNA-mediated Exon 51 skipping in mdx52 mice. Mol. Ther. Methods Clin. Dev. 17, 1037–1047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCourt J. L., et al. , In vitro stability of therapeutically relevant, internally truncated dystrophins. Skelet. Muscle 5, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spitali P., et al. , DMD transcript imbalance determines dystrophin levels. FASEB J. 27, 4909–4916 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Anthony K., et al. , Biochemical characterization of patients with in-frame or out-of-frame DMD deletions pertinent to exon 44 or 45 skipping. JAMA Neurol. 71, 32–40 (2014). [DOI] [PubMed] [Google Scholar]
- 36.van Putten M., et al. , The effects of low levels of dystrophin on mouse muscle function and pathology. PLoS One 7, e31937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudoh H., et al. , A new model mouse for Duchenne muscular dystrophy produced by 2.4 Mb deletion of dystrophin gene using Cre-loxP recombination system. Biochem. Biophys. Res. Commun. 328, 507–516 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data are included in the manuscript and/or SI Appendix.