Significance
Assembly factors play key roles in the biogenesis of mitochondrial protein complexes, regulating their stabilities, activities, and incorporation of essential cofactors. Cytochrome c oxidase assembly factor 7 (COA7) is a metazoan-specific assembly factor, the absence or mutation of which in humans accompanies complex IV assembly defects and neurological conditions. Here, we report the crystal structure of COA7 to 2.4 Å resolution, revealing a banana-shaped molecule composed of five helix-turn-helix (α/α) repeats. COA7 binds heme with micromolar affinity, even though the protein structure does not resemble previously characterized heme-binding proteins. The heme-bound COA7 can redox cycle between oxidation states Fe(II) and Fe(III) and shows disulfide reductase activity toward copper binding assembly factors. We propose that COA7 functions to facilitate the biogenesis of the binuclear copper site (CuA) of complex IV.
Keywords: COA7, mitochondria, cytochrome c oxidase, heme, X-ray crystallography
Abstract
Cytochrome c oxidase (COX) assembly factor 7 (COA7) is a metazoan-specific assembly factor, critical for the biogenesis of mitochondrial complex IV (cytochrome c oxidase). Although mutations in COA7 have been linked to complex IV assembly defects and neurological conditions such as peripheral neuropathy, ataxia, and leukoencephalopathy, the precise role COA7 plays in the biogenesis of complex IV is not known. Here, we show that loss of COA7 blocks complex IV assembly after the initial step where the COX1 module is built, progression from which requires the incorporation of copper and addition of the COX2 and COX3 modules. The crystal structure of COA7, determined to 2.4 Å resolution, reveals a banana-shaped molecule composed of five helix-turn-helix (α/α) repeats, tethered by disulfide bonds. COA7 interacts transiently with the copper metallochaperones SCO1 and SCO2 and catalyzes the reduction of disulfide bonds within these proteins, which are crucial for copper relay to COX2. COA7 binds heme with micromolar affinity, through axial ligation to the central iron atom by histidine and methionine residues. We therefore propose that COA7 is a heme-binding disulfide reductase for regenerating the copper relay system that underpins complex IV assembly.
In eukaryotes, the bulk of ATP generation occurs through oxidative phosphorylation (OXPHOS) that involves five multisubunit protein complexes in the inner mitochondrial membrane, known as complexes I to V (1). Complex IV (cytochrome c oxidase or COX) is the copper and heme a–containing terminal oxidase of the mitochondrial respiratory chain, which catalyzes electron transfer from cytochrome c in the intermembrane space (IMS) to molecular oxygen and contributes to the generation of the proton gradient required to power ATP synthesis. In mammals, complex IV is composed of 14 subunits, including 3 core subunits (COX1, COX2, and COX3) that are encoded by mitochondrial DNA (mtDNA) (2–4).
The assembly of complex IV is complicated and thought to involve the biogenesis of different modules harboring both mtDNA and nuclear encoded subunits (5, 6). A central module is seeded by membrane integration and maturation of COX1, which then incorporates separate modules containing COX2 and COX3. The incorporation of one copper ion (the copper B (CuB) site), heme a, and heme a3 cofactors is necessary for the biogenesis of the COX1 module, which then assembles with the COX2 subunit on incorporation of the binuclear copper site (CuA). Complex IV biogenesis requires the action of more than 30 assembly factors (6). While not part of the final enzyme, these assembly factors have been shown to perform critical roles in subunit maturation, cofactor incorporation, and stabilization of intermediate assemblies of complex IV in humans. These include the COX17, SCO1, SCO2, and COA6 proteins, which function in copper delivery to the CuA site and which bind copper through two cysteine residues, for which redox cycling between disulfide (oxidized) and free/Cu-bound (reduced) states is tightly linked to their functions (7–11). Loss-of-function mutations in both mtDNA and nuclear DNA genes encoding mitochondrial proteins, including complex IV subunits and assembly factors, lead to complex IV deficiency and mitochondrial disease. Complex IV is also found in supercomplexes, which are large assemblies containing complex IV along with complexes I and III, including the complex I-complex III2-complex IV (CI/III2/IV) respirasome (12–14).
The COX assembly factor 7 (COA7, also referred to as RESA1) is a metazoan-specific complex IV assembly factor located in the IMS (15, 16). The function of COA7 in complex IV assembly is unknown. COA7 is a cysteine-rich protein (13 cysteine residues) which, based on sequence analysis, contains five Sel1-like repeat (SLR) domains. SLR proteins are a subclass of the tetratricopeptide repeat (TPR) proteins, which belong to the α/α repeat family of solenoid proteins (17, 18). Available structures of SLR proteins show large solvent accessible surfaces, suitable for the binding of both large and small substrates (17–19). Accordingly, solenoid proteins often function in DNA–, peptide– and protein–protein interactions (17–19), which facilitate cellular processes such as signaling, the cell cycle, and intercompartmental transport (18, 20–22).
Pathogenic mutations in the gene encoding COA7 have been identified in patients exhibiting neurological symptoms of peripheral neuropathy, ataxia, and leukoencephalopathy (23, 24). The first of these studies described an isolated deficiency of complex IV in patient skin fibroblasts and skeletal muscle in the presence of compound heterozygous mutations in COA7 (p.Tyr137Cys and COA7-exon2Δ), resulting in the expression of the COA7 mutant proteins Y137C and an in-frame deletion mutant missing residues 37 to 83. A second study examined a cohort of Japanese patients with recessive mutations in COA7, where skin fibroblasts from these patients showed significant decreases in complex I and complex IV activities. Patients had either homozygous single amino acid p.Asp6Gly (D6G) mutations, compound D6G with p.Ser149Ile (S149I), or a missense p.Gly144fs mutation, and compound p.Arg39Trp (R39W) and COA7-exon2Δ mutations (24). The Arg39, Tyr137, and Ser149 residues are highly conserved among COA7 proteins from different species (SI Appendix, Fig. S1).
In the present study, we used structural and functional approaches to investigate the role of COA7 in mitochondrial complex biogenesis. Loss of COA7 blocks complex IV assembly steps that follow the formation of the COX1 intermediate module, with COX2 being highly labile. We report the crystal structure of human COA7 to 2.4 Å resolution and show that the recombinant COA7 protein binds heme in vitro specifically and with micromolar affinity, through axial His/Met coordination to the iron atom, despite no resemblance between the COA7 structure and well-characterized heme-binding proteins. The heme bound to COA7 has a reduction potential of −353 mV and can reduce the disulfide bonds that compose the copper binding sites in the assembly factors SCO1 and SCO2, enabling regeneration of the copper relay system. These findings point to a role for COA7 in the pathway that leads to the biogenesis of the CuA site of complex IV.
Results and Discussion
Loss of COA7 Results in Reduced Levels of Respiratory Chain Enzymes, with a Severe Complex IV Assembly Defect.
To investigate the function of COA7 in the biogenesis of the respiratory chain, CRISPR-Cas9–mediated gene editing (25) was used to generate COA7 knockout (COA7KO) HEK293T cell lines. Following transfection, cells were sorted to generate individual clones. Two clones (termed COA7KO-1 and COA7KO-2) were found to contain frameshift deletions in the COA7 gene where the single guide RNA (gRNA) was targeted. Immunoblot analysis confirmed complete loss of the COA7 protein from COA7KO mitochondria (SI Appendix, Fig. S2A). Isolated mitochondria were solubilized in digitonin and subjected to blue native (BN) polyacrylamide gel electrophoresis (PAGE) and immunoblotted for components of the respiratory chain complexes (Fig. 1A). Probing for the complex IV subunit COX4 revealed a severe complex IV defect, with no complex IV holoenzyme or higher complex IV-containing supercomplexes (CI/III2/IV or III2/IV) detected (Fig. 1A). Consistent with this, probing for the complex I subunit NDUFA9 revealed the loss of the respirasome (CI/III2/IV), while a faster migrating species corresponding to the CI/III2 supercomplex was seen (Fig. 1A). Probing for the complex III subunit UQCRC1 revealed the loss of the III2/IV supercomplex and the accumulation of the CI/III2 supercomplex (Fig. 1A). Complex V levels were unchanged (Fig. 1A). The defects observed in the supercomplexes and complex IV were rescued upon re-expression of COA7 with a C-terminal Flag epitope (+COA7Flag), indicating the complex IV defect observed was indeed due to the loss of COA7 (Fig. 1A).
Fig. 1.
Loss of COA7 results in complex IV assembly defects. (A and B) Mitochondria isolated from control, COA7KO-1, COA7KO-2, and rescued COA7KO-1 cells were solubilized in 1% digitonin and subjected to BN-PAGE and analyzed using antibodies against subunits in brackets. Coomassie brilliant blue (CBB)–stained membranes showing complex V (CV) are indicative of loading. (C) Cells were pulsed with [35S]-Met for 2 h and chased for the indicated times. Isolated mitochondria were analyzed by SDS-PAGE and phosphorimaging. Immunoblotting (IB) with the complex II subunit SDHA was used as a loading control. (D) Volcano plot showing proteins with altered abundance in COA7KO mitochondria. The SILAC log2-transformed fold change (FC) ratios are indicated on the x-axis. Subunits of the different OXPHOS complexes are color-coded, and complex IV subunits are labeled. The horizontal line indicates P = 0.05, and vertical lines indicate a fold change of 1.5. Topographical heatmap of SILAC log2-transformed fold change (FC) ratios mapped onto the structure of complex IV (PDB 5Z62) (31). Proteins encoded by mtDNA are shown as surface renders, while nuclear-encoded proteins are shown as cartoons. Blue intensity is relative to the decrease in protein abundance. n.d., not detected. (E) Venn diagram summarizing COA7Flag-interacting proteins under native conditions or following chemical cross-linking of cells grown in Glu- or Gal-containing media. Complex IV subunits and assembly factors are in bold. N/A; not applicable.
To delineate the effect of COA7KO on assembly of complex IV, we analyzed representative subunits of each of the three assembly modules: COX1, COX2, and COX3 (detecting COX1, COX2, and COX6A, respectively). As can be seen (Fig. 1B), residual COX1 was present in a faster migrating complex, termed the S2 intermediate (26), while COX2 and COX6A were not present in any observed intermediates. To further investigate the stability of the core mtDNA-encoded proteins (COX1 to 3), we utilized [35S]-methionine pulse-chase labeling in the presence of anisomycin, an inhibitor of cytosolic translation (27). Control and COA7KO-1 cells were labeled for 2 h with [35S]-Met before a chase without labeling and restoration of translation. Sodium dodecyl sulfate (SDS)-PAGE and phosphorimage analyses showed that pulse-labeled COX2 was highly labile in COA7KO cells, with reduced labeling of this subunit at the zero-hour time point and rapid loss over time relative to control cells (Fig. 1C). COX3 also displayed increased turnover during the chase period in COA7KO cells compared with control mitochondria (Fig. 1C). In comparison, the translation and stability of COX1 were similar in both lines (Fig. 1C). The levels of other mtDNA-encoded proteins did not appear to differ between control and COA7KO mitochondria. These results point to a critical role for COA7 in complex IV assembly in steps that follow the assembly of the COX1 module.
We next applied quantitative stable isotope labeling of amino acids in cell culture (SILAC) proteomics (28) to investigate the effect of COA7 loss on the mitoproteome. In agreement with the pulse-chase analysis, a robust decrease in the abundance of COX2 and COX3 was observed (Fig. 1D and SI Appendix, Dataset S1). In addition, the levels of a number of nuclear-encoded complex IV subunits (COX6A1, COX6B1, COX7A2, COX8A, and NDUFA4), which are associated with the COX2 or COX3 modules or later complex IV assembly intermediates (4, 29, 30), were reduced in the absence of COA7 (Fig. 1D and SI Appendix, Dataset S1). Visualization of the relative levels of complex IV subunits on the structure of complex IV using a topographical heatmap (Protein Data Bank [PDB] 5Z62) (31) shows that the COX1 module subunits (COX1, COX4, and COX5A) are largely unaffected by the loss of COA7, in contrast to the other mtDNA-encoded subunits, COX2 and COX3, and their surrounding nuclear-encoded proteins (Fig. 1D).
To understand the complex IV interactome in the absence of COA7, we generated a COA7KO cell line expressing COX4 with a Flag-tag (COA7/COX4KO + COX4Flag) (SI Appendix, Fig. S2B). Affinity-enrichment mass spectrometry analysis (SI Appendix, Fig. S2C and Dataset S2) showed that in the absence of COA7, COX4Flag enriched COX1 module subunits (COX1 and COX5A), COX3 module subunits (COX3 and COX7A2), COX5B (a subunit that associates with COX1 and COX3), (31) and a subunit of the COX2 module (COX6C) (SI Appendix, Fig. S2C). We also identified several complex IV assembly factors, including the CuA copper metallochaperone SCO1, the thiol oxidoreductase SCO2, COA3, the heme a synthase COX15, plus numerous complex I and III subunits (SI Appendix, Fig. S2C and Dataset S2). Given that COX3 is rapidly turned over in the absence of COA7, the S2 subcomplex appears to comprise the mtDNA-encoded subunits COX1 (Fig. 1B), COX4 (Fig. 1A), and likely COX5B. Indeed, in a recent BN-PAGE and complexome profiling analysis of ΔCOX2 cybrids (32), an equivalent subcomplex was identified to harbor COX1, COX4, COX5B, and COA3, but not COX3, COX5A, COX6C, or COX7A2, hinting that these subunits associate after the S2 subcomplex has been assembled. These results further support that in the absence of COX2 (due to the loss of COA7), a subset of complex IV subunits is capable of assembly into a crippled supercomplex, dependent on the presence of COX1 (32). Taken together, these results indicate that COA7 plays a critical role in the assembly of complex IV at a step that involves the maturation of the COX2 and/or COX3 modules onto the COX1 module.
COA7 Interacts Transiently with Complex IV Assembly Factors.
We next performed affinity-enrichment mass spectrometry to identify stable COA7-interacting proteins in an unbiased manner. Under steady-state conditions, we did not enrich COA7Flag with any complex IV subunits or assembly factors (Fig. 1E and SI Appendix, Fig. S2D and Dataset S3). However, COA7Flag did enrich several IMS proteins, including apoptosis-inducing factor 1 (AIFM1) and CHCHD4, which are required for disulfide bond formation in the intermembrane space (33). This correlates with COA7 having already been identified as a client of the CHCHD4 pathway (15). Also enriched were coproporphyrinogen oxidase (CPOX), an enzyme involved in heme biosynthesis (34), the mitochondrial protease LONP1 (35), the disaggregase CLPB and the CLPB-interacting protein HAX1 (36) (Fig. 1E and SI Appendix, Fig. S2D and Dataset S3). RCN2, an endoplasmic reticulum (ER)-luminal protein with potential localization in the mitochondrial IMS (37), was also enriched with COA7Flag. These results suggest that unlike many assembly factors, COA7 does not form stable interactions with components of the respiratory chain.
To identify transient interactions, we performed chemical cross-linking proteomics on control cells and COA7KO + COA7Flag cells grown in standard glucose-containing media or with galactose (where cells are reliant on OXPHOS for ATP) (38). Proteomic analysis of eluted COA7-interacting proteins revealed numerous proteins that were present in the native coimmunoprecipitation (Fig. 1E). In addition, we identified 18 proteins common to both the glucose and galactose samples, including the complex IV assembly factors COA4, SCO2, COA6, and COX17 (Fig. 1E and SI Appendix, Fig. S2 E and F and Datasets S4 and S5). Analysis of the COA7Flag interactome from galactose-grown cells revealed an additional 13 proteins, including the complex IV subunit COX2 (Fig. 1E and SI Appendix, Fig. S2F and Dataset S5). These results suggest that COA7 also interacts in assemblies with several complex IV assembly factors, in addition to the mtDNA-encoded complex IV subunit COX2.
Structural Characterization of COA7.
To uncover further functional information, we sought to determine the molecular structure of COA7. Recombinant COA7 was overexpressed and purified (∼90 to 95% purity), with an intense band at ∼25 kDa, corresponding to the molecular weight of monomeric COA7 (SI Appendix, Fig. S3A). The analytical size exclusion chromatography (SEC) elution profile of the purified COA7 showed a broad elution peak with an elution volume corresponding to a molecular weight of ∼55 kDa, indicating the possible presence of a dimeric species in solution (SI Appendix, Fig. S3B). The quaternary structure of COA7 was further investigated by analytical ultracentrifugation (AUC), using sedimentation velocity experiments. These data were fitted to a continuous size distribution model, which yielded sedimentation coefficients of 1.9 S and 3.5 S, demonstrating that the sample contained a mixture of monomeric and dimeric species (SI Appendix, Fig. S3C and Table S2), with the relative proportions of these species observed to vary with protein concentration, indicating concentration-dependent oligomerization behavior.
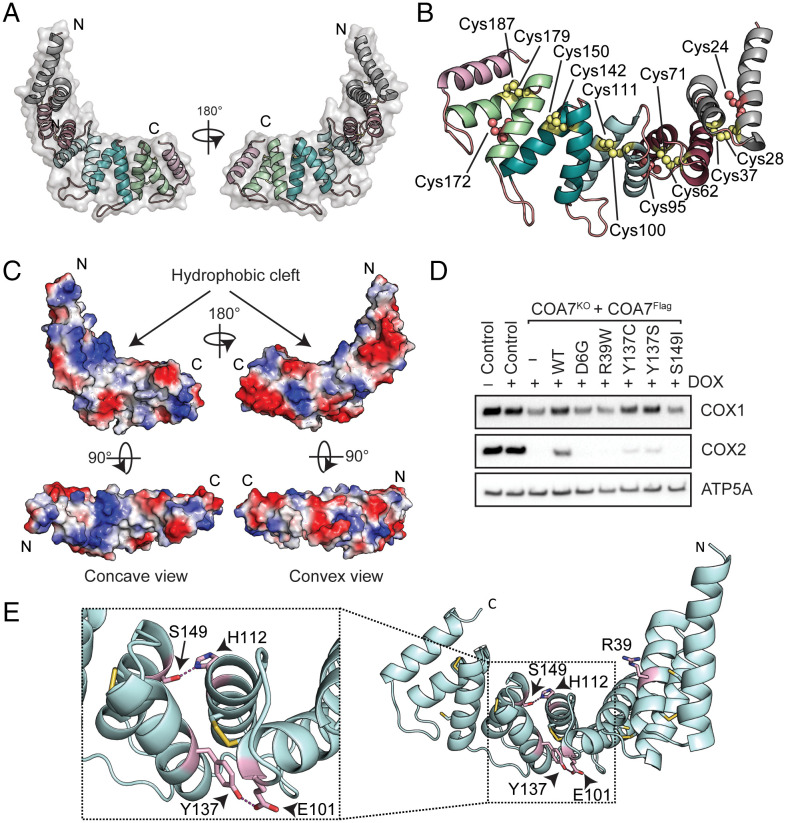
We determined the structure of COA7 by X-ray crystallography. The structure was solved and refined in space group I41 to 2.4 Å resolution, with a single molecule of COA7 per asymmetric unit (Fig. 2A). The final COA7 model includes residues Glu10-His218 and refinement of the model converged with residuals R = 20.5% and Rfree = 25.5% (SI Appendix, Table S3). COA7 is comprised of 11 α-helices, arranged as 5 helix-turn-helix (α/α) repeats (which vary in length from 30 to 36 residues) and an additional C-terminal helix (Fig. 2A). Together, these form an elongated, right-handed superhelix, with a concave and a convex face. The COA7 sequence includes 13 cysteine residues (at positions 24, 28, 37, 62, 71, 95, 100, 111, 142, 150, 172, 179, and 187), with the structure showing five intramolecular disulfide bonds between residues Cys28-Cys37, Cys62-Cys71, Cys100-Cys111, Cys142-Cys150, and Cys179-Cys187 (Fig. 2B), which bridge pairs of helices within each of the five α/α repeats. The position of the disulfide bonds as determined by crystallography is consistent with that proposed for the previously published model of COA7 (15) and with that predicted using AlphaFold (39). The 10 Cys residues that participate in the intramolecular disulfide bonds are conserved in COA7 sequences across all metazoans; however, the cysteine residues at positions 24, 95, and 172 (that do not form disulfide bonds in this structure) are only found in vertebrate COA7 (SI Appendix, Fig. S1) (10).
Fig. 2.
Structure of COA7 and analyses of pathogenic variants. (A) Cartoon representation of the overall structure of COA7. Secondary structures are represented as cartoons with SEL1 repeats colored in gray, raspberry, cyan, teal, and green. The transparent molecular surface is represented in gray. The amino (N) and carboxy (C) termini are indicated. (B) Cysteine residues (labeled) involved in intramolecular disulfide bonds are shown as yellow spheres, and free cysteine residues are shown as salmon spheres. (C) The molecular surface of COA7 is colored according to the electrostatic potentials (red, negatively charged; blue, positively charged; white, uncharged). The Top images show the side view of the COA7 structure. The Left and Right Bottom images show concave and convex views, respectively. The amino (N) and carboxy (C) termini are indicated. (D) Mitochondria isolated from control, COA7KO cells, and COA7KO cells expressing wildtype (WT) COA7Flag and variants in the absence or presence of doxycycline (DOX) as indicated were subjected to SDS-PAGE and immunoblotting with antibodies as shown. ATP5A serves as a loading control. (E) Patient mutations are mapped onto the structure of COA7 and shown as pink sticks and hydrogen bonds between residues (labeled) shown as dashed lines. Inset: Closer view of the interactions formed by residues His112 and Tyr137. The amino (N) and carboxy (C) termini are indicated.
A search against the PDB (40) revealed that COA7 shares significant structural similarity with the Helicobacter cysteine-rich proteins B and C (HcpB [PDB 1KLX] and HcpC [PDB 1OUV], respectively) (41, 42) from Helicobacter pylori (SI Appendix, Fig. S4). Hcp proteins are composed of α/α repeats and belong to the SLR protein family. The presence of disulfide bonds within the repeats for both the Hcps and COA7 is unusual for this protein family (18, 41). Indeed, to our knowledge, COA7 is the first structurally characterized human SLR protein possessing disulfide-bridged α/α repeats.
The outer convex surface of the COA7 molecule is highly negatively charged due to the presence of several Glu and Asp residues (located within repeats 1 and 2 and on the linkers between repeats 3 and 4, 4 and 5 and the C-terminal helix). In addition, the presence of lysine residues 49, 56, 59, 73, 82, 105, and 106 on the inner concave surface of COA7 creates a positively charged surface in this region (Fig. 2C). The residues that comprise both charged surfaces are largely conserved in COA7 proteins (SI Appendix, Fig. S1). Also on the concave surface is a hydrophobic cleft that results from clustering of several nonpolar residues (Ile108, Ala109, Gly144, Gly145, Leu181, and Gly182) (Fig. 2C). Interestingly, the application of crystallographic symmetry operators reveals that in the crystal, this cleft is occupied by the N-terminal helix (residues Glu10-His30) of a neighboring molecule of COA7 (SI Appendix, Fig. S5), such that the crystal is composed of an infinite network of these protein–protein interactions (SI Appendix, Fig. S5). This interaction is mediated by hydrogen bonds and electrostatic interactions, with a total buried surface area of 1,614 Å2, representing ∼7% of the surface areas of the two molecules combined (40). Given the limited extent of this interaction, it is unlikely to represent a persistent oligomeric structure in solution (43), but correlates with the concentration-dependent oligomerization behavior observed by AUC. This contact may indicate a molecular mechanism for an interaction between COA7 and protein partners in the mitochondrial IMS. Similar direct protein–protein interactions have been previously proposed for other TPR proteins, including HcpC (42), the Hsp70/Hsp90 organizing protein (Hop) (PDB 1ELR) (44, 45), and the peroxisomal targeting signal-1 PEX5 (PDB 1FCH) (46). In the case of the related HcpC structure, an interaction between the C terminus of one molecule and the hydrophobic pocket of a neighboring molecule facilitates the formation of a similar extended protein network in the crystal. This was proposed to represent a mechanism for protein–protein interactions in the cell (42).
COA7 Binds Heme with Micromolar Affinity.
A key step in the biogenesis of complex IV is insertion of copper into the CuA site of COX2 (6). Loss-of-function mutations in these assembly factors block complex IV assembly at the level of COX2 maturation, similar to that observed here for COA7 (26). Furthermore, soluble high-affinity Cu(I)-binding proteins (copper metallochaperones) such as ATOX1 (47), COX17 (48), the recently identified Cu(I)-binding protein COA6 (26) and the soluble domains of SCO1 and SCO2 (49) all show CysXnCys sequence motifs and bind Cu(I). Due to the presence of multiple Cys residues within the COA7 sequence, we sought to determine whether COA7 binds copper (specifically Cu[I]) using a competition assay with the [Cu(I)Bcs2]3− (copper bathocuproinedisulfonic acid) complex (26, 50). Cu(I) binding to the COA7 was not detected using this assay; however, Cu(I) binding was observed for the recombinant ATOX1 protein, which was included as a positive control (SI Appendix, Fig. S6).
Given these findings, we investigated whether COA7 might bind heme, the other cofactor required for complex IV biogenesis and activity. Complex IV binds heme a and heme a3, which are essential for function. Heme a is important for the stability of COX1, while insertion of heme a3, which has a farnesyl tail and sits between COX1 and COX2, is required for assembly and stabilization of COX2 (5, 6). We therefore tested whether COA7 might be involved in heme incorporation into complex IV by measuring whether the purified protein was able to bind heme in solution using ultraviolet-visible (UV-Vis) spectrophotometry (51, 52). Upon titration of COA7 with hemin (Fe(III)-PPIX, protoporphyrin IX), difference spectra revealed a Soret band at 416 nm and a broad feature between 500 and 600 nm (Fig. 3A), which increased in intensity with the addition of increasing concentrations of hemin. Fitting of these titration data yielded a dissociation constant KD(heme)= 1.40 ± 0.28 μM (Fig. 3B). This value is similar to those measured for other heme-binding proteins. For instance, Listeria HemQ, which is involved in heme acquisition, binds heme with KD(heme)= 16.2 μM (53), and Pseudomonas Phus binds heme with KD(heme)= 29 μM (54).
Fig. 3.
Heme binding to COA7. (A) Difference absorption spectra of COA7 (1 μM) with increasing concentrations of Fe(III)-PPIX (from 0 to 10 μM) as indicated. (B) The binding curve was fitted to the data from A using an equation describing a single binding site (Y = Bmax * X/KD + X) with GraphPad Prism. (C) UV-Vis absorption spectra of COA7 in the presence of one molar equivalent of Fe(III)-PPIX (black); Fe(II)-PPIX (red), and Zn(II)-PPIX (dotted, black).
Incubation of purified COA7 with one molar equivalent of hemin and reduction with sodium dithionite yielded UV-Vis spectra indicative of Fe(III)- and Fe(II)-heme-COA7, respectively (Fig. 3C). These spectra were very similar to the those reported for the ferric and ferrous states, respectively, of cytochrome b562 from Escherichia coli (55), which shows axial ligation of the heme iron by His and Met residues. Incubation of COA7 with Zn(II)-PPIX also yielded a spectrum very similar to that of Zn(II)-PPIX–loaded cytochrome b562 (Fig. 3C) (55). The incubation of COA7 with excess heme, followed by removal of unbound cofactor, yielded a Fe(III)-heme-COA7 complex with a stoichiometry, as determined by inductively coupled plasma mass spectrometry (ICP-MS), of 1.06 ± 0.03 iron atoms (heme cofactors) per protein molecule (SI Appendix, Fig. S7C). COA7 could not be copurified with PPIX using the same approach (SI Appendix, Fig. S7B).
Examination of the structure of COA7 reveals a possible heme binding site located between repeats 3 and 4, with residues His119 and Met156 in comparable positions to those that act as iron ligands in cytochrome b562 (55) (SI Appendix, Fig. S8). Unfortunately, despite extensive attempts, we were unable to crystallize Fe(III)-heme-COA7. We therefore generated single- and double-mutant proteins (H119ACOA7, M156ACOA7, and H119A/M156ACOA7) to probe the contributions of these residues to heme binding. In addition, we examined the ability of the H119A/M156ACOA7Flag mutant to rescue the assembly of complex IV in COA7KO cells (SI Appendix, Fig. S11). Titration of the H119A/M156ACOA7 variant with hemin yielded no detectable binding (SI Appendix, Fig. S9 B and C), and the UV-Vis spectra of the variant protein following incubation with one equivalent of hemin was significantly different from that observed for COA7 (both in the presence and absence of sodium dithionite; SI Appendix, Fig. S10). To investigate the effect of this mutant in COA7KO cells, we also generated a doxycycline-inducible system with COA7Flag expressed downstream of a GFP and a viral T2A cleavage site to separate the two encoded proteins (SI Appendix, Fig. S11A). In this way, GFP functions as an internal control for controlling equal levels of expression of both wild-type and mutant COA7 (SI Appendix, Fig. S11 A and B). COA7Flag and H119A/M156ACOA7Flag were also detected in whole-cell lysates, migrating slightly slower than endogenous COA7 due to the presence of the Flag epitope (SI Appendix, Fig. S11C). Isolated mitochondria from these cells were analyzed by SDS-PAGE and immunoblotting for COA7 (SI Appendix, Fig. S11D) and the complex IV proteins COX1 and COX2 (SI Appendix, Fig. S11E). While the levels of H119A/M156ACOA7Flag were lower than COA7Flag, both were higher than endogenous COA7 (SI Appendix, Fig. S11D). Analysis of COX1 and COX2 from these mitochondria by SDS-PAGE further revealed that the H119A/M156ACOA7Flag was not able to rescue levels of COX1 and COX2 to the same extent as COA7Flag (SI Appendix, Fig. S11E). To determine the effect on complex IV assembly, isolated mitochondria were analyzed by BN-PAGE, which showed that relative to COA7Flag, H119A/M156ACOA7Flag expression was not able to efficiently rescue complex IV assembly (SI Appendix, Fig. S11F), indicating that residues His119 and Met156 are important for COA7 function.
Finally, we incubated the heme-loaded COA7, H119ACOA7, and M156ACOA7 proteins with sodium cyanide and compared the UV-Vis spectra of the proteins measured before and after incubation (SI Appendix, Fig. S9 D–F). The spectrum of the Fe(III)-heme-COA7 protein showed no change on incubation with CN−. In contrast, for both the Fe(III)-heme-H119ACOA7 and Fe(III)-heme-M156ACOA7 variants, shifts in the positions of the Soret bands and spectral changes in the region between 500 and 600 nm were observed, indicating a change in iron coordination on addition of CN−. These data taken together indicate that heme binding to COA7 is mediated by axial ligation of residues His119 and Met156 to the heme iron. A superposition of the COA7 structure with that of cytochrome b562 (PDB 256B; SI Appendix, Fig. S8B) (56), shows that heme binding in this position, comparable to that seen in the structure of cytochrome b562 (56), would be possible upon slight adjustments to the positioning of the surrounding helices. These data indicate that in vitro COA7 binds heme specifically through axial ligation of the central iron atom with His119 and Met156 protein ligands, which are conserved in the majority of COA7 sequences (SI Appendix, Fig. S1).
COA7 Does Not Facilitate Heme Incorporation into Complex IV.
Given the observed heme binding activity of recombinant COA7, we sought to investigate whether COA7 might act as a heme chaperone and/or function in heme incorporation into complex IV. To that end, we generated and characterized CPOXKO cells by CRISPR-Cas9 genome editing. Sequencing of the CPOX locus revealed disruption at the position of the DNA cleavage (SI Appendix, Fig. S12A). To ensure specificity, we generated a CPOX rescue cell line by reintroducing CPOXFlag into CPOXKO cells that we verified with SDS-PAGE and immunoblotting with CPOX and Flag antibodies (SI Appendix, Fig. S12B). As expected, CPOXKO cells grown in heme-depleted media showed a reduction in the levels of COX1 and COX2, which was reversed to control levels by the expression of CPOXFlag (SI Appendix, Fig. S12C). Additionally, analysis of mitochondria from control, CPOXKO, and CPOXKO expressing CPOXFlag by BN-PAGE showed a defect in complexes II and IV in the CPOXKO cells, which was rescued by the expression of CPOXFlag (SI Appendix, Fig. S12D).
We next compared control, COA7KO, and CPOXKO cells grown in heme-depleted media in the presence or absence of succinylacetone (SA), an inhibitor of 5-aminolevulinic acid dehydratase (ALAD), the second enzyme of the heme biosynthetic pathway (57) (Fig. 4A). We reasoned that if COA7 functions in heme incorporation, the inhibition of heme biosynthesis in COA7KO cells would not result in further decreases in the levels of COX1. Mitochondria isolated from control, COA7KO, and CPOXKO cells grown in heme-depleted serum in the presence or absence of SA were subjected to SDS-PAGE and immunoblotting (Fig. 4A and SI Appendix, Fig. S13 A and C). Interestingly, we found a significant increase in the levels of COA7 (SI Appendix, Fig. S13 A and B) in control cells where heme biosynthesis was inhibited (+SA) and in CPOXKO cells, both in the presence and absence of SA, suggesting that levels of COA7 may be modulated by heme. CPOX levels were relatively similar in control and COA7KO cells, but for the complex II subunit SDHC, which is part of the heme-binding region of succinate dehydrogenase, levels were reduced in samples where heme biosynthesis was inhibited and in CPOXKO cells as expected (SI Appendix, Fig. S13A). We next investigated whether levels of COX1 were perturbed by the inhibition of heme biosynthesis. Following SA treatment, COX1 levels were reduced in control cells, and interestingly further reduced in COA7KO cells. The levels of COX1 and COX2 in CPOXKO mitochondria were unaffected by SA treatment (Fig. 4B and SI Appendix, Fig. S13C). Furthermore, analysis by BN-PAGE revealed a robust decrease in the levels of complex IV in control cells following SA treatment (SI Appendix, Fig. S13D). In addition, the S2 subcomplex present in COA7KO cells was not observed in cells treated with SA (SI Appendix, Fig. S13D), suggesting that this subcomplex requires heme incorporation for stability and that it is therefore hemylated in COA7KO cells. Taking these data together, we conclude that COA7 does not function in the delivery or incorporation of heme into complex IV.
Fig. 4.
COA7 functions as a disulfide reductase in complex IV assembly. (A) Schematic of the heme biosynthetic pathway. Succinylacetone (SA) inhibits the second enzyme in the pathway, 5-aminolevulinic acid dehydratase (ALAD). 5-aminolevulinic acid synthase 1 (ALAS1), 5-aminolevulinic acid (5-ALA), porphobilinogen (PBG), coproporphyrinogen oxidase (CPOX), ferrochelatase (FECH). (B) The relative abundance of COX1 in mitochondria isolated from the cells indicated were quantified by densitometry. n = 4, mean ± SEM, ordinary one-way ANOVA, using Tukey’s multiple comparisons test, ****P < 0.0001; n.s., not significant. (C) Analysis of the reduction of a single disulfide bond in SCO1 by Fe(II)-heme-COA7 via electrospray ionization mass spectrometry. The Top spectrum shows a deconvoluted experimental molecular weight of oxidized SCO1 (S-S) following iodoacetamide labeling (19,766.52 Da). The Bottom spectrum shows a deconvoluted experimental molecular weight of reduced SCO1 following reduction by Fe(II)-heme-COA7 and iodoacetamide labeling (S-IAA, 19,881.15 Da). (D) Analysis of the reduction of a single disulfide in SCO2 by Fe(II)-heme-COA7 via electrospray ionization mass spectrometry. The Top spectrum shows a deconvoluted experimental molecular weight of oxidized SCO2 (S-S) following iodoacetamide labeling (S-IAA, 19,543.48 Da). The Bottom spectrum shows a deconvoluted experimental molecular weight of reduced SCO2 following reduction by Fe(II)-heme-COA7 and iodoacetamide labeling (19,660.03 Da). (E) Mitochondria isolated from control, COA7KO, COA7KO+COA7Flag (+COA7Flag), and HIGD2AKO cells were subjected to redox analysis by treatment with DTT, iodoacetamide (IAA), and AMS and analyzed by SDS-PAGE and immunoblotting (IB) with SCO1 and SCO2 antibodies as indicated. RED, reduced SCO1/SCO2; OX, oxidized SCO1/SCO2.
Heme-COA7 Is Redox Active and Can Reduce Disulfide Bonds in Assembly Factor Proteins that Function in Copper Incorporation into Complex IV.
Given the lack of evidence that COA7 facilitates heme incorporation into complex IV and the apparent spectroscopic similarity of heme-loaded COA7 to E. coli cytochrome b562 (49), we investigated whether heme-bound COA7 showed redox activity. The oxidation of Fe(II)-heme-COA7 to Fe(III)-heme-COA7 with a mixture of ferri/ferrocyanide was monitored spectrophotometrically and fit to the Nernst equation to yield a reduction potential for Fe(III)-heme-COA7 of −353 mV (SI Appendix, Fig. S14A). This reduction potential is lower than those reported for the disulfide bonds that form the copper coordination sites in the assembly factor proteins COX17 (−198 mV) (58), SCO1 (−277 mV) (59), SCO2 (less than −300 mV) (60), and COA6 (−349 mV) (10), some of which were detected as interacting partners to COA7 in the cross-linked proteomics analysis (Fig. 1E and SI Appendix, Fig. S2 E and F). Reduction of the disulfide bonds between specific pairs of cysteine residues in these proteins allows Cu(I) coordination, creating a pathway for insertion of this cofactor into COX2. The functions of SCO1 and SCO2 have been particularly well characterized. SCO1 binds copper through cysteine residues 169 and 173 and acts as a metallochaperone for metal transfer to the CuA site (7, 61, 62). SCO2 acts as a thiol:disulfide oxidoreductase of SCO1 in vivo and COX2 in vitro (7, 8). These activities are dependent on Cu binding between cysteine residues 133 and 137 (7, 60).
Given the reduction potential of Fe(III)-heme-COA7 is lower than those reported for SCO1 and SCO2, we tested whether Fe(II)-heme-COA7 could be oxidized by these proteins and whether there was a concomitant reduction of the disulfide bonds that form their copper binding sites. Incubation of Fe(II)-heme-COA7 with the oxidized (disulfide-containing) SCO1 and SCO2 proteins led to oxidation of the Fe(II)-heme-COA7 to Fe(III)-heme-COA7 as indicated by the disappearance of the alpha peaks at 530 and 558 nm and appearance of a feature at 540 nm in the UV-Vis spectrum of the protein (SI Appendix, Fig. S15 A and B). This observation could not be replicated by the addition of buffer alone (SI Appendix, Fig. S15C). In addition, the substitution of Fe(II)-cytochrome c (with a reduction potential higher than the SCO1 and SCO2 proteins at +256 to 266 mV) (63) for Fe(II)-heme-COA7 showed no spectral changes, indicating no change in the oxidation state of Fe(II)-cytochrome c (SI Appendix, Fig. S15D).
To test whether the spectrophotometrically observed oxidation of Fe(II)-heme-COA7 corresponded to the reduction of disulfide bonds within the SCO1 and SCO2 proteins, we analyzed the reaction mixtures before and after incubation with iodoacetamide (which adds 57 Da per cysteine residue though the generation of S-carbamidomethylcysteine adducts) by electrospray mass spectrometry (Fig. 4 C and D and SI Appendix, Figs. S16–S18). These experiments revealed the reduction of a single disulfide bond in both the SCO1 and SCO2 proteins in the presence of Fe(II)-heme-COA7, demonstrating that heme-COA7 has disulfide reductase activity. This activity could not be observed with hemin alone, apo-COA7, Fe(II)-heme-H119A/M156ACOA7, or Fe(II)-cytochrome c (SI Appendix, Figs. S17 and S18).
The observed disulfide reductase activity of heme-COA7 suggests that heme-COA7 functions in the reduction of copper binding sites of these proteins and therefore biogenesis of the CuA site, which is essential for complex IV assembly. We therefore examined the redox status of SCO1 and SCO2 in control, COA7KO, and COA7 rescue cells using 4-acetoamido-4’-maleimidylstilbene-2,2’-disulfonic acid (AMS) as described previously (8) (Fig. 4E). Relative to control cells, SCO1 and SCO2 in COA7KO cells appeared to be predominantly in the oxidized states, as they were not able to be modified by AMS in the absence of dithiothreitol (DTT) (Fig. 4E). Re-expression of COA7Flag was able to restore this defect to what was observed in control cells. The defect observed in COA7KO mitochondria is likely not due to a general block in complex IV assembly, as the oxidation state of SCO1 and SCO2 in the HIGD2AKO mitochondria (that also shows a complex IV assembly defect) (30) resembled control mitochondria (Fig. 4E). In cells, the absence of COA7 therefore shifts the redox states of the SCO1 and SCO2 proteins toward the oxidized and therefore Cu-free forms, which agrees with the disulfide reductase activity of heme-COA7 observed in solution.
COA7 Pathogenic Mutations.
Pathogenic mutations in the COA7 gene producing the mutant variants D6GCOA7, R39WCOA7, Y137CCOA7, and S149ICOA7 have been identified in patients exhibiting neurological symptoms of peripheral neuropathy, ataxia, and leukoencephalopathy (23, 24). Analysis of the crystal structure of COA7 reveals that residue Arg39 is located on the C-terminal helix of repeat 1 and is exposed on the outer surface of the COA7 molecule (Fig. 2E). Residues Tyr137 and Ser149 are located in the middle of the structure, on repeat 4, and participate in hydrogen bonding interactions with residues Glu101 and His112, respectively, which are both found on repeat 3 (Fig. 2E). Residue Asp6 is at the N terminus of the molecule and could not be resolved in the crystal structure.
To investigate the effects of the pathogenic mutations identified in patients, we generated doxycycline-inducible retroviral plasmids encoding COA7 mutant proteins using the green fluorescent protein-thosea asigna virus self-cleaving 2A peptide (GFP-T2A) system (SI Appendix, Fig. S11A) for inducible expression in COA7KO cells. Equal expression of COA7 and variants was verified by analyzing whole-cell lysates by SDS-PAGE and immunoblotting using GFP antibodies (SI Appendix, Fig. S19A). Analysis of COA7 levels in both whole-cell extracts (SI Appendix, Fig. S19B) and isolated mitochondria (SI Appendix, Fig. S19C) showed that mitochondrial levels of D6GCOA7 and R39WCOA7 were similar to COA7, but the levels of Y137CCOA7 and S149ICOA7 were reduced. Expression of COA7Flag was able to rescue COX1 and COX2 levels (although not to control levels) in the COA7KO line (Fig. 2D). However, upon expression of the COA7 variants D6GCOA7, R39WCOA7, and S149ICOA7, COX1 levels were not rescued, while COX2 was not detected, indicating that these variants were less active in complex IV assembly relative to COA7 (Fig. 2D). Expression of the patient mutation Y137CCOA7 and the analogous Y137SCOA7 protein (containing a similar geometry to the Y137C patient-identified mutation, but unable to form disulfide bridges) partially rescued COX1 and COX2 levels (Fig. 2D).
Recombinant, purified forms of R39WCOA7, Y137CCOA7, S149ICOA7, and Y137SCOA7 were prepared and subjected to differential scanning fluorimetry (SI Appendix, Table S4) to measure their thermal stabilities (64). Reduced protein stabilities relative to COA7 were observed for all variant proteins, consistent with the observed decreased expression in mitochondria (SI Appendix, Table S4). The thermal transition for purified S149ICOA7 could not be observed, indicating the protein was most likely misfolded. The melting temperatures of the Y137CCOA7 and Y137SCOA7 proteins were 16 to 19 °C lower than COA7. Analysis of the quaternary structure of Y137CCOA7 by analytical ultracentrifugation showed evidence of concentration-dependent dimerization (as observed for COA7), in addition to the presence of higher-order oligomeric states indicative of aggregation. These observations may explain the reported aggregation and mislocalization of the Y137CCOA7 protein in cells (15) (SI Appendix, Fig. S20 and Table S2). Interestingly, Y137CCOA7 bound hemin with approximately the same affinity as COA7 (SI Appendix, Fig. S21); however, the reduction potential of Fe(III)-heme-Y137CCOA7 was determined as −256 mV (SI Appendix, Fig. S14B), which is significantly higher than that for COA7. Neither the oxidation of Fe(II)-heme-Y137CCOA7 nor the reduction of the disulfide bonds on incubation with the oxidized forms of the SCO1 or SCO2 proteins could be detected for this COA7 variant (SI Appendix, Figs. S17 and S18). Therefore, the observed pathogenesis of this mutation is likely also linked to defects in the disulfide reductase activity of COA7.
Conclusion
Despite the importance of the assembly of complex IV in health and mitochondrial disease, we have an incomplete understanding of the molecular basis of the biogenesis of this enzyme owing to a lack of knowledge about the identities, structures, and roles of all crucial assembly factors (6). The present study shows that deletion of COA7 resulted in a severe complex IV assembly defect. Crucially, the absence of COA7 causes specific defects in the maturation of the COX2 and COX3 modules, resulting in their degradation and therefore halting complex IV biogenesis.
The high-resolution crystal structure of the human COA7 protein shows the protein is composed of five SLRs, tethered by disulfide bonds. The overall fold of COA7 is similar to other TPRs and SLRs, proposed to function in protein–protein interactions due to their extensive solvent-accessible surfaces (18, 20–22). Fascinatingly, recombinant COA7 was found to bind heme in solution via axial His/Met coordination to the heme iron, despite showing no structural similarity to well-characterized heme-binding folds. However, our data show that COA7 does not function as a heme chaperone or in heme transfer to complex IV. Instead, Fe(II)-heme-COA7 is able to reduce disulfide bonds within the assembly factors SCO1 and SCO2, which function in the pathway for copper delivery to the CuA site in COX2. In the absence of COA7, the relative ratios of the oxidized and reduced forms of these proteins in mitochondria are skewed relative to control samples. The Y137C variant of COA7, associated with mitochondrial disease in patients, shows an altered reduction potential and no disulfide reductase activity.
The redox cycling of the SCO1 and SCO2 proteins is essential for the Cu(I) binding activity of SCO1, which has been proposed to deliver the metal to the CuA site of COX2 and for the thiol disulfide oxidoreductase activity of SCO2, which reduces COX2 for copper binding (7). The fact that Fe(II)-heme-COA7 shows disulfide reductase activity toward both SCO1 and SCO2 is fascinating, given their distinct roles. Given the activity of COA7 overlaps to some extent with that proposed for COA6 (11), the potential for cooperative functions for these proteins will require further investigation.
To date, the functions of assembly factors such as SCO1, SCO2, and others in the pathway, including COA6 and COX17, have been based to a significant extent on in vitro experiments performed and analyzed without consideration for the potential function of other assembly factors, such as COA7. Thus, these studies have arguably painted an incomplete picture of CuA biogenesis. The fact that COA7 shows redox activity and its absence accompanies such a significant complex IV assembly defect reinforce the importance of the of redox status of proteins within the IMS to this process.
Materials and Methods
Cell line generation, culture conditions, affinity enrichment analyses, proteomics analyses, crystallization, X-ray data collection, structure solution, and refinement are described in detail in SI Appendix, SI Materials and Methods. Human HEK293T cells were used for knockout cell line generation as described, using CRISPR-Cas9 gene editing (25, 65). Human COA7 and variants were cloned into the pGEX-6p-1 plasmid with an N-terminal glutathione-S-transferase tag and expressed and purified from E. coli strain SHuffle T7 (New England BioLabs). Published methods for heme-binding studies (51, 52) and measurement of the reduction potential of Fe(III)-heme-COA7 (66, 67) are also given in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was funded by the Australian Research Council (DP140102746 and FT180100397 to M.J.M. and DP220102030 to M.J.M. and M.T.R.), the National Health and Medical Research Council (GNT1165217 to M.T.R. and M.J.M., GNT1140906 and GNT1140851 to D.A.S., and GNT2010149 to L.E.F.), and an Australian Government Research Training Program Scholarship to S.M. L.E.F. also acknowledges support from the Mito Foundation. Part of this study was carried out using the MX2 beamline at the Australian Synchrotron, which is part of the Australian Nuclear Science and Technology Organisation (ANSTO), and made use of the Australian Cancer Research Foundation (ACRF) detector. We thank the beamline staff for their enthusiastic and professional support, Dr. Yee-Foong Mok at the Melbourne Protein characterization facility, the Bio21 Molecular Science and Biotechnology Institute for performing the AUC experiments and data analyses, the Bio21 Mass Spectrometry and Proteomics Facility for the provision of instrumentation, training, and technical support, Daniel Machell for PyMOL and coding assistance, Dr. Katie Ganio and Prof. Christopher McDevitt for the ICP-MS analysis, and Mrs. Mahnaz Dideh Var for her extraordinary support with the structural biology aspects of this study. We thank the reviewers for their constructive comments. Schematics were created using Biorender.com.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110357119/-/DCSupplemental.
Data Availability
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) (68) partner repository (https://www.ebi.ac.uk/pride/archive) with the dataset identifier PXD031589 (69). The atomic coordinates and structure factors have been deposited in the PDB (https://www.rcsb.org; PDB 7MQZ).
References
- 1.Fernández-Vizarra E., Tiranti V., Zeviani M., Assembly of the oxidative phosphorylation system in humans: What we have learned by studying its defects. Biochim. Biophys. Acta 1793, 200–211 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Tsukihara T., et al. , The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Mick D. U., Fox T. D., Rehling P., Inventory control: Cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsa E., et al. , NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 16, 378–386 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Signes A., Fernandez-Vizarra E., Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem. 62, 255–270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timón-Gómez A., et al. , Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 76, 163–178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgada M. N., et al. , Loop recognition and copper-mediated disulfide reduction underpin metal site assembly of CuA in human cytochrome oxidase. Proc. Natl. Acad. Sci. U.S.A. 112, 11771–11776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leary S. C., Sasarman F., Nishimura T., Shoubridge E. A., Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 18, 2230–2240 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Bertini I., Cavallaro G., McGreevy K. S., Cellular copper management—A draft user’s guide. Coord. Chem. Rev. 254, 506–524 (2010). [Google Scholar]
- 10.Maghool S., et al. , Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6. Life Sci. Alliance 2, e201900458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maghool S., Ryan M. T., Maher M. J., What role does COA6 play in cytochrome C oxidase biogenesis: A metallochaperone or thiol oxidoreductase, or both? Int. J. Mol. Sci. 21, 6983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letts J. A., Fiedorczuk K., Sazanov L. A., The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Li Y., et al. , An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 282, 17557–17562 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Diaz F., Fukui H., Garcia S., Moraes C. T., Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26, 4872–4881 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohanraj K., et al. , Inhibition of proteasome rescues a pathogenic variant of respiratory chain assembly factor COA7. EMBO Mol. Med. 11, e9561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozjak-Pavlovic V., et al. , C1orf163/RESA1 is a novel mitochondrial intermembrane space protein connected to respiratory chain assembly. J. Mol. Biol. 426, 908–920 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Andrade M. A., Perez-Iratxeta C., Ponting C. P., Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 134, 117–131 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Mittl P. R., Schneider-Brachert W., Sel1-like repeat proteins in signal transduction. Cell. Signal. 19, 20–31 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Cerveny L., et al. , Tetratricopeptide repeat motifs in the world of bacterial pathogens: Role in virulence mechanisms. Infect. Immun. 81, 629–635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blatch G. L., Lässle M., The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays 21, 932–939 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Kajava A. V., Review: Proteins with repeated sequence—Structural prediction and modeling. J. Struct. Biol. 134, 132–144 (2001). [DOI] [PubMed] [Google Scholar]
- 22.D’Andrea L. D., Regan L., TPR proteins: The versatile helix. Trends Biochem. Sci. 28, 655–662 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Martinez Lyons A., et al. , COA7 (C1orf163/RESA1) mutations associated with mitochondrial leukoencephalopathy and cytochrome C oxidase deficiency. J. Med. Genet. 53, 846–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi Y., et al. , Mutations in COA7 cause spinocerebellar ataxia with axonal neuropathy. Brain 141, 1622–1636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroud D. A., et al. , COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2. Hum. Mol. Genet. 24, 5404–5415 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Formosa L. E., Hofer A., Tischner C., Wenz T., Ryan M. T., Translation and assembly of radiolabeled mitochondrial DNA-encoded protein subunits from cultured cells and isolated mitochondria. Methods Mol. Biol. 1351, 115–129 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Ong S. E., et al. , Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Pitceathly R. D., et al. , UK10K Consortium, NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep. 3, 1795–1805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hock D. H., et al. , HIGD2A is required for assembly of the COX3 module of human mitochondrial complex IV. Mol. Cell. Proteomics 19, 1145–1160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zong S., et al. , Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobo-Jarne T., et al. , Multiple pathways coordinate assembly of human mitochondrial complex IV and stabilization of respiratory supercomplexes. EMBO J. 39, e103912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hangen E., et al. , Interaction between AIF and CHCHD4 regulates respiratory chain biogenesis. Mol. Cell 58, 1001–1014 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Swenson S. A., et al. , From synthesis to utilization: The ins and outs of mitochondrial heme. Cells 9, 579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshwal S., Fiedler K. U., Langer T., Mitochondrial proteases: Multifaceted regulators of mitochondrial plasticity. Annu. Rev. Biochem. 89, 501–528 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Cupo R. R., Shorter J., Skd3 (human ClpB) is a potent mitochondrial protein disaggregase that is inactivated by 3-methylglutaconic aciduria-linked mutations. eLife 9, e55279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung V., et al. , Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang Y., et al. , Function of hTim8a in complex IV assembly in neuronal cells provides insight into pathomechanism underlying Mohr-Tranebjærg syndrome. eLife 8, e48828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Luthy L., Grutter M. G., Mittl P. R., The crystal structure of Helicobacter pylori cysteine-rich protein B reveals a novel fold for a penicillin-binding protein. J. Biol. Chem. 277, 10187–10193 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Lüthy L., Grütter M. G., Mittl P. R., The crystal structure of Helicobacter cysteine-rich protein C at 2.0 A resolution: Similar peptide-binding sites in TPR and SEL1-like repeat proteins. J. Mol. Biol. 340, 829–841 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Jones S., Thornton J. M., Protein-protein interactions: A review of protein dimer structures. Prog. Biophys. Mol. Biol. 63, 31–65 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Scheufler C., et al. , Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Brinker A., et al. , Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J. Biol. Chem. 277, 19265–19275 (2002). [DOI] [PubMed] [Google Scholar]
- 46.G. J. Gatto, Jr, Geisbrecht B. V., Gould S. J., Berg J. M., Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Xiao Z., et al. , Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: Detection probes and affinity standards. J. Biol. Chem. 286, 11047–11055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banci L., et al. , A structural-dynamical characterization of human Cox17. J. Biol. Chem. 283, 7912–7920 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Horng Y. C., et al. , Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 280, 34113–34122 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Brose J., La Fontaine S., Wedd A. G., Xiao Z., Redox sulfur chemistry of the copper chaperone Atox1 is regulated by the enzyme glutaredoxin 1, the reduction potential of the glutathione couple GSSG/2GSH and the availability of Cu(I). Metallomics 6, 793–808 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Gupta N., Ragsdale S. W., Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erbbeta. J. Biol. Chem. 286, 4392–4403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soto I. C., Fontanesi F., Myers R. S., Hamel P., Barrientos A., A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 16, 801–813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofbauer S., et al. , Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes. Arch. Biochem. Biophys. 574, 36–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deredge D. J., et al. , Ligand-induced allostery in the interaction of the Pseudomonas aeruginosa heme binding protein with heme oxygenase. Proc. Natl. Acad. Sci. U.S.A. 114, 3421–3426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Della Pia E. A., et al. , Redox tuning of cytochrome b562 through facile metal porphyrin substitution. Chem. Commun. (Camb.) 48, 10624–10626 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Lederer F., Glatigny A., Bethge P. H., Bellamy H. D., Matthew F. S., Improvement of the 2.5 A resolution model of cytochrome b562 by redetermining the primary structure and using molecular graphics. J. Mol. Biol. 148, 427–448 (1981). [DOI] [PubMed] [Google Scholar]
- 57.Hanna D. A., et al. , Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. U.S.A. 113, 7539–7544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voronova A., et al. , Oxidative switches in functioning of mammalian copper chaperone Cox17. Biochem. J. 408, 139–148 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banci L., et al. , Human Sco1 functional studies and pathological implications of the P174L mutant. Proc. Natl. Acad. Sci. U.S.A. 104, 15–20 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banci L., et al. , A structural characterization of human SCO2. Structure 15, 1132–1140 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Abajian C., Rosenzweig A. C., Crystal structure of yeast Sco1. J. Biol. Inorg. Chem. 11, 459–466 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Banci L., et al. , A hint for the function of human Sco1 from different structures. Proc. Natl. Acad. Sci. U.S.A. 103, 8595–8600 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber C., Michel B., Bosshard H. R., Spectroscopic analysis of the cytochrome c oxidase-cytochrome c complex: Circular dichroism and magnetic circular dichroism measurements reveal change of cytochrome c heme geometry imposed by complex formation. Proc. Natl. Acad. Sci. U.S.A. 84, 6687–6691 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seabrook S. A., Newman J., High-throughput thermal scanning for protein stability: Making a good technique more robust. ACS Comb. Sci. 15, 387–392 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Stroud D. A., et al. , Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Theodorakis J. L., et al. , A chemical modification of cytochrome-c lysines leading to changes in heme iron ligation. Biochim. Biophys. Acta 1252, 103–113 (1995). [DOI] [PubMed] [Google Scholar]
- 67.Wada K., Okunuki K., Studies on chemically modified cytochrome c. II. The trinitrophenylated cytochrome c. J. Biochem. 66, 249–262 (1969). [DOI] [PubMed] [Google Scholar]
- 68.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.L. E. Formosa et al., Data from Mitochondrial COA7 is a heme-binding protein with disulphide reductase activity, which acts in the early stages of complex IV assembly. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD031589. Deposited 11 February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) (68) partner repository (https://www.ebi.ac.uk/pride/archive) with the dataset identifier PXD031589 (69). The atomic coordinates and structure factors have been deposited in the PDB (https://www.rcsb.org; PDB 7MQZ).