Abstract
Testing is one of the important methodologies used by various countries in order to fight against COVID-19 infection. The infection is considered as one of the deadliest ones although the mortality rate is not very high. COVID-19 infection is being caused by SARS-CoV2 which is termed as severe acute respiratory syndrome coronavirus 2 virus. To prevent the community, transfer among the masses, testing plays an important role. Efficient and quicker testing techniques helps in identification of infected person which makes it easier for to isolate the patient. Deep learning methods have proved their presence and effectiveness in medical image analysis and in the identification of some of the diseases like pneumonia. Authors have been proposed a deep learning mechanism and system to identify the COVID-19 infected patient on analyzing the X-ray images. Symptoms in the COVID-19 infection is well similar to the symptoms occurring in the influenza and pneumonia. The proposed model Inception Nasnet (INASNET) is being able to separate out and classify the X-ray images in the corresponding normal, COVID-19 infected or pneumonia infected classes. This testing method will be a boom for the doctors and for the state as it is a way cheaper method as compared to the other testing kits used by the healthcare workers for the diagnosis of the disease. Continuous analysis by convolutional neural network and regular evaluation will result in better accuracy and helps in eliminating the false-negative results. INASNET is based on the combined platform of InceptionNet and Neural network architecture search which will result in having higher and faster predictions. Regular testing, faster results, economically viable testing using X-ray images will help the front line workers to make a win over COVID-19.
Keywords: Chest X-ray images, Novel coronavirus, COVID-19, Transfer learning, Dense model
1. Introduction
COVID-19 is one of the deadliest and infectious diseases caused by Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV2) [1]. It has been firstly identified in December 2019 in Wuhan, Hubei province, China. And the first case has been reported in November, 2019 [2]. The virus initially named as novel coronavirus but later it has been changed to SARS-CoV2 after studying its internal structure by world health organization. Virus has created so much panic among the masses [3]. We are not seeing it for the first time as SARS and MERS are the virus from coronavirus family which affects the humans severely. We still had not been developed the efficient vaccine or particular effective cure. SARS and MERS were the previous outbreak caused by the members of the coronavirus family in 2002 and 2012 respectively and still showing their terror at regular time intervals [4], [5]. Symptoms in the infected persons are similar to the patient getting infected with common cold, influenza, and pneumonia which makes the identification of the infection based on the symptoms. And fever, cold, fatigue, loss of appetite, loss of smell and taste, lung fibrosis are the major symptom seeing in a COVID-19 infected patient [6], [7]. In severe cases, the person is feeling shortness of breath, uneasiness in breathing, acute respiratory syndrome, and even may have severe pneumonia. The incubation period is ranging from two to fourteen days. WHO has declared the diseases as pandemic as it has taken so many lives all around the world and the death toll is still increasing. The infection is mainly transferred through water droplets coming out of the mouth of an infected person while talking, sneezing, and coughing but it may remain in the air. As a result, to which, COVID-19 infection is considered as an airborne disease. According to the world meters, this disease has taken the lives of about 3,566,512 persons and the toll is still rising. Approximately 171,528,463 persons have got infected with the SARS-CoV2 virus around the world [8]. These numbers are very scary and make us to feel the severity of the disease.
Testing is the main weapon in order to prevent the spread of COVID-19 infection. All countries are working on finding a suitable testing method which will be both efficient and cheaper [9]. There are various testing methods that are present which are not as efficient and are taking very much time in giving the results [10]. RTPCR testing is the most commonly used testing where the sample has been collected from the nasal and throat swabs. In this type of testing, RNA has been first converted into DNA [11]. This amplified DNA has been analyzed to predict the infection in the patient [12], [13]. Antigen based testing method is also available where the antigens are being analyzed in the sample. This testing would not be beneficial in giving the results of predicting whether the patient is currently infected with the virus or not and is also generating false-negative results [14], [15]. Testing plays an important part in combating the infection as if a state tests more and more people, after that only they will be able to identify the infected patients. COVID-19 infected patients are isolated and then their symptoms can only be treated as still there is no particular treatment is available in the market. Separation and isolation of infected person reduce the risk of community transfer which in result reduce the burden over the healthcare system.
Artificial Intelligence has already proved its ability in testing various diseases like tuberculosis [16], pneumonia [17], and cancer. Artificial Intelligence is flying its flag in solving medical and healthcare problems like disease detection, drug detection, affected site detection, and many more [18], [19]. Deep learning techniques and neural networks are being used for detecting pneumonia and tuberculosis using X-ray images [20]. Similar to which, we have developed a testing system which will detect the COVID-19 infected patient using X-ray images. The X-ray images are being analyzed and the system will be able to predict whether the concerned person is COVID-19 positive or not. It is very necessary to separate out the COVID-19 patient from the patients having pneumonia and influenza as symptoms are pretty much similar. We have taken care of this issue and our system will be able to identify the difference between the X-ray image of COVID-19 infected person and the pneumonia affected person. We have been proposing a neural architecture that will be able to give efficient results in no time and is a way cheaper than any other testing method. This method does not require extra specialists and can be easily operable without any requirement of additional material.
2. Related work
There is an invisible competition going among various big research labs and institutes for finding a testing and treatment mechanism for curing the COVID-19 infected person. Digital image analysis has been made significant progress in the past recent years. There are various software and image processing techniques that are available to analyze the image and being able to gain the specific knowledge from those captured images. Deep learning techniques involving image analysis using a convolutional neural network is very significant in gaining the key features from the images. Convolutional neural networks have been used in various healthcare applications like detection of cancerous cells, pneumonia detection, localization of pneumonia patches in the X-ray image of lungs, detection of tumors in the brain, and heart disease detection [21]. Researchers and scientists are working on finding the best suitable method for testing the COVID-19 infection to get the results in less amount of the time. Serological and Molecular testing are the two main testing techniques used for identifying whether the person is infected with COVID-19 infection or not. Antibodies have been searched in the blood sample for confirming the presence of the viral infection in serological testing whereas a genetic material or we can say RNA has been looking for in the sample in molecular testing [22]. Antibody testing is a best-suited example for serological testing where the blood sample has been collected from the patient’s body and checked whether the antibodies are being present in the serum sample or not [23]. Based on the analysis of whether the person has been distinguished as COVID-19 infected patient or not. RT-PCR testing is one of the examples of the molecular testing method. Another testing method is, Reverse transcription-polymerase testing method is used for finding out the person having some COVID-19 related symptoms that whether he is COVID-19 positive or not [24]. In this method, they are analyzing the genetic part of the viral structure that has been present in the sample taken from the suspected person. On determining the desired genetic material, the person has been confirmed as COVID-19 infected patient [25]. There are other testing methods like Quidel test, qPCR testing, Antigen based testing, Isothermal amplification testing, plasmonic photothermal sensing-based testing, and CRISPER based testing [26] method which have been used by the doctors in order to test the COVID-19 suspected patients.
Having all of these different types of testing methods available in the market, why is there a need to developing a new testing mechanism for COVID-19. Innovation is everything and we should have to optimize every part of the process. There are both pros and cons of these testing mechanisms. But cons surpass over pros in terms of economy, efficiency, and time taken to give the results. RT-PCR testing method takes more time and results are not very efficient. Antibody testing is not giving the confirmation whether the person is still carrying the virus. Researchers are working on optimizing the testing mechanism in terms of reducing the time taken to provide the results which helps in early and accurate identification of the infected patients and indirectly will help in contact tracing. Deep learning has been considered as the best suitable option opted by the research fellows working in the computer vision field. CheXnet has been used for the classification of various pulmonary activities including pneumonia. Mohapatra et al. have proposed a method for the detection of leukemia in the stained blood smear and bone marrow microscopic images [27]. Fei shan et al. worked on quantification of lung infection in COVID-19 infected patients based on the analysis over CT scan images [28]. Maghdid et al. has been working on creating an AI framework for detecting COVID-19 patients using smartphone embedded sensors [29]. We are proposing a deep learning model which will be based on InceptionNet and Nasnet combination where we are analyzing the X-ray images of the COVID-19 suspects. Our model is successfully predicting the COVID-19 suspect’s persons whether they are normal or have been infected with the SARS-CoV2 virus. In the next section, we are discussing our proposed mechanism in detail.
3. INASNET method
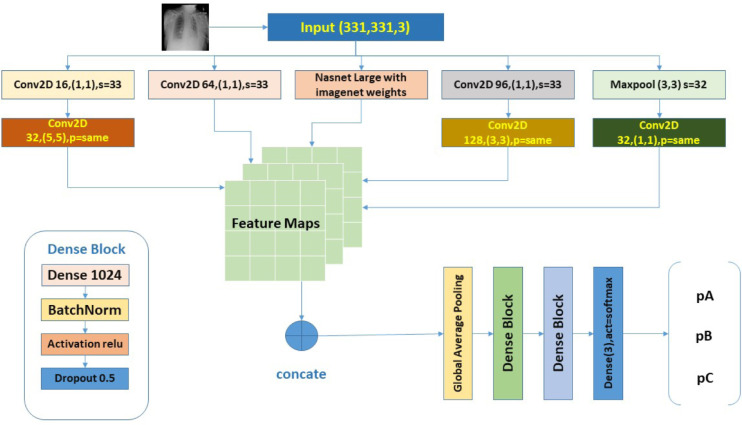
Deep Learning techniques are widely used in the detection of various diseases like pneumonia. Digital image processing has proved its strength in healthcare, bioinformatics, drug design, and medical image analysis. These techniques have been automated most of the process in the medical area. We are using supervised deep learning techniques where we first train our system using a dataset. Based on the experience and the knowledge received from the training process, the system will work in finding the infection in the person when a new X-ray image has been supplemented to it. The artificial neural network used in our system is mainly based on the visual human cortex that can be used in gaining the information. In this work, we are proposing a model for classification of X-ray images from a person having COVID-19 infection, pneumonia infected person, and normal person. Our model is based on deep learning functionalities and is following supervised learning along with reinforcement learning. In supervised learning, the model has been trained over the labeled dataset so that the network is able to learn the features gaining from the corresponding input. We have trained our model over the dataset containing X-ray images of suspects which include COVID-19 infected patients, pneumonia infected patients, and normal persons. These datasets have been provided by the various health institutes and hospitals and are available at open source. The proposed hybrid inception model has been based on the combined result of both NAS structure and InceptionNet. As the X-ray images can easily be analyzed using an ordinary convolution neural network. The overall architecture of the proposed model has been shown in Fig. 1.
Fig. 1.
Overview of the proposed INASNET method.
3.1. Inception block
The CNNs can be stacked together to get a deeper network which may increase the performance of predicting the disease. Gradient upgrades are very difficult to transverse throughout the entire network. Deeper convolutional neural networks have a huge affinity towards over-fitting as the kernel size should be large due to the more global spread of information. And choosing the right kernel is seriously a tedious job. As in the COVID-19 pandemic and because of a large population, speedy inhibition on the infection spread rate, and large randomized dataset availability, inception block will be the best which has provided speedy, efficient and accurate results as compared to other deep learning models. The use of the inception block is computationally less expensive in computing the results due to much wider network. Therefore, instead of having a deeper network, we are creating a wider network applying varied size kernels over the image simultaneously at a particular level. The representation bottleneck has been removed which will reduce the computational expenses and also helps in preventing the loss of information. Label smoothing will be incorporated which will help in preventing over-fitting problems.
3.2. NAS block
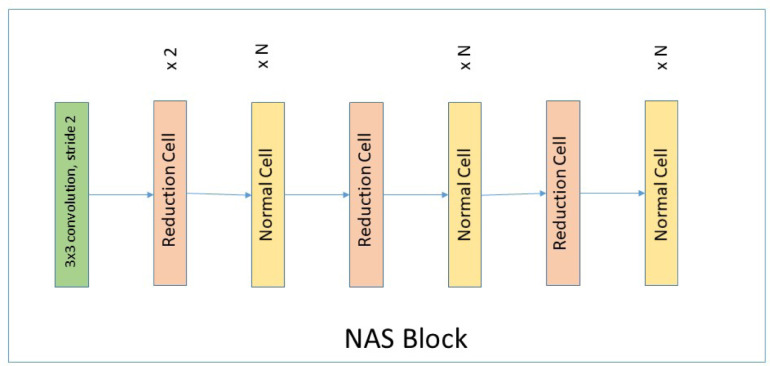
The reinforcement learning supports the neural architecture search space technique which will help in optimizing the regularization parameters and hyper-parameters. The resulting concatenated output from the inception blocks and the NAS layer then be further supplemented to the dense blocks which will help in learning the features and be able to give accurate results. NAS has been added in automating the convolution features in the inception layer. Adding NAS block will help in optimizing the hyper-parameters. Nasnet will helps in learning the features from the large dataset by applying two featured convolution cells [30]. These convolution cells are the normal cells and the reduced cells which return the feature maps. Normal cells return the feature maps of the same height and width whereas the reduced cells return the feature maps whose height and width can be reduced by some factor. The Nas block architecture for imagenet has been shown in Fig. 2.
Fig. 2.
NAS block for imagenet.
Hill climbing technique has been used that applies network morphism where the network has been optimized based on the local search by finding the best solution based on an incremental approach. As we should have to be fully confident about the results that the system will be able to accurately identify. We have included one of the NAS blocks parallel to the inception layer for getting an accurate result. The NAS network has been introduced in parallel to the convolutional blocks and max pool layer as it will add more CNN feature maps to the network and is acting like a skip connection from input to fully connected layer. Doing a stride of 3 × 3 helps the network in gathering the wider function approximation connecting distant features in the image. Max pooling layer in the network will compliment with the locally converged features. All of this aids Nasnet in improving the metrics.
The output is fed to the dense layered block and then the soft-max classifier which will then predict the person category from analyzing the corresponding X-ray image. The dense block consists of a dense layer, batch normalization layer, relu and finally the dropout layer. For increasing the training speed and reduction in co-variance shift of the values, batch normalization layer and dense layer has been used. The dense layer has also been used for reducing the parameters used in the convolution layer. The dropout layer has been added in order to improve the generalization error where we are disabling some of the neurons in the network randomly. Using all these specific layers and developing a hybrid model introduced in this paper that has been used for giving better and faster results in testing on COVID-19 suspects.
3.3. Dense block
Dense block is used in the hybrid model in order to extract the features. It is used for to deepen the network without using any additional parameters. All layers with their matching feature map sizes have been directly connected to each other with the help of dense block. Each layer is obtaining extra inputs from all foregoing layers and the resulting feature map or we can say its own feature map has been passed to its following layers. This property will maintain the feed forward nature in the network. This will help in the reduction of the usage of number of extra parameters. Dense Block consists of dense layer, Batch Normalization layer, relu as an activation function, dropout layer.
3.3.1. Dense layer
Dense layer used is a deep network of neural layers. The output of the dense layer depends on the number of units specified in the dense layer. Using the dense layer will allow the largest potential function approximation in the limit of the provided width of the given layer. This will be used to avail the space under limited function approximations. In dense layer, each hidden unit in one layer has been mapped to every other hidden unit in the subsequent layer. On increasing the width of the network, the network could simultaneously approximate more functions resulting in the expansion of the solution space. Network could use those parallel approximations resulting in making of more informed decisions by increasing the depth of the network and dense layer will help in that in order to increase the depth at the needed time.
3.3.2. Batch normalization
Batch Normalization layer can be used to increase the training speed of the network. Using batch normalization we are actually decreasing the co-variate shift. Co-variate shift will help each layer of the network in learning on a more stable distribution of inputs. It is actually the variation in the dispersion of input values to a learning algorithm [31]. In hidden layers this is called internal co-variate shift which makes the training slow and will require very small learning rate and a good parameter initialization. Batch Normalization is a process by which the layer’s inputs have been normalized over a mini batch. Mini batch consists of sample of random images from all the three class labels. Higher learning rate, regularization effect, usage of saturating non-linearities are advantages achieved using batch normalization.
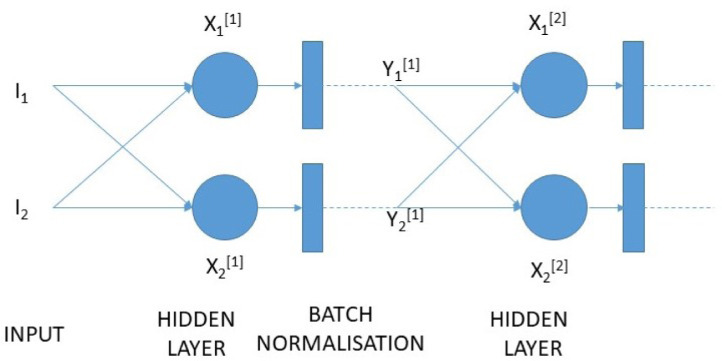
The mathematical formulation for Batch Normalization in a neural network has been given in the equations from 2–5. Let X be the input before batch normalization whereas Y be the output after batch normalization as shown in Fig. 3
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Batch normalization adds only two extra parameters in the neural network. According to the series of above equations and normal assumptions behind batch normalization for understanding purpose, the mean () & variance () have been calculated for the mini batch. The normalized values can be obtained by subtracting mean form and subsequently dividing by standard deviation (). We have calculated by multiplying with the scale () and adding a shift () and placing as a non linearity’s input.
Fig. 3.
Batch normalization on a simple network.
3.3.3. Dropout
To regularize our network we are using an additional dropout layer in the hybrid network. When hidden units or neurons of a specific layer have been always become influenced by the output of a particular hidden unit in the previous layer, it will reduce the efficiency of our model. To prevent this indirect biasing, we are make use of dropout layer which will helps in better training of our hybrid model ultimately leads to better efficiency and accuracy. The layer will reduce the extra dependent nature of particular hidden units in the neural network. Use of dropout layer finally results in reducing the over-fitting problem.
4. Experimental results
4.1. Dataset
In this work, we use the open-source and publicly available chest X-ray dataset. In this, dataset consists of three different class image data such as normal, pneumonia, and COVID-19. The COVID-19 is a new pandemic disease, and the availability of this virus related images are very less.
For experimental analysis, we merged and modified three distinct datasets consisting of COVID-19 images. These three datasets are: 1) COVID-19 Chest X-ray Dataset Initiative [32], RSNA Pneumonia Detection Challenge dataset [33] and COVID-19 Image Data Collection [34]. In this combined dataset contains a very less amount of COVID-19 infection images available compare with the other two classes. The number of COVID-19 images are 183 CXR images that are collected from 121 corona virus-infected patients. And the total number of normal and pneumonia patient cases is 8066 and 5538 respectively. The paper related code is available publicly for open access at https://github.com/murukessanap/inceptionNasnet.
4.2. Results
The performance of our model have been measured on metrics like accuracy (AC), precision, recall and F1 score. For measuring the fruition of our model we have calculated True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN). The results which has been correctly predicted by the model have been included in the True Positive list. The results of a COVID-19 suspects whose results are negative and actually are not infected with the SARS-CoV2 virus have been listed under True Negative list. False positive is an error in the prediction where the person without having COVID-19 infection have been confirmed as COVID-19 patient. False negative results are the error in the results where the COVID-19 infected person have been predicted as COVID-19 negative. The metrics have been calculated by the following equations as mentioned:
| (7) |
The F1-score in terms of precision and recall is being calculated as follows:
| (8) |
| (9) |
| (10) |
Our novel hybrid approach has proved its capability and has made a step forward in becoming the best testing method in COVID-19 pandemic. The results are promising and has been approved by various doctors and have been recognized by front line workers. The calculated parameters for various different models and for INASNET have been shown in the Table 1.
Table 1.
Test metrics comparison among different models on COVID X-ray data (Normal, Pneumonia, COVID-19) where Precision, Recall and F1-score are weighted average across classes.
| S. No. | Model | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| 1 | VggNet | 0.896 | 0.90 | 0.90 | 0.89 |
| 2 | Resnet | 0.904 | 0.91 | 0.90 | 0.90 |
| 3 | InceptionNet | 0.900 | 0.91 | 0.90 | 0.90 |
| 4 | Nasnet | 0.861 | 0.87 | 0.86 | 0.86 |
| 6 | INASNET | 0.943 | 0.94 | 0.94 | 0.94 |
No additional equipment’s are necessary for the proposed testing method. In X-ray testing mechanism, the main problem that the doctors and the states are facing is about the availability of the specialized radiologist who have the necessary skills to identify the confirmed cases on analyzing the X-ray report of a suspect. This manual monitoring is very difficult and there are likely high chances of getting infection to that person due to the continuous exposure with the COVID-19 infected persons. Our proposed hybrid model is handling all these concerns by automating the testing process where no further manual monitoring is required. The results of our proposed method have also been compared with the other well known deep learning models. These models include VGGNet, Resnet, InceptionNet and Nasnet.
We are getting a quite impressive results from our model as compared to other ones. Comparison between the results of our proposed hybrid model with the other deep learning models have been shown in the Table 1. INASNET is working very efficiently and have achieved an accuracy score of 0.943 as compared to any other deep learning model. The accuracy achieved by INASNET over test set is a way higher than the other model and the accuracy results are shown in Table 1.
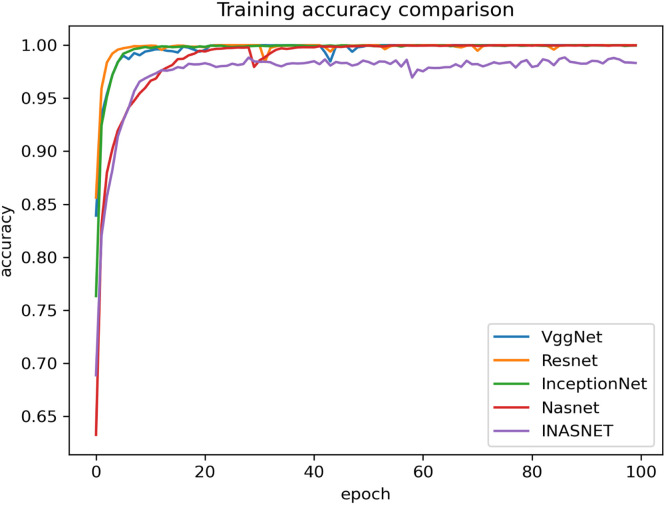
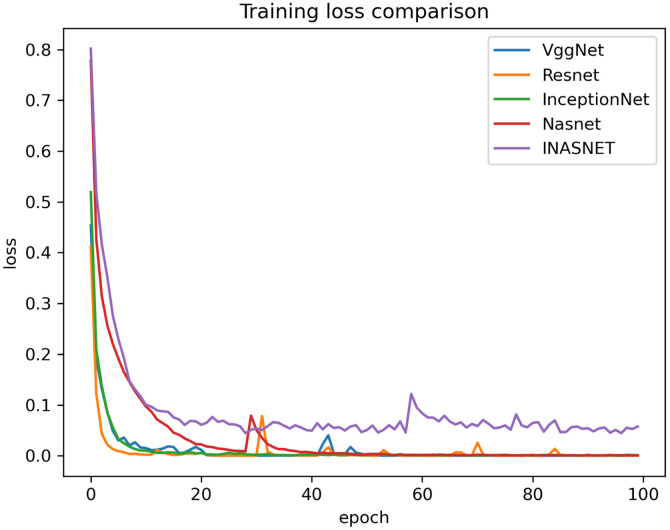
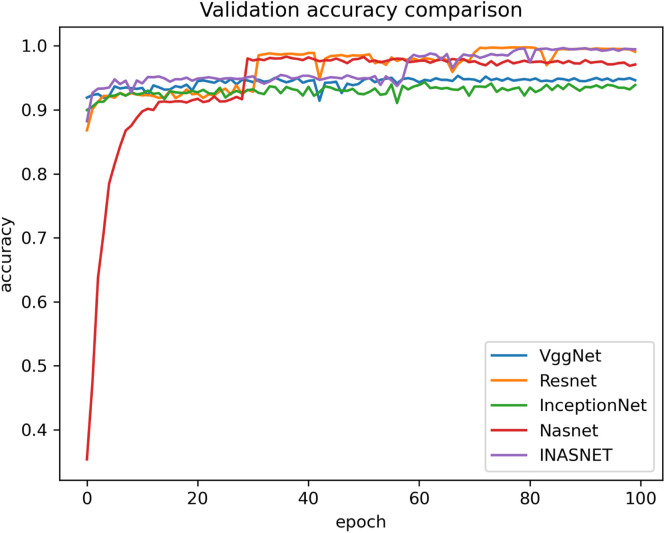
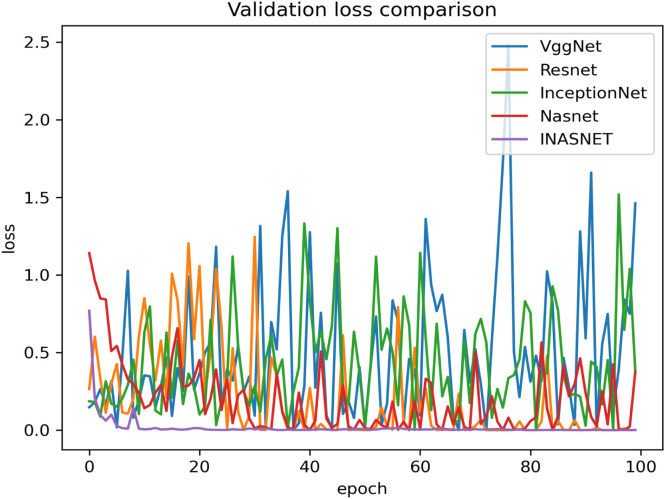
Our proposed model is suitable for the real time classification task and can be used on real time dataset which backs our model to stand uniquely. The other models have gone into over-fitting condition and is not suitable over the real time images. Our model is having excellent training accuracy and have surpassed the over-fitting problem which makes it the only model to use for analyzing the X-ray images from the COVID-19 suspects. The training accuracy graph of the hybrid model along with comparison with the pre-existing ones has been shown in Fig. 4. The proposed model has achieved training accuracy of about 98.84 which is quite remarkable. The information loss during training phase is also not very high which will help in the better training of our hybrid approach. Lower training loss helps in the better classification at modeling the relationship between the input data and the output labels. The training loss is calculated as a moving average over all processed batches. The loss in early training stage when it sinks rapidly the first batch of an epoch will have a much higher loss as compared to the previous one. When the epoch is finished, the resulting training loss will not symbolize the loss at the end of the epoch but the average training loss from start to end of the epoch. The graph showing the training loss comparison of the proposed model with the other deep learning models have been shown in Fig. 5. Validation accuracy is the accuracy calculated over validation dataset or we can say in general it have been computed on the images that have not been used for the training purposes. The images be used for validating the generalization ability of our model. Our model is having the best validation accuracy as compared to the other deep learning models like VGGNet, Resnet, InceptionNet and Nasnet. We are also taking care of the over-fitting problem by regularization mechanism used in the NAS block. The validation accuracy graph showing the validation accuracy of all the models including the hybrid model has been shown in Fig. 6. The graph showing the validation loss of the proposed model comparing it with the other deep models has been shown in Fig. 7.
Fig. 4.
Comparison of proposed method training accuracy with different models.
Fig. 5.
Comparison of proposed method training Loss with different models.
Fig. 6.
Comparison of proposed method validation accuracy with different models.
Fig. 7.
Comparison of proposed method validation loss with different models.
The proposed model is having better validation loss with respect to other showing that our model has been performed well on the validation set as well. The average validation loss for the different models have been shown in the Table 2 for better understanding of our proposed model’s efficiency.
Table 2.
Comparison of the INASNET model along with the different deep learning model over validation loss average.
| S. No. | Method | Average Validation Loss |
|---|---|---|
| 1 | VggNet | 0.508 |
| 2 | Resnet | 0.202 |
| 3 | InceptionNet | 0.423 |
| 4 | Nasnet | 0.197 |
| 6 | INASNET | 0.019 |
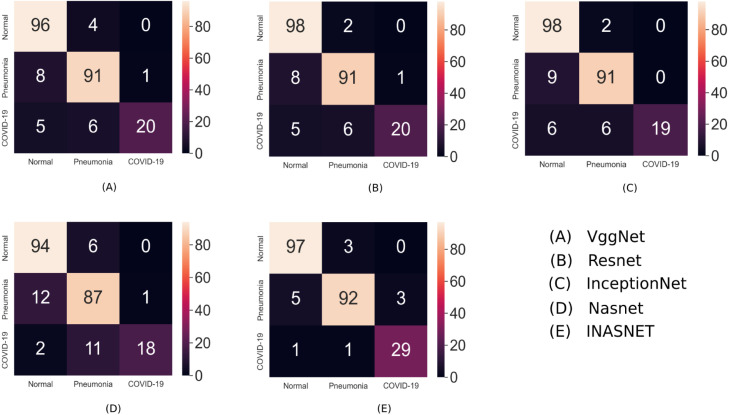
The performance of our hybrid proposed model for COVID-19 detection using X-ray images can be visualized using confusion matrix. The matrix will give a clear understanding of our results and will help in analyzing the statistics of our classification task. Instances in a predicted class has been represented by the rows whereas the columns represents the instances of the actual class. Besides, we have analyzed the performance of the other deep learning models using the confusion matrix. The comparison results and the confusion matrix of INASNET and other models has been shown in Fig. 8. Based on this we are proposing that our hybrid model works best on the COVID-19 detection and is way more efficient than any other deep learning model.
Fig. 8.
Confusion matrix and comparison results of INASNET with the other deep learning models.
5. Conclusion
We have developed a hybrid inception model which will help the doctors and the COVID-19 frontline workers in screening the COVID-19 infected person at community level and is way cheaper as compared to the current detecting techniques. Corona testing kits which are available in the market are too expensive to be afford by a common people which will ultimately lead an economic burden on the patient as well as on the state. INASNET model have been trained on complex datasets and is optimized by having batch normalization and dense layer which will help in increasing the efficiency for detection. This will also helps the hospitals in having a better understanding of COVID-19 by having a significant amount of X-ray’s for a person which makes them to have a regular check on the area that have been affected in the lungs due to SARS-CoV2.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank the anonymous reviewers for their thorough review and valuable comments. This work was supported in part by grants Government of India, Ministry of Human Resource Development and NIT Warangal under NITW/CS/CSE-RSM/2018/908/3118 project.
References
- 1.Zhu Na, Zhang Dingyu, Wang Wenling, Li Xingwang, Yang Bo, Song Jingdong, et al. A novel coronavirus from patients with pneumonia in China 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenjie Tan, Wenjie Tan, Zhao Xiang, Ma Xuejun, Wang Wenling, Niu Peihua, et al. China 2019– 2020, Vol. 2. China CDC weekly; 2020. A novel coronavirus genome identified in a cluster of pneumonia cases– Wuhan; pp. 61–62. [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya Alexander E., Baker Susan C., Baric Ralph, de Groot Raoul J., Drosten Christian, Gulyaeva Anastasia A., et al. 2020. Severe acute respiratory syndrome-related coronavirus: The species and its viruses–a statement of the coronavirus study group. [Google Scholar]
- 4.Heymann David L., Rodier Guénal. Global surveillance, national surveillance, and SARS. Emerg Infect Diseases. 2004;10(2):173. doi: 10.3201/eid1002.031038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Killerby Marie E., Biggs Holly M., Midgley Claire M., Gerber John T. Middle east respiratory syndrome coronavirus transmission. Emerg Infect Diseases. 2020;26(2):191. doi: 10.3201/eid2602.190697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.COVID, CDC, Response Team, Jorden Michelle A., Rudman Sarah L., Villarino Elsa, et al. Evidence for limited early spread of COVID-19 within the United States january–2020. Morb Mortal Wkly Rep. 2020;69(22):680. doi: 10.15585/mmwr.mm6922e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadnytskyi Valentyn, Bax Christina E., Bax Adriaan, Anfinrud Philip. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc Natl Acad Sci. 2020;117(22):11875–11877. doi: 10.1073/pnas.2006874117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.https://www.worldometers.info/coronavirus/.
- 9.Centers for Disease Control and Prevention . 2020. How to protect yourself & others. centers for disease control and prevention website. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/prevention-H.pdf. [Google Scholar]
- 10.World Health Organization . 2020. Laboratory testing of 2019 novel coronavirus (2019-nCoV) in suspected human cases: interim guidance; p. 2020. https://www.who.int/publications/i/item/10665-331501. [Google Scholar]
- 11.2019 Novel Coronavirus (2019-nCoV) Situation Summary . 2020. Centers for disease control and prevention. Archived from the original on 26 2020. Retrieved 30 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200130-sitrep-10-ncov.pdf. [Google Scholar]
- 12.Real-Time RT-PCR Panel for Detection 2019-nCov . 2020. Centers for disease control and prevention. Archived from the original on 30 2020. Retrieved 1 2020. https://www.fda.gov/media/134922/download. [Google Scholar]
- 13.Brueck Hilary. There’s only one way to know if you have the coronavirus, and it involves machines full of spit and mucus. Bus Insider. 2020;28(3804):31–46. [Google Scholar]
- 14.FDACoronavirus U S. Update:FDA authorizes first antigen test to help in the rapid detection of the virus that causes COVID-19 in patients 2020 US Food Drug Administration (9 May).
- 15.FDA issues emergency approval of new antigen test that is cheaper . 2020. Faster and simpler. Washington post.9 may. https://www.washingtonpost.com/health/2020/05/09/fda-issues-emergency-approval-new-antigen-test-that-is-cheaper-faster-simpler/ [Google Scholar]
- 16.Raju Manoj, Aswath Arun, Kadam Amrit, Pagidimarri Venkatesh. Advances in analytics and applications. Springer; Singapore: 2019. Automatic detection of tuberculosis using deep learning methods; pp. 119–129. [Google Scholar]
- 17.Abiyev Rahib H., Mohammad Khaleel Sallam Ma’aitaH. Deep convolutional neural networks for chest diseases detection. J Healthcare Eng. 2018;2018 doi: 10.1155/2018/4168538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadav Milind, Perumal Murukessan, Srinivas M. Analysis on novel coronavirus (COVID-19) using machine learning methods. Chaos Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivas M., Naidu R Ramu, Sastry Challa S., Krishna Mohan C. Content based medical image retrieval using dictionary learning. Neurocomputing. 2015;168:880–895. [Google Scholar]
- 20.Pranav Rajpurkar, Irvin Jeremy, Zhu Kaylie, Yang Brandon, Mehta Hershel, Duan Tony, et al. 2017. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225. [Google Scholar]
- 21.Miao Kathleen H., Miao Julia H. Coronary heart disease diagnosis using deep neural networks. Int J Adv Comput Sci Appl. 2018;9(10):1–8. [Google Scholar]
- 22.Vogel Gretchen. New blood tests for antibodies could show true scale of coronavirus pandemic. Science. 2020 [Google Scholar]
- 23.Abbasi Jennifer. The promise and peril of antibody testing for COVID-19. JAMA. 2020;323(19):1881–1883. doi: 10.1001/jama.2020.6170. [DOI] [PubMed] [Google Scholar]
- 24.Vincent Myriam, Xu Yan, Kong Huimin. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendel Ayal, Bak Rasmus O., Clark Joseph T., Kennedy Andrew B., Ryan Daniel E., Roy Subhadeep, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature Biotechnol. 2015;33(9):985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broughton James P., Deng Xianding, Yu Guixia, Fasching Clare L., Servellita Venice, Singh Jasmeet, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohapatra Subrajeet, Patra Dipti, Satpathy Sanghamitra. An ensemble classifier system for early diagnosis of acute lymphoblastic leukemia in blood microscopic images. Neural Comput Appl. 2014;24(7):1887–1904. [Google Scholar]
- 28.Shan Fei, Gao Yaozong, Wang Jun, Shi Weiya, Shi Nannan, Han Miaofei, et al. 2020. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655. [Google Scholar]
- 29.Maghded Halgurd S., Ghafoor Kayhan Zrar, Sadiq Ali Safaa, Curran B., Rabie Khaled. 2020 IEEE 21st international conference on information reuse and integration for data science. IEEE; 2020. A novel AI-enabled framework to diagnose coronavirus COVID-19 using smartphone embedded sensors: design study; pp. 180–187. [Google Scholar]
- 30.Zoph Barret, Vasudevan Vijay, Shlens Jonathon, Le Quoc V. Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2018, p. 8697–710.
- 31.Ioffe Sergey, Szegedy Christian. International conference on machine learning. PMLR; 2015. Batch normalization: Accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- 32.Pan Ian., Cadrin-Chênevert Alexandre., Cheng Phillip M. Tackling the radiological society of north America pneumonia detection challenge. Am J Roentgenol. 2019;213(3):568–574. doi: 10.2214/AJR.19.21512. [DOI] [PubMed] [Google Scholar]
- 33.Cohen Joseph Paul, Morrison Paul, Dao Lan, Roth Karsten, Duong Tim Q., Ghassemi Marzyeh. 2020. Covid-19 Image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006.11988. [Google Scholar]
- 34.COVID, Actualmed Chung A. 2020. Chest x-ray data initiative. URL https://github com/agchung/Actualmed-COVID-chestxray-dataset (19) [Google Scholar]