Significance
Modifications of RNA, including methylations of cytosine (m5C) and adenosine (m6A), have important roles in RNA metabolism, cellular responses to stress, and biological processes of differentiation and development. We report on the profiles of m5C messenger RNA (mRNA) modifications in malaria parasites that infect rodents (Plasmodium yoelii) and humans (Plasmodium falciparum). These parasites have genes that encode homologs of human and plant NSUN2 methyltransferases (m5C “writers”). We show that one of these homologs, termed PyNSUN2, stabilizes mRNA transcripts in P. yoelii and mediates m5C-associated development of the parasite sexual stages (gametocytes). Further research on m5C and other epitranscriptomic modifications may yield new insights into molecular pathways of gametocyte development and mosquito infectivity that can be exploited to interrupt malaria transmission.
Keywords: RNA methyltransferase, gametocytogenesis, epitranscriptomic modifications, RNA bisulfite sequencing, gene knockout
Abstract
5-methylcytosine (m5C) is an important epitranscriptomic modification involved in messenger RNA (mRNA) stability and translation efficiency in various biological processes. However, it remains unclear if m5C modification contributes to the dynamic regulation of the transcriptome during the developmental cycles of Plasmodium parasites. Here, we characterize the landscape of m5C mRNA modifications at single nucleotide resolution in the asexual replication stages and gametocyte sexual stages of rodent (Plasmodium yoelii) and human (Plasmodium falciparum) malaria parasites. While different representations of m5C-modified mRNAs are associated with the different stages, the abundance of the m5C marker is strikingly enhanced in the transcriptomes of gametocytes. Our results show that m5C modifications confer stability to the Plasmodium transcripts and that a Plasmodium ortholog of NSUN2 is a major mRNA m5C methyltransferase in malaria parasites. Upon knockout of P. yoelii nsun2 (pynsun2), marked reductions of m5C modification were observed in a panel of gametocytogenesis-associated transcripts. These reductions correlated with impaired gametocyte production in the knockout rodent malaria parasites. Restoration of the nsun2 gene in the knockout parasites rescued the gametocyte production phenotype as well as m5C modification of the gametocytogenesis-associated transcripts. Together with the mRNA m5C profiles for two species of Plasmodium, our findings demonstrate a major role for NSUN2-mediated m5C modifications in mRNA transcript stability and sexual differentiation in malaria parasites.
Malaria is a mosquito-borne infectious disease caused by unicellular apicomplexan protozoa of the genus Plasmodium. Transmission of these parasites in 2020 caused an estimated 241 million infections and 627,000 deaths globally, a 12% increase over malaria deaths in 2019 (1). Among the species infecting humans, Plasmodium falciparum is the most virulent and accounts for most of these deaths. Plasmodium parasites have a complex life cycle that alternates between mosquito and vertebrate hosts, in which the parasites complete numerous rounds of asexual proliferation in the haploid state and pass through a brief diploid period following obligatory mating of male and female gametes in the mosquito midgut (2). During the proliferation of intraerythrocytic parasites in the vertebrate bloodstream, a small fraction of the asexual population commits to gametocytogenesis, producing sexual-stage gametocytes that are taken up by mosquitoes in which they emerge from red blood cells as gametes (3, 4). These cellular developments are associated with stage-specific and highly dynamic transcriptomes under the hierarchical control of transcription factors and epigenetic regulators (3, 5, 6).
Epigenetic regulation at the transcriptional level plays a critical role in genome expression events and their global outcome. For example, heterochromatin protein 1 (HP1)-dependent heterochromatin modification provides a transcriptionally repressive microenvironment for the silencing of antigenically variant genes and ap2-g, a master regulator of sexual commitment, thus determining parasite adaptation and development in the human host and transmission into mosquitoes (5, 7, 8). Regulatory pathways, such as RNase-mediated nascent decay, m6A modification-mediated messenger RNA (mRNA) stability, and RNA helicase-associated translational repression confer other important mechanisms for fine-tuning posttranscriptional regulation in malaria parasites (9–11). These findings demonstrate that the highly dynamic transcriptome of malaria parasites is controlled by a complex but well-organized multilayered regulatory network.
While DNA methylation has been widely studied as an epigenetic phenomenon, characterizations of mRNA methylation in the modulation of transcriptome stability and expression are more recent (12–14). Reversible mRNA modifications are now known to be involved in the posttranscriptional regulation of gene expression in eukaryotes (10). For example, N6-methyladenosine (m6A) is the most prevalent RNA modification and has been investigated extensively in many eukaryotic organisms, particularly in its association with various cellular processes, such as cell differentiation, embryonic development, and stress responses. In these processes, m6A epitranscriptomic modifications are involved in regulation of mRNA metabolism, translation, decay, and microRNA biogenesis (10, 15, 16). In malaria parasites, the epitranscriptome has likewise been recognized as an important modulator of posttranscriptional gene expression. Baumgarten et al. (14) described highly regulated features of m6A mRNA methylation in P. falciparum, found that these features are associated with stage-specific fine-tuning of the transcriptional cascade, and suggested that widely distributed m6A modifications may shape transcriptome expression through effects on mRNA stability during blood-stage development.
In addition to m6A, RNA modifications such as N1-methyladenosine (m1A), m5C, 5-hydroxymethylcytosine (hm5C), pseudouridine (Ψ), adenosine-to-inosine (I), cytosine-to-uridine (U), and methylation of ribose at the 2′ position (2′-O-Me) may affect RNA processes, including pre-mRNA splicing, nuclear export, transcript stability, and translation initiation (16). Modification of mRNA by m5C is important for diverse biological processes in bladder cancer, HIV-1 infection, and developmental progressions of zebrafish, Arabidopsis thaliana, and rice (12, 17–19), suggesting that cytosine methylation is a powerful regulator at the epitranscriptomic level (15). In other studies, pathways involving m5C-modified transcripts have been found to include DNA repair and homologous recombination (20). Specific methyltransferases (m5C writers) in various species include NSUN2 in humans, TRM4B in A. thaliana, and OsNSUN2 in rice. NSUN2 can drive human urothelial carcinoma pathogenesis by targeting the m5C methylation site in the HDGF 3′ untranslated region (UTR) (18). TRM4B, by promoting mRNA stability through m5C modifications, influences the transcriptional levels of genes involved in the root development of A. thaliana (12). Mutation of OsNSUN2 affects photosynthesis efficiency in rice (19).
Among binding proteins (m5C “readers”) that recognize m5C-modified transcripts are the Aly/REF export factor (ALYREF) in humans and the Y-box binding protein 1 (Ybx1) in zebrafish. ALYREF is an adaptor that regulates the transport of m5C-modified transcripts from the nucleus to the cytoplasm (13), whereas Ybx1 promotes the stability of its target mRNAs in an m5C-dependent manner (21). A potential m5C demethylase, ALKBH (an AlkB homolog), was reported more recently from A. thaliana and is attracting study for its possible role as an m5C “eraser” (22, 23).
In P. falciparum, the DNA methyltransferase homolog TRDMT1 has been shown to methylate position 38 of aspartic acid transfer RNA (tRNA), where the m5C modification may modulate translation of proteins involved in pathogenesis, parasite stress response, and gametocytogenesis (24, 25). More generally, a number of RNA (cytosine-5)-methyltransferase (RCMT) orthologs have been predicted from genome analysis of Plasmodium species, including P. falciparum and the rodent malaria parasite Plasmodium yoelii (26–28).
The present study was designed to characterize the overall transcriptome profile and stage-specific dynamics of m5C mRNA modifications in the asexual replicating forms and gametocytes of P. yoelii and P. falciparum. Our results demonstrate comparatively high levels of m5C mRNA methylation in the gametocytes of both species. Enhanced stability of m5C transcripts is likely to play an important role in the processes of sexual stage development and transmission. P. yoelii and P. falciparum each carry an ortholog of NSUN2 (PyNSUN2; PfNSUN2) that functions as a methyltransferase with a major role in these m5C modifications of the transcriptome. We show that P. yoelii gametocytogenesis is disrupted by knockout (KO) of the Pynsun2 gene and can be reestablished by restoration of Pynsun2 expression.
Results
Global Profiles of m5C Modification in the P. yoelii and P. falciparum Transcriptomes.
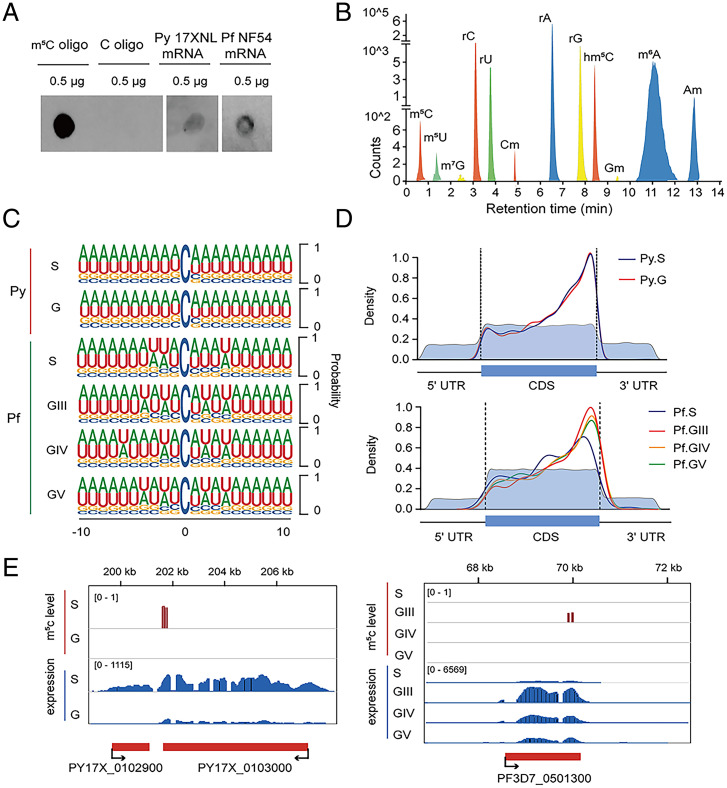
To test for detectable m5C modifications, dot blot assays were performed with mRNA preparations from P. yoelii and P. falciparum mRNAs. Using an anti-m5C antibody previously shown to detect mRNA m5C modifications in Oryza sativa (19), our experiments demonstrated the presence of m5C modifications in schizont-stage parasites of both Plasmodium species (Fig. 1A).
Fig. 1.
Features of m5C-modified mRNA in malaria parasites. (A) Dot blot assays show detection of m5C modifications in schizont-stage mRNAs (enriched from total RNA) from P. falciparum and P. yoelii. m5C-modified and unmodified RNA oligonucleotides served as positive and negative controls, respectively. (B) LC-MS/MS signals indicating modified nucleotides from P. yoelii schizont-stage mRNA. Am, 2′-O-methyladenosine; Cm, 2′-O-methylcytosine; Gm, 2′-O-methylguanosine; hm5C, 5-Hydroxymethylcytosine; m5C, 5-methylcytosine; m5U, 5-methyluridine; m6A, N6-methyladenosine; m7G, N7-methyl-2′-guanosine; rA, adenosine; rC, cytosine; rG, guanosine; rU, uracil. (C) Frequency logo displays of the nucleotides proximal to mRNA m5C sites in the schizonts (S) and gametocytes (G) of P. yoelii (Py), and the schizonts (S) and stage III, IV, and V gametocytes (GIII, GIV, GV) of P. falciparum (Pf). (D) Density distribution of the m5C sites in mRNA transcripts of P. yoelii and P. falciparum. The moving averages (10-bp window) of percentage mRNA cytosine content (light blue) are lower in the 5′ and 3′ UTR regions than in the CDS, as expected for these species. (E) Integrative Genomics Viewer displays of example transcript sequence levels and their m5C sites from genes in P. yoelii (Left) and P. falciparum (Right). Vertical red bars indicate the m5C levels detected in specific parasite stages. The m5C levels were normalized to their corresponding transcript fragment abundance levels before comparison.
For quantitative assessments of modified nucleotides in the P. yoelii transcriptome, purified mRNA preparations from P. yoelii were completely digested to mononucleotides, which were then dephosphorylated and subjected to analysis by triple quadrupole liquid chromatography-mass spectrometry (QQQ LC-MS). Results identified well-known mRNA modifications, including m6A, the most abundant mRNA modification in eukaryotes including P. falciparum (14, 29). Other modifications, including m5C, hm5C, m5U, and m7G were detected at lower levels (Fig. 1B).
To obtain transcriptome-wide maps of m5C modification at single-base resolution, we performed RNA bisulfite sequencing (BisSeq) on mRNA preparations from asexual schizont (S) and sexual gametocyte (G) stages of P. yoelii, and from synchronized schizonts and three stages of induced gametocytes (5 to 6 d stage III, 8 to 9 d stage IV, and 12 to 13 d stage V) of P. falciparum. For each stage, two biological replicates were used for high-confidence site calling (Dataset S1). The m5C sites identified between the independent replicates displayed variation and reproducibility comparable to those of high-quality studies from other organisms, such as zebrafish, human, and mouse (21, 30). About one-third of m5C sites were shared between replicates (SI Appendix, Fig. S1A). The methylation levels of these shared (common) m5C sites were significantly higher than those of the m5C sites unique to one replicate or the other (SI Appendix, Fig. S1A, box plots). Median fractional methylation levels at the shared sites were 0.77 for P. yoelii schizonts and 0.15 for P. falciparum schizonts. In comparison, the median m5C methylation levels were 0.39 for P. yoelii day 3 gametocytes, and 0.2, 0.48, and 0.39 for the stage III, IV, and V P. falciparum gametocytes, respectively (SI Appendix, Fig. S1B).
Examination of complete transcripts harboring m5C modifications showed that individual transcripts more often carried these modifications at two or more sites than at just one site alone, and that a greater fraction of P. yoelii transcripts carried multiple m5C modifications than did P. falciparum transcripts (SI Appendix, Fig. S1C and Dataset S1). Frequency analyses of neighboring nucleotides downstream of the m5C modifications found that most modifications were at CHH sequences (where H = A, C, U) (SI Appendix, Fig. S1D) and often in the context of AU-rich segments (Fig. 1C). We also analyzed the bases upstream of the methylated sites and found that motifs of GHC, HHC, and HGC predominated (SI Appendix, Fig. S1D), which is similar to the case for rice (19).
The large majority of m5C modifications were detected within the transcript coding sequences (CDSs) (SI Appendix, Fig. S1E), where most of these m5C sites were located upstream of stop codons (Fig. 1D) in a different pattern than in human HeLa cells (13) and zebrafish embryos (21). Fig. 1E presents representative examples of P. yoelii and P. falciparum transcripts that carry m5C modifications in the 3′ regions of their CDSs.
Profiles of m5C Modifications in the Asexual and Sexual Stages of P. yoelii and P. falciparum.
Results of RNA-BisSeq analysis identified 7,409 m5C modifications in 527 mRNA transcripts from the P. yoelii schizont set (Py17X_S) (Dataset S2) and 9,168 m5C modifications in 796 transcripts from the P. yoelii gametocyte set (Py17X_G) (Dataset S2). In contrast, results from the P. falciparum datasets showed lower levels of 335 m5C modifications in 220 asexual-stage transcripts, and 781, 419, and 891 m5C modifications in 422 GIII, 224 GIV, and 438 GV transcripts, respectively (Dataset S2).
To further investigate the potential roles of mRNA m5C modifications in the Plasmodium life cycle, we performed gene ontology (GO) analysis on the sets of methylated transcripts from P. yoelii and P. falciparum. For P. yoelii gametocytes, results from this analysis identified enriched numbers of transcripts relating to metabolic/cellular processes and sexual development. Correspondingly, for P. falciparum gametocytes, transcripts relating to cellular and metabolic processes were enriched in GIII stages and transcripts relating to sexual development were enriched in GV stages (Table 1). In the asexual-stage parasites including schizonts, m5C modifications were enriched with transcripts having assigned GO terms of nucleoside transport, nucleic acid metabolic processes, and chromatin organization (Table 1). These features were also evident in heatmaps of the m5C-modified transcripts, which identified similarities as well as distinct differences between the transcript methylations in asexual stages and gametocytes of the P. yoelii and P. falciparum populations (SI Appendix, Fig. S2).
Table 1.
Gene ontology (GO) assignments for m5C-modified transcripts in schizonts and gametocytes of P. yoelii and P. falciparum
| ID | Biological processes | Result count | P value | Stage |
| GO:0007049 | Cell cycle | 5 | 9.12E-03 | Py.S |
| GO:0015698 | Inorganic anion transport | 2 | 1.18E-02 | Py.S |
| GO:0090304 | Nucleic acid metabolic process | 49 | 1.99E-02 | Py.S |
| GO:0006352 | DNA-templated transcription, initiation | 4 | 4.45E-02 | Py.S |
| GO:0006720 | Isoprenoid metabolic process | 3 | 4.73E-02 | Py.S |
| GO:0016310 | Phosphorylation | 28 | 9.44E-04 | Py.G |
| GO:0006796 | Phosphate-containing compound metabolic process | 46 | 1.31E-03 | Py.G |
| GO:0009166 | Nucleotide catabolic process | 7 | 2.20E-03 | Py.G |
| GO:0046031 | ADP metabolic process | 6 | 2.27E-03 | Py.G |
| GO:0046939 | Nucleotide phosphorylation | 6 | 7.41E-03 | Py.G |
| GO:0006468 | Protein phosphorylation | 21 | 1.24E-02 | Py.G |
| GO:GNF0004 | Sexual development | 78 | 1.48E-08 | Py.G |
| GO:0006325 | Chromatin organization | 5 | 6.59E-03 | Pf.S |
| GO:0043543 | Protein acylation | 4 | 7.73E-03 | Pf.S |
| GO:0000280 | Nuclear division | 3 | 2.06E-03 | Pf.S |
| GO:0015858 | Nucleoside transport | 2 | 4.82E-03 | Pf.S |
| GO:0031365 | N-terminal protein amino acid modification | 2 | 9.38E-03 | Pf.S |
| GO:0015858 | Nucleoside transport | 3 | 4.94E-04 | Pf.GIII |
| GO:0009987 | Cellular process | 159 | 1.18E-03 | Pf.GIII |
| GO:0031365 | N-terminal protein amino acid modification | 3 | 1.86E-03 | Pf.GIII |
| GO:0006325 | Chromatin organization | 8 | 1.78E-03 | Pf.GIII |
| GO:1901642 | Nucleoside transmembrane transport | 2 | 6.26E-03 | Pf.GIII |
| GO:0008152 | Metabolic process | 140 | 4.68E-03 | Pf.GIII |
| GO:0042273 | Ribosomal large subunit biogenesis | 5 | 8.46E-03 | Pf.GIII |
| GO:0007155 | Cell adhesion | 9 | 1.65E-03 | Pf.GIV |
| GO:0006811 | Ion transport | 9 | 9.05E-03 | Pf.GIV |
| GO:0048870 | Cell motility | 6 | 4.01E-03 | Pf.GV |
| GO:0051674 | Localization of cell | 6 | 4.01E-03 | Pf.GV |
| GO:0000737 | DNA catabolic process, endonucleolytic | 2 | 5.92E-03 | Pf.GV |
| GO:0006415 | Translational termination | 3 | 7.60E-03 | Pf.GV |
| GO:0043624 | Cellular protein complex disassembly | 3 | 7.60E-03 | Pf.GV |
| GO:GNF0004 | Sexual development | 43 | 2.76E-05 | Pf.GV |
m5C-Mediated mRNA Stability Correlates with Gametocyte Development.
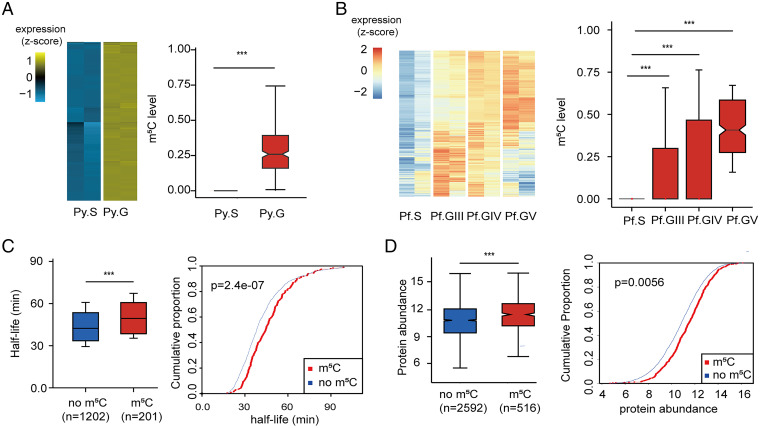
Having found that m5C methylated transcripts were significantly enriched for GO identifications of sexual development in P. yoelii and P. falciparum gametocytes, we focused next on genes involved in gametocyte development. As previously reported, genes with higher transcript abundances in gametocytes relative to asexual parasites have important roles during gametocyte development (31–33). Our search of the P. yoelii data identified 873 genes with transcript abundances more than threefold (log2 ≥ 1.585) higher in gametocytes relative to asexual parasites (Dataset S3). Similarly, 462 gametocyte development-related genes were identified with greater than or equal to threefold higher transcript abundance in P. falciparum gametocytes (stage GIII) vs. asexual stages (Dataset S3). On comparative analysis of these greater than or equal to threefold more abundant gametocyte transcripts, significantly increased m5C levels were observed relative to the schizont transcripts of P. yoelii and P. falciparum, particularly in the comparison using m5C-modified stage V gametocyte transcripts (Fig. 2 A and B).
Fig. 2.
Association of m5C methylation levels with transcript longevity and protein expression in gametocytes. (A and B) Heatmaps indicate the expression of transcripts whose levels are greater than or equal to threefold higher in gametocytes than in schizonts of P. yoelii and P. falciparum. Boxplots show corresponding m5C levels of those genes at different stages (***P < 0.001, Wilcoxon test). The m5C levels were normalized to their corresponding transcript fragment abundance levels before comparison. (C) Boxplot and cumulative fraction plots indicate the longer mRNA half-lives of m5C methylated (red) relative to nonmethylated (blue) transcripts in P. yoelii gametocytes (***P < 0.001, Wilcoxon test). (D) Boxplot and cumulative fraction plots compare the levels of proteins translated from m5C-methylated (red) and nonmethylated (blue) transcripts in P. yoelii gametocytes (***P < 0.001, Wilcoxon test).
Our results confirmed the presence of m5C modifications in transcripts with well-established roles in sexual commitment (3, 5, 6, 34). For example, m5C methylation of AP2-O (PY17X_0907300) transcripts was not detected in schizonts but was present at a level of 0.34 along with higher expression of these transcripts in gametocytes (SI Appendix, Fig. S3A). In another example, the DEAD/DEAH helicase (PY17X_0313200), an AP2-G–induced transcript involved in male gametocyte development in Plasmodium berghei (5), was found to be m5C methylated in both asexual and sexual stages of P. yoelii. Similarly, data from P. falciparum showed gametocyte-stage m5C methylation of three transcripts that possibly regulate sexual commitment and gametocytogenesis (2, 3, 23), including genes whose disruption is known to lead to greatly reduced gametocyte numbers (GIG, AP2-G2, Pfs16) (2, 3) (SI Appendix, Fig. S3A). However, distinct from AP2-O or AP2-G2, no detectable m5C modification was found for AP2-G transcripts in either the P. yoelii or P. falciparum malaria parasites.
In view of the above results and m5C stabilization of transcripts known from other systems (18, 21), we asked if m5C might be associated with mRNA stability in Plasmodium parasites. To address this question, we first assessed the abundance of mRNAs whose m5C levels differed between sexual and asexual parasites, relative to the abundance of mRNAs that showed no change of m5C levels between these stages. Results from these comparisons suggested a positive correlation between m5C level and mRNA abundance in malaria gametocytes relative to schizonts (SI Appendix, Fig. S3B).
We next studied the half-lives of mRNAs in P. yoelii gametocytes that were treated with actinomycin D as a transcription inhibitor (18). Results from RNA-sequencing (RNA-seq) analysis indicated that mRNAs with m5C modification had detectably longer half-lives than those without m5C modification (Fig. 2C and Dataset S4). Greater protein abundance was likewise found to be associated with corresponding transcript m5C modifications in quantification experiments with tandem mass tags (Fig. 2D and Dataset S5). Although these differences were overall modest between the methylated and unmethylated populations, we found that markedly higher transcript m5C levels in gametocytes were in important instances associated with 5 to 10× greater abundance of their corresponding proteins. For example, transcripts for actin-depolymerizing factor (ADF2, PY17X_113890) were significantly m5C-methylated in gametocytes but not in schizont stages, and this methylation was associated with approximately seven times greater protein abundance in the gametocytes; ADF2 is pivotal to the developmental progression of ookinetes to sporozoites (35). Likewise, m5C modifications of transcripts for the PhIL1 interacting protein PIP2 [putative, PY17X_1015100, involved in Plasmodium sexual stage development (36)] and for the secreted ookinete protein PSOP1 [putative, PY17X_0621900, involved in ookinete development (37)] were detected only in gametocytes and were associated with four to five times greater protein abundance in these stages (Dataset S5).
PyNSUN2 Functions as an mRNA m5C Methyltransferase in P. yoelii.
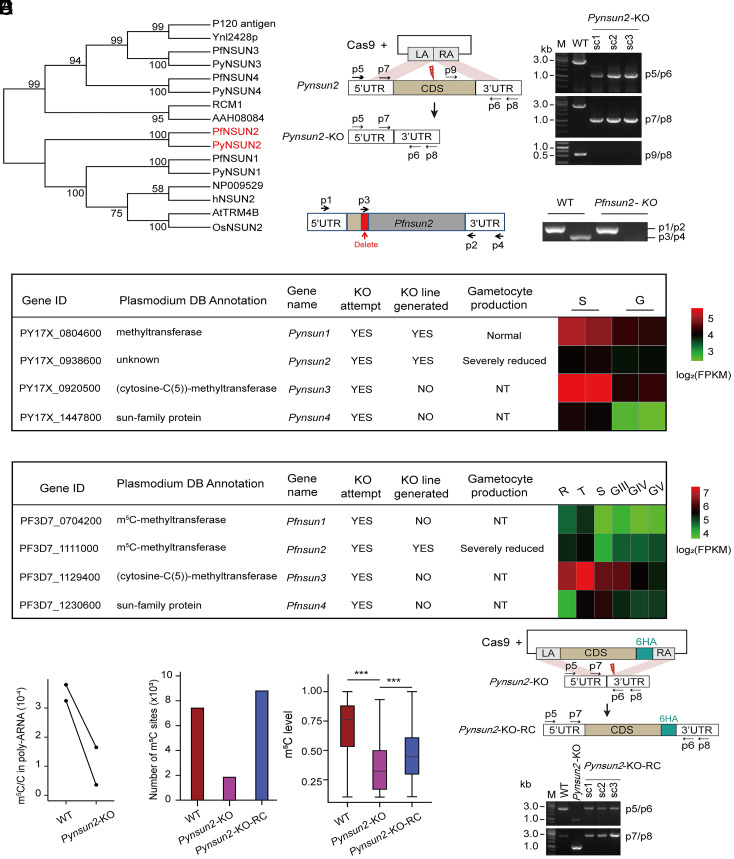
To identify potential mRNA m5C methyltransferases in P. yoelii, we searched for orthologs of hNSUN2 and identified four candidate sequences: PY17X_0804600, PY17X_0938600, PY17X_0920500, PY17X_1447800 (which we named PyNSUN1 to PyNSUN4, respectively) (Fig. 3A). Corresponding searches of the PlasmoDB database (http://www.plasmoDB.org) identified PF3D7_0704200, PF3D7_1111000, PF3D7_1129400, and PF3D7_1230600 (PfNSUN1 to PfNSUN4) as candidate RNA methyltransferases in P. falciparum (Pf3D7_0704200 and Pf3D7_1111000 have been annotated in PlasmoDB as tRNA methyltransferases and Pf3D7_1129400 as a ribosome RNA methyltransferase). Sequence comparisons showed that individual members of this family are highly conserved between P. yoelii and P. falciparum, and that the NSUN1 and NSUN2 sequences have close affinities to orthologs in humans, A. thaliana, Saccharomyces cerevisiae, and O. sativa (Fig. 3A).
Fig. 3.
Identification and verification of PyNSUN2 as a m5C methyltransferase. (A) Phylogenetic analysis of candidate NSUN m5C methyltransferases in P. yoelii and P. falciparum. Sequences were aligned using ClustalX 2.1. The neighbor-joining phylogeny was performed using MEGA 5.2.2 with 1,000 replicates. (B) Strategies to generate Pynsun2-KO and Pfnsun2-KO parasites. For CRISPR/Cas9-mediated deletion of the Pynsun2 CDS, the left arm (LA) and right arm (RA) were designed to match sequences in the 5′ UTR and 3′ UTR of Pynsun2, respectively. For disruption of Pfnsun2, sgRNA targeting was used to delete a portion of the CDS (red box) and introduce multiple stop codons downstream. Red thunderbolt indicates the site for sgRNA targeting. PCR products with indicated primer pairs confirmed the expected differences between the WT and allelically manipulated parasites (single clones: sc1–3, with the expected amplicon sizes; p5/p6: 1.1 kb for the Pynsun2-KO vs. 2.7 kb for WT parasites; p7/p8: 1 kb for Pynsun2-KO vs. 2.6 kb for WT). Sequence data confirming the deletion and stop codons in the Pfnsun2-KO parasites are shown in SI Appendix, Fig. S4A. (C) Summary results from experiments to disrupt homologs of the NSUN family in P. yoelii and P. falciparum. NT: not tested. Heatmaps show transcript fragment abundance levels from each gene in the WT parasites, as determined by RNA-seq. (D) LC-MS/MS-determined levels of m5C/C in the poly(A)-selected transcripts of P. yoelii WT and Pynsun2-KO parasites (schizont stage). Paired points represent the results of each of two biological replicates showing an average 72% reduction (P = 0.046 by paired t test). (E) Histogram shows numbers of mRNA m5C sites in schizont stages of WT, Pynsun2-KO and Pynsun2-KO-RC P. yoelii clones. Box plot shows the corresponding m5C levels in WT, Pynsun2-KO, and Pynsun2-KO-RC lines (Right) (***P < 0.001, Wilcoxon test). Only m5C sites detected in both replicates were used for the graphs. (F) Strategy for genetic complementation repair of the Pynsun2-KO with the gene CDS fused with a C-terminal 6HA. (Upper) The design for the CRISPR/Cas9-mediated gene knockin. LA and RA match sequences in the 5′ UTR and 3′ UTR of Pynsun2, respectively. Red thunderbolt indicates the site for sgRNA targeting. (Lower) The expected PCR products from three Pynsun2-KO-RC clones (sc1–3) by the primer sets p5/p6 (1.1 kb for the Pynsun2-KO line vs. 2.7 kb for the WT and Pynsun2-KO-RC lines) and p7/p8 (1 kb for the Pynsun2-KO vs. 2.6 kb for the WT and Pynsun2-KO-RC lines).
To study the role of these putative m5C methyltransferases in Plasmodium, we performed experiments to disrupt Pynsun2 in the P. yoelii 17XNL line and Pfnsun2 in the P. falciparum NF54 line by CRISPR-Cas9 gene editing (Fig. 3B). Functional disruption attempts of the sun1, sun3, and sun4 homologs were likewise made. After a minimum of three independent attempts with two to three different single-guide RNA (sgRNA) sequences for each individual homolog (Dataset S6), we obtained KO lines for three of the eight genes (Pynsun1, Pynsun2, and Pfnsun2) (Fig. 3C and SI Appendix, Fig. S4A). We note that some of these results differ from those of other studies (38, 39), which found the PF3D7_0704200 (Pfnsun1) gene to be dispensable and PF3D7_1111000 (Pfnsun2) gene to be potentially essential. Among possible explanations for these differences are the use of alternative genetic manipulation strategies, including the choices of selection agent and sequence targets for replacement or deletion.
We used LC-MS/MS and RNA-BisSeq to compare m5C modification levels in the schizont-stage transcripts of the 17XNL wild-type (WT) and Pynsun2-KO lines (sexual stage production by Pynsun2-KO parasites was inadequate for gametocyte comparisons; see below). By LC-MS/MS, m5C/C levels in the Pynsun2-KO schizonts were reduced by an average of 72% (Fig. 3D). Consistent with these results, there was a marked decrease in the m5C transcript modifications detected by RNA-BisSeq analysis. In the 17XNL WT schizonts, 7,409 m5C sites were detected in 527 mRNAs, whereas this number was reduced to 1,845 m5C sites in 265 mRNAs of Pynsun2-KO schizonts (75% and 50% reductions, respectively) (Fig. 3E and Dataset S2). Corresponding experiments with P. falciparum WT and Pfnsun2-KO schizonts did not yield adequate signals for reliable comparisons, either by LC-MS/MS or RNA-BisSeq, presumably because of the much lower WT m5C modification levels in P. falciparum relative to P. yoelii schizonts (SI Appendix, Fig. S1A).
In the case of the Pynsun1-KO line, moderately reduced m5C sites and levels were observed in schizont-stage transcripts (SI Appendix, Fig. S4B). For transcripts of the ApiAP2, AROM, SNF2L, and MSRO2 genes known to be involved in gametocytogenesis (3, 4, 34), no significant reductions of m5C levels were observed for any of these transcripts in Pynsun1-KO schizonts, in contrast to the markedly lower m5C levels in transcripts from three of these genes (ApiAP2, SNF2L, and MSRO2) in Pynsun2-KO relative to WT schizonts (SI Appendix, Fig. S4C and Dataset S2).
To examine the NSUN2 subcellular localization, P. yoelii and P. falciparum parasites were engineered to express NSUN2 as an HA-tagged form: as either PyNSUN2-HA×6 (six HA tags in tandem) or PfNSUN2-3HA-3Ty1 (three HA followed by three Ty1 tags) (SI Appendix, Fig. S5 A and B). Immunofluorescence assay (IFA) and Western blot analyses of these parasites with anti-HA antibody showed signals indicating the presence of PyNSUN2 and PfNSUN2 in nuclear extracts throughout the intraerythrocytic cycle (SI Appendix, Fig. S5 C and D), consistent with its localization in previous studies of other organisms (17, 19).
By IFA and Western blot analyses with HA-tagged enzymes, we detected the signal of PyNSUN2 in the cytoplasmic extracts of P. yoelii, but we did not find a clear PfNSUN2 signal in the cytoplasmic extracts of P. falciparum (SI Appendix, Fig. S5 C and D). This difference may help to explain the higher median fractional shared-site mRNA methylation level of 0.77 in P. yoelii schizonts vs. only 0.15 in P. falciparum schizonts (SI Appendix, Fig. S1B). We note that levels of NSUN2 can vary widely in cell cytoplasms, as has been observed in mouse and human keratinocytes, SZ95 immortalized human sebocytes (40), and osteosarcoma cells (41).
To confirm the evidence from KO parasites that Pynsun2 functions as a major m5C methyltransferase, we used CRISPR/Cas9 allelic modification to reintroduce Pynsun2 to its original locus in the Pysun2-KO parasite (Fig. 3F and SI Appendix, Fig. S5E). Comparative analysis of the restored line (Pynsun2-KO-RC) demonstrated the return of substantial m5C levels, reflecting restored methylation activity (Fig. 3E).
Gametocytogenesis Is Impaired by Disruption of Plasmodium nsun2 and Can Be Restored by Genetic Complementation.
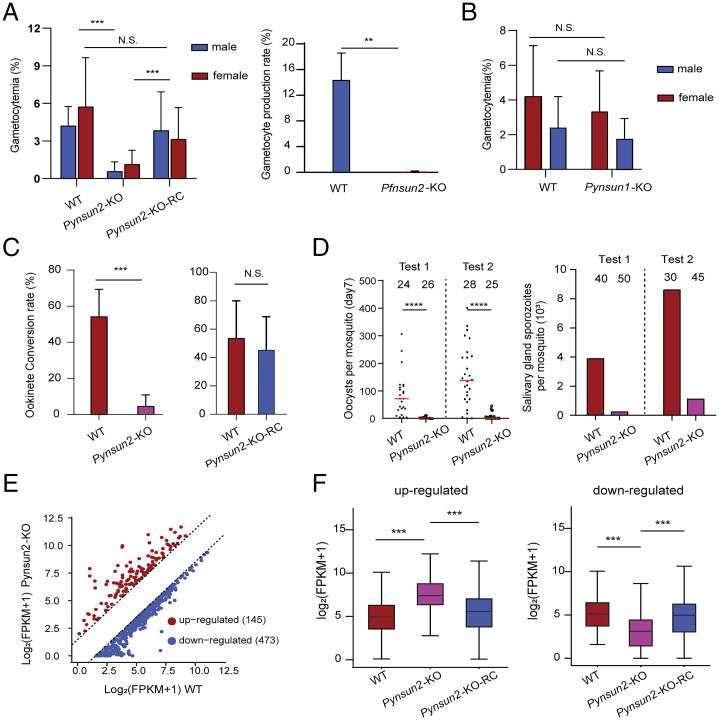
Considering the differentially greater m5C levels in gametocyte vs. schizont mRNA transcripts and the successful KOs of Pynsun1, Pynsun2, and Pfnsun2 in blood-stage parasites, we asked if expression of these genes might be associated with phenotypes of gametocyte production or development of parasite stages in the mosquito. Comparative experiments with the KO and WT lines demonstrated that disruptions of Pynsun2 in P. yoelii and Pfnsun2 in P. falciparum resulted in dramatically reduced gametocyte production (Fig. 4A). In contrast, gametocyte production by the Pynsun1-KO parasites line was not significantly reduced (Fig. 4B). The disruption of Pynsun2 affected the production of male and female gametocytes similarly (Fig. 4A), while it showed no detectable effect on the propagation of asexual parasites (SI Appendix, Fig. S6A). As with m5C modification of the mRNA transcripts, gametocyte production was successfully rescued by the restoration of Pynsun2 expression in the Pynsun2-KO-RC line (Fig. 4A).
Fig. 4.
Reduction of gametocytogenesis in Plasmodium NSUN2 KO lines and its restoration by gene complementation. (A) Gametocytogenesis is markedly decreased in Pynsun2-KO and Pfnsun2-KO parasites relative to high levels in the original WT lines. Repair of the KO in Pynsun2-KO parasites restores gametocyte production. Error bars represent median and 95% confidence intervals (CI) (***P < 0.001; **P < 0.01; N.S.: not significant, Wilcoxon test). (B) No significant effect on gametocyte production was detected after knockout of the Pynsun1 gene. Error bars represent median and 95% CIs (N.S.: no significant difference, Wilcoxon test). (C) Markedly decreased ookinete conversion rate in Pynsun2-KO relative to WT parasites as well as Pynsun2-KO-RC parasites carrying the restored gene. Data were obtained from two independent replicates. Error bars represent median and 95% CIs (***P < 0.001; N.S.: not significant, Wilcoxon test). (D) Number of oocysts in mosquito midguts 7 d after blood feeding with WT or Pynsun2-KO gametocytes (Left), and number of sporozoites in mosquito salivary glands 14 d after blood feeding in the same experiments (Right). The number of mosquitoes dissected in each group is indicated. Medians and 95% CIs are indicated (****P < 0.0001, Wilcoxon test). (E) Identification of transcripts that are differentially expressed between Pynsun2-KO and WT parasites at the schizont stage. Transcripts that were found to have greater than or equal to threefold higher or lower levels in Pynsun2-KO vs. WT parasites are marked in red or blue, respectively. (F) Higher or lower levels of transcripts in the KO clone are reversed by restoration of Pynsun2 expression in the Pynsun2-KO-RC clone (***P < 0.001, Wilcoxon test).
Consistent with the effect of gene disruption on gametocyte production, in vitro ookinete conversion rates to mature forms were significantly reduced in the Pynsun2-KO relative to WT parasites; these rates were then rescued by gene complementation in the Pynsun2-KO-RC parasites (Fig. 4C). Also, the numbers of midgut oocysts at day 7 and salivary gland sporozoites at day 14 were markedly decreased with Pynsun2-KO parasites (Fig. 4D), suggesting that Pynsun2-KO effects may impair the development of parasite stages in the mosquito.
To further investigate the association of decreased numbers of gametocytes with diminished mRNA m5C modification, we identified transcripts whose m5C levels were significantly reduced in Pynsun2-KO relative to WT schizonts. Many of these transcripts were more than threefold changed in Pynsun2-KO vs. WT P. yoelii parasites, including 145 instances of increased transcript abundance and 473 instances of decreased transcript abundance (Fig. 4E); these changes were largely reversed with restoration of Pynsun2 expression in the Pynsun2-KO-RC line (Fig. 4F and Dataset S7). Among the instances of reduced abundance, we identified transcripts that have been reported to be AP2-G–mediated and involved in gametocyte development (5) (SI Appendix, Fig. S6B). These included transcripts of PY17X_0313200 (DEAD/DEAH), disruption of which can greatly reduce the number of gametocyte parasites in P. berghei (5). Interestingly, no significant change of AP2-G expression itself was evident with the Pynsun2-KO (SI Appendix, Fig. S6C).
Discussion
The results of this study illuminate a landscape of m5C mRNA transcriptome methylations in the schizonts and gametocytes of two Plasmodium species, P. yoelii and P. falciparum, and show that these epitranscriptomic modifications are linked to the development of gametocytes. Furthermore, the searches for possible RNA methyltransferases and findings from experiments to KO and repair the candidate genes indicate that a homolog of the NSUN2 family plays a major role in these m5C mRNA modifications. Although little or no effect on the growth of asexual blood-stage parasites was detected in these experiments, markedly decreased gametocyte production was found after disruption of the Pynsun2 homolog in P. yoelii (Pynsun2-KO) and disruption of the Pfnsun2 homolog in P. falciparum (Pfnsun2-KO). Moreover, in evaluations of Pynsun2-KO infectivity, the conversion rate of ookinetes from immature to mature forms was also decreased, suggesting that m5C mRNA modifications may be important both to gametocyte production and to parasite development following zygote formation in the mosquito.
m5C modifications can stabilize mRNA transcripts and increase half-life, and they may also influence the export of the transcripts for translation in the cytoplasm (13, 21, 42, 43). In the present work, several transcripts known to be involved in sexual commitment and gametocyte development were found to be m5C-modified, suggesting a role for this form of posttranscriptional methylation in the timing and control of their expression. Interestingly, no m5C modification was found for AP2-G, a master regulator of sexual commitment and development (44–46), nor was there any evidence for a change of AP2-G expression in NSUN2-disrupted parasites. These observations suggest that pathways downstream of AP2-G are epigenetically modulated at the posttranscriptional level. Transcripts for gametocyte development protein 1 (GDV1, PF3D7_0935400) showed high levels of m5C in the GIII gametocyte stage (m5C level, 0.941) (Dataset S2), but not in other stages. GDV1 has been reported to work as an upstream activator of sexual commitment, with peak transcript abundance in schizont-stage parasites (34). We speculate that the presence of these m5C-methylated transcripts in GIII stages reflects additional functions of GDV1 (after sexual commitment) that remain to be discovered. KOs of NSUN2 and loss of m5C methylation may adversely affect the stability and translation of mRNA transcripts in these and other pathways, resulting in dysregulation and impairment of effective gametocytogenesis.
As in other organisms (12, 13, 21), the m5C sites of transcripts modified by NSUN2 are found predominantly in the protein coding regions of Plasmodium mRNAs. However, the profiles of m5C distributions within these regions differ among organisms. In the two Plasmodium species studied here, a peak of m5C levels in CDSs occurs just upstream of the stop codon. In contrast, the m5C levels in cells from human and mouse cells or from zebrafish embryos are more uniformly distributed across the CDSs or are enriched near the translation initiation sites. A. thaliana shows two peaks of m5C enrichment that flank the stop codon. Differences in the consensus site logos for m5C modification are also present between Plasmodium spp. and other eukaryotes (18, 19, 47). In P. yoelii and P. falciparum, the target C nucleotide is flanked on each side by a highly AU-rich sequence, which may in part reflect the correspondingly high AT content of the genomes and transcriptomes of these two species.
Our homology searches for sequences related to NSUN2 identified other potential methylases, termed Plasmodium NSUN1, NSUN3, and NSUN4 in this work. Multiple attempts with CRISPR/Cas9 strategies to KO these different homologs in P. yoelii and P. falciparum yielded only one parasite line with a disruption of Pynsun1. However, unlike the Pynsun2 and Pfnsun2 KOs, the Pynsun1-KO line showed little or no reduction of gametocyte production relative to that of WT P. yoelii. Further investigations are needed to establish phenotypes associated with Plasmodium NSUN1, as well as NSUN3 and NSUN4, which, in light of the multiple unsuccessful KO attempts, are likely to be critical for parasite survival. We note that the observed reduction of m5C methylation in Pynsun2-KO schizonts was to ∼30% of the WT level, as measured by LC-MS/MS and supported by RNA-BisSeq. Thus, writers other than NSUN2 may also be involved in m5C transcript modifications of asexual- as well as sexual-stage parasites. In addition to its major activity as an mRNA m5C methyltransferase in eukaryotic cells (13), NSUN2 can also target tRNA and vault RNA (48–50). These possibilities, in addition to the potential activities of the NSUN-type m5C methyltransferases on other RNAs, such as tRNA and noncoding RNAs (51), remain to be explored.
We asked if m5C modifications of schizonts and gametocytes might be subject to an eraser (demethylase) activity, but to our knowledge no eraser has been reported from any eukaryote in which m5C mRNA modifications have been studied (52). In the absence of an identified eraser, it is possible that m5C levels simply rise and fall due to different rates of m5C writer activity and subsequent degradation of m5C-modified transcripts. This possibility is supported by the positive correlation between m5C status and mRNA stability (half-life) in gametocytes, as well as the greater average m5C modification to transcript ratios in gametocytes relative to shorter-lived schizonts (Dataset S2).
m5C-modified mRNA trafficking in mammalian cells is mediated by ALYREF and other RNA binding proteins (13, 53), and m5C modifications have also been found to have an important role in RNA transport and function in plants (54, 55). Our searches using the human ALYREF sequence identified seven genes in P. yoelii and P. falciparum with e-values less than 0.0004, and searches for Ybx1 homology identified one predicted protein in each of these species with an e-value less than 0.01 (SI Appendix, Fig. S7). Further studies of these candidate genes may help to determine if an ALYREF-like reader in Plasmodium recognizes m5C-modified mRNAs and facilitates the transport of the methylated transcripts for expression from bound complexes in the cytoplasm.
Control of mRNA turnover and translational repression is critical to Plasmodium sexual development and is mediated by molecules, including DOZI (development of zygote inhibited), a member of the DDX6-family of DEAD-box RNA helicases (56). In the comparative analysis of our data with the DOZI-bound P. berghei mRNAs reported by Guerreiro et al. (57), we identified 50 of 246 (20%) homologs of the DOZI-bound mRNAs that show m5C methylation in WT P. falciparum schizont and gametocytes (SI Appendix, Fig. S8A) as well as 122 of 449 (27%) homologs of the DOZI-bound mRNAs that show m5C methylation in WT P. yoelii schizont and gametocytes (SI Appendix, Fig. S8B). The transcript abundances and m5C levels of these homologs have similar dynamics during the gametocyte development of the two species (SI Appendix, Fig. S8 A and B), suggesting that m5C modifications may function in concert with DOZI complexes to sequester mRNAs for translational repression. Intriguingly, among the 51 DOZI-bound mRNAs with m5C methylation in WT P. yoelii schizont stages, 50 mRNAs showed reduced m5C levels in Pynsun2-KO parasites, including DEAD/DEAH, an AP2-G–induced transcript involved in the male gametocyte development of P. berghei (5) (SI Appendix, Fig. S8C). Translational repression also occurs in mature sporozoites and is associated with their quiescence in mosquito salivary glands until blood feeding and salivation (58–60).
Dynamic m5C modifications are vital to gene regulatory networks in a diverse variety of biological processes, including embryogenesis in zebrafish (21) and stages of development in A. thaliana (12). Global transcriptome fluctuations of 30% in m5C methylation occur with altered photosynthesis efficiency and heat tolerance in rice (19); and m5C fluctuations have likewise been associated with the maternal-to-zygotic transition and the development of bladder carcinoma (18, 21). The m5C modification patterns observed in the present study, including increased levels of methylation in gametocytes and their loss following KO of Plasmodium NSUN2, are also associated with developmental processes of male and female gametocytogenesis and progression to mature ookinetes following fertilization. Further investigation of these m5C-mediated processes and those of other epitranscriptomic modifications, such as m6A (14), in both mRNA and other types of RNA, may yield new insights into gametocytogenesis, mosquito infectivity, and molecular pathways that might be exploited to interrupt malaria transmission.
Methods
Animal Use and Ethics Statement.
All animal experiments were performed in accordance with approved protocols (XMULAC20140004) by the Committee for Care and Use of Laboratory Animals of Xiamen University. ICR mice (female, 5- to 6-wk old) were purchased and housed in the Animal Care Center of Xiamen University and kept at room temperature under a 12-h light/dark cycle at a constant relative humidity of 45%, where they supported parasite propagation, drug selection, parasite cloning, and mosquito feedings.
Parasite Culture and Mouse Infections.
P. falciparum strain 3D7 (NF54) parasites were cultured and synchronized as previously reported (14). Human erythrocytes were obtained under approved clinical protocol at the Shanghai Blood Center. Microscopy of Giemsa-stained thin blood films was used to monitor parasite development.
P. yoelii 17XNL strain parasites were obtained from the Malaria Research and Reference Reagent Resource Center (MR4) (https://www.beiresources.org/About/MR4.aspx). The parasites were injected intravenously or intraperitoneally into mice for infection, and parasitemia was monitored by Giemsa-stained thin blood films. Single clones were obtained by the limiting dilution method (61).
Gametocyte Induction.
For the P. yoelii 17XNL strain, gametocyte induction in mice was performed as previously described (62). Briefly, phenylhydrazine (80 μg/g mouse body weight) was administered to the ICR mice through intraperitoneal injection. The mice were infected 3 d later with 3.0 × 106 parasites through tail vein injection. Gametocytemia typically peaked at day 3 postinjection. Giemsa-stained blood smears were used to count the number of male and female gametocytes. Gametocytemia (male, female, or both) was determined by the ratio of gametocytes over total infected erythrocytes. All experiments were repeated three times independently.
For P. falciparum, gametocyte induction was performed as described previously with minor modifications (63, 64). The P. falciparum population was synchronized and expanded to 8% parasitemia in culture (4% hematocrit). Medium changes were performed daily without dilution of the culture and without disturbing the red blood cell layer at the bottom of the culture dish. N-acetylglucosamine (50 mM; Sigma-Aldrich, A3286) was used for 5 d to eliminate asexual parasites (46). Giemsa-stained thin blood smears were used to count the number of gametocytes from day 3. Stage III gametocytes were harvested on days 5 to 6, stage IV on days 8 to 9, and stage V on days 12 to 13. Gametocytemia determined on days 8 to 9 was used to calculate sexual commitment (%). The coding regions of the Pfap2-g and Pfgdv1 genes were sequenced in the WT- and Pfsun2-KO parasites and confirmed that no mutations or deletions occurred during the KO process (file GSE159126_qPCR_of_gdv1_ap2g.xlsx, available under GEO accession no. GSE159127).
Ookinete Conversion Rate Determinations.
Conversion rates to mature ookinetes were determined as described previously (65). Briefly, 100 µL of mouse blood with 3 to 10% gametocytemia from the WT line or 1 to 3% gametocytemia from the KO line was collected and immediately transferred to a 10-cm cell culture dish (Corning, cat# 801002) containing ookinete culture medium (RPMI 1640, 25 mM Hepes, 10% FCS, 100 mM xanthurenic acid, pH 8.0) to allow gametogenesis, fertilization, and ookinete development at 22 °C (66). For ookinete conversion analysis, samples from 12-h culture were collected for thin blood films stained by Giemsa solution (Sigma, cat# GS80). The conversion rate to mature ookinetes was calculated from the counts ratio of mature ookinetes (stage V) to total ookinetes (stages I to V). Three biological replicates of these experiments were performed with identical volumes of infected blood.
Mosquito Maintenance and Mosquito Feeding.
Sucrose solution (10%) with 0.5 g/L PABA was used to feed Anopheles stephensi mosquitoes (strain Hor) (67), which were reared at 28 °C, 80% relative humidity, in a 12-h light/dark cycle. Blood feeding assays were performed at 23 °C. Each anesthetized mouse with 4 to 6% gametocytemia was used to feed 80 female mosquitoes for 30 min. For oocyst detection, mosquito midguts were dissected 7 d after blood feeding and stained with 0.1% mercurochrome. For salivary gland sporozoite counting, salivary glands from 30 to 50 mosquitoes were dissected on day 14 after blood feeding, and the number of sporozoites per mosquito was calculated.
Gene Homolog Identification and Phylogenetic Analysis of m5C Writer Candidates.
Candidate proteins of the malaria parasite m5C writer complex were identified by blast search against PlasmoDB database (https://plasmodb.org/plasmo/) using the amino acid sequence of human mRNA m5C methylase NSUN2 as a query. The amino acid sequences of human, rice, and A. thaliana RCMT domain family members were obtained from the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/). ClustalX software (68) was used to construct the neighbor-joining phylogenetic tree with 1,000 bootstrap replicates.
GO Analysis.
GO analysis of specific genes were performed using EnrichGO tool from PlasmoDB (https://plasmodb.org/). Only terms in the biological process category were shown. GO terms with P ≤ 0.05 were considered as statistically significant terms. Genes enriched for terms sexual development (GO: GNF0004) were obtained from a previous study (32) and used for enrichment analysis.
Detailed Methods.
Extended descriptions of the dot blot experiments, LC-MS/MS determinations, transcriptome sequencing, mRNA half-life assessments, RNA-BisSeq analyses, parasite transfections, proteomics analyses, Western blotting, immunofluorescence assays, and statistics are provided in SI Appendix, Detailed Methods.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 81630063 and 81971959 (to Q.Z.), 31972882, 31771419, and 31721003 (to C.J.), and 31772443 and 31970387 (to J.Y.); National Key R&D Program of China Grants 2018YFA0507300 (to Q.Z.) and 2019YFA0110001 (to C.J.); the Shanghai Blue Cross Brain Hospital Co., Ltd. and Shanghai Tongji University Education Development Foundation (Q.Z.); and the Division of Intramural Research at the US National Institute of Allergy and Infectious Diseases (J.M. and T.E.W.).
Footnotes
Reviewers: K.D., Weill Cornell Medicine; and S.L., The Pennsylvania State University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110713119/-/DCSupplemental.
Data Availability
In addition to the datasets provided in SI Appendix, the raw and processed high-throughput sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE159127).
References
- 1.World Health Organization, World Malaria Report 2021 (World Health Organization, Geneva, Switzerland, 2021). https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021. Accessed December 8, 2021. [Google Scholar]
- 2.McKenzie F. E., Ferreira M. U., Baird J. K., Snounou G., Bossert W. H., Meiotic recombination, cross-reactivity, and persistence in Plasmodium falciparum. Evolution 55, 1299–1307 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Josling G. A., Llinás M., Sexual development in Plasmodium parasites: Knowing when it’s time to commit. Nat. Rev. Microbiol. 13, 573–587 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Venugopal K., Hentzschel F., Valkiūnas G., Marti M., Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat. Rev. Microbiol. 18, 177–189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent R. S., et al. , Inducible developmental reprogramming redefines commitment to sexual development in the malaria parasite Plasmodium berghei. Nat. Microbiol. 3, 1206–1213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Josling G. A., Williamson K. C., Llinás M., Regulation of sexual commitment and gametocytogenesis in malaria parasites. Annu. Rev. Microbiol. 72, 501–519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kafsack B. F., et al. , A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuda M., Iwanaga S., Kaneko I., Kato T., Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc. Natl. Acad. Sci. U.S.A. 112, 12824–12829 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortés A., Deitsch K. W., Malaria epigenetics. Cold Spring Harb. Perspect. Med. 7, a025528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B. S., Roundtree I. A., He C., Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammam E., et al. , Discovery of a new predominant cytosine DNA modification that is linked to gene expression in malaria parasites. Nucleic Acids Res. 48, 184–199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui X., et al. , 5-methylcytosine RNA methylation in Arabidopsis thaliana. Mol. Plant 10, 1387–1399 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Yang X., et al. , 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27, 606–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumgarten S., et al. , Transcriptome-wide dynamics of extensive m6A mRNA methylation during Plasmodium falciparum blood-stage development. Nat. Microbiol. 4, 2246–2259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trixl L., Lusser A., The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 10, e1510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H., Chai P., Jia R., Fan X., Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer 19, 78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courtney D. G., et al. , Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26, 217–227.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., et al. , 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Tang Y., et al. , OsNSUN2-mediated 5-methylcytosine mRNA modification enhances rice adaptation to high temperature. Dev. Cell 53, 272–286.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Chen H., et al. , m5C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat. Commun. 11, 2834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y., et al. , RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell 75, 1188–1202.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Esteve-Puig R., Bueno-Costa A., Esteller M., Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 474, 127–137 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Huong T. T., Ngoc L. N. T., Kang H., Functional characterization of a putative RNA demethylase ALKBH6 in Arabidopsis growth and abiotic stress responses. Int. J. Mol. Sci. 21, 6707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govindaraju G., Jabeena C. A., Sethumadhavan D. V., Rajaram N., Rajavelu A., DNA methyltransferase homologue TRDMT1 in Plasmodium falciparum specifically methylates endogenous aspartic acid tRNA. Biochim. Biophys. Acta. Gene Regul. Mech. 1860, 1047–1057 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Hammam E., et al. , Malaria parasite stress tolerance is regulated by DNMT2-mediated tRNA cytosine methylation. MBio 12, e0255821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlton J. M., et al. , Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii. Nature 419, 512–519 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Gardner M. J., et al. , Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlopoulou A., Kossida S., Phylogenetic analysis of the eukaryotic RNA (cytosine-5)-methyltransferases. Genomics 93, 350–357 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Meyer K. D., Jaffrey S. R., Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang T., Chen W., Liu J., Gu N., Zhang R., Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 26, 380–388 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Hall N., et al. , A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Young J. A., et al. , The Plasmodium falciparum sexual development transcriptome: A microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143, 67–79 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Prajapati S. K., et al. , The transcriptome of circulating sexually committed Plasmodium falciparum ring stage parasites forecasts malaria transmission potential. Nat. Commun. 11, 6159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filarsky M., et al. , GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359, 1259–1263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doi Y., Shinzawa N., Fukumoto S., Okano H., Kanuka H., ADF2 is required for transformation of the ookinete and sporozoite in malaria parasite development. Biochem. Biophys. Res. Commun. 397, 668–672 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Parkyn Schneider M., et al. , Disrupting assembly of the inner membrane complex blocks Plasmodium falciparum sexual stage development. PLoS Pathog. 13, e1006659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tachibana M., Iriko H., Baba M., Torii M., Ishino T., PSOP1, putative secreted ookinete protein 1, is localized to the micronemes of Plasmodium yoelii and P. berghei ookinetes. Parasitol. Int. 84, 102407 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Bushell E., et al. , Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M., et al. , Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360, eaap7847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frye M., Watt F. M., The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 16, 971–981 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Van Haute L., et al. , NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 47, 8720–8733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caro F., Ahyong V., Betegon M., DeRisi J. L., Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. eLife 3, e04106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Painter H. J., et al. , Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat. Commun. 9, 2656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ralph S. A., Cortés A., Plasmodium sexual differentiation: How to make a female. Mol. Microbiol. 112, 1627–1631 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Josling G. A., et al. , Dissecting the role of PfAP2-G in malaria gametocytogenesis. Nat. Commun. 11, 1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llorà-Batlle O., et al. , Conditional expression of PfAP2-G for controlled massive sexual conversion in Plasmodium falciparum. Sci. Adv. 6, eaaz5057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo G., et al. , Disease activity-associated alteration of mRNA m(5)C methylation in CD4(+) T cells of systemic lupus erythematosus. Front. Cell Dev. Biol. 8, 430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuorto F., et al. , RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Hussain S., et al. , NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco S., et al. , Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sibbritt T., Patel H. R., Preiss T., Mapping and significance of the mRNA methylome. Wiley Interdiscip. Rev. RNA 4, 397–422 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Nombela P., Miguel-López B., Blanco S., The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol. Cancer 20, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan J., et al. , ALYREF links 3′-end processing to nuclear export of non-polyadenylated mRNAs. EMBO J. 38, e99910 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T., et al. , RNA motifs and modification involve in RNA long-distance transport in plants. Front. Cell Dev. Biol. 9, 651278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang L., et al. , m(5)C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 29, 2465–2476.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Mair G. R., et al. , Regulation of sexual development of Plasmodium by translational repression. Science 313, 667–669 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guerreiro A., et al. , Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte. Genome Biol. 15, 493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang M., et al. , The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 207, 1465–1474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindner S. E., et al. , Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat. Commun. 10, 4964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bogale H. N., et al. , Transcriptional heterogeneity and tightly regulated changes in gene expression during Plasmodium berghei sporozoite development. Proc. Natl. Acad. Sci. U.S.A. 118, e2023438118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffmann E. J., Weidanz W. P., Long C. A., Susceptibility of CXB recombinant inbred mice to murine plasmodia. Infect. Immun. 43, 981–985 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y., et al. , An intracellular membrane protein GEP1 regulates xanthurenic acid induced gametogenesis of malaria parasites. Nat. Commun. 11, 1764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delves M. J., et al. , Routine in vitro culture of P. falciparum gametocytes to evaluate novel transmission-blocking interventions. Nat. Protoc. 11, 1668–1680 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Fan Y., et al. , Rrp6 regulates heterochromatic gene silencing via ncRNA RUF6 decay in malaria parasites. MBio 11, e01110–e01120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X., Qian P., Cui H., Yao L., Yuan J., A protein palmitoylation cascade regulates microtubule cytoskeleton integrity in Plasmodium. EMBO J. 39, e104168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao H., et al. , ISP1-anchored polarization of GCβ/CDC50A complex initiates malaria ookinete gliding motility. Curr. Biol. 28, 2763–2776.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Wang M., Wang J., Glucose transporter GLUT1 influences Plasmodium berghei infection in Anopheles stephensi. Parasit. Vectors 13, 285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K., et al. , MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In addition to the datasets provided in SI Appendix, the raw and processed high-throughput sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE159127).