Significance
Inflammatory diseases collectively account for numerous deaths and morbidity worldwide. New treatment approaches are needed. A central feature of inflammatory diseases is the recruitment of leukocytes to the affected tissues, which is stimulated by secreted proteins called chemokines. Effective suppression of leukocyte recruitment could be achieved by simultaneously targeting multiple chemokines, a natural molecular strategy used by tick salivary proteins called evasins. Here, we describe the structural and molecular features of a tick evasin that control its ability to bind and block a limited set of chemokines. By modifying these features, we demonstrate that evasins can be engineered to alter the array of chemokines that they target. Thus, this study establishes a structure-based paradigm for the development of antiinflammatory therapeutics.
Keywords: inflammatory diseases, chemokines, tick evasins, protein engineering
Abstract
As natural chemokine inhibitors, evasin proteins produced in tick saliva are potential therapeutic agents for numerous inflammatory diseases. Engineering evasins to block the desired chemokines and avoid off-target side effects requires structural understanding of their target selectivity. Structures of the class A evasin EVA-P974 bound to human CC chemokine ligands 7 and 17 (CCL7 and CCL17) and to a CCL8-CCL7 chimera reveal that the specificity of class A evasins for chemokines of the CC subfamily is defined by conserved, rigid backbone–backbone interactions, whereas the preference for a subset of CC chemokines is controlled by side-chain interactions at four hotspots in flexible structural elements. Hotspot mutations alter target preference, enabling inhibition of selected chemokines. The structure of an engineered EVA-P974 bound to CCL2 reveals an underlying molecular mechanism of EVA-P974 target preference. These results provide a structure-based framework for engineering evasins as targeted antiinflammatory therapeutics.
While some proteins exhibit absolute specificity for a unique binding partner, many others display “multispecificity,” whereby they interact with several, but not all, members of a partner protein family (1, 2). Understanding how proteins achieve such selectivity provides a basis for rational engineering to regulate alternative targets. In this study, we investigated the structural basis for multispecific recognition of human proinflammatory chemokines by tick evasin proteins.
Chemokines are the master regulators of leukocyte-trafficking, the unifying feature of immune homeostasis and all inflammatory diseases (3). Chemokines stimulate leukocyte migration via activation of chemokine receptors, G protein–coupled receptors expressed on the surfaces of leukocytes. Chemokines are classified into two major families (CCL and CXCL) and two minor families (XCL and CX3CL) based on the arrangement of conserved cysteine residues near the N termini of their amino acid sequences. Chemokine receptors are classified (CCR, CXCR, XCR, and CX3CR) based on their chemokine selectivity. The types of leukocytes recruited to specific tissues depend on the array of chemokines expressed in those tissues and the selectivity of those chemokines for the receptors expressed on different leukocyte subsets. For example, in vascular inflammation associated with hypertension, elevated levels of the chemokines CCL2, CCL7, and CCL8 act via the receptor CCR2 (and possibly also CCR1) to stimulate migration of monocytes into the blood vessel wall (4).
To suppress leukocyte recruitment in inflammatory diseases, numerous antagonists of specific chemokine receptors have been evaluated in clinical trials. However, these trials have not yielded any new antiinflammatory therapeutics (5), in part because most leukocytes can utilize multiple chemokine receptors, thus circumventing the specific antagonists. The alternative approach of targeting chemokines has not generally been favored, because it would require agents that bind with high affinity to multiple chemokines. However, the natural chemokine-binding proteins of ticks, helminths, and viruses (6–8) display multispecificity for mammalian chemokines, suggesting that they could potentially be deployed as antiinflammatory therapeutics.
Evasins are two families of chemokine-binding, antiinflammatory proteins from tick saliva (6). Class A evasins each inhibit multiple CC chemokines of their mammalian hosts but none of the closely related CXC chemokines. Conversely, class B evasins are specific for CXC over CC chemokines but exhibit variable selectivity among CXC chemokines. Typically, each tick species secretes a mixture of evasins, thereby accomplishing broad-spectrum suppression of the host inflammatory response, presumably enabling the tick to feed on the host for extended periods.
The in vivo antiinflammatory activity of tick evasins has been demonstrated using a variety of animal models of inflammatory diseases, including lung fibrosis, skin inflammation, arthritis, colitis, pancreatitis, ischemic reperfusion injury, postinfarction myocardial injury, and Leishmania major infection (9–13). However, deploying evasins as effective antiinflammatory therapeutics in humans would require engineering the natural evasins to selectively target the relevant array of chemokines for any given indication while minimizing off-target inhibition (14). Such engineering requires understanding both the specificity of evasins for a single chemokine subfamily and their target preference among chemokines within that subfamily.
Previously, only a single structure has been reported for an evasin:chemokine complex, class A evasin EVA-1 bound to CCL3 (15), so it has not been possible to identify the conserved and variable features of the interactions. Nevertheless, the structure revealed that EVA-1 binds to several receptor recognition elements of CCL3, explaining its inhibitory activity. Moreover, limited mutational data (15, 16) have confirmed that residues in the N- and C-terminal regions of EVA-1 and the homologous EVA-4, respectively, contribute to binding affinity, raising the question of whether the structural basis of CC chemokine recognition varies across the class A evasin family.
To establish a structure-based platform for engineering the chemokine selectivity of class A evasins, we now report the structures of EVA-P974 (previously called ACA-01) (17, 18), from the Cayenne tick (Amblyomma cajennense), bound to each of two wild-type chemokines and one chimeric CC chemokine. Structural comparisons and extensive evasin and chemokine mutational data revealed the structural basis for CC chemokine specificity and identified several “hotspots” that define target preference among CC chemokines. These insights enabled EVA-P974 to be engineered to modify its target preference. We further verified the molecular basis of the modified selectivity by solving the chemokine-bound structure of the engineered evasin. Finally, by inhibiting a chemokine mixture, we provide proof of principle for applying engineered evasins as multichemokine inhibitors.
Results and Discussion
Chemokine Selectivity of EVA-P974.
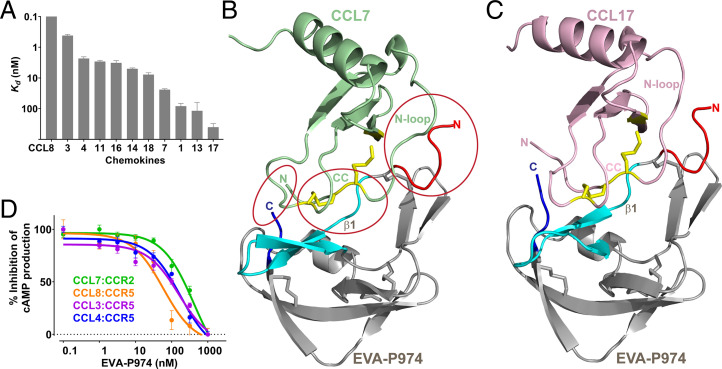
Evasins have previously been produced in either bacterial or eukaryotic expression systems, with both yielding functional proteins (15, 17–19). To avoid the heterogeneity and extensive glycosylation resulting from eukaryotic expression, we overexpressed recombinant EVA-P974 in Escherichia coli and purified it to homogeneity. Mass spectrometry confirmed the presence of the four expected disulfide bonds. EVA-P974 was then immobilized, via a biotinylated Avi-tag, on a surface plasmon resonance (SPR) chip and screened for binding to all available human chemokines. EVA-P974 bound to 11 of the 22 human CC chemokines tested (equilibrium dissociation constants; Kd ∼0.05 to 400 nM) but did not bind to any of the 16 CXC, one CX3C, and two XC chemokines tested (at 500 nM) (Fig. 1A and SI Appendix, Fig. S1 and Table S1). Thus, like other class A evasins, EVA-P974 is specific for CC chemokines over other chemokine families but displays limited selectivity among CC chemokines.
Fig. 1.
EVA-P974 binds and inhibits multiple CC chemokines. (A) Affinities of EVA-P974 for CC chemokines, measured using SPR. (B) Structure of EVA-P974 (gray: N terminus, red: first β-sheet, cyan: C terminus, blue: Cys residues as sticks) bound to CCL7 (green: Cys residues, yellow sticks), with key recognition regions circled and labeled. (C) Structure of EVA-P974 (shown as in B) bound to CCL17 (pink: Cys residues, yellow sticks). (D) EVA-P974 inhibits activation of chemokine receptors by their cognate CC chemokines.
Structure of EVA-P974 in Complex with Chemokines.
We crystallized complexes of EVA-P974 (truncated by eight N-terminal and four C-terminal residues) with human CC chemokine ligands 7 and 17 (CCL7 and CCL17) and solved their structures to resolutions of 1.82 Å and 1.64 Å, respectively (Fig. 1 B and C and SI Appendix, Table S2). EVA-P974 binds to both chemokines with 1:1 stoichiometry. Chemokine-bound EVA-P974 comprises extended N- and C-terminal regions and a structured core (three β-sheets and an α-helix) with four disulfide bonds, which are conserved in other class A evasins (15, 20). CCL7 and CCL17 adopt the canonical chemokine architecture, in which the critical receptor-binding regions (21)—the N terminus, CC motif, and “N-loop”—are linked to the secondary structure core via two conserved disulfide bonds. These chemokine regions interact directly with EVA-P974 (Fig. 1B), consistent with the ability of EVA-P974 to inhibit chemokine stimulation of cognate receptors (Fig. 1D). Specifically: the chemokine CC motif binds to the first β-sheet of EVA-P974; the chemokine N-loop interacts with the EVA-P974 N-terminal region and with a hydrophobic pocket formed by noncontiguous elements from the first two β-sheets of EVA-P974; and the chemokine N terminus interacts with the EVA-P974 C-terminal tail.
Structural Basis of CC Chemokine Specificity of EVA-P974.
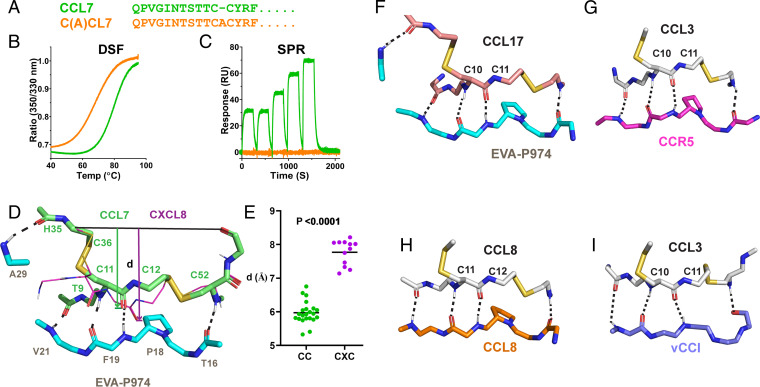
Why do class A evasins target CC chemokines with absolute specificity? CC and CXC chemokines differ not only in the spacing of N-terminal Cys residues but also in the amino acid sequences of their N-terminal and N-loop regions. To probe the direct role of the CC motif, we constructed a variant of CCL7 with an alanine inserted within the CC motif to mimic the CXC equivalent (Fig. 2A). This mutant, C(A)CL7, underwent cooperative thermal denaturation, as shown using differential scanning fluorimetry (Fig. 2B), with a denaturation midpoint of ∼68 °C, compared to ∼78 °C for wild-type CCL7. These data indicated that C(A)CL7 was fully folded at ambient temperature, albeit slightly destabilized relative to the wild-type protein. Nevertheless, C(A)CL7 was unable to bind to immobilized EVA-P974, as shown by SPR (Fig. 2C). These results demonstrate that interactions with the CC motif itself determine the specificity of EVA-P974 (and probably other class A evasins) for CC chemokines.
Fig. 2.
Conserved backbone–backbone interactions dictate specificity of class A evasins for CC chemokines. (A) Sequence alignment of CCL7 and the CCL7 mutant, C(A)CL7, with Ala inserted in the CC motif. (B) Differential scanning fluorometry (DSF) traces show that CCL7 (green) and C(A)CL7 (orange) undergo cooperative thermal unfolding. (C) SPR traces show that CCL7 (green) binds to EVA-P974, whereas C(A)CL7 (orange) does not. (D) EVA-P974 (cyan sticks) recognizes the CC motif of CCL7 (lime green sticks) but sterically clashes with the CXC motif of CXCL8 (thin magenta sticks); the indicated distance, d, is defined as the distance from the line between the Cα atom of the third conserved Cys and the CO atom of the residue after the fourth conserved Cys to the OC atom immediately preceding the second conserved Cys. (E) The key structural distance, d (defined in D), is conserved within CC and CXC chemokine subfamilies but differs between these two subfamilies. (F–I) Conserved mode of CC chemokine recognition by EVA-P974, chemokine receptor CCR5 (Protein Data Bank [PDB]: 7F1T), chemokine CCL8 (PDB: 7S5A) within a homodimer, and viral CC chemokine inhibitor vCCI (PDB: 4ZLT).
EVA-P974 interacts with the cysteine residues (or adjacent residues) of both CCL7 and CCL17 via five identical backbone–backbone hydrogen bonds (Fig. 2 D and F), four of which are conserved in the only previous structure of an evasin:chemokine complex (SI Appendix, Fig. S2) (15). This hydrogen-bonding results in the relative positions of the two Cys residues in the CC motif being highly constrained. We postulated that insertion of even a single residue between them would cause the inserted residue to protrude from the chemokine surface, sterically clashing with the evasin and preventing evasin binding. To systematically assess this hypothesis, we determined the distance, d (defined in Fig. 2D), for all CC and CXC chemokines whose structures were available. Indeed, this analysis showed that the distance is greater in all CXC chemokines than in all CC chemokines (Fig. 2E), supporting the proposal that CXC chemokines cannot be accommodated within the structural constraints of the evasin:chemokine complex. We conclude that the conserved backbone–backbone interactions, rather than directly favoring CC chemokines, are incompatible with binding to CXC chemokines, a phenomenon previously termed “negative selection” (1). This phenomenon likely accounts for the binding specificity of all characterized class A evasins for CC chemokines.
Although our data demonstrate the requirement of the CC motif for recognition of class A evasins, we note that it is not the only required chemokine region. This is clear because not all CC chemokines bind to any given class A evasin. The roles of several additional chemokine elements are also supported by mutational data (see the next section). Thus, simply removing the X residue may not necessarily convert CXC chemokines into evasin binders.
In addition to class A evasins, several other families of proteins selectively recognize CC chemokines over other chemokine families, including CC chemokine receptors (21, 22), viral CC chemokine inhibitors (23, 24), and CC chemokines themselves (by homo- or heterodimerization) (25). Structural comparisons revealed that the mode of CC motif recognition is almost identical in all these protein families (Fig. 2 G–I), with a structurally constrained (rigid) β-strand being the central recognition element in each case. This is a remarkable example of convergent evolution and structural mimicry of a protein recognition motif.
Interactions with the Chemokine N-Loop Dominate the Target Preference of EVA-P974.
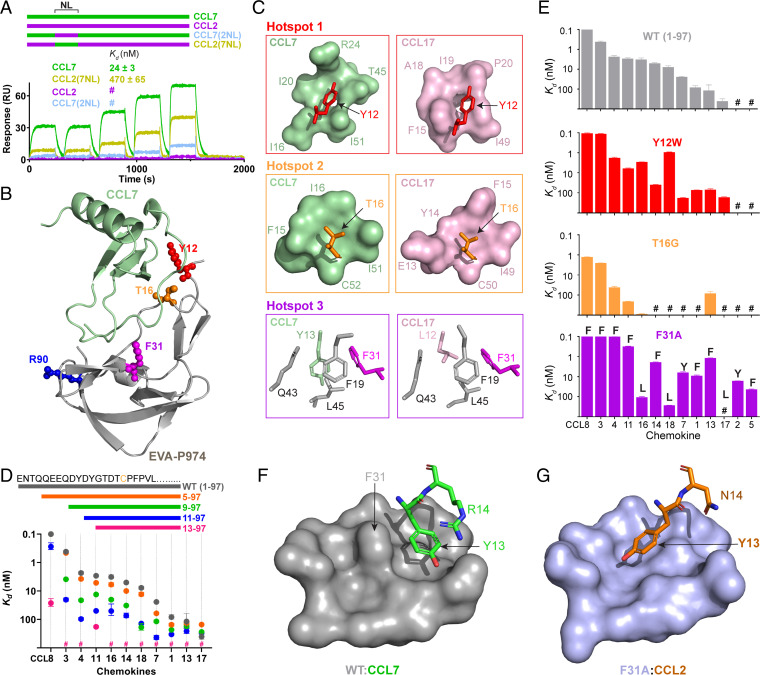
What controls the target preference of class A evasins among CC chemokines? Side-chain interactions are limited to the chemokine N-loop, adjacent β3 strand, and N terminus. Considering that EVA-P974 binds with high affinity to CCL7 (Kd ∼ 24 nM) but not measurably to the 71% identical chemokine CCL2 (at a concentration of 500 nM), we used a set of CCL2–CCL7 chimeras (26) to determine the contributions of these three chemokine regions to the preference of EVA-P974 for CCL7. Swapping of the N terminus or β3 region between these two chemokines had no effect on binding to EVA-P974 (SI Appendix, Fig. S3). However, replacing the N-loop of CCL7 with that of CCL2 (chimera CCL7(2NL)) dramatically reduced binding to EVA-P974 (affinity was not measurable at 500 nM chemokine), whereas replacing the N-loop of CCL2 with that of CCL7 (chimera CCL2(7NL)) substantially enhanced binding to EVA-P974 (Kd = 470 ± 65 nM) (Fig. 3A and SI Appendix, Fig. S3). These data demonstrate that the flexible N-loop of the chemokine is critical for EVA-P974 to bind CCL7 selectively over CCL2.
Fig. 3.
Hotspots between the EVA-P974 N terminus and the chemokine N-loop (NL) dominate target preference. (A) Swapping the NL between CCL7 and CCL2 substantially switches their ability to bind to EVA-P974, as shown by SPR. Chimeric chemokines are shown schematically (color-coded) and named to indicate the parental chemokine and the origin of the inserted NL region; for example, CCL7(2NL) consists of the CCL7 sequence with the NL replaced by that of CCL2. (B) EVA-P974:CCL7 complex, highlighting the EVA-P974 hotspots for target chemokine selectivity. (C) Close-up views of EVA-P974 hotspot residues (colored as in B) interacting with CCL7 (green) and CCL17 (pink). (D) N-terminal truncation of EVA-P974 substantially decreases chemokine-binding affinity. (E) Hotspot mutations alter the chemokine-binding profile of EVA-P974. For F31A, each data point is labeled with the one-letter code for the first residue in the NL. (F) Wild-type EVA-P974 binds to the CCL7 Tyr13 side chain in an orientation stabilized by a cation–π interaction with Arg14. (G) Removal of the bulky phenyl ring in the EVA-P974(F31A) variant enables binding to the CCL2 Tyr13 side chain in an alternative orientation not possible in the wild-type evasin. All Kd values were measured using SPR (n = 3). #, no measurable binding at 500-nM chemokine concentration.
Mutational analysis identified three N-loop interaction “hotspots” within EVA-P974, focused on Tyr12, Thr16, and Phe31, respectively (Fig. 3B). These hotspot interactions differ between the CCL7 and CCL17 complexes and each hotspot could be modified to alter chemokine target preference.
Hotspot 1 represents the interaction of EVA-P974 residue Tyr12 with a shallow pocket created by several hydrophobic and positively charged residues from the chemokine N-loop and β3 regions (Fig. 3C). The side chains forming this pocket differ substantially between CCL7 and CCL17. Several lines of evidence support the importance of Tyr12 for chemokine recognition. First, in a series of N-terminal truncation mutants of EVA-P974, deletion of residues Asp11 and Tyr12 resulted in the largest reduction in binding affinity to most chemokines (Fig. 3D). Second, we have previously observed that sulfation of Tyr12, which occurs posttranslationally in eukaryotic cells, enhances affinity and alters selectivity for target chemokines (19). Third, substitution of Tyr12 by Trp altered the target preference of EVA-P974, enhancing affinity for CCL3 and CCL18 (Fig. 3E).
Hotspot 2 is the interaction between Thr16 of EVA-P974 and residues in the chemokine N-loop and β3 strand (Fig. 3 B and C). Thr16 is conserved in evasins with broad target preference but substituted by Gly, in EVA-1, which has a more-restricted chemokine-binding preference (27). Indeed, the EVA-P974 T16G mutant displayed more-focused target preference than wild-type EVA-P974 (Fig. 3E), demonstrating that broad-spectrum chemokine-binding is dependent on the Thr16 side-chain interactions and/or the effects of this residue on the conformation of the evasin N-terminal region.
Hotspot 3 involves binding of the first residue of the chemokine N-loop, a large hydrophobic residue, into a hydrophobic pocket in the core of EVA-P974 (Fig. 3 B and C). The orientation of this hydrophobic side chain and the composition of the pocket differ substantially between the EVA-P974:CCL7 and EVA-1:CCL3 structures (SI Appendix, Fig. S4). Moreover, mutations in this pocket alter the target preference of EVA-P974, with the F31A mutant exhibiting a profound preference for chemokines in which the first residue of the N-loop is aromatic (Fig. 3E and SI Appendix, Fig. S5).
To explore the structural basis of this altered selectivity, we crystallized EVA-P974(F31A) bound to CCL2 and solved the structure of the complex at 1.74-Å resolution (SI Appendix, Fig. S6 and Table S2). Comparison to the CCL7-bound structure of wild-type EVA-P974 reveals the likely reason for the modified chemokine selectivity of the mutant. In wild-type EVA-P974, the bulky side chain of residue Phe31 narrows the hydrophobic pocket, so the Tyr13 side chain of CCL7 is orientated toward an alternative binding site on the evasin surface (Fig. 3F). This orientation appears to be stabilized by a cation–π interaction (28) with the following residue, Arg14. CCL2 lacks such a stabilizing interaction, explaining the preference of wild-type EVA-P974 to bind CCL7 but not CCL2. In EVA-P974(F31A), deletion of the bulky phenyl ring opens up the hydrophobic pocket, enabling it to readily accommodate the Tyr13 side chain of CCL2 in an extended conformation (Fig. 3G). Indeed, this same extended conformation is observed in the solution structures of all free chemokines in which this residue is aromatic (SI Appendix, Fig. S7), indicating that it is the most stable conformation and explaining why all these chemokines bind more tightly to the F31A mutant. Thus, we conclude that wild-type EVA-P974 cannot accommodate most chemokines for which the first residue of the chemokine N-loop is aromatic without an energetically unfavorable conformational rearrangement, whereas EVA-P974(F31A) binds favorably to the preferred conformation of all these chemokines. By contrast, the F31A mutant showed reduced affinity to chemokines with an aliphatic residue at the same position, apparently due to loss of a favorable interaction with the Phe31 side chain (SI Appendix, Fig. S8).
Interaction of the EVA-P974 C Terminus with Chemokine N Terminus Fine-Tunes Binding Selectivity.
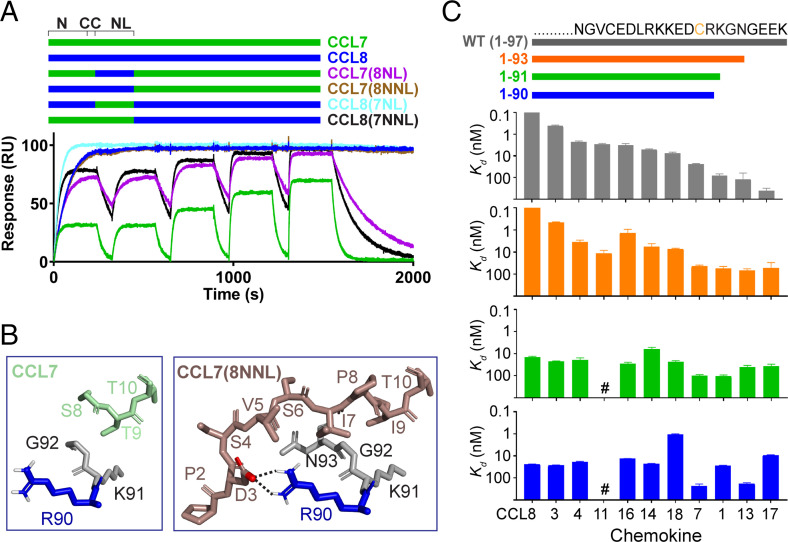
Notwithstanding the importance of N-loop interactions, exchanging the N-loop sequences of CCL8 (the strongest EVA-P974 binder) and CCL7 (a relatively weak binder) did not account for their ∼1,000-fold affinity difference (chimeras CCL7(8NL) and CCL8(7NL) in Fig. 4A and SI Appendix, Fig. S9). However, swapping the N-terminal (in addition to N-loop) sequences, giving the chimeras CCL7(8NNL) and CCL8(7NNL), demonstrated that interactions of the N-terminal region were largely responsible for the affinity difference (Fig. 4A and SI Appendix, Fig. S9). Despite several attempts, the EVA-P974:CCL8 complex failed to crystallize. However, the complex between EVA-P974 and CCL7(8NNL) crystallized, and the structure was solved to 2.39-Å resolution (SI Appendix, Fig. S10 and Table S2). Whereas the N-terminal structures in the CCL7 and CCL17 complexes were only partially resolved, the CCL7(8NNL) complex revealed a set of well-defined hydrophobic interactions between CCL8 N-terminal residues and EVA-P974 C-terminal residues Gly92 and Asn93 and a salt bridge between CCL8 Asp3 and EVA-P974 Arg90 (Fig. 4B). We postulated that these interactions were responsible for the relatively high affinity of CCL8. Consistent with this hypothesis, truncation of the EVA-P974 C terminus past residue Gly92 (EVA-P974(1-91) and (1-90) in Fig. 4C) resulted in complete loss of CCL8 preference and very limited selectivity among chemokine ligands.
Fig. 4.
Interactions of the EVA-P974 C terminus fine-tune chemokine-binding selectivity. (A) Swapping the N terminus (N) and N-loop (NL) between CCL7 and CCL8 substantially switches their ability to bind to EVA-P974, but swapping the NL alone does not, as shown by SPR. Chimeric chemokines are shown schematically (color-coded) and named to indicate the parental chemokine and the origin of the inserted NL or inserted N terminus plus NL (NNL) region; for example, CCL7(8NNL) consists of the CCL7 sequence with both the N-terminal and NL regions replaced by those of CCL8. (B) Detailed interactions between the C terminus of EVA-P974 (R90, blue; K91-N93, gray) and the N termini of CCL7 (left, green) and the chimera CCL7(8NNL) (right, beige). Residues shown in the CCL7(8NNL) but not the CCL7 structure were not resolved in the latter data set. (C) C-terminal truncation of EVA-P974 alters chemokine-binding affinity, with variants 1 through 90 and 1 through 91 exhibiting reduced selectivity among chemokines. All Kd values were measured using SPR (n = 3). #, no measurable binding at 500-nM chemokine concentration.
Application of Engineered EVA-P974 in Chemokine Functional Inhibition.
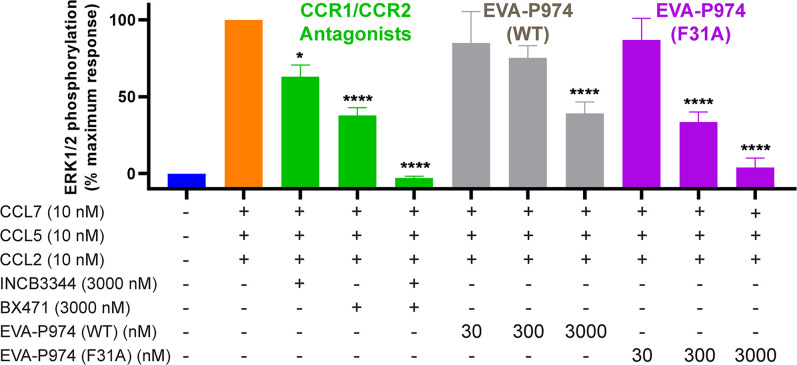
To demonstrate the utility of engineered evasins as chemokine inhibitors, we compared the inhibitory activity of wild-type and F31A mutant EVA-P974 (Fig. 5). THP-1 cells, an immortalized monocytic cell line, express two major proinflammatory chemokine receptors, CCR1 and CCR2. The chemokines CCL2, CCL5, and CCL7 are agonists of CCR2, CCR1, and both receptors, respectively. As expected, treatment of THP-1 cells with a mixture of these three chemokines induced intracellular ERK phosphorylation. Selective small-molecule antagonists of CCR1 (BX471) and CCR2 (INCB3344) confirmed that ERK phosphorylation was mediated via combined activation of these two receptors. Wild-type EVA-P974, which bound with high affinity to CCL7 but not to CCL2 or CCL5, only reduced the combined CCR1/CCR2 activation of these three chemokines by ∼50% at 3,000 nM. In contrast, the F31A mutant bound to all three chemokines and completely abrogated the combined receptor activation. When the cells were treated with each chemokine individually, rather than the mixture, wild-type EVA-P974, at 300 nM, significantly inhibited the response to CCL7 but not to CCL2 or CCL5, whereas the F31A mutant significantly inhibited the responses to all three chemokines (SI Appendix, Fig. S11). These, data demonstrate that it is possible to engineer an evasin with superior ability, relative to the wild-type evasin, to block the cellular response to multiple chemokines acting via multiple receptors.
Fig. 5.
Engineered EVA-P974(F31A) inhibits receptor activation by multiple chemokines. Combined activation of CCR1 and CCR2 in THP-1 monocytes by the chemokine mixture CCL2+CCL5+CCL7 (orange) is fully inhibited by CCR1 inhibitor BX471 plus CCR2 inhibitor INCB3344 (green), partially inhibited by wild-type EVA-P974 (gray), and fully inhibited by EVA-P974(F31A) (magenta). Data points represent the average ± SEM of three independent experiments, each conducted in duplicate. *P < 0.05 and ****P < 0.0001 relative to CCL2+CCL5+CCL7, one-way ANOVA.
Concluding Remarks
Our results show that the absolute specificity of class A evasins for chemokines of the CC subfamily is defined by structurally conserved, rigid backbone–backbone interactions that cannot be accommodated by other chemokine subfamilies. However, the target preference of class A evasins for distinct subsets of CC chemokines is controlled by side chain–side chain interactions at several key hotspots in relatively flexible parts of the structure. Thus, the evasin:chemokine system establishes a structural paradigm by which proteins can achieve binding selectivity within a restricted target family.
The structural basis of chemokine recognition defined here establishes a foundation for engineering class A evasins to selectively target subsets of chemokines involved in inflammatory diseases. The enhanced chemokine-inhibitory activity of EVA-P974(F31A) provides proof of principle for this approach. Our results suggest that through a combination of rational engineering and directed in vitro evolution, it may be feasible to tailor the selectivity of evasins for applications as antiinflammatory therapeutics.
Materials and Methods
Expression and purification of EVA-P974 (or variants thereof) and chemokines (CCL2, CCL7, CCL8, CCL11, CCL16, CCL17, CCL18, or variants thereof), chemokine-binding by SPR, crystallization of complexes between EVA-P974 (or variants thereof) and chemokines, structure solution and refinement, nano differential scanning fluorometry, cAMP inhibition assay, and ERK1/2 phosphorylation assay are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Monash Macromolecular Crystallisation Facility staff and the Monash Proteomics and Metabolomics Facility staff (David Steer) for assistance, and Martin Scanlon and Nathan Companez for advice and discussions. This research was supported by National Health and Medical Research Council Project Grants APP1140867 (M.J.S., R.J.P., and M.C.J.W.) and APP1140874 (M.J.S.) and Investigator Grant APP1174941 (R.J.P.) and by a Bridging Postdoctoral Fellowship from Monash University (R.P.B.). This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122105119/-/DCSupplemental.
Data Availability
The coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: 7S58, 7S4N, 7S59, and 7SO0. All other study data are included in the article and/or SI Appendix.
References
- 1.Schreiber G., Keating A. E., Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 21, 50–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sergeeva A. P., et al. , DIP/Dpr interactions and the evolutionary design of specificity in protein families. Nat. Commun. 11, 2125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zlotnik A., Yoshie O., Chemokines: A new classification system and their role in immunity. Immunity 12, 121–127 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Moore J. P., et al. , M2 macrophage accumulation in the aortic wall during angiotensin II infusion in mice is associated with fibrosis, elastin loss, and elevated blood pressure. Am. J. Physiol. Heart Circ. Physiol. 309, H906–H917 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Zhao S., Wu B., Stevens R. C., Advancing chemokine GPCR structure based drug discovery. Structure 27, 405–408 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Bhusal R. P., et al. , Evasins: Tick salivary proteins that inhibit mammalian chemokines. Trends Biochem. Sci. 45, 108–122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maizels R. M., Smits H. H., McSorley H. J., Modulation of host immunity by Helminths: The expanding repertoire of parasite effector molecules. Immunity 49, 801–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Sanchez J., Stone M. J., Payne R. J., Sulfation of the human cytomegalovirus protein UL22A enhances binding to the chemokine RANTES. Angew. Chem. Int. Ed. Engl. 56, 8490–8494 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Braunersreuther V., et al. , Treatment with the CC chemokine-binding protein Evasin-4 improves post-infarction myocardial injury and survival in mice. Thromb. Haemost. 110, 807–825 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Déruaz M., et al. , Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. J. Exp. Med. 205, 2019–2031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montecucco F., et al. , Treatment with Evasin-3 abrogates neutrophil-mediated inflammation in mouse acute pancreatitis. Eur. J. Clin. Invest. 44, 940–950 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Russo R. C., et al. , Therapeutic effects of evasin-1, a chemokine binding protein, in bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 45, 72–80 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Vieira A. T., et al. , Treatment with a novel chemokine-binding protein or eosinophil lineage-ablation protects mice from experimental colitis. Am. J. Pathol. 175, 2382–2391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lounkine E., et al. , Large-scale prediction and testing of drug activity on side-effect targets. Nature 486, 361–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dias J. M., et al. , Structural basis of chemokine sequestration by a tick chemokine binding protein: The crystal structure of the complex between Evasin-1 and CCL3. PLoS One 4, e8514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonvin P., et al. , Identification of the pharmacophore of the CC chemokine-binding proteins Evasin-1 and -4 using phage display. J. Biol. Chem. 289, 31846–31855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward J., et al. , Ticks from diverse genera encode chemokine-inhibitory evasin proteins. J. Biol. Chem. 292, 15670–15680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh K., et al. , Yeast surface display identifies a family of evasins from ticks with novel polyvalent CC chemokine-binding activities. Sci. Rep. 7, 4267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franck C., et al. , Semisynthesis of an evasin from tick saliva reveals a critical role of tyrosine sulfation for chemokine binding and inhibition. Proc. Natl. Acad. Sci. U.S.A. 117, 12657–12664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denisov S. S., et al. , Structural characterization of anti-CCL5 activity of the tick salivary protein evasin-4. J. Biol. Chem. 295, 14367–14378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhusal R. P., Foster S. R., Stone M. J., Structural basis of chemokine and receptor interactions: Key regulators of leukocyte recruitment in inflammatory responses. Protein Sci. 29, 420–432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kufareva I., Salanga C. L., Handel T. M., Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol. Cell Biol. 93, 372–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo N. W., et al. , Structural insights into the interaction between a potent anti-inflammatory protein, viral CC chemokine inhibitor (vCCI), and the human CC chemokine, Eotaxin-1. J. Biol. Chem. 289, 6592–6603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau E. K., Allen S., Hsu A. R., Handel T. M., Chemokine-receptor interactions: GPCRs, glycosaminoglycans and viral chemokine binding proteins. Adv. Protein Chem. 68, 351–391 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Miller M. C., Mayo K. H., Chemokines from a structural perspective. Int. J. Mol. Sci. 18, 2088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huma Z. E., et al. , Key determinants of selective binding and activation by the monocyte chemoattractant proteins at the chemokine receptor CCR2. Sci. Signal. 10, eaai8529 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Frauenschuh A., et al. , Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus. J. Biol. Chem. 282, 27250–27258 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Kumar K., et al. , Cation-π interactions in protein-ligand binding: Theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. (Camb.) 9, 2655–2665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors have been deposited in the Protein Data Bank under the following accession codes: 7S58, 7S4N, 7S59, and 7SO0. All other study data are included in the article and/or SI Appendix.