Significance
Cellular protein synthesis relies on faithful selection of the translation start codon by the ribosome. Initiation at alternative sites on a messenger RNA (mRNA) impairs production of native proteins, triggers synthesis of junk proteins, and enables regulation of translation. The nucleotide sequence flanking a start codon controls its efficiency of selection. We identify mRNAs containing start codons in conserved poor sequence contexts, including several Hox mRNAs encoding regulators of the body plan. Other Hox mRNAs contain conserved upstream open reading frames with poor start codon contexts that sensitize translation to changes in start codon selection stringency. Thus, alterations in start codon selection stringency has the potential to regulate global gene expression programs, including Hox gene-directed body plan formation in animals.
Keywords: Hox genes, start codon selection stringency, uORF, eIF1, eIF5
Abstract
Translation start site selection in eukaryotes is influenced by context nucleotides flanking the AUG codon and by levels of the eukaryotic translation initiation factors eIF1 and eIF5. In a search of mammalian genes, we identified five homeobox (Hox) gene paralogs initiated by AUG codons in conserved suboptimal context as well as 13 Hox genes that contain evolutionarily conserved upstream open reading frames (uORFs) that initiate at AUG codons in poor sequence context. An analysis of published cap analysis of gene expression sequencing (CAGE-seq) data and generated CAGE-seq data for messenger RNAs (mRNAs) from mouse somites revealed that the 5′ leaders of Hox mRNAs of interest contain conserved uORFs, are generally much shorter than reported, and lack previously proposed internal ribosome entry site elements. We show that the conserved uORFs inhibit Hox reporter expression and that altering the stringency of start codon selection by overexpressing eIF1 or eIF5 modulates the expression of Hox reporters. We also show that modifying ribosome homeostasis by depleting a large ribosomal subunit protein or treating cells with sublethal concentrations of puromycin leads to lower stringency of start codon selection. Thus, altering global translation can confer gene-specific effects through altered start codon selection stringency.
Translation start codon selection is crucial for proper gene expression. The selection of the incorrect start codon can lead to the synthesis of junk peptides from alternate reading frames, the production of N-terminally extended or truncated versions of the native proteins from the correct reading frame, or the reduced synthesis of the native protein. In eukaryotes, the translation start codon is selected by a scanning ribosome. A 43S preinitiation complex (PIC) composed of the small (40S) ribosomal subunit, the eukaryotic translation initiation factor 2 (eIF2)-GTP-methionyl initiator tRNA (Met-tRNAiMet) ternary complex, eIF1, eIF1A, eIF3, and eIF5 binds near the 5′-m7G cap of a messenger RNA (mRNA) (reviewed in ref. 1). The PIC then scans down the mRNA, and base pairing interactions between the anticodon of the Met-tRNAiMet in the PIC and a start codon in the mRNA trigger conformational changes in the PIC, leading to the eIF5-dependent completion of GTP hydrolysis by eIF2 and the release of eIF1. Following this selection of the start codon by the PIC and the dissociation of additional factors, the large (60S) ribosomal subunit joins, and the resulting 80S ribosome enters the elongation phase of protein synthesis and begins synthesizing the protein encoded in the reading frame of the selected start codon.
In most cases, eukaryotic translation initiation occurs at the 5′-most AUG codon with favorable flanking nucleotide context. In mammals, the consensus initiation context is GCC(A/G)CCAUGG. The nucleotides in italics and underlined at positions −3 and +4, relative to the first nucleotide of the AUG codon, play the most important role in determining the efficiency of initiation (2). Additionally, although translation usually initiates at an AUG codon, near-cognate codons that differ from AUG by a single nucleotide change such as CUG or UUG can also be selected, albeit at lower efficiency, by the scanning PIC (1, 3–5). The efficiency of near-cognate start codon selection by a scanning ribosome is even more sensitive to the flanking context nucleotides than is AUG start codon selection (5). At both AUG and near-cognate start codons, favorable sequence contexts reduce leaky scanning, which is defined as an instance in which a ribosome scans over a start codon without initiating translation. In addition to sequence context, the stringency of start codon selection is controlled by two important translation initiation factors, eIF1 and eIF5. eIF1 binds near the ribosomal P site and promotes PIC scanning and the skipping of weak start sites, while eIF5, which binds in the ribosomal P site following eIF1 release, has the opposite effect and increases initiation at these weak start sites (4, 6–9).
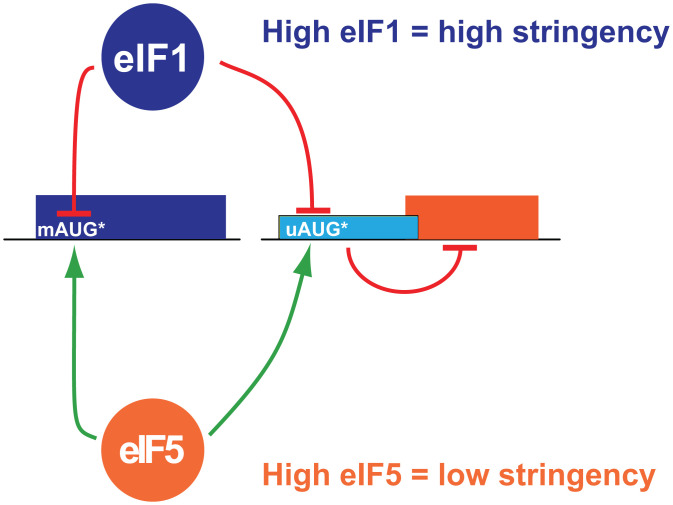
The opposing actions of eIF1 and eIF5 on start codon selection stringency can have differential impacts on translation depending on whether the main open reading frame (mORF) or, alternatively, an inhibitory upstream ORF (uORF) (10) initiates at an AUG codon in weak sequence context (Fig. 1). When the mORF start codon is in weak context, high eIF1 levels relative to eIF5 (Fig. 1, Upper) will result in increased start codon selection stringency and increased scanning past the mORF start codon, leading to a decrease in protein production. In contrast, if the inhibitory uORF start codon is in weak context, high eIF1 levels relative to eIF5 will result in increased leaky scanning past the inhibitory uORF, increased translation of the mORF, and increased protein production (Fig. 1, Right). The opposite effects are expected when eIF5 levels are high relative to eIF1: more ribosomes initiate at weak start sites, resulting in the increased translation of mORFs with start codons in weak context and, conversely, the repression of mORF translation on mRNAs containing inhibitory uORFs with start codons in weak context (Fig. 1, Bottom).
Fig. 1.
A schematic representation of the role of eIF1 and eIF5 in regulating stringency of start codon selection in eukaryotes. (Left) mRNA with the mORF AUG start codon in suboptimal context (mAUG*). (Right) mRNA with inhibitory uORF initiated by an AUG codon in suboptimal context (uAUG*). High levels of eIF1 confer high stringency of start codon selection leading to 1) repressed expression of genes initiated by mAUG* codons and 2) increased leaky scanning of uAUG* codons resulting in the derepression of mORF translation. Low stringency of start codon selection in cells with high levels of eIF5 has the opposite effects, stimulating translation of ORFs starting with mAUG* and repressing the translation of the mORF on mRNAs with inhibitory uORFs initiating at uAUG* codons.
Interestingly, the opposing actions of eIF1 and eIF5 on stringency of start codon selection is exploited to control the relative expression of the two factors within the cell in an autoregulatory feedback loop (4). The AUG start codon (mAUG) of eIF1 is in conserved poor sequence context (6, 11) (like Fig. 1, Left), and increased leaky scanning upon eIF1 overexpression accounts for the negative autoregulation of eIF1 expression (6, 8, 12). By contrast, when eIF5 levels rise, more ribosomes initiate at the eIF1 weak start site, resulting in the production of more eIF1 (4). eIF5 expression is under the inhibitory control of uORFs in its 5′ untranslated region (5'UTR), which themselves are initiated by AUG codons in poor sequence context (4) (like Fig. 1, Right). When eIF1 levels are high, more ribosomes scan over the inhibitory uORFs and synthesize eIF5; whereas, when eIF5 levels are high, more ribosomes translate the uORFs and fail to synthesize eIF5. Thus, auto- and cross-regulation of eIF1 and eIF5 mRNA translation establishes a paradigm for the stringency control of translation. Importantly, however, it is currently not known if perturbations in the stringency of start codon selection might more broadly control the translation of other groups of cellular mRNAs.
In this work, we use a broad search of mammalian mRNA sequences to identify mRNAs containing start codons in conserved poor sequence contexts. Interestingly, mRNAs encoding homeobox (Hox) proteins, ubiquitous regulators of body plan formation (13), are enriched among the mRNAs with conserved poor start codons. Whereas several Hox mRNA leaders were previously proposed to be under the translational control of an internal ribosome entry site (IRES) in the 5′ leader of their mRNA (14), our mapping of the 5′ leaders of mouse Hox mRNAs reveals that the leaders are much shorter than previously proposed and lack the putative IRES elements. In contrast, we identify conserved uORFs that initiate at weak start codons in some of these Hox mRNAs and show that these conserved uORFs control the translation of reporters containing the Hox mRNA leaders. We further show that the alteration of the stringency of translation start site selection through modulating eIF1 or eIF5 levels or through inhibiting general translation affects the translation of the Hox mRNA reporters. Thus, modulators of general translation might have gene-specific effects with consequences for key developmental regulators.
Results
Mammalian Genes with Conserved Poor Context AUG Start Codons.
Previous studies have identified several human genes with conserved suboptimal initiation sites, including either a conserved near-cognate start codon (UUG or CUG) or an AUG codon in conserved suboptimal context (15–17). In some cases, the suboptimal codons provide alternative translation initiation sites to generate N-terminally extended protein isoforms from the same transcript (17–22). However, in other cases, the architecture of the mRNA is such that the suboptimal start codon appears to represent the sole initiation codon; for example, when the next available AUG codon is in frame but near the 3′ end of the coding sequence (CDS) and thus unlikely to initiate a functional protein or when the next available AUG codon is out of frame and will thus lead to nonproductive translation (SI Appendix, Fig. S1).
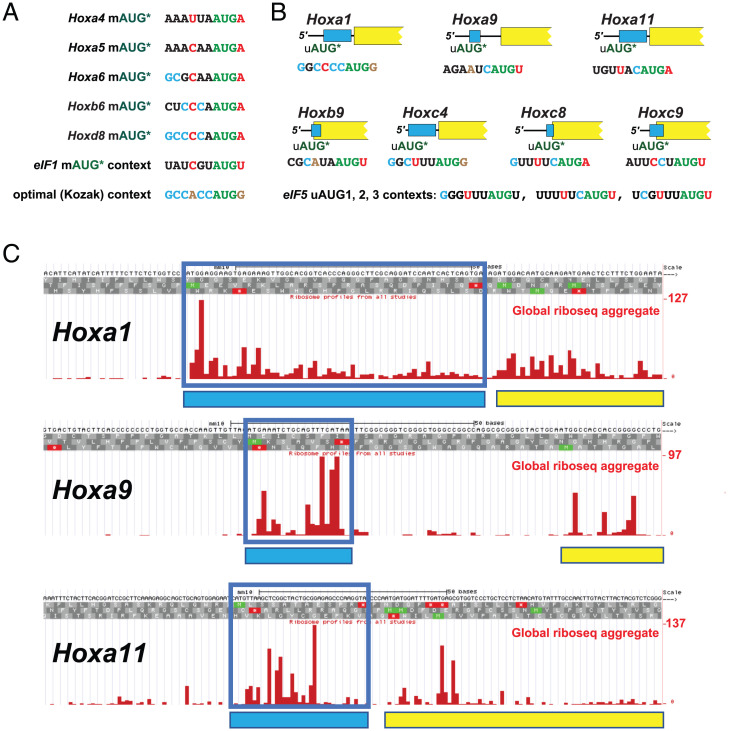
We conducted a search for mammalian mRNAs in which the AUG codon initiating the annotated mORF (mAUG) is found in an evolutionarily conserved poor context. We defined start sites as having a poor context when there is a pyrimidine at the −3 position and no G at the +4 position, as this class has previously been shown to be initiated with an efficiency as low as 3% relative to perfect context (A at −3, G at +4) (2, 4). Of the 394 poor context human mRNAs identified that fulfilled our conservation criteria (Dataset S1, also see Materials and Methods), at least 122 are likely to use the poor context AUG codon as the primary or sole mORF initiation site (“obligatory poor context”) due to the lack of downstream in-frame start codons or the presence of one or more downstream out-of-frame nonpoor context AUG codons before the next in-frame AUG codon (Dataset S2). Four of the human mRNAs—EIF1, EIF1B, BZW1 (also known as 5MP2), and BZW2 (also known as 5MP1)—were previously identified as having conserved poor translation start sites. As all four of these proteins play a critical role in determining start codon selection stringency, the suboptimal mAUG start codon enables these proteins to feedback to inhibit their own mRNA translation in an autoregulatory manner (6, 23). In addition to Eif1/Eif1B and Bzw1/Bzw2, eight other paralogous groups were identified with more than one member initiating at a conserved obligatory poor context start codon—Ext1/Extl1, Hoxa5/Hoxa6/Hoxb5/Hoxb6/Hoxd8 (Fig. 2A and SI Appendix, Figs. S1 and S2), Jph1/Jph3/Jph4, Kdm6a/Kdm6b, Pdgfc/Pdgfd, Smad6/Smad7/Smad9, Tcf19/Tcf25, and Zbtb10/Zbtb18/Zbtb20 (Dataset S1). The presence of five Hox paralogs on this list was striking and suggested the possibility that Hox genes could be candidates for investigating translational regulation conferred by changes in start codon selection stringency.
Fig. 2.
Hox genes with conserved poor context mAUG start codon or conserved uORF initiated by suboptimal uAUG codon. (A) The initiation context for the mAUG start codon of five Hox genes in which the mAUG is the main or only start codon. The suboptimal context of human eIF1 and the consensus optimal (Kozak) context are also shown. (B) A cartoon representation, approximately to scale, of the seven Hox leaders containing uORFs (light blue) conserved from mammals to fish. The suboptimal context of the three uAUG start codons of human eIF5 are also shown. The initiating AUG codon is shown in green; critical consensus optimal nucleotides at position −3 and +4 are shown in brown; nonoptimal residues at positions 3 and +4 are shown in red; consensus residues in positions −6, −5, −4, −2, and −1 are shown in cyan; nonconsensus residues in the same positions are shown in black. (C) Screenshot from the GWIPS-viz (https://gwips.ucc.ie/) browser (31) of global aggregate ribosome footprint profiling (riboseq) coverage in the leaders of mouse Hoxa1, Hoxa9, and Hoxa11. Reads mapping to the uORFs are boxed in blue and represented in the cartoons by with blue rectangles. The mORFs are depicted by yellow rectangles. The first nucleotide of each sequence matches the 5′ end of the mRNA as determined by CAGE-seq of somites (Fig. 3 and SI Appendix, Figs. S1 and S2).
Hox Genes Containing Conserved uORFs with Start Codons in Suboptimal Context.
The 39 HOX genes in the human genome represent ∼0.2% of the total 20,352 protein-coding genes (24). In our analysis, Hox genes represented 31 of the 12,467 genes passing our conservation selection criteria (Dataset S3 and see also Materials and Methods) but 5 of 122 genes with conserved obligatory poor context AUG start codons. This 16.5-fold enrichment (P = 2.8 × 10−5, two-tailed Fisher's exact test) suggests that the evolutionarily conserved Hox genes might have evolved features that make them specifically responsive to changes in global start codon selection stringency. To investigate this possibility, all 39 Hox genes were examined for evolutionarily conserved features in their mRNA leaders that could make them responsive to changes in stringency. In addition to examining the start codon of the mORFs, the start codons of potential uORFs were also examined. Similar to the analysis of mORF start codons, suboptimal upstream AUG (uAUG) context was defined as no purine at the −3 position and/or no G at the +4 position. Importantly, while a suboptimal mAUG confers increased protein expression under conditions of low start site selection stringency (6), the presence of a suboptimal uAUG confers decreased protein expression under conditions of low start site selection stringency because of increased initiation at the inhibitory uORF (4) (Fig. 1).
During the preliminary stages of searching for evolutionarily conserved uORFs in the leaders of Hox mRNAs, we observed discrepancies between annotated transcripts, as currently reported in GenBank, and transcription start sites, as defined by publicly available cap analysis of gene expression sequencing (CAGE-seq) data [Fantom5 (25) on the University of California, Santa Cruz (UCSC) browser (26)]. To resolve these discrepancies and to concentrate on mRNA features that might be relevant during mammalian development regulated by the Hox genes, we independently performed CAGE-seq on total RNA isolated from mouse somites, a mesoderm-derived collection of cells flanking the spinal cord during development (27). Hox gene expression in somites regulates the anterior–posterior patterning of the developing embryo. Thus, we reasoned that the Hox mRNA leaders expressed in mouse somites would be the relevant isoforms for an analysis of start codon selection stringency conferred by Hox gene uORFs and would allow us to directly address previous studies in these tissues (14).
No-amplification nontagging cap analysis of gene expression (nAnT-iCAGE) (28) was performed on total RNA isolated from the somites and neural tubes of three embryonic day 11.5 (E11.5) mice (see Materials and Methods for details on somite/neural tube dissection and RNA extraction). In brief, CAGE-seq utilizes selective oxidation and biotinylation of the 5′ methyl-7-guanosine cap of mRNAs followed by streptavidin immunoprecipitation to isolate mRNA 5′ ends followed by adapter ligation and next-generation sequencing. Following sequencing, the first nucleotide of each read was aligned to the mouse genome (mm10). In total, 15,707,020 reads were mapped from mouse 1, 16,446,419 from mouse 2, and 18,053,549 from mouse 3. Based on these mapping data, the 5′ leaders of Hox genes were defined as the genomic region spanning from the major 5′ UTR peak as determined by CAGE data downstream to the annotated mORF start codon; any uORFs present within this region are considered to be expressed in E11.5 somites. The genomic coordinates of the closest major CAGE peak 5′ to the annotated start codon for all 39 Hox genes are presented in SI Appendix, Table S1. Additionally, leaders in which there are multiple CAGE peaks, in which the peaks are broadly distributed, or in which the peak is uniquely downstream of the annotated start codon are noted in SI Appendix, Table S1.
Based on the CAGE data, 13 Hox mRNAs contain uORFs that met our criteria for conservation (SI Appendix, Table S2)—see Materials and Methods for details of the criteria used. Of those, seven Hox mRNAs—Hoxa1, Hoxa9, Hoxa11, Hoxb9, Hoxc4, Hoxc8, and Hoxc9—contain uORFs conserved from mammals to fish (Fig. 2B and SI Appendix, Figs. S3–S5), four—Hoxa6, Hoxc10, Hoxd10, and Hoxd11—contain uORFs conserved from mammals to tetrapods, and two—Hoxc13 and Hoxd1—contain uORFs conserved in mammals only.
The leaders of three Hoxa mRNAs, which contain uORFs conserved from mammals to fish, Hoxa1, Hoxa9, and Hoxa11, were subjected to further analysis. The leaders of the Hoxa1, Hoxa9, and Hoxa11 mRNAs in mouse somites are relatively short, with lengths of 64 to 98, 83 to 85, and 53 to 91 nucleotides (nts), respectively (Fig. 3 and SI Appendix, Figs. S6 and S8), and their uORFs are 21, 7, and 12 codons in length, respectively (SI Appendix, Figs. S3–S5 and S8). Previous studies of uORFs in mammalian cells demonstrated that the efficiency of ribosome reinitiation following the translation of a uORF is inversely correlated with the length of the uORF (29, 30). Based on the 7 to 21 codon length of the uORFs in these three Hox mRNAs, the translation of these uORFs is predicted to be mildly to moderately inhibitory for the translation of the mORF. Additionally, the context of the initiating uAUG in all three mRNAs was deemed suboptimal: the Hoxa1 uAUG does not have a purine at −3, although it has a G at position +4; Hoxa9 uAUG has neither a purine at the −3 position nor a G at +4; and Hoxa11 uAUG has a preferred A at −3 but does not have the preferred G at +4. Notably, the suboptimal contexts for these three Hox mRNA uORFs are also conserved from mammals to fish (SI Appendix, Figs. S3–S5), and aggregate data from ribosome footprint profiling experiments in mouse cells (31) demonstrate read density of ribosome-protected fragments in all three uORFs, consistent with them being translated in vivo (Fig. 2C).
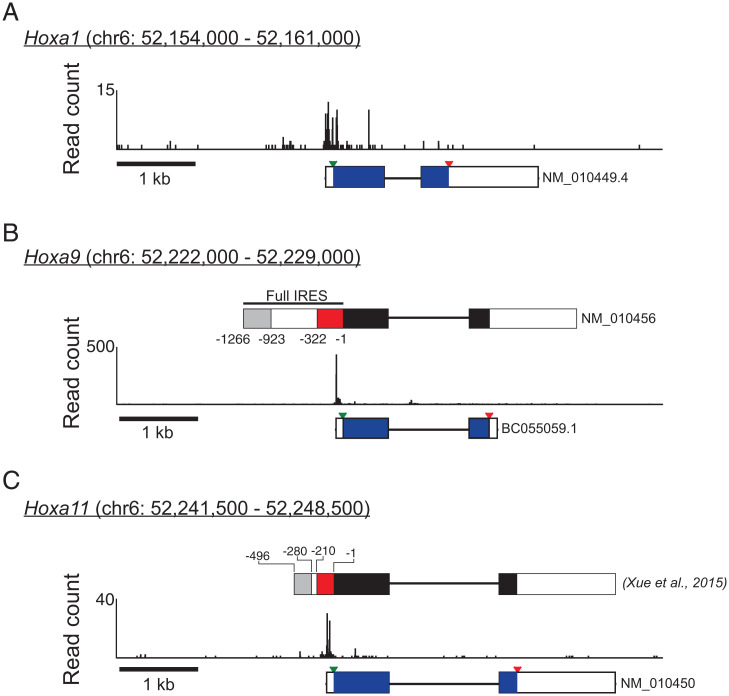
Fig. 3.
The CAGE-seq analysis of mouse somites reveals relatively short leaders on the Hoxa1, Hoxa9, and Hoxa11 mRNAs. Combined reads of three replicates of CAGE-seq analysis mapping the transcription start sites in mouse somites for (A) Hoxa1, (B) Hoxa9, and (C) Hoxa11. For Hoxa9 and Hoxa11, the schematic at Top shows proposed “IRES” elements (red box) (red box, 14) in the transcript drawn to scale. The schematics at Bottom display the National Center for Biotechnology Information (NCBI) annotated transcripts, shown to scale; exons are shown as rectangles, introns as lines; the mORF coding region is shown as a black rectangle, with the start and stop codons indicated by inverted green or red triangles, respectively. Chromosomal coordinates (GRCm38/mm10) of the mapping data and 1-kb scale bar are shown above and below, respectively, each figure. Complementary qRT-PCR analyses reveal that the Hoxa9 CDS is at least 200-fold more abundant in total somite mRNAs that the putative IRES (SI Appendix, Fig. S7).
The Hoxa1, Hoxa9, and Hoxa11 uORFs Confer Stringency-Dependent Control of Gene Expression.
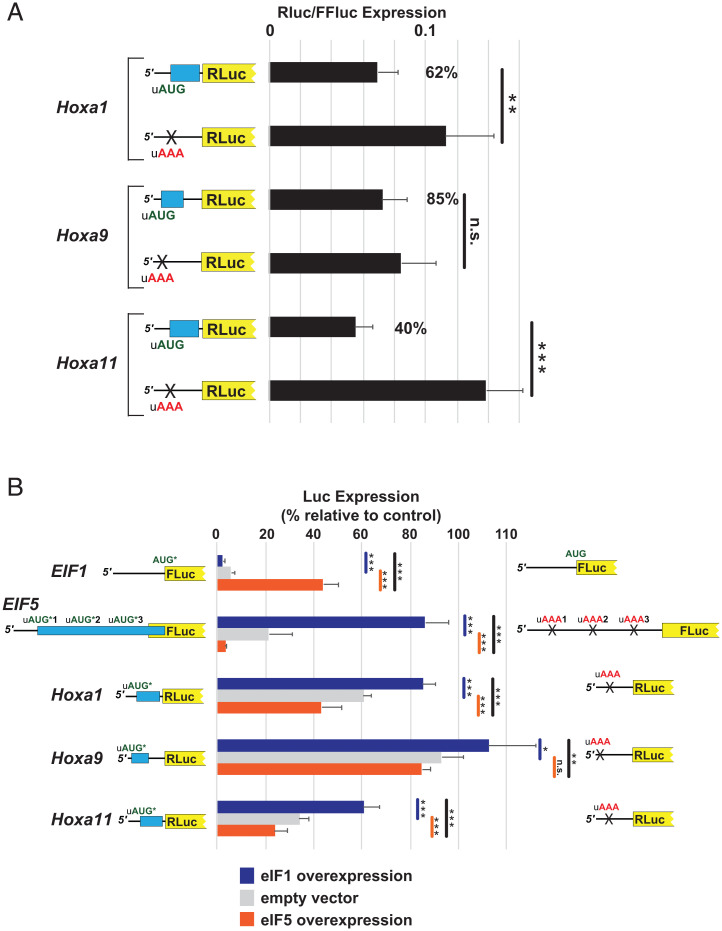
If the conserved Hox uORFs are translated, then they would be expected to repress translation from the downstream mORF (10). To test this prediction, we employed a luciferase reporter in which the conserved leaders of Hoxa1 (96 nts), Hoxa9 (83 nts), and Hoxa11 (91 nts) were placed upstream of the luciferase mORF. Pairs of reporters with the intact uORF or with the uAUG changed to a noninitiating AAA codon were transfected into cultured mammalian U2OS cells. As expected, for Hoxa1 and Hoxa11, eliminating the uORF by mutating the uAUG led to significant derepression of luciferase expression (a greater than twofold increase for Hoxa11 and a more modest increase for Hoxa1) (Fig. 4A). The removal of the uORF of Hoxa9 led to a modest 1.2-fold derepression, which did not meet the criteria for statistical significance (Fig. 4A). Of note, the modest 1.2- to 2.5-fold effects associated with removing these uORFs are consistent with the modest 2.7-fold effect of removing the inhibitory non-AUG–initiated uORF (uCC) in the human AZIN1 mRNA (32) or the fivefold effect associated with removing all three inhibitory uORFs in the EIF5 mRNA (4).
Fig. 4.
The uORFs of Hoxa1, Hoxa9, and Hoxa11 confer translation regulation following global changes in the stringency of start codon selection. (A) U2OS cells were transfected with luciferase reporters containing the leaders of Hoxa1, Hoxa9, and Hoxa11 or mutant versions in which the uAUG codon was mutated to a noninitiating AAA codon. The relative activity of Renilla luciferase reporter to firefly luciferase transfection control is plotted. The percent activity of wild-type versus mutant reporter is indicated on Right. (B) U2OS cells were cotransfected with luciferase reporters described in A, a reporter initiated by an AUG codon in poor EIF1 context (residues −6 to +4) or a reporter containing the leader of EIF5, and with plasmids overexpressing eIF1 (blue), eIF5 (orange), or with empty vector (gray). Activities of the test reporter, shown schematically on Left, are presented as the percentage of the control reporter, shown schematically on Right. The control reporter for eIF1 is initiated by an AUG codon in optimal context; for other controls, the uAUG codons are mutated to noninitiating AAA codons. For Hox reporters, test Renilla luciferase activities were normalized to firefly luciferase activity expressed from the same plasmid. For EIF1 and EIF5 reporters, test firefly activity was normalized to activity from cotransfected Renilla n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s two-tailed t test; n = 8, assayed in duplicate).
Since the uAUGs of the Hoxa1, Hoxa9, and Hoxa11 mRNAs are present in conserved suboptimal initiation context and the uORFs which they initiate are inhibitory to downstream translation, these uORFs might be expected to respond to changes in stringency in a manner similar to the uAUGs of eIF5. To investigate this possibility, the Hoxa1, Hoxa9, and Hoxa11 luciferase reporters were cotransfected into U2OS cells with eIF5-overexpressing (low stringency) or eIF1-overexpressing (high stringency) plasmids or with an empty vector (natural stringency). As a control, the overexpression of eIF1 and eIF5 had the anticipated effect on luciferase reporters fused to the eIF1 or eIF5 mRNA leaders as previously reported (4); the overexpression of eIF1 by ∼2.3-fold (SI Appendix, Fig. S9) repressed the eIF1 reporter and derepressed the eIF5 reporter, while the overexpression of eIF5 by ∼3.1-fold (SI Appendix, Fig. S9) had the opposite effects (Fig. 4B). With all three Hoxa reporters, increasing global stringency by the overexpression of eIF1 resulted in the significant derepression of reporter activity (Fig. 4B), while decreasing global stringency by the overexpression of eIF5 led to the significant repression of the Hoxa1 and Hoxa11 reporters and a modest reduction in Hoxa9 reporter expression (Fig. 4B). The measurement of the reporter mRNA levels under the conditions tested did not explain the observed changes in luciferase expression (SI Appendix, Fig. S10A). Notably, when directly comparing high stringency (eIF1 overexpression) to low stringency (eIF5 overexpression), all three Hox gene reporters showed a statistically significant change in expression (Fig. 4B), demonstrating that Hox protein production is subject to stringency control.
Inhibition of Translation Leads to Decreased Stringency of Start Codon Selection.
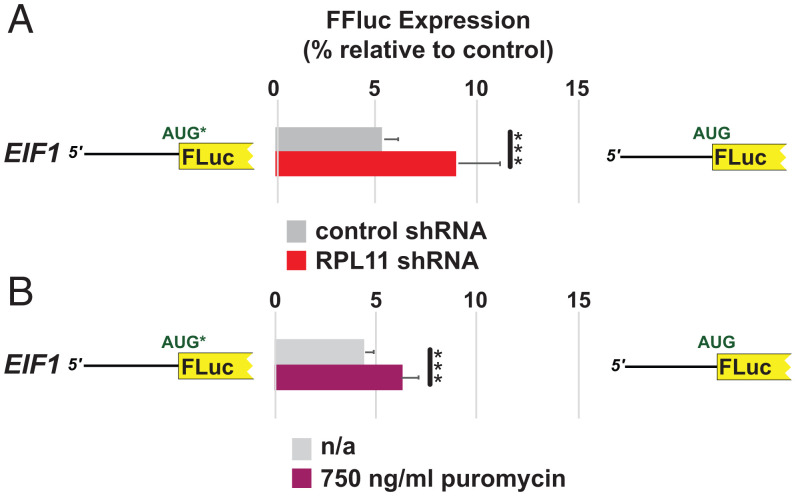
The haploinsufficiency of ribosomal protein Rpl38 in mice leads to homeotic transformation (33). However, since the loss of individual ribosomal proteins typically reduces total ribosome levels and thus global translation (see, for example, refs. 34 and 35), we hypothesized that reduced translation might perturb the stringency of start codon selection, perhaps by impacting the relative levels of eIF1 and eIF5. General perturbation of start site selection stringency could, in turn, alter Hox mRNA translation and explain at least some of the homeotic effects associated with the Rpl38 mutation. To investigate this possibility, we first examined the effects of inhibiting global translation on the stringency of start codon selection using the highly responsive eIF1-luciferase reporter (Fig. 4B). Two approaches were used to impair translation. First, the cells were treated with 250 to 750 ng/mL puromycin, an antibiotic which, at this concentration, causes the stochastic premature termination of protein synthesis and a partial inhibition of overall translation (36). As predicted, treating cells with puromycin for 24 h resulted in a greater increase in eIF1-luciferease expression from the reporter with the native eIF1 poor context AUG start codon than that observed for the reporter with the AUG codon in optimal context (Fig. 5A). Second, the cells were cotransfected with short hairpin RNA (shRNA) targeting expression of the ribosomal protein Rpl11. The knockdown of Rpl11 expression was previously shown to reduce overall ribosome levels and impair cellular protein synthesis (37, 38); we observed an ∼64% reduction in Rpl11 protein levels in cells treated with shRNA targeting Rpl11 (SI Appendix, Fig. S11). A significant increase in eIF1-luciferase expression was observed 72 h after cotransfection from the reporter with poor eIF1 AUG start codon context relative to the reporter with the AUG start codon in an optimal context (Fig. 5B). These data establish that impairing global translation could relax start codon selection stringency.
Fig. 5.
Inhibition of global translation reduces stringency of start codon selection. A comparison of luciferase activities of U2OS cells transfected with luciferase reporters initiated by AUG codons in poor EIF1 context (residues −6 to +4), shown schematically on Left, or optimal context, shown schematically on Right. (A) Cells cotransfected with an empty vector or a plasmid expressing an shRNA targeting RPL11. (B) Untreated cells or cells treated with 750 ng/mL puromycin. Firefly activity was normalized to Renilla activity from a cotransfected plasmid initiating from an AUG codon in optimal context. Error bars denote SD. ***P < 0.001 (Student’s two-tailed t test; n = 8, assayed in duplicate). No statistically significant changes in reporter mRNA levels were detected in these experiments (SI Appendix, Fig. S10B).
We next wondered how impairing global translation could impact stringency and whether this could be directly related to the activity of factors eIF1 and eIF5, the best-characterized regulators of stringency (4, 6). Previous studies demonstrated that simultaneous overexpression of eIF1 and eIF5 cancels out the heightened and relaxed, respectively, effects on start codon selection stringency associated with overexpressing either factor alone (4). Thus, it is the perturbation of the eIF5:eIF1 ratio in cells that is critical for altering start codon selection stringency. Interestingly, an investigation of protein half-lives in primary human and mouse cells reported that eIF1 has the shortest half-life of all the translation factors and that the half-life of eIF1 is roughly one-third to one-half that of eIF5 (39); analogous results were also found in a study of protein half-lives in mouse brain (40). Given its shorter half-life, eIF1 levels are expected to drop more rapidly than eIF5 levels upon inhibition of global translation. We propose that a more rapid turnover of eIF1 relative to eIF5 upon inhibition of general translation mimics eIF5 overexpression and results in a relaxation of start codon selection stringency. Consistent with this hypothesis, the treatment of cells with 250 ng/mL puromycin or with an shRNA targeting Rpl11 modestly increased the eIF5:eIF1 ratio in U2OS cells (SI Appendix, Fig. S12).
Discussion
Over the past decade, the study of genes with evolutionarily conserved mRNA features, like the uORFs in the eIF5 and Azin1 mRNAs and the weak mAUG start codon of eIF1, have established that the stringency of start codon selection can be altered locally in cis (32) or globally in trans (4, 6). More recent studies have revealed additional examples in which translation is regulated by local or global changes in start codon selection stringency. For example, as found for eIF1, the weak start codon of BZW1/BZW2 sensitizes their translation to changes in global stringency (23), and the knockdown of eIF1 was found to regulate eIF1 and eIF5 mRNA translation as well as the translation of many uORFs (12). In addition, the impaired function of translation factor eIF5A during meiosis or because of high levels of polyamines in yeast or upon depletion of the factor in mammalian cells altered mORF translation start site selection or translation of inhibitory uORFs in a manner dependent on specific features in the regulated mRNAs (41–43). In this paper, we identified Hox genes whose translation is sensitive to changes in start codon selection stringency. The translation of the subset of Hox genes with conserved poor mORF start codon contexts will be inhibited under conditions of high stringency as has been shown for the eIF1 mRNA (6), whereas the translation of the subset of Hox mRNAs, including the Hoxa1, Hoxa9, and Hoxa11 mRNAs, with conserved inhibitory uORFs with poor start codons will be induced under conditions of high stringency (Fig. 4B).
Although many eukaryotic genes initiate translation at conserved near-cognate start codons, either as the main start site or to produce alternative protein isoforms with distinct N termini, mammalian mRNAs that initiate translation on an AUG codon in conserved suboptimal context have not been thoroughly investigated. Here, we identified 394 mammalian genes that initiate on AUG codons in conserved poor contexts as defined by the lack of the preferred A residue at position −3 and/or a G residue at position +4 relative to the A of the AUG codon. For 122 of the genes, the conserved poor context AUG start codon is predicted to represent the sole obligatory initiation site. Interestingly, 22 of these 122 genes can be grouped into eight paralogous clusters. Since two of the paralogous clusters Eif1 and Bzw had previously been shown to use the conserved poor context AUG start codon as a sensor for global stringency of start codon selection (6, 23), we proposed that the genes in the other paralogous clusters might have similarly evolved to take advantage of changes in the stringency of mORF start codon selection for translational control.
The significant enrichment of conserved suboptimal initiation contexts in 5 of 39 mammalian Hox mRNAs, with the conservation extending from human to fish for four of the genes, suggests that this group of genes, important for embryo development and body plan specification, might have evolved this feature specifically to sense the stringency of start codon selection. Potential biological significance for stringency-mediated control of Hox expression is reinforced by the presence of conserved potentially inhibitory uORFs initiated by AUG codons in less-than-optimal context in the leaders of at least seven other Hox genes, making them analogous to the leader of the EIF5 mRNA in the EIF1/EIF5 regulatory paradigm (4). As shown in Fig. 4, at least for the three tested Hox leaders Hoxa1, Hoxa9, and Hoxa11, the conserved uORF regulates translation of the main ORF in response to altered stringency of start codon selection caused by the overexpression of eIF1 or eIF5. The sensitivity of Hox mRNA translation to the levels of eIF1 and eIF5 provides support for the idea that the conserved suboptimal mORF and uORF start codons on at least 12 of 39 Hox genes renders Hox expression sensitive to changes in the global stringency of start codon selection. Why the Hox gene family evolved to sense start codon selection stringency and how this feature may be exploited to regulate Hox protein expression during mammalian development will be grounds for further study.
The observation that some Hox genes have poor context mAUG start codons while others have poor context uAUG start codons for inhibitory uORFs indicates that different Hox genes will respond in distinct, and even opposite, manners to changes in start codon selection stringency. Based on the combinatorial model for Hox protein function, the gene dosages or levels of gene expression of different Hox genes plays a critical role in determining the patterning of a region (13). Thus, the proposed distinct translational expression patterns for different Hox genes may play a critical role in directing developmental programs. Interestingly, in addition to the Hox genes with conserved suboptimal mAUG start codons, such as Hoxa5, Hoxa6, Hoxb5, Hoxb6, and Hoxd8, other Hox genes have poor mAUG context in some but not all organisms. These suboptimal AUGs might provide species-specific regulation of Hox expression to control unique developmental trajectories. Likewise, several Hox genes, other than those shown in Fig. 2B, have nonconserved uORFs initiated by uAUGs in suboptimal context. Differential translational control of these latter Hox genes could provide an additional layer of species-specific regulation of development.
It has been proposed that several Hoxa mRNAs expressed in mouse somites contain long 5′ leaders containing multiple translational control features including translation inhibitory elements (TIE) and IRES elements (14). It has further been proposed that the IRES elements partially or fully mediate the changes in the translation of these mRNAs in Rpl38 haploinsufficient mice (14). Our CAGE-seq experiments in mouse somites revealed that many of these specific Hox mRNAs have short 5′ leaders and do not contain the regions corresponding to the proposed IRES elements (Fig. 3 and SI Appendix, Fig. S2 and Table S1). This finding is supported by qRT-PCR analyses of somite mRNAs showing much greater abundance for the Hoxa9 CDS relative to the proposed IRES elements in vivo, while a synthetic Hoxa9 mRNA designed to contain the proposed IRES elements showed that the IRES-directed primers amplify equally well to the CDS-directed primers when the IRES and CDS regions exist in 1:1 stoichiometry (SI Appendix, Fig. S7). Indeed, according to our CAGE-seq data [as well as the results of CAGE-seq studies reported on the UCSC genome browser (25, 26)], 5′ leaders long enough to encode the proposed IRES elements are either not expressed or are expressed at extremely minimal levels in E11.5 mouse somites (Fig. 3 and SI Appendix, Fig. S2).
Of the proposed Hoxa IRES elements, the Hoxa9 element has been studied in the greatest detail (14, 44, 45). The 1,266 nts immediately 5′ of the Hoxa9 start codon are proposed to contain two translational control elements: First, an upstream TIE that prevents ribosomes scanning from the cap from translating the mORF (14). Recently, the TIE was proposed to contain an inhibitory uORF (45). Second, downstream of the TIE, an IRES element is proposed to specifically recruit ribosomes to the mRNA (14, 44) (SI Appendix, Fig. S13A). However, our CAGE-seq results on E11.5 mouse somites (Fig. 3 and SI Appendix, Figs. S6 and S8) as well as the CAGE-seq and Fantom5 data from multiple mouse tissues reported on the UCSC mouse genome browser (25, 26) mapped two prominent transcription start sites corresponding to much shorter leaders of 83 nts and 85 nts on the Hoxa9 mRNA. The original description of the long leader of the mouse Hoxa9 mRNA was based on a “complementary DNA (cDNA) walking” procedure and not from sequencing an intact single cDNA (46). Northern analyses reported by Fujimoto et al. (46) detected a longer ∼2.4-kilobase (kb) transcript that retained the first intron and a shorter ∼1.9-kb transcript that is consistent with the short leader identified in our studies linked to the CDS and 1,472-nt 3′ UTR. Notably, the annotated Hoxa9 mRNA in the Mammalian Gene Collection (47) has this latter structure with the short leader and long 3′ UTR. Based on these data as well as our results showing that the abundance of the Hoxa9 CDS is at least 200-fold higher than that of the putative IRES (SI Appendix, Fig. S7), we hypothesized that much of the putative long Hoxa9 leader sequence examined in previous studies is part of the Hoxa9 promoter. To test this idea, the previously explored 1,266-nt sequence encompassing the putative Hoxa9 leader sequence (with IRES) was inserted upstream of a promoter-less firefly luciferase reporter and transfected into U2OS, human embryonic kidney 293T (HEK293T), and Chinese hamster ovary (CHO) cells. As shown in SI Appendix, Fig. S13B, compared to a control construct lacking the insert, the proposed Hoxa9 leader sequence resulted in a 21- to 30-fold increase in firefly luciferase expression. In light of this result, we propose that the previously characterized IRES of the Hoxa9 locus is in fact part of a promoter sequence, which could readily explain the observed luciferase activity of this sequence when inserted in the spacer of a dual-luciferase DNA vector as well as the expression phenotypes observed within cell culture and mice (14, 44).
Both physiological (meiosis) and environmental (polyamines) factors are known to alter local (cis) stringency of start codon selection depending on features within the regulated mRNAs (32, 42, 43). Finding physiological and environmental factors that alter global (trans) stringency of start codon selection has been more elusive. Considering the central role of eIF1 and eIF5 in controlling global stringency for start site selection, it seems plausible that altering their ratio in cells might provide an entry point for such regulation. Currently, there are no known conditions that alter the transcriptional expression of eIF1 and eIF5; however, studies of global protein turnover have shown that eIF1 and eIF5 have different half-lives (39, 40), with eIF1 displaying a consistently shorter half-life than eIF5. Given this fact, the inhibition of global protein synthesis might be expected to deplete eIF1 faster than eIF5, resulting in decreased stringency of start codon selection. Notably, such a mechanism exploiting differences in protein half-lives resembles the activation of NF-κB upon global inhibition of translation. The NF-κB inhibitor IκB has a much shorter half-life than NF-κB, and upon inhibition of translation, for example by the Integrated Stress Response (ISR), the levels of IκB fall faster than the levels of NF-κB, resulting in NF-κB release and activation (48, 49). Consistent with such a model, a Western blot analysis of cells following the inhibition of translation revealed a potential small increase in the eIF5:eIF1 ratio (SI Appendix, Fig. S12). Whether this modest change in the relative levels of eIF1 and eIF5 accounts for the relaxed stringency of start codon selection upon inhibition of translation remains an open question.
Like eIF1 and eIF5, the proteins BZW1 and BZW2 control start codon selection stringency by counteracting the effect of eIF5 on global stringency of start codon selection (23, 50). In addition, their human homologs have conserved poor initiation contexts that are used for autoregulation (23). Mutations in the Drosophila BZW homolog called Krasavietz (Kra) result in defects in neuronal development and long-term memory (51), reportedly because of mis-regulation of midline axon repulsion (52), and the knockdown of BZW/5MP expression in the red flour beetle Tribolium castaneum impaired larval development (53). These results are consistent with the idea that altering the stringency of start codon selection can impact developmental pathways, and they raise the possibility that start codon selection stringency could be a natural target for regulating gene expression in a tissue-specific manner during development.
Materials and Methods
Identification of Genes with Conserved Poor Initiation Context.
Starting with a reference database of human protein–coding transcripts downloaded from https://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Homo_sapiens/all_assembly_versions/GCF_000001405.37_GRCh38.p11/ on January 12, 2018, and a subject database of vertebrate transcripts downloaded from https://ftp.ncbi.nlm.nih.gov/refseq/release/vertebrate_mammalian/ and https://ftp.ncbi.nlm.nih.gov/refseq/release/vertebrate_other/ on January 12, 2018, only transcripts with annotated coding regions of at least 100 codons in length were selected. Furthermore, for each species and for each gene name, only the transcript with the longest coding sequence was selected. For each human reference sequence, the most closely related sequence in each other organism was found using a reciprocal best hit (RBH) approach, based on pairwise comparisons, as follows. For a given human gene A, the best hit gene Bs in each other species s was found. Then, for each Bs, the best hit gene in human was found. If the best hit in human is gene A, then Bs is an RBH of A. In many cases, Bs will be an ortholog of A (54, 55).
The best hits were defined using tblastn (56) with default parameters. The annotated coding sequence in each human reference database gene was translated into amino acids and used as a query in tblastn, with the annotated coding sequences derived from the database of vertebrate transcripts as the subject. Hits with less than 95% coverage or less than 65% amino acid identity to the query sequence were discarded, and where multiple sequences in one taxon (e.g., paralogs or alternative splice forms) remained, only the hit with highest identity to the query was retained. These best hits in each taxon were then translated to amino acid sequences and used as queries in tblastn, with the annotated coding sequences derived from the database of reference human transcripts as the subject, and the same selection criteria were applied to the hits. RBH were retained as putative orthologs of the initial query human reference sequence. Then, all sequences in which the coding sequence was annotated as being incomplete at the 5′ end or where there were fewer than 3 nts of annotated 5′ UTR were discarded, as both situations preclude identification of the initiation context.
Beginning with 16,636 human reference genes, the genes in which putative orthologs were identified according to the parameters described in the preceding paragraph in at least 30 placental mammals were selected, giving 12,467 genes (including 31 Hox genes). For each gene in each taxon, the context of the annotated initiation codon and the presence or absence of downstream in-frame and/or out-of-frame AUG codons and their contexts were identified. An initiation context was defined to be poor if there is a U or C at the −3 position and an A, C, or U at the +4 position. Moreover, an initiation codon was defined to be an obligatory poor context initiation codon if either there is no in-frame downstream AUG codon anywhere within the annotated coding sequence or if there is at least one downstream out-of-frame AUG codon that is not in a poor context (i.e., A or G at −3 and/or G at +4) between the initiation codon and the next downstream in-frame AUG codon. In total, 394 and 122 of the 12,467 genes were found to have, respectively, poor context and obligatory poor context in at least 90% of the placental mammals in which putative orthologs were identified. The latter set includes Bzw1, Bzw2, Eif1, Eif1B, Hoxa5, Hoxa6, Hoxb5, Hoxb6, and Hoxd8, and the former set additionally includes Hoxa4, Hoxa11, Hoxc4, and Hoxd3.
Identification of Hox Genes with Conserved uORFs.
The leaders of all 39 mouse Hox genes were analyzed for the presence of uORFs. Transcription start sites were based on CAGE-seq data as described in the main text (and shown in SI Appendix, Table S1). After obtaining the coordinates of the corresponding human sequences, the CodAlignView online tool (57) was used to generate alignments from different organisms (as shown in SI Appendix, Figs. S3–S5). An AUG-initiated uORF was deemed conserved if it was present in at least 90% of the placental mammals with available high-quality sequence from the locus under examination. The translation of each scored uORF was confirmed by examining aggregate ribosome profiling data as collated on the Genome Wide Information on Protein Synthesis visualized (GWIPS-viz) (https://gwips.ucc.ie/) browser (31).
Cell Culture and Transfections.
U2OS cells were obtained from Nancy Kedersha (Harvard University, Cambridge, MA). HEK-293T cells were obtained from American Type Culture Collection. Both U2OS and HEK-293T cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Corning) supplemented with 1 mM L-glutamine, 10% fetal bovine serum (FBS) (Gibco), and penicillin/streptomycin (Quality Biological). CHO cells were obtained from David Ron [Cambridge University, Cambridge, United Kingdom (58)] and were grown in DMEM/F-12 media (Thermo Fisher) supplemented with 10% FBS and penicillin/streptomycin. All cells were maintained at 37 °C in 5% CO2.
For testing the Hoxa luciferase reporter alone (Fig. 4A), U2OS cells were grown overnight in 10-cm plates to ∼70% confluence, washed, treated with trypsin, and then transfected using Lipofectamine 2000 reagent (Invitrogen) in a 1-d protocol in which suspension cells were added directly to the DNA mixtures in white 96-well half-area plates (Costar)—0.2 µl Lipofectamine and 2.5 ng reporter plasmid were mixed in 25 µl Opti-MEM (Gibco) and then dispensed to each well along with 104 cells suspended in 25 µl DMEM. The transfection was terminated by removing the media and lysing the cells using 25 µl 1× Passive Lysis Buffer (Promega). For the eIF1 and eIF5 overexpression experiments (Fig. 4B), a similar protocol was followed with the following modifications: 2.5 ng Hoxa dual-luciferase reporter plasmid was mixed with 25 ng eIF1oe or eIF5oe expression vector, corresponding to plasmids “eIF1 good*” and “eIF5 AAA” (4), respectively, or with an empty vector [phRL lacking the Renilla CDS (4)]. In parallel, 5 ng EIF1 or EIF5 firefly reporter was first mixed with 5 ng normalizing Renilla reporter (4), and then the eIF1 or eIF5 expression vector or empty vector was added to the mixture. The cells were transfected and incubated for 22 h before being lysed and assayed for luciferase activity. For the Rpl11 depletion experiment (Fig. 5A), U2OS cells were transfected as described for the other transfection experiments with the following modifications: 5 ng EIF1 or control (Kozak context) firefly reporter was mixed with 5 ng normalizing Renilla reporter and either 25 ng scrambled control or 25 ng Rpl11 shRNA plasmid (38). The cells were transfected and incubated for 48 h before being lysed and assayed for luciferase activity. For the puromycin treatment experiment (Fig. 5B), U2OS cell were transfected as described for the other transfection experiments with the following modifications: 10 ng EIF1 or control (Kozak context) firefly reporter was mixed with 5 ng normalizing Renilla reporter. At the time of transfection, puromycin was added to a final concentration of 750 ng/mL to half of the wells. The cells were incubated for 22 h before being lysed and assayed for luciferase activity. For the Hoxa9 promoter experiment (SI Appendix, Fig. S13B), 200 ng promoterless phRL Renilla luciferase vector or the same vector containing the 1,266-nt putative Hoxa9 IRES-containing mRNA leader upstream of Renilla luciferase (14) was transfected in U2OS, CHO-K1, or HEK293T cells using a protocol similar to those described for the other transfection experiments with the following modifications: 104 U2OS cells, 2 × 104 CHO-K1 cells, and 2 × 104 HEK293T cells were used per well. In total, 1 ng control firefly luciferase (6) was mixed with 200 ng test Renilla reporter per well.
Plasmid Construction.
The Hoxa1, Hoxa9, and Hoxa11 Renilla luciferase reporters were constructed using the vector phRL-CMV (Promega). Synthetic DNA fragments from commercial vendors were designed to start at the SpeI restriction site of the vector, extend 577 base pairs (bp) downstream to include the entire CMV promoter, and then include the entire leader of Hoxa1, Hoxa9, or Hoxa11 followed by the ATG start codon and first seven codons of Renilla luciferase and end with an AvaI restriction site. The Hoxa leader sequences start at their natural transcription start sites as depicted in SI Appendix, Fig. S8 and as determined by CAGE-seq analysis of mouse somites. The DNA fragments were cloned between the SpeI and AvaI sites of phRL. To insert the firefly luciferase (Fluc) control reporter into the Hoxa–Renilla reporter plasmids, a 2.8-kb DNA fragment containing the CMV promoter and Fluc sequence was amplified by PCR using the plasmid PSF-CMV-FLUC (Sigma) as a template and primers designed to introduce SpeI sites at both ends of the PCR product. Following digestion with SpeI, the DNA fragment was inserted into the SpeI site of the Hoxa-Renilla reporter plasmids. Plasmids in which the CMV-Fluc insert was in the opposite orientation relative to Renilla (divergent promoters) were selected for the assays.
The reporters for testing promoter activity of the putative IRES of Hoxa9 were constructed using vector phRL. A commercially synthesized DNA fragment with a 5′ SpeI restriction site and a 3′ AvaI restriction site and containing the 1,266-nt leader of NM_010456 followed by the first seven codons of phRL Renilla was inserted between the same sites of phRL in place of the natural CMV promoter. For the control plasmid, the sequence actagtACTGCAATGGCTTCCAAGGTGTACGACcccgag (restriction sites in lowercase, and Renilla ATG start codon underlined) was inserted between the SpeI and AvaI sites of phRL.
All plasmids were confirmed by DNA sequencing.
Luciferase Assays.
Luciferase activities were determined as described previously (32). For the Hoxa luciferase reporters, the Renilla luciferase activity was normalized relative to firefly luciferase expressed from the same plasmid. The firefly luciferase activity of the EIF1 and EIF5 reporters was normalized relative to the activity of a cotransfected pSV40-Renilla plasmid expressing Renilla luciferase (4). For the Hoxa9 promoter experiments, the Renilla luciferase activity was normalized to firefly luciferase activity expressed from a separate p2luc-based reporter (6).
Western Analysis.
For the analysis of eIF1 and eIF5 levels in the puromycin experiment, 250,000 U2OS cells were seeded in 4 mL DMEM + 10% FBS in triplicate in a 6-well tissue-culture plate. After allowing 1 d for cells to adhere, the cells were treated with puromycin to a final concentration of 250 ng/mL or with an equivalent volume of water (vehicle) as a control. After 22 to 24 h of treatment, the cells were lysed in 100 µl radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher 89901) supplemented with 2.5 mM MgCl2, 1× Halt protease and phosphatase inhibitor mixture (Thermo Fisher 78443), and 50 U/mL Benzonase (MilliporeSigma E1014) by scraping and pipetting up and down. Cell lysates were spun at 10,000 × g for 5 min and the clarified lysate transferred to a new tube.
For the analysis of eIF1 and eIF5 levels in shRpl11-treated samples and for the analysis of shRpl11 knockdown efficiency, the same culturing and lysis protocol was followed except that U2OS cells were transfected with either the shRpl11 or shScramble plasmid using Lipofectamine 2000 reagent according to the manufacturer’s instructions and then lysed after 48 h using RIPA lysis buffer as described in the opening paragraph of this section.
For the analysis of eIF1 and eIF5 overexpression, the same culturing and lysis protocol was followed except that U2OS cells were transfected with either the eIF1oe, eIF5oe, or empty vector plasmids using Lipofectamine 2000 reagent according to the manufacturer’s instructions and then lysed after 24 h using RIPA lysis buffer.
The RNA content of cell lysates was measured using the Nanodrop UV-Vis spectrophotometer A260 channel with background lysis buffer subtraction (Thermo Scientific), normalized to equal RNA content in 1× sodium dodecyl sulfate (SDS) loading dye, and boiled for 5 min at 95 °C. For Western blots, normalized samples were subjected in triplicate to electrophoresis on 4 to 12% bis-Tris polyacrylamide gels (Criterion Bio-Rad) in 2-(N-morpholino)ethanesulfonic acid (MES) running buffer at 150 V for 1 h. The gels were transferred to polyvinylidene fluoride (PVDF) membranes using the Trans-Blot Turbo Transfer System (Bio-Rad), membranes cut to the appropriate size corresponding to either eIF1, eIF5, Rpl11, or Actin, and blocked in 5% milk for 30 min at room temperature (RT) followed by overnight incubation at 4 °C with rabbit anti-eIF1 (Cell Signaling 12496), rabbit anti-eIF5 (Cell Signaling 2480), rabbit anti-Rpl11 (Cell Signaling 18163), or rabbit anti-Actin (Cell Signaling 4967) antibodies at a 1:1,000 dilution in Tris-buffered saline containing 1 mL/l Tween 20 (TBST) and supplemented with 5% milk. The next day, the membranes were washed 3 × 10 min at RT in TBST, incubated with secondary anti-rabbit IgG–HRP (Santa Cruz 2357) 1:10,000 in TBST supplemented with 5% milk for 45 min at RT, and washed 3 × 10 min in TBST. Western blots were visualized by horseradish peroxidase (HRP) chemiluminescence using Super Signal West HRP substrate (Thermo Fisher). The films were developed in a dark room at multiple exposures to ensure quantitation of protein levels within the dynamic range. The bands were quantitated using Fiji (v2.1.0/1.53c) using equal field sizes and background subtraction. For the analysis of eIF1 and eIF5 levels, eIF1:eIF5 ratios were calculated within each sample. For the analysis of shRpl11 knockdown efficiency and eIF1 and eIF5 overexpression efficiency, each signal was normalized to the Actin signal from the same lane of the gel. Each sample was analyzed in single, duplicate, or triplicate on each gel (technical replicates), and each data point therefore reflects the average from one to three measurements. In addition, two or three biological replicates each containing one to three technical replicates were performed. The anti-eIF1 and anti-eIF5 antibodies were tested and shown to be specific to eIF1 and eIF5 by small interfering RNA (siRNA) knockdown of eIF1 or eIF5, respectively.
Mouse Dissection and Preparation of Somites.
Vertebrate animal protocol was approved by the Carnegie Institutional Animal Care and Use Committee (IACUC). C56BL/6J mice were used for timed mating; the date when a vaginal pug is present is assigned as embryonic day (E) 0.5 per convention. At E11.5, embryos were used for somite/neural tube isolation as in ref. 14. The dissection procedure mainly followed that described in ref. 33. Briefly, embryos were placed in dissection media (DMEM/F-12 1:1, 10% FBS, and 1% penn-strep; GIBCO) and chilled on ice in a 60-mm Petri dish (Falcon) until dissection. Each embryo was sequentially transferred to a small dissection dish with Sylgard bottom (Living Systems Instrumentation) in the same media, pinned facing down by small dissection pins (Living Systems Instrumentation), and the neural tube and somites were dissected out using two angled scissors (2.5- and 4-mm cutting edge) from the first somite to the tail end. Prior to transferring the dissected somite/neural tube to a 1.5-mL RNase-free microfuge tube (Thermo Fisher Scientific), any remaining ventral endoderm-derived tissue was removed. To the dissected somite/neural tube (∼100 µl, after removing as much media as possible), 400 µl TRIzol reagent (Invitrogen) was added. The tube was vortexed until the tissue was dissolved and then immediately frozen on dry ice and stored at −80 °C until RNA isolation. Total RNA was isolated using the Direct-zol RNA Miniprep Kit (Zymogen) followed by quantification and analyses using Nanodrop One (Thermo Fisher Scientific) and Bioanalyzer (Agilent), respectively, and stored at −80 °C until use.
Preparation of CAGE Libraries.
nAnT-iCAGE libraries were prepared using the cap-trap method as described in a detailed protocol (28). In brief, 5 µg total mouse somite RNA was reverse transcribed (Superscript III, Thermo Fisher) using random hexamer primers. Following cleanup with Agencourt RNAClean XP (Beckman Coulter), the RNA ends of RNA:cDNA hybrids were oxidized with NaIO4 (11.3 mM) on ice in the dark for 45 min, cleaned with Agencourt RNAClean XP, and biotinylated with biotin hydrazide (0.83 mM) at RT overnight. Single-stranded RNA was cleaved using Ribonuclease I (RNAseI) ribonuclease, and biotinylated RNA:cDNA hybrids were purified by incubation with Dynabeads M-270 streptavidin for 30 min followed by washing and elution on a magnetic stand as previously described (28). Hybrid RNA was degraded with Ribonuclease H (RNAseH), and any remaining free RNA was degraded with RNAseI as previously described (28). cDNA was concentrated in a centrifugal concentrator to ∼5 to 10 µl and ligated to the preannealed double-stranded 5′linker containing a 6-nt barcode using ligation Mighty Mix at 16 °C for 16 h. Following cDNA purification with Agencourt AMPure XP, cDNA was ligated to the preannealed 3′ linker using ligation Mighty Mix at 16 °C for 16 h. cDNA was treated with shrimp alkaline phosphatase and Uracil-Specific Excision Reagent (USER) followed by cleanup, second-strand synthesis using DeepVent DNA polymerase, primer degradation with Exonuclease I, and a final AMPure XP cleanup. Total cDNA was analyzed for quality and quantified using a Bioanalyzer (Agilent). DNA was pooled to a final concentration of 2 nM and sequenced on two lanes of a HiSeq 2500 FlowCell (Illumina) with 50-bp read length.
Analysis of CAGE Libraries.
CAGE libraries were analyzed using the spliced transcripts alignment to a reference (STAR) package v2.7.3a (https://github.com/alexdobin/STAR). Total reads from the two flow cells were concatenated into a single file, and barcoded reads representing reads from the somites of each of three mice were retrieved into individual files. In total, there were 15,707,020 reads from mouse 1, 16,446,419 reads from mouse 2, and 18,053,549 reads from mouse 3. The most up-to-date genome annotation for mouse at the time of analysis was downloaded as a Gene Transfer Format (GTF) file from the Gencode database (vM24), and the most up-to-date genome for mouse at the time analysis was downloaded as a FAST-All (FASTA) file from the Gencode database (GRCm38). The STAR index genome was compiled as described in the STAR manual. For each mouse dataset, the 5′ end of all reads were mapped to the index genome as described in the STAR manual and output as wiggle (WIG) and binary alignment map (BAM) file formats. Mapped reads were visualized from WIG files using the Integrative Genome Browser software (v2.8.0), and plots were constructed from BAM files using Python 2. Data were deposited in the Gene Expression Omnibus (GEO) database under accession number GSE184515.
In Vitro Transcription of Hoxa9 mRNA.
A synthetic 2,232–base pair gene block corresponding to the Mus musculus Hoxa9 mRNA annotation NM_010456.3 was ordered from Integrated DNA Technlogies (IDT). In total, the long synthetic DNA molecule contained the entire annotated 5′ UTR (1,266 nts), the entire spliced coding sequence (816 nts), and the CDS-proximal 150 nts of the 3′ UTR with slight sequence modifications at three sites (64 base pairs mutated in total) in order to decrease guanine and cytosine (GC) content and allow it to be feasibly synthesized. Importantly, no nucleotide modifications were introduced at sites corresponding to qPCR primers or amplicons. In total, the sequence of the entire construct is shown in Dataset S4, with modified regions depicted in capital letters.
The synthetic construct was PCR amplified and inserted into the pSP64 in vitro transcription vector (Promega P1241) at the SmaI restriction site using Gibson assembly according to the manufacturer’s instructions (New England Biolabs, E2611). The colonies were sequenced to confirm accurate cloning of the construct into the pSP64 vector. Synthetic Hoxa9 mRNA was in vitro transcribed from the pSP64-Hoxa9 construct followed by RQ1 deoxyribonuclease (DNase) treatment and phenol:chloroform extraction and ethanol precipitation according to the manufacturer’s instructions (Promega P1280).
qPCR.
All qPCR experiments on RNA isolated from cultured cells followed identical cell culture and transfection protocols as described in the section "Cell Culture and Transfections". U2OS cells were grown in 10-cm plates overnight to 70% confluence and trypsinized, and 2 × 105 cells were seeded in each well of a 12-well plate (Corning). Transfection reagents and volumes were scaled up 20× relative to the transfections using 96-well half-area plates. Incubation times were identical to the incubation times in 96-well plates. At the end of the incubation, the media was removed, and the cells were washed 1× with 500 µl phosphate buffered saline (PBS) and then lysed directly from 12-well plates by adding 100 μL TRIzol reagent followed by repeated pipetting and scraping. Lysed cells in TRIzol reagent were extracted in phenol-chloroform according to the manufacturer’s instructions and resuspended in 100 μL 1× TURBO DNase buffer containing 2 U TURBO DNase (AM2238). Following DNase digestion for 45 min at 37 °C, samples were treated with 200 µg proteinase K (AM2546) for 15 min at 37 °C to degrade DNase, then extracted using phenol-chloroform (AM9720) according to the manufacturer’s instructions, and ethanol-precipitated with 2 volumes 100% EtOH at −80 °C overnight followed by two washes in 70% EtOH. RNA pellets were resuspended in 25 μL nuclease-free water. Reverse transcription was performed using the Protoscript II First Strand cDNA synthesis kit (E6560) using a random hexamer primer with the following cycling times: 2’ at 95 °C to denature followed by addition of reagents (in master mix) and incubation at 25 °C for 5 min, 42 °C for 1 h, 80 °C for 5 min, and then hold at 4 °C. qPCR was performed using SYBR green master mix (Bio-Rad 1725122) using the following cycle times: 95 °C for 2 min, [95 °C for 10 s, 60 °C 30 s] × 40 cycles, and then 60 °C for 3 min on a QuantStudio 6 RT-PCR system (Thermo). Raw cycle threshold (Ct) values for FLuc and RLuc were internally normalized to actin control. All measurements were performed in triplicate and averaged for two or three biological replicates per sample. The following qPCR primers were used:
hActin B (F): ACGTTGCTATCCAGGCTGTG
hActin B (R): GAGGGCATACCCCTCGTAGA
Firefly luciferase single plasmid (F): GTGTTGGGCGCGTTATTTATC
Firefly luciferase single plasmid (R): TAGGCTGCGAAATGTTCATACT
Firefly luciferase dual plasmid (F): GCCCTTCTTCGAGGCTAAGG
Firefly luciferase dual plasmid (R): CCCAGTGTCTTACCGGTGTC
Renilla luciferase (F): TCCAGATTGTCCGCAACTAC
Renilla luciferase (R): CTTCTTAGCTCCCTCGACAATAG.
All qPCR experiments on RNA isolated from mouse somites (related to SI Appendix, Fig. S7) followed identical somite preparation and RNA extraction protocols as described in the section "Mouse Dissection and Preparation of Somites." The RT and qPCR followed the identical protocol as described above except that all reactions were additionally performed with a nonreverse transcribed (“no-RT”) sample in triplicate in order to control for DNA elements or nonspecific background reactions that might lead to a background qPCR amplification signal for each primer set. Then, the raw Ct values for the no-RT IRES- and CDS-directed primer sets were directly subtracted from the Ct values for their respective reverse-transcribed counterparts. As such, primer sets that are directed toward expressed RNA elements will have low Ct values in the reverse-transcribed sample relative to high Ct values due to background amplification in the no-RT control. All primer sequences and amplicon information for both IRES and CDS amplicons are listed in SI Appendix, Table S3. The same qPCR procedure was followed for the synthetic Hoxa9 transcript.
Supplementary Material
Acknowledgments
We thank members of our laboratories as well as Alan Hinnebusch, Jon Lorsch, Nick Guydosh, and members of their laboratories for helpful discussions. We thank Shrey Bhatt for help preparing experiments and reagents. This research was supported, in part, by the Intramural Research Program of the NIH, (T.E.D.). J.A.S. is supported by a National Research Service Award (NRSA) F30 Fellowship from the National Cancer Institute (F30CA260910). R.G. is supported by the HHMI. C.-M.F. is supported by Grants AR060042, AR071976, and AR072644 from National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of NIH, and A.E.F is supported by a Wellcome Trust Grant (220814).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117226119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. CAGE data were deposited in the Gene Expression Omnibus (GEO) under accession number GSE184515.
Note Added in Proof.
A recently posted study has likewise reported promoter activity for the proposed Hoxa9 and other Hox gene IRESes (59).
References
- 1.Hinnebusch A. G., Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak M., Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 (1986). [DOI] [PubMed] [Google Scholar]
- 3.Peabody D. S., Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264, 5031–5035 (1989). [PubMed] [Google Scholar]
- 4.Loughran G., Sachs M. S., Atkins J. F., Ivanov I. P., Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 40, 2898–2906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M., Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 9, 5073–5080 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov I. P., Loughran G., Sachs M. S., Atkins J. F., Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U.S.A. 107, 18056–18060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llácer J. L., et al. , Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife 7, e39273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Marcos P., Cheung Y. N., Hinnebusch A. G., Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 31, 4814–4831 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanda J. S., et al. , eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 394, 268–285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch A. G., Ivanov I. P., Sonenberg N., Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyasaka H., Endo S., Shimizu H., Eukaryotic translation initiation factor 1 (eIF1), the inspector of good AUG context for translation initiation, has an extremely bad AUG context. J. Biosci. Bioeng. 109, 635–637 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Fijalkowska D., et al. , eIF1 modulates the recognition of suboptimal translation initiation sites and steers gene expression via uORFs. Nucleic Acids Res. 45, 7997–8013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander T., Nolte C., Krumlauf R., Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 25, 431–456 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Xue S., et al. , RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kearse M. G., Wilusz J. E., Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 31, 1717–1731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M., Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov I. P., Firth A. E., Michel A. M., Atkins J. F., Baranov P. V., Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 39, 4220–4234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia N. T., Lareau L. F., Weissman J. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritsch C., et al. , Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 22, 2208–2218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Na C. H., et al. , Discovery of noncanonical translation initiation sites through mass spectrometric analysis of protein N termini. Genome Res. 28, 25–36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzani I., et al. , Systematic analysis of the PTEN 5′ leader identifies a major AUU initiated proteoform. Open Biol. 6, 150203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Damme P., Gawron D., Van Criekinge W., Menschaert G., N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men. Mol. Cell. Proteomics 13, 1245–1261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loughran G., Firth A. E., Atkins J. F., Ivanov I. P., Translational autoregulation of BZW1 and BZW2 expression by modulating the stringency of start codon selection. PLoS One 13, e0192648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pertea M., et al. , CHESS: A new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 19, 208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi S., et al. , FANTOM5 CAGE profiles of human and mouse samples. Sci. Data 4, 170112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent W. J., et al. , The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallo M., Wellik D. M., Deschamps J., Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344, 7–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murata M., et al. , Detecting expressed genes using CAGE. Methods Mol. Biol. 1164, 67–85 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Jackson R. J., Hellen C. U., Pestova T. V., Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 86, 45–93 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Luukkonen B. G., Tan W., Schwartz S., Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 69, 4086–4094 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel A. M., Kiniry S. J., O’Connor P. B. F., Mullan J. P., Baranov P. V., GWIPS-viz: 2018 update. Nucleic Acids Res. 46, D823–D830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov I. P., et al. , Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol. Cell 70, 254–264.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondrashov N., et al. , Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khajuria R. K., et al. , Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173, 90–103.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robledo S., et al. , The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14, 1918–1929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nathans D., Puromycin inhibition of protein synthesis: Incorporation of puromycin into peptide chains. Proc. Natl. Acad. Sci. U.S.A. 51, 585–592 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng T., Mercer C. A., Hexley P., Thomas G., Fumagalli S., Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. Mol. Cell. Biol. 33, 4660–4671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei J., et al. , Ribosomal proteins regulate MHC class I peptide generation for immunosurveillance. Mol. Cell 73, 1162–1173.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathieson T., et al. , Systematic analysis of protein turnover in primary cells. Nat. Commun. 9, 689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fornasiero E. F., et al. , Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 9, 4230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manjunath H., et al. , Suppression of ribosomal pausing by eIF5A is necessary to maintain the fidelity of start codon selection. Cell Rep. 29, 3134–3146.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenberg A. R., et al. , Translation initiation site profiling reveals widespread synthesis of non-AUG-initiated protein isoforms in yeast. Cell Syst. 11, 145–160.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vindu A., et al. , Translational autoregulation of the S. cerevisiae high-affinity polyamine transporter Hol1. Mol. Cell 81, 3904–3918.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leppek K., et al. , Gene- and species-specific Hox mRNA translation by ribosome expansion segments. Mol. Cell 80, 980–995.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alghoul F., Laure S., Eriani G., Martin F., Translation inhibitory elements from Hoxa3 and Hoxa11 mRNAs use uORFs for translation inhibition. eLife 10, e66369 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujimoto S., et al. , Analysis of the murine Hoxa-9 cDNA: An alternatively spliced transcript encodes a truncated protein lacking the homeodomain. Gene 209, 77–85 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Temple G., et al. , MGC Project Team, The completion of the Mammalian Gene Collection (MGC). Genome Res. 19, 2324–2333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng J., et al. , Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell. Biol. 24, 10161–10168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu S., et al. , Ultraviolet light activates NFκB through translational inhibition of IκBα synthesis. J. Biol. Chem. 279, 34898–34902 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Tang L., et al. , Competition between translation initiation factor eIF5 and its mimic protein 5MP determines non-AUG initiation rate genome-wide. Nucleic Acids Res. 45, 11941–11953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubnau J., et al. , The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13, 286–296 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Lee S., et al. , The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development 134, 1767–1777 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Hiraishi H., et al. , Essential role of eIF5-mimic protein in animal development is linked to control of ATF4 expression. Nucleic Acids Res. 42, 10321–10330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huynen M. A., Bork P., Measuring genome evolution. Proc. Natl. Acad. Sci. U.S.A. 95, 5849–5856 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatusov R. L., Koonin E. V., Lipman D. J., A genomic perspective on protein families. Science 278, 631–637 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jungreis I., Lin M. F., Chan C. S., Kellis M., “CodAlignView: The Codon Alignment Viewer” CodAlignView (2016). https://data.broadinstitute.org/compbio1/cav.php. Accessed 2 December 2021.
- 58.Crespillo-Casado A., Chambers J. E., Fischer P. M., Marciniak S. J., Ron D., PPP1R15A-mediated dephosphorylation of eIF2α is unaffected by Sephin1 or Guanabenz. eLife 6, e26109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.C. Akirtava, G. E. May, C. J. McManus, False-Positive IRESes from Hoxa9 and other genes resulting from errors in mammalian 5’ UTR annotations. bioRxiv [Preprint] (2022). https://biorxiv.org/cgi/content/short/2022.02.10.479744v1. Accessed 10 February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. CAGE data were deposited in the Gene Expression Omnibus (GEO) under accession number GSE184515.