Abstract
The health of the kidney filtration barrier requires communication among podocytes, endothelial cells, and mesangial cells. Disruption of these cell-cell interactions is thought to contribute to disease progression in chronic kidney diseases (CKDs). Podocyte ablation via doxycycline-inducible deletion of an essential endogenous molecule, CTCF [inducible podocyte-specific CTCF deletion (iCTCFpod−/−)], is sufficient to drive progressive CKD. However, the earliest events connecting podocyte injury to disrupted intercellular communication within the kidney filter remain unclear. Single-cell RNA sequencing of kidney tissue from iCTCFpod−/− mice after 1 week of doxycycline induction was performed to generate a map of the earliest transcriptional effects of podocyte injury on cell-cell interactions at single-cell resolution. A subset of podocytes had the earliest signs of injury due to disrupted gene programs for cytoskeletal regulation and mitochondrial function. Surviving podocytes up-regulated collagen type IV ɑ5, causing reactive changes in integrin expression in endothelial populations and mesangial cells. Intercellular interaction analysis revealed several receptor-ligand-target gene programs as drivers of endothelial cell injury and abnormal matrix deposition. This analysis reveals the earliest disruptive changes within the kidney filter, pointing to new, actionable targets within a therapeutic window that may allow us to maximize the success of much needed therapeutic interventions for CKDs.
Chronic kidney diseases (CKDs) affect 700 million people globally, yet specific therapies are lacking.1 Many kidney diseases originate in the glomerulus, the filtration unit of the kidney. The glomerulus consists of (i) podocytes, specialized postmitotic cells with elaborate foot processes that interdigitate forming slit diaphragms and wrapping around glomerular capillaries; (ii) endothelial cells, that lie opposite podocytes on a shared glomerular basement membrane (GBM); (iii) mesangial cells, that form a matrix that provides structural support for the glomerulus; and (iv) parietal epithelial cells (PECs), that line the Bowman capsule.2 Podocyte injury, in particular, leads to many highly prevalent, progressive kidney diseases, including diabetic kidney disease (DKD), focal segmental glomerulosclerosis (FSGS), and nephrotic syndrome (both idiopathic and genetic). The canonical pattern of injury results in the loss of interdigitating podocyte foot processes, known as foot process effacement, caused by a rearrangement of the actin cytoskeleton. This effacement leads to a disruption of the slit diaphragm, the physical barrier that functions as a filter, followed by podocyte detachment or death.3,4 On the other hand, in addition to intact podocytes, the formation and maintenance of the glomerular filtration barrier require intraglomerular communication, tightly controlled by a series of autocrine and paracrine signaling mechanisms. For example, vascular endothelial growth factor A (VEGFA) is a prosurvival signal for endothelial cells secreted by podocytes, and platelet-derived growth factor B (PDGFB) is a prosurvival signal for mesangial cells secreted by endothelial cells.5 Disruption of these cell-cell interactions are frequently observed in a host of glomerular diseases, including FSGS and DKD.6 Therefore, identifying the earliest disruptive changes to the glomerulus may offer novel targets and the opportunity to optimize therapeutic success for the treatment of kidney diseases.
Single-cell RNA sequencing (scRNAseq) has revolutionized the ability to study individual cell types of complex tissues and cell states after specific perturbations.7,8 Recent scRNAseq studies in kidney have provided insight into the transcriptional profiles of kidney cell types in healthy mice and human samples as well as in some disease states, including DKD and lupus nephritis.9, 10, 11, 12 However, detailed studies of glomerular cell states have been limited by the small number of cells available for analysis. Furthermore, the earliest cell type–specific changes that occur in all glomerular cells on podocyte injury have not been monitored and are yet to be defined.
Podocyte ablation via podocyte-specific inducible deletion of an essential endogenous protein, CTCF, leads to progressive proteinuric kidney disease and CKD.13 Historically, CKD mouse models were generated by inducing kidney injury by exogenous toxins or surgical interventions, suboptimal systems that fall short of recapitulating sequential mechanistic changes. Deletion of CTCF, an essential endogenous molecule, leads to rapid podocyte loss, severe progressive albuminuria, bone mineral metabolism changes, kidney failure, and premature death.13 The inducible podocyte-specific CTCF deletion (iCTCFpod−/−) model provides the unique opportunity to study changes in intraglomerular cell-cell interactions as a consequence of induced podocyte injury. Podocyte CTCF expression is undetectable at the earliest timepoint, at 1 week of doxycycline-mediated Cre induction in iCTCFpod−/− mice.13 Significant and progressive podocyte loss, as measured by histologic analysis, starts at 2 weeks after Cre induction. In the current study, a detailed analysis was performed at the 1-week time point to identify the earliest changes in intraglomerular cell-cell interactions driven by podocyte injury that ultimately lead to CKD.
Materials and Methods
Animal Care
This study was approved by the Animal Care and Use Committee at Brigham and Women's Hospital, Harvard Medical School. All animal studies were performed in accordance with guidelines established and approved by the Animal Care and Use Committee at Brigham and Women's Hospital, Harvard Medical School. iCrepod-Ctcfwt/fl mice were generated as previously described13 and inbred to generate iCrepod-Ctcffl/fl (iCTCFfl/fL) and iCrepod-Ctcfwt/wt [wild-type (WT)] mice. Both male and female mice were used in this study. Doxycycline (4 g/L) (Sigma D9891) was continuously administered in drinking water that contained sucrose (50 g/L) (VWR BDH0308) to both iCTCFfl/fL (generates iCTCFpod−/− mice) and WT littermate control mice (6 to 8 weeks of age at the start of doxycycline use) to drive Cre expression specifically in podocytes.
Glomerular Enrichment from Whole Kidney
iCTCFpod−/− and WT mice were sacrificed after 1 week of doxycycline treatment, and kidneys were quickly dissected and washed with ice-cold Hanks' balanced salt solution (catalog number 14170112, Thermo Scientific, Waltham, MA). After removing the kidney capsules, glomeruli were enriched at 4°C using the sieving technique14 with 180-mm, 75-mm and 53-mm sieves. Glomerular-enriched fractions collected from the 53-mm sieve were rinsed with ice-cold 1× Hanks' balanced salt solution and placed on ice.
Preparation of Single-Cell Suspensions
Glomerular-enriched fractions were centrifuged at 350 × g for 5 minutes at room temperature. After removing most of the Hanks' balanced salt solution, 1 mL of liberase TH digestion buffer (catalog number 5401135001, Sigma-Aldrich, St. Louis, MO) that contained 50 U/mL of DNase I (catalog number 90083, Thermo Scientific) was added to the glomerular pellet and incubated at 37°C for 60 minutes on an orbital shaker (56 × g). The suspension was passed through a 27-gauge needle twice after 20 minutes. Digested glomerular fractions were added to 9 mL of RPMI 1640 medium (catalog number 11875119, Thermo Scientific) containing 10% fetal bovine serum (catalog number 16000044, Thermo Scientific) and centrifuged at 500 × g for 5 minutes at room temperature. Then 1 mL of red blood cell lysis buffer (catalog number A1049201, Thermo Scientific) was added to the glomerular pellet and mixed for 1 minute at room temperature. Next 9 mL of RPMI 1640 medium (10% fetal bovine serum) was added, and the suspension was centrifuged at 500 × g for 5 minutes at room temperature. The media was removed from the pellet and 200 mL of Accumax (catalog number 07921, Stem Cell Technologies, Vancouver, BC, Canada) was added and incubated for 20 minutes at 37°C. Then 1.8 mL 1× phosphate-buffered saline (PBS) (catalog number 14190250, Thermo Scientific) plus 0.04% bovine serum albumin (BSA) (catalog number A1933, Sigma-Aldrich) was added, and the suspension was centrifuged at 500 × g for 8 minutes. The digested glomeruli were washed with 750 mL of 1× PBS plus 0.04% BSA and filtered using a 40-m Flowmi Tip Strainer (catalog number BAH136800040, Sigma-Aldrich). Then 1.25 mL 1× PBS plus 0.04% BSA was added, and the suspension was centrifuged at 500 × g for 8 minutes. The pellet was resuspended in a small volume of 1× PBS plus 0.04% BSA.
Library Preparation and Single-Cell Sequencing
Single cells were processed through the 10× Chromium 3′ Single Cell Platform using the Chromium Single Cell 3′ Library, Gel Bead, and Chip Kits (10× Genomics, Pleasanton, CA), following the manufacturer's protocol. Briefly, 10,000 cells were added to each channel of a chip to be partitioned into Gel Beads in Emulsion in the Chromium instrument, followed by cell lysis and barcoded reverse transcription of RNA in the droplets. Breaking of the emulsion was followed by amplification, fragmentation, and addition of adapter and sample index. Libraries were pooled together and sequenced on Illumina HiSeq.
Hybridization Chain Reaction
All hybridization chain reaction (HCR) v3 reagents (probes, hairpins, and buffers) were purchased from Molecular Technologies (Pasadena, CA). Thin sections of tissue (10 m) were mounted in 24-well glass bottom plates (catalog number 82050-898, VWR, Radnor, PA) coated with a 1:50 dilution of (3-aminopropyl)triethoxysilane (catalog number 440140, Sigma-Aldrich). The following solutions were added to the tissue: 10% formalin (catalog number 100503-120, VWR) for 15 minutes, two washes of 1× PBS (catalog number AM9625, Thermo Fisher Scientific), ice-cold 70% EtOH at −20°C for 2 hours (to overnight), three washes of 5× saline sodium citrate with 0.2% Tween-20 (SSCT) (catalog number 15557044, Thermo Fisher Scientific), hybridization buffer (Molecular Technologies) for 10 minutes, probes in hybridization buffer overnight, four washes of wash buffer (Molecular Technologies) for 15 minutes, three washes of 5× SSCT, amplification buffer (Molecular Technologies) for 10 minutes, and three washes of 15 minutes with 5× SSCT (1:10,000 DAPI, catalog number TCA2412-5MG, VWR) in the second wash. Hairpins were heat denatured in amplification buffer overnight. Samples were stored and imaged in 5× SSCT. Imaging was performed on a spinning disk confocal (Yokogawa W1 on Nikon Eclipse Ti) operating NIS-elements AR software. Image analysis and processing was performed on ImageJ Fiji software version 2.1.0/1.53c (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Data Processing and Quality Control
A Cellranger toolkit (version 2.1.1) was used to perform demultiplexing using the cellranger mkfastq command and the cellranger count command for alignment to the mouse transcriptome, cell barcode partitioning, collapsing unique molecular identifier (UMI) to transcripts, and gene-level quantification. Cells were filtered to include cells expressing a minimum of 500 genes and a maximum of 4000 genes. Furthermore, the percentage of reads mapping to mitochondrial genes was capped at 12%. DoubletFinder was used to identify potential doublets.15 Clusters with >75% of cells classified as high-confidence doublets were removed from further analysis. The remaining cells classified as high-confidence doublets were also removed from further analysis. The analysis involved downsampling the number of cells to account for this difference. For example, if there were fewer WT cells in a given cell type, a random sample of iCTCF cells equal to the number of WT cells was used for the differential expression analysis.
Unsupervised Clustering and Dimensionality Reduction
The default settings in the Seurat R package16 version 3.0 were used for normalization (NormalizeData) of the gene expression counts, identifying variable genes (FindVariableGenes), finding integration anchors (FindIntegrationAnchors), and integrating the samples (IntegrateData). Unwanted variation occurred because of the number of UMIs and ratio of reads mapping to mitochondrial genes (ScaleData). Dimensionality reduction was performed using principal component analyses (RunPCA) on the highly variable genes. The PCElbowPlot() function was used to distinguish principal components for further analysis. For clustering all cells, the first 40 principal components sufficiently captured all of the variance. Molecularly distinct clusters were identified using the default parameters (FindClusters) and a resolution of 0.4 (FindNeighbors). The data were processed and scaled as described above after subsetting glomerular cells. A total of 30 principal components were used for downstream clustering at a resolution of 0.3 (FindNeighbors).
Cell Type Classification
Cluster-enriched or marker genes were computed using the Wilcoxon rank sum test (FindAllMarkers) for differential expression of genes in the cluster cells versus all other cells and selecting those genes that pass the adjusted P value (false discovery rate) cutoff of 0.05 as cluster representative. Cluster identity was assigned by comparing data-driven genes with a list of literature-curated genes for mature kidney cell types.
Differential Gene Expression Analysis
Pairwise differential expression analysis in Seurat (FindMarkers) was used with the log fold change threshold set to 0.01 and default parameters to analyze differential expression between iCTCFpod−/− and WT cells in a specific cluster. Within FindMarkers, ident.1 was set to cluster-specific iCTCFpod−/− cells, and ident.2 was set to the corresponding cluster-specific WT cells. Genes with an adjusted P < 0.05 were considered significant.
Overrepresentation Analysis
Overrepresentation analysis was used to determine whether any known biological functions or processes were overrepresented or enriched in the list(s) of differentially expressed genes.17 The R package clusterProfiler,18 specifically the enrichGO function, was used for the overrepresentation analysis. Default parameters were used, with the exceptions of ont = BP, pvaluecutoff = 0.25, and qvaluecutoff = 0.25. The R package enrichplot (GitHub, https://github.com/YuLab-SMU/enrichplot, last accessed August 30, 2021) was then used, specifically the emapplot function, to visualize the enrichment results by plotting the top enrichment terms.
Ligand Receptor Analysis
iTALK19 was used to map receptors to ligands that were differentially expressed in iCTCFpod−/− podocytes. Specifically, rawParse was used to calculate the mean expression of each gene, using scaled data from Seurat. FindLR using datatype = meancount was used to identify ligand and receptor pairs. Ligand and receptor pairs of interest were plotted using default parameters of LRPlot. The thickness of the lines indicates the relative mean expression of the ligand, and the size of the arrowhead indicates the relative mean expression of the receptor.
NicheNet Analysis
The R package NicheNet20 was used to predict ligand-receptor interactions that might drive gene expression changes in the cell type of interest. All podocyte clusters and all endothelial clusters were combined for this analysis. All default parameters were used with the exception of setting a lower cutoff threshold of 0.11 for prepare_ligand_target_visualization.
HCR Image Processing and Quantification
Images were processed using ImageJ2 software (NIH, Bethesda, MD; http://imagej.nih.gov/ij). For ImageJ files, version 2.1.0/1.53c was used (http://imagej.net/Contributors (last accessed August 2, 2020). Raw ND2 files were background subtracted using the Rolling Ball method (rolling = 50 sliding stack). Mean intensities of the Z-stack images were then projected, and image channels were split and saved separately. CellProfiler version 3.1.5 (Broad Institute of MIT and Harvard, Cambridge, MA)21 was used for cell segmentation based on the fluorescence intensity of DAPI channel and for measuring integrated fluorescence intensity in the rest of the channels. Four WT and four iCTCFpod−/− mice were evaluated for statistical analysis. All glomeruli from three 40× images were quantified. A Welch-corrected two-tailed t-test was performed.
Results
Single-Cell Profiling of >29,000 Glomerular-Enriched Kidney Cells
Current single-cell protocols for whole kidney identify <2.5% glomerular cells,9 and although a purified glomerular preparation using magnetic beads enriches for this population,22 it fails to capture other cell types of the kidney that may be of interest. A sieving method was used to simultaneously enrich glomeruli and capture additional kidney cell types to extend these findings and develop a detailed understanding of cell-cell interactions within the glomerulus in the context of the entire cellular landscape of the kidney, before and after podocyte injury.
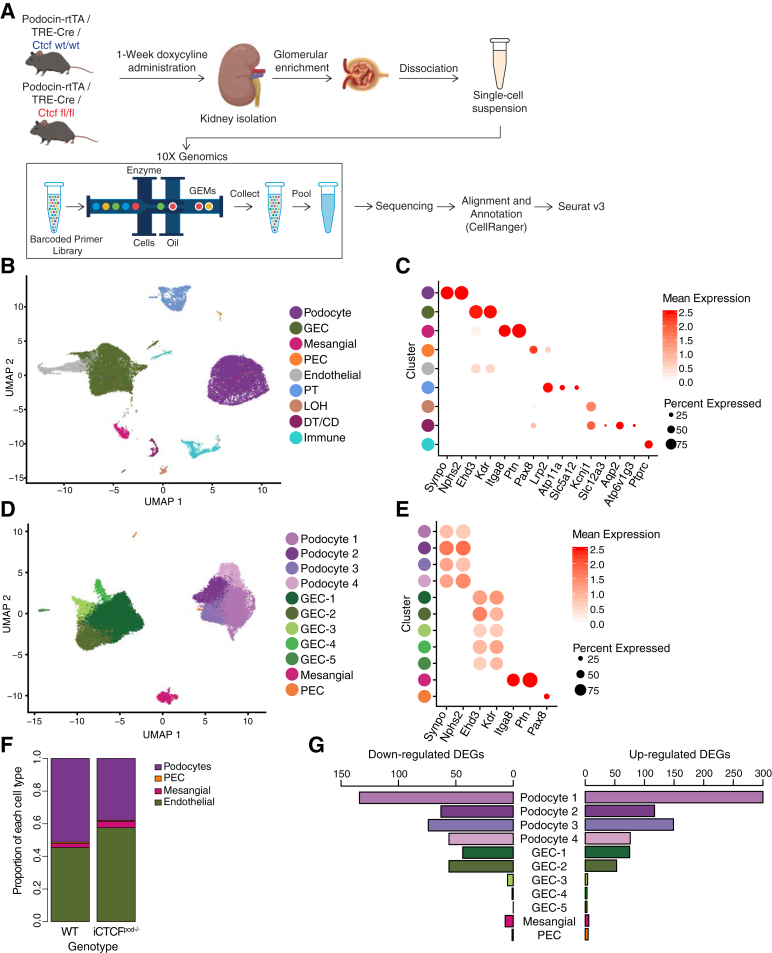
To identify the early transcriptional effects of podocyte ablation in a cell type–specific manner, scRNAseq was performed with kidney tissue from WT and iCTCFpod−/− mice collected by serial sieving after 1 week of doxycycline treatment (Figure 1A). Kidney tissue from four WT animals (four biological replicates) and four iCTCFpod−/− animals yielded a total of 14,783 WT and 14,727 iCTCFpod−/− cells profiled after filtering (Supplemental Figure S1, A and B; Supplemental Tables S1 and S2). Data were normalized to remove effects due to the number of UMIs and percentage of mitochondrial reads. After integration of WT and iCTCFpod−/− samples, a low resolution of clustering was used to detect nine clusters (Figure 1B). Biological replicates of WT and iCTCFpod−/− samples were distributed among all clusters (Supplemental Figure S2, A-C). All cell types of the glomerulus, as well as additional kidney cell types, were identified using established and data-derived markers (Figure 1C; Supplemental Table S3; Supplemental Figure S3A).
Figure 1.
Endothelial cells are most affected by podocyte injury, as indicated by single-cell RNA sequencing of wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) kidneys. A: Experimental design. WT and iCTCFpod−/− mice were treated with doxycycline for 1 week to induce deletion of Ctcf specifically in podocytes. After 1 week, kidneys were isolated, and glomeruli were enriched using serial sieving. Enzymatic digestion was used to obtain a single-cell suspension that was subsequently sequenced on the 10× Genomics platform. Downstream data analysis was performed using CellRanger and Seurat. B: Uniform manifold approximation and projection (UMAP) plot of all cells isolated from WT and iCTCFpod−/− glomerular-enriched kidney fractions. C: Expression of cell type–specific markers used for cluster identification in panel A presented as a dot plot. Color represents the mean expression level of the gene in the specified cluster. WT and iCTCFpod−/− cells split into 10 clusters, with all major cell types of the kidney identified. D: UMAP plot of cells of glomerular origin that were isolated and reclustered. E: Expression of cell type–specific markers used for cluster identification in panel C. WT and iCTCFpod−/− glomerular cells split into 11 clusters. F: Proportion of WT or iCTCFpod−/− cells per glomerular cell type. G: Bar plot of the number of differentially expressed genes (DEGs) in each cluster comparing iCTCFpod−/− and WT cells. Genes were defined as differentially expressed if they were expressed in at least 10% of cells, had a log fold change of 0.01, and had an adjusted P < 0.05. DT/CD, distal tubule/collecting duct; GEC, glomerular endothelial cell; LOH, loss of heterozygosity; PEC, parietal endothelial cell; PT, proximal tubule; rtTA, reverse tetracycline-controlled transactivator.
Glomerular cells, representing a total 82.1% of all recovered cells [36.9% podocytes, 42.1% glomerular endothelial cells (GECs), 2.7% mesangial cells, and 0.36% PECs] were isolated and reclustered (Figure 1D). Four clusters of podocytes, five clusters of GECs, and one cluster each of mesangial cells and PECs, expressing canonical cell-type markers, were identified (Figure 1E; Supplemental Figure S3B). Biological replicates of WT and iCTCFpod−/− samples were distributed throughout all clusters (Supplemental Figure S2, D-F; Supplemental Table S4). As anticipated, given that podocyte-specific CTCF deletion leads to histologically detectable podocyte loss at 2 weeks,13 iCTCFpod−/− samples contained a lower percentage of podocytes than WT samples (Figure 1F; Supplemental Table S5). iCTCFpod−/− samples had 13.1% fewer podocytes, 11.9% more GECs, and 1.5% more mesangial cells than WT samples (Supplemental Table S5). PECs contributed <1% to either of the iCTCFpod−/− or WT samples. This finding suggests that disruption of transcriptional programs critical for podocyte survival precedes histologically detectable podocyte injury and loss and highlights the power of scRNAseq in discerning subtle changes that can be missed by histologic analysis.
Podocyte-Specific Inducible CTCF Deletion Leads to Gene Expression Changes in All Glomerular Cell Types
Differential expression analysis was performed in each of 11 clusters comparing iCTCFpod−/− and WT cells (Supplemental Table S6). Genes were considered differentially expressed if found in at least 10% of cells in a given cluster, with a minimum absolute log fold change of 0.1 and an adjusted P < 0.05. Ctcf was differentially expressed in each of the four podocyte clusters, with mean log fold changes of −0.390, −0.189, −0.331, and −0.303, respectively. The numbers of cells in each cluster were downsampled and the differential expression analysis repeated to compare the number of differentially expressed genes among the 11 clusters. The podocyte clusters had the most differentially expressed genes, followed by GEC-1 and GEC-2 (Figure 1G). The remaining three clusters of GECs, along with the mesangial cells and PECs, had far fewer differentially expressed genes (Figure 1G), suggesting that GEC-1 and GEC-2, among all glomerular clusters, were most affected by the sequelae of CTCF deletion–driven podocyte injury.
Disease-Associated Gene Programs Identified in Specific Podocyte and Endothelial Cell Clusters
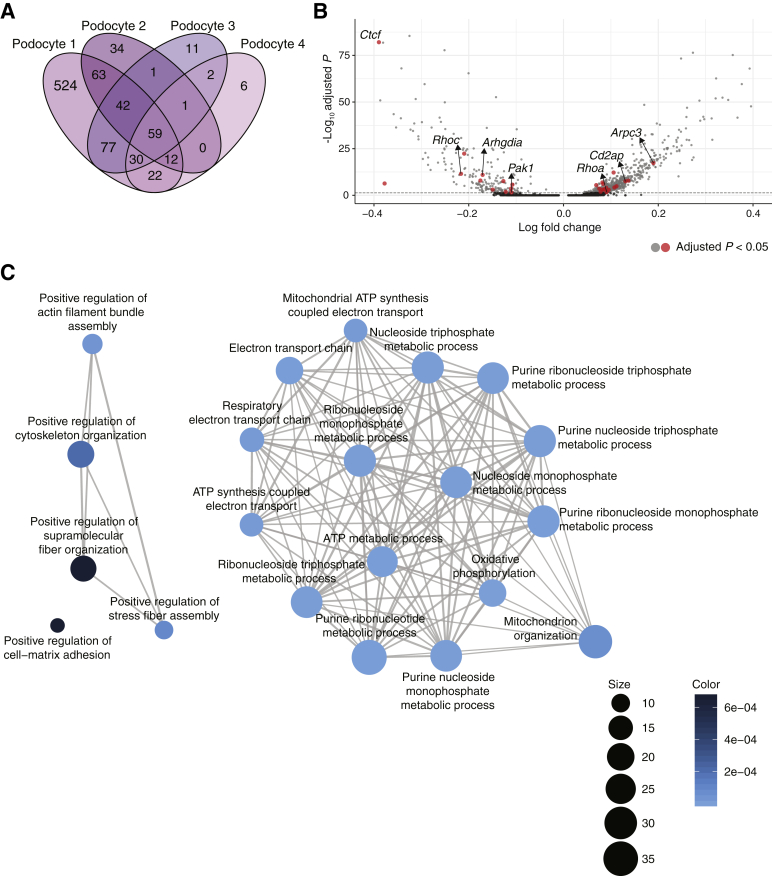
This study first sought to examine how the individual podocyte clusters respond to injury. A Venn diagram of the differentially expressed genes in each of the four podocyte clusters revealed that podocyte 1 had the most uniquely differentially expressed genes of the four podocyte clusters (Figure 2A; Supplemental Table S7). The observation that one of four podocyte clusters was more prominently affected is in agreement with prior histologic data indicating that not all podocytes are affected with equal severity in the face of injury and highlights the power of scRNAseq to molecularly characterize the heterogeneity of cell states within the same cell type.23
Figure 2.
Gene programs of focal adhesions, cytoskeleton organization, and mitochondrial functions enriched in podocytes. A: Venn diagram of the differentially expressed genes identified in each of the four podocyte subclusters. B: Volcano plot of differentially expressed genes comparing inducible podocyte-specific CTCF deletion (iCTCFpod−/−) and wild-type (WT) cells in podocyte 1. All genes from adhesion and cytoskeleton organization gene programs are colored in red, with genes of interest labeled. Significantly differentially expressed genes are represented by light gray and red dots. Genes in dark gray are not significantly differentially expressed. The y axis has been limited to 100. Dashed line indicates default parameters: cutoff P = 0.25. C: Visualization of the overrepresentation analysis of genes uniquely differentially expressed in podocyte 1 presented as a network or enrichment map. Overlapping gene sets cluster together.
An overrepresentation analysis was performed to identify gene programs enriched as a consequence of CTCF loss in the podocyte 1 cluster (Supplemental Table S8). The top enriched terms were visualized with an enrichment map to cluster mutually overlapping gene sets (Figure 2B). A prominent group of enriched terms was mitochondrial functions, including ATP synthesis, mitochondrial organization, electron transport chain, and oxidative phosphorylation (Figure 2B). These data extend recent work pointing to mitochondrial dysfunction as a sign of podocyte injury.24 In addition, human genetics have pointed to the importance of mitochondrial functions in podocytes, including several mutations in the CoQ biosynthesis pathway (PDSS1, PDSS2, COQ2, COQ6, and ADCK4) that cause nephrotic syndrome, mainly in children.25,26 It has therefore been postulated that podocyte mitochondrial dysfunction may represent a prominent cell state associated with all diseases that stem from podocyte loss.24 The data provide support for this notion at single-cell resolution, suggesting that mitochondrial dysfunction may represent the earliest injury state in a specific population of podocytes, leading to podocyte loss.
One of the main groups of enriched terms from the overrepresentation analysis (Figure 2B) was cytoskeletal organization as well as a related single enriched term for cell-matrix adhesion. The actin cytoskeleton plays an essential role in maintaining the podocytes' unique and complex structure, and adhesion to the GBM is essential for podocyte function.27 Mature focal adhesions contain hundreds of proteins that link the actin cytoskeleton, receptor matrix binding, intracellular signal transduction, and actin polymerization. One of the most down-regulated genes in podocyte 1 was Rhpn1 (Supplemental Figure S4A), an essential component for establishing podocyte cytoskeleton dynamics and maintaining podocyte foot process architecture.28 The Arp2/3 complex component Arpc3, a driver of actin polymerization, was up-regulated in the podocyte 1 cluster (Figure 2C; Supplemental Figure S4B). A gene for an additional component of the complex, Actr2, was also up-regulated in both podocyte 1 and podocyte 4. In addition, Wasl, whose protein product, N-WASP, activates the Arp2/3 complex and is required for the maintenance of podocyte foot processes in vivo29, was also up-regulated in podocyte 1, podocyte 3, and podocyte 4. The cytoskeletal regulator Arhgdia, whose deletion is associated with nephrotic syndrome in mice,30 was down-regulated in podocyte 1 (Figure 2C). The expression levels of two Rho GTPases that are well established regulators of the actin cytoskeleton and cell adhesion dynamics31 were also altered: RhoA was up-regulated and RhoC was down-regulated, as was a central downstream effector of Rac1, Pak1, consistent with a prior study.23 Furthermore, Cd2ap, a critical podocyte actin cytoskeleton component,27,32 was up-regulated in podocyte 1. Together, these changes suggest that one of the earliest podocyte responses to CTCF deletion–driven injury is to alter critical components of the actin cytoskeleton in a struggle to maintain attachment to the GBM and thus survive the injury. In addition, scRNAseq identified specific mediators of this response matched to a specific population of podocytes, pointing to putative targets for early therapeutic intervention.
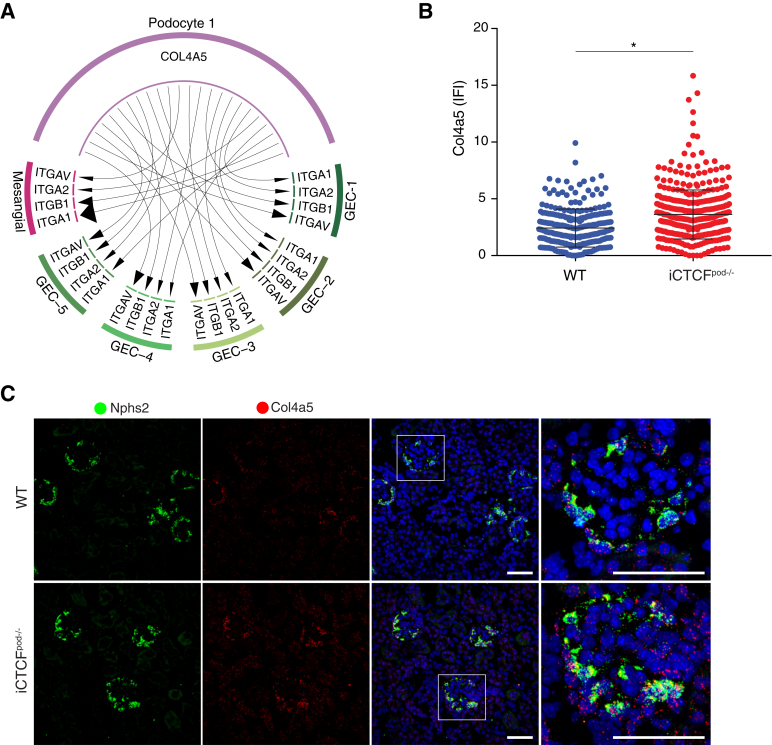
The GBM is a meshwork of extracellular matrix proteins situated between podocytes and GECs that provides structural support for the glomerular capillaries, harbors ligands for receptors on the surface of the adjacent GECs, podocytes, and mesangial cells, and contributes to glomerular filter selectivity.33 One of the critical components of the GBM is collagen type IV ɑ5 (Col4a5); mutations in this gene cause Alport syndrome and FSGS in humans.33,34 Variants in Col4a3, another structural component of the GBM, were recently identified by a comprehensive genome-wide association study in DKD.35 Therefore, changes to GBM components as a consequence of podocyte injury are broadly relevant to many kidney diseases. Whether expression of genes encoding structural components of the GBM were disrupted in the model was queried. Col4a5 was up-regulated in iCTCFpod−/− podocytes (with a log fold change of 0.130) (Supplemental Figure S4C; Supplemental Table S6). Ligand receptor analysis suggested that Col4a5 upregulation in podocyte 1 leads to increased interactions with cells of all five GEC clusters and mesangial cells through several integrins (Figure 3A). The expression of Col4a5 in Nphs2-expressing podocytes was significantly increased (P < 0.0001; Welch-corrected two-tailed t-test) (Figure 3B). The observed Col4a5 up-regulation, with spatial resolution, was validated by HCR, a method that generates single-molecule fluorescence via in situ hybridization36 (Figure 3C; Supplemental Figure S5). This result confirmed that individual gene data derived from the single-cell transcriptomic experiment could be independently validated, with spatial resolution, bolstering the validity of the conclusions drawn by computational analyses. Furthermore, these data highlight that the key response to injury by a specific population of podocytes is to up-regulate collagen production, which may account for the prominent or thick GBM observed in progressive diseases, such as DKD.35
Figure 3.
Podocyte cluster up-regulates Col4a5. A:Col4a5 was significantly up-regulated in inducible podocyte-specific CTCF deletion (iCTCFpod−/−) podocyte 1 cells compared with wild-type (WT) podocyte 1 cells. Collagen type IV ɑ5 (COL4A5) receptors were strongly expressed in mesangial cells and glomerular endothelial cell (GEC) subclusters 1 to 4. Size of the arrowhead represents the relative expression of the receptor. B: Quantification of hybridization chain reaction (HCR). Col4a5 was significantly up-regulated in iCTCFpod−/−Nphs2-expressing cells compared with WT Nphs2-expressing cells. Four WT and four iCTCFpod−/− mice were evaluated. All glomeruli from three images were quantified. C: Col4a5 expression in glomeruli of WT and iCTCFpod−/−animals as evaluated by HCR. ∗P < 0.0001 (two-tailed t-test with Welch correction). Original magnification, ×40; ×63 (enlargements of boxed areas) (C). Scale bars = 50 μm (C). IFI, immunofluorescent imaging.
GEC-1 and GEC-2 had nearly as many differentially expressed genes as podocyte clusters 2 to 4, whereas the remaining GEC clusters, mesangial cells, and PECs had fewer differentially expressed genes. Therefore, these two endothelial clusters were most affected by podocyte injury. GEC-1 expressed >600 uniquely differentially expressed genes (Supplemental Figure S6A; Supplemental Table S9). Overrepresentation analysis indicated that the uniquely differentially expressed genes in GEC-1 were enriched in gene programs for cell migration and adhesion (Supplemental Figure S6, B and C; Supplemental Table S10). These analyses revealed the earliest transcriptional changes that occur in GECs as a response to podocyte injury.
Modeling Intercellular Communication Reveals Key Interactions between Cell Types in Response to Early Podocyte Injury
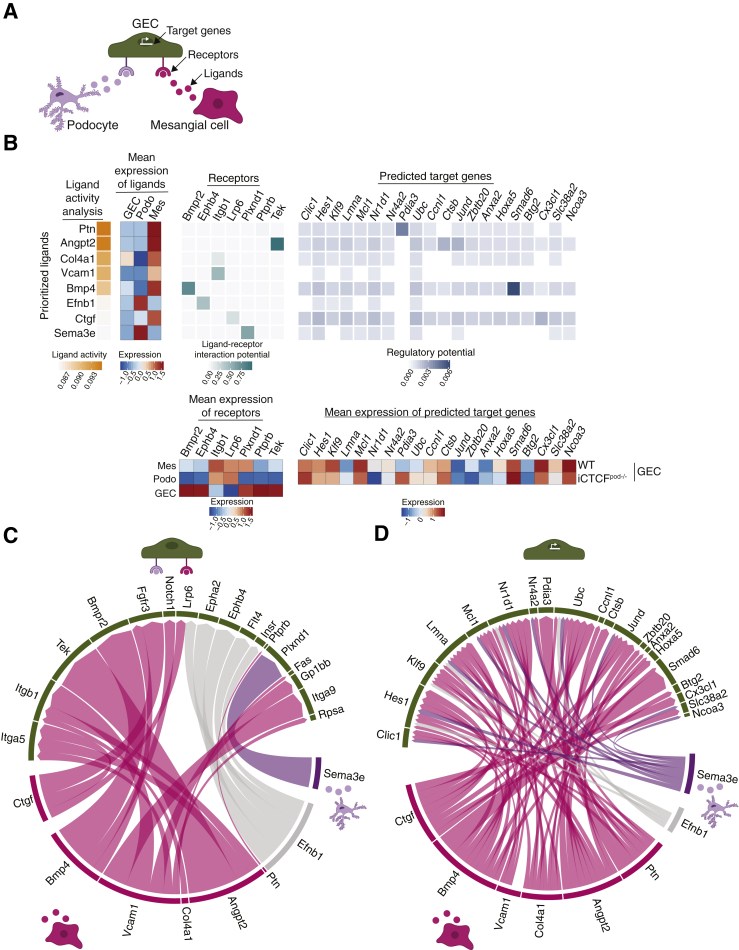
Having identified disrupted gene programs in several distinct cell clusters, the study next sought to understand how glomerular cell-cell crosstalk was influenced by podocyte injury. The study first probed the list of differentially expressed genes in podocyte, GEC, and mesangial cell clusters. The expression of two key autocrine prosurvival ligands, Vegfa and Pdgfb, as well as the expression of the PDGFB receptor, Pdgfrb, were disrupted (Supplemental Table S6). Vegfa expression was decreased in all podocyte clusters (mean log fold change between −0.034 and −0.149). In contrast, Pdgfb expression was increased in all GEC clusters (mean log fold change between 0.031 and 0.140), and the receptor Pdgfrb was up-regulated in mesangial cells (mean log fold change of 0.155). This analysis suggested that podocyte injury leads to decreased expression of the prosurvival ligand Vegfa, negatively affecting GECs. To compensate, GECs may up-regulate the prosurvival ligand Pdgfb, triggering mesangial cells to up-regulate the receptor Pdgfrb.
NicheNet, a novel algorithm that infers how ligand-receptor interactions derived from expression data may affect specific targets by integrating preexisting knowledge of signaling and regulatory networks, was used to more deeply investigate ligand-receptor interactions and their putative target genes.20 NicheNet was applied to model interactions between podocytes and GECs as well as mesangial cells and GECs that could potentially induce differentially expressed genes (target genes) in GECs in the setting of podocyte injury (Figure 4A). For this analysis, all clusters of each cell type were combined into a single cluster. The top predicted ligands expressed by mesangial cells and/or podocytes were pleiotrophin (Ptn), angiopoietin 2 (Angpt2), Col4a1, vascular cell adhesion molecule 1, bone morphogenetic protein 4 (Bmp4), Ephrin B1, connective tissue growth factor, and semaphoring 3e (Sema3e) (Figure 4B). The expression pattern of GEC receptors through which these ligands are known to act were mapped next to the ligand activity analysis. In addition, the predicted target genes that were differentially expressed in iCTCFpod−/− GECs compared with WT controls were also mapped. The results in Figure 4B are summarized as circular plots in Figure 4, C and D.
Figure 4.
NicheNet analysis reveals ligands, receptors, and target genes that contribute to transcriptional changes in glomerular endothelial cells (GECs) as a consequence of podocytes injury. A: Model for the NicheNet analysis. Potential target genes for inducible podocyte-specific CTCF deletion (iCTCFpod−/−) GECs were defined as differentially expressed [compared with wild-type (WT) GECs] if they were expressed in at least 10% of cells, had a log fold change of 0.1, and had an adjusted P < 0.05. Potential ligand-receptor pairs that lead to differentially expressed genes in iCTCFpod−/− GECs were then identified. B: Results of the NicheNet analysis. Ligands expressed in mesangial cells and/or podocytes were ranked by the likelihood that the ligand would affect gene expression changes in GECs. Receptors, expressed in GECs, were selected based on their known potential to interact with the prioritized ligands. Finally, target genes were selected based on their differential expression in GECs and their potential to be regulated by the ligand-receptor interactions identified between GECS-podocytes and GECs–mesangial cells. C: Summary of the ligand-receptor interactions identified in panel B. D: Summary of the ligand-target interactions identified in panel B.
Several ligands, receptors, and target genes were notable from this analysis. Pleiotrophin, a ligand highly expressed in mesangial cells (Figure 4B), is a secreted growth factor that can bind and inhibit protein tyrosine phosphatase receptor type B, stimulating endothelial cell migration via increased Tek (Tie2)/Angpt137 and Kdr (Vegfr2)/Vegfa38 signaling. Accordingly, Tek was significantly up-regulated in GECs (Figure 4B). Angpt2, highly expressed in mesangial cells, is an antagonistic ligand of Tek, inhibiting the binding of Angpt1.39 Because Angpt1 signaling is known to promote podocyte survival and a disruption of the Angpt1/Angpt2 ratio contributes to the development of DKD,40 this ligand-receptor analysis appeared to point to maladaptive Tek signaling in GECs as one of the earliest consequences of podocyte injury.
This study also found prominent changes in type IV collagen expression in several glomerular cells (Figure 4B). Normally produced by healthy podocytes, collagen IV heterotrimers transition from ɑ1ɑ1ɑ2 to ɑ3ɑ4ɑ5 during glomerular development, which is necessary for proper GBM formation and function.33,34 Up-regulation of Col4a1, along with expansion of the mesangial matrix, is frequently observed in DKD.41,42 In the current analysis, Col4a1 was expressed by both mesangial cells and GECs (mean expression of ligand heatmap) (Figure 4B) and was specifically up-regulated in iCTCFpod−/− GECs compared with WT controls. These data suggest that GECs up-regulate Col4a1 in the setting of podocyte injury, which may lead to a stiffer, fibrotic GBM that contributes to the development of segmental sclerosis.33
Bmp4 was a highly expressed mesangial ligand identified by NicheNet analysis (Figure 4, B-D). BMPs play key roles in kidney development and disease.43 Bmp4 is up-regulated in the setting of diabetic nephropathy in rats,42 and treatment of mice with diabetes with an anti-BMP4 antibody prevents the up-regulation of Col4a1 and mesangial matrix expansion.44 Of particular interest, mesangial Bmp4 was found to trigger GEC Smad6 expression (Figure 4B). Expression of Smad6 is induced by BMPs and in a negative-feedback loop, Smad6 specifically inhibits BMPs, including BMP4.45 Smad6 was down-regulated in iCTCFpod−/− GECs compared with WT controls, suggesting increased BMP signaling in endothelial cells in the setting of podocyte injury.
One of the prioritized ligands identified in podocytes was Sema3e (Figure 4, B-D). Class 3 semaphorins are secreted proteins that function in a variety of biological processes, including angiogenesis, lymphangiogenesis, and disease.46 Sema3a is up-regulated in human DKD,47 and Sema3g was recently identified as a podocyte-specific gene that protects podocytes from inflammation in vivo.48 The role of Sema3e in the podocyte remains unclear. The current analysis revealed a previously unrecognized receptor-ligand pair whereby podocyte Sema3e interacts with plexinD1 on GECs to trigger several downstream gene targets in the setting of podocyte injury (Figure 4, C and D).
Among several newly identified GEC target genes, Hes1 and Cx3cl1 were also found in the adhesion gene programs enriched in GEC-1 (Figure 4, B-D; Supplemental Figure S6). Hes1 is a target of Notch signaling, which plays an important role in the developing kidney, but reactivation can lead to fibrosis.49 Cx3cl1 is a chemokine mainly produced by glomerular endothelium that acts as a chemoattractant and adhesion molecule for its receptor, Cx3cr1, which is ubiquitously expressed on mononuclear and circulatory lymphatic leukocytes.50 Cx3cl1 has been implicated in a variety of kidney diseases, including DKD, IgA nephropathy, and glomerulonephritis.50 Hes1 and Cx3cl1 were both up-regulated in iCTCFpod−/− GECs compared with WT controls, suggesting that GECs respond to podocyte injury by up-regulating proinflammatory and profibrosis programs.
Finally, because many of the ligands and targets identified in the NicheNet analysis are implicated in DKD, in both rodent models as well as humans, the data set was compared with a single nuclei transcriptomic data set of early human DKD.10 A specific comparison was made of differentially expressed genes between human and mouse cell clusters. Of the 138 differentially expressed genes in human DKD GECs, 25 genes were also identified in mouse iCTCFpod−/− endothelial cells, including Col4a1 (Supplemental Figure S7; Supplemental Table S11). Together, these results suggest that endothelial cell programs frequently disrupted in human DKD can be partially recapitulated in the mouse model of early glomerular injury via selective podocyte ablation. In addition, these results suggest that, in addition to podocytes, there may be potential endothelial-specific targets and therapeutic strategies to halt or slow the progression of complex and highly prevalent kidney diseases, such as DKD.
Discussion
Podocyte dysfunction and loss are the cause of many progressive kidney diseases. This study used scRNAseq to dissect the transcriptional networks and cellular crosstalk in the glomerular compartment shortly after induction of selective podocyte injury. The study took advantage of a mouse model of iCTCFpod−/− at 1 week after induction to generate a single-cell map of the kidney filter at the earliest stages of podocyte injury. This comprehensive study of glomerular cells, the first to evaluate the effects of a specific and selective genetic perturbation, has revealed four important insights about the kidney filter in health and disease.
First, scRNAseq was performed after 1 week of doxycycline treatment to induce CTCF deletion in iCTCFpod−/− animals to detect the earliest transcriptional changes that occur after podocyte injury. Although podocyte quantification by image analysis revealed a statistical difference in the number of podocytes in WT and iCTCFpod−/− glomeruli after 2 weeks of doxycycline treatment,13 a reduction of podocytes in iCTCFpod−/− glomeruli was observed even after 1 week of doxycycline induction. This finding suggests that disruption of transcriptional programs critical for podocyte survival precedes histologically detectable podocyte injury and loss and highlights the power of scRNAseq in discerning subtle changes that can be missed by histologic analysis. At a high level, this finding suggests that efforts to develop podocyte-protective therapies should focus on preventing podocyte injury as early as possible during the disease.
Second, enriching for glomeruli from whole kidneys using the sieving method was used to increase the proportion of isolated glomerular cells that were recovered, including a rare population of PECs that have not been captured in previous scRNAseq studies.9,22,51 In addition, all major cell types of the kidney were identified. This identification provides the opportunity for future studies to evaluate transcriptional changes driven by podocyte injury in the entire kidney, which will deepen understanding of kidney-wide transcriptional events that lead to the progression of CKD.
Third, the data suggest that the iCTCFpod−/− mouse model recapitulates many aspects of human disease at the transcriptional level, including differential expression of genes critical for podocyte adherence to the GBM and autocrine prosurvival ligands. Because adherence of podocytes to the GBM is critical for their function, the up-regulation of Col4a5 and other genes in cell adhesion gene programs support a model whereby podocytes respond to initial injury by increasing adherence to the GBM. Human mutations in podocyte focal adhesion genes, including actin cytoskeleton genes, type IV collagen, and integrins, have all been implicated in FSGS and/or nephrotic syndrome.52,53 Furthermore, ligand-receptor analysis revealed that signaling molecules implicated in human DKD are also disrupted in the CTCF mouse model (eg, BMP4 and Col4a1).41,42,44 We also observed the dysregulation of two key autocrine prosurvival ligands, Vegfa and Pdgfb, whose roles have been well established in vitro and in vivo, and alterations in their expression and/or activity have been associated with human kidney diseases, such as thrombotic microangiopathies and DKD.54 Specifically, anti-VEGF therapy, a common cancer treatment, leads to renal thrombotic microangiopathy,55 and Pdgfb expression is increased in human kidney biopsy samples of patients with DKD and mesangial proliferative glomerulonephritis.56, 57, 58 Taken together, these data support the notion that the iCTCFpod−/− mouse model may offer a reasonably faithful transcriptional profile of early glomerular changes with relevance to several human kidney diseases.
The detailed modeling of ligand-receptor-target gene interactions in all glomerular cells after podocyte injury led to several key observations with important therapeutic implications. GECs were most affected by podocyte injury, consistent with the physical proximity between the two cell types. This result emphasizes the importance of moving beyond reductionist studies that focus on one cell type alone by understanding intercellular communication patterns to gain deeper insights into mechanisms of kidney disease progression. The integrated analysis revealed several previously unrecognized molecules in GECs, such as BMP4 and PlexinD1, and in podocytes, such as Sema3E, that may be further explored for their potential to be targeted for therapeutic benefit. In this context, the detailed single-cell map of the mouse glomerulus after early podocyte injury generated in this study, made openly available to all investigators, may be used to generate hypotheses about actionable targets for much needed therapies.
Acknowledgments
We thank Aviv Regev, Orit Rosenblatt-Rosen, and members of the Regev group for ongoing advice on single-cell technology and computational approaches, and the Broad GP platform for sequencing services.
Footnotes
Supported by NIHNational Institute of Diabetes and Digestive and Kidney Diseases grants DK103658, DK099465, and DK095045 (A.G.).
Disclosures: A.G. has a financial interest in Goldfinch Biopharma, which was reviewed and is managed by Brigham and Women's Hospital and Partners HealthCare and the Broad Institute of MIT and Harvard in accordance with their conflict of interest policies.
Current address of Y.Z., Medical Research Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, Guangdong, China; of M.S.M., Morphic Therapeutic, Waltham, MA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2021.11.004.
Author Contributions
A.R.C., J.M., and A.G. designed the experiments; A.R.C. performed animal experiments, assisted by Y.Z. and M.S.M. A.R.C. performed computational analysis of single-cell RNA sequencing data; L.N. generated and sequenced 10× libraries; J.M. performed HCR experiments; H.C. performed HCR computational analysis for quantification; A.G. directed the project; and A.R.C. and A.G. wrote the manuscript. All authors read, commented on, and approved the manuscript.
Supplemental Data
Quality control metrics of single-cell RNA sequencing data. A and B: Violin plots showing the distribution of the number of unique molecular identifiers (counts), number of genes (features), and percentage of mitochondrial reads (% mito) in each sample before (A) and after (B) filtering.
Clustering of wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) cells by sample and genotype. A: Distribution of all profiled WT and iCTCFpod−/− cells per cluster. B: Distribution of WT and iCTCFpod−/− glomerular cells per cluster. C: Clustering of all WT and iCTCFpod−/− cells colored by number. D: Clustering of WT and iCTCFpod−/− glomerular cells colored by sample number. E: Clustering of all WT and iCTCFpod−/− cells colored by genotype. F: Clustering of WT and CTCFpod−/− glomerular cells colored by genotype.
Expression of cell-type specific markers in wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) cells used for cluster identification in Figure 1. Violin plots of cell type–specific markers in WT and iCTCFpod−/− cells used for cluster identification for all cells (A) or glomerular cells only (B).
Extension of Figure 2. A: Full volcano plot presented in Figure 2, highlighting Rhpn1 and Ctcf. Significantly differentially expressed genes are represented by light gray and red dots. Genes in dark gray are not significantly differentially expressed. Dashed line indicates default parameters: cutoff P = 0.25. B: Dot plot showing the expression of all labeled genes in volcano plots of podocyte 1 cells. Size of the dot represents the percentage of cells expressing the genes. Color represents the mean expression level of the gene in the specified cluster. C: Dot plot showing the expression of Nphs2 and Col4a5 in all clusters.
Extended validation of genes presented in Figure 3. Additional hybridization chain reaction (HCR) images used for quantification of Nphs2 and Col4a5 expression in wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) animals. A–D: Replicates of HCR images used for quantification of Nphs2 and Col4a5. White box denotes the section of the 40× image used in the Zoom panel. Scale bars = 50 μm.
Endothelial cells enriched for gene programs important for adhesion and migration. A: Venn diagram of the differentially expressed genes identified in glomerular endothelial cells (GECs) 1 and 2. B: Visualization of the overrepresentation analysis of genes uniquely differentially expressed in GEC-1 presented as a network or enrichment map. Overlapping gene sets cluster together. C: Heat map of all genes from the adhesion and migration gene programs from panel B.
Comparison of inducible podocyte-specific CTCF deletion (iCTCFpod−/−) glomerular endothelial cells (GECs) with human diabetic kidney disease (DKD) GECs. Venn diagram of the differentially expressed genes found in all iCTCFpod−/− GEC clusters and human DKD GEC clusters.
References
- 1.Webster A.C., Nagler E.V., Morton R.L., Masson P. Chronic kidney disease. Lancet. 2017;389:1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 2.Scott R.P., Quaggin S.E. Review series: the cell biology of renal filtration. J Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greka A. Human genetics of nephrotic syndrome and the quest for precision medicine. Curr Opin Nephrol Hypertens. 2016;25:138–143. doi: 10.1097/MNH.0000000000000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriz W., Shirato I., Nagata M., LeHir M., Lemley K.V. The podocyte's response to stress: the enigma of foot process effacement. Am J Physiol Ren Physiol. 2013;304:F333–F347. doi: 10.1152/ajprenal.00478.2012. [DOI] [PubMed] [Google Scholar]
- 5.Eremina V., Cui S., Gerber H., Ferrara N., Haigh J., Nagy A., Ema M., Rossant J., Jothy S., Miner J.H., Quaggin S.E. Vascular endothelial growth factor a signaling in the podocyte-endothelial compartment is required for mesangial cell migration and survival. J Am Soc Nephrol. 2006;17:724–735. doi: 10.1681/ASN.2005080810. [DOI] [PubMed] [Google Scholar]
- 6.Dimke H., Maezawa Y., Quaggin S.E. Crosstalk in glomerular injury and repair. Curr Opin Nephrol Hypertens. 2015;24:231–238. doi: 10.1097/MNH.0000000000000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Stuart T., Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 9.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson P.C., Wu H., Kirita Y., Uchimura K., Ledru N., Rennke H.G., Welling P.A., Waikar S.S., Humphreys B.D. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci. 2019;116:19619–19625. doi: 10.1073/pnas.1908706116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young M.D., Mitchell T.J., Vieira Braga F.A., Tran M.G.B., Stewart B.J., Ferdinand J.R., et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark A.R., Greka A. The power of one: advances in single-cell genomics in the kidney. Nat Rev Nephrol. 2020;16:73–74. doi: 10.1038/s41581-019-0237-1. [DOI] [PubMed] [Google Scholar]
- 13.Christov M., Clark A.R., Corbin B., Hakroush S., Rhee E.P., Saito H., Brooks D., Hesse E., Bouxsein M., Galjart N., Jung J.Y., Mundel P., Jüppner H., Weins A., Greka A. Inducible podocyte-specific deletion of CTCF drives progressive kidney disease and bone abnormalities. JCI Insight. 2018;3 doi: 10.1172/jci.insight.95091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreisberg J.I., Hoover R.L., Karnovsky M.J. Isolation and characterization of rat glomerular epithelial cells in vitro. Kidney Int. 1978;14:21–30. doi: 10.1038/ki.1978.86. [DOI] [PubMed] [Google Scholar]
- 15.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Wang R., Zhang S., Song S., Jiang C., Han G., Wang M., Ajani J., Futreal A., Wang L. iTALK: an R package to characterize and illustrate intercellular communication. bioRxiv. 2019:507871. [Google Scholar]
- 20.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2019;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 21.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., Sabatini D.M. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karaiskos N., Rahmatollahi M., Boltengagen A., Liu H., Hoehne M., Rinschen M., Schermer B., Benzing T., Rajewsky N., Kocks C., Kann M., Müller R.-U. A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol. 2018;29:2060–2068. doi: 10.1681/ASN.2018030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greka A., Mundel P. Cell biology and pathology of podocytes. Annu Rev Physiol. 2012;74:299–323. doi: 10.1146/annurev-physiol-020911-153238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkkoetter P.T., Bork T., Salou S., Liang W., Mizi A., Özel C., Koehler S., Hagmann H.H., Ising C., Kuczkowski A., Schnyder S., Abed A., Schermer B., Benzing T., Kretz O., Puelles V.G., Lagies S., Schlimpert M., Kammerer B., Handschin C., Schell C., Huber T.B. Anaerobic glycolysis maintains the glomerular filtration barrier independent of mitochondrial metabolism and dynamics. Cell Rep. 2019;27:1551–1566.e5. doi: 10.1016/j.celrep.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emma F., Bertini E., Salviati L., Montini G. Renal involvement in mitochondrial cytopathies. Pediatr Nephrol. 2012;27:539–550. doi: 10.1007/s00467-011-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akchurin O., Reidy K.J. Genetic causes of proteinuria and nephrotic syndrome: impact on podocyte pathobiology. Pediatr Nephrol. 2015;30:221–233. doi: 10.1007/s00467-014-2753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schell C., Huber T.B. The evolving complexity of the podocyte cytoskeleton. J Am Soc Nephrol. 2017;28:3166–3174. doi: 10.1681/ASN.2017020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lal M.A., Andersson A.-C., Katayama K., Xiao Z., Nukui M., Hultenby K., Wernerson A., Tryggvason K. Rhophilin-1 is a key regulator of the podocyte cytoskeleton and is essential for glomerular filtration. J Am Soc Nephrol. 2015;26:647–662. doi: 10.1681/ASN.2013111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schell C., Baumhakl L., Salou S., Conzelmann A.-C., Meyer C., Helmstädter M., Wrede C., Grahammer F., Eimer S., Kerjaschki D., Walz G., Snapper S., Huber T.B. N-wasp is required for stabilization of podocyte foot processes. J Am Soc Nephrol. 2013;24:713–721. doi: 10.1681/ASN.2012080844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gee H.Y., Saisawat P., Ashraf S., Hurd T.W., Vega-Warner V., Fang H., Beck B.B., Gribouval O., Zhou W., Diaz K.A., Natarajan S., Wiggins R.C., Lovric S., Chernin G., Schoeb D.S., Ovunc B., Frishberg Y., Soliman N.A., Fathy H.M., Goebel H., Hoefele J., Weber L.T., Innis J.W., Faul C., Han Z., Washburn J., Antignac C., Levy S., Otto E.A., Hildebrandt F. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123:3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodge R.G., Ridley A.J. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 32.Shih N.Y., Li J., Karpitskii V., Nguyen A., Dustin M.L., Kanagawa O., Miner J.H., Shaw A.S. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 1999;286:312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 33.Suh J.H., Miner J.H. The glomerular basement membrane as a barrier to albumin. Nat Rev Nephrol. 2013;9:470–477. doi: 10.1038/nrneph.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunwar S., Ballester F., Noelken M.E., Sado Y., Ninomiya Y., Hudson B.G. Glomerular basement membrane: identification of a novel disulfide-cross-linked network of alpha3, alpha4, and alpha5 chains of type IV collagen and its implications for the pathogenesis of Alport syndrome. J Biol Chem. 1998;273:8767–8775. doi: 10.1074/jbc.273.15.8767. [DOI] [PubMed] [Google Scholar]
- 35.Salem R.M., Todd J.N., Sandholm N., Cole J.B., Chen W.-M., Andrews D., et al. SUMMIT Consortium, DCCT/EDIC Research Group, GENIE Consortium Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J Am Soc Nephrol. 2019;30:2000–2016. doi: 10.1681/ASN.2019030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi H.M.T., Schwarzkopf M., Fornace M.E., Acharya A., Artavanis G., Stegmaier J., Cunha A., Pierce N.A. Third-generation hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145:dev165753. doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yacyshyn O.K., Lai P.F.H., Forse K., Teichert-Kuliszewska K., Jurasz P., Stewart D.J. Tyrosine phosphatase beta regulates angiopoietin-Tie2 signaling in human endothelial cells. Angiogenesis. 2009;12:25–33. doi: 10.1007/s10456-008-9126-0. [DOI] [PubMed] [Google Scholar]
- 38.Mellberg S., Dimberg A., Bahram F., Hayashi M., Rennel E., Ameur A., Westholm J.O., Larsson E., Lindahl P., Cross M.J., Claesson-Welsh L. Transcriptional profiling reveals a critical role for tyrosine phosphatase VE-PTP in regulation of VEGFR2 activity and endothelial cell morphogenesis. FASEB J. 2009;23:1490–1502. doi: 10.1096/fj.08-123810. [DOI] [PubMed] [Google Scholar]
- 39.Maisonpierre P.C., Suri C., Jones P.F., Bartunkova S., Wiegand S.J., Radziejewski C., Compton D., McClain J., Aldrich T.H., Papadopoulos N., Daly T.J., Davis S., Sato T.N., Yancopoulos G.D. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 40.Fu J., Lee K., Chuang P.Y., Liu Z., He J.C. Glomerular endothelial cell injury and cross talk in diabetic kidney disease. Am J Physiol Ren Physiol. 2015;308:F287–F297. doi: 10.1152/ajprenal.00533.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alicic R.Z., Rooney M.T., Tuttle K.R. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol. 2017;12:2032–2045. doi: 10.2215/CJN.11491116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C., Lin J., Li L., Zhu T., Gao L., Wu W., Liu Q., Ou S. The role of the BMP4/Smad1 signaling pathway in mesangial cell proliferation: a possible mechanism of diabetic nephropathy. Life Sci. 2019;220:106–116. doi: 10.1016/j.lfs.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 43.Nishinakamura R., Sakaguchi M. BMP signaling and its modifiers in kidney development. Pediatr Nephrol. 2014;29:681–686. doi: 10.1007/s00467-013-2671-9. [DOI] [PubMed] [Google Scholar]
- 44.Matsubara T., Araki M., Abe H., Ueda O., Jishage K.-I., Mima A., Goto C., Tominaga T., Kinosaki M., Kishi S., Nagai K., Iehara N., Fukushima N., Kita T., Arai H., Doi T. Bone morphogenetic protein 4 and Smad1 mediate extracellular matrix production in the development of diabetic nephropathy. Diabetes. 2015;64:2978–2990. doi: 10.2337/db14-0893. [DOI] [PubMed] [Google Scholar]
- 45.Wang R.N., Green J., Wang Z., Deng Y., Qiao M., Peabody M., Zhang Q., Ye J., Yan Z., Denduluri S., Idowu O., Li M., Shen C., Hu A., Haydon R.C., Kang R., Mok J., Lee M.J., Luu H.L., Shi L.L. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toledano S., Nir-Zvi I., Engelman R., Kessler O., Neufeld G. Class-3 semaphorins and their Receptors: potent multifunctional modulators of tumor progression. Int J Mol Sci. 2019;20:556. doi: 10.3390/ijms20030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aggarwal P.K., Veron D., Thomas D.B., Siegel D., Moeckel G., Kashgarian M., Tufro A. Semaphorin3a promotes advanced diabetic nephropathy. Diabetes. 2015;64:1743–1759. doi: 10.2337/db14-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishibashi R., Takemoto M., Akimoto Y., Ishikawa T., He P., Maezawa Y., Sakamoto K., Tsurutani Y., Ide S., Ide K., Kawamura H., Kobayashi K., Tokuyama H., Tryggvason K., Betsholtz C., Yokote K. A novel podocyte gene, semaphorin 3G, protects glomerular podocyte from lipopolysaccharide-induced inflammation. Sci Rep. 2016;6:25955. doi: 10.1038/srep25955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edeling M., Ragi G., Huang S., Pavenstädt H., Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol. 2016;12:426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhuang Q., Cheng K., Ming Y. CX3CL1/CX3CR1 Axis, as the therapeutic potential in renal diseases: friend or foe? Curr Gene Ther. 2017;17:442–452. doi: 10.2174/1566523218666180214092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lennon R., Randles M.J., Humphries M.J. The importance of podocyte adhesion for a healthy glomerulus. Front Endocrinol. 2014;5:160. doi: 10.3389/fendo.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vivante A., Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016;12:133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartlett C.S., Jeansson M., Quaggin S.E. Vascular growth factors and glomerular disease. Annu Rev Physiol. 2016;78:437–461. doi: 10.1146/annurev-physiol-021115-105412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eremina V., Jefferson J.A., Kowalewska J., Hochster H., Haas M., Weisstuch J., Richardson C., Kopp J.B., Kabir M.G., Backx P.H., Gerber H.-P., Ferrara N., Barisoni L., Alpers C.E., Quaggin S.E. VEGF inhibition and renal thrombotic microangiopathy. N Engl J Med. 2008;358:1129–1136. doi: 10.1056/NEJMoa0707330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Floege J., Eitner F., Alpers C.E. A new look at platelet-derived growth factor in renal disease. J Am Soc Nephrol. 2008;19:12–23. doi: 10.1681/ASN.2007050532. [DOI] [PubMed] [Google Scholar]
- 57.Matsuda M., Shikata K., Makino H., Sugimoto H., Ota K., Akiyama K., Hirata K., Ota Z. Gene expression of PDGF and PDGF receptor in various forms of glomerulonephritis. Am J Nephrol. 1997;17:25–31. doi: 10.1159/000169067. [DOI] [PubMed] [Google Scholar]
- 58.Uehara G., Suzuki D., Toyoda M., Umezono T., Sakai H. Glomerular expression of platelet-derived growth factor (PDGF)-A, -B chain and PDGF receptor-alpha, -beta in human diabetic nephropathy. Clin Exp Nephrol. 2004;8:36–42. doi: 10.1007/s10157-003-0265-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality control metrics of single-cell RNA sequencing data. A and B: Violin plots showing the distribution of the number of unique molecular identifiers (counts), number of genes (features), and percentage of mitochondrial reads (% mito) in each sample before (A) and after (B) filtering.
Clustering of wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) cells by sample and genotype. A: Distribution of all profiled WT and iCTCFpod−/− cells per cluster. B: Distribution of WT and iCTCFpod−/− glomerular cells per cluster. C: Clustering of all WT and iCTCFpod−/− cells colored by number. D: Clustering of WT and iCTCFpod−/− glomerular cells colored by sample number. E: Clustering of all WT and iCTCFpod−/− cells colored by genotype. F: Clustering of WT and CTCFpod−/− glomerular cells colored by genotype.
Expression of cell-type specific markers in wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) cells used for cluster identification in Figure 1. Violin plots of cell type–specific markers in WT and iCTCFpod−/− cells used for cluster identification for all cells (A) or glomerular cells only (B).
Extension of Figure 2. A: Full volcano plot presented in Figure 2, highlighting Rhpn1 and Ctcf. Significantly differentially expressed genes are represented by light gray and red dots. Genes in dark gray are not significantly differentially expressed. Dashed line indicates default parameters: cutoff P = 0.25. B: Dot plot showing the expression of all labeled genes in volcano plots of podocyte 1 cells. Size of the dot represents the percentage of cells expressing the genes. Color represents the mean expression level of the gene in the specified cluster. C: Dot plot showing the expression of Nphs2 and Col4a5 in all clusters.
Extended validation of genes presented in Figure 3. Additional hybridization chain reaction (HCR) images used for quantification of Nphs2 and Col4a5 expression in wild-type (WT) and inducible podocyte-specific CTCF deletion (iCTCFpod−/−) animals. A–D: Replicates of HCR images used for quantification of Nphs2 and Col4a5. White box denotes the section of the 40× image used in the Zoom panel. Scale bars = 50 μm.
Endothelial cells enriched for gene programs important for adhesion and migration. A: Venn diagram of the differentially expressed genes identified in glomerular endothelial cells (GECs) 1 and 2. B: Visualization of the overrepresentation analysis of genes uniquely differentially expressed in GEC-1 presented as a network or enrichment map. Overlapping gene sets cluster together. C: Heat map of all genes from the adhesion and migration gene programs from panel B.
Comparison of inducible podocyte-specific CTCF deletion (iCTCFpod−/−) glomerular endothelial cells (GECs) with human diabetic kidney disease (DKD) GECs. Venn diagram of the differentially expressed genes found in all iCTCFpod−/− GEC clusters and human DKD GEC clusters.