Bacteria do not have organelles. This textbook statement is as simple as it is false. An organelle is defined as a specialized structure inside a cell that has specific functions, such as the mitochondrium, the chloroplast, and the Golgi apparatus in a eukaryotic cell. Bacteria were believed to be devoid of such structures, but this dogma was overturned within the past decades. Many bacteria and archaea do have subcellular structures with a defined function. Compartmentalization of cellular processes as found in bacteria and archaea is achieved by lipid- or protein-bounded organelles (1, 2). Photosynthetic structures such as the thylakoid and chlorosome that have been found by electron microscopy some time ago are probably the best-studied specimens (3). Other examples are the gas vesicles in halophilic archaea (4), the halobacteria; magnetosomes in magnetotactic bacteria (5); intracellular membranes in methylotrophs that harbor the key enzyme of their metabolism, the methane monooxygenase (6); and compartments of largely unknown function in planctomyces (7) as well as in the hyperthermophilic archaeon Ignicoccus hospitalis (8).
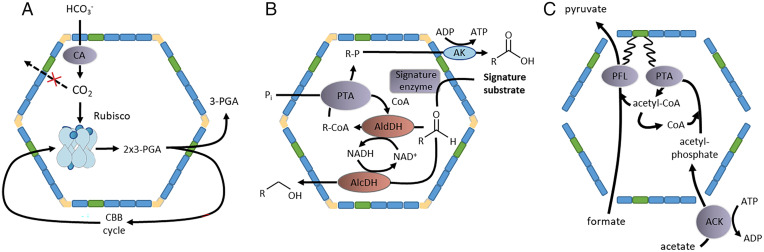
Whereas the aforementioned compartments are found in specialized cells and are not widely distributed over the phylogenetic tree, bacterial microcompartments (BMCs) are. Interestingly, they were discovered already some time ago in cyanobacteria by electron microscopy, and functional analyses revealed them as site of the first step in CO2 fixation. The key enzyme, ribulose-bisphosphate carboxylase (Rubisco), requires CO2 as a substrate, and the cell is faced with the problem of concentrating the gas inside to fit the catalytic demands of Rubisco. This is achieved by compartmentalization of the entire process into one class of BMCs, the carboxysomes. Bicarbonate is transported into the carboxysome, then converted into CO2 by the enzyme carbonic anhydrase, and CO2 is fixed by Rubisco (Fig. 1A). The product of this enzymatic reaction, 3-phosphoglycerate, is transported out of the carboxysome and further metabolized by the reactions of the Calvin cycle (9). Although subcellular structures and carboxysomes have been known for more than half a century, the field has moved with very high speed within the last two decades. Carboxysomes have been found in lithoautotrophic bacteria as well, and in addition to these anabolic BMCs catabolic BMCs with different functions have been described in many different organoheterotrophic bacteria. There, the BMCs also serve to concentrate substrates or to protect the cytoplasm from toxic intermediates of metabolic processes. They are specifically involved in the metabolism of propanediol, ethanolamine, fucose, and rhamnose. The catabolic metabolosomes involved in propanediol utilization (Pdu) or ethanolamine utilization (Eut) house substrate-specific signature enzymes like propanediol dehydratase or ethanolamine ammonia lyase that degrade their corresponding substrates to generate a cytotoxic aldehyde that is separated from the cytoplasm in the BMCs. The aldehyde is further disproportionated by aldehyde-processing enzymes to the corresponding alcohol and carbonic acids in the BMC (Fig. 1B). The genes encoding the BMC structure as well as the metabolic enzyme are usually clustered on the genome and typically bacteria have one such BMC gene cluster that is induced by one substrate (10, 11). Recently, anaerobic acetogenic bacteria were shown to produce BMCs while growing on very different substrates such as organic sources but also during lithotrophic growth on H2 + CO2 (12).
Fig. 1.
Carboxysomes (A), metabolosomes (B), and a synthetic formate- and acetate-utilizing microcompartment (C). The shell is made by BMC-T (green), BMC-H (blue), and BMC-P (yellow). Within the carboxysome, carboanhydrase (CA) catalyzes conversion of bicarbonate to carbon dioxide, which is then fixed by Rubisco to yield two molecules of 2 phyosphoglycerate (PGA). In the metabolosome, an aldehyde is disproportionated. One part is reduced with NADH by an alcohol dehydrogenase (AlcDH) to an alcohol and the other part is oxidized by a CoA-dependent acetaldehyde dehydrogenase (AldDH) to the carboxylic acid with rereduction of NAD. The CoA ester is then converted by PTA to yield the phosphorylated fatty acid, which is then dephosphorylated by a kinase to yield the free fatty acid and ATP by substrate level phosphorylation. The signature substrate is taken up by the BMC and converted by the signature enzyme to the aldehyde. An example would be the dehydration of 1,2 propanediol (signature substrate) by the B12-dependent enzyme propanediol dehydratase (signature enzyme), which yields propionaldehyde that is further disproportionated to propanol and propionic acid. The synthetic formate- and acetate-utilizing BMC engineered by Kirst et al. (16) converts formate and acetyl-CoA by the PFL to pyruvate. Acetate is activated by acetate kinase outside the BMC, and acetyl phosphate enters the BMC and is converted to acetyl-CoA by the PTA. PFL and PTA are directed into the BMC by fusion to BMC-T; therefore, a Spy-tag and Snoop-tag are fused to BMC-T and the PFL and PTA are engineered with a Spy and Snoop catcher, respectively. Acetate is activated by acetate kinase (ACK) outside the BMC. Adapted from refs. 11 and 16.
Synthetic BMCs for Challenging Metabolic Reactions
The proteinaceous shell of BMCs is assembled by mainly three types of shell proteins, BMC-H (hexamers), BMC-T (pseudohexamers), and BMC-P (pentamers) that assemble into an icosahedral shell with a diameter of ca. 400 Å (Fig. 1). Shell proteins are highly conserved across the phylogenetic tree, but the function of the metabolic enzymes and their nature are very different. Shell proteins are selectively permeable and, therefore, the question is how specific transport of metabolites as well as enzymes is achieved. This has not been answered, but signature sequences in the enzymes may be responsible (13).
BMCs protect cells from toxic metabolic intermediates and concentrate substrates inside and allow bacteria to conquer different ecological niches. They have also attracted considerable interest in recent years due to their potential use as small nanoreactors in biotechnology. Despite the many basic questions that remain, several groups have started to engineer synthetic BMCs carrying different signature enzymes (14, 15). In PNAS, Kirst et al. (16) have chosen to engineer a formate-oxidizing BMC. Formate is a highly interesting compound in biotechnology. It can be produced enzymatically or chemically from CO2 and further converted to valuable products by metabolic engineering (17). Naturally formatotrophic aerobic and anaerobic bacteria that are genetically tractable are known. For example, acetogenic bacteria are of special interest. Some can grow on formate and produce added-value compounds from it. Some can also produce formate from H2 + CO2 and thus can be used to store CO2, to store molecular hydrogen, or simply produce formate as feedstock from CO2 (18, 19). However, metabolic engineering in these organisms is hampered by the fact that they grow at thermodynamic equilibrium, which restricts the synthesis of valuable, energy-demanding compounds (20). An alternative is to engineer the metabolism of a versatile bacterium such as Escherichia coli to use formate. Kirst et al. (16) have used an even different approach: to synthesize pyruvate, the central intermediate of bacterial metabolism, from formate and acetate in a synthetic BMC.
Several groups have started to engineer synthetic BMCs carrying different signature enzymes. In PNAS, Kirst et al. have chosen to engineer a formate-oxidizing BMC.
A Synthetic Formate- and Acetate-Utilizing BMC
Three enzymes are required for pyruvate formation from formate and acetate: an acetate-activating enzyme, a phosphotransacetylase (PTA) that forms acetyl-CoA from acetyl-phosphate, and the pyruvate formate lyase (PFL) that condenses acetyl-CoA and formate to pyruvate. PFL uses a glycyl radical for catalysis which is very sensitive to inactivation by oxidation, but the production of an oxygen-sensitive enzyme (hydrogenase) in a BMC was recently shown to be feasible (21). Reactivation after spontaneous oxidation of the PFL is achieved by a PFL-activating enzyme that requires S-adenosylmethionine (SAM) for activity. Therefore, the authors also cloned the activating enzyme as well as a SAM-synthesizing enzyme. To prove function of the enzymes containing tags in vivo they produced these proteins in a mutant of E. coli devoid of PFL and the activating enzymes and indeed observed growth of this strain on acetate and formate. This is a nice physiological study that demonstrates an active system expressed from the plasmids to convert acetate and formate. Also, the backward reaction, oxidation of pyruvate to formate, was possible in E. coli.
The major challenge in this work was successful targeting of PFL and PTA in the microcompartment in an active form. Furthermore, it was essential to clearly demonstrate the activity within the BMCs. To encapsulate both enzymes, the authors fused two different tags to the shell protein BMC-T, a Spy-Tag and a Snoop-Tag, in a way that resulted in an “inward” orientation of the tags, i.e., to the lumen of the BMCs. The corresponding Spy and Snoop catcher domains were fused to PFL and PTA that retained activity. They then expressed together in E. coli the BMC-T variant, BMC-H, but not the pentameric BMC-P. This trick resulted in BMCs devoid of BMC-P, leaving behind little pores in the BMC planned to enhance exchange between the BMC and the cytoplasm (Fig. 1C). Indeed, that was possible, and the resulting structures were called wiffleballs. These were purified by affinity chromatography and shown to also contain the cargo proteins PFL and PTA fused to BMC-T. Then, the critical experiment was done. The proteins were indeed active and produced formate from pyruvate, but yields and activities were low. The only drawback is that pyruvate synthesis from formate and acetyl-CoA was not demonstrated. At any rate, these studies show how intelligent design of microcompartments using two different tags can be used for targeting of the proteins in such a microcompartment. This is exciting and paves the road to use the strategy to engineer BMCs for the production of various compounds from cheap substrates such as formate and acetate. In contrast to the synthetic BMCs first pioneered, which were used for ethanol production (22), polyphosphate storage (23), and hydrogen production (21), the system described here can be used as a platform in ambitious engineering projects to compartmentalize entire metabolic pathways for the production of a biomolecule of interest.
Acknowledgments
Work from the V.M. laboratory on BMCs is funded by an Advanced Grant of the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program (Grant Agreement No. 741791).
Footnotes
The author declares no competing interest.
See companion article, “Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate,” 10.1073/pnas.2116871119.
References
- 1.Greening C., Lithgow T., Formation and function of bacterial organelles. Nat. Rev. Microbiol. 18, 677–689 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Murat D., Byrne M., Komeili A., Cell biology of prokaryotic organelles. Cold Spring Harb. Perspect. Biol. 2, a000422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saer R. G., Blankenship R. E., Light harvesting in phototrophic bacteria: Structure and function. Biochem. J. 474, 2107–2131 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer F., Distribution, formation and regulation of gas vesicles. Nat. Rev. Microbiol. 10, 705–715 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Uebe R., Schüler D., Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 14, 621–637 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hanson R. S., Netrusov A. I., Tsuji K., “The obligate methanotrophic bacteria Methylococcus, Methylomonas and Methylosinus” in The Prokaryotes, Balows E., Trüper H. G., Dworkin M., Harder W., Schleifer K.-H., Eds. (Springer, New York, NY, ed. 2, 1991), vol. 3, pp. 2350–2364. [Google Scholar]
- 7.Wiegand S., Jogler M., Jogler C., On the maverick Planctomycetes. FEMS Microbiol. Rev. 42, 739–760 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Küper U., Meyer C., Müller V., Rachel R., Huber H., Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U.S.A. 107, 3152–3156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L. N., Advances in the bacterial organelles for CO2 fixation. Trends Microbiol., 10.1016/j.tim.2021.10.004 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Bobik T. A., Lehman B. P., Yeates T. O., Bacterial microcompartments: Widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol. Microbiol. 98, 193–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M., Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury N. P., Alberti L., Linder M., Müller V., Exploring bacterial microcompartments in the acetogenic bacterium Acetobacterium woodii. Front. Microbiol. 11, 593467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobik T. A., Stewart A. M., Selective molecular transport across the protein shells of bacterial microcompartments. Curr. Opin. Microbiol. 62, 76–83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M. J., Palmer D. J., Warren M. J., Biotechnological advances in bacterial microcompartments technology. Trends Biotechnol. 37, 325–336 (2019). [DOI] [PubMed] [Google Scholar]
- 15.McDowell H. B., Hoiczyk E., Bacterial nanocompartments: Structures, functions and applications. J. Bacteriol., 10.1128/JB.00346-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirst H., et al. , Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate. Proc. Natl. Acad. Sci. U.S.A. 119, e2116871119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yishai O., Lindner S. N., Gonzalez de la Cruz J., Tenenboim H., Bar-Even A., The formate bio-economy. Curr. Opin. Chem. Biol. 35, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Moon J., et al. , Formate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 23, 4214–4227 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Müller V., New horizons in acetogenic conversion of one-carbon substrates and biological hydrogen storage. Trends Biotechnol. 37, 1344–1354 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Katsyv A., Müller V., Overcoming energetic barriers in acetogenic C1 conversion. Front. Bioeng. Biotechnol. 8, 621166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., et al. , Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production. Nat. Commun. 11, 5448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M. J., et al. , Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm. Nat. Chem. Biol. 14, 142–147 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Liang M., Frank S., Lünsdorf H., Warren M. J., Prentice M. B., Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol. J. 12, 1600415 (2017). [DOI] [PubMed] [Google Scholar]