Significance
Acute myeloid leukemias (AMLs) with NUP98-NSD1 or MLL abnormality are generally aggressive, demanding a better understanding of the underlying oncogenic mechanisms. We show that these AMLs rely on a regulatory axis involving PRC2–|Kdm5b–|stemness genes for sustaining an oncogenic program. The H3K27 methylase activity of polycomb repressive complex 2 (PRC2) is crucial for repressing Kdm5b, a corepressor carrying a H3K4me3 reader domain, that antagonizes the AML oncoproteins by directly binding to and down-regulating the AML stemness genes, thereby suppressing acute leukemogenesis. Such an AML-suppressing role of Kdm5b is not dependent on its intrinsic demethylase activity but requires its scaffold and/or chromatin association functions. The findings of this study shall aid in potential therapeutics of the affected AML patients.
Keywords: PRC2, Kdm5b, stemness, tumorigenicity, AML
Abstract
Acute myeloid leukemias (AMLs) with the NUP98-NSD1 or mixed lineage leukemia (MLL) rearrangement (MLL-r) share transcriptomic profiles associated with stemness-related gene signatures and display poor prognosis. The molecular underpinnings of AML aggressiveness and stemness remain far from clear. Studies with EZH2 enzymatic inhibitors show that polycomb repressive complex 2 (PRC2) is crucial for tumorigenicity in NUP98-NSD1+ AML, whereas transcriptomic analysis reveal that Kdm5b, a lysine demethylase gene carrying “bivalent” chromatin domains, is directly repressed by PRC2. While ectopic expression of Kdm5b suppressed AML growth, its depletion not only promoted tumorigenicity but also attenuated anti-AML effects of PRC2 inhibitors, demonstrating a PRC2–|Kdm5b axis for AML oncogenesis. Integrated RNA sequencing (RNA-seq), chromatin immunoprecipitation followed by sequencing (ChIP-seq), and Cleavage Under Targets & Release Using Nuclease (CUT&RUN) profiling also showed that Kdm5b directly binds and represses AML stemness genes. The anti-AML effect of Kdm5b relies on its chromatin association and/or scaffold functions rather than its demethylase activity. Collectively, this study describes a molecular axis that involves histone modifiers (PRC2–|Kdm5b) for sustaining AML oncogenesis.
Acute myeloid leukemia (AML) is a common form of childhood cancer (affecting patients age 0 to 15 y), and despite improvement, its prognosis remains generally poor. In particular, chromosomal abnormality of NUP98-NSD1 (1–3) and MLL rearrangements (MLL-r) (4, 5) account, respectively, for ∼5 to 16% and 15 to 20% of pediatric AML cases. Acute lymphoblastic leukemia with MLL-r is more prevalent in pediatric patients. These patients with NUP98-NSD1 or MLL-r generally show particularly poor outcomes in the clinic, demanding more effective therapeutics. There is a need to better understand the molecular underpinnings of tumorigenesis and aggressiveness in these AMLs. The translocation of NUP98-NSD1 generates a chimeric protein that fuses the N-terminal segment of NUP98, enriched in the intrinsically disorganized region causing liquid–liquid phase separation (6), with a C-terminal region of NSD1, which contains histone-methyltransferase and chromatin-binding domains (1–3, 7), whereas MLL-r chimeras fuse an N-terminal MLL segment that interacts with Menin and LEDGF with a C-terminal part of an MLL-r fusion partner such as AF9, ENL, or AF4 (4, 5, 8). Previous studies using murine and human AMLs have collectively shown that both NUP98-NSD1 and MLL-r oncoproteins (such as MLL-AF9 and MLL-AF4) directly bind and activate a set of stemness- and proliferation-related gene targets, notably the Hox cluster genes, thereby potentiating AML tumorigenesis (1–3, 5, 7, 8). It remains to be fully understood how these AMLs sustain tumorigenicity.
Chromatin modulation is increasingly appreciated to be pivotal for malignant development (9–12). Previously, it has been shown that MLL-r+ AMLs rely partly on polycomb repressive complex 2 (PRC2) for maintaining tumorigenesis, implicative of an attractive therapeutic target (13–17). However, it remains far from clear which molecular pathways are responsible for oncogenic actions of PRC2 in AML. In this study, we show that NUP98-NSD1+ AML cells are exquisitely sensitive to an inhibitor of PRC2 enzymatic activity, thus extending PRC2 dependency to multiple subtypes of genetically defined AMLs displaying poor prognosis. Our transcriptomic profiling in AML further identified Kdm5b (also known as Jarid1b, Plu-1, or RBP2-H1), a multifunctional demethylase that can remove histone H3 lysine 4 tri/di-methylation (H3K4me3/2), to be a critical downstream target repressed by PRC2. Depletion of Kdm5b not only promoted AML development but also significantly desensitized AML cells to the PRC2 inhibitor, thus uncovering an involvement of the PRC2–|Kdm5b axis for AML tumorigenesis. Genomic profiling by chromatin immunoprecipitation followed by sequencing (ChIP-seq), Cleavage Under Targets & Release Using Nuclease (CUT&RUN), and RNA-seq further showed that both Kdm5b and NUP98-NSD1 are targeted to stemness- and proliferation-related genes and that Kdm5b functions to suppress a suite of AML proto-oncogenes. Systematic mutagenesis showed Kdm5b’s intrinsic demethylase activity to be dispensable for AML suppression, whereas its chromatin-binding functions are essential, indicating a scaffolding role of Kdm5b in assembling the chromatin-bound complex for transcriptional repression. Together, this study unveils a PRC2–|Kdm5b–|stemness axis that operates to promote AML oncogenesis and aggressiveness, which sheds light on potential therapeutic means.
Results
NUP98-NSD1+ AML Cells Are Sensitive to Enzymatic Inhibitor of PRC2.
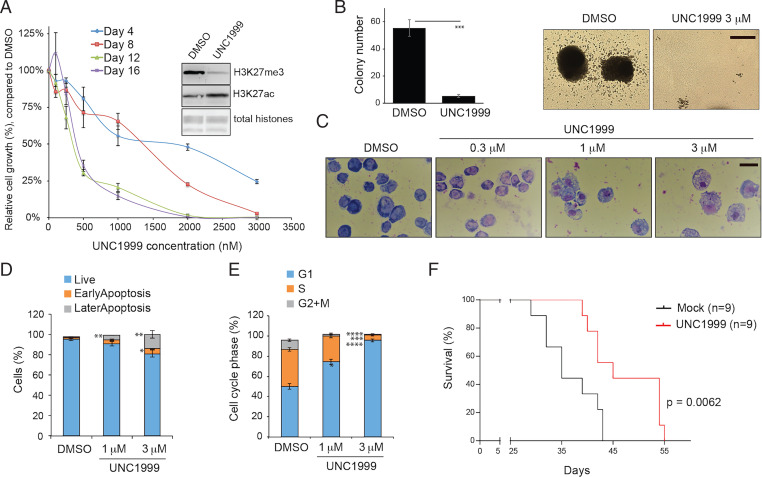
Previously, it has been shown that UNC1999, a selective inhibitor of the PRC2 catalytic subunit (EZH2 or EZH1), suppressed growth of MLL-r AML cells (17). Here, we treated murine AML cells established by NUP98-NSD1 (7) with UNC1999. Following the depletion of global H3K27me3 (Fig. 1A, Inset), we observed a robust response and sensitivity to UNC1999, comparable to what was seen with MLL-r AML (17). Relative to mock treatment, UNC1999 dramatically suppressed the in vitro proliferation (Fig. 1A) and colony formation of NUP98-NSD1+ AML cells (Fig. 1B). Treatment with UNC1999 also resulted in enhanced cell differentiation (Fig. 1C), increased apoptosis (Fig. 1D and SI Appendix, Fig. S1), and slower cell cycle progression (Fig. 1E) in AML cells. Furthermore, when compared to vehicle treatment, UNC1999 treatment also significantly delayed tumor progression in the murine NUP98-NSD1+ AML model (Fig. 1F). Taken together, these results demonstrate a PRC2 dependency for the NUP98-NSD1–associated AML growth.
Fig. 1.
PRC2 sustains tumorigenesis in the NUP98-NSD1+ AML. (A) Proliferation of NUP98-NSD1–transformed murine AML cells after treatment with UNC1999, an enzymatic inhibitor of EZH2/1, for the indicated duration. The y-axis represents the relative percentage of cell numbers in cultures, normalized to dimethyl sulfoxide (DMSO) treatment (n = 3 independent experiments; shown as mean ± SD). Inset shows immunoblotting of global H3K27me3 and H3K27ac after a 24-h treatment with 3 μM of UNC1999, relative to DMSO. (B) Quantification of CFUs (Left) and images of representative single-cell colonies (Right) formed by NUP98-NSD1+ AML cells, cultured in the semisolid medium containing DMSO or 3 μM of UNC1999 for 10 d (n = 3 independent experiments; mean ± SD; unpaired two-tailed Student’s t test). ***P < 0.001. (Scale bar, 2 mm.) (C) Wright–Giemsa staining images showing the differentiation status of NUP98-NSD1+ AML cells, treated with DMSO or the indicated concentration of UNC1999 for 4 d. (Black bar, 10 μm.) (D and E) Summary of apoptotic cells, analyzed by propidium iodide (PI), and annexin-V staining (D), and cell cycle progress, analyzed by PI-based DNA staining (E), in the NUP98-NSD1+ AML cells, following a 2-d treatment with DMSO or the indicated concentration of UNC1999 (n = 3 independent experiments; mean ± SD; unpaired two-tailed Student’s t test). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (F) Kaplan–Meier curve showing kinetics of NUP98-NSD1-induced AML in a transplantation model using syngeneic mice. Starting from day 7 posttransplantation, mice received oral administration of either vehicle (blue) or 100 mg/kg UNC1999 (red) once per day. n, cohort size. Statistical significance was determined by log-rank test.
Transcriptomic Profiling Identified Kdm5b to Be a PRC2-Repressed Gene in AML with NUP98-NSD1 or MLL-r.
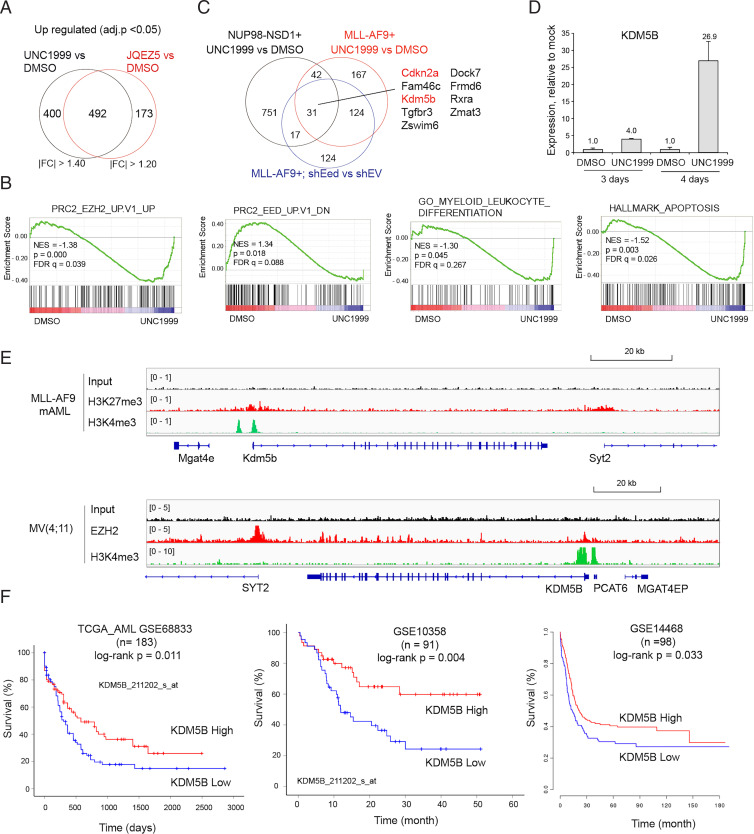
In order to dissect downstream mediators through which PRC2 sustains AML oncogenesis, we conducted transcriptomic profiling of NUP98-NSD1+ AML cells posttreatment with PRC2 inhibitors. To rule out a potential off-target effect, we treated NUP98-NSD1+ AML cells either with UNC1999 or with a second PRC2 inhibitor, JQEZ5 (18, 19), and then performed RNA-seq analysis (Dataset S1 and SI Appendix, Fig. S2A). Replicated RNA-seq profiles within the same treatment group were highly consistent (SI Appendix, Fig. S2B), and transcripts showing the increased expression after treatment with either of the two inhibitors also overlapped significantly (Fig. 2A and SI Appendix, Fig. S2C), although the overall effect of JQEZ5 was somewhat milder (please note a milder overall change in gene expression caused by JQEZ5, compared to UNC1999, as revealed by principal component analysis, PC1 of SI Appendix, Fig. S2B). Gene set enrichment analysis (GSEA) showed the blockade of PRC2’s enzymatic activity by either inhibitor to be correlated with up-regulation of transcripts repressed by PRC2 or those related either to leukocyte differentiation or apoptosis (Fig. 2B and SI Appendix, Fig. S2 D and E), consistent with the observed cell phenotypes posttreatment (Fig. 1). To further identify genes commonly repressed by PRC2 in NUP98-NSD1+ and MLL-r AMLs, we compared UNC1999-derepressed transcripts identified in the two AMLs and, to further enhance the rigor, also included our previous transcriptomic data for knockdown of the essential component of PRC2, Eed (17) (Fig. 2C). This analysis identified a 31-gene signature that is commonly up-regulated among all three comparisons (Fig. 2C and SI Appendix, Table S1) and included Cdkn2a, a cell cycle inhibitor gene known to be directly repressed by PRC2 (17). In addition, Kdm5b, encoding a histone demethylase shown to have a tumor-suppressive role in MLL-r AML (20), was also found to be significantly activated upon PRC2 inhibition (Fig. 2C). The qRT-PCR assays verified the marked up-regulation of Kdm5b following either PRC2 enzymatic inhibition (Fig. 2D) or EZH2 depletion (SI Appendix, Fig. S2F) in independent AML cell lines.
Fig. 2.
Kdm5b, encoding a tumor suppressor of AML, is a direct target of PRC2 and H3K27me3 in AML. (A) Venn diagram showing the overlap between DEGs, as determined by RNA-seq to be up-regulated in NUP98-NSD1+ AML cells after a 4-d treatment with 3 μM of independent PRC2 inhibitors, either UNC1999 (Left) or JQEZ5 (Right), relative to DMSO (n = 3 replicated samples). Threshold of DEG is set as adjusted DESeq calculated probability value (adj.p) < 0.05, the indicated fold-change (FC) cutoff, and mean tag counts >10. (B) GSEA revealing that enzymatic inhibition of PRC2 by UNC1999 is positively correlated to activation of the indicated PRC2-repressed, myeloid differentiation–related, or apoptosis-related gene signature in the NUP98-NSD1+ AML cells. NES, normalized enrichment score; FDR, false discovery rate. (C) Venn diagram showing overlap among the DEGs up-regulated posttreatment of NUP98-NSD1+ (black) or MLL-AF9+ AML cells (red) with UNC1999, or postknockdown of Eed, an essential PRC2 component, in the latter cells, relative to the respective controls. (D) qRT-PCR for Kdm5b in MLL-AF9+ murine AML cells treated with 3 μM of UNC1999 for the indicated duration, relative to DMSO (n = 3 independent experiments; shown as mean ± SD). The y-axis represents PCR signals relative to DMSO-treated cells after normalization to GAPDH. (E) ChIP-seq profiles of H3K27me3 or EZH2 and H3K4me3 at the Kdm5b gene in MLL-AF9+ murine AML (mAML) cells (Upper) and MV4;11 cells, an MLL-AF4+ human AML line (Lower). (F) Kaplan–Meier survival curve for KDM5B expression in the indicated AML patients, including the TCGA cohort (Left). Statistical significance was determined by log-rank test. n, cohort size.
To assess whether Kdm5b is a direct target of PRC2 and H3K27me3, we examined H3K27me3 and H3K4me3 ChIP-seq data that we previously generated in MLL-AF9–transformed murine AML cells (17) and found the Kdm5b promoter to be demarcated with both histone marks (Fig. 2E), which is a characteristic of bivalent domains in chromatin. We also conducted ChIP-seq in MV4;11 cells, a human AML line harboring MLL-AF4, and also found KDM5B to be directly bound by EZH2 and H3K4me3 (Fig. 2E). Interrogation of multiple clinical outcome datasets of human AML patients, including the TCGA AML cohort, consistently displayed a positive correlation between the lower KDM5B expression and the significantly poorer prognosis (Fig. 2F). Collectively, these observations support the view that KDM5B is dynamically regulated, with PRC2-catalyzed H3K27me3 serving as a critical repressor, and that KDM5B expression is a predictive marker of AML prognosis.
Kdm5b Depletion not only Promotes AML Oncogenesis but also Significantly Desensitizes AML Cell Response to PRC2 Inhibition.
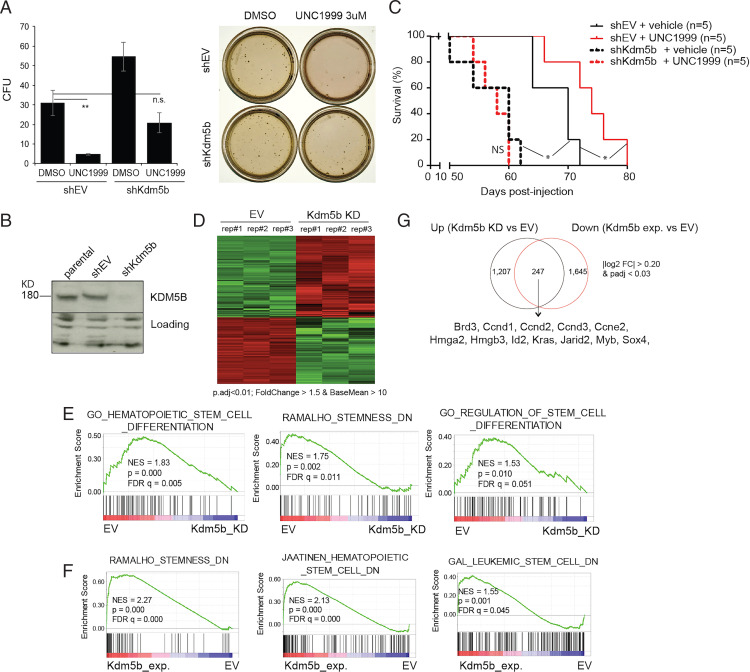
Kdm5b depletion in the NUP98-NSD1+ AML cells resulted in the significantly accelerated proliferation (SI Appendix, Fig. S3A) and increased colony formation in the in vitro assays (Fig. 3 A and B; see DMSO, Right versus Left). Compared to mock treatment, Kdm5b depletion also significantly accelerated AML development in syngeneic mice (Fig. 3C; see black lines, dashed versus solid), consistent with what was previously observed with MLL-r AML (20). Importantly, Kdm5b depletion significantly attenuated the AML cell sensitivity to UNC1999 in both liquid cultures (SI Appendix, Fig. S2B) and colony formation assays (Fig. 3A). Relative to mock treatment, Kdm5b depletion completely abrogated the UNC1999-mediated suppression of AML growth in mice (Fig. 3C; see dashed lines, black versus red). These results demonstrated a tumor-suppressive role for Kdm5b and an oncogenic axis involving PRC2–|Kdm5b in NUP98-NSD1+ AMLs.
Fig. 3.
Depletion of Kdm5b not only renders AML cells unresponsive to PRC2 inhibition but also promotes aggressiveness of AML. (A) Quantification of CFU (Left; n = 3 independent experiments; mean ± SD; unpaired two-tailed Student’s t test) and image of single-cell colonies (Right) formed by NUP98-NSD1+ murine AML cells, transduced with either EV (shEV) or Kdm5b-targeting shRNA (shKdm5b), after a 10-d cultivation in the semisolid medium containing either DMSO or 3 μM of UNC1999. **P < 0.01; n.s., not significant. (B) Immunoblotting of Kdm5b after Kdm5b depletion. (C) Survival curve showing the kinetics of murine AMLs induced by NUP98-NSD1+ cells, which were stably transduced with vector (shEV; solid lines) or Kdm5b-targeting shRNA (shKdm5b; dashed lines), in syngeneic mice (n = 5) treated with either vehicle (black) or UNC1999 (red). Statistical significance was determined by log-rank test. *P < 0.05; NS, not significant. (D) Heatmap of RNA-seq results showing relative expression of DEGs in NUP98-NSD1+ murine AML cells after Kdm5b knockdown (KD), relative to mock (EV). Threshold of DEG is indicated. n = 3 biological replicates (rep.). (E) GSEA shows a negative correlation between Kdm5b knockdown (Kdm5b_KD) and the indicated differentiation-related gene sets in NUP98-NSD1+ murine AML cells (n = 3 replicated samples). (F) GSEA shows a positive correlation between ectopic expression of Kdm5b (Kdm5b_exp.) and the indicated differentiation-related gene sets in MLL-AF9+ murine AML cells (n = 2 replicated samples). (G) Venn diagram showing overlap between the up-regulated transcripts after Kdm5b KD (Left) and the down-regulated transcripts after ectopic Kdm5b expression (Kdm5b exp.) (Right) relative to their respective controls, as determined by RNA-seq in AML cells. AML-related oncogenes and the DEG threshold are labeled. padj, adjusted P values.
RNA-seq Profiling Shows a Role of Kdm5b in Suppressing a Stemness-Related Gene-Expression Program in AML.
To examine the gene-regulatory role for Kdm5b in NUP98-NSD1+ AMLs, we carried out RNA-seq profiling after Kdm5b depletion, which revealed transcripts showing the altered expression due to Kdm5b loss (Fig. 3D and Dataset S2). GSEA using RNA-seq profiles revealed a negative correlation between Kdm5b depletion and the differentiation-related gene sets (Fig. 3E). Furthermore, GSEA of RNA-seq profiles obtained after ectopic Kdm5b expression in MLL-AF9+ AML cells (Dataset S3) revealed a positive correlation between Kdm5b expression and gene sets related to differentiation, apoptosis, or slowed cell proliferation (Fig. 3F and SI Appendix, Fig. S3 D and E). Despite the cell background difference, there is a significant overlap between the Kdm5b-repressed transcripts in the two AML models, including genes, such as Sox4, Id2, Myb, Brd3, Hmga2, and Hmgb3, known to be essential for AML stemness or differentiation arrest (Fig. 3G and SI Appendix, Table S2). Interestingly, oncogenes such as Kras and cell cycle–related genes that include Ccnd1, Ccnd2, and Ccne2 were also found to be repressed by Kdm5b (Fig. 3G). These results thus link Kdm5b to AML-related gene-expression programs.
ChIP-seq Data Establish Direct Cobinding of Kdm5b and NUP98-NSD1 to AML-Related Target Genes, Thus Providing a Molecular Explanation for Antagonism between KDM5B and AML Oncoproteins.
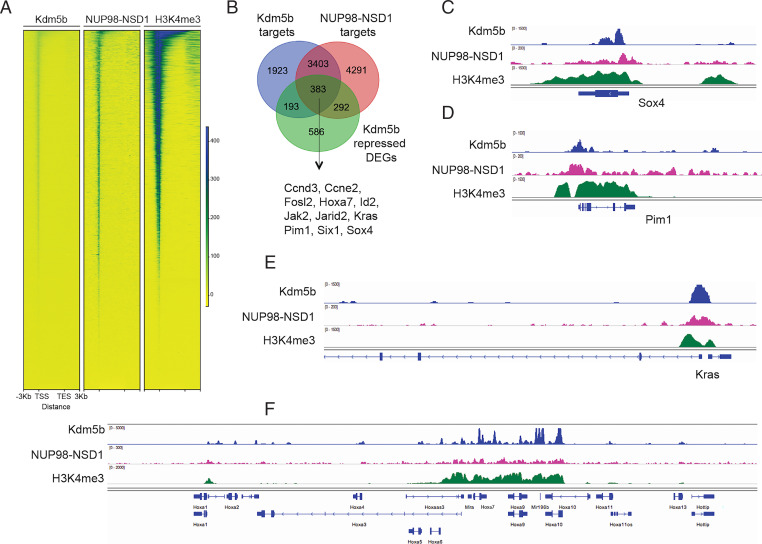
To further address whether Kdm5b directly regulates the AML-related gene program, we conducted ChIP-seq for Kdm5b, NUP98-NSD1 (flag-tagged), and H3K4me3 in NUP98-NSD1+ AML cells (Datasets S4 and S5). Notably, our results revealed a significant coexistence of the three factors at gene promoters (Fig. 4 A and B), which is in good agreement with previous reports that the C-terminal PHD finger domain of Kdm5b directly binds H3K4me3 (21, 22) and that Kdm5b binding sites overlap H3K4me3 in different cell models (23, 24); also, chimeric NUP98 fusion oncoproteins (NUP98-NSD1 or NUP98-PHD) were found targeted to promoters for oncogene activation (7, 25–27). Integrated analyses of our ChIP-seq and RNA-seq datasets in NUP98-NSD1+ AML cells further defined genes that are cotargeted by both Kdm5b and NUP98-NSD1 and also transcriptionally repressed by Kdm5b (Fig. 4B and SI Appendix, Table S3), which again included a suite of AML proliferative and stemness genes such as Brd3, Ccnd3, Kras, Sox4, and Hoxa7 (Fig. 4 C–F and SI Appendix, Fig. S4). These results demonstrate a direct suppressing effect by Kdm5b on the oncogenic gene-expression program, thereby antagonizing AML-promoting effects by oncoproteins.
Fig. 4.
Both Kdm5b and the leukemic oncoprotein, NUP98-NSD1, directly bind to stemness-related and AML-causing genes. (A) Heatmap showing ChIP-seq signals of Kdm5b, Flag-tagged NUP98-NSD1, and H3K4me3 at promoter-proximal regions (±3 kb from transcriptional start site [TSS] of all genes) in NUP98-NSD1+ AML cells. TES, transcriptional end site. (B) Venn diagram showing the overlap among gene targets directly bound by Kdm5b (blue) or NUP98-NSD1 (red) and the Kdm5b-repressed genes (green; based on RNA-seq) in NUP98-NSD1+ AML cells. (C–F) Integrative genomics viewer (IGV) views showing ChIP-seq profiles (input depth–normalized) of Kdm5b, NUP98-NSD1, and H3K4me3 at genes related to AML cell proliferation and stemness such as Sox4 (C), Pim1 (D), Kras (E), and Hox-a (F).
Kdm5b’s Chromatin Association Domains Are Essential for Its AML Suppressive Role, whereas Its Demethylase Activity Is Dispensable.
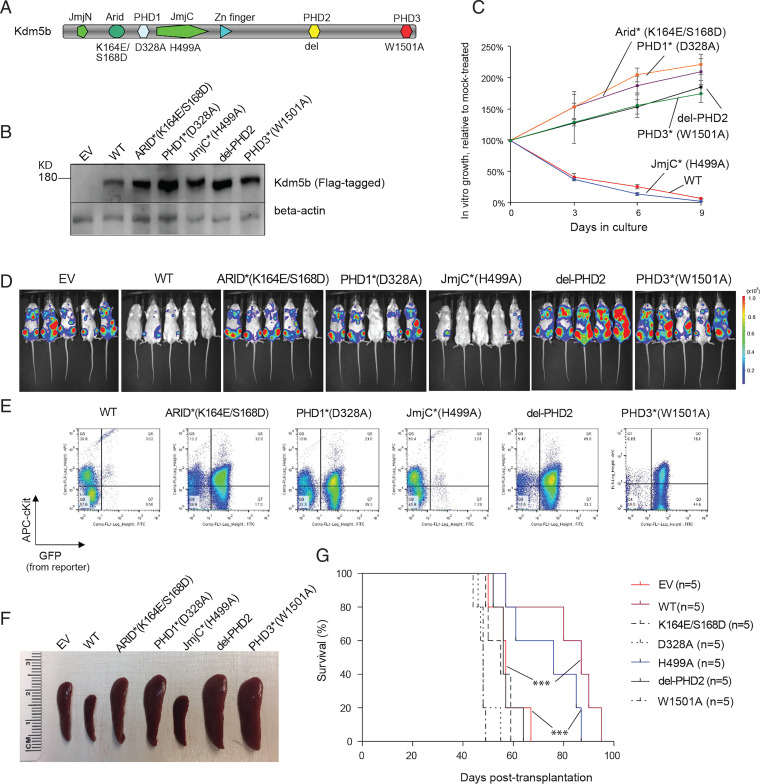
Kdm5b is a large protein with multiple functional motifs. Besides a Jumonji (Jmj) domain, essential for lysine demethylation, Kdm5b contains three PHD fingers, with the first and third ones specifically binding nonmodified (H3K4me0) and highly methylated (e.g., H3K4me3) H3K4, respectively, as well as an ARID domain previously shown to bind CG-rich DNA (28) (Fig. 5A). To examine potential involvement of these functional domains in Kdm5b-mediated AML suppression, we ectopically expressed Kdm5b in the NUP98-NSD1+ AML cells. Here, we employed a set of Kdm5b mutants exhibiting the abolished activity in either chromatin binding or demethylation (Fig. 5A). These mutants included K164E/S168D (in the Arid domain) (28), D328A (in PHD1) (21, 22), H499A (demethylase dead) (29, 30), ΔPHD2 (PHD2 deletion), and W1501A (in PHD3) (22, 26). Wild-type (WT) and mutant forms of Kdm5b showed comparable levels of expression (Fig. 5B). Compared to mock, WT Kdm5b robustly suppressed AML growth in vitro (Fig. 5C; see WT) and in mice, as assessed by assessment of the in vivo expansion of luciferase/GFP-labeled AML cells via live animal imaging and fluorescence-activated cell sorting (FACS) with the isolated cells from killed mice (Fig. 5 D and E; see WT). As a result, the typical splenomegaly seen with the NUP98-NSD1–induced murine AML was suppressed by WT Kdm5b compared to mock, and leukemogenesis was significantly delayed (Fig. 5 F and G; see WT versus empty vector [EV]). These results are in agreement with the positive correlation between the higher KDM5B expression and improved clinical outcomes of AML patients (Fig. 2F). In contrast, all of the tested chromatin-binding mutations, including K164E/S168D (ARID), D328A (PHD1), and W1501A (PHD3), almost completely abolished Kdm5b’s anti-AML effects both in vitro (Fig 5 B and C) and in vivo (Fig. 5 D–G; see chromatin-binding mutants versus WT). Unexpectedly, the Kdm5b H499A mutant, which lacks the H3K4me3 demethylase activity (29, 30), retained the tumor-suppressive activity at a level comparable to what was observed for WT (Fig. 5 C–G; H499A versus WT). These observations in the NUP98-NSD1+ AML model indicate essential requirements for Kdm5b’s chromatin association and/or scaffolding functions for suppressing leukemogenesis, whereas its intrinsic demethylase activity is dispensable.
Fig. 5.
The chromatin association and/or scaffolding function of Kdm5b is essential for its tumor-suppressive effect in AML, whereas its intrinsic demethylase activity is dispensable. (A) Protein architecture of Kdm5b, with its functional domains and used mutations labeled on the Top and Bottom, respectively. del, deletion. (B) Anti-Flag immunoblotting of WT and mutant Flag-tagged Kdm5b after stable expression in cells. (C) Proliferation of NUP98-NSD1+ murine AML cells after stable transduction of WT or mutant Kdm5b, relative to EV. The y-axis represents relative percentage of cell numbers normalized to EV-expressing controls (n = 3 independent experiments; presented as mean ± SD). Star indicates the domain mutation. (D–G) Effect of ectopically expressed Kdm5b, either WT or mutant, on AML leukemogenesis. NUP98-NSD1+ murine AML cells carrying a luciferase-IRES-GFP reporter were stably transduced with the indicated Kdm5b and then transplanted intravenously into syngeneic mice. Leukemogenesis was monitored by chemiluminescence imaging of live animals (30 d posttransplantation; D), FACS (E) of the isolated bone marrow cells (cKit+GFP+ indicates in vivo expansion of GFP-labeled NUP98-NSD1+ AML cells), measurement (F) of spleen size (50 d posttransplantation when leukemic mice were close to their terminal stage), and survival curves (G) of mice transplanted with the indicated NUP98-NSD1+ AML cells (n = 5 per cohort). ***P < 0.001. KD, knockdown.
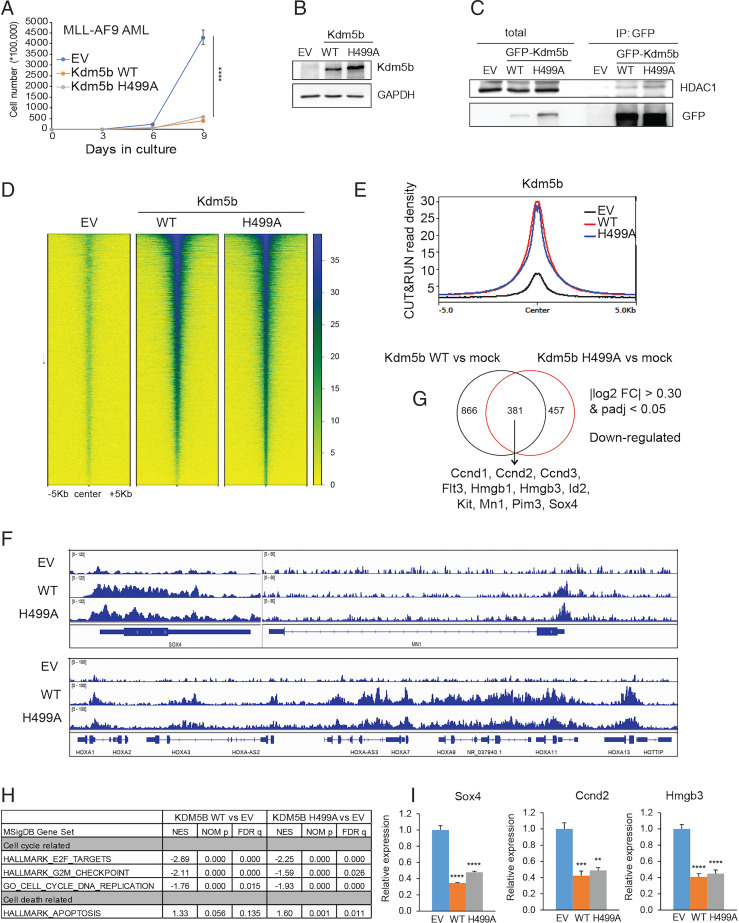
To further assess the role of the Kdm5b demethylase activity during leukemogenesis, we employed MLL-AF9–transformed AML as a second model and confirmed comparable suppressive effects by WT Kdm5b and its H499A mutant on AML cell proliferation when compared to mock (Fig. 6 A and B). It has been reported that Kdm5b or related Kdm5a associates with the nucleosome remodeling and deacetylase (NuRD) complex, which has histone deacetylase (such as HDAC1) and gene-repressive activities (22, 31, 32). Coimmunoprecipitation (CoIP) showed that both WT and H499A-mutated Kdm5b associated with HDAC1 (Fig. 6C), a strong transcriptional corepressor. These results indicate a scaffolding role, rather than an intrinsic demethylase function, for Kdm5b in suppressing AML. Furthermore, we compared the genome-targeting and gene-modulatory effects by WT Kdm5b versus an enzymatic-dead mutant (H499A). First, we introduced either WT or mutant forms of Kdm5b into HEK293 cells because this cell line is generally insensitive to Kdm5b expression and thus allows comparison of WT versus mutant Kdm5b without being affected by cell status changes (such as differentiation and proliferation seen with AML). CUT&RUN results for WT and H499A-mutated Kdm5b revealed comparable binding at target genes (Fig. 6 D and E), as exemplified by Kdm5b peaks at AML-related genes such as MN1, HOX-A cluster genes, SOX4, and CCND2/3 (Fig. 6F and SI Appendix, Fig. S5A). Complementary to the genomic binding results, RNA-seq analyses further confirmed that gene-modulatory effects of WT and H499A-mutated Kdm5b are largely comparable in MLL-AF9+ murine AML cells (Dataset S6), with there being a significant overlap between differentially expressed genes (DEGs), either down- or up-regulated, following stable expression of either form of Kdm5b (Fig. 6G and SI Appendix, Fig. S5B). Again, GSEA of RNA-seq datasets confirmed that both WT Kdm5b and its H499A mutant show a positive correlation with the slowed cell cycle progression and increased cell death (Fig. 6H and SI Appendix, Fig. S5C). Equally repressive effects of WT and H499A-mutated Kdm5b on the tested AML oncogenes (Sox4, Ccnd2, and Hmgb3) were also verified by qRT-PCR (Fig. 6I).
Fig. 6.
Kdm5b suppresses AML-related gene targets in a demethylase-independent manner. (A) Proliferation of MLL-AF9+ murine AML cells transduced with EV or Flag-tagged Kdm5b, either WT or H499A-mutated. The y-axis represents accumulative cell numbers (n = 3 independent experiments; mean ± SD). ****P < 0.0001. (B) Immunoblotting for exogenously expressed Kdm5b using cells in (A). (C) CoIP for HDAC1 interaction with GFP-tagged Kdm5b, WT, or H499A-mutated, in HEK293 cells. IP, immunoprecipitation. (D and E) Heatmap (D) and averaged plotting (E) of GFP CUT&RUN signals for WT and H499A-mutated Kdm5b (GFP-tagged) at the peak-centered genomic regions (± 5 kb) in the HEK293 stable expression cells. Non–GFP-tagged cells (EV) act as a background control. (F) IGV views showing WT and H499A-mutated Kdm5b CUT&RUN signals at the indicated gene in the HEK293 stable expression cells. (G) Venn diagram showing the overlap of DEGs, as determined by RNA-seq to be down-regulated in MLL-AF9+ murine AML cells posttransduction of WT (Left) or H499A-mutated (Right) Flag-Kdm5b, relative to EV (n = 2 replicated samples). AML-related oncogenes and the DEG threshold are indicated. (H) GSEA reveals that relative to EV, expression of WT (Left) or H499A-mutated (Right) Kdm5b is negatively correlated to activation of proliferation-related gene sets in MLL-AF9+ AML cells. NOM p, normalized p-values. (I) qRT-PCR of the indicated AML-related genes in MLL-AF9+ murine AML cells after ectopic expression of WT or H499A-mutated Kdm5b. The qRT-PCR signals are relative to EV-transduced cells after being normalized to GAPDH (n = 3 independent experiments; presented as the mean ± SD). **P < 0.01; ***P < 0.001; ****P < 0.0001. FC, fold-change.
Taken together, our results demonstrate a striking similarity between WT Kdm5b and its demethylase-dead mutant in terms of AML growth suppression, chromatin binding, cofactor interaction (such as HDAC1), and gene-expression modulation.
Discussion
AML is an aggressive disease, demanding a better appreciation of the underlying oncogenic mechanisms and improved therapy. Expression of the stem-like gene-expression program in AML is known to be well correlated with poor prognosis (33, 34). How such a transcriptomic feature is maintained in AML has remained unclear. Chromatin modulation is increasingly appreciated as a major molecular determinant for the gene-expression patterns and cellular identities, and its dysregulation is frequent in cancer, including AML (9–12, 26, 35). Among various chromatin-modulatory players, PRC2 catalyzes formation of H3K27me3, leading to the silenced chromatin state. Involvement of PRC2 and H3K27me3 in tumorigenesis has been demonstrated with several cancer models, in which the overexpression and/or activating mutation of EZH2 or PRC2 cofactor promotes development of cancers such as AML (13–17, 36, 37), B cell lymphoma (38), melanoma (18), lung cancer (19), and multiple myeloma (39). Here, we report that, in AML subtypes with adverse outcomes (NUP98-NSD1+ or MLL-r+), a PRC2–|Kdm5b axis functions to sustain AML tumorigenicity. Integrated profiling showed that Kdm5b and oncogenic AML proteins cobind target genes related to stemness and proliferation, where Kdm5b acts to repress such an oncogenic program. A role for Kdm5b in regulating stemness genes in normal hematopoietic stem/progenitor cells has been reported (40). Consistent with our AML animal data, interrogation of multiple clinical datasets of AML patients supported a strong correlation of the reduced KDM5B expression with poorer prognosis. This work not only uncovered a PRC2–|Kdm5b pathway underlying AML tumorigenicity and aggressiveness but also provides an explanation for the anti-AML effect of the PRC2 catalytic inhibitors. It is noteworthy to mention that this axis of PRC2–|Kdm5b–|oncogenic/stemness transcripts can cooperate with the previously reported pathways enforced by PRC2 and H3K27me3 readers, such as silencing of tumor suppressors (41) and immunity-related genes (42), which collectively establish aggressive features seen in AML (Fig. 7).
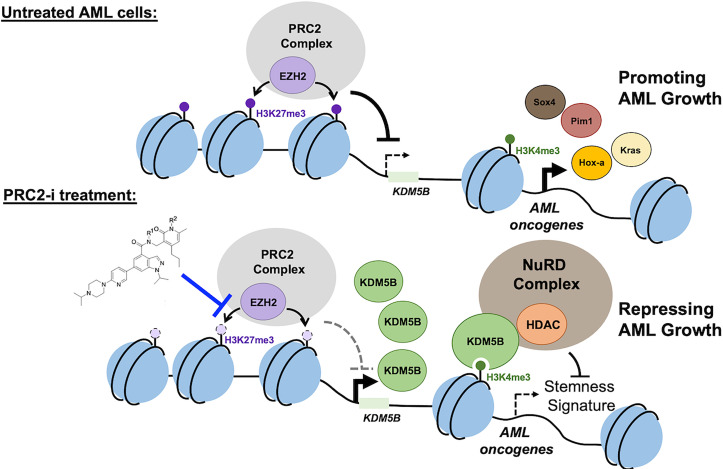
Fig. 7.
Scheme showing a role for the PRC2-|KDM5B axis in mediating AML tumorigenesis. Kdm5b is subject to epigenetic repression by PRC2 and H3K27me3, and such a Kdm5b repression is required for sustaining the expression of AML-causing and stemness-related genes, promoting the leukemogenesis initiated by NUP98-NSD1 and MLL-r. Upon treatment with PRC2 inhibitors (PRC2-i) such as UNC1999, the Kdm5b level is elevated. Then, Kdm5b assembles a transcriptional repressive machinery (with HDAC1 and others), leading to the down-regulation of stemness genes and suppression of AML growth. In this scenario, Kdm5b’s gene-repressing and anti-AML effects are largely demethylase-independent.
While Kdm5b’s chromatin association domains (ARID, PHD1, and PHD3 domains) are essential for suppressing AML tumorigenesis, its demethylation activity is unexpectedly dispensable for the anti-AML effects of Kdm5b both in vitro and in vivo. In this regard, a recent investigation pointed to a role for KDM5B in suppressing the expression of endogenous retroelements such as MMVL30 in mouse melanoma models (43), with the immunity-inhibiting effect of KDM5B being mediated by recruitment of the H3K9 methyltransferase SETDB1 and not by its intrinsic demethylase activity. Thus, the target-repressing activities of KDM5B are often demethylase-independent, as seen at endo-retrovirus elements in melanoma (43) and stemness genes in AML. We further show that Kdm5b’s genome-targeting and cofactor (HDAC1) interaction are unaffected by an enzymatic-dead mutation (H499A). We thus favor a model in which Kdm5b functions as a scaffold protein to mediate the recruitment and/or assembly of a gene-repressive complex (HDAC-containing SIN3B and NuRD or other gene-repressive complexes) at AML oncogenes, thereby attenuating the leukemogenesis.
KDM5’s PHD1 and PHD3 carry histone tail-binding activities (22, 26). It has been suggested that the KDM5 PHD2 may serve as a part of the structural linker in forming a “dumbbell shaped and curved” KDM5B architecture (44), although its exact role merits further investigation. The PHD3-containing fragment of KDM5A (a functionally related KDM5B family protein) was found to be recurrently fused to NUP98 in a subset of AMLs, driving AML development (26, 45). In this scenario, the resultant NUP98-KDM5A(PHD3) oncoproteins (or similar chimeras) bind H3K4me3 (via the H3K4me3-binding PHD3) and can form phase-separated protein condensate (6), leading to maintenance of high expression of a suite of stemness and leukemogenic genes (25–27). Thus, the H3K4me3/2-binding activity harbored within PHD3 is important for both NUP98-KDM5A fusion and normal KDM5B. It is very likely that disruption of one normal KDM5A allele by a NUP98-KDM5A chromosomal translocation contributes to the aggressive features of AML, which requires additional investigation. Interestingly, interrogation of KDM5B and KDM5A using cBioPortal revealed low rates of deletion or somatic mutation among patients of myeloid malignancy and AML (SI Appendix, Fig. S6A). Mutations of these functionally related genes show a tendency of enrichment at a region encoding the chromatin-binding and structural domains (SI Appendix, Fig. S6 B and C), such as PHD3 and a PLU1 domain suggested to be a structural “tower region” (44).
Deregulation of lysine demethylases is a recurrent theme in cancers (12, 46). KDMs may have both tumor-promoting and -suppressive roles. On the one hand, KDM5 family proteins can be oncogenic in solid tumors, including breast cancer and melanoma (47–49). As an example, KDM5B can silence tumor immunity-associated pathways (43). On the other hand, tumor-suppressing roles for KDM5 have been demonstrated in solid tumors such as breast cancer (22, 31), consistent with its AML-suppressing function shown in this work. The functions of KDM5 proteins are multifaceted and subject to further investigation.
Materials and Methods
Plasmid construction, tissue culture and cell lines, antibodies and Western blotting, chemicals, phenotypic assays of hematological cells, such as colony-forming units (CFUs), Wright-Giemsa staining and proliferation assays, assays of cell cycle progression and apoptosis, the in vivo leukemogenic studies, RNA sequencing and data analysis, ChIP-seq and data analysis, CUT&RUN and data analysis, analysis of publicly available patient datasets, qRT-PCR, and statistics and reproducibility can be found in SI Appendix, Materials and Methods. All animal experiments were performed according to the protocol approved by Institutional Animal Care and Use Committee, University of North Carolina UNC).
Supplementary Material
Acknowledgments
We thank Drs. Qin Yan and David L. Bentley for providing reagents used in the study and all laboratory members for discussion and technical support. We sincerely thank UNC facilities, including the High-Throughput Sequencing Facility, Bioinformatics Core, Flow Cytometry Core, Tissue Culture Facility, and Animal Studies Core, for their professional assistance of this work. The cores affiliated with the UNC Cancer Center are supported in part by the UNC Lineberger Comprehensive Cancer Center Core Support Grant P30-CA016086. The publicly available The Cancer Genome Atlas (TCGA) dataset used here is in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. This work was supported by NIH (CA178765, to R.G.R.), the Leukemia and Lymphoma Society Specialized Center of Research Grant (17403-19, to R.G.R.), the Gilead Sciences Research Scholars Program in hematology/oncology (to G.G.W.), When Everyone Survives Leukemia Research Foundation (to G.G.W.), the Ministry of Science and Technology in Taiwan (MOST 107-2320-B-010-024-MY3 and 110-2320-B-A49A-533-MY3, to W.-Y.C.), and the Cancer Progression Research Center, National Yang Ming Chiao Tung University, from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan (to W.-Y.C.).
Footnotes
Reviewers: H.J., University of Virginia; and Z.Q., University of Florida.
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122940119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. RNA-seq, ChIP-seq, and CUT&RUN datasets related to this work have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE179826.
References
- 1.Shiba N., et al. , NUP98-NSD1 gene fusion and its related gene expression signature are strongly associated with a poor prognosis in pediatric acute myeloid leukemia. Genes Chromosomes Cancer 52, 683–693 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Hollink I. H., et al. , NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 118, 3645–3656 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ostronoff F., et al. , NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: A COG and SWOG report. Blood 124, 2400–2407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balgobind B. V., Zwaan C. M., Pieters R., Van den Heuvel-Eibrink M. M., The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia 25, 1239–1248 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Rao R. C., Dou Y., Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn J. H., et al. , Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G. G., Cai L., Pasillas M. P., Kamps M. P., NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 9, 804–812 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Krivtsov A. V., Armstrong S. A., MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7, 823–833 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Shih A. H., Abdel-Wahab O., Patel J. P., Levine R. L., The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer 12, 599–612 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Xu B., Konze K. D., Jin J., Wang G. G., Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp. Hematol. 43, 698–712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi P., Allis C. D., Wang G. G., Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S., Allis C. D., Wang G. G., The language of chromatin modification in human cancers. Nat. Rev. Cancer 21, 413–430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J., et al. , The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene 32, 930–938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neff T., et al. , Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. U.S.A. 109, 5028–5033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W., et al. , Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 9, 643–650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka S., et al. , Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 120, 1107–1117 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Xu B., et al. , Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 125, 346–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souroullas G. P., et al. , An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 22, 632–640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H., et al. , Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov. 6, 1006–1021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong S. H., et al. , The H3K4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell 28, 198–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., et al. , The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B. Protein Cell 5, 837–850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein B. J., et al. , The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 6, 325–335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto S., et al. , JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell 25, 762–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidder B. L., Hu G., Zhao K., KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol. 15, R32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., et al. , Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion. Nat. Commun. 11, 3339 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G. G., et al. , Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gough S. M., et al. , NUP98-PHF23 is a chromatin-modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD histone reader function. Cancer Discov. 4, 564–577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu S., et al. , The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 15, 419–421 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Seward D. J., et al. , Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 14, 240–242 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Wong P. P., et al. , Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A). Mol. Cell. Biol. 32, 1633–1644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q., et al. , Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 71, 6899–6908 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Nishibuchi G., et al. , Physical and functional interactions between the histone H3K4 demethylase KDM5A and the nucleosome remodeling and deacetylase (NuRD) complex. J. Biol. Chem. 289, 28956–28970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eppert K., et al. , Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17, 1086–1093 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Shlush L. I., et al. , Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Dawson M. A., Kouzarides T., Huntly B. J., Targeting epigenetic readers in cancer. N. Engl. J. Med. 367, 647–657 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Danis E., et al. , Inactivation of Eed impedes MLL-AF9-mediated leukemogenesis through Cdkn2a-dependent and Cdkn2a-independent mechanisms in a murine model. Exp. Hematol. 43, 930–935.e6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiel A. T., Feng Z., Pant D. K., Chodosh L. A., Hua X., The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia. Haematologica 98, 918–927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Béguelin W., et al. , EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren Z., et al. , PHF19 promotes multiple myeloma tumorigenicity through PRC2 activation and broad H3K27me3 domain formation. Blood 134, 1176–1189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cellot S., et al. , RNAi screen identifies Jarid1b as a major regulator of mouse HSC activity. Blood 122, 1545–1555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K. H., Roberts C. W., Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burr M. L., et al. , An evolutionarily conserved function of polycomb silences the MHC Class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell 36, 385–401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S. M., et al. , KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 598, 682–687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorosz J., et al. , Molecular architecture of the Jumonji C family histone demethylase KDM5B. Sci. Rep. 9, 4019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Rooij J. D., et al. , NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 27, 2280–2288 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Klose R. J., Kallin E. M., Zhang Y., JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Hinohara K., et al. , KDM5 Histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 34, 939–953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinogradova M., et al. , An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12, 531–538 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Roesch A., et al. , Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23, 811–825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. RNA-seq, ChIP-seq, and CUT&RUN datasets related to this work have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE179826.