Significance
The mitotic checkpoint system is essential for the prevention of mistakes in the segregation of chromosomes in mitosis. As long as chromosomes are not attached correctly to the mitotic spindle, a mitotic checkpoint complex (MCC) is assembled and inhibits the action of ubiquitin ligase APC/C (anaphase-promoting complex/cyclosome) to initiate anaphase. When the checkpoint is turned off, MCC is disassembled, allowing anaphase initiation. The mechanisms of MCC disassembly have been studied, but the regulation of this process remained obscure. We found that a second ubiquitin ligase, UBR5 (ubiquitin-protein ligase N-recognin 5), ubiquitylates MCC components and stimulates the disassembly of MCC from APC/C, as well as the dissociation of a subcomplex of MCC.
Keywords: cell cycle, mitosis, ubiquitin
Abstract
The mitotic (or spindle assembly) checkpoint system ensures accurate chromosome segregation in mitosis by preventing the onset of anaphase until correct bipolar attachment of sister chromosomes to the mitotic spindle is attained. It acts by promoting the assembly of a mitotic checkpoint complex (MCC), composed of mitotic checkpoint proteins BubR1, Bub3, Mad2, and Cdc20. MCC binds to and inhibits the action of ubiquitin ligase APC/C (anaphase-promoting complex/cyclosome), which targets for degradation regulators of anaphase initiation. When the checkpoint system is satisfied, MCCs are disassembled, allowing the recovery of APC/C activity and initiation of anaphase. Many of the pathways of the disassembly of the different MCCs have been elucidated, but the mode of their regulation remained unknown. We find that UBR5 (ubiquitin-protein ligase N-recognin 5) is associated with the APC/C*MCC complex immunopurified from extracts of nocodazole-arrested HeLa cells. UBR5 binds to mitotic checkpoint proteins BubR1, Bub3, and Cdc20 and promotes their polyubiquitylation in vitro. The dissociation of a Bub3*BubR1 subcomplex of MCC is stimulated by UBR5-dependent ubiquitylation, as suggested by observations that this process in mitotic extracts requires UBR5 and α−β bond hydrolysis of adenosine triphosphate. Furthermore, a system reconstituted from purified recombinant components carries out UBR5- and ubiquitylation-dependent dissociation of Bub3*BubR1. Immunodepletion of UBR5 from mitotic extracts slows down the release of MCC components from APC/C and prolongs the lag period in the recovery of APC/C activity in the exit from mitotic checkpoint arrest. We suggest that UBR5 may be involved in the regulation of the inactivation of the mitotic checkpoint.
The accurate segregation of chromosomes in mitosis is ensured by a surveillance mechanism called the mitotic (or spindle assembly) checkpoint system (reviewed in 1–5). This system monitors the lack of correct bipolar attachment of sister chromatids by their kinetochores to the mitotic spindle and thus prevents premature or faulty chromosome separation. Its action is mediated by a mitotic checkpoint complex (MCC), consisting of mitotic checkpoint proteins BubR1, Bub3, Mad2, and Cdc20. MCC inhibits the action of the ubiquitin ligase anaphase promoting complex/cyclosome (APC/C), which targets for degradation inhibitors of anaphase initiation, cyclin B, and securin (6). A sequence of events, which leads to MCC assembly when the checkpoint is turned on, is initiated by phosphorylation of specific sites on unattached kinetochores, which then serve as docking sites for Bub3. This promotes the recruitment to kinetochores of Bub3-binding proteins BubR1 and Bub1. In turn, Bub1 recruits to kinetochores downstream mitotic checkpoint protein components such as Mad1 and Mad2. An important step in the assembly of MCC is the conversion of Mad2 from an inactive, open conformation (O-Mad2) to an active closed conformation (C-Mad2), the latter of which binds to Cdc20 in the assembled MCC (3–5).
After all chromosomes are correctly attached to the mitotic spindle in metaphase, the checkpoint is satisfied and MCC is disassembled to allow APC/C activation, which is necessary for anaphase initiation. We have been studying the mechanisms of MCC disassembly and checkpoint inactivation by the use of a cell-free system that recapitulates these processes in vitro. In this system, soluble extracts derived from nocodazole-arrested cells are incubated with adenosine triphosphate (ATP), leading to increased APC/C activity after a time lag that corresponds to the disassembly of MCC (7, 8). MCC exists in free and APC/C-bound pools, both of which have to be disassembled for checkpoint inactivation (9). The disassembly of MCC in both pools requires ATP (8), but their pathways are different. Furthermore, different moieties of free MCC are dissociated by different processes. Thus, in free MCC, Mad2 is released from Cdc20 by the joint action of the Mad2-binding protein p31comet and the AAA-ATPase TRIP13 (10, 11). In this process, C-Mad2 is converted back to inactive O-Mad2, which is incapable of assembly into MCC (10, 12). BubR1 is dissociated from Cdc20 in free MCC by the phosphorylation of Cdc20 (13) and by the action of the CCT/TRiC chaperonin (14). By contrast, the disassembly of APC/C-bound MCC requires APC/C-catalyzed ubiquitylation of its Cdc20 (15, 16) and BubR1 (16) components. In this process, Cdc20 and BubR1 are dissociated from each other and from APC/C (16).
More recently, we have turned our attention to the problem of how the different pathways of MCC disassembly are regulated. Thus, the disassembly of free MCC appears to be subject to negative regulation by phosphorylation of p31comet by Polo-like kinase 1 (17). In the present investigation, we searched for possible regulatory factors that may influence the disassembly of APC/C-bound MCC and therefore followed up the observation that UBR5 (ubiquitin protein ligase N-recognin 5) (18), an evolutionarily conserved enzyme involved in a variety of cellular processes (reviewed in 19), is associated with APC/C-MCC. UBR5 binds to and ubiquitylates mitotic checkpoint proteins BubR1, Bub3, and Cdc20 and the disassembly of Bub3*BubR1 subcomplex is stimulated by UBR5-mediated ubiquitylation. The removal of UBR5 from mitotic extracts slows down the release of MCC components from APC/C and prolongs the lag period of APC/C activation in exit from the mitotic checkpoint. It thus seems that UBR5 may be involved in the regulation of the inactivation of the mitotic checkpoint.
Results
UBR5 Interacts with Mitotic Checkpoint Proteins and Promotes Their Ubiquitylation.
Initially, we have searched for factors that interact with the APC/C*MCC complex and thus may regulate its action in the mitotic checkpoint. For this purpose, the APC/C*MCC complex was immunopurified from extracts of nocodazole-arrested HeLa cells with an antibody against the Cdc27 subunit of APC/C and was subjected to mass spectrometry analysis. In addition to expected proteins, such as APC/C subunits and MCC components, we noted the presence of UBR5 associated with APC/C (SI Appendix, Table S1). UBR5 is an evolutionarily conserved, N-degron recognition-domain containing enzyme (18) in metazoans that has a variety of essential roles in cellular regulation (reviewed in 19). A possible role of UBR5 in mitosis was suggested by the observation that UBR5 coimmunoprecipitated in mammalian cells with MCC components, such as Bub3, BubR1, and Cdc20 (20). It was furthermore reported that small interfering RNA-mediated knockdown of UBR5 abrogated the accumulation of G2/M cells in response to nocodazole, suggesting a role in G2/M or mitotic checkpoint (20). In contrast, the results of another study showed that UBR5 knockdown prolonged metaphase-anaphase transition, consistent with a role of this enzyme in the inactivation of the mitotic checkpoint (21). The latter authors also reported that UBR5 interacts directly with Bub3 and promotes its ubiquitylation and degradation in cells (21).
To examine the role of UBR5 in mitotic checkpoint, we raised a polyclonal antibody directed against UBR5 and used it to immunoprecipitate this enzyme, along with associated proteins, from extracts of HeLa cells arrested in mitosis. We confirmed that in mitotic extracts, UBR5 associates with all MCC components, BubR1, Cdc20, Bub3, and Mad2 (Fig. 1A, lane 3). Some of these interactions could be indirect, due to the association of MCC components with each other and with APC/C in mitosis. However, we furthermore found that in extracts from asynchronous cells, in which MCC levels are much lower than in mitosis, the extent of coimmunoprecipitation of all MCC components was quite similar to that in mitotic extracts (Fig. 1A, lane 6). This raised the possibility that some MCC components may interact directly with UBR5 and not indirectly through their association with MCC.
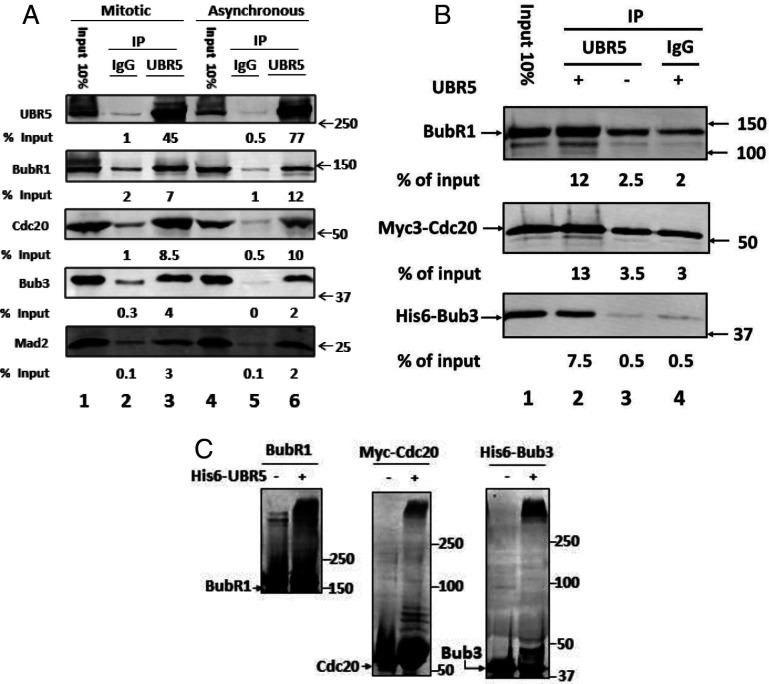
Fig. 1.
UBR5 binds to mitotic checkpoint proteins and promotes their ubiquitylation. (A) Binding of mitotic checkpoint proteins to UBR5 in extracts from mitotic and asynchronous HeLa cells. Extracts from nocodazole-arrested (“mitotic”) or logarithmically growing (“asynchronous”) HeLa cells were prepared as described previously (7). Immunoprecipitation (IP) of extracts with an anti-UBR5 antibody or with nonimmune rabbit IgG was carried out as described in SI Appendix, SI Materials and Methods. Results are expressed as the percentage of immunoprecipitated proteins relative to the corresponding inputs. Inputs show 10% of the amounts of the indicated proteins in mitotic or asynchronous extracts used for immunoprecipitation. Numbers on the Right indicate the electrophoretic migration of marker proteins (in kDa). (B) Binding of UBR5 to individual recombinant mitotic checkpoint proteins. The indicated recombinant mitotic checkpoint proteins (100 nM) were incubated with recombinant his6-UBR5 (at a quantity similar to that in 40 μg protein of mitotic HeLa cell extract) in a buffer containing: 50 mM Tris⋅HCl (pH 7.2), 20% (vol/vol) glycerol, 1 mg/mL bovine serum albumin (BSA), and 1 mM dithiothreitol (DTT). Following incubation for 1 h at 23 °C, the samples were immunoprecipitated with anti-UBR5 polyclonal antibody or with nonimmune rabbit IgG. Immunoprecipitated material was resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted for the indicated proteins. Results are expressed as the percentage of immunoprecipitated material relative to input. Numbers on the Right (kDa) indicate the electrophoretic migration position of marker proteins. (C) Ubiquitylation of individual mitotic checkpoint proteins by recombinant UBR5. The indicated purified recombinant checkpoint proteins (100 nM, each) were incubated in a volume of 30 μL with a ubiquitylation mixture consisting of 40 mM Tris⋅HCl (pH 7.6), 5 mM MgCl2, 1 mg/mL BSA, 1 mM DTT, 150 nM E1 (Enzo BML-UW9410), 600 nM UbcH5ɑ/UBE2D1 (Boston Biochem E2-616), 100 μM ubiquitin, and 2 mM adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP), in the presence or absence of recombinant his6-UBR5 (supplemented at a quantity similar to its amount in 40 μg protein of HeLa cell extract). Following incubation at 37 °C for 1 h, samples were subjected to SDS-PAGE and immunoblotting for the indicated proteins. Numbers on the Right indicate the electrophoretic migration position of marker proteins.
To examine this problem more directly, we expressed and purified full-length recombinant his6-UBR5; incubated it with individual recombinant BubR1, Bub3, and Cdc20 proteins; and estimated their association with UBR5 by immunoprecipitation with anti-UBR5. As shown in Fig. 1B, lane 2, all the separate mitotic checkpoint proteins significantly coimmunoprecipitated with recombinant UBR5. Control immunoprecipitations showed that extents of nonspecific binding observed without UBR5 (lane 3) or with nonimmune immunoglobulin G (IgG) (lane 4) were much lower than those obtained in the presence of UBR5 and its specific antibody (lane 2). These results suggested that UBR5 may directly interact with each of these mitotic checkpoint proteins.
The observed binding of BubR1 and Cdc20 to UBR5 raised the question of whether they are substrates for UBR5-mediated ubiquitylation, as has been shown previously for Bub3 (21). We tested this possibility by incubation of the recombinant checkpoint proteins with a ubiquitylation mixture, in the absence or presence of recombinant UBR5. As shown in Fig. 1C, the formation of high-molecular-weight, polyubiquitylated products of BubR1, Cdc20, and Bub3, dependent upon the supplementation of UBR5, could be seen.
UBR5 Stimulates the Release of Bub3 from BubR1.
The interaction of mitotic checkpoint proteins with UBR5 and their ubiquitylation by this enzyme raised the question about whether it may affect the assembly-disassembly state of different complexes of mitotic checkpoint proteins. We first examined whether UBR5 is involved in the release of Bub3 from its binding partner, BubR1, which has been reported to take place in exit from mitosis (22; Discussion), by immunodepletion of UBR5 from extracts of HeLa cells. As shown in SI Appendix, Fig. S1A, the immunodepletion procedure effectively removed most of UBR5 from extract, as compared to sham depletion with nonimmune IgG. UBR5-depleted or sham-depleted mitotic extracts were incubated with a recombinant his6-Bub3*BubR1 subcomplex in the presence of ATP, samples were immunoprecipitated with anti-BubR1, and the release of his6-Bub3 from BubR1 was determined. Recombinant his6-Bub3*BubR1 was supplemented at a level ∼sixfold lower than that of endogenous Bub3 (see SI Appendix, Fig. S1B) to minimize the reassociation of released his6-Bub3 with BubR1 by competition with endogenous free Bub3. There was no significant decrease in the levels of total his6-Bub3 in the course of incubation (SI Appendix, Fig. S1B), indicating that his6-Bub3 is not subject to proteolytic degradation under these conditions. By contrast, immunoprecipitated his6-Bub3 was rapidly released from BubR1 in sham-treated extracts, but the rate of release was noticeably slowed down by immunodepletion of UBR5 (Fig. 2 A and B). These findings suggested that UBR5 may have a role in the dissociation of the Bub3*BubR1 subcomplex, although we cannot rule out the possibility that some other factor, coimmunodepleted with UBR5, is involved in this process.
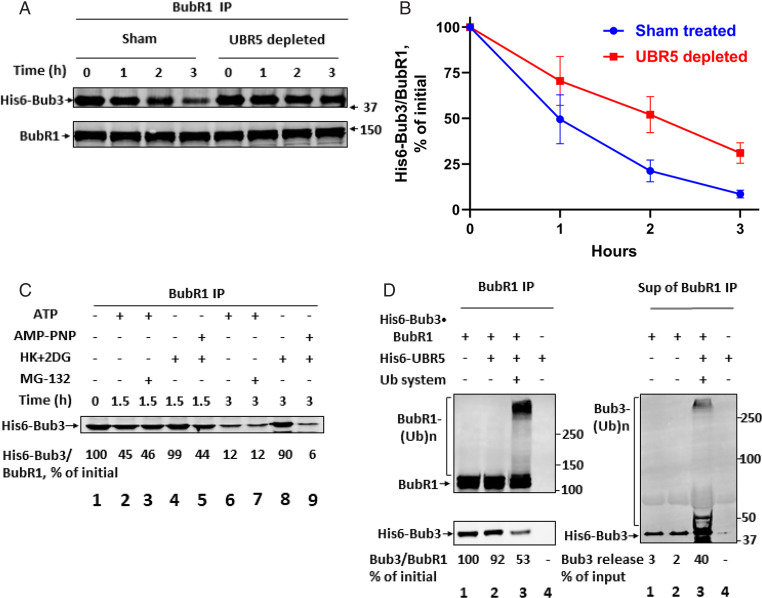
Fig. 2.
UBR5 stimulates the release of Bub3 from BubR1. (A) Effect of immunodepletion of UBR5 from extracts on the release of Bub3 from BubR1. Samples of UBR5-depleted or sham-treated mitotic extracts (1.6 mg of protein) were incubated in a reaction volume of 100 μL containing 40 nM his6-Bub3*BubR1 subcomplex, 40 mM Tris HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, 2 mM ATP, 10 mM phosphocreatine, and 100 μg/mL creatine phosphokinase. Samples were incubated at 23 °C for the indicated time periods. Subsequently, samples were immunoprecipitated with anti-BubR1- beads (10 μL, packed), with rotation for 2 h at 4 °C. Beads were then washed and treated with lambda phosphatase as described in SI Appendix, SI Materials and Methods. The release of His6-Bub3 from anti-BubR1 beads was determined by immunoblotting of immunoprecipitated material with anti-Bub3. Numbers on the Right indicate the position of molecular size marker proteins (kDa). (B) Quantitation of data from A. Ratios of his6-Bub3/BubR1 were determined for each lane and were expressed as percentage of this ratio at time 0. (C) The release of Bub3 from BubR1 in HeLa cell extracts requires ATP but not proteasome activity. The His6-Bub3*BubR1 subcomplex was incubated with sham-treated extracts as described in A. Where indicated, the following additions were made: 2 mM ATP together with 10 mM phosphocreatine and 100 μg/mL creatine phosphokinase; 2 mM adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP); 10 μM MG-132; a mixture of hexokinase (Roche 11426362001, 0.4 mg/mL) and 2-deoxyglucose (10 mM) (“HK+DG”). Reaction mixtures were incubated at 23 °C for the indicated time periods. Samples were subjected to immunoprecipitation with anti-BubR1, and the release of His6-Bub3 from anti-BubR1 beads was analyzed as in A. Results (shown at Bottom) were expressed as the percentage of His6-Bub3 bound to BubR1 relative to the initial value. (D) The dissociation of BubR1*Bub3 subcomplex in a purified system is stimulated by UBR5-dependent ubiquitylation. Recombinant BubR1*Bub3 subcomplex (100 nM) and his6-UbR5 (at a quantity similar to that in 40-μg HeLa cell extract) were supplemented as indicated to a reaction mixture containing the following: 40 mM Tris HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, and 1 mg/mL BSA. Where indicated, a ubiquitylation system consisting of 2 mM AMP-PNP, 150 nM E1, 600 nM UbcH5ɑ, and 100 μM ubiquitin was added. Following incubation at 37 °C for 1 h, Nonidet P-40 (0.2%) and NaCl (150 mM) were added and samples were subjected to immunoprecipitation with anti-BubR1 beads. Supernatants were collected and beads were washed as described under SI Appendix, SI Materials and Methods. Samples of immunoprecipitated material (Left) and of supernatants (Right) were immunoblotted for the indicated proteins. Numbers below blots for Bub3 express the percentage of Bub3/BubR1 ratio relative to the initial value (Lower Left) and the percentage of Bub3 released to the supernatant (Right). Electrophoretic migration positions of molecular size marker proteins are indicated on the Right (kDa).
The properties of the dissociation of the Bub3*BubR1 subcomplex by checkpoint extract were examined in the experiment shown in Fig. 2C. Incubations were carried out for different time periods in the presence of ATP (Fig. 2C, lanes 2 and 6) or without ATP and with added hexokinase and deoxyglucose to remove endogenous ATP (lanes 4 and 8). As seen, the removal of ATP inhibited completely the dissociation of the Bub3*BubR1 subcomplex. The dependence on ATP was not due to the requirement for proteasome action for this process, as shown by the lack of influence of the proteasome inhibitor MG-132 (lanes 3 and 7). When ATP was removed with hexokinase and deoxyglucose, but replaced by the β−γ nonhydrolyzable analog AMPPNP, rapid dissociation of Bub3-BubR1 took place (lanes 5 and 9). These findings suggested that the α−β bond hydrolysis of ATP is required for the dissociation of Bub3*BubR1. Since the action of the ubiquitin-activating enzyme E1 requires the α−β bond hydrolysis of ATP (23), a possible explanation is that ubiquitylation may be necessary for Bub3*BubR1 dissociation.
We examined the possibility that UBR5-mediated ubiquitylation may be involved in the dissociation of the Bub3*BubR1 subcomplex by the use of a system reconstituted from purified components. In the experiments shown in Fig. 2D and SI Appendix, Fig. S2, the recombinant his6-Bub3*BubR1 subcomplex was incubated with recombinant his6-UBR5, in the presence or absence of a ubiquitylation system consisting of E1, an E2 (UbcH5α), ubiquitin, and AMPPNP. An immunoblot analysis of samples taken from the total reaction mixture (SI Appendix, Fig. S2) showed significant ubiquitylation of both BubR1 and Bub3 components of the complex, similar to that described above for the free proteins (Fig. 1C). Other samples from this experiment were subjected to immunoprecipitation with anti-BubR1 and to analysis by immunoblotting. BubR1 immunoblots of immunoprecipitates showed the expected presence of both free and polyubiquitylated BubR1 following incubation of the complex with UBR5 and ubiquitylation mixture (Fig. 2 D, Left, Top, lane 3). Immunoblotting of Bub3 in these immunoprecipitates showed a marked decrease of BubR1-bound Bub3 following incubation with UBR5 and ubiquitylation mixture (Fig. 2 D, Left, Bottom, lane 3 vs. lanes 1 and 2). Examination of Bub3 in the supernatants of BubR1 immunoprecipitates showed a prominent increase of both free and polyubiquitylated forms of Bub3 in this incubation (Fig. 2 D, Right, lane 3 vs. lanes 1 and 2). These results indicated that UBR5-dependent ubiquitylation causes dissociation of the Bub3*BubR1 subcomplex. It is notable that a greater part of Bub3 was released in a free, nonubiquitylated form. This suggested that the ubiquitylation of Bub3 is not obligatory for its release from BubR1 and raised the possibility that BubR1 ubiquitylation may be responsible for this process (Discussion).
UBR5 Stimulates the Release of MCC from APC/C and Advances the Activation of APC/C.
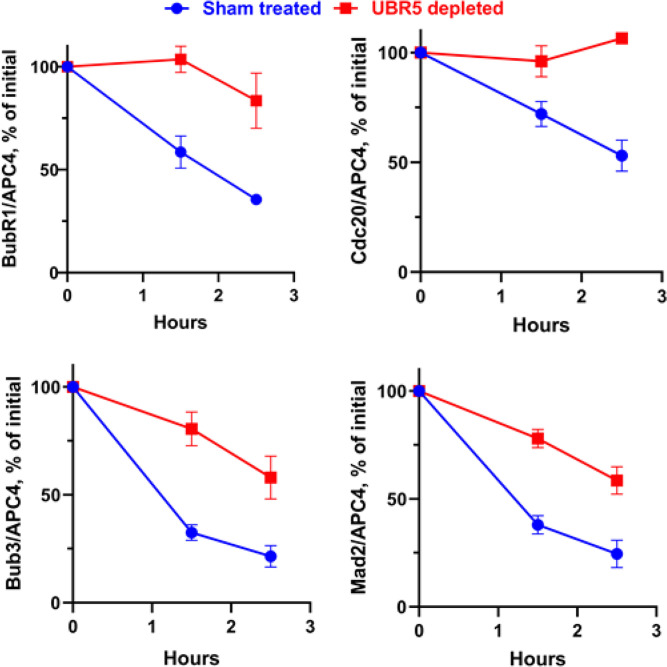
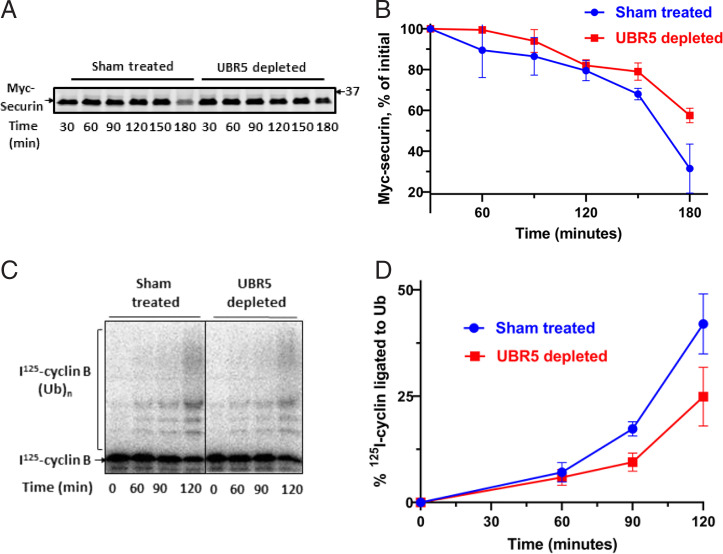
Our observation that UBR5 promotes the ubiquitylation of Cdc20 and BubR1 components of MCC raised the possibility that these activities are related to similar reactions catalyzed by APC/C, which is known to stimulate the disassembly of APC/C-bound MCC (6, 15, 16). It is possible, for example, that UBR5 has an auxiliary or regulatory role in these processes. We therefore examined the influence of immunodepletion of UBR5 on the release of MCC components from APC/C in checkpoint extracts. In the experiment shown in Fig. 3 and SI Appendix, Fig. S3, UBR5-depleted or sham-treated extracts were incubated with ATP, samples were immunoprecipitated with anti-Cdc27, and the amounts of MCC components bound to APC/C were determined by quantitative immunoblotting. Results were normalized to the amounts of the APC4 subunit of APC/C. As shown in Fig. 3, immunodepletion of UBR5 slowed down the release of all MCC components from APC/C. UBR5 depletion also prolonged the lag in the degradation of securin, an APC/C substrate, in checkpoint extracts incubated with ATP (Fig. 4 A and B). This was due to a delay in the release of APC/C activity from checkpoint inhibition, as shown by an assay of activity of immunopurified APC/C in the ubiquitylation of cyclin B (Fig. 4 C and D). These data suggest that UBR5 has actions similar to those of APC/C in promoting the disassembly of APC/C-bound MCC and the activation of APC/C in the exit from mitotic checkpoint and raise the possibility that these processes may be regulated by the joint action of these two ubiquitin ligases (Discussion).
Fig. 3.
Effect of UBR5 depletion on the release of MCC components from APC/C. Quantitation of immunoblots shown in SI Appendix, Fig. S3. The ratios of the different mitotic checkpoint proteins to APC4 were determined for each lane and were expressed as the percentage of corresponding ratios at time 0. All values of quantitation of APC4 immunoblots were in the linear range of the assay, and thus, APC4 blots could serve as valid loading controls for the estimation of the levels of the other proteins shown in the figure.
Fig. 4.
UBR5 shortens the lag period in the activation of APC/C in exit from mitotic checkpoint. (A) Influence of UBR5 on the degradation of myc-securin in checkpoint extracts incubated with ATP. Recombinant Myc-securin (2.5 ng) was added to samples of UBR5-depleted or sham-treated HeLa cell extracts (150 μg of protein) and was incubated in a reaction volume of 10 μL containing 40 mM Tris⋅HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, 2 mM ATP, 10 mM phosphocreatine, and 100 μg/mL creatine phosphokinase. Following incubation at 23 °C for the time periods indicated, samples were resolved by SDS-PAGE and were blotted with Myc-tag antibody. (B) Quantitation of data from A. Results were expressed as the percentage of Myc-securin at time 0. (C) Influence of immunodepletion of UBR5 on activation of APC/C in checkpoint extract incubated with ATP. Checkpoint extracts subjected to UBR5-depletion or to sham depletion were incubated with ATP as described under A, for the time periods indicated. Samples were mixed with anti-Cdc27 beads for 2 h at 4 °C, and then beads were washed three times with a buffer consisting of 50 mM Tris⋅HCl (pH 7.2), 20% (vol/vol) glycerol, 1 mg/mL BSA, and 1 mM DTT. APC/C activity was determined in samples of 1-μL packed beads contained by the ubiquitylation of an 125I-labeled N-terminal fragment of cyclin B, as described previously (41). (D) Quantitation of data from C. The data were expressed as the percentage of I125-cyclin B-(Ub)n conjugates formed relative to the total amount of I125-cyclin B in each lane.
Discussion
This study was initiated by a search for factors that may regulate the action of the MCC on the ubiquitin ligase APC/C. Mass spectrometry analysis detected the association of the ubiquitin ligase UBR5 with immunopurified APC/C-MCC. We have followed up this observation because of the apparent importance of UBR5 in cellular regulation. UBR5 (also called EDD for E3 identified by differential display) is an essential and evolutionarily conserved HECT (homologous to E6AP C-terminus)-type ubiquitin ligase, homologous to Drosophila tumor suppressor Hyd (hyperplastic discs). It is frequently mutated in human cancers and has been implicated in a variety of cellular processes, such as DNA damage response, apoptosis, cell cycle control, transcriptional control, and regulation of gluconeogenesis (reviewed in 19), as well as in protein quality control (24, 25). UBR5 has been reported to act on diverse cellular proteins such as katanin p60 subunit of microtubule-severing ATPase (26), TOBP1 topoisomerase-binding protein (27), ATMIN mediator of ATM signaling (28), TERT catalytic subunit of telomerase (29), proapoptotic protein MAOP-1 (30), deubiquitylating enzyme DUBA (31), pregnane X receptor (32), and gluconeogenetic enzyme PEPCK (33). In some cases, UBR5 recognizes protein substrates directly through different substrate-binding regions, such as the binding of PEPCK to the N-lobe of the HECT domain (34), binding of Bub3 to HECT domain (21), or binding of PAIP2 to an MLLE/PABC interaction region (described in 35). In other cases, UBR5 binds to protein substrates via adaptor complexes, such as the EVDP complex composed of EDD/UBR5, DDB1, VPRBP proteins bound to protein kinase DYRK2 (26, 29, 30, 32). However, in many other cases, the mode of recognition of substrate proteins by UBR5 is not known. UBR5 has a binding domain recognizing N-degrons of the Arg/N-degron pathway (18, 36), but to our knowledge, cellular N-degron substrates of this ubiquitin ligase have not yet been described.
We have confirmed previous reports that UBR5 coimmunoprecipitates with mitotic checkpoint proteins (20) and that it binds and ubiquitylates Bub3 (21). We furthermore found that it also directly binds to and ubiquitylates mitotic checkpoint proteins Cdc20 and BubR1 (Fig. 1 B and C). It is surprising that the three different mitotic checkpoint proteins are substrates for UBR5, and it remains to be investigated how the different mitotic checkpoint proteins are recognized by this ubiquitin ligase. Since ubiquitylation of MCC components has been shown previously be involved in MCC disassembly (6, 15, 16), these observations led us to examine the possible roles of UBR5 in the disassembly of complexes of mitotic checkpoint proteins. We first examined whether UBR5 influences the dissociation of the Bub3*BubR1 subcomplex. Assembly-disassembly of Bub3*BubR1 apparently takes place in the cell cycle since it has been reported that during the interphase these proteins have different subcellular locations; Bub3 is localized in the nucleus, while BubR1 is mostly cytosolic (37). In prometaphase, Bub3 has an important role in the initiation of the assembly of MCC by binding to sites on kinetochore phosphorylated by Mps1 protein kinase and then recruiting to the kinetochore its interacting partners BubR1 and Bub1 (reviewed in 3, 5). In the mitotic checkpoint, Bub3 is a component of MCC and was shown to be necessary for the action of MCC to inhibit APC/C (38). It has been recently reported that when MCC is disassembled upon exit from mitosis, Bub3 is also dissociated from BubR1 (22). This is in contrast to previous observations suggesting that Bub3 is constitutively bound to BubR1 throughout the cell cycle (39), which have been attributed to methods of cell lysis that disrupt the sequestration of Bub3 in the nucleus (22). It thus seems that the dissociation of Bub3 from BubR1 at the end of mitosis is necessary to reset the interphase subcellular sequestration of these checkpoint proteins.
We found that the release of Bub3 from BubR1 requires ATP but not proteasome action (Fig. 2C). These results are reminiscent of the properties of the disassembly of MCC by APC/C-driven ubiquitylation, which is also not affected by proteasome inhibition (8, 9), but are different from a report suggesting that UBR5 targets Bub3 for degradation (21). In the presently investigated system, ATP is presumably required for ubiquitylation. By the use of a purified reconstituted system, we showed that UBR5-catalyzed ubiquitylation causes disassembly of the BubR1*Bub3 subcomplex (Fig. 2D). Remarkably, a great part of Bub3 released in the reconstituted system was not ubiquitylated, raising the possibility that the observed ubiquitylation of its partner, BubR1, may elicit a structural change that decreases its interaction with Bub3.
The activity of UBR5 to ubiquitylate Cdc20 and BubR1 components of MCC resembles similar activities of APC/C, which are known to be involved in MCC disassembly and checkpoint inactivation (9, 15, 16). Immunodepletion of UBR5 slowed down the release of MCC components from APC/C in checkpoint extracts incubated with ATP (Fig. 3 and SI Appendix, Fig. S3) and prolonged the lag in the activation of APC/C (Fig. 4 A–D). However, UBR5 depletion did not abolish these processes, and at later times of incubation, APC/C was activated in the virtual absence of UBR5. It thus seems that MCC disassembly and APC/C activation in checkpoint inactivation are driven by a basic APC/C-catalyzed process and that UBR5 acts to modify or to regulate this basic process by augmenting the rate of ubiquitylation of MCC components. It is also possible that UBR5 cooperates with APC/C to produce specific mixed branches of polyubiquitin chains on Cdc20 and BubR1, which may affect their release from APC/C. Another possibility is that the activity of UBR5 itself may be regulated by the mitotic checkpoint. Many HECT-type ubiquitin ligases, to which UBR5 belongs, are subject to autoinhibition, usually by intramolecular interactions between regions of the HECT domain responsible for activity (reviewed in 40). Autoinhibition can be relieved by a variety of regulatory processes, such as posttranslational modifications or interaction with regulatory proteins (40). In the case of UBR5, potential autoinhibitory interactions in its HECT domain have been described (41), but its effects on the regulation of enzymatic activity have not been reported. It remains to be investigated whether the activity of UBR5 is regulated by the state of the mitotic checkpoint system and, if so, how it regulates the disassembly of MCCs in the inactivation of the mitotic checkpoint.
Materials and Methods
Further experimental details are provided in SI Appendix, SI Materials and Methods.
Preparations.
Recombinant his6-UBR5 was prepared by transfection of HEK293T cells (ATCC) with pCMV-Tag2B EDD, coding for the full-length human UBR5 fused to his6-tag (kindly provided by Dr. Darren N. Saunders, Garvan Institute of Medical Research), using the Lipofectamine 2000 reagent (Invitrogen). Following incubation for 48 h at 37 °C, cells were harvested and lysed, and his6-UBR5 was purified on Ni-nitriloacetic acid agarose (Qiagen). Recombinant MCC and BubR1*Bub3 subcomplex were formed by coinfection of SF9 cells with baculoviruses expressing the appropriate proteins (streptavidin-binding peptide-BubR1, myc3-Cdc20, flag3-Mad2, and his6-Bub3), followed by purification procedures as described previously (42).
Supplementary Material
Acknowledgments
We thank Dr. Tamar Ziv, Smoler Proteomics Center, Technion, for mass spectrometry analysis and Dr. Darren N. Saunders, Garvan Institute of Medical Research, Sydney, Australia, for pCMV-Tag2B plasmid coding for the full-length human UBR5. We are grateful to Dr. Michael Fry for helpful comments on the manuscript. This work was supported by grants from the Israel Science Foundation, the Israel Cancer Research Fund, and the Gitta and Saul Kurlat Foundation.
Footnotes
Reviewers: M.P., HHMI and NYU School of Medicine; and A.V., California Institute of Technology.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121478119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or in SI Appendix.
References
- 1.Musacchio A., Salmon E. D., The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Lara-Gonzalez P., Westhorpe F. G., Taylor S. S., The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Musacchio A., The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25, R1002–R1018 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Watson E. R., Brown N. G., Peters J. M., Stark H., Schulman B. A., Posing the APC/C E3 ubiquitin ligase to orchestrate cell division. Trends Cell Biol. 29, 117–134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.London N., Biggins S., Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15, 736–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfieri C., et al. , Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 536, 431–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein I., Miniowitz S., Moshe Y., Hershko A., Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc. Natl. Acad. Sci. U.S.A. 104, 4870–4875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miniowitz-Shemtov S., Teichner A., Sitry-Shevah D., Hershko A., ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc. Natl. Acad. Sci. U.S.A. 107, 5351–5356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eytan E., Sitry-Shevah D., Teichner A., Hershko A., Roles of different pools of the mitotic checkpoint complex and the mechanisms of their disassembly. Proc. Natl. Acad. Sci. U.S.A. 110, 10568–10573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eytan E., et al. , Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet). Proc. Natl. Acad. Sci. U.S.A. 111, 12019–12024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K., et al. , Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J. Biol. Chem. 289, 23928–23937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Q., et al. , TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife 4, e07367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miniowitz-Shemtov S., et al. , Role of phosphorylation of Cdc20 in p31(comet)-stimulated disassembly of the mitotic checkpoint complex. Proc. Natl. Acad. Sci. U.S.A. 109, 8056–8060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaisari S., Sitry-Shevah D., Miniowitz-Shemtov S., Teichner A., Hershko A., Role of CCT chaperonin in the disassembly of mitotic checkpoint complexes. Proc. Natl. Acad. Sci. U.S.A. 114, 956–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy S. K., Rape M., Margansky W. A., Kirschner M. W., Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446, 921–925 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Sitry-Shevah D., Kaisari S., Teichner A., Miniowitz-Shemtov S., Hershko A., Role of ubiquitylation of components of mitotic checkpoint complex in their dissociation from anaphase-promoting complex/cyclosome. Proc. Natl. Acad. Sci. U.S.A. 115, 1777–1782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaisari S., et al. , Role of Polo-like kinase 1 in the regulation of the action of p31comet in the disassembly of mitotic checkpoint complexes. Proc. Natl. Acad. Sci. U.S.A. 116, 11725–11730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasaki T., et al. , A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25, 7120–7136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shearer R. F., Iconomou M., Watts C. K. W., Saunders D. N., Functional roles of the E3 ubiquitin ligase UBR5 in cancer. Mol. Cancer Res. 13, 1523–1532 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Scialpi F., Mellis D., Ditzel M., EDD, a ubiquitin-protein ligase of the N-end rule pathway, associates with spindle assembly checkpoint components and regulates the mitotic response to nocodazole. J. Biol. Chem. 290, 12585–12594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H., He X., Feng D., Zhu X., Zheng Y., RanGTP aids anaphase entry through Ubr5-mediated protein turnover. J. Cell Biol. 211, 7–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirnekhi H. K., Herman J. A., Paddison P. J., DeLuca J. G., BuGZ facilitates loading of spindle assembly checkpoint proteins to kinetochores in early mitosis. J. Biol. Chem. 295, 14666–14677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hershko A., Ciechanover A., The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Yau R. G., et al. , Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171, 918–933.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyuncu S., et al. , The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington’s disease patients. Nat. Commun. 9, 2886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddika S., Chen J., Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11, 409–419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda Y., et al. , Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem. 277, 3599–3605 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Zhang T., Cronshaw J., Kanu N., Snijders A. P., Behrens A., UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proc. Natl. Acad. Sci. U.S.A. 111, 12091–12096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung H.-Y., Wang X., Jun S., Park J. I., Dyrk2-associated EDD-DDB1-VprBP E3 ligase inhibits telomerase by TERT degradation. J. Biol. Chem. 288, 7252–7262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuura K., Huang N. J., Cocce K., Zhang L., Kornbluth S., Downregulation of the proapoptotic protein MOAP-1 by the UBR5 ubiquitin ligase and its role in ovarian cancer resistance to cisplatin. Oncogene 36, 1698–1706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutz S., et al. , Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Ong S. S., et al. , Stability of the human pregnane X receptor is regulated by E3 ligase UBR5 and serine/threonine kinase DYRK2. Biochem. J. 459, 193–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong Y., Lei Q.-Y., Zhao S., Guan K. L., Regulation of glycolysis and gluconeogenesis by acetylation of PKM and PEPCK. Cold Spring Harb. Symp. Quant. Biol. 76, 285–289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Q., Qiu Z., Wu W., Zheng J., Jia Z., Characterization of interaction and ubiquitination of phosphoenolpyruvate carboxykinase by E3 ligase UBR5. Biol. Open 7, bio037366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida M., et al. , Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 25, 1934–1944 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshavsky A., N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor S. S., Ha E., McKeon F., The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142, 1–11 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overlack K., et al. , BubR1 promotes Bub3-dependent APC/C inhibition during spindle assembly checkpoint signaling. Curr. Biol. 27, 2915–2927.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan G. K., Jablonski S. A., Sudakin V., Hittle J. C., Yen T. J., Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146, 941–954 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah S. S., Kumar S., Adaptors as the regulators of HECT ubiquitin ligases. Cell Death Differ. 28, 455–472 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muñoz-Escobar J., Matta-Camacho E., Kozlov G., Gehring K., The MLLE domain of the ubiquitin ligase UBR5 binds to its catalytic domain to regulate substrate binding. J. Biol. Chem. 290, 22841–22850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaisari S., Sitry-Shevah D., Miniowitz-Shemtov S., Hershko A., Intermediates in the assembly of mitotic checkpoint complexes and their role in the regulation of the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 113, 966–971 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or in SI Appendix.