Summary
Background
Alterations in the respiratory microbiome are common in chronic lung diseases, correlate with decreased lung function, and have been associated with disease progression. The clinical significance of changes in the respiratory microbiome after lung transplant, specifically those related to development of chronic lung allograft dysfunction (CLAD), are unknown. The aim of this study was to evaluate the effect of lung microbiome characteristics in healthy lung transplant recipients on subsequent CLAD-free survival.
Methods
We prospectively studied a cohort of lung transplant recipients at the University of Michigan (Ann Arbor, MI, USA). We analysed characteristics of the respiratory microbiome in acellular bronchoalveolar lavage fluid (BALF) collected from asymptomatic patients during per-protocol surveillance bronchoscopy 1 year after lung transplantation. For our primary endpoint, we evaluated a composite of development of CLAD or death at 500 days after the 1-year surveillance bronchoscopy. Our primary microbiome predictor variables were bacterial DNA burden (total 16S rRNA gene copies per mL of BALF, quantified via droplet digital PCR) and bacterial community composition (determined by bacterial 16S rRNA gene sequencing). Patients’ lung function was followed serially at least every 3 months by spirometry, and CLAD was diagnosed according to International Society of Heart and Lung Transplant 2019 guidelines.
Findings
We analysed BALF from 134 patients, collected during 1-year post-transplant surveillance bronchoscopy between Oct 21, 2005, and Aug 25, 2017. Within 500 days of follow-up from the time of BALF sampling, 24 (18%) patients developed CLAD, five (4%) died before confirmed development of CLAD, and 105 (78%) patients remained CLAD-free with complete follow-up. Lung bacterial burden was predictive of CLAD development or death within 500 days of the surveillance bronchoscopy, after controlling for demographic and clinical factors, including immunosuppression and bacterial culture results, in a multivariable survival model. This relationship was evident when burden was analysed as a continuous variable (per log10 increase in burden, HR 2·49 [95% CI 1·38–4·48], p=0·0024) or by tertiles (middle vs lowest bacterial burden tertile, HR 4·94 [1·25–19·42], p=0·022; and highest vs lowest, HR 10·56 [2·53–44.08], p=0·0012). In patients who developed CLAD or died, composition of the lung bacterial community significantly differed to that in patients who survived and remained CLAD-free (on permutational multivariate analysis of variance, p=0·047 at the taxonomic level of family), although differences in community composition were associated with bacterial burden. No individual bacterial taxa were definitively associated with CLAD development or death.
Interpretation
Among asymptomatic lung transplant recipients at 1-year post-transplant, increased lung bacterial burden is predictive of chronic rejection and death. The lung microbiome represents an understudied and potentially modifiable risk factor for lung allograft dysfunction.
Introduction
Survival after lung transplantation is poorer than for other solid organ transplants,1 with the leading cause of death in patients who survive 1 year after transplant being chronic rejection.2 Chronic rejection is manifested by fibrotic infiltration of the lung allograft, resulting in irreversible pulmonary dysfunction, termed chronic lung allograft dysfunction (CLAD).2 Inflammatory and immune-mediated events, such as primary graft dysfunction, acute rejection, and the development of donor-specific antibodies, have been associated with subsequent development of CLAD.3 However, the factors that predispose to the development of CLAD are incompletely understood.
In the past decade, the use of culture-independent microbiological techniques has revealed that the lower respiratory tract, classically believed to be sterile, harbors diverse and dynamic communities of bacteria.4 Lung bacteria are appreciable in healthy subjects,5,6 altered in chronic lung diseases,4,7 and correlate with altered immune responses.6,8,9 Several studies in recent years have revealed that the respiratory microbiome is independently predictive of outcomes in diverse lung diseases, both acute and chronic. For instance, increased lung bacterial burden has been shown to predict poor clinical outcomes in pulmonary fibrosis10,11 and critically ill patients receiving mechanical ventilation.12 Decreased diversity of sputum bacteria predicts mortality in chronic obstructive pulmonary disease,13 and altered respiratory communities predict exacerbations in bronchiectasis14 and respiratory infections in children.15
Lung transplant recipients have several unique factors that affect the balance between bacterial immigration and elimination, which collectively shape the respiratory microbiome. Immunosuppressive medications decrease baseline immune surveillance, prophylactic antibiotics exert exogenous selection pressures, increased rates of reflux lead to increased immigration of particular taxa, and surgical anastomoses and denervation result in reduced bacterial clearance.4,16 Consequentially, compared with healthy individuals, lung transplant recipients have altered respiratory microbiome characteristics, including increased bacterial burden and altered lung communities.17,18 The clinical significance of these differences in lung microbiota on outcomes after lung transplant is, to date, undetermined.
We hypothesised that the lung microbiome is an independent risk factor for chronic rejection after lung transplant, representing an understudied and targetable source of clinical variation in this at-risk population. To test this hypothesis, we did a prospective cohort study of asymptomatic, CLAD-free lung transplant recipients undergoing per-protocol surveillance bronchoscopy 1 year after transplant. We evaluated the association of bacterial burden and community composition in bronchoalveolar lavage fluid (BALF) with subsequent development of chronic rejection and death.
Methods
Study design and participants
We prospectively studied a cohort of lung transplant recipients at the University of Michigan (Ann Arbor, MI, USA). The aim of this study was to evaluate the effect of lung microbiome characteristics in healthy lung transplant recipients on subsequent CLAD-free survival. Thus, we limited our cohort to asymptomatic patients undergoing per-protocol surveillance bronchoscopy 1 year after transplant, without age restrictions. Enrolled participants permitted the use of any BALF surplus to clinical requirements for microbiome analysis. Exclusion criteria were reported respiratory symptoms, decline in pulmonary function, or new radiographic opacities at the time of the 1-year post-transplant bronchoscopy. Enrolled patients without surplus BALF available from bronchoscopy were subsequently excluded from our analyses. All lung transplant recipients received immunosuppression and antimicrobial prophylaxis per institutional protocol. Specific treatments are provided in the appendix (p 1). This study was approved by the University of Michigan institutional research board (reference number HUM00043357) and written informed consent was obtained for participation in the study.
Specimen collection and processing
Bronchoscopy was done per institutional protocol. Briefly, after patients received conscious sedation and nebulised lidocaine, the bronchoscope was advanced via the mouth or nose and through the vocal cords. After an airway exam, the bronchoscope was wedged in the right middle lobe or lingula of the allograft. Bronchoalveolar lavage was done with instillation of between 120 mL and 300 mL of sterile isotonic saline. Samples were stored on ice, filtered, centrifuged at 1000 rpm for 5 min to remove eukaryotic cells, and stored at –80°C until the time of DNA extraction. DNA was extracted, amplified, and sequenced according to previously published protocols.12,19,20 Bacterial DNA burden was measured with a QX200 Droplet Digital PCR System (BioRad, Hercules, CA, USA). Sequencing was done with the Illumina MiSeq platform (San Diego, CA, USA). Additional details on sample and DNA processing are provided in the appendix (p 2).
Low biomass microbiome studies are vulnerable to contamination from bacterial DNA present in reagents used for DNA extraction and library preparation.21 We therefore compared the bacterial signal detected in our cell-free BALF specimens with numerous negative control specimens (n=46; 26 in droplet digital PCR and 20 in 16S rRNA sequencing) representing potential sources of contamination (appendix pp 2–3).
Outcomes and predictor variables
Lung transplant recipients are monitored at least every 3 months with spirometry according to American Thoracic Society and European Respiratory Society guidelines,22 and CLAD was diagnosed in accordance with the International Society of Heart and Lung Transplant guidelines, as summarised in the appendix (p 1).2 For our primary endpoint, we elected to evaluate a composite of development of CLAD or death at 500 days after the 1-year surveillance bronchoscopy. This timepoint was selected because it met our criteria of, first, allowing sufficient time for patients to develop CLAD and, second, because we had complete follow-up data on all patients, thus avoiding the potential bias associated with censored data. For patients who died, the date and cause of death were determined by medical record review. We prespecified that our primary microbiome predictors of CLAD development or death would be: bacterial DNA burden (quantified as total 16S rRNA gene copies per mL of BALF via droplet digital PCR), bacterial community diversity within specimens (α-diversity, quantified according to the Shannon diversity index), bacterial community richness (quantified as number of unique operational taxonomic units [OTUs] per 100000 sequences), and differences in community composition between specimens (β-diversity) at different taxonomic levels. We assessed differences in each predictor between patients who remained CLAD-free and those who developed CLAD or died. Similar to previous studies,10,23,24 we also stratified patients into tertiles of bacterial burden (lowest, middle, and highest bacterial burden) and compared between these groups the occurrence of CLAD development or death. Demographic predictor variables of the composite outcome were age, sex, pretransplant diagnosis, and bilateral versus single lung transplant. Clinical predictor variables were current lung function (FEV1) and immunosuppression regimen, severity of any primary graft dysfunction immediately after transplant based on the irst blood gas available25 (graded per International Society of Heart and Lung Transplant guidelines26), cumulative acute rejection scores (per International Society of Heart and Lung Transplant schema27) in the first year of transplant, presence of donor-specific antibodies in the first year of transplant, BALF cell counts, current and previous BALF culture results, history of either cytomegalovirus (CMV) pneumonitis or community-acquired respiratory viral infections between transplant and 1-year surveillance, antibiotic use in the 30 days before bronchoscopy, and azithromycin use for CLAD prevention at the time of bronchoscopy. Further details on outcome variables are provided in the appendix (p 1).
Statistical analysis
The sequence data from BALF and control specimens were processed and analysed with the software mothur (version 1.38.0), according to the standard operating procedure for MiSeq sequence data.28 We analysed microbial ecology parameters using the vegan package and mvabund in R (version 3.6.1).29,30 We visualised the relative community composition of specimens using principal component analysis, and interrogated which taxa drove clustering via biplot analysis. We also visualised differences in community composition across groups with rank abundance plots. We determined statistical significance in community composition comparisons firstly using the adonis function of vegan,29 which performs permutational multivariate analysis of variance (PERMANOVA) via 10000 permutations; and secondly using mvabund,30 a model-based approach to analysis of multivariable abundance data. We used the adonis function with default parameters, including the Bray-Curtis index to compare β-diversity. Analyses unrelated to microbial ecology were done with Stata (version 12.0). Categorical variables were summarised as percentages and compared with the χ2 test. Continuous data were summarised as means (with SD) and compared with two-sample t test, ANOVA, and simple linear regression (when data were normally distributed), or summarised as medians (with IQR) and compared with the Wilcoxon rank-sum test, Kruskal–Wallis test, or Spearman’s correlation (when at least one variable was not normally distributed). Time-to-event analysis for primary microbiome predictor variables of our composite outcome (CLAD development or death) were done with the log-rank test31,32 and results presented as Kaplan–Meier curves.33 To evaluate the association of lung microbiome variables, and our demographic and clinical predictor variables, with the composite outcome, we first did univariable Cox proportional hazards regression analyses.34 Significant univariable associations (at p<0·05) were further evaluated in a multivariable Cox proportional hazards regression model that included known risk factors for CLAD and variables associated with relevant microbiome predictors (appendix p 4). Missing data were omitted in univariable analyses and included as a categorical variable in multivariable analyses. Additionally, we did exploratory analyses investigating the potential association of lung microbiome characteristics with demographic and clinical predictor variables. In a supplementary analysis, we evaluated whether culture-independent detection of Pseudomonas aeruginosa was associated with development of our composite outcome. For this we identified the OTU representing P aeruginosa via comparison of its representative nucleotide sequence to sequences of speciated bacterial strains. We subsequently evaluated whether the culture-independent presence and relative abundance of P aeruginosa was associated with our composite outcome using the log-rank test and univariable Cox regression, respectively. As a post-hoc analysis, we evaluated potential interactions between bacterial burden, relevant predictor variables, and our composite outcome.
A p value of less than 0·05 was considered to indicate significance in all analyses. Our statistical methods are outlined further in the appendix (pp 2–5).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
160 lung transplant recipients were enrolled in our prospective study between Sept 29, 2005, and Oct 13, 2017. Of these patients, 13 were excluded due to reported respiratory symptoms, pulmonary function decline, or new radiographic opacities at the time of 1-year post-transplant bronchoscopy. An additional 13 patients did not have any BALF surplus to clinical requirements for microbiome analysis. The remaining 134 samples comprised our cohort of asymptomatic 1-year surveillance BALF samples, collected between Oct 21, 2005, and Aug 25, 2017. The demographic and clinical characteristics of patients contributing these samples are shown in table 1. Within 500 days of follow-up from the point of BALF sampling, 24 (18%) of 134 patients developed CLAD and five (4%) died before developing CLAD (four patients died of respiratory failure before CLAD could be established via spirometry and one died of metastatic adenocarcinoma from the native lung). The remaining 105 (78%) patients remained CLAD-free with complete follow-up. Patients who remained CLAD-free did not differ from patients who died or developed CLAD with regard to most demographic and clinical characteristics (table 1). Compared with patients who did not meet the primary endpoint, those who remained CLAD-free were more likely to be receiving mycophenolate for antiproliferative immunosuppression (63 [60%] of 105 vs 9 [31%] of 29) and less likely to have immunosuppression held (eight [8%] vs seven [24%]; χ2 p=0·0064) at the time of 1-year surveillance bronchoscopy. All BALF samples were used for bacterial quantification and 16S rRNA gene sequencing.
Table 1:
Demographic and clinical variables of 1-year post-transplant surveillance cohort
| Total (n=134) | CLAD-free* (n=105) | Developed CLAD or died* (n=29) | p value† | |
|---|---|---|---|---|
| Age, years | 50·2 (13·4) | 50·0 (13·3) | 50·8 (14·2) | 0·79 |
| Female patients | 53 (40%) | 38 (36%) | 15 (52%) | 0·13 |
| Pretransplant diagnosis | ||||
| Chronic obstructive pulmonary disease | 43 (32%) | 31 (30%) | 12 (41%) | ·· |
| Interstitial lung disease | 46 (34%) | 34 (32%) | 12 (41%) | ·· |
| Cystic fibrosis | 30 (22%) | 27 (26%) | 3 (10%) | ·· |
| Other | 15 (11%) | 13 (12%) | 2 (7%) | 0.21 |
| Double lung transplant | 97 (72%) | 77 (73%) | 20 (69%) | 0·64 |
| FEV1, L | 2·35 (0·81) | 2·41 (0·82) | 2·11 (0·71) | 0·074 |
| Induction immunosuppression | ||||
| No induction | 125 (93%) | 97 (92%) | 28 (97%) | ·· |
| Basiliximab | 9 (7%) | 8 (8%) | 1 (3%) | 0·43 |
| Calcineurin inhibition‡ | ||||
| Cyclosporine | 29 (22%) | 21 (20%) | 8 (28%) | ·· |
| Tacrolimus | 105 (78%) | 84 (80%) | 21 (72%) | 0·38 |
| Antiproliferative immunosuppression‡ | ||||
| Azathioprine | 47 (35%) | 34 (32%) | 13 (45%) | ·· |
| Mycophenylate | 72 (54%) | 63 (60%) | 9 (31%) | ·· |
| None or held | 15 (11%) | 8 (8%) | 7 (24%) | 0·0064 |
| Primary graft dysfunction immediately (<24 h) after transplant | ||||
| Grade 0 | 41 (31%) | 31 (30%) | 10 (34%) | ·· |
| Grade 1 | 25 (19%) | 20 (19%) | 5 (17%) | ·· |
| Grade 2 | 28 (21%) | 24 (23%) | 4 (14%) | ·· |
| Grade 3 | 40 (30%) | 30 (29%) | 10 (34%) | 0·71 |
| Cumulative grade A rejection score§ | 0·3 (0·4) | 0·3 (0·4) | 0·3 (0·5) | 0·7 |
| Cumulative grade B rejection score§ | 0·1 (0·2) | 0·1 (0·2) | 0·1 (0·1) | 0·57 |
| DSA present¶ | 17/70 (24%) | 12/53 (23%) | 5/17 (29%) | 0·57 |
| BALF neutrophil percentage|| | 12·3% (16·6) | 12·2% (16·9) | 12·6% (16·0) | 0·88 |
| BALF lymphocyte percentage|| | 8·0 (9·8) | 7·3 (9·1) | 10·4 (12·2) | 0·77 |
| BALF bacterial culture|| | ||||
| Negative; no growth | 38 (28%) | 32 (30%) | 6 (21%) | ·· |
| Oral flora | 78 (58%) | 60 (57%) | 18 (62%) | ·· |
| Positive; speciated result | 18 (13%) | 13 (12%) | 5 (17%) | 0·53 |
| Number of previous positive BALF bacterial cultures**†† | ||||
| None | 67 (50%) | 51 (49%) | 16 (55%) | ·· |
| One | 21 (16%) | 19 (18%) | 2 (7%) | ·· |
| Two or more | 46 (34%) | 35 (33%) | 11 (38%) | 0·34 |
| History of community-acquired respiratory viral infection**‡‡ | 8/79 (10%)§§ | 5/62 (8%)§§ | 3/17 (18%)§§ | 0·26 |
| History of CMV pneumonitis**¶¶ | 13 (10%) | 10 (10%) | 3 (10%) | 0·90 |
| Antibiotics in the 30 days before bronchoscopy|||| | 18 (13%) | 15 (14%) | 3 (10%) | 0·58 |
| Azithromycin for CLAD prevention at time of bronchoscopy | 7 (5%) | 6 (6%) | 1 (3%) | 0·63 |
Data are mean (SD) or number of patients (%). Percentages might not always add to 100% due to rounding. CLAD=chronic lung allograft dysfunction. DSA=donor-specific antibodies. BALF=bronchoalveolar lavage fluid. CMV=cytomegalovirus.
Within 500 days of follow-up.
Calculated with the Student’s t test (when normally distributed) or the Wilcoxon rank-sum test (when not normally distributed) for continuous variables, and the χ2 test for categorical variables.
As determined at the time of 1-year post-transplant surveillance bronchoscopy only; all patients were on corticosteroids per protocol, although the exact immunosuppression regimen during the 500 days of follow up, including decisions to stop or start immunosuppression, transition within class, and add other drugs was at the discretion of the transplant pulmonologist.
Calculated by adding the ordinal values of each biopsy specimen’s A or B score (per International Society of Heart and Lung Transplant schema27) divided by the total number of biopsies done in the first year post-transplant.
Defined as a mean fluorescence intensity of ≥3000 via single antigen bead testing of the patient’s serum before or at the time of 1-year post-transplant surveillance (64 patients had no DSA information available: 52 among those who remained CLAD-free and 12 among those who developed CLAD or died).
Identified in the 1-year surveillance BALF (regarding cell counts, five patients had no BALF cell count information available: four among those who remained CLAD-free and one among those who developed CLAD or died).
At any point from transplant to 1-year surveillance bronchoscopy.
Positive BALF culture was defined as any culture for which a bacterial species was identified per clinical microbiology laboratory protocol (culture described as oral flora was not considered positive).
Defined as any positive respiratory pathogen panel in the presence of respiratory symptoms, transient decline in spirometry, or radiographic infiltrate (55 patients had no respiratory viral PCR results available: 43 among those who remained CLAD-free and 12 among those who developed CLAD or died).
Denominators represent patients with available viral PCR data.
Defined as any CMV detected on transbronchial biopsy, CMV culture from BALF, or CMV shell antigen from BALF.
Excluding routine Pneumocystis carinii prophylaxis.
In comparing bacterial DNA burden, BALF specimens contained significantly more bacterial DNA than did negative control specimens (median 3563 copies per mL [IQR 2377–8316] vs 776 copies per mL [IQR 678–934], p<0·0001; appendix p 8). In comparing the identity of detected bacteria, the bacterial taxa in cell-free BALF specimens were significantly distinct from those found in negative sequencing controls (p<0·0001; appendix p 8). The dominant bacterial family in negative control specimens (Comamonadaceae) comprised 42·6% of bacterial sequences in negative controls, but only 2·3% of sequences in BALF specimens. Additional taxonomic comparisons between cell-free BALF specimens and negative controls are provided in the appendix (p 8). We concluded that our BALF specimens contained a bacterial signal that was greater than and distinct from that of negative control specimens. No specimens or taxa were excluded from subsequent analyses.
We aimed to determine whether lung bacterial burden predicts the composite outcome of CLAD development or death in the 500 days after 1-year surveillance bronchoscopy. As shown in figure 1A, the median bacterial burden in patients with the composite outcome was approximately 50% higher than in CLAD-free survivors (5·85 × 103 [IQR 2·17 × 103–5·03 × 104] vs 3·32 × 103 [2·17 × 103–5·52 × 103] copies/mL; p=0·0018). In univariable survival analysis (table 2), increased bacterial burden predicted occurrence of the composite outcome (per log10 increase in bacterial burden, HR 1·89 [95% CI 1·30–2·75], p=0·0008). Subsequently, we stratified all patients into tertiles of bacterial burden and compared their times to either CLAD development or death. As shown in figure 1B, patients in the lowest bacterial burden tertile had significantly longer CLAD-free survival than patients in the middle tertile and highest tertile. We further tested whether lung bacterial burden independently predicts CLAD development or death in a multivariable survival model. As detailed in the appendix (p 4), our model included known risk factors for CLAD and variables associated with either our primary outcome in univariable analyses or bacterial burden. As shown in table 2, increasing lung bacterial burden was an independent predictor of CLAD development or death, whether analysed as a continuous variable (per log10 increase in burden, HR 2·49 [1·38–4·48], p=0·0024) or as tertiles (middle vs lowest bacterial burden tertile, HR 4·94 [1·25–19·42], p=0·022; and highest vs lowest, HR 10·56 [2·53–44·08], p=0·0012). We thus concluded that lung bacterial burden is an independent predictor of CLAD-free survival.
Figure 1: Increased lung bacterial burden predicts chronic rejection and death after lung transplantation.
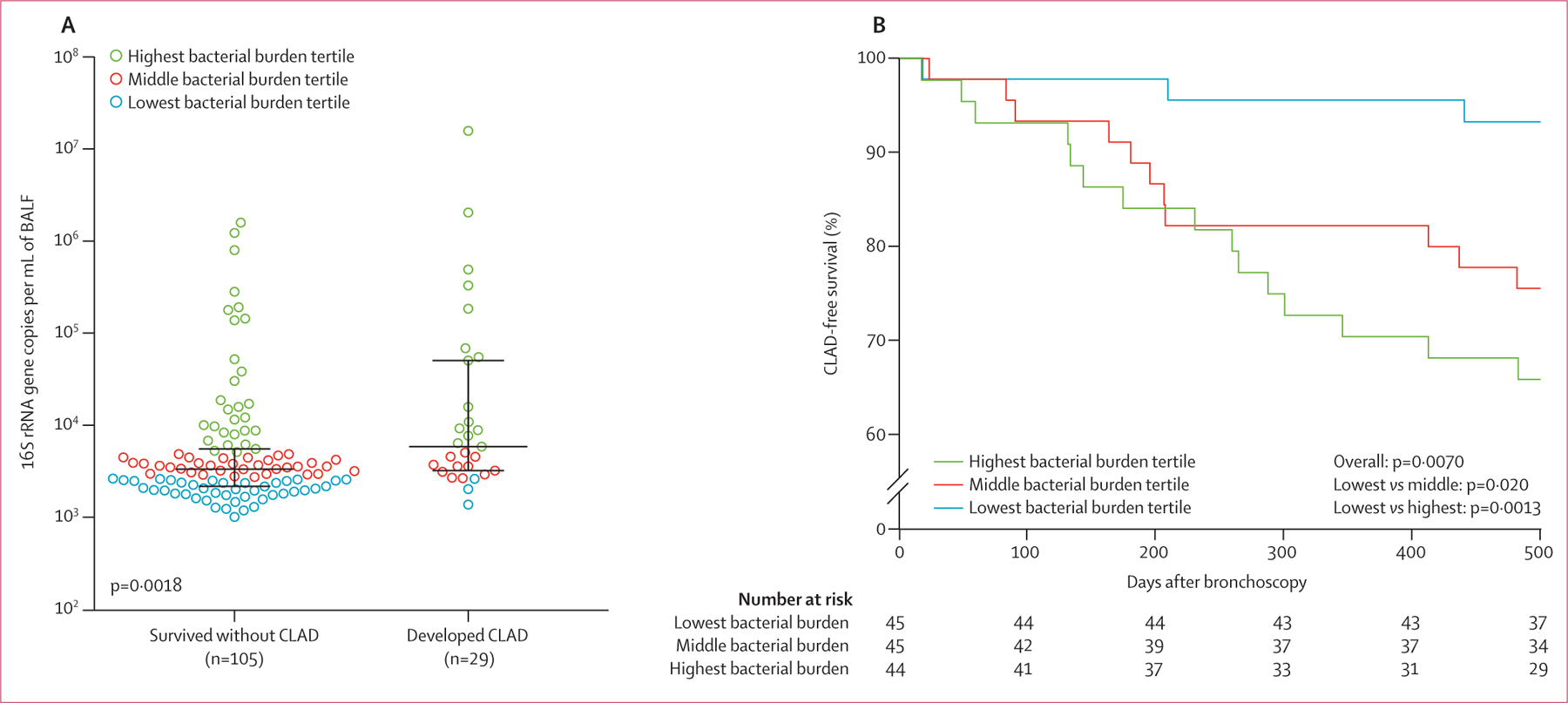
(A) BALF bacterial burden in lung transplant recipients who developed CLAD or died within 500 days after 1-year surveillance bronchoalveolar lavage was higher than in CLAD-free survivors. Lines indicate the median and IQR (log10 scale). Hypothesis testing was done with the Wilcoxon rank-sum test. (B) Kaplan–Meier curves illustrating the time to CLAD development or death, stratified by lung bacterial burden in BALF. Patients with the lowest bacterial burden had a decreased risk of developing CLAD or death compared with those with the higher bacterial burdens. Hypothesis testing was done with univariable log-rank tests. CLAD=chronic lung allograft dysfunction. BALF=bronchoalveolar lavage.
Table 2:
Predictors of CLAD development or death in the 500 days after 1-year post-transplant surveillance
| Univariable |
Multivariable |
|||
|---|---|---|---|---|
| HR (95% CI) | p value* | HR (95% CI) | p value* | |
| Age, per 1-year increase | 1·00 (0·97–1·03) | 0·91 | ·· | ·· |
| Female sex | 1·62 (0·73–3·61) | 0·24 | ·· | ·· |
| Pretransplant diagnosis | ||||
| Chronic obstructive pulmonary disease | 1 (ref) | ·· | 1 (ref) | ·· |
| Interstitial lung disease | 0·93 (0·42–2·07) | 0·86 | 1·16 (0·42–3·20) | 0·77 |
| Cystic fibrosis | 0·33 (0·09–1·17) | 0·086 | 0·27 (0·07–1·10) | 0·068 |
| Other | 0·44 (0·10–1·95) | 0·28 | 0·68 (0·12–3·68) | 0·65 |
| Double lung transplant | 0·82 (0·37–1·81) | 0·63 | ·· | ·· |
| Mean FEV1, per 1-L increase | 0·81 (0·11–5·98) | 0·83 | ·· | ·· |
| Induction Immunosuppression | ||||
| No induction | 1 (ref) | ·· | 1 (ref) | ·· |
| Basiliximab | 0·58 (0·08–4·33) | 0·60 | 0·45 (0·04–4·89) | 0·51 |
| Calcineurin inhibition | ||||
| Cyclosporine | 1 (ref) | ·· | ·· | ·· |
| Tacrolimus | 0·70 (0·31–1·58) | 0·39 | ·· | ·· |
| Antiproliferative immunosuppression | ||||
| Azathioprine | 1 (ref) | ·· | 1 (ref) | ·· |
| Mycophenylate | 0·43 (0·18–0·99) | 0·049 | 0·30 (0·11–0·85) | 0·022 |
| None or held | 1·92 (0·77–4·82) | 0·16 | 2·24 (0·53–9·39) | 0·27 |
| Primary graft dysfunction immediately (<24 h) after transplant | ||||
| Grade 0 | 1 (ref) | ·· | 1 (ref) | ·· |
| Grade 1 | 0·82 (0·28–2·41) | 0·72 | 1·03 (0·28–3·83) | 0·96 |
| Grade 2 | 0·55 (0·17–1·74) | 0·31 | 0·40 (0·11–1·44) | 0·16 |
| Grade 3 | 1·09 (0·46–2·63) | 0·84 | 1·52 (0·57–4·11) | 0·40 |
| Cumulative A rejection score, per 1-point increase | 1·01 (0·40–2·54) | 0·98 | 0·53 (0·17–1·67) | 0·28 |
| Cumulative B rejection score, per 1-point increase | 0·57 (0·07–4·40) | 0·59 | 0·44 (0·04–4·34) | 0·48 |
| DSA present† | 1·33 (0·47–3·78) | 0·59 | 1·29 (0·40–4·15) | 0·67 |
| BALF neutrophil percentage, per 10% increase‡ | 1·03 (0·83–1·27) | 0·79 | ·· | ·· |
| BALF lymphocyte percentage, per 10% increase‡ | 1·25 (0·93–1·68) | 0·14 | ·· | ·· |
| BALF bacterial culture | ||||
| Negative; no growth | 1 (ref) | ·· | 1 (ref) | ·· |
| Oral flora | 1·53 (0·61–3·86) | 0·37 | 0·94 (0·34–2·60) | 0·91 |
| Positive; speciated result | 2·02 (0·62–6·61) | 0·25 | 0·73 (0·17–3·06) | 0·66 |
| Number of previous positive BALF bacterial cultures | ||||
| None | 1 (ref) | ·· | ·· | ·· |
| One | 0·50 (0·11–2·25) | 0·37 | ·· | ·· |
| Two or more | 1·29 (0·56–2·98) | 0·55 | ·· | ·· |
| History of community-acquired respiratory viral infection§ | 1·87 (0·56–6·18) | 0·31 | 0·72 (0·11–4·62) | 0·73 |
| History of CMV pneumonitis | 1·14 (0·34–3·75) | 0·84 | 1·82 (0·39–8·54) | 0·45 |
| Antibiotics in the 30 days before bronchoscopy | 0·75 (0·23–2·48) | 0·64 | ·· | ·· |
| Azithromycin for CLAD prevention | 0·81 (0·11–5·98) | 0·83 | ·· | ·· |
| Lung bacterial burden, per log10 increase¶ | 1·89 (1·30–2·75) | 0·0008 | 2·49 (1·38–4·48) | 0·0024 |
| Lung bacterial burden tertiles¶ | ||||
| Lowest bacterial burden tertile | 1 (ref) | ·· | 1 (ref) | ·· |
| Middle bacterial burden tertile | 4·06 (1·13–14·54) | 0·032 | 4·94 (1·25–19·42) | 0·022 |
| Highest bacterial burden tertile | 5·98 (1·73–20·68) | 0·0047 | 10·56 (2·53–44·08) | 0·0012 |
The multivariable model included known risk factors for CLAD and variables associated with either our primary outcome or bacterial burden in univariable analyses (appendix p 4). HR=hazard ratio. DSA=donor-specific antibodies. BALF=bronchoalveolar lavage fluid. CMV=cytomegalovirus. CLAD=chronic lung allograft dysfunction.
Calculated with Cox proportional hazards regression models.
64 patients had no DSA information available and were analysed as a unique category (not reported).
Five patients had no BALF cell count information available and were analysed as a unique category (not reported).
Among 55 patients without respiratory viral PCR results at the 1-year surveillance bronchoscopy, 53 had no evidence of a CARV within the first year and two had a previous CARV that cleared by 1 year; no patients with excluded or analysed as missing for this variable.
Distinct multivariable analyses were done for bacterial burden as a continuous variable and bacterial burden tertiles as categorical variables.
We next asked if the community composition of lung bacteria predicts CLAD-free survival. We compared both α-diversity (within-specimen diversity) and β-diversity (across-specimen diversity of identified taxa). In our analysis of α-diversity, the lung communities of patients who developed CLAD or died did not differ from those in CLAD-free survivors, when measured either by the Shannon diversity index (mean 2·10 [SD 0·75] vs 2·06 [SD 0·61], p=0·78) or by community richness (unique bacterial taxa per sample; mean 32·5 per 1000 reads [SD 16·5] vs 27·7 per 1000 reads [13·4], p=0·11; appendix p 9). To characterise β-diversity, we visualised the community composition of lung bacteria using principal component analysis (figure 2A). Although we observed considerable overlap between specimens from patients who developed CLAD or died and from those who remained CLAD-free, a detectable difference was evident between the groups. This difference in community composition was confirmed statistically via permutation testing (figure 2A, appendix p 32). We thus concluded that the community composition of lung bacteria is associated with CLAD-free survival in lung transplant recipients.
Figure 2: Community composition of lung bacteria differs between patients who develop chronic rejection and those who do not after lung transplant.
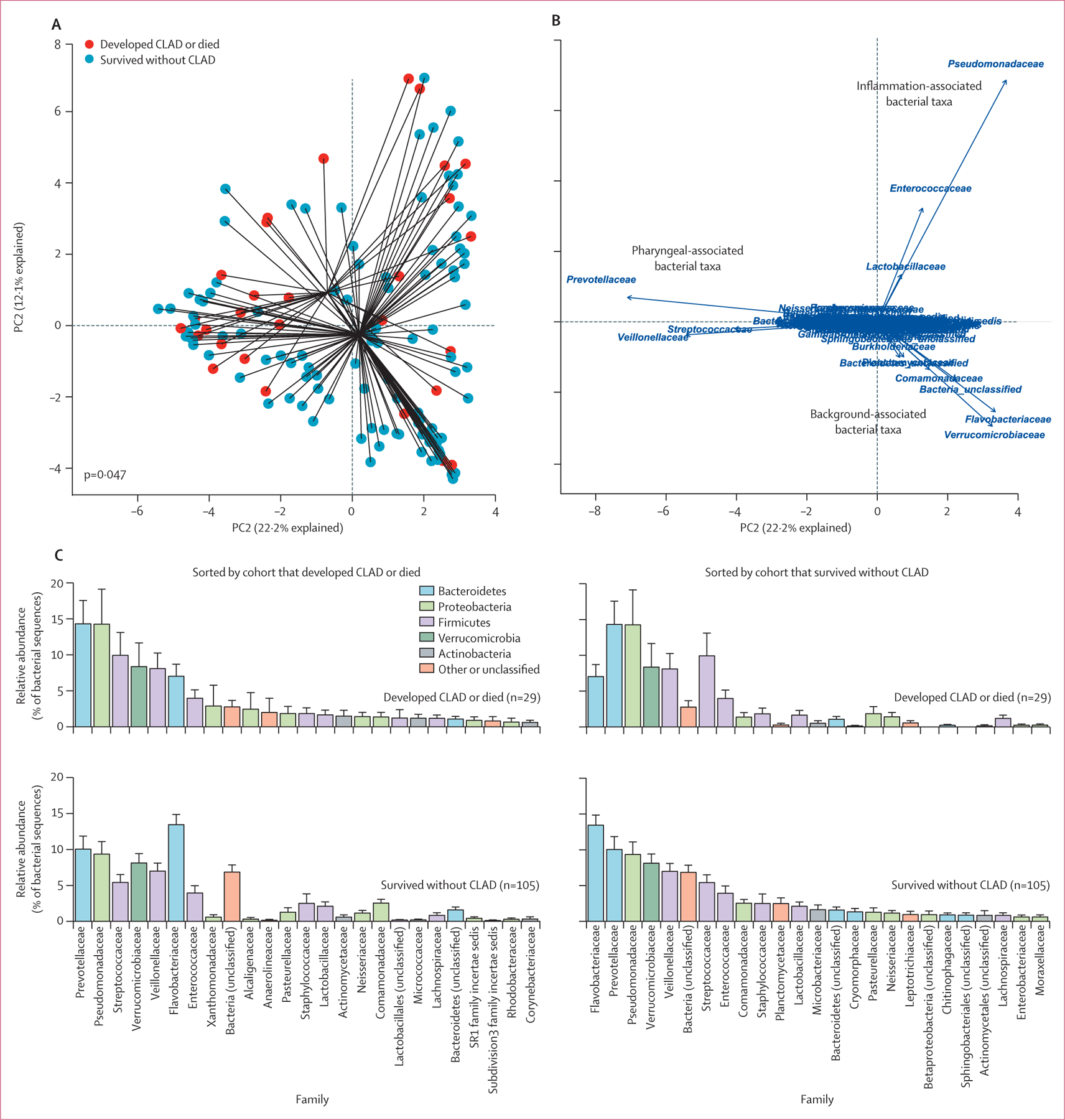
(A) The overall community composition of BALF microbiota differed between transplant recipients who developed CLAD or died and those who did not, both via permutational multivariate analysis of variance (appendix p 32) and visually via principal component analysis. (B) Biplot analysis revealed that this difference was likely to be driven by background (contaminant) taxa disproportionately abundant in transplant recipients who survived without CLAD development. (C) Rank abundance analysis identified probable contaminants (eg, Flavobacteriaceae) enriched within transplant recipients who survived without CLAD development, whereas pharyngeal-associated and infection-associated taxa (eg, Prevotellaceae and Pseudomonadaceae) were enriched in those who developed CLAD or died. CLAD=chronic lung allograft dysfunction. PC1=principal component 1.
We used complementary techniques to determine bacterial taxa responsible for the difference in community composition across outcome groups. Using a biplot analysis (figure 2B), we observed three clusters of taxa driving the separation of specimens: a cluster of pharyngeal-associated taxa commonly found in the lungs of healthy adults5,35 (eg, Prevotellaceae, Veillonellaceae, and Streptococcaceae); a cluster of inflammation-associated taxa commonly found in the lungs of patients with acute and chronic respiratory disease4,8 (eg, Pseudomonadaceae and Enterococcaceae); and a cluster of taxa associated with so-called background signal from reagent contamination21,36 (eg, Flavobacteriaceae and Verrucomicrobiaceae). The microbiota of samples from patients who developed CLAD or died were, overall, more likely to resemble the pharyngeal-associated cluster and the inflammation-associated cluster, whereas samples from patients who survived CLAD-free were more likely to resemble the background-associated cluster. We subsequently used rank abundance analysis to identify specific taxa enriched within cohorts (figure 2C). No taxa at the family level or other taxonomic levels was responsible for the difference in community composition between outcome groups (mvabund p>0·05 for individual comparisons). However, compared with patients who developed CLAD or died, CLAD-free survivors had decreased relative abundance of Prevotellaceae and Pseudomonadaceae families, with increased enrichment of the background-associated Flavobacteriaceae family. The relative abundance of the Flavobacteriaceae family was inversely associated with total bacterial burden across specimens (mvabund p=0·0010), and overall bacterial community composition of specimens was significantly associated with variation in bacterial burden (mvabund p<0·0001). We thus concluded that although community composition predicts CLAD-free survival, differences in community composition between outcome groups might be secondary to differences in total bacterial burden and susceptibility to sequencing contamination.
Given the differences in antiproliferative immunesuppression at the time of 1-year surveillance bronchoscopy between our outcome groups (table 1) and the observed protective effect of receiving mycophenolate at the time of 1-year surveillance bronchoscopy in multivariable analysis (for the composite outcome of developing CLAD or death, HR 0·30 [95% CI 0·11–0·85], p=0·022; table 2), we did exploratory and post-hoc analyses to evaluate the potential interactions between immunosuppression, lung microbiota variables, and the composite outcome (appendix pp 3–4). Differences in immunosuppression were not associated with differences in bacterial burden, community diversity, community richness, or community composition (appendix p 19). We also observed no interaction between bacterial burden and immunosuppression in survival analyses (appendix p 40). We thus concluded that antiproliferative immunosuppression at the time of 1-year surveillance bronchoscopy is not associated with lung microbiome characteristics, and the predictive significance of the microbiome in lung transplant recipients is independent of variation in immunosuppression at the time of 1-year surveillance bronchoscopy. Additionally, although previous research has shown that primary graft dysfunction,25 acute cellular rejection,27 lymphocytic bronchiolitis,37 presence of donor-specific antibodies,38,39 CMV pneumonitis,40,41 and community-acquired respiratory viral infections42 are known risk factors for CLAD, we found no association between these and characteristics of the lung microbiome (appendix pp 20–23, 28–29). As might be expected, BALF culture results were associated with bacterial burden and overall community composition (appendix p 26). However, bacterial culture results were not associated with our primary outcome in either univariable or multivariable analyses. We thus concluded that the lung microbiome represents a novel risk factor for CLAD development.
Culture-dependent studies have shown that colonisation with P aeruginosa is a risk factor for CLAD development.43–46 We therefore investigated whether the association of the lung microbiome with our composite outcome of developing CLAD or death could be attributed to P aeruginosa. In our cohort of 134 asymptomatic patients, seven (5%) had BALF cultures from the 1-year surveillance bronchoscopy that were positive for P aeruginosa; all but one of these patients had previous evidence of P aeruginosa in BALF culture in the first year of transplant consistent with colonisation (appendix p 33). Identification of P aeruginosa on BALF culture at the 1-year surveillance point was not predictive of our composite outcome (log-rank p=0·56). On 16S rRNA gene sequencing of BALF samples, 58 (43%) of 134 patients had detectable bacteria from the family Pseudomonadaceae, with relative abundance in terms of percentage of bacterial sequences in the range 1·4–97·4% (mean 24·1% [SD 24·5%]). We observed no association between presence of Pseudomonadaceae and our composite outcome when analysed either dichotomously (present vs absent, log-rank p=0·32) or continuously (per 10% increase in relative abundance, univariable Cox regression p=0·19). Finally, as described in the appendix (p 5), we identified OTU 0006 (number assigned arbitrarily) as representative of P aeruginosa, with 16 (12%) samples in our cohort having sequences identified as OTU 0006 (relative abundance range 1·4–98·6%, mean 30·9% [SD 37·8%]). We found no association between presence of OTU 0006 and CLAD development or death (present vs absent, log-rank p=0·68; per 10% increase in relative abundance, univariable Cox regression p=0·14). We thus concluded that the association between increased bacterial DNA burden and risk of CLAD development is not attributable to the presence or relative abundance of Pseudomonas spp.
Discussion
In this study, we showed that among healthy lung transplant recipients, the lung microbiome predicts the development of chronic rejection or death. Lung bacterial burden, measured 1 year after transplant, independently predicted CLAD development or death, even when controlled for other potential predictors and known risk factors for CLAD. Patient outcomes were predicted both by lung bacterial burden and by lung bacterial community composition, although not by conventional culture-based microbiological assessment. Our findings suggest that the lung microbiome is a novel and potentially modifiable risk factor for the development of chronic rejection after lung transplantation.
These results reveal the inadequacy in the current understanding of CLAD pathogenesis. Previously identified predictors of CLAD have either represented disordered host immune function (eg, primary graft dysfunction, acute cellular rejection, and donor specific antibodies25,27,37,38,47) or acute infectious episodes (eg, CMV pneumonitis and viral pneumonia40,42,48). Our results suggest that lung microbiota, even in healthy and asymptomatic patients, might participate in the pathogenesis of CLAD. Given the well established correlation between lung microbiota and lung immune tone, even in a healthy state,6,49 we believe lung bacteria are a plausible key driver of post-transplant lung inflammation and allograft dysfunction, independent of acute infections. An alternative interpretation is that lung bacteria are instead an innocent bystander in CLAD pathogenesis, reflecting some underlying host derangement. Arguing against this interpretation, we found no association between lung microbiota and indices of host dysfunction (immunosuppression, acute rejection, or lung immune cellularity), and the lung microbiome has been causally implicated in the pathogenesis of experimental models of acute and chronic lung diseases.31,50 Further research, including interventional human studies and translational animal modeling, will be necessary to confirm the causal significance of the lung microbiome in lung transplant rejection.
Our study design and analytical approach address several key limitations to previous studies of the lung microbiome in lung transplant recipients. To date, most studies have been retrospective analyses of convenience samples, limited by small sample sizes, cohort heterogeneity (eg, varied time since transplant), and confounding by acute respiratory infections.17,51–58 Our prospective study followed a large cohort of asymptomatic lung transplant recipients undergoing per-protocol surveillance bronchoscopy 1-year after transplant, minimising the potential confounding of varying time post-transplant and concurrent respiratory events that might alter the lung microbiome. Additionally, most lung transplant microbiome studies to date have focused only on the identity of lung microbiota (determined by 16S rRNA gene amplicon sequencing),51,56 whereas we incorporated ultrasensitive quantification of lung bacterial burden (determined via droplet digital PCR of the 16S rRNA gene). Although several studies have similarly reported disordered microbiota within the lungs of transplant recipients,17,51,52,57,58 to our knowledge we have shown the prognostic significance of this respiratory dysbiosis in the first adequately powered prospective study. Most previous studies have included both asymptomatic patients (undergoing surveillance bronchoscopy) and symptomatic patients (with suspected infection). Given the confounding effects of acute pneumonia on assessment of the lung microbiome,58 we believe future studies should stratify patients by the presence of clinically detected respiratory infections.
Based on culture-dependent methods, a number of studies have correlated colonisation of P aeruginosa in the airways of lung transplant recipients with subsequent development of CLAD.43–46 However, to date, no studies have shown this association with culture-independent techniques. In our study, we found no association between the presence or relative abundance of Pseudomonas spp and development of CLAD. Some studies have found that the association between airway colonisation with pathological bacteria after transplant and CLAD development is strongest in cases of de-novo colonisation,44,45,59 prompting some to suggest that recolonisation of the airways with bacteria present before transplant might be protective.53,59 We lacked pretransplant microbiological data for our cohort, and were thus unable to evaluate whether the clinical significance of detecting P aeruginosa via culture-independent methods differed among patients in whom this finding might represent recolonisation or de-novo colonisation. Future studies that use culture-independent techniques to compare the microbiota before and after transplant might clarify the undetermined role of Pseudomonas spp on transplant outcomes.
Our finding that lung bacterial burden predicts clinical outcomes in lung transplantation aligns with multiple studies in the past decade in other acute and chronic pulmonary conditions. In one prospective cohort of critically ill patients receiving mechanical ventilation, increased bacterial burden predicted fewer ventilator-free days.12 In several studies, lung bacterial burden has been found to predicted disease progression and mortality in idiopathic pulmonary fibrosis, a disease which, like CLAD, involves fibrotic remodeling of the lung.10,23,24 Prophylactic antibiotics might reduce mortality and hospitalisations in patients with idiopathic pulmonary fibrosis,60,61 and in a murine model of pulmonary fibrosis, mortality was diminished in germ-free animals.24 Although these data suggest that modification of the lung microbiome might impact disease progression, whether modification can be accomplished clinically is undetermined. In one study in lung transplant patients, azithromycin three times a week versus placebo did not result in any significant differences in lung bacterial burden or the relative abundance of key lung taxa.52 Similarly, in patients with chronic obstructive pulmonary disease, chronic azithromycin therapy did not alter lung bacterial burden.62 However, clinical response to chronic antibiotic therapy is highly variable; for instance, patients with BALF neutrophilia appear to have markedly improved pulmonary function and survival.63 Whether these azithromycin responders and non-responders differ with regard to pretreatment microbiome characteristics is not yet known. Variation in lung microbiota has recently been shown to predict patient response to inhaled antibiotic therapy in bronchiectasis.64 Future studies should establish if lung bacterial burden is modifiable with antibiotics or other interventions, and whether variation in lung microbiota explains variation in patient response to therapy after lung transplantation.
In our multivariable model, we observed a protective effect of receiving mycophenolate versus azathioprine, in terms of preventing CLAD development or death, at the time of 1-year surveillance bronchoscopy. Single centre trials of lung transplant recipients have reported lower rates of acute rejection in patients treated with mycophenolate than in those treated with azathioprine,65,66 and mycophenolate was associated with improved graft survival in a meta-analysis of renal transplant recipients.67 However, a multicentre, randomised study of lung transplant recipients found no difference between mycophenolate and azathioprine with regard to CLAD development or survival.68 As our study was not designed nor powered to evaluate the effect of immunosuppression regimen on CLAD development, we are cautious not to overinterpret the association of mycophenolate and CLAD-free survival in our study.
Our study has some limitations that warrant further investigation. First, although we detected a distinct bacterial signal in our specimens, some overlap was observed with background sequencing noise. Future studies of whole BALF, which has increased biomass and permits detection of cell-associated bacteria,57 might find stronger bacterial signal than our acellular samples and further clarify the relationship of lung bacteria and lung transplant outcomes. Additionally, although lung bacterial burden was predictive of CLAD development or death in multivariable models that accounted for confounding factors, such as differences in immunosuppression regimen at the time of bronchoalveolar lavage, residual confounding was probable. Future randomised trials investigating immunomodulatory or antimicrobial therapy in lung transplant patients could address this issue by investigating lung microbiota as a potential mediator of treatment efficacy. Finally, 16S-based sequencing cannot provide species-level discrimination or detect virulence factors. Complementary approaches, such as metagenomics, could characterise lung microbiota with improved taxonomic resolution and inform understanding of how lung bacteria affect clinical outcomes.
In summary, increased lung bacterial burden is predictive of chronic rejection and death in healthy lung transplant recipients. The lung microbiome is an important and understudied risk factor for lung allograft dysfunction. Future studies should establish whether the lung microbiome is a therapeutic target for the prevention and reversal of chronic rejection after lung transplantation.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed from database inception to March 17, 2020, for reports in any language using the search terms (“lung transplantion/lung transplant” AND “chronic rejection” OR “chronic lung allograft dysfunction” OR “bronchiolitis obliterans syndrome” AND “microbiome/microbiota”), which yielded 19 articles. Of these, nine were research articles, five were reviews, one was a case report, one was a case series, one investigated only the respiratory virome, one was an animal study, and one investigated rejection after haemopoietic stem cell transplant. Among the research articles, only two investigated the effect of the lung microbiome on subsequent CLAD development, and no articles were limited to patients without evidence of chronic rejection, lung function decline, or respiratory symptoms.
Added value of this study
Among asymptomatic lung transplant recipients undergoing 1-year post-transplant bronchoscopy, we found increased lung bacterial burden, measured in the bronchoalveolar lavage fluid, to be predictive of chronic rejection and death. Lung bacterial community composition differed between patients who remained CLAD-free and patients who developed CLAD or died, although no particular bacterial taxon—including Pseudomonas aeruginosa—was responsible for these differences.
Implications of all the available evidence
The lung microbiome generally, and bacterial burden in particular, are novel and potentially modifiable risk factors for chronic lung allograft dysfunction. Interventional studies are needed to determine if lung bacterial burden is modifiable, or whether variation in lung microbiota explains variation in patient response to therapy after lung transplantation
Acknowledgments
This study was supported by US NIH grants T32 HL007749 (to MPC and DSW), R01 HL118017 and R01 HL094622 (to VNL), and K23 HL130641 and R01 HL144599 (to RPD); by Cystic Fibrosis Foundation grant LAMA16XX0 (to VNL); and by the Brian and Mary Campbell and Elizabeth Campbell Carr research gift fund (to VNL). We are grateful to the lung transplant patients at the University of Michigan who participated in this study.
Funding
US National Institutes of Health, Cystic Fibrosis Foundation, Brian and Mary Campbell and Elizabeth Campbell Carr research gift fund.
Footnotes
Declaration of interests
MPC, DSW, VNL, and RPD report grants from the US National Institutes of Health (NIH). VNL reports grants from the Cystic Fibrosis Foundation. All other authors declare no competing interests.
Data sharing
Sequences are available via the NCBI Sequence Read Archive (accession number PRJNA615630). Operational taxonomic unit tables, taxonomy classification of sequences, and metadata tables are available online.
Contributor Information
Michael P Combs, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
David S Wheeler, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
Jenna E Luth, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
Nicole R Falkowski, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
Natalie M Walker, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
John R Erb-Downward, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
Vibha N Lama, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA.
Robert P Dickson, Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, MI, USA; Department of Microbiology and Immunology, University of Michigan, Ann Arbor, MI, USA; Michigan Center for Integrative Research in Critical Care, Ann Arbor, MI, USA.
References
- 1.Rana A, Gruessner A, Agopian VG, et al. Survival benefit of solid-organ transplant in the United States. JAMA Surg 2015; 150: 252–59. [DOI] [PubMed] [Google Scholar]
- 2.Verleden GM, Glanville AR, Lease ED, et al. Chronic lung allograft dysfunction: definition, diagnostic criteria, and approaches to treatment-a consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant 2019; 38: 493–503. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KC, Raghu G, Verleden GM, et al. An international ISHLT/ATS/ERS clinical practice guideline: diagnosis and management of bronchiolitis obliterans syndrome. Eur Respir J 2014; 44: 1479–503. [DOI] [PubMed] [Google Scholar]
- 4.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol 2016; 78: 481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson RP, Erb-Downward JR, Freeman CM, et al. Bacterial topography of the healthy human lower respiratory tract. MBio 2017; 8: e02287–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal LN, Clemente JC, Tsay JC, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 2016; 1: 16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Nuñez M, Millares L, Pomares X, et al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol 2014; 52: 4217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol 2016; 1: 16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Dwyer DN, Zhou X, Wilke CA, et al. Lung dysbiosis, inflammation, and injury in hematopoietic cell transplantation. Am J Respir Crit Care Med 2018; 198: 1312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molyneaux PL, Cox MJ, Willis-Owen SA, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2014; 190: 906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Dwyer DN, Ashley SL, Gurczynski SJ, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med 2019; 199: 1127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson RP, Schultz MJ, van der Poll T, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med 2020; 201: 555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leitao Filho FS, Alotaibi NM, Ngan D, et al. Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am J Respir Crit Care Med 2019; 199: 1205–13. [DOI] [PubMed] [Google Scholar]
- 14.Rogers GB, Zain NM, Bruce KD, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc 2014; 11: 496–503. [DOI] [PubMed] [Google Scholar]
- 15.Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med 2017; 196: 1582–90. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell AB. The lung microbiome and transplantation. Curr Opin Organ Transplant 2019; 24: 305–10. [DOI] [PubMed] [Google Scholar]
- 17.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med 2012; 186: 536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickson RP, Erb-Downward JR, Freeman CM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS One 2014; 9: e97214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason KL, Erb-Downward JR, Mason KD, et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 2012; 80: 3371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79: 5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014; 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J 2005; 26: 319–38. [DOI] [PubMed] [Google Scholar]
- 23.Invernizzi R, Barnett J, Rawal B, et al. Bacterial burden in the lower airways predicts disease progression in idiopathic pulmonary fibrosis and is independent of radiological disease extent. Eur Respir J 2020; 55: 1901519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Dwyer DN, Ashley SL, Gurczynski SJ, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med 2019; 199: 1127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daud SA, Yusen RD, Meyers BF, et al. Impact of immediate primary lung allograft dysfunction on bronchiolitis obliterans syndrome. Am J Respir Crit Care Med 2007; 175: 507–13. [DOI] [PubMed] [Google Scholar]
- 26.Snell GI, Yusen RD, Weill D, et al. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction, part I: definition and grading—a 2016 consensus group statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant 2017; 36: 1097–103. [DOI] [PubMed] [Google Scholar]
- 27.Burton CM, Iversen M, Carlsen J, et al. Acute cellular rejection is a risk factor for bronchiolitis obliterans syndrome independent of post-transplant baseline FEV1. J Heart Lung Transplant 2009; 28: 888–93. [DOI] [PubMed] [Google Scholar]
- 28.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75: 7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oksanen J, Blanchet FG, Friendly M, et al. vegan: Community Ecology Package. Sept 26, 2005. https://CRAN.R-project.org/package=vegan (accessed Nov 6, 2018).
- 30.Wang Y, Naumann U, Wright ST, Warton DI. mvabund-an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol 2012; 3: 471–74. [Google Scholar]
- 31.Harrington DP, Fleming TR. A class of rank test procedures for censored survival data. Biometrika 1982; 69: 553–66. [Google Scholar]
- 32.Savage IR. Contributions to the theory of rank order statistics-the two-sample case. Ann Math Statist 1956; 27: 590–615. [Google Scholar]
- 33.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958; 53: 457–81. [Google Scholar]
- 34.Cox DR. Regression models and life-tables. J R Stat Soc B 1972; 34: 187–202. [Google Scholar]
- 35.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013; 1: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weyrich LS, Farrer AG, Eisenhofer R, et al. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour 2019; 19: 982–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glanville AR, Aboyoun CL, Havryk A, Plit M, Rainer S, Malouf MA. Severity of lymphocytic bronchiolitis predicts long-term outcome after lung transplantation. Am J Respir Crit Care Med 2008; 177: 1033–40. [DOI] [PubMed] [Google Scholar]
- 38.Tikkanen JM, Singer LG, Kim SJ, et al. De novo DQ donor-specific antibodies are associated with chronic lung allograft dysfunction after lung transplantation. Am J Respir Crit Care Med 2016; 194: 596–606. [DOI] [PubMed] [Google Scholar]
- 39.Safavi S, Robinson DR, Soresi S, Carby M, Smith JD. De novo donor HLA-specific antibodies predict development of bronchiolitis obliterans syndrome after lung transplantation. J Heart Lung Transplant 2014; 33: 1273–81. [DOI] [PubMed] [Google Scholar]
- 40.Paraskeva M, Bailey M, Levvey BJ, et al. Cytomegalovirus replication within the lung allograft is associated with bronchiolitis obliterans syndrome. Am J Transplant 2011; 11: 2190–96. [DOI] [PubMed] [Google Scholar]
- 41.Snyder LD, Finlen-Copeland CA, Turbyfill WJ, Howell D, Willner DA, Palmer SM. Cytomegalovirus pneumonitis is a risk for bronchiolitis obliterans syndrome in lung transplantation. Am J Respir Crit Care Med 2010; 181: 1391–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalifah AP, Hachem RR, Chakinala MM, et al. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. Am J Respir Crit Care Med 2004; 170: 181–87. [DOI] [PubMed] [Google Scholar]
- 43.Vos R, Vanaudenaerde BM, Geudens N, Dupont LJ, Van Raemdonck DE, Verleden GM. Pseudomonal airway colonisation: risk factor for bronchiolitis obliterans syndrome after lung transplantation? Eur Respir J 2008; 31: 1037–45. [DOI] [PubMed] [Google Scholar]
- 44.Botha P, Archer L, Anderson RL, et al. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation 2008; 85: 771–74. [DOI] [PubMed] [Google Scholar]
- 45.Gottlieb J, Mattner F, Weissbrodt H, et al. Impact of graft colonization with gram-negative bacteria after lung transplantation on the development of bronchiolitis obliterans syndrome in recipients with cystic fibrosis. Respir Med 2009; 103: 743–49. [DOI] [PubMed] [Google Scholar]
- 46.Gregson AL, Wang X, Weigt SS, et al. Interaction between Pseudomonas and CXC chemokines increases risk of bronchiolitis obliterans syndrome and death in lung transplantation. Am J Respir Crit Care Med 2013; 187: 518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brugière O, Roux A, Le Pavec J, et al. Role of C1q-binding anti-HLA antibodies as a predictor of lung allograft outcome. Eur Respir J 2018; 52: 1701898. [DOI] [PubMed] [Google Scholar]
- 48.Weigt SS, Derhovanessian A, Liao E, et al. CXCR3 chemokine ligands during respiratory viral infections predict lung allograft dysfunction. Am J Transplant 2012; 12: 477–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am J Respir Crit Care Med 2018; 198: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashley SL, Sjoding MW, Popova AP, et al. Lung and gut microbiota are altered by hyperoxia and contribute to oxygen-induced lung injury in mice. Sci Transl Med 2020; 12: eaau9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schott C, Weigt SS, Turturice BA, et al. Bronchiolitis obliterans syndrome susceptibility and the pulmonary microbiome. J Heart Lung Transplant 2018; 37: 1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spence CD, Vanaudenaerde B, Einarsson GG, et al. Influence of azithromycin and allograft rejection on the post-lung transplant microbiota. J Heart Lung Transplant 2020; 39: 176–83. [DOI] [PubMed] [Google Scholar]
- 53.Willner DL, Hugenholtz P, Yerkovich ST, et al. Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome. Am J Respir Crit Care Med 2013; 187: 640–47. [DOI] [PubMed] [Google Scholar]
- 54.Sharma NS, Wille KM, Athira S, et al. Distal airway microbiome is associated with immunoregulatory myeloid cell responses in lung transplant recipients. J Heart Lung Transplant 2017; 37: 206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernasconi E, Pattaroni C, Koutsokera A, et al. Airway microbiota determines innate cell inflammatory or tissue remodeling profiles in lung transplantation. Am J Respir Crit Care Med 2016; 194: 1252–63. [DOI] [PubMed] [Google Scholar]
- 56.Mouraux S, Bernasconi E, Pattaroni C, et al. Airway microbiota signals anabolic and catabolic remodeling in the transplanted lung. J Allergy Clin Immunol 2018; 141: 718–29.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dickson RP, Erb-Downward JR, Prescott HC, et al. Cell-associated bacteria in the human lung microbiome. Microbiome 2014; 2: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dickson RP, Erb-Downward JR, Freeman CM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS One 2014; 9: e97214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orfanos S, Gomez C, Baron S, et al. Impact of gram negative bacteria airway recolonization on the occurrence of chronic lung allograft dysfunction after lung transplantation in a population of cystic fibrosis patients. BMC Microbiol 2018; 18: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macaluso C, Maritano Furcada J, Alzaher O, et al. The potential impact of azithromycin in idiopathic pulmonary fibrosis. Eur Respir J 2019; 53: 1800628. [DOI] [PubMed] [Google Scholar]
- 61.Shulgina L, Cahn AP, Chilvers ER, et al. Treating idiopathic pulmonary fibrosis with the addition of co-trimoxazole: a randomised controlled trial. Thorax 2013; 68: 155–62. [DOI] [PubMed] [Google Scholar]
- 62.Segal LN, Clemente JC, Wu BG, et al. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 2017; 72: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vos R, Vanaudenaerde BM, Ottevaere A, et al. Long-term azithromycin therapy for bronchiolitis obliterans syndrome: divide and conquer? J Heart Lung Transplant 2010; 29: 1358–68. [DOI] [PubMed] [Google Scholar]
- 64.Sibila O, Laserna E, Shoemark A, et al. Airway bacterial load and inhaled antibiotic response in bronchiectasis. Am J Respir Crit Care Med 2019; 200: 33–41. [DOI] [PubMed] [Google Scholar]
- 65.Speich R, Schneider S, Hofer M, et al. Mycophenolate mofetil reduces alveolar inflammation, acute rejection and graft loss due to bronchiolitis obliterans syndrome after lung transplantation. Pulm Pharmacol Ther 2010; 23: 445–49. [DOI] [PubMed] [Google Scholar]
- 66.Ross DJ, Waters PF, Levine M, Kramer M, Ruzevich S, Kass RM. Mycophenolate mofetil versus azathioprine immunosuppressive regimens after lung transplantation: preliminary experience. J Heart Lung Transplant 1998; 17: 768–74. [PubMed] [Google Scholar]
- 67.Wagner M, Earley AK, Webster AC, Schmid CH, Balk EM, Uhlig K. Mycophenolic acid versus azathioprine as primary immunosuppression for kidney transplant recipients. Cochrane Database Syst Rev 2015; 12: CD007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNeil K, Glanville AR, Wahlers T, et al. Comparison of mycophenolate mofetil and azathioprine for prevention of bronchiolitis obliterans syndrome in de novo lung transplant recipients. Transplantation 2006; 81: 998–1003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


