Abstract
Enzymes boost protein engineering, directed evolution, and the biochemical industry and are also the cornerstone of metabolic engineering. Basidiomycetes are known to produce a large variety of terpenoids with unique structures. However, basidiomycetous terpene synthases remain largely untapped. Therefore, we provide a modeling method to obtain specific terpene synthases. Aided by bioinformatics analysis, three γ-cadinene enzymes from Ganoderma lucidum and Ganoderma sinensis were accurately predicted and identified experimentally. Based on the highly conserved amino motifs of the characterized γ-cadinene enzymes, the enzyme was reassembled as model 1. Using this model as a template, 67 homologous sequences of the γ-cadinene enzyme were screened from the National Center for Biotechnology Information (NCBI). According to the 67 sequences, the same gene structure, and similar conserved motifs to model 1, the γ-cadinene enzyme model was further improved by the same construction method and renamed as model 2. The results of bioinformatics analysis show that the conservative regions of models 1 and 2 are highly similar. In addition, five of these sequences were verified, 100% of which were γ-cadinene enzymes. The accuracy of the prediction ability of the γ-cadinene enzyme model was proven. In the same way, we also reanalyzed the identified Δ6-protoilludene enzymes in fungi and (−)-α-bisabolol enzymes in plants, all of which have their own unique conserved motifs. Our research method is expected to be used to study other terpenoid synthases with a similar or the same function in basidiomycetes, ascomycetes, bacteria, and plants and to provide rich enzyme resources.
Introduction
Basidiomycetes are a strong division of higher fungi because of their 30000 + species richness and are the key players in the global ecosystem carbon cycle and the production of small molecular bioactive compounds.1,2 The natural products of mushrooms have a breathtaking structural diversity, especially sesquiterpenoids, diterpenoids, and triterpenoids, which are formed by terpene synthases (TSs) catalyzing precursors geranyl diphosphate (GPP), (2E,6E)-farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP) in a process of biosynthesis.3−6 Subsequently, the modification of terpene gene clusters further increases the structural diversity of terpenoids, including many reported active natural products, such as the antimicrobial compound lagopodin B from Coprinopsis cinerea,7,8 antitumor compounds illudin M and S9,10 of Omphalotus olearius and O. illudens, and antibiotic melleolides.11,12 Terpenoids are natural products with extremely diversified structures, but just a small level of them is explored thus far.13
Sesquiterpenoids are the largest terpenoid group in Basidiomycetes, and a considerable number of compounds with chemical structures different from those produced by other microbes have been identified. However, because Basidiomycetes are difficult to culture under laboratory conditions and because of the complexity of genetic domestication, compared with plants, bacteria, and ascomycetes, there are few studies on the sesquiterpene synthase (STS) biosynthesis pathway.5,14−16 Each basidiomycete has an average of 10–20 hypothetical STS homologues,10 indicating that basidiomycetous sesquiterpenoids and STSs represent rich but largely unexploited natural resources. Over the past decade, with the continuous development of sequencing technology and the emergence of a large amount of genomic data, a certain basis for the study of STSs has been provided but has also brought challenges for gene characterization. The groundbreaking work, identification of five STSs (Cop1–4 and Cop6) from C. cinerea,7 was carried out in 2009. In 2012, the STSs (Omp1–10) from O. olearius and 40 available basidiomycete genomes provided a prediction framework for sesquiterpene biosynthesis in basidiomycetes, which can predict the products of the corresponding cyclization mechanism according to the five classes of the phylogenetic tree.10 In 2013, the cloning and characterization of STSs [Stehi1_64702 (now ShSTS15), Stehi1_73029 (now ShSTS16), and Stehi1_25180 (now ShSTS18)] from S. hirsutum proved the accuracy of the prediction framework,17 but this method cannot be used to predict the specific structures of sesquiterpenoids. Until 2020, when STSs (Agr1-9) from Agrocybe aegerita were identified, sequence similarity networks (SSNs) were established by using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST, http://efi.igb.illinois.edu/efi-est/)18 to probe similar or isofunctional fungal STSs and products, which further broadened the knowledge of basidiomycetous STSs, but Galma_104215 from Ganoderma marginata and Pilcr_825684 from P. croceum were not correctly predicted,19 indicating that there are some limitations to predicting the functions of STSs by SSNs. Therefore, this study puts forward a new point of view to accurately identify the novel STSs we need and provides some additions for the STS prediction framework.
In the recently reported and sequenced genomes of G. lucidum(20) and G. sinensis,21 we have identified three highly selective γ-cadinene synthases (γ-CSs). Ganoderma, a world-famous medical macrofungus in Basidiomycetes, has been used to treat a variety of diseases for more than 2000 years. Ganoderma is one of the most deeply studied medicinal model organisms22 and has also been used as bonsai, which represents a great commercial value. Triterpenes and polysaccharides are the most studied bioactive components in Ganoderma.23 In addition, sesquiterpene oxygen derivatives with antifungal activity have also been reported,24 but there are few studies on Ganoderma sesquiterpenoids, which are worthy of further study. Three STSs (GS11330, GS14272, and GS02363) from G. sinensis(25,26) and 1 STS (GL26009) from G. lucidum(27) have been identified previously. In this study, three interesting STSs from their STS family with the same gene structure and sequence characteristics were cloned and characterized in Escherichia coli chassis strains. The homology model of γ-CS was established according to the conservatism of sequences, and five sequences verified the accuracy of this model used to identify novel γ-CSs. When we used this modeling method to analyze the identified Δ6-protoilludene synthases19 in Basidiomycetes and (−)-α-bisabolol synthases (BOSs)28 in plants, we found that these highly conserved sequences can still be used to model and discover more new and valuable enzymes and gene clusters. In the future, this modeling method is expected to be extended to Basidiomycetes, ascomycetes, plants, and bacteria, providing unlimited enzyme resources for the production of target products and intermediates.
Results and Discussion
Identification of γ-CSs in G. lucidum and G. sinensis
Over the past decade, more than 80 kinds of TSs in Basidiomycetes have been characterized.1 By retrieving the literature and searching the JGI Basidiomycetous genome database,29 the number of Basidiomycetes has increased from 113 in 201414 to 605 thus far. The influx of sequenced and annotated genes has brought new opportunities and challenges to the research of natural products. In the early stage of this study, by analyzing the genome sequencing data of G. lucidum and G. sinensis, we found two particularly interesting STS genes, GlSTS6 and GsSTS43, whose exon number was the highest in the family, and the exon size and splicing pattern of each gene were consistent (Table 1). We preliminarily speculate that the gene containing this nine-exon gene structure may have evolved a more conserved similar function, so next, this study aimed to predict its function by bioinformatics and carry out experimental characterization.
Table 1. Comparison of Exon/Intron Size in the Subtree.
| exon1 | intron1 | exon2 | intron2 | exon3 | intron3 | exon4 | intron4 | exon5 | intron5 | exon6 | intron6 | exon7 | intron7 | exon8 | intron8 | exon9 | length | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpSTS18 | 131 | 52 | 139 | 52 | 137 | 58 | 48 | 63 | 134 | 53 | 107 | 55 | 18 | 52 | 244 | 58 | 224 | 1625 |
| STC9 | 131 | 57 | 139 | 53 | 137 | 48 | 48 | 50 | 134 | 48 | 107 | 56 | 18 | 49 | 244 | 55 | 122 | 1496 |
| ShSTS5 | 140 | 55 | 139 | 57 | 137 | 59 | 48 | 48 | 134 | 54 | 107 | 58 | 18 | 52 | 244 | 57 | 86 | 1493 |
| GsSTS45b | 140 | 67 | 136 | 64 | 137 | 51 | 48 | 54 | 134 | 59 | 107 | 62 | 18 | 72 | 244 | 102 | 269 | 1764 |
| GISTS6 | 140 | 80 | 136 | 66 | 137 | 51 | 48 | 62 | 134 | 49 | 107 | 62 | 18 | 55 | 244 | 76 | 296 | 1761 |
| GsSTS43 | 140 | 80 | 136 | 66 | 137 | 53 | 48 | 63 | 134 | 46 | 107 | 62 | 18 | 56 | 244 | 85 | 293 | 1768 |
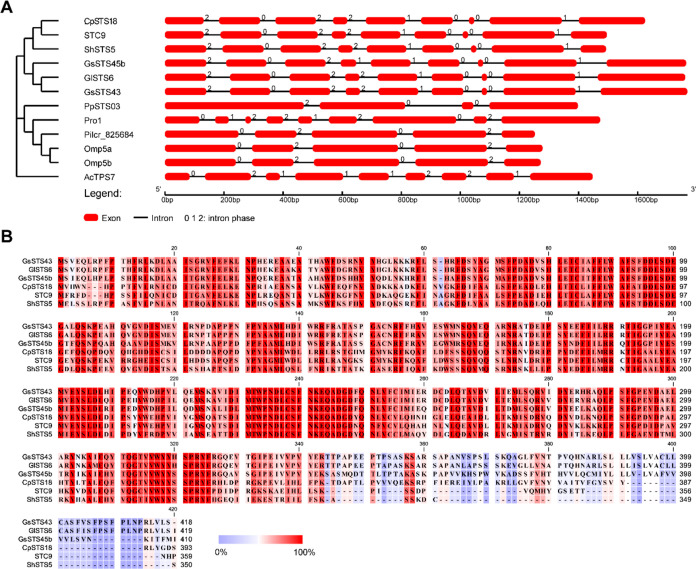
We sorted out the STS genes of Basidiomycetes that had been characterized in the last 10 years, marked their exon number, and found that the average exon number was 5.53 (see the Supporting Information), indicating that nine exons were obviously dominant in number and may have a specific relationship with function. The phylogenetic tree of the characterized STS genes was constructed with GlSTS6 and GsSTS43. The STS gene was in the same small branch as STC9,30 ShSTS5, and CpSTS1831 (see the Supporting Information), which are all specific enzymes coding a single product of γ-cadinene. Therefore, we further speculated that GlSTS6 and GsSTS43 may also be γ-CSs. To improve the accuracy of prediction, we selected genes containing γ-cadinene products and nine exons to reconstruct the gene structure-phylogenetic tree (Figure 1A). The gene structures of GlSTS6 and GsSTS43 were highly similar to the gene structures of STC9, ShSTS5, and CpSTS18, while PpSTS03,32 Pilcr_825684,19 and Omp5a/b10 were all γ-CSs, but their genetic relationships were distant, and their gene structures were also different, which may result from their nonspecific enzymes. Although Pro111 and AcTPS733 have nine exons, the structure and function of the genes are very different (see Figure 1A and the Supporting Information). The size comparison of exons 1–9 of GlSTS6, GsSTS43, STC9, ShSTS5, and CpSTS18 showed that exons 3–8 were the same except exons 1, 2, and 9, indicating that they had evolved a neat gene structure. In addition, multiple sequence alignments were carried out, and the highest sequence similarity was found to be 51.8%, and the lowest was 3.8% (see the Supporting Information). Although the overall sequence similarity was not high, many motifs were highly conserved (Figure 1B). Therefore, according to the high conservation of gene structure and amino acid residue sites, we are more convinced that GlSTS6 and GsSTS43 are γ-CSs.
Figure 1.
Gene structure-phylogenetic tree and sequence alignment of γ-CSs. (A) Gene structure-phylogenetic tree of γ-CS sequences and nine-exon genes. (B) Amino acid sequence alignments of six γ-CSs. Amino acids range from low (blue) to high (red) conservation.
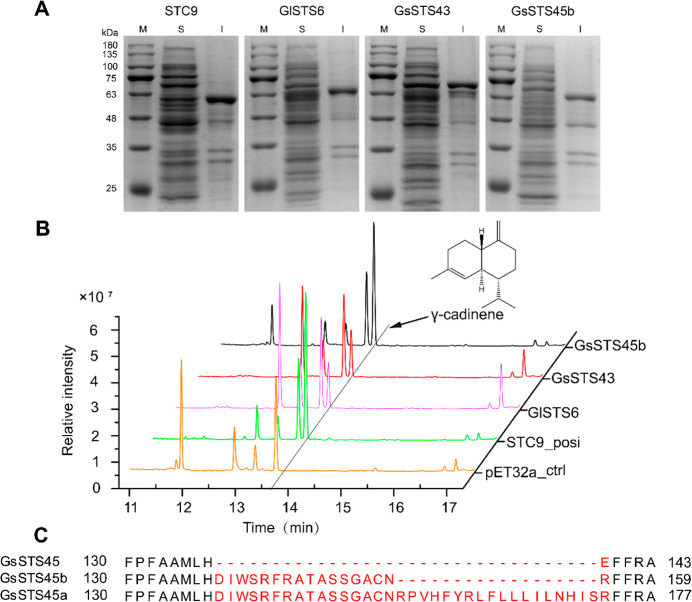
Through the extraction of RNA from G. lucidum and G. sinensis, GlSTS6 and GsSTS43 were cloned by cDNA, and pGEM T Easy-GlSTS6 and pGEM T Easy-GsSTS43 clone vectors and pET32a-GlSTS6 and pET32a-GsSTS43 fusion protein expression vectors were all successfully constructed. The expression of the soluble protein was low, and most of them formed inclusion bodies (Figure 2A). Since there is no market standard for γ-cadinene, we synthesized the STC9 gene. The expression vectors of fusion proteins for pET32a-GlSTS6, pET32a-GsSTS43, and pET32a-STC9 were transferred into the hosts of E. coli Rosetta (DE3), and the products were detected by HS-SPME-GC-MS in the headspace of E. coli culture medium. The results showed that E. coli cells expressing GlSTS6 and GsSTS43 produced a single peak of γ-cadinene (the MS spectra for γ-cadinene can be found in the Supporting Information), consistent with the major product of STC9. There are the minor products of low response value for STC9, together accounting for ≈3.4% of (relative peak area) the total sesquiterpenes in the headspace of E. coli cultures expressing STC9. In addition, when we cloned GsSTS45, a member of the STS family of G. sinensis, two correct transcripts, GsSTS45a and GsSTS45b, were also obtained by manual reannotation, in which GsSTS45b is a functional single-product γ-CS and has gene structure and amino acid residues similar to that of GlSTS6, GsSTS43, STC9, ShSTS5, and CpSTS18 (Figure 2B,C). Therefore, the function of genes can be initially predicted by bioinformatics analysis, such as comparison of exon size, gene structure-phylogenetic tree, sequence alignment, and so forth.
Figure 2.
Characterization of γ-CSs from G. lucidum and G. sinensis in E. coli. (A) Expression of various γ-CSs from G. lucidum and G. sinensis. Lane M, molecular weight marker protein; S, soluble fraction of total cell extracts; and I, insoluble fraction of total cell extracts. (B) GC–MS chromatograms of cultural supernatants of strains GlSTS6, GsSTS43, and GsSTS45b and the control strains pET32a _ctrl and the positive STC9_posi. (C) Prediction and cloning of a gene in the G. sinensis STS family. The predicted gene was GsSTS45. The cloned and reannotated genes were GsSTS45a and GsSTS45b, and only GsSTS45b revealed to be functional.
Homology Search for Unidentified γ-CSs in Basidiomycetes
To explore whether other highly conserved genes with this special gene structure would have the same function in Basidiomycetes, we established homology model 1 (see the Supporting Information) of γ-CS based on the conserved regions of GlSTS6, GsSTS43, GsSTS45b, STC9, ShSTS5, and CpSTS18, with amino acid residues with high similarity extracted and recombined into a new γ-CS protein sequence. Using this sequence as a template for protein BLASTA searching in the NCBI nonredundant protein sequence (nr), 67 genes were obtained by manually deleting the sequences in which exon introns were too long or too short or the number difference and the lack of terpenoid-conserved domains DDXXD and NSE/DTE. Finally, 67 genes were obtained, which were distributed among 31 genera and 58 species (see the Supporting Information).
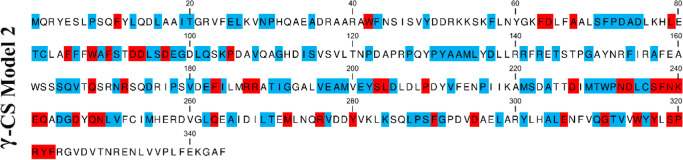
To improve the accuracy of prediction model 1, we combine the following three strategies: (1) comparison of gene structure, (2) multiple sequence alignment, and (3) homologous modeling. First, 67 sequences were used to construct the gene structure-phylogenetic tree, which was divided into approximately three classes, and as a whole, the intron phase was conserved (see the Supporting Information). The gene structure of branch I is the most similar, in which branch Ia is almost the species of Suillus, which may contain rich γ-cadinene. The exon 9 length of branch II is larger than the exon 9 length of branches I and III, indicating that the C-terminus of branches I and III is truncated or that the C-terminus of branch II is lengthened to varying degrees. To accurately analyze their structure, we counted the number of bases for each exon of each gene, and the average is as follows: exon1: 134 bp, exon2: 139 bp, exon3: 137 bp, exon4: 48 bp, exon5: 134 bp, exon6: 107 bp, exon7: 18 bp, exon8: 244 bp, and exon9: 119 bp, among which most of exon 9 was 74 bp (see the Supporting Information). The average value of exons 3–8 is the same as the average value of homologous model 1, so we infer that the same splicing pattern of exons 3–8 may be related to the function of STSs. Then, a multisequence alignment of 67 amino acid sequences was carried out, and most of the amino acid residues were found to be highly conserved. In addition to the conserved motifs DDXXD and NSE/DTE shared by terpenoids, they also contained many unique conserved motifs, especially upstream and downstream of these two conserved motifs, such as FFXWAFSXDDLSDEGXLQXFP and DXMTWPNDLCSFNKEQXDGDXQNLV, as well as several conserved regions, such as FDXXAXLSFPDAD, PYAAMLXD, FIXXRR, and QGTVXWYYXSPRYF (see the Supporting Information). These conserved regions are almost the same as homologous model 1. Based on this knowledge, we established homology model 2, in which the amino acid residues with high similarity in 67 sequences were extracted and reassembled into a new protein sequence (Figure 3). Our goal is to use this sequence and >80% of the highly conserved regions as templates to identify γ-CSs in Basidiomycetes and to provide a new method for active search and accurate identification of some important STSs.
Figure 3.
Homology model 2 of γ-CS was constructed. Model 2 based on conservative regions of 67 sequences (see the Supporting Information) retrieved by model 1 (see the Supporting Information) in NCBI. Red represented 100% conservatism, and blue represents >80% conservatism of amino acid.
Verification of Predictive Ability for Homology Model 2
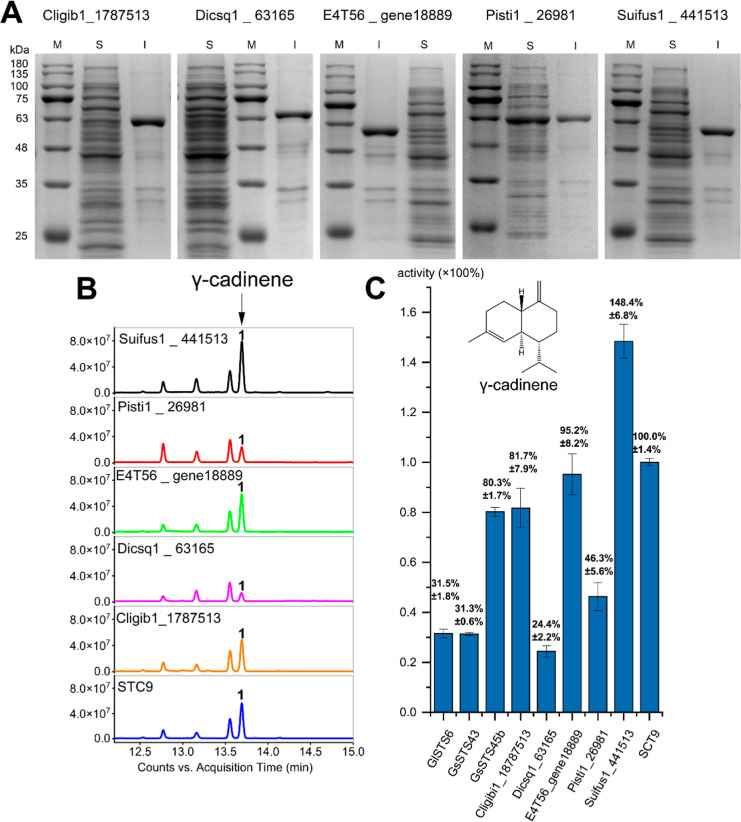
By establishing the connection between sequence conservation and products, it is particularly important for us to obtain more accurate prediction models to explore or design enzymes with better yield, activity, and selectivity. To test the prediction ability of homologous model 2 from each small class, we manually selected five sequences with high similarity to the conservative region of model 2 for functional verification: Cligib1_1787513 from Infundibulicybe gibba, Dicsq1_63165 from Dichomitus squalens LYAD-421 SS1, E4T56_gene18889 from Termitomyces sp. T112, Pisti1_26981 from Pisolithus tinctorius Marx 270, and Suifus1_441513 from Suillus fuscotomentosus (see the Supporting Information). Each sequence was chemically synthesized and connected with pET32a vectors to construct a fusion protein expression vector and cloned into E. coli chassis strains to express the product. Each protein was approximately 65 kDa, and the soluble protein content of Pisti1_26981 was the highest (Figure 4A). Fortunately, the E. coli clones expressing Cligib1_1787513, Dicsq1_63165, E4T56_gene18889, Pisti1_26981, and Suifus1_441513 all produced γ-cadinene (Figure 4B), which was consistent with STC9, ShSTS5, and CpSTS18. Similar to GlSTS6, GsSTS43, and GsSTS45b, Dicsq1_63165 and Pisti1_26981 produced only a single γ-cadinene product, while Cligib1_1787513, E4T56_gene18889, and Suifus1_441513, like STC9, produced the main product γ-cadinene > 95%, as well as the two same small product peaks, with a total content of <5%. The yield of Suifus1_441513 is 6.08 times higher than the yield of Dicsq1_63165 (Figure 4C), which proves the accuracy of the prediction ability of homology model 2 constructed in this experiment, and homology model 2 can also be used to find more active enzymes. When we used homology model 2 to search homologous proteins among 605 Basidiomycetes from the JGI Genome database, we found that the sequences with gene structures similar to homology model 2 were endless, which provided abundant candidate genes for accurate identification of γ-CS. At the same time, our work method is also expected to be used to find other TSs with potential application value and better selectivity.
Figure 4.
Verification of predictive ability for homology model 2, and γ-CSs were selected according to a principle that the amino acid sequence in each branch of the phylogenetic tree is most similar to the red and blue amino acids of model 2. (A) Expression of five selected γ-CSs. Lane M, molecular weight marker protein; S, soluble fraction of total cell extracts; and I, insoluble fraction of total cell extracts. (B) GC–MS chromatograms of cultural supernatants of five γ-CS strains. (C) Production of γ-cadinene for five γ-CSs. STC9 production of γ-cadinene is set to 100%; error bars indicate standard deviations determined from triplicates.
Study on the Functions of Other TSs
In previous research work, the characterized TSs were used to predict the possible TS genes in the sequenced and annotated genome databases, and then these genes were directly cloned or synthesized after cyclization mechanisms, or products of TSs were predicted by constructing evolutionary trees. SSNs were constructed by using the Enzyme Function Initiative-Enzyme Similarity Tool to predict or characterize new TSs, such as 15 characterized Δ6-protoilludene synthases.19 Novel enzymes with higher activity and yield were also found by multiple sequence alignments, such as (−)-α-bisabolol synthase,28 or combining two strategies: (1) full-sequence alignment and (2) comparison of predicted active sites to identify linalool synthases.34 One thing is in common in the discovery of all TSs: the conservation of sequences is used to predict and identify TSs, which coincides with the central point of view of this study. The establishment of a highly conservative model can be used to find the required enzymes. Therefore, we want to use this method to reanalyze some identified TSs to establish the relationship between sequence conservation and specific products.
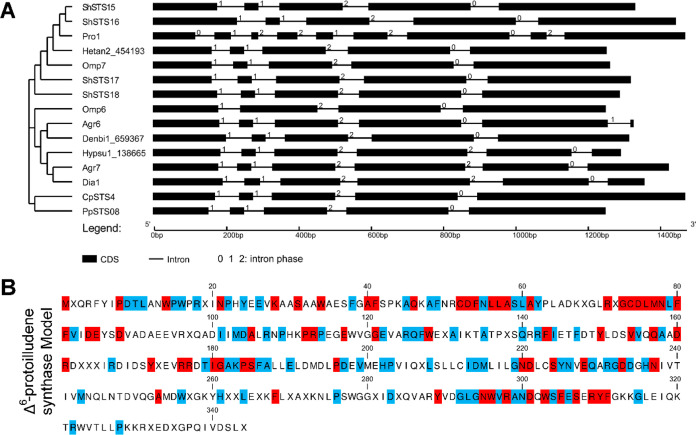
After 15 gene sequences of characterized Δ6-protoilludene synthases cloned by cDNA or synthesized by reannotating were rearranged and used to reconstruct the gene structure-phylogenetic tree, we found that the splicing patterns of the other 14 genes were similar except Pro1 (Figure 5A). Based on the intron/exon pattern, Pro1 may have evolved from a distant ancestor plant terpene synthase containing 12 introns and 13 exons.35 Dia1, a Δ6-protoilludene synthase from Diaporthe sp. in ascomycetes, a cross20 phyla horizontal gene transfer event between Basidiomycota and Ascomycota of BR109, has been identified.36−39 The small Omp7 cluster is a gene duplication from the large Omp6 cluster to improve the rate-limiting steps in the biosynthesis of illudin compounds.10 Then, through multiple sequence alignments, we further found that 15 Δ6-protoilludene synthases also contain many unique conserved motifs, such as RXGCDLMNLFFVXDEXXD and GNDXXSYNXEQXRGDDXHN upstream and downstream of the traditional conserved motif of terpene synthase, as well as CDFNLLASLAY, VVXQAXDR, YXXXRRXTIGAKPSFA, and GLGNWVRANDXWSFESXRYF (see the Supporting Information), not one by one. Finally, the Δ6-protoilludene synthase homology model protein sequence containing 344 amino acids was established and assembled by using the method of this study (Figure 5B). We can use this model to look for more Δ6-protoilludene synthase candidate genes and corresponding gene clusters in the JGI genome database, which can bring rich enzyme resources for the study of biosynthesis pathways of illudin with antitumor and antimicrobial effects and may even bring better titer intermediates.
Figure 5.
Gene structure-phylogenetic tree and homology model of Δ6-protoilludene synthases. (A) Gene structure-phylogenetic tree of 15 characterized Δ6-protoilludene synthases (see the Supporting Information). (B) Homology model of Δ6-protoilludene synthases. The modeling method is the same as the γ-CS; red represents 100% conservatism, and blue represents >80% conservatism of the amino acid.
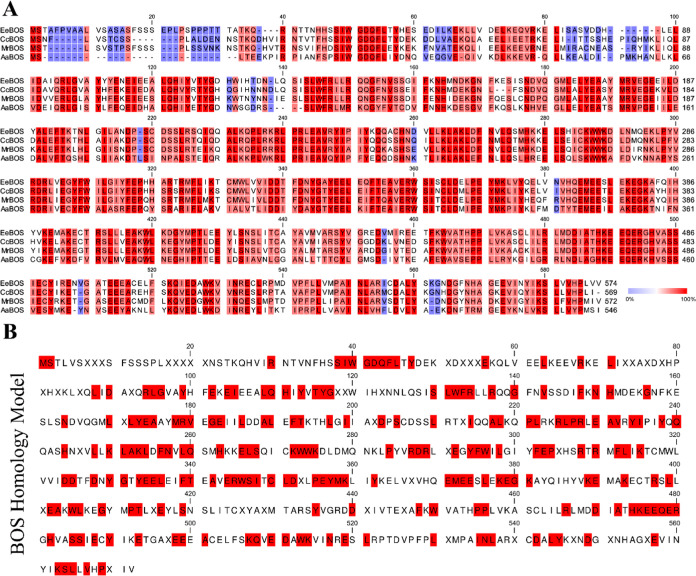
In addition, as one of the four stereoisomers of α-bisabolol, (+)-α-bisabolol,40 (−)-α-bisabolol,28,41−44 (+)-epi-α-bisabolol,45 and (−)-epi-α-bisabolol,46 (−)-α-bisabolol, which is a monocyclic sesquiterpene alcohol, is highly valued and widely used as an active ingredient in the cosmetics and pharmaceutical industries.28,47 The productivity of (−)-α-bisabolol has been improved by using engineered microbes and high-efficiency BOS for the industrial production. To obtain BOS with better activity and selectivity, the researcher combined three strategies: (1) use the identified enzyme homology search, (2) build a phylogenetic tree to lock a smaller one, and (3) multiple sequence alignments and finally obtained a higher yield of CcBOS from Cynara cardunculus var. scolymus.28 The identified process for the new BOS in this study is similar to our research method. Therefore, we can construct a BOS homology model to discover more novel and more active BOS enzyme resources. Then, the BOS homology model (Figure 6B) was built by sequence alignment of BOS (Figure 6A), and we can find that BOS also has unique motifs, such as SIWGDCFL, DDXXDXYGXYEELXXFTXAXERWSIXCLDXXPEYMK, HKEEQER, and so forth. At the same time, we can make use of the differences in the conserved sites of amino acid sequences to study the key active sites of the BOS enzyme and develop more selective and active BOS by site-directed mutagenesis. Finally, more detailed information on the BOS homology model will be provided, and combined with bioinformatics technology, the method of BOS identification will also be simplified. The discovery of a new type of BOS indicates that there may be more highly conserved motifs of TS in the plant kingdom than in Basidiomycetes,28 and further studies will be needed. Moreover, the results also show that the method of this study is applicable in the plant kingdom. In the future, with the increasing number of TS characterizations, there are believed to be an increasing number of homology models, which will make it easier for us to identify the enzymes we need. At the same time, the increasing number of TS characterizations can also promote the development of bioinformatics technology and make the characterization of new TSs more convenient.
Figure 6.
Sequence alignment and homology model of BOSs. (A) Amino acid sequence alignments of four BOSs. MrBOS, Matricaria recutita (AIG92846.1); AaBOS, Artemisia annua (AFV40969.1); EeBOS, E. erythropappus (DC) McLeisch (AYJ71561.1); and CcBOS, C. cardunculus var. scolymus (XP_024994640.1). Amino acids range from low (blue) to high (red) conservation. (B) Homology model of BOSs. The modeling method is the same as the γ-CS, red represents 100% conservatism.
Conclusions
Through gene structure, phylogenetic tree construction, and multisequence alignment analysis, we predicted and screened three interesting genes in G. lucidum and G. sinensis for cloning and characterization. GC–MS detection results showed that the three genes were consistent with previous research results, all of which were highly specific γ-CSs. Based on the high conservation of the sequences, we constructed the γ-CS homology model 1 and established the one-to-one correspondence between the conserved residue sites of the sequence and γ-cadinene products. In order to make the verification method more convincing, we used homologous model 1 to retrieve 61 highly conserved sequences in NCBI, and based on the principle of being as similar as possible to conservative residues, we selected five genes for functional verification, 100% of which were γ-CSs. This study is to hope that this method of SEQUENCE homology modeling can be used to find more efficient enzymes, such as BOS, Δ6-protoilludene synthase, and so forth. We also hope that γ-CS can have new applications in future research and encourage more scientific researchers to construct more other sequence homology models to improve the productivity of enzymes.
Methods
Strains and Growth Conditions
G. lucidum and G. sinense strain sources and growth conditions were carried out as described previously.20,21E. coli DH5α was used to amplify genes and clone plasmids. E. coli Rosetta (DE3) was used to express proteins and produce sesquiterpenoids. Lysogeny broth (LB) medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with the appropriate antibiotic(s) ampicillin (50 μg mL–1), chloramphenicol (25 μg mL–1), and kanamycin (30 μg mL–1) was used to culture the cells. Isopropyl-β-D-1-thiogalactopyranoside (IPTG) induced the expression of genes involved in sesquiterpenoid synthesis.
Gene Predictions and Selection
Family STS gene annotation, alignment, structure prediction, and manual correction were performed as described previously.20,21 Briefly, four software programs were used for ab initio gene identification, including Augustus, GeneMark, Fgenesh, and SNAP. STS gene predictions were manually corrected using Apollo software. Two STS families were aligned with functionally characterized basidiomycetous STSs, and a neighbor-joining phylogenetic tree was subsequently produced using MEGAX.48 Moreover, a gene structure-phylogenetic tree was constructed by using GSDS2.049 (http://gsds.gao-lab.org/) to select interesting genes.
mRNA Extraction and cDNA Preparation
The mycelium material wrapped in tin foil from 7, 14, 21, and 28 days of plate culture was frozen with liquid nitrogen and then stored in a −80 °C refrigerator. Then, total RNA was extracted by using the Universal Plant Total RNA Extraction Kit (Bioteke, Beijing) according to the manufacturer’s procedures without DNase and further purified using Recombinant DNase I alone (RNase-free, Takara) to remove genomic DNA. Equal quantities of RNA at the four different ages were mixed and then used for RT-PCR by using a PrimerScript II first Strand cDNA Synthesis Kit (Takara). Single-stranded cDNA was synthesized utilizing Oligo dT Primers.
Cloning of STS Genes
According to the predicted candidate gene sequences, three target-encoding sequences were amplified using 5′ and 3′ end-specific primers designed with restriction sites (see the Supporting Information) and Pyrobest DNA Polymerase (Takara) according to the manufacturer’s procedures. Hard-to-obtain genes were further amplified using Q5 hot-activated ultrafidelity DNA polymerases (BioLabs) to improve the efficiency of amplification. The PCR products were purified, attached to the pGEM-Teasy vectors (Promega), and transformed into competent E. coli DH5α cells (TIANGEN), which grew for 12–16 h on LB solid plates containing 50 μg/mL ampicillin antibiotic at 37 °C. Through blue–white screening, at least three positive clones of each gene were selected and sent to GENEWIZ in Tian Jin for sequencing by the Sanger method. The sequencing results were assembled with the predicted sequences by SeqMan software, and Apollo software was used to manually correct the incomplete matching sequences to obtain the correct open-reading frame obtained in the experiment.
Heterologous Expression of STSs
The STS genes were ligated to the pET32a (+) expression vectors through the restriction sites. The successfully constructed plasmids were transformed into E. coli Rosetta (DE3) (TIANGEN) hosts to heterologously express proteins. When the OD600 value of the bacterial liquid reached 0.8, 0.5 mM (final concentration) IPTG was added to the culture medium to induce transient protein expression at 18 °C for 24 h. The pET32a (+) vectors with a thioredoxin tag can promote STS expression and solubility. The Rosetta (DE3)50 strain contains a pRARE plasmid, which can provide tRNA of AUA, AGG, AGA, CUA, CCC, and GGA rare codons and enhance the expression level of heterologous proteins.
GC–MS Analysis of Terpenoids
After centrifugation of 8 mL of culture, the upper medium was quickly placed in 20 mL SPME Flasche (GERSTEL). The whole sampling process is automatically completed by the MPS Multipurpose Sampler (GERSTEL). The program was set as follows: incubation at 50 °C for 20 min, extraction for 15 min by SPME fiber (50/30 μm DVB/CAR/PDMS, Supelco, USA), desorption for 5 min, and cleanup for 3 min before and after injection. Volatile terpenoids were analyzed by a 7890B-7000D (Agilent Technologies). Samples were desorbed in a 250 °C injection port and eluted with helium into an HP-5MS column (30 m × 250 μm × 0.25 μm) splitless. The following GC oven temperature program was applied: 60 °C hold for 2 min, 10 °C/min ramped to 150 °C, 2 °C/min ramped to 160 °C, 15 °C/min ramped to 250 °C, and hold for 3 min (total GC program time: 25 min). Mass spectra were scanned over the range of 30–500 Da for 300 ms. The compounds were identified by electron ionization mode mass spectrometry compared with the standard spectra of terpenoids in the National Institute of Standards and Technology (NIST) database, mass spectra reported in the literature, a C8–C20 alkane mix compared to the published retention indices, and the characterized STC9.
Model Building and Homology Search
The STSs from this study were compared with the protein sequences of the same published functional STS using CLC Genomics Workbench 12 software, and the conserved regions were analyzed by a Clustal Omega Multiple Sequence Alignment (https://www.ebi.ac.uk/Tools/msa/clustalo/) and modified by Jalview software. Then, the amino acid residues with the highest frequency were recombined into a novel protein sequence. Finally, more similar sequences were found in NCBI Protein BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi? PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), and the Joint Genome Institute under the Fungal Genomics Program (http://genome.jgi-psf.org/programs/fungi/index.jsf) was searched to verify the homologous model.
Acknowledgments
This research was funded by the National Natural Science Foundation of China, grant no 81603221. Thanks for the 7890B-7000D (Agilent Technologies) instrument support of College of Pharmaceutical Engineering of Traditional Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin 301617, P. R. China.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c06792.
Products, protein ID, and exon number of published STSs from Basidiomycetes; phylogenetic tree of characterized STSs in Basidiomycetes; protein sequence similarity of the STSs in the small red branch; MS spectra for γ-cadinene; γ-CS model 1; 67 γ-CS homology sequences in NCBI; gene structure-phylogenetic tree of 67 sequences; comparison of exon/intron size for 67 sequences; amino acid sequence alignments of 67 sequences; amino acid sequence alignments of 15 characterized Δ6-protoilludene synthases; primers and plasmids; and cloned coding sequences from G. lucidum and G. sinensis (PDF)
Author Contributions
R.C., L.W., and C.S. conceived the projects, developed the methods, curated the data, and wrote the manuscript. R.C., X.W., Q.W., P.Q., and Y.Z. did the experiments. All authors contributed to the discussion and approved the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Gressler M.; Löhr N. A.; Schäfer T.; Lawrinowitz S.; Seibold P. S.; Hoffmeister D. Mind the mushroom: natural product biosynthetic genes and enzymes of Basidiomycota. Nat. Prod. Rep. 2021, 38, 702–722. 10.1039/d0np00077a. [DOI] [PubMed] [Google Scholar]
- Blackwell M. The fungi: 1, 2, 3 ... 5.1 million species?. Am. J. Bot. 2011, 98, 426–438. 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Davis E. M.; Croteau R.. Cyclization Enzymes in the Biosynthesis of Monoterpenes, Sesquiterpenes, and Diterpenes; Biosynthesis, Leeper F. J., Vederas J. C., Eds.; Academic PressSpringer: Berlin, Heidelberg, 2000; pp 53–95. [Google Scholar]
- Zhou Z.-Y.; Liu J.-K. Pigments of fungi (macromycetes). Nat. Prod. Rep. 2010, 27, 1531–1570. 10.1039/c004593d. [DOI] [PubMed] [Google Scholar]
- Lin H.-C.; Hewage R. T.; Lu Y.-C.; Chooi Y.-H. Biosynthesis of bioactive natural products from Basidiomycota. Org. Biomol. Chem. 2019, 17, 1027–1036. 10.1039/c8ob02774a. [DOI] [PubMed] [Google Scholar]
- Keller N. P.; Turner G.; Bennett J. W. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Agger S.; Lopez-Gallego F.; Schmidt-Dannert C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 2009, 72, 1181–1195. 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckli M.; Morinaka B. I.; Lackner G.; Kombrink A.; Sieber R.; Margot C.; Stanley C. E.; deMello A. J.; Piel J.; Künzler M. Bacteria-induced production of the antibacterial sesquiterpene lagopodin B in Coprinopsis cinerea. Mol. Microbiol. 2019, 112, 605–619. 10.1111/mmi.14277. [DOI] [PubMed] [Google Scholar]
- Nord C.; Menkis A.; Broberg A. Cytotoxic Illudane Sesquiterpenes from the Fungus Granulobasidium vellereum (Ellis and Cragin) Jülich. J. Nat. Prod. 2015, 78, 2559–2564. 10.1021/acs.jnatprod.5b00500. [DOI] [PubMed] [Google Scholar]
- Wawrzyn G. T.; Quin M. B.; Choudhary S.; López-Gallego F.; Schmidt-Dannert C. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem. Biol. 2012, 19, 772–783. 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels B.; Heinig U.; Grothe T.; Stadler M.; Jennewein S. Cloning and characterization of an Armillaria gallica cDNA encoding protoilludene synthase, which catalyzes the first committed step in the synthesis of antimicrobial melleolides. J. Biol. Chem. 2011, 286, 6871–6878. 10.1074/jbc.m110.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörfer M.; Gressler M.; Hoffmeister D. Diversity and bioactivity of Armillaria sesquiterpene aryl ester natural products. Mycol. Prog. 2019, 18, 1027–1037. 10.1007/s11557-019-01508-z. [DOI] [Google Scholar]
- Bisht B. S.; Bankoti H.; Bharti T. A Review on Therapeutic Uses of Terpenoids. J. Drug Deliv. Therapeut. 2021, 11, 182–185. 10.22270/jddt.v11i1-s.4523. [DOI] [Google Scholar]
- Schmidt-Dannert C. Biosynthesis of terpenoid natural products in fungi. Adv. Biochem. Eng. Biotechnol. 2014, 148, 19–61. 10.1007/10_2014_283. [DOI] [PubMed] [Google Scholar]
- Abraham W.-R. Bioactive sesquiterpenes produced by fungi: are they useful for humans as well?. Curr. Med. Chem. 2001, 8, 583–606. 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- Quin M. B.; Flynn C. M.; Schmidt-Dannert C. Traversing the fungal terpenome. Nat. Prod. Rep. 2014, 31, 1449–1473. 10.1039/c4np00075g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quin M. B.; Flynn C. M.; Wawrzyn G. T.; Choudhary S.; Schmidt-Dannert C. Mushroom hunting by using bioinformatics: application of a predictive framework facilitates the selective identification of sesquiterpene synthases in basidiomycota. Chembiochem 2013, 14, 2480–2491. 10.1002/cbic.201300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A.; Bouvier J. T.; Davidson D. B.; Imker H. J.; Sadkhin B.; Slater D. R.; Whalen K. L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037. 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Chen X.; Orban A.; Shukal S.; Birk F.; Too H.-P.; Rühl M. Agrocybe aegerita Serves As a Gateway for Identifying Sesquiterpene Biosynthetic Enzymes in Higher Fungi. ACS Chem. Biol. 2020, 15, 1268–1277. 10.1021/acschembio.0c00155. [DOI] [PubMed] [Google Scholar]
- Chen S.; Xu J.; Liu C.; Zhu Y.; Nelson D. R.; Zhou S.; Li C.; Wang L.; Guo X.; Sun Y.; Luo H.; Li Y.; Song J.; Henrissat B.; Levasseur A.; Qian J.; Li J.; Luo X.; Shi L.; He L.; Xiang L.; Xu X.; Niu Y.; Li Q.; Han M. V.; Yan H.; Zhang J.; Chen H.; Lv A.; Wang Z.; Liu M.; Schwartz D. C.; Sun C. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; Xu J.; Sun C.; Zhou S.; Xu H.; Nelson D. R.; Qian J.; Song J.; Luo H.; Xiang L.; Li Y.; Xu Z.; Ji A.; Wang L.; Lu S.; Hayward A.; Sun W.; Li X.; Schwartz D. C.; Wang Y.; Chen S. Chromosome-level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense. Sci. Rep. 2015, 5, 11087. 10.1038/srep11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.; Luo H.; Xu J.; Guo H.; Li C.; Hu Y.; Song J.; Chen S. Ganoderma lucidum : an Emerging Medicinal Model Fungus for Study of the Biosynthesis of Natural Medicines. Sci. Sin. Vitae 2013, 43, 447. 10.1360/052012-425. [DOI] [Google Scholar]
- Boh B.; Berovic M.; Zhang J.; Zhi-Bin L. Ganoderma lucidum and its pharmaceutically active compounds. Biotechnol. Annu. Rev. 2007, 13, 265–301. 10.1016/s1387-2656(07)13010-6. [DOI] [PubMed] [Google Scholar]
- Wang L.; Li J.-q.; Zhang J.; Li Z.-m.; Liu H.-g.; Wang Y.-z. Traditional uses, chemical components and pharmacological activities of the genus Ganoderma P. Karst.: a review. RSC Adv. 2020, 10, 42084–42097. 10.1039/d0ra07219b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Wang H.; Pu X.; Sun S.; Wei J.; Sun C. Cloning and characterization of sesquiterpene synthase genes from the Ganoderma sinense genome. Sci. Sin. Vitae 2018, 48, 447–454. [Google Scholar]
- Chu L.; Wang L.; Chen S.; Zeng X.; Xu J.; Li Y.; Sun C. Functional Identification of a Multi-product Sesquiterpene Synthase from Ganoderma sinense. Chin. J. Exp. Tradit. Med. Formulae 2019, 25, 151–157. [Google Scholar]
- Wang L.; Pu X.; Tan S.; Sun S.; Bi Y.; Sun C.; Chen S.; Wang H. Cloning and expression of the sesquiterpene synthase gene from Ganoderma lucidum. J. Agric. Univ. Hebei 2017, 40, 67–72. [Google Scholar]; (in Chinese)
- Lim H. S.; Kim S. K.; Woo S.-G.; Kim T. H.; Yeom S.-J.; Yong W.; Ko Y.-J.; Kim S.-J.; Lee S.-G.; Lee D.-H. (−)-α-Bisabolol Production in Engineered Escherichia coli Expressing a Novel (−)-α-Bisabolol Synthase from the Globe Artichoke Cynara cardunculus var. Scolymus. J. Agric. Food Chem. 2021, 69, 8492–8503. 10.1021/acs.jafc.1c02759. [DOI] [PubMed] [Google Scholar]
- Martin F.; Cullen D.; Hibbett D.; Pisabarro A.; Spatafora J. W.; Baker S. E.; Grigoriev I. V. Sequencing the fungal tree of life. New Phytol. 2011, 190, 818–821. 10.1111/j.1469-8137.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- Burkhardt I.; Kreuzenbeck N. B.; Beemelmanns C.; Dickschat J. S. Mechanistic characterization of three sesquiterpene synthases from the termite-associated fungus Termitomyces. Org. Biomol. Chem. 2019, 17, 3348–3355. 10.1039/c8ob02744g. [DOI] [PubMed] [Google Scholar]
- Nagamine S.; Liu C.; Nishishita J.; Kozaki T.; Sogahata K.; Sato Y.; Minami A.; Ozaki T.; Schmidt-Dannert C.; Maruyama J. I.; Oikawa H. Ascomycete Aspergillus oryzae Is an Efficient Expression Host for Production of Basidiomycete Terpenes by Using Genomic DNA Sequences. Appl. Environ. Microbiol. 2019, 85, e00409-19 10.1128/AEM.00409-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H.; Kitaoka T. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism. Microb. Biotechnol. 2018, 11, 952–965. 10.1111/1751-7915.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-L.; Ma L.-T.; Lee Y.-R.; Shaw J.-F.; Wang S.-Y.; Chu F.-H. Differential Gene Expression Network in Terpenoid Synthesis of Antrodia cinnamomea in Mycelia and Fruiting Bodies. J. Agric. Food Chem. 2017, 65, 1874–1886. 10.1021/acs.jafc.6b05386. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Chen X.; Lee R. T. C.; Rehka T.; Maurer-Stroh S.; Rühl M. Bioinformatics-aided identification, characterization and applications of mushroom linalool synthases. Commun. Biol. 2021, 4, 223. 10.1038/s42003-021-01715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp S. C.; Croteau R. B. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 2001, 158, 811–832. 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J. C.; Hibbett D. S. Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS One 2007, 2, e1097 10.1371/journal.pone.0001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J. C.; Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr. Biol. 2011, 21, 134–139. 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Wisecaver J. H.; Slot J. C.; Rokas A. The evolution of fungal metabolic pathways. PLoS Genet. 2014, 10, e1004816 10.1371/journal.pgen.1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sena Filho J. G.; Quin M. B.; Spakowicz D. J.; Shaw J. J.; Kucera K.; Dunican B.; Strobel S. A.; Schmidt-Dannert C. Genome of Diaporthe sp. provides insights into the potential inter-phylum transfer of a fungal sesquiterpenoid biosynthetic pathway. Fungal Biol. 2016, 120, 1050–1063. 10.1016/j.funbio.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangphrom P.; Seki H.; Suzuki M.; Komori A.; Nishiwaki M.; Mikawa R.; Fukushima E. O.; Muranaka T. Functional Analysis of Amorpha-4,11-Diene Synthase (ADS) Homologs from Non-Artemisinin-Producing Artemisia Species: The Discovery of Novel Koidzumiol and (+)-α-Bisabolol Synthases. Plant Cell Physiol. 2016, 57, 1678–1688. 10.1093/pcp/pcw094. [DOI] [PubMed] [Google Scholar]
- Son Y.-J.; Kwon M.; Ro D.-K.; Kim S.-U. Enantioselective microbial synthesis of the indigenous natural product (-)-α-bisabolol by a sesquiterpene synthase from chamomile (Matricaria recutita). Biochem. J. 2014, 463, 239–248. 10.1042/bj20140306. [DOI] [PubMed] [Google Scholar]
- Han G. H.; Kim S. K.; Yoon P. K.-S.; Kang Y.; Kim B. S.; Fu Y.; Sung B. H.; Jung H. C.; Lee D.-H.; Kim S.-W.; Lee S.-G. Fermentative production and direct extraction of (-)-α-bisabolol in metabolically engineered Escherichia coli. Microb. Cell Factories 2016, 15, 185. 10.1186/s12934-016-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertti L. A. G.; Delatte T. L.; Souza de Farias K.; Galdi Boaretto A.; Verstappen F.; van Houwelingen A.; Cankar K.; Carollo C. A.; Bouwmeester H. J.; Beekwilder J. Identification of the Bisabolol Synthase in the Endangered Candeia Tree (Eremanthus erythropappus (DC) McLeisch). Front. Plant Sci. 2018, 9, 1340. 10.3389/fpls.2018.01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-J.; Kim S. K.; Seong W.; Woo S.-G.; Lee H.; Yeom S.-J.; Kim H.; Lee D.-H.; Lee S.-G. Enhanced (−)-α-Bisabolol Productivity by Efficient Conversion of Mevalonate in Escherichia coli. Catalysts 2019, 9, 432. 10.3390/catal9050432. [DOI] [Google Scholar]
- Attia M.; Kim S.-U.; Ro D.-K. Molecular cloning and characterization of (+)-epi-α-bisabolol synthase, catalyzing the first step in the biosynthesis of the natural sweetener, hernandulcin, in Lippia dulcis. Arch. Biochem. Biophys. 2012, 527, 37–44. 10.1016/j.abb.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Nakano C.; Kudo F.; Eguchi T.; Ohnishi Y. Genome mining reveals two novel bacterial sesquiterpene cyclases: (-)-germacradien-4-ol and (-)-epi-α-bisabolol synthases from Streptomyces citricolor. Chembiochem 2011, 12, 2271–2275. 10.1002/cbic.201100418. [DOI] [PubMed] [Google Scholar]
- Maurya A.; Singh M.; Dubey V.; Srivastava S.; Luqman S.; Bawankule D. α-(-)-bisabolol reduces pro-inflammatory cytokine production and ameliorates skin inflammation. Curr. Pharm. Biotechnol. 2014, 15, 173–181. 10.2174/1389201015666140528152946. [DOI] [PubMed] [Google Scholar]
- Saitou N.; Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Hu B.; Jin J.; Guo A.-Y.; Zhang H.; Luo J.; Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegel H.; Tourle S.; Ottosson J.; Persson A. Increased levels of recombinant human proteins with the Escherichia coli strain Rosetta (DE3). Protein Expr. Purif. 2010, 69, 159–167. 10.1016/j.pep.2009.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.