Abstract
The development over the past two decades of molecular methods for manipulation of RNA and DNA has afforded molecular virologists the ability to study viral genomes in detail that has heretofore not been possible. There are many molecular techniques now available for typing and subtyping of viruses. The available methods include restriction fragment length polymorphism analysis, Southern blot analysis, oligonucleotide fingerprint analysis, reverse hybridization, DNA enzyme immunoassay, RNase protection analysis, single-strand conformation polymorphism analysis, heteroduplex mobility assay, nucleotide sequencing, and genome segment length polymorphism analysis. The methods have certain advantages and disadvantages which should be considered in their application to specific viruses or for specific purposes. These techniques are likely to become more widely used in the future for epidemiologic studies and for investigations into the pathophysiology of virus infections.
The major methods for detection of viruses in the clinical laboratory today include (i) identification of cytopathic effects (CPE) in cell cultures, (ii) use of fluorescent antibodies directly on specimen material, (iii) enzyme immunoassays for antigen detection, and (iv) amplification techniques with viral genomes as targets. These methods are used routinely as the first means of detection. In most cases, presumptive identification of the virus is made in conjunction with detection. Thus, if a viral culture from a nasopharyngeal swab shows positive CPE which is characteristic of respiratory syncytial virus (RSV), then a virus is detected and identified at the same time. The above-mentioned methods allow identification of the virus. Typing and subtyping require additional investigation employing either serologic techniques to identify unique antigenic epitopes or molecular techniques to dissect the genome of the virus and compare, directly or indirectly, the nucleotide compositions of different isolates. Today’s molecular techniques have been instrumental in viral subtype analysis that has gone well beyond the realm of antigen-antibody interaction. Many of the methods discussed here are capable of identifying a single base change in a viral genome of several hundred kilobases. Molecular characterization for the purpose of subtyping is not relevant to treatment (except in the case of hepatitis C virus [HCV]) but is useful mainly for epidemiologic purposes and for investigations into pathogenesis and disease progression.
Two decades ago, the major method of identification of viruses was growth in cell cultures and observation for CPE. The use of fluorescent antibodies (initially polyclonals, later monoclonals, and, still later, mixtures of monoclonals) was just making its way into the clinical virology lab. Restriction endonucleases had recently been discovered and were being put to use in various techniques for characterization of viral genomes at the molecular level. Nucleic acid probe technology was being developed for use in the clinical laboratory. However, probes could not be radioactively labeled to a high enough specific activity to be used as devices for sensitive direct detection of viral genomes in clinical specimens. The reality of their use fell far short of the expectations at the time. Probes would later find their greatest utility in research laboratories with such techniques as Southern blotting and RNase protection assays.
The middle 1980s brought the PCR to the virology laboratory and with it the ability to amplify femtogram amounts of DNA or RNA to levels that could be easily detected in ethidium bromide-stained agarose gels or by enzyme-linked immunosorbent assay or hybrid capture assays. PCR has arguably been the single most important development in the past two decades with regard to the ability to characterize and compare the genomes of viruses. It has not only been at the center of numerous assays that have been aimed at the detection of minute levels of virus has but also allowed detection of viruses that had previously been very difficult to detect (e.g., those causing cerebrospinal fluid infections), thus enabling a diagnosis in cases that would otherwise have gone undiagnosed. In addition to sensitivity and specificity, another major advantage that PCR provides is the ability to sample (amplify) a relatively small portion of the viral genome (a few hundred to a few thousand base pairs) that may have been subjected to evolutionary pressures and to enter that portion into a comparison or subtype analysis while excluding the remainder of the genome. Thus, relatively small variable regions that actually confer the subtype can be compared without the background clutter of the much larger genome. Numerous techniques for identification and comparison of viruses, many of which are discussed here, have been spun off of PCR.
The classical method for typing and subtyping viruses is serotyping. Long before molecular methods were available, identifying differences among viruses was accomplished by the use of antibodies that could define antigenic differences. Some virus species, such as human cytomegalovirus (CMV) and measles virus, cannot be divided into different types or subtypes because significant antigenic differences do not exist. On the other hand, serologic methods have been used to define major viral groups (i.e., types), such as influenza virus types A to C, parainfluenza virus types 1 to 4, poliovirus types 1 to 3, and herpes simplex virus types 1 and 2 (HSV-1 and -2, respectively). In some cases, and with the use of monoclonal antibodies, finer distinctions can be made. In general, serotyping has been useful for making relatively large distinctions among viruses but does not have the ability to distinguish individual isolates within a serotype simply because the major antigenic epitopes that define the serotype are highly resistant to change. The need for this capability has provided the impetus for the development of molecular methods described in this review.
Viral genomes can vary somewhat (i.e., mutate) at the nucleotide level and yet maintain their essential characteristics at the protein and virion levels. This variability is the basis for molecular characterization and subtype classification of viruses. Some regions of the viral genome may be extremely stable and resistant to mutation, while other regions may be hypervariable. If less than the whole genome is being studied, the investigator must choose the region for analysis with care so as to observe a reasonable number of mutations. The changes at the nucleotide level may or may not result in amino acid changes, but it is irrelevant for these genotypic methods. Nucleotide changes in a viral isolate from an infected patient may allow that isolate to be uniquely identified or grouped with similar isolates of the same virus by molecular techniques. It is also noteworthy that RNA viruses have much higher rates of spontaneous mutation than DNA viruses, primarily due to the fact that viral RNA polymerases do not have 3′-to-5′ exonuclease activity and thus cannot edit mistakes made during replication of the genome.
Tables 1 and 2 summarize the techniques and applications discussed in this review. Table 1 describes the methods and their requirements, such as the use of restriction enzymes, other enzymes, and probes, as well as the ability of the technique to sample a large number of bases and detect point mutations. One must consider these attributes of a particular assay when deciding which to use for an application. Table 2 identifies the practical uses of the methods and gives examples of viruses that have been studied by a particular method as discussed in this review.
TABLE 1.
Molecular methods for characterization of viruses
| Method | Restriction enzyme | Probe | Basis for distinctions | Level of resolution | Advantage(s) | Disadvantage(s) |
|---|---|---|---|---|---|---|
| Nucleotide sequencing | No | No | Nucleotide sequence | Single genome (if cloned) | Wide applicability; can identify single nucleotide mutations | Technically complex; produces large amounts of data; automated sequencing requires expensive equipment |
| RFLP analysis | Yes | No | Restriction sites | Subtypes | Simple; wide applicability | Samples only the restriction sites; difficult to use with RNA viruses |
| Southern blotting | Yes | Yes | Restriction sites, probe sites | Subtypes | Wide applicability; ability to analyze complex genomes | Samples only the restriction sites and probe sites; technically complex; may require radioisotopes |
| Oligonucleotide fingerprint analysis | No | No | RNase T1 cleavage sites | Subtypes, quasispecies | Directly applicable to RNA viruses; can detect point mutations | Involves complex electrophoresis procedure |
| RH | No | Yes | Probe sites | Subtypes | Simple; commercially available | May not correctly identify all subtypes of HCV |
| DEIA | No | Yes | Probe sites | Subtypes | Simple; commercially available | May not correctly identify all subtypes of HCV |
| RNase protection assay | No | Yes | Probe sites | Subtypes | Readily applicable to RNA viruses | Requires a radioactive probe; technically difficult |
| SSCP analysis | No | No | Mobility differences | Subtypes | Detects a few mutations in a large number of bases; simple procedure | Identifies the presence but not the location of mutations |
| HMA | No | No | Mobility of hetero- vs homoduplexes | Quasispecies | Can visualize a large number of quasispecies | Cannot distinguish quasispecies with <2–3% nucleotide differences; technically difficult and complex |
| Genome segment length polymorphism analysis | No | No | Segment length | Subtypes | Easy technique for use with segmented genomes | Cannot detect mutations (detects only variations in segment length); applicable only to viruses with segmented genomes |
TABLE 2.
Application of molecular subtyping methods to specific viruses
| Virus | Method(s) | Reference(s) |
|---|---|---|
| Adenovirus | RFLP analysis | 34, 36, 53 |
| CMV | RFLP analysis, PCR-RFLP analysis, Southern blotting, glycoprotein B genotyping | 38, 45, 72, 77, 81 |
| Dengue virus | Nucleotide sequencing | 19 |
| Epstein-Barr virus | RFLP analysis, analysis of termini | 24, 42, 66, 69 |
| Enterovirus | RFLP analysis, RT-PCR–RFLP analysis, oligonucleotide fingerprint analysis, SSCP analysis, nucleotide sequencing | 25, 47, 57–59, 62 |
| HBV | Southern blotting, SSCP analysis, PCR-SSCP analysis, nucleotide sequencing | 30 |
| HCV | SSCP analysis, PCR-SSCP analysis, RH, DEIA, nucleotide sequencing | 7, 11, 48, 49, 65 |
| HIV | HMA, HTA, nucleotide sequencing | 18, 55, 61, 64 |
| HSV | RFLP analysis, PCR-RFLP analysis, Southern blotting | 8, 9 |
| Influenza virus | RNase protection assay, HMA, nucleotide sequencing | 54, 75, 86 |
| JC virus | Nucleotide sequencing | 2 |
| Measles virus | RT-PCR–RFLP analysis | 41 |
| Parainfluenza virus | Serotyping, sequencing | 6, 46 |
| Parvovirus B19 | SSCP analysis, PCR-SSCP analysis | 43 |
| RSV | RNase protection assay, PCR-RFLP analysis, sequencing | 15, 74 |
| Rhinovirus | Serotyping, nucleotide sequencing | 14, 23, 37, 39, 68 |
| Rotavirus | Genome segment length polymorphism analysis | 5, 21, 26 |
| Small round-structured viruses (Norwalk-like viruses) | Southern blotting, nucleotide sequencing | 3, 52, 83 |
| Varicella-zoster virus | RFLP analysis, long PCR-RFLP analysis | 1, 76 |
NUCLEOTIDE SEQUENCING
Introduction and General Approach
The experimental determination of the linear arrangement of bases in a viral genome is the ultimate form of subtyping. Sequencing of the genome has the potential to distinguish even between parent and progeny if a single mutation has occurred in the replicative process. Sequencing generally requires a commitment of time and resources beyond those of the other methods discussed herein, and it produces a volume of data that may exceed what the investigator had hoped for and is capable of dealing with. However, if only small portions or specific variable regions of a viral genome are sequenced for the purpose of subtyping, the volume of sequencing data can be kept to a minimum. Generally this is the case. Only the portions of the genome that confer the subtype need be compared, but in the beginning this information may not be known. In most cases the comparison will include only the regions that are subjected to immunological pressure by the host (i.e., the major antigenic epitopes of the surface proteins). However, in other cases the untranslated regions of the genomes are unique to a subtype because they contain random mutations that may persist indefinitely in the complete absence of immunologic pressure. The use of PCR amplification in conjunction with sequencing and recent developments in automated sequencing (with cycle labeling of the oligonucleotides) have removed much of the tedium from this process and streamlined it to the point that one can obtain a fully analyzed sequence of a small (e.g., 1-kb) portion of a viral genome within about 3 days after obtaining a clinical isolate or, in some cases, after receiving a clinical specimen. Of course, all RNA genomes must first be converted to DNA prior to sequencing, so the use of reverse transcriptase PCR (RT-PCR) is a natural fit for this process and leads directly to a sequencable product (Fig. 1).
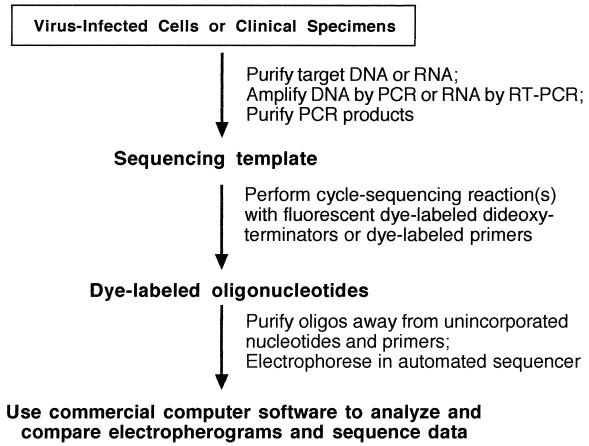
FIG. 1.
Automated cycle sequencing as a rapid and convenient method for sequence analysis of PCR products. The target DNA was extracted and amplified and the PCR products were purified away from unincorporated nucleotides and primers. The sequencing template was “amplified” in another PCR mixture containing the sequencing primers, a mixture of deoxy- and dideoxynucleotides, and modified Taq polymerase. Dideoxynucleotides were coupled to fluorescent dyes, a different color for each of the four bases; these act as terminators of the growing oligonucleotides. Thus, the color of the dye is the key to which dideoxynucleotide is at the 3′ terminus of each oligonucleotide. The large array of oligonucleotides was separated in a polyacrylamide gel which was continuously scanned by a laser during the run, and the colors were detected as they passed the scan point. Each oligonucleotide in the array, as they move through the gel, is only one base shorter than the oligonucleotide above it in the gel. The detection information was fed directly into a computer for analysis and construction of the four-color electropherogram, which was translated into the linear sequence by commercially available software.
Application to Human Immunodeficiency Virus
The complete sequence of the human immunodeficiency virus type 1 (HIV-1) genome was published in 1985 (67). Since that time, there have been numerous reports of the use of sequencing to demonstrate the genetic variability of the virus. Early reports showed a high degree of variation over time in specific genes within an individual patient (28). Other investigators using HIV sequencing attempted to establish a correlation between sequence heterogeneity in the V3 region of the envelope in longitudinal specimens and the stage of HIV disease as determined by CD4+-cell counts (55). These studies led to the conclusion that only a highly restricted subset of virus quasispecies are apparently transmitted and establish infection in a new host. Sequence divergence between an HIV-infected asymptomatic blood donor and three pediatric recipients was also studied by sequencing the immunodominant Env V3 region (56). Mutations occur during natural selection within the host in specific locations surrounding the V3 loop but rarely, if ever, within the loop structure.
Globally circulating strains of HIV-1 are classified according to their phylogenetic relationships into genetic groups (termed M for “major” and O for “outlier”) and subtypes (currently A through J). Serotyping may identify the majority of HIV-1 isolates, but highly divergent isolates and recombinant isolates are difficult to classify. Two regions of the HIV genome have been used for these phylogenetic analyses, gag and env. Due to the highly variable nature of the HIV-1 genome, the definitive method for subtype classification is to sequence one or both of these regions and then build phylogenetic trees which group the isolates with others of the same subtype. An epidemiologic study in which HIV-1 env DNA from dried blood spots was PCR amplified and sequenced demonstrated that the technique worked and showed the presence of subtypes A, B, C, and E in Asia (10). The investigators amplified a 389-bp segment of the region of env from C2 to V3, from which 287 bases were aligned and used in the tree analysis.
Others have sequenced full-length proviral clones of “subtype E” virus from Thailand in an effort to demonstrate intersubtype recombination as the evolutionary origin of this virus (27). Previous analyses of gag and env regions had shown the Thailand subtype E virus to be a hybrid of subtypes A (the gag region) and E (the env region). The sequence of the complete genome demonstrated convincingly that there were several crossover points between the two subtypes. The gp120 portion of env, parts of vif and vpr, and most of the long terminal repeat were derived from subtype E, and the remainder of the genome was derived from subtype A (27). This study again points out the amazingly plastic nature of the HIV genome and the usefulness of nucleotide sequencing in analyzing this genome.
Molecular Forensics of HIV
The power of sequencing for determination of molecular epidemiology related to a forensic inquiry was demonstrated in a highly publicized investigation of the possible transmission of HIV in a dental practice (64). In this extensive investigation, a PCR-amplified product of 680 bp from the envelope gene of proviral DNA from lymphocytes of the dentist, seven of his HIV-positive patients, and 35 local HIV-infected controls was sequenced. From the primary sequence, three analyses were performed: analysis of DNA distance, construction of phylogenetic trees, and analysis of amino acid signature patterns. The results showed that the viruses from the dentist and five of the patients were closely related, and taken with the epidemiologic results, it was concluded that these patients had been infected (via an unknown mechanism) while receiving care at the dentist’s office. Obviously the nature of this particular investigation necessitated a highly sophisticated analysis of the viral genomes. Investigators using the various techniques discussed in this review must determine the ability of a particular method to provide the level of sophistication appropriate for the application at hand.
Use with Dengue, JC, and Influenza Viruses
Dengue virus is an RNA virus of the flavivirus genus that causes a mild flu-like illness in millions of people each year and severe to life-threatening hemorrhagic fever and shock syndrome in some individuals. A molecular epidemiologic study which was performed by direct sequencing of PCR products revealed six genotypic groups among 28 dengue virus type 2 isolates (19). The investigation was undertaken because of the suggestion that certain topotypes (i.e., genetically related viruses that circulate in a particular geographic region) might be more likely to cause severe disease than others. The authors were unable to demonstrate this possibility because of many confounding variables but nonetheless generated surveillance data that may be useful in vaccine development.
Another investigation with a similar hypothesis was reported for JC virus (2). JC virus is found in four different genotypes in 70 to 90% of the adult population worldwide and is the cause of progressive multifocal leukoencephalopathy (PML) in about 5% of autopsied AIDS patients. By direct sequencing of specific regions of the genome, type determination was made and compared among 50 PML patients and 103 control subjects. Brain tissues from the PML patients had a significantly higher proportion of JC virus type 2 than urine isolates from the controls, thus indicating a biologic difference between the JC virus genotypes and perhaps demonstrating a propensity for type 2 to cause PML.
A very recent application of sequencing for the purpose of molecular characterization is the case of avian influenza A (H5N1) virus isolated from a child in Hong Kong with fatal influenza (75). The isolate was obtained from a tracheal aspirate specimen. The hemagglutinin (HA) and neuraminidase genes were amplified by RT-PCR and sequenced to confirm the H5N1 genotype. The sequence analysis revealed the presence of a multiple basic amino acid insertion upstream from the trypsin cleavage site (75). This insertion has previously been found in highly pathogenic avian influenza virus strains and is thought to extend the tissue range of the virus by allowing proteases other than trypsin to cleave the HA protein into HA1 and HA2 domains and thus enable systemic spread of the virus.
RESTRICTION FRAGMENT LENGTH POLYMORPHISM ANALYSIS
Review of the Method
The site of cleavage of DNA by a restriction endonuclease is sequence dependent. The presence of mutations at potential cleavage sites in some viral strains results in different patterns of fragments when they are separated in an agarose gel, a phenomenon termed restriction fragment length polymorphism (RFLP) (Fig. 2). The technique requires fairly large amounts of purified or partially purified viral DNA, a set of restriction enzymes to enzymatically cleave the DNA, the ability to separate the resulting DNA fragments by electrophoresis, and a method of documenting the results. The results are displayed as patterns of bands in an ethidium bromide-stained agarose gel. Viruses with large genomes (e.g., CMV) may have 20 to 50 bands, while viruses with smaller genomes (e.g., adenoviruses) have only 5 to 10 bands. Of course, the restriction enzyme used for the digestion is an important factor in the resultant number of bands, and care must be taken to ensure that digestion is complete so that partial digestion products do not obscure the final analysis of bands. Restriction endonucleases have recognition sites usually consisting of either four or six bases in a specific palindromic sequence. If a mutation occurs in the DNA genome to either eliminate or create a restriction site, polymorphisms occur in the pattern of bands in the gel (Fig. 2). There is no easy way, nor is there generally a need, to correlate the pattern (i.e., a missing band or an extra band) with mutations at specific locations in the genome without extensive molecular hybridization studies or even sequencing of the genomes being compared. A distinct limitation of the method is that the presence of a mutation cannot be detected unless that mutation happens to fall within the recognition sequence of the restriction endonuclease being used for digestion of the DNA. The use of different restriction enzymes will optimize the probability of detecting mutations in a particular genome or portion of a genome. Also, the absence of differing restriction patterns may simply be the result of testing with too few restriction enzymes.
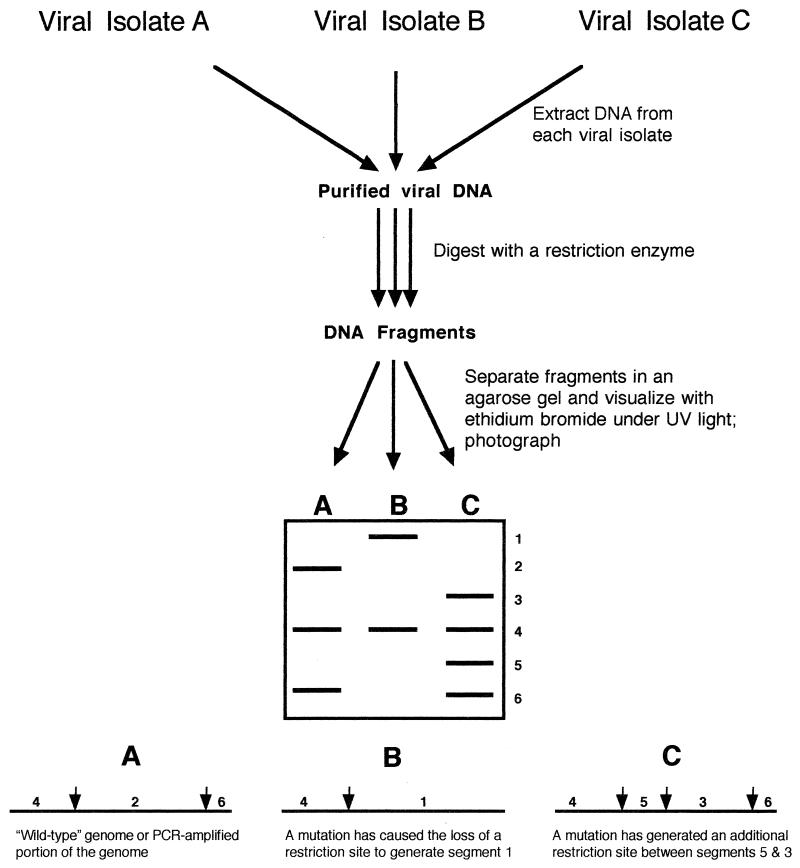
FIG. 2.
Hypothetical comparison of three viral isolates by RFLP analysis. In this case, viral genomic DNA was extracted from cells infected with the different isolates and then was digested with a restriction enzyme to produce DNA fragments, which were separated in an agarose gel containing ethidium bromide to allow subsequent visualization and documentation of patterns. Virus isolate A was the hypothetical wild type, with two restriction sites. Isolates B and C differ from A by the loss and gain, respectively, of a restriction site, which resulted in altered banding patterns in the gel. The method can be modified for the use of PCR products instead of genomic DNA or for the use of RT-PCR to allow the use of genomic RNA.
Examples Involving DNA Viruses: CMV, HSV, and Adenovirus
CMV was the first virus to be subjected to RFLP analysis of viral genomic DNA pursuant to epidemiologic studies. Kilpatrick et al. (45) demonstrated the usefulness of this method by restriction enzyme digestion of 11 human strains of CMV. Each strain had a unique pattern of fragments when separated by electrophoresis in an agarose gel. Subsequent studies have documented a number of interesting observations with regard to the epidemiology of CMV. Five of six congenitally infected babies had the same strain of CMV as their mothers, thus leading to the obvious conclusion that endogenous CMV is the most frequent cause of intrauterine transmission (38). Spector and Spector (72) used the RFLP technique to study the epidemiologic relationships of three CMV isolates from newborn twins and their mother. The infants were infected with two different strains of virus during an extended hospitalization after birth (twin A was infected at 9 weeks of age and twin B was infected at 6 weeks). The mother, who was CMV seronegative prior to and shortly after giving birth, became infected at 6 months postpartum with the strain carried by twin A. In another study among infants in a nursery, RFLP analysis of CMV strains isolated from the urine of eight babies demonstrated that one of the babies had transmitted CMV to two other babies, apparently via fomites. The two babies began shedding CMV on the same day, 22 days after the index patient was transferred to the intensive care nursery.
Other viral genomes have been subjected to RFLP analysis. The method was originally referred to as restriction endonuclease fingerprinting as it was applied to HSV in groundbreaking studies demonstrating exogenous reinfection (9) and nosocomial outbreaks (8). These and other studies found HSV-1 to have 19 of 57 total restriction enzyme cleavage sites that were variable, with four restriction enzymes among the isolates in the studies (8, 9). From this information it was estimated (9) that the smallest number of potential differentiable strains of HSV-1 is 524,288, thus lending theoretical support to the observation that epidemiologically unlinked isolates have different RFLP patterns. An RFLP analysis of the isolates from the earlier study (8) led to the conclusion that there had been two independent introductions of HSV-1 into a pediatric intensive care unit. An outbreak of HSV encephalitis in Boston, Mass., engendered an RFLP analysis of the isolates, and all seven had different restriction patterns, thus proving that they were not epidemiologically related (29).
The molecular epidemiology of adenoviruses based on RFLP analysis has been extensively investigated over the past 20 years. Wadell assembled a compendium on the subject (78), and Wigand and Adrian (80) devised a system for classification of the genome types. Others have followed with articles focused on more local aspects of the topic (4, 40) to describe populations of adenovirus genotypes of particular interest and to gain insight into the possibility that the pathology of adenovirus disease is related to the genotype of the virus (53). The AIDS epidemic has provided a unique opportunity to study adenoviruses. Because they are spread easily and persist for long periods of time in immunocompromised patients, the conditions are good for widespread exchange of genetic material among and within serotypes through recombination. Hierholzer et al. (33–36) found numerous antigenically intermediate strains and DNA variants. They concluded that restriction enzyme analysis of adenoviruses from AIDS patients thus had only limited usefulness for typing but was helpful in identifying groups of isolates with similar properties (34).
RT-PCR for RFLP Analysis of RNA Viruses: Enteroviruses and Measles Virus
Restriction endonucleases are capable of cleaving only DNA, but with the help of RT-PCR, portions of the genomes of RNA viruses can be converted to DNA, amplified, and subjected to RFLP analysis. Many of the enteroviruses have been analyzed by this method for the purposes of both detection and differentiation of the numerous serotypes by Kuan (47). In this study, a 297-bp amplicon from the 5′ untranslated region (UTR) of the virus was cleaved by restriction enzymes and the fragments were analyzed on an agarose gel. Fragment patterns were characteristic of the serotype, as shown by comparison with prototype strains. Molecular epidemiology of measles virus, another RNA virus, has recently been used to document the changing distribution of measles virus genotypes in Japan. This investigation was accomplished by amplifying the HA and nucleoprotein genes and then subjecting the amplified product to digestion by restriction enzymes, which allowed strain typing of the isolate (41). Measles virus type 2 represented 80% of the isolates in western Japan in 1985, but by 1990 the majority were type 1 and by 1995 all strains studied were type 1 as determined by RFLP analysis.
RFLP analysis is a simple yet powerful technique. It has been widely used because of its ability to detect point mutations if they occur in a restriction site. It can be used to compare a large number of isolates in epidemiologic studies and can even be adapted for use with RNA viruses after an RT-PCR step.
SOUTHERN BLOT ANALYSIS
Description of Method
Classical Southern blotting (70) is a modification of RFLP analysis in which the viral DNA is cut with a restriction enzyme and then fragments are subjected to electrophoresis in an agarose or polyacrylamide gel, transferred onto a nitrocellulose sheet, and then hybridized with a labeled probe from the entire genome or against a specific region of the genome.
Use with CMV
This method (22) and RFLP analysis (71) have been used to demonstrate that an AIDS patient can be infected with multiple strains of CMV. In a landmark study on the transmission of CMV in cadaveric renal transplantation (12), 15 distinct strains (genotypes) of CMV were isolated from 19 organ recipients (i.e., four pairs [8 patients] of the 19 had paired isolates with the same genotype). In all four pairs of patients who had received kidneys from the same cadaver, both recipients shed the same strain of CMV, suggesting that both had acquired it from the donor and that seropositive recipients can be reinfected by a new strain of CMV after transplantation (12).
OLIGONUCLEOTIDE FINGERPRINT ANALYSIS
Method
Oligonucleotide fingerprint analysis, a technically complex but powerful method of analyzing randomly distributed noncontiguous segments of an RNA molecule or RNA virus genome, was first described by DeWachter and Fiers (20). The method employs digestion of radioactively labeled genomic RNA with RNase T1, an endonuclease purified from Aspergillus oryzae that cleaves single-stranded RNA on the 3′ side of G residues, and two-dimensional electrophoresis. The electrophoresis procedure is performed on 8% polyacrylamide in 6 M urea at 4°C in the first direction and then on 22% acrylamide in Tris-borate buffer at room temperature in the second direction after a 90° rotation from the first direction. 32P-labeled RNA oligonucleotides are detected by autoradiography as a fan-shaped array of spots and are distributed according to size and composition (20). The method allows comparison of the larger T1-resistant oligonucleotides present in the RNA being tested (representing about 10% of the genome of certain enteroviruses [44]). This procedure was considered to be the most sensitive method available, short of nucleotide sequencing, for analysis of RNA genetic relatedness.
Application to Enteroviruses
The oligonucleotide fingerprint method was first applied to poliovirus RNA by Lee et al. (50) and Lee and Wimmer (51) and soon thereafter was used in the analysis of other RNA viruses, including influenza virus (84), vesicular stomatitis virus (13), and enteroviruses (31, 57, 62). The most widely used application has been in the study of molecular variation among the enteroviruses, particularly polioviruses. T1 fingerprinting was a powerful tool in the early epidemiologic studies of the distribution of poliovirus subpopulations among infected human communities and in studies undertaken to clarify the molecular evolution of poliovirus during infection and passage through the human intestine. Using T1 fingerprinting, it was observed that the poliovirus genome was quite plastic, since the genome underwent continual change during natural epidemic transmission. Within a single epidemic in a confined geographic area, small differences between patterns were detected and no two patterns were exactly alike (62). New spots appeared on the autoradiograms and others disappeared; even the patterns among infected family members were different (62).
Other practical applications were also discovered. Isolates of vaccine origin could easily be distinguished from circulating “wild” strains of poliovirus by this method, proving it a timely method for detection of wild-type infections and for tracking possible vaccine revertants of the type 1 oral polio vaccine (62).
Oligonucleotide fingerprinting showed that in contrast to the extreme variability of the poliovirus genome, the genome of enterovirus 70 (EV 70), the cause of epidemic acute hemorrhagic conjunctivitis around the world, is quite stable (44). Only one basic genotype of EV 70 appears to be in circulation worldwide, and isolates from one year to the next and from one geographic region to the next have very similar patterns. Numerous intervening infections that linked different isolates did not result in major changes in the fingerprint patterns of EV 70 (44).
REVERSE HYBRIDIZATION
Technique
The reverse hybridization (RH) method is based on a system in which PCR products amplified from test specimens are hybridized to probes bound in parallel lines to membrane strips. An assay employing this technology for type and subtype analysis of HCV is commercially available (Inno-LiPA HCV; Innogenetics, Zwijnaarde, Belgium, and, in the United States, Norcross, Ga.). HCV RNA is extracted from serum and amplified by a nested RT-PCR with biotinylated primers directed against the HCV 5′ noncoding (NC) region or 5′ UTR. These products are hybridized to probes on the strips and hybrids are detected by alkaline phosphatase-conjugated streptavidin. Reaction of the amplified fragment with specific probes results in the formation of visible lines on the strip, thus presumably allowing identification of at least six of the nine currently known major HCV genotypes and also the respective subtype.
Application to HCV
This assay has been evaluated for its usefulness in genotyping of HCV. HCV RNA isolated from the sera of 61 HCV-positive patients was analyzed by sequencing of the 5′ NC region and the less well conserved nonstructural-5 region and by RH (85). The RH assay was able to correctly identify all of the HCV genotypes (only types 1, 2, and 3 were found in these patients). However, the RH assay incorrectly identified the subtype of 1 of 11 HCV subtype 1a genotypes as subtype 1b and identified 3 of 31 subtype 1b genotypes as subtype 1a, and it also identified 4 of 4 subtype 2c genotypes as subtype 2a. All HCV subtype 2a, 2b, and 3a genotypes in this study were correctly identified by the RH assay. A subsequent evaluation by the same group (49) and by another group (65) led to the conclusion that this particular RH assay misinterpreted 2 to 10% of the subtype 1a and 1b genotypes and also that the method could not be used for differentiation of HCV subtypes 2a and 2c. The shortcomings of this assay are apparently the result of the choice of probes. The distinction between subtypes 1a and 1b is dependent on a single nucleotide difference at position −99 in the 5′ UTR, and subtypes 2a and 2c are identical in the region targeted by the Inno-LiPA probes.
DNA ENZYME IMMUNOASSAY
Description of the Commercial Assay
A commercial DNA enzyme immunoassay (DEIA) product is available for identification of HCV types and subtypes. RNA from serum is extracted and amplified by RT-PCR with nested primers directed against the HCV core region. Heat-denatured PCR products are hybridized to probes bound to multiwell plates. Hybrids are detected with a murine monoclonal antibody against double-stranded DNA followed by rabbit anti-mouse antibody conjugated to horseradish peroxidase. A substrate is then added. The DEIA reagents contain six different probes directed against the HCV core region.
Use for HCV and Comparison to RH Assay
In a recent evaluation and direct comparison of the RH assay (Inno-LiPA HCV II) and the DEIA (65), 112 of 120 evaluable samples (93.3%) yielded concordant results. Of the eight subtyping discrepancies, all involved subtype 1 and 2 viruses that were misassigned by the RH assay apparently because of the suboptimal probes discussed above.
Thus, both the RH assay and the DEIA appear to be excellent choices for the determination of HCV type and reasonable choices for the determination of subtype. However, the DEIA has a higher accuracy for subtyping, especially for identifying the subtypes of HCV-1 and -2.
RNASE PROTECTION ANALYSIS
Development of the Method and Application to Viruses
RNase protection analysis was developed by Myers et al. (60) for the purpose of demonstrating the presence and approximate locations of point mutations in DNA. The technique involves the use of a relatively short, in vitro-synthesized, radioactive RNA probe that is transcribed from wild-type genomic DNA (or cloned DNA) from a standard strain or the type strain of an organism. In the original method, this RNA probe was then hybridized to DNA from test strains prior to digestion of the heteroduplex with RNase A. The resulting fragments were separated in a polyacrylamide gel containing 8 M urea (Fig. 3). RNase A cleaves the radioactive RNA probe at positions of mismatch with the DNA. Test genomes with mutations that are not present in the probe or with mutations at different positions than the probe will have altered banding patterns in the gel. The method is able to detect about half of all single-base mutations (60). It is not clear why all mutations are not detected, but this is perhaps related to the secondary structure of the probe-test heteroduplex.
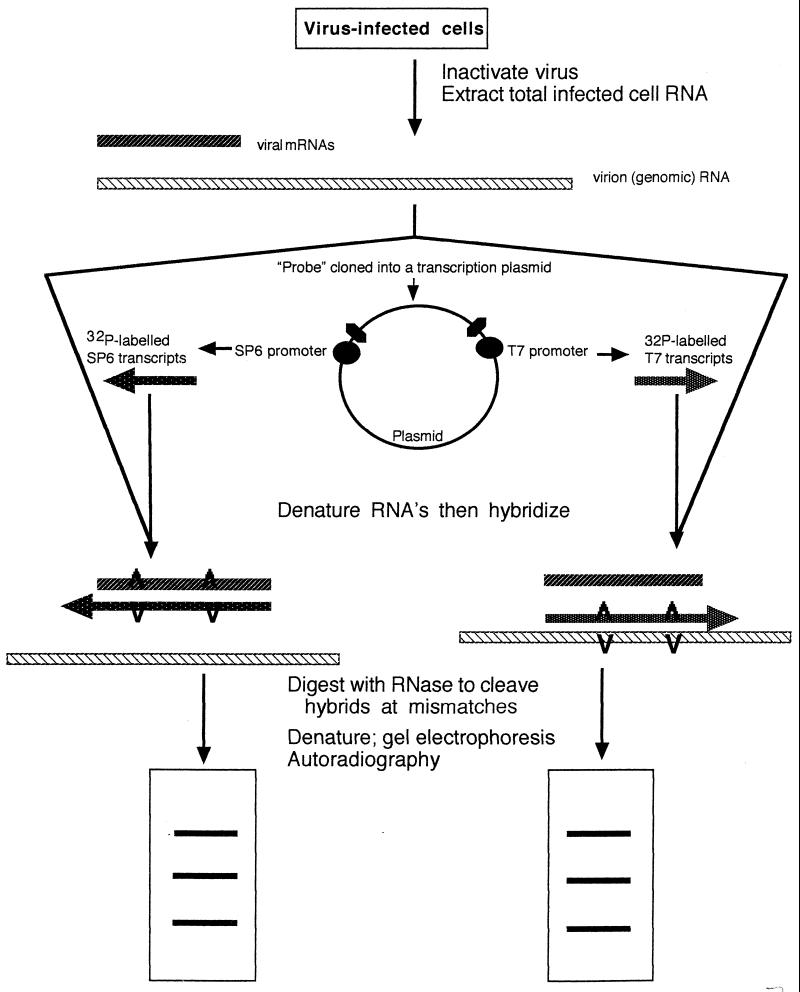
FIG. 3.
RNase protection analysis. RNA from virus-infected cells was extracted and purified. This mixture of mRNA and genomic RNA was mixed with transcripts, labeled in vitro with 32P, from a cloned probe. The cloned probe was transcribed in both directions, from an SP6 promoter at one end and a T7 promoter at the other end. The mixtures were denatured and reannealed to allow the formation of hybrids which would contain “bubbles” at the sites of mismatches. RNase digests all the single-stranded regions and cleaves the double-stranded regions at the bubbles or mismatches. After RNase digestion, the products were subjected to electrophoresis in polyacrylamide gels and bands were detected by autoradiography. Viral isolates with different mutations relative to the probe differ from each other in their banding patterns. Adapted from reference 74 with permission of the publisher.
Influenza Virus and RSV Analysis
A modification of ribonuclease protection, sometimes called the RNase A mismatch cleavage method, has allowed its use with the RNA genomes of viruses, including influenza virus and RSV. By using a technique that had been developed to demonstrate the presence of point mutations in RNA transcripts from the c-ras gene (82), the genetic relatedness and evolution of field isolates of influenza virus were investigated (54). This method was also employed in an epidemiologic study of RSV in which it was found, using a probe made from the G glycoprotein of the A2 strain of RSV, that all epidemiologically linked isolates (from coinfected twins, infants infected during a nosocomial outbreak, and institutionalized adults infected during an outbreak) had identical band patterns (74). The same method was used to study the genetic variability and evolution of the different subtypes of RSV circulating in Spain (15). Mutations which appeared in early isolates were retained in later isolates, and viruses isolated during a short time span showed highly similar band patterns. Additionally, different isolates from the same winter outbreak of RSV showed considerable heterogeneity and the RSV G gene accumulated mutations at a faster rate than other genes. Although the method is technically difficult, it has considerable power to define epidemiologically related isolates in the same way that RFLP analysis has for DNA viruses.
SINGLE-STRAND CONFORMATION POLYMORPHISM ANALYSIS
Development of the Assay and Application to Viruses
Single-strand conformation polymorphism (SSCP) analysis is the term applied to the method developed by Orita et al. (63) with which they demonstrated that a single nucleotide substitution was sufficient to cause a mobility shift of a fragment of single-stranded DNA in a neutral polyacrylamide gel. Initially, the general procedure was to use RFLP fragments from genomic DNA (one from the wild-type genome and one from a possible mutant), denature the fragments by alkali treatment, subject them to electrophoresis in a neutral polyacrylamide gel, and compare their mobilities. More recently, modifications have been made to accommodate PCR amplification of a specific region of wild-type or mutant genomes prior to denaturation and separation on a neutral gel (32). In either case, if mutations are present in the segment of the mutant genome being tested, that segment will likely run at a different position in the gel than the same segment from the wild-type genome. The altered electrophoresis pattern is apparently due to the mutation-altered secondary structure of the restriction fragment or the PCR amplicon (63). The separation of the wild-type and mutant fragments is dependent on several environmental factors, including the temperature of the gel during electrophoresis, the concentration and composition of the electrophoresis buffer, and the presence of denaturing agents in the gel. Several sets of conditions should be tried empirically to optimize mutation detection. One major advantage of this method is that it can “sample” the genetic makeup of several hundred base pairs of DNA, whereas RFLP analysis can sample only a few bases (the restriction sites).
Analysis of Parvovirus, Hepatitis B Virus, and HCV
An analysis of the genetic variability in the nonstructural gene of human parvovirus B19 by PCR-SSCP analysis revealed the presence of six genotypes among 50 samples of virus from several countries (43). Sequencing of this region confirmed the presence of mutations in the different genotypes, and all were silent mutations. There was a good correlation between the SSCP type and the country from which the virus was obtained. Within Japan, genotypes 1, 2, 3, and 4 circulated between 1981 and 1987 in about equal numbers. However, between 1990 and 1994, 90% of the samples tested were type 3.
An epidemiologic investigation into an outbreak of hepatitis B virus (HBV) infection in a pediatric oncology unit was based on the ability to distinguish genotypes of the virus by PCR-SSCP analysis (30). PCR was used to amplify a 189-bp product from the hypervariable (pre-S1) region of the genome, and this product was denatured and subjected to electrophoresis in neutral polyacrylamide gels. Forty unrelated controls all had distinct patterns in gels, and all but 6 of 58 oncology patients had patterns that fell into five different groups: one shared by 16 patients, one shared by 19 patients, one shared by 9 patients, one shared by 5 patients, and one shared by 3 patients. Thus, there had been several independent introductions of HBV into the unit, and some of these isolates had also been spread extensively within the unit.
Genotyping of HCV is useful because of marked differences in pathogenesis and the ability to treat different strains. A method for rapid and sensitive genotyping of HCV by PCR-SSCP analysis has been described (48). A nested RT-PCR assay was used to amplify a 289-bp amplicon from the conserved 5′ NC region, which was analyzed in nondenaturing polyacrylamide gels (Fig. 4). This method was able to correctly identify the strain types of 73 HCV-positive samples which had been genotyped by sequencing and dideoxy fingerprinting. The PCR-SSCP method was more rapid and considerably cheaper than either of the established methods and yet provided the same information with regard to strain typing.
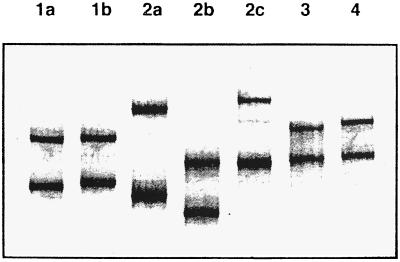
FIG. 4.
SSCP analysis. RT-PCR products from the 5′ NC regions from various HCV genotypes were electrophoresed in 11.5% polyacrylamide gel and visualized by silver staining. HCV types and subtypes are shown across the top. Reprinted from reference 48 with permission of the publisher.
HETERODUPLEX MOBILITY ASSAY AND HETERODUPLEX TRACKING ASSAY
HIV Heteroduplex Mobility Assay
The effect of a single base deletion on the electrophoresis of heteroduplex DNA in cross-linked gels was studied by Wang and Griffith (79) using synthetic 30-bp duplexes. All DNAs containing bulges due to a single base deletion in one strand which resulted in a bulge in the other strand were electrophoretically retarded in comparison to DNAs with no bulges. These observations have been the basis for the technique of heteroduplex mobility assays (HMAs), which are now used to investigate the genetic variability (at the molecular level) of various strains or quasispecies of viruses.
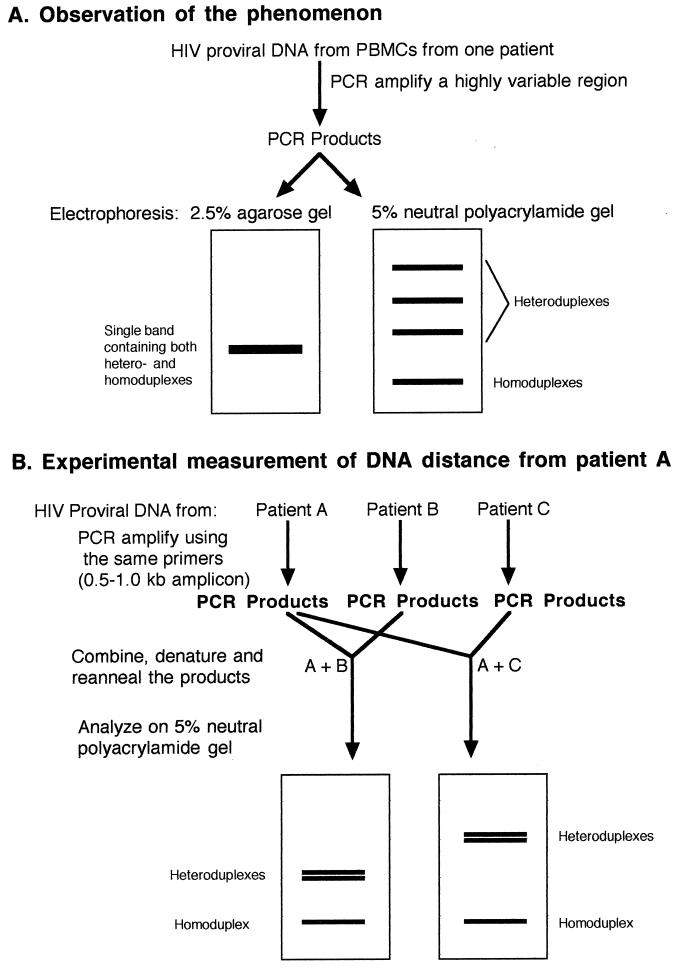
Following PCR amplification of HIV proviral DNA from peripheral blood mononuclear cells, a slowly migrating species of product appeared if analyzed in neutral 5% polyacrylamide gels (18). The same product mixtures migrated as a single band in a 2.5% agarose gel (Fig. 5A). The fact that the slowly migrating products in polyacrylamide gels were heteroduplexes was proven when they were eliminated either by performing one additional round of PCR in the presence of excess Taq polymerase and excess primers or by reducing the number of divergent proviral genotypes by dilution of the input target proviral DNA (18). The former method produces all homoduplexes, since with excess polymerase and primers all targets are replicated; if not denatured, they will remain as homoduplexes. The latter method simply results in a less divergent population of targets in the PCR, with the final products therefore being predominantly homoduplexes.
FIG. 5.
HMA. (A) Illustration of the phenomenon and how HMA was initially observed. PCR products generated from a highly variable region of HIV contain many heteroduplexes which are not resolved in agarose gels but may be resolved and appear as distinct bands in polyacrylamide gels. The heteroduplexes migrate more slowly due to conformational changes which result from the bulged regions and can be converted to homoduplexes as described in the text. PBMCs, peripheral blood mononuclear cells. (B) Illustration of the use of the technique to determine the relative relatedness of virus from two patients to that from patient A. Purified amplicons from the test patients were mixed with amplicons from patient A, denatured, reannealed, and subjected to electrophoresis in a neutral polyacrylamide gel. The migration of the heteroduplex bands was retarded in proportion to their genetic distance from the virus from patient A. See the text for details.
Further demonstration of the usefulness of the HMA technique to investigate the diversity of HIV strains was provided by denaturing and reannealing mixtures of amplified DNA from divergent HIV strains with known sequences. If two diverse amplicons of the same size were mixed, denatured, reannealed, and subjected to electrophoresis in neutral polyacrylamide gels, three bands were observed: one band consisted of homoduplexes and the other two were the slower-migrating heteroduplexes (Fig. 5B). If three diverse amplicons were mixed, denatured, and reannealed, there were six slower-migrating bands in the gel (18), indicating the presence of all combinations of heteroduplexes. The relative retardation of the heteroduplexes was proportional to the DNA distance (i.e., the percent nucleotide difference) between the strains being tested (Fig. 5B). In Fig. 5B, strain C is more divergent from A than strain B is divergent from A since the heteroduplex bands are further from the homoduplex band, and the relative distances can be calculated (18) (see below). The technique can detect genetic differences of as little as 2% in an amplicon of several hundred base pairs but is not reliable at detection of differences less than that. The major advantage of this technique is that it can be used to screen isolates and determine genetic relatedness without the laborious task of DNA sequencing.
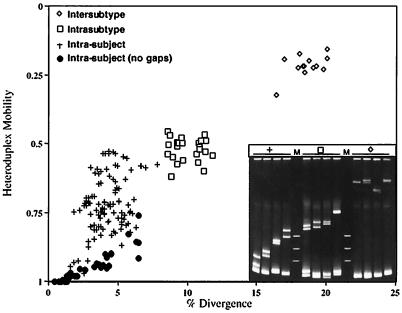
The HMA method has been used efficiently to analyze the relatedness of various HIV strains (quasispecies) from a single individual and from around the world. Delwart et al. (18) showed that intrasubject relatedness of DNA from the env gene (excluding gaps in the sequences) was about 1 to 5%, with corresponding heteroduplex mobilities of 1.0 to 0.9 (the ratio of the mobility of the heteroduplex to that of the homoduplex). If gaps caused by insertions and deletions were considered in the distance calculations, then the DNA distances ranged from 2 to 8% and the heteroduplex mobilities ranged from 1.0 to about 0.45. At the other end of the spectrum was a comparison of U.S. and African isolates in which the DNA distances were about 18 to 23% and the heteroduplex mobilities were about 0.18 to 0.32. Similar data are presented in Fig. 6. In this case, HIV-1 specimens known to be of subtype B were compared by HMA with others of the same subtype and with others of different subtypes (subtypes A, C, D, and E) as well as within individual patients. Heteroduplex mobilities of about 0.9 to 0.5 correlated with up to an 8% divergence in the intrasubject analysis (2 to 8% with and 1 to 7% without gaps), 0.6 to 0.4 correlated with an 8 to 12% divergence in the intrasubtype comparison, and 0.3 to 0.2 correlated with a 7 to 20% divergence in the intersubtype analysis. Thus, the method allows an experimental estimation of DNA distance simply by performing the HMA, and in fact, one can quickly determine the subtype of an HIV strain by using this assay (18).
FIG. 6.
Use of HMA to determine HIV relatedness. Heteroduplex mobilities versus DNA distance of sequences of the region of HIV-1 Env from V3 to V5 from the same individuals and from individuals infected with the same (intrasubtype) or different (intersubtype) subtypes of HIV-1. Mobilities were determined by dividing the average distance of migration of the heteroduplex bands by the distance of migration of the homoduplex band. Cloned fragments of the patient proviral DNA were used as starting material. DNA distances (i.e., divergence) were determined by counting mismatches in the best alignment of the sequences, disregarding bases that are unpaired due to insertions or deletions. (Inset) Polyacrylamide (5%) gel stained with ethidium bromide showing representative heteroduplexes from the comparisons discussed above. Courtesy of E. L. Delwart and J. I. Mullins.
In a separate analysis, Delwart et al. (18) plotted pairwise comparisons between viruses from a given geographic region. A very high degree of viral genetic diversity was shown for isolates from Africa and was consistent with the notion that the HIV-1 epidemic began on that continent. The divergence of HIV-1 isolates from North America and Europe was limited (i.e., the DNA distances formed a single cluster), whereas the experimentally determined DNA distances for isolates from Brazil and from Thailand revealed two distinct clusters which indicated the recent introduction of virus from two divergent sources. Delwart and others (16, 17) have continued to use HMA in heroic efforts to study HIV-1 evolution in individual patients during progression to AIDS.
HIV Heteroduplex Tracking Assay
A variation of the HMA methodology is to amplify by RT-PCR products that represent an entire pool of quasispecies from an individual. This pool of PCR products is then hybridized to a radioactively labeled probe (made from a cloned region of HIV DNA), and the resulting heteroduplexes are detected by electrophoresis and autoradiography. This is called the heteroduplex tracking assay (HTA). Since the heteroduplexes displayed in the gel represent the relatedness of many (perhaps all) of the quasispecies, HTA may provide the best overall analysis of genetic distances between quasispecies generated within a single individual. Thus, HMA detects all of the heteroduplexes that are formed by viewing them in ethidium bromide-stained gels, whereas HTA detects only the heteroduplexes that form between the radioactive probe and the PCR products.
Recently, by using a gp120 V3-specific HTA in which an HIV subtype B consensus Env V3 sequence was used as a probe, evolution was measured by divergence from the consensus probe and was correlated with the syncytium-inducing (SI) phenotype of the isolates (61). The V3 HTA was able to detect 96% of the SI variants tested, and the overall correspondence for identifying SI and non-SI variants was 88%. Thus, this assay may be useful for the identification of certain phenotypic traits in HIV-1 and preclude the use of costly and time-consuming cultures to distinguish SI and non-SI isolates.
Use of HMA To Type and Subtype Influenza Virus
A practical application of the HMA has been described in which multiplex reverse transcription and multiplex PCR were employed to differentiate the HA genes of influenza viruses (86). The subtype-specific primers in the multiplex-reverse transcription–multiplex PCR method differentiated the strains by type and subtype, because the amplified products of type A (subtypes H1 and H3), B (HA gene), and C (HA, esterase, and fusion [HEF] gene) were different sizes. In the subsequent HMA analysis, the amplified product of a clinical strain was mixed with the corresponding amplicon of a reference strain, and the resulting mobility shift pattern after electrophoresis indicated the divergence of the clinical amplicon. This process can be performed in 2 days and thus is a rapid and simple method for identification of clinical influenza virus strains.
GENOME SEGMENT LENGTH POLYMORPHISM ANALYSIS (ELECTROPHEROTYPING)
Application to Viruses with Segmented Genomes
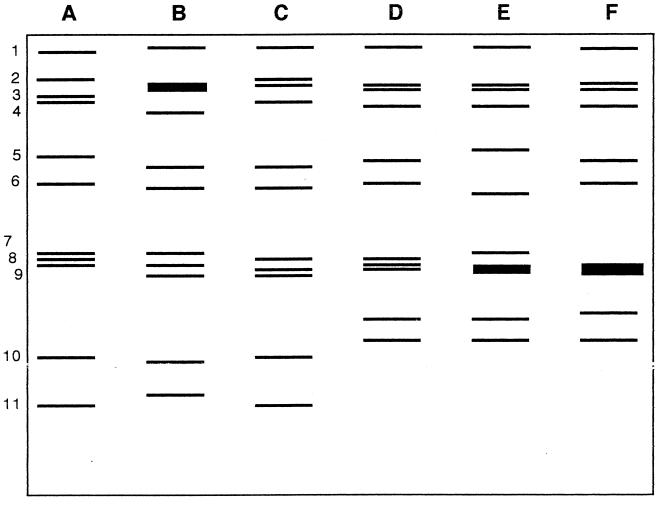
Certain groups of viruses are known to have segmented genomes. For example, the influenza viruses have segmented negative-strand RNA genomes, and the reoviruses (of which rotavirus is the most clinically significant member) have double-stranded RNA genomes. Influenza A and B viruses have eight different genome segments and influenza C virus has seven RNA segments. Each segment is of nearly constant length (the segments vary in length for influenza A virus from 1,263 to 2,320 nucleotides), and no length polymorphisms exist. Rotaviruses that infect humans fall into groups A, B, and C (there are also groups D through G, which are not known to infect humans), all of which have 11 segments of double-stranded RNA; some of these segments are of variable length. When total RNA is extracted from a clinical sample and subjected to electrophoresis on a polyacrylamide gel (5), the classical electropherotype pattern of group A rotaviruses reveals four size classes, with segments 1 to 4 in the largest class, 5 and 6 in the second class, 7 to 9 in the third class, and 10 to 11 in the smallest class (Fig. 7). Patterns that spread out more during gel electrophoresis are referred to as long genomes, and others that spread out less are short genomes.
FIG. 7.
Electropherotyping of rotaviruses. Stylized drawing of 10% polyacrylamide gel analysis of representative RNAs (A through F) extracted from stool specimens from children. The gels were stained with ethidium bromide to visualize the RNA bands (genome segments) numbered 1 to 11 at the left. Samples A through C are the long electropherotype and samples D through F are the short electropherotype. Adapted from reference 73.
The observation that the electropherotypes of epidemiologically unrelated rotaviruses may vary due to small differences in the lengths of some segments has been used to study the epidemiology and transmission of rotavirus. A simplified method for these analyses was developed by Dolan et al. (21), in which RNA was extracted directly from clinical specimens, subjected to electrophoresis in Laemmli stacking polyacrylamide gels, and detected with the extremely sensitive silver stain technique. They found 10 different electropherotypes among 68 rotavirus-containing specimens and noted that the predominant strain was different in each of three consecutive winter outbreaks. This same basic technique had been used to study the molecular epidemiology of rotavirus infection in children in South Africa (73) (Fig. 7) and to demonstrate the nosocomial transmission of rotavirus in hospitalized patients in Santiago, Chile (26).
CONCLUDING REMARKS
The methods described here have all been developed over the past 20 years. They find usefulness in our efforts to further subdivide and characterize the broad groups of viruses that are isolated in cell culture, identified by fluorescent-antibody reagents, and detected by PCR. This activity by molecular virologists in subtyping of viruses has arisen mainly out of interest in epidemiologic studies. Direct analysis of the viral genome is a reasonable and practical way to approach these issues. Many of the methods described here can be performed with very simple equipment and in relatively unsophisticated laboratory environments, thus making them useful to clinical laboratories as well as research laboratories. Other methods discussed, particularly nucleotide sequencing, are becoming more widely used as tools in clinical laboratories because of major advances in the technology. Some of these methods were considered quite esoteric only a few years ago but have been catapulted into common use by the development of automated or computerized instrumentation, PCR technology, and commercial development of kits. The HIV/AIDS epidemic has also driven some of this molecular genotyping technology in the search for greater understanding of the mechanisms of disease transmission and progression and the characterization of isolates resistant to antiretroviral agents. In the future, these methods for the molecular analysis of viral genomes will become more widely available and will be used more frequently in practical applications in clinical laboratories.
REFERENCES
- 1.Adams S G, Dohner D E, Gelb L D. Restriction fragment differences between the genomes of the Oka varicella vaccine virus and American wild-type varicella-zoster virus. J Med Virol. 1989;29:38–45. doi: 10.1002/jmv.1890290108. [DOI] [PubMed] [Google Scholar]
- 2.Agostini H T, Ryschkewitsch C F, Mory R, Singer E J, Stoner G L. JC virus (JCV) genotypes in brain tissue from patients with progressive multifocal leukoencephalopathy (PML) and in urine from controls without PML: increased frequency of JCV type 2 in PML. J Infect Dis. 1997;176:1–8. doi: 10.1086/514010. [DOI] [PubMed] [Google Scholar]
- 3.Ando T, Jin Q, Gentsch J R, Monroe S S, Noel J S, Dowell S F, Cicirello H G, Kohn M A, Glass R I. Epidemiologic applications of novel molecular methods to detect and differentiate small round structured viruses (Norwalk-like viruses) J Med Virol. 1995;47:145–152. doi: 10.1002/jmv.1890470207. [DOI] [PubMed] [Google Scholar]
- 4.Arens M, Dilworth V. Remarkably homogeneous population of adenovirus type 3 and 7 genome types. J Clin Microbiol. 1988;26:1604–1608. doi: 10.1128/jcm.26.8.1604-1608.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arens M, Swierkosz E M. Detection of rotavirus by hybridization with a nonradioactive synthetic DNA probe and comparison with commercial enzyme immunoassays and silver-stained polyacrylamide gels. J Clin Microbiol. 1989;27:1277–1279. doi: 10.1128/jcm.27.6.1277-1279.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beraud F, Kessler N, Aymard M. Contribution of monoclonal antibodies to the study of parainfluenza virus antigens. Dev Biol Stand. 1984;57:257–267. [PubMed] [Google Scholar]
- 7.Biasin M R, Fiordalisi G, Zanella I, Cavicchini A, Marchelle G, Infantolino D. A DNA hybridization method for typing hepatitis C virus genotype 2c. J Virol Methods. 1997;65:307–315. doi: 10.1016/s0166-0934(97)02202-7. [DOI] [PubMed] [Google Scholar]
- 8.Buchman T G, Roizman B, Adams G, Stover B H. Restriction endonuclease fingerprinting of herpes simplex virus DNA: a novel epidemiological tool applied to a nosocomial outbreak. J Infect Dis. 1978;138:488–498. doi: 10.1093/infdis/138.4.488. [DOI] [PubMed] [Google Scholar]
- 9.Buchman T G, Roizman B, Nahmias A J. Demonstration of exogenous genital reinfection with herpes simplex virus type 2 by restriction endonuclease fingerprinting of viral DNA. J Infect Dis. 1979;140:295–304. doi: 10.1093/infdis/140.3.295. [DOI] [PubMed] [Google Scholar]
- 10.Cassol S, Weniger B G, Babu G, Salminen M O, Zheng X, Htoon M T, Delaney A, O’Shaughnessy M, Ou C-Y. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res Hum Retroviruses. 1996;70:7013–7029. doi: 10.1089/aid.1996.12.1435. [DOI] [PubMed] [Google Scholar]
- 11.Castelain S, Khorsi H, Zawadzki P, Sueur J-M, Capron J-P, Eb F, Duverlie G. Direct blotting electrophoresis for sequencing and genotyping hepatitis C virus. J Virol Methods. 1997;65:237–243. doi: 10.1016/s0166-0934(97)02184-8. [DOI] [PubMed] [Google Scholar]
- 12.Chou S. Acquisition of donor strains of cytomegalovirus by renal-transplant recipients. N Engl J Med. 1986;314:1418–1423. doi: 10.1056/NEJM198605293142205. [DOI] [PubMed] [Google Scholar]
- 13.Clewley J P, Bishop D H L, Kang C-Y, Coffin J, Schnitzlein W M, Reichmann M E, Shope R E. Oligonucleotide fingerprints of RNA species obtained from rhabdoviruses belonging to the vesicular stomatitis virus subgroup. J Virol. 1977;23:152–166. doi: 10.1128/jvi.23.1.152-166.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooney M K, Fox J P, Kenny G E. Antigenic groupings of 90 rhinovirus serotypes. Infect Immun. 1982;37:642–647. doi: 10.1128/iai.37.2.642-647.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristina J, López J A, Albó C, García-Barreno B, García J, Melero J A, Portela A. Analysis of genetic variability in human respiratory syncytial virus by the RNase A mismatch cleavage method: subtype divergence and heterogeneity. Virology. 1990;174:126–134. doi: 10.1016/0042-6822(90)90061-u. [DOI] [PubMed] [Google Scholar]
- 16.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 19.Deubel B, Nogueira R M, Drouet M T, Zeller H, Reynes J M, Ha D Q. Direct sequencing of genomic cDNA fragments amplified by the polymerase chain reaction for molecular epidemiology of dengue-2 viruses. Arch Virol. 1993;129:197–210. doi: 10.1007/BF01316895. [DOI] [PubMed] [Google Scholar]
- 20.De Wachter R, Fiers W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32P-labeled RNA. Anal Biochem. 1972;49:184–197. doi: 10.1016/0003-2697(72)90257-6. [DOI] [PubMed] [Google Scholar]
- 21.Dolan K T, Twist E M, Horton-Slight P, Forrer C, Bell L M, Plotkin S A, Clark H F. Epidemiology of rotavirus electropherotypes determined by a simplified diagnostic technique with RNA analysis. J Clin Microbiol. 1985;21:753–758. doi: 10.1128/jcm.21.5.753-758.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drew W L, Sweet E S, Miner R C, Mocarski E S. Multiple infections by cytomegalovirus in patients with acquired immunodeficiency syndrome: documentation by Southern blot hybridization. J Infect Dis. 1984;150:952–953. doi: 10.1093/infdis/150.6.952. [DOI] [PubMed] [Google Scholar]
- 23.Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, Blaas D, Kuechler E. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2, and 14. Proc Natl Acad Sci USA. 1987;84:2605–2609. doi: 10.1073/pnas.84.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falk K, Gratama J W, Rowe M, Zou J Z, Khanim F, Young L S, Oosterveer M A, Ernberg I. The role of repetitive DNA sequences in the size variation of Epstein-Barr virus (EBV) nuclear antigens, and the identification of different EBV isolates using RFLP and PCR analysis. J Gen Virol. 1995;76:779–790. doi: 10.1099/0022-1317-76-4-779. [DOI] [PubMed] [Google Scholar]
- 25.Fujioka S, Koide H, Kitaura Y, Duguchi H, Kawamura K. Analysis of enterovirus genotypes using single-strand conformation polymorphisms of polymerase chain reaction products. J Virol Methods. 1995;51:253–258. doi: 10.1016/0166-0934(94)00112-t. [DOI] [PubMed] [Google Scholar]
- 26.Gaggero A, Avendaño L F, Fernández J, Spencer E. Nosocomial transmission of rotavirus from patients admitted with diarrhea. J Clin Microbiol. 1992;30:3294–3297. doi: 10.1128/jcm.30.12.3294-3297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn B H, Shaw G M, Taylor M E, Redfield R R, Markham P D, Salahuddin S Z, Wong-Staal F, Gallo R C, Parks E S, Parks W P. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 29.Hammer S M, Buchman T G, D’Angelo L J, Karchmer A W, Roizman B, Hirsch M S. Temporal cluster of herpes simplex encephalitis: investigation by restriction endonuclease cleavage of viral DNA. J Infect Dis. 1980;141:436–440. doi: 10.1093/infdis/141.4.436. [DOI] [PubMed] [Google Scholar]
- 30.Hardie D R, Kannemeyer J, Stannard L M. DNA single strand conformation polymorphism identifies five defined strains of hepatitis B virus (HBV) during an outbreak of HBV infection in an oncology unit. J Med Virol. 1996;49:49–54. doi: 10.1002/(SICI)1096-9071(199605)49:1<49::AID-JMV8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 31.Harris T J R, Robson K J H, Brown F. A study of the level of nucleotide sequence conservation between the RNAs of two serotypes of foot and mouth disease virus. J Gen Virol. 1980;50:403–418. doi: 10.1099/0022-1317-50-2-403. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. Genome Res. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 33.Hierholzer J C. Adenoviruses in the immunocompromised host. Clin Microbiol Rev. 1992;5:262–274. doi: 10.1128/cmr.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hierholzer J C, Adrian T, Anderson L J, Wigand R, Gold J W. Analysis of antigenically intermediate strains of subgenus B and D adenoviruses from AIDS patients. Arch Virol. 1988;103:99–115. doi: 10.1007/BF01319812. [DOI] [PubMed] [Google Scholar]
- 35.Hierholzer J C, Barme M. Counterimmunoelectrophoresis with adenovirus type-specific anti-hemagglutinin sera as a rapid diagnostic method. J Immunol. 1974;112:987–995. [PubMed] [Google Scholar]
- 36.Hierholzer J C, Wigand R, Anderson L J, Adrian T, Gold J W. Adenoviruses from patients with AIDS: a plethora of serotypes and a description of five new serotypes of subgenus D (types 43–47) J Infect Dis. 1988;158:804–813. doi: 10.1093/infdis/158.4.804. [DOI] [PubMed] [Google Scholar]
- 37.Horsnell C, Gama R E, Hughes P J, Stanway G. Molecular relationships between 21 human rhinovirus serotypes. J Gen Virol. 1995;76:2549–2555. doi: 10.1099/0022-1317-76-10-2549. [DOI] [PubMed] [Google Scholar]
- 38.Huang E-S, Alford C A, Reynolds D W, Stagno S, Pass R F. Molecular epidemiology of cytomegalovirus infections in women and their infants. N Engl J Med. 1980;303:958–962. doi: 10.1056/NEJM198010233031702. [DOI] [PubMed] [Google Scholar]
- 39.Hughes P J, North C, Jellis C H, Minor P D, Stanway G. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J Gen Virol. 1988;69:49–58. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- 40.Johansson M E, Andersson M A, Thörner P Å. Adenoviruses isolated in the Stockholm area during 1987–1992: restriction endonuclease analysis and molecular epidemiology. Arch Virol. 1994;137:101–115. doi: 10.1007/BF01311176. [DOI] [PubMed] [Google Scholar]
- 41.Katayama Y, Shibahara K, Kohama T, Homma M, Hotta H. Molecular epidemiology and changing distribution of genotypes of measles virus field strains in Japan. J Clin Microbiol. 1997;35:2651–2653. doi: 10.1128/jcm.35.10.2651-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katz B Z, Niederman J C, Olson B A, Miller G. Fragment length polymorphisms among independent isolates of Epstein-Barr virus from immunocompromised and normal hosts. J Infect Dis. 1988;157:299–308. doi: 10.1093/infdis/157.2.299. [DOI] [PubMed] [Google Scholar]
- 43.Kerr J R, Curran M D, Moore J E, Erdman D D, Coyle P V, Nunoue T, Middleton D, Ferguson W P. Genetic diversity in the non-structural gene of parvovirus B19 detected by single-stranded conformational polymorphism assay (SSCP) and partial nucleotide sequencing. J Virol Methods. 1995;53:213–222. doi: 10.1016/0166-0934(95)00017-o. [DOI] [PubMed] [Google Scholar]
- 44.Kew O M, Nottay B K, Hatch M H, Hierholzer J C, Obijeski J F. Oligonucleotide fingerprint analysis of enterovirus 70 isolates from the 1980 to 1981 pandemic of acute hemorrhagic conjunctivitis: evidence for a close genetic relationship among Asian and American strains. Infect Immun. 1983;41:631–635. doi: 10.1128/iai.41.2.631-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilpatrick B A, Huang E-S, Pagano J S. Analysis of cytomegalovirus genomes with restriction endonucleases HinD III and EcoR-1. J Virol. 1976;18:1095–1105. doi: 10.1128/jvi.18.3.1095-1105.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Komada H, Klippmark E, Orvell C, Randall R E, Ito Y, Norrby E. Immunological relationships between parainfluenza virus type 4 and other paramyxoviruses studied by use of monoclonal antibodies. Arch Virol. 1991;116:277–283. doi: 10.1007/BF01319249. [DOI] [PubMed] [Google Scholar]
- 47.Kuan M M. Detection and rapid differentiation of human enteroviruses following genomic amplification. J Clin Microbiol. 1997;35:2598–2601. doi: 10.1128/jcm.35.10.2598-2601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lareu R R, Swanson N R, Fox S A. Rapid and sensitive genotyping of hepatitis C virus by single-strand conformation polymorphism. J Virol Methods. 1997;64:11–18. doi: 10.1016/s0166-0934(96)02134-9. [DOI] [PubMed] [Google Scholar]
- 49.Lee J-H, Roth W K, Zeuzem S. Evaluation and comparison of different hepatitis C virus genotyping and serotyping assays. J Hepatol. 1997;26:1001–1009. doi: 10.1016/s0168-8278(97)80108-0. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y F, Kitamura N, Nomoto A, Wimmer E. Sequence studies of poliovirus RNA. IV. Nucleotide sequence complexities of poliovirus type 1, type 2 and two type 1 defective interfering particles RNAs, and fingerprint of the poliovirus type 3 genome. J Gen Virol. 1979;44:311–322. doi: 10.1099/0022-1317-44-2-311. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y F, Wimmer E. Fingerprinting high molecular weight RNA by two-dimensional electrophoresis: application to poliovirus RNA. Nucleic Acids Res. 1976;3:1647–1658. doi: 10.1093/nar/3.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levett P N, Gu M, Luan B, Fearon M, Stubberfield J, Jamieson F, Petric M. Longitudinal study of molecular epidemiology of small round-structured viruses in a pediatric population. J Clin Microbiol. 1996;34:1497–1501. doi: 10.1128/jcm.34.6.1497-1501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Zheng Q, Liu Y, Wadell G. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J Med Virol. 1996;49:170–177. doi: 10.1002/(SICI)1096-9071(199607)49:3<170::AID-JMV3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 54.Lopez-Galindez C, Lopez J A, Melero J A, De La Fuente L, Martinez C, Ortin J, Perucho M. Analysis of genetic variability and mapping of point mutations in influenza virus by the RNase mismatch cleavage method. Proc Natl Acad Sci USA. 1988;85:3522–3526. doi: 10.1073/pnas.85.10.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McNearney T, Hornickova Z, Kloster B, Birdwell A, Storch G A, Polmar S H, Arens M, Ratner L. Evolution of sequence divergence among human immunodeficiency virus type 1 isolates derived from a blood donor and a recipient. Pediatr Res. 1993;33:36–42. doi: 10.1203/00006450-199301000-00008. [DOI] [PubMed] [Google Scholar]
- 57.Minor P D, Schild G C, Ferguson M, Mackay A, Magrath D I, John A, Yates P J, Spitz M. Genetic and antigenic variation in type 3 polioviruses: characterization of strains by monoclonal antibodies and T1 oligonucleotide mapping. J Gen Virol. 1982;61:167–176. doi: 10.1099/0022-1317-61-2-167. [DOI] [PubMed] [Google Scholar]
- 58.Mulders M N, Lipskaya G Y, van der Avoort H G A M, Koopmans M P G, Kew O M, van Loon A M. Molecular epidemiology of wild poliovirus type 1 in Europe, the Middle East, and the Indian subcontinent. J Infect Dis. 1995;171:1399–1405. doi: 10.1093/infdis/171.6.1399. [DOI] [PubMed] [Google Scholar]
- 59.Mulders M N, van Loon A M, van der Avoort H G A M, Reimerink J H J, Ras A, Bestebroer T M, Drebot M A, Kew O M, Koopmans M P G. Molecular characterization of a wild poliovirus type 3 epidemic in The Netherlands (1992 and 1993) J Clin Microbiol. 1995;33:3252–3256. doi: 10.1128/jcm.33.12.3252-3256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myers R M, Larin Z, Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985;230:1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- 61.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nottay B K, Kew O M, Hatch M H, Heyward J T, Obijeski J F. Molecular variation of type 1 vaccine-related and wild polioviruses during replication in humans. Virology. 1981;108:405–423. doi: 10.1016/0042-6822(81)90448-7. [DOI] [PubMed] [Google Scholar]
- 63.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ou C-Y, Ciesielski C A, Myers G, Bandea C I, Luo C-C, Korber B T M, Mullins J I, Schochetman G, Berkelman R L, Economou A N, Witte J J, Furman L J, Satten G A, MacInnes K A, Curran J W, Jaffe H W. Laboratory Investigation Group, Epidemiologic Investigation Group: molecular epidemiology of HIV transmission in a dental practice. Science. 1992;256:1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 65.Pogam S L, Dubois F, Christen R, Raby C, Cavicchini A, Goudeau A. Comparison of DNA enzyme immunoassay and line probe assays (Inno-LiPA HCV I and II) for hepatitis C virus genotyping. J Clin Microbiol. 1998;36:1461–1463. doi: 10.1128/jcm.36.5.1461-1463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rabb-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 67.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski F A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 68.Rivera V M, Welsh J D, Maizel J V. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology. 1988;165:42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- 69.Sidagis J, Ueno K, Tokunaga M, Ohyama M, Eizuru Y. Molecular epidemiology of Epstein-Barr virus (EBV) in EBV-related malignancies. Int J Cancer. 1997;72:72–76. doi: 10.1002/(sici)1097-0215(19970703)72:1<72::aid-ijc11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 70.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 71.Spector S A, Hirata K K, Neuman T R. Identification of multiple cytomegalovirus strains in homosexual men with acquired immunodeficiency syndrome. J Infect Dis. 1984;150:953–955. doi: 10.1093/infdis/150.6.953. [DOI] [PubMed] [Google Scholar]
- 72.Spector S A, Spector D H. Molecular epidemiology of cytomegalovirus infections in premature twin infants and their mother. Pediatr Infect Dis. 1982;1:405–409. doi: 10.1097/00006454-198211000-00009. [DOI] [PubMed] [Google Scholar]
- 73.Steele A D. Shift in genomic RNA patterns of human rotaviruses isolated from white children in South Africa. SAMJ. 1991;79:143–145. [PubMed] [Google Scholar]
- 74.Storch G A, Park C S, Dohner D E. RNA fingerprinting of respiratory syncytial virus using ribonuclease protection. J Clin Investig. 1989;83:1894–1902. doi: 10.1172/JCI114096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 76.Takayama M, Takayama N, Inoue N, Kameoka Y. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J Clin Microbiol. 1996;34:2869–2874. doi: 10.1128/jcm.34.12.2869-2874.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tolpin M D, Stewart J A, Warren D, Mojica B A, Collins M A, Doveikis S A, Cabradilla C, Schauf V, Raju T N K, Nelson K. Transfusion transmission of cytomegalovirus confirmed by restriction endonuclease analysis. J Pediatr. 1985;107:953–956. doi: 10.1016/s0022-3476(85)80201-8. [DOI] [PubMed] [Google Scholar]
- 78.Wadell G. Molecular epidemiology of human adenoviruses. Curr Top Microbiol Immunol. 1984;110:191–220. doi: 10.1007/978-3-642-46494-2_7. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y-H, Griffith J. Effects of bulge composition and flanking sequence on the kinking of DNA by bulged bases. Biochemistry. 1991;30:1358–1363. doi: 10.1021/bi00219a028. [DOI] [PubMed] [Google Scholar]
- 80.Wigand R, Adrian T. A rational system for classifying and denominating adenovirus genome types. Virus Res. 1991;142:47–56. doi: 10.1016/0923-2516(91)90027-z. [DOI] [PubMed] [Google Scholar]
- 81.Winston D J, Huang E-S, Miller M J, Lin C-H, Ho W G, Gale R P, Champlin R E. Molecular epidemiology of cytomegalovirus infections associated with bone marrow transplantation. Ann Intern Med. 1985;102:16–20. doi: 10.7326/0003-4819-102-1-16. [DOI] [PubMed] [Google Scholar]
- 82.Winter E, Yamamoto F, Almoguera C, Perucho M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc Natl Acad Sci USA. 1985;82:7575–7579. doi: 10.1073/pnas.82.22.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamazaki K, Oseto M, Seto Y, Utagawa E, Kimoto T, Minekawa Y, Inouye S, Yamazaki S, Okuno Y, Oishi I. Reverse transcription-polymerase chain reaction detection and sequence analysis of small round-structured viruses in Japan. Arch Virol Suppl. 1996;12:271–276. doi: 10.1007/978-3-7091-6553-9_29. [DOI] [PubMed] [Google Scholar]
- 84.Young J F, Desselberger U, Palese P. Evolution of human influenza A viruses in nature: sequential mutations in the genomes of new H1N1 isolates. Cell. 1979;18:73–83. doi: 10.1016/0092-8674(79)90355-6. [DOI] [PubMed] [Google Scholar]
- 85.Zeuzem S, Rüster B, Lee J-H, Stripf T, Roth W K. Evaluation of a reverse hybridization assay for genotyping of hepatitis C virus. J Hepatol. 1995;23:654–661. doi: 10.1016/0168-8278(95)80030-1. [DOI] [PubMed] [Google Scholar]
- 86.Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]