Abstract
Non-coding RNAs (ncRNAs) are a large family of RNA molecules with no capability in encoding proteins. However, they participate in developmental and biological processes and their abnormal expression affects cancer progression. These RNA molecules can function as upstream mediators of different signaling pathways and enhancer of zeste homolog 2 (EZH2) is among them. Briefly, EZH2 belongs to PRCs family and can exert functional roles in cells due to its methyltransferase activity. EZH2 affects gene expression via inducing H3K27me3. In the present review, our aim is to provide a mechanistic discussion of ncRNAs role in regulating EZH2 expression in different cancers. MiRNAs can dually induce/inhibit EZH2 in cancer cells to affect downstream targets such as Wnt, STAT3 and EMT. Furthermore, miRNAs can regulate therapy response of cancer cells via affecting EZH2 signaling. It is noteworthy that EZH2 can reduce miRNA expression by binding to promoter and exerting its methyltransferase activity. Small-interfering RNA (siRNA) and short-hairpin RNA (shRNA) are synthetic, short ncRNAs capable of reducing EZH2 expression and suppressing cancer progression. LncRNAs mainly regulate EZH2 expression via targeting miRNAs. Furthermore, lncRNAs induce EZH2 by modulating miRNA expression. Circular RNAs (CircRNAs), like lncRNAs, affect EZH2 expression via targeting miRNAs. These areas are discussed in the present review with a focus on molecular pathways leading to clinical translation.
Keywords: EZH2, MiRNA, LncRNA, CircRNA, SiRNA, ShRNA, Cancer therapy
Introduction
Over the past decades, attention has been directed towards revealing the role of signaling networks in cancer, as this disease causes high mortality and morbidity worldwide [1]. It is believed that abnormal proliferation and metastasis of cancer cells result from alterations in molecular pathways [2–4]. In fact, these tumor-promoting molecular pathways drive cancer progression via activating positive factors for cancer survival [5–7]. In contrast, tumor-suppressor pathways sensitize cancer cells to death and prevent their progression and migration [8]. Due to progress in sequencing and bioinformatics, such molecular pathways have been identified and by continuing research, other novel signaling networks with potential roles in cancer progression/inhibition are revealed [9–11]. The importance of revealing such molecular pathways paves the way for developing novel therapeutics for effective cancer treatment. These therapeutics can be based on designing genetic tools for targeting molecular pathways or using small molecules as drugs for suppressing cancer progression [12–15]. Furthermore, plant derived-natural products have also demonstrated capacity in targeting molecular pathways for cancer therapy [16–19]. According to newly published article by Siegel and colleagues, cancer is still a major challenge for public health and more research should be directed towards basic and clinical understanding of cancer [20].
In the present review, our aim is to focus on a special signaling pathway known as enhancer of zeste homolog 2 (EZH2) in cancer, and its regulation by major upstream mediators, called non-coding RNAs (ncRNAs). The regulatory impact of these ncRNAs on EZH2 signaling, both induction and inhibition are discussed. Furthermore, to ease the understanding of the miRNA roles, we divided the sections based on cancer types. Each section is discussed with a focus on molecular pathways, and to pave the way for clinical translation, the role of these signaling networks, related biomarkers, and their application in clinic are discussed.
EZH2 signaling in cancer
Signaling pathways
There is a large protein family, known as Polycomb Repressive Complexes (PRCs) capable of modifying lysine residues on histones [21, 22]. In mammals, there are two primary PRCs including PRC1 and PRC2. EZH2 is a member of PRCs that exerts functional roles in cells [23]. In PCR2 complex, EZH2 is considered as a catalytic subunit and is responsible for methyltransferase activity. It is noteworthy that EZH proteins can be phosphorylated. However, there are differences among them. The phosphorylation of EZH1 results in its degradation and significantly diminishes its function [24, 25]. The regulatory impact of EZH2 on other genes is related to its impact on histone H3 lysine 27 (H3K27me3). H3K27me3 can be identified by WD40-repeat domain of EED, contributing to the spread of H3K27me3 in genome through the formation of feedforward loop. As a catalytic subunit of PRC2, EZH2 suppresses the transcription of target genes via triggering the trimethylation (Lysine-27) of H3K27me [26]. By regulating gene transcription, EZH2 participates in biological mechanisms including cell fate, cell lineage specification and tumorigenesis [27–30]. EZH2 can also affect cell cycle progression, autophagy, apoptosis, DNA damage repair, and cellular senescence [31–33]. Furthermore, EZH2 can interact with non-histone targets or directly interact with proteins to influence downstream targets in a PRC2-independent manner [34–36]. Overall, there are three different actions for EZH2 including PRC2-dependent H3K27m3, PRC2-dependent non-histone protein methylation, and PRC2-independent gene transactivation [37].
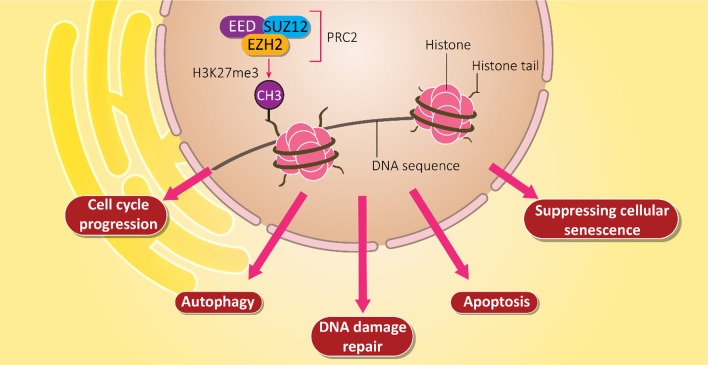
Structurally, EZH2 gene is located on chromosome 7q35 and contains 20 exons with 746 amino acid residues [38]. It has been reported that EZH2 has five distinct domains that are responsible for its regulatory effects. These domains include EED-interaction domain (EID), Domain I and Domain II cysteine-rich domain (CXC domain), C-terminal suppressor of variegation 39 enhancer of zeste, and trithorax domain (SET domain) [39, 40]. Each domain has its own activities. For instance, SET domain cooperates with CXC domain N-terminal for histone methyltransferase activity of EZH2 through its N-terminal domains [39]. Figure 1 demonstrates role of EZH2 in regulation of vital biological mechanisms in cell.
Fig. 1.
EZH2 signaling and its role in regulating downstream signaling pathways. Epigenetic regulation of molecular mechanisms in cells such as DNA repair, cell cycle, apoptosis, autophagy and senescence are regulated by EZH2, showing vital functions of this signaling pathway in cells
The roles of EZH2 in cancer
With respect to the role of EZH2 in regulation of biological mechanisms, studies have focused on revealing its role in cancer. For exerting its tumorigenesis impact, we need to consider EZH2 stability. It has been reported that protein arginine methyltransferase 1 (PRMT1) can promote EZH2 stability via its methylation. On the other hand, EZH2 enhances breast cancer metastasis via epithelial-to-mesenchymal transition (EMT) induction [41]. It is noteworthy that genome stability is affected by EZH2 in cancer cells. EZH2 overexpression is associated with instability of ribosomal DNA and an increase in ribosome synthesis that is in favor of increasing proliferation and invasion of breast cancer cells [42]. Therefore, EZH2 stabilization leads to cancer progression and silencing EZH2 diminishes cancer proliferation and metastasis [43]. It was showed that EZH2 stimulates EMT and leads to an increase in cancer metastasis. It seems that EZH2 can affect metabolism of cancer cells in increasing their growth. EZH2 uses its methyltransferase activity to silence expression of aldehyde oxidase (AOX1). These results in the activation of tryptophan-kynurenine pathway that subsequently promotes NADP levels to provide cancer growth [44]. It is noteworthy that EZH2 can regulate genes involved in cell cycle progression of cancer cells [45]. The Yes-associated protein (YAP) activates EZH2 to transcriptionally suppress expression level of p27, as a cell cycle kinase inhibitor [46]. EZH2 signaling can affect immune cells in favor of cancer progression. In this way, EZH2 stimulates chemokine ligand 5 (CCL5) to provide macrophage chemotaxis, leading to metastasis of lung cancer cells [47]. EZH2 also regulates the response of cancer cells to therapy. Low expression of EZH2 mediates better responses to chemotherapy compared to high expression. Additionally, EZH2 can be considered as a prognostic factor, so that low expression of EZH2 leads to a good prognosis and improve progression-free survival rate of ovarian cancer patients [48]. Furthermore, suppressing EZH2 signaling can prevent the development of drug resistance in small cell lung cancer [49].
Noteworthy, EZH2 signaling can regulate response of cancer cells to immunotherapy. The checkpoint inhibitors are commonly utilized in prostate cancer therapy. It has been reported that suppressing EHZ2 signaling enhances number of M1 macrophages and induces infiltration of CD8 + T cells, increasing response of prostate cancer cells to checkpoint inhibitors [50]. Furthermore, knock-down of EZH2 elevates IFN-γ-mediated cytokines, confirming modulatory impacts of EZH2 signaling on immune responses [51]. Clinical studies have confirmed the role of EZH2 in cancer prognosis. For instance, moderate and high expression levels of EZH2 are associated with unfavorable prognosis of patients with ovarian cancer [52]. Overall, EZH2 is a critical player in cancer [53–56] and can affect proliferation, metastasis, and therapy response of cancer cells. However, there are studies showing that EZH2 can also function as a tumor-suppressor. For instance, SOX4 overexpression is in favor of breast cancer progression and EZH2 reduces its expression by binding to the promoter, leading to a decrease in invasion and migration of breast cancer cells [57]. Furthermore, EZH2 has been shown to suppress immune evasion of hepatocellular carcinoma cells by recruiting H3K27me3 to promoter and negatively affect programmed death-ligand 1 (PD-L1) expression [58]. Table 1 provides a summary of EZH2’s role in different cancers.
Table 1.
A summary of EZH2’s role in different cancers
| Cancer type | Signaling network | Remarks | References |
|---|---|---|---|
| Breast cancer | EZH2/PP2A |
EZH2 reduces expression level of PP2A via triggering histone modification Conferring resistance to HER2 inhibitors |
[59] |
| Breast cancer | PRMT1/EZH2 | Tumor-associated macrophages stimulate PRMT1 expression to enhance EZH2 stability and expression, leading to breast cancer invasion | [60] |
| Triple-negative breast cancer | EZH2/DLC1 |
Curcumin impairs metastasis and proliferation of cancer cells Curcumin induces apoptosis Reducing EZH2 expression and subsequent upregulation of DLC1 |
[61] |
| Non-small cell lung cancer | EZH2/TGFBR2 | Synergistic impact between EZH2 and YAP/TAZ in transcription repression of TGFBR2 and promoting cancer progression | [62] |
| Lung cancer | – |
Association of EZH2 overexpression with cancer proliferation, metastasis, and therapy resistance Providing poor prognosis |
[63] |
| Colorectal cancer | DUXAP8/EZH2/EMT |
Enhancing metastasis of cancer cells via EMT induction Activation of EZH2 by DUXAP8 is vital for EMT stimulation |
[64] |
| Colon cancer | – |
Overexpression of EZH2 in colon cancer compared to normal colonic mucosa Reduced tumor differentiation, and lymph node metastasis Association with lower survival Therefore, EZH2 can be considered as a prognostic factor |
[65] |
| Ovarian cancer | EZH2/DAB2IP |
DAB2IP overexpression suppresses cancer stem cell features in ovarian cancer EZH2 down-regulates DAB2IP expression to induce Wnt signaling, leading to ovarian cancer progression |
[66] |
| Gastric cancer | EZH2/Rho/ROCK/EMT |
EZH2 promotes cancer metastasis via inducing Rho/ROCK-mediated EMT Diosgenin and GSK126 synergistically down-regulate EZH2 in suppressing cancer metastasis |
[67] |
| Prostate cancer | AR/EZH2 |
The interaction between AR and EZH2 leads to prostate cancer progression EZH2 inhibition enhances anti-tumor activity of metformin |
[68] |
EZH2 inhibitors: an overview
In according to critical role of EZH2 signaling in vital biological mechanisms and modulating cancer progression as well as affecting immune system, significant efforts have been made in developing novel EZH2 inhibitors. Different kinds of EZH2 inhibitors have been developed and some of them are being applied in clinical trials for treatment of patients [69–73]. The S-adenosylmethionine (SAM)-competitive inhibitors are among the common EZH2 inhibitors in cancer therapy. The GSK126 is such compound that suppresses methyltransferase activity of EZH2 and diminishes H3K27me3 levels. These activities pave the way for stimulating silenced PRC2 downstream targets and preventing cancer growth in xenografts [74]. The EED226 is another EZH2 inhibitor capable of repressing PRC2 activity. There is a binding site on the H3K27me3, known as EED, that EED226 can bind to it, triggering conformational alterations and inhibiting PRC2 activity [75]. In addition to pre-clinical experiments, clinical studies have also evaluated role of EZH2 inhibitors in treatment of cancer patients. The intravenous administration of GSK1816126 demonstrates moderate anti-tumor activity in solid tumors and lymphoma [76]. The previous clinical trial was in phase I and highlighted that low half-life survival of EZH2 inhibitors limits their tumor-suppressor activity. Therefore, methods for targeted delivery of EZH2 inhibitors such as nanotechnological approaches can be developed in near future.
The first small molecule with capacity of EZH2 inhibition was carbocyclic adenosine analog 3-deazaneplanocin (DZNep) that was derived from neplanocin-A as a natural antibiotic [77]. However, DZNep lacks specificity and selectivity and it may negatively affect physiological processes such as regeneration [78]. Furthermore, DZNep had poor bilability and low half-life due to its hydrophilic nature [79]. Therefore, more selective EZH2 inhibitors have been developed. The EPZ005687 was developed as a high selective and potent small molecule capable of EZH2 inhibition. The EZP005687 demonstrated potential in preventing H3K27 methylation in a concentration-dependent manner and suppressed progression of various tumors including lymphoma, breast and prostate tumors [80]. The GSK126 can affect both mutant and WT EZH2 with high selectivity and it prevents the growth of lymphoma in mice and currently, it has been introduced to clinic (phase I) for treatment of lymphoma [81–83]. GSK343 is also an EZH2 inhibitor, but it has a different structure from GFSK126 and instead of indole, GSK343 has indazole nucleus [84].
It has been reported that EZH2 inhibitors are effective in suppressing metastasis and neovascularization in human tumors. The clinical aspect of EZH2 inhibitors is also attributed to improving ability of oncologists in suppressing metastasis and angiogenesis in cancer. Interestingly, tumor cells respond more likely to EZH2 inhibitors and they have been applied in treatment of colon cancer (stage II and III). A similar approach can be utilized in treatment of others cancers such as breast and gastric cancers. Furthermore, EZH2 inhibitors can be utilized as adjuvant in synergistic cancer therapy in clinical course. As out knowledge in oncology and EZH2 signaling enhances, more novel therapeutics for EZH2 targeting are introduced. Furthermore, since various complicated signaling networks are involved in cancer progression, EZH2 inhibitors and other anti-cancer agents should be used in combination in effective cancer therapy [85].
The administration route of EZH2 inhibitors in clinical course is also different. For instance, EPZ6438 and CPI-1205 are administered via oral route, while GSJ126 is administered via intravenous route [86]. There are some limitations associated with EZH2 inhibitors that should be considered for their clinical application. The poor efficacy, large molecular weight and poor bioavailability are most important drawbacks of EZH2 inhibitors [87]. For instance, TAZVERIK™ is a commercial EZH2 inhibitor that should be used with dose of 800 mg twice daily. Recently, efforts have been made in developing covalent inhibitors for EZH2 in cancer therapy. These kinds of inhibitors develop covalent bonds with EZH2 and they have improved pharmacodynamic duration. Therefore, there is no need of continuous exposure to EZH2 inhibitors [88]. Increasing evidence has shown benefits of covalent EZH2 inhibitors including high potency, good pharmacokinetics and unique selectivity [89–91]. However, a few covalent EZH2 inhibitors have been developed. For instance, GNA002 can covalently bind to EZH2 for inhibiting its function [92]. It is worth mentioning that covalent EZH2 inhibitors have also their drawbacks such as complex structure and low bioavailability that limits their clinical application [93]. Hence, more investigation should be made in developing novel and effective EZH2 inhibitors. Table 2 provides a summary of EZH2 inhibitors.
Table 2.
An overview of EZH2 inhibitors based on pre-clinical and clinical studies
| EZh2 inhibitor | In vitro/in vivo/clinical trial | Remarks | References |
|---|---|---|---|
| GSK2816126 | Clinical trial (phase I) |
Preventing the progression of solid tumors and lymphoma Exerting mild anti-tumor activity Low half-life restricts its anti-tumor activity Intravenous administration |
[76] |
| GSK126 |
In vitro (DLBCL cell line) In vivo (xenografts) |
Preventing methyltransferase activity of EZH2 Decreasing H2K27me3 levels Stimulating expression of PCR2 target genes |
[74] |
| EED226 | In vivo (human lymphoma xenograft tumors) |
Triggering conformational changes in EED site of H3K27me3 Suppressing PRC2 activity Preventing tumor growth |
[75] |
|
GSK926 GSK343 |
In vitro (HCC1806 breast cancer cells) |
Reducing nuclear H3K27me3 levels in a concentration-dependent manner Acting like other SAM compounds in suppressing EZH2 activity |
[94] |
| EPZ-6438 |
In vitro (lymphoma cells) In vivo (EZH2-mutant NHL xenograft-bearing mice) |
Acting in a time- and concentration-dependent manner Preventing lysine 27 methylation of H3K27me3 Suppressing EZH2 signaling Exerting anti-tumor activity |
[95] |
| SAH-EZH2 (a peptide) | In vitro (MLL-AF9 leukemia cells) |
Proliferation inhibition Inducing monocyte-macrophage differentiation Inhibiting EZH2 signaling by impairing EZH2-EED complex |
[96] |
| AZD9291 | In vitro (lymphoma and breast cancer cells) |
Inhibiting PRC2 activity by disrupting EZH2-EED interaction Reducing EZH2 expression at mRNA and protein levels via miRNA-34a overexpression |
[97] |
| Astemizole | In vitro (SU-DHL6, Toledo, DB, SU-DHL4, and Pfeiffer lymphoma cell lines) |
Suppressing growth of cancer cells Inhibiting EZH2 signaling via preventing interaction between PRC2 and EZH2-EED complex |
[98] |
| Wedelolactone | In vitro (HepG2, K562 and 293T cells) |
Binding to EED and inhibiting EED and EZH2 interaction Mediating PRC2 degradation Suppressing cancer proliferation |
[99] |
Non-coding RNAs and EZH2 signaling
MiRNAs and EZH2 signaling
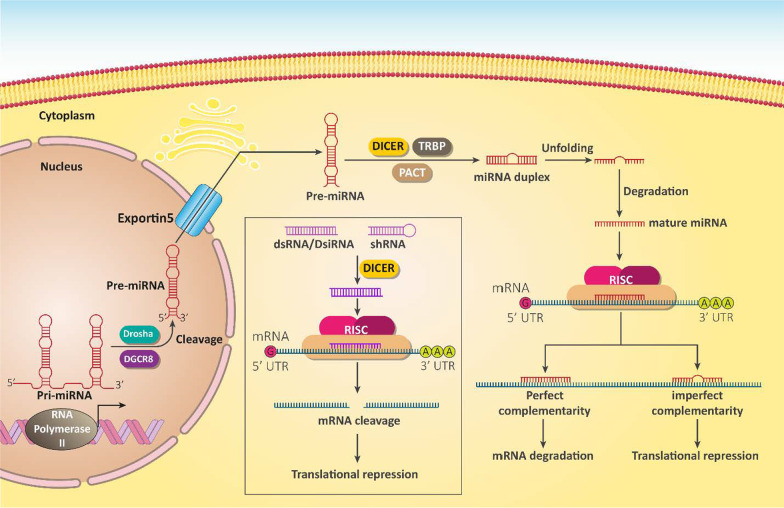
In 1993, the discovery of microRNAs (miRNAs) improved our knowledge of biological mechanisms and their regulations in cells [100–103]. These short ncRNAs have a length of 22 nucleotides and can bind to complementary target messenger RNA (mRNA) via 3′-untranslated region (3′-UTR) [104]. Before targeting mRNA, miRNAs form a complex with Argonaute protein to be embedded in RNA-induced silencing complex (RISC) and exerts its regulatory function on gene expression (Fig. 2) [105, 106]. The expression of miRNAs is tissue-specific and is categorized in two groups including tumor-promoting and tumor-suppressor miRNAs [107]. Regardless of physiological function of miRNAs in cells, increasing evidence highlights their role in cancer. For instance, modulating their expression using transfection strategies can be beneficial in cancer treatment [108, 109]. In the next sections, we will provide a summary of miRNA and EZH2 interaction in different cancers and how this interaction can affect the malignancy of cancer cells.
Fig. 2.
The biogenesis route and functions of miRNAs, siRNA and shRNA in cells [110, 111]
Brain tumors
Brain tumors are responsible for high mortality and morbidity worldwide. Here, we cover the role of miRNAs in cancer progression and inhibition [112, 113]. Glioma is a well-known brain tumor with an upregulated EZH2 expression [114]. On the other hand, miRNA-32 is a tumor-suppressor factor undergoing down-regulation in glioma cells and tissues. Enhancing miRNA-32 expression significantly impairs proliferation and migration of glioma cells via EZH2 down-regulation [115]. The association between miRNA and EZH2 expression has been investigated in glioma [116]. MiRNA-340 is considered as a tumor-suppressor factor in which its upregulation increases anti-tumor immunity and promotes macrophage phagocytosis [117]. Furthermore, miRNA-340 can suppress cancer metastasis via down-regulating RhoA expression [118]. In glioblastoma multiforme, miRNA-340 stimulates apoptosis and cell cycle arrest while suppressing cancer cell metastasis. Furthermore, this miRNA induces autophagy and differentiation. It is thus suggested that EZH2 down-regulation by miRNA-340 is involved in anti-tumor activities [99].
The LIM-only protein 3 (LMO3) was first identified in glioma and is considered as a DNA methylation gene [119]. LMO3 has been implicated in brain tumors and can enhance cancer proliferation and tumorigenesis in neuroblastoma [120, 121]. By inhibiting EZH2, miRNA-101 diminishes promoter occupation of LMO3 by H3K27me3 and prevents its methylation, resulting in glioma suppression [122]. In fact, miRNAs target EZH2 to affect the methylation condition of a specific gene that is of importance for its activation or inhibition [123].
Therefore, proliferation and invasion of brain tumors are affected by miRNA/EZH2 axis [124]. In the tumor microenvironment, there are competitions among rapidly dividing and proliferating cancer cells, leading to hypoxia and subsequent induction of angiogenesis for promoting cancer progression [125, 126]. EZH2 can induce angiogenesis and promote cancer growth [127]. MiRNA-137 can bind to 3′-UTR of EZH2 to diminish its expression, impairing glioblastoma proliferation and angiogenesis [128]. Down-regulation of tumor-suppressor miRNAs results in EZH2 activation and subsequent induction of angiogenesis and growth of glioblastoma cells [129].
Thoracic tumors
Lung cancer
Among thoracic cancers, lung cancer is a common type and recent studies have highlighted the role of miRNAs in regulating its malignancy and aggressive behavior. Increasing evidence suggests that EZH2 upregulation is in favor of lung cancer progression [130]. On the other hand, there are miRNAs capable of regulating EZH2 expression in lung cancer. A recent experiment has shown that miRNA-92b as a tumor-suppressor role and binds to 3′-UTR of EZH2 to reduce its expression, impairing proliferation and metastasis of lung cancer cells [114]. Furthermore, miRNAs can regulate the behavior of cancer stem cells (CSCs) in lung cancer. Briefly, CSCs are responsible for cancer progression and inducing resistance to therapy. Furthermore, the presence of CSCs is associated with cancer recurrence [131, 132]. Therefore, it is of importance to delineate the relationship between miRNA/EZH2 axis and CSCs for developing novel therapeutics. It has been reported that miRNA-21 promotes the expression level of EZH2 and enhances the progression of CSCs to provide resistance to radiotherapy and chemotherapy in lung cancer cells. The apoptosis and cell cycle regulators including Cdc2, cyclin B1 and Bcl-2 are regulated by miRNA-21/EZH2 axis in CSCs. Silencing miRNA-21/EZH2 axis enhances the potential of radiotherapy and chemotherapy suppressing lung cancer by 39.2% and 69.7% [133]. The downregulation of EZH2 in lung cancer cells by miRNAs is mediated by its binding to 3′-UTR [134].
Methotrexate is a potent anti-tumor agent with a capacity of inhibiting dihydrofolate reductase to suppress cancer progression [135, 136]. This agent is extensively applied in lung cancer therapy, however, the development of drug resistance in cancer cells can limit its anti-tumor activity. To overcoming resistance, nanoparticles have been developed for targeted delivery of methotrexate with some degrees of success [137, 138]. Another tool that is commonly used to reduce the development of drug resistance is gene therapy. This requires an in-depth understanding of signaling networks that are involved in methotrexate resistance and the resulting genetic modulations in cellular responses. In A549 cells, EZH2 overexpression diminishes sensitivity to methotrexate therapy and prevents apoptosis and cell cycle arrest. For potentiating methotrexate-mediated cell death and cell cycle arrest, transfection of A549 cells with miRNA-200c results in down-regulation of EZH2 [139].
One of the mechanisms responsible for tumor metastasis is EMT. At morphological level, epithelial cells are transformed to mesenchymal cells that have high motility rate. At molecular level, a number of factors known as EMT-inducing transcription factors (EMT-TFs) such as ZEB1/2, Snail, Slug and TGF-β result in EMT stimulation via E-cadherin down-regulation, and N-cadherin and vimentin upregulation. These processes are reversible and its reversible type, known as MET can be induced. As EMT leads to metastasis and poor prognosis, its suppression is of importance in cancer therapy [140–144]. A recent experiment has shown how miRNA/EZH2 axis can modulate EMT in lung cancer cells. The expression of miRNA-124 undergoes down-regulation in lung adenocarcinoma compared to normal cells. MiRNA-124 transfection results in down-regulation of EZH2 and subsequent inhibition of EMT, impairing lung cancer migration [145].
Breast cancer
Breast cancer is one of the most encountered malignancies among women with miRNAs and EZH2 playing a significant role in proliferation, metastasis, and therapy response [146, 147]. This section is dedicated to discussing miRNA and EZH2 interaction in breast cancer cells. As a dynamic process, autophagy activation occurs when damaged proteins and organelles accumulate and their degradation is vital for preserving cell homeostasis [148, 149]. Increasing evidence demonstrates the role of autophagy induction in reducing proliferation of breast cancer cells [112]. As a tumor-suppressor factor, miRNA-92b stimulates autophagy and diminishes viability and growth of breast cancer cells. In this way, miRNA-92b binds to 3′-UTR of EZH2 to reduce its expression, resulting in the upregulation of light chain-3 (LC3) and SQSTM1 degradation [150]. However, it should be mentioned that autophagy possesses a pro-survival role and its activation may enhance breast cancer progression [151–153]. Thus, the dual role of autophagy in cancer should also be considered.
It is worth mentioning that miRNAs can exert a synergistic impact with anti-tumor agents in regulating EZH2 expression and affecting breast cancer progression. Venom peptides derived from predatory marine cone snails are considered as promising agents in cancer therapy. It has been reported that Conus peptides have a high stability, are resistance to proteases, and can selectivity target receptors [134, 154, 155]. A recent experiment has applied Syn-Cal14.1a, a synthetic peptide isolated from Californiconus californicus (Cal14.1a), in addition to miRNA-101-3p transfection to suppress breast cancer progression. This combination synergistically down-regulates EZH2 expression and inhibit proliferation and migration of breast cancer cells [156]. As more experiments are performed, more miRNAs involved in regulating EZH2 expression are revealed. MiRNA-340 is a tumor-suppressor and significantly diminishes proliferation and metastasis of breast cancer cells via regulating different molecular pathways such as inhibiting Wnt and ROCK1 [157, 158]. In vitro and in vivo experiments have shown that miRNA-340 transfection significantly inhibits tumor growth in vitro and in vivo. In this way, miRNA-340 down-regulates EZH2 expression to decrease DNMT1, H3K27me3, β-catenin and signal transducer and activator of transcription 3 (STAT3), an oncogenic transcription factor. This axis results in down-regulation of miRNA-21 and upregulation of miRNA-200a/b, which are of key importance in breast cancer therapy [159].
MiRNA-33a is another tumor-suppressor that impairs breast cancer metastasis and growth while promoting sensitivity to doxorubicin chemotherapy via inhibiting EMT, eukaryotic translation initiation factor 5A2 (eIF5A2)[160, 161]. In triple-negative breast cancer cells, miRNA-33a reduces growth and induces cell cycle arrest (G1 phase). Enhancing miRNA-33a expression is associated with EZH2 down-regulation and suppression of breast cancer progression [162]. It has been reported that miRNA/EZH2 axis can regulate the therapy response of breast cancer cells. Doxorubicin (DOX) is a well-known anti-tumor agent that participates in cancer suppression by inhibiting the activity of topoisomerase II and inducing cell cycle arrest [163, 164]. However, recent experiments have shown the development of DOX resistance and the need for using complementary strategies in suppressing resistance [165, 166]. One of the potential mechanisms that can be used is miRNA replacement therapy to increase the expression levels of tumor-suppressing miRNAs. In breast cancer cells, enhancing expression levels of miRNA-15a and miRNA-16 reduces the EZH2 expression and partially prevents DNA repair and provides DOX sensitivity [167].
Gastrointestinal tumors
Gastric cancer
Gastric cancer is a lethal disease and EZH2 upregulation is in favor of gastric carcinogenesis. As a tumor-suppressor, miRNA-26 suppresses EZH2 expression to impair gastric cancer progression. Investigation of molecular pathways demonstrates that TET, a member of DNA demethylases family, can provide sequestration of miRNA-26, paving the way for EZH2 overexpression and gastric tumorigenesis [168]. To enhance our capacity in enhancing miRNA expression, delivery methods including viral vectors have been used. A recent experiment uses lentivirus for delivery of miRNA-124 to down-regulate EZH2 expression and successfully inhibit to gastric cancer progression [169].
Gastrokine 1 (GKN1) has been isolated from gastric mucosa cells and is also expressed in autocrine/paracrine gastric mucosa [170, 171]. GKN1 is involved in the process of healing and protecting antral mucosa [172]. GKN1 can suppress gastric cancer proliferation via upregulating miRNA-185. EZH2 down-regulation, on the other hand, leads to inhibition of gastric cancer progression, and promotion of sensitivity to 5-fluorouracil chemotherapy [173].
Liver cancer
Liver cancer is a leading cause of cancer related mortality worldwide. MiRNA-26a is a tumor-suppressor factor capable of suppressing cancer progression via down-regulating matrix metalloproteinases (MMPs) [174]. Furthermore, miRNA-26a induces cancer cell apoptosis via E2F7 down-regulation [175]. In hepatocellular carcinoma, miRNA-26a stimulates cell growth inhibition via inhibiting EZH2 expression [176]. EZH2 can function as upstream mediator of STAT3 by providing methylation [177]. In hepatocellular carcinoma, miRNA-137 inhibits migration and metastasis via targeting EZH2/STAT3 signaling. In this way, miRNA-137 inhibits EZH2 and its downstream target STAT3 to provide E-cadherin upregulation and Snail down-regulation in favor of metastasis inhibition [178]. Similar to STAT3, Wnt/β-catenin is also a downstream pathway of EZH2 [179]. Many miRNAs affect Wnt signaling via targeting EZH2. Among them, miRNA-98 suppresses EZH2 expression to inhibit Wnt signaling, resulting in a decrease in hepatocellular carcinoma proliferation [180]. Interferon-alpha (IFN-γ) is an inflammatory factor and can induce the expression of miRNAs in cancer cells. In hepatocellular carcinoma, IFN-γ enhances the expression level of miRNA-26a that subsequently, down-regulates EZH2 to suppress metastasis [181]. Besides, miRNA/EZH2 axis can regulate the response of hepatocellular carcinoma cells to therapy. It has been reported that EMT induction can lead to cisplatin resistance [182]. MiRNA-138 reduces EZH2 expression to inhibit EMT, resulting in an increase in cisplatin sensitivity in hepatocellular carcinoma cells [183]. Overall, these studies agree with the fact that miRNA/EZH2 axis can affect proliferation, metastasis, and therapy response of hepatocellular carcinoma cells, and in this way, different molecular pathways such as Wnt, STAT3 and EMT are affected.
Colon cancer
Like other kinds of gastrointestinal tumors, EZH2 overexpression enhances colon cancer progression and its inhibition by anti-tumor agents such as salinomycin is associated with activation of death receptors [184]. MiRNA and EZH2 interaction is a determining factor for colon cancer progression. It has been reported that colon cancer metastasis is impaired following miRNA-101 overexpression and is attributed to EZH2 down-regulation and subsequent inhibition of cancer invasion [185]. It is suggested that the inhibitory impact of miRNA-101 on colon cancer metastasis is due to EMT inhibition. Overexpression of miRNA-101 inhibits EZH2 signaling post-transcriptional to suppress EMT, leading to colon cancer metastasis impairment [186].
Reproductive tumors
Prostate cancer
Prostate cancer is a common tumor among men and EZH2 has been shown to play a role in its malignancy affecting growth and senescence [187]. Furthermore, EZH2 can enhance prostate cancer metastasis via Twist upregulation and increasing N-cadherin levels [188]. The expression of miRNA-605-3p as a tumor-suppressor factor is down-regulated in prostate cancer cells, which increases the expression of EZH2 to disrupt prostate cancer growth and metastasis. These results have been shown in both in vitro and in vivo [189]. It is noteworthy that miRNAs can indirectly affect EZH2 via targeting other upstream mediators. Hypoxia diminishes miRNA-137 expression to promote migration of prostate cancer cells via EMT induction [190]. A recent experiment has shown that miRNA-137-3p is down-regulated in prostate cancer cells, and its low expression is associated with tumor stage. MiRNA-137-3p down-regulates EZH2 expression via c-Jun N-terminal kinase 3 (JNK3) inhibition which significantly suppresses prostate cancer proliferation and metastasis, while stimulating apoptosis [191].
The therapeutic targeting of miRNA/EZH2 axis has been shown to suppress prostate cancer progression. The Aristeromycin (a derivative of 3-deazaneplanocin A (DZNeP) has been effective at upregulating the expression level of miRNA-26a. In turn, miRNA-26a binds to 3′-UTR of EZH2 to reduce its expression and impair prostate cancer proliferation [192]. There is a close relationship between miRNAs and androgen receptor (AR) signaling in enhancing prostate cancer progression. Briefly, AR signaling plays a key role in prostate cancer development and progression, and androgen deprivation therapy (ADT) is used to suppress prostate cancer progression [193]. As a tumor-promoting factor, AR signaling enhances EZH2 expression to inhibit apoptosis and provide enzalutamide resistance. Intracellular delivery of miRNA-124 reduces AR splice variants and suppresses EZH2 as a downstream target to prevent prostate cancer progression [194].
Gynecological tumors
Ovarian cancer
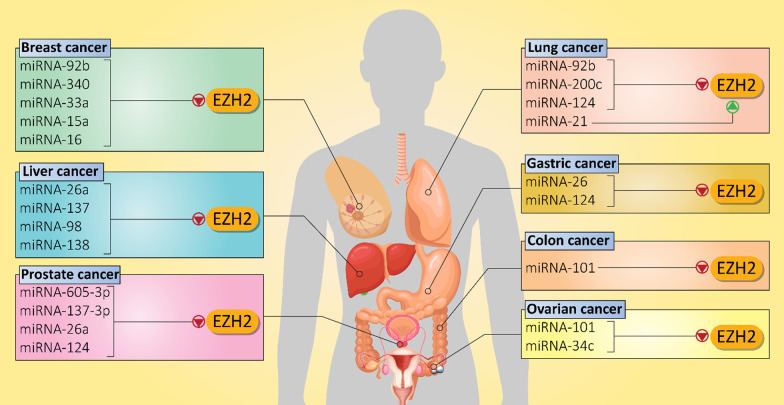
Ovarian cancer is a common gynecologic cancer with a high global mortality rate. Both miRNAs and EZH2 demonstrate abnormal expressions in ovarian cancer [195–198]. Indeed, the miRNA/EZH2 interaction determines progression and chemoresistance of ovarian cancer cells. MiRNA-101 is observed at low levels in ovarian cancer. Enhancing miRNA-101 expression is associated with EZH2 down-regulation and a subsequent decrease in proliferation and migration of ovarian cancer cells. Furthermore, as aggressive behavior of these malignant cells decreases, their sensitivity to cisplatin enhances [199]. It has been reported that EZH2 increases ovarian cancer growth and promotes cell cycle progression through transcriptional activation of CDKN1C. It is noteworthy that miRNA-34c inhibits EZH2 in favor of reducing ovarian cancer proliferation (Table 3) [200]. A few studies have examined the role of miRNA/EZH2 interaction in ovarian cancer and more experiments are needed to delineate the mechanisms of action. Furthermore, studies on regulation of miRNA/EZH2 axis by lncRNAs are discussed below. Figure 3 shows regulation of EZH2 signaling by miRNAs in different cancers.
Table 3.
MiRNAs as potential upstream mediators of EZH2 signaling in cancers
| MiRNA | Signaling network | Cancer type | In vitro/in vivo | Cell line/Animal model | Remarks | References |
|---|---|---|---|---|---|---|
| MiRNA-876-3p | SPRR3/EZH2 | Non-small cell lung cancer |
In vitro In vivo |
H1299, PC9, HCC827 and A549 cells NOS/SCID mice |
Exerting an anti-tumor function Inhibiting SPRR3/EZH2 axis Apoptosis induction Disrupting metastasis |
[201] |
| MiRNA-21 | – | Lung cancer | In vitro | A549 cell line |
MiRNA-21 promotes proliferation and therapy response of cancer cells Silencing miRNA-21 and its downstream target EZH2 enhance therapy sensitivity |
[133] |
| MiRNA-200c | – | Lung cancer | In vitro | A549 cells | Inhibiting migration and invasion of cancer cells via enhancing E-cadherin levels and decreasing EZH2 expression | [139] |
| MiRNA-124 | – | Pancreatic cancer | In vitro | AsPC-1, PANC1, BxPC-3 and SW1990 cells |
Impairing metastasis and proliferation of cancer cells Delivery to tumor cells via exosomes EZH2 overexpression disrupts anti-tumor activity of miRNA-124 |
[202] |
| MiRNA-137 | EZH2/LSD1 | Endometrial cancer |
In vitro In vivo |
AN3CA, HEC1A, KLE, RL-95-2 cells Xenografts |
Suppressing proliferation of cancer cells Inhibiting EZH2 and LSD1 expression levels |
[203] |
| MiRNA-494 | MYC/EZH2 | Burkitt lymphoma | In vitro | BL cell lines |
The MYC can increase EZH2 expression in maintaining malignancy of lymphoma cells MiRNA-494 inhibits MYC/EZH2 axis |
[204] |
| MiRNA-26a | – | Bladder cancer | In vitro | EJ cells |
Apoptosis induction Decreasing proliferation of cancer cells MiRNA-26a inhibits EZH2 signaling |
[205] |
| MiRNA-98 | EZH2/Wnt/β-catenin | Hepatocellular carcinoma | In vitro | HCCLM3, HepG2, SMMC7721, Hep3 B cell lines |
Binding to 3′-UTR of EZH2 and reducing its expression Inactivating Wnt signaling Suppressing cancer growth |
[180] |
|
MiRNA-378a-3p MiRNA-378d |
EZH2/STAT3 | Breast cancer |
In vitro In vivo |
CAL51, MDA-MB-231 and MCF-7 cells Xenografts |
Upregulation of EZH2 and subsequent induction of STAT3 signaling in increasing chemoresistance feature and stemness of breast cancer cells | [206] |
| MiRNA-124 | EZH2/STAT3 | Cholangiosarcoma |
In vitro In vivo |
HuCCT1, KMBC, and MZChA1 cells Mouse xenograft model |
MiRNA-124 inhibits EZH2 and its downstream target STAT3 Inducing autophagy-related cell death via ATG5 upregulation Reducing miRNA-124 expression enhances disease progression |
[207] |
| MiRNA-26a | EZH2 | Skin cancer | In vitro | HaCaT cells |
Reducing EZH2 expression to mediate UV-induced apoptosis Using EZH2 inhibitors aggravates apoptosis |
[208] |
Fig. 3.
MiRNAs regulating EZH2 in different cancers
EZH2 as upstream mediator of miRNA expression
The previous section highlighted the role of miRNAs as upstream mediators of EZH2 in cancer regulation. It is noteworthy that there are studies demonstrating that EZH2 can regulate miRNAs and result in cancer progression. Glycolysis or Warburg effect is responsible for increased glucose uptake and higher metabolism ensures rapid proliferation of cancer cells. With respect to the role of glycolysis in cancer progression, various inhibitory therapeutic agents have been developed [16, 209]. EZH2 upregulation in prostate cancer cells results in the induction of hexokinase-2 (HK2) to increase glycolysis. MiRNA-181b as a tumor-suppressor factor binds to 3′-UTR of HK2 to inhibit glycolysis and progression of prostate cancer cells. However, transcriptional knockdown of EZH2 activates HK2-mediated glycolysis [210]. In addition, miRNA regulation by EZH2 can affects cell cycle progression. It has been reported that miRNA-200c binds to 3′-UTR of E2F3 to reduce its expression, inducing cell cycle arrest. In turn, EZH2 as an upstream mediator, down-regulates miRNA-200c expression to induce E2F3-mediated cell cycle progression in prostate cancer [211].
EZH2 can also interact with other tumor-promoting factors such as YY1 to down-regulate miRNA expression. YY1 is a zinc finger transcriptional factor that exerts a variety of biological activities including cell growth, differentiation, apoptosis, embryonic development, and carcinogenesis [212–214]. YY1 recruits EZH2 in prostate cancer cells to down-regulate miRNA-146a expression. Silencing YY1 enhances miRNA-146a expression to induce apoptosis in cancer cells [215]. SOX4 is considered as a tumor-promoting factor in ovarian cancer and its upregulation by lncRNA FEZF1-AS1 results in cancer inhibition [216]. It is noteworthy that SOX4 can function as an upstream mediator of EZH2 to induce H3K27me3. This axis leads to down-regulation of miRNA-212 and 132 and activating EMT for ovarian cancer metastasis [217].
It has been reported that EZH2 can recruit factors responsible for DNA methylation such as DNA methyltransferases enzymes (DNMTs) that induce hypermethylation of CpG islands [218]. In cervical cancer cells, EZH2 recruits DNMT1 to silence miRNA-484 via triggering methylation of its promoter. Subsequently, activation of membrane-bound matrix metalloproteinase (MMP14) and the hepatocyte nuclear factor 1A (HNF1A) drive cancer metastasis and invasion [219].
The miRNA-125a is a tumor-suppressor and its upregulation by propofol results in reduced growth and invasion of gastric cancer cells [220]. MiRNA-125a can be considered as a diagnostic factor for gastric cancer [221], enhancing expression level of breast cancer metastasis suppressor 1 (BRMS1) to inhibit metastasis and migration of gastric cancer cells. EZH2 as an upstream mediator down-regulates miRNA-125a and BRMS1 to enhance gastric cancer metastasis [222].
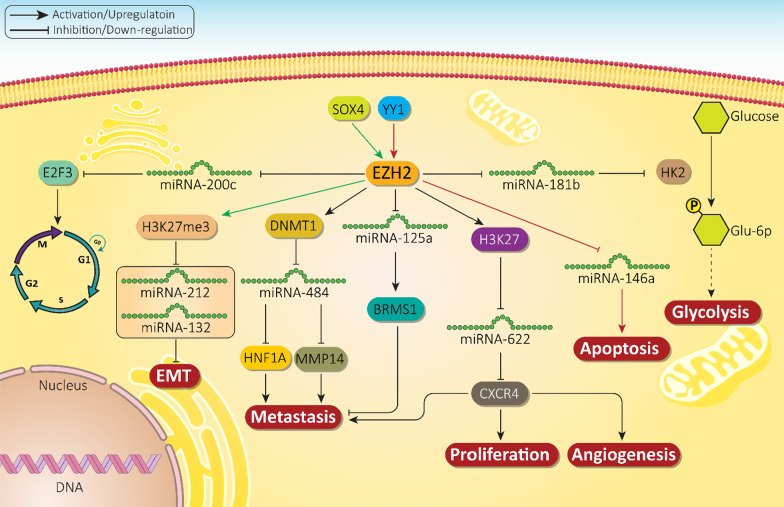
The CXC chemokine receptor 4 (CXCR4) is a receptor for chemokine ligand 12 (CXCL12) and regulates various biological events in cells including angiogenesis, EMT, dissemination, migration, and cell stemness [223, 224]. By binding to 3′-UTR of CXCR4, miRNA-622 down-regulates its expression. EZH2 activates trimethylation of miRNA-622 promoter at H3K27 to diminish its expression, leading to CXCR4 upregulation and increased progression of hepatocellular carcinoma [225]. Overall, owing to the trimethylation activity of EZH2 at H3K27 site, the expression of miRNAs can be regulated through promoter transcriptional suppression [226–236]. Figure 4 depicts role of EZH2 in regulation of miRNAs in different cancers.
Fig. 4.
EZH2 as upstream mediator of miRNA expression in cancers. Due to transcriptional role of EZH2, this pathway can target miRNAs in affecting proliferation and invasion of cancer cells that subsequently, determine response of cancer cells to therapy
Synthetic short non-coding RNAs
SiRNAs
The small interfering RNA (siRNA) is a kind of RNA interference (RNAi) consisting of double-stranded RNA and has a homologous sequence to target gene [237, 238]. SiRNAs can down-regulate the expression of target genes (silencing), a mechanism that begins in the cytoplasm with the aid of Dicer enzymes that cleaves double-stranded RAN to produce shorter RNA molecules, known as siRNA with length of 21–25 nucleotides. For this purpose, the stranded guide is loaded in RISC and after matching with a target mRNA, cleaves it and results in gene silencing. As gene deregulation is a common phenomenon in different diseases, especially cancer, a wide variety of experiments have applied siRNA for gene silencing to suppress cancer progression [239]. Despite significant progress in gene identification and further application of siRNA, the use of siRNA in clinical course is limited. There are several reasons for limitations in the clinical application of siRNA. The first reason is that siRNA should overcome physiological and cellular barriers that prevent entrance of siRNA into cytoplasm of cells. Furthermore, as RNAses are present in different locations in cells and tissues, they can degrade siRNAs and inhibit their potential in gene silencing [240]. For overcoming the aforementioned challenges, delivery systems have been designed to improve the potential of siRNA in gene silencing [241]. In this section, we discuss the role of siRNAs and delivery systems to silence EZH2 and suppress cancer malignancy.
With respect to the tumor-promoting role of EZH2, its inhibition should significantly diminish the survival of cancer cells. In this way, siRNA-EZH2 reduces the proliferation of bladder cancer cells by 37.9% and migration by 67% [242]. Furthermore, EZH2 upregulation is in favor of chemoresistance and reducing the sensitivity of cancer cells to chemotherapy [206, 243]. EZH2 down-regulation by siRNA is associated with apoptosis induction (caspase 3/8 activation) and cell cycle arrest (G0/G1 phase). Besides, EZH2 silencing by siRNA results in the downregulation of tumor-promoting factors including cyclin D1 and multidrug resistance 1 (MDR1) at protein and mRNA levels. On the other hand, EZH2 silencing upregulates tumor-suppressor factors such as p15, p21, p27 and miRNA-218 that can in turn suppress the progression of lung and gastric cancers and enhance their sensitivity to cisplatin chemotherapy [244]. However, the efficacy of siRNAs needs to be improved to gain better results in gene silencing. Polymeric nanoparticles can promote the intracellular accumulation of siRNA-EZH2 up to 98%. Due to their low size (35.6 nm), nanoparticles demonstrate high stability as confirmed by their zeta potential of 36.7 mV. Both in vitro and in vivo experiments reveal the role of siRNA-EZH2-loaded polymeric nanoparticles in gene silencing and subsequent apoptosis induction in cancer cells [245]. Co-delivery of siRNAs with other chemotherapeutic agents can be provided via nanocarriers. This leads to their synergistic impact and improvement in cancer elimination. Furthermore, surface modification of nanoparticles, for instance by RGD peptide, can enhance the selectivity of nanoparticles towards cancer cells and in EZH2 silencing [246]. Compared to siRNA-EZH2 or cisplatin alone, cisplatin- and siRNA-EZH2-loaded nanoparticles induce more toxicity against cancer cells, showing a synergistic impact capable of reversing chemoresistance [247]. As siRNA-EZH2 nanoparticles are administered systemically for cancer suppression, a special attention should be directed towards their biocompatibility and low toxicity towards normal cells [248]. Future experiments need to focus on developing green-synthesized nanocarriers for siRNA-EZH2 delivery and examine downstream targets of EZH2 in normal cells (Table 4).
Table 4.
The role of SiRNAs and nanoscale delivery systems in regulating EZH2 in cancer therapy
| Cancer type | Nanocarrier | Co-delivery | In vitro/in vivo | Cell line/Animal model | Remarks | References |
|---|---|---|---|---|---|---|
| Bladder cancer | – | – | In vitro | T24 cells |
EZH2 down-regulation by siRNA Reducing growth up to 37.9% Decreasing metastasis up to 67% |
[242] |
|
Non-small cell lung cancer Gastric cancer |
– | – | In vitro | AGS and A549 cells |
Inducing cell cycle arrest at G0/G1 phase after siRNA-EZH2 application Apoptosis stimulation Caspase-3/8 activation Down-regulation of cyclin D1 and MDR1 Upregulation of p15, p21, p27 and miRNA-218 as tumor-suppressor factors |
[244] |
| Non-small cell lung cancer | Multifunctional nanoparticles |
SiRNA-EZH2 Etoposide |
In vitro In vivo |
A549 cells Orthotopic lung cancer model |
Reducing mRNA and protein levels of EZH2 Decreasing proliferation and invasion of cancer cells Selective targeting tumor cells via RGD modification Synergistic impact |
[246] |
| Glioma | Polymeric nanoparticles | – |
In vitro In vivo |
U87 cells Tumor-bearing mice |
High transfection efficiency (up to 98%) Zeta potential of 36.7 demonstrates high stability Particle size of 35.6 nm Providing gene silencing and suppressing cancer progression |
[176] |
| Ovarian cancer | Iron nanoparticles |
Platinum siRNA |
In vitro In vivo |
A2780 cells Tumor-bearing mice |
Synergistic impact for overcoming drug resistance Cancer elimination Apoptosis induction |
[247] |
ShRNA
The short-hairpin RNA (shRNA) is like siRNA in gene silencing. ShRNAs are 19–20 nucleotides in length with a short hairpin loop of 4–11 nucleic acids. ShRNA is transcribed from DNA with the aid of RNA polymerase II and III. Pre-shRNAs translocate to cytoplasm by Exportin-5, where they form complexes with Dicer enzymes and are loaded in RISC for cleavage [249]. To date, few experiments have investigated the role of shRNAs in EZH2 silencing and suppressing cancer progression. These studies have applied delivery systems including nanocarriers and biological carriers for shRNA silencing of EZH2. Polymeric nanoparticles and adenoviruses for shRNA-EZH2 delivery have high transfection efficiency, great potential in gene silencing and lead to effective suppression of proliferation and invasion of prostate cancer cells, reduce expression of EZH2 and its downstream targets including Ki67 and CCND1 [250, 251]. Although no absolute conclusions can be made from these experiments, they demonstrate the efficiency of shRNA in EZH2 silencing and further studies can provide a comparative investigation of the role of shRNA and siRNA in EZH2 silencing and their potential.
LncRNAs and EZH2 signaling
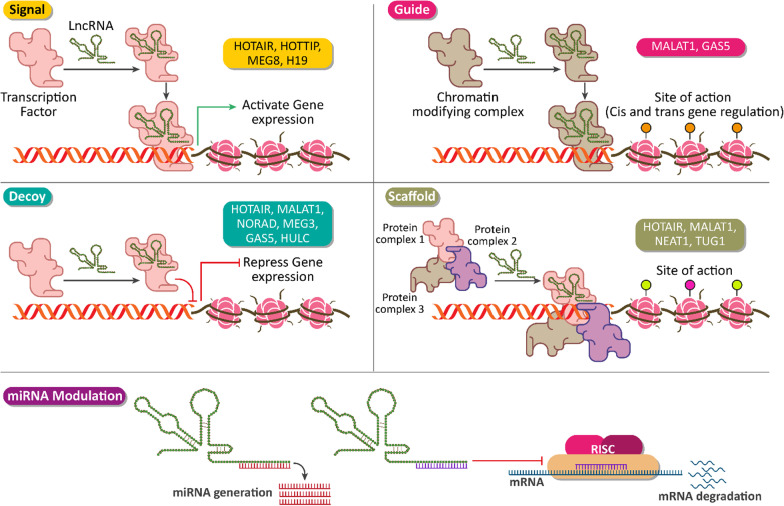
Long non-coding RNAs (lncRNAs) are important members of ncRNAs with a length more than 200 nucleotides and do not encode any proteins (Fig. 5) [230, 252]. As most part of genome is formed by ncRNA encoded by junk DNA, they are not transcribed to proteins [253, 254]. However, this concept was changed after the discovery of potential roles of lncRNAs in biological events [255]. LncRNAs can affect gene expression at various levels including chromatin, transcriptional and post-transcriptional levels [256]. Increasing evidence has revealed the role of lncRNAs in differentiation, cell cycle, and stem cell pluripotency [257–260]. It is suggested that lncRNAs are important in cancer biology and can affect proliferation, metastasis, and therapy response of cancer cells [10, 261–264]. By affecting other ncRNAs or proteins, lncRNAs can function as signals, guides, decoys or scaffolds to change cellular functions [265, 266]. Similar to miRNAs, lncRNAs can have both tumor-suppressing and tumor-promoting functions as suggested by been different experiments [263]. Single-cell sequencing has shown that lncRNAs have heterogeneous roles in individual cancer cells [267]. Therefore, it is of interest to explore the role of lncRNAs in different types of cancer. In the next section, we examine the role of lncRNAs in regulating EZH2 expression in various cancers.
Fig. 5.
The lncRNA function in cells [268]
LncRNAs inducing EZH2
The lncRNA small nucleolar RNA host gene 1 (SNHG1) is a tumor-promoting factor and recent studies have revealed its role in cancer progression. This lncRNA induces phosphoinositide 3-kinase (PI3K)/protein kinase-B (Akt) to enhance bladder cancer proliferation and metastasis [269]. Furthermore, SNHG1 can regulate miRNA expression to provide chemoresistance [270]. In colorectal cancer cells and tissues, SNHG1 is increased in expression and is associated with decreased patient survival. Investigation of molecular pathways reveals down-regulation of miRNA-154-5p by SNHG1 via sponging and EZH2 induction which increases colorectal cancer progression [271]. Another study reveals the role of SNHG1 in bladder cancer progression via regulating EZH2. In this way, SNHG1 enhances EZH2 expression in the nucleus to down-regulate CDH1, resulting in a decrease in E-cadherin levels and promoting metastasis and migration of bladder cancer cells [272].
LncRNA SNHG6 is another factor with a potential role in cancer. SNHG6 upregulation is associated with poor prognosis of patients with colorectal cancer [273]. SNHG6 can also enhance radio-resistance of cervical cancer cells [274]. In colorectal cancer cells, SNHG6 overexpression occurs due to SP1 stimulation and DNA copy number gains. SNHG6 enhances both migration and proliferation of cancer cells. Mechanistically, SNHG6 down-regulates the expression levels of miRNA-214 and miRNA-26a/b via sponging to enhance EZH2 expression, leading to colorectal cancer progression [275]. Interestingly, most of the studies have focused on the role of miRNA sponging by lncRNAs and subsequent induction of EZH2 [276]. However, lncRNAs can also interact with miRNAs to regulate EZH2. For instance, lncRNA plasmacytoma variant translocation I (PVT1) interacts with EZH2 to down-regulate miRNA-214 expression, leading to an increase in proliferation and metastasis of ovarian cancer cells [277]. Furthermore, lncRNA LINC00114 can induce EZH2/DNMT1 to reduce miRNA-133b expression and promote colorectal cancer progression [278]. Such interactions are in favor of cancer metastasis. EZH2 upregulation can promote cancer metastasis via EMT induction [279]. The lncRNA taurine upregulated gene 1 (TUG1) cooperates with EZH2 to down-regulate miRNA-382 and aid in sponging, leading to EMT induction and pancreatic cancer invasion [280].
Therapeutic targeting of lncRNAs has been tested in cancer therapy. Curcumin is a phytochemical isolated from root and rhizome of Curcuma longa and reveals anti-tumor activities via regulating the expression level of ncRNAs [281–285]. It has been reported that curcumin administration can promote sensitivity of pancreatic cancer cells to gemcitabine via regulating the expression level of PVT1 and its interaction with EZH2. In this way, curcumin suppresses the expression of EZH2 as a subunit of PRC2 and its related lncRNA PVT1 to prevent pancreatic cancer progression and increase gemcitabine sensitivity [286]. The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) can promote gastric cancer progression via reducing miRNA-124-3p expression and subsequent induction of EZH2 signaling. It has been reported that exposing gastric cancer cells to hydrogen is associated with down-regulation of MALAT1, and subsequent upregulation of miRNA-124-3p as a tumor-suppressor, resulting in EZH2 inhibition and suppression of proliferation and invasion [287].
Exosomes are minute structures with sizes of 40–150 nm that are secreted by most cells [288]. The formation process of exosomes includes plasma membrane budding inward to produce early endosomes. Subsequently, late endosomes, known as multivesicular bodies (MVBs) form and are secreted to extracellular space [289–292]. It is noteworthy that exosomes can function as carriers within cells to contain proteins, lipids, and nucleic acids. Exosomes can also transfer lncRNAs in different cancers and depending on the function of lncRNAs exert cancer progression or inhibition effects [293, 294]. For instance, lncRNA UFC1 is suggested to promote lung cancer progression. UFC1 is upregulated in lung cancer tissues, serum, and serum exosomes of patients with non-small cell lung cancer and is suggested to mediate their proliferation and invasion. Silencing UFC1 is associated with cell cycle arrest and apoptosis induction. Investigation of molecular pathways demonstrates that lncRNA UFC1 binds to EZH2 to increase its accumulation at promoter of phosphatase and tensin homolog (PTEN). This leads to the trimethylation of H3K27 which subsequently down-regulates PTEN expression and promotes lung cancer progression. This suggests the key role of exosomes in facilitating the transfer of UFC1 to non-small cell lung cancer cells [295].
It was reported that miRNA/EZH2 axis affects EMT and metastasis of cancer cells. Importantly, lncRNAs can affects EMT via EZH2 regulation. The lncRNA H19 is a tumor-promoting factor capable of enhancing cancer proliferation and invasion via p53 down-regulation and subsequent induction of TNFAIP8 [274]. Silencing H19 is in favor of apoptosis induction and impairing breast cancer proliferation [296]. On the other hand, lncRNA H19 induces STAT3 expression to upregulate EZH2, leading to EMT induction in esophageal cancer cells [297]. A similar pathway by lncRNA NRON occurs to increase bladder cancer metastasis. Mechanistically, NRON stimulates EZH2 signaling to provide EMT induction and bladder cancer migration [298]. Overall, these studies agree with the regulatory role of EZH2/EMT axis by lncRNAs in different cancers. Interestingly, there is a close relationship between EMT and chemoresistance. It has been reported that EMT induction can provide paclitaxel (PTX) resistance in cancer cells [196]. The lncRNA PVT1 interacts with EZH2 to recruit it at promoter region of miRNA-195 and reduce its expression, resulting in PTX-mediated EMT. Silencing PVT1 is associated with increased sensitivity of cervical cancer cells to chemotherapy [299]. Overall, experiments agree with the fact that lncRNAs can induce cancer progression via inducing EZH2 expression. In most cases, lncRNAs stimulate EZH2 via miRNA sponging. Furthermore, it has been reported that lncRNAs recruit EZH2 to the promoter of miRNAs and other factors to reduce their expression, resulting in cancer malignancy and progression. Finally, proliferation and metastasis of cancer cells are regulated by lncRNA/EZh2 axis [296, 300–311].
LncRNAs inhibiting EZH2
It is noteworthy that most of studies have focused on revealing the role of tumor-promoting lncRNAs in cancer progression via EZH2 regulation. However, there are evidence showing that lncRNAs can also inhibit EZH2 signaling to function as tumor-suppressor factors. In this section, we will discuss lncRNAs capable of inhibiting EZH2. Previously, it was mentioned that chemoresistance occurs in cancer cells due to activation of tumor-promoting factors. It seems that lncRNA/EZH2 axis can also regulate the response of cancer cells to radiotherapy. The lncRNA MAGI2-AS3 is down-regulated in esophageal cancer cells. Enhancing MAGI2-AS3 expression is associated with recruitment of EZH2 to induce H3K27me3, leading to HOXB7 down-regulation and increasing radio-sensitivity [312]. This study demonstrates the tumor-suppressing role of EZH2 in cancer. Due to the methyltransferase role of EZH2, its recruitment is key in regulating the expression of other genes. For instance, as a tumor-suppressor, lncRNA MEG3 binds to EZH2 resulting in H3K27-mediated trimethylation of Engrailed-2 (EN-2) to suppress prostate cancer progression [313]. Therefore, EZH2 can also function as transcriptional regulators.
Although previous experiments demonstrate the anti-tumor activity of EZH2 in cancers via reducing expression levels of downstream targets, it appears that its down regulation by lncRNAs can reduce cancer progression. The lncRNA ANCR plays a dual role as tumor-suppressor or tumor-promoter, demonstrating that more experiments are required to reveal the role of lncs in cancer [314–317]. Specifically, ANCR exerts anti-tumor activity in breast cancer via regulating EZH2 expression. In this way, ANCR increases CDK1 and EZH2 interaction that is vital for enhancing EZH2 phosphorylation at threonine-345 and threonine-487. This phosphorylation provides the conditions for EZH2 ubiquitination and its subsequent degradation, resulting in a significant decrease in metastasis and migration of breast cancer cells [318].
Glycogen synthase kinase-3beta (GSK-3β) is considered as a driver of cancer progression and is a downstream target of PI3K/Akt axis in increasing pancreatic cancer growth and migration [319, 320]. Increased phosphorylation of GSK-3β is in favor of chemoresistance induction [321]. On the other hand, lncRNA brain-derived neurotrophic factor antisense (BDNF-AS) is suggested to impair the malignancy of colorectal cancer cells. BDNF-AS recruits EZH2 to down-regulate GSK-3β [322] and silence the tumor-promoting gene GSK-3β.
LncRNAs can affect stem cell regulation and progression of cancer cells [323]. On the other hand, CSCs play a significant role in mediating therapy resistance [324, 325]. Therefore, it is of importance to reveal how lncRNA and EZH2 interactions affect CSC’s behavior. The lncRNA STXBP5-AS1 significantly diminishes proliferation and survival of cancer cells and is associated with favorable prognosis of pancreatic cancer. This lncRNA reduces stemness of pancreatic cancer via suppressing stem cell-like features. Mechanistically, this anti-tumor activity is mediated via recruiting EZH2, suggesting its role in tumor suppression [326]. Suppressing the expression of such lncRNAs increases self-renewal capacity of CSCs in favor of cancer progression [327]. Although a few studies have examined tumor-suppressor lncRNAs, more experiments are needed to reveal lncRNA and EZH2 interaction in cancer.
EZH2 in lncRNA regulation
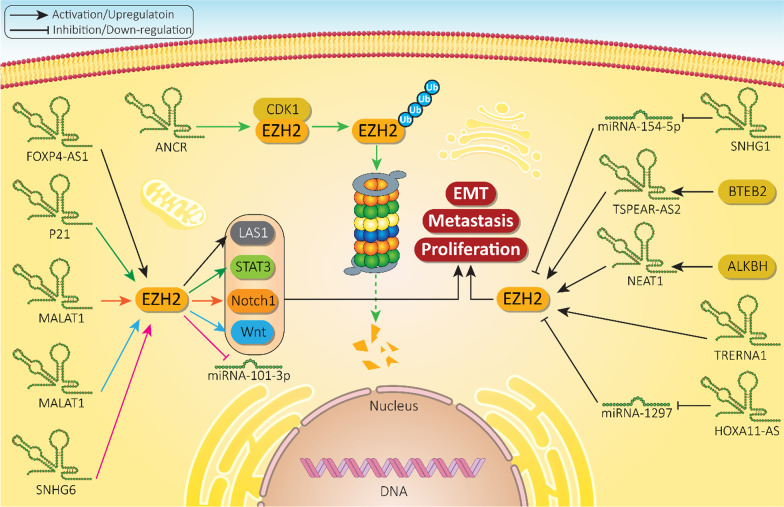
Like miRNAs, it has been reported that lncRNA expression can be regulated by EZH2 in cancer cells. Although a few studies have investigated this interaction, future experiments will shed more light on their relationship. A recent experiment has shown that lncRNA SVUGP2 is downregulated by EZH2 in lung cancer cells. Increasing expression level of lncRNA SVUGP2 is associated with impairing proliferation and invasion of lung cancer cells. Mechanistically, EZH2 down-regulates SVUGP2 to induce Wnt/β-catenin signaling to exert its tumorigenesis role (Fig. 6 and Table 5) [301].
Fig. 6.
Regulation of EZH2 by lncRNAs. LncRNAs not only affect EZH2, but also its downstream targets including LAS1, STAT3, Notch1 and Wnt are affected, leading to a significant change in EMT, metastasis and growth of cancer cells
Table 5.
The role of lncRNAs in regulating EZH2 in different cancers
| LncRNA | Signaling network | Cancer type | Remarks | References |
|---|---|---|---|---|
| NEAT1 | ALKBH/NEAT1/EZH2 | Gastric cancer |
Overexpression of ALKBH in cancer cells and tissues Demethylation of NEAT1 for its activation Upregulation of EZH2 and enhancing cancer progression |
[328] |
| TSPEAR-AS2 | BTEB2/TSPEAR-AS2/EZH2/GJA1 | Gastric cancer |
Upregulation of lncRNA by BTEB2 Driving cancer progression via enhancing EZH2 expression and down-regulating GJA1 |
[329] |
| TRERNA1 | EZH2/CDH1 | Gastric cancer | Enhancing cancer metastasis via upregulating EZH2 and subsequent inhibition of CDH1, leading to EMT induction | [330] |
| SNHG6 | EZH2/miRNA-101-3p/ZEB1 | Gastric cancer |
Transcriptional inhibition via recruiting EZH2 Inducing EMT via miRNA-101-3p down-regulation and subsequent stimulation of ZEB1 expression Increasing cancer metastasis |
[331] |
| LINC00460 | EZH2/LSD1/CCNG2 | Gastric cancer |
Overexpression of lncRNA in cancerous tissues compared to normal tissues Inducing EZH2/LSD1 axis to down-regulate CCNG2 expression Enhancing cancer progression |
[332] |
| HOXA11-AS | MiRNA-1297/EZH2 | Gastric cancer |
Reducing miRNA-1297 expression via sponging Inducing EZH2 expression and its complex formation with histone demethylase LSD1 or DNMT1 Increasing cancer cell growth |
[333] |
| FOXP4-AS1 | EZH2/LSD1 | Gastric cancer |
Facilitating proliferation and metastasis of cancer cells Inducing EZH22/LAS1 axis |
[334] |
| P21 | EZH2/STAT3 | Prostate cancer |
Increased transcription of lncRNA p21 by enzalutamide through activating androgen signaling Activating non-histone methyltransferase activity of EZH2 STAT3 methylation and subsequent induction of NED |
[335] |
| MALAT1 | – | Prostate cancer | Recruitment of EZH2 and enhancing its tumorigenesis activity | [336] |
| MALAT1 | EZH2/Notch1 | Esophageal cancer | Inducing EZH2/Notch1 axis to promote metastasis via EMT induction | [337] |
| HERES | EZH2/Wnt | Esophageal squamous cell carcinoma |
Interaction of HERES with EZH2 through G-quadruple structure-like motif Activating Wnt signaling Enhancing growth, migration and colony formation capacity of cancer cells |
[338] |
| CASC9 | EZH2/PDCD4 | Esophageal squamous cell carcinoma |
Association with poor survival of cancer patients Increasing cancer growth Enriching EZH2 Reducing PDCD4 expression after binding of EZH2 to its promoter |
[339] |
| HOXA-AS2 | EZH2/LSD1 | Pancreatic cancer |
Increasing cancer growth and survival Apoptosis inhibition Enhancing cell cycle progression Inducing EZH2/LSD1 axis |
[340] |
| AGAP2-AS1 | RREB1/AGAP-AS1/EZH2 | Pancreatic cancer |
Overexpression of lncRNA by RREB1 Transcriptional repression of ANKRD1 and ANGPTL4 to activate EZH2 signaling Increasing cancer progression and metastasis |
[341] |
| BLACAT1 | EZH2/CDKN1C | Pancreatic cancer |
BLACAT1 recruits EZH2 to provide trimethylation of CDKN1C promoter via H3K27 Enhancing proliferation and inducing glycolysis |
[342] |
| HOTAIRM1 | HOXA1/EZH2 | Glioblastoma multiforme |
Upregulation of HOXA1 by HOTAIRM1 Demethylation and sequestering EZH2 Increasing cancer proliferation and metastasis |
[343] |
| LINC00115 | TGF-β/LINC00115/EZH2 | Glioma |
MiRNA-200s down-regulation by TGF-β-mediated LINC00115 upregulation Inducing ZNF596 transcription Triggering EZH2/STAT3 axis for cancer progression |
[344] |
| PVT1 | EZH2/Hippo/Notch1 | Non-small cell lung cancer |
PVT1 stimulates Hippo/Notch1 axis via upregulating EZH2 Increasing cancer metastasis |
[345] |
| FOXC2 | EZH2/p15 | Non-small cell lung cancer |
Apoptosis inhibiting Preventing cell cycle arrest P53 down-regulation via activating EZH2 signaling |
[346] |
| UCA1 | EZH2/CDKN1A | Non-small cell lung cancer |
Epigenetic silencing of CDKN1A via recruiting EZH2 Enhancing proliferation and inhibiting apoptosis |
[347] |
| PVT1 | MiRNA-526b/EZH2 | Non-small cell lung cancer |
Association with poor prognosis Down-regulation of miRNA-526b and subsequent induction of EZH2 |
[348] |
| MSTO2P | – | Lung cancer | Enhancing EZH2 expression and promoting proliferation and invasion of cancer cells | [228] |
| HOTAIR | – | Lung cancer | Silencing HOTAIR/EZH2 axis increases potential of atractylenolide 1 and erlotinib in lung cancer suppression | [349] |
| HOTAIR | – | Breast cancer |
Apoptosis inhibition Inducing cell cycle progression Increasing cancer proliferation EZH2 recruitment Increasing DNA repair Inducing radio-resistance |
[350] |
| TUG1 | EZH2/miRNA-194-5p/CCND2 | Bladder cancer |
Recruiting EZH2 to down-regulate miRNA-194-5p Inhibiting CCND2 expression Silencing TUG1 increases cisplatin sensitivity of cancer cells |
[351] |
| SPRY4-IT1 | MiRNA-101-3p/EZH2 | Bladder cancer |
Increasing cancer proliferation and metastasis Reducing miRNA-101-3p expression via sponging Increasing EZH2 expression |
[352] |
| AWPPH | EZH2/Smad4 | Bladder cancer |
Recruitment of EZH2 by AWPPH Subsequent upregulation of Smad4 and enhancing cancer proliferation and progression |
[353] |
| CACS15 | EZH2/APC | Ovarian cancer |
Overexpression of CACS15 is associated with poor survival of cancer patients Increasing proliferation and metastasis Recruiting EZH2 to promoter of APC to inhibit it |
[354] |
| SUMO1P3 | EZH2/CPEB3 | Colorectal cancer |
Apoptosis inhibition and increasing proliferation upon SUMO1P3 overexpression Recruiting EZH2 to promoter of CREB3 Epigenetic repression of CREB3 by EZH2 |
[355] |
| DUXAP8 | EZH2/LSD1 | Colorectal cancer |
DUXAP8 enhances EZH2 and LSD1 levels in providing cancer progression Association with tumor size and tumor grade |
[356] |
| LL22NC03-N64E9.1 | EZH2/KLF2 | Colorectal cancer |
Enhancing cancer proliferation and colony formation capacities Apoptosis inhibition Exerting carcinogenesis impact LncRNA binds to EZH2 to down-regulate KLF4 and provide its tumorigenesis impact |
[357] |
| MALAT1 | MiRNA-363-3p/EZH2 | Colorectal cancer |
Down-regulating miRNA-363-3p via sponging Inducing EZH2 signaling Enhancing cancer progression in vitro and in vivo |
[358] |
| FAM83C | EZH2/SEMA3F | Colorectal cancer |
Promoting malignant transformation of colorectal cancer Stabilizing EZH2 and increasing methylation of SEMA3F |
[359] |
| SNHG6 | MiRNA-26a/EZH2 | Colorectal cancer |
MiRNA-26a inhibition and subsequent upregulation of EZH2 Enhancing metastasis via EMT induction Increasing growth and survival of cancer cells |
[341] |
| PRADX | EZH2/NF-κB | Colon adenocarcinoma |
Recruiting EZH2 by PRADX in promoter of UBXN1 gene NF-κB activation and increasing cancer progression |
[360] |
| CASC11 | STAT3/CASC11/EZH2/PTEN | Hepatocellular carcinoma |
Overexpression of CASC11 by STAT3 Recruiting EZH2 and subsequent down-regulation of PTEN Enhancing cancer migration and invasion via EMT induction |
[361] |
| HOXD-AS1 | MiRNA-130a-3p/SOX4/EZH2 | Liver cancer |
Protecting SOX4 against degradation via miRNA-130a-3p down-regulation Enhancing EZH2 expression and paving the way for cancer progression |
[362] |
| PVT1 | EZH2/MYC | Liver cancer |
Recruitment of EZH2 by PVT1 and subsequent induction of MYC expression Increasing cancer progression |
[363] |
| SNHG8 | EZH2/RECK | Cervical cancer |
Apoptosis inhibition Facilitating proliferation Recruiting EZH2 for transcriptional repression of RECK |
[364] |
| LINC01535 | MiRNA-214/EZH2 | Cervical cancer |
Reverse relationship between LINC01535 and miRNA-214 Inducing EZH2 signaling Promoting growth in vitro and in vivo |
[365] |
| PVT1 | EZH2/miRNA-200b | Cervical cancer |
Binding to EZH2 and recruiting it at promoter of miRNA-200b Enhancing proliferation and migration |
[366] |
CircRNAs and EZH2 signaling
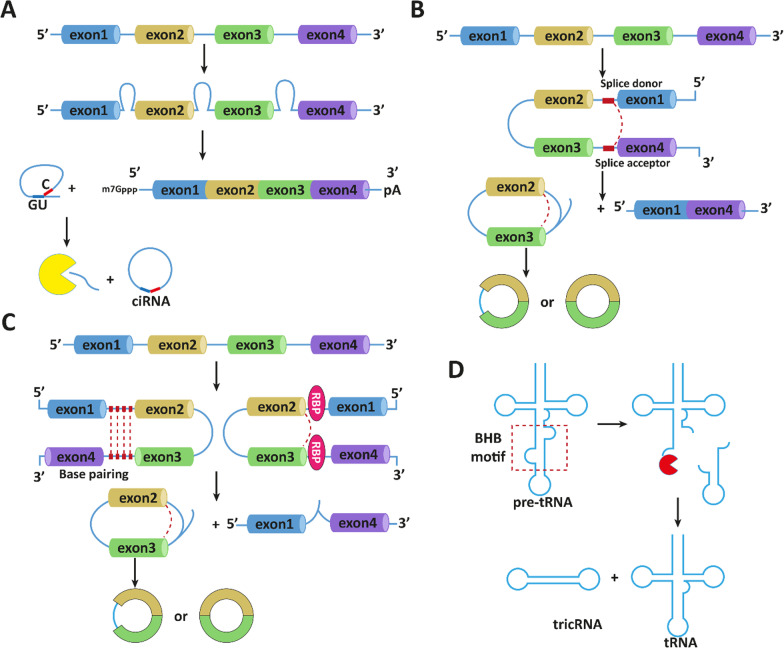
Circular RNAs (circRNAs) were first discovered in plant viroids and Sendai viruses in 1976 using electron microscopy [367]. Subsequently, attempts have been made to identify them in eukaryotic cells which occurred by 1979 [368–371]. However, it was believed that circRNAs result from splicing errors with low abundance [372]. Later, various kinds of circRNAs were discovered with progress in bioinformatics and sequencing technologies. CircRNAs have a loop structure attached through covalent bonds and lack 5′ cap and 3′ poly (A) tails (Fig. 7) [373]. The biogenesis of circRNAs starts with the precursor mRNA back-splicing of exons. In this way, a downstream 5/ splice site of an exon attaches to upstream 5/ splice site to form a loop structure [374–376]. Reverse complementary sequences and RNA binding proteins aid with exon skipping and circRNA formation [377–379]. It has been reported that expression of circRNAs is cell- and tissue-specific [380]. Recently, much attention has been directed towards the role of circRNAs in cancer as diagnostic, prognostic, and therapeutic tools [174, 381–384]. Furthermore, circRNAs can regulate miRNA expression via sponging like lncRNAs [385]. In this section, we provide a discussion of EZH2 regulation by circRNAs in cancer.
Fig. 7.
The circRNA biogenesis in cells [386]. A canonical splicing; B lariat-driven circularization; C intron-pairing-driven circularization; D bulge–helix–bulge (BHB) motif recognition
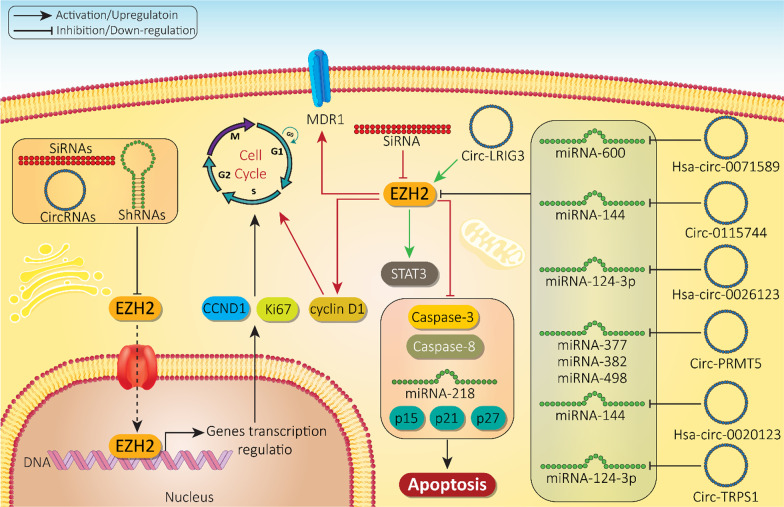
Most studies have focused on revealing the role of tumor-promoting circRNAs in cancer. Based on EZH2’s role in cancer progression, such circRNAs can enhance EZH2 expression. For instance, hsa-circ-0071589 undergoes upregulation in colorectal cancer cells and tissues. Mechanistically, this circRNA down-regulates miRNA-600 expression via sponging to enhance EZH2 expression [387]. Therefore, silencing such circRNAs may suppress tumorigenesis. For instance, silencing hsa-circ-0026123 is a promising strategy that enhances the expression level of miRNA-124-3p, leading to EZH2 down-regulation in ovarian cancer cells to inhibit their migration and growth [388]. In hepatocellular carcinoma, circ-LRIG3 enhances EZH2 expression and induces STAT3 methylation to promote cancer progression. Positive feedback loops between STAT3 and circ-LRIG3 occur through STAT3 binding to promoter of circ-LRIG3 and enhancing STAT3 expression, to subsequently lead to hepatocellular carcinoma progression [389]. These feedback loops are in favor of cancer malignancy and further complicates molecular pathways related to circRNA/EZH2 axis.
New evidences demonstrate that circRNAs can target more than one miRNA when targeting EZH2. MiRNA-377, miRNA-382 and miRNA-498 are considered as tumor-suppressor factors in non-small cell lung cancer and their overexpression suppresses lung cancer growth via EZH2 down-regulation. On the other hand, circ-PRMT5 can down-regulate the expression levels of miRNA-377, -382 and -498 to enhance EZH2 expression, leading to an increase in proliferation of non-small cell lung cancer cells [390]. Therefore, competitive binding between circRNAs and miRNAs is of importance for regulating EZH2 expression and subsequent impact on proliferation and invasion of cancer cells [391]. One of the limitations of experiments related to circRNA/EZH2 axis is the lack of in vivo studies in many cancers. Most of studies have focused on in vitro models and we have still a long way before translation to clinic. It is noteworthy that the most the circRNAs investigated to date are tumor-promoting factors and their mechanism of action is suggested by sponging miRNAs to enhance EZH2 expression (Fig. 8 and Table 6) [392–394]. Further experiments need to focus on revealing the role of tumor-suppressor circRNAs in cancer.
Fig. 8.
Regulation of EZH2 by siRNA, shRNA and circRNAs in cancer
Table 6.
The role of CircRNAs in regulating EZH2 expression in different cancers
| CircRNA | Signaling network | Cancer type | Signaling network | References |
|---|---|---|---|---|
| Hsa-circ-0071589 | MiRNA-600/EZH2 | Colorectal cancer |
Reverse association between circRNA and miRNA-600 Reducing miRNA-600 expression via sponging Inducing EZH2 expression and promoting cancer progression |
[387] |
| Circ-0115744 | MiRNA-144/EZH2 | Colorectal cancer |
Reducing miRNA-144 expression and subsequent induction of EZH2 signaling Enhancing cancer metastasis |
[392] |
| Hsa-circ-0026123 | MiRNA-124-3p/EZH2 | Ovarian cancer |
Elevating cancer proliferation and migration Sponging miRNA-124-3p and increasing EZH2 expression |
[206] |
| Circ-PRMT5 | MiRNA-377/382/498/EZH2 | Non-small cell lung cancer |
Decreasing expression levels of miRNAs with tumor-suppressing role Paving the way for EZH2 upregulation Accelerating cancer progression |
[390] |
| Hsa-circ-0020123 | MiRNA-144/EZH2 | Non-small cell lung cancer | Competitive binding with miRNA-144 and paving the way for EZH2 upregulation in elevating cancer progression | [391] |
| Circ-TRPS1 | MiRNA-124-3p/EZH2 | Prostatic cancer |
Promoting stemness of cancer cells Reverse relationship between circRNA and miRNA Inducing EZH2 signaling |
[393] |
| Circ-LRIG3 | EZH2/STAT3 | Hepatocellular carcinoma | Promoting expression level of STAT3 via activating EZH2 to ensure cancer survival | [389] |
| Hsa-circ-0000129 | – | Breast cancer |
Overexpression of EZH2 as a downstream target of circRNA Enhancing tumor progression Considering as a biomarker |
[394] |
Non-coding RNA and EZH2 interaction in tumor microenvironment
The tumor microenvironment (TME) is a complex environment that not only comprises of cancer cells, abut also has immune cells, secreted proteins, adipocytes, fibroblasts and hematopoietic derived cells, among others. The interaction between cancer cells and TME affects and determines progression of tumor cells. The interactions occurring in TME are responsible for proliferation, invasion, angiogenesis and survival of cancer cells [395]. The EZH2 signaling affects TME to regulate cancer progression. For instance, EZH2 mutation affects infiltration of T cells in TME [396]. The EZH2 enhances viability and function of CD4 + and CD8 + T cells and can suppress differentiation of Th1 and Th2 cells [397]. The poor expression of EZH2 in CD8 + T cells leads to undesirable prognosis of cancers. Besides, EZH2 is responsible for preserving T-cell memory precursors in tumor suppression [398]. The T-regulatory cells, natural killer cells and dendritic cells are also affected by EZH2 signaling [399]. A recent experiment revealed that EZH2 upregulation by circPVT1 results in high infiltration of macrophages in TME and enhancing tumor progression [400]. Notably, immune checkpoint inhibitors are applied for improving cancer immunotherapy. However, resistance to immune checkpoint inhibitors has been observed in various tumors and EZH2 inhibition enhances anti-cancer immunity and prevents checkpoint inhibitor resistance via improving T regulatory cell trafficking and elevated antigen presentation [401].
The interaction between ncRNAs and EZH2 signaling in TME determines tumor progression. The down-regulation of miRNA-144/451a occurs in hepatocellular carcinoma and they are associated with anti-tumor immunity. This miRNA cluster negatively regulates EZH2 expression to elevate M1 polarization of macrophages and to enhance anti-tumor immunity. There is a negative feedback loop in which EZH2 can reduce miRNA expression in hepatocellular carcinoma to mediate immune evasion [402]. However, EZH2 functions in TME like a double-edged sword. The EZH2 signaling is vital for appropriate function of cytotoxic CD8 + T cells in TME and preventing tumor progression. The overexpression of miRNA-26a impairs function of T cells and promotes tumor progression, while miRNA-26a down-regulation is in favor of improving T cell function and inducing tumor growth suppression. The miRNA-26a has negative correlation with EZH2 in cytotoxic T cells and inhibits EZH2 signaling to disrupt T cell function and to enhance lung cancer progression [403]. An important feature of TME is hypoxia that can enhance both growth and metastasis of cancer cells [404, 405]. The expression level of lncRNA HITT undergoes down-regulation in hypoxic TME that promotes progression of cervical and colorectal tumor cells. The lncRNA HITT directs EZH2 to promoter of HIF-1α to form RNA–DNA triplex, leading to HIF-1α down-regulation and subsequent tumor suppression [406]. These studies highlight the fact that ncRNAs are capable of regulating EZH2 signaling in TME and affecting cancer progression.
Biomarker application
As cancer claims the second leading cause of death after cardiovascular diseases, there have been efforts in identification of biomarkers for diagnosis, prognosis, and prediction. NcRNAs can be considered as reliable and non-invasive biomarkers for cancer diagnosis. It has been reported that upregulation of lncRNAs in lung cancer are associated transcript 1 (LUCAT1) which enhances cancer proliferation via EZH2 overexpression. Silencing LUCAT1 remarkably diminishes cancer progression and malignancy. With respect to its tumor-promoting role, LUCAT1 is considered as a prognostic factor as its upregulation provides undesirable prognosis for patients with thyroid cancer [407]. A similar phenomenon occurs in gastric cancer with lncRNA urothelial cancer-associated 1 (UCA1) which binds to EZH2 and induces the down-regulation of tumor-suppressor factors including p21 and Sprouty RTK signaling antagonist 1 (SPRY1). The lncRNA UCA1 is a prognostic factor in gastric cancer with its upregulation being correlated to unfavorable prognosis [408]. As another example, lncRNA SNHG1/EZH2 axis, a prognostic factor, contributes to rectal cancer metastasis and initiation [409]. These experiments highlight the fact that ncRNAs regulate EZH2 expression and can be considered as reliable and non-invasive biomarkers for cancer prognosis and diagnosis.
Pre-clinical studies are in agreement with the fact that EZH2 signaling is tightly regulated by ncRNAs. These findings are advantageous, when they are translated to clinic, paving the way for effective treatment of cancer patients. Throughout this review article, we report that lncRNAs, circRNAs and miRNAs can regulate EZH2 expression in cancer cells. It is noteworthy that most experiments have focused on revealing the role of tumor-promoting factors. It has been shown that siRNA and shRNA can be applied to down-regulate lncRNAs [410, 411]. Thus, next experiments can focus on the targeting of ncRNAs and translating these findings in clinical settings. Furthermore, as conditions in pre-clinical and clinical studies are different and elicit different responses in vivo and in vitro, strategies that increase the efficiency of therapeutics such as biocompatible, safe and well-tolerated nanocarriers for delivery of therapeutics in targeting ncRNA/EZH2 axis need be considered [412, 413]. Further experiments can help shed more light on the efficacy of nanocarriers at cellular and systemic levels.
Conclusion and remarks
The present review provides a comprehensive discussion of EZH2 signaling role in cancer, and its regulation by upstream mediators, such as ncRNAs. The introduction section demonstrated the dual role of EZH2 in cancer. In most cases, EZH2 functions as a tumor-promoting factor, while owing to its methyltransferase activity, EZH2 may also function as tumor-suppressor. Tumor suppression occurs through recruitment of H3K27 to promoter of target genes and reducing their expression, paving the way for cancer elimination. Among ncRNAs, miRNAs and lncRNAs play a key regulatory impact on EZH2 signaling compared to other kinds of ncRNAs. MiRNAs can indirectly affect the expression of target gene by recruiting EZH2 to promoter. It is noteworthy that EZH2 can form a feedback and function as upstream mediator of miRNAs. In most cases, lncRNAs regulate the expression of EZH2 via sponging miRNAs. CircRNAs also regulate the expression of EZH2 signaling, but similar to lncRNAs, they regulate EZH2 expression mainly via targeting miRNAs. We also discuss clues for the development of siRNA and shRNA for targeting EZH2 and their clinical applications. However, efficacy of siRNA and shRNA is limited in vivo which will affect their potential in clinical course. Thus, we explored the use of nanocarriers for their targeted delivery in cancer patients which warrants additional research.
Acknowledgements
Not applicable.
Abbreviations
- EZH2
Enhancer of zeste homolog 2
- ncRNAs
Non-coding RNAs
- PRCs
Polycomb repressive complexes
- PRMT1
Protein arginine methyltransferase 1
- EMT
Epithelial-to-mesenchymal transition
- AOX1
Aldehyde oxidase 1
- YAP
Yes-associated protein
- CCL5
Chemokine ligand 5
- miRNAs, microRNAs, 3′-UTR
3′-Untranslated region
- mRNA
Messenger RNA
- RISC
RNA-induced silencing complex
- LMO3
LIM-only protein 3
- CSCs
Cancer stem cells
- LC3
Light chain-3
- STAT3
Signal transducer and activator of transcription 3
- DOX
Doxorubicin
- GKN1
Gastrokine 1
- MMPs
Matrix metalloproteinases
- IFN-γ
Interferon-γ
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- MVBs
Multivesicular bodies
- PTEN
Phosphatase and tensin homolog
- PTX
Paclitaxel
- EN-2
Engrailed-2
- GSK-3β
Glycogen synthase kinase-3beta
- BDNF-AS
Brain-derived neurotrophic factor antisense
- circRNAs
Circular RNAs
- LUCAT1
Lung cancer associated transcript 1
- CXCL12
Chemokine ligand 12
- siRNA
Small interfering RNA
- RNAi
RNA interference
- MDR1
Multidrug resistance 1
- shRNA
Short-hairpin RNA
- lncRNAs
Long non-coding RNAs
- SNHG1
Small nuclear RNA host gene 1
- Akt
Protein kinase-B
- PI3K
Phosphoinositide 3-kinase
- PVT1
Plasmacytoma variant translocation I
- TUG1
Taurine upregulated gene 1
- JNK3
C-Jun N-terminal kinase 3
- DZNeP
3-Deacaneplanocin A
- AR
Androgen receptor
- ADT
Androgen deprivation therapy
- HK2
Hexokinase-2
- DNMT
DNA methyltransferases enzymes
- MMP14
Matrix metalloproteinase 14
- BRMS1
Breast cancer metastasis suppressor 1
- CXCR4
CXC chemokine receptor 4
- UCA1
Urothelial cancer-associated 1
- SPRY1
Sprouty RTK signaling antagonist 1
Authors' contributions
APK, MA and YW conceptualized this idea. SM, MHG, KH, FH, AZ and MA, NR and PM participated in literature review and writing of manuscript. AZ performed software work and drew the various schematic figures. APK, YW, MA, IC, ARA, FC and NN participated in English editing and supervising manuscript. All authors read and approved the final manuscript.
Funding
This research was supported in part by the Canadian Institutes of Health Research (#141635, #144159, #153081, #173338) (YW), Terry Fox Research Institute (#1062) (YW), Singapore Ministry of Education (MOE-T2EP30120-0016), the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Center of Excellence initiative to Cancer Science Institute of Singapore, and National University of Singapore (APK).
Availability of data and materials
Not applicable.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
All authors have read the manuscript and given their consent for publication.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuzhuo Wang, Email: ywang@bccrc.ca.
Milad Ashrafizadeh, Email: milad.ashrafizadeh@sabanciuniv.edu.
Alan Prem Kumar, Email: apkumar@nus.edu.sg.
References
- 1.Abadi AJ, Zarrabi A, Hashemi F, Zabolian A, Najafi M, Entezari M, Hushmandi K, Aref AR, Khan H, Makvandi P, et al. The role of SOX family transcription factors in gastric cancer. Int J Biol Macromol. 2021;180:608–624. doi: 10.1016/j.ijbiomac.2021.02.202. [DOI] [PubMed] [Google Scholar]
- 2.Ashrafizadeh M, Delfi M, Hashemi F, Zabolian A, Saleki H, Bagherian M, Azami N, Farahani MV, Sharifzadeh SO, Hamzehlou S, et al. Biomedical application of chitosan-based nanoscale delivery systems: Potential usefulness in siRNA delivery for cancer therapy. Carbohydr Polym. 2021;260:117809. doi: 10.1016/j.carbpol.2021.117809. [DOI] [PubMed] [Google Scholar]
- 3.Ang HL, Yuan Y, Lai X, Tan TZ, Wang L, Huang BB, Pandey V, Huang RY, Lobie PE, Goh BC, et al. Putting the BRK on breast cancer: From molecular target to therapeutics. Theranostics. 2021;11:1115–1128. doi: 10.7150/thno.49716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohan CD, Bharathkumar H, Dukanya, Rangappa S, Shanmugam MK, Chinnathambi A, Alharbi SA, Alahmadi TA, Bhattacharjee A, Lobie PE, et al. N-substituted pyrido-1,4-oxazin-3-ones induce apoptosis of hepatocellular carcinoma cells by targeting NF-κB signaling pathway. Front Pharmacol. 2018;9:1125. [DOI] [PMC free article] [PubMed]
- 5.Cai W, Xiong Chen Z, Rane G, Satendra Singh S, Choo Z, Wang C, Yuan Y, Zea Tan T, Arfuso F, Yap CT, et al. Wanted DEAD/H or alive: helicases winding up in cancers. J Natl Cancer Inst. 2017;109. [DOI] [PubMed]
- 6.Puar YR, Shanmugam MK, Fan L, Arfuso F, Sethi G, Tergaonkar V. Evidence for the involvement of the master transcription factor NF-κB in cancer initiation and progression. Biomedicines. 2018;6. [DOI] [PMC free article] [PubMed]
- 7.Arora L, Kumar AP, Arfuso F, Chng WJ, Sethi G. The role of signal transducer and activator of transcription 3 (STAT3) and its targeted inhibition in hematological malignancies. Cancers (Basel). 2018;10. [DOI] [PMC free article] [PubMed]
- 8.Mirzaei S, Zarrabi A, Asnaf SE, Hashemi F, Zabolian A, Hushmandi K, Raei M, Goharrizi MASB, Makvandi P, Samarghandian S, et al. The role of microRNA-338-3p in cancer: growth, invasion, chemoresistance, and mediators. Life Sci. 2021;268:119005. [DOI] [PubMed]
- 9.Delfi M, Sartorius R, Ashrafizadeh M, Sharifi E, Zhang Y, De Berardinis P, Zarrabi A, Varma RS, Tay FR, Smith BR, Makvandi P. Self-assembled peptide and protein nanostructures for anti-cancer therapy: Targeted delivery, stimuli-responsive devices and immunotherapy. Nano Today. 2021;38:101119. doi: 10.1016/j.nantod.2021.101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashrafizaveh S, Ashrafizadeh M, Zarrabi A, Husmandi K, Zabolian A, Shahinozzaman M, Aref AR, Hamblin MR, Nabavi N, Crea F, et al. Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett. 2021;508:104–114. doi: 10.1016/j.canlet.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Mirzaei S, Mohammadi AT, Gholami MH, Hashemi F, Zarrabi A, Zabolian A, Hushmandi K, Makvandi P, Samec M, Liskova A, et al. Nrf2 signaling pathway in cisplatin chemotherapy: potential involvement in organ protection and chemoresistance. Pharmacol Res. 2021;167:105575. doi: 10.1016/j.phrs.2021.105575. [DOI] [PubMed] [Google Scholar]
- 12.Halim CE, Xinjing SL, Fan L, Bailey Vitarbo J, Arfuso F, Tan CH, Narula AS, Kumar AP, Sethi G, Ahn KS. Anti-cancer effects of oxymatrine are mediated through multiple molecular mechanism(s) in tumor models. Pharmacol Res. 2019;147:104327. doi: 10.1016/j.phrs.2019.104327. [DOI] [PubMed] [Google Scholar]
- 13.Kirtonia A, Gala K, Fernandes SG, Pandya G, Pandey AK, Sethi G, Khattar E, Garg M. Repurposing of drugs: an attractive pharmacological strategy for cancer therapeutics. Semin Cancer Biol. 2021;68:258–278. doi: 10.1016/j.semcancer.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Ashrafizadeh M, Ahmadi Z, Kotla NG, Afshar EG, Samarghandian S, Mandegary A, Pardakhty A, Mohammadinejad R, Sethi G. Nanoparticles Targeting STATs in Cancer Therapy. Cells. 2019;8. [DOI] [PMC free article] [PubMed]
- 15.Kashyap D, Tuli HS, Yerer MB, Sharma A, Sak K, Srivastava S, Pandey A, Garg VK, Sethi G, Bishayee A. Natural product-based nanoformulations for cancer therapy: opportunities and challenges. Semin Cancer Biol. 2021;69:5–23. doi: 10.1016/j.semcancer.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Brockmueller A, Sameri S, Liskova A, Zhai K, Varghese E, Samuel SM, Büsselberg D, Kubatka P, Shakibaei M. Resveratrol’s anti-cancer effects through the modulation of tumor glucose metabolism. Cancers. 2021;13:188. doi: 10.3390/cancers13020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubatka P, Kello M, Kajo K, Samec M, Liskova A, Jasek K, Koklesova L, Kuruc T, Adamkov M, Smejkal K. Rhus coriaria L. (Sumac) demonstrates oncostatic activity in the therapeutic and preventive model of breast carcinoma. Int J Mol Sci. 2021;22:183. doi: 10.3390/ijms22010183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai K, Brockmüller A, Kubatka P, Shakibaei M, Büsselberg D. Curcumin’s beneficial effects on neuroblastoma: mechanisms, challenges, and potential solutions. Biomolecules. 2020;10:1469. doi: 10.3390/biom10111469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liskova A, Koklesova L, Samec M, Smejkal K, Samuel SM, Varghese E, Abotaleb M, Biringer K, Kudela E, Danko J. Flavonoids in cancer metastasis. Cancers. 2020;12:1498. doi: 10.3390/cancers12061498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 21.Stairiker CJ, Thomas GD, Salek-Ardakani S. EZH2 as a regulator of CD8+ T cell fate and function. Front Immunol 2020;11. [DOI] [PMC free article] [PubMed]
- 22.Wang J, Wang GG. No easy way out for EZH2: its pleiotropic, noncanonical effects on gene regulation and cellular function. Int J Mol Sci. 2020;21:9501. doi: 10.3390/ijms21249501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaballa JM, Braga Neto MB, Ramos GP, Bamidele AO, Gonzalez MM, Sagstetter MR, Sarmento OF, Faubion WA., Jr The role of histone methyltransferases and long non-coding RNAs in the regulation of T cell fate decisions. Front Immunol. 2018;9:2955. doi: 10.3389/fimmu.2018.02955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Shuaib M, Zhang H, Nadeef S, Orlando V. Ubiquitin ligases HUWE1 and NEDD4 cooperatively control signal-dependent PRC2-Ezh1α/β-mediated adaptive stress response pathway in skeletal muscle cells. Epigenetics Chromatin. 2019;12:78. doi: 10.1186/s13072-019-0322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JR, Lee CH, Oksuz O, Stafford JM, Reinberg D. PRC2 is high maintenance. Genes Dev. 2019;33:903–935. doi: 10.1101/gad.325050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Mierlo G, Veenstra GJC, Vermeulen M, Marks H. The complexity of PRC2 subcomplexes. Trends Cell Biol. 2019;29:660–671. doi: 10.1016/j.tcb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Mas G, Di Croce L. The role of Polycomb in stem cell genome architecture. Curr Opin Cell Biol. 2016;43:87–95. doi: 10.1016/j.ceb.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Nutt SL, Keenan C, Chopin M, Allan RS. EZH2 function in immune cell development. Biol Chem. 2020;401:933–943. doi: 10.1515/hsz-2019-0436. [DOI] [PubMed] [Google Scholar]
- 32.Yao Y, Hu H, Yang Y, Zhou G, Shang Z, Yang X, Sun K, Zhan S, Yu Z, Li P, et al. Downregulation of enhancer of zeste homolog 2 (EZH2) is essential for the induction of autophagy and apoptosis in colorectal cancer cells. Genes (Basel). 2016;7. [DOI] [PMC free article] [PubMed]
- 33.Ito T, Teo YV, Evans SA, Neretti N, Sedivy JM. Regulation of cellular senescence by polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep. 2018;22:3480–3492. doi: 10.1016/j.celrep.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Lee Y, Lu X, Song B, Fong KW, Cao Q, Licht JD, Zhao JC, Yu J. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 2018;25:2808–2820.e2804. doi: 10.1016/j.celrep.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13:1–12. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardoso C, Mignon C, Hetet G, Grandchamps B, Fontes M, Colleaux L. The human EZH2 gene: genomic organisation and revised mapping in 7q35 within the critical region for malignant myeloid disorders. Eur J Hum Genet. 2000;8:174–180. doi: 10.1038/sj.ejhg.5200439. [DOI] [PubMed] [Google Scholar]
- 39.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the polycomb-group gene enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Wang D, Lu J, Huang B, Wang Y, Dong M, Fan D, Li H, Gao Y, Hou P, et al. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 2020;27:3226–3242. doi: 10.1038/s41418-020-00615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu W, Zhang X, Qi L, Fu Y, Wang P, Zhao W, Du J, Zhang J, Zhan J, Wang Y, et al. The EZH2-PHACTR2-AS1-ribosome axis induces genomic instability and promotes growth and metastasis in breast cancer. Cancer Res. 2020;80:2737–2750. doi: 10.1158/0008-5472.CAN-19-3326. [DOI] [PubMed] [Google Scholar]
- 43.Lee JE, Park CM, Kim JH. USP7 deubiquitinates and stabilizes EZH2 in prostate cancer cells. Genet Mol Biol. 2020;43:e20190338. doi: 10.1590/1678-4685-GMB-2019-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vantaku V, Putluri V, Bader DA, Maity S, Ma J, Arnold JM, Rajapakshe K, Donepudi SR, von Rundstedt FC, Devarakonda V, et al. Epigenetic loss of AOX1 expression via EZH2 leads to metabolic deregulations and promotes bladder cancer progression. Oncogene. 2020;39:6265–6285. doi: 10.1038/s41388-019-0902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biswas A, Mukherjee G, Kondaiah P, Desai KV. Both EZH2 and JMJD6 regulate cell cycle genes in breast cancer. BMC Cancer. 2020;20:1159. doi: 10.1186/s12885-020-07531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoxha S, Shepard A, Troutman S, Diao H, Doherty JR, Janiszewska M, Witwicki RM, Pipkin ME, Ja WW, Kareta MS, Kissil JL. YAP-mediated recruitment of YY1 and EZH2 represses transcription of key cell-cycle regulators. Cancer Res. 2020;80:2512–2522. doi: 10.1158/0008-5472.CAN-19-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia L, Zhu X, Zhang L, Xu Y, Chen G, Luo J. EZH2 enhances expression of CCL5 to promote recruitment of macrophages and invasion in lung cancer. Biotechnol Appl Biochem. 2020;67:1011–1019. doi: 10.1002/bab.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun S, Yang Q, Cai E, Huang B, Ying F, Wen Y, Cai J, Yang P. EZH2/H3K27Me3 and phosphorylated EZH2 predict chemotherapy response and prognosis in ovarian cancer. PeerJ. 2020;8:e9052. doi: 10.7717/peerj.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, Ni A, Khodos I, de Stanchina E, Nguyen T, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, Qiu X, Cañadas I, Roehle K, Heckler M, et al. EZH2 inhibition activates a dsRNA–STING–interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat Cancer. 2021;2:444–456. doi: 10.1038/s43018-021-00185-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, Campisi M, Kuang Y, Zhang Y, Gjini E, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med. 2018;24:1143–1150. doi: 10.1038/s41591-018-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leitner K, Tsibulak I, Wieser V, Knoll K, Reimer D, Marth C, Fiegl H, Zeimet AG. Clinical impact of EZH2 and its antagonist SMARCA4 in ovarian cancer. Sci Rep. 2020;10:20412. doi: 10.1038/s41598-020-77532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rezaei S, Hosseinpourfeizi MA, Moaddab Y, Safaralizadeh R. Contribution of DNA methylation and EZH2 in SRBC down-regulation in gastric cancer. Mol Biol Rep. 2020;47:5721–5727. doi: 10.1007/s11033-020-05619-9. [DOI] [PubMed] [Google Scholar]
- 54.Tian JH, Mu LJ, Wang MY, Zeng J, Long QZ, Guan B, Wang W, Jiang YM, Bai XJ, Du YF. FOXM1-dependent transcriptional regulation of EZH2 induces proliferation and progression in prostate cancer. Anticancer Agents Med Chem 2020. [DOI] [PubMed]
- 55.Huang B, Mu P, Yu Y, Zhu W, Jiang T, Deng R, Feng G, Wen J, Zhu X, Deng Y. Inhibition of EZH2 and activation of ERRγ synergistically suppresses gastric cancer by inhibiting FOXM1 signaling pathway. Gastric Cancer. 2021;24:72–84. doi: 10.1007/s10120-020-01097-x. [DOI] [PubMed] [Google Scholar]
- 56.Yu J, Xie Y, Liu Y, Wang F, Li M, Qi J. MBD2 and EZH2 regulate the expression of SFRP1 without affecting its methylation status in a colorectal cancer cell line. Exp Ther Med. 2020;20:242. doi: 10.3892/etm.2020.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song Z, Zhang X, Lin Y, Wei Y, Liang S, Dong C. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J Cell Mol Med. 2019;23:7554–7565. doi: 10.1111/jcmm.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao G, Jin LL, Liu CQ, Wang YC, Meng YM, Zhou ZG, Chen J, Yu XJ, Zhang YJ, Xu J, Zheng L. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J Immunother Cancer. 2019;7:300. doi: 10.1186/s40425-019-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bao Y, Oguz G, Lee WC, Lee PL, Ghosh K, Li J, Wang P, Lobie PE, Ehmsen S, Ditzel HJ, et al. EZH2-mediated PP2A inactivation confers resistance to HER2-targeted breast cancer therapy. Nat Commun. 2020;11:5878. doi: 10.1038/s41467-020-19704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, Wang D, Wang W, Chen X, Tang A, Hou P, Li M, Zheng J, Bai J. Macrophages-stimulated PRMT1-mediated EZH2 methylation promotes breast cancer metastasis. Biochem Biophys Res Commun. 2020;533:679–684. doi: 10.1016/j.bbrc.2020.10.037. [DOI] [PubMed] [Google Scholar]
- 61.Zhou X, Jiao D, Dou M, Zhang W, Lv L, Chen J, Li L, Wang L, Han X. Curcumin inhibits the growth of triple-negative breast cancer cells by silencing EZH2 and restoring DLC1 expression. J Cell Mol Med. 2020;24:10648–10662. doi: 10.1111/jcmm.15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo Sardo F, Pulito C, Sacconi A, Korita E, Sudol M, Strano S, Blandino G. YAP/TAZ and EZH2 synergize to impair tumor suppressor activity of TGFBR2 in non-small cell lung cancer. Cancer Lett. 2021;500:51–63. doi: 10.1016/j.canlet.2020.11.037. [DOI] [PubMed] [Google Scholar]
- 63.Cao Z, Wu W, Wei H, Zhang W, Huang Y, Dong Z. Downregulation of histone-lysine N-methyltransferase EZH2 inhibits cell viability and enhances chemosensitivity in lung cancer cells. Oncol Lett. 2021;21:26. doi: 10.3892/ol.2020.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He W, Yu Y, Huang W, Feng G, Li J. The Pseudogene DUXAP8 promotes colorectal cancer cell proliferation, invasion, and migration by inducing epithelial-mesenchymal transition through interacting with EZH2 and H3K27me3. Oncol Targets Ther. 2020;13:11059–11070. doi: 10.2147/OTT.S235643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdel Raouf SM, Ibrahim TR, Abdelaziz LA, Farid MI, Mohamed SY. Prognostic value of TWIST1 and EZH2 expression in colon cancer. J Gastrointest Cancer. 2021;52:90–98. doi: 10.1007/s12029-019-00344-4. [DOI] [PubMed] [Google Scholar]
- 66.Zong X, Wang W, Ozes A, Fang F, Sandusky GE, Nephew KP. EZH2-mediated downregulation of the tumor suppressor DAB2IP maintains ovarian cancer stem cells. Cancer Res. 2020;80:4371–4385. doi: 10.1158/0008-5472.CAN-20-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu S, Rong G, Li X, Geng L, Zeng Z, Jiang D, Yang J, Wei Y. Diosgenin and GSK126 produce synergistic effects on epithelial-mesenchymal transition in gastric cancer cells by mediating EZH2 via the Rho/ROCK signaling pathway. Oncol Targets Ther. 2020;13:5057–5067. doi: 10.2147/OTT.S237474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kong Y, Zhang Y, Mao F, Zhang Z, Li Z, Wang R, Liu J, Liu X. Inhibition of EZH2 enhances the antitumor efficacy of metformin in prostate cancer. Mol Cancer Ther. 2020;19:2490–2501. doi: 10.1158/1535-7163.MCT-19-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan R, Du W, Guo WJJoH, Oncology. EZH2: a novel target for cancer treatment. 2020;13:1–12. [DOI] [PMC free article] [PubMed]
- 70.Kang N, Eccleston M, Clermont P-L, Latarani M, Male DK, Wang Y, Crea FJE. EZH2 inhibition: a promising strategy to prevent cancer immune editing. 2020;12:1457–76. [DOI] [PMC free article] [PubMed]
- 71.Villanueva MTJNRDD: All roads lead to EZH2 inhibition. 2017;16:239–9. [DOI] [PubMed]
- 72.Eich M-L, Athar M, Ferguson JE, Varambally SJCR. EZH2-targeted therapies in cancer: hype or a reality. 2020;80:5449–58. [DOI] [PMC free article] [PubMed]
- 73.Stazi G, Zwergel C, Mai A, Valente S. EZH2 inhibitors: a patent review (2014–2016). 2017;27:797–813. [DOI] [PubMed]
- 74.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. [DOI] [PubMed]
- 75.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, Zhang M, Zhang J, Yu Z, Li L, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol. 2017;13:381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 76.Yap TA, Winter JN, Giulino-Roth L, Longley J, Lopez J, Michot JM, Leonard JP, Ribrag V, McCabe MT, Creasy CL, et al. Phase I study of the novel enhancer of zeste homolog 2 (EZH2) inhibitor GSK2816126 in patients with advanced hematologic and solid tumors. Clin Cancer Res. 2019;25:7331–7339. doi: 10.1158/1078-0432.CCR-18-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glazer RI, Knode MC, Tseng CK, Haines DR, Marquez VE. 3-Deazaneplanocin A: a new inhibitor of S-adenosylhomocysteine synthesis and its effects in human colon carcinoma cells. 1986;35:4523–7. [DOI] [PubMed]
- 78.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. 2009;8:1579–88. [DOI] [PMC free article] [PubMed]
- 79.Coulombe R, Sharma RP, Huggins JJ. Pharmacokinetics of the antiviral agent 3-deazaneplanocin A. 1995;20:197–202. [DOI] [PubMed]
- 80.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song JJ. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. 2012;8:890–6. [DOI] [PubMed]
- 81.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). 2012;109:2989–94. [DOI] [PMC free article] [PubMed]
- 82.Morera L, Lübbert M, Jung MJ. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. 2016;8:1–16. [DOI] [PMC free article] [PubMed]
- 83.Yap TA, Winter JN, Leonard JP, Ribrag V, Constantinidou A, Giulino-Roth L, Michot J-M, Khan TA, Horner T, Carver JJB. A phase I study of GSK2816126, an enhancer of zeste homolog 2 (EZH2) inhibitor, in patients (pts) with relapsed/refractory diffuse large B-cell lymphoma (DLBCL), other non-Hodgkin lymphomas (NHL), transformed follicular lymphoma (tFL), solid tumors and multiple myeloma (MM). 2016;128:4203.
- 84.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. 2012;3:1091–6. [DOI] [PMC free article] [PubMed]
- 85.Crea F, Fornaro L, Bocci G, Sun L, Farrar WL, Falcone A, Danesi R. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 2012;31:753–761. doi: 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- 86.Gulati N, Béguelin W, Giulino-Roth L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma. 2018;59:1574–1585. doi: 10.1080/10428194.2018.1430795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Q, Chen X, Hu X, Duan X, Wan G, Li L, Feng Q, Zhang Y, Wang N, Yu L. Covalent inhibitors of EZH2: Design, synthesis and evaluation. Biomed Pharmacother. 2022;147:112617. doi: 10.1016/j.biopha.2022.112617. [DOI] [PubMed] [Google Scholar]
- 88.Singh J, Petter RC, Baillie TA, Whitty AJ. The resurgence of covalent drugs. 2011;10:307–17. [DOI] [PubMed]
- 89.Sanderson KJ. Irreversible kinase inhibitors gain traction: the approval of boehringer ingelheim's anticancer drug afatinib highlights the growing enthusiasm for once-shunned irreversible kinase inhibitors. 2013;12:649–52. [DOI] [PubMed]
- 90.Lonsdale R, Ward RA. Structure-based design of targeted covalent inhibitors. 2018;47:3816–30. [DOI] [PubMed]
- 91.Wang L, Zhao J, Yao Y, Wang C, Zhang J, Shu X, Sun X, Li Y, Liu K, Yuan HJ. Covalent binding design strategy: a prospective method for discovery of potent targeted anticancer agents. 2017;142:493–505. [DOI] [PubMed]
- 92.Wang X, Cao W, Zhang J, Yan M, Xu Q, Wu X, Wan L, Zhang Z, Zhang C, Qin XJ. A covalently bound inhibitor triggers EZH 2 degradation through CHIP‐mediated ubiquitination. 2017;36:1243–60. [DOI] [PMC free article] [PubMed]
- 93.Rodrigues T, Reker D, Schneider P, Schneider GJ. Counting on natural products for drug design. 2016;8:531–41. [DOI] [PubMed]
- 94.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 96.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang K-l, Shen Q-q, Fang Y-f, Sun Y-m, Ding J, Chen YJ. AZD9291 inactivates the PRC2 complex to mediate tumor growth inhibition. 2019;40:1587–95. [DOI] [PMC free article] [PubMed]
- 98.Kong X, Chen L, Jiao L, Jiang X, Lian F, Lu J, Zhu K, Du D, Liu J, Ding H, et al. Astemizole arrests the proliferation of cancer cells by disrupting the EZH2-EED interaction of polycomb repressive complex 2. J Med Chem. 2014;57:9512–9521. doi: 10.1021/jm501230c. [DOI] [PubMed] [Google Scholar]
- 99.Chen H, Gao S, Li J, Liu D, Sheng C, Yao C, Jiang W, Wu J, Chen S, Huang W. Wedelolactone disrupts the interaction of EZH2-EED complex and inhibits PRC2-dependent cancer. Oncotarget. 2015;6:13049–13059. doi: 10.18632/oncotarget.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cao Z, Qiu J, Yang G, Liu Y, Luo W, You L, Zheng L, Zhang T. MiR-135a biogenesis and regulation in malignancy: a new hope for cancer research and therapy. Cancer Biol Med. 2020;17:569. doi: 10.20892/j.issn.2095-3941.2020.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ashrafizadeh M, Ang HL, Moghadam ER, Mohammadi S, Zarrin V, Hushmandi K, Samarghandian S, Zarrabi A, Najafi M, Mohammadinejad R, Kumar AP. MicroRNAs and their influence on the ZEB family: mechanistic aspects and therapeutic applications in cancer therapy. Biomolecules. 2020;10. [DOI] [PMC free article] [PubMed]
- 102.Ashrafizadeh M, Zarrabi A, Hushmandi K, Kalantari M, Mohammadinejad R, Javaheri T, Sethi G. Association of the epithelial–mesenchymal transition (EMT) with cisplatin resistance. Int J Mol Sci. 2020;21. [DOI] [PMC free article] [PubMed]
- 103.Ashrafizadeh M, Hushmandi K, Hashemi M, Akbari ME, Kubatka P, Raei M, Koklesova L, Shahinozzaman M, Mohammadinejad R, Najafi M, et al. Role of microRNA/epithelial-to-mesenchymal transition axis in the metastasis of bladder cancer. Biomolecules 2020;10. [DOI] [PMC free article] [PubMed]
- 104.Panoutsopoulou K, Avgeris M, Magkou P, Mavridis K, Dreyer T, Dorn J, Obermayr E, Reinthaller A, Michaelidou K, Mahner S. miR-181a overexpression predicts the poor treatment response and early-progression of serous ovarian cancer patients. Int J Cancer. 2020;147:3560–3573. doi: 10.1002/ijc.33182. [DOI] [PubMed] [Google Scholar]
- 105.Bartel DP. Metazoan micrornas. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gebert LF, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pourdavoud P, Pakzad B, Mosallaei M, Saadatian Z, Esmaeilzadeh E, Alimolaie A, Shaygannejad A. MiR-196: emerging of a new potential therapeutic target and biomarker in colorectal cancer. Mol Biol Rep. 2020:1–8. [DOI] [PubMed]
- 108.Mirzaei S, Zarrabi A, Asnaf SE, Hashemi F, Zabolian A, Hushmandi K, Raei M, Goharrizi MASB, Makvandi P, Samarghandian S. The role of microRNA-338–3p in cancer: growth, invasion, chemoresistance, and mediators. Life Sci. 2021:119005. [DOI] [PubMed]
- 109.Ashrafizadeh M, Zarrabi A, Hushmandi K, Hashemi F, Moghadam ER, Owrang M, Hashemi F, Makvandi P, Goharrizi MASB, Najafi M. Lung cancer cells and their sensitivity/resistance to cisplatin chemotherapy: Role of microRNAs and upstream mediators. Cell Signal. 2020:109871. [DOI] [PubMed]
- 110.Faraoni I, Antonetti FR, Cardone J, Bonmassar EJ. miR-155 gene: a typical multifunctional microRNA. 2009;1792:497–505. [DOI] [PubMed]
- 111.Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang X-JJ. Therapeutic siRNA: state of the art. 2020;5:1–25. [DOI] [PMC free article] [PubMed]
- 112.Yang S, Zheng Y, Zhou L, Jin J, Deng Y, Yao J, Yang P, Yao L, Wu Y, Zhai Z, et al. miR-499 rs3746444 and miR-196a-2 rs11614913 are associated with the risk of glioma, but not the prognosis. Mol Ther Nucleic Acids. 2020;22:340–351. doi: 10.1016/j.omtn.2020.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhen J, Zhang H, Dong H, Tong X. miR-9-3p inhibits glioma cell proliferation and apoptosis by directly targeting FOXG1. Oncol Lett. 2020;20:2007–2015. doi: 10.3892/ol.2020.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cheng C, Dong Y, Ru X, Xia Y, Ji Y. LncRNA ANCR promotes glioma cells invasion, migration, proliferation and inhibits apoptosis via interacting with EZH2 and repressing PTEN expression. Cancer Gene Ther. 2020. [DOI] [PubMed]
- 115.Zhang Y, Wang J, An W, Chen C, Wang W, Zhu C, Chen F, Chen H, Zheng W, Gong J. MiR-32 inhibits proliferation and metastasis by targeting EZH2 in glioma. Technol Cancer Res Treat. 2019;18:1533033819854132. doi: 10.1177/1533033819854132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuan DH, Zhao J, Shao GF. Circular RNA TTBK2 promotes the development of human glioma cells via miR-520b/EZH2 axis. Eur Rev Med Pharmacol Sci. 2019;23:10886–10898. doi: 10.26355/eurrev_201912_19792. [DOI] [PubMed] [Google Scholar]
- 117.Xi Q, Zhang J, Yang G, Zhang L, Chen Y, Wang C, Zhang Z, Guo X, Zhao J, Xue Z, et al. Restoration of miR-340 controls pancreatic cancer cell CD47 expression to promote macrophage phagocytosis and enhance antitumor immunity. J Immunother Cancer. 2020;8. [DOI] [PMC free article] [PubMed]
- 118.Algaber A, Al-Haidari A, Madhi R, Rahman M, Syk I, Thorlacius H. MicroRNA-340-5p inhibits colon cancer cell migration via targeting of RhoA. Sci Rep. 2020;10:16934. doi: 10.1038/s41598-020-73792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Z, Tang H, Wang Z, Zhang B, Liu W, Lu H, Xiao L, Liu X, Wang R, Li X. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol Cancer. 2011;10:1–16. doi: 10.1186/1476-4598-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Can Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- 121.Isogai E, Ohira M, Ozaki T, Oba S, Nakamura Y, Nakagawara A. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS ONE. 2011;6:e19297. doi: 10.1371/journal.pone.0019297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu X, Lei Q, Yu Z, Xu G, Tang H, Wang W, Wang Z, Li G, Wu M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget. 2015;6:7930–7943. doi: 10.18632/oncotarget.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lei Q, Liu X, Fu H, Sun Y, Wang L, Xu G, Wang W, Yu Z, Liu C, Li P, et al. miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget. 2016;7:5007–5022. doi: 10.18632/oncotarget.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo P, Lan J, Ge J, Nie Q, Mao Q, Qiu Y. miR-708 acts as a tumor suppressor in human glioblastoma cells. Oncol Rep. 2013;30:870–876. doi: 10.3892/or.2013.2526. [DOI] [PubMed] [Google Scholar]
- 125.Olejarz W, Kubiak-Tomaszewska G, Chrzanowska A, Lorenc T. Exosomes in angiogenesis and anti-angiogenic therapy in cancers. Int J Mol Sci. 2020;21. [DOI] [PMC free article] [PubMed]
- 126.Unterleuthner D, Neuhold P, Schwarz K, Janker L, Neuditschko B, Nivarthi H, Crncec I, Kramer N, Unger C, Hengstschläger M, et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis. 2020;23:159–177. doi: 10.1007/s10456-019-09688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liang W, Wu J, Qiu X. LINC01116 facilitates colorectal cancer cell proliferation and angiogenesis through targeting EZH2-regulated TPM1. J Transl Med. 2021;19:45. doi: 10.1186/s12967-021-02707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun J, Zheng G, Gu Z, Guo Z. MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J Neurooncol. 2015;122:481–489. doi: 10.1007/s11060-015-1753-x. [DOI] [PubMed] [Google Scholar]
- 129.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao Y, Wang XX, Wu W, Long H, Huang J, Wang Z, Li T, Tang S, Zhu B, Chen D. EZH2 regulates PD-L1 expression via HIF-1α in non-small cell lung cancer cells. Biochem Biophys Res Commun. 2019;517:201–209. doi: 10.1016/j.bbrc.2019.07.039. [DOI] [PubMed] [Google Scholar]
- 131.Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50:117–125. doi: 10.5483/BMBRep.2017.50.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Najafi M, Mortezaee K, Majidpoor J. Cancer stem cell (CSC) resistance drivers. Life Sci. 2019;234:116781. doi: 10.1016/j.lfs.2019.116781. [DOI] [PubMed] [Google Scholar]
- 133.Xia H, Zhang W, Zhang B, Zhao Y, Zhao Y, Li S, Liu Y. miR-21 modulates the effect of EZH2 on the biological behavior of human lung cancer stem cells in vitro. Oncotarget. 2017;8:85442–85451. doi: 10.18632/oncotarget.20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang H, Zhang H, Zhao M, Lv Z, Zhang X, Qin X, Wang H, Wang S, Su J, Lv X, et al. MiR-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell Physiol Biochem. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- 135.Brandt JV, Piazza RD, Dos Santos CC, Vega-Chacón J, Amantéa BE, Pinto GC, Magnani M, Piva HL, Tedesco AC, Primo FL, et al. Synthesis and colloidal characterization of folic acid-modified PEG-b-PCL Micelles for methotrexate delivery. Colloids Surf B Biointerfaces. 2019;177:228–234. doi: 10.1016/j.colsurfb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 136.Katiyar SS, Kushwah V, Dora CP, Jain S. Lipid and TPGS based novel core-shell type nanocapsular sustained release system of methotrexate for intravenous application. Colloids Surf B Biointerfaces. 2019;174:501–510. doi: 10.1016/j.colsurfb.2018.11.053. [DOI] [PubMed] [Google Scholar]
- 137.Rudnik LAC, Farago PV, Manfron Budel J, Lyra A, Barboza FM, Klein T, Kanunfre CC, Nadal JM, Bandéca MC, Raman V, et al. Co-loaded curcumin and methotrexate nanocapsules enhance cytotoxicity against non-small-cell lung cancer cells. Molecules 2020, 25. [DOI] [PMC free article] [PubMed]
- 138.Coutinho AJ, Costa Lima SA, Afonso CMM, Reis S. Mucoadhesive and pH responsive fucoidan–chitosan nanoparticles for the oral delivery of methotrexate. Int J Biol Macromol. 2020;158:180–188. doi: 10.1016/j.ijbiomac.2020.04.233. [DOI] [PubMed] [Google Scholar]
- 139.Shan W, Zhang X, Li M, Deng F, Zhang J. Over expression of miR-200c suppresses invasion and restores methotrexate sensitivity in lung cancer A549 cells. Gene. 2016;593:265–271. doi: 10.1016/j.gene.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 140.Ashrafizadeh M, Mirzaei S, Hashemi F, Zarrabi A, Zabolian A, Saleki H, Sharifzadeh SO, Soleymani L, Daneshi S, Hushmandi KJB. New insight towards development of paclitaxel and docetaxel resistance in cancer cells: EMT as a novel molecular mechanism and therapeutic possibilities. 2021, 141:111824. [DOI] [PubMed]
- 141.Ashrafizadeh M, Ang HL, Moghadam ER, Mohammadi S, Zarrin V, Hushmandi K, Samarghandian S, Zarrabi A, Najafi M, Mohammadinejad R. MicroRNAs and their influence on the ZEB family: mechanistic aspects and therapeutic applications in cancer therapy. 2020;10:1040. [DOI] [PMC free article] [PubMed]
- 142.Lee JH, Chinnathambi A, Alharbi SA, Shair OH, Sethi G, Ahn KS. Farnesol abrogates epithelial to mesenchymal transition process through regulating Akt/mTOR pathway. 2019;150:104504. [DOI] [PubMed]
- 143.Hwang ST, Yang MH, Kumar AP, Sethi G, Ahn KS. Corilagin represses epithelial to mesenchymal transition process through modulating Wnt/β-catenin signaling cascade. 2020;10:1406. [DOI] [PMC free article] [PubMed]
- 144.Lee JH, Mohan CD, Deivasigamani A, Jung YY, Rangappa S, Basappa S, Chinnathambi A, Alahmadi TA, Alharbi SA, Garg MJ. Brusatol suppresses STAT3-driven metastasis by downregulating epithelial-mesenchymal transition in hepatocellular carcinoma. 2020;26:83–94. [DOI] [PMC free article] [PubMed]
- 145.Wu J, Li L, Zhang Y, Zhu J. Decreased miR-124 contributes to the epithelial-mesenchymal transition phenotype formation of lung adenocarcinoma cells via targeting enhancer of zeste homolog 2. Pathol Res Pract. 2020;216:152976. doi: 10.1016/j.prp.2020.152976. [DOI] [PubMed] [Google Scholar]
- 146.Pappas K, Martin TC, Wolfe AL, Nguyen CB, Su T, Jin J, Hibshoosh H, Parsons R. NOTCH and EZH2 collaborate to repress PTEN expression in breast cancer. Commun Biol. 2021;4:312. doi: 10.1038/s42003-021-01825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou K, Wang L, Cao Y, Sun P, Wang T. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond) 2020;40:484–500. doi: 10.1002/cac2.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 149.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 150.Liu F, Sang M, Meng L, Gu L, Liu S, Li J, Geng C. miR-92b promotes autophagy and suppresses viability and invasion in breast cancer by targeting EZH2. Int J Oncol. 2018;53:1505–1515. doi: 10.3892/ijo.2018.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hao M, Yeo SK, Turner K, Harold A, Yang Y, Zhang X, Guan JL. Autophagy blockade limits HER2+ breast cancer tumorigenesis by perturbing HER2 trafficking and promoting release via small extracellular vesicles. Dev Cell. 2021;56:341–355.e345. doi: 10.1016/j.devcel.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 152.Deng S, Shanmugam MK, Kumar AP, Yap CT, Sethi G, Bishayee A. Targeting autophagy using natural compounds for cancer prevention and therapy. Cancer. 2019;125:1228–1246. doi: 10.1002/cncr.31978. [DOI] [PubMed] [Google Scholar]
- 153.Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 154.Akondi KB, Muttenthaler M, Dutertre S, Kaas Q, Craik DJ, Lewis RJ, Alewood PF. Discovery, synthesis, and structure-activity relationships of conotoxins. Chem Rev. 2014;114:5815–5847. doi: 10.1021/cr400401e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Olivera BM. Conus venom peptides: correlating chemistry and behavior. J Comp Physiol A. 1999;185:353–359. doi: 10.1007/s003590050394. [DOI] [PubMed] [Google Scholar]
- 156.Jiang H, Li L, Zhang J, Wan Z, Wang Y, Hou J, Yu Y. MiR-101–3p and Syn-Cal14.1a synergy in suppressing EZH2-induced progression of breast cancer. Oncol Targets Ther. 2020;13:9599–9609. doi: 10.2147/OTT.S264600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Maskey N, Li D, Xu H, Song H, Wu C, Hua K, Song J, Fang L. MicroRNA-340 inhibits invasion and metastasis by downregulating ROCK1 in breast cancer cells. Oncol Lett. 2017;14:2261–2267. doi: 10.3892/ol.2017.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mohammadi-Yeganeh S, Paryan M, Arefian E, Vasei M, Ghanbarian H, Mahdian R, Karimipoor M, Soleimani M. MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway. Tumour Biol. 2016;37:8993–9000. doi: 10.1007/s13277-015-4513-9. [DOI] [PubMed] [Google Scholar]
- 159.Shi Z, Li Y, Qian X, Hu Y, Liu J, Zhang S, Zhang J. MiR-340 inhibits triple-negative breast cancer progression by reversing EZH2 mediated miRNAs dysregulated expressions. J Cancer. 2017;8:3037–3048. doi: 10.7150/jca.19315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guan X, Gu S, Yuan M, Zheng X, Wu J. MicroRNA-33a-5p overexpression sensitizes triple-negative breast cancer to doxorubicin by inhibiting eIF5A2 and epithelial-mesenchymal transition. Oncol Lett. 2019;18:5986–5994. doi: 10.3892/ol.2019.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang C, Zhang Y, Ding W, Lin Y, Huang Z, Luo Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6:881–889. doi: 10.1007/s13238-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weihua Z, Guorong Z, Xiaolong C, Weizhan L. MiR-33a functions as a tumor suppressor in triple-negative breast cancer by targeting EZH2. Cancer Cell Int. 2020;20:85. doi: 10.1186/s12935-020-1160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hortobágyi G. Anthrazykline in der Krebstherapie: Ein Überblick. Drugs. 1997;54:1–7. [Google Scholar]
- 164.Yang F, Teves SS, Kemp CJ, Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Oo KK, Kamolhan T, Soni A, Thongchot S, Mitrpant C, Thuwajit C, Thuwajit P. Development of an engineered peptide antagonist against periostin to overcome doxorubicin resistance in breast cancer. BMC Cancer. 2021;21:65. doi: 10.1186/s12885-020-07761-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kubiliute R, Januskeviciene I, Urbanaviciute R, Daniunaite K, Drobniene M, Ostapenko V, Daugelavicius R, Jarmalaite S. Nongenotoxic ABCB1 activator tetraphenylphosphonium can contribute to doxorubicin resistance in MX-1 breast cancer cell line. Sci Rep. 2021;11:6556. doi: 10.1038/s41598-021-86120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Patel N, Garikapati KR, Pandita RK, Singh DK, Pandita TK, Bhadra U, Bhadra MP. miR-15a/miR-16 down-regulates BMI1, impacting Ub-H2A mediated DNA repair and breast cancer cell sensitivity to doxorubicin. Sci Rep. 2017;7:4263. doi: 10.1038/s41598-017-02800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Deng M, Zhang R, He Z, Qiu Q, Lu X, Yin J, Liu H, Jia X, He Z. TET-mediated sequestration of miR-26 drives EZH2 expression and gastric carcinogenesis. Cancer Res. 2017;77:6069–6082. doi: 10.1158/0008-5472.CAN-16-2964. [DOI] [PubMed] [Google Scholar]
- 169.Pan Y, Wu A, Xu F, Chen C, Jiang L, Jin R. Lentivirus-mediated overexpression of miR-124 suppresses growth and invasion by targeting JAG1 and EZH2 in gastric cancer. Oncol Lett. 2018;15:7450–7458. doi: 10.3892/ol.2018.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Martin TE, Powell CT, Wang Z, Bhattacharyya S, Walsh-Reitz MM, Agarwal K, Toback FG. A novel mitogenic protein that is highly expressed in cells of the gastric antrum mucosa. Am J Physiol Gastrointest Liver Physiol. 2003;285:G332–G343. doi: 10.1152/ajpgi.00453.2002. [DOI] [PubMed] [Google Scholar]
- 171.Xing R, Li W, Cui J, Zhang J, Kang B, Wang Y, Wang Z, Liu S, Lu Y. Gastrokine 1 induces senescence through p16/Rb pathway activation in gastric cancer cells. Gut. 2012;61:43–52. doi: 10.1136/gut.2010.230623. [DOI] [PubMed] [Google Scholar]
- 172.Toback FG, Walsh-Reitz MM, Musch MW, Chang EB, Del Valle J, Ren H, Huang E, Martin TE. Peptide fragments of AMP-18, a novel secreted gastric antrum mucosal protein, are mitogenic and motogenic. Am J Physiol Gastrointest Liver Physiol. 2003;285:G344–G353. doi: 10.1152/ajpgi.00455.2002. [DOI] [PubMed] [Google Scholar]
- 173.Yoon JH, Choi YJ, Choi WS, Ashktorab H, Smoot DT, Nam SW, Lee JY, Park WS. GKN1-miR-185-DNMT1 axis suppresses gastric carcinogenesis through regulation of epigenetic alteration and cell cycle. Clin Cancer Res. 2013;19:4599–4610. doi: 10.1158/1078-0432.CCR-12-3675. [DOI] [PubMed] [Google Scholar]
- 174.Zheng W, Li ZY, Zhao DL, Li XL, Liu R. microRNA-26a directly targeting MMP14 and MMP16 inhibits the cancer cell proliferation, migration and invasion in cutaneous squamous cell carcinoma. Cancer Manag Res. 2020;12:7087–7095. doi: 10.2147/CMAR.S265775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 175.Cheng C, Guo L, Ma Y, Wang Z, Fan X, Shan Z. Up-Regulation of miR-26a-5p inhibits E2F7 to regulate the progression of renal carcinoma cells. Cancer Manag Res. 2020;12:11723–11733. doi: 10.2147/CMAR.S271710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhao WT, Lin XL, Liu Y, Han LX, Li J, Lin TY, Shi JW, Wang SC, Lian M, Chen HW, et al. miR-26a promotes hepatocellular carcinoma invasion and metastasis by inhibiting PTEN and inhibits cell growth by repressing EZH2. Lab Invest. 2019;99:1484–1500. doi: 10.1038/s41374-019-0270-5. [DOI] [PubMed] [Google Scholar]
- 177.Xu T, Yan S, Wang M, Jiang L, Ma P, Lu B, Chen Q, Wei C, Wang Z. LncRNA UCA1 induces acquired resistance to gefitinib by epigenetically silencing CDKN1A expression in non-small-cell lung cancer. Front Oncol. 2020;10:656. doi: 10.3389/fonc.2020.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Huang B, Huang M, Li Q. MiR-137 suppresses migration and invasion by targeting EZH2-STAT3 signaling in human hepatocellular carcinoma. Pathol Res Pract. 2018;214:1980–1986. doi: 10.1016/j.prp.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 179.Jiang Q, Lei YH, Krishnadath DC, Zhu BY, Zhou XW. Curcumin regulates EZH2/Wnt/β-Catenin pathway in the mandible and femur of ovariectomized osteoporosis rats. Kaohsiung J Med Sci. 2021. [DOI] [PubMed]
- 180.Zhang JJ, Chen JT, Hua L, Yao KH, Wang CY. miR-98 inhibits hepatocellular carcinoma cell proliferation via targeting EZH2 and suppressing Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;85:472–478. doi: 10.1016/j.biopha.2016.11.053. [DOI] [PubMed] [Google Scholar]
- 181.Wang G, Sun Y, He Y, Ji C, Hu B, Sun Y. miR-26a promoted by interferon-alpha inhibits hepatocellular carcinoma proliferation and migration by blocking EZH2. Genet Test Mol Biomarkers. 2015;19:30–36. doi: 10.1089/gtmb.2014.0245. [DOI] [PubMed] [Google Scholar]
- 182.Ashrafizadeh M, Zarrabi A, Hushmandi K, Kalantari M, Mohammadinejad R, Javaheri T, Sethi G. Association of the epithelial–mesenchymal transition (EMT) with cisplatin resistance. Int J Mol Sci. 2020;21:4002. doi: 10.3390/ijms21114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zeng T, Luo L, Huang Y, Ye X, Lin J. Upregulation of miR-138 increases sensitivity to cisplatin in hepatocellular carcinoma by regulating EZH2. Biomed Res Int. 2021;2021:6665918. doi: 10.1155/2021/6665918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Singh AK, Verma A, Singh A, Arya RK, Maheshwari S, Chaturvedi P, Nengroo MA, Saini KK, Vishwakarma AL, Singh K, et al. Salinomycin inhibits epigenetic modulator EZH2 to enhance death receptors in colon cancer stem cells. Epigenetics. 2021;16:144–161. doi: 10.1080/15592294.2020.1789270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang Z, Wu X, Li J. miR-101 suppresses colon cancer cell migration through regulation of EZH2. Rev Esp Enferm Dig. 2020. [DOI] [PubMed]
- 186.Jiang M, Xu B, Li X, Shang Y, Chu Y, Wang W, Chen D, Wu N, Hu S, Zhang S, et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene. 2019;38:301–316. doi: 10.1038/s41388-018-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Burkhart DL, Morel KL, Wadosky KM, Labbé DP, Galbo PM, Dalimov Z, Xu B, Loda M, Ellis L. Evidence that EZH2 deregulation is an actionable therapeutic target for prevention of prostate cancer. Cancer Prev Res (Phila) 2020;13:979–988. doi: 10.1158/1940-6207.CAPR-20-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jin L, Zhou Y, Chen G, Dai G, Fu K, Yang D, Zhu J. EZH2-TROAP pathway promotes prostate cancer progression via TWIST signals. Front Oncol. 2020;10:592239. doi: 10.3389/fonc.2020.592239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pan MZ, Song YL, Gao F. MiR-605-3p inhibits malignant progression of prostate cancer by up-regulating EZH2. Eur Rev Med Pharmacol Sci. 2019;23:8795–8805. doi: 10.26355/eurrev_201910_19274. [DOI] [PubMed] [Google Scholar]
- 190.Zhang H, Liang F, Yue J, Liu P, Wang J, Wang Z, Li H, Cheng D, Du J, Zhang K, Du P. MicroRNA-137 regulates hypoxia-mediated migration and epithelial-mesenchymal transition in prostate cancer by targeting LGR4 via the EGFR/ERK signaling pathway. Int J Oncol. 2020;57:540–549. doi: 10.3892/ijo.2020.5064. [DOI] [PubMed] [Google Scholar]
- 191.Zang Y, Zhu J, Li Q, Tu J, Li X, Hu R, Yang D. miR-137-3p modulates the progression of prostate cancer by regulating the JNK3/EZH2 axis. Oncol Targets Ther. 2020;13:7921–7932. doi: 10.2147/OTT.S256161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Uchiyama N, Tanaka Y, Kawamoto T. Aristeromycin and DZNeP cause growth inhibition of prostate cancer via induction of mir-26a. Eur J Pharmacol. 2017;812:138–146. doi: 10.1016/j.ejphar.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 193.Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: adaptive responses in the androgen axis. Cancer Treat Rev. 2014;40:426–433. doi: 10.1016/j.ctrv.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 194.Shi XB, Ma AH, Xue L, Li M, Nguyen HG, Yang JC, Tepper CG, Gandour-Edwards R, Evans CP, Kung HJ, deVere White RW. miR-124 and androgen receptor signaling inhibitors repress prostate cancer growth by downregulating androgen receptor splice variants, EZH2, and Src. Cancer Res. 2015;75:5309–5317. doi: 10.1158/0008-5472.CAN-14-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu L, Ning Y, Yi J, Yuan J, Fang W, Lin Z, Zeng Z. miR-6089/MYH9/β-catenin/c-Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomed Pharmacother. 2020;125:109865. doi: 10.1016/j.biopha.2020.109865. [DOI] [PubMed] [Google Scholar]
- 196.Gasca J, Flores ML, Jiménez-Guerrero R, Sáez ME, Barragán I, Ruíz-Borrego M, Tortolero M, Romero F, Sáez C, Japón MA. EDIL3 promotes epithelial-mesenchymal transition and paclitaxel resistance through its interaction with integrin α(V)β(3) in cancer cells. Cell Death Discov. 2020;6:86. doi: 10.1038/s41420-020-00322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Y, Ai H, Fan X, Chen S, Wang Y, Liu L. Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting miR-138-5p-regulated EZH2 and SIRT1. Biol Res. 2020;53:18. doi: 10.1186/s40659-020-00286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Huo X, Sun H, Qian Q, Ma X, Peng P, Yu M, Zhang Y, Yang J, Cao D, Gui T, Shen K. CYP27B1 downregulation: a new molecular mechanism regulating EZH2 in ovarian cancer tumorigenicity. Front Cell Dev Biol. 2020;8:561804. doi: 10.3389/fcell.2020.561804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liu L, Guo J, Yu L, Cai J, Gui T, Tang H, Song L, Wang J, Han F, Yang C, et al. miR-101 regulates expression of EZH2 and contributes to progression of and cisplatin resistance in epithelial ovarian cancer. Tumour Biol. 2014;35:12619–12626. doi: 10.1007/s13277-014-2585-6. [DOI] [PubMed] [Google Scholar]
- 200.Yu Z, Kim J, He L, Creighton CJ, Gunaratne PH, Hawkins SM, Matzuk MM. Functional analysis of miR-34c as a putative tumor suppressor in high-grade serous ovarian cancer. Biol Reprod. 2014;91:113. doi: 10.1095/biolreprod.114.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li Q, Wang Y, Hu R, Yang G. Dysregulation of SPRR3/miR-876-3p axis contributes to tumorigenesis in non-small-cell lung cancer. Oncol Targets Ther. 2020;13:2411–2419. doi: 10.2147/OTT.S245422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xu Y, Liu N, Wei Y, Zhou D, Lin R, Wang X, Shi B. Anticancer effects of miR-124 delivered by BM-MSC derived exosomes on cell proliferation, epithelial mesenchymal transition, and chemotherapy sensitivity of pancreatic cancer cells. Aging (Albany NY) 2020;12:19660–19676. doi: 10.18632/aging.103997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang W, Chen JH, Shan T, Aguilera-Barrantes I, Wang LS, Huang TH, Rader JS, Sheng X, Huang YW. miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation. Lab Invest. 2018;98:1397–1407. doi: 10.1038/s41374-018-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Białopiotrowicz E, Noyszewska-Kania M, Kachamakova-Trojanowska N, Łoboda A, Cybulska M, Grochowska A, Kopczyński M, Mikula M, Prochorec-Sobieszek M, Firczuk M, et al. Serine biosynthesis pathway supports MYC-miR-494-EZH2 feed-forward circuit necessary to maintain metabolic and epigenetic reprogramming of burkitt lymphoma cells. Cancers (Basel). 2020;12. [DOI] [PMC free article] [PubMed]
- 205.Zhou B, Wei E, Shi H, Huang J, Gao L, Zhang T, Wei Y, Ge B. MiR-26a inhibits cell proliferation and induces apoptosis in human bladder cancer through regulating EZH2 bioactivity. Int J Clin Exp Pathol. 2017;10:11234–11241. [PMC free article] [PubMed] [Google Scholar]
- 206.Yang Q, Zhao S, Shi Z, Cao L, Liu J, Pan T, Zhou D, Zhang J. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2021;40:120. doi: 10.1186/s13046-021-01901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ma J, Weng L, Wang Z, Jia Y, Liu B, Wu S, Cao Y, Sun X, Yin X, Shang M, Mao A. MiR-124 induces autophagy-related cell death in cholangiocarcinoma cells through direct targeting of the EZH2-STAT3 signaling axis. Exp Cell Res. 2018;366:103–113. doi: 10.1016/j.yexcr.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 208.Zhang T, Qian H, Hu C, Lu H, Li JB, Wu YF, Li W. MiR-26a mediates ultraviolet B-induced apoptosis by targeting histone methyltransferase EZH2 depending on Myc expression. Cell Physiol Biochem. 2017;43:1188–1197. doi: 10.1159/000481759. [DOI] [PubMed] [Google Scholar]
- 209.Varghese E, Samuel SM, Líšková A, Samec M, Kubatka P, Büsselberg D. Targeting glucose metabolism to overcome resistance to anticancer chemotherapy in breast cancer. Cancers. 2020;12:2252. doi: 10.3390/cancers12082252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tao T, Chen M, Jiang R, Guan H, Huang Y, Su H, Hu Q, Han X, Xiao J. Involvement of EZH2 in aerobic glycolysis of prostate cancer through miR-181b/HK2 axis. Oncol Rep. 2017;37:1430–1436. doi: 10.3892/or.2017.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tao T, Liu D, Liu C, Xu B, Chen S, Yin Y, Ang L, Huang Y, Zhang X, Chen M. Autoregulatory feedback loop of EZH2/miR-200c/E2F3 as a driving force for prostate cancer development. Biochim Biophys Acta. 2014;1839:858–865. doi: 10.1016/j.bbagrm.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 212.Bhalla SS, Robitaille L, Nemer M. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J Biol Chem. 2001;276:11439–11445. doi: 10.1074/jbc.M100208200. [DOI] [PubMed] [Google Scholar]
- 213.Lee MY, Lu A, Gudas LJ. Transcriptional regulation of Rex1 (zfp42) in normal prostate epithelial cells and prostate cancer cells. J Cell Physiol. 2010;224:17–27. doi: 10.1002/jcp.22071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Luo J, Zhou X, Ge X, Liu P, Cao J, Lu X, Ling Y, Zhang S. Upregulation of Y ing Y ang 1 (YY 1) suppresses esophageal squamous cell carcinoma development through heme oxygenase-1. Cancer Sci. 2013;104:1544–1551. doi: 10.1111/cas.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang Y, Tao T, Liu C, Guan H, Zhang G, Ling Z, Zhang L, Lu K, Chen S, Xu B, Chen M. Upregulation of miR-146a by YY1 depletion correlates with delayed progression of prostate cancer. Int J Oncol. 2017;50:421–431. doi: 10.3892/ijo.2017.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sun Z, Gao S, Xuan L, Liu X. Long non-coding RNA FEZF1-AS1 induced progression of ovarian cancer via regulating miR-130a-5p/SOX4 axis. J Cell Mol Med. 2020;24:4275–4285. doi: 10.1111/jcmm.15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lin L, Wang Z, Jin H, Shi H, Lu Z, Qi Z. MiR-212/132 is epigenetically downregulated by SOX4/EZH2-H3K27me3 feedback loop in ovarian cancer cells. Tumour Biol. 2016. [DOI] [PubMed]
- 218.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 219.Hu Y, Wu F, Liu Y, Zhao Q, Tang H. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin Epigenet. 2019;11:186. doi: 10.1186/s13148-019-0786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zeng J, Li YK, Quan FF, Zeng X, Chen CY, Zeng T, Zou J, Tong WJ. Propofol-induced miR-125a-5p inhibits the proliferation and metastasis of ovarian cancer by suppressing LIN28B. Mol Med Rep. 2020;22:1507–1517. doi: 10.3892/mmr.2020.11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Dos Santos MP, Pereira JN, De Labio RW, Carneiro LC, Pontes JC, Barbosa MS, Smith MAC, Payão SLM, Rasmussen LT. Decrease of miR-125a-5p in gastritis and gastric cancer and its possible association with H. pylori. J Gastrointest Cancer 2020. [DOI] [PubMed]
- 222.Xiong J, Tu Y, Feng Z, Li D, Yang Z, Huang Q, Li Z, Cao Y, Jie Z. Epigenetics mechanisms mediate the miR-125a/BRMS1 axis to regulate invasion and metastasis in gastric cancer. Oncol Targets Ther. 2019;12:7513–7525. doi: 10.2147/OTT.S210376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zlotnik A. New insights on the role of CXCR4 in cancer metastasis. J Pathol. 2008;215:211–213. doi: 10.1002/path.2350. [DOI] [PubMed] [Google Scholar]
- 224.Jung MJ, Rho JK, Kim YM, Jung JE, Jin YB, Ko YG, Lee JS, Lee SJ, Lee JC, Park MJ. Upregulation of CXCR4 is functionally crucial for maintenance of stemness in drug-resistant non-small cell lung cancer cells. Oncogene. 2013;32:209–221. doi: 10.1038/onc.2012.37. [DOI] [PubMed] [Google Scholar]
- 225.Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. 2015;6:8494. doi: 10.1038/ncomms9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen S, Pu J, Bai J, Yin Y, Wu K, Wang J, Shuai X, Gao J, Tao K, Wang G, Li H. EZH2 promotes hepatocellular carcinoma progression through modulating miR-22/galectin-9 axis. J Exp Clin Cancer Res. 2018;37:3. doi: 10.1186/s13046-017-0670-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Feng GX, Li J, Yang Z, Zhang SQ, Liu YX, Zhang WY, Ye LH, Zhang XD. Hepatitis B virus X protein promotes the development of liver fibrosis and hepatoma through downregulation of miR-30e targeting P4HA2 mRNA. Oncogene. 2017;36:6895–6905. doi: 10.1038/onc.2017.291. [DOI] [PubMed] [Google Scholar]
- 228.Wang LJ, Sun GZ, Chen YF. LncRNA MSTO2P promotes proliferation and autophagy of lung cancer cells by up-regulating EZH2 expression. Eur Rev Med Pharmacol Sci. 2019;23:3375–3382. doi: 10.26355/eurrev_201904_17701. [DOI] [PubMed] [Google Scholar]
- 229.Chien YC, Chen JN, Chen YH, Chou RH, Lee HC, Yu YL. Epigenetic silencing of miR-9 promotes migration and invasion by EZH2 in glioblastoma cells. Cancers (Basel). 2020;12. [DOI] [PMC free article] [PubMed]
- 230.Vinchure OS, Sharma V, Tabasum S, Ghosh S, Singh RP, Sarkar C, Kulshreshtha R. Polycomb complex mediated epigenetic reprogramming alters TGF-β signaling via a novel EZH2/miR-490/TGIF2 axis thereby inducing migration and EMT potential in glioblastomas. Int J Cancer. 2019;145:1254–1269. doi: 10.1002/ijc.32360. [DOI] [PubMed] [Google Scholar]
- 231.Jiang X, Hu C, Arnovitz S, Bugno J, Yu M, Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun. 2016;7:11452. doi: 10.1038/ncomms11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yin H, Wang Y, Wu Y, Zhang X, Zhang X, Liu J, Wang T, Fan J, Sun J, Yang A, Zhang R. EZH2-mediated epigenetic silencing of miR-29/miR-30 targets LOXL4 and contributes to tumorigenesis, metastasis, and immune microenvironment remodeling in breast cancer. Theranostics. 2020;10:8494–8512. doi: 10.7150/thno.44849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Dou D, Ge X, Wang X, Xu X, Zhang Z, Seng J, Cao Z, Gu Y, Han M. EZH2 contributes to cisplatin resistance in breast cancer by epigenetically suppressing miR-381 expression. Oncol Targets Ther. 2019;12:9627–9637. doi: 10.2147/OTT.S214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ihira K, Dong P, Xiong Y, Watari H, Konno Y, Hanley SJ, Noguchi M, Hirata N, Suizu F, Yamada T, et al. EZH2 inhibition suppresses endometrial cancer progression via miR-361/Twist axis. Oncotarget. 2017;8:13509–13520. doi: 10.18632/oncotarget.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Miele E, Po A, Mastronuzzi A, Carai A, Besharat ZM, Pediconi N, Abballe L, Catanzaro G, Sabato C, De Smaele E, et al. Downregulation of miR-326 and its host gene β-arrestin1 induces pro-survival activity of E2F1 and promotes medulloblastoma growth. Mol Oncol. 2021;15:523–542. doi: 10.1002/1878-0261.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xu S, Li X, Li L, Wang Y, Geng C, Guo F, Zhang T, Du A, Lu Z, Hui H, Wang Q. CTCF-silenced miR-137 contributes to EMT and radioresistance in esophageal squamous cell carcinoma. Cancer Cell Int. 2021;21:155. doi: 10.1186/s12935-020-01740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Roscigno G, Scognamiglio I, Ingenito F, Chianese RV, Palma F, Chan A, Condorelli G. Modulating the crosstalk between the tumor and the microenvironment using SiRNA: a flexible strategy for breast cancer treatment. Cancers. 2020;12:3744. doi: 10.3390/cancers12123744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mirzaei S, Mahabady MK, Zabolian A, Abbaspour A, Fallahzadeh P, Noori M, Hashemi F, Hushmandi K, Daneshi S, Kumar AP, et al. Small interfering RNA (siRNA) to target genes and molecular pathways in glioblastoma therapy: current status with an emphasis on delivery systems. Life Sci. 2021;275:119368. doi: 10.1016/j.lfs.2021.119368. [DOI] [PubMed] [Google Scholar]
- 239.Anguela XM, High KA. Entering the modern era of gene therapy. Annu Rev Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 240.Mainini F, Eccles MR. Lipid and polymer-based nanoparticle siRNA delivery systems for cancer therapy. Molecules. 2020;25:2692. doi: 10.3390/molecules25112692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hattab D, Bakhtiar A. Bioengineered siRNA-based nanoplatforms targeting molecular signaling pathways for the treatment of triple negative breast cancer: preclinical and clinical advancements. Pharmaceutics. 2020;12:929. doi: 10.3390/pharmaceutics12100929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang HF, Yang H, Hu LB, Lei YH, Qin Y, Li J, Bi CW, Wang JS, Huo Q. Effect of siRNA targeting EZH2 on cell viability and apoptosis of bladder cancer T24 cells. Genet Mol Res. 2014;13:9939–9950. doi: 10.4238/2014.November.27.23. [DOI] [PubMed] [Google Scholar]
- 243.Wen Y, Hou Y, Yi X, Sun S, Guo J, He X, Li T, Cai J, Wang Z. EZH2 activates CHK1 signaling to promote ovarian cancer chemoresistance by maintaining the properties of cancer stem cells. Theranostics. 2021;11:1795–1813. doi: 10.7150/thno.48101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhou W, Wang J, Man WY, Zhang QW, Xu WG. siRNA silencing EZH2 reverses cisplatin-resistance of human non-small cell lung and gastric cancer cells. Asian Pac J Cancer Prev. 2015;16:2425–2430. doi: 10.7314/apjcp.2015.16.6.2425. [DOI] [PubMed] [Google Scholar]
- 245.Wang X, Hua Y, Xu G, Deng S, Yang D, Gao X. Targeting EZH2 for glioma therapy with a novel nanoparticle-siRNA complex. Int J Nanomed. 2019;14:2637–2653. doi: 10.2147/IJN.S189871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yuan ZQ, Chen WL, You BG, Liu Y, Yang SD, Li JZ, Zhu WJ, Zhou XF, Liu C, Zhang XN. Multifunctional nanoparticles co-delivering EZH2 siRNA and etoposide for synergistic therapy of orthotopic non-small-cell lung tumor. J Control Release. 2017;268:198–211. doi: 10.1016/j.jconrel.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 247.Yu C, Ding B, Zhang X, Deng X, Deng K, Cheng Z, Xing B, Jin D, Ma P, Lin J. Targeted iron nanoparticles with platinum-(IV) prodrugs and anti-EZH2 siRNA show great synergy in combating drug resistance in vitro and in vivo. Biomaterials. 2018;155:112–123. doi: 10.1016/j.biomaterials.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 248.McMahon KM, Plebanek MP, Thaxton CS. Properties of native high-density lipoproteins inspire synthesis of actively targeted in vivo siRNA delivery vehicles. Adv Funct Mater. 2016;26:7824–7835. doi: 10.1002/adfm.201602600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Acharya R. The recent progresses in shRNA-nanoparticle conjugate as a therapeutic approach. Mater Sci Eng C. 2019;104:109928. doi: 10.1016/j.msec.2019.109928. [DOI] [PubMed] [Google Scholar]
- 250.Wu Y, Yu J, Liu Y, Yuan L, Yan H, Jing J, Xu G. Delivery of EZH2-shRNA with mPEG-PEI nanoparticles for the treatment of prostate cancer in vitro. Int J Mol Med. 2014;33:1563–1569. doi: 10.3892/ijmm.2014.1724. [DOI] [PubMed] [Google Scholar]
- 251.Xu SG, Yu JJ, Shi Q, Niu Q, Guo Z, Guo BY, Zhou GC, Gu X, Wu YX. Conditionally replicative adenovirus carrying shRNA targeting EZH2 inhibits prostate cancer growth and invasion. Oncol Rep. 2019;42:273–282. doi: 10.3892/or.2019.7157. [DOI] [PubMed] [Google Scholar]
- 252.Ong MS, Cai W, Yuan Y, Leong HC, Tan TZ, Mohammad A, You ML, Arfuso F, Goh BC, Warrier S, et al. 'Lnc'-ing Wnt in female reproductive cancers: therapeutic potential of long non-coding RNAs in Wnt signalling. Br J Pharmacol. 2017;174:4684–4700. doi: 10.1111/bph.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Javed Z, Khan K, Sadia H, Raza S, Salehi B, Sharifi-Rad J, Cho WC. LncRNA & Wnt signaling in colorectal cancer. Cancer Cell Int. 2020;20:1–10. doi: 10.1186/s12935-020-01412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mishra S, Verma SS, Rai V, Awasthee N, Chava S, Hui KM, Kumar AP, Challagundla KB, Sethi G, Gupta SC. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell Mol Life Sci. 2019;76:1947–1966. doi: 10.1007/s00018-019-03053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Jiang N, Zhang X, Gu X, Li X, Shang L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021;7:1–11. doi: 10.1038/s41420-021-00407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, Tai S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 258.Yang Z, Jiang S, Shang J, Jiang Y, Dai Y, Xu B, Yu Y, Liang Z, Yang Y. LncRNA: shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res Rev. 2019;52:17–31. doi: 10.1016/j.arr.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 259.Liao K, Xu J, Yang W, You X, Zhong Q, Wang X. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol Immunol. 2018;101:182–188. doi: 10.1016/j.molimm.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 260.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Can Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shermane Lim YW, Xiang X, Garg M, Le MTN, Li-Ann Wong A, Wang L, Goh B-C. The double-edged sword of H19 lncRNA: Insights into cancer therapy. Cancer Lett. 2021;500:253–262. doi: 10.1016/j.canlet.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 262.Bhardwaj V, Tan YQ, Wu MM, Ma L, Zhu T, Lobie PE, Pandey V. Long non-coding RNAs in recurrent ovarian cancer: theranostic perspectives. Cancer Lett. 2021;502:97–107. doi: 10.1016/j.canlet.2020.12.042. [DOI] [PubMed] [Google Scholar]
- 263.Wu M, Zhang X, Han X, Pandey V, Lobie PE, Zhu T. The potential of long noncoding RNAs for precision medicine in human cancer. Cancer Lett. 2021;501:12–19. doi: 10.1016/j.canlet.2020.11.040. [DOI] [PubMed] [Google Scholar]
- 264.Shen C, Yang C, Xia B, You M. Long non-coding RNAs: emerging regulators for chemo/immunotherapy resistance in cancer stem cells. Cancer Lett. 2021;500:244–252. doi: 10.1016/j.canlet.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 265.Barth DA, Juracek J, Slaby O, Pichler M, Calin GA. lncRNA and mechanisms of drug resistance in cancers of the genitourinary system. Cancers. 2020;12:2148. doi: 10.3390/cancers12082148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Han T-S, Hur K, Cho H-S, Ban HS. Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers. 2020;12:2622. doi: 10.3390/cancers12092622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hu W, Wang T, Yang Y, Zheng S. Tumor heterogeneity uncovered by dynamic expression of long noncoding RNA at single-cell resolution. Cancer Genet. 2015;208:581–586. doi: 10.1016/j.cancergen.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 268.Pandya G, Kirtonia A, Sethi G, Pandey AK, Garg MJ. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. 2020;1874:188423. [DOI] [PubMed]
- 269.Du Q, Chen J. SNHG1 promotes proliferation, migration and invasion of bladder cancer cells via the PI3K/AKT signaling pathway. Exp Ther Med. 2020;20:110. doi: 10.3892/etm.2020.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Pei ML, Zhao ZX, Shuang T. Dysregulation of lnc-SNHG1 and miR-216b-5p correlate with chemoresistance and indicate poor prognosis of serous epithelial ovarian cancer. J Ovarian Res. 2020;13:144. doi: 10.1186/s13048-020-00750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, Xu X, Xu T, Hu X, Sun L, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17:141. doi: 10.1186/s12943-018-0894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Xiang W, Lyu L, Huang T, Zheng F, Yuan J, Zhang C, Jiang G. The long non-coding RNA SNHG1 promotes bladder cancer progression by interacting with miR-143-3p and EZH2. J Cell Mol Med. 2020;24:11858–11873. doi: 10.1111/jcmm.15806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Yao X, Lan Z, Lai Q, Li A, Liu S, Wang X. LncRNA SNHG6 plays an oncogenic role in colorectal cancer and can be used as a prognostic biomarker for solid tumors. J Cell Physiol. 2020;235:7620–7634. doi: 10.1002/jcp.29672. [DOI] [PubMed] [Google Scholar]
- 274.Liu J, Liu X, Li R. LncRNA SNHG6 enhances the radioresistance and promotes the growth of cervical cancer cells by sponging miR-485-3p. Cancer Cell Int. 2020;20:424. doi: 10.1186/s12935-020-01448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X, Pan B, Xu T, Sun L, He B, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12:3. doi: 10.1186/s13045-018-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wang W, Ge L, Xu XJ, Yang T, Yuan Y, Ma XL, Zhang XH. LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the miR-144-3p/EZH2 axis. Radiol Oncol. 2019;53:434–442. doi: 10.2478/raon-2019-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. 2018;15:238–250. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lv L, He L, Chen S, Yu Y, Che G, Tao X, Wang S, Jian Z, Zhang X. Long non-coding RNA LINC00114 facilitates colorectal cancer development through EZH2/DNMT1-induced miR-133b suppression. Front Oncol. 2019;9:1383. doi: 10.3389/fonc.2019.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wu J, Sun L, Liu T, Dong G. Ultrasound-targeted microbubble destruction-mediated downregulation of EZH2 inhibits stemness and epithelial-mesenchymal transition of liver cancer stem cells. Onco Targets Ther. 2021;14:221–237. doi: 10.2147/OTT.S269589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The Lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging Mir-382. Cell Physiol Biochem. 2017;42:2145–2158. doi: 10.1159/000479990. [DOI] [PubMed] [Google Scholar]
- 281.Cai J, Sun H, Zheng B, Xie M, Xu C, Zhang G, Huang X, Zhuang J. Curcumin attenuates lncRNA H19-induced epithelial-mesenchymal transition in tamoxifen-resistant breast cancer cells. Mol Med Rep. 2021;23:1. doi: 10.3892/mmr.2020.11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhou C, Hu C, Wang B, Fan S, Jin W. Curcumin suppresses cell proliferation, migration, and invasion through modulating miR-21-5p/SOX6 axis in hepatocellular carcinoma. Cancer Biother Radiopharm. 2020. [DOI] [PubMed]
- 283.Ashrafizadeh M, Zarrabi A, Hashemi F, Zabolian A, Saleki H, Bagherian M, Azami N, Bejandi AK, Hushmandi K, Ang HL, et al. Polychemotherapy with curcumin and doxorubicin via biological nanoplatforms: enhancing antitumor activity. Pharmaceutics. 2020;12. [DOI] [PMC free article] [PubMed]
- 284.Moballegh Nasery M, Abadi B, Poormoghadam D, Zarrabi A, Keyhanvar P, Khanbabaei H, Ashrafizadeh M, Mohammadinejad R, Tavakol S, Sethi G. Curcumin delivery mediated by bio-based nanoparticles: a review. Molecules. 2020, 25. [DOI] [PMC free article] [PubMed]
- 285.Tewari D, Nabavi SF, Nabavi SM, Sureda A, Farooqi AA, Atanasov AG, Vacca RA, Sethi G, Bishayee A. Targeting activator protein 1 signaling pathway by bioactive natural agents: possible therapeutic strategy for cancer prevention and intervention. Pharmacol Res. 2018;128:366–375. doi: 10.1016/j.phrs.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 286.Yoshida K, Toden S, Ravindranathan P, Han H, Goel A. Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis. 2017;38:1036–1046. doi: 10.1093/carcin/bgx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zhu B, Cui H, Xu W. Hydrogen inhibits the proliferation and migration of gastric cancer cells by modulating lncRNA MALAT1/miR-124-3p/EZH2 axis. Cancer Cell Int. 2021;21:70. doi: 10.1186/s12935-020-01743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Fan T, Sun N, He J. Exosome-derived lncRNAs in lung cancer. Front Oncol. 2020;10:1728. doi: 10.3389/fonc.2020.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 291.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [DOI] [PMC free article] [PubMed]
- 293.Piao HY, Guo S, Wang Y, Zhang J. Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin Transl Oncol. 2021;23:246–256. doi: 10.1007/s12094-020-02412-9. [DOI] [PubMed] [Google Scholar]
- 294.Li Q, Wang X, Jiang N, Xie X, Liu N, Liu J, Shen J, Peng T. Exosome-transmitted linc00852 associated with receptor tyrosine kinase AXL dysregulates the proliferation and invasion of osteosarcoma. Cancer Med. 2020;9:6354–6366. doi: 10.1002/cam4.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zang X, Gu J, Zhang J, Shi H, Hou S, Xu X, Chen Y, Zhang Y, Mao F, Qian H, et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11:215. doi: 10.1038/s41419-020-2409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Zhong G, Lin Y, Wang X, Wang K, Liu J, Wei W. H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-130a-3p/SATB1 in breast cancer cells. Oncol Targets Ther. 2020;13:12501–12513. doi: 10.2147/OTT.S280142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Chen MJ, Deng J, Chen C, Hu W, Yuan YC, Xia ZK. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol. 2019;113:27–36. doi: 10.1016/j.biocel.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 298.Xiong T, Huang C, Li J, Yu S, Chen F, Zhang Z, Zhuang C, Li Y, Zhuang C, Huang X, et al. LncRNA NRON promotes the proliferation, metastasis and EMT process in bladder cancer. J Cancer. 2020;11:1751–1760. doi: 10.7150/jca.37958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25:637–644. doi: 10.1080/1061186X.2017.1307379. [DOI] [PubMed] [Google Scholar]
- 300.Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J, Shi Y, Ren J, Ji G, Wang K. A novel lncRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther Nucleic Acids. 2018;12:684–697. doi: 10.1016/j.omtn.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wei S, Liu J, Li X, Liu X. Repression of lncRNA-SVUGP2 mediated by EZH2 contributes to the development of non-small cell lung cancer via brisking Wnt/β-catenin signal. Artif Cells Nanomed Biotechnol. 2019;47:3400–3409. doi: 10.1080/21691401.2019.1648279. [DOI] [PubMed] [Google Scholar]
- 302.Lian Y, Xiao C, Yan C, Chen D, Huang Q, Fan Y, Li Z, Xu H. Knockdown of pseudogene derived from lncRNA DUXAP10 inhibits cell proliferation, migration, invasion, and promotes apoptosis in pancreatic cancer. J Cell Biochem. 2018;119:3671–3682. doi: 10.1002/jcb.26578. [DOI] [PubMed] [Google Scholar]
- 303.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC) Biomed Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 304.Wang XY, Jian X, Sun BQ, Ge XS, Huang FJ, Chen YQ. LncRNA ROR1-AS1 promotes colon cancer cell proliferation by suppressing the expression of DUSP5/CDKN1A. Eur Rev Med Pharmacol Sci. 2020;24:1116–1125. doi: 10.26355/eurrev_202002_20162. [DOI] [PubMed] [Google Scholar]
- 305.Xu Y, Yao Y, Jiang X, Zhong X, Wang Z, Li C, Kang P, Leng K, Ji D, Li Z, et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37:81. doi: 10.1186/s13046-018-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zeng XY, Jiang XY, Yong JH, Xie H, Yuan J, Zeng D, Dou YY, Xiao SS. lncRNA ABHD11-AS1, regulated by the EGFR pathway, contributes to the ovarian cancer tumorigenesis by epigenetically suppressing TIMP2. Cancer Med. 2019;8:7074–7085. doi: 10.1002/cam4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 307.Dai ZY, Jin SM, Luo HQ, Leng HL, Fang JD. LncRNA HOTAIR regulates anoikis-resistance capacity and spheroid formation of ovarian cancer cells by recruiting EZH2 and influencing H3K27 methylation. Neoplasma. 2021. [DOI] [PubMed]
- 308.Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol Ther Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Xu JL, Hua T, Ding J, Fan Y, Liu ZJ, Lian JW. FOXF2 aggravates the progression of non-small cell lung cancer through targeting lncRNA H19 to downregulate PTEN. Eur Rev Med Pharmacol Sci. 2019;23:10796–10802. doi: 10.26355/eurrev_201912_19782. [DOI] [PubMed] [Google Scholar]
- 310.Kim CY, Oh JH, Lee JY, Kim MH. The LncRNA HOTAIRM1 promotes tamoxifen resistance by mediating HOXA1 expression in ER+ breast cancer cells. J Cancer. 2020;11:3416–3423. doi: 10.7150/jca.38728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Zhang W, Huang X, Shi J. EZH2-mediated lncRNA ABHD11-AS1 promoter regulates the progression of ovarian cancer by targeting miR-133a-3p. Anticancer Drugs. 2021;32:269–277. doi: 10.1097/CAD.0000000000001039. [DOI] [PubMed] [Google Scholar]
- 312.Cheng W, Shi X, Lin M, Yao Q, Ma J, Li J. LncRNA MAGI2-AS3 overexpression sensitizes esophageal cancer cells to irradiation through down-regulation of HOXB7 via EZH2. Front Cell Dev Biol. 2020;8:552822. doi: 10.3389/fcell.2020.552822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhou Y, Yang H, Xia W, Cui L, Xu R, Lu H, Xue D, Tian Z, Ding T, Cao Y, et al. LncRNA MEG3 inhibits the progression of prostate cancer by facilitating H3K27 trimethylation of EN2 through binding to EZH2. J Biochem. 2020;167:295–301. doi: 10.1093/jb/mvz097. [DOI] [PubMed] [Google Scholar]
- 314.Yang ZY, Yang F, Zhang YL, Liu B, Wang M, Hong X, Yu Y, Zhou YH, Zeng H. LncRNA-ANCR down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017;18:95–104. doi: 10.3233/CBM-161715. [DOI] [PubMed] [Google Scholar]
- 315.Li Z, Dong M, Fan D, Hou P, Li H, Liu L, Lin C, Liu J, Su L, Wu L, et al. LncRNA ANCR down-regulation promotes TGF-β-induced EMT and metastasis in breast cancer. Oncotarget. 2017;8:67329–67343. doi: 10.18632/oncotarget.18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Xie C, Guo Y, Lou S. LncRNA ANCR promotes invasion and migration of gastric cancer by regulating FoxO1 expression to inhibit macrophage M1 polarization. Dig Dis Sci. 2020;65:2863–2872. doi: 10.1007/s10620-019-06019-1. [DOI] [PubMed] [Google Scholar]
- 317.Wang S, Lan F, Xia Y. lncRA ANCR inhibits non-small cell lung cancer cell migration and invasion by inactivating TGF-β pathway. Med Sci Monit. 2018;24:6002–6009. doi: 10.12659/MSM.911492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Li Z, Hou P, Fan D, Dong M, Ma M, Li H, Yao R, Li Y, Wang G, Geng P, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71. doi: 10.1038/cdd.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Jiang J, Bai J, Qin T, Wang Z, Han L. NGF from pancreatic stellate cells induces pancreatic cancer proliferation and invasion by PI3K/AKT/GSK signal pathway. J Cell Mol Med. 2020;24:5901–5910. doi: 10.1111/jcmm.15265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ong PS, Wang LZ, Dai X, Tseng SH, Loo SJ, Sethi G. Judicious toggling of mTOR activity to combat insulin resistance and cancer: current evidence and perspectives. Front Pharmacol. 2016;7:395. doi: 10.3389/fphar.2016.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Chen S, He Z, Zhu C, Liu Y, Li L, Deng L, Wang J, Yu C, Sun C. TRIM37 mediates chemoresistance and maintenance of stemness in pancreatic cancer cells via ubiquitination of PTEN and activation of the AKT-GSK-3β-β-catenin signaling pathway. Front Oncol. 2020;10:554787. doi: 10.3389/fonc.2020.554787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Zhi H, Lian J. LncRNA BDNF-AS suppresses colorectal cancer cell proliferation and migration by epigenetically repressing GSK-3β expression. Cell Biochem Funct. 2019;37:340–347. doi: 10.1002/cbf.3403. [DOI] [PubMed] [Google Scholar]
- 323.Yan P, Lu JY, Niu J, Gao J, Zhang MQ, Yin Y, Shen X. LncRNA Platr22 promotes super-enhancer activity and stem cell pluripotency. J Mol Cell Biol. 2020. [DOI] [PMC free article] [PubMed]
- 324.Jung J, Kim S, An HT, Ko J. α-Actinin-4 regulates cancer stem cell properties and chemoresistance in cervical cancer. Carcinogenesis. 2020;41:940–949. doi: 10.1093/carcin/bgz168. [DOI] [PubMed] [Google Scholar]
- 325.Iwamoto K, Takahashi H, Okuzaki D, Osawa H, Ogino T, Miyoshi N, Uemura M, Matsuda C, Yamamoto H, Mizushima T, et al. Syntenin-1 promotes colorectal cancer stem cell expansion and chemoresistance by regulating prostaglandin E2 receptor. Br J Cancer. 2020;123:955–964. doi: 10.1038/s41416-020-0965-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Chen S, Huang L, Li G, Qiu F, Wang Y, Yang C, Pan J, Wu Z, Chen J, Tian Y. LncRNA STXBP5-AS1 suppresses stem cell-like properties of pancreatic cancer by epigenetically inhibiting neighboring androglobin gene expression. Clin Epigenetics. 2020;12:168. doi: 10.1186/s13148-020-00961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Li F, Xu Y, Xu X, Ge S, Zhang F, Zhang H, Fan X. lncRNA HotairM1 depletion promotes self-renewal of cancer stem cells through HOXA1-nanog regulation loop. Mol Ther Nucleic Acids. 2020;22:456–470. doi: 10.1016/j.omtn.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ma ZH, Shuai Y, Gao XY, Yan Y, Wang KM, Wen XZ, Ji JF. BTEB2-activated lncRNA TSPEAR-AS2 drives GC progression through suppressing GJA1 expression and upregulating CLDN4 expression. Mol Ther Nucleic Acids. 2020;22:1129–1141. doi: 10.1016/j.omtn.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wu H, Hu Y, Liu X, Song W, Gong P, Zhang K, Chen Z, Zhou M, Shen X, Qian Y, Fan H. LncRNA TRERNA1 function as an enhancer of SNAI1 promotes gastric cancer metastasis by regulating epithelial–mesenchymal transition. Mol Ther Nucleic Acids. 2017;8:291–299. doi: 10.1016/j.omtn.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 332.Yang J, Lian Y, Yang R, Lian Y, Wu J, Liu J, Wang K, Xu H. Upregulation of lncRNA LINC00460 facilitates GC progression through epigenetically silencing CCNG2 by EZH2/LSD1 and indicates poor outcomes. Mol Ther Nucleic Acids. 2020;19:1164–1175. doi: 10.1016/j.omtn.2019.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 333.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 334.Chen RY, Ju Q, Feng LM, Yuan Q, Zhang L. The carcinogenic complex lncRNA FOXP4-AS1/EZH2/LSD1 accelerates proliferation, migration and invasion of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23:8371–8376. doi: 10.26355/eurrev_201910_19148. [DOI] [PubMed] [Google Scholar]
- 335.Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, Xu W, Niu Y, Cheng L, Maity SN, et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10:2571. doi: 10.1038/s41467-019-09784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, Zhang J, Huang H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6:41045–41055. doi: 10.18632/oncotarget.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chen M, Xia Z, Chen C, Hu W, Yuan Y. LncRNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anticancer Drugs. 2018;29:767–773. doi: 10.1097/CAD.0000000000000645. [DOI] [PubMed] [Google Scholar]
- 338.You BH, Yoon JH, Kang H, Lee EK, Lee SK, Nam JW. HERES, a lncRNA that regulates canonical and noncanonical Wnt signaling pathways via interaction with EZH2. Proc Natl Acad Sci USA. 2019;116:24620–24629. doi: 10.1073/pnas.1912126116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X, Meng H, Guan X, Yang K, Bai Y. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150. doi: 10.1186/s12943-017-0715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Lian Y, Li Z, Fan Y, Huang Q, Chen J, Liu W, Xiao C, Xu H. The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex promotes cell proliferation in pancreatic cancer. Am J Transl Res. 2017;9:5496–5506. [PMC free article] [PubMed] [Google Scholar]
- 341.Hui B, Ji H, Xu Y, Wang J, Ma Z, Zhang C, Wang K, Zhou Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019;10:207. doi: 10.1038/s41419-019-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Zhou X, Gao W, Hua H, Ji Z. LncRNA-BLACAT1 facilitates proliferation, migration and aerobic glycolysis of pancreatic cancer cells by repressing CDKN1C via EZH2-induced H3K27me3. Front Oncol. 2020;10:539805. doi: 10.3389/fonc.2020.539805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37:265. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Tang J, Yu B, Li Y, Zhang W, Alvarez AA, Hu B, Cheng SY, Feng H. TGF-β-activated lncRNA LINC00115 is a critical regulator of glioma stem-like cell tumorigenicity. EMBO Rep. 2019;20:e48170. doi: 10.15252/embr.201948170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zeng SHG, Xie JH, Zeng QY, Dai SHH, Wang Y, Wan XM, Liu JCH. lncRNA PVT1 promotes metastasis of non-small cell lung cancer through EZH2-mediated activation of hippo/NOTCH1 signaling pathways. Cell J. 2021;23:21–31. doi: 10.22074/cellj.2021.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sun Z, He C, Xiao M, Wei B, Zhu Y, Zhang G, Zhou H, Yuan J, Hu X, Yi Y. LncRNA FOXC2 antisense transcript accelerates non-small-cell lung cancer tumorigenesis via silencing p15. Am J Transl Res. 2019;11:4552–4560. [PMC free article] [PubMed] [Google Scholar]
- 347.Li Y, Ma HY, Hu XW, Qu YY, Wen X, Zhang Y, Xu QY. LncRNA H19 promotes triple-negative breast cancer cells invasion and metastasis through the p53/TNFAIP8 pathway. Cancer Cell Int. 2020;20:200. doi: 10.1186/s12935-020-01261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Qiu C, Li S, Sun D, Yang S. lncRNA PVT1 accelerates progression of non-small cell lung cancer via targeting miRNA-526b/EZH2 regulatory loop. Oncol Lett. 2020;19:1267–1272. doi: 10.3892/ol.2019.11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Xiao Q, Zheng F, Tang Q, Wu JJ, Xie J, Huang HD, Yang XB, Hann SS. Repression of PDK1- and LncRNA HOTAIR-mediated EZH2 gene expression contributes to the enhancement of atractylenolide 1 and erlotinib in the inhibition of human lung cancer cells. Cell Physiol Biochem. 2018;49:1615–1632. doi: 10.1159/000493497. [DOI] [PubMed] [Google Scholar]
- 350.Qian L, Fei Q, Zhang H, Qiu M, Zhang B, Wang Q, Yu Y, Guo C, Ren Y, Mei M, et al. lncRNA HOTAIR promotes dna repair and radioresistance of breast cancer via EZH2. DNA Cell Biol. 2020. [DOI] [PubMed]
- 351.Yu G, Zhou H, Yao W, Meng L, Lang B. lncRNA TUG1 promotes cisplatin resistance by regulating CCND2 via epigenetically silencing miR-194-5p in bladder cancer. Mol Ther Nucleic Acids. 2019;16:257–271. doi: 10.1016/j.omtn.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, Wang M, Huang C, Wang L, Zeng F, Jiang G. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 353.Zhu F, Zhang X, Yu Q, Han G, Diao F, Wu C, Zhang Y. LncRNA AWPPH inhibits SMAD4 via EZH2 to regulate bladder cancer progression. J Cell Biochem. 2018;119:4496–4505. doi: 10.1002/jcb.26556. [DOI] [PubMed] [Google Scholar]
- 354.Liu Y, Sun J, Yu J, Ge W, Xiao X, Dai S, Xiang Q. LncRNA CACS15 accelerates the malignant progression of ovarian cancer through stimulating EZH2-induced inhibition of APC. Am J Transl Res. 2019;11:6561–6568. [PMC free article] [PubMed] [Google Scholar]
- 355.Lin H, Guo Q, Lu S, Chen J, Li X, Gong M, Tang L, Wen J. LncRNA SUMO1P3 promotes proliferation and inhibits apoptosis in colorectal cancer by epigenetically silencing CPEB3. Biochem Biophys Res Commun. 2019;511:239–245. doi: 10.1016/j.bbrc.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 356.Gong A, Huang Z, Ge H, Cai Y, Yang C. The carcinogenic complex lncRNA DUXAP8/EZH2/LSD1 accelerates the proliferation, migration and invasion of colorectal cancer. J buon. 2019;24:1830–1836. [PubMed] [Google Scholar]
- 357.Lian Y, Yan C, Ding J, Xia R, Ma Z, Hui B, Ji H, Zhou J, Wang K. A novel lncRNA, LL22NC03-N64E9.1, represses KLF2 transcription through binding with EZH2 in colorectal cancer. Oncotarget. 2017;8:59435–59445. doi: 10.18632/oncotarget.19738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Xie JJ, Li WH, Li X, Ye W, Shao CF. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33:331–343. [PubMed] [Google Scholar]
- 359.Xue W, Wang F, Han P, Liu Y, Zhang B, Gu X, Wang Y, Li M, Zhao Y, Cui B. The oncogenic role of LncRNA FAM83C-AS1 in colorectal cancer development by epigenetically inhibits SEMA3F via stabilizing EZH2. Aging (Albany NY) 2020;12:20396–20412. doi: 10.18632/aging.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Li Y, Liu X, Cui X, Tan Y, Wang Q, Wang Y, Xu C, Fang C, Kang C. LncRNA PRADX-mediated recruitment of PRC2/DDX5 complex suppresses UBXN1 expression and activates NF-κB activity, promoting tumorigenesis. Theranostics. 2021;11:4516–4530. doi: 10.7150/thno.54549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Han Y, Chen M, Wang A, Fan X. STAT3-induced upregulation of lncRNA CASC11 promotes the cell migration, invasion and epithelial-mesenchymal transition in hepatocellular carcinoma by epigenetically silencing PTEN and activating PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2019;508:472–479. doi: 10.1016/j.bbrc.2018.11.092. [DOI] [PubMed] [Google Scholar]
- 362.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Jiang B, Yang B, Wang Q, Zheng X, Guo Y, Lu W. lncRNA PVT1 promotes hepatitis B virus-positive liver cancer progression by disturbing histone methylation on the c-Myc promoter. Oncol Rep. 2020;43:718–726. doi: 10.3892/or.2019.7444. [DOI] [PubMed] [Google Scholar]
- 364.Qu X, Li Y, Wang L, Yuan N, Ma M, Chen Y. LncRNA SNHG8 accelerates proliferation and inhibits apoptosis in HPV-induced cervical cancer through recruiting EZH2 to epigenetically silence RECK expression. J Cell Biochem. 2020;121:4120–4129. doi: 10.1002/jcb.29646. [DOI] [PubMed] [Google Scholar]
- 365.Song H, Liu Y, Jin X, Liu Y, Yang Y, Li L, Wang X, Li G. Long non-coding RNA LINC01535 promotes cervical cancer progression via targeting the miR-214/EZH2 feedback loop. J Cell Mol Med. 2019;23:6098–6111. doi: 10.1111/jcmm.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124:649–658. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 367.Liu B, Zhao N, Zhou Y, Lu Y, Chen W, Huang Z, Wang D, Xu Y, Yam JWP, Cui Y. Circular RNA circ_ABCB10 in cancer. Clinica Chimica Acta. 2021. [DOI] [PubMed]
- 368.Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer. 2020;19:1–18. doi: 10.1186/s12943-019-1121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Patop IL, Wüst S, Kadener S. Past, present, and future of circ RNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Hsu M-T, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 372.Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, Li H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 373.Li Z, Chen X, Xu D, Li S, Chan MT, Wu WK. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52:e12704. doi: 10.1111/cpr.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Chen L-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 377.Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2017;14:1000–1006. doi: 10.1080/15476286.2016.1227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14:1007–1017. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, Tu J, Zhi Z, Qin Y, Sun LN, Li JM. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer. 2020;19:164. doi: 10.1186/s12943-020-01272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Zhang X, Lu N, Wang L, Wang Y, Li M, Zhou Y, Yan H, Cui M, Zhang M, Zhang L. Circular RNAs and esophageal cancer. Cancer Cell Int. 2020;20:362. doi: 10.1186/s12935-020-01451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Cai Z, Li H. Circular RNAs and bladder cancer. Oncol Targets Ther. 2020;13:9573–9586. doi: 10.2147/OTT.S268859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, Dai X, Zhou H, Zhu J, Zhang H, Jiang Y. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y, Ma Z, Chen Y. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111:2824–2836. doi: 10.1111/cas.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Li R, Jiang J, Shi H, Qian H, Zhang X, Xu WJC, Sciences ML. CircRNA: a rising star in gastric cancer. 2020;77:1661–80. [DOI] [PMC free article] [PubMed]
- 387.Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother. 2018;102:1188–1194. doi: 10.1016/j.biopha.2018.03.085. [DOI] [PubMed] [Google Scholar]
- 388.Yang X, Wang J, Li H, Sun Y, Tong X. Downregulation of hsa_circ_0026123 suppresses ovarian cancer cell metastasis and proliferation through the miR-124-3p/EZH2 signaling pathway. Int J Mol Med. 2021;47:668–676. doi: 10.3892/ijmm.2020.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Sun S, Gao J, Zhou S, Li Y, Wang Y, Jin L, Li J, Liu B, Zhang B, Han S, et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2020;39:252. doi: 10.1186/s13046-020-01779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi: 10.1016/j.gene.2019.144099. [DOI] [PubMed] [Google Scholar]
- 391.Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8:1387–1402. [PMC free article] [PubMed] [Google Scholar]
- 392.Ma X, Lv L, Xing C. Circ_ 0115744 acts as miR-144 sponge to promote and predict the metastasis of colorectal cancer. Aging (Albany NY) 2021;13:5892–5905. doi: 10.18632/aging.202513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Sha J, Xia L, Han Q, Chi C, Zhu Y, Pan J, Huang Y, Xia W, Dong B, Xue W, Yang C. Downregulation of circ-TRPS1 suppressed prostatic cancer prognoses by regulating miR-124-3p/EZH2 axis-mediated stemness. Am J Cancer Res. 2020;10:4372–4385. [PMC free article] [PubMed] [Google Scholar]
- 394.Zhang Z, Shi Z, Zhang S, Lu Q, Wei H, Wu X, Han L. Upregulated hsa_circ_0000129 expression promotes proliferation and migration of breast cancer cells. Oncol Lett. 2021;21:239. doi: 10.3892/ol.2021.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Kiran AVR, Kumari GK, Krishnamurthy PT, Khaydarov RR. Tumor microenvironment and nanotherapeutics: intruding the tumor fort. 2021. [DOI] [PubMed]
- 396.Rai S, Inoue H, Sakai K, Hanamoto H, Matsuda M, Maeda Y, Haeno T, Watatani Y, Kumode T, Serizawa K, et al. Decreased expression of T-cell-associated immune markers predicts poor prognosis in patients with follicular lymphoma. Cancer Sci. 2021. [DOI] [PMC free article] [PubMed]
- 397.Karantanos T, Chistofides A, Barhdan K, Li L, Boussiotis VA. Regulation of T cell differentiation and function by EZH2. Front Immunol. 2016;7:172. doi: 10.3389/fimmu.2016.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.He S, Liu Y, Meng L, Sun H, Wang Y, Ji Y, Purushe J, Chen P, Li C, Madzo J, et al. Ezh2 phosphorylation state determines its capacity to maintain CD8(+) T memory precursors for antitumor immunity. Nat Commun. 2017;8:2125. doi: 10.1038/s41467-017-02187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Wang X, Brea LT, Yu J. Immune modulatory functions of EZH2 in the tumor microenvironment: implications in cancer immunotherapy. Am J Clin Exp Urol. 2019;7:85–91. [PMC free article] [PubMed] [Google Scholar]
- 400.Liu Y, Li L, Song X. Exosomal circPVT1 derived from lung cancer promotes the progression of lung cancer by targeting miR-124-3p/EZH2 axis and regulating macrophage polarization. Cell Cycle. 2022:1–17. [DOI] [PMC free article] [PubMed]
- 401.Kim H-J, Cantor H, Cosmopoulos K. Overcoming immune checkpoint blockade resistance via EZH2 inhibition. Trends Immunol. 2020;41:948–963. doi: 10.1016/j.it.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 402.Zhao J, Li H, Zhao S, Wang E, Zhu J, Feng D, Zhu Y, Dou W, Fan Q, Hu J, et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol Cancer. 2021;20:46. doi: 10.1186/s12943-021-01343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Long H, Xiang T, Luo J, Li F, Lin R, Liu S, Jiang S, Hu C, Chen G, Wong E, et al. The tumor microenvironment disarms CD8(+) T lymphocyte function via a miR-26a-EZH2 axis. Oncoimmunology. 2016;5:e1245267. doi: 10.1080/2162402X.2016.1245267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Ma Z, Wang LZ, Cheng J-T, Lam WST, Ma X, Xiang X, Wong AL-A, Goh BC, Gong Q, Sethi GJA. Targeting Hypoxia-Inducible Factor-1-Mediated Metastasis for Cancer Therapy. 2021;34:1484–97. [DOI] [PubMed]
- 405.Yu L, Li J, Peng B, Cai P, Zhao B, Chen Y, Zhu H. CircASXL1 knockdown restrains hypoxia-induced DDP resistance and NSCLC progression by sponging miR-206. Cancer Manag Res. 2021;13:5077–5089. doi: 10.2147/CMAR.S276964. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 406.Wang X, Wang Y, Li L, Xue X, Xie H, Shi H, Hu Y. A lncRNA coordinates with Ezh2 to inhibit HIF-1α transcription and suppress cancer cell adaption to hypoxia. Oncogene. 2020;39:1860–1874. doi: 10.1038/s41388-019-1123-9. [DOI] [PubMed] [Google Scholar]
- 407.Luzón-Toro B, Fernández RM, Martos-Martínez JM, Rubio-Manzanares-Dorado M, Antiñolo G, Borrego S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Sci Rep. 2019;9:14374. doi: 10.1038/s41598-019-50913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.He X, Wang J, Chen J, Han L, Lu X, Miao D, Yin D, Geng Q, Zhang E. lncRNA UCA1 predicts a poor prognosis and regulates cell proliferation and migration by repressing p21 and SPRY1 expression in GC. Mol Ther Nucleic Acids. 2019;18:605–616. doi: 10.1016/j.omtn.2019.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Qi X, Lin Y, Liu X, Chen J, Shen B. Biomarker discovery for the carcinogenic heterogeneity between colon and rectal cancers based on lncRNA-associated ceRNA network analysis. Front Oncol. 2020;10:535985. doi: 10.3389/fonc.2020.535985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Wu B, Yuan Y, Han X, Wang Q, Shang H, Liang X, Jing H, Cheng W. Structure of LINC00511-siRNA-conjugated nanobubbles and improvement of cisplatin sensitivity on triple negative breast cancer. FASEB J. 2020;34:9713–9726. doi: 10.1096/fj.202000481R. [DOI] [PubMed] [Google Scholar]
- 411.Yan C, Wei S, Han D, Wu L, Tan L, Wang H, Dong Y, Hua J, Yang W. LncRNA HULC shRNA disinhibits miR-377-5p to suppress the growth and invasion of hepatocellular carcinoma in vitro and hepatocarcinogenesis in vivo. Ann Transl Med. 2020;8:1294. doi: 10.21037/atm-20-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Delfi M, Sartorius R, Ashrafizadeh M, Sharifi E, Zhang Y, De Berardinis P, Zarrabi A, Varma RS, Tay FR, Smith BR. Self-assembled peptide and protein nanostructures for anti-cancer therapy: targeted delivery, stimuli-responsive devices and immunotherapy. Nano Today. 2021;38:101119. doi: 10.1016/j.nantod.2021.101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Ashrafizade M, Delfi M, Hashemi F, Zabolian A, Saleki H, Bagherian M, Azami N, Farahani MV, omid Sharifzadeh S, Hamzehlou S. Biomedical application of chitosan-based nanoscale delivery systems: potential usefulness in siRNA delivery for cancer therapy. Carbohydr Polym. 2021:117809. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.