Abstract
Ethyl carbamate is a group of 2A carcinogen ubiquitously existed in fermented foods. The monitoring of its residues was important for evaluating the potential risk to human beings. Immunoassays with good accuracy and simplicity are great analytical tools for small molecule contaminants. However, it is typically confined in a competitive mode for small molecules with drawback of the sensitivity curbing. In this work, three different phages displayed peptides with capability of identifying the xanthyl ethyl carbamate immunocomplex were isolated from phage library. The binding mechanism of peptides and immunocomplex was studied by computer-assisted simulation. Results indicated that the xanthydrol group of xanthyl ethyl carbamate and the Asn-32 and Asn-92 residues of the antibody light chain were mainly responsible for binding. Simultaneously, a sensitive non-competitive immunoassay for detecting ethyl carbamate in wine samples was developed. The established method exhibited a limit of detection of 5.4 ng/mL and a linear range from 8.7 ng/mL to 32 ng/mL for wine samples. In comparison with the conventional competitive immunoassay, the sensitivity of the proposed non-competitive immunoassay was improved by 17-fold. The results of the immunoassay were validated by a standard ultra-performance liquid chromatography-quadrupole/orbitrap high-resolution mass spectrometry, which illustrated good reliability of the proposed assay.
Keywords: Non-competitive ELISA, Immune complex, Computer simulation, Peptide displayed phage, Ethyl carbamate
1. Introduction
Small molecules contaminants (molecular weight less than 1000 Da) such as toxins and pesticides in environment and food may cause a series of public safety issues (Chen et al., 2020; Li et al., 2019; Liu et al., 2020). Hence, the reports of analytical methods for monitoring the small molecules contaminants showed remarkable growing in recent years (Chamkasem et al., 2013; Lehotay et al., 2010; Liu et al., 2020; Saito-Shida et al., 2020). Among all kinds of detection methods, immunoassays with advantages of sensitivity, rapidness, high-throughput and simplicity are considered as a promising test tool (Cheng et al., 2019; Fu et al., 2018; Li et al., 2018; Zhang et al., 2017; Zhang et al., 2018). Technically, sandwich-type non-competitive immunoassay in two site binding format is a preferred strategy with better sensitivity. The detection can be achieved upon reagent excess, owing to measured signal only from the occupancy binding sites (Akter et al., 2016; Vanrell et al., 2012). Moreover, double sites recognition can identify analytes more accurately (Farka et al., 2018; Ji et al., 2020; Ricciardi et al., 2018). Nevertheless, most of the small molecules will be buried into the pockets of antibody deeply and resulted in insufficient space for binding with a secondary antibody. Thus, the immunoassay for small molecules is typically confined in a competitive mode (Bu et al., 2019; Fu et al., 2020). Therefore, researchers have made some attempts to implement detection of small molecule analytes by non-competitive format, such as antibody variable region-based open sandwich immunoassay (Cong et al., 2019; Kanai et al., 2019; Ohmuro-Matsuyama and Ueda, 2018), anti-metatype antibody-based immunoassay (Akter et al., 2019); (Arola et al., 2017), anti-idiotypic antibody-based immunoassay (Mares et al., 1995) and Giraudi’s method (Acharya and Dhar, 2008). However, the difficulty of antibody production, case-specific dependency and preparation of appropriate blocking reagent have limited their application. The small peptide loops displayed on the coat protein of M13 phage was able to recognize conformational change of antibody binding pocket after binding with target. It has become an appropriate substitution of producing anti-immune complex antibody. Up to now, several immunoassays by using phage displayed peptides for the determination of atrazine (González-Techera et al., 2007), phenoxybenzoic acid (González-Techera et al., 2007), clomazone (Rossotti et al., 2010), malachite green (Dong et al., 2014) and brominated diphenyl ether 47 (Hua et al., 2015) have been developed. However, the identification of key amino acid residues of phage displayed peptide was obtained generally by analysis of the consensus motif of the large amount positive sequences. For the case of a few positive phages obtained, it is difficult to forecast effective binding sites and analyze the reason for activity difference.
Ethyl carbamate (EC, C2H5OCONH2, Mwt: 89.1 Da) exists ubiquitously in daily fermented foods. It could induce cancer in mammals due to the genotoxicity (Gowd et al., 2018; Miller et al., 2003). It is categorized as probably carcinogenic to humans (group 2A) by the International Agency for Research on Cancer. Many countries such as Japan, France, Canada and the United States have set the maximum residue level (MRL) of EC ranging from 15 to 1000 ng/mL in different alcoholic beverages (Ajtony et al., 2013; Tu et al., 2018). Notably, as red wine is one of the most popular alcoholic beverages worldwide, the United States has set the MRL of EC at 15 ng/mL for red wine. It is a challenge to detect such low concentration of EC in red wine due to the occurrence of interferences. Previously, we prepared polyclonal and monoclonal antibodies against xanthyl derivatives of EC (xanthyl ethyl carbamate, XEC) and developed competitive ELISAs for EC (Luo et al., 2017; Luo et al., 2018). However, the dilution of wine sample to eliminate matrix effects resulted in loss of sensitivity. Consequently, the sensitivity of traditional competitive immunoassay methods could not meet the requirement of EC screening in red wine samples unless complicated ratiometric fluorescence approach was involved (Luo et al., 2018). As an alternative, ultra-sensitive non-competitive immunoassay mode may provide the possibility for achieving the rapid screening of EC in a large number of wine samples.
In this work, we first obtained three different anti XEC-monoclonal antibody immune complex binding peptides via phage display selection technique. We systematically investigated the difference of recognition between the obtained peptides and immune complex by molecular modeling and computer simulation techniques. Based on the peptide with the best performance, non-competitive immunoassay was developed for analyzing EC in wine samples. The results of proposed method were then validated by a standard ultra-performance liquid chromatography-quadrupole/orbitrap high-resolution mass spectrometry (UPLC-Q-Orbitrap HRMS).
2. Materials and methods
2.1. Reagents
Ethyl carbamate, methyl carbamate and acrylamide were purchased from Aladdin Chemical Technology Co., Ltd. (Shanghai, China). XEC and anti-XEC monoclonal antibody (mAb) were produced in our laboratory (Luo et al., 2018). Bovine serum albumin (BSA), 9-xanthydrol, Tween 20, polyethylene glycol 8000 (PEG 8000), isopropyl-β-D-thiogalactopyranoside (IPTG), Xgal and 3, 3′, 5, 5′-tetramethylbenzidine (TMB) were supplied by Sigma (Shanghai, China). Ph.D.-C7C Peptide Library Kit, ER2738 host strain and 96 gIII sequencing primer (5’-CCC TCA TAG TTA GCG TAA CG-3’) were obtained from New England Biolabs, Inc. (Ipswich, MA, USA). Mouse anti-M13 mAb-HRP was purchased from GE Healthcare (Piscataway, NJ, U.S.A.). Tryptone and yeast extract were supplied by Fisher Scientific Co. (Fair Lawn, NJ, U.S.A.).
2.2. Biopanning
The procedure of phage biopanning (Fig. 1) was carried out following the manufacturer’s instructions with some modifications. Briefly, three wells of plate were coated with 100 μL of purified anti-XEC mAb (10 μg/mL) in phosphate buffered saline (PBS, 0.01 mol/mL) at 4 °C overnight. After two times of washing with PBST (PBS solution with 0.1% Tween-20), plate was subsequently blocked by 3% skimmed milk (in PBS) at 37 °C for 1 h. Then, 100 μL of XEC (10 μg/mL) was added to form the XEC mAb immune complex (XEC-mAb). After 1 h incubation at 37 °C, the excess XEC was removed by rinsing with 300 μL of cold PBS twice. The library was diluted into concentration of 1010 pfu/mL using PBS containing 5% skimmed milk and then dispensed into three microtiter wells (100 μL/well). After incubating for 2 h at 4 °C, the three wells were washed 10 times with cold PBST (0.1% Tween-20) and 10 times with cold PBS to ensure the removal of unbound phage. The 0.2 mol/L glycine-HCl solution (pH 2.2) was added to 96-well plate (100 μL/well) with gently shaking for 10 min to elute bound phages twice. Then 4.5 μL of Tris-HCl solution (1 mol/L, pH 9.1) was immediately added for neutralization. To avoid the eluted phage of clones adsorbing uncombined antibody, a postselection-adsorption step was included. Briefly, the eluted solution was incubated in the wells coated with 10 μg/mL uncombined antibody and 5% BSA at 37 °C for 1 h. The eluted solution (600 μL) was collected for amplification and titration. For amplification, the collected eluent was used to infect 20 mL E. coli ER2738 (OD600=0.05) and incubated at 37 °C with vigorous aeration (250 rpm) for 4.5 h. Phage was first obtained from precipitating supernatant with 20% PEG 8000 (v/v) contained 2.5 mol/L NaCl in ice bath overnight after ER2738 culture centrifugation (12, 000 rpm for 10 min at 4°C). With re-centrifugation in the same conditions, phages were collected with 1 mL of PBS and then titrated using E. coli ER2738 (OD600=0.5). After amplification, the obtained phage was subsequently applied for the next panning round. In the following two rounds of panning, the mAb coating concentration was used with 5 and 1 μg/mL respectively to remove weak binder, as well as other procedures. After the third round biopanning was finished, individual blue plaques were picked from titrated plates to test the XEC-mAb immune complex binding activity.
Fig. 1.
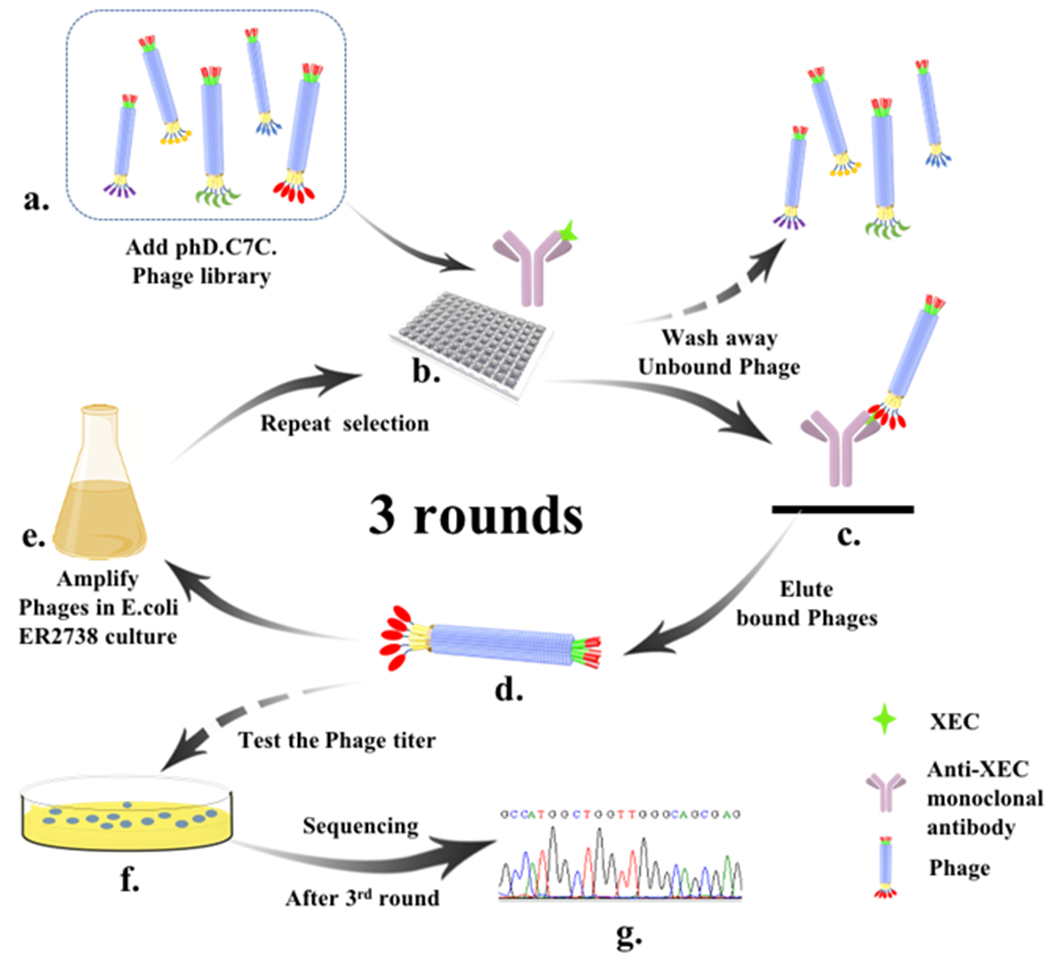
Schematic of biopanning of XEC-mAb immune complex binding peptides using the Ph.D.-C7C phage library. (a) Dilute library to 1010 pfu/mL. (b) The phage library is added into plate and incubated with immune complex. (c) Immune complex-bound phages remain in the well after the 20 times washing and the unbound phages phages are removed. (d) The immune complex-binding phages are eluted from immunecomplex upon glycine-HCl. (e) The eluted phages are amplified by infecting the E. coli ER2738 then as a sub-phage library for the following round selection (b→c→d→e). (f) Titer the eluted phages. (g) After 3rd round, positive phages were identified by sequencing with the primer.
2.3. Screening of phage eluate for positive clones by phage ELISA.
To screen for the positive clones, diluted phage elutes of the third panning (10 μL) was added to 200 μL of E. coli ER2738 (OD600=0.5). Then, the cells were dispensed to 3 mL of top agar (45 °C) and immediately spread on a prepared LB/IPTG/Xgal plate with incubation overnight at 37 °C. Twenty random blue plaques were picked and amplified by 1 mL of E. coli ER2738 (OD600=0.05). After 10-min centrifugation at 12000 rpm, the obtained supernatants (50 μL/well) and an equal volume of 1 μg/mL XEC in PBS or PBS itself were added into 10 μg/mL of mAb-coated wells. Simultaneously, the ability of non-specific binding of each clone was detected by 1 μg/mL of BSA. After 1 h incubation at room temperature, the plate was washed seven times with 0.05% PBST. Then 100 μL of anti-M13 phage antibody-HRP (diluted 5000-fold with PBST) was added to each well for 30 min incubation at 37 °C. After five-time washes, TMB substrate buffer was added (100 μL/well) for 10-min incubation at 37 °C. Finally, the absorbance (450 nm) was measured after the reaction was terminated with 50 μL of H2SO4 (10%, v/v) for each well. The selected positive clones should possess both high binding ability to the immune complex-coated wells and weak binding ability to antibody and BSA. The DNA sequence of the target peptide displayed by positive phages was identified by sequencing with the primer-96 gIII.
2.4. The Selection of phage-borne peptide for non-competitive phage ELISA
To choose the phage particle with the best performance, the XEC was diluted with PBS to 10 ng/mL and added into wells (50 μL/well) coated with the anti-XEC mAb (10, 5 and 2 μg/mL). Then serial dilutions of corresponding purified phage suspension were added subsequently. After incubation 1 h at room temperature and washing 7 times, the phage ELISA was established as described above. The phage-borne peptide with the best performance was selected and used to develop the non-competitive calibration curve at optimal working conditions.
2.5. Molecular docking and simulations
The fragment of antigen binding (Fab) model was built via homology modeling (details were shown in supporting information). Energy minimization of the obtained Fab was proceeded via molecular dynamics (MD) using Gromacs 2016.4 (http://www.gromacs.org/) software with the GROMOS96 43a2 force field. A cubic box for MD was created with a minimum distance of 1.5 Å from protein to the edge and the SPC solvent model was subsequently filled in the box. For production MD, the structure energy was minimized after 15 ns simulation (7500000 runs, 2 fs/run) at the temperature of 310 K. The molecular structures of XEC and peptides (ligands) were created via Gauss View 3.09 software (http://gaussian.com). LeadIT 2.1.8 software (http://www.biosolvet.de/LeadIT/) was used for the docking simulations of XEC and Fab by using Enthalpy and Entropy (hybrid approach) ligand binding setting (other settings were default). The obtained semi-flexible Fab-XEC-peptide docking complex was also analyzed by MD method as described above. All three-dimensional (3D) and two-dimensional (2D) maps were created by PyMOL (version 2.4.0a0, open-source) and Ligplot v.21 software respectively.
2.6. Sensitivity and specificity of non-competitive phage ELISA
Sensitivity was assessed by analyte producing 50% saturation of the signal (SC50), obtained from the calibration curve for detection of EC using phage-borne peptide. A serial of EC standards with different concentrations (0-20 ng/mL in 1% alcohol-PBS) were prepared via the following derivatization reaction (Fig. S1). First, the derivatizing agent 9-xanthydrol (0.4 mL of 4 mg/mL) solved in acetonitrile was mixed with 0.6 mL a serial concentration of EC standards. Then the derivatization was triggered using 0.1 mL of HCl (1.5 mol/L) and terminated with 0.1 mL of NaOH (1.5 mol/L) after 5-min reaction. The mixture was subsequently diluted 5-fold with PBS, which was used for phage ELISA analysis. Calibration curves were normalized as binding rate (B/B0) between the phage and antibody (or immunocomplex). The obtained calibration curves were calculated according to the formula: B/B0=(B-Bmin)/(Bmax-Bmin)×100% (B represents measured absorbance value, Bmax represents absorbance value at the maximum EC concentration, and Bmin represents absorbance value at no EC). The phage ELISA calibration curves were fitted with a four-parameter logistic function by plotting the binding rate (B/B0) against the logarithm of the EC concentration. The limit of detection (LOD) for assay was determined as SC10 with the lower and upper limits of quantitative concentration as SC20~SC80, respectively.
The structurally related EC compounds, acrylamide and methyl carbamate (Luo et al., 2017) were also derived to xanthyl acrylamide (XAA) and xanthyl methyl carbamate (XMC) for evaluating the specificity of the non-competitive assay. The concentration of SC50 of the curve was used to calculate the cross-reactivity (CR) of the assay. The CR formula as follows: CR (%)=SC50 (XEC)/SC50 (cross-reactive compounds) ×100.
2.7. Sample analysis and validation
Ten wine samples were purchased from several local supermarkets in Guangzhou, China. The samples were first analyzed using UPLC-Q-Orbitrap HRMS to obtain the background level of EC. One sample was chosen to perform the recovery test. Briefly, the sample was spiked with three concentrations of EC (10, 15 and 25 ng/mL). To eliminate the effect of alcoholic strength on derivatization, the samples were diluted 10-fold by distilled water (Luo et al. 2017). Then the samples were submitted for derivatization: 0.6 mL of sample was mixed with 0.4 mL of 9-xanthydrol (4 mg/mL, in acetonitrile), then 0.1 mL of HCl (1.5 mol/L) was added to trigger the reaction. After 5 min, the reaction was terminated by adding 0.1mL of NaOH (1.5 mol/L). After 5-fold dilution with PBS, the final mixture solution was used for non-competitive phage ELISA.
UPLC-Q-Orbitrap HRMS (Thermo Fisher) was performed according to the previous report (Wang et al., 2016). Before analysis, the wines were diluted 5-fold with distilled water and filtered through 0.22 μm microporous membrane. The detail of the procedure was described in Supporting Information. EC was analyzed by high-resolution mass spectrometry in positive mode.
3. Results and discussion
3.1. Biopanning the phage library with monoclonal antibody against XEC
The Ph.D.-C7C phage display peptide library was panned as described in section 2.2. The isolated phage-borne peptide for developing immunoassay should recognize the XEC-antibody immunocomplex region and simultaneously avoid nonspecific binding with other regions of mAb. Twenty clones were picked randomly and further analyzed in the presence or absence of XEC with mAb coated plates by phage ELISA. As shown in Fig. 2, 14 of the 20 clones exhibited strong binding affinity to the XEC-antibody immunocomplex and negligible affinity to the mAb and BSA (1 μg/mL). The results indicated that the phage-borne peptides could recognize the XEC-antibody immune complex specifically. After sequencing of the selected clones, three different types of sequences were obtained. The amino acid sequences (named as N.1, N.2, and N.3) are presented in Table 1.
Fig. 2.
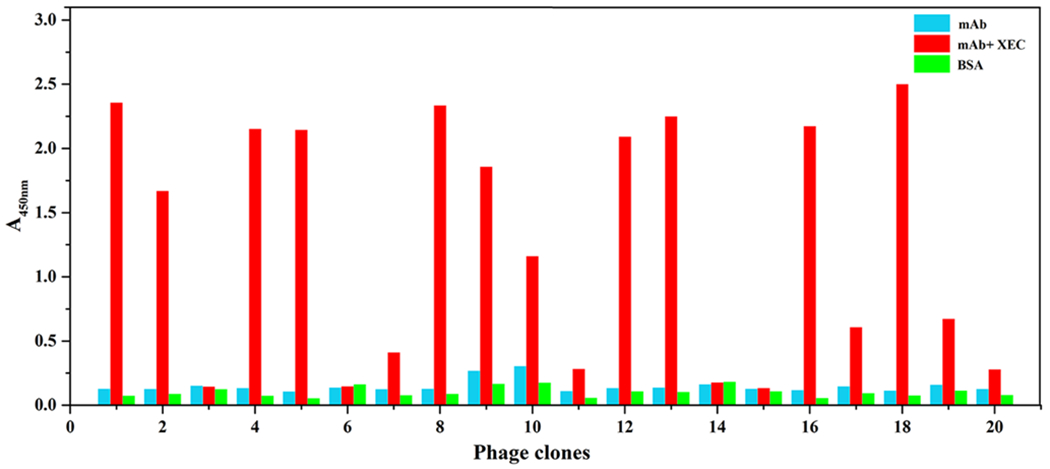
The results of positive phage clones screening obtained from phage ELISA with the mAb concentration of 10 μg/mL in the absence (blue) or presence (red) of XEC (1 μg/mL). Non-specific binding was evaluated upon 1 μg/mL BSA. Clones 1, 2, 4, 5, 7, 8, 9, 10, 12, 13, 16, 17, 18, 19 reacting specifically with the XEC-mAb immune complex shows negligible binding to the unbound antibody and BSA.
Table 1.
The peptide sequences from the isolation of XEC-mAb immunecomplex.
| Phage clone | Amino acid sequence |
|---|---|
| N.1 | CTMHLSVYC |
| N.2 | CTPRSANYC |
| N.3 | CEKMVATHC |
3.2. Selection of phage-borne peptides
Checkerboard procedure was carried out to determine the appropriate concentration of coating mAb and phage-borne peptides for developing sensitive assay. The obtained three representative phage-borne peptides (N.1, N.2 and N.3) with different sequences were serially diluted respectively. Then the peptides were added to the 96-well plates which coated with three concentration levels of mAb (10, 5 and 2 μg/mL) with the presence of XEC (10 ng/mL). Two common results (Fig. S2A~2C) were observed for all the three phages-borne peptides. One was that with a higher concentration of coated mAb, the signal differences produced by the presence and absence of XEC were all more obvious. Another was that the signal differences all gradually decreased when the phage dilution gradually increased. The maximal signal difference was observed at the high level of antibody concentration both for N.1 and N.2. The working affinity of N.1 and N.2 were evaluated to ~109 pfu/mL and ~1010 pfu/mL, respectively. As a result, N.1 was chosen and further estimated by making standard curves in PBS containing 1% ethanol. The highest sensitivity against XEC (SC50 1.6 ng/mL) was achieved with the phage-borne peptide N.1 of 2.3×109 pfu/mL and mAb concentration of 10 μg/mL, respectively (Fig. S2D). These concentrations were then applied for following non-competitive immunoassay of EC detection.
3.3. Analysis of interaction between immunocomplex and peptides
Computer-assisted simulation is an effective tool for identification of the interactions between protein and other molecules (Lape et al., 2010). The key amino acid residues of peptide for binding was first studied by computer-assisted simulation. The three-dimensional (3D) Fab model was created by SWISS MODEL (https://swissmodel.expasy.org/) and PyMOL (Chen et al., 2018). The results were showed in Fig. S3A and B. The model reliability for heavy chain and light chain were assessed with PROCHECK (http://services.mbi.ucla.edu/PROCHECK/). The Ramachandran plot map (Fig. S3C, D) showed that more than 90% of the residues of heavy chain and light chain located in the most favored and additional allowed region. However, atom overlap and the other unreasonable conformation problems resulted in the rough combined 3D model. Molecular dynamic (MD) is an effective method to obtain reasonable minimized conformation and thereby was applied to correct the unreasonable docking models. The root-mean-square deviation (RMSD, Fig. S4A) indicated that the conformation was minimized after 15 ns MD and the final confirmation of 3D Fab model was obtained (Fig. S4B). All the data suggested that the created 3D Fab model was reasonable.
In order to build Fab-XEC-peptide complex interaction models, the Fab model was used for XEC docking firstly (Fig. 3A1-2, the details of analysis were in Supporting information). Three peptide models were applied for the following docking with the prepared Fab-XEC complex docking model and minimized by MD. After 15 ns MD, the conformation reached equilibrium (Fig. S5) and their docking maps were show in Fig. 3B, C and D. For the Fab-XEC-peptide N.1 complex model (Fig. 3B1-3), the length of the hydrogen bond between Tyr-94L and NH group of XEC was shorter (1.5 Å, Fig. 3B2) than that of XEC docking (Fig. 3A2). Meanwhile, xanthydrol group of XEC formed a 2.4 Å-long hydrogen bond to the oxygen atom of and the NH group of Cys-9P. The residues of peptide N.1 including Cys-1P, Thr-2P, Ser-6P, Val-7P and Tyr-8P wrapped one of the benzene rings of xanthydrol group and thereby formed strong hydrophobic interaction. Therefore, the xanthydrol group was the main binding site for peptide N.1 to antibody-XEC complex, while hydrophobic interaction was the main binding interaction. By contrast, hydrophobic interaction did not play a leading role in the interaction between Fab and peptide N.1 (Fig. 3B3). Instead of that, the binding activity was mainly contributed by hydrogen bond from Fab. The oxygen atom of amide between Thr-2P and Met-3P formed a 2.4 Å and 1.7 Å-long hydrogen bond to the side chain amidogen (ND2) of Asn-32L and Asn-92L, respectively (Fig. 3B3). Meanwhile, the side chain hydroxyl (OG) of Ser-6P formed a 2.4 Å-long hydrogen bond to Asn-32L. The three hydrogen bonds between Fab and peptide N.1 may provide a strong binding affinity. Nevertheless, according to the results of non-competitive ELISA (Fig. 2), the absence of XEC significantly decreased the binding activity of peptide N.1 to antibody, indicating that the xanthydrol group was the determinant binding site for peptide N.1. For peptide N.2 (Fig. 3C1-3), only a 2.4 Å-long hydrogen bond was formed between the oxygen atom of amide of Asn-92L and the amidogen (NE) of side chain of Arg-4P (Fig. 3C3). Meanwhile, eight residues of peptide N.2 formed hydrophobic interaction to Fab, which was more than the six residues of peptide N.1. For peptide N.3, Thr-7P and Cys-1P formed hydrogen bonds to Tyr-91L, Asn-58H and Asp-99H, respectively, and eleven residues formed hydrophobic interaction to Fab, which was more than that of peptide N.1 and N.2. However, no significant blank background value was observed from the three peptides in the non-competitive ELISA. The results indicated that the number difference of hydrogen bond and hydrophobic interaction residue between Fab and the peptides did not mainly influence their binding activities in ELISA. The main reason may be that the interaction region between the Fab and the three peptides was not mainly in the CDR pocket. Peptides only covered the surface of antibody near the CDR, and did not form a reasonable docking conformation. Hence, the key binding site between the peptides and antibody-XEC complex is the XEC molecule. For the interaction between peptide N.2 and XEC, the amidogen of Asn-7P side chain formed a shorter hydrogen bond to oxygen atom of xanthydrol than peptide N.1 (Fig. 3C2). However, the ELISA results showed that the binding activity of peptide N.2 to antibody-XEC complex was lower than that of peptide N.1. It showed that hydrogen bond was not the main factor that influence the docking activity. Only two residues (Ala-6P and Tyr-8P) of peptide N.2 formed hydrophobic interaction to xanthydrol of XEC. Few residues resulted in a weaker hydrophobic interaction and therefore causing a lower binding activity of peptide N.2 to antibody-XEC complex. In the peptide N.3 docking model (Fig. 3D1-3), no hydrogen bond was formed between XEC and peptide. Two residues (Lys-3P and His-8P) of peptide N.3 formed hydrophobic interaction to xanthydrol of XEC, just like the situation of peptide N.2. However, the hydrophobic interactions for N. 3 were not strong enough to provide a bind activity due to the higher polarity of the two residues than that of peptide N.2. All the above analysis explained that the weakest hydrophobic interaction resulted the lowest affinity of peptide N.3 to immunocomplex. In summary, the xanthydrol group of XEC was the key binding site for peptide docking and mainly contributed to hydrophobic interaction. The different activities of the three peptides were clearly explained, which providing guidance for subsequent evolution to change the peptide performance for detecting EC in different samples.
Fig. 3.
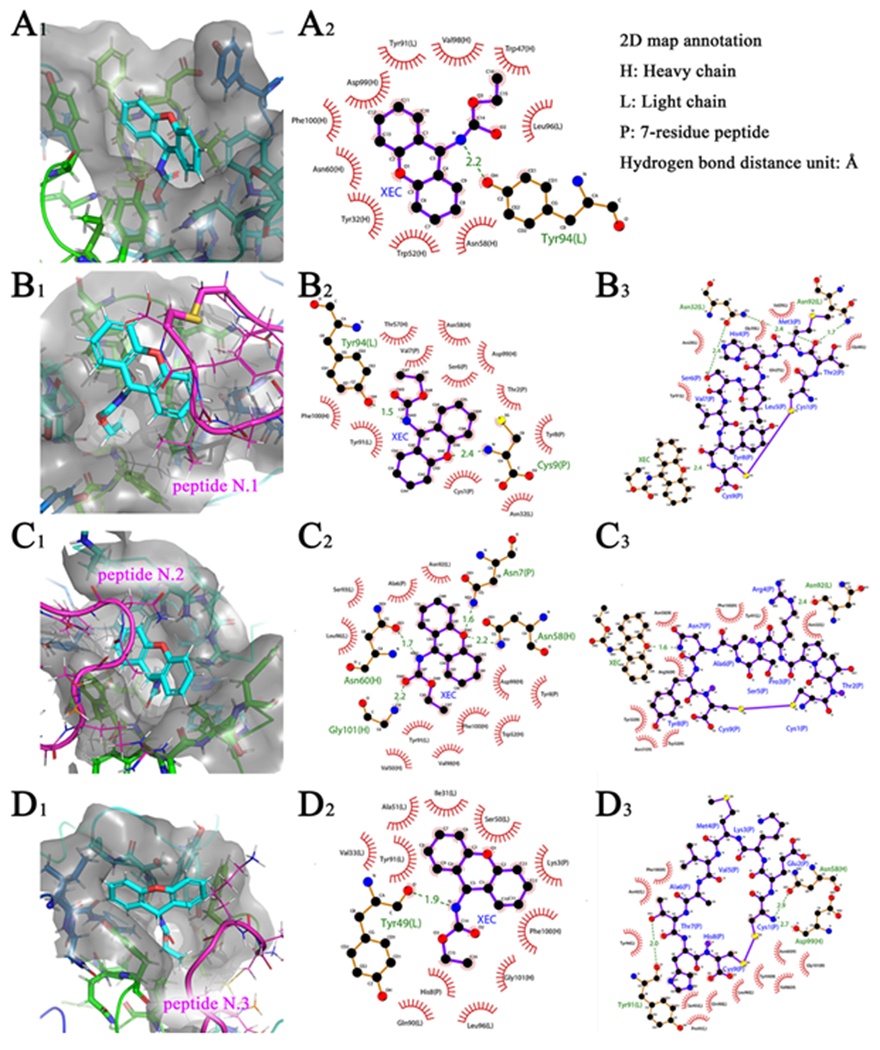
3D docking map (subscript1), 2D docking map which selected XEC as ligand (subscript2) and 2D docking map which selected peptide as ligand (subscript3). (A) Fab-XEC complex; (B) Fab-XEC-peptide N.1 complex; (C) Fab-XEC-peptide N.2 complex; (D) Fab-XEC-peptide N.3 complex.
3.4. Sensitivity and specificity of non-competitive phage ELISA
The analytical capability of non-competitive phage ELISA was evaluated after EC derivatization as described in the experimental section. The optimized concentration of antibody and phage was 10 μg/mL and 2.3×109 pfu/mL, respectively. The calibration and equation for EC determination under 1% (v/v) alcohol solution was obtained. As shown in Fig. 4A, the non-competitive ELISA assay exhibited a SC50 of 1.66 ng/mL and LOD of 0.54 ng/mL for EC. The linear range (SC20~SC80) was 0.87-3.20 ng/mL (y=1.080lgx +0.2961). Although it was found that the linear range of this non-competitive method was narrower (Fig. 4A) than that of competitive one (16.8-127.92 ng/mL), the sensitivity of non-competitive method has a 17-fold improvement in comparison with the competitive immunoassay (LOD of 9.28 ng/mL). The proposed assay was mainly used as a screening purpose, samples with EC concentration higher than the upper LOQ can be diluted and tested again or confirmed by a standard instrumental method.
Fig. 4.
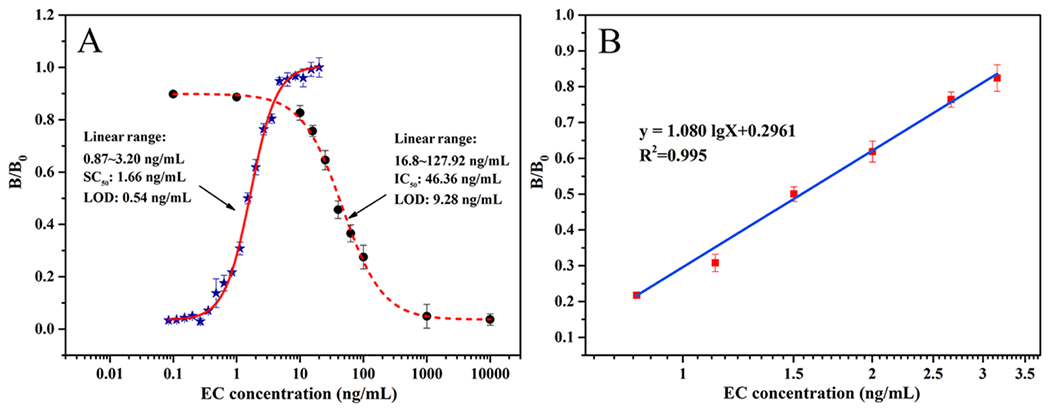
(A) Calibration curves for EC determination using phage-displayed peptide based non-competitive ELISA (full line) and competitive ELISA (dotted line). (B) The linear region of non-competitive ELISA standard curve for determining EC concentration in PBS with 1% alcohol (n=3).
To confirm the reusability of the proposed non-competitive phage ELISA, under the same condition, eight concentration levels of EC (0.8, 0.9, 1.0, 1.2, 1.6, 1.8, 2.0 and 3.2 ng/mL) were analyzed by five tests, respectively. The results showed that the coefficients of variation (CVs) were 10.3%, 8.7%, 8.1%, 5.2%, 8.2%, 7.0%, 6.4% and 10.1%, respectively. The results indicated good repeatability of this non-competitive immunoassay based on peptide displayed phage. Furthermore, the stability of the phage for developing competitive immunoassay was also investigated. When phage screened from the library was stored at 4 °C for 30 days, the sensitivity (SC50, 1.71ng/mL) showed no obvious decrease. The peptide displayed phage was stable for further application.
For purpose of investigating the specificity of the method, the cross-reactivity (CR) for XEC structurally related compounds under same conditions was analyzed. As shown in Table 2, a little CR (7.4%) for XMC as observed, while negligible CR for XAA and 9-xanthydrol was obtained. The specificity of the proposed assay was similar to that of mAb-based competitive immunoassay for EC (Luo et al., 2018). It could be inferred that ethyl carbamate was a key group for antibody recognition and entirely inserted into antigen binding cavity of antibody due to its diminutive size (Mwt: 89.09 Da). Consequently, the aromatic rings of assembled XEC-antibody immunocomplex are dominating part for phage-displayed peptide recognizing. In order to verify the specificity, we further applied the surface plasmon resonance (SPR) to further analyze affinity difference of anti-XEC mAb against XEC and its structurally related compounds. As shown in Fig. S6 and Table S1, the KD of mAb to XEC was 6.42×10−6 mol/L, which was significantly higher than other ligands. The strong affinity and weak dissociation (Kd, 8.47×10−3 s−1) of mAb to XEC simultaneously illustrated the possibility of the immunocomplex forming for phage screening. The results were consistent with the experimental results. Since methyl carbamate was seldom found in red wine sample and the level was less than 1% of EC content (Luo et al., 2017), the little CR would not affect the specific determination of EC by the assay.
Table 2.
Performance of specificity of phage-displayed peptide based non-competitive phage ELISA (n=3).
| Compound | Structure | CRa(%) |
|---|---|---|
| XEC |
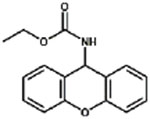
|
100% |
| XMC |
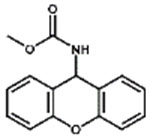
|
7.4% |
| XAA |
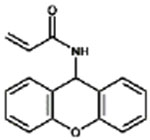
|
0.3% |
| 9-Xanthydrol |
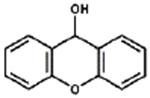
|
< 0.1% |
CR, Cross reactivity.
CR(%)=SC50 (XEC)/SC50(cross-reactive compound)×100.
3.5. Validation of non-competitive ELISA
To evaluate the practicality of the proposed non-competitive ELISA, one red wine sample (background level of EC was 1.8 ng/mL) was spiked with three concentrations of EC (10, 15 and 25 ng/mL) and determined by both non-competitive ELISA and UPLC-Q-Orbitrap HRMS. For non-competitive ELISA, good recovery was obtained ranging from 87.2% to 94.4% with CVs below 10.7% (Table 3). For UPLC-Q-Orbitrap HRMS, the detection range of calibration curves (y=−0.0042+0.0124x) was from 1 to 30 ng/mL (R2=0.995) and the limit of detection (LOD) was 0.05 ng/mL (S/N=3). Recovery ranging from 89.3% to 97.1% were obtained for UPLC-Q-Orbitrap HRMS.
Table 3.
Results of spiked wine samples analyzed by non-competitive ELISA and UPLC-Q-Orbitrap HRMS (n=3).
| Non-competitive ELISA | UPLC-Q-Orbitrap HRMS | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Original EC conc.(ng/mL) | Spiked level (ng/mL) | Mean± SDa, (ng/mL) | Recovery (%) | CVb (%) | Mean± SDa, (ng/mL) | Recovery (%) | CVb (%) | P valuec |
| 1.8 | 10 | 10.5±1.1 | 87.2 | 10.4 | 11.5±0.2 | 97.1 | 1.7 | 0.07 |
| 15 | 14.9±1.6 | 87.3 | 10.7 | 15.2±0.2 | 89.3 | 1.3 | 0.58 | |
| 25 | 25.4±2.4 | 94.4 | 9.4 | 25.2±0.4 | 93.6 | 1.6 | 0.85 | |
SD, Standard deviation
CV, Coefficient of variance
P value (>0.05) shows no significant differences between two methods for t-Test result.
To further validate the accuracy of the proposed method, all the ten red wine samples were randomly selected and determined by both non-competitive ELISA and UPLC-Q-Orbitrap HRMS (see Table S2). Bland-Altman method was used for statistically analyzing the difference between the results of two methods. As shown in Fig. 5, the scatter points almost distributed within 95% limits of agreement. The results indicated good accuracy and reliability of developed method. All of the aforementioned results demonstrated the proposed method based phage-displayed peptide can be promising for application in real sample detection.
Fig. 5.
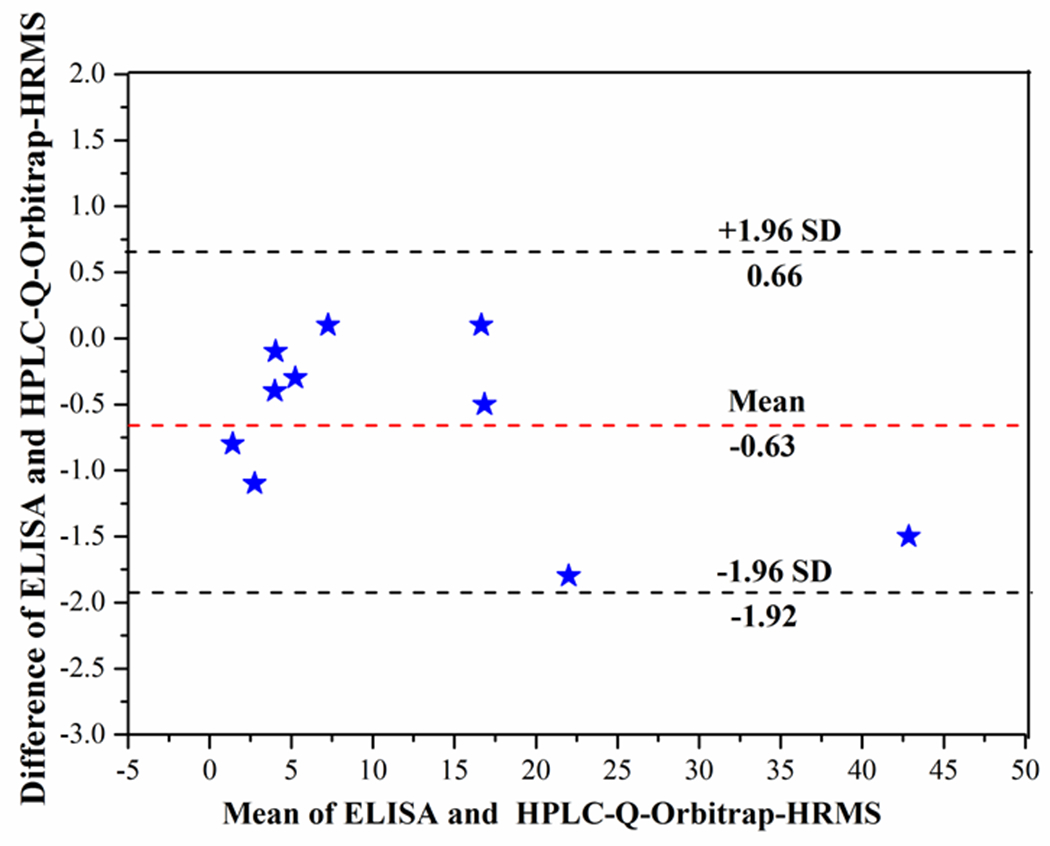
The results of Bland-Altman analysis for EC concentration in blind samples between non-competitive phage ELISA and UPLC-Q-Orbitrap HRMS (n=3).
4. Conclusions
In conclusion, three phage-displayed peptides with different binding activities to immunocomplex were isolated from the phage display library. Computer-assisted simulation analysis revealed that the xanthydrol of xanthyl ethyl carbamate and the residues (Asn-32 and Asn-92) at light chain of antibody were mainly responsible for binding. The activity differences of the peptides mainly attributed to the hydrophobic interaction between the peptides and xanthydrol. The binding mechanism provided a basis for further implementing directed evolution to improve performance of peptide. Based on the peptide with the best performance, non-competitive immunoassay for sensitive determination of ethyl carbamate was developed. The assay sensitivity was improved about 17-fold in comparison with traditional competitive immunoassay. The proposed assay also showed good specificity and consistency with that of standard UPLC-Q-Orbitrap HRMS. Therefore, the non-competitive immunoassay developed has great potential for other small molecules monitoring in the future.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31822039), the Guangdong Special Support Program (2019TX05N052), the Key Project of Guangdong Provincial High School (2019KJDXM002, 2019SCAUGH03) and the National Institutes of Environmental Health Sciences Superfund Research Program (P42ES04699) and RIVER Award (R35 ES030443-01).
Footnotes
The authors declare no conflict of interest.
References
- Acharya D, Dhar TK, 2008. A novel broad-specific noncompetitive immunoassay and its application in the determination of total aflatoxins. Anal. Chim. Acta 630, 82–90. [DOI] [PubMed] [Google Scholar]
- Ajtony Z, Szoboszlai N, Bencs L, Viszket E, Mihucz VG, 2013. Determination of ethyl carbamate in wine by high performance liquid chromatography. Food Chem. 141, 1301–1305. [DOI] [PubMed] [Google Scholar]
- Akter S, Kustila T, Leivo J, Muralitharan G, Vehniäinen M, Lamminmäki U, 2019. Noncompetitive chromogenic lateral-flow immunoassay for simultaneous detection of microcystins and nodularin. Biosensors 9, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Vehniäinen M, Spoof L, Nybom S, Meriluoto J, Lamminmäki U, 2016. Broad-spectrum noncompetitive immunocomplex immunoassay for cyanobacterial peptide hepatotoxins (microcystins and nodularins). Anal. Chem 88, 10080–10087. [DOI] [PubMed] [Google Scholar]
- Arola H, Tullila A, Nathanail A, Nevanen T, 2017. A simple and specific noncompetitive ELISA method for HT-2 toxin detection. Toxins 9, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu T, Zhang M, Sun X, Tian Y, Bai F, Jia P, Bai Y, Zhe T, Wang L, 2019. Gold nanoparticles-functionalized microorganisms assisted construction of immunobiosensor for sensitive detection of ochratoxin A in food samples. Sens. Actuators B Chem 299, 126969. [Google Scholar]
- Chamkasem N, Ollis LW, Harmon T, Lee S, Mercer G, 2013. Analysis of 136 pesticides in avocado using a modified QuEChERS method with LC-MS/MS and GC-MS/MS. J. Agr. Food Chem 61, 2315–2329. [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu X, Xiao Z, Fu H, Huang Y, Huang S, Shen Y, He F, Yang X, Hammock B, Xu Z, 2020. Production of a specific monoclonal antibody for 1-naphthol based on novel hapten strategy and development of an easy-to-use ELISA in urine samples. Ecotox. Environ. Safe 196, 110533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang X, Wang B, Rao M, Wang H, Lei H, Liu H, Zhang Y, Sun Y, Xu Z, 2018. Production of antigen-binding fragment against O,O-diethyl organophosphorus pesticides and molecular dynamics simulations of antibody recognition. Int. J. Mol. Sci 19, 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Shi Q, Zhu C, Li S, Lin Y, Du D, 2019. Pt–Ni(OH)2 nanosheets amplified two-way lateral flow immunoassays with smartphone readout for quantification of pesticides. Biosens. Bioelectron 142, 111498. [DOI] [PubMed] [Google Scholar]
- Cong Y, Dong H, Wei X, Zhang L, Bai J, Wu J, Huang JX, Gao Z, Ueda H, Dong J, 2019. A novel murine antibody and an open sandwich immunoassay for the detection of clenbuterol. Ecotox. Environ. Safe 182, 109473. [DOI] [PubMed] [Google Scholar]
- Dong J, Xu C, Wang H, Xiao Z, Gee SJ, Li Z, Wang F, Wu W, Shen Y, Yang J, Sun Y, Hammock BD, 2014. Enhanced sensitive immunoassay: noncompetitive phage anti-immune complex assay for the determination of malachite green and leucomalachite green. J. Agr. Food Chem 62, 8752–8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farka Z, Čunderlová V, Horáčková V, Pastucha M, Mikušová Z, Hlaváček A, Skládal P, 2018. Prussian blue nanoparticles as a catalytic label in a sandwich nanozyme-linked immunosorbent assay. Anal. Chem 90, 2348–2354. [DOI] [PubMed] [Google Scholar]
- Fu H, Wang Y, Xiao Z, Wang H, Li Z, Shen Y, Lei H, Sun Y, Xu Z, Hammock BD, 2020. A rapid and simple fluorescence enzyme-linked immunosorbent assay for tetrabromobisphenol A in soil samples based on a bifunctional fusion protein. Ecotox. Environ. Safe 188, 109904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Yuan L, Shen Y, Liu Y, Liu B, Zhang S, Xie Z, Lei H, Sun Y, Xu Z, 2018. A full-automated magnetic particle-based chemiluminescence immunoassay for rapid detection of cortisol in milk. Anal. Chim. Acta 1035, 129–135. [DOI] [PubMed] [Google Scholar]
- González-Techera A, Kim HJ, Gee SJ, Last JA, Hammock BD, González-Sapienza G, 2007. Polyclonal antibody-based noncompetitive immunoassay for small analytes developed with short peptide loops isolated from phage libraries. Anal. Chem 79, 9191–9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Techera A, Vanrell L, Last JA, Hammock BD, González-Sapienza G, 2007. Phage anti-immune complex assay: general strategy for noncompetitive immunodetection of small molecules. Anal. Chem 79, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowd V, Su H, Karlovsky P, Chen W, 2018. Ethyl carbamate: an emerging food and environmental toxicant. Food Chem. 248, 312–321. [DOI] [PubMed] [Google Scholar]
- Hua X, Zhou L, Feng L, Ding Y, Shi H, Wang L, Gee SJ, Hammock BD, Wang M, 2015. Competitive and noncompetitive phage immunoassays for the determination of benzothiostrobin. Anal. Chim. Acta 890, 150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Li X, Lu Y, Guo P, Zhang G, Wang Y, Zhang Y, Zhu W, Pan J, Wang J, 2020. Nanobodies based on a sandwich immunoassay for the detection of staphylococcal enterotoxin b free from interference by protein A. J. Agr. Food Chem 68, 5959–5968. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Ohmuro-Matsuyama Y, Tanioku M, Ushiba S, Ono T, Inoue K, Kitaguchi T, Kimura M, Ueda H, Matsumoto K, 2019. Graphene field effect transistor-based immunosensor for ultrasensitive noncompetitive detection of small antigens. ACS Sens. 5, 24–28. [DOI] [PubMed] [Google Scholar]
- Lape M, Paula S, Ball WJ, 2010. A molecular model for cocaine binding by the immunotherapeutic human/mouse chimeric monoclonal antibody 2E2. Eur. J. Med. Chem 45, 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehotay SJ, Son KA, Kwon H, Koesukwiwat U, Fu W, Mastovska K, Hoh E, Leepipatpiboon N, 2010. Comparison of QuEChERS sample preparation methods for the analysis of pesticide residues in fruits and vegetables. J. Chromatogr. A 1217, 2548–2560. [DOI] [PubMed] [Google Scholar]
- Li H, Ma S, Zhang X, Li C, Dong B, Mujtaba MG, Wei Y, Liang X, Yu X, Wen K, Yu W, Shen J, Wang Z, 2018. Generic hapten synthesis, broad-specificity monoclonal antibodies preparation, and ultrasensitive ELISA for five antibacterial synergists in chicken and milk. J. Agr. Food Chem 66, 11170–11179. [DOI] [PubMed] [Google Scholar]
- Li Y, Ye X, Zheng X, Chen W, 2019. Transcription factor EB (TFEB)-mediated autophagy protects against ethyl carbamate-induced cytotoxicity. J. Hazard. Mater 364, 281–292. [DOI] [PubMed] [Google Scholar]
- Liu H, Huang R, Zeng G, Xu Z, Sun Y, Lei H, Sheng Y, Wang H, Xu B, Wei X, 2020. Discrimination of reconstituted milk in China market using the content ratio of lactulose to furosine as a marker determined by LC-MS/MS. LWT-Food Sci. Technol 117, 108648. [Google Scholar]
- Liu Z, Hua Q, Wang J, Liang Z, Li J, Wu J, Shen X, Lei H, Li X, 2020. A smartphone-based dual detection mode device integrated with two lateral flow immunoassays for multiplex mycotoxins in cereals. Biosens. Bioelectron 158, 112178. [DOI] [PubMed] [Google Scholar]
- Luo L, Lei H, Yang J, Liu G, Sun Y, Bai W, Wang H, Shen Y, Chen S, Xu Z, 2017. Development of an indirect ELISA for the determination of ethyl carbamate in Chinese rice wine. Anal. Chim. Acta 950, 162–169. [DOI] [PubMed] [Google Scholar]
- Luo L, Song Y, Zhu C, Fu S, Shi Q, Sun Y, Jia B, Du D, Xu Z, Lin Y, 2018. Fluorescent silicon nanoparticles-based ratiometric fluorescence immunoassay for sensitive detection of ethyl carbamate in red wine. Sens. Actuators B Chem 255, 2742–2749. [Google Scholar]
- Mares A, De Boever J, Osher J, Quiroga S, Barnard G, Kohen F, 1995. A direct non-competitive idiometric enzyme immunoassay for serum oestradiol. J. Immunol. Methods 181, 83–90. [DOI] [PubMed] [Google Scholar]
- Miller YE, Dwyer-Nield LD, Keith RL, Le M, Franklin WA, Malkinson AM, 2003. Induction of a high incidence of lung tumors in C57BL/6 mice with multiple ethyl carbamate injections. Cancer Lett. 198, 139–144. [DOI] [PubMed] [Google Scholar]
- Ohmuro-Matsuyama Y, Ueda H, 2018. Homogeneous noncompetitive luminescent immunodetection of small molecules by ternary protein fragment complementation. Anal. Chem 90, 3001–3004. [DOI] [PubMed] [Google Scholar]
- Ricciardi C, Santoro K, Stassi S, Lamberti C, Giuffrida MG, Arlorio M, Decastelli L, 2018. Microcantilever resonator arrays for immunodetection of β-lactoglobulin milk allergen. Sens. Actuators B Chem 254, 613–617. [Google Scholar]
- Rossotti MA, Carlomagno M, González-Techera A, Hammock BD, Last J, González-Sapienza G., 2010. Phage Anti-immunocomplex assay for clomazone: two-site recognition increasing assay specificity and facilitating adaptation into an on-site format. Anal. Chem 82, 8838–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Shida S, Kashiwabara N, Shiono K, Nemoto S, Akiyama H, 2020. Development of an analytical method for determination of total ethofumesate residues in foods by gas chromatography-tandem mass spectrometry. Food Chem. 313, 126132. [DOI] [PubMed] [Google Scholar]
- Tu Q, Qi W, Zhao J, Zhang L, Guo Y, 2018. Quantification ethyl carbamate in wines using reaction-assisted-extraction with 9-xanthydrol and detection by heart-cutting multidimensional gas chromatography-mass spectrometry. Anal. Chim. Acta 1001, 86–92. [DOI] [PubMed] [Google Scholar]
- Vanrell L, Gonzalez-Techera A, Hammock BD, Gonzalez-Sapienza G, 2012. Nanopeptamers for the development of small-analyte lateral flow tests with a positive readout. Anal. Chem 85, 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Huang QT, Qi P, 2016. Determination and Identification of ethyl carbamate in alcoholic drinks by UPLC-quadrupole/electrostatic field orbitrap high-resolution mass spectrometry. Liquor-Making Sci. Technol 7, 112–115. [Google Scholar]
- Zhang Y, Wang L, Shen X, Wei X, Huang X, Liu Y, Sun X, Wang Z, Sun Y, Xu Z, Eremin SA, Lei H, 2017. Broad-Specificity Immunoassay for simultaneous detection of ochratoxins A, B, and C in millet and maize. J. Agr. Food Chem 65, 4830–4838. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu Z, Wang F, Cai J, Dong J, Zhang J, Si R, Wang C, Wang Y, Shen Y, Sun Y, Wang H, 2018. Isolation of bactrian camel single domain antibody for parathion and development of one-step dc-FEIA method using VHH-alkaline phosphatase fusion protein. Anal. Chem 90, 12886–12892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


