Abstract
Background
Delirium (an acute change in cognition) is a common, morbid, and costly syndrome seen primarily in aging adults. Despite increasing knowledge of its epidemiology, delirium remains a clinical diagnosis with no established biomarkers to guide diagnosis or management. Advances in proteomics now provide opportunities to identify novel markers of risk and disease progression for postoperative delirium and its associated long-term consequences (eg, long-term cognitive decline and Alzheimer’s disease [AD]).
Methods
In a nested matched case–control study (18 delirium/no-delirium pairs) within the Successful Aging after Elective Surgery study (N = 556), we evaluated the association of 1305 plasma proteins preoperatively [PREOP] and on postoperative day 2 [POD2]) with delirium using SOMAscan. Generalized linear models were applied to enzyme-linked immunosorbant assay (ELISA) validation data of one protein across the full cohort. Multi-protein modeling included delirium biomarkers identified in prior work (C-reactive protein, interleukin-6 [IL6]).
Results
We identified chitinase-3-like-protein-1 (CHI3L1/YKL-40) as the sole delirium-associated protein in both a PREOP and a POD2 predictor model, a finding confirmed by ELISA. Multi-protein modeling found high PREOP CHI3L1/YKL-40 and POD2 IL6 increased the risk of delirium (relative risk [95% confidence interval] Quartile [Q]4 vs Q1: 2.4[1.2–5.0] and 2.1[1.1–4.1], respectively).
Conclusions
Our identification of CHI3L1/YKL-40 in postoperative delirium parallels reports of CHI3L1/YKL-40 and its association with aging, mortality, and age-related conditions including AD onset and progression. This highlights the type 2 innate immune response, involving CHI3L1/YKL-40, as an underlying mechanism of postoperative delirium, a common, morbid, and costly syndrome that threatens the independence of older adults.
Keywords: Inflammation, Postoperative, Proteomics
Delirium affects 25% of older adults undergoing major elective surgery and is associated with greater nosocomial complications (1), longer hospitalizations (2), higher rates of discharge to nursing homes (3), increased risk of long-term cognitive and functional decline (4–6), incident dementia (7), and mortality (8,9). In Alzheimer’s disease (AD) patients, delirium is associated with accelerated cognitive decline (10–12). Annual U.S. health care costs attributable to delirium are $182 billion (13). Despite increasing knowledge of its epidemiology, delirium pathophysiology and etiopathogenesis remain poorly understood due to its multifactorial and heterogeneous etiology.
Among proposed pathophysiologies (14), recent work has supported an inflammatory model of postoperative delirium. Specifically, a systemic low-grade preinflammatory state is linked to an increased risk of developing delirium (15,16), which aligns with the literature on inflammation and immune activation in AD (17). Specifically, we found: (i) interleukin (IL)-6 levels measured on postoperative day 2 (POD2) were elevated in delirium cases relative to matched no-delirium controls using a Luminex cytokine panel (18), and (ii) C-reactive protein (CRP) was the strongest delirium-associated protein preoperatively (PREOP) and at POD2 using unbiased proteomics (Isobaric tags for relative and absolute quantitation [iTRAQ]–based relative quantitation mass spectrometry) followed by enzyme-linked immunosorbant assay (ELISA) validation (15). Hence, there is emerging evidence that blood-based protein biomarkers for delirium might play a role in elucidating pathophysiological mechanisms and laying the foundation for noninvasive, cost-effective immunoassays to guide prediction, diagnosis, and monitoring of delirium.
Our prior Luminex and mass spectrometry-based approaches were limited to a restricted set of measurable plasma proteins. Advances in proteomics platforms now provide opportunities to further identify new biological pathways of delirium through measurement of exponentially more proteins with minimal sample volume: from approximately 100 proteins using mass spectrometry based-iTRAQ to now 1305 proteins measured across the entire dynamic range using aptamer-based SOMAscan (SOMALogic). We hypothesize that a more comprehensive analysis of the proteome using this innovative proteomics platform may enhance understanding of delirium pathophysiology. Therefore, using banked samples from the well-characterized Successful Aging after Elective Surgery (SAGES) cohort, we: (i) analyzed the plasma proteome at PREOP and POD2 using SOMAscan to discover markers of postoperative delirium in a nested matched case–control study (18 delirium/no delirium pairs), (ii) conducted ELISA validation of one identified delirium-associated protein in the full SAGES cohort (PREOP N = 553, POD2 N = 556), and (iii) used multi-protein modeling to examine the independent associations of the SOMAscan-identified and ELISA-validated protein with previously discovered plasma markers for delirium (CRP and IL6) (15,16,18,19).
Method
Study Population
The SAGES study, designed to understand novel risk factors and long-term outcomes of delirium, enrolled 560 patients ≥70 years old scheduled for major noncardiac surgery. Study details, including major inclusion and exclusion criteria have been previously published (20,21). Notably, patients underwent a detailed screening process to exclude dementia based on patient or family report of dementia diagnosis, medical record review, capacity assessment, and cognitive testing using the Modified Mini-Mental State Examination (22) and detailed neuropsychological battery that enabled calculation of a General Cognitive Performance (GCP) measure (23).
Informed consent for study participation was obtained from all subjects according to procedures approved by institutional review boards at Beth Israel Deaconess Medical Center and Brigham and Women’s Hospital, the 2 surgical sites, and Hebrew SeniorLife, the study coordinating center, all located in Boston, Massachusetts.
Specimen Collection
Patients underwent phlebotomy at PREOP, post-anesthesia care unit, POD2, and 1 month post-surgery. Based on previous findings, this study focused on the PREOP and POD2 time points due to their paramount importance for identifying risk markers (PREOP) and disease markers (POD2) of delirium, respectively (18). Details about quality control measures, blood processing, and storage have been published (15,18).
Delirium and Delirium Severity
Postoperative delirium was determined from daily interviews during hospitalization, supplemented with a validated chart review method, used to maximize sensitivity (24). The presence of delirium was determined by the Confusion Assessment Method (CAM) diagnostic algorithm (25). Patients were considered delirious if delirium was present on either the CAM or the chart review method on any postoperative day.
Delirium severity was quantified using the CAM-Severity long form (CAM-S LF) score (24). The CAM-S LF includes severity ratings of 10 CAM features (range 0–19, 19 most severe); each scored as 0 (absent), 1 (present, mild), and 2 (present, marked)—except for fluctuating course, which is scored 0 (absent) and 1 (present). Our primary outcome for delirium severity was defined as the sum of CAM-S scores (sum CAM-S) across all postoperative days until hospital discharge, which considers both intensity and duration and thus reflects the total “burden” of delirium features across the entire hospitalization. We previously described sum CAM-S and found it to be the delirium feature severity measure most strongly associated with clinical outcomes (26). For analysis, we categorized the sum CAM-S into 3 groups: no delirium (regardless of CAM-S score), mild delirium (sum CAM-S ≤17, the median sum CAM-S value among patients with delirium), and severe delirium (sum CAM-S >17).
Selection of the Nested Matched Case–Control Sample for SOMAscan Discovery and ELISA Validation
For biomarker discovery, we created a nested, matched case–control sample from the full SAGES cohort. Delirium cases were defined as participants with either: (i) delirium on POD2; or (ii) delirium on POD1 and subsyndromal delirium on POD2 or POD3 (see Supplementary Material for details). No-delirium controls were defined as patients without delirium or without subsyndromal delirium on any postoperative day. Cases and controls were matched on: age within 5 years; baseline GCP score within 5 points [0–100 scale]; and an exact match for sex, surgery type, presence of vascular comorbidity, and Apolipoprotein E ε4 status (see Supplementary Materials for description of variables). Details on the inclusion/exclusion criteria and matching methods have been described (15,18,27). Plasma samples from 18 matched case–control pairs at PREOP and POD2 were considered in the SOMAscan biomarker discovery phase (Aim 1). ELISA validation was performed on the entire SAGES cohort with plasma available (N = 556).
SOMAscan Assay
Heparin plasma samples (50 μL) were analyzed using the SOMAscan Assay Kit for human serum 1.3k (cat. #900-00012), which measures expression of 1305 human proteins using highly selective single-stranded modified Slow Off-rate Modified DNA Aptamers (SOMAmer) according to the manufacturer’s standard protocol (SomaLogic; Boulder, CO). See Supplementary Materials for details. Data quality control, calibration, and normalization were done according to the manufacturer’s protocol as previously described (28).
Sample Processing and Statistical Analysis
Figure 1 illustrates the overall methodologic steps that are described in detail below.
Figure 1.
Overall workflow and methodologic steps for protein identification of delirium protein expression profiles and ELISA validation of proteins. Proteins significantly differentially expressed in delirium samples at PREOP and POD2 were used for hierarchical clustering and PCA to investigate their potential to distinguish the 2 classes of samples. Both PREOP and POD2 delirium-specific protein sets were used for System Biology Analysis to identify their biological implications using Ingenuity Pathway Analysis. Ten thousand subsamples of the data at both PREOP and POD2 were generated to calculate the frequency with which a protein remains delirium-specific. Proteins with high-frequency and fold change, i.e. robust differential expression, were evaluated as predictors of delirium using all possible combinations. For each combination, 1000 4-fold cross-validations were performed to identify predictor performance using support vector machines. ELISA validation of CHI3L1/YKL-40 as the sole delirium-associated protein in both a PREOP and a POD2 predictor model was confirmed by ELISA in a larger cohort of 554 samples. CHI3L1/YKL-40 and markers previously associated with Delirium (CRP [PREOP, POD2] and IL6 [POD2]) were analyzed in a combined model. CHI3L1/YKL-40 = Chitinase-3-like protein 1; CRP = C-reactive protein; ELISA = Enzyme-linked immunosorbent assay; PCA = Principal component analysis; POD2 = Postoperative day 2; PREOP = Preoperative; SVM = Support vector machine.
Step 1: SOMAscan proteomics discovery, systems biology, and development of delirium predictor models
Identification of a proteomic signature of postoperative delirium.
—Differential expression analysis compared delirium (DEL) and control (CNT) samples at PREOP and POD2 using paired t tests. A protein was considered to be differentially expressed if it was significantly dysregulated (p < .05). Fold change (FC) of protein expression was calculated by applying the one-step Tukey’s biweight algorithm on the FC values (DEL/CNT) for each paired sample (tFC) (29). This provides a robust estimation of the FC for each individual protein that is unaffected by outliers. Discriminatory power of the protein expression was established using hierarchical clustering and principal components analysis (PCA) (30). Sample classification was completed using support vector machines (SVM) (31) on the PCA results using Gaussian, polynomial (degree = 2, 3, 4), and linear kernels. See Supplementary Materials for additional details.
Systems biology analysis.
—To acquire new insights into potential pathophysiological pathways underlying delirium-specific plasma protein signatures at PREOP and POD2 and to better understand the interactions between the differentially expressed proteins and candidate upstream regulators, we performed functional category, canonical pathway, interactive network, upstream regulator, and regulator effect analyses of all dysregulated proteins with a p < .05 using the Ingenuity Pathway Analysis (IPA) software tool (QIAGEN, Redwood City, CA). IPA is a repository of biological interactions and functions created from millions of individually modeled relationships ranging from molecular (proteins, genes) to organism (diseases) level (31).
Machine learning to develop diagnostic classifiers using support vector machines (SVM).
—To assess the robustness in differential expression, 10 000 subsampled data sets were obtained at each time point. In each subsampled data set, 75% of the delirium and control samples were retained and differential expression analysis was performed. The frequency of occurrence in the differentially expressed protein list (p < .05) for each protein was calculated. Twelve proteins each that were differentially expressed in at least 50% of the subsampled data sets and that were linked to delirium were selected as candidate predictors at PREOP and POD2 based on the frequency count (top one-half) and p-value (p < .05). All combinations (1–12) of the 12 proteins were evaluated as predictor sets, resulting in 212–1 = 4095 candidate predictor protein sets.
To develop risk predictor models, we developed classifiers with a minimal number of proteins that could serve as risk and disease biomarkers of delirium. For each candidate predictor set, 1000 splits based on 4-fold cross-validation was performed to assess the performance of the predictor set by assessing its accuracy, sensitivity, and specificity in each split. Prediction was performed by applying SVM-based machine learning to the samples projected on the PCA bases to develop high accuracy delirium risk and disease predictors. In contrast to the average prediction accuracies for the individual proteins, combinations of multiple proteins significantly increased average accuracy, sensitivity, and specificity.
Step 2: ELISA validation
ELISA validation of proteomic data.
—We selected one of the proteins included in predictors at both PREOP and POD2 for validation across the entire SAGES cohort. Expression levels of this protein were determined by ELISA (R&D Systems, Minneapolis, MN) according to the manufacturer’s protocol (PREOP, N = 553; POD2, N = 556). All samples were run in duplicate. Each 96-well plate contained the standard curve and 19 participant samples at 2 timepoints (PREOP and POD2). Duplicate coefficient of variations (CVs) were typically ≤5%, and samples with more than 10% CVs were repeated. Two internal samples were run on every 96-well ELISA plate for calibration. Inter-plate variation ranged from 5% to 10% based on the internal calibration value. ELISA plates were read using a BioTek MX plate reader at optical density = 450 nm. A 4-parameter logistic curve was used for optimizing the best-fit model.
Step 3: Multivariable analysis and modeling of ELISA data
For validation (PREOP N = 553; POD2 N = 556), we used ELISA measurement of the SOMAscan-identified protein to determine the association between the protein and postoperative delirium using generalized linear models with a log-link and binomial error term (relative risks [RR]). Using multinomial logistic regression models, we determined the association between the protein and the 3-category delirium severity measure (no delirium, mild delirium, severe delirium). All models adjusted for age, sex, baseline GCP, surgery type, Charlson comorbidity index, and medical complications.
Using generalized linear models, we conducted multi-protein modeling to determine the independent association of the SOMAscan-identified protein, along with plasma markers previously associated with delirium (CRP at PREOP and POD2, IL6 at POD2 only). Using multinomial logistic regression, we conducted multi-protein modeling to determine the association of each protein with delirium severity. All models adjusted for age, sex, baseline GCP, surgery type, Charlson comorbidity index, and medical complications. Statistical analyses for Steps 2 and 3 were performed using SAS, Version 9.4.
Results
Sample Characteristics
Supplementary Table S1 shows the sample characteristics of the SOMAscan sample (n = 36; 18 matched pairs) and the ELISA validation sample of the SAGES cohort (N = 556). No substantial differences between the 2 samples were observed for age and sex, though the SOMAscan sample included a greater proportion of patients undergoing orthopedic surgery, having a Charlson comorbidity score ≥2, having higher vascular comorbidity, and developing postoperative delirium (50% given the matched case–control study design).
SOMAscan proteomics for delirium biomarker discovery
Paired t tests identified 85 proteins at PREOP (Table 1 showing the top 20 up- and downregulated out of 85 proteins, based on tFC) and 128 proteins at POD2 (Table 2 showing the top 20 up- and downregulated out of 128 proteins, based on tFC) that were significantly differentially expressed between delirium cases and matched no-delirium controls (p < .05). Forty-four of these proteins were dysregulated at both timepoints. These results indicated that a portion of the proteins linked to delirium presence are dysregulated prior to surgery, enhancing the risk for postoperative delirium. See Supplementary Table S2A and S2B for a complete list of differentially expressed proteins at PREOP and POD2 (respectively).
Table 1.
Top 10 Up-Regulated and Down-Regulated Proteins Between Delirium Cases and Controls at PREOPa
| Protein Name | Entrez Gene ID |
Entrez Gene Symbol | p Value | tFC (DEL/CNT) |
|---|---|---|---|---|
| Up-Regulated/Increased in Delirium | ||||
| Chitinase-3-like protein 1 | 1116 | CHI3L1 | .0045 | 1.99 |
| Platelet factor 4 | 5196 | PF4 | .0088 | 1.83 |
| MHC class I polypeptide-related sequence A | 4276 | MICA | .0082 | 1.69 |
| Pituitary adenylate cyclase-activating polypeptide 27 | 116 | ADCYAP1 | .0090 | 1.66 |
| Resistin | 56729 | RETN | .0388 | 1.64 |
| CMRF35-like molecule 6 | 10871 | CD300C | .0059 | 1.61 |
| Programmed cell death 1 ligand 1 | 29126 | CD274 | .0251 | 1.53 |
| Low affinity immunoglobulin gamma Fc region receptor III-B | 2215 | FCGR3B | .0312 | 1.51 |
| Pappalysin-1 | 5069 | PAPPA | .0036 | 1.48 |
| Tumor necrosis factor receptor superfamily member 1A | 7132 | TNFRSF1A | .0013 | 1.48 |
| Down-Regulated/Decreased in Delirium | ||||
| N-acylethanolamine-hydrolyzing acid amidase | 27163 | NAAA | .0458 | −1.34 |
| Acidic leucine-rich nuclear phosphoprotein 32 family member B | 10541 | ANP32B | .0140 | −1.34 |
| Plasma kallikrein | 3818 | KLKB1 | .0423 | −1.35 |
| Bone morphogenetic protein 1 | 649 | BMP1 | .0106 | −1.37 |
| Complement C4b | 720 721 | C4A C4B | .0315 | −1.40 |
| C-C motif chemokine 27 | 10850 | CCL27 | .0157 | −1.42 |
| Cathepsin L2 | 1515 | CTSV | .0259 | −1.43 |
| Tumor necrosis factor ligand superfamily member 9 | 8744 | TNFSF9 | .0066 | −1.44 |
| Eotaxin | 6356 | CCL11 | .0420 | −1.59 |
| Interleukin-25 | 64806 | IL25 | .0169 | −1.60 |
Note: CNT = No delirium control; DEL = Delirium case; tFC = Tukey’s fold change.
aTop proteins selected based on fold change.
Table 2.
Top 10 Up-Regulated and Down-Regulated Proteins Between Delirium Cases and Controls at POD2a
| Protein Name | Entrez Gene ID |
Entrez Gene Symbol | p Value | tFC (DEL/CNT) |
|---|---|---|---|---|
| Up-Regulated/Increased in Delirium | ||||
| Phospholipase A2; membrane associated | 5320 | PLA2G2A | .0450 | 1.94 |
| Interleukin-6 | 3569 | IL6 | .0047 | 1.90 |
| MHC class I polypeptide-related sequence A | 4276 | MICA | .0128 | 1.79 |
| Metalloproteinase inhibitor 1 | 7076 | TIMP1 | .0031 | 1.74 |
| Thrombospondin-1 | 7057 | THBS1 | .0220 | 1.69 |
| CD177 antigen | 57126 | CD177 | .0092 | 1.68 |
| Pituitary adenylate cyclase-activating polypeptide 27 | 116 | ADCYAP1 | .0142 | 1.63 |
| Chitinase-3-like protein 1 | 1116 | CHI3L1 | .0049 | 1.60 |
| Creatine kinase M-type | 1158 | CKM | .0448 | 1.54 |
| CMRF35-like molecule 6 | 10871 | CD300C | .0183 | 1.53 |
| Down-Regulated/Decreased in Delirium | ||||
| Protein amnionless | 81693 | AMN | .0364 | −1.33 |
| Syntaxin-1A | 6804 | STX1A | .0455 | −1.33 |
| C-C motif chemokine 16 | 6360 | CCL16 | .0450 | −1.34 |
| Choline/ethanolamine kinase | 1120 | CHKB | .0328 | −1.34 |
| Plasma protease C1 inhibitor | 710 | SERPING1 | .0033 | −1.35 |
| Melanoma-derived growth regulatory protein | 8190 | MIA | .0190 | −1.37 |
| C-C motif chemokine 27 | 10850 | CCL27 | .0366 | −1.38 |
| Cadherin-1 | 999 | CDH1 | .0047 | −1.39 |
| Angiostatin | 5340 | PLG | .0043 | −1.39 |
| Leucine-rich repeats and immunoglobulin-like domains protein 3 | 121227 | LRIG3 | .0024 | −1.39 |
Note: CNT = No delirium control; DEL = Delirium case, tFC = Tukey’s fold change.
aTop proteins selected based on Tukey’s fold change.
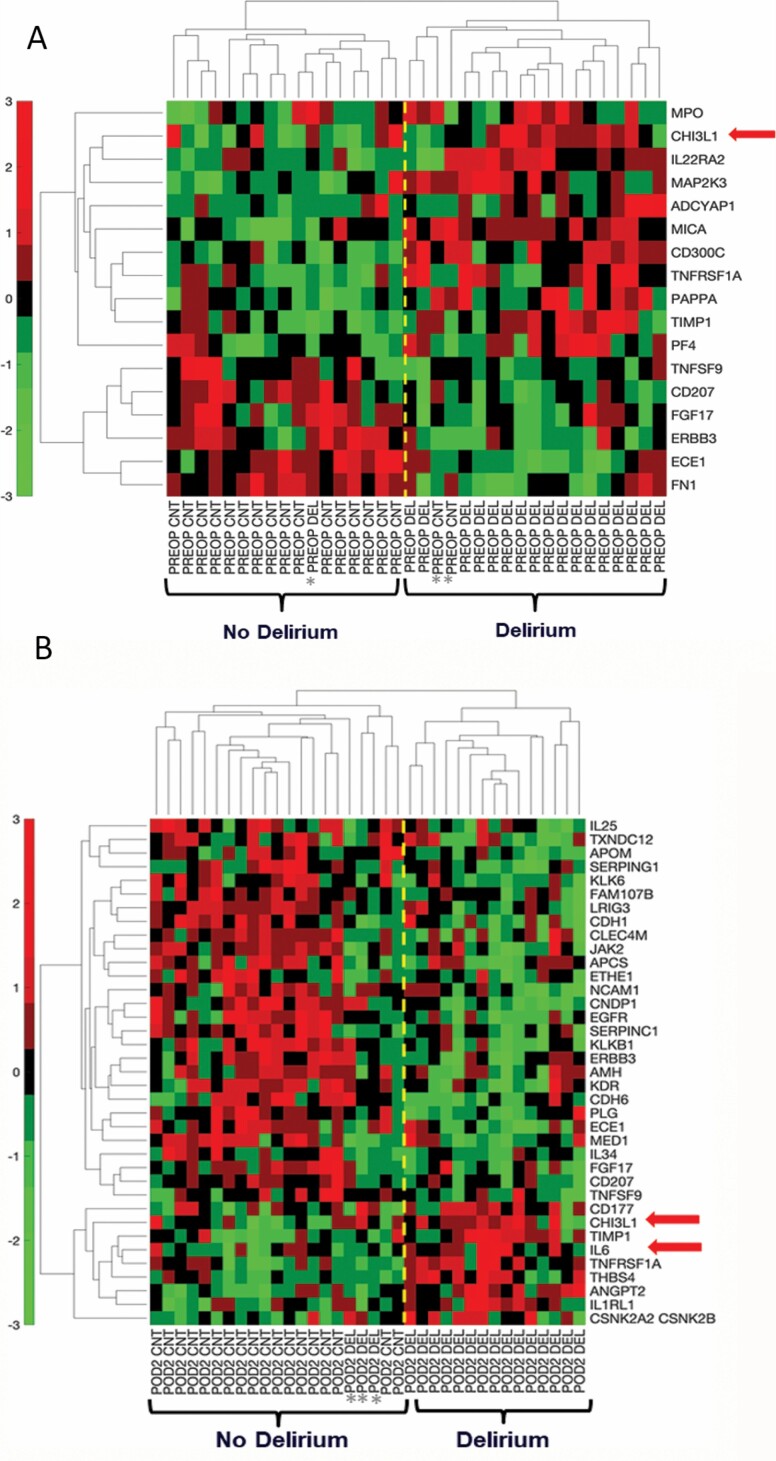
Figure 2A shows hierarchical clustering analysis of the top 17 proteins with the most significant differential expression (p < .01) between cases and controls at PREOP: 11 proteins were increased and 6 proteins were decreased in patients with postoperative delirium. There was excellent discrimination between the delirium cases and controls, except for 1 of 18 delirium cases that clustered with the no-delirium controls and 2 of 18 controls that clustered with the delirium cases. Similarly, at POD2, hierarchical clustering of the most significantly differentially expressed 37 proteins (p < .01) separated delirium cases from no delirium controls, with 3 of 18 delirium cases clustering with the controls, and no controls clustering with the delirium cases (Figure 2B). Among these 37 proteins, 9 proteins were increased and 28 proteins were decreased in patients who developed delirium compared to their matched no-delirium controls. Excellent discrimination at PREOP and POD2 was observed when performing PCA using these sets of proteins as visualized by the SVM classification decision line (Supplementary Figure S1A and S1B, respectively).
Figure 2.
Hierarchical clustering of top proteins differentially expressed between delirium cases and matched controls at PREOP and POD2. (A) Hierarchical clustering of delirium cases and matched non-delirium controls using the top 17 plasma proteins at PREOP (p < .01), reflecting relative minimum and maximum expression levels for each protein as quantified by SOMAscan. (B) Hierarchical clustering of delirium cases and matched non-delirium controls using the top 37 plasma proteins at POD2 (p < .01). In the hierarchical clustering colormap, red denotes relatively higher level and green denotes relatively lower level. CHI3L1/YKL-40 = Chitinase 3-like protein 1; IL6 = Interleukin-6; POD2 = Postoperative day 2; PREOP = Preoperative.
Functional Annotation and Systems Biology Analysis
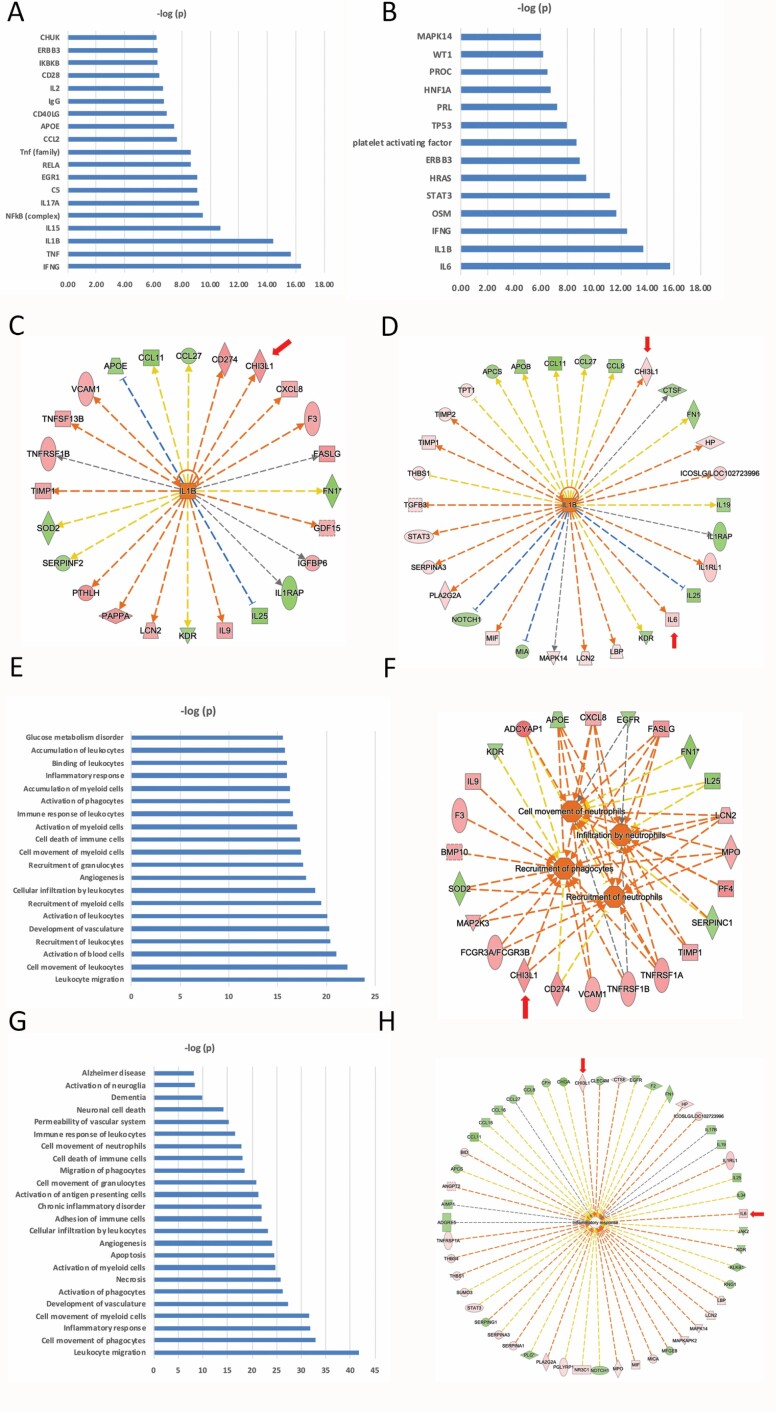
We performed IPA using the 85 PREOP and 128 POD2 proteins (p < .05 threshold for both). Modeling the links between delirium-associated proteins and their established associations with upstream regulatory proteins was particularly informative. The predicted upstream regulators with highest statistical significance converged on activation of pro-inflammatory cytokines (IFNG, TNF, IL1B, IL15, IL17A, CCL2, and IL2) and the NF-κB pathway (NFKB, RELA, and IKBKB) at PREOP (Figure 3A) and on a similar set of cytokines (IL6, IL1B, IFNG, and OSM) at POD2 (Figure 3B).
Figure 3.
(A) PREOP Upstream regulators (ie, a protein/gene that can affect the expression of another protein/gene) with highest statistical significance that best explain the observed expression changes in the input 85 protein list as their targets. The x-axis indicates the -log p-values. (B) POD2 Upstream regulators (ie, a protein/gene that can affect the expression of another protein/gene) with highest statistical significance that best explain the observed expression changes in the input 128 protein list as their targets. The x-axis indicates the -log p-values. (C) PREOP Downstream targets of IL1B (ie, proteins whose expression is affected by IL1B) from among the 85-protein list. Red indicates upregulation and green denotes downregulation in delirium. Proteins are coded by shape; square: cytokine, vertical rhombus: enzyme, horizontal rhombus: peptidase, trapezoid: transporter, ellipse: transmembrane receptor, circle: other. Links are color-coded as red: leads to activation, blue: leads to inhibition, yellow: findings inconsistent with state of downstream protein, black: effect not predicted. (D) POD2 Downstream targets of IL1B (ie, proteins whose expression is affected by IL1B) from among the 128-protein list. The color, shape, and link coding are the same as part C. (E) PREOP Biological functions that are significantly enriched (ie, statistically relative high number of proteins dysregulated in cases versus controls) by the 85-input protein list. The x-axis indicates the -log p-values. (F) PREOP proteins among the input list that are linked to immune cell movement. The color, shape, and link coding are the same as part C. (G) POD2 Biological functions that are significantly enriched by the 128-input protein list. The x-axis indicates the -log p-values. (H) POD2 proteins among the input list that are linked to the “Inflammatory Response” biological functions. The color, shape, and link coding are the same as part B. In parts B and D, proteins that are validated by ELISA are indicated by a red arrow. The orange color of the target upstream regulator and biological function shown in the center implies ‘‘activation,’’ and the shade of the color implies confidence in activation with darker shades implying more confidence in the prediction. See text for more details. ELISA = enzyme-linked immunosorbent assay, POD2 = postoperative day 2, PREOP = preoperative.
The pro-inflammatory cytokines common to both PREOP and POD2 are IL1B and IFNG (Figure 3A and B). Twenty-four of the 85 PREOP proteins (Figure 3C) and 29 of the 128 POD2 proteins were predicted to be regulated by IL1B (Figure 3D). Among the proteins downstream of IL1B is chitinase-3-like protein 1 (CHI3L1/YKL-40). This was of particular interest given its association with AD (32). At POD2, but not PREOP, IL6 was elevated in delirium and was the most significant upstream regulator of 28 proteins. STAT3, which is activated by IL6, is another significant upstream regulator at POD2. Complement C5 is predicted to be active and upstream of a set of proteins linked to delirium at PREOP (Supplementary Figure S2A).
Among the significantly enriched disease and biological functions at PREOP, the immune regulatory pathways were most prominently represented (Figure 3E). This included pathways related to migration, movement, and activation of immune cells, the inflammatory response, and vasculogenesis. Figure 3F highlights in detail the 23 proteins linked to a predicted enhancement in movement, recruitment, and infiltration of neutrophils among the 85 PREOP proteins. This pathway analysis underscores a preinflammatory state in patients who develop postoperative delirium.
Inflammatory processes are also prominently represented in the significantly enriched disease and biological functions at POD2 (Figure 3G). The 51 proteins linked to the inflammatory response, including CHI3L1/YKL-40 and IL6, are highlighted in Figure 3H (Supplementary Figure S2B). Additional relevant pathways are linked to cell death, vascular function, and neurological functions (Figure 3G), including proteins involved in vascular function downstream of platelet activating factor (PAF) such as ANGPT2, THBS1, TIMP1, and TIMP2 (Supplementary Figure S2C). PAF has been demonstrated to induce transient blood–brain barrier dysfunction.
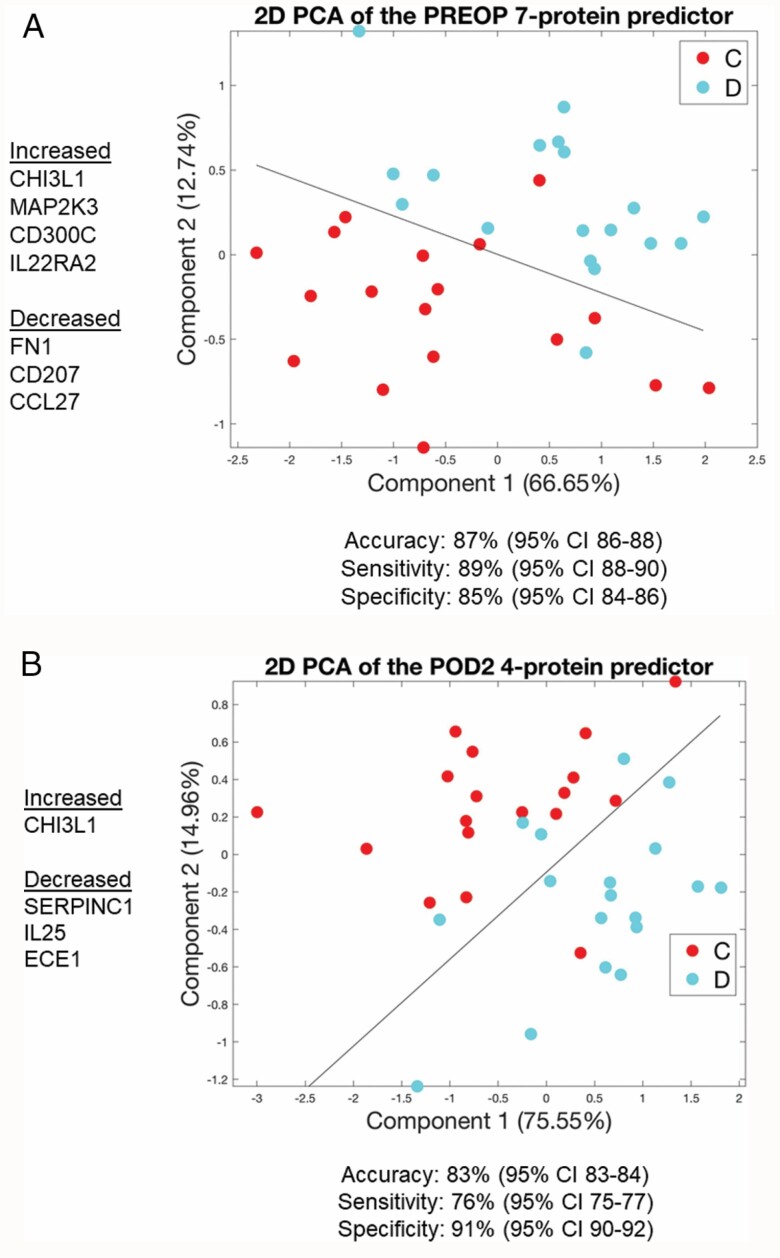
Risk Predictor Models
At PREOP, a 7-protein delirium risk predictor was identified with 86.9% average accuracy (95% confidence interval [CI): [86.1, 87.6]), 89.0% average sensitivity (95% CI: [88.0, 90.0]), and 84.7% average specificity (95% CI: [83.5, 85.9]) and an area under the curve (AUC) of 0.94 (Supplementary Figure S3). This 7-protein delirium risk predictor model comprised 4 proteins that were increased (CHI3L1/YKL-40, MAP2K3, IL22RA2, and CD300C) and 3 proteins that were decreased (CCL27, CD207, and FN1) in patients with delirium. PCA using this 7-protein panel separated the delirium cases from the controls into 2 clusters as seen by 2D visualization of the SVM classification line (Figure 4A).
Figure 4.
PCA plot using the 7 proteins from the PREOP Delirium Classifier (A) and the 4 proteins from the POD2 Delirium Classifier (B) demonstrates discrimination between delirium cases and non-delirium controls with the Support Vector Machines separating line based on a linear kernel. Blue circles, delirium [D]; Red circles, no-delirium controls [C]. POD2 = Postoperative day 2; PREOP = Preoperative.
At POD2, a 4-protein delirium disease predictor was identified with a 4-fold-cross-validation of 83.4% average accuracy (95% CI: [82.6, 84.1]), 76.0% average sensitivity (95% CI: [74.7, 77.2), and 90.8% average specificity (95% CI: [89.9, 91.7]) and AUC of 0.94 (Supplementary Figure S4). This 4-protein predictor was comprised of 1 protein that was increased (CHI3L1/YKL-40) and 3 proteins that were decreased (SERPINC1, IL25, and ECE1) in patients with delirium. PCA using this 4-protein panel effectively separated the delirium cases from the controls into 2 clusters as seen by 2D visualization of the SVM classification line (Figure 4B). See Supplementary Figures S5 and S6 for box and whisker plots showing the distribution of SOMAscan expression levels for each of these 7 PREOP and 4 POD2 proteins, respectively.
ELISA Validation
CHI3L1/YKL-40 is among the proteins significantly increased in delirium cases at both PREOP and POD2 and is the only one shared by the predictor models for both timepoints. Given this and its known link to AD and neuroinflammation, we selected CHI3L1/YKL-40 for independent ELISA validation in the entire SAGES cohort (N = 556). As visualized in Supplementary Figure S7, median CHI3L1/YKL-40 levels were significantly higher in patients with delirium relative to patients without delirium at PREOP and POD2 (p < 10–4 for both).
Multivariable Analysis
Table 3 reports the adjusted RR of postoperative delirium for each sample-based quartile of CHI3L1/YKL-40 (PREOP, POD2) along with markers previously associated with delirium (CRP [PREOP, POD2] and IL6 [POD2]). These are considered in separate, individual analytic models (Models 1) and in a single multi-protein model (Model 2). Although each of the markers was associated with postoperative delirium in the individual models (Models 1), when all proteins were considered together only CHI3L1/YKL-40 PREOP and IL6 POD2 remained significant. At PREOP, patients in quartiles 3 and 4 (Q3, Q4) of CHI3L1/YKL-40 had more than a 2-fold increased risk of postoperative delirium (RR [95% CI]: 2.2[1.1, 4.3] and 2.4[1.2, 5.0], respectively) compared to those in Q1. At POD2, those in Q4 of IL6 had over a 2-fold increased risk of delirium compared to those in Q1 (RR [95% CI]: 2.1 [1.1–4.1], respectively).
Table 3.
Modeling of CHI3L1/YKL-40 With and Without IL-6 and CRP (a comparison of models) and Their Association With Delirium
| Marker | Models 1: Individual Models | Model 2: Single Multivariable Model |
|---|---|---|
| RR (95% CI) | RR (95% CI) | |
| PREOP: | ||
| YKL-40 | ||
| Q1 | Reference | Reference |
| Q2 | 2.0 (1.1–3.9) | 1.9 (1.0a–3.6) |
| Q3 | 2.5 (1.4–4.7) | 2.2 (1.1–4.3) |
| Q4 | 2.9 (1.6–5.5) | 2.4 (1.2–5.0) |
| p-for trend | <.01 | .03 |
| CRP | ||
| Q1 | Reference | Reference |
| Q2 | 1.2 (0.7–2.2) | 1.3 (0.7–2.3) |
| Q3 | 1.6 (1.0b–2.8) | 1.5 (0.8–2.6) |
| Q4 | 1.8 (1.0 c –3.0) | 1.4 (0.8–2.4) |
| p-for trend | .02 | .23 |
| POD2: | ||
| YKL-40 | ||
| Q1 | Reference | Reference |
| Q2 | 1.2 (0.7–2.2) | 0.9 (0.5–1.7) |
| Q3 | 1.8 (1.0 d –3.1) | 1.2 (0.6–2.1) |
| Q4 | 2.0 (1.1–3.4) | 1.1 (0.5–2.1) |
| p-for trend | <.01 | .66 |
| CRP | ||
| Q1 | Reference | Reference |
| Q2 | 1.1 (0.7–2.0) | 1.0 (0.6–1.8) |
| Q3 | 1.5 (0.9–2.5) | 1.2 (0.7–2.0) |
| Q4 | 1.5 (0.9–2.5) | 1.0 (0.6–1.7) |
| p-for trend | .06 | .99 |
| IL-6 | ||
| Q1 | Reference | Reference |
| Q2 | 1.5 (0.8–2.8) | 1.5 (0.8–2.7) |
| Q3 | 2.0 (1.1–3.5) | 1.8 (0.9–3.3) |
| Q4 | 2.3 (1.3–4.1) | 2.1 (1.1–4.1) |
| p-for trend | <.01 | .02 |
Notes: All models adjusted for age, sex, surgery type, Charlson comorbidity index, medical complications, and baseline general cognitive performance.
Bold indicates significant at p < .05.
CHI3L1/YKL-40 (ng/l): PREOP - Q1 ≤38.97, Q2 38.98–70.80, Q3 70.81–122.69, Q4 ≥122.70; POD2 – Q1 ≤286.74, Q2 286.75–542.84, Q3 54.85–1040.66, Q4 ≥1040.67.
CRP (mg/l): POD2 - Q1 ≤127.53, Q2 127.54–177.05, Q3 177.06–235.73, Q4 ≥235.74.
IL-6 (mg/l): POD2 – Q1 ≤110.12, Q2 110.13–127.53, Q3 127.54–169.22, Q4 ≥265.4.
Model 1: CHI3L1/YKL-40 PREOP (N = 553), CHI3L1/YKL-40 POD2 (N = 556), CRP PREOP (N = 553), CRP POD2 (N = 556), IL6 POD2 (N = 555).
Model 2: N = 547.
CHI3L1/YKL-40 = Chitinase-3-like protein 1; CI = Confidence interval; CRP = C-reactive protein; IL6 = Interleukin-6; POD2 = Postoperative day 2; PREOP = Preoperative; RR = Relative risk.
Actual values: a0.97, b0.96, c1.04, d1.02.
Supplementary Table 3 reports the adjusted odds ratios (OR) of delirium severity for each sample-based quartile of CHI3L1/YKL-40, CRP, and IL6 considered in separate, individual analytic models (Model 1) and a single multi-protein model (Model 2). A strong association was observed for delirium severity and CHI3L1/YKL-4 (PREOP and POD2) and IL6 POD2. Relative to patients in Q1 of CHI3L1/YKL-40 at PREOP, patients in Q2, Q3, and Q4 had an increased odds of having severe delirium as opposed to no delirium (OR [95% CI]; Model 1): 3.7 [1.3–9.9], 3.2 [1.1–8.8], and 5.7 [2.1–15.6]), respectively. Relative to patients in Q1 of CHI3L1/YKL-40 at POD2, patients in Q3 and Q4 had an increased odds of having severe delirium as opposed to no delirium (OR [95% CI]; Model 1): 2.9 [1.2–7.3] and 3.8 [1.6–9.5], respectively. When all proteins were considered together (Model 2), CHI3L1/YKL-40 PREOP and IL-6 POD2 remained significant: relative to patients in Q1 of CHI3L1/YKL-40 PREOP, patients in Q2, Q3, and Q4 had an increased odds of having severe delirium compared to no delirium (OR [95% CI]): 3.2 [1.1–9.1], 2.5 [0.8–7.6], and 4.2[1.3–13.5], respectively).
Conclusions
Using SOMAscan proteomics for discovery followed by ELISA validation, we identified a novel, potentially clinically relevant delirium risk and disease plasma biomarker: CHI3L1/YKL-40. The following evidence provides support for the involvement of CHI3L1/YKL-40 in delirium: (i) strong and consistent associations of elevated CHI3L1/YKL-40 at PREOP and POD2 in the SOMAscan proteomics discovery phase; (ii) the only protein in common to PREOP and POD2 delirium predictor models; (iii) independent ELISA validation across the full SAGES cohort with significant adjusted RR in individual models at PREOP and POD2, and in a PREOP multi-protein model. Taken together, these findings indicate the potential value of CHI3L1/YKL-40 as a risk marker of delirium (ie, elevated before onset of delirium) and as a disease marker on POD2 (ie, rises with delirium onset, and may fall with delirium recovery).
Our study contributes important new information to the previous literature on inflammation and delirium by more specifically highlighting the involvement of the type 2 immune response. CHI3L1/YKL-40, a member of the glycosylhydrolase family, drives type 2 immunity by eliciting lymphocytic and eosinophilic inflammatory responses, spurring alternative macrophage responses (M2 response), shutting down innate immunity, and driving repair (33,34). Elevated blood levels of CHI3L1/YKL-40 have been associated with chronological age (35), multiple inflammatory conditions (eg, diseases associated with tissue remodeling, stroke, and traumatic brain injury) (36), AD (37), and all-cause mortality in octogenerarians (38).
CHI3L1/YKL-40 is expressed by different cell types, including macrophages, neutrophils, vascular smooth muscle cells, and chondrocytes, and plays a critical role in the type 2 immune response (39). Specifically, CHI3L1/YKL-40 mediates the differentiation of mature macrophages from monocytes, suggesting that the dysregulated levels of CHI3L1/YKL-40 expression are actively involved in chronic inflammation and immune disorders (40) and that CHI3L1/YKL40 is involved in diseases that are characterized by tissue remodeling and/or inflammation. CHI3L1/YKL-40 is a central regulator of adaptive and anti-pathogen T helper 2 (Th2) cell responses (33,39). Independently, infusions of both Th1 and Th2 mediating T cells have reported benefits in brain diseases, which include fewer plaque-related microglia in the hippocampus and restoration of near normal working memory in mice with the human transgenes for both the APP Swedish mutation and Presenilin 1 mutation (41). Taken together, our findings shed new light on the potential role of the type 2 immune activation in delirium pathophysiology.
Our identification of CHI3L1/YKL-40 in delirium pathophysiology aligns well with prior work demonstrating that CHI3L1/YKL-40, among several other proteins, increases post-surgery and that its relative abundance fold change (as opposed to absolute levels) is associated with postoperative outcomes (medical complications, longer length of stay, and discharge to a nursing home) (42). In contrast, our current study is the first, to our knowledge, to show that elevated absolute levels of plasma CHI3L1/YKL-40 are associated with risk and presence of postoperative delirium, as well as delirium severity. Our CHI3L1/YKL-40 finding is particularly interesting given the known epidemiologic links between delirium and AD (3,7,11,43,44). As the search for potential pathophysiologic mechanisms linking delirium and AD continues, our identification of CHI3L1/YKL-40 in delirium incidence and severity highlights a potential promising new avenue for investigation on delirium and its relationship with AD.
Our study has several notable strengths. First, the use of SOMAscan deep proteomics enabled measurement of over 1300 proteins for detecting and conducting high-dimensional functional characterization of pathophysiological correlates of delirium—a platform that has been used to identify proteins associated with additional outcomes (eg, biliary atresia (45)). Functional enrichment analysis using IPA highlighted key upstream regulators and biological functions that appear to be associated with delirium. This includes inflammation and other immune pathways (eg, migration, adhesion, and activation of immune cells) at PREOP and POD2, and vascular functions at POD2.
We acknowledge some limitations of our study. First, for the discovery proteomics phase, we used a subsample of 18 matched delirium/no-delirium pairs, which may not be representative of the full SAGES cohort. Despite this, our CHI3L1/YKL-40 findings were validated in the full cohort. This finding supports the feasibility and efficiency of using the matched case–control design for the initial biomarker discovery phase. Second, the SOMAscan cohort is a subset of the ELISA validation cohort, thus, the validation set is not entirely independent, although the overlap is small (36 out of 554). Third, our cohort included only patients undergoing major elective surgery, mostly orthopedic, in a relatively healthy, older population without dementia. Thus, our results may not be generalizable to patients undergoing other types of surgeries, delirium in nonsurgical settings (eg, ICU), or to older adults with dementia.
In summary, the identification of CHI3L1/YKL-40 using SOMAscan combined with ELISA validation and multi-protein modeling places CHI3L1/YKL-40 on a growing list of promising delirium biomarkers. CHI3L1/YKL-40 is unique in its specific role in immune activation and in its links to aging and AD (33,34,37,38). The emergence of CHI3L1/YKL-40 from a comprehensive proteomic analysis points to a potential link to etiopathogenic mechanisms of delirium. Although clinical utility of plasma CHI3L1/YKL-40 concentration as a stand-alone biomarker for delirium requires further investigation in additional, more diverse populations, our data reveal a robust peripheral signature of this highly specific, immune activating protein in delirium pathophysiology and potentially provides a link between delirium, aging, and AD.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the contributions of the patients, family members, nurses, physicians, staff members, and members of the Executive Committee who participated in the Successful Aging after Elective Surgery (SAGES) Study.
Funding
This work was supported by the National Institute on Aging grants (R01AG051658 to E.R.M. and T.A.L., P01AG031720 to S.K.I., K01AG057836 to S.M.V., R03AG061582 to S.M.V., R24AG054259 to S.K.I., R21AG057955 to T.G.F., R01AG041274 to Z.X., R21AG048600 to Z.X., and K24AG035075 to E.R.M.); and the Alzheimer’s Association (AARF-18-560786 to S.M.V.). S.I. holds the Milton and Shirley F. Levy Family Chair.
Author Contributions
S.M.V., E.R.M., and T.A.L. conceived of the idea for this project. S.M.V. and T.A.L. drafted the manuscript. S.T.D., N.Y.C., M.J., S.K.I., and E.R.M. acquired the data. S.M.V., L.H.N., B.T., and H.H.O. analyzed the data. All authors interpreted the data, and critically revised and approved the final submission for important intellectual content.
Conflict of Interest
The reports report no conflicts of interest.
Data Availability
A limited data set of the SAGES study can be requested through the NIH biobank: https://agingresearchbiobank.nia.nih.gov/
References
- 1. Martin BJ, Buth KJ, Arora RC, Baskett RJ. Delirium as a predictor of sepsis in post-coronary artery bypass grafting patients: a retrospective cohort study. Crit Care. 2010;14:R171. doi:10.1186/cc9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rudolph JL, Jones RN, Rasmussen LS, Silverstein JH, Inouye SK, Marcantonio ER. Independent vascular and cognitive risk factors for postoperative delirium. Am J Med. 2007;120:807–813. doi:10.1016/j.amjmed.2007.02.026 [DOI] [PubMed] [Google Scholar]
- 3. Witlox J, Eurelings LS, de Jonghe JF, Kalisvaart KJ, Eikelenboom P, van Gool WA. Delirium in elderly patients and the risk of postdischarge mortality, institutionalization, and dementia: a meta-analysis. JAMA. 2010;304:443–451. doi:10.1001/jama.2010.1013 [DOI] [PubMed] [Google Scholar]
- 4. Koster S, Hensens AG, van der Palen J. The long-term cognitive and functional outcomes of postoperative delirium after cardiac surgery. Ann Thorac Surg. 2009;87:1469–1474. doi:10.1016/j.athoracsur.2009.02.080 [DOI] [PubMed] [Google Scholar]
- 5. Marcantonio ER, Flacker JM, Michaels M, Resnick NM. Delirium is independently associated with poor functional recovery after hip fracture. J Am Geriatr Soc. 2000;48:618–624. doi:10.1111/j.1532-5415.2000.tb04718.x [DOI] [PubMed] [Google Scholar]
- 6. Saczynski JS, Marcantonio ER, Quach L, et al. Cognitive trajectories after postoperative delirium. N Engl J Med. 2012;367:30–39. doi:10.1056/NEJMoa1112923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis DH, Muniz Terrera G, Keage H, et al. Delirium is a strong risk factor for dementia in the oldest-old: a population-based cohort study. Brain. 2012;135(Pt 9):2809–2816. doi:10.1093/brain/aws190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marcantonio ER, Goldman L, Mangione CM, et al. A clinical prediction rule for delirium after elective noncardiac surgery. JAMA. 1994;271:134–139. doi:10.1001/jama.1994.03510260066030 [PubMed] [Google Scholar]
- 9. Robinson TN, Raeburn CD, Tran ZV, Angles EM, Brenner LA, Moss M. Postoperative delirium in the elderly: risk factors and outcomes. Ann Surg. 2009;249:173–178. doi:10.1097/SLA.0b013e31818e4776 [DOI] [PubMed] [Google Scholar]
- 10. Fong TG, Jones RN, Shi P, et al. Delirium accelerates cognitive decline in Alzheimer disease. Neurology. 2009;72:1570–1575. doi:10.1212/WNL.0b013e3181a4129a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fong TG, Davis D, Growdon ME, Albuquerque A, Inouye SK. The interface between delirium and dementia in elderly adults. Lancet Neurol. 2015;14:823–832. doi:10.1016/S1474-4422(15)00101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weiner WF. Impact of delirium on the course of Alzheimer disease. Arch. Neurol. 2012;69:1639–1640. doi:10.1001/archneurol.2012.2703 [DOI] [PubMed] [Google Scholar]
- 13. Leslie DL, Inouye SK. The importance of delirium: economic and societal costs. J Am Geriatr Soc. 2011;59Suppl 2:S241–S243. doi:10.1111/j.1532-5415.2011.03671.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maldonado JR. Neuropathogensis of delirium: review of current etiologic theories and common pathways. Am. J. Geriatr. Psychiatry 2013;12:1190–222. doi:10.1016/j.jagp.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 15. Dillon ST, Vasunilashorn SM, Ngo L, et al. Higher C-reactive protein levels predict postoperative delirium in older patients undergoing major elective surgery: a longitudinal nested case-control study. Biol. Psychiatry 2017;15:145–153. doi:10.1016/j.biopsych.2016.03.2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vasunilashorn SM, Dillon ST, Inouye SK, et al. High C-reactive protein predicts delirium incidence, duration, and severity after major non-cardiac surgery. J. Am. Geriatr. Soc. 2017;65:e109–e116. doi:10.1111/jgs.14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holmes C. Review: systemic inflammation and Alzheimer’s disease. Neuropathol Appl Neurobiol. 2013;39:51–68. doi:10.1111/j.1365-2990.2012.01307.x [DOI] [PubMed] [Google Scholar]
- 18. Vasunilashorn SM, Ngo L, Inouye SK, et al. Cytokines and postoperative delirium in older patients undergoing major elective surgery. J Gerontol A Biol Sci Med Sci. 2015;70:1289–1295. doi:10.1093/gerona/glv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vasunilashorn SM, Ngo LH, Chan NY, et al. Development of a dynamic multi-protein signature of postoperative delirium. J Gerontol A Biol Sci Med Sci. 2019;74:261–268. doi:10.1093/gerona/gly036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmitt EM, Marcantonio ER, Alsop DC, et al. ; SAGES Study Group . Novel risk markers and long-term outcomes of delirium: the successful aging after elective surgery (SAGES) study design and methods. J Am Med Dir Assoc. 2012;13:818.e1–818.10. doi:10.1016/j.jamda.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmitt EM, Saczynski JS, Kosar CM, et al. The successful aging after elective surgery study: cohort description and data quality procedures. J. Am. Geriatr. Soc. 2015;63:2463–2471. doi:10.1111/jgs.13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teng EL, Chui HC. The modified mini-mental state (3MS) examination. J Clin Psychiatry. 1987;48:314–318. [PubMed] [Google Scholar]
- 23. Jones RN, Rudolph JL, Inouye SK, et al. Development of a unidimensional composite measure of neuropsychological functioning in older cardiac surgery patients with good measurement precision. J Clin Exp Neuropsychol. 2010;32:1041–1049. doi:10.1080/13803391003662728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saczynski JS, Kosar CM, Xu G, et al. A tale of two methods: chart and interview methods for identifying delirium. J Am Geriatr Soc. 2014;62:518–524. doi:10.1111/jgs.12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inouye SK, van Dyck CH, Alessi CA, Balkin S, Siegal AP, Horwitz RI. Clarifying confusion: the confusion assessment method. A new method for detection of delirium. Ann Intern Med. 1990;113:941–948. doi:10.7326/0003-4819-113-12-941 [DOI] [PubMed] [Google Scholar]
- 26. Vasunilashorn SM, Marcantonio ER, Gou Y, et al. Quantifying the severity of a delirium episode throughout hospitalization: the combined importance of intensity and duration. J Gen Int Med. 2016;31:1164. doi:10.1007/s11606-016-3671-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ngo LH, Inouye SK, Jones RN, et al. Methodologic considerations in the design and analysis of nested case-control studies: association between cytokines and postoperative delirium. BMC Med Res Methodol. 2017;17:88. doi:10.1186/s12874-017-0359-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hathout Y, Brody E, Clemens PR, et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2015;112:7153–7158. doi:10.1073/pnas.1507719112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoaglin DC, Mosteller F, Tukey JW.. Understanding Robust and Exploratory Data Analysis. New Jersey: Wiley; 2000. [Google Scholar]
- 30. Pearson KL III. On lines and planes of closest fit to systems of points in space. London Edinburgh Dublin Philosoph. Mag. J. Sci. 1901;2:559–572. [Google Scholar]
- 31. Hsu CW, Lin CJ. A comparison of methods for multiclass support vector machines. IEEE Trans Neural Netw. 2002;13:415–425. doi:10.1109/72.991427 [DOI] [PubMed] [Google Scholar]
- 32. Hampel H, Toschi N, Baldacci F, et al. Alzheimer’s disease biomarker-guided diagnostic workflow using the added value of six combined cerebrospinal fluid candidates: Aβ1-42, total-tau, phosphorylated-tau, NFL, neurogranin, and YKL-40. Alzheimers Dement. 2018;14:492–501. doi:10.1016/j.jalz.2017.11.015 [DOI] [PubMed] [Google Scholar]
- 33. Lee CG, Da Silva CA, Dela Cruz CS, et al. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011;73:479–501. doi:10.1146/annurev-physiol-012110-142250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee CG, Dela Cruz CS, Herzog E, Rosenberg SM, Ahangari F, Elias JA. YKL-40, a chitinase-like protein at the intersection of inflammation and remodeling. Am J Respir Crit Care Med. 2012;185:692–694. doi:10.1164/rccm.201202-0203ED [DOI] [PubMed] [Google Scholar]
- 35. Bojesen SE, Johansen JS, Nordestgaard BG. Plasma YKL-40 levels in healthy subjects from the general population. Clin Chim Acta. 2011;412:709–712. doi:10.1016/j.cca.2011.01.022 [DOI] [PubMed] [Google Scholar]
- 36. Bonneh-Barkay D, Zagadailov P, Zou H, et al. YKL-40 expression in traumatic brain injury: an initial analysis. J Neurotrauma. 2010;27:1215–1223. doi:10.1089/neu.2010.1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Villar-Piqué A, Schmitz M, Hermann P, et al. Plasma YKL-40 in the spectrum of neurodegenerative dementia. J Neuroinflammation. 2019;16:145. doi:10.1186/s12974-019-1531-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johansen JS, Pedersen AN, Schroll M, Jørgensen T, Pedersen BK, Bruunsgaard H. High serum YKL-40 level in a cohort of octogenarians is associated with increased risk of all-cause mortality. Clin Exp Immunol. 2008;151:260–266. doi:10.1111/j.1365-2249.2007.03561.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee CG, Hartl D, Lee GR, et al. Role of breast regression protein 39 (BRP039)/chitinase 3-like-1 in th2 and IL-13-induced tissue responses and apoptosis. J. Exp. Med. 2009;206:1149–1166. doi:10.1084/jem.20081271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rehli M, Niller HH, Ammon C, et al. Transcriptional regulation of CHI3L1, a marker gene of late stages of macrophage differentiation. J. Biol. Chem. 2003;278:44058–44067. doi:10.1074/jbc.M306792200 [DOI] [PubMed] [Google Scholar]
- 41. Cao C, Arendash GW, Dickson A, Mamcarz MB, Lin X, Ethell DW. Abeta-specific Th2 cells provide cognitive and pathological benefits to Alzheimer’s mice without infiltrating the CNS. Neurobiol Dis. 2009;34:63–70. doi:10.1016/j.nbd.2008.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fong TG, Chan NY, Dillon ST, et al. Identification of plasma proteome signatures associated with surgery using SOMAscan. Ann. Surg. 2019. Epub ahead of print. doi:10.1097/SLA.0000000000003283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Inouye SK, Marcantonio ER, Kosar CM, et al. The short-term and long-term relationship between delirium and cognitive trajectory in older surgical patients. Alzheimers Dement. 2016;12:766–775. doi:10.1016/j.jalz.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fong TG, Vasunilashorn SM, Libermann T, Marcantonio ER, Inouye SK. Delirium and Alzheimer disease: a proposed model for shared pathophysiology. Int J Geriatr Psychiatry. 2019;34:781–789. doi:10.1002/gps.5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lertudomphonwanit C, Mourya R, Fei L, et al. Large-scale proteomics identifies MMP-7 as a sentinel of epithelial injury of biliary arterisa. Sci. Transl. Med. 2017;9:eaan8462. doi:10.1126/scitranslmed.aan8462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A limited data set of the SAGES study can be requested through the NIH biobank: https://agingresearchbiobank.nia.nih.gov/