Abstract
The transparency of Caenorhabditis elegans provides a unique window to observe and study the function of germ granules. Germ granules are specialized ribonucleoprotein (RNP) assemblies specific to the germline cytoplasm, and they are largely conserved across Metazoa. Within the germline cytoplasm, they are positioned to regulate mRNA abundance, translation, small RNA production, and cytoplasmic inheritance to help specify and maintain germline identity across generations. Here we provide an overview of germ granules and focus on the significance of more recent observations that describe how they further demix into sub-granules, each with unique compositions and functions.
Keywords: C. elegans, germline, germ granules, P granules, Mutator foci, Z granules, piRNAs, WormBook
Overview of germ granules
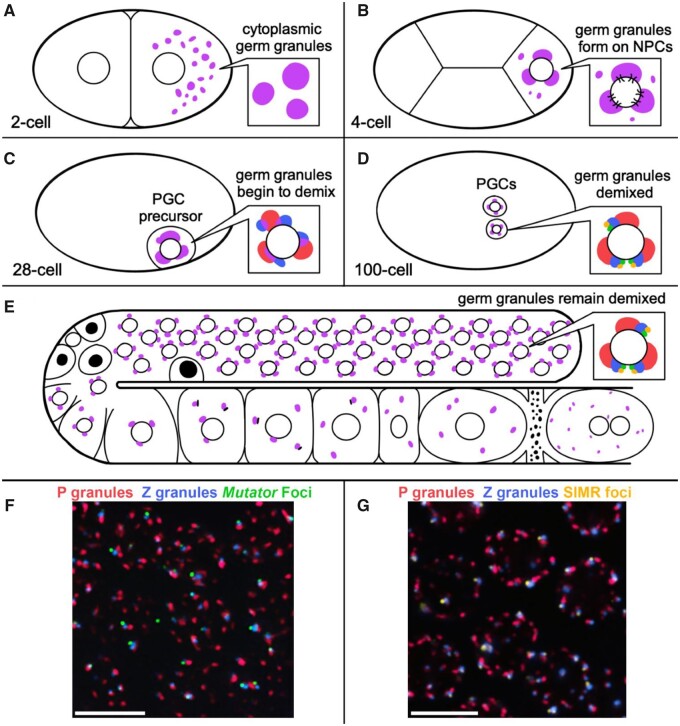
Early studies of animal germline specification noted visibly dense assemblies on the cytoplasmic surface of germline nuclei [reviewed in Eddy (1975)]. These assemblies have been called by various names across species and throughout germline development, but in recent years a consensus has emerged to collectively refer to them as germ granules. In C. elegans, the first observed germ granules were called P granules because they segregate with the P cell lineage (germline blastomeres) during embryogenesis [Figure 1, A–D; reviewed in Strome (2005), Hubbard and Greenstein (2005)]. Today, the term “germ granule” defines a collection of dynamic and germline-specific perinuclear assemblies that can be distinguished through high-resolution microscopy. Other cytoplasmic assemblies, such as P bodies, stress granules, and those which form in arrested oocytes will not be covered here. In early embryos, the terms germ granule and P granule are used interchangeably, but as development progresses, germ granules further demix into sub-granules that, along with P granules, include Mutator foci, Z granules, SIMR-1 foci, and likely other perinuclear assemblies yet to be defined (Figure 1, C–G). Caenorhabditis elegans provide a window to observe the dynamics of these sub-granules, revealing their function in the germline and specific contributions of their individual components. These findings can then be extended to better define the roles of germ-granule proteins in other animals.
Figure 1.
Germ granule distribution and demixing during development. (A) Posteriorly localized germ granules (purple) in the 2-cell embryo are dispersed in the cytoplasm. (B) Germ granules in the 4-cell embryo begin to adhere to the cytoplasmic surface of the nuclear envelope and cluster NPCs. (C) Germ granule demixing begins at the 28-cell stage in primordial germ cell (PGC) precursors. (D) Germ granules in the PGCs of 100-cell embryos have demixed into adjacent P granules, Z granules, SIMR foci and Mutator foci. (E) Germ granules remain demixed in adult germ cells (inset). PGL proteins, but not GLH proteins, are cleared from P granules during physiological apoptosis (black nuclei). P granules disperse into the cytoplasm of oocytes prior to fertilization, initially with part of the nuclear envelope attached. (F, G) Immunofluorescence image of adult germ cells (pachytene) with demixed sub-granules. (images Celja Uebel. scale bars = 5 μm).
Germ granules and their subtypes are heterogeneous ribonucleoproteins. Of the ∼90 C. elegans proteins currently known to be germ-granule enriched (Table 1), almost all have RNA binding domains. They include RNA (mostly DEAD-box) helicases, mRNA stabilizing and destabilizing proteins, translation initiation factors, small RNA-binding Argonautes, RNA polymerases, mRNA export and nuclear pore complex components, and many proteins harboring KH, CCCH zinc-finger, Tudor, and LOTUS domains. Together, assemblies of these germline proteins at the nuclear periphery position them to regulate mRNA abundance, translation, small RNA production, and cytoplasmic inheritance through cell divisions and across generations. In the proceeding paragraphs, we explore the function of germ granules, and what the composition and modularity of known germ-granule subtypes reveal about their function.
Table 1.
Germ granule proteins (P granules, Z granules, Mutator foci, SIMR foci, and unknown)
| Germ granule | Protein | Description | References |
|---|---|---|---|
| P | ALG-3 | Argonaute expressed during spermatogenesis | Conine et al. (2010) |
| P | ALG-4 | Argonaute expressed during spermatogenesis | Conine et al. (2010) |
| P | ALG-5 | Argonaute associated with miRNAs | Brown et al. (2017) |
| P | CAR-1 | Cytokinesis, apoptosis, and RNA-binding 1 TRAL/Lsm14 | Audhya et al. (2005), Boag et al. (2005), Squirrell et al. (2006) |
| P | CCF-1 | CCR4/NOT deadenylase complex | Gallo et al. (2008) |
| P | CDE-1 | Uracil nucleotidyltransferase | van Wolfswinkel et al. (2009) |
| P | CGH-1 | Dhh1/DDX6 DEAD-box helicase | Navarro et al. (2001) |
| P | CSR-1 | Argonaute required for endo-siRNA | Claycomb et al. (2009) |
| P | DCAP-1 | mRNA decapping enzyme | Squirrell et al. (2006) |
| P | DCAP-2 | mRNA decapping enzyme | Lall et al. (2005) |
| P | DCR-1 | Dicer-related RNAse | Beshore et al. (2011) |
| P | DDX-19 | DDX19 DEAD-box helicase | Sheth et al. (2010) |
| P | DEPS-1 | Defective P granules and Sterile | Spike et al. (2008a) |
| P | DRH-3 | Dicer-related DEAD-box helicase | Claycomb et al. (2009) |
| P | EGO-1 | RNA-directed RNA polymerase (RdRP) | Claycomb et al. (2009) |
| P | ERH-2 | 21U-RNA maturation | Cordeiro Rodrigues et al. (2019) and Zeng et al. (2019) |
| P | FBF-2 | PUF-domain fem-3 mRNA 3′UTR-binding factor | Voronina (2012) |
| P | GLD-1 | RNA-binding KH domain | Jones et al. (1996) |
| P | GLD-2 | Poly(A) polymerase | Wang et al. (2002) |
| P | GLD-3 | RNA-binding KH domain | Eckmann et al. (2002) |
| P | GLD-4 | Poly(A) polymerase | Schmid et al. (2009) |
| P | GLH-1 | Vasa DEAD-box helicase | Gruidl et al. (1996) |
| P | GLH-2 | Vasa DEAD-box helicase | Gruidl et al. (1996) |
| P | GLH-3 | Vasa DEAD-box helicase | Kuznicki et al. (2000) |
| P | GLH-4 | Vasa DEAD-box helicase | Kuznicki et al. (2000) |
| P | GLS-1 | GLD-3/4 interacting protein | Rybarska et al. (2009) |
| P | HENN-1 | 3′ RNA methyltransferase | Kamminga et al. (2012) |
| P |
|
Heritable RNAi deficient | Spracklin et al. (2017) and Lewis et al. (2020) |
| P | IFE-1 | eIF4E mRNA cap-binding | Amiri et al. (2001) |
| P | IFE-3 |
|
Cordeiro Rodrigues et al. (2019), Zeng et al. (2019), and Huggins et al. (2020) |
| P | IFET-1 | eIF4E transporter | Sengupta et al. (2013) |
| P | LAF-1 | DDX3 DEAD-box helicase | Hubert and Anderson (2009) |
| P | MBK-2 | DYRK3 YAK-related kinase | Stitzel et al. (2007) |
| P | MEG-1 | Maternal effect germ cell defective | Leacock and Reinke (2008) |
| P | MEG-2 | Maternal effect germ cell defective | Leacock and Reinke (2008) |
| P |
|
Maternal effect germ cell defective GCNA-IDR | Wang et al. (2014) |
| P | MEG-4 | Maternal effect germ cell defective GCNA-IDR | Wang et al. (2014) |
| P | MEX-1 | CCCH-type zinc-finger protein | Guedes and Priess (1997) |
| P | MEX-3 | RNA-binding KH domain | Draper et al. (1996) |
| P | MIP-1 | LOTUS-containing MEG-3 interacting protein | Cipriani et al. (2021) |
| P | MIP-2 | LOTUS-containing MEG-3-interacting protein | Cipriani et al. (2021) |
| P | NOS-2 | Nanos-related protein | Subramaniam and Seydoux (1999) |
| P | NPP-8 | NUP155 NPC protein | Voronina and Seydoux (2010) |
| P | NPP-10 | NUP98 NPC protein | Voronina and Seydoux (2010) |
| P | NXF-1 | NXF1/TAP-like mRNA export factor | Sheth et al. (2010) |
| P | OMA-1 | CCCH-type zinc-finger protein | Shimada et al. (2002) |
| P | OMA-2 | CCCH-type zinc-finger protein | Shimada et al. (2002) |
| P | PAB-1 | Poly(A)-binding protein 1 | Gallo et al. (2008) |
| P | PAN-1 | LRRTM4 DEAD-box helicase | Gao et al. (2012) |
| P | PARN-1 | Poly(A)-specific 3′–5′-exoribonuclease | Tang et al. (2016) |
| P | PATR-1 | Pat1 decapping cofactor | Gallo et al. (2008) |
| P | PGL-1 | RGG-containing P granule endoribonuclease | Kawasaki et al. (1998) |
| P | PGL-2 | PGL-1 related | Kawasaki et al. (2004) |
| P | PGL-3 | RGG-containing P granule endoribonuclease | Kawasaki et al. (2004) |
| P | PID-1 | piRNA-induced silencing defective | Cordeiro Rodrigues et al. (2019) and Zeng et al. (2019) |
| P |
|
piRNA-induced silencing defective | Cordeiro Rodrigues et al. (2019) and Zeng et al. (2019) |
| P | PID-4 | piRNA-induced silencing defective | Placentino et al. (2021) |
| P | PID-5 | Aminopeptidase, piRNA-induced silencing defect | Placentino et al. (2021) |
| P | PIE-1 | CCCH-type zinc-finger protein | Mello et al. (1996) |
| P | PLP-1 | Pur alpha-like protein | Witze et al. (2009) |
| P | POS-1 | CCCH-type zinc-finger protein | Tabara et al. (1999a) |
| P | PRG-1 | Argonaute required for piRNA synthesis | Batista et al. (2008) |
| P | PUF-8 | PUF (Pumilio/FBF) domain 3′UTR-binding factor | Ariz et al. (2009) |
| P | RDE-12 | RNAi defective DEAD-box helicase | Sheth et al. (2010) |
| P | RNP-8 | RRM poly(G) RNA binding | Kim et al. (2009) |
| P | SIR-2.2 | Sirtuin 4-like protein deacetylase | Jedrusik-Bode et al. (2013) |
| P | Sm prot. | Splicing factors | Barbee et al. (2002) |
| P | SPN-2 | eIF4E-binding protein | Li et al. (2009) |
| P | SPN-4 | RNP-type RNA-binding domain | Ogura et al. (2003) |
| P | TIA-1 | TIA-1 RNP-type RNA-binding domain | Gallo et al. (2008) |
| P | TOFU-6 | 21U-RNA fouled up | Cordeiro Rodrigues et al. (2019) and Zeng et al. (2019) |
| P | VBH-1 | Vasa Belle-like DEAD-box helicase | Salinas et al. (2007) |
| P | WAGO-1 | Argonaute required for endo-siRNA | Gu et al. (2009) |
| P | WAGO-3 | Argonaute present in sperm | Schareier et al. (2021) |
| P | Y51F10.2 | TRIM32 E3 ubiquitin-protein ligase | Lee et al. (2020) |
| M | MUT-2 RDE-3 | Mutator with predicted nucleotidyltransferase activity | Phillips et al. (2012) |
| M | MUT-7 | Mutator with predicted 3′–5′ exoRNAse activity for miRNA end processing | Phillips et al. (2012) |
| M | MUT-8 RDE-2 | Mutator RNAi defective | Phillips et al. 2012) |
| M | MUT-14 | Mutator resembling the DDX3 DEAD-box helicase | Phillips et al. (2012) |
| M | MUT-15 RDE-5 | Mutator RNAi defective | Phillips et al. (2012) |
| M |
|
Mutator RNAi defective | Phillips et al. (2012) |
| M | NYN-1 | NYN domain ribonuclease homolog | Uebel et al. (2018) |
| M | NYN-2 | NYN domain ribonuclease homolog | Uebel et al. (2018) |
| M | RDE-8 | mRNA-binding endo-RNAse that positively regulates RdRP activity | Tsai et al. (2015) |
| M | RRF-1 | RNA-directed RNA polymerase (RdRP) | Phillips et al. (2012) |
| M | SMUT-1 | Synthetic mutator DDX3 DEAD-box helicase-like | Phillips et al. (2014) |
| Z | LOTR-1 | LOTUS and Tudor domain protein | Marnik et al. (2021) |
| Z |
|
piRNA-induced silencing defective | Placentino et al. (2021) and Wan et al. (2021) |
| Z | WAGO-4 | Argonaute required for RNAi inheritance | Wan et al. (2018) |
| Z | ZNFX-1 | NFX1-type zinc finger-containing protein | Ishidate et al. (2018) and Wan et al. (2018) |
| S | HPO-40 | SIMR-1-like Tudor-domain protein | Manage et al. (2020) |
| S | RSD-2 | RNAi spreading defective | Manage et al. (2020) |
| S | SIMR-1 | siRNA defective and mortal germline Tudor-domain protein | Manage et al. (2020) |
| ? | FBF-1 | PUF-domain fem-3 mRNA 3′UTR-binding factor that docks next to PGL-1 | Voronina et al. (2012) |
| ? | MINA-1 | RNA-binding KH protein that docks next to PGL-1 | Sendoel et al. (2019) |
Organization and function of P granules
Overview of P granules
The discovery of P granules in C. elegans (Strome and Wood 1982) followed the discovery of asymmetric segregation of germ granules that had been previously described during early embryonic cleavages in several invertebrates and amphibians (Ritter 1890; Hegner 1911; Penners 1922; Smith 1966; Mahowald et al. 1976). It was recognized that partitioning of P granules in early embryos could be used to understand how asymmetry is established prior to the first cell division [(Kemphues et al. 1988; reviewed in Rose and Gonczy (2014)]. Moreover, the accessibility of P granules provided a way to identify their core components and observe their formation, condensation, and dissolution in early embryogenesis (reviewed in Seydoux 2018).
Structural composition of P granules
The list of P-granule-associated proteins continues to expand (Table 1). Many of these proteins associate transiently with P granules in the early embryo or at other stages of germline development and gametogenesis. A smaller contingent can be described as constitutive proteins that are found in P granules at all stages of the C. elegans lifecycle. Among these are a few novel proteins (DEPS-1, PGL-1, and PGL-3), the Vasa DEAD-box germline helicases (GLH-1, GLH-2, GLH-3, and GLH-4), other related DEAD-box helicases (RDE-12, LAF-1, VBH-1), and some of the Argonaute proteins (CSR-1, PRG-1, and worm-specific Argonaute (WAGO)-1). PGL-1 and PGL-3 are scaffolding proteins that interact through two dimerization domains, exhibit RNA endonuclease activity in vitro, and have the ability to repress tethered mRNA expression in vivo (Aoki et al. 2016, 2021). Ectopic expression of PGL-1 and PGL-3, either in the soma of C. elegans or in mammalian cell culture, is sufficient to nucleate P-granule-like condensates (Hanazawa et al. 2011; Updike et al. 2011). Interestingly, different proteins nucleate germ-granule formation in other animals, supporting the idea that germ-granule nucleators evolved through convergent evolution [reviewed in Kulkarni and Extavour (2017)]. Recombinant PGL-3 can self-assemble under physiological conditions in vitro (Saha et al. 2016). In vivo, the posterior condensation of PGL-1 and PGL-3 in the zygote is facilitated by a MEG-3 and MEG-4 scaffold to coat PGL assemblies, and LOTUS-domain MEG-3 interacting proteins MIP-1 and MIP-2 (Wang et al. 2014; Chen et al. 2016; Smith et al. 2016; Putnam et al. 2019; Folkmann et al. 2021; Price et al. 2021; Schmidt et al. 2021; Cipriani et al. 2021). During spermatogenesis, PGL-1 and PGL-3 are cleared from P granules while GLH proteins are retained, suggesting that nucleators are not always needed to maintain P granules after they are formed [discussed in Updike and Strome (2010)].
The RNA composition of P granules is better defined in embryos than in adults. Early studies demonstrated that embryonic P granules retain both maternally expressed and developmentally regulated mRNAs (Seydoux and Fire 1994; Subramaniam and Seydoux 1999; Schisa et al. 2001). More recently, MEG-3 iCLIPs identified approximately 500 specific mRNAs that are enriched in embryonic P granules (Lee et al. 2020). In situ hybridization studies revealed that nascent mRNAs pass into and through P granules, and that their perinuclear enrichment may be a result of their slowed diffusion during the transit (Sheth et al. 2010). In contrast to mRNAs, ribosomal RNAs are not enriched in P granules and even appear excluded (Schisa et al. 2001; Marnik et al. 2019), suggesting that P granules are devoid of translation. In fact, some P-granule localized transcripts are correlated with low translational status and ribosome coverage, and global translation inhibition directs numerous transcripts to embryonic P granules in a MEG-3 dependent but nonsequence specific manner (Lee et al. 2020; Parker et al. 2020). These results suggest embryonic P granules are a way to maintain a pool of maternal mRNAs in germline precursors until they resume zygotic transcription.
Association of P granules with the nuclear pore complex
The association of C. elegans P granules on the cytoplasmic surface of the nuclear periphery reflects the general distribution of germ granules across species. In the C. elegans germline, P granules cluster nuclear pore complexes (NPCs), and 75% of NPCs are covered by P granules (Figure 1B;Pitt et al. 2000). NPCs not covered by P granules are only found where a prominent lobe from the nucleolus contacts the nuclear envelope, suggesting that rRNAs enter the cytoplasm without passing through P granules (Sheth et al. 2010). Nuclear export factors, such as NXF-1 and DDX-19, and peripheral nucleoporins, such as NPP-8, NPP-9, and NPP-10, localize to the base of P granules (Sheth et al. 2010; Voronina and Seydoux 2010). Upon the cytoplasmic dispersal of P granules during oogenesis, some P granules retain attached NPCs (Pitt et al. 2000). In addition, RNAi depletion of a number of nucleoporins cause P granules to detach from the nuclear periphery and disperse into the cytoplasm (Updike and Strome 2009; Voronina and Seydoux 2010). These findings demonstrate the tight association between P granules and NPCs.
P granules and NPC contacts are likely mediated through glycine-rich FG-repeat domains present in several nucleoporins (FG-Nups) and P-granule proteins such as GLH-1, GLH-2, GLH-4, DDX-19, RDE-12 (Sheth et al. 2010). Unstructured FG-repeat domains fill up the pore of NPCs to establish the size exclusion barrier between the nucleus and cytoplasm. A current model is that the regularly spaced phenylalanines form weak hydrophobic interactions to create a mesh or smart sieve (Schmidt and Görlich 2016). Proteins under 40 kD can diffuse freely through the sieve, while larger proteins require a karyopherin for import and export. P granules extend the 40 kD size exclusion barrier beyond the pore and into the cytoplasm, and weak concentrations of hexanediol capable of disrupting the hydrophobic interactions within the pore also disperse P granules (Updike et al. 2011). P-granule FG-repeat proteins form hydrophobic tethers with FG-Nups to maximize coverage of NPCs, positioning P granules to receive nascent transcripts that exit the nucleus. Deleting FG-repeats from GLH-1 and GLH-2 increases P-granule size and sphericity as they lose contact with the nuclear periphery (Marnik et al. 2019; Chen et al. 2020).
P granules regulate germline apoptosis
Over half of the oogenic germ cells undergo physiological apoptosis (Gartner et al. 2008). Germ cells are connected to a cytoplasmic syncytium, and excess germ cells function as nurse cells and dump their mitochondria and other cytoplasmic contents into the shared cytoplasm as apoptosis is initiated (Raiders et al. 2018). PGL-1 and PGL-3 disappear from these apoptotic cells, suggesting that P granules and their components not only play a role in the formation of germ cells, but also their preservation (Pitt et al. 2000; Sheth et al. 2010). In response to UV damage, pgl-1 and pgl-3 mutants show elevated levels of apoptosis; the pro-apoptotic factor Apaf1/CED-4 accumulates, and Sirtuin/SIR-2.1, which functions as an antiapoptotic factor in the nucleus, is translocated into the cytoplasm. These findings suggest that P granules (or the presence of PGL proteins in P granules) suppress these pro-apoptotic activities (Min et al. 2016). Supporting these findings, somatic programmed cell death is suppressed in synMuvB mutants that express somatic P granules; similarly, ectopic expression of PGL-1 or PGL-3 from transgenes is sufficient to repress apoptosis in the soma in a SIR-2.1-dependent manner (Al-Amin et al. 2016). PGL proteins are cleared by autophagy in somatic blastomeres during embryogenesis (Zhang et al. 2009, 2018a). Similarly, PGL proteins in the adult germline are cleared by autophagy following UV-induced DNA damage, linking the requirement of autophagy machinery to UV-induced apoptosis in the germline (Min et al. 2019).
P granules promote germline gene expression
Given the complex composition of P granules and the redundancy involved in their nucleation, depleting single P-granule components does not clear P granules from the adult germline. However, core P-granule proteins and electron-dense P-granule assemblies are no longer detected in mex-3 gld-1 double mutants, and this absence correlates with germ-to-soma transdifferentiation (Ciosk et al. 2006). The simultaneous RNAi depletion of multiple core P-granule proteins inhibits fertility and also causes germ-to-soma transdifferentiation (Updike et al. 2014), and an increase in somatic expression in the germline of older adults (Knutson et al. 2017). These results suggest that P granules safeguard germline development through mRNA surveillance mechanisms that repress the accumulation and translation of somatic transcripts that become stochastically expressed. The analysis of deps-1, glh-1, glh-2, glh-4, pgl-1, and pgl-3 single mutants has revealed underproliferated germlines and defects in gametogenesis at restrictive temperatures, while sterility in double mutants can make it difficult to distinguish primary from secondary effects on gene expression (Kawasaki et al. 2004; Spike et al. 2008a, 2008b). Before the onset of these defects in deps-1 and glh-1 single mutants, expression profiling reveals only subtle germline expression changes (Spike et al. 2008a, 2008b). The same is the case in healthy germlines of young adults after simultaneous RNAi depletion of multiple transcripts (pgl-1, pgl-3, glh-1, glh-4), except for a global increase of spermatogenic transcripts in proximal germ cells slated to undergo oogenesis (Campbell and Updike 2015; Knutson et al. 2017).
Mechanisms used by P granules to recognize and suppress somatic and spermatogenic expression in the adult germline are areas of continuing focus. During spermatogenesis PGL proteins are cleared from secondary spermatocytes (Amiri et al. 2001), while GLH proteins, which are necessary for the completion of spermatogenesis (Kuznicki et al. 2000), are retained in P granules until they are deposited in the residual body near the completion of spermatogenesis (Gruidl et al. 1996). The expression of P-granule associated Argonaute proteins like CSR-1, WAGO-1 and WAGO-3 is accompanied by transient expression of Argonautes ALG-3 and ALG-4 during spermatogenesis; while all of these Argonautes are implicated in paternal inheritance, CSR-1, WAGO-1 and WAGO-3 persist in sperm while ALG-3 and ALG-4 become deposited in residual bodies (Conine et al. 2010, 2013; Schreier et al. 2021). How the temporospatial expression of each these factors impact spermatogenesis still needs to be resolved. Recent studies have shown that CSR-1 has both long (a) and short (b) isoforms, but the long CSR-1a isoform is selectively expressed during spermatogenesis in L4 hermaphrodites, where it primarily targets spermatogenic genes (Nguyen and Phillips 2021; Charlesworth et al. 2021). Dimethylarginine modifications to the CSR-1a isoform are necessary for this target specificity (Nguyen and Phillips 2021). Translational initiation may also be involved; for example, the PGL-associated isoform of the m7G cap-binding eIF4E initiation factor IFE-1 promotes sperm translation—an activity likely repressed with the occurrence of PGLs in the first wave of oogenesis (Amiri et al. 2001; Henderson et al. 2009; Friday et al. 2015). In contrast, a second isoform of the m7G cap eIF4E, IFE-3, associates with eIF4E transporter IFET-1 to drive oocyte translation and the sperm-to-oocyte switch but is not required for spermatogenesis (Sengupta et al. 2013; Huggins et al. 2020).
P granules as sites of mRNA surveillance
The tight association with the NPC positions P granules to survey transcripts for foreign or somatic sequences as they exit the nucleus. This epigenetic memory of germline expression is conferred through the small RNA machinery within P granules. The PIWI-class Argonaute, PRG-1, is a constitutive P-granule component that associates with more than 10,000 distinct Piwi-interacting RNAs (piRNAs), also known as 21U RNAs in C. elegans due to their 21-nt length and 5′ bias for uracil, to form piRNA-induced silencing complexes (piRISCs). These piRISCs use imperfect complementarity to engage and surveil the entire germline transcriptome, including mRNAs, non-coding RNAs, and transposable elements (Ruby et al. 2006; Wang and Reinke 2008; Batista et al. 2008; Lee et al. 2012; Bagijn et al. 2012; Shen et al. 2018; Zhang et al. 2018b). piRNA targeting can initiate heritable epigenetic silencing that, in many cases, bypasses the need for piRNAs to maintain that silencing in subsequent generations (Shirayama et al. 2012; Ashe et al. 2012; Luteijn et al. 2012). This heritable silencing is mediated by WAGO 22G-RNAs, 22-nt siRNAs with a 5′ guanosine bias, that are bound by a class of WAGO proteins. WAGO 22G-RNAs are synthesized by RNA-dependent RNA polymerases (RdRPs) associated with the mutator complex (described in more detail below; Gu et al. 2009; Phillips et al. 2012).
The biogenesis of piRNAs requires both 5′ and 3′ processing. First, the 5′ piRNA end maturation is carried out by the PETISCO/PICS complex, made up of PID-1, PID-3/PICS-1, ERH-2, TOFU-6, and IFE-3 (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019; Perez-Borrajero et al. 2021). This maturation involves decapping and removal of two nucleotides from the 5′ end of the piRNA precursor (Gu et al. 2012). The PETISCO/PICS complex is also P-granule associated, though the co-localization of this complex with known P-granule factors may be imperfect (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019; Perez-Borrajero et al. 2021). For example, perinuclear IFE-3 granules appear to dock next to PGL-1 labeled granules (Huggins et al. 2020). 5′ end maturation is followed by loading of the piRNA precursor into PRG-1 where the 3′ end is trimmed by the P granule-localized exonuclease PARN-1 (Tang et al. 2016). Finally, the 3′ end of the piRNA is 2′O-methylated by the methyltransferase HENN-1, which may also localize to P granules (Billi et al. 2012; Montgomery et al. 2012; Kamminga et al. 2012). While initially the mobilization of transposable elements was assumed to cause the transgenerational decline in fertility in prg-1 mutants, prg-1 sterility correlates more with perturbed P-granule structure and may be independent of genomic stability (Spichal et al. 2021). Also correlating with the progressive loss of fertility in prg-1 mutants, is the accumulation of 22G-RNAs antisense to replicative histone genes and ribosomal RNA genes and a corresponding reduced expression of histone mRNAs (Barucci et al. 2020; Montgomery et al. 2020; Reed et al. 2020; Wahba et al. 2021). Whether reduced expression of rRNAs and histones, disruption of P granules, or another unknown factor is underlying cause of transgenerational sterility in prg-1 mutant animals is a matter that will need further investigation.
Opposing piRNA-mediated silencing, 22G-RNAs bound by the P granule-associated Argonaute CSR-1 (CSR-1 22G-RNAs) license protein-encoding transcripts for germline expression (Claycomb et al. 2009; Gu et al. 2009; Seth et al. 2013; Wedeles et al. 2013; Cecere et al. 2014; Tu et al. 2014). CSR-1 22G-RNAs are produced through the activity of the EGO-1 RdRP (Claycomb et al. 2009), yet the mechanism by which EGO-1 activity is initiated on CSR-1 target transcripts has been a bit of an enigma. While WAGO 22G-RNAs can be initiated by piRNAs or other classes of primary siRNAs, there has been no known primary siRNA class in the CSR-1 pathway. However, recent work has demonstrated that CSR-1 slicer activity is necessary to trigger biogenesis of CSR-1 22G-RNAs within the coding region of CSR-1 target genes. Interestingly, CSR-1 slicer activity is not required for initiation of EGO-1 RdRP activity in CSR-1 target 3′UTRs. Thus, while it is unknown what triggers the recruitment of EGO-1 to target 3′UTRs, these new data indicate that within target gene bodies EGO-1 22G-RNA synthesis may be initiated following cleavage by 22G-RNA-bound CSR-1, independent of any primary siRNAs (Singh et al. 2021). Furthermore, it appears that CSR-1 22G-RNA biogenesis occurs on actively translated mRNAs in the cytoplasm, in contrast to other 22G-RNAs which are mostly synthesized in germ granules (Singh et al. 2021). Lastly, CSR-1 22G-RNAs can be uridylated by the P granule-localized nucleotidyl transferase CDE-1, to restrict their accumulation (van Wolfswinkel et al. 2009).
Through CSR-1 22G-RNA/WAGO 22G-RNA opposition, P granules have the capacity to retain a memory of germline expression. How CSR-1 promotes the expression of its 22G-RNA targets is unclear; however, one model is that CSR-1 may secure the passage of its targets through P granules and into the cytoplasm where they can ultimately be translated. Compromising CSR-1 and select components upstream of CSR-1 22G-RNA synthesis cause a very distinct enlarged P-granule phenotype (Vought et al. 2005; Claycomb et al. 2009; Updike and Strome 2009; Campbell and Updike 2015; Andralojc et al. 2017). This could reflect the pooling of CSR-1 22G-RNA target transcripts that can no longer make their way through and into the cytoplasm. Interestingly, while CSR-1 22G-RNA target mRNA levels change very little in dissected germlines and whole worm lysates, expression profiling in early embryos suggest a P-granule independent role for the slicer activity of CSR-1 in clearing its maternally deposited targets from somatic blastomeres (Quarato et al. 2021). Determining whether germ granules in other animals confer a similar mRNA surveillance system to retain an epigenetic memory of germline expression will be a critical next step.
Organization and function of Mutator foci
Overview of Mutator foci
The term “mutator” was first used to describe spontaneous mutations caused by insertions of the transposon Tc1 (Eide and Anderson 1985; Collins et al. 1987). Later experiments linked the phenomenon of transposon activation to the disruption of RNA interference pathways via two parallel genetic screens, one for defects in RNAi and the other for germline mobilization of transposons (Ketting et al. 1999; Tabara et al. 1999b). The overlap of these two screens provided some of the first evidence that the RNAi pathway is required for transposon silencing. Since then, nearly a dozen mutator genes have been identified, with their protein products found to interact to form the mutator complex and function in the production of WAGO 22G-RNAs and ERGO-1 26G-RNAs (see Table 1; Ketting et al. 1999; Tabara et al. 1999b; Tijsterman et al. 2002; Vastenhouw et al. 2003; Tops et al. 2005; Chen et al. 2005; Grishok et al. 2005; Robert et al. 2005; Kim et al. 2005; Phillips et al. 2012; Tsai et al. 2015). Consequently, loss of the mutator complex results in defects in the production of WAGO 22G-RNAs and ERGO-1 26G-RNAs but not other classes of small RNAs such as piRNAs, microRNAs (miRNAs), CSR-1 22G-RNAs, and ALG-3/4 26G- and 22G-RNAs (Gu et al. 2009; Zhang et al. 2011; Lee et al. 2012; Phillips et al. 2014; Tsai et al. 2015). Interestingly, the loss of ERGO-1 26G-RNAs in mutator mutants can be attributed to the disruption of a homeostatic feedback loop mediated by mutator-dependent 22G-RNAs at the eri-6/7 gene locus rather than the direct involvement of the mutator proteins in ERGO-1 26G-RNA biogenesis (Rogers and Phillips 2020b). Therefore, the mutator complex is thought to function primarily for the amplification of WAGO 22G-RNAs through the activity of RdRPs.
Mutations in genes from this group not only have active germline transposition and defects in exogenous RNAi, but they also are temperature-sensitive sterile and have more male progeny, suggestive of chromosome segregation defects (Ketting et al. 1999; Tabara et al. 1999b; Zhang et al. 2011; Wallis et al. 2019). The fertility defects at elevated temperature are present in spermatogenic and oogenic cells, though more severe during spermatogenesis, manifesting within a single generation at elevated temperature (Rogers and Phillips 2020a). In contrast, the oogenic defect accumulates over several generations, becoming progressively sterile over 2–3 generations at elevated temperature (Rogers and Phillips 2020a). While the spermatogenic defect has not been fully characterized, the oogenic defect can be attributed to changes in germline chromatin accessibility resulting in the aberrant expression of somatic and spermatogenic genes in oogenic nuclei (Rogers and Phillips 2020a).
Composition and assembly of Mutator foci
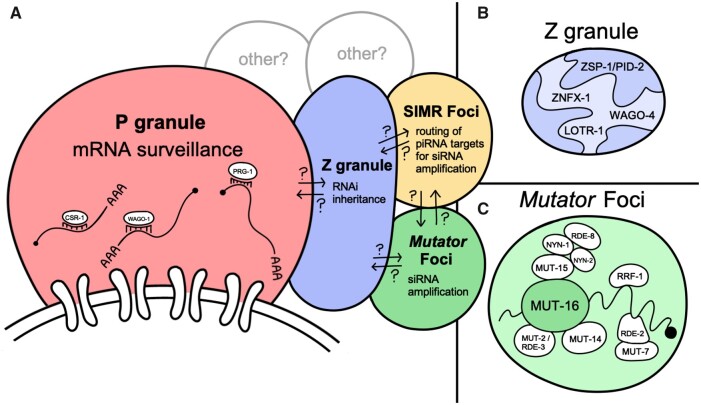
Assembly of mutator complex and accumulation of the mutator complex into visible perinuclear germline foci, referred to as Mutator foci, depend on the scaffolding properties of MUT-16 (Figures 1F and 2, A and C; Phillips et al. 2012; Uebel et al. 2018). Both structured and unstructured/disordered regions of MUT-16 are required for Mutator foci assembly and for the recruitment of other mutator complex members (Uebel et al. 2018). Specifically, the most C-terminal region of MUT-16 (∼275 amino acids), is both necessary and sufficient for the germline Mutator foci assembly (Phillips et al. 2012). MUT-2/RDE-3, a nucleotidyltransferase that adds untemplated poly(UG) tails (pUG tails) to the 3′ termini of cleaved siRNA-targeted mRNAs, is recruited to the complex through a structured region near the N-termius of MUT-16 (Chen et al. 2005; Phillips et al. 2012; Uebel et al. 2018; Shukla et al. 2020). The same region also recruits a DEAD-box helicase MUT-14, which acts redundantly with its paralog SMUT-1 in the initiation of WAGO 22G-RNA amplification (Tijsterman et al. 2002; Phillips et al. 2014), and MUT-15, a protein with no known domains that recruits NYN-1 and NYN-2 to Mutator foci (Uebel et al. 2018). NYN-1 and NYN-2, two NYN-domain proteins, in turn recruit to Mutator foci the NYN-domain endoribonuclease RDE-8, which associates with MUT-2/RDE-3 and cleaves siRNA-targeted mRNAs for MUT-2 pUGylation (Tsai et al. 2015; Uebel et al. 2018).
Figure 2.
Physical and functional compartmentalization of germ granules. (A) P granules are positioned to receive and scan nascent transcripts as they exit the nucleus. This is accomplished through CSR-1, PRG-1, and WAGO-1 Argonautes and their associated small RNAs, which can distinguish foreign transcripts from those licensed for germline expression. Z granules promote amplification of siRNAs for memory storage and transgenerational silencing. SIMR foci route piRNA targets for siRNA amplification. Mutator foci are sites of siRNA amplification. The trajectory of small RNAs and mRNAs through the granules is still unknown and possible routes are indicated by arrows. (B) Substructure within Z granules is mediated through ZSP-1/PID-2. (C) Mutator foci are nucleated by MUT-16, which contains both structured and disordered regions. Additional mutator complex proteins are recruited to Mutator foci through either direct or indirect interactions with MUT-16.
The disordered central region of MUT-16 recruits the RdRP RRF-1 and RDE-2, a protein with no known domains, which then recruit the 3′–5′ exonuclease MUT-7 to Mutator foci (Ketting et al. 1999; Tops et al. 2005; Phillips et al. 2012; Uebel et al. 2018). The RNAs targeted by the MUT-7 exonuclease are currently unknown. It is also worth noting that the RdRPs EGO-1 and RRF-1 are redundantly required for mutator complex-dependent small RNA amplification; however, only RRF-1 localizes to Mutator foci and co-IPs with mutator complex proteins (Gu et al. 2009; Phillips et al. 2012; Manage et al. 2020). Perhaps the distinct localization of the two RdRPs can be attributed to their activity in two distinct 22G-RNA pathways: EGO-1 is also required for biogenesis of CSR-1 22G-RNAs (Claycomb et al. 2009), suggesting that EGO-1 may act primarily with CSR-1 in P granules or the cytoplasm for CSR-1 22G-RNA biogenesis, whereas RRF-1 acts with the mutator complex for WAGO 22G-RNA biogenesis. Perhaps then, only in the absence of RRF-1, EGO-1 can compensate for the loss by also generating WAGO 22G-RNAs with the Mutator complex. Despite the distinction, both RdRPs, EGO-1 and RRF-1, and MUT-16 co-IP with the Dicer-related helicase DRH-3 and the Tudor-domain protein EKL-1 (Gu et al. 2009; Thivierge et al. 2011; Manage et al. 2020). DRH-3 and EKL-1 are required for both CSR-1 22G-RNA and WAGO 22G-RNA biogenesis making them likely members of the mutator complex (Claycomb et al. 2009; Gu et al. 2009), though it has not been demonstrated that they colocalize with mutator complex proteins at Mutator foci.
Regulation of Mutator foci
Several variables have been shown to modulate the presence and intensity of Mutator foci. Early observations of Mutator foci indicated that they are present in germ cells but not somatic cells, which was initially perplexing due to the necessity of the mutator complex to promote RNAi and WAGO 22G-RNA production in somatic cells (Chen et al. 2005; Kim et al. 2005; Gent et al. 2010; Phillips et al. 2012; Uebel et al. 2018). Interestingly, increasing the concentration of MUT-16 in somatic cells can lead to the formation of ectopic Mutator foci that are capable of nucleating other mutator complex proteins (Uebel et al. 2018). This result suggests that somatic cells are not lacking any factors required for Mutator foci formation, but rather that somatic cells do not possess MUT-16 at high enough concentration to nucleate visible Mutator foci. In addition to protein concentration, Mutator foci are also regulated by environmental temperature, as elevated temperatures promote their dissolution (Uebel et al. 2018). Examination of Mutator foci suggested that their intensity is greatest in the mitotic region, transition zone, and late pachytene/diplotene (Phillips et al. 2012; Uebel et al. 2020). This association of foci intensity with germ cell progression can be monitored through the examination of mitotic and meiotic mutants, demonstrating that extended regions of mitosis but not the transition zone can expand the region of intense Mutator foci and establishing that foci intensity is modulated by germline cell cycle stage (Uebel et al. 2020). Finally, similar to work demonstrating that RNA is integral to P-granule stability (Sheth et al. 2010), injection of the transcriptional inhibitor α-amanitin results in dispersal of Mutator foci and indicating that RNA is a critical component of this granule (Uebel et al. 2020).
Organization and function of Z granules
Z granules are defined by the localization of ZNFX-1, a superfamily one (SF1) RNA helicase and zinc-finger domain protein, and WAGO-4, a WAGO-clade Argonaute protein (Ishidate et al. 2018; Wan et al. 2018). Loss of ZNFX-1 and WAGO-4 disrupt RNAi inheritance from one generation to the next but do not disable the response to double-stranded RNA within a single generation (Ishidate et al. 2018; Wan et al. 2018; Xu et al. 2018). Curiously, in a transgene expression assay, mutations in znfx-1 do not simply disrupt the inheritance of silencing signals but rather alter the stability of transgene expression, with the transgenes switching back and forth between silencing and expressed states across generations (Ishidate et al. 2018). Furthermore, ZNFX-1 co-immunoprecipitates with Argonaute proteins in activating and silencing pathways, including PRG-1, WAGO-1, and CSR-1 (Ishidate et al. 2018). These data suggest that Z granules may not solely be required for RNAi inheritance of silencing signals but instead for the inheritance of both expressed and silent epigenetic states.
There is some discrepancy regarding the small RNA that associates with WAGO-4, with separate reports indicating that it primarily associates with CSR-1 22G-RNAs or WAGO 22G-RNAs (Xu et al. 2018; Wan et al. 2021). However, these data, along with data indicating that WAGO-4 binds small RNAs antisense to genes targeted by exogenous RNAi, suggest that, like ZNFX-1, WAGO-4 may act with both activating and silencing small RNA pathways (Xu et al. 2018). Yet, precisely how ZNFX-1 and WAGO-4 promote balanced epigenetic inheritance is still unclear. One clue comes from the sequencing of small RNAs in a znfx-1 mutant. This mutant shows a shift in the distribution of both CSR-1 and WAGO 22G-RNAs across target mRNAs and toward the 5′ end, indicating that one role for ZNFX-1 may be the redistribution of RdRPs from the 5′ end to the 3′ end of target mRNAs (Ishidate et al. 2018). Another Z-granule protein called LOTR-1 contributes to this function, as lotr-1 mutants exhibit a similar loss of small RNAs from the 3′ ends of WAGO and mutator targets and impacts transgenerational RNAi inheritance (Marnik et al. 2021). The Tudor domain of LOTR-1 positions ZNFX-1 and LOTR-1 in perinuclear Z granules, while the LOTUS domain of LOTR-1 appears to associate with cytoskeletal and 3′UTR-binding components. While the LOTUS-domain proteins MIP-1 and MIP-2 interact with the helicase GLH-1 to help nucleate P-granule assembly, LOTR-1 functions similarly by interacting with the helicase ZNFX-1 to help nucleate Z-granule assembly (Marnik et al. 2021; Cipriani et al. 2021).
More recently, PID-2/ZSP-1, a protein with intrinsically disordered regions (IDRs), has been shown to localize to the surface of Z granules where it regulates Z-granule size, number, and fluidity (Figure 2B;Placentino et al. 2021; Wan et al. 2021). PID-2/ZSP-1 is required for germline RNAi and heritable silencing downstream of piRNAs, and, similarly to znfx-1, mutations in pid-2/zsp-1 display only modest alterations in overall small RNA levels or levels of small RNAs mapping to previously defined categories of 22G-RNAs (Ishidate et al. 2018; Placentino et al. 2021; Wan et al. 2021). Furthermore, like znfx-1, pid-2/zsp-1 mutants alter the distribution of both WAGO 22G-RNAs and CSR-1 22G-RNAs along mRNA targets (Ishidate et al. 2018; Placentino et al. 2021). However, pid-2/zsp-1 mutants have reduced small RNAs at the 5′ end of target mRNA transcripts, which is opposite to the effect of a znfx-1 mutation and suggests a possible role for PID-2/ZSP-1 in the processivity of the RdRPs (Placentino et al. 2021). Nonetheless, the fact that most small RNA target genes still produce small RNAs, including germline genes targeted by RNAi, yet pid-2/zsp-1 mutants have strong desilencing of a piRNA sensor and fail to silence RNAi-targeted germline mRNAs, suggests that PID-2/ZSP-1 may somehow fail to couple siRNA production to gene silencing (Placentino et al. 2021; Wan et al. 2021). Altogether, there are clearly still questions to be answered regarding the precise role Z granules play in RNA silencing and RNA inheritance.
Organization and function of SIMR foci
SIMR-1, an extended Tudor domain protein, was initially identified through a MUT-16 immunoprecipitation followed by mass spectrometry of interacting proteins (Manage et al. 2020). Loss of simr-1 has no RNAi-defective (Rde) or Enhanced RNAi (Eri) phenotypes, though the mutation causes a depletion of piRNA-dependent but not piRNA-independent Mutator targets. Further, simr-1 mutants can desilence a piRNA sensor and prevent sterility after reestablishing WAGO-class 22G-RNA production, a phenotype previously only associated with piRNA pathway mutants (de Albuquerque et al. 2015; Phillips et al. 2015; Manage et al. 2020). The piRNAs themselves are unaffected in a simr-1 mutant, so together, this data suggests that SIMR-1 acts in the piRNA pathway, downstream of piRNA biogenesis but upstream of the mutator complex. Interestingly, SIMR-1 also forms germline foci, but these foci are discrete from P granules, Z granules, and Mutator foci (Figures 1G and 2A; Manage et al. 2020). Rather, SIMR-1 colocalizes with RSD-2, a factor required for exogenous RNAi introduced at low doses and production of 22G-RNAs at ERGO-1 target mRNAs (Tijsterman et al. 2004; Han et al. 2008; Zhang et al. 2012; Sakaguchi et al. 2014) and that has no known association with the piRNA pathway. Thus, the current hypothesis is that the SIMR-1 foci function to promote interactions between primary and secondary small RNA pathways, RSD-2 for exogenous RNAi and ERGO-1 target genes and SIMR-1 for piRNA target genes. Also, likely co-localizing with SIMR-1 and RSD-2 is the SIMR-1 paralog, HPO-40. While no function has yet been attributed to HPO-40, it has a similar localization pattern as SIMR-1 and fails to colocalize completely with MUT-16 (Manage et al. 2020). Since no other components of the SIMR foci are currently known, further study will be necessary to reveal the molecular details of this compartment.
Interactions between germ-granule compartments
The current model of germ granule organization in the C. elegans germline proposes sequential stacking of P granules, Z granules, and Mutator foci at the nuclear periphery (Figure 2A;Wan et al. 2018). SIMR foci are also found in tripartite structures, adjacent to Z granules and opposite P granules (Manage et al. 2020), however the orientation of all four condensates relative to one another is still undetermined. Nonetheless, the consistency of which these granules are found in this stacked organization suggests that the interaction between condensates may promote efficient small RNA-based silencing and inheritance. In the germline progenitor cells of early embryos, Z-granule proteins, ZNFX-1 and WAGO-4, co-localize to P granules rather than forming discrete structures; however, after the 100-cell stage of embryonic development, the Z granules demix into discrete condensates (Figure 1C;Wan et al. 2018). Similarly, the Mutator foci and SIMR foci are first observed as robust granules around the 100-cell stage of embryonic development in the Z2/Z3 progenitor germ cells, but unlike Z granules, the Mutator foci and SIMR foci appear to nucleate de novo from cytoplasmically localized proteins rather than through demixing of proteins from the P granule (Figure 1D;Uebel et al. 2018, 2021; Wan et al. 2018). These events roughly coincide with initiation of transcription from embryonic germ cells, suggesting that the presence of newly synthesized mRNAs traversing the nuclear pores and their recognition by various small RNA pathway proteins may drive changes in germline condensate morphology (Seydoux and Dunn 1997; Wan et al. 2018).
Based on their protein composition, Mutator foci are considered hubs of WAGO 22G-RNA biogenesis, positioned near P granules and nuclear pores to capture recently-transcribed target mRNAs for small RNA amplification. Simultaneous RNAi knockdown of four core components of the P granule is sufficient to disrupt Mutator foci formation, while Mutator foci are not required for P-granule assembly (Phillips et al. 2012; Singh et al. 2021). SIMR foci also do not require Mutator foci for formation and, reciprocally, Mutator foci do not require SIMR foci; however, interactions between SIMR foci and other germ-granule compartments have not been thoroughly tested (Manage et al. 2020). Similar to Mutator foci, ZNFX-1 appears to require P granules for localization, as csr-1 and glh-1 mutants, which disrupt P granules, also disrupt ZNFX-1 localization (Ishidate et al. 2018). Interestingly, loss of the P-granule protein DEPS-1, alters P-granule, Z-granule and Mutator foci morphology, while loss of mutator complex proteins disrupt DEPS-1 localization, suggesting there may be a more complex interplay between condensates (Wan et al. 2018; Suen et al. 2020).
Similar to P granules, Z granules and Mutator foci have properties associated with phase-separated condensates, canonically characterized by rapid internal recovery after photobleaching (Brangwynne et al. 2009; Uebel et al. 2018; Wan et al. 2018). Furthermore, Mutator foci assemble after reaching a critical concentration threshold, which may explain their presence in germ cells but not somatic cells. They are disrupted by elevated temperature and, like P granules, they dissolve in low concentrations of an aliphatic alcohol that is thought to only affect weak hydrophobic interactions (Updike et al. 2011; Uebel et al. 2018). These liquid-like properties may allow a continuation of RNA exchange through distinct granule compartments, coordinating RNA silencing between granules.
Proteins and RNAs not yet assigned to specific germ-granule compartments
While the above-described sub-granules constitute the most well-characterized of the germ-granule compartments, several other proteins have been associated with germ granules but whether they form distinct compartments or overlap with a known compartment is unclear. For example, the Tudor domain protein RSD-6 localizes to foci near P granules referred to as R2 bodies (Yang et al. 2014). RSD-6, along with the DEAD box helicase RDE-12, associate with mRNAs targeted by RNAi and are required for production of secondary siRNAs at ERGO-1 and exogenous RNAi targets (Zhang et al. 2012; Shirayama et al. 2014; Yang et al. 2014). Interestingly RDE-12 localizes to both the R2 bodies and to P granules, possibly shuttling between the two; it has been proposed that RDE-12 may carry primary siRNA-targeted mRNAs from P granules to R2 bodies for mutator-dependent siRNA amplification (Yang et al. 2014). Given the overlap in phenotypes and likely function between rsd-2, in SIMR foci, with rsd-6 and rde-12, in R2 bodies, a likely conclusion is that SIMR foci and R2 bodies are one in the same; however, this inference has yet to be proven.
Another example of a germ granule-associated protein which has not been precisely localized to one of the known germ-granule compartments is FBF-1. FBF-1, and its paralog FBF-2, are PUF-family RNA binding proteins that are required in germline stem cells to silence the expression of mRNAs required for meiosis (Voronina et al. 2012). While FBF-2 seems to associate with P granules, FBF-1 forms both cytoplasmic and perinuclear foci, the majority of which do not overlap with P granules though many are immediately adjacent (Voronina et al. 2012). Two additional PUF-family proteins, PUF-3 and PUF-11, in combination with FBF-1 and FBF-2, are required for germline proliferation and have been localized to unidentified cytoplasmic and perinuclear foci germline foci (Haupt et al. 2020).
Similarly, the KH protein MINA-1, identified in a screen for apoptosis regulators, binds the 3′UTRs of genes associated with germ cell development and localizes to a germ-granule compartment adjacent to P granules (Sendoel et al. 2019). Curiously, MINA-1 down-regulates the Z-granule Argonaute protein WAGO-4, and loss of mina-1 leads to enhanced exogenous RNAi (Sendoel et al. 2019). Further work will be necessary to uncover the precise role of MINA-1 in small RNA and gene regulatory pathways.
Finally, in addition to proteins that have yet to be localized to specific germ granules, mRNAs have also been localized to the perinuclear germ-granule regions. Specifically, mRNAs are differentially regulated by miRNAs in germ cells compared to somatic cells, where germline miRNA-targeted mRNAs are stabilized while somatic miRNA-targeted mRNAs are destabilized (Dallaire et al. 2018). These miRNA-targeted mRNAs localize adjacent to P granules dependent on GLH-1, but not to any specific, known compartment (Dallaire et al. 2018). Thus, the localization of RSD-6, FBF-1, PUF-3, PUF-11, MINA-1, and miRNA-targeted mRNAs to undetermined perinuclear germ-granule compartments will necessitate further co-localization studies to determine whether these proteins are components of known germ-granule compartments or whether they are the defining members of a newly identified compartments.
Germ granules likely collaborate in heritable gene silencing
Overview of transgenerational inheritance
In many eukaryotes, including C. elegans, endogenous siRNAs can transmit epigenetic information, including responses to environmental stress, from parents to offspring (Rechavi and Lev 2017). When siRNAs carried by either egg or sperm are deposited into the embryo, they are amplified through the activity of RdRPs. mRNA targets of the deposited siRNAs become RdRP templates, leading to further production of abundant secondary siRNAs and enhanced silencing. Loss of many RNAi pathway and germ-granule components lead to a germline mortal (Mrt) phenotype, where the animals become progressively less fertile over generations (Buckley et al. 2012; Simon et al. 2014; Sakaguchi et al. 2014; Spracklin et al. 2017; Ishidate et al. 2018; Wan et al. 2018, 2021; Manage et al. 2020; Placentino et al. 2021). This phenotype hints at the possibility that sterility arises from changes in germ cell gene expression that become exacerbated by the transgenerational small RNA amplification cycle.
Phenotypic variation can depend on parental or ancestral genotype
It has been observed that mutations in multiple germ-granule components, including pgl-1, glh-1, and meg-3; meg-4, are either RNAi defective or RNAi inheritance defective (Robert et al. 2005; Spike et al. 2008b; Wang et al. 2014; Spracklin et al. 2017; Ouyang et al. 2019; Dodson and Kennedy 2019). The implication being that germ granules are essential for RNAi. However, this RNAi-defective phenotype can be uncoupled, or “transgenerationally disconnected,” from the germ granule-defective genotype. For example, wild-type animals with meg-3; meg-4 mutant ancestors can exhibit RNAi defects and meg-3; meg-4 mutants from wild-type ancestors can exhibit a wild-type RNAi response (Lev et al. 2019; Ouyang et al. 2019; Dodson and Kennedy 2019). This phenotypic disconnect can last >8 generations, indicating that loss of germ granules can have transgenerational repercussions. It is worth noting, however, that the expected coupling of phenotype and genotype ultimately returns, suggesting the information needed to coordinate small RNA gene silencing is genome-encoded (Dodson and Kennedy 2019).
P granules and piRNAs coordinate siRNA biogenesis and transgenerational inheritance
If embryonic P granules are not required, per se, for germline RNAi, what role do they play in coordinating siRNA production and RNA silencing (Dodson and Kennedy 2019)? P granules house multiple RNAi pathway proteins, including Argonaute proteins and RdRPs, and in their absence, aberrant siRNAs are generated at some genes while siRNAs are lost at others. Changes in small RNA poly-uridylation have also been observed, which could affect small RNA stability or sorting into distinct Argonaute proteins, ultimately altering RNA silencing efficacy (van Wolfswinkel et al. 2009; Xu et al. 2018; Lev et al. 2019). Some of the aberrantly targeted genes include genes required for RNAi. For example, rde-4 mRNA expression is reduced when P granules are dispersed in deps-1 mutants (Spike et al. 2008a). In addition, sid-1 and rde-11 have increased small RNAs and reduced mRNA expression when embryonic P granules are dispersed in meg-3; meg-4 double mutants, presumably reducing RNAi’s efficacy in this mutant (Ouyang et al. 2019; Dodson and Kennedy 2019). Curiously, the misexpressed small RNAs tend to be very consistent between replicates and experiments, leading to the question of why specific genes are mistargeted and if certain sequences or features drive aberrant small RNA targeting following the loss of germ granules. One clue comes from the fact that mutants in prg-1, as well as transgenerational RNA silencing mutants hrde-1 and znfx-1, can restore RNAi competence to meg-3; meg-4 mutants (Ouyang et al. 2019). Both sid-1 and rde-11 appear to be direct targets of PRG-1, as their siRNA levels are reduced and their mRNA expression increased in a prg-1 mutant (McMurchy et al. 2017; Shen et al. 2018; Ouyang et al. 2019). This comes as a surprise, as these genes are expressed in wild-type animals and required for an effective response to exogenous RNAi; however, the findings invoke a “safe harbor” model where P granules can protect some transcripts from transgenerational piRNA-mediated silencing (Ouyang et al. 2019).
While it is generally understood that piRNAs promote siRNA-mediated gene silencing, it may not be that simple. For example, PRG-1 is required maternally to coordinate siRNA biogenesis and prevent siRNA-mediated silencing of essential genes (de Albuquerque et al. 2015; Phillips et al. 2015). In fact, following the loss of PRG-1 and piRNAs, siRNA-mediated silencing of the rDNA locus directly contributes to transgenerational sterility (Wahba et al. 2021), while siRNA-mediated silencing at histone genes can become permanent (Barucci et al. 2020; Reed et al. 2020; Shukla et al. 2021). While the signal that predisposes some genes to silencing in prg-1 mutants is unknown, it has been hypothesized that the lack of a poly(A) tail common in histone transcripts and rRNAs (Montgomery et al. 2020; Reed et al. 2020), or the susceptibility of transcripts to perpetual poly-UG (pUG) RNA/siRNA cycling (Shukla et al. 2020, 2021; Wahba et al. 2021), could make specific genes more permissive for siRNA biogenesis. Like the “safe harbor” model above, PRG-1 targeted transcripts may be protected from permanent RDE-3-mediated siRNA silencing of Mutator foci if they are sequestered within P granules and away from RDE-3 silencing in Mutator foci (Shukla et al. 2021). As these details are worked out, the roles of germ-granule demixing are sure to become more apparent.
Outlook and outstanding questions
The occurrence of germ-granule demixing into distinct sub-granules leaves a number of questions unanswered. To start with, what are the events in germline blastomeres that trigger demixing? Is demixing a passive phenomenon caused by the localized synthesis of particular small RNAs, the clustering of similar RNA modifications and their binding proteins, or the condensation of phases similar to those that partition nucleolar sub-compartments (Feric et al. 2016; Lafontaine et al. 2021)? Instead, is demixing an active process that requires energy metabolism and the cytoskeleton? Is there directionality or progression from the emergence of one sub-granule to the next? Then once formed, how is the separation of these compartments maintained?
Determining what constitutes a P granule, Z granule, Mutator focus, or SIMR focus, their upper and lower size limits, and how each are defined is left to an investigator’s discretion. This requires us to ask whether distinct compositions of germ-granule compartments exist and whether this indicates functional specialization. At any given time are the RNA and protein constituents of the individual sub-granules the same, and if not, does the association, loss, or exchange of RNAs and proteins constitute a new compartment with distinct functions? If the answer is yes, it will be important to understand the extent that proteins are shared across sub-granules, and if proteins moving between compartments are trafficking RNAs or other substrates from one location to the next. If so, what then is the trajectory of RNAs between sub-granules? Does this trajectory reflect changing functions for these RNAs, or an assembly line for their progressive modifications? And then, finally, how are these activities coordinated to ensure germline integrity? The answer to these and other questions should be attainable with further study and the increasing availability of new tools and improved imaging resolution.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript.
Conflicts of interest
The authors declare that there is no conflict of interest. Conclusions, implications and opinions expressed within the content are solely the authors.
Literature cited
- Al-Amin M, Min H, Shim Y-H, Kawasaki I.. 2016. Somatically expressed germ-granule components, PGL-1 and PGL-3, repress programmed cell death in C. elegans. Sci Rep. 6:33884.doi:10.1038/srep33884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, et al. 2001. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 128:3899–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andralojc KM, Campbell AC, Kelly AL, Terrey M, Tanner PC, et al. 2017. ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLoS Genet. 13:e1006611.doi:10.1371/journal.pgen.1006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki ST, Kershner AM, Bingman CA, Wickens M, Kimble J.. 2016. PGL germ granule assembly protein is a base-specific, single-stranded RNase. Proc Natl Acad Sci U S A. 113:1279–1284. doi:10.1073/pnas.1524400113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki ST, Lynch TR, Crittenden SL, Bingman CA, Wickens M, et al. 2021. C. elegans germ granules require both assembly and localized regulators for mRNA repression. Nat Commun. 12:996. doi:10.1038/s41467-021–21278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariz M, Mainpal R, Subramaniam K.. 2009. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev Biol. 326:295–304. doi:10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 150:88–99. doi:10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, et al. 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 171:267–279. doi:10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, et al. 2012. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 337:574–578. doi:10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Lublin AL, Evans TC.. 2002. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr Biol. 12:1502–1506. doi:10.1016/S0960-9822(02)01111-9.[pii] [DOI] [PubMed] [Google Scholar]
- Barucci G, Cornes E, Singh M, Li B, Ugolini M, et al. 2020. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat Cell Biol. 22:235–245. doi:10.1038/s41556-020–0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 31:67–78. doi:10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshore EL, McEwen TJ, Jud MC, Marshall JK, Schisa JA, et al. 2011. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev Biol. 350:370–381. doi:10.1016/j.ydbio.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, et al. 2012. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 8:e1002617.doi:10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag PR, Nakamura A, Blackwell TK.. 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 132:4975–4986. doi:10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, et al. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. doi:10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brown KC, Svendsen JM, Tucci RM, Montgomery BE, Montgomery TA.. 2017. ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res. 45:9093–9107. doi:10.1093/nar/gkx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, et al. 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 489:447–451. doi:10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AC, Updike DL.. 2015. CSR-1 and P granules suppress sperm-specific transcription in the C. elegans germline. Development. 142:1745–1755. doi:10.1242/dev.121434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Hoersch S, O'Keeffe S, Sachidanandam R, Grishok A.. 2014. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. NatStruct Mol Biol. 21:358–65. 10.1038/nsmb.2801. 24681887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth AG, , Seroussi U, LehrbachNJ, , RenaudMS, , SundbyAE, , et al. 2021, . Two isoforms of the essential C. elegans Argonaute CSR-1 differentially regulate sperm and oocyte fertility. Nucleic Acids Res. 49:8836–8865. 10.1093/nar/gkab619. 34329465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-CG, Simard MJ, Tabara H, Brownell DR, McCollough JA, et al. 2005. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol. 15:378–383. doi:10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Chen J-X, Cipriani PG, Mecenas D, Polanowska J, Piano F, et al. 2016. In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into P granule dynamics. Mol Cell Proteom. 15:1642–1657. doi:10.1074/mcp.M115.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Hu Y, Lang CF, Brown JS, Schwabach S, et al. 2020. The dynamics of P granule liquid droplets are regulated by the Caenorhabditis elegans germline RNA helicase GLH-1 via its ATP hydrolysis cycle. Genetics. 215:421–434. doi:10.1534/genetics.120.303052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR.. 2006. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 311:851–853. doi:10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- Cipriani PG, Bay O, Zinno J, Gutwein M, Gan HH, et al. 2021. Novel LOTUS-domain proteins are organizational hubs that recruit C. elegans Vasa to germ granules. Elife. 10:e60833. doi:10.7554/eLife.60833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 139:123–134. doi:10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J, Saari B, Anderson P.. 1987. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature. 328:726–728. doi:10.1038/328726a0. [DOI] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, et al. 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 107:3588–3593. doi:10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, et al. 2013. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell. 155:1532–1544. doi:10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro Rodrigues RJ, de Jesus Domingues AM, Hellmann S, Dietz S, de Albuquerque BFM, et al. 2019. PETISCO is a novel protein complex required for 21U RNA biogenesis and embryonic viability. Genes Dev. 33:857–870. doi:10.1101/gad.322446.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallaire A, Frédérick P-M, Simard MJ.. 2018. Somatic and germline microRNAs form distinct silencing complexes to regulate their target MRNAs differently. Dev Cell. 47:239–247.e4. doi:10.1016/j.devcel.2018.08.022. [DOI] [PubMed] [Google Scholar]
- de Albuquerque BFM, Placentino M, Ketting RF.. 2015. Maternal piRNAs are essential for germline development following de novo establishment of endo-siRNAs in Caenorhabditis elegans. Dev Cell. 1–9. doi:10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Dodson AE, Kennedy S.. 2019. Germ granules coordinate RNA-based epigenetic inheritance pathways. Dev Cell. 50:704–715.e4. doi:10.1016/j.devcel.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR.. 1996. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 87:205–216. doi: 10.1016/S0092-8674(00)81339-2.[pii] [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J.. 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell. 3:697–710. doi:S1534580702003222.[pii] [DOI] [PubMed] [Google Scholar]
- Eddy EM. 1975. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 43:229–280. doi:10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Eide D, Anderson P.. 1985. Transposition of Tc1 in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 82:1756–1760. doi:10.1073/pnas.82.6.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, et al. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 165:1686–1697. doi:10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann A, Putnam A, Lee CF, Seydoux G.. 2021. Pickering stabilization of a dynamic intracellular emulsion. bioRxiv. doi:10.1101/2021.06.22.449249. [Google Scholar]
- Friday AJ, Henderson MA, Morrison JK, Hoffman JL, Keiper BD.. 2015. Spatial and temporal translational control of germ cell mRNAs mediated by the eIF4E isoform IFE-1. J Cell Sci. 128:4487–4498. doi:10.1242/jcs.172684. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G.. 2008. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol. 323:76–87. doi:10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Gao G, Deeb F, Mercurio JM, Parfenova A, Smith PA, et al. 2012. PAN-1, a P-granule component important for C. elegans fertility, has dual roles in the germline and soma. Dev Biol. 364:202–213. doi:10.1016/j.ydbio.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK.. 2008. Germline survival and apoptosis. WormBook. Sep 4;1–20. doi:10.1895/wormbook.1.145.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, et al. 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 37:679–689. doi:10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA.. 2005. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 19:683–696. doi:10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruidl ME, Smith P. A, Kuznicki K. A, McCrone JS, Kirchner J, et al. 1996. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 93:13837–13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 36:231–244. doi:10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee H-C, Chaves D, Youngman EM, Pazour GJ, et al. 2012. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 151:1488–1500. doi:10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes S, Priess JR.. 1997. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. 124:731–739. [DOI] [PubMed] [Google Scholar]
- Han W, Sundaram P, Kenjale H, Grantham J, Timmons L.. 2008. The Caenorhabditis elegans rsd-2 and rsd-6 genes are required for chromosome functions during exposure to unfavorable environments. Genetics. 178:1875–1893. doi:10.1534/genetics.107.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa M, Yonetani M, Sugimoto A.. 2011. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J Cell Biol. 192:929–937. doi:10.1083/jcb.201010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt KA, Law KT, Enright AL, Kanzler CR, Shin H, et al. 2020. A PUF hub drives self-renewal in Caenorhabditis elegans germline stem cells. Genetics. 214:147–161. doi:10.1534/genetics.119.302772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegner RW. 1911. The germ cell determinants in the eggs of chrysomelid beetles. Science. 33:71–72. doi:10.1126/science.33.837.71. [DOI] [PubMed] [Google Scholar]
- Henderson MA, Cronland E, Dunkelbarger S, Contreras V, Strome S, et al. 2009. A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J Cell Sci. 122:1529–1539. doi:10.1242/jcs.046771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJA, Greenstein D.. 2005. Introduction to the germ line. WormBook. Sep 1;1–4. doi:10.1895/wormbook.1.18.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert A, AndersonP. 2009. The C. elegans sex determination gene laf-1 encodes a putative DEAD-box RNA helicase. Dev Biol. 330:358–67. 10.1016/j.ydbio.2009.04.003. 19361491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins HP, Subash JS, Stoffel H, Henderson MA, Hoffman JL, et al. 2020. Distinct roles of two eIF4E isoforms in the germline of Caenorhabditis elegans. J Cell Sci. 133:jcs237990. doi:10.1242/jcs.237990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishidate T, Ozturk AR, Durning DJ, Sharma R, Zhi Shen E, et al. 2018. ZNFX-1 functions within perinuclear nuage to balance epigenetic signals. Mol Cell. 70:639–649.e6. doi:10.1016/j.molcel.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik-Bode M, Studencka M, Smolka C, Baumann T, Schmidt H, et al. 2013. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J Cell Sci. 126:5166–5177. doi:10.1242/jcs.130708. [DOI] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T.. 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 180:165–183. doi:10.1006/dbio.1996.0293 [DOI] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJT, Bagijn MP, et al. 2012. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 8:e1002702.doi:10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, et al. 1998. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 94:635–645. doi:S0092-8674(00)81605-0.[pii] [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, et al. 2004. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 167:645–661. doi:10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS.. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 52:311–320. doi:S0092-8674(88)80024-2.[pii] [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HGA, Plasterk RH.. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 99:133–141. doi:10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, et al. 2005. Functional genomic analysis of RNA interference in C. elegans. Science. 308:1164–1167. doi:10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Kim KW, Nykamp K, Suh N, Bachorik JL, Wang L, et al. 2009. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell. 16:723–733. doi:10.1016/j.devcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson AK, Egelhofer T, Rechtsteiner A, Strome S.. 2017. Germ granules prevent accumulation of somatic transcripts in the adult Caenorhabditis elegans Germline. Genetics. 206:163–178. doi:10.1534/genetics.116.198549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Extavour CG.. 2017. Convergent evolution of germ granule nucleators: a hypothesis. Stem Cell Res. 24:188–194. doi:10.1016/j.scr.2017.07.018. [DOI] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, et al. 2000. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 127:2907–2916. [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP.. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 22:165–182. doi:10.1038/s41580-020–0272-6. [DOI] [PubMed] [Google Scholar]
- Lall S, Piano F, Davis RE.. 2005. Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol Biol Cell. 16:5880–5890. doi: 10.1091/mbc.E05-07-0622.[pii]10.1091/mbc.E05-07–0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leacock SW, Reinke V.. 2008. MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics. 178:295–306. doi:10.1534/genetics.107.080218.[pii]10.1534/genetics.107.080218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-C, Gu W, Shirayama M, Youngman E, Conte D, et al. 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 150:78–87. doi:10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-YS, Putnam A, Lu T, He S, Ouyang JPT, et al. 2020. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife. 9:e52896. doi:10.7554/eLife.52896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev I, Toker IA, Mor Y, Nitzan A, Weintraub G, et al. 2019. Germ granules govern small RNA inheritance. Curr Biol. 29:2880–2891.e4. doi:10.1016/j.cub.2019.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Berkyurek AC, Greiner A, Sawh AN, Vashisht A, et al. 2020. A family of argonaute-interacting proteins gates nuclear RNAi. Mol Cell. 78:862–875.e8. doi:10.1016/j.molcel.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, DeBella LR, Guven-Ozkan T, Lin R, Rose LS.. 2009. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J Cell Biol. 187:33–42. doi:10.1089/jcb.200903003.[pii]10.1083/jcb.200903003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJT, Almeida MV, Roovers EF, et al. 2012. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. Embo J. 31:3422–3430. doi:10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP, Illmensee K, Turner FR.. 1976. Interspecific transplantation of polar plasm between Drosophila embryos. J Cell Biol. 70:358–373. doi:10.1083/jcb.70.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manage KI, Rogers AK, Wallis DC, Uebel CJ, Anderson DC, et al. 2020. A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans. Elife. 9:e56731. doi:10.7554/eLife.56731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnik EA, Fuqua JH, Sharp CS, Rochester JD, Xu EL, et al. 2019. Germline maintenance through the multifaceted activities of GLH/Vasa in Caenorhabditis elegans P granules. Genetics. 213:923–939. doi:10.1534/genetics.119.302670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnik EA, Almeida MV, Cipriani PG, Chung G, Caspani E, et al. 2021. The Caenorhabditis elegans TDRD5/7-like protein. LOTR-1, interacts with the helicase ZNFX-1 to balance epigenetic signals in the germline. bioRxiv. doi:10.1101/2021.06.18.448978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchy AN, Stempor P, Gaarenstroom T, Wysolmerski B, Dong Y, et al. 2017. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. Elife. 6:e21666. doi:10.7554/eLife.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, et al. 1996. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 382:710–712. doi:10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Min H, Shim Y, Kawasaki I.. 2016. Loss of PGL-1 and PGL-3, members of a family of constitutive germ-granule components, promotes germline apoptosis in C. elegans. J Cell Sci. 129:341–353. doi:10.1242/jcs.174201. [DOI] [PubMed] [Google Scholar]
- Min H, Lee Y-U, Shim Y-H, Kawasaki I.. 2019. Autophagy of germ-granule components, PGL-1 and PGL-3, contributes to DNA damage-induced germ cell apoptosis in C. elegans, (M. P. Colaiácovo, Ed. ). PLoS Genet. 15:e1008150.doi:10.1371/journal.pgen.1008150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim Y-S, Zhang C, Dowen RH, Phillips CM, et al. 2012. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8:e1002616.doi:10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BE, Vijayasarathy T, Marks TN, Reed KJ, Montgomery TA.. 2020. piRNAs prevent runaway amplification of siRNAs from ribosomal RNAs and histone mRNAs. bioRxiv. doi:10.1101/2020.06.15.153023. [Google Scholar]
- Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK.. 2001. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. 128:3221–3232. [DOI] [PubMed] [Google Scholar]
- Nguyen DAH, Phillips CM.. 2021. Arginine methylation promotes siRNA-binding specificity for a spermatogenesis-specific isoform of the Argonaute protein CSR-1. Nat Commun. 12:4212.doi:10.1038/s41467-021–24526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y.. 2003. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. 130:2495–2503. [DOI] [PubMed] [Google Scholar]
- Ouyang JPT, Folkmann A, Bernard L, Lee C-Y, Seroussi U, et al. 2019. P granules protect RNA interference genes from silencing by piRNAs. Dev Cell. 50:716–728.e6. doi:10.1016/j.devcel.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DM, Winkenbach LP, Boyson S, Saxton MN, Daidone C, et al. 2020. mRNA localization is linked to translation regulation in the Caenorhabditis elegans germ lineage. Development. 147:dev186817. doi:10.1242/dev.186817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penners A. 1922. Die Furchung von Tubifex rivulorum Lam. Verh Deutsch Zool Ges. 29:69–73. [Google Scholar]
- Perez-Borrajero C, Podvalnaya N, Holleis K, Lichtenberger R, Karaulanov E, et al. 2021. Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans. Genes Dev. 35:1304–1323., doi:10.1101/gad.348648.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery T. A, Breen PC, Ruvkun G.. 2012. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26:1433–1444. doi:10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery BE, Breen PC, Roovers EF, Rim Y-S, et al. 2014. MUT-14 and SMUT-1 DEAD box RNA helicases have overlapping roles in germline RNAi and endogenous siRNA formation. Curr Biol. 24:839–844. doi:10.1016/j.cub.2014.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Brown KC, Montgomery BE, Ruvkun G, Montgomery TA.. 2015. piRNAs and piRNA-dependent siRNAs protect conserved and essential C. elegans genes from misrouting into the RNAi pathway. Dev Cell. 34:457–465. doi:10.1016/j.devcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR.. 2000. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 219:315–333. doi:10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- Placentino M, de Jesus Domingues AM, Schreier J, Dietz S, Hellmann S, et al. 2021. Intrinsically disordered protein PID-2 modulates Z granules and is required for heritable piRNA-induced silencing in the Caenorhabditis elegans embryo. Embo J. 40:e105280.doi:10.15252/embj.2020105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IF, Hertz HL, Pastore B, Wagner J, Tang W.. 2021. Proximity labeling identifies LOTUS domain proteins that promote the formation of perinuclear germ granules in C. elegans. Elife. 10. doi:10.7554/eLife.72276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam A, Cassani M, Smith J, Seydoux G.. 2019. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat Struct Mol Biol. 26:220–226. doi:10.1038/s41594-019–0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarato P, Singh M, Cornes E, Li B, Bourdon L, et al. 2021. Germline inherited small RNAs facilitate the clearance of untranslated maternal mRNAs in C. elegans embryos. Nat Commun. 12:1441.doi:10.1038/s41467-021–21691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiders SA, Eastwood MD, Bacher M, Priess JR.. 2018. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLoS Genet. 14:e1007417.doi:10.1371/journal.pgen.1007417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Lev I.. 2017. Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr Biol. 27: R720–R730. doi:10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]
- Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, et al. 2020. Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48:1811–1827. doi:10.1093/nar/gkz1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter R. 1890. Die Entwicklung der Geschlechtsorgane und des Darmes bei Chiromomus. Zeit Furr Wiss Zool Bd. 50:408–427. [Google Scholar]
- Robert VJP, Sijen T, van Wolfswinkel J, Plasterk RHA.. 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19:782–787. doi:10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AK, Phillips CM.. 2020a. RNAi pathways repress reprogramming of C. elegans germ cells during heat stress. Nucleic Acids Res. 48:4256–4273. doi:10.1093/nar/gkaa174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AK, Phillips CM.. 2020b. A small-RNA-mediated feedback loop maintains proper levels of 22G-RNAs in C. elegans. Cell Rep. 33:108279.doi:10.1016/j.celrep.2020.108279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Gönczy P.. 2014. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook. Dec 30;1–43. doi:10.1895/wormbook.1.30.2. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 127:1193–1207. doi:10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Rybarska A, Harterink M, Jedamzik B, Kupinski AP, Schmid M, et al. 2009. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet. 5:e1000494.doi:10.1371/journal.pgen.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, et al. 2016. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell. 166:1572–1584.e16. doi:10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A, Sarkies P, Simon M, Doebley A-L, Goldstein LD, et al. 2014. Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc Natl Acad Sci U S A. 111: E4323–E4331. doi:10.1073/pnas.1406131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas LS, Maldonado E, Macias-Silva M, Blackwell TK, Navarro RE.. 2007. The DEAD box RNA helicase VBH-1 is required for germ cell function in C. elegans. Genesis. 45:533–546. doi:10.1002/dvg.20323. [DOI] [PubMed] [Google Scholar]
- Schisa JA, Pitt JN, Priess JR.. 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development. 128:1287–1298. [DOI] [PubMed] [Google Scholar]
- Schmid M, Kuchler B, Eckmann CR.. 2009. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 23:824–836. doi:10.1101/gad.494009. [pii]10.1101/gad.494009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Görlich D.. 2016. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci. 41:46–61. doi:10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Putnam A, Rasoloson D, Seydoux G.. 2021. Protein-based condensation mechanisms drive the assembly of RNA-rich P granules. Elife. 10:e63698. doi:10.7554/eLife.63698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier J, Dietz S, Jesus Domingues A. D, Seistrup A-S, Nguyen DAH, et al. 2021. A membrane-associated condensate drives paternal epigenetic inheritance in C. elegans. bioRxiv. doi:10.1101/2020.12.10.417311v1. [Google Scholar]
- Sendoel A, Subasic D, Ducoli L, Keller M, Michel E, et al. 2019. MINA-1 and WAGO-4 are part of regulatory network coordinating germ cell death and RNAi in C. elegans. Cell Death Differ. 26:2157–2178. doi:10.1038/s41418-019–0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta MS, Low WY, Patterson JR, Kim H-M, Traven A, et al. 2013. ifet-1 is a broad-scale translational repressor required for normal P granule formation in C. elegans. J Cell Sci. 126:850–859. doi:10.1242/jcs.119834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D, et al. 2013. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell. 27:656–663. doi:10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Fire A.. 1994. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 120:2823–2834. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA.. 1997. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 124:2191–2201. [DOI] [PubMed] [Google Scholar]
- Seydoux G. 2018. The P granules of C. elegans: a genetic model for the study of RNA–protein condensates. J Mol Biol. 430:4702–4710. doi:10.1016/j.jmb.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E, Chen H, Ozturk AR, Tu S, Shirayama M, et al. 2018. Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell. 172:937–951.e18. doi:10.1016/j.cell.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Pitt J, Dennis S, Priess JR.. 2010. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 137:1305–1314. doi:10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Kawahara H, Doi H.. 2002. Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells. 7:933–947. doi:10.1046/j.1365–2443.2002.00570.x.[pii] [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, et al. 2012. PiRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 150:65–77. doi:10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Stanney W, Gu W, Seth M, Mello CC.. 2014. The Vasa Homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr Biol. 24:845–851. doi:10.1016/j.cub.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Yan J, Pagano DJ, Dodson AE, Fei Y, et al. 2020. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature. 582:283–288. doi:10.1038/s41586-020-2323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Perales R, KennedyS. 2021. piRNAs coordinate poly(UG) tailing to prevent aberrant and perpetual gene silencing. Curr Biol. CB. 31:4473–4485.e3. 10.1016/j.cub.2021.07.076. 34428467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley A-L, Goldstein LD, et al. 2014. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7:762–773. doi:10.1016/j.celrep.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Cornes E, Li B, Quarato P, Bourdon L, et al. 2021. Translation and codon usage regulate Argonaute slicer activity to trigger small RNA biogenesis. Nat Commun. 12:3492.doi:10.1038/s41467-021-23615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LD. 1966. The role of a “germinal plasm” in the formation of primordial germ cells in Rana pipiens. Dev Biol. 14:330–347. doi:10.1016/0012–1606(66)90019-4. [DOI] [PubMed] [Google Scholar]
- Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, et al. 2016. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife. 5:e21337.doi:10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichal M, Heestand B, Billmyre KK, Frenk S, Mello CC, et al. 2021. Germ granule dysfunction is a hallmark and mirror of Piwi mutant sterility. Nat Commun. 12:1420.doi:10.1038/s41467-021-21635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Bader J, Reinke V, Strome S.. 2008a. DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development. 135:983–993. doi:10.1242/dev.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, et al. 2008b. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics. 178:1973–1987. doi:10.1534/genetics.107.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G, Fields B, Wan G, Becker D, Wallig A, et al. 2017. The RNAi inheritance machinery of Caenorhabditis elegans. Genetics. 206:1403–1416. doi:10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, et al. 2006. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 17:336–344. doi:10.1091/mbc.e05-09–0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Cheng KC-C, Seydoux G.. 2007. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr Biol. 17:1545–1554. doi:10.1016/j.cub.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB.. 1982. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 79:1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. 2005. Specification of the germ line. WormBook. Jul 28:1–10. doi:10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G.. 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 126:4861–4871. [DOI] [PubMed] [Google Scholar]
- Suen KM, Braukmann F, Butler R, Bensaddek D, Akay A, et al. 2020. DEPS-1 is required for piRNA-dependent silencing and PIWI condensate organisation in Caenorhabditis elegans. Nat Commun. 11:4242.doi:10.1038/s41467-020-18089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y.. 1999a. pos-1 Encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 126:1–11. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, et al. 1999b. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 99:123–132. doi:10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee H-C, Weng Z, Mello CC.. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell. 164:974–984. doi:10.1016/j.cell.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, et al. 2011. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol. 19:90–97. doi:10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RHA.. 2002. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 295:694–697. doi:10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RHA.. 2004. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol. 14:111–116. doi:10.1016/j.cub.2003.12.029. [DOI] [PubMed] [Google Scholar]
- Tops BBJ, Tabara H, Sijen T, Simmer F, Mello CC, et al. 2005. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33:347–355. doi:10.1093/nar/gki183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-Y, Chen C-CG, Conte D, Moresco JJ, Chaves DA, et al. 2015. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell. 160:407–419. doi:10.1016/j.cell.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Wu MZ, Wang J, Cutter AD, Weng Z, et al. 2014. Comparative functional characterization of the CSR-1 22G-RNA pathway in Caenorhabditis nematodes. Nucleic Acids Res. 1–17. doi:10.1093/nar/gku1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebel CJ, Anderson DC, Mandarino LM, Manage KI, Aynaszyan S, et al. 2018. Distinct regions of the intrinsically disordered protein MUT-16 mediate assembly of a small RNA amplification complex and promote phase separation of Mutator foci. PLoS Genet. 14:e1007542.doi:10.1371/journal.pgen.1007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebel CJ, Agbede D, Wallis DC, Phillips CM.. 2020. Mutator foci are regulated by developmental stage, RNA, and the germline cell cycle in Caenorhabditis elegans. G3 (Bethesda). 10:3719–3728. doi:10.1534/g3.120.401514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebel CJ, Manage KI, Phillips CM.. 2021. SIMR foci are found in the progenitor germ cells of C. elegans embryos. microPubl Biol. Feb 22;2021. doi:10.17912/micropub.biology.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Strome S.. 2009. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics. 183:1397–1419. doi:10.1534/genetics.109.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D, Strome S.. 2010. P granule assembly and function in Caenorhabditis elegans germ cells. J Androl. 31:53–60. doi:10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Hachey SJ, Kreher J, Strome S.. 2011. P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol. 192:939–948. doi:10.1083/jcb.201010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Knutson AK, Egelhofer TA, Campbell AC, Strome S.. 2014. Germ-granule components prevent somatic development in the C. elegans germline. Curr Biol. 24:970–975. doi:10.1016/j.cub.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, et al. 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 139:135–148. doi:10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Fischer SEJ, Robert VJP, Thijssen KL, Fraser AG, et al. 2003. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol. 13:1311–1316. [DOI] [PubMed] [Google Scholar]
- Voronina E, Paix A, Seydoux G.. 2012. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development. 139:3732–3740. doi:10.1242/dev.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G.. 2010. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development. 137:1441–1450. doi:10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vought VE, Ohmachi M, Lee M-H, Maine EM.. 2005. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics. 170:1121–1132. doi:10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Hansen L, Fire AZ.. 2021. An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans. Dev Cell. 56:2295–2312.e6. doi:10.1016/j.devcel.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis DC, Nguyen DAH, Uebel CJ, Phillips CM.. 2019. Visualization and quantification of transposon activity in Caenorhabditis elegans RNAi pathway mutants. G3 (Bethesda). 9:3825–3832. doi:10.1534/g3.119.400639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Fields BD, Spracklin G, Shukla A, Phillips CM, et al. 2018. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature. 557:679–683. doi:10.1038/s41586-018–0132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Bajaj L, Fields B, Dodson AE, Pagano D, et al. 2021. ZSP-1 is a Z granule surface protein required for Z granule fluidity and germline immortality in Caenorhabditis elegans. Embo J. 40:e105612.doi:10.15252/embj.2020105612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J.. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 419:312–316. doi:10.1038/nature01039nature01039.[pii] [DOI] [PubMed] [Google Scholar]
- Wang G, Reinke V.. 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 18:861–867. doi:10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen B-C, Schmidt H, Rasoloson D, et al. 2014. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically-disordered proteins in C. elegans. Elife. 3:e04591. doi:10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, Claycomb JM.. 2013. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell. 27:664–671. doi:10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Witze ES, Field ED, Hunt DF, Rothman JH.. 2009. C. elegans pur alpha, an activator of end-1, synergizes with the Wnt pathway to specify endoderm. Dev Biol. 327:12–23. doi:10.1016/j.ydbio.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Feng X, Chen X, Weng C, Yan Q, et al. 2018. A Cytoplasmic Argonaute protein promotes the inheritance of RNAi. Cell Rep. 23:2482–2494. doi:10.1016/j.celrep.2018.04.072. [DOI] [PubMed] [Google Scholar]
- Yang H, Vallandingham J, Shiu P, Li H, Hunter CP, et al. 2014. The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. elegans. Curr Biol. 24:832–838. doi:10.1016/j.cub.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Weng C, Wang X, Yan Y-H, Li W-J, et al. 2019. Functional proteomics identifies a PICS complex required for piRNA maturation and chromosome segregation. Cell Rep. 27:3561–3572.e3. doi:10.1016/j.celrep.2019.05.076. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321. doi:10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SEJ, Phillips CM, et al. 2011. Mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 108:1201–1208. doi:10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Fischer SEJ, Garcia SMDA, Riedel CG, et al. 2012. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr Biol. 22:881–890. doi:10.1016/j.cub.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang Z, Du Z, Zhang H.. 2018a. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 174:1492–1506.e22. doi:10.1016/j.cell.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu W-S, Huang W-C, et al. 2018b. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 359:587–592. doi:10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript.