Abstract
Introduction
In the cell cycle, cyclin-dependent kinases (CDKs) play a positive regulatory role, which is essential for normal cell growth, but the expression pattern and prognostic significance of the CDK family in colorectal cancer (CRC) have not been systematically investigated.
Methods
In our study, we analyzed and visualized the expression of CDKs in CRC using TCGA, GEPIA, GSCALite, TIMER, HPA database, and R language CDKs risk model was constructed.
Results
Overall, CDKs (CDK1, CDK2, CDK3, CDK4, CDK5, CDK6, CDK7 and CDK8) were differentially expressed between normal controls and colorectal cancer. Three CDKs genes (CDK3, CDK5 and CDK8) associated with prognosis were obtained by univariate and multivariate Cox and LASSO regression analysis. In CRC, CDK3, CDK5 and CDK8 are significantly associated with expression levels of recognized immune infiltrates.
Conclusion
CDK3, CDK5 and CDK8 are potential diagnostic markers for CRC; meanwhile, CDK3, CDK5 and CDK8 are potential prognostic markers for CRC; studying the relationship between CDKs and tumor immunology may be helpful for immunotherapy of CRC, and more studies are needed to confirm these results.
Keywords: colorectal cancer, cyclin-dependent kinases family, mRNA expression, prognosis, risk score model
Introduction
Colorectal cancer (CRC), which includes both colon and rectal cancers, is one of the most common malignant tumors of the gastrointestinal system. In recent years, the incidence and mortality rate of CRC in China have been among the top 5 of all malignant tumors, and the trend has continued to increase in the past decade.1 Early treatment is still based on surgical resection supplemented with radiotherapy, but the efficacy is poor. Although the pathogenesis of CRC is still unclear, its etiology is multifactorial, as both internal and external factors contribute to the disease. Therefore, a better understanding of the molecular mechanisms of CRC can help in the development of new therapeutic strategies. To improve CRC patient survival, it is important to use novel biomarkers in the diagnosis and prognosis of colorectal cancer.2
CDKs are serine/threonine (Ser/Thr) protein kinases and are central molecules that regulate the cell cycle network. In mammals, the CDK family consists of 20 members (CDK1 to CDK20). Among them, CDK1 to CDK6 are associated with cell cycle regulation, while CDK7 to CDK9, CDK12 and CDK13 are involved in transcriptional processes.3 Studies have confirmed that the abnormal activation of CDKs can result in disorder of cell cycle process, which can lead to abnormal proliferation of tumor cells. In the process of carcinogenesis of breast, lung, esophagus and other cells, it is often accompanied by abnormal increase of CDKs activity and out of control of cell cycles.4 More studies have been published on inhibitors of interphase CDKs, such as CDK2,5 CDK4/CDK6 and CDK5.6,7 Therefore, CDKs are a focus in anti-tumor research field. At present, several CDKs small molecule inhibitors are undergoing preclinical or clinical trials for the treatment of lung cancer, leukemia, ovarian cancer and other diseases.8–10 Nevertheless, some fundamental research data in regards to the role of CDKs in colorectal cancer are available. For example, CDK5 silencing via transfection can directly reduce the proliferation of human HCT116 and SW480 tumor cell lines.11 Dale et al showed that stable silencing of CDK8 in Colo205 human colon cancer cells reduced β-catenin/TCF-dependent transcription.12 Furthermore, CDK3, CDK5 and CDK8 have been less studied in colorectal cancer. Therefore, this study comprehensively analyzed the expression levels of CDKs in colorectal cancer and its association with clinical prognosis of colorectal cancer patients, and constructed a predictive model of CRC that can be applied for clinical practice. In addition, we investigated the correlation between CDKs and different immune cell infiltration levels to provide a theoretical basis for basic research and clinical treatment.
Materials and Methods
RNA-Seq Data Source
The mRNA expression profiles and clinical characteristics of 620 CRC patients are available at the Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/projects). Fragments per kilobase million (FPKM) values were transformed into transcripts per kilobase million (TPM) value, and were further log transformed for better comparisons between samples.
Expression of CDK Gene in Pan Cancer
Next, we used the R package “ggplot2” to analyse the expression of CDKs in various cancer types and to analyse the expression of CDK1-CDK8 in different tumour cell lines.13–15 The expression of CDKs in pan-cancer was also analyzed in normal and tumor tissues.
Expression of CDKs in CRC
To evaluate the mRNA transcript levels of CDKs in CRC, we analyzed the differential expression of CDKs in CRC using Oncomine (https://www.oncomine.org/resource/login.html),16 GEPIAdata (http://gepia.cancerpku.cn/),17 GSCALite data (http://bioinfo.life.hust.edu.cn/web/GSCALite/) and ComplexHeatmap R package.18,19 In addition, the pheatmap R package was used to explore correlations between CDK1,2,3,4,5,6,7 and 8 levels in CRC. The AUC values of CDKs were evaluated using pROC R package and ggplot2 r package.
Construction of “CDKs Risk Score”Predicting System
Univariate Cox proportional hazards regression analysis was first performed on the expression matrix of RNA processing factors to estimate the relationship between RNA processing factors and prognosis (OS) in the TCGA-CRC cohort. RNA processing factors of p-value < 0.2 were selected as the potential prognosis-related RNA processing factors. Then, to further delineate the impact on CDKs and its related genes on the prognosis of patients, we applied the following protocol: First, Pearson correlation analysis and univariate cox analysis was implemented to screen CDKs related prognostic genes. Then, least absolute shrinkage and selection operator (LASSO) Cox regression, implemented by R package “glmnet”, was conducted to perform dimension reduction.20–22 After these procedures, a CDKs-related gene set composed of three genes was successfully developed, the three genes were “CDK3”,“CDK5”, and“CDK8”. The risk score of the signature was calculated for each sample based on the following formula:
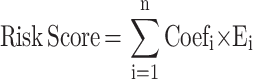 |
where Coefi is the coefficient and Ei is the normalized expression value of each selected gene by log2 and z-score transformations. Risk scores were computed for all patients included in our study. To further obtain a clinical model with predicting values, another multivariate Cox regression was applied to establish a nomogram,23–25 integrating all clinical characteristics that had HR >1 (or < −1) and p-value < 0.05. The calibration plots show the prognostic predictive accuracy of the nomogram.
TIMER Database Analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a web server for comprehensive analysis of tumor-infiltrating immune cells. TIMER was used to analyze the correlation between CDK expression level with immune cell infiltration level or immune checkpoint expression level in CRC.26 p-value<0.05 was considered as statistically significant.
The Human Protein Atlas Database Analysis
The Human Protein Atlas (HPA) (https://www.proteinatlas.org/) is a database of immunohistochemistry (IHC)‐based protein expression profiles in different cancers,27 normal tissue as well as cell lines. In this study, we used the HPA database for protein expression profiling.
Statistics and Data Analysis
All of the statistical analysis and charts are carried out using R language (version 3.6.3). In this study, the Wilcoxon Rank Sum Test was used to compare the mRNA expression of CDKs in CRC and normal colorectal tissues. The log rank test was then used to analyze the survival and prognosis of CRC patients. The pathological stage of CRC was determined using Kruskal–Wallis Test. In all tests, p-value<0.05 was considered statistically significant.
Results
mRNA Expression in Pan Cancer in Patients with Colorectal Cancer
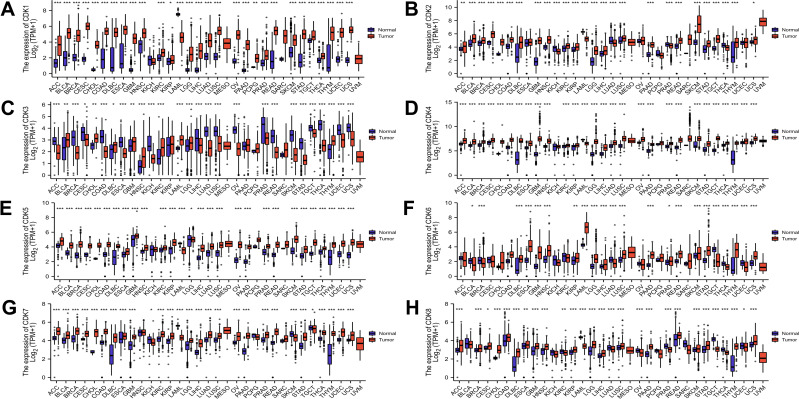
The mRNA levels of CDKs in different cancer cell lines were obtained from the CCLE database. The results showed that colorectal cancer cell lines maintained high mRNA expression of CDKs from 3–7 in 18 cancer cell lines, and CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, and CDK8 were relatively high compared with other cancer cell lines except for CDK3, which was relatively low (Supplement Figure 1). Using the R data visualization package (“ggplot2”), we analyzed CDK mRNA expression of pan-cancers. The results showed that CDK was different in different tumors. CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, and CDK8 were highly expressed in colorectal cancer compared with normal tissues, whereas CDK3 was expressed at lower levels in colorectal cancer (Figure 1).
Figure 1.
Expression of CDK family mRNA in different types of tumors. (A-H) Expression levels of CDK1-8 in multiple cancers. Blue represents normal tissue and red represents tumor tissue.*p<0.05, **p<0.01, and ***p<0.001.
Relationship Between mRNA Levels of CDKs and Clinicopathologic Characteristics of CRC Patients
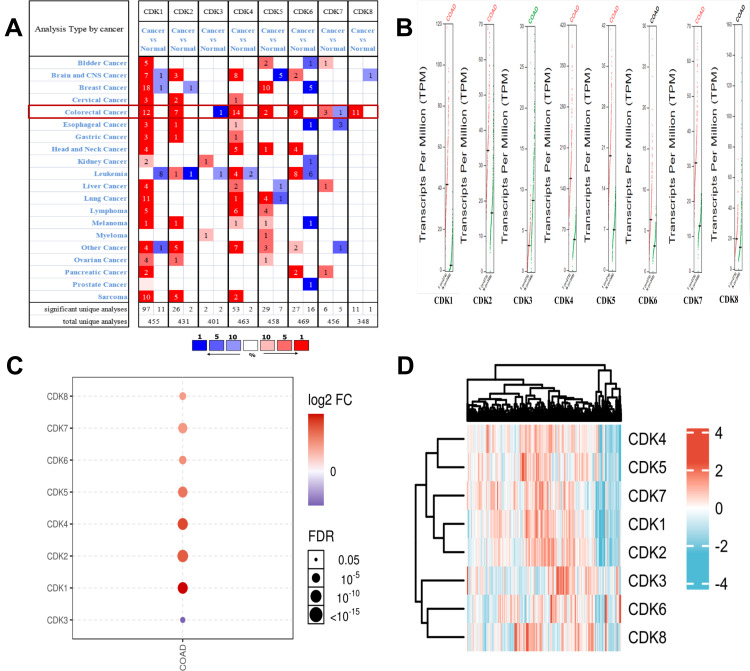
We utilized the public Oncomine database to analyze CDK expression of CRC and normal tissue. We found that the transcriptional levels of the target genes CDK1, CDK2, CDK4 and CDK5, CDK6, CDK7, CDK8 were significantly increased in CRC, while CDK3 transcriptional levels were reduced compared with normal tissue (Figure 2A). Using the GSCALite online tool to analyze the differential expression of CDK genes (Figure 2C), we found that CDK1, CDK2, CDK4, CDK5, CDK6, CDK7 and CDK8 were highly expressed, while CDK3 was expressed at low levels in CRC. In addition, we verified the mRNA expression of CDK1, CDK2, CDK3, CDK4, CDK5, CDK6, CDK7 and CDK8 using the online database GEPIA, and the results were consistent with the above findings (Figure 2B). The heatmap illustrates the expression of CDKs in tissues from the TCGA database (colon adenocarcinoma (COAD); Figure 2D).
Figure 2.
Expression levels of CDK genes in CRC.(A) Analysis of CDKs in the Oncomine database. (B) CDK expression levels in CRC (GEPIA).(C) CDKs are differentially expressed (GSCALite). The dots indicate the FDR from multiple t-tests; the larger the circle, the smaller the FDR.(D) Hierarchical cluster analysis of the CDK family (ComplexHeatmap package).
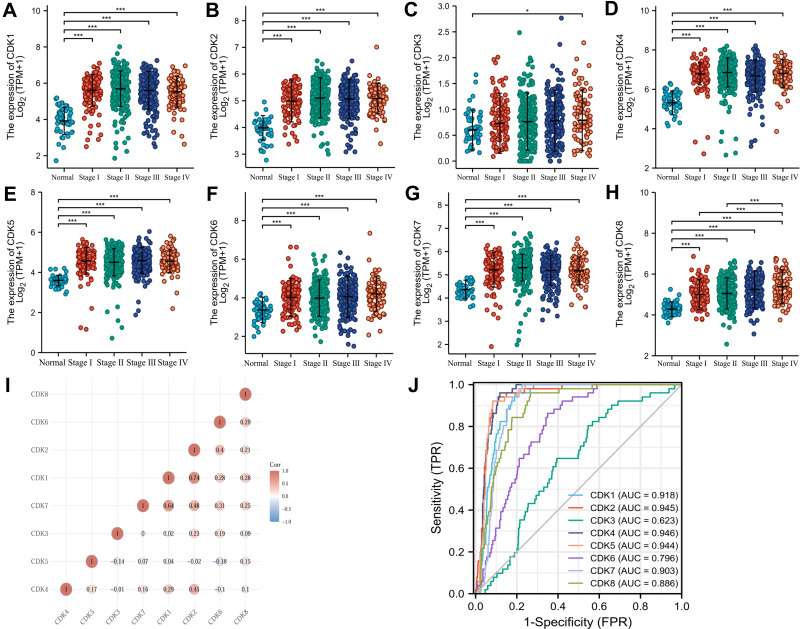
The clinical stage expression status of CDKs in CRC tissues was shown by Kruskal–Wallis Test, and the expression of CDK1-CDK8 was significantly higher in CRC tissues than in normal tissues (P<0.05) (Figure 3A–H). CDK1-CDK8 was significantly expressed in CRC (P<0.05), revealing that the expression of CDK1-CDK8 was associated with colorectal carcinogenesis. As shown in Figure 3I, there was a significant correlation among CDK1-CDK8 gene levels. The AUC values of CDK1-8 were 0.918 (CI:0.895–0.942), 0.945 (CI:0.923–0.967), 0.623 (CI:0.555–0.692), 0.946 (CI:0.929–0.963), 0.946 (CI:0.929–0.963), 0.944 (CI:0.925–0.963), 0.796 (CI:0.750–0.843), 0.903 (CI:0.880–0.926) and 0.886 (0.852–0.920)(Figure 3J).
Figure 3.
CDK1-8 expression levels in CRC from TCGA data. (A–H) Expression levels of CDK1-8 in CRC and normal tissues; (I) Correlation between CDK1-8 members; (J) Receiver operating characteristic analysis (ROC) of CDK1-8 in CRC. *p<0.05 and ***p<0.001.
Construction of Risk Scores and their Predictive Value
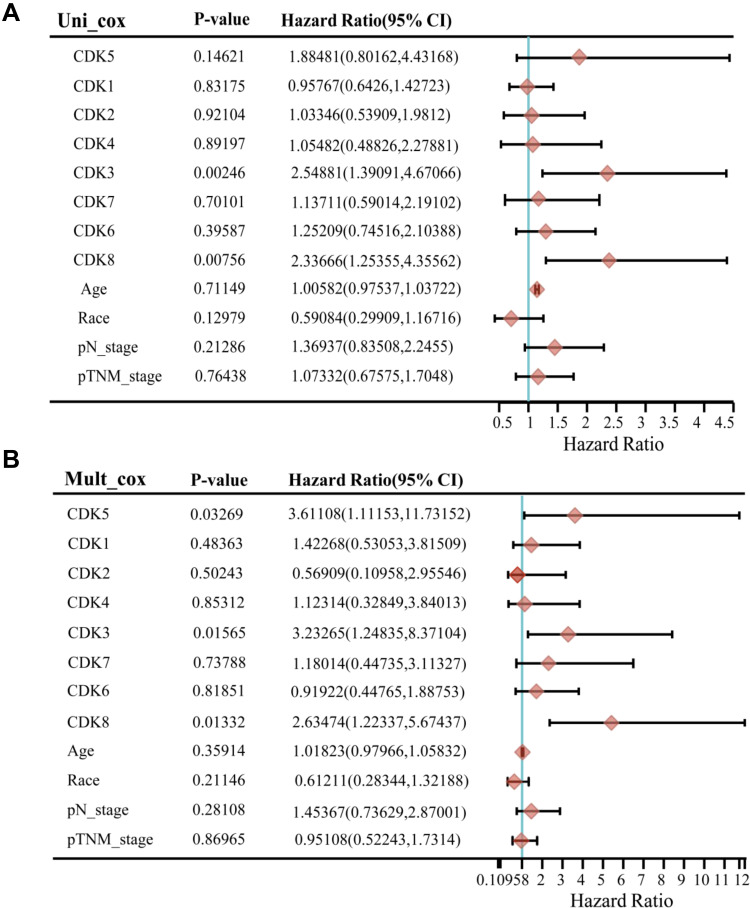
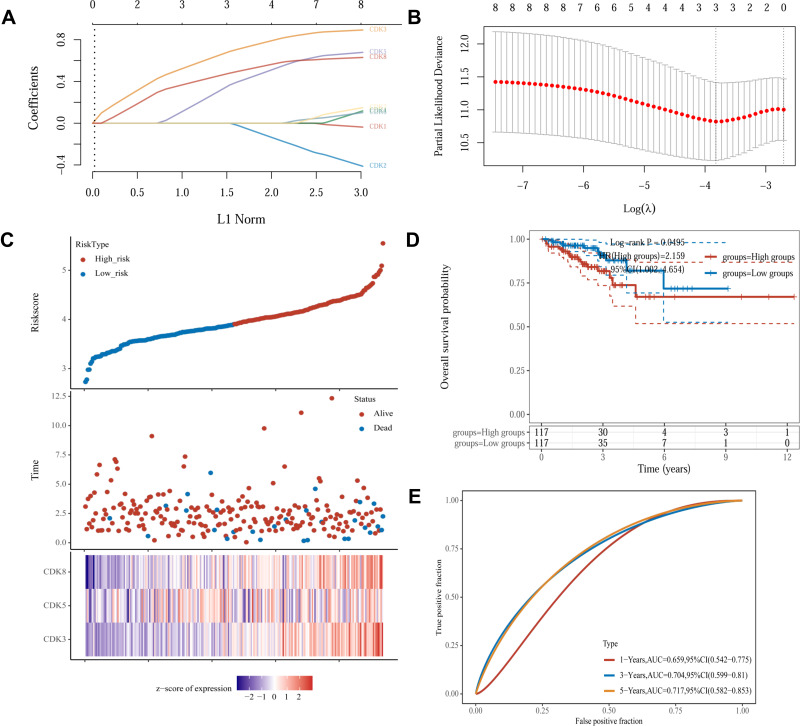
To investigate whether CDKs can be used as an effective biomarker to predict the prognosis of CRC, we further performed LASSO regression analysis of CDKs. Univariate Cox regression analysis was used for the initial screening of survival-related genes. Three genes (CDK3, CDK5 and CDK8) that met the p-value< 0.2 criteria were retained for further analysis, with CDK3, CDK5 and CDK8 genes associated with increased risk (HR>1) (Figure 4A and B). Subsequently, LASSO regression analysis was performed. Combining the results of Figure 5A and B, three genes were constructed based on the optimal λ value. The risk score is as follows: Riskscore = (0.598)×CDK3 + (0.2248)×CDK5 + (0.4186)×CDK8. Based on the score calculated by the risk scoring formula, 620 colorectal cancer patients were equally divided into low-risk and high-risk groups (Figure 5C). Compared with patients in the low-risk group, patients in the high-risk group had more deaths and shorter survival time. A significant difference in OS time was detected between the low and high risk groups (P<0.05, Figure 5D). Applying time-dependent receiver operating characteristic (ROC) analysis to evaluate the sensitivity and specificity of the prognostic model, we found an area under the ROC curve (AUC) of 0.659 at 1 year, 0.704 at 3 years, and 0.717 at 5 years (Figure 5E).
Figure 4.
Univariate and multivariate analysis of clinical characteristics and risk scores of DFS patients. CDKs expression and other clinicopathologic factors with DFS in CRC were calculated via univariate (A) and multivariate (B) regression analysis. Three genes were obtained with p-value < 0.2.
Figure 5.
Construction of risk characteristics in the TCGA queue.(A) LASSO coefficient profiles.(B) Ten-time cross-validation for tuning parameter selection in the LASSO mode.(C) The risk score, survival status, and heat map of CDK3, CDK5 and CDK8 in patients with CRC.(D) Kaplan-Meier curves for the OS of patients in the high- and low-risk groups.(E) ROC curves demonstrated the predictive efficiency of the risk score.
Prognostic Model Development Based on CDK3/5/8 and Clinicopathological Factors
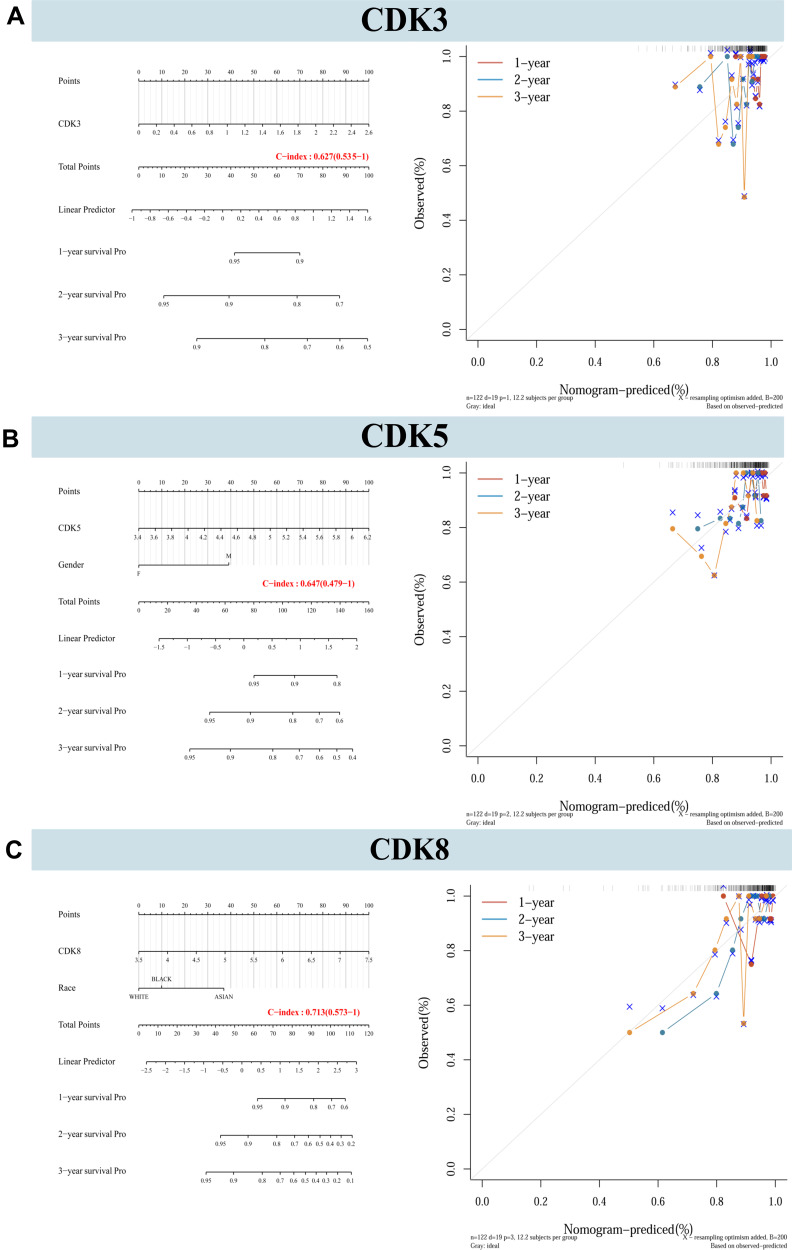
Nomograms were constructed for CDK3/5/8 expression and independent clinical risk factors (age and pathological stage) (Figure 6). Higher total number of points in the nomogram represents worse prognosis. In addition, the C index values of CDK3, CDK5 and CDK8 were 0.627, 0.647 and 0.713, respectively. The deviation correction line in the calibration chart is close to the ideal curve (ie, 45°line), indicating that the prediction results are good agreement with the observation results.
Figure 6.
Nomograms and calibration plots predicting the probability of 1-year, 2-year, and 3-year DFS in CRC patients. (A–C) Integration of nomograms and calibration plots for CDK3, CDK5 and CDK8, and other prognostic factors for CRC from TCGA data.
Correlation of Characteristic Immune-Related Genes with the Immune Microenvironment
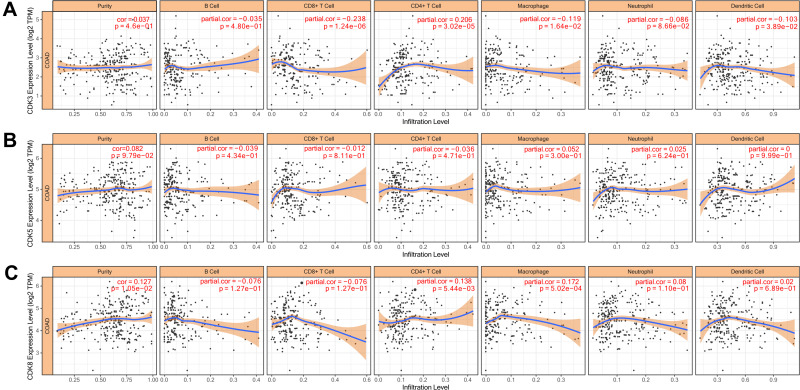
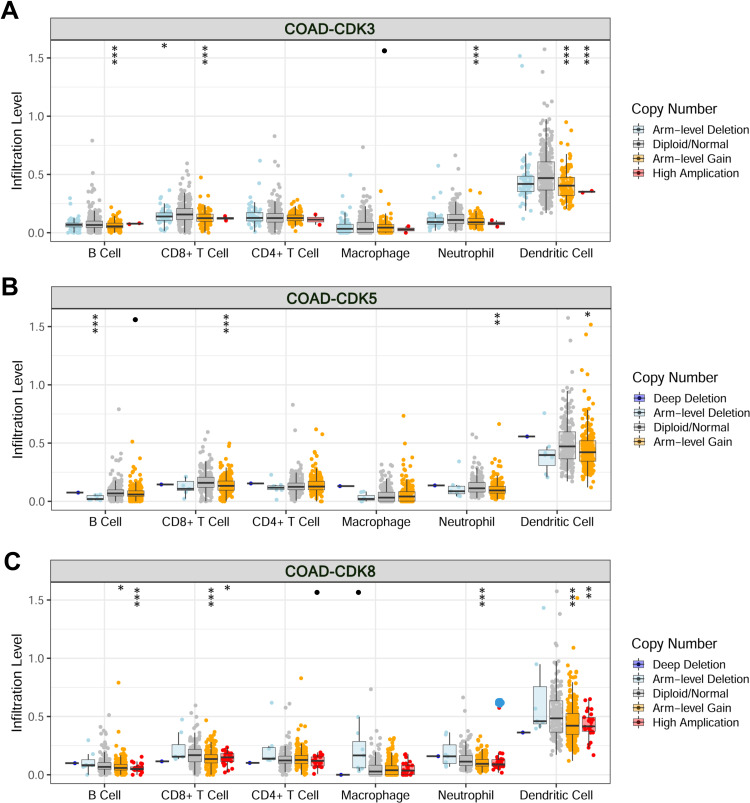
The level of immune cell infiltration is related to the proliferation and development of cancer cells. To assess the potential relationship between CDK expression and the level of immune infiltration in CRC, we used TIMER to perform the following analysis of the correlation between CDK3, CDK5 and CDK8 members and immune cell infiltration. We explored whether CDK3, CDK5 and CDK8 expression from the TIMER database could influence the level of various immune cell infiltrations in COAD. The results showed that in COAD, CDK3 expression was significantly negatively correlated with CD8+ T Cell (r=−0.238, p=1.24e-06), Macrophage (r=−0.0119, p=1.64e-02) and Dendritic Cell (r=−0.103, p=3.89e-02), and was positively correlated with the infiltration level of CD4+T Cell (r=0.206, p=3.02e-05), and was not correlated with B Cell (r=−0.035, p=4.80e-01) and Neutrophil (Figure 7A). CDK8 expression was positively correlated with levels of CD4 + T Cell (r = 0.138, p = 5.44e-03) and Macrophage (r = 0.172, p = 5.02e-04) infiltration (Figure 7C). In addition, CDK5 expression did not significantly correlate with the infiltration levels of B Cell, CD8+ T Cell, CD4+ T Cell, Macrophage, Neutrophil and Dendritic Cell (Figure 7B). In addition, CDK3, CDK5 and CDK8 CNA significantly correlated with the level of major immune cell infiltration (Figure 8). These results strongly suggest that CDK3, CDK5 and CDK8 could serve as a major tumor immune infiltration regulator in COAD.
Figure 7.
Correlation of CDKs levels of immune cell infiltration in COAD.Correlations between CDK3 (A), CDK5 (B) and CDK8 (C) expression and tumor purity and dominant immune cells infiltration levels (TIMER). p<0.05 considered statistically significant.
Figure 8.
Changes in somatic copy number of CDKs in COAD.Correlations between CDK3 (A), CDK5 (B) and CDK8 (C) somatic copy number alterations and dominant immune cells infiltration levels. •p<0.1, *p<0.05, **p<0.01, ***p<0.001.
Prognostic Gene Validation Using Clinical Tissue Samples
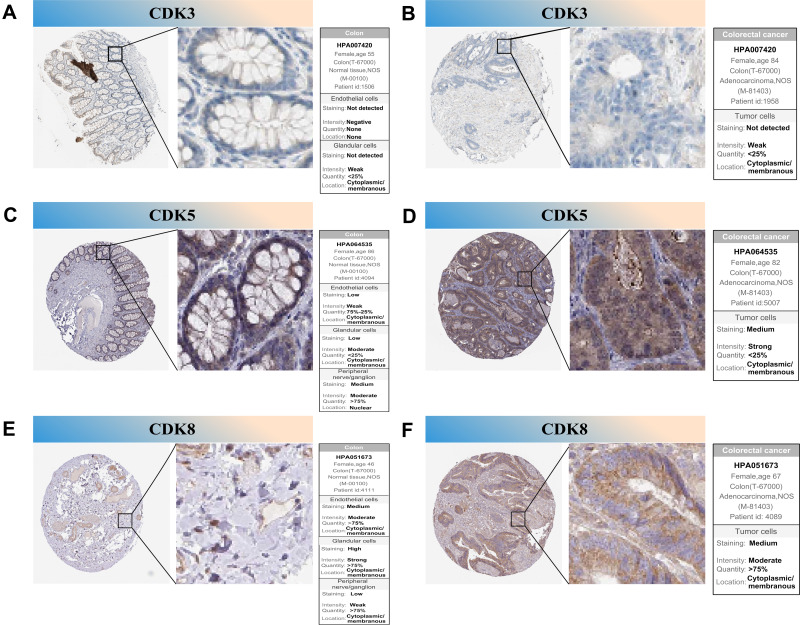
To further confirm the prognostic value of the hub genes with prognostic values, we used immunohistochemical (IHC) stain to detect the protein expression of CDK3, CDK5 and CDK8 in normal tissues and tumor tissues. The results showed that CDK3 was significantly under-expressed in primary CRC tissues compared with normal tissues and CDK5, CDK8 were significantly highly expressed in primary colorectal cancer tissues (Figure 9), which is consistent with our findings.
Figure 9.
IHC analysis of CDK3, CDK5 and CDK8 with prognostic values. (A–F) Differentially expressed proteins of CDK3, CDK5 and CDK8 with prognostic values in colorectal normal tissues and colorectal cancer in The Human Protein Atlas database.
Discussion
CRC is the consequence of a progressive accumulation of genetic and epigenetic changes in normal colonic epithelial cells, which leads to colorectal adenomas and invasive adenocarcinomas.28 CRC has a high incidence and is one of the leading causes of tumor-related mortality.29 CDKs regulate cell cycle transcription and progression, and aberrant expression of these genes has been observed in different human tumors. Impaired regulation of cell cycle progression is associated with intestinal (small and large intestine) diseases, particularly colon and rectal cancers, which account for 1/5 of all cancers worldwide.30 Previous data clearly show that epigenetic changes are important molecular features that occur early and frequently. Epigenetic changes are common in cancer and include aberrant DNA methylation and aberrant histone modifications. Proteins in the CDK family function in the cell cycle and may also be involved in epigenetic processes. While CDKs have been shown to play a key role in breast and liver cancers, the different roles of CDKs in CRC remain to be elucidated. Therefore, the identification of new molecular mechanisms is necessary to develop therapeutic targets in CRC.
Our results suggest that CDK3, CDK5 and CDK8 may be a prognostic biomarker in CRC. low CDK3 expression and high CDK5 and CDK8 expression are associated with poor survival. CDK3 has been reported to promote myoblast cell proliferation;31 Yue et al showed that CDK5 promotes the proliferation of medullary thyroid carcinoma cells;32 in addition, Chen et al showed that the promotion of OSCC cell proliferation by direct binding of miR-567 leads to increased CDK8 expression.33 These previous findings suggest that CDK3, CDK5 and CDK8 is an oncogene.
Similarly, our results demonstrated CDK5 and CDK8 was upregulated in CRC tissues, supporting its potential role in the development of CRC. In addition, several studies suggest that CDK3, CDK5 and CDK8 may be a target for cancer therapy.31,34,35 However, the association between CDK3, CDK5 and CDK8, and CRC has not been explored so far. Therefore, our results provide the first evidence for its therapeutic and prognostic value in CRC.
The tumor microenvironment (TME) affects the occurrence and development of tumors through circulating cells and lymphoid systems containing peripheral cells; therefore, the TME has been widely involved in tumors. Some TMEs contain cytotoxic CD8+ memory T cells that are capable of killing tumor cells, which leads to a good prognosis.36 Cytotoxic CD8+ memory T cells kill tumor cells by identifying specific antigens on those cells and by inducing an immune response. Our research shows that CDK3, CDK5 and CDK8 expression may be significantly associated with infiltration of the TME by six immune cell types, which indicates that CDK3, CDK5 and CDK8 may also reflect the immune response in addition to the prognosis of the disease. This study provides detailed information on immune cells in CRC that may help in the design of new immunotherapies.
Inevitably, our research also has some limitations that need to be pointed out. First, The signature was developed based on public databases and retrospective cohort studies. Second, this is a retrospective study that may display statistical bias, and the bulk sequence transcriptome data were used, in which there is a lack of comprehensive exploration for intratumoral heterogeneity. Finally, we need to perform in vitro and in vivo experiments regarding these genes to further validate the silico results and the response of immunotherapy should be further verified in randomized clinical trials.
Conclusion
This CDK3, CDK5 and CDK8 signature may be a useful biomarker for predicting CRC patient survival, and could expand the application of targeted therapy and improve treatment efficacy through patient stratification.
Funding Statement
This work was funded by grants from the Medical School of Facial Features of Hubei University of Science and Technology (No.2020XZ35), the High-level Talent Research Project of Xianning City (No.202001), the Xianning Municipal Science and Technology Plan Project (No.2021ZRKX025), and the Hubei Provincial Colleges and Universities Provincial Teaching Research Project (No.2020641).
Abbreviations
CDKs, cyclin-dependent kinases; CRC, colorectal cancer; TCGA, the Cancer Genome Atlas; FPKM, fragments per kilobase million; TPM, transcripts per kilobase million; LASSO, least absolute shrinkage and selection operator; HPA, the human protein atlas; ROC, receiver operating characteristic; AUC, the area under the curve; IHC, immunohistochemical; TME, the tumor microenvironment.
Ethics Approval and Consent to Participate
This study was conducted with approval from the Ethics Committee of Hubei University of Science and Technology. TCGA belong to public databases. Users can download relevant data for free for research and publish relevant articles. Our study is based on open-source data and therefore has no other conflicts of interest.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, agreed to the submitted journal, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhu J, Tan Z, Hollis-Hansen K, et al. Epidemiological trends in colorectal cancer in china: an ecological study. Dig Dis Sci. 2017;62(1):235–243. doi: 10.1007/s10620-016-4362-4 [DOI] [PubMed] [Google Scholar]
- 2.Xu M, Guo X, Wang RD, et al. Long non-coding RNA HANR as a biomarker for the diagnosis and prognosis of colorectal cancer. Medicine. 2020;99(7):e19066. doi: 10.1097/MD.0000000000019066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solaki M, Ewald JC. Fueling the cycle: cDKs in carbon and energy metabolism. Front Cell Dev Biol. 2018;6:93. doi: 10.3389/fcell.2018.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2011;137(10):1409–1418. doi: 10.1007/s00432-011-1039-4 [DOI] [PubMed] [Google Scholar]
- 5.Somarelli JA, Roghani RS, Moghaddam AS, et al. A precision medicine drug discovery pipeline identifies combined CDK2 and 9 inhibition as a novel therapeutic strategy in colorectal cancer. Mol Cancer Ther. 2020;19(12):2516–2527. doi: 10.1158/1535-7163.MCT-20-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Q, Guo X, Wang M, et al. The application and prospect of CDK4/6 inhibitors in malignant solid tumors. J Hematol Oncol. 2020;13(1):41. doi: 10.1186/s13045-020-00880-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz de Porras V, Bystrup S, Cabrero-de Las Heras S, et al. Tumor expression of cyclin-dependent kinase 5 (Cdk5) Is a prognostic biomarker and predicts outcome of oxaliplatin-treated metastatic colorectal cancer patients. Cancers. 2019;11(10):1540. doi: 10.3390/cancers11101540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sánchez-Martínez C, Lallena MJ, Sanfeliciano SG, et al. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs: recent advances (2015–2019). Bioorg Med Chem Lett. 2019;29(20):126637. doi: 10.1016/j.bmcl.2019.126637 [DOI] [PubMed] [Google Scholar]
- 9.Whittaker SR, Mallinger A, Workman P, et al. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther. 2017;173:83–105. doi: 10.1016/j.pharmthera.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marak BN, Dowarah J, Khiangte L, et al. A comprehensive insight on the recent development of cyclic dependent kinase inhibitors as anticancer agents. Eur J Med Chem. 2020;203:112571. doi: 10.1016/j.ejmech.2020.112571 [DOI] [PubMed] [Google Scholar]
- 11.Zhuang K, Zhang J, Xiong M, et al. CDK5 functions as a tumor promoter in human colorectal cancer via modulating the ERK5-AP-1 axis. Cell Death Dis. 2016;7:e2415. doi: 10.1038/cddis.2016.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale T, Clarke PA, Esdar C, et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat Chem Biol. 2015;11:973–980. doi: 10.1038/nchembio.1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big bioMedical data analyses. Nat Biotechnol. 2017;35(4):314–316. doi: 10.1038/nbt.3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou T, Cai Z, Ma N, et al. Novel ten-gene signature predicting prognosis in hepatocellular carcinoma. Front Cell Dev Biol. 2010;8:629. doi: 10.3389/fcell.2020.00629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin Y, Conley AP, Grimm EA, et al. tool for discovering drug sensitivity and gene expression associations in cancer cells. PLoS One. 2017;12(4):e0176763. doi: 10.1371/journal.pone.0176763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Lin E, Zhuang H, et al. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int. 2020;20:27. doi: 10.1186/s12935-020-1113-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Lin W, Weng Y, et al. Characterization of hypoxia signature to evaluate the tumor immune microenvironment and predict prognosis in glioma. J Clin Oncol. 2020;38(15_suppl):e14534–e14534. doi: 10.1200/JCO.2020.38.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu F, Huang X, Li Y, et al. (2021). m6A-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD. Mol Ther Nucleic Acids. 2021;(24):780–791. doi: 10.1016/j.omtn.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y, Yuan L, Xiong J, et al. An outcome model for human bladder cancer: a comprehensive study based on weighted gene co-expression network analysis. J Cell Mol Med. 2020;24(3):2342–2355. doi: 10.1111/jcmm.14918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Mi M, Li X, et al. A lncRNA prognostic signature associated with immune infiltration and tumour mutation burden in breast cancer. J Cell Mol Med. 2020;24(21):12444–12456. doi: 10.1111/jcmm.15762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong SH, Kim RB, Park SY, et al. Nomogram for predicting gastric cancer recurrence using biomarker gene expression. Eur J Surg Oncol. 2020;46(1):195–201. doi: 10.1016/j.ejso.2019.09.143 [DOI] [PubMed] [Google Scholar]
- 26.Li T, Fan J, Wang B, et al. TIMER A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asplund A, Edqvist PH, Schwenk JM, et al. Antibodies for profiling the human proteome-the human protein atlas as a resource for cancer research. Proteomics. 2012;12(13):2067–2077. doi: 10.1002/pmic.201100504 [DOI] [PubMed] [Google Scholar]
- 28.Jung G, Hernández-Illán E, Moreira L, et al. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17(2):111–130. doi: 10.1038/s41575-019-0230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. Ca Cancer J Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601 [DOI] [PubMed] [Google Scholar]
- 30.Costa NR, Gil da costa RM, Medeiros R. A viral map of gastrointestinal cancers. Life Sci. 2018;199:188–200. doi: 10.1016/j.lfs.2018.02.025 [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Xi Y, Sun C, et al. CDK3 is a major target of miR-150 in cell proliferation and anti-cancer effect. Exp Mol Pathol. 2017;102(2):181–190. doi: 10.1016/j.yexmp.2017.01.008 [DOI] [PubMed] [Google Scholar]
- 32.Do PA, Lee CH. The role of CDK5 in tumours and tumour microenvironments. Cancers. 2020;13(1):101. doi: 10.3390/cancers13010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Wang X, Zhou S, et al. LncRNA HOXA-AS2 promotes tumor progression by suppressing miR-567 expression in oral squamous cell carcinoma. Cancer Manag Res. 2021;13:5443–5455. doi: 10.2147/CMAR.S305946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Yue L, Fan C, et al. Mechanism of Cdk5-synaptophysin-SNARE pathway in acute and chronic inflammatory pain. Am J Transl Res. 2021;13(3):1075–1084. PMID: 33841641. [PMC free article] [PubMed] [Google Scholar]
- 35.Liang J, Chen M, Hughes D, et al. CDK8 selectively promotes the growth of colon cancer metastases in the liver by regulating gene expression of TIMP3 and matrix metalloproteinases. Cancer Res. 2018;78(23):6594–6606. doi: 10.1158/0008-5472.CAN-18-1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hivroz C, Chemin K, Tourret M, et al. Crosstalk between T lymphocytes and dendritic cells. Crit Rev Immunol. 2012;32(2):139–155. doi: 10.1615/critrevimmunol.v32.i2.30 [DOI] [PubMed] [Google Scholar]