Abstract
Background and Objectives
We report the pathogenic sequence variant m.5789T>C in the anticodon stem of the mitochondrial tRNA for cysteine as a novel cause of neuropathy, ataxia, and retinitis pigmentosa (NARP), which is usually associated with pathogenic variants in the MT-ATP6 gene.
Methods
To address the correlation of oxidative phosphorylation deficiency with mutation loads, we performed genotyping on single laser-dissected skeletal muscle fibers. Stability of the mitochondrial tRNACys was investigated by Northern blotting. Accompanying deletions of the mitochondrial genome were detected by long-range PCR and their breakpoints were determined by sequencing of single-molecule amplicons.
Results
The sequence variant m.5789T>C, originating from the patient's mother, decreases the stability of the mitochondrial tRNA for cysteine by disrupting the anticodon stem, which subsequently leads to a combined oxidative phosphorylation deficiency. In parallel, we observed a prominent cluster of low-abundance somatic deletions with breakpoints in the immediate vicinity of the m.5789T>C variant. Strikingly, all deletion-carrying mitochondrial DNA (mtDNA) species, in which the corresponding nucleotide position was not removed, harbored the mutant allele, and none carried the wild-type allele.
Discussion
In addition to providing evidence for the novel association of a tRNA sequence alteration with NARP syndrome, our observations support the hypothesis that single nucleotide changes can lead to increased occurrence of site-specific mtDNA deletions through the formation of an imperfect repeat. This finding might be relevant for understanding mechanisms of deletion generation in the human mitochondrial genome.
Neuropathy, ataxia, and retinitis pigmentosa (NARP) syndrome is characterized by sensory neuropathy, ataxia, and retinitis pigmentosa and is typically caused by mitochondrial DNA (mtDNA) sequence variants leading to amino acid substitutions in the MT-ATP6 gene that codes for a subunit of the mitochondrial ATP synthase, complex V of oxidative phosphorylation (OXPHOS).1,2 In addition, a combination of ataxia and retinitis pigmentosa in a young adult was described to be associated with the m.1606G>A variant in the mitochondrial tRNA for valine.3
Here, we demonstrate that the novel m.5789T>C sequence variant in the tRNACys gene of the mtDNA is also associated with late-onset NARP syndrome. Previously reported cases of pathogenic tRNACys variants are very rare and are usually associated with mitochondrial encephalomyopathy4,5 and/or hearing loss6-8 or MERRF syndrome (myoclonic epilepsy with ragged-red fibers).9 Furthermore, we show that the m.5789T>C variant, in combination with a silent homoplasmic allelic variant, contributes to an increased de novo generation of somatic mtDNA deletions by stabilizing an imperfect repeat.
Methods
Enzymatic Analysis
Activity of citrate synthase, NADH:ubiquinone oxidoreductase (complex I of the respiratory chain), and the cytochrome c oxidase (complex IV) in skeletal muscle homogenate was measured using a common spectrophotometry protocol reported before.10
DNA Isolation
Biopsy specimen from M. vastus lateralis, urine sediment, blood, and cultured skin fibroblasts were collected from the patient. Furthermore, blood and urine sediment samples were collected from all 3 unaffected siblings. No material was available from the deceased parents. DNA was extracted from tissue samples using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany).
mtDNA Sequencing
Four large (4.5–5.8 kb) overlapping fragments of the mitochondrial genome were amplified, and PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Sanger sequencing of the entire mitochondrial genome was performed by a commercial service (Eurofins, Ebersberg, Germany) using 30 sequencing primers.
Whole-Exome Sequencing
Whole-exome sequencing (WES) was performed on DNA isolated from the patient's cultured fibroblast using an Agilent SureSelect V6 enrichment. Paired-end 100-bp reads were aligned to the GRCh38 assembly of the human genome. Mean coverage of 92-fold (30-fold coverage for 83% and 10-fold coverage for 95.7% of target sequences) was achieved. Filtering and variant prioritization was performed using the VARBANK database and analysis tool at the Cologne Center for Genomics. In particular, we filtered for high-quality (coverage > 15-fold; PHRED-scaled quality > 25), rare variants (minor allele frequency ≤ 0.01, according to the gnomAD database v2.1.1,11 last accessed on November 15, 2021) with predicted effects on protein sequence or splicing. We scored potential functional effects of variants using the Combined Annotation-Dependent Depletion (CADD) v1.612 (last accessed November 15, 2021). Only variants with a CADD score of >14 were considered. To identify previously reported pathogenic or likely pathogenic sequence variants we checked the list of variants against the ClinVar databank13 (last accessed on November 10, 2021). To exclude pipeline-related artifacts, we filtered (minor allele frequency ≤ 0.01) against variants from in-house WES data sets from 511 patients with epilepsy.
Mutation Detection by Restriction Fragment Length Polymorphism Analysis
The m.5789T>C variant was detected by amplifying a 186-bp PCR product using primers 5694F (5′-AACAGCTAAGCACCCTAATCAACT-3') and 5879R (5′-GAGTGAAGCATTGGACTGTAAATCT-3′). The PCR mix contained 10 ng of DNA, 0.2 U JumpStart Taq DNA polymerase, 0.1 mM of each dNTP, 1× PCR buffer (Sigma-Aldrich, St. Louis), 2.5 mM MgCl2, and 250 nM of each primer in a total volume of 25 µL. Amplification conditions were 95°C for 10 minutes; 35 cycles of 95°C for 15 seconds, 55°C for 30 seconds, and 72°C for 40 seconds; and finally 72°C for 7 minutes. Amplification products were digested with the restriction enzymes TaqαI or Hpy188I (New England Biolabs, Ipswich, MA) using the producer's protocol. TaqαI cuts the wild-type molecules into 2 fragments of 89 bp and 97 bp length, whereas it does not cut the mtDNA molecules carrying the m.5789T>C variant. Hyp188I cuts the wild-type mtDNA into 2 fragments of 58 bp and 128 bp length and the mutated mtDNA into 3 fragments of 58 bp, 31 bp, and 97 bp length. Restriction digestion fragments were separated on a 10% polyacrylamide gel and visualized by SYBR Green I staining (Sigma-Aldrich) and fluorescence detection using a GelDoc system (Bio-Rad Laboratories, Hercules). Proportions of wild-type and mutant mtDNA were estimated from band intensities using ImageJ software. Heteroplasmy values from TaqαI and Hpy188I restriction digestions were averaged.
Laser Microdissection and Single-Fiber Genotyping
Single muscle fibers were dissected using the P.A.L.M. MicroBeam system, as previously described.14 Briefly, unstained air-dried 10-µm skeletal muscle sections flanked on both sides by cytochrome c oxidase (COX)/succinate dehydrogenase (SDH)-stained sections were mounted on a membrane slide (Zeiss, Oberkochen, Germany) and immersed sequentially in 70% ethanol, 95% ethanol, absolute ethanol, and xylene.
Skeletal muscle fibers were cut using a P.A.L.M. MicroBeam system and catapulted into 0.2-mL microcentrifuge tube caps containing 20 µL of magnesium-free PCR buffer, 10× diluted Tris-EDTA buffer, 0.5% Tween 20, and 1 mg/mL Proteinase K. After incubation at 55°C for 30 minutes and inactivation of the Proteinase K at 95°C for 10 minutes, we used 8 µL of each sample to quantify mutation loads by PCR and restriction fragment length polymorphisms (RFLP) analysis as described above. Identical fibers from 3 consecutive sections were quantified and their values averaged.
Detection of mtDNA Deletions
We investigated the presence of deletions of the mtDNA by long-range PCR. Primers 5462F (5′-CCTTACCACGCTACTCCTACCTATCTCC-3') and 638R (5′-GGTGATGTGAGCCCGTCTAAAC-3′) were used for amplification. Reaction mixtures contained 20 ng DNA, 0.25 mM dNTPs, 0.2 µM of each primer, 0.6 U JumpStart AccuTaq LA DNA Polymerase, and 1× AccuTaq PCR buffer (Sigma-Aldrich). Amplification conditions were 95°C for 2 minutes 30 seconds; 10 cycles of 92°C for 20 seconds and 68°C for 14 minutes; 20 cycles of 92°C for 25 seconds and 68°C for 16 minutes; and finally 72°C for 10 minutes. Amplification products were separated on an ethidium bromide–containing 1% agarose gel and visualized by fluorescence using a GelDoc system (Bio-Rad Laboratories, Hercules).
Single-Molecule Genotyping
We used a single-molecule dilution technique for genotyping mtDNA molecules.15 Before PCR, DNA samples were diluted to a degree at which not more than 30% of multiple identical PCR amplifications resulted in a product. Under these conditions, the majority of amplicons are expected to originate from a single molecule according to the Poisson distribution. The PCR mix contained 0.12 U of Takara HS DNA polymerase, 1× PCR buffer, 0.2 mM dNTPs (TaKaRa Bio, Kusatsu, Japan), 400 nM of each primer, and 0.5 µL of diluted DNA in a final volume of 12.5 µL. Amplification conditions were 95°C for 2 minutes 30 seconds; 10 cycles of 92°C for 20 seconds and 68°C for 14 minutes; 32 cycles of 92°C for 25 seconds and 68°C for 16 minutes; and finally 72°C for 10 minutes. Single mtDNA deletions were amplified using primers 5462F and 638R. Deletion breakpoints were mapped by reamplification and direct sequencing of single-deletion amplicons.
To detect low amounts of the m.5789T>C variant in DNA extracted from urine and blood of the siblings, samples were digested with TaqαI restriction endonuclease before single-molecule PCR. This step was introduced to cut wild-type mtDNA molecules, thus rendering them nonamplifiable. Single-molecule amplification was then performed using primers 5462F and 5985R (5′-CTCCAGCTCATGCGCCGAATAATAG-3′) and amplification conditions: 95°C for 2 minutes 30 seconds; 10 cycles of 92°C for 20 seconds and 68°C for 2 minutes; 32 cycles of 92°C for 25 seconds and 68°C for 2.5 minutes; and finally 72°C for 10 minutes. PCR products were sequenced by a commercial sequencing service (Eurofins, Ebersberg, Germany).
Determination of mtDNA Copy Number by Quantitative PCR
Quantitative PCR was performed using the Bio-Rad iCycler iQ Real-Time PCR Detection System to determine the mtDNA copy number in bulk skeletal muscle DNA of the patient. Ten or 20 ng of template DNA was used for amplification together with SYBR Green qPCR Master Mix (Bimake) and 250 nM of each primer. Primers 3150F (5′-TACTTCACAAAGCGCCTTCC-3') and 3246R (5′-GGCTCTGCCATCTTAACAAACC-3′) were used to amplify a short region in the MT-ND1 gene in the minor arc of the mtDNA. Part of the single-copy nuclear gene KCNJ10 was amplified using primers KirF (5′-GCGCAAAAGCCTCCTCATT-3') and KirR (5′-CCTTCCTTGGTTTGGTGGG-3′). Amplification conditions were 95°C for 15 minutes, 45 cycles of 95°C for 15 seconds, and 62.5°C for 1 minute. Experimental data were fitted to a 4-parameter Chapman-Richards growth curve, and the x parameter of the inflection point of the sigmoidal curve was defined as CT value. MtDNA copy number per nucleus was calculated using the formula 2 ^ (CT[nuclear] − CT[mitochondrial]) * 2. Quantification of deleted mtDNA molecules was performed by comparing CT values obtained by primers 3150F and 3246R for total mtDNA and primers 12062F (5′-ACCCTCATGTTCATACACCTA-3′) and 12135R (5′-GAGGAAAACCCGGTAATGATGTC-3′) for nondeleted mtDNA.
Northern Blot Analysis
RNA samples from the patient's and 3 controls' skeletal muscle pieces as well as from 106 human osteosarcoma 143B cells were extracted using a common TRIzol protocol. RNA pellets were dissolved in 0.3 M sodium acetate (pH 5.0) + 1 mM EDTA buffer. Part of the 143B RNA sample was treated with 0.5 M Tris-HCl (pH 9.0) at 75°C for 15 minutes to deacylate tRNA molecules. Gel electrophoresis and Northern blotting was performed as previously described.16 Briefly, aliquots of 2 µg total RNA were run on a denaturing 6.5% polyacrylamide gel containing 8 M urea in 0.1 M sodium acetate buffer. After electroblotting to a nylon membrane (GeneScreen, NEN Life Science Products, Boston, MA), samples were hybridized with [γ-32P]-ATP 5′ end labeled oligonucleotides 5761F (5′-AAGCCCCGGCAGGTTTGAAGCTGC-3′) for tRNACys and 7478F (5′-GGTTTCAAGCCAACCCCATGGCCTC-3′ for tRNASerUCN. Hybridization was performed overnight at 65°C in a buffer containing 1 M NaCl, 10% dextran sulfate, and 1% sodium dodecyl sulfate (SDS). The membrane was then washed twice for 10 minutes in 2× SSC + 0.1% SDS buffer and once in 0.1× SSC + 0.1% SDS buffer at 65°C. Signals were detected using a phosphoimager device and quantified with ImageJ software.
Standard Protocol Approvals, Registrations, and Patient Consents
Experiments on samples of human individuals were approved by the ethical committees of the University of Bonn (128/09) and the University of Magdeburg (06/09). Written informed consent was obtained from all participants in the study.
Data Availability
The patient's complete mtDNA sequence can be accessed from GenBank (accession number OM132208).
Results
Patient Description
A 59-year-old Caucasian male patient presented with marked walking and visual difficulties. The previously healthy man with an uninformative family history (Figure 1A) noticed a rapidly progressive gait instability at age 51 years. An inflammatory polyneuropathy (Guillain-Barré syndrome variant, acute motor axonal neuropathy) was diagnosed. The patient was subsequently admitted to an intensive care unit because mechanical respiratory support was needed for some days. The muscle strength was graded 1–2/5 according to the Medical Research Council (MRC) scale for the proximal and 2–3/5 for the distal upper and lower limbs. Tendon stretch reflexes were absent. The motor strength gradually recovered in the following months, and the patient was able to walk without support more than 1,000 m. He did not require specific neurologic support in the following years.
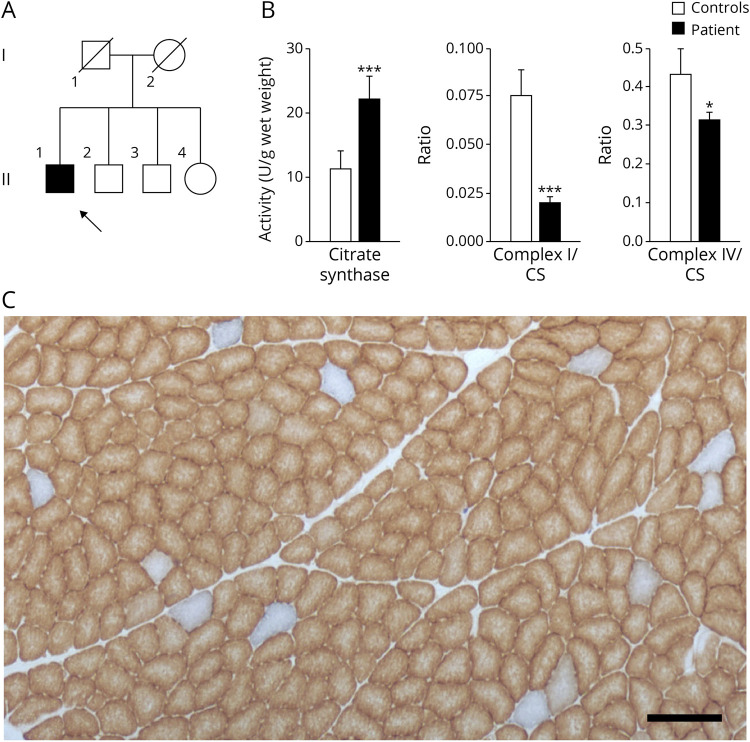
Figure 1. Mosaic Mitochondrial Impairment in Skeletal Muscle of a Patient Having Neuropathy, Ataxia, and Retinitis Pigmentosa Syndrome.
(A) Pedigree of the patient's family. The patient is indicated by an arrow. (B) Spectrophotometric measurements of mitochondrial enzyme activities in skeletal muscle homogenate of the patient and controls. NADH:ubiquinone oxidoreductase (complex I) and cytochrome c oxidase (complex IV) activities were normalized to citrate synthase activities. Mean values of 3 independent measurements are shown for the patient, whereas control values indicate averages of 3 healthy individuals. Error bars represent standard deviations. For significance, the t test was performed; *p < 0.05; ***p < 0.001. (C) COX/SDH double staining of the patient's skeletal muscle. Brown color indicates functional COX, whereas blue color results from SDH activity in the absence of COX activity. Scale bar, 200 µm. COX = cytochrome c oxidase; SDH = succinate dehydrogenase.
At age 58 years, he developed a progressively broad-based gait with a tendency to fall as well as a slurred speech. He observed mild memory deficits, which, however, did not significantly interfere with daily life and usual activities. In addition, he experienced visual problems, first noticed through the inability to pick up small objects from the table. With increasing physical impairment, his condition was classified grade III out of V in the state long-term care insurance. Clinical examination at age 59 years revealed mild dysarthria with occasional word repetitions, swallowing difficulties especially for crumbling foods, and slowed saccadic horizontal and vertical eye movements but no ophthalmoparesis or ptosis. Visual acuity was reduced to the examiner's finger movements at 20 cm distance, and confrontation visual field testing was not feasible. The patient had severe cerebellar limb ataxia more pronounced in the legs and mild muscle weakness (arms 4+/5 and legs 4−/5 proximally and 4+/5 distally on the MRC scale) but no marked muscular atrophy. Tendon stretch reflexes were preserved. Apart from bilateral slightly reduced vibratory sense at the ankles (4/8), examination of sensation was entirely normal, and no autonomic symptoms like edema, discoloring of skin, or skin temperature differences were present. Furthermore, the patient showed marked postural tremor of the head and the arms. He walked with a wide base and used a walker; also, he was unable to stand with feet together.
A detailed neuropsychological examination revealed executive function deficits consistent with a diagnosis of mild cognitive impairment. Deficits included reduced verbal fluency and flexibility performances (Regensburg Verbal Fluency Test17), a reduced working memory capacity (revised Wechsler Memory Scale18), and an encoding deficit while learning an unstructured word list (episodic memory; Rey Auditory Verbal Learning Test19). The retrieval of previously encoded information (late and delayed recall, and recognition), as well as semantic memory of past and current events were intact. The personal, spatial, and temporal orientation was preserved. A self-rating scale assessment of psychiatric symptoms (Geriatric Depression Scale20) revealed minor depressive signs consistent with the patient's report of sleep disturbance and rumination. The complete neuropsychological profile including percentiles can be found in eFigure 1, links.lww.com/NXG/A515.
Laboratory tests focusing on metabolic or inflammatory disease gave no evidence for abnormalities. Serum creatine kinase activity (CK) was normal. Repeated CSF examinations showed normal cell counts but markedly increased protein (2.7–3.4 g/L; reference value < 0.5 g/L) and lactate levels (4.5–6.4 mmol/L; reference value < 2.5 mmol/L) in comparison to serum lactate (1.5–2 mmol/L; reference value < 2.5 mmol/L). An MRI (1.5 T) scan of the brain revealed posterior predominant white matter changes and pontocerebellar atrophy (eFigure 2, links.lww.com/NXG/A515). 1H-MR-spectroscopy confirmed elevation of lactate accompanied with decreased N-acetylaspartate within the affected brain regions. Nerve conduction studies (NCSs) were compatible with a mild to moderate chronic axonal motor neuropathy (eTable 1). Needle EMG studies of clinically affected muscles demonstrated typical chronic neuropathic changes consisting of an increased proportion of long-high-amplitude motor unit potentials with weak voluntary contraction but no spontaneous electrical activity in a relaxed resting muscle. EEG showed diffuse continuous slowing consisting of fast theta activity. Ophthalmologic examination revealed a mottled macular pigment epithelium and midperipheral salt-and-pepper retinopathy compatible with retinitis pigmentosa.
Impairment of Mitochondrial Function
Skeletal muscle biopsy from M. vastus lateralis at age 59 years showed only little variations in fiber size and only sporadically clumps of nuclei but no significant neurogenic or myopathic changes. COX/SDH double staining of skeletal muscle slices demonstrated that 3% of fibers were COX deficient (Figure 1C). No fibers with intense SDH staining were observed, indicating the lack of mitochondrial proliferation. In accordance with this, no ragged-red fibers were detected on Gomori trichrome staining.
Spectrophotometric measurements of mitochondrial enzyme activities in skeletal muscle homogenates indicated elevated levels of the mitochondrial marker enzyme citrate synthase (CS) and strongly reduced complex I/CS and complex IV/CS ratios compatible with a combined OXPHOS impairment (Figure 1B).
Heteroplasmic Variant m.5789T>C Leads to COX Deficiency and Destabilizes tRNACys
Whole-exome sequencing of DNA isolated from the patient's cultured fibroblast revealed no pathogenic or likely pathogenic sequence variant in nuclear genes that have been associated with disorders compatible with the phenotype of the patient. Particularly, no pathogenic variants in known nuclear-encoded mitochondrial proteins (as listed in MitoCarta 3.021), ataxia-associated gene, or genes associated with retinitis pigmentosa were observed. Sanger sequencing of the patient's skeletal muscle mtDNA revealed the presence of the heteroplasmic variant m.5789T>C in the MT-TC gene coding for the mitochondrial tRNACys (Figure 2A). This sequence variant is neither present in Mitomap's collection of 52,633 human mtDNA sequences nor listed there as known pathogenic variant.22 RFLP analysis confirmed the presence of the mutation and displayed heteroplasmy levels of 44% in skeletal muscle, 11% in blood, 28% in urine sediment, and 12% fibroblasts (Figure 2B). Position 5789 of the human mitochondrial genome corresponds to nucleotide 43 of the tRNACys and the alteration m.5789T>C turns a perfect base-pairing between nucleotides 27 and 43 in the anticodon stem into a wobble pair (Figure 2C). Although position 43 of the mitochondrial tRNA for cysteine itself is not highly conserved in mammals, a perfect Watson-Crick pair with nucleotide 27 can be found in most species. All species harbor at least 1 perfect pair at the first 2 base pairs of the anticodon stem. The alteration m.5789T>C results in 2 wobble pairs in human, thus potentially decreasing the stability of the anticodon stem (eFigure3, links.lww.com/NXG/A515).
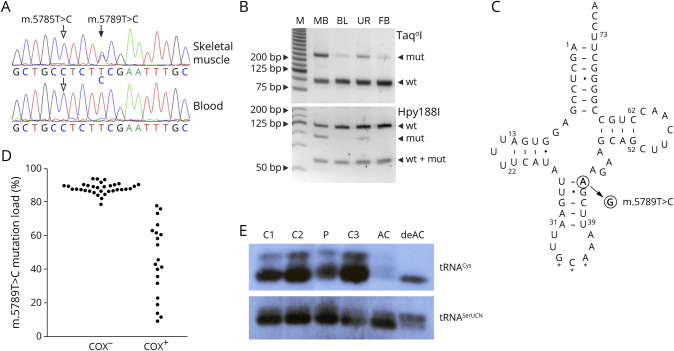
Figure 2. Detection and Functional Effects of the m.5789T>C Sequence Variant.
(A) Sequencing chromatograms from the patient's skeletal muscle and blood. Filled arrow indicates the heteroplasmic m.5789T>C variant, whereas empty arrows show the homoplasmic m.5785T>C variant. (B) Restriction fragment length polymorphism (RFLP) analysis of the patient's skeletal muscle biopsy sample (MB), blood (BL), urine (UR), and fibroblasts (FB). M, molecular weight marker. wt, wild type; mut, mutant. (C) Secondary structure of the human mitochondrial tRNA cysteine adapted from the Mamit-tRNA database.23 Filled arrow indicates the position of the novel m.5789T>C variant. Note that the replacement of a Watson-Crick base pair by a U-G base pair generates 2 adjacent wobble base pairs in the anticodon stem. Stars indicate the anticodon. Dash, Watson-Crick base pair; dot, wobble base pair. (D) Bee swarm diagram of m.5789T>C mutation loads in COX-negative and COX-positive single skeletal muscle fibers as determined by RFLP. Note that fibers with 78% mutation load or less show COX activity. (E) Northern blot analysis of skeletal muscle RNA of the patient (P) and 3 controls (C1–3) as well as aminoacylated (AC) and deaminoacylated (deAC) human osteosarcoma cells 143B RNA. The overall intensity of the tRNACys signal is decreased in comparison to tRNASerUCN, whereas the proportions of aminoacylated and deaminoacylated RNA species are similar in the patient and in controls. COX = cytochrome c oxidase.
No mutant m.5789C allele was detectable in urine sediments and blood samples from the patient's siblings by RFLP, but increasing the sensitivity using single-molecule sequencing of mutant-enriched mtDNA demonstrated the presence of the sequence alteration in all healthy siblings, thus indicating that the variant originated from the maternal germ line (eFigure 4, links.lww.com/NXG/A515). In addition, the previously reported22 homoplasmic variant m.5785T>C, located 4 nucleotides upstream of the heteroplasmic variant, was detected in all available family members.
Mutation loads of the m.5789T>C variant showed a strong correlation with COX deficiency in single skeletal muscle fibers (Figure 2D). Fibers harboring at least 78% mutant mtDNA displayed a loss of COX activity. This threshold value is comparable with other pathogenic tRNA sequence variants.14
Because the m.5789T>C variant turns a Watson-Crick base pair in the anticodon stem of the mitochondrial tRNACys into a wobble pair (Figure 2C), it is predicted to alter tRNA structure and stability. Therefore, we performed Northern blotting experiments and found that the amount of the mitochondrial tRNACys in the patient's skeletal muscle was 56% of the mean value of 3 control samples when normalized to the mitochondrial tRNASerUCN (p < 0.01). No alteration of amino acid charging of tRNACys was observed (Figure 2E).
m.5789T>C Colocalizes With Low-Abundance mtDNA Deletions and Alters Hypothetical Annealing
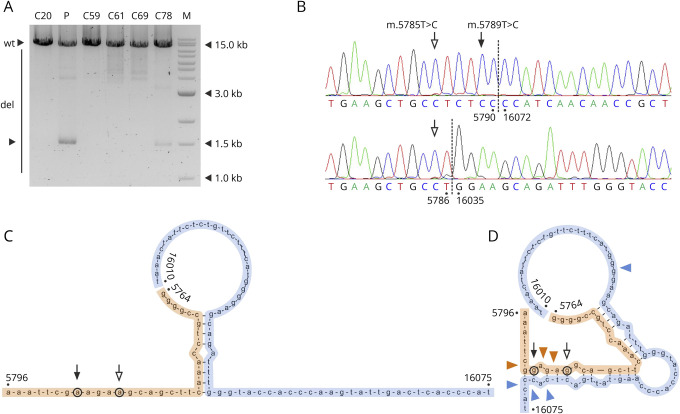
Long-range PCR showed the presence of a prominent mtDNA deletion cluster in the patient's skeletal muscle (Figure 3A, lane 2). These deletions, however, represented less than 5% of mtDNA molecules as determined by qPCR. Sequencing of single mtDNA deletion species revealed that 5′ breakpoints of these deletions were located in the immediate vicinity of the m.5789T>C variant, near to the replication origin oriL (Figure 3B and eTable 2, links.lww.com/NXG/A515). 3′ breakpoints clustered at a well-known deletion hotspot around position 16070, near to the replication-relevant D-loop region. In 30% of deletion-carrying molecules, the position 5789 was retained (eTable 2). Strikingly, all these molecules carried the m.5789C mutated allele and none the wild-type allele. The Fisher exact test indicated that the m.5789C allele and deletions occur not independently (p = 0.003). In silico alignment of np5764–np5796 to np16010–np16075 displayed a short hypothetical stretch of hybridization in the presence of the wild-type m.5785T and m.5789T alleles (Figure 3C). The mutant alleles m.5785C and m.5879C together resulted in a substantial extension of the hybridizing region. Breakpoints of the observed prominent deletions clustered at the 2 ends of the extended hybridization stretch (Figure 3D).
Figure 3. Prominent Cluster of mtDNA Deletion Detected in the Patient's Skeletal Muscle.
(A) Multiple mtDNA deletions in the patient's skeletal muscle and in 5 controls. M indicates a molecular weight marker. Note the single major band in the patient's DNA sample. (B) Sequencing chromatograms of the 2 most abundant deletions species in the patient's skeletal muscle, m.5791_16071 and m.5787_16034. Empty arrows point at the m.5785T>C homoplasmic variant, whereas the filled arrow marks the position of the m.5789T>C variant (upper panel), which is lost in the second deletion (lower panel). (C and D) In silico prediction of hybridization between nucleotides 5764–5796 (orange) and 16010–16075 (blue) in the presence of wild-type alleles (C) and mutant alleles (D) at positions m.5789 (filled arrow) and position m.5785 (empty arrow). Colored arrowheads indicate deletion breakpoints clustering at the ends of the extended hybridizing stretch. Annealing of the H-strand of the 5,790 region to the L-strand of the 16070 region was simulated using the Hybrid-min operator in UNAFold 3.7 software24 at default settings for DNA, as previously described.25 C = controls (numbers indicate ages in years); del = deletion-carrying mtDNA species; mtDNA = mitochondrial DNA; P = patient; wt = full-length wild-type mtDNA.
Discussion
In accordance with previously published pathogenicity scores,26 we provide evidence for the causative role of the novel heteroplasmic m.5789T>C variant in a patient having a mitochondrial disorder phenotypically resembling NARP syndrome. The following findings support the pathogenicity of the sequence variant: (1) the variant was present in heteroplasmic state (>10%) in all investigated tissues of the patient but was only detected in traces (<1%) in the unaffected siblings. (2) Combined oxidative phosphorylation deficiency was confirmed in skeletal muscle homogenate, and (3) COX deficiency was restricted to fibers with high (>78%) mutation loads. (4) Northern blot analysis showed reduced steady state amounts of tRNACys. The total score of 14 classifies the sequence variant as definitely pathogenic.26 Using a sensitive single-molecule sequencing technique, we confirmed its presence in family members, which has important implications for genetic counseling.
Previously reported pathogenic sequence variants in the mitochondrial tRNACys were found to be present in skeletal muscle samples of patients at homoplasmic or nearly homoplasmic levels (representing >90% of total mitochondrial genomes).4-9 The fact that the mutation load was much lower in the patient described in this study is in accordance with the late onset of the disease. Very high mutation loads in other reported patients often lead to a disease manifestation in early childhood; the 44% mutation load in our patient's skeletal muscle was associated with an onset in the sixth decade of life. Notably, in previously reported cases of pathogenic mitochondrial tRNACys sequence variants, high mutation loads were detected also in blood samples of patients4-9 and even in maternal relatives with mild unspecific symptoms or no symptoms at all.5,7,8 Accordingly, many of these mutations have also been described as mitochondrial haplogroup-defining polymorphisms present in the general population (m.5814T>C, haplogroups L2b and B4a5; m.5783G>A, haplogroup M3a2; m.5780G>A, haplogroup H1ap).27 In opposite, the m.5789T>C variant was present to a much less extent in proliferative tissues of the patient than in skeletal muscle, only in traces in unaffected family members, and absent in human mtDNA sequences reported so far.22 These features are typical for many heteroplasmic pathogenic mitochondrial tRNA sequence variants.3,14
NARP syndrome is caused by pathogenic point sequence variants of the mtDNA, and it is characterized by a variable combination of peripheral neuropathy, ataxia, pigmentary retinopathy, epilepsy, and dementia.1 Except epilepsy, all these features were present in the patient described in this study. Of interest, the most prominent abnormality on MRI was pontocerebellar atrophy and bilateral occipital leukoencephalopathy. A similar pattern of tissue injury has been previously reported in a 49-year-old man with NARP syndrome and the most common pathogenic mtDNA sequence variant m.8993T>G.28 However, in this patient, minor putaminal signal changes were detectable in the basal ganglia on one side only, which were not clearly evident in our case at the time of MRI examination. Heterogeneous areas of neuronal loss in NARP syndrome have been also reported in other brain imaging studies.28 Cognitive impairment in mitochondrial disorders is increasingly recognized. Similar to our patient, reduced processing speed, memory, verbal fluency, and affective symptoms have been reported in NARP syndrome.29 EMG-NCSs revealed evidence of a chronic motor axonal neuropathy generally compatible with NARP syndrome, but also with the previous history of inflammatory polyneuropathy. Of interest, sural sensory nerve action potential amplitudes in our case were relatively well preserved for NARP syndrome (eTable 1, links.lww.com/NXG/A515). However, patterns of peripheral nerve involvement have been reported to be heterogeneous even between members of the same NARP family.29
Ataxia and neuropathy are also prominent features of another group of mitochondrial diseases that are characterized by the presence of multiple mtDNA deletions. In these disorders, disturbed maintenance of mtDNA is due to pathogenic sequence variants in nuclear genes that play a role in replication of the mitochondrial genome, most prominently the mtDNA polymerase γ.30 The general term ataxia neuropathy spectrum includes a number of syndromic manifestations of disturbed mtDNA maintenance, among others SANDO (sensory ataxic neuropathy with dysarthria and opthalmoparesis). Although dysarthria was present in our patient, ataxia was predominantly cerebellar, and no opthalmoparesis was observed. Furthermore, we did not detect a typical heterogeneous mixture of multiple mtDNA deletions that is a hallmark of maintenance disorders. Instead, we observed a single prominent cluster of deletions with one of the breakpoints located in the close vicinity of the heteroplasmic sequence variant. The low overall amount of deletions and the lack of the typical pattern of multiple mtDNA deletions suggest that mtDNA deletions do not play a causative role in the patient's condition.
The class of somatic mtDNA deletions that were identified in the patient's skeletal muscle spanned the whole major arc of the mitochondrial genome between the oriL region and the D-loop. Misannealing between separate regions of the mitochondrial genome due to sequence similarities is a key element of present models of mitochondrial deletion generation.31 In concert with this, in silico analysis suggested that the heteroplasmic m.5789T>C variant in combination with the homoplasmic m.5785T>C variant may facilitate a misannealing between the oriL and D-loop regions through creating an extended imperfect repeat. The colocalization of the mutated m.5879C allele with deletions on the same mtDNA molecules further supports this hypothesis. The effect of a single-base exchange on deletion formation was demonstrated in the mitochondrial haplogroup N1b.25 In this haplogroup, the m.8472C>T variant interrupts the 13-nucleotide-long direct repeat that flanks the frequently observed common deletion (8470–13447), which results in a reduced occurrence of this deletion. In contrast to this situation, we propose that the m.5789T>C variant (in combination with the m.5785T>C variant) increases the probability of annealing between oriL and D-loop, thus promoting mtDNA deletion generation.
In conclusion, we provide an example of phenotypical overlap between pathogenic mtDNA sequence variants that act through different molecular mechanisms. The novel MT-TC m.5789T>C variant destabilizes tRNACys structure, causes a combined oxidative phosphorylation defect, and leads to similar symptoms as amino acid substitutions in the protein-coding MT-ATP6 gene, which are typical causes of the NARP syndrome. Deletions that were observed in association with the m.5789T>C variant are unlikely to substantially contribute to the pathogenicity, but their presence strengthens the hypothesis that the generation of specific somatic mtDNA deletions might depend on hybridization between different regions of the mitochondrial genome.
Acknowledgment
The authors thank Prof. Frank Hochholdinger at the Institute of Crop Sciences and Resources Conservation, University of Bonn, for providing access to the P.A.L.M. Laser Dissection System. They thank Susanne Beyer, Karin Kappes-Horn (Bonn), Maria Bust (Cologne), and Dr. Daniel Behme (Magdeburg) for excellent technical assistance.
Glossary
- CADD
Combined Annotation-Dependent Depletion
- COX
cytochrome c oxidase
- CS
citrate synthase
- MRC
Medical Research Council
- mtDNA
mitochondrial DNA
- NARP
neuropathy, ataxia, and retinitis pigmentosa
- NCS
nerve conduction study
- RFLP
restriction fragment length polymorphisms
- SDH
succinate dehydrogenase
- SDS
sodium dodecyl sulfate
- WES
whole-exome sequencing
Appendix. Authors
Contributor Information
Marius Hippen, Email: marius141095@aol.com.
Gábor Zsurka, Email: gabor.zsurka@ukbonn.de.
Viktoriya Peeva, Email: vik_peeva@hotmail.com.
Judith Machts, Email: judith.wesenberg@med.ovgu.de.
Kati Schwiecker, Email: kati.schwiecker@med.ovgu.de.
Grazyna Debska-Vielhaber, Email: grazyna.debska-vielhaber@med.ovgu.de.
Rudolf J. Wiesner, Email: rudolf.wiesner@uni-koeln.de.
Stefan Vielhaber, Email: stefan.vielhaber@med.ovgu.de.
Study Funding
Deutsche Forschungsgemeinschaft KU 911/21-2 and KU 911/22-1 to W.S.K. Deutsche Forschungsgemeinschaft ZS 99/3-2 and ZS 99/4-1 to G.Z.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosures.
References
- 1.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet . 1990;46(3):428-433. [PMC free article] [PubMed] [Google Scholar]
- 2.Stendel C, Neuhofer C, Floride E, et al. . Delineating MT-ATP6-associated disease: from isolated neuropathy to early onset neurodegeneration. Neurol Genet . 2020;6(1):e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacconi S, Salviati L, Gooch C, Bonilla E, Shanske S, DiMauro S. Complex neurologic syndrome associated with the G1606A mutation of mitochondrial DNA. Arch Neurol . 2002;59(6):1013-1015. [DOI] [PubMed] [Google Scholar]
- 4.Manfredi G, Schon EA, Bonilla E, Moraes CT, Shanske S, DiMauro S. Identification of a mutation in the mitochondrial tRNA(Cys) gene associated with mitochondrial encephalopathy. Hum Mutat . 1996;7(2):158-163. [DOI] [PubMed] [Google Scholar]
- 5.Scuderi C, Borgione E, Musumeci S, et al. . Severe encephalomyopathy in a patient with homoplasmic A5814G point mutation in mitochondrial tRNACys gene. Neuromuscul Disord . 2007;17(3):258-261. [DOI] [PubMed] [Google Scholar]
- 6.Lehtonen MS, Moilanen JS, Majamaa K. Increased variation in mtDNA in patients with familial sensorineural hearing impairment. Hum Genet . 2003;113(3):220-227. [DOI] [PubMed] [Google Scholar]
- 7.Feigenbaum A, Bai RK, Doherty ES, et al. . Novel mitochondrial DNA mutations associated with myopathy, cardiomyopathy, renal failure, and deafness. Am J Med Genet A . 2006;140(20):2216-2222. [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Sun D, Yang L, et al. . Mitochondrial ND5 T12338C, tRNA(Cys) T5802C, and tRNA(Thr) G15927A variants may have a modifying role in the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese pedigrees. Am J Med Genet A . 2008;146A(10):1248-12458. [DOI] [PubMed] [Google Scholar]
- 9.Kawazoe T, Tobisawa S, Sugaya K, et al. . Myoclonic epilepsy with ragged-red fibers with intranuclear inclusions. Intern Med. 2022;61(4):547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedemann FR, Vielhaber S, Schröder R, Elger CE, Kunz WS. Evaluation of methods for the determination of mitochondrial respiratory chain enzyme activities in human skeletal muscle samples. Anal Biochem . 2000;279(1):55-60. [DOI] [PubMed] [Google Scholar]
- 11.Karczewski KJ, Francioli LC, Tiao G, et al. . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature . 2020;581(7809):434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med . 2021;13(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landrum MJ, Chitipiralla S, Brown GR, et al. . ClinVar: improvements to accessing data. Nucleic Acids Res . 2020;48(D1):D835-D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlapakow E, Peeva V, Zsurka G, et al. . Distinct segregation of the pathogenic m.5667G>A mitochondrial tRNA(Asn) mutation in extraocular and skeletal muscle in chronic progressive external ophthalmoplegia. Neuromuscul Disord . 2019;29(5):358-367. [DOI] [PubMed] [Google Scholar]
- 15.Nicholls TJ, Zsurka G, Peeva V, et al. . Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum Mol Genet . 2014;23(23):6147-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Möllers M, Maniura-Weber K, Kiseljakovic E, et al. . A new mechanism for mtDNA pathogenesis: impairment of post-transcriptional maturation leads to severe depletion of mitochondrial tRNASer(UCN) caused by T7512C and G7497A point mutations. Nucleic Acids Res . 2005;33(17):5647-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aschenbrenner S, Tucha O, Lange KW. RWT. Regensburger Wortflüssigkeits-Test. Hogrefe; 2000. [Google Scholar]
- 18.Haerting C, Markowitsch HJ, Neufeld H, Calabrese P, Deisinger K, Kessler J. Wechsler Memory Scale—Revised Edition, German Edition. Huber; 2000. [Google Scholar]
- 19.Helmstaedter C, Lendt M, Lux S. Verbaler Lern-und Merkfähigkeitstest: VLMT, Manual. Beltz Test; 2001. [Google Scholar]
- 20.Sheikh JI, Yesavage JA. Geriatric Depression Scale (GDS): recent evidence and development of a shorter version. Clin Gerontol . 1986;5:165-173. [Google Scholar]
- 21.Rath S, Sharma R, Gupta R, et al. . MitoCarta3.0: an updated inventory of the mitochondrial proteome, now with sub-organelle localization and pathway annotations. Nucleic Acids Res . 2021;49(D1):D1541-D1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lott MT, Leipzig JN, Derbeneva O, et al. . mtDNA variation and analysis using MITOMAP and MITOMASTER. Curr Protoc Bioinformatics . 2013;44(123):1.23.1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pütz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA . 2007;13(8):1184-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res . 2005;33:W577-W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, Popadin KY, Markuzon N, et al. . Repeats, longevity and the sources of mtDNA deletions: evidence from 'deletional spectra'. Trends Genet . 2010;26(8):340-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarham JW, Al-Dosary M, Blakely EL, et al. . A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat . 2011;32(11):1319-1325. [DOI] [PubMed] [Google Scholar]
- 27.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat . 2009;30(2):E386-E394. [DOI] [PubMed] [Google Scholar]
- 28.Renard D, Labauge P. Posterior leukoencephalopathy in NARP syndrome. Acta Neurol Belg . 2012;112(4):417-418. [DOI] [PubMed] [Google Scholar]
- 29.Gelfand JM, Duncan JL, Racine CA, et al. . Heterogeneous patterns of tissue injury in NARP syndrome. J Neurol. 2011;258(3):440-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman S, Copeland WC. POLG-related disorders and their neurological manifestations. Nat Rev Neurol . 2019;15(1):40-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nissanka N, Minczuk M, Moraes CT. Mechanisms of mitochondrial DNA deletion formation. Trends Genet . 2019;35(3):235-244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The patient's complete mtDNA sequence can be accessed from GenBank (accession number OM132208).