Abstract
Microbial pathogens use a number of genetic strategies to invade the host and cause infection. These common themes are found throughout microbial systems. Secretion of enzymes, such as phospholipase, has been proposed as one of these themes that are used by bacteria, parasites, and pathogenic fungi. The role of extracellular phospholipase as a potential virulence factor in pathogenic fungi, including Candida albicans, Cryptococcus neoformans, and Aspergillus, has gained credence recently. In this review, data implicating phospholipase as a virulence factor in C. albicans, Candida glabrata, C. neoformans, and A. fumigatus are presented. A detailed description of the molecular and biochemical approaches used to more definitively delineate the role of phospholipase in the virulence of C. albicans is also covered. These approaches resulted in cloning of three genes encoding candidal phospholipases (caPLP1, caPLB2, and PLD). By using targeted gene disruption, C. albicans null mutants that failed to secrete phospholipase B, encoded by caPLB1, were constructed. When these isogenic strain pairs were tested in two clinically relevant murine models of candidiasis, deletion of caPLB1 was shown to lead to attenuation of candidal virulence. Importantly, immunogold electron microscopy studies showed that C. albicans secretes this enzyme during the infectious process. These data indicate that phospholipase B is essential for candidal virulence. Although the mechanism(s) through which phospholipase modulates fungal virulence is still under investigations, early data suggest that direct host cell damage and lysis are the main mechanisms contributing to fungal virulence. Since the importance of phospholipases in fungal virulence is already known, the next challenge will be to utilize these lytic enzymes as therapeutic and diagnostic targets.
To aid in the invasion of host tissues, microbial cells possess constitutive and inducible hydrolytic enzymes that destroy or derange constituents of host cell membranes, leading to membrane dysfunction and/or physical disruption (167). Since membranes are made up of lipids and proteins, these biochemicals constitute the target of enzyme attack. Pathogenic fungi (e.g., Candida albicans) secrete enzymes which are considered to be integral to their pathogenesis; these are categorized into two main types, proteinases (72, 73), which hydrolyze peptide bonds, and phospholipases (75), which hydrolyze phospholipids. In this review, we focus on fungal phospholipases. It should be emphasized that this field is evolving rapidly and that most of the data reviewed in this article were published recently. New insights into the biology and contribution of these enzymes to fungal virulence will be forthcoming in the near future.
DEFINITION OF PHOSPHOLIPASES AND RATIONALE IN CONSIDERING THEM FOR A ROLE IN VIRULENCE
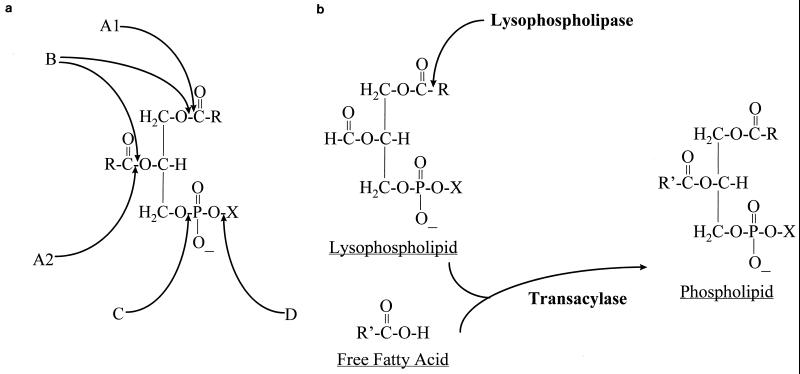
The term “phospholipases” refers to a heterogeneous group of enzymes that share the ability to hydrolyze one or more ester linkage in glycerophospholipids. Although all phospholipases target phospholipids as substrates, each enzyme has the ability to cleave a specific ester bond (Fig. 1a). Thus, qualifying letters, such as A, B, C, and D, are used to differentiate among phospholipases and to indicate the specific bond targeted in the phospholipid molecule (4). For example, phospholipase A1 (PLA1) hydrolyzes the fatty acyl ester bond at the sn-1 position of the glycerol moiety, while phospholipase A2 (PLA2) removes the fatty acid at the sn-2 position of this molecule. The action of PLA1 (EC 3.1.1.32) and PLA2 (EC 3.1.1.4) results in the accumulation of free fatty acids and 2-acyl lysophospholipid or 1-acyl lysophospholipid, respectively. The fatty acid still linked to the lysophospholipid is, in turn, cleaved by other enzymes termed lysophospholipases (Lyso-PL) (EC 3.1.1.5) (Fig. 1b). Phospholipase C (PLC) (EC 3.1.4.3) hydrolyzes the phosphodiester bond in the phospholipid backbone to yield 1,2-diacylglycerol and, depending on the specific phospholipid species involved, phosphatidylcholine, phosphatidylethanolamine, etc. The second phosphodiester bond is cleaved by phospholipase D (PLD) (EC 3.1.4.4) to yield phosphatidic acid and choline or ethanolamine, again depending on the phospholipid class involved (4).
FIG. 1.
Sites of action of various phospholipases. (a) A1 and A2, PLA1 and PLA2, respectively; B, PLB; C, PLC; D, PLD. (b) Lyso-PL and Lyso-PL transacylase.
Although phospholipase B (PLB) (synonyms: lysophospholipase, lysophospholipase-transacylase) refers to an enzyme that can remove both sn-1 and sn-2 fatty acids, its nomenclature is confusing (4). This confusion arises because PLB has both hydrolase (fatty acid release) and lysophospholipase-transacylase (LPTA) activities. The hydrolase activity allows the enzyme to cleave fatty acids from both phospholipids (PLB activity) and lysophospholipids (lysophospholipase [Lyso-PL] activity), while the transacylase activity allows the enzyme to produce phospholipid by transferring a free fatty acid to a lysophospholipid (Fig. 1b). The finding that PLB has both hydrolase and acyltransferase activities has been reported for enzymes purified from C. albicans, Penicillium notatum, and Saccharomyces cerevisiae (100, 122, 164, 224, 225). This complex pattern of activity led to difficulties in the nomenclature of the enzyme, with some authors naming it PLB (as in the case of S. cerevisiae and P. notatum) and others naming it LPTA (10, 122, 193). Recent cloning and disruption of genes encoding PLB provided further evidence that this enzyme has both PLB and Lyso-PL activities. Deletion of caPLB1, the gene encoding PLB from C. albicans, showed that both PLB and Lyso-PL activities were reduced in the PLB1-deficient mutant relative to the wild type, confirming that caPLB1 encodes an enzyme responsible for the PLB and Lyso-PL activities (75, 101). Similar findings were reported for PLB deletion in S. cerevisiae (100).
Invasion of host cells by microbes entails penetration and damage of the outer cell envelope. This transmigration process is mediated, most probably, by either physical or enzymatic means or a combination of the two. Phospholipids and proteins represent the major chemical constituents of the host cell envelope. Therefore, enzymes capable of hydrolyzing these chemical classes, such as phospholipases and proteinases, are likely to be involved in the membrane disruption processes that occur during host cell invasion. By cleaving phospholipids, phospholipases destabilize the membrane and cell lysis results (166). Evidence implicating phospholipases in host cell penetration, injury, and lysis by microorganisms has been reported for Rickettsia rickettsii (207), Toxoplasma gondii (160, 161), Entamoeba histolytica (154), and C. albicans (101). Consequently, phospholipases have been included among the virulence factors that damage host cells (166).
PHOSPHOLIPASES IN ORGANISMS OTHER THAN FUNGI
Besides fungi, which are the subject of this review, extracellular phospholipases have been implicated as pathogenicity factors for bacteria (Table 1). Since bacterial phospholipases have been the subject of at least two recent reviews (183, 194), to avoid repetition, only a brief description of these enzymes and their role in virulence is covered in this article. Furthermore, including some information about phospholipases from organisms other than fungi will show a global perspective of microbial phospholipases.
TABLE 1.
Phospholipases from microorganisms other than fungi
| Microbial class | Enzymea | Gene cloned | Substrate specificitya | Reference(s) |
|---|---|---|---|---|
| Bacteria | ||||
| Clostridium perfringens | Alpha-toxin | Yes | PC, SM, PS, LPC | 103, 136, 162, 195, 197 |
| Clostridium novyi | Gamma-toxin | No | PC, SM, LPC, PE, PI, PG | 191 |
| PI-PLC | No | |||
| Clostridium bifermentans | PLC | Yes | Unknown | 197 |
| Listeria monocytogenes | PLC-A | Yes | PI | 102, 119, 203 |
| PLC-B | Yes | PC, PE, PS, SM | ||
| Pseudomonas aeruginosa | PLC-H | Yes | PC, LPC, SM | 13, 29, 137 |
| PLC-N | ||||
| Pseudomonas cepacia | PLC | Yes | 202 | |
| Staphylococcus aureus | Beta-toxin | Yes | SM, LPC | 38, 76, 148, 157, 216, 217 |
| PI-PLC | No | PI, LPI | ||
| Bacillus cereus | PC-PLC | Yes | PC, PE | 77, 85, 96, 104 |
| SMase | Yes | SM | ||
| PI-PLC | Yes | PI, LPI | ||
| Bacillus thuringiensis | PI-PLC | Yes | PI, LPI | 68, 78, 192 |
| Mycobacterium tuberculosis | MPC-Ac | SM, PC | 83 | |
| MPC-Bc | SM, PC | |||
| PLD | PC | |||
| PLA | 177 | |||
| Rickettsia prowazekii | PLA | |||
| corynebacterium pseudotuberculosis | PLD | Yes | 118 | |
| Arcanobacterium haemolyticum | PLD | Yes | 118 | |
| Legionella pneumophila | PLC | No | PC | 8 |
| Ureaplasma urealyticum | PLC | No | 15, 36 | |
| Protozoa | ||||
| Toxoplasma gondii | PLA | No | 160, 161 | |
| Entamoeba histolytica | PLA | No | 106, 153 |
Abbreviations: PI, phosphatidylinositol; LPI, lysophosphatidylinositol; PG, phosphatidylglycerol; SM, sphingomyelin; LPC, lysophosphatidylcholine; LPG, lysophosphatidylglycerol; PLC-N, nonhemolytic PLC; PLC-H, hemolytic PLC.
Bacterial Phospholipases
Clostridium perfringens.
PLC (synonyms: alpha-toxin, lecithinase), which is one of the most active bacterial phospholipases, is 1 of at least 12 different soluble antigens referred to as toxins, produced by Clostridium perfringens, that may be involved in pathogenesis. The findings that clostridial PLC was a potent toxin (109, 110, 123) with hemolytic (162, 195), lethal (196), dermonecrotic (115), vascular permeabilization (186), and platelet-aggregating (134, 185) properties attracted a large number of investigators to study this secreted protein, making it the most extensively studied bacterial toxin. Thus, it is not surprising that extensive reviews have been written about this enzyme (67, 79, 194), its application to the study of cell membranes (3, 5, 48, 126, 159, 229), and its role in virulence (183).
Over the past 60 years, extensive data have accumulated concerning both the physiology of toxin production by C. perfringens (179) and some of the physical and biological properties of the enzyme (115). However, many of the earlier data have been difficult to interpret because of the lack of purity of the toxin preparations used (127). In these earlier studies, many authors used preparations of the enzyme from commercial or other sources with insufficient or no regard for the fact that these preparations, unless produced under rigorous purification standards, may contain as many as 11 contaminating substances (115). Furthermore, purification of this enzyme is difficult. Fortunately, recent molecular biology techniques allowed the cloning and sequencing of the gene encoding clostridial PLC (195, 227) and subsequent synthesis of purified recombinant enzyme (227). Availability of the gene and the purified protein made studies aimed at defining the role of PLC in the virulence of C. perfringens feasible.
To study the pathogenesis of C. perfringens-mediated gas gangrene, Awad et al. (6) used reverse genetics to construct a suicide plasmid in which the plc gene, encoding PLC, was inactive. Using this plasmid, they isolated mutants that had lost their ability to produce detectable PLC. Comparing the mutants and their parent in a mouse virulence model showed that the mutants were markedly less pathogenic than the parent strain (6). Unlike animals infected with the phospholipase-deficient mutants, which showed minimal swelling, muscle destruction, inflammation, or necrosis of the infected tissues and remained active with an otherwise healthy appearance, animals challenged with the parent strain had extensive swelling of the infected foot, leg, and hip, as well as a severely necrotic infected foot, demonstrable signs of hematuria, and extensive muscle destruction. This study provided definitive genetic evidence of the essential role of PLC in gas gangrene or clostridial myonecrosis. For a more in-depth review of clostridial PLC with regard to its substrate specificity, molecular architecture, interaction with phospholipids and membranes, and role in disease, the reader is referred to excellent reviews by Titball (194) and Songer (183).
Listeria monocytogenes.
Similar to C. perfringens, L. monocytogenes secretes a number of extracellular enzymes including listeriolysin (encoded by hly) and two phospholipases: i) PI-PLC or PLC-A, encoded by plcA, which is an inositol-specific PLC (21), and (ii) PC-PLC or PLC-B, encoded by plcB, which is a broadly active PLC with the ability to hydrolyze most cellular phospholipids (203). Although each phospholipase may contribute to the virulence of L. monocytogenes by itself (21, 119, 203), elegant work by Portnoy and colleagues showed that the two phospholipases play overlapping roles in the pathogenesis of L. monocytogenes (178). In their study, mutants harboring a deletion in each PLC, as well as a double mutant lacking both enzymes, were characterized with regard to virulence. Abolishing PI-PLC and PC-PLC resulted in strains that were 2- and 20-fold less virulent than the parent strain, respectively. In addition, these strains were defective in escape from the vacuole and in cell-to-cell spread, depending on which enzyme was deleted. Interestingly, the mutant lacking both PLCs was 500-fold less virulent in mice and was severely diminished in its ability to escape from the vacuole and to spread from cell to cell (178). These findings are consistent with the two enzymes having overlapping functions throughout the course of infection. Importantly, deleting a single gene resulted in only a modest reduction in virulence, while simultaneous deletion of the two genes in the same strain led to a highly significant decrease in the virulence, suggesting that the two enzymes have a synergistic effect on the ability of the organism to invade host tissues. For more details on entry of L. monocytogenes into phagocytic cells and the role of secreted enzymes, including phospholipases, in the ability of the organism to escape from the vacuole and transmigrate cell-to-cell, the reader is referred to references 144, 183, and 194.
Pseudomonas aeruginosa.
Two distinct PLCs are produced by Pseudomonas aeruginosa: PLC-N (nonhemolytic) and PLC-H (hemolytic) (183). The genes (plcN and plcS) encoding these enzymes have been cloned (137). Although the expression and secretion of both enzymes are phosphate regulated, each enzyme has a distinct substrate specificity, with PLC-H hydrolyzing sphingomyelin in addition to phosphatidylcholine (PC), while PLC-N is active in phosphatidylserine and PC degradation (137). This difference in substrate specificity has a bearing on the efficiency with which these enzymes degrade eukaryotic membranes. Ostroff et al. (138) proposed that the two enzymes could work sequentially and synergistically to lyse host cells. PLC-H initiates membrane lysis by hydrolyzing PC and sphingomyelin (the major constituents of the membrane outer leaflet), and this is followed by the action of PLC-N which cleaves phosphatidylserine (the major constituent of the membrane inner leaflet). Furthermore, substrate specificity studies have shown that PLC-H preferentially cleaves phospholipids containing quaternary ammonium groups, such as phosphatidylcholine, which are found primarily in eukaryotic membranes and lung surfactants, but that it has minimal activity toward phospholipids such as phosphatidylethanolamine, which are found in the prokaryotic membrane (14). This selective ability may explain why the invading organism can lyse the host cells without damaging its own membrane.
Correlation between PLC-H and P. aeruginosa pathogenicity can be derived from studies showing that the enzyme is secreted in vivo and in experiments comparing the wild type with mutants in which this gene is deleted in animal models. Three lines of evidence suggest that PLC-H is secreted in vivo: (i) clinical isolates from the lungs produce PLC (13), (ii) PLC is produced when the bacterium is grown in bronchial washings (105), and (iii) high titers of antibody against PLC-H are detected in patients with chronic P. aeruginosa infection (62).
Mutants disrupted in the plcS gene were generated and used to determine the role of PLC-H in pseudomonal virulence (138). In this study, the virulence of wild-type strains and plcS-disrupted mutants was compared in a mouse burn model of infection. A reduction was observed in the ability of the disruptant grown under phosphate-limiting conditions to kill mice (200-fold increase in the 50% lethal dose compared with the wild-type strain). Since virulence in these studies was phosphate dependent, mutants disrupted in plcR (a phosphate-regulatory gene) were generated to investigate its effect alone and combined with plcS. These studies showed that when the strains were grown under phosphate-limiting conditions, the virulence of the ΔplcS ΔplcR strain was 200- to 10,000-fold lower than that of the parent strain (138). Interestingly, mutants with only plcR deleted were also attenuated in virulence even though they produced greater amounts of PLC and hemolysin (138). In spite of this apparent enigma, which suggests a role for plcR in pseudomonal pathogenicity, the significantly greater reduction in the pathogenicity of the double disruptant suggests that PLC-H is associated with virulence (194).
Bacillus cereus.
Several PLCs, including phosphatidylinositol-specific and PC-preferring enzymes, as well as sphingomyelinase, are produced by Bacillus cereus (80, 95, 198, 199). The genes encoding the three different proteins have been cloned and sequenced (84, 85, 96). PC-preferring and sphingomyelinase-encoding genes form a gene cluster (tandemly located) (59), which is not positioned in close proximity to the gene encoding phosphatidylinositol-specific PLC (59, 96). The PC-preferring enzyme has structural homology to the C. perfringens alpha-toxin (103), while the phosphatidylinositol-specific enzyme has stretches of sequence homology to other eukaryotic phospholipases (97).
The contribution of phospholipases to the virulence of B. cereus has not been investigated in animals. Although PLC of this bacterium is considered to be nontoxic (126), data suggestive of its involvement in host cell lysis have been reported. Gilmore et al. (59) suggested that by creating a duplex hemolysin named cereolysin AB, the PC-preferring phospholipase and the sphingomyelinase act in concert to cause hemolysis. Others (212) showed that B. cereus strains producing PLC cause degranulation of human neutrophils with a dose-dependent release of lysosomal enzymes, which may mediate tissue damage. Finally, it has been suggested that B. cereus protects itself against phagocytosis by releasing phospholipases (152).
Rickettsia.
Two species of the genus Rickettsia (Rickettsia rickettsii and Rickettsia prowazekii) were shown to possess PLA that was suggested to mediate host cell lysis (63, 177, 220, 222). Previously, Winkler and Miller (221) reported that rickettsiae enter cells through a mechanism involving a PLA. Their hypothesis was based on the demonstration that host cells exposed to large numbers of R. prowazekii release considerable amounts of lysophospholipids and free fatty acids into the culture medium. Involvement of phospholipases in the penetration and damage of host cells by R. rickettsii was first suggested by Walker et al. (207). More recently, Silverman et al. (177) confirmed these findings and provided suggestive evidence that phospholipase activity associated with internalization of this intracellular parasite lies directly with the infecting organism rather than with the host cell. However, as stated by the authors, definitive evidence that the phospholipase originates from these organisms could be provided only by isolation and cloning of specific rickettsial gene(s) involved in the internalization process (177).
Corynebacterium pseudotuberculosis.
Corynebacterium pseudotuberculosis, the agent of caseous lymphadenitis in small ruminants and ulcerative lymphangitis in horses, produces extracellular PLD, which possibly plays a role in the pathogenicity of this and similar bacteria (Corynebacterium ulcerans and Arcanobacterium haemolyticum) (1, 34, 118, 184). The genes encoding the PLD in C. pseudotuberculosis and A. haemolyticum have been cloned and sequenced (118). Targeted mutagenesis of PLD in C. pseudotuberculosis reduced the ability of this bacterium to establish a primary infection or cause chronic abscess formation in regional lymph nodes (117). These results indicate that PLD is a virulence determinant of C. pseudotuberculosis, increasing the persistence and spread of the bacteria within the host (117).
Protozoan Phospholipases
Phospholipase A facilitates host cell penetration by the two protozoan species Toxoplasma gondii and Entamoeba histolytica.
Indirect evidence implicating a calcium dependent-PLA2 in host cell invasion by T. gondii was obtained by Saffer et al. (160), who showed that incorporation of exogenous PLA2 from snake venom increases host cell penetration by T. gondii. Furthermore, penetration of host fibroblasts by the parasite was inhibited following preincubation with PLA2 inhibitors, such as p-bromophenacyl bromide and nordihydroguaiaretic acid, or antisera to this enzyme (160). In another study, the same group extended these findings and demonstrated that treating fibroblasts with fractions of disrupted T. gondii led to accumulation of degradation products of the phospholipids (fatty acids and lysophospholipids). The data suggest that PLA may be associated with T. gondii cells (161). Moreover, fractions of T. gondii that had PLA enzymatic activity also increased host cell penetration (161). Although the findings of Saffer and Schwartzman (161) implicate phospholipases in the process of penetration of host cells by T. gondii, these workers used a crude enzyme preparation, which makes interpretation of the results quite difficult. To unequivocally establish a correlation between PLA and T. gondii virulence and to elucide how the enzyme may influence the penetration process of host cells by this parasite, gene cloning and enzyme purification are required.
E. histolytica, similar to T. gondii, possesses PLA (106, 153). Ravdin and coworkers (106) reported that this ameba has two PLA enzymes: a calcium-dependent protein which is associated with the plasma membrane and is most active at an alkaline pH, and a calcium-independent enzyme that is localized predominantly to soluble subcellular fractions of E. histolytica and is optimally active at an acidic pH. Amebic cytolytic activity, and thereby virulence, is associated with the calcium-dependent PLA. However, as in the case with T. gondii, definitive demonstration of the role of amebic PLA enzymes in the cytolytic event awaits their purification, gene cloning, and disruption.
PHOSPHOLIPASES IN PATHOGENIC FUNGI
Phospholipases of Candida albicans
The overall incidence of Candida bloodstream infections has increased significantly in the last two decades (9, 140, 143), ranging from a 75% increase in small hospitals to an over 400% increase in some large tertiary-care centers (140). This increase led to a tremendous interest in the study of candidal pathogenesis and strategies for control and prevention of this clinically important fungus. Candidal virulence factors have also attracted interest as a possible means for developing novel therapeutic interventions against candidiasis (33, 52, 139). Such virulence factors include adherence (20, 55, 90), germination (180), extracellular proteinases (71, 72) and phospholipases (75), and phenotypic switching (181, 182).
Early work on candidal phospholipases.
(i) Candidal phospholipase.
The secretion of extracellular phospholipases by C. albicans was first reported in the 1960s by Costa et al. (A. Costa, A. Misefari, and A. Amaro, Abstr. Atti XIV Congr. Naz. Microbiol. Messina, abstr. P35 and P36 1967) and Werner (213) by growing the yeast on solid media containing egg yolk or lecithin and analyzing the lipid breakdown products. Later, phospholipase activity was found in many pathogenic C. albicans strains by using media containing blood serum and sheep erythrocytes (30). The observation that C. albicans secretes phospholipase prompted Pugh and Cawson (150) to develop a lecithin-based cytochemical method to detect this enzyme. In a subsequent study, these authors used this method (149) in conjunction with a chicken chorioallantoic membrane model to evaluate ultrastructural details of candidal invasion and to determine the site of phospholipase production. Invasion was initiated by placing stationary-phase blastospores of C. albicans on the membrane, which stimulated cellular changes in the blastospores. Many of the blastospores developed hyphae with phospholipase activity concentrated at the growing tip. The activity was highest where the hyphae were in direct contact with the membrane (149). In general, only hyphae invaded the membrane successfully. Based on these results, the investigators proposed that extracellular phospholipases were important in the invasion of tissue by C. albicans.
Subsequent studies by other groups were directed at developing simple methods to detect candidal phospholipases. Odds and Abbott (132) described a biochemical assay to measure intracellular PLA and Lyso-PL activity in C. albicans. By using this assay, it was found that PC was hydrolyzed to give Lyso-PC. A small amount of this degradation product was always detected (132). One disadvantage of this assay is that it is time-consuming and therefore unsuitable for testing a large number of isolates. Price et al. (147) described a plate method for the detection of phospholipase activity in C. albicans which circumvents this disadvantage. Since egg yolk contains large amounts of phospholipids, predominantly PC and phosphatidylethanolamine, it was incorporated into a Sabouraud dextrose agar-based medium. When grown on this medium, phospholipase-positive candidal isolates produce a distinct, well-defined, dense white zone of precipitation around the colony (Fig. 2). This white zone is probably due to the formation of calcium complex with the fatty acids released by the action of phospholipase on the phospholipids present in the egg yolk (110). In this assay, phospholipase activity (expressed as a Pz value) is defined as the ratio of colony diameter to the diameter of the dense white zone of precipitation around phospholipase positive colonies. This easy plate method became the traditional screening method for phospholipase activity for Candida species (66, 99, 168, 215) and other fungi such as Cryptococcus neoformans (23). However, because egg yolk contains substrates for both phospholipases (phospholipids) and lipases (triglycerides), the egg yolk-based assay is not specific, and therefore its use should be limited to initial screens only (49). Furthermore, the assay is not suitable for the screening of fungal isolates that produce low levels of phospholipase. Confirmation of phospholipase activity necessitates the use of a specific radiometric (10) or colorimetric (122, 176) assay and the use of concentrated culture filtrate, particularly in poorly phospholipase-producing strains. Awareness of this caveat is especially relevant for researchers attempting to determine the phospholipase activity of genetically manipulated mutants, e.g., phospholipase-deficient clones generated by site-specific mutagenesis (49, 101).
FIG. 2.
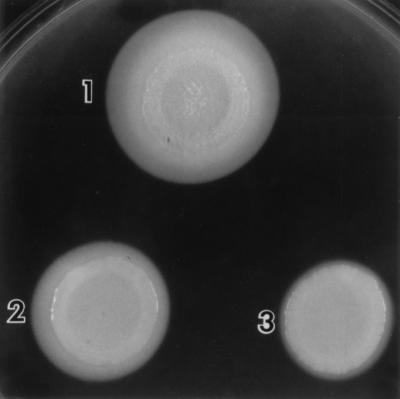
Production of phospholipase by three different clinical C. albicans isolates. Note the difference in the precipitation zones around the three colonies.
(ii) Production of phospholipase by various candidal species.
Early studies in which the egg yolk-based assay was used to evaluate the ability of different Candida species to produce phospholipase showed that only C. albicans and not other species of Candida produce extracellular phospholipase. Samaranayake et al. (168) screened 41 Candida isolates for phospholipase activity by using a plate assay method and found that 79% of the C. albicans strains tested produced extracellular phospholipases whereas no strains of C. tropicalis, C. glabrata, and C. parapsilosis produced the enzymes. The quantity of phospholipase produced by C. albicans varied with the specific isolate and correlated with the site of infection. Blood isolates generally produce much higher levels than do isolates from wounds or urine (147). In contrast, Clancy et al. (C. J. Clancy, M. A. Ghannoun, and M. H. Nguyen, Programs Abstr. 36th Annu. Meet. Infect. Dis. Soc. Am., abstr. 317, 1998) recently showed that non-albicans Candida species produce extracellular phospholipases as determined by both egg yolk-based and colorimetric assays. In this study, 41% of C. glabrata, 50% of C. parapsilosis, 70% of C. tropicalis, 80% of C. lusitaniae, and 100% of C. krusei strains produced detectable phospholipase activity. However, it is important to emphasize that relative to C. albicans, the non-albicans species secreted significantly smaller amounts of phospholipase (for example, C. krusei has approximately 10 times less phospholipase activity [P. Mukherjee and M. Ghannoum, unpublished data]). Although Clancy et al. demonstrated that non-albicans species can secrete phospholipase similar to C. albicans, it is important to point out that the number of candidal isolates tested was limited (n = 51). Therefore, the percentages reported in this study may have been significantly different if a larger number of isolates had been examined. Furthermore, the discrepancy observed by different workers in the phospholipase activity for the non-albicans species may be attributed to strain-to-strain variation or differences in the preparation of the egg yolk agar plates used to detect phospholipase secretion in these species.
Lane and Garcia (99), using an egg yolk-based assay, tested the ability of C. albicans switching variants (wild type, star, stipple, and ring) to produce phospholipase (181). Star and ring variants produced a similar amount of phospholipase to the wild type. In contrast, the stipple variant produced between 27 and 34% more phospholipase than did the other three strains tested. Although candidal switching is considered to be a virulence factor (181), the relevance of the findings of Lane and Garcia to candidal pathogenicity remains to be shown through in vivo experimentation.
(iii) Types of candidal phospholipases.
The literature contains contradicting reports on the number and specific types of phospholipase enzymes secreted by C. albicans. Costa et al. (Abstr. Atti XIV Cong. Naz. Microbiol. Messina, abstr. P35 and P36, 1967) reported the secretion of both PLA and PLC by this clinically important yeast. Their results were based on the isolation of palmitic acid and phosphatidylcholine from the proximity of candidal colonies cultured on Sabouraud agar supplemented with serum and sheep erythrocytes. Since this medium is not chemically defined, the sources of the hydrolysis products are uncertain. Banno et al. (10) performed a crude fractionation of the proteins in culture filtrates of C. albicans by using DEAE-Sephadex and then assayed the fractions for phospholipase activity. Their data suggested that C. albicans secreted three types of phospholipases: Lyso-PL, LPTA, and PLB. Takahashi et al. (193) purified two distinct forms of LPTA from culture filtrates of C. albicans. These candidal enzymes differed from mammalian enzymes in amino acid composition; however, the substrate specificities were not determined (193).
To determine the type and substrate specificity of phospholipases secreted by C. albicans, a high-phospholipase-producing strain was grown to late log phase and the supernatant was concentrated and assayed for phospholipase activity by using two complementary assays: a specific-substrate radial diffusion assay capable of differentiating between PLA, PLB, and PLC activities (64) and a colorimetric acyl coenzyme A-oxidase system (101). Only PLB activity was observed in the diffusion-based assay (128). In the colorimetric assay, the concentrated supernatant was incubated with a substrate that is specific for PLB (PC) or Lyso-PL (Lyso-PC) activities. Both PLB and Lyso-PL activities were detected in the supernatant, suggesting that there are two enzymes with individual activity, or one enzyme with two activities (see “Definition of phospholipases and rationale in considering them for a role in virulence” above). To resolve this dilemma, protein purification and gene cloning were undertaken.
In collaboration with Yoshinori Nozawa (Gifu University, Gifu, Japan), we purified to homogeneity the protein responsible for candidal extracellular phospholipase activities. This enzyme is a glycoprotein with a molecular mass of 84 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The specific activities of the enzyme were 117 μmol/min/mg of protein for fatty acid release (hydrolase), and 459 μmol/min/mg of protein for PC formation (LPTA activity). The apparent Km of the hydrolase activity of the enzyme for 1-palmitoyl-sn-glycero-3-phosphocholine was 60.6 μM. PLB activity was optimal at pH 6.0. The activity of the purified enzyme was not dependent on divalent cations (Ca2+ and Mg2+) and was not inhibited by the addition of EDTA or EGTA (122).
To characterize the functional activity of the purified enzyme, Mirbod et al. (122) used both the radiometric and colorimetric methods described above. In these assays the enzyme was incubated with a substrate that is specific for PLB activity or Lyso-PL activities. In addition, to determine the LPTA activity, Lyso-PC was incubated with the purified enzyme and the rate of production of PC was monitored. These studies revealed that the purified C. albicans enzyme has both hydrolase (PLB and Lyso-PL) and LPTA activities. The finding that one enzyme has two hydrolase activities is not unique to C. albicans. A similar observation has been reported for PLB secreted by Penicillium notatum. Saito et al. (164) reported that P. notatum PLB is a glycoprotein with intrinsic Lyso-PL and PLB activities. Similarly, Lee et al. (100) showed that the Saccharomyces cerevisiae PLB1 gene encodes a protein required for Lyso-PL and PLB activities. These data clearly demonstrate that the purified enzyme is a phospholipase with dual activities. Similar to the candidal enzyme, PLB from S. cerevisiae is reported to have an acyltransferase activity (224, 225). This complex pattern of activity led to difficulties in the nomenclature of the fungal enzyme (see above). Further evidence that candidal PLB has both PLB and Lyso-PL activities was obtained by assaying phospholipase activities in supernatants from the parent and PLB1-deleted mutants (see below for construction and characterization of mutants). Unlike the parent, deletion of caPLB1 led to loss of both PLB and Lyso-PL activities.
The phospholipase activities described so far are due to secreted candidal enzymes. Recently, McLain and Dolan (116) reported on a membrane-associated enzyme in C. albicans. This enzyme has a PLD activity and is capable of performing a transphosphatidylation reaction in the presence of primary alcohols (see below for cloning of candidal PLD).
Evidence correlating phospholipases and virulence.
Although the contribution of phospholipases to the pathogenesis of bacteria and protozoa has been known for some time, investigations into whether these enzymes are associated with fungal virulence have only recently been undertaken. Barrett-Bee et al. (11) was the first to evaluate the role of extracellular candidal phospholipases in virulence by using a murine model of candidiasis. When phospholipase activity was measured in six yeasts (four strains of C. albicans and a single strain each of C. parapsilosis and S. cerevisiae), a correlation was found between phospholipase activity and two potential parameters of pathogenicity. The C. albicans isolates which adhered most strongly to buccal epithelial cells and were most pathogenic in mice had the highest phospholipase activities. Less pathogenic isolates of C. albicans, C. parapsilosis, and S. cerevisiae were less adherent to epithelial cells and less lethal to mice and had lower phospholipase activities (11). Although these findings suggest a correlation between phospholipase activity and candidal virulence, the strains used were not genetically related, and therefore the differences observed in the virulence of the tested strains could be attributed to factors other than phospholipase secretion.
Two strategies were followed by Ibrahim et al. (75) to determine the role of phospholipase in candidal virulence: (i) the ability of blood isolates of C. albicans from patients and oral isolates from healthy volunteers to produce phospholipase were compared, and (ii) the pathogenicity of clinical isolates with different levels of phospholipase secretion was compared in a murine model of disseminated candidiasis (75). In the first comparative study, 22 isolates of C. albicans were tested (11 were blood isolates obtained from patients with disseminated candidiasis, and 11 were commensal isolates recovered from the oral cavities of healthy volunteers). Marked differences among strains from the two sources were observed. Significantly higher levels of phospholipase production were found in the blood isolates than in the commensals (P = 0.0081). In addition, the blood isolates had significantly higher rates of germination (P = 0.03), and their germ tubes were longer than the germ tubes formed by the commensal strains (P = 0.016). These findings suggest that blood isolates, unlike commensals, may have enhanced the expression of several virulence factors, including both germination and phospholipase production, to enable them to invade host tissues (75).
In the same study, Ibrahim et al. (75) prospectively examined nine blood isolates for expression of virulence factors, including phospholipase and proteinase production, adherence, germination, growth rate, and ability to damage endothelial cells. Additionally, the mortality of mice infected with each of these isolates was determined and the predictive value of each virulence factor for mortality was determined by Cox proportional-hazards analysis. Of the virulence factors studied, only extracellular phospholipase activity was predictive of mortality (75).
To obtain further evidence of the contribution of phospholipases to candidal pathogenicity, the virulence of two C. albicans isolates, CA30 and CA87, was compared in an infant-mouse model (28). Strain CA87 failed to cross the bowel wall of the infant mouse after oral-intragastric challenge and did not cause disseminated infection, while strain CA30 was able to cross the bowel wall and to disseminate hematogenously. A variety of putative virulence factors of these two strains were measured in vitro to determine the relationship of these factors to virulence (75). The only apparent difference in expression of virulence factors between the two strains was in the superior ability of CA30 to produce phospholipase (75). Since this strain was distinguished from CA87 by its ability to invade the bowel wall and undergo subsequent hematogenous dissemination and tissue invasion, these data further suggest that phospholipase is involved in the invasion process of C. albicans.
Although these studies provide evidence that implicates phospholipases in the pathogenesis of C. albicans, they do not prove this association. As in the study of Barrett-Bee et al. (11), the candidal isolates used were not genetically related. The use of such strains does not rule out the possibility that differences observed in the virulence of these strains could be attributed to factors other than phospholipases. Thus, the correlation found between phospholipase and candidal virulence should be confirmed by the use of an isogenic strain pair that differs only in phospholipase production. Molecular cloning of the candidal gene(s) encoding the extracellular phospholipases is the essential first step in the molecular genetic dissection of the role of these enzymes in pathogenesis.
Cloning of the gene(s) encoding candidal phospholipases.
Efforts to clone genes encoding candidal phospholipases have resulted in the cloning of two genes encoding PLB (caPLB1 and caPLB2) and one gene (caPLD) coding for PLD.
(i) Candidal phospholipases B.
(a) caPLB1. The C. albicans PLB1 gene was cloned by using a PCR-based approach relying on degenerate oligonucleotide primers designed on the basis of the amino acid sequences of two peptide fragments obtained from a purified candidal enzyme displaying phospholipase activity (122). Sequence analysis of a 6.7-kb EcoRI-ClaI genomic clone revealed a single open reading frame of 1,818 bp that predicts a preprotein of 605 residues. The size of the candidal PLB1 protein is similar to those reported for other fungal PLBs, which ranged between 612 and 664 amino acids (Table 2). The genomic DNA sequence encodes 17 amino acid residues that are absent from the NH2 terminus of the mature protein. This stretch of residues represents a possible signal sequence (111, 169). The predicted protein contains seven Asn-X-Ser/Thr motifs (residues 199, 261, 399, 451, 465, 492, and 573) that could potentially be N glycosylated. One possible tyrosine phosphorylation site, Lys-Ser-Asn-Ile-Asp-Val-Ser-Ala-Tyr (residues 369 to 377), was also identified (101). Hydropathy analysis of the predicted protein sequence (98) revealed the presence of a single stretch of hydrophobic amino acids present at the amino terminus (residues 1 to 18). This segment of amino acids most probably functions as a single peptide which targets the protein to the endoplasmic reticulum for subsequent processing and, ultimately, secretion. Comparison of the putative candidal phospholipase with other proteins in the redundant database (BLASTP program) revealed significant homology to known fungal PLBs from S. cerevisiae (45%), P. notatum (42%), Torulaspora delbrueckii (48%), and Schizosaccharomyces pombe (38%) (Fig. 3). This gene, designated caPLB1, was mapped to chromosome 6 (101). Consistent with our findings, Hoover et al. (69a) used degenerate oligonucleotide primers derived from conserved regions of PLB1 genes from S. cerevisiae and other fungi to clone the corresponding candidal homolog. Their results confirm our findings and the identity of caPLB1.
TABLE 2.
Fungal phospholipases
| Fungus | Enzyme | Gene cloned (name) | Substrate specificitya | Protein size (amino acid residues) | Molecular mass (kDa) | Potential GPI anchor sequence for attachment | Reference(s) |
|---|---|---|---|---|---|---|---|
| Pathogenic | |||||||
| Candida albicans | PLB1 | Yes (caPLB1) | PC, 1-palmitoyl-Lyso-PC, 1-arachidonyl-Lyso-PC, 1-olejoyl-Lyso-PC, f1-Palmitoyl-Lyso-PE, 1-Palmitoyl-Lyso-PI | 605 | 84 | No | 101, 122, 193 |
| PLB2 | Yes (caPLB2) | 608 | 67b | No | 188 | ||
| PLD | Yes (caPLD) | 1,710 | 196.4b | 87 | |||
| Candida glabrata | PLB | No | PC, Lyso-PC | Clancy et al., Programs Abstr. 36th Annu. Meet. Infect. Dis. Soc. Am., abstr. 316 and 317 | |||
| Cryptococcus neoformans | PLB | Yes (PLB) | PC | 617 | 23, 24; Gottfredsson et al., Abstr. 97th Gen. Meet. Am. Soc. Microbiol. 1997 | ||
| Aspergillus fumigatus | PLB | No | PC | 16; Koul et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998 | |||
| PLC | No | ||||||
| PLD | No | 16 | |||||
| Aspergillus flavus | PLB | No | PC, Lyso-PC | 16; Koul et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998 | |||
| PLA | No | Unknown | |||||
| PLC | No | Unknown | |||||
| Nonpathogenic | |||||||
| Saccharomyces cerevisiae | Yes (PLB1)e | 664 | Yes | 100, 223–225 | |||
| PLBc | 200–280 | ||||||
| PLB1d | 220 | ||||||
| PLB2d | 145 | 87, 206 | |||||
| PLD | Yes (SPO14) | 1,683 | 195 | ||||
| Penicillium notatum | PLB | Yes (PLB) | PC and Lyso-PC | 603 | 95 | Yes | 113, 163 |
| Schizosaccharomyces pombe | Yes | Yes | |||||
| Torulaspora delbrueckii | Yes (PLB) | Unknown | 649 | 72 | Yes | 211 | |
| Neurospora crassa | Yesf | ||||||
| Kluyveromyces lactus | Yesf |
For abbreviations, see Table 1, footnote a.
Predicted from the amino acid sequence and may vary due to posttranslational modifications.
Secreted enzyme.
Membrane-associated enzyme.
Coding for a core protein moiety of the three isoforms of PLB present in S. cerevisiae.
Gene sequence available in GenBank but not published.
FIG. 3.
Sequence alignment of fungal PLBs. The amino acid sequences predicted by caPLB1 and caPLB2 (C. alb.1 and C. alb.2) were aligned with PLBs from S. cerevisiae (S. cer.), P. notatum (P. not.), S. pombe (S. pom.), and T. delbrueckii (T. del.). Shaded areas indicate positions where the amino acids are identical.
A deeper analysis of the PLB1 protein sequence revealed a feature that distinguishes the candidal enzyme from the other known fungal PLBs. Unlike the PLBs of the nonpathogenic fungi, S. cerevisiae, S. pombe, T. delbrueckii, and P. notatum, candidal PLB1 is characterized by the absence of a hydrophobic COOH terminus (Table 2). Such a hydrophobic COOH-terminal region may ultimately be replaced by a glycosylphatidylinositol (GPI) anchor. Indeed, potential GPI anchor sites, immediate to the hydrophobic COOH-terminus, were identified in fungal PLBs from S. cerevisiae, S. pombe, T. delbrueckii, and P. notatum (22, 53). Proteins modified with a GPI anchor may be transiently tethered to the plasma membrane or ultimately cross-linked to the insoluble glucan component of the cell wall (43, 107, 108). Release of proteins associated with the plasma membrane would require the action of a GPI-specific phospholipase. In this view, a GPI anchor may serve to regulate the release of the enzyme to the surroundings. Unlike PLBs from nonpathogenic fungi, the candidal PLB, escaping GPI anchoring, would probably be directly secreted. Such a characteristic may enhance the virulence of C. albicans. Further characterization of PLBs from each fungal species will be necessary to clarify whether any are GPI anchored, and what effect this modification may have on the function and subcellular localization of PLBs, as well as on the pathogenicity of these fungi.
(b) caPLB2. Attempts to clone caPLB2 were prompted by evidence suggesting that C. albicans may possess more than one gene encoding PLB. This evidence is summarized as follows: (i) two proteins which share phospholipase activity were purified by Takahashi et al. (193); (ii) similarly, three enzymes with PLB activity have been purified and characterized from S. cerevisiae (224, 225); (iii) the completion of the sequencing of the S. cerevisiae genome revealed that this organism has at least three genes encoding PLB that are highly homologous (GenBank accession no. L23089, S53035, and S66693); (iv) the S. pombe genome has two sequences which potentially encode PLB (D89183 and D89204); and (v) deletion of caPLB1 did not lead to 100% loss of phospholipase activity, suggesting that the residual activity (about 1% and 10% PLB and Lyso-PL activities, respectively) may be due to a second gene.
To clone caPLB2, Sugiyama et al. (188) used a PCR-based approach similar to the one used to clone caPLB1. A number of similarities are observed between caPLB1 and caPLB2 in size, availability of N-glycosylation sites, the presence of a single stretch of hydrophobic amino acids at the amino terminus, and the absence of GPI attachment site (Table 2). The nucleotide sequence of caPLB2 contained a single open reading frame encoding a putative 608-amino-acid protein with an estimated molecular mass of about 67 kDa. The predicted amino acid sequence contains six potential N-glycosylation sites [Asn-X-(Ser/Thr) motifs] at residues 259, 365, 450, 464, 491, and 572. The deduced amino acid sequence of caPLB2 was homologous to that of caPLB1 (65% identity). caPLB2 was also similar to PLBs from S. cerevisiae, T. delbrueckii, and P. notatum (42, 46, and 42% identity, respectively) (Fig. 3). Hydropathy analysis of the predicted protein (98) revealed the presence of a cluster of hydrophobic amino acids at the N terminus. Similar to S. cerevisiae and T. delbrueckii PLBs (100, 211), caPLB2 possesses a potential signal sequence at the N-terminal region of caPLB2, where two polar amino acids (Gln-Ser) are followed by a cluster of six hydrophobic amino acids (Ile-Leu-Leu-Phe-Val-Val). Such sequence may guide proteins to the secretory pathway. Like caPLB1, caPLB2 lacks the GPI attachment site (a cluster of hydrophobic amino acids at the carboxy terminal) found in the PLBs from the nonpathogenic fungi such as S. cerevisiae, P. notatum, and T. delbrueckii PLBs (100, 113, 211) (see above). Therefore, caPLB2, as with caPLB1, is probably not GPI anchored. Since fungal PLBs and mammalian PLA2 have lysophospholipase activities, it is likely that these enzymes also share common conserved amino acid regions. In this regard, three amino acid residues essential for the catalytic function have been identified in PLA2 (200Arg, 228Ser, and 549Asp) (142). Among them, the serine residue participates in the catalytic function. Interestingly, three regions surrounding these amino acids are also conserved in PLBs from fungi. The deduced amino acid sequence of caPLB2 contains the motifs SGGGX97RA(M/L), GL133SG(G/S), and 381D(S/G)G(E/L)XXXN, which may have a catalytic function (188).
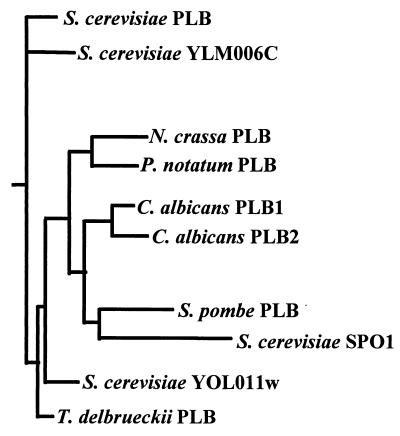
It is clear that fungal PLBs have certain features that are common and others in which they differ. To determine the phylogenetic relationship among various fungal PLBs, Sugiyama et al. (188) constructed a phylogenetic tree of PLBs by using the neighbor-joining method (165). Sequence data from S. cerevisiae PLB (GenBank accession no. L23089), S. cerevisiae SP01 (P53541), S. cerevisiae YLM006c (S53035), S. cerevisiae YOLO11w (S66693), T. delbrueckii PLB (D32134), P. notatum PLB (P39457), S. pombe (Z99258), Neurospora crassa (AF045575), and C. albicans caPLB1 and caPLB2 were used in the tree construction. Figure 4 shows that PLBs and potential PLB analogues are contained in a large cluster of PLB family members. caPLB1 and caPLB2 are closely related to each other, and caPLB genes are more closely related to PLB genes from S. pombe and P. notatum than to PLBs genes from S. cerevisiae and T. delbrueckii.
FIG. 4.
Phylogenetic tree analysis of fungal PLBs. S. cerevisiae PLB (GenBank accession no. L 23089), S. cerevisiae SPO1 (P 53541), S. cerevisiae YLM006c (S53035), S. cerevisiae YOL011w (S66693), T. delbrueckii PLB (D 32134), P. notatum PLB (P 39457), S. pombe PLB (Z 99258), N. crassa PLB (AF045575), and C. albicans caPLB1 and caPLB2.
(ii) Candidal phospholipase D.
PLD catalyzes the hydrolysis of PC to produce phosphatidic acid and choline (Fig. 1a above). Both mammalian and fungal (S. cerevisiae) genes encoding PLD have been cloned and characterized (130, 206). Mammalian PLD has emerged as one of the key enzymes in intracellular signaling (44), while PLD from S. cerevisiae (encoded by SPO14) is essential for meiosis (41, 158, 206). The absence of meiosis in C. albicans, which, unlike S. cerevisiae, exists as a diploid, indicates that PLD in this clinically important yeast may play other roles than meiosis. McLain and Dolan (116) have recently shown that PLD may be an important regulator of the dimorphic transition. Since this yeast-hypha transformation is thought to be an important virulence determinant in C. albicans (33, 54, 133), cloning and disruption of candidal PLD may contribute to our understanding of the biological role of this phospholipase.
For cloning candidal PLD, Kanoh et al. (87) designed two oligonucleotide primers based on the conserved amino acid sequences in human PLD1a and SPO14. These primers were used in a PCR-based approach to clone the full-length gene. The cloned PLD sequence had a potential open reading frame encoding a protein of 1,710 amino acids with a calculated molecular mass of 196.4 kDa. The putative ATG codon was surrounded by the consensus Kozak sequence for translation initiation (94), and the in-frame stop codon and the putative TATA box sequence (TATATAA) were found 6 and 172 bases upstream from the start codon, respectively. The deduced amino acid sequence contained the four conserved regions (I, II, III, and IV) defined by the primary structures of plant, yeast, and human PLDs (125). Furthermore, the HKD motif (HxKxxxxD) in regions I and IV and a serine residue in the GSRS motif in region IV, which are critical for PLD biochemical activity (189), were also completely conserved.
The number of amino acids and the calculated molecular mass of C. albicans PLD closely resembled these of SPO14 protein (1,683 amino acids; 195 kDa). Comparison of the primary structure with those of other PLDs showed that it had the highest homology to the SPO14 protein. The overall homology between the amino acid sequences of C. albicans PLD and the SPO14 protein was 42%. At the four conserved regions, the homology between C. albicans PLD and the SPO14 protein ranged from 65 to 71%, while that between C. albicans PLD and rat PLDs was 42 to 55%. In addition to the four conserved regions found in both fungal and mammalian PLDs, candidal PLD has seven regions (A to G) of 10 to 91 amino acids that are highly homologous to SPO14 protein. Of the seven regions, only two, F and G, are conserved in mammalian PLDs. Therefore, Kanoh et al. (87) speculated that these five regions (A to E) may compose some functional domains specific for fungi.
A phylogenetic tree for PLDs of various species showed three major clusters (87). The first cluster is composed of mammalian PLD1 and plant (151, 209), nematode, and Streptomyces PLDs. The second cluster was composed of mammalian PLD2, and the third cluster was composed of fungal PLDs, including C. albicans PLD and the SPO14 protein. Because fungal PLDS are in a separate grouping from mammalian and other PLDs, it is tempting to speculate that this enzyme may serve a fungus-specific function(s). Unfortunately, elucidation of the biological function of candidal PLD awaits the disruption of the gene encoding it.
Disruption of phospholipase B1.
Gene disruption (deletion) is frequently used to determine the functionality of a specific gene. This is particularly relevant in assessing the contribution of a given gene to microbial virulence (45). Several workers have disrupted a number of genes to evaluate their contribution to the virulence of C. albicans (37, 61, 74, 170, 228). Most recently, C. albicans strains with deletions of the INT1 gene, which encodes a cell surface protein with similarity to mammalian integrins, were constructed (50). Adhesion, hyphal formation, and virulence were subsequently found to be correlated with the expression of this gene.
The successful cloning of caPLB1 allowed us to construct PLB-deficient mutants by targeted gene disruption by using the ura-blaster technique (47) (see reference 101 for details of the method used to disrupt caPLB1). Initial disruption of caPLB1 was complicated by the finding that three rounds of transformation were required to delete the gene, suggesting that strain CAI-4 (a candidal strain derived from C. albicans SC5314 by deletion of the ura3 gene) was triploid for this locus. This finding seemed plausible since disruption of the C. albicans chitin synthase (CHS2) and catalase (CAT1) genes also revealed that they are triploid (61, 226). However, chromosome-blotting experiments suggested that the triallelic state of the caPLB1 genetic locus probably originated from a gene duplication/translocation event rather than an inherent triploid state in the parental strain.
Given the above finding, we proceeded to disrupt the caPLB1 gene a second time, starting with the original CAI-4 strain. Chromosomal analysis was performed at every stage of this second disruption as a means of screening newly constructed strains for genetic translocation or incorrect targeting of transforming DNA. Southern hybridization, chromosomal analysis, and measurement of functional activity confirmed the successful disruption of caPLB1 and demonstrated that Candida is diploid for this locus (101). Although we cannot discount the fact that some candidal genes may be aneuploid (61, 226), our study with caPLB1 highlights the importance of fully characterizing the nature of a suspected triploidy even if mutant strains are constructed by the generally reliable technique of targeted gene disruption. Similar experience has been encountered by a number of Candida researchers (personal communications); therefore, intensive molecular genetic analysis of the constructed mutants should be undertaken so as to disclose potential mutations that otherwise go undetected, especially if disruption of the gene in question produces no obvious phenotypic defect.
Phenotypic characterization of phospholipase B1-deficient mutants.
Phenotypic characterization of the parent and deletion mutants is necessary prior to evaluating the virulence of the isogenic strain pair. Such characterization may provide some measure of assurance that deleting the caPLB1 gene results in only loss of PLB production and not other unrelated phenotypic properties. Comparison of the growth rates and germination capabilities of PLB-deficient mutant with that of the parental strain revealed that disruption of caPLB1 did not affect growth and germination of C. albicans (101), indicating that caPLB1 is not essential for these processes.
Disruption of caPLB1 reduced the ability of C. albicans to secrete the enzyme. When examined by Western blot analysis, PLB was found only in culture filtrates produced by parental and plb1-Δ1 strains but not in the supernatant produced by strain plb1-Δ2 (null mutant). Furthermore, assay of supernatants collected from the parent and the PLB-deficient mutants for PLB and Lyso-PL activities by using specific substrates revealed that the two activities were reduced in the PLB-deficient mutant by approximately 99 and 80% respectively, relative to the wild type. The residual phospholipase activities secreted by the caPLB1-deficient strain may be the result of an additional candidal phospholipase-encoding gene. In this regard, a second gene, caPLB2, which has significant homology to caPLB1, was recently cloned by our group (see “Candidal phospholipases B” above). Experiments to delete this second gene are under way. Characterization of mutants with caPLB1 and caPLB2 deleted may clarify the source of the residual phospholipase activity observed in caPLB1-deficient mutant.
Testing of phospholipase B1-deficient mutants in murine models of candidiasis.
Two different models of candidiasis, representing different clinical settings, were used to determine the role of caPLB1 in the virulence of C. albicans: (i) a hematogenous-dissemination murine model (58) and (ii) an oral-intragastric infant mouse model (28).
(i) Hematogenous-dissemination model.
Leidich et al. (101) challenged BALB/c mice with 5 × 105 yeast cells of either the parent or the PLB-deficient strains intravenously through the lateral tail vein. Both survival and tissue fungal burden were used to assess the pathogenicity of the infecting strains. Their data showed that all mice infected with parental strain SC5314 succumbed to candidal infection within 9 days. In contrast, 50 and 60% of mice challenged with either the PLB-deficient strain plb1-Δ1 or plb1-Δ2, respectively, were alive at day 15. The mean survival time ± standard deviation for mice infected with parent was 4.4 ± 2.1 days, compared to 12.7 ± 2.7 and 13.3 ± 2.6 days for mice infected with plb1-Δ1 or plb1-Δ2, respectively. Statistical analyses revealed that mice infected with either strain plb1-Δ1 or plb1-Δ2 survived significantly longer than did mice infected with strain SC5314 (P < 0.0001 for both comparisons).
Gross inspection of the kidneys showed that numerous visible candidal foci covered the renal cortex of mice infected with the parental strain. In contrast, no candidal foci were visible on renal surfaces of mice infected with either of the PLB-deficient strains. Tissue fungal burden experiments were carried out to assess the severity of infection caused by the wild-type and mutant strains (101). Consistent with the results of the survival experiment, candidal strains plb1-Δ1 and plb1-Δ2 were cleared significantly faster from the kidneys and brain than was parental strain SC5314. For example, the mean fungal burden in the kidneys ± standard deviation was 6.13 ± 0.05 and 4.33 ± 0.35 CFU/g of tissue for the parent and plb1-Δ2 mutant, respectively (P < 0.004). The relative rates of clearance were as follows: plb1-Δ2 > plb1-Δ1 > SC5314.
Taken together, these findings demonstrate that caPLB1 is associated with candidal virulence. Survival and tissue fungal burden data showed that mortality of and tissue invasion in mice infected with strain plb1-Δ1, which harbors only a single deleted caPLB1 allele, was also significantly reduced compared to those for the wild type, suggesting that there may be a dose-dependent relationship between PLB and virulence. Thus, a threshold level of PLB may be required to effectively enhance candidal virulence. In this regard, analysis of culture filtrates for PLB activity revealed that the levels of free fatty acid activity, relative to the parent, released from dipalmitoyl-PC following incubation with culture filtrates obtained from CAI-4, plb1-Δ1, and plb-Δ2 were 100, 54, and 1%, respectively. Moreover, the fact that deletion of caPLB1 did not render C. albicans strains completely avirulent underscores the notion that candidal pathogenicity is multifactorial and is regulated by more than one determinant (33).
(ii) Oral-intragastric infant-mouse model.
Seshan et al. (174) used an oral-intragastric infant-mouse model to examine the effect of caPLB1 on the ability of C. albicans to traverse the gastrointestinal barrier and colonize systemic target organs, such as the kidneys and liver. This model simulates candidal migration across the gastrointestinal tract, one of the major routes for contracting disseminated candidiasis (131). In these experiments, inbred infant mice [crl:CFW (SW) BR] were inoculated intragastrically with 2 × 108 blastospores of either the wild-type or caPLB1-deficient strain. Transmigration of candidal cells across the gastrointestinal tract was monitored microscopically as well as by determining tissue fungal burden of target organs.
The histopathologic appearance of the gastric mucosa of mice infected with the parental strain differed markedly from that of mice infected with the PLB-deficient mutant. Light and transmission electron microscopic examination revealed yeast and hyphal elements in the gastric mucosa 14 days postchallenge. Fungal elements were observed after challenge with both the parental and PLB-deficient strains. However, mucosal invasion in mice challenged with the PLB-deficient strain was confined to the stomach lumen and inner layers of the gastric mucosa, and a minimal neutrophil-dependent inflammatory response was elicited. Furthermore, several areas of the stomach revealed no demonstrable infection. In contrast, the parental strain invaded the submucosal tissue of the stomach, elicited a neutrophil-dependent inflammatory response, and produced systemic candidiasis to a far greater extent. Additionally, the parental strain was able to transverse the vasculature, since numerous hyphal elements were observed within blood vessel lumens (see below) (174).
The number of mice that developed systemic candidal infections differed markedly following challenge with either the parental or PLB-deficient strain. Of the mice challenged with the parental strain, 90 and 70%, respectively, exhibited liver and kidney colonization. In contrast, only 45 and 27% of mice infected with the PLB-deficient mutant exhibited liver and kidney involvement, respectively (174). This difference was reflected in the number of candidal cells (CFU) recovered from these organs. The relative candidal cells recovered from the kidneys and liver of mice challenged with the parental or caPLB1-deficient strain were 2.05 ± 0.4 and 0.77 ± 0.4 CFU/g for the kidney and 2.87 ± 0.4 and 1.33 ± 0.4 CFU/g for the liver, respectively. These differences were statistically significant (P values for kidneys and liver were 0.042 and 0.014, respectively).
The findings of Seshan et al. (174) that the parental strain was more efficient at crossing the gastrointestinal tract and invading internal organs than was its PLB-deficient counterpart suggest a role for PLB in candidal transmigration across the gastrointestinal tract and subsequent dissemination to target organs. Furthermore, their study presents further evidence in support of PLB as a virulence determinant for C. albicans. Finally, the results of the studies of Leidich et al. (101) and Seshan et al. (174) indicate that candidal PLB may be critical for dissemination of C. albicans by the gastrointestinal and hematogenous routes.
Expression of phospholipase B1 during host tissue invasion.
One of the criteria to prove that a particular gene or its product plays an important role in the disease process is to show that it is expressed by the microorganism during the infectious process (45, 167). Both the hematogenous-dissemination (101) and oral-intragastric mouse (174) models for candidiasis were used to determine whether PLB is secreted in vivo.
In the hematogenous-dissemination model, Leidich et al. (101) challenged mice with either the parental or the caPLB1-deleted mutant. Kidneys were harvested, and sections were processed for immunoelectron microscopy by being incubated with either PLB antiserum or goat serum, which served as a negative control. The data revealed that while PLB was secreted from the parental strain during the infectious process, as evidenced by the formation of immunogold complexes following incubation of the tissue sections with PLB antiserum, it was not secreted from the PLB-deficient mutant. In other experiments, C. albicans cells were recovered from the kidneys of mice infected with either the parental or the PLB-deficient strain and prepared for immunogold electron microscopy. Sections examination showed that immunogold labeling was observed only with cells recovered from mice infected with the wild type strain and not with cells recovered from animals infected with the PLB-deficient mutant (J. Vitullo, S. D. Leidich, C. J. Jessup, and M. A. Ghannoum, Program Abstr. 98th Gen. Meet. Am. Soc. Med., abstr. F-39, 1998). These data demonstrate that C. albicans secretes PLB during the invasion of target organs. Moreover, deletion of PLB abrogate the fungal cell ability to secrete this lytic enzyme (see “Phenotypic and genotypic characterization of phospholipase B1-deficient mutants” above).
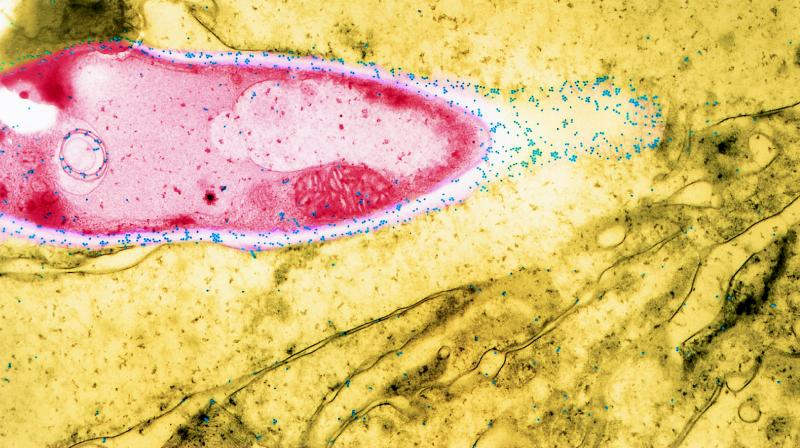
Using indirect-immunofluorescence microscopy, Seshan et al. (174) investigated the expression of PLB during the course of candidal transmigration across the gastrointestinal tract of infant mice. Following challenge with either the parent or the PLB-deficient strain, mice were sacrificed and their gastric mucosal tissues were removed and prepared for indirect-immunofluorescence microscopy. The data showed that the vast majority of candidal cells secrete PLB. Both hyphal and yeast forms of C. albicans secreted the enzyme. Although the periphery of most cells exhibited low-level fluorescence suggestive of PLB secretion, intense fluorescence was observed at the growing tips of mature and developing hyphae, suggesting that PLB secretion is concentrated at the invading hyphal tip. This phenomenon was strikingly clear when immunogold transmission electron microscopy was used to localize PLB secretion in the stomach of infected mice (Fig. 5).
FIG. 5.
Expression of PLB during candidal invasion of gastrointestinal tract of an infant mouse. Inbred infant mice [cr1: CFW (CSW) BR] were inoculated intragastrically with 2 × 108 cells of the phospholipase-producing parental strain (SC5314). The stomach was harvested, and sections were prepared for immunogold microscopy. The candidal cell appears in red, and the stomach tissue appears in greenish-yellow. Immunogold complexes that formed following incubation of the sections with PLB antiserum are shown in blue. Magnification, ×10,000.
The observation that PLB is expressed by C. albicans in murine models prompted us to investigate whether C. albicans also secretes this enzyme during human infection. Sera from patients with proven invasive candidal infection (as determined by a positive blood culture) were obtained from Leo Mendoza (Michigan State University, Lansing, Mich.). Using Western blot analysis, we showed that sera from human patients with systemic candidiasis contain antibodies that reacted with purified PLB (data not shown), suggesting that PLB is secreted during an infectious episode. Taken together, the above findings, using two fundamentally different candidiasis murine models and sera from patients with systemic candidiasis, suggest that PLB is secreted during the course of C. albicans infection and thereby may be involved in virulence.
Phospholipases of Candida glabrata
C. glabrata is recognized increasingly as an important nosocomial pathogen (2, 89, 92, 146, 156, 201, 210, 214, 218, 219); D. W. Warnock, J. Burke, N. J. Cope, E. M. Johnson, N. A. Von Frauenhofer, and E. W. Williams, Letter, Lancet ii:310, 1988). It ranks third among Candida species in the number of reported cases of candidemia, and in one study, C. glabrata surpassed C. tropicalis to become the most common nonalbicans species isolated (51). C. glabrata is important clinically, not only because of the increase in its frequency but also because it is associated with high complication rates (51) and high mortality rates (7, 12). In one study, the mortality rate of 83% exceeded the observed rates from any other Candida species (92). Therefore, identification of factors that contribute to the virulence of this organism may provide new attractive therapeutic targets. Recently, Clancy et al. (Programs Abstr. 36th Annu. Meet. Infect. Dis. Soc. Am., abstr. 317, 1998) reported on the detection of extracellular phospholipase activity in C. glabrata and provided data implicating it as a virulence factor in patients with candidemia caused by this yeast. In their study, phospholipase activity (as determined by opacity around colonies growing on egg yolk-containing media) was investigated in 51 non-albicans Candida species (C. glabrata, 22; C. parapsilosis, 12; C. tropicalis, 10; C. lusitaniae, 5; C. krusei, 2) recovered from patients in a multicenter study of candidemia. Phospholipase activity was detected in 53% of isolates. An assay involving agar containing specific phospholipid substrates was used to identify the type of phospholipase activity secreted by C. glabrata (64). As with C. albicans, PLB and Lyso-PL accounted for 100% of phospholipase activity. No PLA or PLC activities were detected in supernatants collected from various C. glabrata isolates tested. To determine whether a correlation exists between phospholipase activity and clinical virulence, the authors classified the C. glabrata isolates into two groups: persistent isolates were strains that remained in the blood despite therapy with antifungal agents effective in vitro and despite removal of all intravenous catheters, and nonpersistent isolates were strains that were cleared. Among non-albicans Candida species tested, the association between phospholipase activity and persistent candidemia was strongest for C. glabrata (P = 0.004). Moreover, the median PLB and Lyso-PL activities were higher for persistent isolates (9.5 and 24 pmol/mg, respectively) than for nonpersistent isolates (0 and 5.8 pmol/mg) (P < 0.0001 and P = 0.02, respectively).
To establish the role of PLB in the pathogenesis of C. glabrata, an isogenic strain pair of this yeast which differed only in PLB activity was created (C. J. Clancy, A. Leuin, M. A. Ghannoum, and M. H. Nguyen, Programs Abstr. 36th Annu. Meet. Infect. Dis. Soc. Am., abstr. 316, 1998). Conserved sequences of scPLB1, the gene responsible for PLB activity in S. cerevisiae, were used to design guessmer primers for PCR with DNA from a clinical C. glabrata isolate. A 1,030-bp PCR product that encoded part of a putative protein with 78% homology to scPLB1 protein was generated. To create an isogenic mutant with disruption of the PLB gene, Clancy et al. first created a ura3− auxotroph of the parent C. glabrata strain. S. cerevisiae URA3, which is >80% identical to C. glabrata URA3, was amplified by PCR, BamHI restriction sites were added to either end, and the resulting product was inserted in the unique BamHI site of the 1,030-bp PCR fragment. The disrupted product was liberated from the plasmid by EcoRI digestion and was then used to transform C. glabrata ura3− auxotrophs. The resulting mutants were selected by growth on Ura− media. Targeted disruption was confirmed by Southern blot analysis. Colorimetric assay of phospholipase activity revealed >90% elimination of both PLB and Lyso-PL activities in the mutant isolate (URA3+ plb−) compared to the parent C. glabrata (URA3+ plb+), confirming that the cloned gene was responsible for the vast majority of PLB and Lyso-PL activities. Comparative in vivo pathogenicity studies are currently being performed to determine the role of the cloned gene in C. glabrata virulence.
Phospholipases of Cryptococcus neoformans
Cryptococcus neoformans is the cause of the most common life-threatening fungal infection in patients with AIDS. Depending on the study, estimates of the frequency of cryptococcosis among AIDS patients range from 5% to slightly greater than 10% (25, 42, 172). Although the occurrence has decreased in the last couple of years in the developed world due to the introduction of triple HIV therapy (B. Dupont, Abstr. 4th Congr. Eur. Confed. Med. Mycol., abstr. S1, 1998), this incidence is still high, particularly in developing countries such as Uganda. Given the high rate of relapse after initial antifungal therapy, the current management of C. neoformans infections includes lifelong suppressive therapy with antifungals.
Correlation of phospholipase and virulence.
The ability of C. neoformans to secrete phospholipase was first reported by Vidotto et al. (204). Their data showed that 22 of 23 cryptococcal isolates tested produced phospholipase. As with Candida, a wide variation was observed in the ability of various strains to secrete phospholipase (Pz values ranged between 0.271 and 0.49). Although these authors found a correlation between phospholipase production and the size of the capsule in the strains isolated from AIDS patients, the number of isolates analyzed for these factors was small. Therefore, more isolates should be examined to confirm this notion. In another study, Sorrell and coworkers (23) examined 50 C. neoformans isolates for extracellular phospholipase activity by using an egg yolk-based agar assay. Forty-nine of these isolates produced a pericolonial precipitate indicative of phospholipase activity. No difference in phospholipase production was observed between environmental and clinical isolates of C. neoformans var. gattii. Quantitation of cryptococci in the lungs and brains of BALB/c mice inoculated intravenously with four strains expressing high, intermediate, or low phospholipase activity revealed a correlation between phospholipase activity and virulence (23). Based on this finding, the authors proposed that phospholipases secreted by C. neoformans may be associated with cryptococcal virulence.
In a second study, Chen et al. (24), using 1H and 31P nuclear magnetic resonance spectroscopy combined with thin-layer chromatography analyses, extended their work to define the nature of the phospholipase activity produced by C. neoformans. Nuclear magnetic resonance spectroscopy revealed that the sole phospholipid degradation product of the reaction between the substrate PC and cryptococcal culture supernatants was glycerophosphocholine, indicating the presence of PLB. No products indicative of PLA, PLC, PLD, or other lipase activity were detected (24). Thin-layer chromatography analysis confirmed that PLB and Lyso-PL activities were detected in C. neoformans supernatants. Additionally, LPTA activity was identified, by the formation of radioactive PC from Lyso-PC, in the supernatants. The Lyso-PL activity was 10- to 20-fold greater than the PLB activity in these supernatants, with mean specific activities ± standard deviation of 34.9 ± 7.9 and 3.18 ± 0.2 μmol of substrate hydrolyzed/min/mg of protein, respectively. Enzyme activities were stable at acidic pHs (pH 3.8), with pH optima of 3.5 to 4.5. Activities were unchanged in the presence of exogenous serine protease inhibitors, divalent cations, and EDTA. Thus, C. neoformans secretes phospholipases with similar activity (PLB, Lyso-PL, and LPTA) to these observed in C. albicans.
Cloning of the gene encoding phospholipase B.
A PCR-based approach similar to the one used to clone other fungal phospholipases was used to clone cryptococcal PLB (M. Gottfredsson, G. M. Cox, M. Ghannoum, and J. R. Perfect, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. F-71, 1998). Multiple degenerate PCR primers were designed to conserved regions in known fungal PLB genes. From one of the primer combinations, a 1.2-kb amplicon was obtained and cloned into plasmid SK. This fragment was sequenced to confirm its identity and used to screen an EMBL3 C. neoformans genomic library to isolate the entire gene. The data showed that the cryptococcal PLB gene exists in a single copy of approximately 2.4 kb and the putative protein is made up of 617 amino acids. Comparison of the cloned C. neoformans gene to other fungal PLB genes revealed 37, 36, and 36% homology to S. cerevisiae, P. notatum, and C. albicans PLB, respectively, at the amino acid level.
The importance of PLB production for virulence properties in C. neoformans is still unknown and awaits the construction of an isogenic strain pair with specific deletion of PLB. Construction of this pair is currently being accomplished.
Phospholipases of Aspergillus Species and Their Relation to Virulence
Aspergillus fumigatus is the most pathogenic member of its genus, being responsible for 90% of Aspergillus infections. In spite of tremendous efforts, no specific virulence factors have been identified in this genus. Only two studies attempted to determine whether Aspergillus strains secrete phospholipase. Birch et al. (16) investigated the ability of A. fumigatus to produce extracellular phospholipases. The use of fast atom bombardment (which compares lipid-containing media, before and at intervals during A. fumigatus growth, for the presence of anions corresponding to major classes of phospholipids and fatty acids) enabled the detection of several anions corresponding to phospholipid breakdown products. Based on specific degradation products Birch et al. (16) suggested that A. fumigatus secretes multiple extracellular phospholipases, including PLA, PLB, PLC, and PLD. Further characterization of PLC revealed that this activity was initially observed after 30 h of growth and accumulated in broth cultures up to 50 h. Maximal PLC activity coincided with fungal cultures entering the stationary phase, with greater activity occurring at 37°C than at lower temperatures.
Koul et al. studied eight strains of A. fumigatus and A. flavus (A. fumigatus, n = 5; A. flavus, n = 3) for their ability to produce phospholipase (A. Koul, C. J. Jessup, D. J. Deluca, C. J. Elnicky, M. Nunez, R. G. Washburn, and M. A. Ghannoum, Abstr. 987. Gen. Meet. Am. Soc. Microbiol. 1998, abstr. F-78, 1998). Following growth for 3 days at 30 or 37°C in Sabouraud dextrose broth supplemented with 4% glucose, supernatants were concentrated and assayed for phospholipase activity by using a specific substrate radial-diffusion assay capable of differentiating between PLA, PLB, and PLC. Phospholipase activity was detected in all strains regardless of temperature. For both species, PLB was the predominant enzyme. Two of three A. flavus strains also secreted PLA and PLC. For A. fumigatus grown at 30 and 37°C, quantitative PLB activity was 76 to 98 U/100 ml (mean ± standard deviation = 84 ± 9) and 67 to 90 U/100 ml (mean 79 ± 11), respectively. Similarly, for A. flavus, activities were 50 to 81 U/100 ml (mean 62 ± 17) and 33 to 68 U/100 ml (mean, 45 ± 20), respectively. Unlike Birch et al. (16), Koul et al. did not detect enzymatic activity indicative of PLD. Furthermore, the latter group detected only PLB activity in A. fumigatus. This discrepancy could be due to strain-to strain variation. Alternatively, the discrepancy in the detected activities could be because Birch et al. deduced the type of phospholipase secreted by A. fumigatus based on phospholipid degradation products and not assays incorporating specific substrates as utilized by Koul et al.
Although the role of phospholipase in Aspergillus virulence must await cloning and disruption of the gene(s) encoding these enzymes, Koul et al. (Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998) showed that reagent-grade PLB produced extensive cytolysis of cultured pneumocytes, offering the possibility that this enzyme enhances the virulence of Aspergillus by causing damage to host cells.
MECHANISMS BY WHICH PHOSPHOLIPASE AUGMENTS CANDIDAL VIRULENCE
The mechanisms through which the secreted PLB contributes to candidal virulence are currently unknown. Intuitively, it is reasonable to assume that this enzyme is likely to be involved in the early steps of host invasion (adherence, penetration, and damage).
Influence of Phospholipase B1 on Candidal Adherence to Host Cells
A number of investigators provided evidence implicating phospholipases in microbial adherence to host cells. Silverman et al. (177) suggested that phospholipases mediate adhesion to and lysis of host cell membranes during infections by Rickettsia. Prakobphol et al. (145) presented evidence suggesting that enzymes which use lysophospholipids as substrates, e.g., PLB and Lyso-PL, mediate adhesive interactions between host cells and C. tropicalis. Additionally, Barrett-Bee et al. (11) found a correlation between the ability of yeasts, including C. albicans, to adhere to buccal epithelial cells and phospholipase activity. In contrast, comparison of two C. albicans isolates that differed in their ability to produce phospholipase showed that the low phospholipase producer adhered more avidly to the gastrointestinal tract of infant mice than did the high phospholipase producer (28). These contradictory findings could be because the above studies used genetically unrelated strains. Alternatively, phospholipases may facilitate adherence in some organisms and not in others or in certain specific settings. To determine whether caPLB1 influences candidal adherence, the abilities of PLB-deficient mutant and parental strain to adhere to epithelial and endothelial cells were compared by using a standard in vitro assay (57). Our data showed that the percent adherence for the PLB-deficient mutant and parent strain to epithelial cells (HT-29) were 36% ± 3.9% and 35% ± 2.2%, respectively. Similarly, no significant difference was noted in the ability of the isogenic strain pairs to adhere to human umbilical vein endothelial cells (HUVEC) (101). These data suggest that PLB does not appear to directly affect the ability of C. albicans to adhere to host cells. Susan Fisher (University of California, San Francisco), in collaboration with our group, is currently comparing the adherence capabilities of the parent and mutant strains by using more sensitive adherence assays developed by her group. These investigations should determine if subtle differences exist in the adherence of these strains.
Involvement of Phospholipase B1 in Host Cell Injury and Penetration
Since phospholipase targets membrane phospholipids and digests these components, leading to cell lysis (166), direct host cell damage and lysis has been proposed as a major mechanism contributing to microbial virulence. Such host cell injury would be expected to facilitate the penetration of the infecting agent. In this context, Klotz et al. (91), using an in vitro model depicting the earliest events of metastatic Candida infection, showed that Candida organisms first adhere to and then penetrate the endothelium. During transmigration, endothelial-cell continuity was disrupted by the yeasts. As destruction of the endothelium progressed, the fungus penetrated deeper into the substance of the vascular tissue. These authors attributed the dissolution of a portion of the endothelial cells to phospholipase activity (91). Similar suggestive evidence for enzymatic activity by C. albicans in the penetration of the glossal epithelium in both rats (70) and humans (124) has been described, and studies that document this mechanism in murine cervical tissue and skin have been reported (171).
More recently, evidence implicating phospholipase in host cell penetration and injury has been derived from in vitro and in vivo studies. The in vitro studies compared the ability of the parent and PLB-deficient strains to penetrate epithelial and endothelial cell monolayers (101). Also, the abilities of supernatants from cultures of these strains to cause damage to epithelial cells were compared (P. K. Mukhergee, S. D. Leidich, J. Vitulla and M. A. Ghannoum, Program Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. J-81, 1998). Leidich et al. (101), using scanning electron microscopy, compared the abilities of the parent and caPLB1-deficient mutant to penetrate both HUVEC and HT-29 epithelial cells. Their data showed that both the parent and caPLB1-deficient mutant formed germ tubes which penetrated HUVEC and HT-29 cells. However, the capacity of the caPLB1-deficient strain to penetrate HUVEC and HT-29 host cells was significantly lower than that of the parent. The percentage of penetrating parental hyphae was 66.7% ± 1.7% for the endothelial cell line and 57.8% ± 2.5% for the epithelial cell line. In contrast, the percentages of penetrating PLB-deficient hyphae were 37.3% ± 1.3% and 29.0% ± 2.9% for the HUVEC and HT-29 cell lines, respectively (P < 0.0002 for HUVEC; P < 0.0003 for HT-29) (101).
The ability of supernatant from cultures of the parental or PLB-deficient strains to cause damage to epithelial cells was compared by using a thiazolium blue-based assay (Mukhergee et al., 38th ICAAC). Supernatant from the parental culture was about twofold more efficient in causing cell damage than that from the caPLB1-deficient culture (supernatant from the parental strain caused 62.33% ± 1.15% damage, while supernatant from the caPLB1-deficient strain caused 34.67% ± 0.58% damage [P < 0.0001). In the same experiments, damage caused by supernatants obtained from C. albicans and the nonpathogenic yeast S. cerevisiae was compared. In contrast to the significant damage caused by C. albicans, only minor injury was caused by S. cerevisiae (3.95% ± 0.48%). The above data suggest that PLB may enhance candidal virulence by directly damaging host cell membranes.
Seshan et al. (174) provided in vivo data demonstrating that candidal PLB facilitates candidal penetration of host tissues. Using an oral-intragastric infant-mouse model of candidiasis, these authors showed that the phospholipase-producing parental strain penetrate deep into the gastric mucosal and submucosal tissues following intragastric challenge. In contrast, the caPLB1-deficient mutant was not as invasive and was generally sequestered to the stomach lumen. The invasiveness of the parental strain is likely to have increased its access to the gastric vasculature, allowing the organism to hematogenously disseminate more efficiently than the PLB-deficient mutant. Consistent with this notion, more hyphal elements were observed in blood vessel lumens following challenge with the parental strain. These differences in penetration and dissemination were reflected by the number of candidal cells recovered from the livers and kidneys of mice infected with either the parental or caPLB1-deficient strains. Organs (kidneys and liver) from mice infected with the parental strain had significantly higher candidal CFU than did organs from mice infected with the PLB-deficient mutant (174) (see above).
Therefore, the decrease in the capability of PLB1-deficient strains to cross the gastrointestinal tract and cause systemic infection, combined with their decreased capability to penetrate host cell monolayers, and the ability of the enzymatic preparation to lyse the host cell indicate that PLB may play a role in candidal virulence by causing direct damage to host cell membranes. Such injury would allow fungal hyphal elements to more effectively traverse the vascular endothelium, ultimately increasing the rapidity of dissemination to and invasion of target organs.
Do Fungal Extracellular Phospholipases Have Other Functions That Facilitate Virulence in Addition to Cell Lysis and Damage?
Evidence accumulated so far suggests that fungal phospholipases enhance virulence by damaging host cell membranes. However, based on findings derived from studies of phospholipases from bacteria, it is likely that these hydrolytic enzymes may have other functions that facilitate virulence, in addition to causing direct tissue damage (183, 194). Clues to the functions and mechanisms of phospholipases during fungal infection will be provided by the availability of purified fungal phospholipases and isogenic strains from different pathogenic fungi. While C. albicans strains differing in phospholipase production became available only recently and were tested in vivo for their virulence (101, 174), sets from other pathogenic fungi such as C. neoformans are still being constructed, and in some cases (e.g., A. fumigatus) the genes encoding phospholipases await cloning. Thus, mechanistic studies represent important areas of future inquiry.
A number of potential investigational areas relevant to the phospholipase mechanism(s) could be proposed, including (but not limited to) the contributions by phospholipases to signal transduction, stimulation of host cells to release cytokines, and the host inflammatory response.
Many lipids and lipid-derived products generated by phospholipases acting on phospholipids present in host cell membranes are implicated as mediators and second messengers in signal transduction (35, 173). Given the substrate specificity of the enzyme and the lipid degradation products, candidal PLB could also be involved in an as yet unidentified signal transduction pathway potentiated at the lysophospholipid levels (187). In this regard, by-products of PLB degradation, such as lysophospholipids, are reported to induce the activation of protein kinase C (135). Activation of protein kinase C could play a role in the deregulation of cell signaling through local effects or as a consequence of global impacts on host cell physiology (178, 183).
Stimulation of host cells to produce cytokines in response to soluble factors of microbial origin is well documented. For example, PLC from Clostridium perfringens induces the expression of interleukin-8 (IL-8) synthesis by endothelial cells (18, 19). Entamoeba histolytica, which utilizes phospholipase to injure host cells, also stimulates epithelial cells to secrete IL-6 and IL-8 (40). Stimulation of cytokine production in response to lytic enzymes of microbial origin has also been demonstrated in vivo (114). Since phospholipase production is closely associated with candidal host cell injury (see above and reference (101), it is conceivable that this enzyme may directly or indirectly stimulate host cells to produce specific cytokines. A possible mechanism responsible for the expression of genes encoding IL-6 and IL-8 by epithelial cells following microbial injury was proposed by Eckemann et al. (40). These authors showed that E. histolytica injures epithelial cells and causes them to release IL-1α into the medium. This cytokine stimulates adjacent epithelial cells to produce IL-8 and IL-6. Similarly, Kaplanski et al. (88) reported that injury of endothelial cells following exposure to Rickettsia conorii led to the synthesis of IL-6 and IL-8 via a cell-associated IL-1α-dependent pathway. Thus, it is feasible that fungal phospholipases may mediate cytokine production via a similar mechanism.
Microbial phospholipases have been demonstrated to be potent inflammatory agents, inducing the accumulation of inflammatory cells and plasma proteins and the release of various inflammatory mediators in vivo (120). Moreover, microbial phospholipases are able to mobilize arachidonic acid and subsequent prostaglandin synthesis (208). The availability of purified fungal phospholipases and isogenic strain pairs that differ in phospholipase production only will make it feasible to probe the above exciting areas of investigation.
Role of Fungal Phospholipases in the Antifungal Activity of Amphotericin B Lipid Complex
New lipid formulations of amphotericin B with improved safety and efficacy profiles were approved in the United States in the mid-1990s (175). It has been hypothesized that the enhanced therapeutic index of amphotericin B lipid complex (ABELCET) relative to conventional amphotericin B is due, in part, to selective release of active amphotericin B at sites of fungal infection (81). Moreover, this release may occur through the action of phospholipases that are released by the fungus itself or by activated host cells (81, 200). To test this hypothesis, Minassian et al. generated phospholipase-deficient C. albicans mutants by nitrous acid treatment and evaluated them for extracellular phospholipase activity and killing kinetics in the presence of both amphotericin B and amphotericin B lipid complex (ABLC) (B. Minassian, R. K. Flamm, R. R. Whitney, L. Kurselman, L. Bender, R. Summerill, and D. P. Bonner, Program Abstr. 30th Intersci. Conf. Antimicrob. Agents Chemother, abstr. 847, 1990). Compared to the parent the mutants were phospholipase-deficient with increased ABLC MICs and slightly elevated amphotericin B values (121). When tested in vivo, ABLC and conventional amphotericin B were equally efficacious (80 to 90% animals survived following treatment with either agent). Based on these findings, the authors concluded that in vitro release of amphotericin B from ABLC in the case of C. albicans may be due to the breakdown of phospholipids by fungal phospholipases. Furthermore, reduced phospholipase may account for the elevated MICs of ABLC compared to amphotericin B. Moreover, the in vitro activity of amphotericin B but not of ABLC correlates with in vivo efficacy of ABLC against certain phospholipase-deficient Candida. Finally, it was suggested that standard in vitro susceptibility tests with ABLC itself (rather than conventional amphotericin B) may not accurately predict in vivo activity. More recently, Swenson et al. performed an in-depth analysis of the role of candidal and host phospholipases in the antifungal activity of ABLC (190). To evaluate the contribution of fungal phospholipases, these authors used the same candidal mutants created by Minassian et al. (30th ICAAC) and reached similar conclusions. One drawback to these elegant studies is that they were performed with genetically unrelated strains that were created by chemical mutagenesis and were performed prior to the availability of isogenic candidal strains that differ only in phospholipase activity. It is recognized that chemical mutagenesis may have resulted in other changes besides abrogation of phospholipase and thereby may have complicated the data interpretation.
Recently, our group cloned and disrupted the gene caPLB1, which encodes PLB, the dominant phospholipase in C. albicans, and constructed an isogenic strain pair which differ only in phospholipase secretion (101). Next, we used the isogenic strain pair to determine whether a difference exists in the susceptibility of ABLC and conventional amphotericin B in vitro. Our data showed that both conventional amphotericin B and ABLC had similar MICs (0.125-0.25 μg/ml) for the phospholipase-producing wild-type strain (SC5314) and the phospholipase-deficient mutant (plb-1Δ2). This finding was consistent even when different media (RPMI 1640, Sabouraud's dextrose broth, and yeast nitrogen base) were used to assay the in vitro activities of these antifungals (C. Jessup and M. A. Gharroum, unpublished data). Similarly, no difference was observed in the in vitro susceptibility of parent and phospholipase-deficient C. neoformans strains (constructed by Gottfredsson, et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998) against amphotericin B and ABLC (Jessup and Ghannoum, unpublished). Therefore, data derived from our group and that of Perfect clearly demonstrate that deletion of fungal phospholipases does not influence antifungal susceptibility of ABLC. Furthermore, our data support the conclusion of Minassian et al. (30th ICAAC), who suggested that in vivo efficacy of ABLC may be more closely correlated with host phospholipase activity and not with yeast or fungal phospholipase activity. It is conceivable that the effects seen in the studies of Minassian et al. (30th ICAAC) and Swenson et al. (190) could be due to a second phospholipase beside PLB. However, this is unlikely since PLB is the dominant phospholipase and is responsible for about 90% of the extracellular activity of the candidal phospholipase.
FUNGAL PHOSPHOLIPASES AS A THERAPEUTIC AND DIAGNOSTIC TARGET
The validation of PLB as a virulence factor in C. albicans by using animal models of hematogenously disseminated (101) and gastrointestinal (174) candidiasis and the detection of this hydrolytic enzyme in other pathogenic fungi including C. neoformans, A. fumigatus, and A. flavus makes it a potential therapeutic target. The finding that phospholipases are secreted by different clinically important fungal genera and evidence that these fungi share the same class of phospholipase, namely, PLB, suggest that these hydrolytic enzymes may represent a common theme utilized by pathogenic fungi as a universal virulence factor (56). Moreover, demonstration that phospholipase secretion is not limited to one fungal genus increases the potential of fungal phospholipases as a therapeutic target.
A number of drug development approaches could be pursued which may lead to the discovery of novel agents targeting fungal phospholipases. These approaches include the development of vaccines, identification of agents that inhibit the production or release of fungal phospholipases (medicinal chemistry synthetic programs, screening of chemical or combinatorial libraries), and rational inhibitor design (based on the tertiary structure of the protein).
The use of microbial phospholipases as vaccines in protection against diseases has been investigated. Clostridial alpha-toxin (PLC) was reported by Kameyama et al. (86) to induce protection against C. perfringens-mediated gas gangrene. Williamson and Titball (214a) were able to protect sheep against experimentally induced Clostridium novyi and C. perfringens gas gangrene by using a toxoid mixture from C. perfringens-C. septicum-C. novyi as a vaccine. Instead of using PLD to develop a vaccine against C. pseudotuberculosis, Hodgson et al. (69) constructed a PLD-negative strain (called Toxminus), which was incapable of inducing caseous lymphadenitis, and used it to vaccinate sheep. Sheep vaccination and challenge trials with this mutant showed that it elicited strong humoral and cell-mediated immune response and protected the animals from wild-type challenge (69).
Preliminary attempts to identify inhibitors to candidal phospholipases have been previously undertaken by Hänel et al. (65), who screened for synthetic phospholipase inhibitory substances by using in vitro and in vivo models. Their efforts resulted in the identification of lead structures capable of inhibiting phospholipase activity in vitro (65). Additionally, prolongation of animal survival was observed when these compounds were used in combination with fluconazole (65). The above studies provide some support to the concept of targeting fungal phospholipases for drug discovery and warrant a systemic approach for drug discovery based on these lytic enzymes.
Another potential use of fungal extracellular phospholipases, particularly candidal PLB, is as a diagnostic tool. Since it has been demonstrated that candidal PLB is released during the progression of candidal infection in murine models of candidiasis (101, 174) and that sera from patients with systemic candidiasis contain antibodies which react with purified candidal PLB (see above), it is conceivable that this particular antigen could be applied in an antibody detection assay. PLB possesses a number of advantages which make it attractive for development as a diagnostic tool: (i) it is a naturally secreted protein and not a cell wall or cytoplasmic component as is the case with all fungal antigens, with the exception of aspartyl proteinases (46), under development (for reviews, see references 112 and 155); (ii) it is a purified well-defined antigen rather than a crude extract containing cell wall polysaccharide (112); and (iii) blood isolates generally secrete much higher levels of enzyme than do isolates from the oral cavity of healthy individuals (75) or isolates from wounds or urine (147).
Secretion of PLB by a number of pathogenic fungi, including Candida, Cryptococcus neoformans, and Aspergillus, may pose a challenge for the development of a highly specific and sensitive diagnostic test. However, this challenge could be met as more information on genes encoding PLB from specific pathogens become available and these proteins are purified. As stated by Martinez et al. (112), “with their amino acid sequences readily available and the application of new specific epitope-mapping technique, it is now easier to identify specific epitopes within these antigens and to adapt this knowledge to development of improved serologic tests for the diagnosis of candidiasis”.
CONCLUSION AND OUTLOOK
The aim of this review was to evaluate our current knowledge of fungal phospholipases, with special emphasis on their possible role in virulence. It is clear that we are at an early stage in our investigations and that exciting areas for future studies lie ahead. Among the pathogenic fungi, cloning, disruption, and virulence evaluation of genes encoding extracellular phospholipases have been completed only for C. albicans. It is also clear that efforts to clone the genes encoding these enzymes in other pathogenic species are being pursued in earnest. Investigations aimed at determining the role of phospholipases in the virulence of different pathogenic fungi will reveal whether these enzymes are universal virulence factor in pathogenic fungi. Elucidation of the mechanism(s) by which phospholipases contribute to virulence is another area that warrants attention. In addition to gene cloning, enzyme purification should be pursued. Purification of fungal phospholipases will affect our mechanistic studies and, in the long term, may help in rational drug design of novel antifungal targets. A word of caution: purification of these enzymes is not a trivial matter. They are produced in small quantities, and therefore large volumes of medium are required to produce reagent amounts of protein. Moreover, the high lytic potency of these enzymes may interfere with recombinant approaches to purify the gene products. Finally, purification procedures could be complicated further by organism-related factors. For example, in C. neoformans, the presence of a large capsule may make it difficult to purify the enzyme. There is a lack of knowledge of phospholipase gene regulation in vivo, as well as whether this class of enzymes acts in concert with other secreted proteins, such as aspartic proteinases, to augment fungal virulence. Research into these areas will further our understanding of the pathobiology of fungal infection. Once the importance of phospholipases in fungal virulence is proven, the next challenge will be to use these enzymes in drug discovery efforts to identify and design inhibitors and/or develop a vaccine. The utility of phospholipases as diagnostic markers of fungal infections is yet another area of study that may prove to be fruitful.
ACKNOWLEDGMENTS
I thank Steven Leidich for useful discussions and Pranab Mukherjee for his assistance in figure preparation and putting the manuscript together. Also, I thank Kathleen Smith for her secretarial help and K. Seshan, Medical College of Ohio, for providing Fig. 5.
The work reviewed in this article which originated from the my laboratory was partly supported by grants from the National Institutes of Health (AI35097), and Pfizer Pharmaceutical Group, New York, N.Y.
REFERENCES
- 1.Abrehem K, Zamiri I. Purification of a protein toxin from Corynebacterium ulcerans. J Med Microbiol. 1980;13:587–592. doi: 10.1099/00222615-13-4-587. [DOI] [PubMed] [Google Scholar]
- 2.Aisner J, Schimpff S C, Sutherland J C, Young V M, Wiernik P H. Torulopsis glabrata infections in patients with cancer: increasing incidence and relationship to colonization. Am J Med. 1976;61:23–28. doi: 10.1016/0002-9343(76)90026-7. [DOI] [PubMed] [Google Scholar]
- 3.Alouf J E. Cell membranes and cytolytic bacterial toxins. In: Cuatrecasas P, editor. The specificity and action of animal, bacterial and plant toxins. Receptors and recognition, series B. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1977. pp. 219–270. [Google Scholar]
- 4.Ansell G B, Hawthorne J N. Catabolism. In: Ansell G B, Hawthorne J N, editors. Phospholipids. Amsterdam, The Netherlands: Elsevier Publishing Co.; 1964. pp. 152–174. [Google Scholar]
- 5.Avigad G. Microbial phospholipases. In: Bernheimer A W, editor. Mechanisms in bacterial toxinology. New York, N.Y: John Wiley & Sons, Inc.; 1976. pp. 99–167. [Google Scholar]
- 6.Awad M M, Bryant A E, Stevens D L, Rood J I. Virulence studies on chromosomal α-toxin and θ-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of α-toxin in Clostridium perfringens-mediated gas gangrene. Mol Microbiol. 1995;15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. [DOI] [PubMed] [Google Scholar]
- 7.Bailey J E, Kliegman R M, Annable W L, Dahms B B, Fanaroff A A. Torulopsis glabrata sepsis appearing as necrotizing enterocolitis and endophthalmitis. Am J Dis Child. 1984;183:965–966. doi: 10.1001/archpedi.1984.02140480067020. [DOI] [PubMed] [Google Scholar]
- 8.Baine W B. A phospholipase C from the Dallas 1E strain of Legionella pneumophila serogroup 5: purification and characterization of conditions for optimal activity with an artificial substrate. J Gen Microbiol. 1988;134:489–498. doi: 10.1099/00221287-134-2-489. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee S N, Emori T G, Culver D H, Gaynes R P, Jarvis W R, Horan T, Edwards J E, Tolson J, Henderson T, Martone and National Nosocomial Infections Surveillance System W J. Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1989. In: Rex J, Anderson M D, editors. Serious Candida infections: risk factors, treatment, and prevention. Selected readings: focus on fluconazole. New York, N.Y: Medical Information Press, Pfizer, Inc.; 1995. pp. 3–8. [Google Scholar]
- 10.Banno Y, Yamada T, Nozawa Y. Secreted phospholipases of the dimorphic fungus, Candida albicans; separation of three enzymes and some biological properties. Sabouraudia. 1985;23:47–54. doi: 10.1080/00362178585380081. [DOI] [PubMed] [Google Scholar]
- 11.Barrett-Bee K, Hayes Y, Wilson R G, Ryley J F. A comparison of phospholipase activity, cellular adherence and pathogenicity of yeasts. J Gen Microbiol. 1985;131:1217–1221. doi: 10.1099/00221287-131-5-1217. [DOI] [PubMed] [Google Scholar]
- 12.Beck-Sagué C M, Jarvis W R National Nosocomial Infections Surveillance System. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 13.Berka R, Gray G, Vasil M. Studies of phospholipase C (heat-labile haemolysin) in Pseudomonas aeruginosa. Infect Immun. 1981;34:1071–1074. doi: 10.1128/iai.34.3.1071-1074.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berka R, Vasil M. Phospholipase C (heat-labile hemolysin) of Pseudomonas aeruginosa: purification and preliminary characterization. J Bacteriol. 1982;152:239–245. doi: 10.1128/jb.152.1.239-245.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernheimer A W, Bey R F. Copurification of Leptospira interrogans serovar pomona hemolysin and sphingomyelinase C. Infect Immun. 1986;54:262–264. doi: 10.1128/iai.54.1.262-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birch M, Robson G, Law D L, Denning D W. Evidence of multiple extracellular phospholipase activities of Aspergillus fumigatus. Infect Immun. 1996;64:751–755. doi: 10.1128/iai.64.3.751-755.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman M H, Ottolenghi A C, Mengel C E. The effects of phospholipase C on human erythrocytes. J Med Microbiol. 1972;5:467–472. doi: 10.1007/BF02431968. [DOI] [PubMed] [Google Scholar]
- 18.Bryant A E, Stevens D L. Phospholipase C and perfringolysin O from Clostridium perfringens upregulate endothelial cell-leukocyte adherence molecule 1 and intercellular leukocyte adherence molecule 1 expression and induce interleukin-8 synthesis in cultured human umbilical vein endothelial cells. Infect Immun. 1996;64:358–362. doi: 10.1128/iai.64.1.358-362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunting M, Lorant D E, Bryant A E, Zimmerman G A, McIntyre T M, Stevens D L, Prescott S M. Alpha toxin from Clostridium perfringens induces proinflammatory changes in endothelial cells. J Clin Investig. 1997;100:565–574. doi: 10.1172/JCI119566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calderone R, Braun P. Adherence and receptor relationships of Candida albicans. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camilli A, Goldfine H, Portnoy D A. Listeria monocytogenes mutants lacking phosphatidylinositol-specific phospholipase C are avirulent. J Exp Med. 1991;173:751–754. doi: 10.1084/jem.173.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caro L H P, Tettelin H, Vossen J H, Ram A F, van den Ende H, Klis F M. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Muller M, Zhou Z, Wright L, Sorrell T. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J Infect Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Wright L C, Santangelo R T, Muller M, Moran V R, Kuchel P W, Sorrell T C. Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect Immun. 1997;65:405–411. doi: 10.1128/iai.65.2.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuck S L, Sande M A. Infections with Cryptococcus neoformans in the acquired immunodeficiency syndrome. N Engl J Med. 1989;321:794–799. doi: 10.1056/NEJM198909213211205. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Reference deleted.
- 28.Cole G T, Lynn K T, Seshan K R. An animal model for oropharyngeal, esophageal and gastric candidosis. Mycoses. 1990;33:7–19. doi: 10.1111/myc.1990.33.1.7. [DOI] [PubMed] [Google Scholar]
- 29.Coleman K, Dougan G, Arbuthnott J P. Cloning and expression in Escherichia coli K12, of the chromosomal hemolysin (phospholipase C) determinant of Pseudomonas aeruginosa. J Bacteriol. 1983;153:909–915. doi: 10.1128/jb.153.2.909-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa A, Costa C, Misefari A, Amato A. On the enzymatic activity of certain fungi. VII. Phosphatidase activity on media containing sheep's blood of pathogenic strains of Candida albicans. Atti Soc Sci Fis Mat Nat. 1968;XIV:93–101. [Google Scholar]
- 31.Reference deleted.
- 32.Reference deleted.
- 33.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 34.Cutter D, Kreger A. Cloning and expression of the damselysin gene from Vibrio damsela. Infect Immun. 1990;58:266–268. doi: 10.1128/iai.58.1.266-268.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis E A, Rhee S G, Billah M M, Hannun Y A. Role of phospholipases in generating lipid second messengers in signal transduction. FASEB J. 1991;5:2068–2077. doi: 10.1096/fasebj.5.7.1901288. [DOI] [PubMed] [Google Scholar]
- 36.De Silva N S, Quinn P A. Rapid screening for phospholipase C activity in mycoplasmas. J Clin Microbiol. 1987;25:729–731. doi: 10.1128/jcm.25.4.729-731.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diez-Orejas R, Molero G, Navarro-Garcia F, Pla J, Nombela C, Sanchez-Perez M. Reduced virulence of Candida albicans MKC1 mutants: a role for mitogen-activated protein kinase in pathogenesis. Infect Immun. 1997;65:833–837. doi: 10.1128/iai.65.2.833-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doery H M, Magnusson B J, Gulasekharam J, Pearson J E. The properties of phospholipase enzymes in staphylococcal toxins. J Gen Microbiol. 1965;40:283–296. doi: 10.1099/00221287-40-2-283. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Eckmann L, Reed S L, Smith J R, Kagnoff M F. Entamoeba histolytica trophozoites induce an inflammatory cytokine response by cultured human cells through the paracrine action of cytolytically released interleukin-1α. J Clin Investig. 1995;96:1269–1279. doi: 10.1172/JCI118161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ella K M, Dolan J W, Qi C, Meier K E. Characterization of Saccharomyces cerevisiae deficient in expression of phospholipase D. Biochem J. 1996;314:15–19. doi: 10.1042/bj3140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eng R H K, Bishburg E, Smith S M. Cryptococcal infections in patients with acquired immune deficiency syndrome. Am J Med. 1986;81:19–23. doi: 10.1016/0002-9343(86)90176-2. [DOI] [PubMed] [Google Scholar]
- 43.Englund P T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 44.Exton J H. New developments in phospholipase D. J Biol Chem. 1997;272:15579–15582. doi: 10.1074/jbc.272.25.15579. [DOI] [PubMed] [Google Scholar]
- 45.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 46.Flahaut M L, Sanglard D, Monod M, Bille J, Rossier M. Rapid detection of Candida albicans in clinical sample by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J Clin Microbiol. 1998;36:395–401. doi: 10.1128/jcm.36.2.395-401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fonzi F, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1998;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freer J H, Arbuthnott J P. Biochemical and morphologic alterations of membranes by bacterial toxins. In: Bernheimer A W, editor. Mechanisms of bacterial toxicology. New York, N.Y: John Wiley & Sons, Inc.; 1976. pp. 169–193. [Google Scholar]
- 49.Fu Y, Ibrahim A, Fonzi F, Zhou X, Ramos C, Ghannoum M. Cloning and characterization of a gene (LIP1) which encodes a lipase from the pathogenic yeast Candida albicans. Microbiology. 1997;143:340. doi: 10.1099/00221287-143-2-331. [DOI] [PubMed] [Google Scholar]
- 50.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 51.Gassama-Diagne A, Rogalle P, Fauvel J, Willson M, Klaébè A, Chaps H. Substrate specificity of phospholipase B from guinea pig intestine. J Biol Chem. 1992;267:13418–13424. [PubMed] [Google Scholar]
- 52.Georgopapadakou N H, Walsh T J. Antifungal Agents: chemotherapeutic targets and immunologic strategies. Antimicrob Agents Chemother. 1996;40:279–291. doi: 10.1128/aac.40.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerber L D, Kodukula K, Udenfriend S. Phosphatidylinositol glycan (PI-G) anchored membrane proteins. Amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J Biol Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- 54.Ghannoum M, Abu-Elteen K H. Pathogenicity determinants in Candida: a review. J Mycol Med. 1990;33:265–282. doi: 10.1111/myc.1990.33.6.265. [DOI] [PubMed] [Google Scholar]
- 55.Ghannoum M, Radwan S. Candida adherence to epithelial cells. Boca Raton, Fla: CRC Press, Inc.; 1990. [Google Scholar]
- 56.Ghannoum M A. Extracellular phospholipases as universal virulence factor in pathogenic fungi. Jpn J Med Mycol. 1998;39:55–59. doi: 10.3314/jjmm.39.55. [DOI] [PubMed] [Google Scholar]
- 57.Ghannoum M A, Filler S G, Ibrahim A S, Fu Y, Edwards J E., Jr Modulation of interactions of Candida albicans and endothelial cells by fluconazole and amphotericin B. Antimicrob Agents Chemother. 1992;36:2239–2244. doi: 10.1128/aac.36.10.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghannoum M A, Spellberg B J, Saporito-Irwin S M, Fonzi W. Reduced Virulence of Candida albicans PHR1 mutants. Infect Immun. 1995;63:4528–4530. doi: 10.1128/iai.63.11.4528-4530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilmore M S, Cruz-Rodgz A L, Leimeister-Wachter M, Kreft J, Goebel W. A Bacillus cereus cytolytic determinant, cereolysin AB, which comprises the phospholipase C and sphingomyelinase genes: nucleotide sequence and genetic linkage. J Bacteriol. 1989;171:744–753. doi: 10.1128/jb.171.2.744-753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reference deleted.
- 61.Gow N A R, Robbins P W, Lester J, Brown A, Fonzi F, Chapman T, Kinsman O. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granstrom M, Erickson A, Strandvik B, Wretlind B, Pavlovskis O, Berka R, Vasil M. Relationship between antibody response to Pseudomonas aeruginosa exoproteins and colonization/infection in patients with cystic fibrosis. Acta Paediatr Scand. 1984;73:772–777. doi: 10.1111/j.1651-2227.1984.tb17774.x. [DOI] [PubMed] [Google Scholar]
- 63.Graybill J R, Montalbo E, Kirkpatrick W R, Luther M F, Revankar S G, Patterson T F. Fluconazole versus Candida albicans: a complex relationship. Antimicrob Agents Chemother. 1998;42:2938–2942. doi: 10.1128/aac.42.11.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Habermann E, Hardt K L. A sensitive and specific plate test for the quantitation of phospholipases. Anal Biochem. 1972;50:163–173. doi: 10.1016/0003-2697(72)90495-2. [DOI] [PubMed] [Google Scholar]
- 65.Hänel H, Kirsch R, Schmidts H-L, Kottmann H. New systematically active antimycotics from the beta-blocker category. Mycoses. 1995;38:251–264. doi: 10.1111/j.1439-0507.1995.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 66.Hänel H, Menzel I, Holzmann H. High phospholipase A activity of Candida albicans isolated from the intestine of psoriatic patients. Mycoses. 1988;31:451–453. . (In German.) [PubMed] [Google Scholar]
- 67.Hauschild A H W, Walcroft M J, Campbell W. Emesis and diarrhea induced by enterotoxin of Clostridium perfringens type A in monkeys. Can J Microbiol. 1971;17:1141–1143. doi: 10.1139/m71-180. [DOI] [PubMed] [Google Scholar]
- 68.Henner D J, Yang M, Chen E, Hellmiss R, Rodriguez H, Low M G. Sequence of the Bacillus thuringiensis phosphatidylinositol-specific phospholipase C. Nucleic Acids Res. 1988;16:10383. doi: 10.1093/nar/16.21.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hodgson A L M, Krywult J, Corner L A, Rothel J S, Radford A J. Rational Attenuation of Corynebacterium pseudotuberculosis: potential cheesy gland vaccine and live delivery vehicle. Infect Immun. 1992;60:2900–2905. doi: 10.1128/iai.60.7.2900-2905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69a.Hoover C I, Jantapour M J, Newport G, Agabian N, Fischer S J. Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol Lett. 1998;167:163–169. doi: 10.1111/j.1574-6968.1998.tb13223.x. [DOI] [PubMed] [Google Scholar]
- 70.Howlett J A, Squier C A. Candida albicans ultrastructure: colonization and invasion of oral epithelium. Infect Immun. 1980;29:252–260. doi: 10.1128/iai.29.1.252-260.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hube B. Candida albicans secreted aspartyl proteinases. Curr Top Med Mycol. 1996;7:55–69. [PubMed] [Google Scholar]
- 72.Hube B. Possible role of secreted proteinases in Candida albicans infections. Rev Iberoam Micol. 1998;15:65–68. [PubMed] [Google Scholar]
- 73.Hube B, Monod M, Monod M, Sanglard D, Odds F. Functional aspects of secreted Candida proteinases. In: James M N G, editor. Aspartic proteinases. New York, N.Y: Plenum Press; 1998. pp. 339–344. [DOI] [PubMed] [Google Scholar]
- 74.Hube B, Sanglard D, Odds F, Hess D, Monod M, Schafer W, Brown A J P, Gow N A R. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ibrahim A S, Mirbod F, Filler S G, Banno Y, Cole G T, Kitajima Y, Edwards J E, Jr, Nozawa Y, Ghannoum M A. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect Immun. 1995;63:1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ikezawa H. The physiological action of bacterial phosphatidylinositol-specific phospholipase C. The release of ectoenzymes and other effects. J Toxicol Toxin Rev. 1986;5:1–24. [Google Scholar]
- 77.Ikezawa H, Matsushita M, Tomita M, Taguchi R. Effects of metal ions on sphingomyelinase activity of Bacillus cereus. Arch Biochem Biophys. 1986;249:588–595. doi: 10.1016/0003-9861(86)90037-8. [DOI] [PubMed] [Google Scholar]
- 78.Ikezawa H, Nakabayashi T, Suzuki K, Nakajima M, Taguchi T, Taguchi R. Complete purification of a phosphatidylinositol-specific phospholipase C from a strain of Bacillus thuringiensis. J Biochem. 1983;93:1717–1719. doi: 10.1093/oxfordjournals.jbchem.a134315. [DOI] [PubMed] [Google Scholar]
- 79.Ispolatovskaya M V. Type A Clostridium perfringens toxin. In: Kadis S, Montie T C, Ajl S J, editors. Microbial toxins. IIA. Bacterial protein toxins. New York, N.Y: Academic Press, Inc.; 1971. pp. 109–158. [Google Scholar]
- 80.Jackson S G. Development of a fluorescent immunodot assay for Bacillus cereus enterotoxin. J Immunol Methods. 1989;120:215–220. doi: 10.1016/0022-1759(89)90245-7. [DOI] [PubMed] [Google Scholar]
- 81.Janoff A S, Perkins W R, Saletan S L, Swenson C E. Amphotericin B lipid complex (ABLC®): a molecular rationale for the attenuation of amphotericin B related toxicities. J Liposome Res. 1993;3:451–471. [Google Scholar]
- 82.Reference deleted.
- 83.Johansen K A, Gill R E, Vasin M L. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect Immun. 1996;64:3259–3266. doi: 10.1128/iai.64.8.3259-3266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johansen T, Haugli H, Ikezawa H, Little C. Bacillus cereus strain SE-1: nucleotide sequence of the sphingomyelinase C gene. Nucleic Acids Res. 1988;16:10370. doi: 10.1093/nar/16.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johansen T, Holm T, Guddal P H, Sletten K, Haugli F B, Little C. Cloning and sequencing of the gene encoding the phosphatidylcholine-preferring phospholipase C of Bacillus cereus. Gene. 1988;65:293–304. doi: 10.1016/0378-1119(88)90466-0. [DOI] [PubMed] [Google Scholar]
- 86.Kameyama S, Sato H, Murata R. The role of α-toxin of Clostridium perfringens in experimental gas gangrene in guinea pigs. Jpn J Med Sci Biol. 1975;25:200. [PubMed] [Google Scholar]
- 87.Kanoh H, Nakashima S, Zhao Y, Sugiyama Y, Kitajima Y, Nozawa Y. Molecular cloning of a gene encoding phospholipase D from the pathogenic and dimorphic fungus, Candida albicans. Biochim Biophys Acta. 1998;1398:359–364. doi: 10.1016/s0167-4781(98)00067-0. [DOI] [PubMed] [Google Scholar]
- 88.Kaplanski G, Teysseire N, Farnarier C, Kaplanski S, Lissitzky J, Durand J, Soubeyrand J, Dinarello C A, Bongard P. IL-6 and IL-8 production from cultured human endothelial cells stimulated by infection with Rickettsia conorii via a cell-associated IL-1 alpha-dependent pathway. J Clin Investig. 1995;96:2839–2844. doi: 10.1172/JCI118354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaufmann C S, Merz W G. Electrophoretic karyotypes of Torulopsis glabrata. J Clin Microbiol. 1989;27:2165–2168. doi: 10.1128/jcm.27.10.2165-2168.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klotz S. Fungal adherence to the vascular compartment: a critical step in the pathogenesis of disseminated candidiasis. Clin Infect Dis. 1992;14:340–347. doi: 10.1093/clinids/14.1.340. [DOI] [PubMed] [Google Scholar]
- 91.Klotz S A, Drutz D J, Harrison J L, Huppert M. Adherence and penetration of vascular endothelium by Candida yeasts. Infect Immun. 1983;42:374–384. doi: 10.1128/iai.42.1.374-384.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Komshian S V, Uwaydah A K, Sobel J D, Crane L R. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;11:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 93.Reference deleted.
- 94.Kozak M. An analysis of 5′-encoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kramer J M, Gilbert G. Bacillus cereus and other Bacillus species. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 21–70. [Google Scholar]
- 96.Kuppe A, Evans L M, McMillen D A, Griffith O H. Phosphatidylinositol-specific phospholipase C of Bacillus cereus: cloning, sequencing, and relationship to other phospholipases. J Bacteriol. 1989;171:6077–6083. doi: 10.1128/jb.171.11.6077-6083.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuppe A, Hedberg K K, Volwerk J J, Griffith O H. Inhibition of the phosphatidylinositol-specific phospholipase C from Bacillus cereus by a monoclonal antibody binding to a region with sequence similarity to eukaryotic phospholipases. Biochim Biophys Acta. 1990;1047:41–48. doi: 10.1016/0005-2760(90)90258-y. [DOI] [PubMed] [Google Scholar]
- 98.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 99.Lane T, Garcia J R. Phospholipase production in morphological variants of Candida albicans. Mycoses. 1991;34:217–220. doi: 10.1111/j.1439-0507.1991.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 100.Lee K S, Patton J L, Fido M, Hines L K, Kohlwein S D, Paltauf F, Henry S A, Levin D E. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J Biol Chem. 1994;269:19725–19730. [PubMed] [Google Scholar]
- 101.Leidich S D, Ibrahim A S, Fu Y, Koul A, Jessup C, Vitullo J, Fonzi W, Mirbod F, Nakashima S, Nozawa Y, Ghannoum M A. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem. 1998;273:26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- 102.Leimeister-Wachter M, Domann E, Chakraborty T. Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is co-ordinately expressed with listeriolysin in Listeria monocytogenes. Mol Microbiol. 1991;5:361–366. doi: 10.1111/j.1365-2958.1991.tb02117.x. [DOI] [PubMed] [Google Scholar]
- 103.Leslie D, Fairweather N, Pickard D, Dougan G, Kehoe M. Phospholipase C and haemolytic activities of Clostridium perfringens alpha-toxin cloned in Escherichia coli: sequence and homology with a Bacillus cereus phospholipase C. Mol Microbiol. 1989;3:383–392. doi: 10.1111/j.1365-2958.1989.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 104.Little C, Aurebekk B, Otnaess A-B. Purification by affinity chromatography of phospholipase C from Bacillus cereus. FEBS Lett. 1975;52:175–179. doi: 10.1016/0014-5793(75)80800-3. [DOI] [PubMed] [Google Scholar]
- 105.Liu P. Toxins of Pseudomonas aeruginosa. In: Dogget R G, editor. Pseudomonas aeruginosa: clinical manifestations of infection and current therapy. New York, N.Y: Academic Press, Inc.; 1979. pp. 63–68. [Google Scholar]
- 106.Long-Krug S, Fischer K, Hysmith R, Ravdin J. Phospholipase A enzymes of Entamoeba histolytica: description and subcellular localization. J Infect Dis. 1985;152:536–541. doi: 10.1093/infdis/152.3.536. [DOI] [PubMed] [Google Scholar]
- 107.Lu C F, Kurjan J, Lipke P N. A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol Cell Biol. 1994;14:4825–4833. doi: 10.1128/mcb.14.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu C F, Montijn R C, Brown J L, Klis F, Kurjan J, Bussey H, Lipke P N. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β-1,6-glucan in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.MacFarlane M G. On the biochemical mechanisms of action of gas-gangrene toxins. In: Howie J W, Ollea A J, editors. Mechanisms of microbial pathogenicity. Cambridge, United Kingdom: Cambridge University Press; 1955. pp. 57–77. [Google Scholar]
- 110.MacFarlane M G, Knight B C J G. The biochemistry of bacterial toxins. I. Lecithinase activity of Cl. welchii toxins. Biochem J. 1941;35:884–902. doi: 10.1042/bj0350884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez J P, Gil M L, Lopez-Ribot J L, Chaffin W L. Serologic response to cell wall mannoproteins and proteins of Candida albicans. Clin Microbiol Rev. 1998;11:121–141. doi: 10.1128/cmr.11.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Masuda N, Kitamura N, Saito K. Primary structure of protein moiety of Penicillium notatum phospholipase B deduced from the cDNA. Eur J Biochem. 1991;202:783–787. doi: 10.1111/j.1432-1033.1991.tb16433.x. [DOI] [PubMed] [Google Scholar]
- 114.May A K, Sawyer R G, Gleason T, Whitworth A, Pruett T L. In vivo cytokine response to Escherichia coli alpha-hemolysin determined with genetically engineered hemolytic and nonhemolytic E. coli variants. Infect Immun. 1996;64:2167–2171. doi: 10.1128/iai.64.6.2167-2171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McDonel J L. Toxins of Clostridium perfringens types A, B, C, D, and E. In: Dorner F, Drews J, editors. Pharmacology of bacterial toxins. Oxford, United Kingdom: Pergamon Press; 1986. pp. 477–517. [Google Scholar]
- 116.McLain N, Dolan J W. Phospholipase D activity is required for dimorphic transition in Candida albicans. Microbiology. 1997;143:3521–3526. doi: 10.1099/00221287-143-11-3521. [DOI] [PubMed] [Google Scholar]
- 117.McNamara P J, Bradley G A, Songer J G. Targeted mutagenesis of the phospholipase D gene results in decreased virulence of Corynebacterium pseudotuberculosis. Mol Microbiol. 1994;12:921–930. doi: 10.1111/j.1365-2958.1994.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 118.McNamara P J, Cuevas W A, Songer J G. Toxic phospholipases D of Corynebacterium pseudotuberculosis, C. ulcerans and Arcanobacterium haemolyticum: cloning and sequence homology. Gene. 1995;156:113–118. doi: 10.1016/0378-1119(95)00002-n. [DOI] [PubMed] [Google Scholar]
- 119.Mengaud J, Barun-Breton C, Cossart P. Identification of a phosphatidylinositol-specific phospholipase C in Listeria monocytogenes: a novel type of virulence factor? Mol Microbiol. 1991;5:367–372. doi: 10.1111/j.1365-2958.1991.tb02118.x. [DOI] [PubMed] [Google Scholar]
- 120.Meyers D J, Berk R S. Characterization of phospholipase C from Pseudomonas aeruginosa as a potent inflammatory agent. Infect Immun. 1990;58:659–666. doi: 10.1128/iai.58.3.659-666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reference deleted.
- 122.Mirbod F, Banno Y, Ghannoum M A, Ibrahim A S, Nakashima S, Kitajima Y, Cole G T, Nozawa Y. Purification and characterization of lysophospholipase-transacylase (h-LPTA) from a highly virulent strain of Candida albicans. Biochim Biophys Acta. 1995;1257:181–188. doi: 10.1016/0005-2760(95)00072-k. [DOI] [PubMed] [Google Scholar]
- 123.Mitsui K, Mitsui N, Hase J. Clostridium perfringens exotoxins. I. Purification and properties of the α-toxin. Jpn J Exp. 1973;43:65–80. [PubMed] [Google Scholar]
- 124.Montes L F, Wilborn W H. Ultrastructural features of host-parasite relationship in oral candidiasis. J Bacteriol. 1981;96:1349–1356. doi: 10.1128/jb.96.4.1349-1356.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morris A J, Engebrecht J, Frohman M A. Structure and regulation of phospholipase D. Trends Pharmacol Sci. 1996;17:182–185. doi: 10.1016/0165-6147(96)10016-x. [DOI] [PubMed] [Google Scholar]
- 126.Möllby R. Bacterial phospholipases. In: Jeljaszewicz J, Wadström T, editors. Bacterial toxins and cell membranes. New York, N.Y: Academic Press, Inc.; 1978. pp. 367–424. [Google Scholar]
- 127.Möllby R, Nord C E, Wadstrom T. Biological activities contaminating preparations of phospholipase C (α-toxin) from Clostridium perfringens. Toxicon. 1973;11:139–147. doi: 10.1016/0041-0101(73)90075-5. [DOI] [PubMed] [Google Scholar]
- 128.Reference deleted.
- 129.Reference deleted.
- 130.Nakashima S, Matsuda Y, Akao Y, Yoshimura S, Sakai H, Hayakawa K, Andoh M, Nozawa Y. Molecular cloning and chromosome mapping of rat phospholipase D genes, Pld1a, Pld1b and Pld2. Cytogenet Cell Genet. 1997;79:109–113. doi: 10.1159/000134694. [DOI] [PubMed] [Google Scholar]
- 131.Odds F. Candida and candidosis. London, United Kingdom: Bailliere Tindall; 1988. Candidosis of the gastrointestinal tract; pp. 156–163. [Google Scholar]
- 132.Odds F, Abbott A. A simple system for presumptive identification of Candida albicans and differentiation of strains within the species. Sabouraudia. 1980;18:301–307. [PubMed] [Google Scholar]
- 133.Odds F C. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- 134.Ohsaka A, Tsuchiya M, Oshio C, Miyari M, Suzuki K, Yamakawa Y. Aggregation of platelets in the mesenteric microcirculation of the rat induced by α-toxin (phospholipase C) of Clostridium perfringens. Toxicon. 1978;16:333–341. doi: 10.1016/0041-0101(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 135.Oishi K, Raynor R L, Charp P A, Kuo J F. Regulation of protein kinase C by lysophospholipids. Potential role in signal transduction. J Biol Chem. 1988;263:6865–6871. [PubMed] [Google Scholar]
- 136.Okabe A, Shimizu T, Hayashi H. Cloning and sequencing of a phospholipase C gene of Clostridium perfringens. Biochem Biophys Res Commun. 1989;160:33–39. doi: 10.1016/0006-291x(89)91616-1. [DOI] [PubMed] [Google Scholar]
- 137.Ostroff R M, Vasil A I, Vasil M L. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J Bacteriol. 1990;172:5915–5923. doi: 10.1128/jb.172.10.5915-5923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ostroff R M, Wretlind B, Vasil M L. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect Immun. 1989;57:1369–1373. doi: 10.1128/iai.57.5.1369-1373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Perfect J R. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob Agents Chemother. 1996;40:1577–1583. doi: 10.1128/aac.40.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pfaller M, Wenzel R. Impact of the changing epidemiology of fungal infections in the 90's. Eur J Clin Microbiol Infect Dis. 1992;11:287–291. doi: 10.1007/BF01962067. [DOI] [PubMed] [Google Scholar]
- 141.Pfaller M A, Zhang J, Messer S A, Brandt M E, Hajjeh R A, Jessup C J. In vitro activities on voriconazole, fluconazole, and itraconazole against 566 clinical isolates of Cryptococcus neoformans from the United States and Africa. Antimicrob Agents Chemother. 1998;43:169–171. doi: 10.1093/oxfordjournals.jac.a020873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pickard R T, Chiou X G, Strifler B A, DeFelippis M R, Hyslop P A, Tebbe A L, Yee Y K, Reynolds L J, Dennis E A, Kramer R M, Sharp J D. Identification of essential residues for the catalytic function of 85-kDa cytosolic phospholipase A2. J Biol Chem. 1996;271:19225–19231. doi: 10.1074/jbc.271.32.19225. [DOI] [PubMed] [Google Scholar]
- 143.Pittet D, Tarrara D, Wenzel R P. Nosocomial bloodstream infection in critically ill patients: excess length of stay, extra costs, and attributable mortality. In: Rex J, Anderson M D, editors. Serious Candida Infections: risk factors, treatment, and prevention. Selected readings: focus on fluconazole. New York, N.Y: Medical Information Press, Pfizer, Inc.; 1985. pp. 17–24. [Google Scholar]
- 144.Portnoy D A. Cellular biology of Listeria monocytogenes infection. In: Miller V L, Kaper J B, Portnoy D A, Isberg R R, editors. Molecular genetics of bacterial pathogenesis. Washington, D.C.: American Society for Microbiology; 1994. pp. 279–293. [Google Scholar]
- 145.Prakobphol A, Leffler H, Fisher S J. Specific adherence of Candida tropicalis to lysophospholipids. Biochemistry. 1998;33:9496–9503. doi: 10.1021/bi00198a015. [DOI] [PubMed] [Google Scholar]
- 146.Price M F, LaRocco M T, Gentry L O. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob Agents Chemother. 1994;38:1422–1424. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Price M F, Wilkinson I D, Gentry L O. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia. 1982;20:7–14. doi: 10.1080/00362178285380031. [DOI] [PubMed] [Google Scholar]
- 148.Projan S J, Kornblum J, Kreiswirth B, Moghazeh S L, Eiser W, Novick R P. Nucleotide sequence of the β-hemolysin gene of Staphylococcus aureus. Nucleic Acids Res. 1989;17:3305. doi: 10.1093/nar/17.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pugh D, Cawson R A. The cytochemical localization of phospholipase in Candida albicans infecting the chick chorio-allantoic membrane. Sabouraudia. 1977;15:29–35. [PubMed] [Google Scholar]
- 150.Pugh D, Cawson R A. The cytochemical localization of phospholipase A and lysophospholipase in Candida albicans. Sabouraudia. 1975;13:110–115. doi: 10.1080/00362177585190181. [DOI] [PubMed] [Google Scholar]
- 151.Qin W, Pappan K, Wang X. Molecular heterogeneity of phospholipase D (PLD): cloning of PLDγ and regulation of plant PLDγ, -β, and -α by polyphosphoinositides and calcium. J Biol Chem. 1997;272:28267–28273. doi: 10.1074/jbc.272.45.28267. [DOI] [PubMed] [Google Scholar]
- 152.Rahmet-Alla M, Rowley A F. Studies on the cellular defense reactions of the madeira cockroach, Leucophaea maderae: in vitro phagocytosis of different strains of Bacillus cereus and their effect on hemocyte viability. J Invertebr Pathol. 1990;55:350–356. doi: 10.1016/0022-2011(90)90078-k. [DOI] [PubMed] [Google Scholar]
- 153.Ravdin J, Murphy C, Guerrant R, Long-Krug S. Effect of antagonists of calcium and phospholipase A on the cytopathogenicity of E. histolytica. J Infect Dis. 1985;152:542–549. doi: 10.1093/infdis/152.3.542. [DOI] [PubMed] [Google Scholar]
- 154.Ravdin J I, Croft B Y, Guerrant R L. Cytopathogenic mechanisms of Entamoeba histolytica. J Exp Med. 1998;152:377. doi: 10.1084/jem.152.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reiss E, Morrison C J. Nonculture methods for diagnosis of disseminated candidiasis. Clin Microbiol Rev. 1993;6:311–323. doi: 10.1128/cmr.6.4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rex J H, Bennett J E, Sugar A M, Pappas P G, van der Horst C M, Edwards J E, et al. A randomized trial comparing fluconazole with amphotericin B for the treatment of candidemia in patients without neutropenia. N Engl J Med. 1994;331:1325–1330. doi: 10.1056/NEJM199411173312001. [DOI] [PubMed] [Google Scholar]
- 157.Rogolsky M. Nonenteric toxins of Staphylococcus aureus. Microbiol Rev. 1979;43:320–360. doi: 10.1128/mr.43.3.320-360.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rose K, Rudge S A, Frohman M A, Morris A J, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rosenberg P. Bacterial and snake venom phospholipases: enzymatic probes in the study of structure and function in bioelectrically excitable tissues. In: Ohsaka A, Hayashi K, Sawai Y, editors. Animal, plant, and microbial toxins. Vol. 2. New York, N.Y: Plenum Press; 1976. pp. 229–262. [Google Scholar]
- 160.Saffer L D, Krug S A L, Schwartzman J D. The role of phospholipase in host cell penetration by Toxoplasma gondii. Am J Trop Med Hyg. 1989;40:145–149. doi: 10.4269/ajtmh.1989.40.145. [DOI] [PubMed] [Google Scholar]
- 161.Saffer L D, Schwartzman J D. A soluble phospholipase of Toxoplasma gondii associated with host cell penetration. J Protozool. 1991;38:454–460. doi: 10.1111/j.1550-7408.1991.tb04816.x. [DOI] [PubMed] [Google Scholar]
- 162.Saint-Joanis B, Garnier T, Cole S. Gene cloning shows the alpha-toxin of Clostridium perfringens to contain both sphingomyelinase and lecithinase activities. Mol Gen Genet. 1989;219:453–460. doi: 10.1007/BF00259619. [DOI] [PubMed] [Google Scholar]
- 163.Saito K, Kates M. Substrate specificity of a highly purified phospholipase B from Penicillium notatum. Biochim Biophys Acta. 1974;369:245–253. doi: 10.1016/0005-2760(74)90255-0. [DOI] [PubMed] [Google Scholar]
- 164.Saito K, Sugatani J, Okumura T. Phospholipase B from Penicillium notatum. Methods Enzymol. 1991;197:446–456. doi: 10.1016/0076-6879(91)97170-4. [DOI] [PubMed] [Google Scholar]
- 165.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 166.Salyers A, Witt D. Virulence factors that damage the host. In: Salyers A, Witt D, editors. Bacterial pathogenesis: a molecular approach. Washington, D.C.: ASM Press; 1994. pp. 47–62. [Google Scholar]
- 167.Salyers A, Witt D. Virulence factors that promote colonization. In: Salyers A, Witt D, editors. Bacterial pathogenesis: a molecular approach. Washington, D.C.: ASM Press; 1994. pp. 30–46. [Google Scholar]
- 168.Samaranayake L P, Raeside J M, MacFarlane T W. Factors affecting the phospholipase activity of Candida species in vitro. Sabouraudia. 1984;22:201–207. [PubMed] [Google Scholar]
- 169.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sanglard D, Hube B, Monod M, Odds F, Gow N A R. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Scherwitz C. Ultrastructure of human cutaneous candidosis. J Investig Dermatol. 1982;78:200–205. doi: 10.1111/1523-1747.ep12506451. [DOI] [PubMed] [Google Scholar]
- 172.Selik R M, Starcher E T, Curran J W. Opportunistic diseases reported in AIDS patients: frequencies, association, and trends. AIDS. 1987;1:175–182. [PubMed] [Google Scholar]
- 173.Serhan C N, Haeggstrom J Z, Leslie C C. Lipid mediator networks in cell signaling: update and impact of cytokines. FASEB J. 1996;10:1147–1158. doi: 10.1096/fasebj.10.10.8751717. [DOI] [PubMed] [Google Scholar]
- 174.Seshan, K. R., J. C. Vitullo, S. D. Leidich, C. Jessup, G. T. Cole, and M. A. Ghannoum. A genetically defined phospholipase B-deficient Candida albicans mutant is less virulent in the oral-intragastric infant mouse model. Submitted for publication.
- 175.Sheehan D J, Hitchcock C A, Sibley C M. Current and emerging azole antifungal agents. Clin Microbiol Rev. 1999;12:40–79. doi: 10.1128/cmr.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shimizu S, Tani Y, Yamada H, Tabata M, Murachi T. Enzymatic determination of serum-free fatty acids: a colorimetric method. Anal Biochem. 1980;107:193–198. doi: 10.1016/0003-2697(80)90511-4. [DOI] [PubMed] [Google Scholar]
- 177.Silverman D J, Santucci L A, Meyers N, Sekeyova Z. Penetration of host cells by Rickettsia rickettsii appears to be mediated by a phospholipase of rickettsial origin. Infect Immun. 1992;60:2733–2740. doi: 10.1128/iai.60.7.2733-2740.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. The two distinct phospholipase C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Smith L D S. Virulence factors of Clostridium perfringens. Rev Infect Dis. 1979;1:254–262. doi: 10.1093/clinids/1.2.254. [DOI] [PubMed] [Google Scholar]
- 180.Soll D. Candida albicans. In: Szaniszlo P, editor. Fungal dimorphism with emphasis on fungi pathogenic to humans. New York, N.Y: Plenum Press; 1985. pp. 167–195. [Google Scholar]
- 181.Soll D. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Soll D R. Gene regulation during high frequency switching in Candida albicans. Microbiology. 1997;143:279–288. doi: 10.1099/00221287-143-2-279. [DOI] [PubMed] [Google Scholar]
- 183.Songer J G. Bacterial phospholipases and their role in virulence. Trends Microbiol. 1997;5:156–161. doi: 10.1016/S0966-842X(97)01005-6. [DOI] [PubMed] [Google Scholar]
- 184.Soucek A, Michalec C, Souckova A. Identification and characterization of a new enzyme of the group ‘phospholipase D’ isolated from Corynebacterium ovis. Biochim Biophys Acta. 1971;227:116–128. doi: 10.1016/0005-2744(71)90173-2. [DOI] [PubMed] [Google Scholar]
- 185.Sugahara T, Takahashi T, Yamaya S, Ohsaka A. In vitro aggregation of platelets induced by α-toxin (phospholipase C) of Clostridium perfringens. Jpn J Med Sci Biol. 1976;29:255–263. [PubMed] [Google Scholar]
- 186.Sugahara T, Takahashi T, Yamaya S, Ohsaka A. Vascular permeability increase by α-toxin (phospholipase C) of Clostridium perfringens. Toxicon. 1977;15:81–87. doi: 10.1016/0041-0101(77)90074-5. [DOI] [PubMed] [Google Scholar]
- 187.Sugimoto H, Yamashita S. Purification, characterization, and inhibition by phosphatidic acid of lysophospholipase transacylase from rat liver. J Biol Chem. 1994;269:6252–6258. [PubMed] [Google Scholar]
- 188.Sugiyama Y, Nakashima S, Mirbod F, Kanoh H, Kitajima Y, Ghannoum M A, Nozawa Y. Molecular cloning of a second phospholipase B gene, caPLB2 from Candida albicans. Med Mycol. 1999;37:61–67. [PubMed] [Google Scholar]
- 189.Sung T-C, Roper R L, Zhang Y, Rudge S A, Temel R, Hammond S M, Morris A J, Moss B, Engebrecht J, Frohman M A. Mutagenesis of phospholipase D defines a super-family including a trans-Golgi viral protein required for pox-virus pathogenicity. EMBO J. 1997;16:4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Swenson C E, Perkins W R, Roberts P, Ahmad I, Stevens R, Stevens D A, Janoff A S. In vitro and in vivo antifungal activity of amphotericin B lipid complex: are phospholipases important? Antimicrob Agents Chemother. 1998;42:767–771. doi: 10.1128/aac.42.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Taguchi R, Ikezawa H. Phospholipase C from Clostridium novyi type A.I. Biochim Biophys Acta. 1975;409:75–85. doi: 10.1016/0005-2760(75)90082-x. [DOI] [PubMed] [Google Scholar]
- 192.Taguchi R, Ikezawa H. Phosphatidylinositol-specific phospholipase C from Clostridium novyi type A. Arch Biochem Biophys. 1978;186:196–201. doi: 10.1016/0003-9861(78)90480-0. [DOI] [PubMed] [Google Scholar]
- 193.Takahashi M, Banno Y, Nozawa Y. Secreted Candida albicans phospholipases: purification and characterization of two forms of lysophospholipase-transacylase. J Med Vet Mycol. 1991;29:193–204. [PubMed] [Google Scholar]
- 194.Titball R W. Bacterial phospholipases C. Microbiol Rev. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Titball R W, Hunter S E C, Martin K L, Morris B C, Shuttleworth A D, Rubidge T, Anderson D W, Kelly D C. Molecular cloning and nucleotide sequence of the alpha-toxin (phospholipase C) of Clostridium perfringens. Infect Immun. 1989;57:367–376. doi: 10.1128/iai.57.2.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Titball R W, Leslie D L, Harvey S, Kelly D. Haemolytic and sphingomyelinase activities of Clostridium perfringens alpha-toxin are dependent on a domain homologous to that of an enzyme from the human arachidonic acid pathway. Infect Immun. 1991;59:1872–1874. doi: 10.1128/iai.59.5.1872-1874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tso J Y, Siebel C. Cloning and expression of the phospholipase C gene from Clostridium perfringens and Clostridium bifermentans. Infect Immun. 1989;57:468–476. doi: 10.1128/iai.57.2.468-476.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Turnbull P C, Kramer J, Melling J. Bacillus. In: Topley W W C, Wilson G S, editors. Topley and Wilson's principles of bacteriology, virology and immunity. 8th ed. Vol. 2. London, United Kingdom: Edward Arnold; 1990. pp. 188–210. [Google Scholar]
- 199.Turnbull P C B. Bacillus cereus toxins. In: Dorner F, Drews J, editors. Pharmacology of bacterial protein toxins. New York, N.Y: Permagon Press; 1986. pp. 397–448. [Google Scholar]
- 200.Vadas P, Browning J, Edelson J, Pruzanski W. Extracellular phospholipase A2 expression and inflammation: the relationship with associated disease states. J Lipid Mediators. 1993;8:1–30. [PubMed] [Google Scholar]
- 201.Vanden Bossche H, Marichal H P, Odds F, Le Jeune L, Coene M-C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Vasil M L, Krieg D P, Kuhns J S, Ogle J W, Shortridge V D, Ostroff R M, Vasil A I. Molecular analysis of haemolytic and phospholipase C activities of Pseudomonas cepacia. Infect Immun. 1990;58:4020–4029. doi: 10.1128/iai.58.12.4020-4029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vazquez-Boland J A, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Vidotto V, Sinicco A, Di Fraia D, Cardaropoli S, Aoki S, Ito-Kuwa S. Phospholipase activity in Cryptococcus neoformans. Mycopathologia. 1997;136:119–123. doi: 10.1007/BF00438916. [DOI] [PubMed] [Google Scholar]
- 205.Reference deleted.
- 206.Waksman M, Eli Y, Liscovitch M, Gerst J E. Identification and characterization of a gene encoding phospholipase D activity in yeast. J Biol Chem. 1998;271:2361–2364. doi: 10.1074/jbc.271.5.2361. [DOI] [PubMed] [Google Scholar]
- 207.Walker D H, Firth W T, Ballard J G, Hegarty B C. Role of phospholipase-associated penetration mechanism in cell injury by Rickettsia rickettsii. Infect Immun. 1983;40:840–842. doi: 10.1128/iai.40.2.840-842.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Walker T S, Brown J S, Hoover C S, Morgan D A. Endothelial prostaglandin secretion: effects of typhus rickettsiae. J Infect Dis. 1990;162:1136–1144. doi: 10.1093/infdis/162.5.1136. [DOI] [PubMed] [Google Scholar]
- 209.Wang X, Xu L, Xheng L. Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L. J Biol Chem. 1994;269:20312–20317. [PubMed] [Google Scholar]
- 210.Reference deleted.
- 211.Watanabe Y, Yashiki Y, Sultana G N-N, Maruyama M, Kangawa K, Tamai Y. Cloning and sequencing of phospholipase B gene from the yeast Torulaspora delbrueckii. FEMS Microbiol Lett. 1994;124:29–34. doi: 10.1111/j.1574-6968.1994.tb07257.x. [DOI] [PubMed] [Google Scholar]
- 212.Wazny T K, Mummaw N, Styrt B. Degranulation of human neutrophils after exposure to bacterial phospholipase C. Eur J Clin Microbiol Infect Dis. 1990;9:830–832. doi: 10.1007/BF01967385. [DOI] [PubMed] [Google Scholar]
- 213.Werner H. Untersuchungen uber die Lipase-Aktivitat bei Hefen und Hefeahnlichen Pilzen. Zentbl Bakteriol Mikrobiol Hyg 1 Abt Orig A. 1966;200:113. [PubMed] [Google Scholar]
- 214.Wickerham L J. Apparent increase in frequency of infections involving Torulopsis glabrata: procedure for its identification. JAMA. 1957;165:47–48. doi: 10.1001/jama.1957.72980190007010b. [DOI] [PubMed] [Google Scholar]
- 214a.Williamson E D, Titball R W. A genetically engineered vaccine against α-toxin of Clostridium perfringens protects mice against experimental gas gangrene. Vaccine. 1993;11:1253–1258. doi: 10.1016/0264-410x(93)90051-x. [DOI] [PubMed] [Google Scholar]
- 215.Williamson M I, Samaranayake L P, MacFarlane T W. Phospholipase activity as a criterion for biotyping Candida albicans. J Med Vet Mycol. 1986;24:415–417. [PubMed] [Google Scholar]
- 216.Willis T A. Clostridia of wound infection. London, United Kingdom: Butterworths; 1969. [Google Scholar]
- 217.Willis T A. Anaerobic bacteriology: clinical and laboratory practice. 3rd ed. London, United Kingdom: Butterworths; 1977. [Google Scholar]
- 218.Wingard J R. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995;20:115–125. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]
- 219.Wingard J R, Merz W G, Rinaldi M G, Miller C B, Karp J E, Saral R. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob Agents Chemother. 1993;37:1847–1849. doi: 10.1128/aac.37.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Winkler H, Miller E. Phospholipase A activity in the hemolysis of sheep and human erythrocytes by Rickettsia prowazekii. Infect Immun. 1980;29:316–321. doi: 10.1128/iai.29.2.316-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Winkler H, Miller E. Immediate cytotoxicity and phospholipase A: the role of phospholipase in the internalization of Rickettsia prowazekii and L-cells. In: Burgdorfer W, Anacker R, editors. Proceedings of the Rocky Mountain Laboratory Conference on Rickettsiae and Rickettsial Diseases. New York, N.Y: Academic Press, Inc.; 1981. pp. 327–333. [Google Scholar]
- 222.Winkler H H. Early events in the interaction of the obligate intracytoplasmic parasite, Rickettsia prowazekii, with eucaryotic cells: entry and lysis. Ann Inst Pasteur Microbiol. 1986;137A:333–336. doi: 10.1016/s0769-2609(86)80044-8. [DOI] [PubMed] [Google Scholar]
- 223.Witt W, Bruller H-J, Falker G, Fuhrmann G F. Purification and properties of a phospholipid acyl hydrolase from plasma membranes of Saccharomyces cerevisiae. Biochim Biophys Acta. 1982;711:403–410. doi: 10.1016/0005-2760(82)90054-6. [DOI] [PubMed] [Google Scholar]
- 224.Witt W, Mertsching A, Konig E. Secretion of phospholipase B from Saccharomyces cerevisiae. Biochim Biophys Acta. 1984;795:117–124. doi: 10.1016/0005-2760(84)90111-5. [DOI] [PubMed] [Google Scholar]
- 225.Witt W, Schweingruber M E, Mertsching A. Phospholipase B from the plasma membrane of Saccharomyces cerevisiae. Separation of two forms with different carbohydrate content. Biochim Biophys Acta. 1984;795:108–116. doi: 10.1016/0005-2760(84)90110-3. [DOI] [PubMed] [Google Scholar]
- 226.Wysong D R, Christin L, Sugar A M, Robbins P W, Diamond R D. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun. 1998;66:1953–1961. doi: 10.1128/iai.66.5.1953-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yun Tso J, Siebel C. Cloning and expression of the phospholipase C gene from Clostridium perfringens and Clostridium bifermentans. Infect Immun. 1989;57:468–476. doi: 10.1128/iai.57.2.468-476.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhao X J, McElhaney-Feser G E, Sheridan M J, Broedel S E., Jr Avirulence of Candida albicans FAS2 mutants in a mouse model of systemic candidiasis. Infect Immun. 1997;65:829–832. doi: 10.1128/iai.65.2.829-832.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zwaal R F A, Roelofsen B, Colley C M. Localization of red cell membrane constituents. Biochim Biophys Acta. 1973;300:159–182. doi: 10.1016/0304-4157(73)90003-8. [DOI] [PubMed] [Google Scholar]