Abstract
Aims: Oral bacteria have been reported to be associated with the pathogenesis of atherosclerosis; however, the relationship between the oral microbiota and atherosclerosis remains unclear. The present study aimed to investigate whether or not salivary microbiota of patients with atherosclerotic cardiovascular disease (ACVD) differs from that of subjects without ACVD, and to characterize the salivary microbiota of patients with ACVD.
Methods: This study included 43 patients with ACVD and 86 age- and sex-matched non-ACVD individuals. 16S rRNA metagenomic analysis were performed using DNA isolated from the saliva samples of the participants. To select unique operational taxonomic unit (OTU) sets of ACVD, we conducted the random forest algorithm in machine learning, followed by confirmation via 10-fold cross-validation
Results: There was no difference in richness or evenness between the ACVD and non-ACVD groups (alpha diversity; observed OTU index, p =0.503; Shannon’s index, p =0.478). However, significant differences were found in the overall salivary microbiota structure (beta diversity; unweighted UniFrac distances, p =0.001; weighted UniFrac distances, p =0.001). The Actinobacteria phylum was highly abundant in patients with ACVD, while the Bacteroidetes phylum was less abundant. The random forest classifier identified 43 OTUs as an optimal marker set of ACVD. In a 10-fold cross validation using the validation data, an area under the curve (AUC) of 0.933 (95% CI, 0.855–1.000) was obtained.
Conclusions: The salivary microbiota in patients with ACVD was distinct from that of non-ACVD individuals, indicating that the salivary microbiota may be related to ACVD.
Keywords: Oral microbiota, Saliva, Atherosclerosis, 16S rRNA metagenomic analysis
Introduction
Growing evidence suggests that oral health plays an important role in the pathogenesis of atherosclerotic cardiovascular disease (ACVD). Several species of oral bacteria that cause periodontitis and dental caries have been found in atherosclerotic plaques 1 - 4) . Animal models infected by oral or intravenous periodontal pathogens, such as Porphyromonas gingivalis , have shown enhanced development of atherosclerosis 5 , 6) . It is possible that the bacteria from the oral cavity might invade the bloodstream, translocate to the blood vessel, cause inflammation, and increase the risk of ACVD 7) . Epidemiological evidence suggests that periodontal disease, an oral infectious disease, occurs in response to the presence of specific bacteria in the oral cavity that are associated with ACVD 8 , 9) . The causal mechanism underlying the association between these two disease conditions is not yet clearly elucidated; periodontopathic bacterial infection and the subsequent inflammatory response are speculated to play a role in the pathogenesis of ACVD 10) . These findings indicate that the oral microbial environment might contribute to the development of ACVD.
More than 700 species of bacteria exist in the human oral cavity and constitute the oral microbiota 11 , 12) . Current advances in molecular methods have revealed that the oral microbiota plays an important role in the human microbial community and health status 13) . Emerging evidence has shown alterations in the oral microbiota of patients with various diseases. Moreover, the microbiota has been identified as a potential diagnostic and prognostic biomarker for some diseases 14 - 16) . Although past cross-sectional studies have identified individual oral bacterial species associated with ACVD 17 - 19) , no studies have been conducted to identify a group of bacteria closely associated with ACVD from the oral microbiota as a whole. There are several lines of evidence suggesting that the gut microbiota plays a role in ACVD development. The gut microbiota might influence atherosclerosis through activation of the immune system, the alteration of cholesterol metabolism, and production of bacterial metabolites such as Trimethylamine N-oxide (TMAO), which contribute to atherosclerosis by increasing foam cell formation in macrophages, decreasing reverse cholesterol transport, and enhancing platelet hyperreactivity 20 , 21) . In addition, several studies indicate that the gut microbiota may be affected by oral bacteria, leading to dysbiosis 22 - 24) . These observations suggest that oral microbiota could be linked to ACVD.
Aim
In the present study, we compared the salivary bacterial diversity and composition in patients with ACVD with those in the age- and sex-matched control subjects, using bacterial 16S ribosomal RNA (16S rRNA) gene sequencing. Furthermore, a group of bacteria was utilized to distinguish patients with ACVD from control subjects through machine learning techniques. The aim of the present study was to investigate whether or not salivary microbiota of patients with ACVD differ from that of subjects without ACVD, and to characterize the salivary microbiota of patients with ACVD.
Methods
Participants
This study was conducted in accordance with the Helsinki Declaration and its latest amendments and was approved by the Ethics Committee of Osaka Medical College, Takatsuki city, Japan (approval no. 2145). Written informed consent was obtained from all participants.
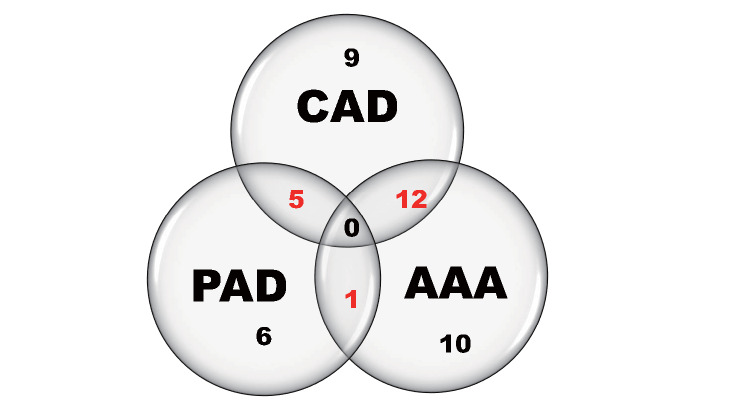
Study participants included 129 Japanese individuals, including 43 patients with ACVD (72.1% male) between 63-86 years of age (mean 74) and 86 age- and sex-matched non-ACVD control subjects. The exclusion criteria in both groups were as follows: participants with ongoing infectious diseases, autoimmune diseases, renal or hepatic failure, under current treatment of cancer, as well as use of antibiotics within one month of sample collection. The ACVD group included patients diagnosed with coronary artery disease (CAD), abdominal aortic aneurysm (AAA), and/or peripheral artery disease (PAD) at the Department of Cardiology of Osaka Medical College Hospital, Takatsuki city, Japan between December 2017 and August 2019. The exclusion criteria were acute coronary syndrome, inflammatory aortic aneurysm, and critical limb ischemia, neither of which can be considered a chronic pathological condition associated with atherosclerosis. The number of patients with each diagnosis is shown using Venn diagrams in Fig.1 . For the non-ACVD control group, we sampled 86 age- and sex-matched control subjects (ratio 1:2, age difference within 3 year) from 277 community-dwelling individuals who participated in the Takatsuki study from March 2018 to November 2019, after excluding individuals with self-reported medical history for coronary or other vascular diseases. Medical history and information about medication were collected from the medical records for patients with ACVD and from a questionnaire for non-ACVD subjects.
Fig.1. Venn diagrams of the group of patients with ACVD.
The patients with ACVD in this study included 26 patients with coronary artery disease (CAD), 12 with peripheral artery disease (PAD), and 23 with abdominal aortic aneurysm (AAA) patients. The unscaled Venn diagrams show sets of patients in the study. There were 26 patients with both CAD and PAD, 12 patients with both CAD and AAA, and one patient with both PAD and AAA. In addition, nine patients only had CAD, and six patients only had PAD, and 10 patients only had AAA, making a total of 43 patients.
Oral Sample Collection and Oral Examination
Saliva samples were collected from participants in the morning (at least 2 h after brushing or consumption of meals) by the cotton swab method, as previously described 25) using the saliva collection system SalivaBio ® Oral Swab and Swab Storage Tube (Salimetrics, State College, PA, USA) prior to oral examination. The samples were immediately frozen after collection and stored at −80℃ prior to DNA extraction.
All participants were subjected to a full-mouth clinical examination by certified dentists. Their periodontal condition was evaluated by measuring the periodontal pocket depth (PPD) and by bleeding on probing (BOP) at six sites for all teeth; the periodontal inflammation surface area (PISA), a periodontal parameter that quantifies the amount of inflamed periodontal tissue 26) , was also calculated. A diagnosis of periodontitis was made following the recent diagnostic criteria for periodontal diseases based on the World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions 27) .
DNA Extraction, 16S rRNA Sequencing, and Taxonomic Classification
For DNA extraction, briefly, a total of 500 µL of sample, including 30 µL of the saliva sample and 470 µL of detergent reagent No. 10 (Kurabo Industries Ltd., Osaka, Japan) was transferred into a 2.0 mL tube. The samples were homogenized with 0.1 mm glass beads using a homogenizer, Disruptor Genie (Scientific Industries Inc., Bohemia, NY, USA), for 90 s. Bacterial genomic DNA from the homogenized samples was extracted using GENE PREP STAR PI-480 (Kurabo Industries Ltd.) according to the manufacturer’s instructions.
The V1–V2 region of the 16S rRNA gene was amplified for 25 cycles with KAPA HiFi Hot Start Ready Mix (Roche, Basel, Switzerland) using the primer set 27Fmod: 5´-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGRGTTTGATYMTGGCTCAG-3´ and 338R: 5´-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGCTGCCTCCCGTAGGAGT-3´.
Each library was prepared in accordance with the 16S metagenomic sequencing library preparation protocol (Illumina, CA, San Diego, USA). DNA was sequenced with 500 cycles using a MiSeq Reagent Kit v2 (Illumina). An average of 37,251 sequence reads with 250 bp paired-ends were denoised and quality filtered using DADA2 in Quantitative Insights Into Microbial Ecology 2 (QIIME2) version 2020.02 28) . After quality filtering, 3,893,333 sequences were obtained with a mean of 30,181 sequences per sample (min: 11,867; max: 66,936). A rarefaction minimum depth cut-off was chosen at 10,000, and this retained all the samples for downstream analysis 29) . The processed sequences were clustered into operational taxonomic units (OTUs) and defined according to a similarity cut-off of 97%. Representative sequences for each OTU were assigned against the curated Silva 132 reference database.
Statistical Analysis
For characteristics of the participants, categorical variables were presented using frequency counts, and intergroup comparisons were analyzed using Fisher’s exact tests or Mann–Whitney U tests. Continuous parametric data were compared using Student’s t -tests. The statistical software JMP pro 15.1.0 (SAS Institute Inc., Cary, NC, USA) was used for database construction and data analysis. A p -value <0.05 was considered significant.
The within-subject alpha diversity of bacterial communities was assessed by Shannon’s index and the observed OTU index, and it was compared between groups using the Kruskal-Wallis test. The between-subjects beta diversity was assessed based on the Bray-Curtis dissimilarity and the unweighted and weighted UniFrac distance metrics 30 , 31) . To visualize the global differences in the microbiome structure in the UniFrac analysis, we performed Principal Coordinate Analysis (PCoA). The significance of compositional differences between groups was assessed by permutational multivariate analysis of variance (PERMANOVA) 32) . QIIME2 was used for these analyses.
The linear discriminant analysis (LDA) effect size (LEfSe) algorithm was used to detect differentially abundant genera among the two groups 33) . All analyses were run with LEfSe’s α parameter for pairwise tests set to 0.05 and the threshold of the logarithmic score for LDA analysis set to 3.0. To characterize specific microbiota of ACVD patients, a model to predict ACVD based on microbial composition at the genus level was constructed using the Random Forest package in R version 3.3.1. The predictive power of the model was evaluated by a 10-fold cross-validation using 70% of the data as training data and 30% as validation data. Following model construction, the variable importance in the projection (VIP) of each genus was calculated and the candidate genera were selected to reflect the differences between the groups. A p -value of less than 0.01 was considered significant. The receiver operating characteristic curve (ROC) and area under the ROC curves (AUCs) of the predictive model were calculated.
Results
Characteristics of the Participants
The characteristics, medications, laboratory data, and oral condition of the participants in the ACVD and non-ACVD groups are summarized in Table 1 . Nearly 70.5% of the participants were male with a mean age of 75 y. The distribution of smoking habits in the following three categories: non-smoker, ex-smoker and current smoker, indicated statistical significance ( p =0.004). The percentage of current smokers was not different between the ACVD and non-ACVD groups ( p =0.083). There was no significant difference in body mass index (BMI) or the percentage of patients with hypertension between the ACVD and non-ACVD groups. The proportions of background medication between the two groups indicated statistical significance ( Table 1 ) . Oral conditions of the participants such as the number of teeth, denture usage, and PISA were significantly different between the two groups.
Table 1. Baseline characteristics and laboratory data of the study population ( n = 129) .
| Variables | ACVD ( n = 43) | Non-ACVD ( n = 86) | p value |
|---|---|---|---|
| Characteristic | |||
| Age (years) | 74.4±5.0 | 75.0±4.8 | 0.527 |
| Sex (male, %) | 72.1 | 69.8 | 0.840 |
| BMI (kg/m 2 ) | 23.6±2.5 | 22.7±2.8 | 0.075 |
| Never smoker (%) | 25.6 | 53.5 | 0.004 |
| Ex-smoker (%) | 60.5 | 41.9 | |
| Current smoker (%) | 14.0 | 4.7 | |
| Dyslipidemia (%) † | 51.2 | 29.1 | 0.020 |
| Hypertension (%) ‡ | 58.1 | 45.4 | 0.194 |
| Diabetes (%) § | 34.9 | 15.1 | 0.013 |
| Oral conditions | |||
| Number of teeth | 19.3±6.5 | 23.6±5.7 | <0.001 |
| Denture wearing (%) | 69.8 | 31.4 | <0.001 |
| Severe periodontitis (%) | 30.2 | 30.2 | 0.638 |
| PISA (cm 2 ) | 296.5±474.7 | 163.2±184.8 | 0.039 |
| Medications | |||
| Anti-diabetes drug (%) | 27.9 | 8.1 | 0.007 |
| Statin (%) | 65.1 | 11.6 | <0.001 |
| ACE-I/ARB (%) | 53.5 | 15.1 | <0.001 |
| Beta-blocker (%) | 34.9 | 1.2 | <0.001 |
| Calcium channel blocker (%) | 58.1 | 17.4 | <0.001 |
| PPI/H2 blocker (%) | 48.8 | 4.7 | <0.001 |
| Anti-coagulant (%) | 9.3 | 2.3 | 0.095 |
| Anti-platelet (%) | 65.1 | 4.7 | <0.001 |
| Laboratory data | |||
| HDL-C (mg/dL) | 53.3±15.7 | 66.6±16.9 | <0.001 |
| LDL-C (mg/dL) | 107.5±36.9 | 117.1±27.1 | 0.150 |
| TG (mg/dL) | 153.3±75.8 | 90.8±62.2 | <0.001 |
| HbA1c (NGSP, %) | 6.1±0.8 | 5.8±0.6 | 0.024 |
| CRP (mg/dL) | 0.55±1.46 | 0.05±0.08 | <0.001 |
Results are expressed as the mean±SD or percentage. p values are the results of comparisons against the non-ACVD group using Mann–Whitney U test, Student’s t -test, or Fisher’s exact test.
† Dyslipidemia is defined as LDL-C > 140 mg/dl, triglycerides > 150 mg/dl or use of anti-dyslipidemic drugs.
‡ Hypertension is defined as blood pressure > 140/90 mmHg or use of anti-hypertensive drugs.
§ Diabetes is defined as HbA1c > 6.5 % (NGSP) or use of oral anti-diabetic drugs or insulin therapy.
PISA, periodontal inflamed surface area;
Microbiota Composition
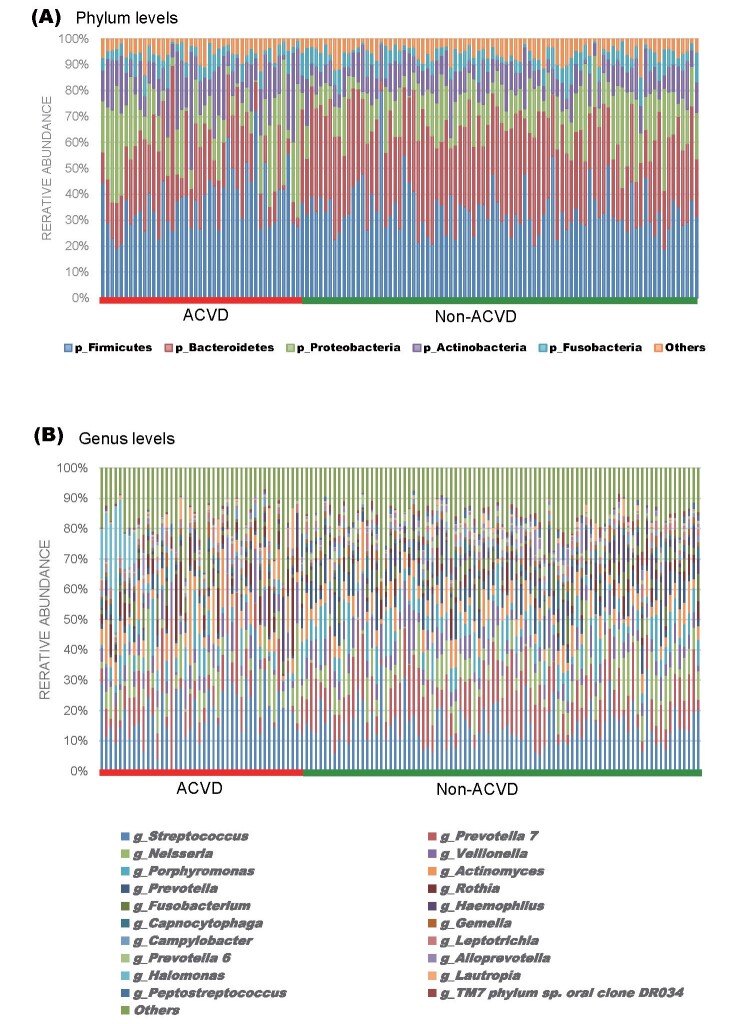
Salivary bacteria with a relative abundance of at least 0.1% in both groups were classified into 11 phyla, 18 classes, 37 orders, 71 families, and 144 genera. The predominant bacteria (>5% of the total sequences in either group) at the phylum level included Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria, which comprised 94.99% and 94.73% of the oral microbiota in ACVD and non-ACVD groups, respectively ( Fig.2A ) . At the genus level, the ACVD group was characterized by 136 genera, of which 22 were absent in the non-ACVD group, while the non-ACVD group included 124 genera, out of which 10 were absent in the ACVD group ( Supplemental Table 1 ) . A total of 39 genera were present in at least 50% of subjects in both groups, of which 37 were common to both the groups. The predominant bacteria at the genus level included Streptococcus with a mean relative abundance of 20.16% and 15.45%, and Prevotella7 with a mean relative abundance of 9.28% and 13.01% in the ACVD and non-ACVD groups, respectively. The 20 most abundant genera in the ACVD and non-ACVD groups accounted for 82.92% and 84.87%, respectively, of the total genera abundance in the two groups ( Fig.2B , Supplemental Table 2 ) .
Fig.2. Taxonomic composition of saliva microbiota in patients with ACVD and non-ACVD subjects.
Vertical bar plot showing the relative abundance of bacterial phyla (A) and genus (B) in each sample.
(A) Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria were the five most abundant phyla in all the subjects comprising 94.99% and 94.72% of the total bacterial communities in patients with ACVD and non-ACVD subjects, respectively.
(B) Bar plot of the 20 most abundant genera in patients with ACVD and non-ACVD subjects, revealing that the majority of genera in the “top 20” groups were common in all the subjects with the overall dominance of Streptococcus (blue) and Prevotella7 (orange).
Supplemental Table 1. Bacterial genus found only in ACVD or non-ACVD group.
| ACVD group | non-ACVD group |
|---|---|
| Genus | Genus |
| Halomonas | Unidentified genus in f_Lentimicrobiaceae |
| Delftia | Desulfovibrio |
| Uncultured bacterium in o_Bacteroidales; f_p-2534-18B5 gut group | Alysiella |
| Cutibacterium | Moraxella |
| Prevotella 9 | Chryseobacterium |
| Anaerobacillus | Streptobacillus |
| Stenotrophomonas | Clostridiales bacterium canine oral taxon 260 |
| Gracilibacteria bacterium oral taxon 871 | Candidatus Tammella |
| Pseudomonas | Acinetobacter |
| Staphylococcus | Faecalibacterium |
| Family XIII AD3011 group | |
| Peptoniphilus | |
| Uncultured bacterium in o_Izimaplasmatales | |
| Lachnospiraceae NK4A136 group | |
| Wolinella | |
| Bacillus | |
| Microbacterium | |
| Jonquetella | |
| Alphaproteobacteria bacterium canine oral taxon 081 | |
| Novosphingobium | |
| Endobacter | |
| Pelospora |
Supplemental Table 2. The 20 most abundant genera in ACVD and non-ACVD group.
| ACVD group | non-ACVD group | ||
|---|---|---|---|
| Genus |
Mean relative abundance (%) |
Genus |
Mean relative abundance (%) |
| Streptococcus | 20.16 | Streptococcus | 15.45 |
| Prevotella 7 | 9.22 | Prevotella 7 | 13.01 |
| Neisseria | 7.31 | Neisseria | 7.95 |
| Actinomyces | 7.00 | Porphyromonas | 6.84 |
| Veillonella | 6.28 | Veillonella | 6.53 |
| Rothia | 5.98 | Prevotella | 6.31 |
| Porphyromonas | 5.09 | Actinomyces | 5.63 |
| Halomonas | 3.35 | Fusobacterium | 3.79 |
| Fusobacterium | 3.07 | Haemophilus | 3.03 |
| Haemophilus | 2.39 | Rothia | 2.81 |
| Prevotella | 2.03 | Capnocytophaga | 1.71 |
| Leptotrichia | 1.70 | Gemella | 1.66 |
| Gemella | 1.57 | Prevotella 6 | 1.65 |
| Lautropia | 1.56 | Campylobacter | 1.50 |
| Campylobacter | 1.54 | Alloprevotella | 1.41 |
| Capnocytophaga | 1.52 | Leptotrichia | 1.38 |
| Corynebacterium | 0.84 | Peptostreptococcus | 1.22 |
| Prevotella 6 | 0.79 | TM7 phylum sp. oral clone DR034 | 1.12 |
| Alloprevotella | 0.78 | Candidatus Saccharimonas | 1.02 |
| Selenomonas 3 | 0.73 | Parvimonas | 0.86 |
Microbial Diversity in ACVD Patients and Non-ACVD Subjects
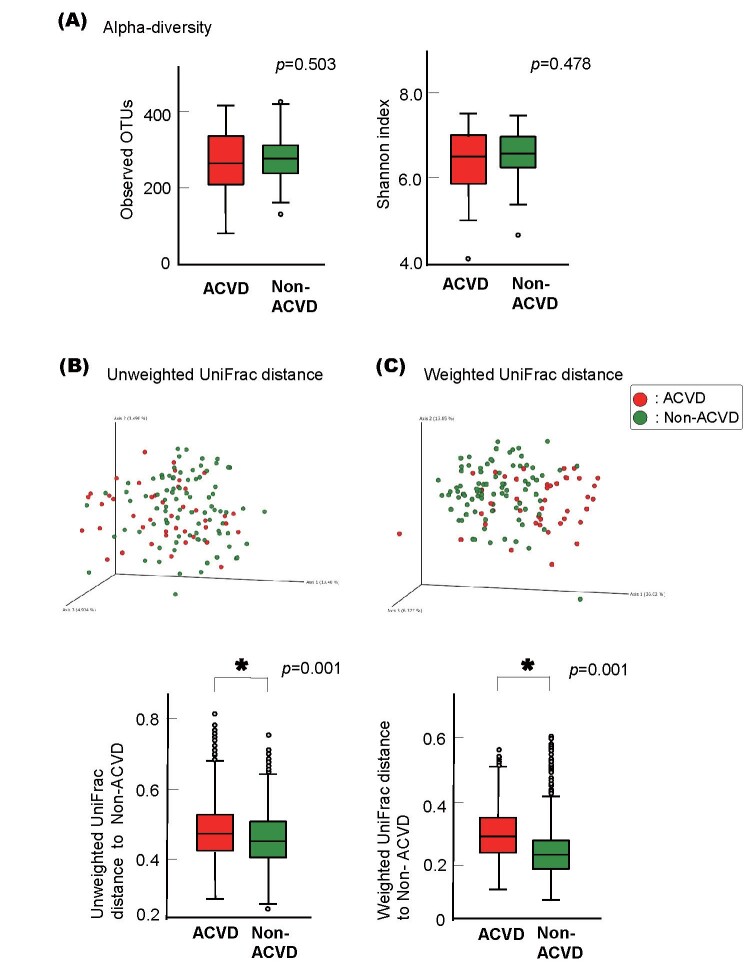
As shown in Fig.3A , no statistically significant differences were observed between the ACVD and non-ACVD groups in terms of alpha diversity (observed OTUs, p =0.503; Shannon’s index, p =0.478).
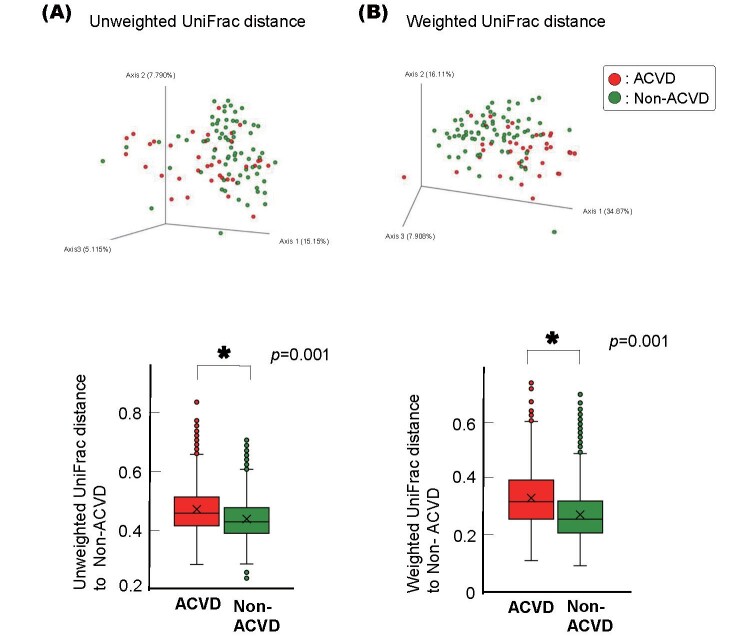
Fig.3. Alpha and Beta diversity of the ACVD and non-ACVD salivary microbiota.
(A) Observed operational taxonomic unit (OTU) index and Shannon index of the patients with ACVD ( n =43, red) compared with those of the non-ACVD subjects ( n =86, green). p values from Wilcoxon rank-sum test are shown.
(B) Unweighted UniFrac distances.
(C) Weighted UniFrac distances.
Principal coordinate analysis (PCoA) plot for the samples from 43 ACVD (red) and 86 non-ACVD (green) subjects. Box plots represent UniFrac distances to non-ACVD subjects from patients with ACVD (red columns) and non-ACVD subjects (green columns). * p <0.05 compared between groups from PERMANOVA, 999 permutations.
PCoA of the unweighted and weighted UniFrac distance metric-based results revealed that although the salivary microbiota showed an interindividual difference, the saliva samples differed between the two groups in 3D space ( Fig.3B, Fig.3C ) . This compositional variation that distinguished the ACVD and non-ACVD groups was verified by PERMANOVA based on the UniFrac data (999 permutations, p =0.001), showing that the bacterial communities of each group were more similar to each other than to the bacterial communities of the other group. PERMANOVA on Bray-Curtis metrics based on relative abundances of bacterial genera also revealed the difference between the two groups (999 permutations, p =0.001).
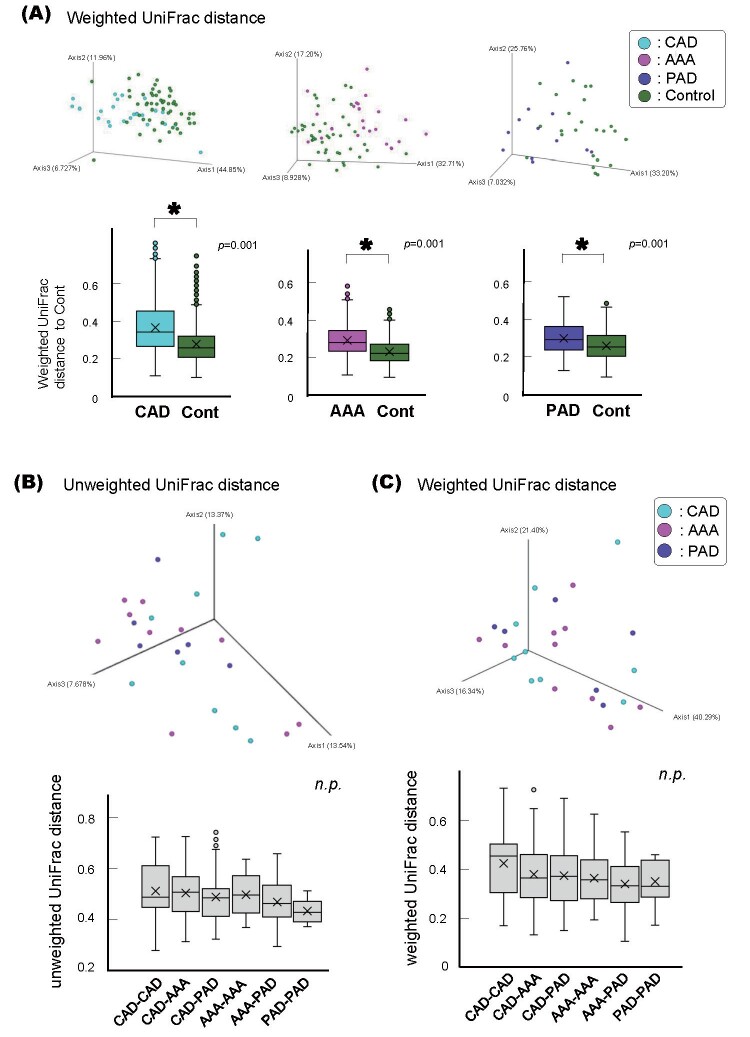
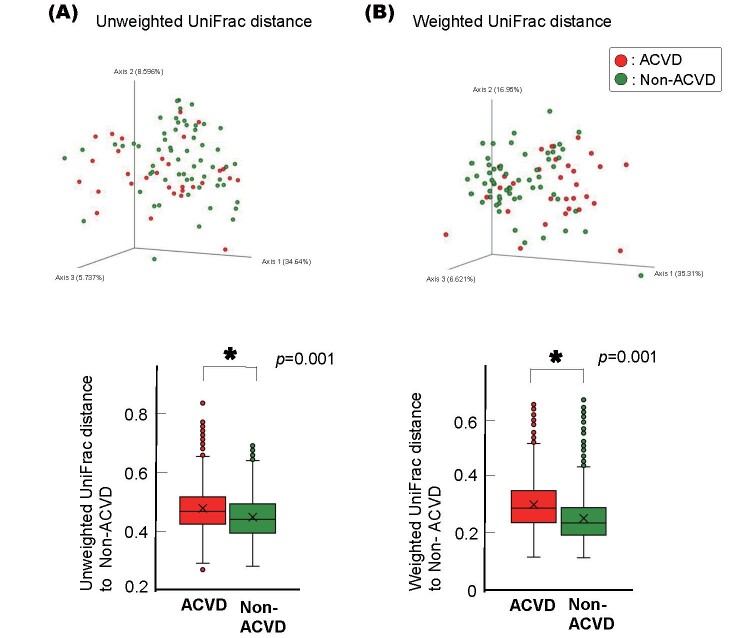
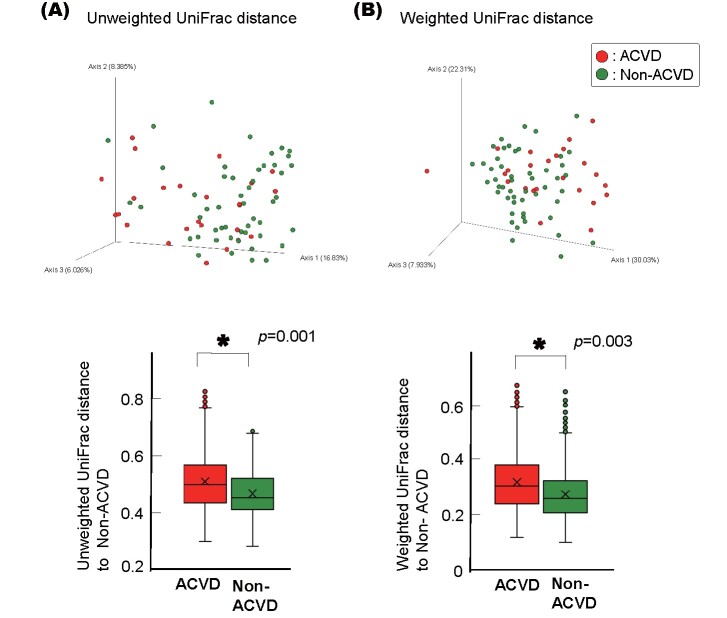
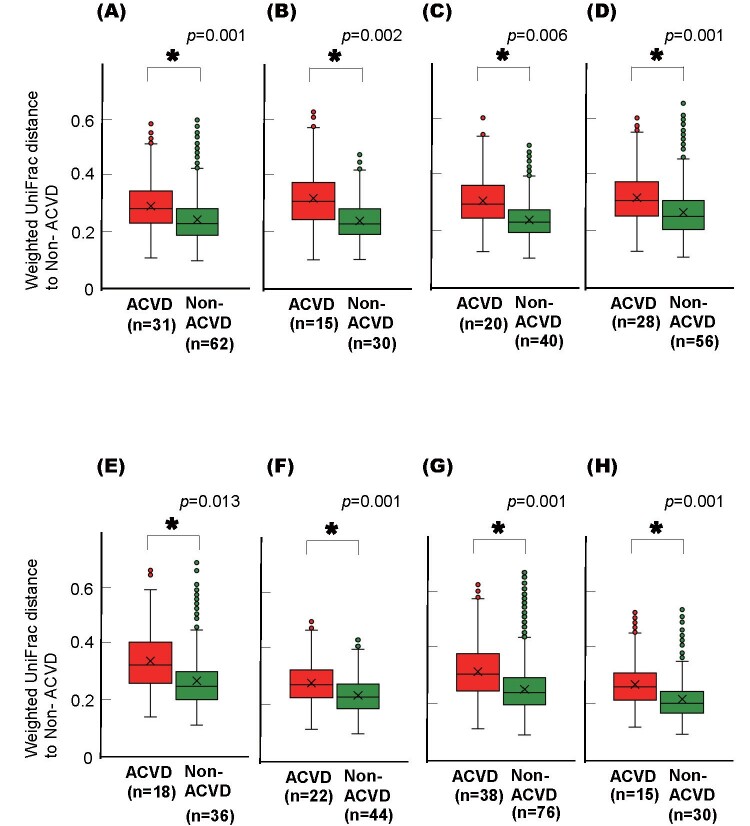
This difference between the ACVD and non-ACVD groups was retained when the ACVD group was first categorized into three sub-groups, i.e., patients with CAD, AAA, or PAD, and then compared with age- and sex-matched non-ACVD control subjects, respectively ( Supplemental Fig.1A ) . There was no difference among the CAD, AAA, and PAD groups ( Supplemental Fig.1B and C ) . The differences between the ACVD and non-ACVD groups were also retained when current smokers or subjects with poorly controlled diabetes mellitus were excluded from either group ( Supplemental Fig.2 and 3 ) . The difference between the ACVD and non-ACVD groups was significant even when the comparison was limited to those with 20 or more teeth or those who were not taking certain medications ( Supplemental Fig.4 and 5 ) .
Supplemental Fig.1. Beta diversities of the salivary microbiotas of the ACVD sub-groups and non-ACVD group.
(A) Weighted UniFrac distances of the ACVD sub-groups and non-ACVD control groups.
PCoA plot for the samples from 26 coronary artery disease (CAD; blue), 23 abdominal aortic aneurysm ( AAA; pink), and 12 peripheral artery disease (PAD; purple) patients, and their corresponding 52, 46, and 46 age- and sex-matched non-ACVD control subjects (green), respectively. Box plots represent the UniFrac distances from the CAD (blue columns), AAA (pink columns), or PAD (purple colums) patients to the non-ACVD control subjects. * p <0.01, compared between groups by using PERMANOVA with 999 permutations.
Unweighted (B) and Weighted (C) UniFrac distances of the ACVD sub-group with CAD, AAA, or PAD are shown.
Principal coordinate analysis (PCoA) plot for the samples from 9 CAD (blue), 10 AAA (pink), and 6 PAD (purple) patients. Box plots represent the UniFrac distances of the sub-groups, CAD, AAA, and PAD. n.s.: not significant, compared between groups by using PERMANOVA with 999 permutations.
Supplemental Fig.2. Beta diversities of the salivary microbiotas of the non-smokers in the ACVD or non-ACVD group.
Unweighted (A) and Weighted (B) UniFrac distances are shown.
Principal coordinate analysis (PCoA) plot for the samples from 37 ACVD (red) and 73 age- and sex-matched non-ACVD (green) subjects excluding current smokers. Box plots represent the UniFrac distances from the ACVD (red columns) or non-ACVD (green columns) subjects to non-ACVD subjects. * p <0.01, compared between groups by using PERMANOVA with 999 permutations.
Supplemental Fig.3. Beta diversities of the salivary microbiotas of the ACVD or non-ACVD subjects excluding those with diabetes mellitus.
Unweighted (A) and Weighted (B) UniFrac distances are shown.
Principal coordinate analysis (PCoA) plot for the samples from 30 ACVD (red) and 59 age- and sex-matched non-ACVD (green) patients excluding non-controlled diabetes patients (FBS <109 mg/dl and HbA1c [NGSP] <6.5%). Box plots represent UniFrac distances from the patients with ACVD (red columns) and non-ACVD (green columns) patients to non-ACVD patients. * p <0.01, compared between groups by using PERMANOVA with 999 permutations.
Supplemental Fig.4. Beta diversities of the salivary microbiotas of the ACVD and non-ACVD subjects excluding those with fewer than 20 teeth.
Unweighted (A) and weighted (B) UniFrac distances are shown. A principal coordinate analysis (PCoA) plot for the samples from 25 ACVD (red) and 50 age- and sex-matched non-ACVD (green) patients excluding those with fewer than 20 teeth is shown. Box plots represent UniFrac distances from the patients with ACVD (red columns) and non-ACVD (green columns) patients to non-ACVD patients. * p <0.01, based on a comparison between groups by PERMANOVA with 999 permutations.
Supplemental Fig.5. Weighted UniFrac distances of the salivary microbiotas of the ACVD and non-ACVD subjects excluding those taking each specific medication.
Box plots represent weighted UniFrac distances of samples from patients of ACVD (red) and age- and sex-matched non-ACVD (green) groups excluding those taking (A) anti-diabetes drugs, (B) statins, (C) ACE-I/ARBs, (D) beta-blockers, (E) calcium channel blockers, (F) PPI/H2 blockers, (G) anti-coagulants, and (H) anti-platelet medication. * p <0.01, based on a comparison between groups by PERMANOVA with 999 permutations.
Linear Discriminant Analysis Effect Size (LEfSe)
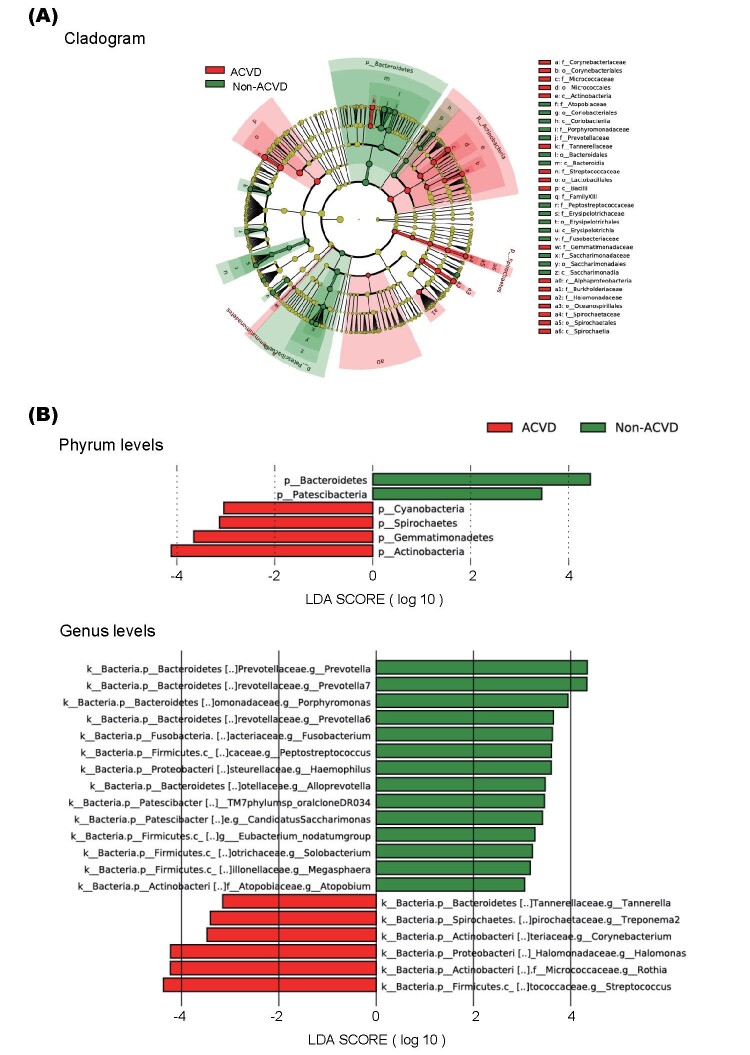
The cladogram in Fig.4A represents the significantly different taxa between the two groups, with a hierarchy reflecting the taxonomic rank from phylum to genus. We found that the ACVD group was associated with a significantly higher abundance of Actinobacteria and lower abundance of Bacteroidetes at the phylum level over four orders of magnitude. At the genus level, LEfSe demonstrated increased abundance of Streptococcus , Rothia , and Halomonas and decreased abundance of Prevotella , Prevotella7 , and Porphyromonas in the ACVD group compared with that in the non-ACVD group ( Fig.4B ) . These results revealed the presence of specific bacteria in the saliva of patients with ACVD and non-ACVD subjects.
Fig.4. The differentially abundant bacterial genera between the ACVD and non-ACVD groups as identified by linear discriminant analysis effect size (LEfSe).
(A) Cladogram of the differentially abundant bacterial taxa where each layer represents different taxonomy. The enriched taxa in ACVD salivary microbiome are represented in the cladogram. The central point represents the root of the tree (Bacteria), and each ring represents the next lower taxonomic level (phylum to genus: p, phylum; c, class; o, order; f, family; g, genus). The diameter of each circle represents the relative abundance of the taxon.
(B) Histogram of the linear discriminant analysis (LDA) scores for differentially abundant bacterial taxa between patients with ACVD and non-ACVD subjects.
LDA score ≥ 3.0 are shown. Red color represents significantly abundant taxa in the ACVD group when compared to those in the non-ACVD group. Green color represents the significantly abundant taxa in the non-ACVD group.
Random Forest (RF) Predictive Models
The RF model identified 43 oral microbiota OTUs ( Fig.5A ) that distinguished individuals with ACVD from non-ACVD controls. Principal component analysis ( Fig.5B ) illustrated that the two groups were distinct from each other. In the 10-fold cross validation with the train-test set, the mean±standard deviation of the AUC was 0.899±0.099. Using the validation data, an AUC of 0.933 (95% CI, 0.855–1.000) was obtained, suggesting that the genera identified for the microbial signature of the patients with ACVD were robust.
Fig.5. Random forest (RF) predictive models.
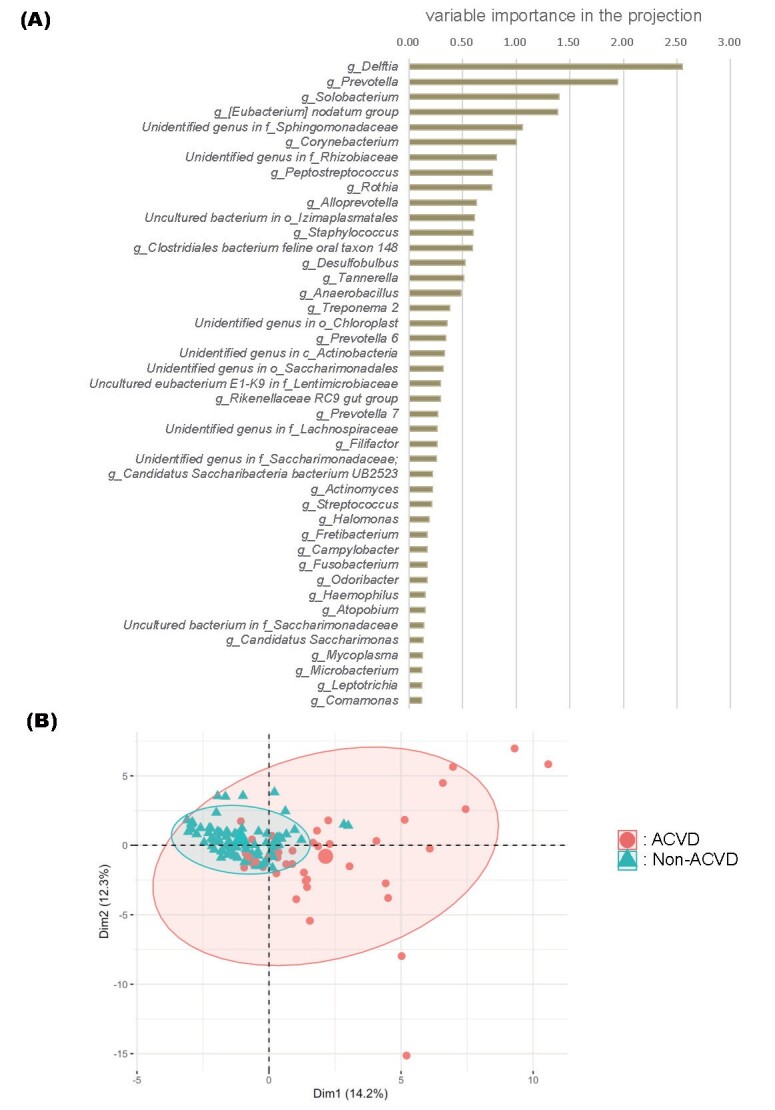
(A) Variable importance plot of the 43 OTUs selected as the optimal marker set.
Following the construction of the RF model, the variable importance in the projection (VIP) of each genus was calculated to select candidate genera that reflected the difference between the two groups. p <0.01 was considered significant.
(B) Principal component analysis of subjects based on the set of OTUs used to train the RF classifier. The centroids and 90% confidence ellipse for each group are shown. The large circle and triangle represent the mean points of the distribution of the ACVD and non-ACVD groups, respectively. Numbers in parentheses indicate the proportion of variance. Dim; dimension
Discussion
In the present study, we compared the salivary microbiota from patients with ACVD with age- and sex-matched non-ACVD individuals using 16S rRNA sequencing analysis. Our data demonstrated that the salivary microbiota in patients with ACVD is distinct from that in the non-ACVD subjects. In addition, we identified a set of bacterial genera that efficiently distinguished the patients with ACVD from the controls. These data indicate the potentiality of the salivary microbiota as an ACVD biomarker.
The overall microbial composition of the oral cavity in both groups was consistent with that of previous findings 34 - 36) . At the genus level, oral bacteria with a relative abundance of at least 0.1% were classified into 136 genera in the ACVD group and into 124 genera in the non-ACVD group; however, alpha diversity, and evenness and richness of the microbial community did not differ between the ACVD and non-ACVD groups. These results were consistent with the results from previous studies 3 , 18) . Beta diversity showed a significant difference between the two groups and revealed a separation of the groups across components of variation, which was contrary to the results from previous studies 3 , 18) . We hypothesize that the inconsistency was caused by the differences in the backgrounds of the participants, such as ethnicity, food-related culture, etc.
Our LEfSe analyses revealed notable distinctive features in the ACVD and non-ACVD groups. We observed a dominance of the bacterial genera Prevotella , Prevotella7 , and Porphyromonas in the non-ACVD group. In the ACVD group, Streptococcus , Rothia , Halomonas , and Corynebacterium were over-represented. In a previous study, the genera Streptococcus , Rothia , and Corynebacterium were detected in both the oral cavity and the atherosclerotic plaques of the same patients who underwent carotid endarterectomy for minor ischemic stroke or transient ischemic attack 3) . Therefore, our study corroborated the possibility of these bacterial genera being related with ACVD. The genus Rothia is an oral commensal; Rothia mucilaginosa has recently been reported to be abundantly present in the buccal and tongue microbiome of smokers 37 , 38) . Since smoking has been recognized as an important factor affecting oral condition, periodontal disease status, and oral microbiota 39) , the frequency and duration of smoking should also be taken into consideration while evaluating the salivary microbiota of patients with ACVD. Interestingly, these findings regarding the oral cavity were partially consistent with another study on ACVD-associated gut microbiota characterized by a relative reduction in Prevotella and enrichment of Streptococcus 40) . Similar to previous studies reporting that the gut microbiome plays a role in ACVD 20 , 21) , our study suggested that the oral microbiota could also be involved in the pathogenesis of ACVD.
Recent reports have shown the potential of salivary microbiota profiles for screening various diseases such as rheumatoid arthritis 41) , primary sclerosing cholangitis 14) , pancreatic cancer 42) , lung cancer 15) , and colorectal cancer 16) . The oral microbiota-based classifiers have been created to distinguish patients from control subjects via RF models through machine learning in several diseases 14 , 16) . To examine the bacterial community characteristics of salivary microbiota of patients with ACVD, we developed an RF model and identified 43 genera that efficiently distinguished the patients with ACVD and non-ACVD subjects. The set of 43 bacteria in the saliva had a high AUC value of 0.933, suggesting that the salivary microbiota of the ACVD patients were unique and may be different from that of non-ACVD subjects. Our results suggest that an entire oral commensal microbiota, rather than a single atherosclerosis-associated bacterium, is associated with ACVD. The growth or the expression of pathogenic factors of a single atherosclerosis-associated bacterium may be regulated by synergistic or antagonistic interactions with surrounding symbiotic bacteria. Furthermore, even bacteria without pathogenic factors may be involved in the inhibition or promotion of ACVD pathogenesis through interactions with related bacteria. In the case of gastric cancer, commensal gastric microbes have been found to interact with Helicobacter pylori and regulate its pathogenicity and carcinogenicity 43) . Thus, the association between salivary microbiota and ACVD requires further investigation.
Although the results of this study have revealed novel microbial determinants of ACVD, our study has the following limitations: First, because of the cross-sectional nature of our data, no inference can be made about a causal relationship between oral microbiota and ACVD. Thus, the next crucial step is to investigate whether the oral microbiota is causally linked to ACVD in a longitudinal setting. Second, all study participants in our study were Japanese elderly individuals living around Takatsuki city and it is possible that the oral microbial association results for ACVD may not be extrapolated to other populations. As the background of participants, such as age, race, and food-related culture have been associated with changes in the oral microbiota 44 , 45) , evaluation of the oral microbiota profile in each setting is necessary. Third, the design of this study does not allow us to match the background medication between the ACVD and non-ACVD groups. Another recent study showed that medication could affect the composition of the gut microbiota in ACVD 40) ; so, the possible effect of drug use on the oral microbiota should be explored in intervention trials with larger cohorts. Lastly, to elucidate the bacterial environment and microbiota changes that affect the pathogenesis of ACVD in detail, not only 16S rRNA metagenomic analysis, but also complex analysis methods such as shotgun sequencing or various omics analyses should be performed. Nevertheless, the present study is a preliminary pilot project with a limited sample size and more extensive studies are required to confirm these observations as well as to thoroughly investigate the causal relationships between ACVD and salivary microbiota.
Conclusion
In summary, we investigated the composition and diversity of the salivary microbiota to distinguish patients with ACVD from non-ACVD subjects. The salivary microbiota could potentially serve as an ACVD marker. As the oral microbiota may be involved in the development of ACVD, further elucidation of the mechanisms underlying the condition could lead to medications or lifestyle changes for reducing the risk of ACVD.
Acknowledgements
We gratefully acknowledge the support of Cykinso, Inc. for their help in conducting microbiome analysis of this research.
This work was partly presented at the AHA Scientific Sessions 2019, Philadelphia, Pennsylvania, November 16 to 18, 2019, and published in the form of an abstract (Circulation 2019;140 Suppl 1).
Conflict of Interest
The authors declare no conflict of interest.
Notice of Grant Support
This research was partially supported by Private University Research Branding Project (2017-2019) of the Ministry of Education, Culture, Sports, Science, and Technology and Japan Society for the Promotion of Science KAKENHI Grant Number 20K18521 and 20K09932
References
- 1).Nakano K, Nemoto H, Nomura R, Inaba H, Yoshioka H, Taniguchi K, Amano A and Ooshima T: Detection of oral bacteria in cardiovascular specimens. Oral Microbiol Immunol, 2009; 24: 64-68 [DOI] [PubMed] [Google Scholar]
- 2).Ishihara K, Nabuchi A, Ito R, Miyachi K, Kuramitsu HK and Okuda K: Correlation between detection rates of periodontopathic bacterial DNA in coronary stenotic artery plaque [corrected] and in dental plaque samples. J Clin Microbiol, 2004; 42: 1313-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE and Backhed F: Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA, 2011; 108: 4592-4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Chhibber-Goel J, Singhal V, Bhowmik D, Vivek R, Parakh N, Bhargava B and Sharma A: Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes, 2016; 2: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Li L, Messas E, Batista EL, Jr., Levine RA and Amar S: Porphyromonas gingivalis infection accelerates the progression of atherosclerosis in a heterozygous apolipoprotein E-deficient murine model. Circulation, 2002; 105: 861-867 [DOI] [PubMed] [Google Scholar]
- 6).Lalla E, Lamster IB, Hofmann MA, Bucciarelli L, Jerud AP, Tucker S, Lu Y, Papapanou PN and Schmidt AM: Oral infection with a periodontal pathogen accelerates early atherosclerosis in apolipoprotein E-null mice. Arterioscler Thromb Vasc Biol, 2003; 23: 1405-1411 [DOI] [PubMed] [Google Scholar]
- 7).Aarabi G, Heydecke G and Seedorf U: Roles of Oral Infections in the Pathomechanism of Atherosclerosis. Int J Mol Sci, 2018; 19: 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Tonetti MS and Van Dyke TE: Periodontitis and atherosclerotic cardiovascular disease: consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J Periodontol, 2013; 84: 24-29 [DOI] [PubMed] [Google Scholar]
- 9).Herrera D, Molina A, Buhlin K and Klinge B: Periodontal diseases and association with atherosclerotic disease. Periodontol 2000, 2020; 83: 66-89 [DOI] [PubMed] [Google Scholar]
- 10).Lockhart PB, Bolger AF, Papapanou PN, Osinbowale O, Trevisan M, Levison ME, Taubert KA, Newburger JW, Gornik HL, Gewitz MH, Wilson WR, Smith SC, Jr. and Baddour LM: Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association?: a scientific statement from the American Heart Association. Circulation, 2012; 125: 2520-2544 [DOI] [PubMed] [Google Scholar]
- 11).Willis JR and Gabaldón T: The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms, 2020; 8: 308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Baker JL, Bor B, Agnello M, Shi W and He X: Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol, 2017; 25: 362-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Gao L, Xu T, Huang G, Jiang S, Gu Y and Chen F: Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell, 2018; 9: 488-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Iwasawa K, Suda W, Tsunoda T, Oikawa-Kawamoto M, Umetsu S, Takayasu L, Inui A, Fujisawa T, Morita H, Sogo T and Hattori M: Dysbiosis of the salivary microbiota in pediatric-onset primary sclerosing cholangitis and its potential as a biomarker. Sci Rep, 2018; 8: 5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Zhang W, Luo J, Dong X, Zhao S, Hao Y, Peng C, Shi H, Zhou Y, Shan L, Sun Q, Li Y and Zhao X: Salivary Microbial Dysbiosis is Associated with Systemic Inflammatory Markers and Predicted Oral Metabolites in Non-Small Cell Lung Cancer Patients. J Cancer, 2019; 10: 1651-1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O’Riordain M, Shanahan F and O’Toole PW: The oral microbiota in colorectal cancer is distinctive and predictive. Gut, 2018; 67: 1454-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Desvarieux M, Demmer RT, Rundek T, Boden-Albala B, Jacobs DR, Jr., Sacco RL and Papapanou PN: Periodontal microbiota and carotid intima-media thickness: the Oral Infections and Vascular Disease Epidemiology Study (INVEST). Circulation, 2005; 111: 576-582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Fak F, Tremaroli V, Bergstrom G and Backhed F: Oral microbiota in patients with atherosclerosis. Atherosclerosis, 2015; 243: 573-578 [DOI] [PubMed] [Google Scholar]
- 19).Hyvarinen K, Mantyla P, Buhlin K, Paju S, Nieminen MS, Sinisalo J and Pussinen PJ: A common periodontal pathogen has an adverse association with both acute and stable coronary artery disease. Atherosclerosis, 2012; 223: 478-484 [DOI] [PubMed] [Google Scholar]
- 20).Tang WH, Kitai T and Hazen SL: Gut Microbiota in Cardiovascular Health and Disease. Circ Res, 2017; 120: 1183-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Jonsson AL and Backhed F: Role of gut microbiota in atherosclerosis. Nat Rev Cardiol, 2017; 14: 79-87 [DOI] [PubMed] [Google Scholar]
- 22).Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, Gotoh K, Motooka D, Nakamura S, Iida T and Yamazaki K: Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep, 2014; 4: 4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Lira-Junior R and Bostrom EA: Oral-gut connection: one step closer to an integrated view of the gastrointestinal tract? Mucosal Immunol, 2018; 11: 316-318 [DOI] [PubMed] [Google Scholar]
- 24).Kato T, Yamazaki K, Nakajima M, Date Y, Kikuchi J, Hase K, Ohno H and Yamazaki K: Oral Administration of Porphyromonas gingivalis Alters the Gut Microbiome and Serum Metabolome. mSphere, 2018; 3: e00460-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Omori M, Kato-Kogoe N, Sakaguchi S, Fukui N, Yamamoto K, Nakajima Y, Inoue K, Nakano H, Motooka D, Nakano T, Nakamura S and Ueno T: Comparative evaluation of microbial profiles of oral samples obtained at different collection time points and using different methods. Clin Oral Investig, 2020; in press, doi: 10.1007/s00784-020-03592-y [DOI] [PubMed] [Google Scholar]
- 26).Nesse W, Abbas F, van der Ploeg I, Spijkervet FK, Dijkstra PU and Vissink A: Periodontal inflamed surface area: quantifying inflammatory burden. J Clin Periodontol, 2008; 35: 668-673 [DOI] [PubMed] [Google Scholar]
- 27).Tonetti MS, Greenwell H and Kornman KS: Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J Periodontol, 2018; 89: 159-172 [DOI] [PubMed] [Google Scholar]
- 28).Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R and Caporaso JG: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol, 2019; 37: 852-857 [Google Scholar]
- 29).Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER and Knight R: Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome, 2017; 5: 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Lozupone C and Knight R: UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol, 2005; 71: 8228-8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Bray JR and Curtis JT: An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr, 1957; 27: 325-349 [Google Scholar]
- 32).Anderson MJ: Permutational Multivariate Analysis of Variance (PERMANOVA). In: Wiley StatsRef: Statistics Reference Online, ed by N. Balakrishnan TC, B. Everitt, W. Piegorsch, F. Ruggeri and J.L. Teugels, pp1-15, John Wiley & Sons, Ltd., UK, 2017 [Google Scholar]
- 33).Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS and Huttenhower C: Metagenomic biomarker discovery and explanation. Genome Biol, 2011; 12: R60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C and Izard J: Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol, 2012; 13: R42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM and Crielaard W: Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res, 2008; 87: 1016-1020 [DOI] [PubMed] [Google Scholar]
- 36).Dong L, Yin J, Zhao J, Ma SR, Wang HR, Wang M, Chen W and Wei WQ: Microbial Similarity and Preference for Specific Sites in Healthy Oral Cavity and Esophagus. Front Microbiol, 2018; 9: 1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, Ma Y, Purdue MP, Jacobs EJ, Gapstur SM, Li H, Alekseyenko AV, Hayes RB and Ahn J: Cigarette smoking and the oral microbiome in a large study of American adults. ISME J, 2016; 10: 2435-2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, Kobayashi W, Sawada K, Tamura Y, Tokuda I, Murashita K, Nakaji S, Imoto S, Yanagita M and Okuno Y: Metagenomic analysis of bacterial species in tongue microbiome of current and never smokers. NPJ Biofilms Microbiomes, 2020; 6: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Jiang Y, Zhou X, Cheng L and Li M: The Impact of Smoking on Subgingival Microflora: From Periodontal Health to Disease. Front Microbiol, 2020; 11: 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen JY, Geng QS, Zhang ZW, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K and Kristiansen K: The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun, 2017; 8: 845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y and Wang J: The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med, 2015; 21: 895-905 [DOI] [PubMed] [Google Scholar]
- 42).Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K and Wong DT: Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut, 2012; 61: 582-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Espinoza JL, Matsumoto A, Tanaka H and Matsumura I: Gastric microbiota: An emerging player in Helicobacter pylori-induced gastric malignancies. Cancer Lett, 2018; 414: 147-152 [DOI] [PubMed] [Google Scholar]
- 44).Belstrom D, Holmstrup P, Nielsen CH, Kirkby N, Twetman S, Heitmann BL, Klepac-Ceraj V, Paster BJ and Fiehn NE: Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J Oral Microbiol, 2014; 6: 23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Yamashita Y and Takeshita T: The oral microbiome and human health. J Oral Sci, 2017; 59: 201-206 [DOI] [PubMed] [Google Scholar]