Graphical abstract
Keywords: KRASG12C, AMG 510, Molecular dynamic simulation, Principal component analysis, Resistant mutation
Abstract
The KRASG12C mutant is often associated with human cancers, and AMG 510 as a promising covalent inhibitor of KRASG12C has achieved surprising efficacy in clinical trials. However, the interaction mechanism between KRASG12C and AMG 510 is not completely understood. Here, we performed all-atom molecular dynamics simulations on the complex of KRASG12C-AMG 510 to explore the influence of this covalent inhibitor on the conformational change of KRASG12C. A PCA (Principal Component Analysis) model was constructed based on known KRAS crystal structures to distinguish different conformations (active, inactive, and other). By mapping simulation trajectories onto the PCA model, we observed that the conformations of KRASG12C bound with AMG 510 were mainly concentrated in the inactive conformation. Further analysis demonstrated that AMG 510 reduced the flexibility of two switch regions to make the complex of KRASG12C-AMG 510 restricted in the inactive conformation. In the meantime, we also identified key interacting residues between KRASG12C and AMG 510 through the calculation of binding energy. Finally, we built a series of KRAS second-site mutation systems (i.e. KRASG12C/mutations) to conduct large-scale screening of potential resistance mutations. By further combining MD simulations and the PCA model, we not only recapitulated the currently known resistance mutations of AMG 510 successfully but also proposed some novel potential resistant mutations. Taken together, these results broaden our insight into the influence of AMG 510 on the conformational change of the KRASG12C mutant at the atomic level, thereby providing crucial hints for the improvement and optimization of drug candidates.
1. Introduction
As an important type of small GTPase, RAS proteins (KRAS, HRAS, and NRAS) function as molecular switches and play indispensable roles in a series of cellular signal transduction processes, including cell proliferation, differentiation, and survival [1], [2]. In normal cells, RAS proteins primarily contain two states: the GTP-bound active state and the GDP-bound inactive state. Although RAS proteins have the ability of intrinsic GTP hydrolysis and nucleotide exchange, the state transition of RAS proteins in the signal transduction process is mainly accelerated by two types of proteins: GTPase-activating proteins (GAPs) that increase the hydrolysis rate of GTP and guanine nucleotide exchange factors (GEFs) that catalyze the GDP release. Activated RAS proteins interact with downstream effectors, such as Raf, PI3K, and RalGDS, to regulate diverse cell signaling pathways [3].
KRAS, NRAS, and HRAS are mutated in approximately 20% of all human malignancies [4]. Among them, KRAS is the most frequently mutated oncogene, participating in 85% of RAS-driven cancers [5], [6]. The KRAS mutants can cause continuous cellular proliferation and cancer cell development, particularly in pancreatic, colorectal, and lung cancers [7]. In general, the residues G12, G13, and Q61 are three mutation hotspots [5], [8]. These oncogenic mutations considerably affect the nucleotide exchange process of KRAS. On the one hand, the mutations impair the intrinsic or GAP-mediated GTP hydrolysis process [7], [9]. On the other hand, the intrinsic exchange is unchanged or accelerated during the exchange of GDP for GTP but the sensitivity to GEF activity can be reduced [10], [11]. Thus, the resulting net effect is the accumulation of GTP-bound form of KRAS. Among KRAS mutants in tumors, oncogenic mutations predominantly occur in G12 [12], [13]. Indeed, the most prevalent mutation is G12D (41%), followed by G12V (28%) and G12C (14%) [5], [14]. Consequently, inhibiting the activity of these mutants and then hindering the transduction of abnormal signals has long been regarded as an ideal strategy to treat KRAS-driven cancers.
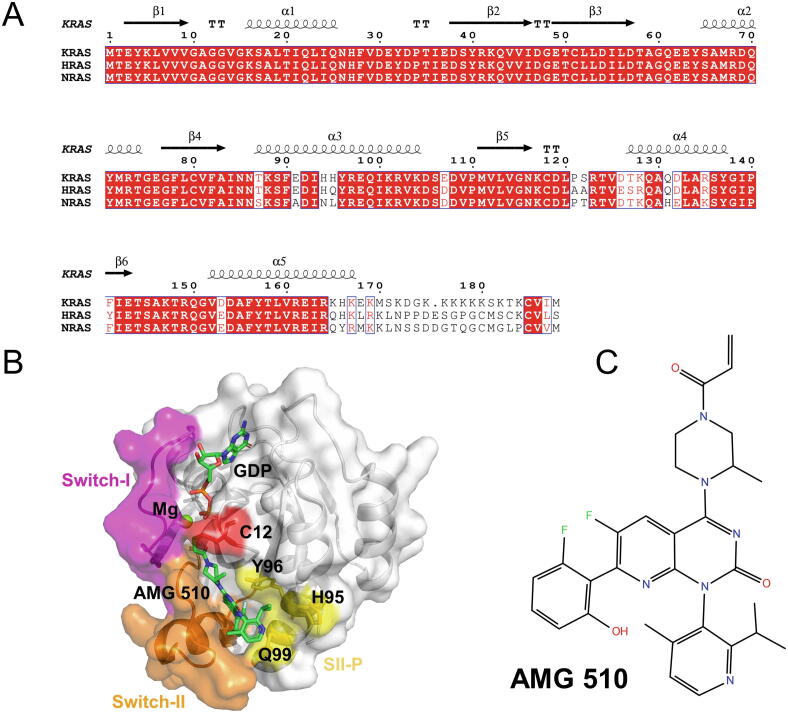
Sharing high sequence identity with HRAS and NRAS, KRAS contains two components: a conserved catalytic domain (residues 1–166) and a membrane-targeting hypervariable region (HVR, residues 167–188 and note that the residues 186–188 are removed when targeting membrane) (Fig. 1A). The catalytic domain, consisting of six β-strands surrounded by five α-helices, participates in the interactions with effectors and regulators, which is influenced by the conformational changes of its two flexible regions: Switch-I (SI, residues 30–38) and Switch-II (SII, residues 59–76) (Fig. 1B). The two switch regions also adopt different conformations in the GTP- and GDP-bound forms. In the active state, both the SI and SII residues are involved in the interaction of GTP. In particular, the two hydrogen bonds formed by T35 and G60 with γ-phosphate in GTP make KRAS in a relatively compact conformation. However, the release of γ-phosphate after the GTP hydrolysis renders KRAS return to the flexible conformation in the inactive state [8], [15]. Thanks to the efforts of structural biologists, a plethora of structures related to KRAS have been experimentally resolved and deposited in Protein Data Bank (PDB), which has provided an essential foundation for deciphering the molecular mechanisms of KRAS as well as conducting structure-based drug discovery [6].
Fig. 1.
Sequence and structure of KRASG12C–AMG 510 complex. (A). Multiple sequence alignment of three RAS proteins (KRAS, HRAS, and NRAS), including the catalytic domains and HVRs. (B) Cartoon and surface representations of the KRASG12C–AMG 510 complex (PDB entry: 6OIM). The SI, SII, HVR, SII-P, and residue C12 are colored as magenta, orange, purple, yellow, and red, respectively. (C) The chemical structure of AMG 510. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Although KRAS mutants have been regarded as drug targets in the past several decades and considerable drug development efforts have been carried out, the strategy of directly targeting the KRASG12C mutant was unsuccessful until the discovery of covalent inhibitors in 2013 [8], [16], [17]. KRAS has been widely considered as an ‘undruggable’ target for a long time due to two reasons [1], [18], [19]. First, the binding affinity of KRAS and GDP/GTP reaches to a picomolar level, seriously affecting the goal to explore the nucleotide-competitive inhibitors [14], [20]. Second, the KRASG12C mutant lacks other reasonable deep hydrophobic pockets, and thus it is challenging to develop high-affinity allosteric inhibitors [14], [17], [20]. Therefore, numerous alternative strategies have been proposed to design the inhibitors of KRAS-driven tumors [9], [13]. For example, the SOS1 and SHP2 inhibitors that block the nucleotide exchange cycle to reduce the number of active KRAS in the cell [9], [21], and the RAF, MEK, and ERK inhibitors that impair the activation of MEPK pathway regulated by KRAS [9], [22] have entered clinical trials. However, these inhibitors developed by targeting KRAS mutants indirectly have not shown satisfactory performance because of lacking the selectivity between mutated and wild-type KRAS [1], [13].
Recently, the strategy of developing covalent inhibitors [23], [24] has received wide attention. The mechanism is to covalently target active site cysteine of KRASG12C because of the inherent reactive nature of cysteine. Shokat and colleagues have focused on the discovery of covalent inhibitors for KRASG12C, and they first identified a novel allosteric pocket behind SII, named the SII pocket (SII-P), and then developed a series of compounds that bind with this novel pocket as well as forming the covalent bond with C12 of KRASG12C. Similar to the SOS inhibitors, these compounds are able to block the processes of SOS-mediated nucleotide exchange and then influence KRASG12C association with RAF [17]. Because the wild-type KRAS lacks active cysteine residue in position 12, these compounds specifically bind to the KRASG12C mutant with low side effects [16]. Therefore, directly targeting receptor protein itself is again a desirable approach for treating KRASG12C-driven tumors. By now, with the continuous optimization, the covalent inhibitors of targeting KRASG12C have entered clinical trials and revealed good antitumor activity, such as AMG 510 (sotorasib), MRTX849 (adagrasib), LY3499446, and ARS-3248 [9], [25], [26], [27], [28]. The detailed information of these four compounds is available in Table S1.
As a promising antitumor compound, AMG 510 (Fig. 1C) is the first KRASG12C covalent inhibitor in clinical development and was approved by FDA in 2021 [26]. The clinical data reported that it was able to make the regression of KRASG12C tumors and enhance the antitumor efficacy of targeted agents [27]. In advanced non-small cell lung cancer (NSCLC) patients with KRASG12C, AMG 510 showed significant efficacy with an objective response rate of 37% and a disease control rate of 81% [29]. However, resistance to small molecular drugs remains a major challenge to realize durable efficacy in patients. Recently, some acquired KRAS mutations have been reported in AMG 510 or MRTX849 treated patients, including R68S, Y96C/D/S, H95D/Q/R and so on [30], [31], [32].
The high-resolution complex structure (PDB entry: 6OIM, X-ray resolution: 1.65 Å) of KRASG12C-AMG 510 reveals that the compound locates at SII-P, which is adjacent to the mutant cysteine residue in the inactive GDP-bound form (Fig. 1B). Meanwhile, a sub-pocket formed by H95/Y96/Q99 offers an additional site to stabilize the binding mode of AMG 510 [14], [27] (Fig. 1B). In general, the complex structure of KRASG12C-AMG 510 has provided a good starting point for employing structural bioinformatics to decipher why AMG 510 could specifically lock KRASG12C in the inactive state and block oncogenic signaling transmission. Although the interaction mechanism of KRASG12C-AMG 510 has been explored through 3D structure determination and other experimental methods as described above [27], [33], there are only a few structural bioinformatics studies on how AMG 510 influences the conformation of KRASG12C at the atomic level. Although Pantsar has utilized molecular dynamics (MD) simulations to characterize the interaction dynamics of the KRASG12C-AMG 510 complex [34], several issues remain to be further addressed through computational methods. For instance, the molecular mechanism of the covalent inhibitor locking the KRASG12C mutant in the GDP-bound state, and the influence of the inhibitor binding on the conformational dynamics change of two switch regions should be further deciphered. In addition, the drug resistance sites raised by AMG 510 have received wide attention, which also deserves further computational investigation. Therefore, further MD studies on the KRASG12C-AMG 510 system are still highly demanded to accelerate KRAS-related drug discovery.
In this study, we performed MD simulations and related computational analyses to investigate the influence of AMG 510 on the KRASG12C structure and to decipher the reason why the mutant is locked in the GDP-bound state. In particular, we constructed a PCA (Principal Component Analysis) model based on known KRAS crystal structures to predict the conformational type of KRAS. By combining MD simulations and the PCA model, the dynamic conformations of KRASG12C bound with and without AMG 510 were characterized. To provide in-depth understanding of the interaction mode of AMG 510 and KRASG12C, a series of key residues involved in the interaction of KRASG12C and AMG 510 were also identified. Moreover, a large-scale in silico mutagenesis experiment was implemented, and potential drug resistance variations were proposed through further combining MD simulations and the PCA model. In addition to recapitulating the known resistance sites of KRASG12C and AMG 510 (K16T, Q61L, Q61R, R68M, R68S, Y96C, Y96D, Y96S, and Q99K), we also predicted some new potential drug resistance variations, which are worthy of further clinical attention.
2. Methods
2.1. Preparation of simulation systems
The crystal structures of KRASG12C-AMG510 (PDB entry: 6OIM) and KRASG12C-apo (PDB entry: 4LDJ) were first downloaded from PDB [27], [35], and then only the catalytic domain (residues 1–166) in each structure was retained for simulation. Note that some residues with missing structural information in the original crystal structure were further modeled using Modeller 9.17 [36], and three engineered mutations (C51S, C80L, C118S) of 6OIM were muated back to native cysteines. The tleap module in the AMBER18 package was employed to add the missing hydrogen atoms of these two crystal structures [37]. Molecular mechanics parameters of proteins were assigned according to the ff14SB force field, while the general AMBER force field (GAFF) was used for AMG 510 and the corresponding RESP charges were assigned using Antechamber [38], [39]. Additionally, the parameters of GDP were downloaded from the AMBER parameter database (www.pharmacy.manchester.ac.uk/bryce/amber). Both systems were solvated in a cubic box of TIP3P water molecules and neutralized with sodium ions. The minimum distance between the protein surface and the boundary of the water box was set to 12 Å.
2.2. MD simulations
Two energy minimization procedures were conducted for each system. To remove wrong contacts in the water box, the first minimization procedure was carried out, which contained 2000 steps of the steepest descent and 3000 steps of the conjugate gradient with a positional restraint of 500 kcal mol−1 Å−2 imposing on all of the protein atoms. Second, the system was further minimized by 5000 steps of steepest descent and 3000 steps of conjugate gradient without any restraint. Subsequently, each system was gradually heated from 0 K to 290 K through 0.9 ns simulation and from 290 K to 300 K through 0.1 ns simulation, in which the protein was restricted in the NVT ensemble with a positional restraint of 2 kcal mol−1 Å−2 for heavy atoms. Moreover, the system was equilibrated without restraint in the NPT ensemble with 300 K and 1 atm for 1 ns. Finally, 800 ns simulation was performed for each system with periodic boundary condition using the NPT ensemble.
During the MD simulation process, the time step was set to 2 fs, and a coordinate trajectory was saved in each 40 ps. The SHAKE algorithm was applied to constrain covalent bonds involving hydrogen atoms [40]. The Langevin thermostat with a collision frequency of 2 ps−1 and Berendsen barostat were used for temperature and pressure regulation, respectively. The particle mesh Ewald method (grid spacing of 1 Å) was used to deal with the long-range electrostatic interactions [41], and the short-range electrostatic and van der Waals interactions cutoff was set to 10 Å.
2.3. Structural analysis of MD simulations
The CPPTRAJ software in AMBER 18 was used to calculate the root mean squared deviation/fluctuation (RMSD/RMSF) of MD simulation trajectories [42], in which the initial structure was used as the reference. The protein–ligand interaction profiler (PLIP) was utilized to detect the interactions between KRAS protein and its ligands (GDP and AMG 510) [43]. Structural visualization was implemented with PyMOL (https://pymol.org/2/).
2.4. Cross-Correlation analysis
To identify the dynamical coupling of the motions between protein segments, the cross-correlation coefficient (Cij) was proposed for measuring the motion correlation between the Cα atom pair in residues i and j, which is defined as:
| (1) |
where and are the displacement from the mean position of the Cα atom pair in residues i and j, considered over the sampled period. Positive Cij denotes lockstep motion in the residue pair under investigation, whereas negative Cij stands for negatively correlated motion. The cross-correlation analysis in this work was implemented through the Bio3D package (http://thegrantlab.org/bio3d).
2.5. Principal component analysis
PCA was often applied to examine the relationship and difference among various structures (e.g., the frames from the MD trajectories). In this study, the Cα atom distance between residues in the SI and SII regions was used as features to represent the conformation of a protein. In more detail, the residue distances from the residues in the SI (residues 30–38) and SII (residues 59–76) regions to the conserved residue K16, and the pairwise residue distances within the SI region were taken into account. Thus, a protein structure can be represented as a 63-dimensional feature vector.
We downloaded 150 KRAS structures, previously compiled by [6], from the PDB database. To precisely extract the structural features of KRAS, the structures whose catalytic domains contain missing residues were deleted. After filtering, 96 structures were reserved and used to conduct PCA in this work (Table S2). According to the type of nucleotide-binding or the literature description [44], [45], [46], these structures can be divided into three groups: the active conformation (GTP-bound, N = 35), the inactive conformation (GDP-bound, N = 48), and other conformation (the SI region presenting open conformation, N = 13). Then, PCA was performed on these structures, and the first three principal components (PCs) were used to build a 3D space for the visualization of these structures. To quantitatively cluster these structures, a simple classifier was established. First, we defined two minimum spheres, Sactive and Sinactive, to represent the clusters of active and inactive conformations. The radius of Sactive (Ractive) represents the maximum distance of the active conformation structures to their geometric center. Likewise, the radius of Sinactive (Rinactive) can be defined. When a KRAS structure or a frame from KRAS MD simulation trajectories is projected onto this 3D space, we can use the distance of the query structure to these two spheres to estimate its conformational type. For example, if the distance of the query structure to the geometric center of Sactive is less than the corresponding distance to Sinactive, and the distance is not greater than Ractive, the query structure can be grouped as the active conformation. Similarly, the determination of an inactive conformational structure can be made. However, when the query structure is grouped neither in the active conformation nor in the inactive conformation, we regard it as in the other conformation. Leave-one-out test was employed to estimate the reliability of the conformational state classifier.
2.6. Binding free energy calculations and energy decomposition
The molecular mechanics energies combined with the Poisson-Boltzmann and surface area continuum solvation (MM/PBSA) approach was used to calculate the binding free energy in this study [47]. The function of the MM/PBSA method is defined as:
| (2) |
where is the change of the gas phase molecular mechanics energy, which includes the van der Waals (), electrostatic (), and internal () terms. represents the electrostatic solvation energy calculated using the Poisson-Boltzmann method [48], and means the non-polar contribution of desolvation, which was approximated by the LCPO method [49]. The entropy term () was ignored in this study because of the computational time and the low accuracy. We further used the MM/PBSA free energy decomposition method in AMBER 18 to identify and quantify the critical residues interacting with the ligand. The last 300 frames of all the simulation trajectories were extracted to conduct the binding free energy calculation and binding energy decomposition.
To calculate the binding affinity between AMG 510 and KRAS mutants, the following modelling steps were adopted: (1) extracting the last frame of the 800 ns MD simulation in each system; (2) breaking up the covalent bond between the C25 atom of AMG 510 and the sulfur atom of C12 in KRASG12C; (3) running another 1 ns production MD simulation; (4) selecting the last 10 frames to calculate the binding free energy between AMG 510 and KRAS mutants.
3. Results and discussion
3.1. PCA model can capture the conformational change of KRASG12C structure
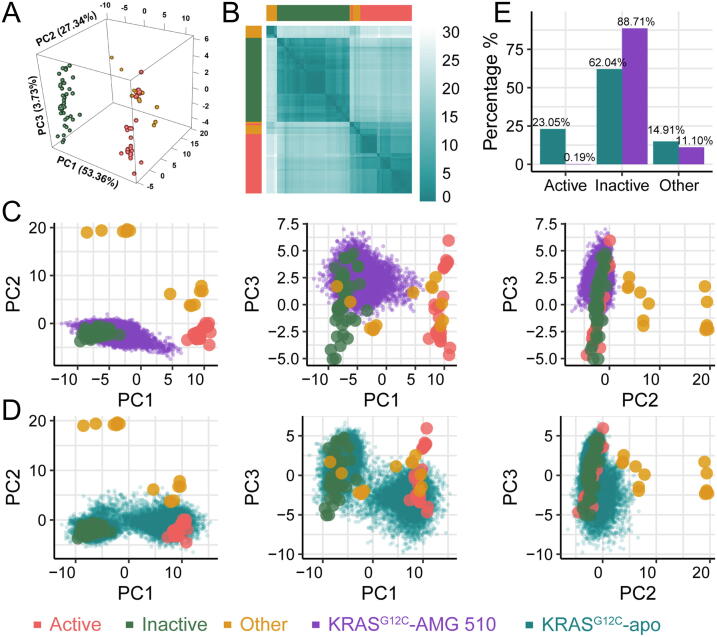
In the PCA result of the 96 experimental structures, the first three components capture more than 85% variance of residue distances related to two switch regions, suggesting that they can provide a clear description of the conformational space of KRAS (Fig. S1A). By projecting the original 96 structures onto the 3D coordinate built by the first three PCs, we observed that these structures were divided into three main groups: two dense clusters that represent the GTP-bound active conformation and GDP-bound inactive conformation, respectively, and a sparse cluster corresponding to the other conformation (Fig. 2A). To supplement the visualization of the structure clustering, the structure distributions measured by (PC1 and PC2) and (PC1 and PC3) are also shown in Fig. S1B and C. Based on the first three PCs, the pairwise Euclidean distances of these 96 structures are also measured and clustered. As shown in Fig. 2B, inactive structures could be clearly distinguished from the other two conformations. However, we also observed that some structures in other conformation type are mixed with active structures (Fig. 2B), implying that the other conformation type tends to be active-like. Meanwhile, we designed a simple rule to classify different conformations of KRAS structures. The result shows that the conformations for 89 out of 96 structures (accuracy = 92.7%) are correctly classified through the Leave-One-Out test, suggesting that the performance of this conformational state classifier is satisfactory.
Fig. 2.
PCA results of the 96 known KRAS structures and the trajectories extracted from MD simulations. (A) Projection of experimental structures onto the 3D space built by the first three PCs. (B) Clustering of the experimental structures based on the pairwise Euclidean distances of the first three PCs. The simulation trajectories from KRASG12C-AMG 510 (C) and KRASG12C-apo (D) are projected onto the 2D spaces built by the first three PCs. (E) The state distribution of structures in the two systems KRASG12C-AMG 510 and KRASG12C-apo, which were extracted from simulation trajectories during the 800 ns all-atom MD simulations.
To further explore the effect of AMG 510 on the conformational state of KRASG12C, we projected the MD simulation trajectories of KRASG12C, which are bound with and without the covalent inhibitor, on the established PCA model to quantitatively characterize the changes of conformational state. Fig. 2C and D show the clustering of the KRASG12C-AMG 510 and KRASG12C-apo trajectories, which provide details to evaluate the structural relationship among these two types of trajectories and the 96 experimentally known KRAS structures. The conformation for each trajectory can be further determined through the proposed conformation classifier. Fig. 2E represents the percentage of three conformational types in the two 800 ns MD simulation trajectories (KRASG12C-AMG 510 and KRASG12C-apo). In general, the majority of KRASG12C-AMG 510 structures (88.71%) tend to be in an inactive conformation during the simulation process, followed by the other conformation (11.10%) and the active conformation (0.19%). Regarding the conformational distribution of KRASG12C not bound with the covalent inhibitor (KRASG12C-apo), the dominant conformation remains inactive, although the proportion decreases to 62.04% compared with the system bound with AMG 510. In addition, 23.05% of frames sampled from simulation trajectories belong to the active conformation, and the other conformation also accounts for 14.91%. As expected, we can know from the above results that the majority of the frames extracted from the 800 ns MD simulation trajectories of two KRASG12C are classified as the inactive conformation, since they are GDP-bound. However, without the restraint exerted by AMG 510, the structures from KRASG12C-apo cluster with active or other conformation during the simulation process. When covalently targeted by AMG 510, the proportion of all the structures of the KRASG12C generated by the MD simulation belonging to the inactive conformation is significantly higher than that of KRASG12C-apo (p < 0.001, Chi-square test). It indicates that AMG 510 can keep KRASG12C continuously in the GDP-bound inactive state and then impede the process of converting it to the active state [14], [27].
3.2. AMG 510 decreases the flexibility of two switch regions of KRASG12C
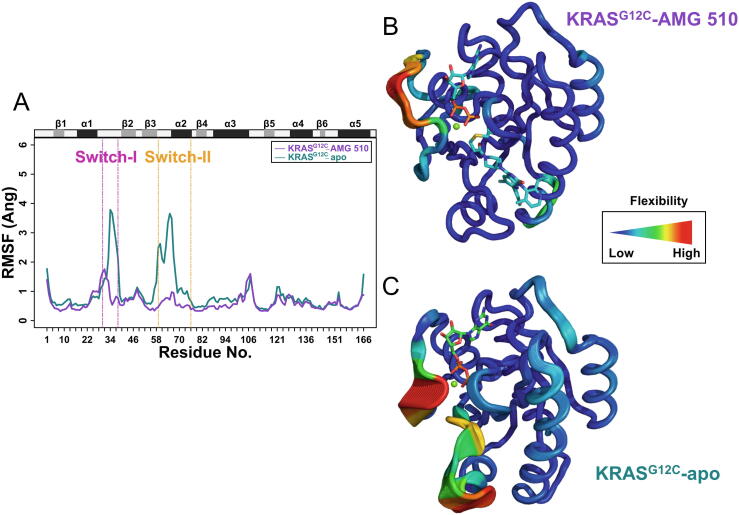
The catalytic domain of the RAS family has a relatively conserved conformation except for the two switch regions (SI and SII), whose conformations influence the interaction with other proteins [44]. To further illustrate the effect of AMG 510 on the structure of KRASG12C, we also performed RMSF analysis to compare the flexibility of the two systems. As shown in Fig. 3A, two peaks with higher fluctuation appear in the SI and SII of the KRASG12C-apo system, which are in accordance with previous observations regarding the dynamic structural feature of those regions. However, as for the KRASG12C-AMG 510, the RMSF values show a significant decrease, especially for the SII region. In the SI region, the fluctuation in the KRASG12C-AMG 510 is less than half compared to the case of KRASG12C-apo (average RMSF values: 1.080 vs. 2.403), while the ratio of the SII region fluctuation in the two systems is close to one-third (average RMSF values: 0.613 vs. 1.854). Fig. 3B and C represent the 3D structures of KRASG12C-AMG 510 and KRASG12C-apo, respectively. It is found that the former structure has lower flexibility in the two switch regions compared to the latter one. Collectively, the RMSF analysis reveals that AMG 510 keeps KRASG12C mutant stably in the inactive state by reducing its flexibility. In 2020, Pantsar explored the dynamic interaction of KRASG12C and AMG 510 through a 10 μs MD simulation, and he also found that AMG 510 reduced the the flexibility of the switch-II region [34]. However, our simulation results further observed the fluctuation of the switch-I reigon decreased during the 800 ns MD simulation, which was failed to be detected in Pantsar’s work. The difference between these two simulation results may be ascribed to different simulation settings.
Fig. 3.
Flexibility analysis of KRASG12C proteins. (A) RMSF values of Cα atoms in two systems (KRASG12C-AMG 510 and KRASG12C-apo). (B) Ribbon representations of the KRASG12C-AMG 510 and KRASG12C-apo structures. Note that the average RMSF value of each residue is annotated by the width of the ribbon as well as the color scheme. Briefly, residues of high fluctuations are denoted by red and thick ribbons, while blue and thin ribbons mean residues with low fluctuations. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Moreover, we calculated the RMSD values along with the simulation time between the two systems. The results indicate that the Cα RMSD values of the whole catalytic domain keep at about 1 Å in the KRASG12C-AMG 510 system during the 800 ns simulation process (Fig. S2A). Comparatively, the RMSD values of KRASG12C-apo are stably around 1 Å in the first 500 ns simulation but show a considerable increase in the 500–800 ns simulation time (Fig. S2A). We further examined the RMSD values of three regions (whole catalytic domain without two switch regions, SI and SII), and it can be inferred that the change of RMSD values between two systems mainly comes from the two switch regions (Fig. S2B-D).
3.3. AMG 510 decreases the correlation between two switch regions
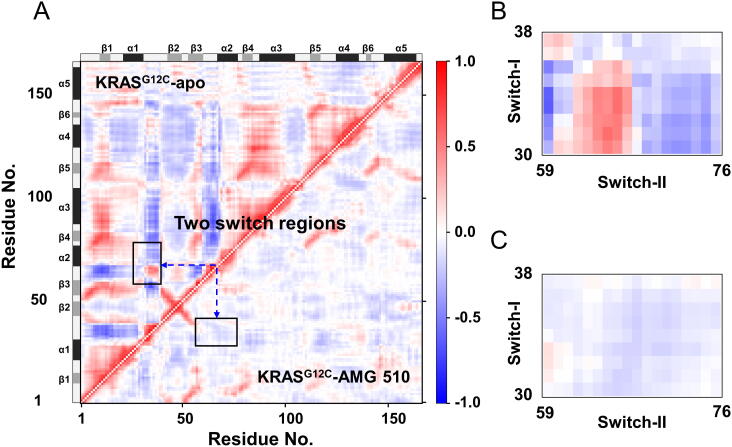
The signal transduction through KRAS follows a hypothetical scenario [44]. First, the upstream regulators are bound with the SII region and induce conformational change, promoting the exchange of GDP and GTP. Subsequently, the conformation of the SI region is changed to be in favor of binding with downstream effector proteins. Thus, although the proteins binding with the two switch regions are different in signal transduction, there is a specific correlation between the conformational changes. To investigate the effect of AMG 510 on the conformational dynamics of two switch regions of the KRASG12C mutant, the dynamics correlation analysis was performed. Fig. 4 displays the residue pair cross-correlations of KRASG12C-apo and KRASG12C-AMG 510.
Fig. 4.
Correlated motions of residue pairs in KRASG12C. (A) Combination of residue pair cross-correlation matrices between KRASG12C-apo (upper left triangle) and KRASG12C-AMG 510 (lower right triangle). The strength of cross-correlation is colored ranging from red (for Cij ≥ 0, lockstep motions) to blue (for Cij ≤ 0, anti-correlated motions). The enlarged plots of residue pair cross-correlations between switch-I (residues 30–38) and switch-II (residues 59–76) in KRASG12C-apo and KRASG12C-AMG 510 are shown in panels B and C, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4A shows the cross-correlation matrices of residue pairs in two systems under investigation, in which the upper triangle represents KRASG12C-apo, and the down triangle denotes KRASG12C-AMG 510. We observed considerable correlations of residue pairs in KRASG12C-apo. In particular, the two switch regions show stronger anti-correlated motion with other parts of the catalytic domain. By contrast, in the system of KRASG12C-AMG 510, the majority of cross-correlation values of residue pairs are close to 0, indicating that residue pairs share a reduced correlation. The dynamic correlation between two switch regions is further zoomed-in in Fig. 4B and C for KRASG12C-apo and KRASG12C-AMG 510, respectively. As shown in Fig. 4B, strongly correlated or anti-correlated motions exist in some residue pairs between the SI and SII regions. However, when KRASG12C binds with AMG 510, the two switch regions almost lose correlated and anti-correlated motions (Fig. 4C). These results indicate that AMG 510 could reduce the correlated motions of KRASG12C, especially the degree of coupling between two switch regions, which impair the conformational change induced by each other.
Regarding the scenario of KRAS signal transduction, the mechanism of AMG 510 inhibiting the propagation of tumor cells may be due to the fact that KRASG12C is bound to the covalent inhibitor, the residue fluctuation in two switch regions is significantly reduced, resulting in the degree of the correlated motion is also weakened. The conformational change induced by each other is hindered, followed by the efficiency of the exchange of GDP and GTP.
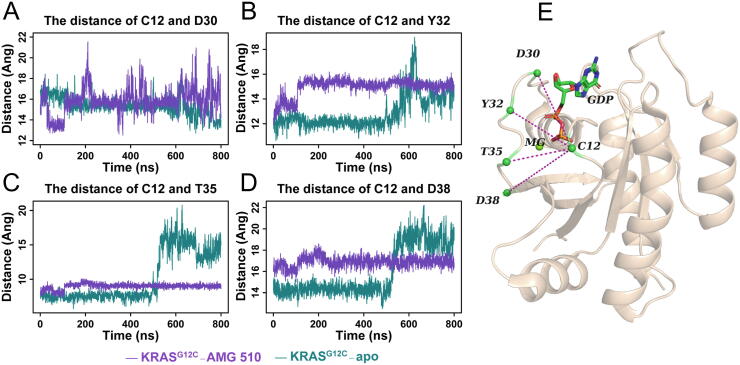
3.4. AMG 510 impedes the process of opening up the nucleotide-binding pocket
The structural diversity of the two highly flexible switch regions offers KRAS the ability to recognize or interact with different proteins, thus mediating multiple signaling pathways in the cells [2], [50]. When converting the inactive state (GDP-bound) to the active state (GTP-bound) of KRAS, the nucleotide-binding pocket opening, including the displacement of the residues in SI [51], is a critical step. Thus, we measured the Cα atom distances between four residues of SI (D30, Y32, T35, and D38) with C12 of P-loop (phosphate-binding loop, residues 10–17), which keep in a stable status during the 800 ns MD simulations, to compare the pocket change in KRASG12C-AMG 510 and KRASG12C-apo.
Fig. 5 shows the distance change of the four residue pairs along the simulation time. Regarding the KRASG12C-apo system, the three residue pairs (C12-Y32, C12-T35, and C12-D38) reveal sharply increased distances in the later period of simulation, although this phenomenon was not applied to the distance of C12-D30. Comparatively, the binding of AMG 510 with KRASG12C eliminates these distance changes. In other words, AMG 510 would stabilize the distance between C12 with the residues of the SI region, especially with the C-terminal residues of SI. Compared with the KRASG12C-apo structure, the nucleotide-binding pocket of KRASG12C-AMG 510 becomes relatively rigid, which is unfavorable to the open of binding pocket followed by the GDP release. In addition, from the statistical results of the non-bond interaction occupancy of AMG 510 with residues of KRASG12C, we observed that the residue T35 was able to form hydrophobic interaction with AMG 510 and had 34.8% occupancy during the 800 ns MD simulation process (Fig. S3). Collectively, it can be inferred that the flexibility of the nucleotide-binding pocket is reduced by the formation of hydrophobic interaction between AMG 510 and T35 of the SI region, which affects the process of pocket opening and then the interaction of KRAS and GEFs [52]. Therefore, the process of conversion from inactive to active state of KRASG12C catalyzed by GEFs may be impeded, resulting in the conformation of KRASG12C continuously kept in the GDP-bound state[10], [51].
Fig. 5.
Fluctuation analysis of the switch-I region revealed by four residue pair distances. Panel A-D shows the time-dependent Cα atom distance of C12-D30, C12-Y32, C12-T35, and C12-D38, respectively. Four monitored residue pairs are shown in the KRASG12C-AMG 510 structure (E).
3.5. AMG 510 enhances the affinity between KRASG12C and GDP
To explore the influence of the affinity of KRASG12C and GDP caused by the covalent binding of AMG 510, we extracted the last 300 frames of the two systems to calculate the binding free energy by MM/PBSA. As listed in Table 1, the calculated ΔGbind values of KRASG12C-AMG 510/GDP and KRASG12C-apo/GDP are −898.832 and −720.251 kcal/mol, respectively. In general, the low binding free energies in both systems are consistent with previous experimental observations that KRAS and nucleotides share high affinities in the cells [14]. The decrease of ΔGbind in the KRASG12C-AMG 510/GDP complex suggests that the binding of AMG 510 would further enhance the affinity between KRASG12C and GDP. In addition, except for the ΔGSA term, the rest three energetic components (ΔEvdw and ΔEele in the gas-phase, ΔGPB) mainly contribute to the ΔGbind differences in these two systems.
Table 1.
Binding free energies and the corresponding energy terms in two systems (KRASG12C-AMG 510/GDP and KRASG12C-apo/GDP).
| KRASG12C-AMG 510/GDP | KRASG12C-apo/GDP | |
|---|---|---|
| ΔEvdw | −17.302 ± 0.110 | −1.145 ± 0.004 |
| ΔEele | −762.164 ± 2.869 | −671.103 ± 2.479 |
| ΔGPB | −116.244 ± 0.711 | −44.689 ± 0.823 |
| ΔGSA | −3.122 ± 0.091 | −3.314 ± 0.501 |
| ΔGbind | −898.832 ± 3.781 | −720.251 ± 2.153 |
All energies in kcal/mol.
The binding energy was also decomposed at the residue level to interrogate the contribution of individual residues. As shown in Fig. S4, the decomposed binding energy results show that the critical residues (binding energy < -0.5 kcal/mol) in these two systems are similar and mainly concentrated in the P-loop and base-binding loops (residues 116–120 and 145–147). However, compared to the KRASG12C-apo/GDP system, the binding of AMG 510 could raise the energy contributions of those critical residues for the interaction of KRASG12C and GDP. In particular, the residue Y32 has an immense contribution to the binding free energy, which appears only in the KRASG12C-AMG 510/GDP system. The above results indicate that AMG 510 reduces the flexibility of the two switch regions and strengthens the affinity of KRASG12C and GDP. Therefore, the influence raised by AMG 510 renders the KRASG12C restricted in the GDP-bound conformation, and then hindering the process of its conversion to the active state.
3.6. Prediction of key residues and potential drug resistance sites of AMG 510
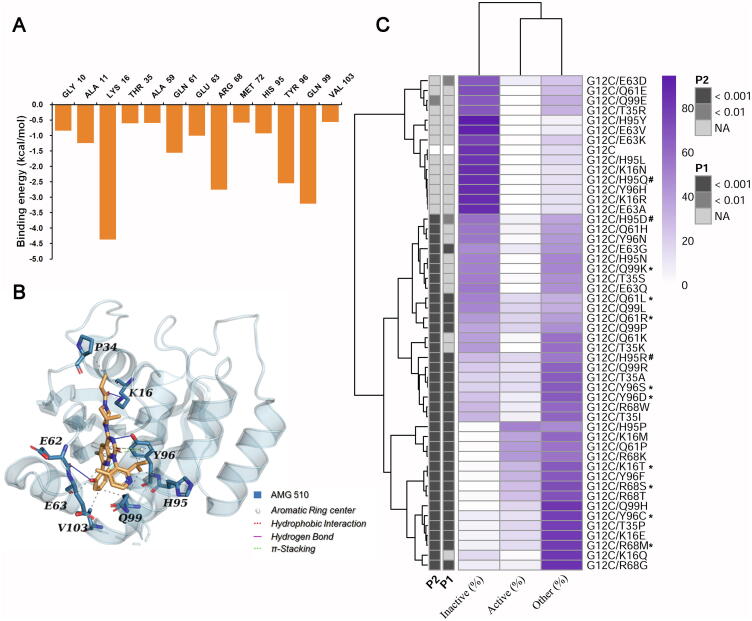
Drug resistance is a common and intractable clinical challenge in cancer therapy. Although AMG 510 presents promising efficacy in KRASG12C-mutant cancers, the drug resistance is very worthy of attention throughout the whole process of clinical trials. The missense mutations frequently cause the emergence of drug resistance, which would impair the interaction of drugs and their targeted proteins. Here, we utilized the strategy of binding energy decomposition based on the 800 ns MD simulation trajectories to predict the key residues contributing to the binding of AMG 510 and KRASG12C.
Fig. 6A shows the key residues (decomposed binding energy < -0.5 kcal/mol) for the binding of AMG 510. Fig. 6B represents the interaction residues and binding mode of KRASG12C with AMG 510 in the crystal structure, which was generated by the online PLIP server. In general, the residues forming non-covalent interactions with the inhibitor molecule are almost consistent with the key residues defined by the energy decomposition. In particular, three residues (H95/Y96/Q99) from SII-P play an essential role in the interaction between AMG 510 and KRASG12C. To explore the importance of these key residues for the affinity of KRASG12C and AMG 510, we performed a series of virtual alanine mutation experiments and compared the binding energy difference of AMG 510 with KRASG12C and KRASG12C/mutations. As shown in Table 2, the mutations at three residue positions (T35, R68, and Y96) considerably impacted the affinity between AMG 510 and KRASG12C (ΔΔGbind greater than 10 kcal/mol). For instance, the mutation Y96A introduced the neutral alanine, disrupted the π-stacking interaction formed by the phenyl ring of Y96 and AMG 510, and resulted in decreased binding affinity. Moreover, the side chains of T35 and R68 formed the hydrophobic interaction and hydrogen bond with AMG 510, respectively, to render the compound fixed in SII-P (Fig. S3). Therefore, the mutations of T35A and R68A would inevitably weaken the interaction between KRASG12C and AMG 510. Based on the above analysis, the mutations at T35/R68/Y96 would influence the affinity of the covalent inhibitor in the SII-P of KRASG12C and then reduce the efficacy of the compound. Thus, we speculated that the residues T35/R68/Y96 would be the potential drug resistance sites. Intriguingly, two sites (R68 and Y96) have been proved by existing clinical data [31]. A possible explanation of the site T35 unseen in clinical data is that the mutation of T35 could compromise the ability of KRAS to bind effector proteins, which precludes its selective advantage as a resistant mutant.
Fig. 6.
Identification of key residues and potential acquired resistance mutations of KRASG12C interacting with AMG 510. (A) The residues with decomposed energy values less than −0.5 kcal/mol for the binding between AMG 510 and KRASG12C. (B) The non-bond interactions formed by AMG 510 with KRASG12C in its crystal structure (PDB entry: 6OIM). (C) The distributions of KRAS conformation states of KRASG12C/mutations simulation systems. The experimentally validated resistant mutations for AMG 510 (sotorasib) were marked with asterisks, and the experimentally validated non-resistant mutations for AMG 510 were marked with “#”. P1 denotes the p value of chi-square test for active and inactive states of KRASG12C and KRASG12C/mutations, while P2 stands for the p value of chi-square test for active-like (active plus other) and inactive states of KRASG12C and KRASG12C/mutations.
Table 2.
Binding energy change of AMG 510 interacting with various KRASG12C mutants.
| △G (kcal/mol)a | △△G (kcal/mol)b | |
|---|---|---|
| KRASG12C | −51.664 | 0.000 |
| KRASG12C/K16A | −50.189 | 1.475 |
| KRASG12C/T35A | −37.934 | 13.730 |
| KRASG12C/Q61A | −43.118 | 8.547 |
| KRASG12C/R68A | −40.793 | 10.872 |
| KRASG12C/H95A | −45.988 | 5.676 |
| KRASG12C/Y96A | −32.879 | 18.786 |
| KRASG12C/Q99A | −48.376 | 3.288 |
| KRASG12C/V103A | −43.114 | 8.550 |
The binding energy of AMG 510 interacting with protein single mutant (i.e. KRASG12C) or double mutants (KRASG12C/mutations).
The binding energy change of double mutants relative to single mutant (i.e. KRASG12C).
To further detect resistant mutations, we have systematically screened all possible missense mutations for the identified key residues based on the genetic codon changes. In other words, for the triplet codon of one selected residue, one nucleotide is mutated each time, and the corresponding missense mutation is recorded. We performed virtual mutation experiments and MD simulations on a total of 50 KRASG12C/mutations systems (Fig. 6C). Then, simulation trajectories from each system were mapped to our PCA model. The distributions of active and inactive conformations of each simulation system were tested with chi-square test. The proportions of three conformation types in the 50 simulation systems and the corresponding p values are listed in Table S3.
As depicted in Fig. 6C, two major clusters were observed in all the 50 virtual mutation systems based on KRAS conformation states. Cluster #1 included 14 virtual mutation systems, which shared similar conformational distribution with KRASG12C, indicating that the introduced secondary mutations in KRASG12C did not affect the interaction of KRASG12C and AMG 510. The remaining 36 virtual mutation systems in Cluster #2 shared common features with the increased percentage of KRAS active and/or other conformation (Fig. 6C). As shown in Fig. 2B, the other conformation tended to be active-like. Therefore, the introduced second mutations in these 36 virtual mutation systems might be potential resistant mutations for AMG 510. In these simulation systems, the inactive conformations remarkably reduced accompanied by the increase of active or other conformations in comparison to the KRASG12C system (p < 0.01, chi-square test), implying that these potential resistant mutations significantly influence the interaction between KRASG12C and AMG 510. Since different KRASG12C inhibitors present distinct resistance mutation profiles [29], [30], [31], in this work we only focused on experimental resistance mutations for AMG 510. We collected 12 experimentally validated mutations associated with our virtual mutation systems, which were obtained from a positive-selection screen experiment in Ba/F3 cells[32], including nine resistance mutations and three non-resistance mutations (i.e., the mutation remains sensitive to AMG 510 according to the current experimental result). Interestingly, all of nine experimentally validated resistance mutations are identified as resistance to AMG 510 with our PCA model (Fig. 6C). One experimentally validated non-resistance mutations (H95Q) is accurately predicted in our PCA model (Fig. 6C). However, the other two experimentally validated non-resistance mutations, H95D and H95R, are incorrectly predicted to be resistance mutations with our PCA model (Fig. 6C). Overall, the aforementioned prediction and structural understanding of resistance mutations should provide crucial hints to develop new therapeutic strategies or drugs for overcoming acquired resistance of AMG 510.
3.7. AMG 510 has the potential efficacy for HRASG12C or NRASG12C
Although KRAS is the most frequently mutated isoform in RAS-driven cancers, some cases are also associated with mutations in HRAS or NRAS [1], [13]. These three RAS proteins share high similarities in sequence and structure (Fig. S5). The pairwise full-length sequence identities reach approximately 85% (Fig. 1A, Fig. S5A). In case the HVRs in their C-terminal are not taken into account, the catalytic domains (residues 1–166) among them share more than 90% sequence identity (Fig. S5A). Although AMG 510 was originally designed to treat the KRASG12C-induced cancers, in this work we also attempted to examine the potential interactions between AMG 510 and HRASG12C or NRASG12C. According to previous analysis results, the three residues 95/96/99 of SII-P play crucial roles in the interaction of AMG 510 and KRASG12C. Although position 95 is one of the five positions around SII-P region containing different residues (Fig. S5B and C), the H95A mutation does not significantly reduce the affinity of AMG 510 and KRASG12C (Table 2). In addition, Table S4 shows the calculated binding energy of AMG 510 and three RAS proteins, and we can find that the ΔGbind values between three mutants and the covalent inhibitor are at a similar level. Comparatively, the values in the HRASG12C or NRASG12C systems are slightly greater than the one in KRASG12C. In addition, the results of energy decomposition also reflect that the key residues of HRASG12C or NRASG12C with AMG 510 are similar to those of KRASG12C (Fig. S5D). Given that these three RAS proteins share sufficient structural similarity as well as similar binding energy with AMG 510, we can infer that AMG 510 may not only target HRASG12C or NRASG12C but also restrict them to an inactive state and impede the step of activation. We hope the above computational experiment can provide some hints for the repurposing of AMG 510 in targeting HRASG12C or NRASG12C, although its actual inhibitory effect still needs further experimental validation.
4. Conclusions
In the present work, we combined multifaceted computational methods, including MD simulation, MM/PBSA calculation, and trajectories analysis, to investigate the molecular interaction mechanism between KRASG12C and AMG 510. The PCA-based structural state classifier quantitatively identified that the frames extracted from the KRASG12C-AMG 510 system have a higher proportion in the inactive conformation during the 800 ns MD simulation compared to those from the KRASG12C-apo system. Then, the trajectory analysis reflected that the reduction in the flexibility of the two switch regions was the primary reason for KRASG12C conformation change. In addition, AMG 510 formed the hydrophobic interactions with T35 to fix the fluctuation of the SI region and impeded the opening of the nucleotide-binding site. Meanwhile, the MM/PBSA calculation demonstrated that AMG 510 enhances the affinity of KRASG12C and GDP. These results suggest that AMG 510 reduced the flexibility of two switch regions in KRASG12C to render it located in the GDP-bound state, and then blocked the activation of KRASG12C.
We predicted the key residues contributing to the interaction between KRASG12C and AMG 510 through the binding energy decomposition. Further large-scale in silico mutagenesis experiment not only provided mechanistic insights into known acquired resistance mutations to AMG 510 but also proposed a series of new potential drug resistance variations, which are deserved further clinical attention. From the similarity of protein sequence and 3D structure between KRAS and HRAS/NRAS, we speculated the potential inhibitory effects of AMG 510 on HRASG12C or NRASG12C and suggested the pursuit of experimental approaches to determine whether AMG 510 targets HRASG12C or NRASG12C in vivo. Taken together, our work provides an atomic-level characterization of the KRASG12C conformational dynamics when bound with the covalent inhibitor AMG 510. It is also hoped that the current work can serve as a proof-of-concept regarding the application of structural bioinformatics in precision oncology.
CRediT authorship contribution statement
Yu Li: Methodology, Formal analysis, Writing - original draft. Lei Han: Conceptualization, Formal analysis, Writing - review & editing. Ziding Zhang: Conceptualization, Resources, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFC1313000, 2018YFC1313001). We thank the support of the high-performance computing platform of the State Key Laboratory of Agrobiotechnology.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.02.018.
Contributor Information
Lei Han, Email: leihan@tmu.edu.cn.
Ziding Zhang, Email: zidingzhang@cau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 3.Dharmaiah S., Bindu L., Tran T.H., Gillette W.K., Frank P.H., Ghirlando R., et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc Natl Acad Sci U S A. 2016;113:E6766–E6775. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karnoub A.E., Weinberg R.A. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pantsar T. The current understanding of KRAS protein structure and dynamics. Comput Struct Biotechnol J. 2020;18:189–198. doi: 10.1016/j.csbj.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo S.P., Coulson J.M., Prior I.A. RAS variant signalling. Biochem Soc Trans. 2018;46:1325–1332. doi: 10.1042/BST20180173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore A.R., Rosenberg S.C., McCormick F., Malek S. RAS-targeted therapies: Is the undruggable drugged? Nat Rev Drug Discov. 2020;19:533–552. doi: 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter J.C., Manandhar A., Carrasco M.A., Gurbani D., Gondi S., Westover K.D. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 11.Gebregiworgis T., Kano Y., St-Germain J., Radulovich N., Udaskin M.L., Mentes A., et al. The Q61H mutation decouples KRAS from upstream regulation and renders cancer cells resistant to SHP2 inhibitors. Nat Commun. 2021;12:6274. doi: 10.1038/s41467-021-26526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan M.B., Corcoran R.B. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol. 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 14.Lanman B.A., Allen J.R., Allen J.G., Amegadzie A.K., Ashton K.S., Booker S.K., et al. Discovery of a covalent inhibitor of KRASG12C (AMG 510) for the treatment of solid tumors. J Med Chem. 2020;63:52–65. doi: 10.1021/acs.jmedchem.9b01180. [DOI] [PubMed] [Google Scholar]
- 15.Gasper R., Wittinghofer F. The Ras switch in structural and historical perspective. Biol Chem. 2019;401:143–163. doi: 10.1515/hsz-2019-0330. [DOI] [PubMed] [Google Scholar]
- 16.Ostrem J.M.L., Shokat K.M. Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat Rev Drug Discov. 2016;15:771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 17.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kessler D., Gmachl M., Mantoulidis A., Martin L.J., Zoephel A., Mayer M., et al. Drugging an undruggable pocket on KRAS. Proc Natl Acad Sci. 2019;116:15823–15829. doi: 10.1073/pnas.1904529116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S., Jang H., Gu S., Zhang J., Nussinov R. Drugging Ras GTPase: A comprehensive mechanistic and signaling structural view. Chem Soc Rev. 2016;45:4929–4952. doi: 10.1039/c5cs00911a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Res A.H.C., Biomol A.J.J., Natl D.M.P., Sci A., John J., Sohmen R., et al. Kinetics of interaction of nucleotides with nucleotide-free H-ras. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 21.Maurer T., Garrenton L.S., Oh A., Pitts K., Anderson D.J., Skelton N.J., et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen I., Shanahan F., Merchant M., Orr C., Hunsaker T., Durk M., et al. Pharmacological induction of RAS-GTP confers RAF inhibitor sensitivity in KRAS mutant tumors. Cancer Cell. 2018;34:611–625. doi: 10.1016/j.ccell.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 23.De Cesco S., Kurian J., Dufresne C., Mittermaier A.K., Moitessier N. Covalent inhibitors design and discovery. Eur J Med Chem. 2017;138:96–114. doi: 10.1016/j.ejmech.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Sutanto F., Konstantinidou M., Dömling A. Covalent inhibitors: A rational approach to drug discovery. RSC Med Chem. 2020;11:876–884. doi: 10.1039/d0md00154f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Kang R, Tang D. The KRAS-G12C inhibitor: Activity and resistance. Cancer Gene Ther 2021;2021. 10.1038/s41417-021-00383-9. [DOI] [PubMed]
- 26.Sunaga N., Miura Y., Kasahara N., Sakurai R. Targeting oncogenic kras in non-small-cell lung cancer. Cancers (Basel) 2021;13:5956. doi: 10.3390/cancers13235956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 28.Hallin J., Sudhakar N., Bowcut V., Baer B.R., Ballard J.A., Burkard M.R., et al. The KRASG12C inhibitor, MRTX849, provides insight toward therapeutic susceptibility of KRAS mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skoulidis F., Li B.T., Dy G.K., Price T.J., Falchook G.S., Wolf J., et al. Sotorasib for lung cancers with KRAS p. G12C mutation. N Engl J Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koga T., Suda K., Fujino T., Ohara S., Hamada A., Nishino M., et al. KRAS secondary mutations that confer acquired resistance to KRAS G12C inhibitors, sotorasib and adagrasib, and overcoming strategies: Insights from in vitro experiments. J Thorac Oncol. 2021;16:1321–1332. doi: 10.1016/j.jtho.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka N., Lin J.J., Li C., Ryan M.B., Zhang J., Kiedrowski L.A., et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11:1913–1922. doi: 10.1158/2159-8290.cd-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awad M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W., et al. Acquired resistance to KRASG12C inhibition in cancer. N Engl J Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasta JD, Peacock DM, Zheng Q, Walker JA, Zhang Z, Zimprich CA, et al. KRAS is vulnerable to reversible switch-II pocket engagement in cells. BioRxiv 2021:2021.10.15.464544. 10.1101/2021.10.15.464544. [DOI] [PMC free article] [PubMed]
- 34.Pantsar T. KRAS(G12C)–AMG 510 interaction dynamics revealed by all-atom molecular dynamics simulations. Sci Rep. 2020;10:11992. doi: 10.1038/s41598-020-68950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter J.C., Gurbani D., Ficarro S.B., Carrasco M.A., Lim S.M., Choi H.G., et al. In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc Natl Acad Sci U S A. 2014;111:8895–8900. doi: 10.1073/pnas.1404639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinforma 2014;2014:5.6.1-5.6.32. 10.1002/0471250953.bi0506s47. [DOI] [PubMed]
- 37.Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the amber biomolecular simulation package. Wiley Interdiscip Rev Comput Mol Sci. 2013;3:198–210. doi: 10.1002/wcms.1121. [DOI] [Google Scholar]
- 38.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J Comput Chem. 2004;56531:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 40.Lambrakos S.G., Boris J.P., Oran E.S., Chandrasekhar I., Nagumo M. A modified shake algorithm for maintaining rigid bonds in molecular dynamics simulations of large molecules. J Comput Phys. 1989;85:473–486. doi: 10.1016/0021-9991(89)90160-5. [DOI] [Google Scholar]
- 41.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 42.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 43.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milburn M.V., Tong L., DeVos A.M., Brünger A., Yamaizumi Z., Nishimura S., et al. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 45.Shima F., Ijiri Y., Muraoka S., Liao J., Ye M., Araki M., et al. Structural basis for conformational dynamics of GTP-bound ras protein. J Biol Chem. 2010;285:22696–22705. doi: 10.1074/jbc.M110.125161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hillig R.C., Sautier B., Schroeder J., Moosmayer D., Hilpmann A., Stegmann C.M., et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS–SOS1 interaction. Proc Natl Acad Sci U S A. 2019;116:2551–2560. doi: 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David L., Luo R., Gilson M.K. Comparison of generalized born and poisson models: Energetics and dynamics of HIV protease. J Comput Chem. 2000;21:295–309. doi: 10.1002/(SICI)1096-987X(200003)21:4<295::AID-JCC5>3.0.CO;2-8. [DOI] [Google Scholar]
- 49.Weiser J., Shenkin P.S., Still W.C. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J Comput Chem. 1999;20:217–230. doi: 10.1002/(SICI)1096-987X(19990130)20:2<217::AID-JCC4>3.0.CO;2-A. [DOI] [Google Scholar]
- 50.Gimple R.C., Wang X. RAS: Striking at the core of the oncogenic circuitry. Front Oncol. 2019;9:965. doi: 10.3389/fonc.2019.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boriack-Sjodin P.A., Margarit S.M., Bar-Sagi D., Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 52.Cherfils J., Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.