Highlights
-
•
Automation led to the localization of FtsZ filaments and constriction mechanism.
-
•
CEMOVIS confirmed the presence of the mycoloic-acid outer membrane in Mycobacteria.
-
•
Cryo-FIB SEM illustrated the sporulation process and asymmetrical cell division.
-
•
Cryo-CLEM was used for the identification of a non-canonical T6SS.
-
•
Subtomogram averaging led to higher resolution by overcoming missing information reveals internal conformation changes and mechanisms.
-
•
Perspective of further coming advancements.
Keywords: Cryo-ET, Data processing, CEMOVIS, FIB-milling, Cryo-CLEM, Subtomogram averaging
Abstract
Advancements in the field of cryo-electron tomography have greatly contributed to our current understanding of prokaryotic cell organization and revealed intracellular structures with remarkable architecture. In this review, we present some of the prominent advancements in cryo-electron tomography, illustrated by a subset of structural examples to demonstrate the power of the technique. More specifically, we focus on technical advances in automation of data collection and processing, sample thinning approaches, correlative cryo-light and electron microscopy, and sub-tomogram averaging methods. In turn, each of these advances enabled new insights into bacterial cell architecture, cell cycle progression, and the structure and function of molecular machines. Taken together, these significant advances within the cryo-electron tomography workflow have led to a greater understanding of prokaryotic biology. The advances made the technique available to a wider audience and more biological questions and provide the basis for continued advances in the near future.
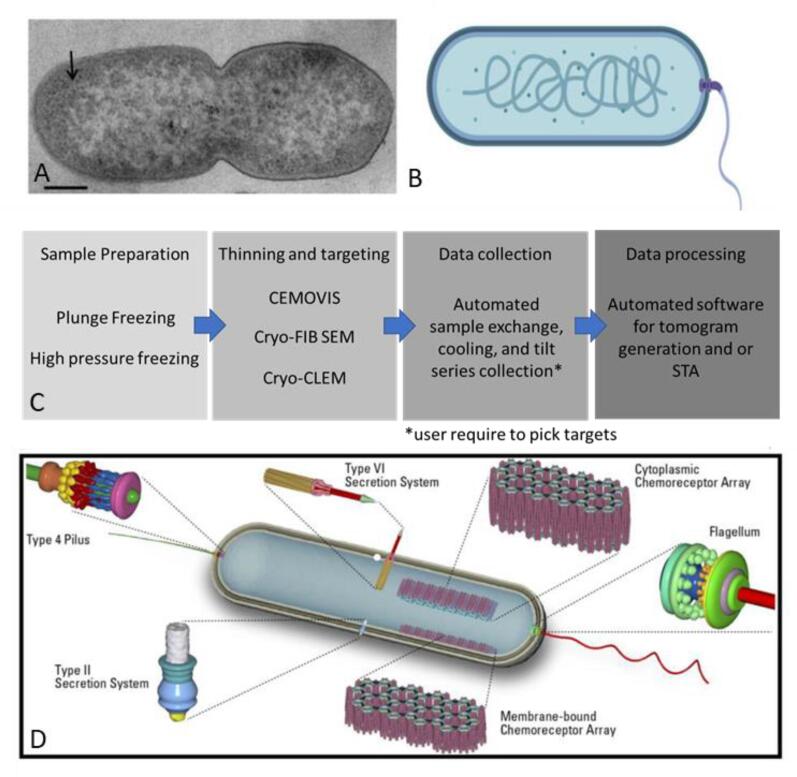
Microscopy has played a crucial role in gaining insights into the hidden world of bacteria. The ability to directly observe live bacterial cells has allowed us to gain understanding into the basic biology of these organisms, such as their shape, growth and division cycles, and motility behaviors. With the advent of traditional electron microscopy (EM), the finer structural details of bacteria became clearer: differences in cell envelope structures could be distinguished, and the intercellular contents, such as the nucleoid and storage granules, could be identified. However, more detailed information could not be gained due to the nature of the sample preparation: chemical fixation, dehydration, plastic embedding, and staining of the sample largely obscured the delicate ultrastructure of the bacterial cells (Fig. 1A). Subsequently, the usefulness of EM was thought to be limited and significant advances in genetic engineering and the development of fluorescent tags rendered EM a niche method. Therefore, despite increasingly detailed insights into the biology of bacterial cells gained by such alternative microbiological methods, many structural aspects of bacteria remained obscure. This lack of insight is still evident in the typical cell architecture comparisons between eukaryotic and bacterial cells in textbooks. While eukaryotic cell types were known to contain a myriad of structures, such as a diverse cytoskeleton, Golgi apparatus, and mitochondria, bacterial cells were often depicted with a generic rod-shaped cell containing only a nucleoid, ribosomes, cell envelope layers, and occasional appendages (Fig. 1B).
Fig. 1.
Changing view of the bacterial cell structure. A; Traditional EM image of Pseudomonas aeruginosa (arrow indicates tightly packed material). Image reproduced from (Latino et al., 2019) with permission. B; Simplified schematic of a bacterial cell, lacking structural detail (made in Biorender.com). C; Workflow of cryo-electron tomography. D; Modern interpretation of a bacterium, containing selected structural features (schematic by Robert van Sluis).
The introduction of cryo-electron tomography (cryo-ET) made it possible to elucidate the finer structural features of bacterial cells in a near-native state, in three dimensions, and at macromolecular resolution (briefly described in Fig. 1C; see Cyrklaff et al., 2017 & Turk and Baumeister, 2020 for more details). To do this, the bacterial cells are flash-frozen on an EM support grid in a liquid-nitrogen-cooled cryogen (Dubochet et al., 1988). This freezing process is so fast that water molecules do not form crystalline ice. Instead, the sample is embedded in vitreous ice which preserves the delicate ultrastructure of the cells (Dubochet, 2007, Dubochet et al., 1988). Vitrified samples are then imaged using a cryogenic transmission electron microscope (cryo-TEM). The cryo-TEM collects a series of 2-dimensional (2D) images while the sample is being tilted with respect to the electron beam (a common tilt range is + 60° to –60° at 2° increments). The resulting set of images, referred to as a tilt-series, can then be computationally back-projected to generate a 3-dimensional (3D) volume of the target (referred to as a tomogram). However, the potential impact of this workflow on the current understandings of bacterial ultrastructure was not immediately realized by microbiologists. A decade after its beginning, the Baumeister group first applied this technology to prokaryotic cells in 1998 (Grimm et al., 1998). Even with this achievement, in the following decade, only a small number of studies used this method on bacterial cells (reviewed in Jensen and Briegel, 2007).
Today, cryo-ET is widely recognized as a powerful tool to unravel the structural aspects of microbes. This method has greatly contributed to our understanding of how microbes are structurally organized, how they grow and divide using a highly complex cytoskeleton, and how they use molecular machines such as secretion systems, cell appendages, and chemotaxis arrays to navigate and interact with the environment (Fig. 1D). In its early days, data collection was practically manual, requiring constant supervision. However, automation of the technique quickly advanced and the data collection process is now largely self-operating (Baldwin et al., 2018, Mastronarde, 2005, Thompson et al., 2016). In addition, improvements in the hardware, such as new types of cameras/detectors, the ability to thin vitrified samples at cryogenic temperatures, the development of correlative techniques with fluorescent light microscopy, and new software packages to analyze the data, aided in raising cryo-ET to a key method in the prokaryotic biology toolbox (Koning et al., 2018, Turk and Baumeister, 2020).
Cryo-ET automation enabled insight into bacterial cytoskeleton
Following its proof of concept, cryo-ET was first practically used to describe novel cell attachment structures (Grimm et al., 1998, Nickell et al., 2003). These studies relied on only a handful of tomograms, since data collection and processing were time-consuming and mostly manual. Specifically, data collection had to be supervised to ensure proper imaging conditions throughout the tilt-series, such as accurate target tracking and maintaining defocus settings of the individual images. The subsequent processing of the 2D images to generate the tomograms was equally taxing. For instance, the early software programs, such as the EM software (Hegerl, 1996), required the user to manually track gold particles in each image. These gold beads are typically added to the sample before freezing to allow for an accurate alignment of the images of a tilt-series, which ensures a precise 3D reconstruction.
This manual workflow resulted in a low throughput of data acquisition and processing, which in turn greatly limited insights into many aspects of bacterial cell biology. The development of robust cryo-ET data collection software provided the means for a higher sample throughput (for example, TOM toolbox, UCSFtomo, FEI software, and serialEM) (Mastronarde, 2005, Nickell et al., 2005, Zheng et al., 2007). In addition, technical improvements such as hardware upgrades that allows maintaining the liquid nitrogen cooling of the samples overnight enables the automation of tilt-series collection without supervision. This advance greatly increased the number of tilt-series collected. In turn, it necessitated the creation of automated data processing routines such as IMOD and Warp (Kremer et al., 1996, Tegunov and Cramer, 2019). These new software allowed the automatic processing of the collected tilt-series, both for quality checks and tomogram generation. A good description of the data processing steps can be found for example in the article of Baldwin et al. (2018).
In addition to the technical developments directly related to instrumentation and software, advances in computational processing power with improved CPUs and GPUs greatly improved data processing times. Furthermore, the advent of direct electron detectors capable of detection of individual electron events with high speed and improved accuracy, have aided in the increased automation of the workflow as well as greatly improved image quality (Baldwin et al., 2018, McMullan et al., 2016). Together, these improvements not only result in more reliable data but also increase the amount of information available for analysis. These advancements directly led to a better understanding of the components of a complex system such as the bacterial cytoskeleton.
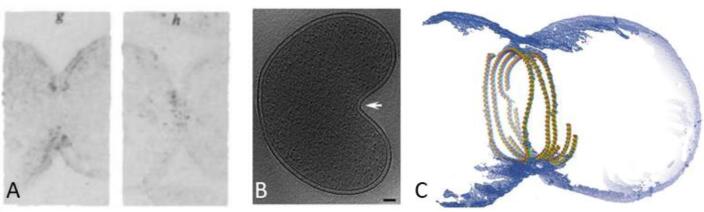
In eukaryotes, it has long been known that the cytoskeleton is an intracellular network system of filaments and tubules that defines the structure of a cell and gives it coherence in its spatial–temporal operations (Szwedziak and Ghosal, 2017). In contrast, bacteria were thought to lack cytoskeletal elements and therefore were devoid of any internal order. Later, it was discovered that bacteria contain homologs of eukaryotic cytoskeletal filaments, but many questions regarding their detailed structure and function remained. Homologs of all three main protein classes of the eukaryotic cytoskeleton were discovered in bacteria, for example, FtsZ (tubulin-like), MreB (actin-like), and CreS (intermediate filament-like). Of these, the first protein of the bacterial cytoskeleton described was FtsZ. This protein was shown to be essential for cell division in most bacteria. Bi & Lutkenhaus (1991) were the first to visualize the FtsZ filaments by immuno-electron microscopy (Fig. 2A). In this study, the FtsZ filament localization at the division site was revealed. With this information, the authors proposed an FtsZ assembly model that proposed a complete ring formation of FtsZ filaments. However, the question if the FtsZ filaments indeed formed such a continuous ring remained.
Fig. 2.
FtsZ structure; (A) comparison from the first EM visualization of the FtsZ structure within Escherichia coli cell (Bi and Lutkenhaus, 1991) with (B) the current models of asymmetric constriction of a Belliella baltica cell (white arrow indicates the asymmetric division site) (Yao et al., 2017) and (C) a visualization of a semi-atomic model of symmetric FtsZ ring constriction in a liposome (Szwedziak et al., 2014).
While cryo-ET is very powerful in resolving structures in intact cells, determining their identity remains a challenge. At present, there are no easily applicable tags comparable to the fluorescent markers used in light microscopy. Instead, the identity of a structure of interest can be indirectly determined by imaging a variety of mutant strains, where the protein of interest is altered in some way. This includes genetic methods such as protein overexpression, deletion or depletion, the generation of non-hydrolyzable variants, or the use of chemicals affecting the protein polymerization. Images collected of these samples are then compared to the wild-type strain to identify the target structure. This requires a large amount of data to be collected and therefore requires a large degree of automation in the workflow to be feasible. This approach was used for example in an early cryo-ET study that compared cryo-ET data of several different FtsZ mutants in Caulobacter crescentus. This research gained insight into the appearance of FtsZ filaments in the cell cycle, its position, orientation, and number and length. The cryo-ET data revealed that FtsZ forms filaments that are relatively short (∼100 nm) and overlapping in vivo. This insight led to the proposition of the ‘iterative pinching model’ (Li et al., 2007).
While early studies proposed that FtsZ does not form a complete ring in C. crescentus, its presence in other bacteria remained disputed (Szwedziak et al., 2014, Yao et al., 2017). The reason for this lies partly in the ‘missing wedge’ artefact created during tilt-series collection because the sample is usually only tilted to +/- 60°due to the decreasing electron transparency of the sample. This in turn results in missing information which could not be obtained from the greater tilt angles, resulting in the absence of discernable features on the top and bottom of a cell. As a consequence, the FtsZ ring could not be visualized as a complete ring (Szwedziak et al., 2014). Recent advancements try to reduce the challenges caused by missing wedge artefacts by using computational algorithms and deep learning based software (Koning et al., 2018, Liu et al., 2021, Penczek et al., 1995, Zhai et al., 2020). Another approach to reduce the missing wedge artefacts is the use of dual-tilt with a rotating specimen stage, where the sample is tilted in the X- and Y-axis (Koning et al., 2018, Penczek et al., 1995). However, dual-tilt cryo-tomography is currently rarely used due to the additional challenge of acquiring and processing this dual-tilt data. Furthermore, the total dose is spread twice over the number of tilts, which decreases the signal-to-noise ratio in each image.
Newer cryo-ET studies support the idea that a complete FtsZ ring is not needed to drive successful cell constriction and division in several species besides C. crescentus (Fig. 2). Yao et al. (2017) visualized various bacterial species in different division stages (pre-, early, mid, and post constriction stage). Additionally, they utilized a cryo-ET database which contained more than 15,000 tomographic 3D images of 88 different species (Ortega et al., 2019, Yao et al., 2017). They categorized the imaged cells in the mentioned stages and ultimately identified 159 dividing cells of 47 species. Using this information, they confirmed that FtsZ filaments can indeed be found in different lengths and forms. Furthermore, they highlighted that the constriction process takes longer than the lifetime of the individual FtsZ filaments (Yao et al., 2017). Despite all these insights, many questions remain on the cell division process, especially in the orchestration of the FtsZ ring formation and constriction in the context of the growing number of proteins that are known to be involved in this process, together called the “Divisome”. Cryo-ET may also help answer these open questions in the future.
Cryogenic electron microscopy of vitreous section (CEMOVIS) as a method to visualize cell envelope layers
A limitation of EM is the thickness of the sample that can be successfully imaged. When a sample is too thick for direct imaging (typically greater than 400–500 nm) the chances of elastic and inelastic scattering increases which causes a reduction of transmission through the sample that in turn leads to a loss of structural information. Therefore, various techniques have been developed to gain insight into the architecture of samples with a thickness beyond the scope of plunge freezing (limited to 10–20 µm in thickness; Dubochet et al., 1988). Instead, these samples are high pressure frozen. This process vitrifies the sample by applying high pressure and rapid cooling to liquid nitrogen temperatures (Moor, 1987). As with plunge freezing, this sample preparation method preserves the delicate structure of the biological material. The frozen sample is subsequently transferred to a cryo-ultramicrotome and sectioned with a diamond knife into cryo-EM suitable sections (Al-Amoudi et al., 2004). This sample thinning technique is referred to as cryogenic electron microscopy of vitreous sections (CEMOVIS).
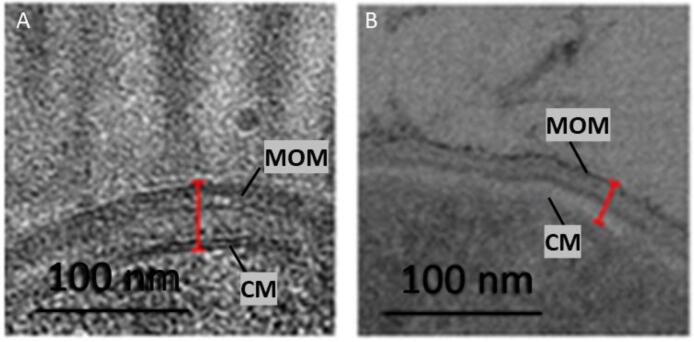
The key difference between traditional EM methods and cryo-EM lies in the sample preparation. The traditional chemical fixation method is strongly dependent on the chemicals used for fixation, staining, and embedding, all of which can lead to significant structural changes. For example, one of the most famous artefacts is the mesosome which was believed to be a compartment of the bacterial cell for many years (Bleck et al., 2010, Chapman and Hillier, 1953, Ebersold et al., 1981, Robertson, 1959). However, with the application of the CEMOVIS technique, mesosomes were found to be artefacts caused by chemical fixation (Bleck et al., 2010, Ebersold et al., 1981). The advantages of CEMOVIS are that it circumvents the artefacts associated with chemical treatment and gives the possibility of serial-sectioning. One disadvantage of CEMOVIS is that it is not possible to combine it with immunogold labeling methods for structures inside of cells. Another disadvantage is the generation of physical artefacts caused by the mechanical stress of sectioning, for example, knife marks and compression. These mechanical artefacts are well known and characterized thus can be considered for interpretation (Alamoudi et al., 2005). Bleck et al. (2010) compared the different sectioning methods with each other and concluded that even though CEMOVIS is not an artefact-free method, it is still the preferable method when compared to chemical fixation. It provides a better understanding of biological structure within a cell and even tissue, which are usually too thick for direct imaging. For example, CEMOVIS was used to image the cell envelope of mycobacteria and to confirm the existence of their unique cell envelope. The cell envelope is a multi-component, layered structure that is responsible for maintaining cell shape as well as protection from environmental stressors. Mycobacteria have an unusual outer membrane that consists mainly of mycolic acid, a lipid that gets easily dissolved by chemical treatment. Thus, due to this characteristic, it was not possible to visualize it with the common sectioning methods and its existence was controversial until 2008 (Fig. 3). CEMOVIS was crucial in confirming the presence of this structure.
Fig. 3.
Comparative micrographs of Mycobacteria smegmatis cell envelope for the different sectioning types; CEMOVIS (A); and classical preparation with chemical fixation and resin embedding (B). Red line indicates the cell wall thickness, whereby the inner and outer membranes are easily detectable in A compared to B. Adapted from (Bleck et al., 2010). CM, cytoplasmic membrane; MOM, mycobacterial outer membrane.
Advances in sample thinning for cryo-ET
Since the introduction of cryo-ET, sample thickness has been a major limitation. The general guideline within the community is that the maximum thickness of the sample is roughly twice the power of the electron beam. Theoretically, this has been predicted to be slightly less, but other factors such as the contents of the sample can also play a role (Yonekura et al., 2006). Thus, working with a 300 keV instrument, the electrons will pass through a sample of up to 600 nm in effective thickness. However, it is important to consider that the increasing thickness of the sample also diminishes the availability of high-resolution information, thus limiting the questions that can be addressed.
About fifteen years ago, a technique typically used in material sciences called focused ion beam scanning electron microscopy (FIB SEM) was adapted to work at cryogenic temperatures and it was shown to be suitable for vitrified biological samples (Marko et al., 2007). Now referred to as cryo-focus ion beam scanning electron microscopy (cryo-FIB SEM), the technique preserves the vitreous nature of the sample while using the focused ion beam to systematically remove material from the sample until it is sufficiently thin. The remaining material, typically with a thickness of 150 – 300 nm, is referred to as a lamella. The SEM beam is used to monitor the progress of the thinning process. Cryo-FIB SEM has evolved into a valuable method to thin plunge frozen samples that are otherwise too thick for cryo-ET, and it has been used to study a variety of samples and biological questions (Melia et al., 2021, Weiss et al., 2019).
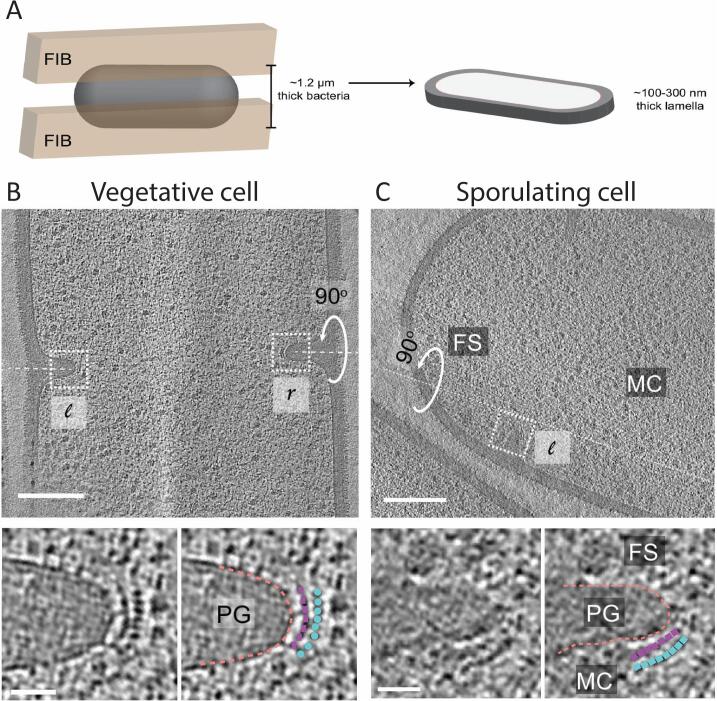
The power of this technique was recently demonstrated by the identification of the structural proteins associated with endospore formation. These spores are highly resistant to environmental stresses and can endure for extended periods due to a thick, protective protein coat. For some bacterial species, like Bacillus subtilis, this dense coat prohibited direct visualization by cryo-ET (Khanna et al., 2021). Previous research had shown that the process of sporulation was significantly different from the division events during vegetative growth, though exactly how was unknown (Tocheva et al., 2013, Tocheva et al., 2011). To solve this problem, Khanna et al. (2021) used cryo-FIB SEM to first thin the cells to approximately 200 nm, which were then subsequently imaged (Fig. 4A) (Khanna et al., 2021). Remarkably, they were able to show that the FtsZA division machinery is evenly distributed along the division septum in vegetative cells (Fig. 4B). Conversely, in a sporulating cell, the FtsZA is not distributed evenly along the septum but rather located only on the mother cell side of the septum (Fig. 4C). Using a combination of genetic and imaging techniques, they had further shown that this asymmetry is likely due to SpoIIE localization on the spore side of the septa, which lead to an exclusion of FtsZA and resulted in a thinner division septum. Cryo-FIB SEM, followed by cryo-ET, was essential to gain this important structural insight into spore formation.
Fig. 4.
A. Schematic representation of cryo-FIB milling of the bacterial cell). B & C. By thinning of Bacillus subtilis using cryo-FIB SEM, Khanna et al. were able to identify the location of FtsZ (blue) and FtsA (purple) at the division septum for vegetative (B) and sporulating cells (C). Adapted from Khanna et al. (Khanna et al., 2021).
Additional advancements in cryo-FIB SEM are currently in development. For instance, most studies using this technique have involved samples that can be prepared by plunge freezing. 200 nm lamellae are routinely produced in samples that are less than 5–10 µm in starting thickness. The next revolution in cryo-FIB SEM will be the processing of large-volume samples that require high pressure freezing, which currently includes thicknesses up to approximately 250 µm. This type of sample includes small multicellular organisms such as Caenorhabditis elegans and bacterial biofilms (Harapin et al., 2015, Melia et al., 2021). Two primary methods in development are the ‘lift-out’ technique using a mechanical arm called microgripper, and focused ion beam milling with oxygen plasma (Kuba et al., 2021, Schaffer et al., 2019, Sergey et al., 2018). The lift-out device allows for the excision of chunks of tissue from a large volume sample. These thinner samples are then transferred to a specialized grid for further thinning to create lamellae for imaging. The oxygen plasma beam has been demonstrated to ablate larger volumes of HPF tissue without damage to the tissue and in less time, thus providing an alternative to the cryo-lift-out and current Ga+ FIB beam (Sergey et al., 2018). These techniques have already been used in large volume samples such as C. elegans. However, they are not yet readily available to the scientific community (Schaffer et al., 2019).
Cryo-electron tomography combined with fluorescence light microscopy (cryo-CLEM) provides the distinction of structure compartments
The absence of widely applicable labels for cryo-ET to confidently identify a feature of interest in a tomogram remains a challenge. However, combining cryo-ET with fluorescence light microscopy (FLM) at both room and cryogenic temperatures (cryo-CLEM) can help to alleviate this issue. This combination allows structures of interest to be tagged with a fluorescent probe, imaged using a light microscope, and then prepared for and imaged with cryo-EM. At room temperature, challenges include the need for the sample to remain static during FLM and cryo-fixation; otherwise, the two images cannot be properly correlated. In addition, the resolution limitation of LM often makes it very difficult to accurately identify the structure of interest. Despite these challenges, this method has been successfully used to determine the location of the chemotaxis arrays, filaments formed by the metabolic enzyme CTP synthase, and the diffusion barriers in the bacterium Caulobacter cresecentus (Briegel et al., 2008, Ingerson-Mahar et al., 2010, Schlimpert et al., 2012).
With the introduction of cryo-CLEM, the sample is vitrified, immobilizing the structures in place, and then imaged using the cryo-FLM and cryo-EM workflow. This technique has been used to identify and describe several prokaryotic structures, including the cytoskeletal MreB filaments in C. crescentus, and the type 6 secretion system (T6SS) (Swulius et al., 2011). First observed in Vibrio cholerae, the T6SS is a syringe-like structure found in some gram-negative bacteria (Basler et al., 2012). The T6SS acts as a molecular needle that contains effector molecules used to stave off offending bacteria or to occupy new environmental niches. It consists of a membrane-bound inner tube and a spike structure surrounded by a sheath assembly that is responsible for contraction and injection. Bound to the membrane at the proximal end, the process of contraction results in a rearrangement of the sheath proteins causing the effectors located inside the tube to be delivered into the neighboring cell. Disassembly quickly follows, allowing the sheath subunits to be re-used in the formation of a new T6SS.
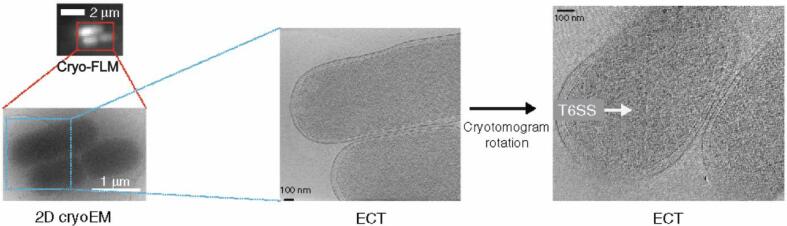
A recent example of the use of cryo-CLEM in the study of T6SS was shown by Szwedziak and Pilhofer (2019). In this study, the authors examined T6SS in enteroaggregative Escherichia coli, identifying a subset of T6SS structures that were connected to the membrane at both the proximal and distal ends of the sheath/tube complex (Fig. 5). Cryo-CLEM was integral in the identification of the T6SS expressing cells in this model, providing adequate data to then model the behavior of contraction at both ends of the needle.
Fig. 5.
cryo-FLM workflow used in the identification of the extended T6SS in enteroaggregative E. coli. The cells were first vitrified and then imaged by cryo-FLM to identify the cells that contained the Hcp1-CouAA labeled T6SS. Subsequent cryo-ET and rotation of the tomogram in the area of interest led to the visualization of the T6SS.
Adapted from Szwedziak and Pilhofer (2019)
Advances in the cryo-FLM portion of the workflow will allow for better correlation of the fluorescent signal with the cryo-ET data. For example, the development of cryogenic photoactivated localization microscopy (cryo-PALM) combined with cryo-ET allowed the identification of the T6SS in the bacterium Myxococcus xanthus (Chang et al., 2014). Without cryo-PALM, the T6SS is challenging to distinguish from other tubular structures present in this bacterium. By tagging the sheath protein VipA with a photoactivatable green fluorescence protein (GFP), the authors were able to locate the structure within the cell in multiple states and with increased precision. This is just one example of the potential power that CLEM can have in identifying and understanding the various molecular machines in prokaryotes. Most recently, the introduction of integrated fluorescent light microscopes into the cryo-FIB SEM will likely significantly improve the combination of the two techniques (Gorelick et al., 2019, Smeets et al., 2021).
Subtomogram averaging to gain insight into molecular machines: Chemotaxis and flagellar motors
As illustrated above, cryo-ET has proven to be a powerful technique to study bacterial cell architecture. However, due to the anisotropic resolution of tomograms, they are typically not sufficient to reliably extract the structure and function of molecular machines in situ. Additionally, if the background noise is equal to or higher than the signal noise, the target structure cannot be differentiated from the background (Lučić et al., 2005). This is mostly the case for cryo-ET datasets since the data is acquired using low dose schemes due to the necessity to distribute the dose that the sample can take among the full tilt-series. Another limitation is the above-described missing wedge artefact. These limitations can be partially overcome for certain cellular content that has a consistent, identical structure. In this case, a method can be applied where the identical structures are computationally extracted in so-called sub-volumes. These are then aligned to match in orientation and averaged. The resulting density map of such an average has a substantially increased signal-to-noise ratio that allows the interpretation of structural details at higher resolution. This technique is referred to as sub-tomogram averaging (STA) (Lučić et al., 2005).
STA also helps to minimize the effect of the missing wedge artefact if the particles within the individual sub-volumes have different orientations. Thus, with more particles of different orientations, this artefact becomes less severe. Several dedicated specialized software packages allow for STA, such as PEET, TOM/AV3 toolbox, Dynamo, EM clarity, M−software, EMAN2, or Relion (Baldwin et al., 2018, Hrabe et al., 2012, Pyle and Zanetti, 2021). While these all have their specific advantages, they all can extract, align and average sub-volumes. Some of the mentioned packages also conduct missing wedge orientation and compensation by interpolating the missing information from other particles (Baldwin et al., 2018). Furthermore, STA can be combined with the molecular dynamics flexible fitting (MDFF). which allows the fitting of high-resolution atomic structures into cryo-EM maps. This combination results in a pseudo-atomic model that in turn can provides structural insight beyond the resolution-limited STA map alone (Trabuco et al., 2009).
This additional processing of cryo-ET data has provided some breakthrough insights into the structure and function of two macromolecular machines that enable the cells to sense their environment and actively move toward beneficial environments: the chemoreceptor arrays and flagellar motors.
Chemotaxis is a system that allows the bacteria to sense their chemical surroundings and control their motility apparatus to seek out their preferred environmental niches and evade harmful ones. The bacterial chemotaxis system in the bacterium E. coli is arguably the best-studied signal-transduction system in biology. It relies on chemoreceptors called methyl-accepting chemotaxis proteins (MCPs), a histidine kinase CheA and the coupling protein CheW. These three proteins cluster together to form extended arrays at the cell poles, which can contain thousands of chemoreceptors. In the presence of repellents or absence of attractants, CheA triggers the response messenger CheY by phosphorylation. CheY-P in turn binds to the flagellar motor and modifies the direction of flagella rotation. Chemoreceptor arrays are highly cooperative, and the activation of one receptor can spread through the array.
While the individual components of this system have been well-studied in the past, the structure and function of the in vivo arrays were unknown until they were imaged by cryo-ET. Insights into the more detailed architecture of the arrays and the changes following activation could only be gained using STA. It revealed the architecture of these arrays, where the hexagonal receptor arrays are networked by rings of alternating CheA and CheW monomers (Briegel et al., 2012, Liu et al., 2012). In addition, STA of in situ arrays in combination with MDFF revealed insights into the structural changes that occur during the activation of CheA.
MDFF is nowadays already implemented in molecular dynamics packages and widely used for the investigation of dynamic simulation, like in GROMACS (Igaev et al., 2019, Lindahl and Hess, 2022). However, the receptor-trimers adopt a more closed conformation at their baseplate interacting tips in the inactivating state, and a more open, splayed conformation in the activating state (Yang et al., 2019). STA further revealed changes in the structure of the kinase between the on– and off states. In the off state, the two CheA domains that contain the phosphoryl group for transfer to the response messenger, as well as the domain responsible for the docking of the CheY protein, are bound to the rest of the protein in an unproductive manner (Fig. 6C). Once activated, these two domains are released, allowing for CheY binding and phosphoryl transfer (Briegel et al., 2013). With ever-increasing resolution of STA, it can be expected that soon further details of the signal-transduction will be revealed. This will undoubtedly help to understand still open questions on how the different components of the chemotaxis system interact with each other and how they are controlled.
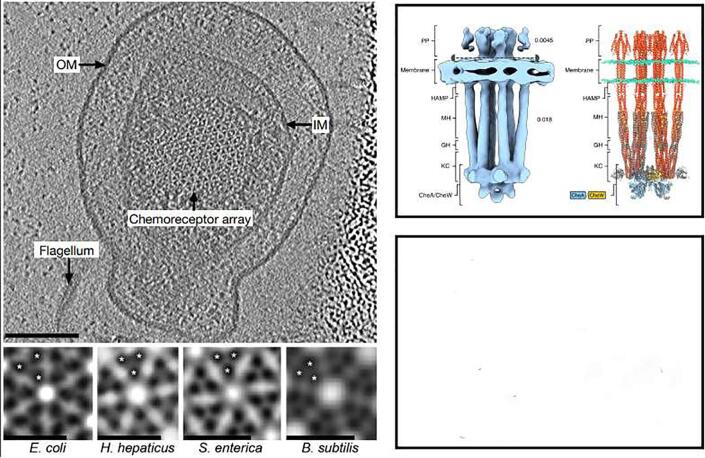
Fig. 6.
A, Top: Top view of a chemotaxis array in a Salmonella enterica minicell. IM: inner membrane, OM: outer membrane. Bottom: subtomogram averages of chemoreceptor arrays in diverse species, highlighting the universal receptor arrangement. Reproduced with permission from Briegel et al., 2012. B, Left: STA of the functional unit. PP: periplasmic ligand-binding domains, MH: methylation-helix bundles, GH: flexible regions containing the glycine hinge, KC: kinase control region Right: Molecular model. Red: receptors, blue: CheA, gold: CheW. Reproduced with permission from Burt et al. (2020). C. Schematic representation of the structural changes between the kinase-off (left) and kinase-on states (right). Courtesy of Dr. Alise Muok (Muok et al., 2020).
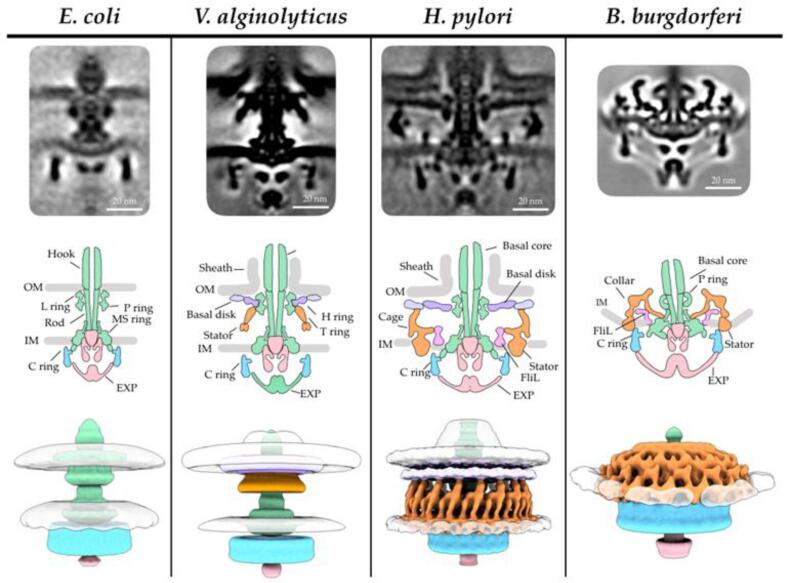
Another well-understood macromolecular complex is the flagella motor which powers flagellar rotation. Flagella are extracellular appendages that provide the cells with the ability to move, either by swimming through a liquid medium or by swarming on surfaces. The flagellum consists of a basal body (motor), a hook (flexible linker), and a propeller (helical filament). It is attached to the flagellar motor, which extends from the cytoplasm, through the inner membrane and the peptidoglycan layer (periplasmic space), to the outer membrane. In general, the core of the flagellar motor is structurally well conserved and shares the same mechanism among species, but the entire motor structure varies greatly (Fig. 7). Depending on the species, the motor is either powered by a proton-motive force or by ion-motive force. In E. coli, depending on the orientation of the filament rotation the cell either swims/runs (counterclockwise rotation, CCW) or tumbles (clockwise rotation, CW). The motor rotation is biased by the binding of the phosphorylated CheY from the chemotaxis system. A more detailed description of the flagellum and the flagellar motor composition can be found in the review of (Carroll and Liu, 2020).
Fig. 7.
Flagellar motor, showing the conserved core elements as well as the species and genera-specific motor adaptations to fulfill their needs in their specific environment. Adapted from (Carroll and Liu, 2020).
In the past years, STA was not just used to reveal species-specific flagellar motor compartments. It also investigated how the components interact with each other. A recently revealed example is the interaction between the messenger protein CheY with the C-ring (FliM, FliN, FliG). CheY binds to FliM and leads to a major conformational change of FliG, while FliN prevents disassociation of the C-ring during the conformational change. In turn, the conformational change of FliG leads to a remodeling of the rotor–stator interaction and a switch from a CCW to a CW tumble rotation of the filament (Liu et al., 2009). The rotor and stator complex are known to be highly dynamic, and the moment of switching is rather volatile, and therefore difficult to visualize. A described solution to improve the visualization is to increase the density of the small switching proteins by GFP labeling and/or using homologs with a higher molecular mass in combination with STA (Rossmann et al., 2020).
In general, with increasing resolution, cryo-ET and STA will likely aid in revealing other aspects of the flagellar motor. For example, binding affinity vs. competition, stator assembly, and relative orientation of c-ring components are still not fully understood yet and might be answered in the coming future.
The future of cryo-ET
As we, and many others, have described, cryo-ET has evolved tremendously over the past twenty years, due in part to the work of Baumeister and his scientific heritage. Advancements in the field will continue in the years to come, significantly enhancing our knowledge of prokaryotic cell biology. Those enhancements include but are not limited to improved sample preparation workflows, advances in hardware and software, and the curation of the vast amount of data into publicly available resources.
Sample preparation is one of the key steps in the cryo-ET workflow. Without a properly prepared sample, no subsequent step is likely to succeed. In the past few years, significant developments in plunge freezing technologies allow the combination of grid preparation, sample application, and freezing into a single device (Dandey et al., 2018, Ravelli et al., 2020). In addition, the development of inkjet-like printing technologies and the introduction of self-blotting grids will enable the study of volume-limited or condition specific samples, which was previously impossible (Dandey et al., 2018, Ravelli et al., 2020, Wei et al., 2018). Both, Dandey et al., 2018, Ravelli et al., 2020, demonstrated that both techniques allow to use smaller sample volumes. As noted in the large volume section, thicker samples are also becoming increasingly accessible. With the application of the cryo-FIB SEM, plunge frozen samples can currently be thinned. The development of tools for larger volume samples such as multicellular tissues will greatly increase our understanding of how microbes interact with their environment.
Hardware and software developments will continue to increase the amount of data collected as well as our ability to process this data for valuable information. Phase plate technology is a promising area due to its ability to increase contrast with minimal defocus. Previously, phase plates had limited implementation in prokaryotic cell biology studies (for examples, see Fukuda et al., 2015, Guerrero-Ferreira and Wright, 2014, Rapisarda et al., 2019, Ultee et al., 2020). The development of the laser phase plate will likely allow a typically challenging technique to be applied to a wider range of samples with increased consistency (Schwartz et al., 2019). Most importantly, the addition of this tool will aid in the analysis of smaller complexes and better interpretation of tomograms to a higher resolution. Software developments include those that enable fast tomography, a collection method that continuously records a movie as the sample is tilted with the electron beam on the sample (Baldwin et al., 2018, Chreifi et al., 2021, Eisenstein et al., 2019). Instead of the repetitive process of tilting, tracking, and imaging, these steps are essentially combined to produce one movie per target. Depending on the collection scheme and chosen magnification, fast tomography could increase collection time per target by 50–75%, vastly multiplying the amount of data that can be collected per sample. While the speed of data collection, and as a result, the amount of data will increase, it is important to note that the biological question determines the required amount of data to answer it. The introduction of new processing programs such as Warp and M provides important information during data collection and a processing pipeline that connects the data collection directly to the tomogram and ultimately structural analysis (Tegunov et al., 2021, Tegunov and Cramer, 2019). For example, these advancements allowed the in-situ determination of bacterial ribosome to 3.5 Å, and 7.4 Å resolution of the hexameric S-Layer lattice, an extracellular paracrystalline layer of gram positive and gram-negative bacteria and most archaea (Bharat et al., 2017, Tegunov et al., 2021). Software improvements are also impacting the sample preparation steps. For instance, for cryo-FIB SEM processed samples, scripts are not available to automate a typically slow and tedious process, which will lead to increased output and improved consistency (Tacke et al., 2021, Zachs et al., 2020).
Once the data are collected, accessibility by the scientific community becomes important. Research groups typically focus on a single part of the data or are looking to answer specific biological questions. Yet, cellular cryo-ET collects an abundance of information about the cell and much of this information is ripe for examination. Resources such as the Caltech Electron Tomography Database (Ortega et al., 2019), EMPIAR (Iudin et al., 2016) and EMDB (Lawson et al., 2011) have laid the groundwork for building comprehensive collections of cryo-ET data. In turn, some of this information has been translated into resources such the Atlas of Bacterial and Archaeal Cell Structure (Oikonomou and Jensen, 2021). This open access, digital resource provides detailed information about the prokaryotic ultrastructure of 85 species, which can be used as a source for comparison of structures in different strains, education, and comparison of sample treatments. Initiatives like this are extremely valuable to the community and should be a goal of the cryo-EM community.
Together, this review highlights the impact and advancements of cryo-ET on prokaryotic biology since its application in 1998. The technique has proved to be a valuable tool that is only now reaching its potential. We look forward to the future where cryo-ET is a standard part of the microbiology toolbox.
Credit authorship contribution statement
Janine Liedtke: Conceptualization, Writing – original draft, Writing – review & editing. Jamie S. Depelteau: Conceptualization, Writing – original draft, Writing – review & editing. Ariane Briegel: Conceptualization, Writing – original draft, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. This work was supported by the OCENW.GROOT.2019.063 and Building Blocks of Life Grant 737.016.004 grants from the Netherlands Organization for Scientific Research (NWO).
References
- Al-Amoudi A., Chang J.-J., Leforestier A., McDowall A., Salamin L.M., Norlén L.P.O., Richter K., Blanc N.S., Studer D., Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23(18):3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamoudi A., Studer D., Dubochet J. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J. Struct. Biol. 2005;150(1):109–121. doi: 10.1016/j.jsb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Baldwin P.R., Tan Y.Z., Eng E.T., Rice W.J., Noble A.J., Negro C.J., Cianfrocco M.A., Potter C.S., Carragher B. Big data in cryoEM: automated collection, processing and accessibility of EM data. Curr. Opin. Microbiol. 2018;43:1–8. doi: 10.1016/j.mib.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M., Pilhofer M., Henderson G.P., Jensen G.J., Mekalanos J.J. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483(7388):182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat T.A.M., Kureisaite-Ciziene D., Hardy G.G., Yu E.W., Devant J.M., Hagen W.J.H., Brun Y.V., Briggs J.A.G., Löwe J. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat Microbiol. 2017;2(7) doi: 10.1038/nmicrobiol.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bleck C.K.E., Merz A., Gutierrez M.G., Walther P., Dubochet J., Zuber B., Griffiths G. Comparison of different methods for thin section em analysis of Mycobacterium smegmatis. J. Microsc. 2010;237(1):23–38. doi: 10.1111/j.1365-2818.2009.03299.x. [DOI] [PubMed] [Google Scholar]
- Briegel A., Ames P., Gumbart J.C., Oikonomou C.M., Parkinson J.S., Jensen G.J. The mobility of two kinase domains in the escherichia coli chemoreceptor array varies with signalling state. Mol. Microbiol. 2013;89:831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel, A., Ding, H.J., Li, Z., Werner, J., Gitai, Z., Dias, D.P., Jensen, R.B., Jensen, G.J., 2008. Location and architecture of the Caulobacter crescentus chemoreceptor array 69. https://doi.org/10.1111/J.1365-2958.2008.06219.X. [DOI] [PMC free article] [PubMed]
- Briegel A., Li X., Bilwes A.M., Hughes K.T., Jensen G.J., Crane B.R. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. 2012;109(10):3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., Cassidy C.K., Ames P., Bacia-Verloop M., Baulard M., Huard K., Luthey-Schulten Z., Desfosses A., Stansfeld P.J., Margolin W., Parkinson J.S., Gutsche I. Complete structure of the chemosensory array core signalling unit in an E. coli minicell strain. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-14350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, B.L., Liu, J., 2020. Structural Conservation and Adaptation of the Bacterial Flagella Motor. Biomol. 2020, Vol. 10, Page 1492 10, 1492. https://doi.org/10.3390/BIOM10111492. [DOI] [PMC free article] [PubMed]
- Chang Y.-W., Chen S., Tocheva E.I., Treuner-Lange A., Löbach S., Søgaard-Andersen L., Jensen G.J. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods. 2014;11(7):737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman G.B., Hillier J. Electron microscopy of ultra-thin sections of bacteria I. : Cellular division in bacillus cereus. J. Bacteriol. 1953;66(3):362–373. doi: 10.1128/jb.66.3.362-373.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chreifi G., Chen S., Jensen G.J. Rapid tilt-series method for cryo-electron tomography: Characterizing stage behavior during FISE acquisition. J. Struct. Biol. 2021;213(2):107716. doi: 10.1016/j.jsb.2021.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M., Frischknecht F., Kudryashev M. Functional insights into pathogen biology from 3D electron microscopy. FEMS Microbiol. Rev. 2017;41:828–853. doi: 10.1093/FEMSRE/FUX041. [DOI] [PubMed] [Google Scholar]
- Dandey V.P., Wei H., Zhang Z., Tan Y.Z., Acharya P., Eng E.T., Rice W.J., Kahn P.A., Potter C.S., Carragher B. Spotiton: New features and applications. J. Struct. Biol. 2018;202(2):161–169. doi: 10.1016/j.jsb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J. The physics of rapid cooling and its implications for cryoimmobilization of cells. Methods Cell Biol. 2007 doi: 10.1016/S0091-679X(06)79001-X. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Adrian M., Chang J.-J., Homo J.-C., Lepault J., McDowall A.W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21(2):129–228. doi: 10.1017/S0033583500004297. [DOI] [PubMed] [Google Scholar]
- Ebersold H.R., Cordier J.-L., Lüthy P. Bacterial mesosomes: Method dependent artifacts. Arch. Microbiol. 1981;130(1):19–22. doi: 10.1007/BF00527066. [DOI] [PubMed] [Google Scholar]
- Eisenstein F., Danev R., Pilhofer M. Improved applicability and robustness of fast cryo-electron tomography data acquisition. J. Struct. Biol. 2019;208(2):107–114. doi: 10.1016/j.jsb.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Laugks U., Lučić V., Baumeister W., Danev R. Electron cryotomography of vitrified cells with a Volta phase plate. J. Struct. Biol. 2015;190(2):143–154. doi: 10.1016/j.jsb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Gorelick S., Buckley G., Gervinskas G., Johnson T.K., Handley A., Caggiano M.P., Whisstock J.C., Pocock R., de Marco A. PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. Elife. 2019;8 doi: 10.7554/eLife.45919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm R., Singh H., Rachel R., Typke D., Zillig W., Baumeister W. Electron tomography of ice-embedded prokaryotic cells. Biophys. J. 1998;74(2):1031–1042. doi: 10.1016/S0006-3495(98)74028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Ferreira R.C., Wright E.R. Zernike phase contrast cryo-electron tomography of whole bacterial cells. J. Struct. Biol. 2014;185(1):129–133. doi: 10.1016/j.jsb.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapin J., Börmel M., Sapra K.T., Brunner D., Kaech A., Medalia O. Structural analysis of multicellular organisms with cryo-electron tomography. Nat. Methods. 2015;12(7):634–636. doi: 10.1038/nmeth.3401. [DOI] [PubMed] [Google Scholar]
- Hegerl R. The EM program package: A platform for image processing in biological electron microscopy. J. Struct. Biol. 1996;116(1):30–34. doi: 10.1006/jsbi.1996.0006. [DOI] [PubMed] [Google Scholar]
- Hrabe T., Chen Y., Pfeffer S., Kuhn Cuellar L., Mangold A.-V., Förster F. PyTom: A python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J. Struct. Biol. 2012;178(2):177–188. doi: 10.1016/j.jsb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Igaev M., Kutzner C., Bock L.V., Vaiana A.C., Grubmüller H. Automated cryo-EM structure refinement using correlation-driven molecular dynamics. Elife. 2019;8 doi: 10.7554/eLife.43542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M., Briegel A., Werner J.N., Jensen G.J., Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12(8):739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iudin A., Korir P.K., Salavert-Torres J., Kleywegt G.J., Patwardhan A. EMPIAR: a public archive for raw electron microscopy image data. Nat Methods. 2016;13(5):387–388. doi: 10.1038/nmeth.3806. [DOI] [PubMed] [Google Scholar]
- Jensen G.J., Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr. Opin. Struct. Biol. 2007;17(2):260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Khanna K., Lopez-Garrido J., Sugie J., Pogliano K., Villa E. Asymmetric localization of the cell division machinery during bacillus subtilis sporulation. Elife. 2021;10 doi: 10.7554/ELIFE.62204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning R.I., Koster A.J., Sharp T.H. Advances in cryo-electron tomography for biology and medicine. Ann. Anat. - Anat. Anzeiger. 2018;217:82–96. doi: 10.1016/J.AANAT.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kuba JAKUB, Mitchels JOHN, Hovorka MILOŠ, Erdmann PHILIPP, Berka LUKÁŠ, Kirmse ROBERT, König JULIA, De bock JAN, Goetze BERNHARD, Rigort ALEXANDER. Advanced cryo-tomography workflow developments – correlative microscopy, milling automation and cryo-lift-out. J. Microsc. 2021;281(2):112–124. doi: 10.1111/jmi.12939. [DOI] [PubMed] [Google Scholar]
- Latino L., Midoux C., Vergnaud G., Pourcel C., Shafer W.M. Investigation of Pseudomonas aeruginosa strain PcyII-10 variants resisting infection by N4-like phage Ab09 in search for genes involved in phage adsorption. PLoS ONE. 2019;14(4):e0215456. doi: 10.1371/journal.pone.0215456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C.L., Baker M.L., Best C., Bi C., Dougherty M., Feng P., van Ginkel G., Devkota B., Lagerstedt I., Ludtke S.J., Newman R.H., Oldfield T.J., Rees I., Sahni G., Sala R., Velankar S., Warren J., Westbrook J.D., Henrick K., Kleywegt G.J., Berman H.M., Chiu W. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39(Database):D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Trimble M.J., Brun Y.V., Jensen G.J. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26(22):4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, Abraham, Hess, Spoel, van der, 2022. GROMACS 2021.5 Manual. https://doi.org/10.5281/ZENODO.5849961.
- Liu J., Hu B., Morado D.R., Jani S., Manson M.D., Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. 2012;109(23):E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lin T., Botkin D.J., McCrum E., Winkler H., Norris S.J. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J. Bacteriol. 2009;191(16):5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-T., Zhang, H., Wang, H., Tao, C.-L., Bi, G.-Q., Zhou, Z.H., 2021. Isotropic Reconstruction of Electron Tomograms with Deep Learning. bioRxiv 2021.07.17.452128. https://doi.org/10.1101/2021.07.17.452128.
- Lučić V., Förster F., Baumeister W. STRUCTURAL STUDIES BY ELECTRON TOMOGRAPHY: From Cells to Molecules. STRUCTURAL STUDIES BY ELECTRON TOMOGRAPHY: From Cells to Molecules. 2005;74(1):833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Marko M., Hsieh C., Schalek R., Frank J., Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat. Methods. 2007;4(3):215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McMullan G., Faruqi A.R., Henderson R. Direct Electron Detectors. Methods Enzymol. 2016;579:1–17. doi: 10.1016/BS.MIE.2016.05.056. [DOI] [PubMed] [Google Scholar]
- Melia C.E., Bolla J.R., Katharios-Lanwermeyer S., Mihaylov D.B., Hoffmann P.C., Huo J., Wozny M.R., Elfari L.M., Böhning J., Morgan A.N., Hitchman C.J., Owens R.J., Robinson C.V., O’Toole G.A., Bharat T.A.M. Architecture of cell-cell junctions in situ reveals a mechanism for bacterial biofilm inhibition. Proc. Natl. Acad. Sci. U. S. A. 2021;118(31) doi: 10.1073/pnas.2109940118. e2109940118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor H. Theory and Practice of High Pressure Freezing. Cryotech. Biol. Electron Microsc. 1987;175–191 doi: 10.1007/978-3-642-72815-0_8. [DOI] [Google Scholar]
- Muok A.R., Briegel A., Crane B.R. Regulation of the chemotaxis histidine kinase CheA: A structural perspective. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2020;1862(1):183030. doi: 10.1016/j.bbamem.2019.183030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S., Förster F., Linaroudis A., Net W.D., Beck F., Hegerl R., Baumeister W., Plitzko J.M. TOM software toolbox: Acquisition and analysis for electron tomography. J. Struct. Biol. 2005;149(3):227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Nickell, S., Hegerl, R., Baumeister, W., Rachel, R., S, N., R, H., W, B., R, R., 2003. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography 141, 34–42. https://doi.org/10.1016/S1047-8477(02)00581-6. [DOI] [PubMed]
- Oikonomou C.M., Jensen G.J. The Atlas of Bacterial & Archaeal Cell Structure : an Interactive Open-Access Microbiology Textbook. J. Microbiol. Biol. Educ. 2021;22 doi: 10.1128/jmbe.00128-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega D.R., Oikonomou C.M., Ding H.J., Rees-Lee P., Jensen G.J., Promponas V.J. ETDB-Caltech: A blockchain-based distributed public database for electron tomography. PLoS ONE. 2019;14(4):e0215531. doi: 10.1371/journal.pone.0215531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P., Marko M., Buttle K., Frank J. Double-tilt electron tomography. Ultramicroscopy. 1995;60(3):393–410. doi: 10.1016/0304-3991(95)00078-X. [DOI] [PubMed] [Google Scholar]
- Pyle E., Zanetti G. Current data processing strategies for cryo-electron tomography and subtomogram averaging. Biochem. J. 2021;478:1827–1845. doi: 10.1042/BCJ20200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapisarda C., Cherrak Y., Kooger R., Schmidt V., Pellarin R., Logger L., Cascales E., Pilhofer M., Durand E., Fronzes R. In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J. 2019;38 doi: 10.15252/EMBJ.2018100886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli R.B.G., Nijpels F.J.T., Henderikx R.J.M., Weissenberger G., Thewessem S., Gijsbers A., Beulen B.W.A.M.M., López-Iglesias C., Peters P.J. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-16392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. The ultrastructure of cell membranes and their derivatives. Biochem. Soc. Symp. 1959;16:3–43. [PubMed] [Google Scholar]
- Rossmann F.M., Hug I., Sangermani M., Jenal U., Beeby M. In situ structure of the Caulobacter crescentus flagellar motor and visualization of binding of a CheY-homolog. Mol. Microbiol. 2020;114(3):443–453. doi: 10.1111/mmi.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M., Pfeffer S., Mahamid J., Kleindiek S., Laugks T., Albert S., Engel B.D., Rummel A., Smith A.J., Baumeister W., Plitzko J.M. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat. Methods. 2019;16(8):757–762. doi: 10.1038/s41592-019-0497-5. [DOI] [PubMed] [Google Scholar]
- Schlimpert S., Klein E., Briegel A., Hughes V., Kahnt J., Bolte K., Maier U., Brun Y., Jensen G., Gitai Z., Thanbichler M. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151(6):1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O., Axelrod J.J., Campbell S.L., Turnbaugh C., Glaeser R.M., Müller H. Laser phase plate for transmission electron microscopy. Nat. Methods. 2019;16(10):1016–1020. doi: 10.1038/s41592-019-0552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergey, G., Denis, K., Ava, H., Gediminas, G., Viola, O., Owen L., K., Ruby H.P., L., Moira O’, B., Roger, P., James C., W., Alex, de M., 2018. Oxygen plasma focused ion beam scanning electron microscopy for biological samples. bioRxiv 457820. https://doi.org/10.1101/457820.
- Smeets M., Bieber A., Capitanio C., Schioetz O., van der Heijden T., Effting A., Piel É., Lazem B., Erdmann P., Plitzko J. Integrated Cryo-Correlative Microscopy for Targeted Structural Investigation In Situ. Micros. Today. 2021;29(6):20–25. doi: 10.1017/S1551929521001280. [DOI] [Google Scholar]
- Swulius M.T., Chen S., Jane Ding H., Li Z., Briegel A., Pilhofer M., Tocheva E.I., Lybarger S.R., Johnson T.L., Sandkvist M., Jensen G.J., Ding H.J., Li Z., Briegel A., Pilhofer M., Tocheva E.I., Lybarger S.R., Johnson T.L., Sandkvist M., Jensen G.J. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem. Biophys. Res. Commun. 2011;407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P., Ghosal D. FtsZ-ring architecture and its control by MinCD. Subcell. Biochem. 2017;84:213–244. doi: 10.1007/978-3-319-53047-5_7. [DOI] [PubMed] [Google Scholar]
- Szwedziak P., Pilhofer M. Bidirectional contraction of a type six secretion system. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P., Wang Q., Bharat T.A.M., Tsim M., Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014;3 doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke S., Erdmann P., Wang Z., Klumpe S., Grange M., Plitzko J., Raunser S. A streamlined workflow for automated cryo focused ion beam milling. J. Struct. Biol. 2021;213(3):107743. doi: 10.1016/j.jsb.2021.107743. [DOI] [PubMed] [Google Scholar]
- Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16(11):1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D., Xue L., Dienemann C., Cramer P., Mahamid J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat. Methods. 2021;18:186–193. doi: 10.1038/s41592-020-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.F., Walker M., Siebert C.A., Muench S.P., Ranson N.A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods. 2016;100:3–15. doi: 10.1016/j.ymeth.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E.I., López-Garrido J., Hughes H.V., Fredlund J., Kuru E., VanNieuwenhze M.S., Brun Y.V., Pogliano K., Jensen G.J. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol. 2013;88(4):673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E., Matson E., Morris D., Moussavi F., Leadbetter J., Jensen G. Peptidoglycan Remodeling and Conversion of an Inner Membrane into an Outer Membrane During Sporulation. Cell. 2011;146(5):799–812. doi: 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco L.G., Villa E., Schreiner E., Harrison C.B., Schulten K. Molecular Dynamics Flexible Fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49(2):174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk M., Baumeister W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 2020;594(20):3243–3261. doi: 10.1002/1873-3468.13948. [DOI] [PubMed] [Google Scholar]
- Ultee E., van der Aart L.T., Zhang L.e., van Dissel D., Diebolder C.A., van Wezel G.P., Claessen D., Briegel A. Teichoic acids anchor distinct cell wall lamellae in an apically growing bacterium. Commun Biol. 2020;3(1) doi: 10.1038/s42003-020-1038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Dandey V.P., Zhang Z., Raczkowski A., Rice W.J., Carragher B., Potter C.S. Optimizing “self-wicking” nanowire grids. J. Struct. Biol. 2018;202(2):170–174. doi: 10.1016/j.jsb.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G.L., Kieninger A.-K., Maldener I., Forchhammer K., Pilhofer M. Structure and Function of a Bacterial Gap Junction Analog. Cell. 2019;178(2):374–384.e15. doi: 10.1016/j.cell.2019.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Cassidy C.K., Ames P., Diebolder C.A., Schulten K., Luthey-Schulten Z., Parkinson J.S., Briegel A., Harwood C.S. In situ conformational changes of the escherichia coli serine chemoreceptor in different signaling states. MBio. 2019;10(4) doi: 10.1128/mBio.00973-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q., Jewett A.I., Chang Y., Oikonomou C.M., Beeby M., Iancu C.V., Briegel A., Ghosal D., Jensen G.J. Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J. 2017;36:1577–1589. doi: 10.15252/EMBJ.201696235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K., Braunfeld M.B., Maki-Yonekura S., Agard D.A. Electron energy filtering significantly improves amplitude contrast of frozen-hydrated protein at 300 kV. J. Struct. Biol. 2006;156(3):524–536. doi: 10.1016/j.jsb.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Zachs T., Schertel A., Medeiros J., Weiss G.L., Hugener J., Matos J., Pilhofer M. Fully automated, sequential focused ion beam milling for cryo-electron tomography. Elife. 2020;9 doi: 10.7554/eLife.52286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Lei D., Zhang M., Liu J., Wu H., Yu Y., Zhang L., Ren G. LoTToR: An Algorithm for Missing-Wedge Correction of the Low-Tilt Tomographic 3D Reconstruction of a Single-Molecule Structure. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-66793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Keszthelyi B., Branlund E., Lyle J.M., Braunfeld M.B., Sedat J.W., Agard D.A. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J. Struct. Biol. 2007;157(1):138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]