Abstract
Monitoring the progression of SARS-CoV-2 outbreaks requires accurate estimation of the unobservable fraction of the population infected over time in addition to the observed numbers of COVID-19 cases, as the latter present a distorted view of the pandemic due to changes in test frequency and coverage over time. The objective of this report is to describe and illustrate an approach that produces representative estimates of the unobservable cumulative incidence of infection by scaling the daily concentrations of SARS-CoV-2 RNA in wastewater from the consistent population contribution of fecal material to the sewage collection system.
Subject terms: Environmental sciences, Infectious diseases
Introduction
Estimating the unobservable fraction of individuals infected in coronavirus outbreaks is of first-order importance in monitoring epidemic progress and evaluating interventions meant to slow transmission. Given the lack of repeated representative COVID-19 testing over time, researchers have attempted to infer SARS-CoV-2 incidence from observable lagging indicators of infection including clinically diagnosed cases, hospitalizations, and deaths1–5. Such indicators present a distorted view of the pandemic due to temporal changes in the rates and coverage of COVID-19 testing, changes in hospital admission and treatment policies, and undercounting of deaths from COVID-19.
Recognizing these difficulties, we initiated daily sampling at a wastewater treatment plant (WWTP) serving a mid-sized US municipality with the objective of obtaining a representative estimate of the unobservable incidence of SARS-CoV-2 infections over time. We previously reported daily SARS-CoV-2 RNA concentrations in this community’s wastewater during the March 2020 epidemic wave, and showed that RNA concentrations followed an epidemic curve while providing an earlier epidemic signal than observed cases or hospitalizations6. Via a mathematical epidemic model, we estimated the associated reproductive number R0 and cumulative incidence of infection in this same community7.
Building upon this previous work, the objective of this report is to develop and illustrate a simple model that directly scales measured RNA concentrations in sewage sludge to the unobservable fraction of the population infected with SARS-CoV-2 over time. The incidence of infection is determined from the start of the pandemic in Connecticut, USA (March 19, 2020) through May 22, 2021, and the results are compared to three independently developed estimates based on observable cases, hospitalizations, and deaths3–5.
Results
Figure 1a reports the total and positive daily number of COVID-19 tests conducted on the residents of the four towns served by the WWTP. These data demonstrate the large increase in testing as the pandemic progressed, while the positive test results illustrate the two major COVID-19 waves experienced in the area with the second wave yielding higher daily case rates over a longer duration compared to the first. Figure 1b plots SARS-CoV-2 RNA concentration measured in sewage sludge over this same time period. This figure also shows two waves of infection. Unlike the number of positive tests, note that for the wastewater data the first wave, though shorter in duration, peaks at concentrations similar to the second wave. This demonstrates that the early lack of testing impacted the accuracy of reported COVID-19 case information.
Figure 1.
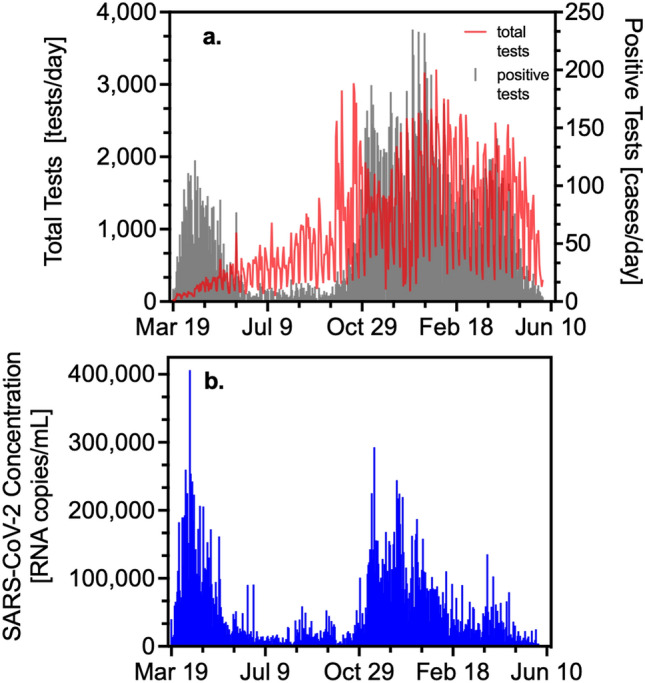
(a) Total and positive COVID-19 tests over time. (b) SARS-CoV-2 RNA wastewater concentration (copies/mL sludge).
Applying Eq. (1) (see “Methods”) to the data contained in Fig. 1b yields the cumulative fraction infected in the population over time (Fig. 2). We estimate that 33.6% (95% CI [24.3%, 42.9%]) of the population was infected by May 22, 2021. By contrast, only 24,296 diagnosed COVID-19 cases were reported in our study population by May 22, 2001, which amounts to 12% of the population. This illustrates the difference between estimating the unobserved incidence of infection and counting the observed number of diagnosed cases of COVID-19. Figure 2 also shows point estimates for the cumulative fraction of the population infected in New Haven County (which subsumes the treatment plant population) produced by three independently developed and computationally intensive statistical models using completely different methods and data sources including COVID-19 cases and deaths (Model 1)3; cases, deaths, hospitalizations, and close-contact measures deduced from cell phone geolocation data (Model 2)4; and deaths alone (Model 3)5. These models demonstrate strikingly similar shapes to and bracket the results of the wastewater-based estimates. Model 13 hugs the wastewater model’s upper 95% confidence limit, Model 24 hugs the wastewater model’s lower 95% confidence limit, and Model5 falls just beneath the point-estimate trajectory of Eq. (1).
Figure 2.
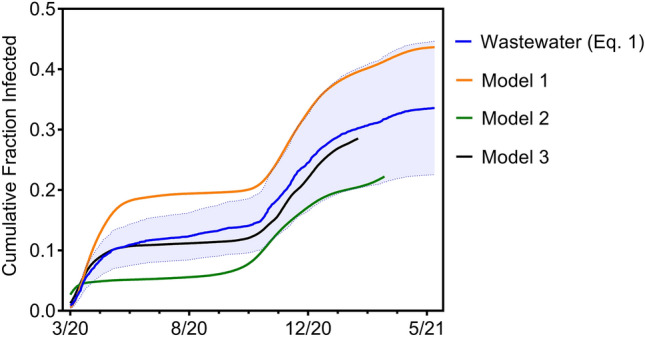
Estimated cumulative incidence of infection with 95% CIs in the population served by the WWTP based on the SARS-CoV-2 RNA concentrations shown in Fig. 1b using Eq. (1). Also shown are cumulative incidence estimates for New Haven County produced by three different statistical models. While all four models show similar trajectories over time, the estimates from Eq. (1) are the middle of the range exhibited by the other models.
Discussion
The utilization of wastewater-based epidemiology surged during the COVID-19 pandemic with applications to outbreak detection and tracking temporal trends8–10. This report presents a major advance for wastewater surveillance by using a simple scaling model to directly estimate the unobservable fraction of persons in a population infected over time from SARS-CoV-2 RNA concentrations in wastewater. This approach circumvents problems with non-representative sampling inherent in observable COVID-19 cases, hospitalizations, or deaths, and in principle can be applied in any location where continuous wastewater sampling over time is possible.
There are some limitations to our study that invite further investigation. The data collection period in our research ended before the emergence of the Delta and Omicron coronavirus variants of concern. It is possible that the mean shedding time differs for such variants, which would imply a different time shift from the historical value assumed in our study, though at least one recent report has estimated that the mean generation times for the Alpha and Delta variants are similar to historical strains11. It is also possible that infection with different variants could change the scaling from SARS-CoV-2 concentrations to infections. Determining whether the mean shedding times and RNA concentrations differ for variants of concern relative to historical strains are topics for future research.
Methods
The number of total tests and confirmed and probable COVID-19 cases was provided by the Connecticut Department of Public Health (CT DPH).
Nucleic acid was extracted from the primary sewage sludge of the New Haven, CT, USA wastewater treatment plant (which serves 200,000 residents), and SARS-CoV-2 RNA concentrations were quantified. Nucleic acid was extracted using commercial kits (Qiagen, RNeasy Powersoil Total RNA kit and Zymo, Quick-RNA Fecal/Soil Microbe Microprep). Nucleic acids were measured by spectrophotometry, the concentration adjusted to 200 ng µL−1 (NanoDrop, Thermo Fisher Scientific) and SARS-CoV-2 RNA concentrations were quantified through one-step qRT-PCR kit (BioRad iTaq™ Universal Probes One-Step Kit) using SARS-CoV-2 N1 and N2 primer sets for quantification in accordance with previously described protocols6,12. SARS-CoV-2 RNA concentrations were quantified daily throughout the study period. Further details regarding the construction of the SARS-CoV-2 RNA concentration dataset appear in the Supplementary Information.
There are two fundamental assumptions in our scaling model: 1. RNA concentrations provide a proportional measure of the extent of infection in the community given the population’s consistent discharge of fecal material into the local sewage collection system, and 2. the concentration of SARS-CoV-2 RNA in sewage sludge lags the population incidence of SARS-CoV-2 in accord with the generation time distribution (also referred to as the shedding load distribution) from infection to transmission, the mean of which is approximately 9 days7,13. Letting πt denote the fraction of the population that is newly infected on day t (the incidence of infection) and ℓ denote the mean generation lag, the SARS-CoV-2 RNA concentration measured on day t, Zt, should approximately reflect the incidence of infection ℓ days earlier, that is, Zt ≈ kπt-ℓ where k is the constant of proportionality, and consequently the cumulative fraction of the population infected by the end of day t, given by , should approximately follow where k′ = 1/k is the constant of proportionality scaling SARS-CoV-2 RNA concentration to infections per person. Given the cumulative number of infections Ct* as of some particular date t*, let denote the cumulative sludge RNA observed through day t. Then the scaling constant k′ can be evaluated from the relation yielding . Substituting back into the equation for cumulative incidence up to an arbitrary day t yields = The cumulative incidence of infection in the 200,000 population served by the WWTP treatment plant was previously estimated as = 9.3% (95% CI [0.0643, 0.1217]) as of t* = May 1, 2020 with a mean generation lag of 8.9 days7 which we round up to ℓ = 9. Substituting yields our scaling of cumulative RNA concentration to cumulative incidence shown in Fig. 2 as
| 1 |
Confidence intervals follow from the variance of estimated via the delta method14.
Supplementary Information
Acknowledgements
We thank Stephen Bart from the Connecticut Department of Public Health for assembling the COVID-19 testing data for the four towns served by the WWTP. We also thank Olga Morozova and Forrest W. Crawford for sharing the cumulative incidence estimates produced by their model for New Haven County. We also acknowledge the Rothberg Cognitive Science Donor Advised Fund at Yale University for supporting this research.
Author contributions
E.H.K., A.Z., and J.P. wrote the manuscript; A.Z. and M.S. performed the laboratory analysis to produce the SARS-CoV-2 RNA concentrations; E.H.K. performed the statistical analysis reported in “Methods” and Supporting Information that led to Fig. 2; J.P. prepared Figs. 1 and 2.
Data availability
All data employed in this report are available in the Excel file contained in the Source Data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07523-7.
References
- 1.Reese H, et al. Estimated incidence of coronavirus disease 2019 (COVID-19) illness and hospitalization—United States, February–September 2020. Clin. Infect. Dis. 2021;72:e1010–e1017. doi: 10.1093/cid/ciaa1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiner R, Jr, et al. Modeling COVID-19 scenarios for the United States. Nat. Med. 2021;27:94–105. doi: 10.1038/s41591-020-1132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitwood MH, et al. Reconstructing the course of the COVID-19 epidemic over 2020 for US states and counties: Results of a Bayesian evidence synthesis model. MedRxiv. 2021 doi: 10.1101/2020.06.17.20133983v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morozova O, Li ZR, Crawford FW. One year of modeling and forecasting COVID-19 transmission to support policymakers in Connecticut. Sci. Rep. 2021;11:20271. doi: 10.1038/s41598-021-99590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gou, Y. About Covid19-projections.com. https://covid19-projections.com/about/ (2021). Accessed 14 July 2021.
- 6.Peccia J, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan EH, et al. Aligning SARS-CoV-2 indicators via an epidemic model: Application to hospital admissions and RNA detection in sewage sludge. Health Care Manage. Sci. 2020 doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polo D, et al. Making waves: Wastewater-based epidemiology for COVID-19—approaches and challenges for surveillance and prediction. Water Res. 2020;186:116404. doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, et al. Early warning of COVID-19 via wastewater-based epidemiology: Potential and bottlenecks. Sci. Total Environ. 2021;767:145124. doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bivins A, et al. Wastewater-based epidemiology: Global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(7754–7757):2021. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- 11.Blanquart F, et al. Selection for infectivity profiles in slow and fast epidemics, and the rise of SARS-CoV-2 variants. MedRxiv. 2021 doi: 10.1101/2021.12.08.21267454v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zulli A, et al. Predicting daily COVID-19 case rates from SARS-CoV-2 RNA concentrations across a diversity of wastewater catchments. FEMS Microb. MedRxiv. 2021 doi: 10.1101/2021.04.27.21256140v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huisman JS, et al. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. MedRxiv. 2021 doi: 10.1101/2021.04.29.21255961v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop YMM, Fienberg SE, Holland PW. Discrete Multivariate Analysis: Theory and Practice. MIT Press; 1975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data employed in this report are available in the Excel file contained in the Source Data.


