Abstract
Circulating cell-free DNA (cfDNA) detection, a non-invasive method, appears promising for genetic analyses as well as quantitative assessment of tumor burden in patients with cancer. Although the analysis of cfDNA for clinical prognosis and monitoring disease burden in multiple myeloma (MM) has been recently studied, the results are unclear. In this meta-analysis, we explored the clinical significance of circulating cfDNA detection in patients with MM. We searched PubMed, Embase, and the Cochrane Library for eligible studies published up until July 25, 2021. Diagnostic accuracy variables were calculated and analyzed using Meta-Disc, and prognostic data were analyzed using Review Manager. Overall, seven studies comprising 235 myeloma patients met our inclusion criteria. The overall sensitivity and specificity of cfDNA to detect minimal residual disease (MRD) were 0.58 and 0.91, respectively. Moreover, higher levels of cfDNA were associated with worse progression-free survival as well as with poor overall survival. Our meta-analysis revealed that ctDNA detection has an obvious advantage in terms of MRD detection specificity, but it showed no superiority over bone marrow assessment in terms of MRD detection sensitivity, and higher levels of cfDNA were indicative of worse prognosis in patients with MM. cfDNA detection is a non-invasive method and thus shows promise as a good alternative to BM biopsies for monitoring clonal evolution and tumor burden so as to guide the treatment of patients with MM.
Keywords: circulating tumor DNA, cell-free DNA, multiple myeloma, minimal residual disease, prognosis, meta-analysis
Introduction
Multiple myeloma (MM), an incurable hematological malignancy, is characterized by recurrent cytogenetic and molecular abnormalities. Malignant plasma cells show typical multifocal distribution in the bone marrow (BM) and occasional extramedullary dissemination (1). Considering the spatial heterogeneity of myeloma, analyzing BM aspirates collected from a single site often does not provide comprehensive insights into the genetic profile of tumors. During disease progression, drug resistance and clonal evolution pose major issues (2, 3). Longitudinal evaluation of mutational landscape and tracking tumor burden can facilitate the identification of early signs of treatment resistance and relapse. However, repeated BM aspiration is impractical as the procedure is invasive and causes discomfort. Although BM biopsy is the current gold standard for MM diagnosis and prognostic stratification, tracking malignant clones remains challenging because of the spatial and temporal limitations of BM biopsies; therefore, there exists an urgent need to identify a novel biomarker to monitor disease progression.
The presence of cell-free nucleic acid fragments in human blood was first described in 1948 by Mandel and Métais (4). Cell-free DNA (cfDNA) is believed to have originated as a consequence of cell apoptosis and necrosis, and possibly also active secretion (5). Increased cfDNA levels were first reported in the serum of cancer patients in 1977 (6). According to several studies, cfDNA is of potential diagnostic and prognostic importance in various cancer types, and its levels during treatment are reportedly correlated with outcome (7–9). With the development of molecular methods, mutant DNA fragments, confirmed to be of tumor origin, have been detected in plasma. Mutations in cfDNA can serve as highly specific markers for cancer, and tumor-derived DNA in cfDNA, also known as circulating tumor DNA (ctDNA) (10), detection provides a non-invasive approach to diagnose cancers. ctDNA carries information pertaining to the dynamics of cancer-specific genetic and epigenetic alterations; moreover, ctDNA represents the entire epitome of mutations present in primary and metastatic tumors (11). In comparison to previously used blood-based biomarkers, cfDNA-based detection methods show higher sensitivity, and thus, they seem to have great potential for both quantitative analysis of tumor burden and genetic analysis in case of patients with cancer.
Relative to other cancer types, cfDNA concentration is higher in MM, and there exists high concordance between mutations found in DNA using BM aspirates and those found in ctDNA (12, 13). Although cfDNA analysis for clinical prognosis and to monitor disease burden in MM has been recently reported (14, 15), the results are unclear. Therefore, herein we performed a meta-analysis to systematically explore the clinical relevance of circulating cfDNA in patients with MM.
Materials and Methods
Data Source and Search Strategy
We searched for eligible studies in PubMed, Embase, and the Cochrane Library using the following keywords: “circulating DNA” OR “cell free DNA” OR “ctDNA” OR “cfDNA” OR “blood DNA” OR “plasma DNA” OR “serum DNA” OR “liquid biopsy” AND “myeloma” OR “plasmacytoma” OR “plasma cell neoplasms” OR “plasma cell dyscrasias”. All the data retrieved were updated to July 25, 2021.
Study Selection
Two investigators independently reviewed all the titles and abstracts obtained on implementing our search strategy. We reviewed potentially relevant articles in full to ensure that they satisfied the following inclusion criteria: (1) clinical studies comprising patients with MM, (2) samples collected from the peripheral blood, (3) availability of information pertaining to the diagnostic and prognostic importance of cfDNA or ctDNA or ability to obtain such information from published data, and (4) clarity regarding techniques and target genes. The exclusion criteria were as follows: (1) reviews, conference abstracts, case reports, and non-English publications; (2) circulating viral DNA; (3) lack of outcomes; and (4) republished articles or samples.
Data Extraction
All relevant studies were assessed by full-text review and those meeting the inclusion criteria were included in final analyses. Both investigators independently extracted the following data from each selected article: first author details, publication year, number of patients, cfDNA detection method, target genes, outcomes, and minimal residual disease (MRD) detection accuracy. If the eligible studies already reported the hazard ratio (HR) of the outcomes [progression-free survival (PFS) and overall survival (OS)] and 95% confidence interval (CI), then the data were directly extracted via full-text review. However, if this information was not available, then the HR was estimated using methods previously reported by Tierney et al. (16).
Study Quality Assessment
The quality of diagnostic studies was assessed using the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) criteria (17) ( Table 1 ). Besides, the quality of prognostic studies was evaluated according to the Newcastle–Ottawa scale (21) ( Table 2 ). Newcastle–Ottawa scale scores of more than five stars were considered to represent high quality.
Table 1.
Assessment of the quality of diagnostic studies using the QUADAS-2 criteria.
| Study | Risk of bias | Concerns about applicability | |||||
|---|---|---|---|---|---|---|---|
| Patients Selection | Index Text | Reference Standard | Flow and Timing | Patients Selection | Index Text | Reference Standard | |
| Mazzotti et al., (18) | U | L | L | L | L | L | L |
| Biancon et al., (19) | U | L | L | L | L | L | L |
| Vrabel et al., (20) | L | L | L | L | L | L | L |
L, low risk of bias; H, high risk of bias; U, unclear risk of bias.
Table 2.
Main characteristics of the included studies.
| NO | Study | Number of patients | Detection method | Detection item | Outcome | Comparison | NOS |
|---|---|---|---|---|---|---|---|
| 1 | Mazzotti C, 2018 (18) | 37 | NGS | MRD (IGH, IGK, IGL rearrangements) | |||
| 2 | Biancon G, 2018 (19) | 22 | NGS | MRD (IGH rearrangement) | PFS | the frequency of clonal IGH <4.7% vs. ≥4.7% | 6 |
| 3 | Vrabel D, 2019 (20) | 12 | ASO-qPCR | MRD (IGH rearrangements) | |||
| 4 | Mithraprabhu S, 2019_1 (22) | 20 | NGS | KRAS, NRAS, CTNNB1, EGFR, TP53, PIK3CA, FOXL2, GNAS, BRAF | OS | FA (%) <1 vs. > 1 | 6 |
| 5 | Mithraprabhu S, 2019_2 (23) | 52 | NGS | KRAS, NRAS, BRAF, TP53 | OS | FA (%) <1 vs. > 1 | 6 |
| 6 | Li Q, 2020 (24) | 17 | ddPCR | KRAS, NRAS, BRAF | OS | undetectable vs. detectable | 6 |
| 7 | Deshpande S, 2021 (25) | 75 | cfDNA quantification | Total cfDNA level | OS, PFS | cfDNA level ≤ 25.2 ng/ml vs. > 25.2 ng/ml | 6 |
MRD, minimal residual disease; FA, fractional abundance.
Statistical Analysis
Diagnostic accuracy variables, such as sensitivity, specificity, likelihood ratios [i.e., positive likelihood ratio and negative likelihood ratio], diagnostic odds ratio, and summary receiver operating characteristic(SROC) curve were calculated and analyzed using Meta-Disc v1.4. Sensitivity and specificity were defined as the proportion of MRD-positive and -negative patients identified via ctDNA detection in plasma among all patients confirmed to be MRD positive and negative, respectively, on BM assessment by multiparametric flow cytometry (MFC) or next-generation sequencing (NGS). The pooled HR and 95% CIs for PFS or OS were analyzed with Review Manager v5.4.1. The I2 statistic was used to quantify heterogeneity among the studies. I2 > 50% represented high heterogeneity, and a random effects model was accordingly used; if I2 was <50%, a fixed effects model was used for analyses.
Results
Study Selection
Our search strategy led to the identification of 588 references, and 94 duplicates were removed ( Figure 1 ). On screening the titles and abstracts, 57 articles were considered worthy of a thorough evaluation. Finally, after full-text review, seven studies comprising 235 myeloma patients were selected for our meta-analysis. Among them, 3 studies were available for calculating the overall sensitivity and specificity of MRD detection. They all used immunoglobulin gene rearrangements in cfDNA to track residual myeloma cells. These three studies used different ctDNA detection methods: Mazzotti (18) and Biancon (19) used NGS, while Vrabel (20) used ASO-qPCR. In addition to Biancon’s study (19) which explore the relationship between the tumor-associated IGH sequence and PFS, the other 4 articles (22–25) included analysis on the association of cfDNA and survival in patients with multiple myeloma. Except the levels of total cfDNA were quantified in Deshpande’s research, the other articles used NGS or PCR to detect the levels of tumor specific DNA. Table 2 summarizes the main characteristics and quality assessment of the included prognostic studies.
Figure 1.
Flow diagram showing the selection of studies for meta-analysis.
Relevance of cfDNA in MRD Detection in MM
Three studies including 71 myeloma patients were pooled for the meta-analysis of MRD detection accuracy. As evident from Figure 2 , the overall sensitivity and specificity were 0.58 (95% CI, 0.43–0.72) and 0.91 (95% CI, 0.72–0.99), respectively. The pooled positive and negative likelihood ratios were 4.82 (95% CI, 1.44–16.12) and 0.31 (95% CI, 0.05–1.82), respectively. The area under the SROC was 0.95, and the diagnostic odds ratio was 16.41 (95% CI, 1.64–164.46).
Figure 2.
Forest plots related to MRD detection accuracy. (A) Forest plots for overall sensitivity, (B) overall specificity, (C) positive likelihood ratio, (D) negative likelihood ratio, (E) SROC curve, and (F) diagnostic odds ratio.
Prognostic Significance of cfDNA in MM
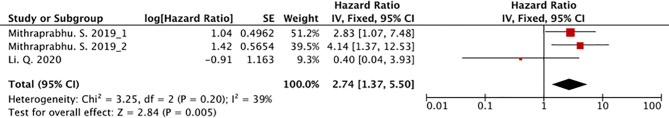
Five studies comprising 186 myeloma patients were pooled for the meta-analysis of survival. Of them, three articles included analysis on the association of cfDNA and OS in patients with MM (22–24), and one article included the information of both PFS and OS (25). Moreover, there was one study which only analyzed the association of cfDNA and PFS (19). In the two studies that included PFS as the outcome indicator, we found that high cfDNA levels in patients with MM were significantly associated with poor PFS (HR, 4.78; 95% CI, 2.00–11.45; P = 0.0004; Figure 3 ). We also found that high cfDNA levels in patients with MM were associated with worse OS (HR, 3.06; 95% CI, 1.66–5.63; P = 0.0003; Figure 4 ). To analyze the relationship between the level of tumor-derived DNA in circulating cfDNA and OS more specifically, we performed subgroup analysis using three studies that detected ctDNA levels. The results showed that high ctDNA levels in myeloma patients were associated with poor OS (HR, 2.74; 95% CI, 1.37–5.50; P = 0.005; Figure 5 ).
Figure 3.
Forest plot showing the prognostic role of cfDNA on PFS.
Figure 4.
Forest plot showing the prognostic role of cfDNA on OS.
Figure 5.
Forest plot showing the prognostic role of ctDNA on OS.
Discussion
To date, myeloma remains an incurable disease, and patients often experience a relapse owing to residual tumor cells. MRD has become one of the most important biomarkers for outcome prediction and therapy optimization. However, longitudinal monitoring of BM MRD remains limited as repetitive BM biopsies are invasive to patients. In addition, in case of some patients with extramedullary disease, BM specimens may not accurately reflect the disease burden. Previous studies based on circulating cfDNA for MRD detection in various hematological malignancies have validated its significance for measuring a small number of residual tumor cells (26–28). Unfortunately, most such studies involving patients with MM either assessed only a few patients or lacking of the comparison with BM MRD assessment; therefore, the conclusions remain controversial.
Our pooled data confirmed that ctDNA detection had an obvious advantage in terms of MRD detection specificity (specificity, 0.91; 95% CI, 0.72–0.99). According to the guidelines for area under the SROC value interpretation (29), ctDNA presence in patients with MM has a relatively high diagnostic ability (AUC > 0.9) and can indicate MRD positive ( Figure 2 ). Clonal immunoglobulin gene rearrangements in plasma samples have been detected by NGS or allele-specific oligonucleotide–qPCR to monitor tumor burden (18–20). At the time of MRD evaluation, the results of ctDNA detection using blood samples were compared with those of NGS or MFC using paired BM samples. Mazzotti (18) demonstrated the absence of a correlation between ctDNA and BM MRD detection by NGS using only immunoglobulin gene rearrangements in patients with MM. Further, Vrabel (20) compared ctDNA and BM MFC, a common method for MRD detection, and the sensitivity of MRD detection using ctDNA was only 66.7%. In contrast, Biancon (19) analyzed MRD by MFC using BM samples and found complete concordance with ctDNA data in all cases, and a high level of correlation was detected between ctDNA and BM MFC data (r = 0.5831, P = 0.0044, Pearson’s correlation test). Finally, regarding the sensitivity of MRD detection, ctDNA detection in plasma was not superior over BM assessment as per our analysis (sensitivity, 0.58; 95% CI, 0.43–0.72).
We believe that further studies with a larger cohort of patients achieving CR are warranted to determine the reasons for inconsistency in MRD detection sensitivity using ctDNA in different studies. The three studies pooled for the meta-analysis of MRD detection accuracy all used immunoglobulin gene rearrangements in cfDNA to track residual myeloma cells. However, MM is characterized by multiple recurrent somatic mutations, copy number variants, and structural alterations (30). Thus, given the extensive heterogeneity in MM, a large targeted sequencing panel may prove useful for improving MRD detection sensitivity using ctDNA.
In other cancers, cfDNA analysis has been used to identify mutations related to drug resistance and to predict therapeutic response, which could influence the choice of treatment (31–33). The prognostic value of cfDNA has been explored in myeloma. Although cfDNA is an admixture of normal and tumor-derived DNA (ctDNA), increased concentration of cfDNA signifies higher tumor burden and is indicative of the prognostic value of survival in many cancers (34, 35). In addition, high cfDNA levels at baseline are significantly associated with poor PFS and OS in patients with myeloma (25). ctDNA represents the entire epitome of mutations present in primary as well as metastatic tumors (36). Plasma-derived ctDNA analysis in patients with MM was found to show good concordance with standard mutation analysis using BM samples. Mithraprabhu (22, 23) analyzed plasma-derived ctDNA as an adjunct to BM biopsy for mutational characterization and tracking disease progression and found that in comparison with BM samples, plasma samples showed a higher proportion of TP53 mutations. Besides, Li (24) found a significantly higher detection rate of BRAF, KRAS, and NRAS mutations in plasma-derived cfDNA samples than in BM samples (53% vs 34%). Plasma evidently reflects mutations originating from all focal sites, both at the intramedullary and extramedullary levels. Therefore, assessing both BM and plasma samples should provide a more comprehensive landscape of tumor mutational burden.
The results of the studies included herein indicate the prognostic value of circulating cfDNA. cfDNA levels in MM are reportedly significantly associated with both PFS and OS. In two studies including PFS as the outcome indicator, higher levels of cfDNA were found to be associated with poor PFS ( Figure 3 ) (19, 25). As evident from Figure 4 , higher level of cfDNA was found to be associated with poorer OS. Because ctDNA only accounts for a small proportion of total cfDNA, we also performed subgroup analysis to assess the usefulness of ctDNA quantification to predict OS; we found that ctDNA-positive patients or those with a high level of ctDNA showed inferior OS ( Figure 5 ).
In the analyzed studies, gene mutations, mainly including RAS-RAF and TP53, were detected, which may predict poor prognosis in patients with MM (37). Mithraprabhu (23) reported that high levels of ctDNA are a prognostic factor in case of relapsed/refractory MM patients but not in case of newly diagnosed MM patients. This could be because a higher frequency of gene mutations in plasma was detected in relapsed/refractory MM patients than in newly diagnosed MM patients. However, Li found a significantly higher detection rate of BRAF, KRAS, and NRAS mutations on using plasma-derived cfDNA samples (53%) than on using BM samples (34%) in case of newly diagnosed MM patients, and patients with these mutations showed shorter OS than those without them (24). These two studies used different ctDNA detection methods: Mithraprabhu used NGS, while Li used droplet digital PCR. Although previous studies have verified that these methods show high concordance in terms of tumor genotype (38, 39), the inconsistency in the data reported by Mithraprabhu and Li suggest that further studies are warranted to compare these methods; furthermore, more patients need to be evaluated to confirm the prognostic significance of plasma-derived ctDNA in newly diagnosed MM patients.
This study has several limitations that need to be addressed. First, the lack of currently recognized ctDNA gene targets in patients with MM might contribute to bias. Second, different ctDNA detection methods were used in the included studies. Finally, owing to the limited number of studies on ctDNA detection in MM, the data included is not very rich, which may contribute to further bias.
Conclusions
To the best of our knowledge, this meta-analysis represents the first comprehensive study to investigate MRD detection and prognostic value of circulating cfDNA in patients with MM. We report that ctDNA detection has an obvious advantage in terms of MRD detection specificity; moreover, higher levels of cfDNA were found to be associated with worse prognosis in patients with MM. cfDNA detection is a non-invasive method and thus shows promise as a good alternative to BM biopsies for monitoring clonal evolution and tumor burden so as to guide the treatment of patients with MM. However, before its wide application in patients with MM, accurate ctDNA gene targets and standardized detection methods need to be established.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
XY conceived and designed the study. XY and WL collected and analyzed the data. LZ and JY analyzed the data. XY wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple Myeloma. Nat Rev Dis Primers (2017) 3:17046. doi: 10.1038/nrdp.2017.46 [DOI] [PubMed] [Google Scholar]
- 2. Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of Genomic Evolution and Mutational Profiles in Multiple Myeloma. Nat Commun (2014) 5:2997. doi: 10.1038/ncomms3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J Clin Oncol (2015) 33:3911–20. doi: 10.1200/JCO.2014.59.1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mandel P, Métais P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil (1948) 142:241–3. [PubMed] [Google Scholar]
- 5. Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, Structures, and Functions of Circulating DNA in Oncology. Cancer Metastasis Rev (2016) 35:347–76. doi: 10.1007/s10555-016-9629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res (1977) 37:646–50. [PubMed] [Google Scholar]
- 7. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med (2014) 6:224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Mattos-Arruda L, Olmos D, Tabernero J. Prognostic and Predictive Roles for Circulating Biomarkers in Gastrointestinal Cancer. Future Oncol (2011) 7:1385–97. doi: 10.2217/fon.11.122 [DOI] [PubMed] [Google Scholar]
- 9. Spindler KL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Circulating Free DNA as Biomarker and Source for Mutation Detection in Metastatic Colorectal Cancer. PloS One (2015) 10:e0108247. doi: 10.1371/journal.pone.0108247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat Rev Cancer (2017) 17:223–38. doi: 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 11. Lo YMD, Lam WKJ. Tracing the Tissue of Origin of Plasma DNA Feasibility and Implications. Ann N Y Acad Sci (2016) 1376:14–7. doi: 10.1111/nyas.13163 [DOI] [PubMed] [Google Scholar]
- 12. Kis O, Kaedbey R, Chow S, Danesh A, Dowar M, Li T, et al. Circulating Tumour DNA Sequence Analysis as an Alternative to Multiple Myeloma Bone Marrow Aspirates. Nat Commun (2017) 8:15086. doi: 10.1038/ncomms15086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerber B, Manzoni M, Spina V, Bruscaggin A, Lionetti M, Fabris S, et al. Circulating Tumor DNA as a Liquid Biopsy in Plasma Cell Dyscrasias. Haematologica (2018) 103:e245–8. doi: 10.3324/haematol.2017.184358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo G, Raje NS, Seifer C, Kloeber J, Isenhart R, Ha G, et al. Genomic Discovery and Clonal Tracking in Multiple Myeloma by Cell Free DNA Sequencing. Leukemia (2018) 32:1838–41. doi: 10.1038/s41375-018-0115-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long X, Xu Q, Lou Y, Li C, Gu J, Cai H, et al. The Utility of Non-Invasive Liquid Biopsy for Mutational Analysis and Minimal Residual Disease Assessment in Extramedullary Multiple Myeloma. Br J Haematol (2020) 189:e45–8. doi: 10.1111/bjh.16440 [DOI] [PubMed] [Google Scholar]
- 16. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical Methods for Incorporating Summary Time-to-Event Data Into Meta-Analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: A Revisedtool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med (2011) 155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 18. Mazzotti C, Buisson L, Maheo S, Perrot A, Chretien ML, Leleu X, et al. Myeloma MRD by Deep Sequencing From Circulating Tumor DNA Does Not Correlate With Results Obtained in the Bone Marrow. Blood Adv (2018) 2:2811–3. doi: 10.1182/bloodadvances.2018025197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biancon G, Gimondi S, Vendramin A, Carniti C, Corradini P. Non-Invasive Molecular Monitoring in Multiple Myeloma Patients Using Cell-Free Tumor DNA: A Pilot Study. J Mol Diagn (2018) 20:859–70. doi: 10.1016/j.jmoldx.2018.07.006 [DOI] [PubMed] [Google Scholar]
- 20. Vrabel D, Sedlarikova L, Besse L, Rihova L, Bezdekova R, Almasi M, et al. Dynamics of Tumor-Specific cfDNA in Response to Therapy in Multiple Myeloma Patients. Eur J Haematol (2020) 104:190–7. doi: 10.1111/ejh.13358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stang A. Critical Evaluation of the Newcastle-Ottawa Scale for the Assessment of the Quality of Nonrandomized Studies in Meta-Analyses. Eur J Epidemiol (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 22. Mithraprabhu S, Morley R, Khong T, Kalff A, Bergin K, Hocking J, et al. Monitoring Tumour Burden and Therapeutic Response Through Analysis of Circulating Tumour DNA and Extracellular RNA in Multiple Myeloma Patients. Leukemia (2019) 33:2022–33. doi: 10.1038/s41375-019-0469-x [DOI] [PubMed] [Google Scholar]
- 23. Mithraprabhu S, Hocking J, Ramachandran M, Choi K, Klarica D, Khong T, et al. DNA-Repair Gene Mutations Are Highly Prevalent in Circulating Tumour DNA From Multiple Myeloma Patients. Cancers (Basel) (2019) 11:917. doi: 10.3390/cancers11070917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Q, Huang HJ, Ma J, Wang Y, Cao Z, Karlin-Neumann G, et al. RAS/RAF Mutations in Tumor Samples and Cell-Free DNA From Plasma and Bone Marrow Aspirates in Multiple Myeloma Patients. J Cancer (2020) 11:3543–50. doi: 10.7150/jca.43729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deshpande S, Tytarenko RG, Wang Y, Boyle EM, Ashby C, Schinke CD, et al. Monitoring Treatment Response and Disease Progression in Myeloma With Circulating Cell-Free DNA. Eur J Haematol (2021) 106:230–40. doi: 10.1111/ejh.13541 [DOI] [PubMed] [Google Scholar]
- 26. Yeh P, Dickinson M, Ftouni S, Hunter T, Sinha D, Wong SQ, et al. Molecular Disease Monitoring Using Circulating Tumor DNA in Myelodysplastic Syndromes. Blood (2017) 129:1685–90. doi: 10.1182/blood-2016-09-740308 [DOI] [PubMed] [Google Scholar]
- 27. Roschewski M, Staudt LM, Wilson WH. Dynamic Monitoring of Circulating Tumor DNA in Non-Hodgkin Lymphoma. Blood (2016) 127:3127–32. doi: 10.1182/blood-2016-03-635219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oberle A, Brandt A, Voigtlaender M, Thiele B, Radloff J, Schulenkorf A, et al. Monitoring Multiple Myeloma by Next-Generation Sequencing of V(D)J Rearrangements From Circulating Myeloma Cells and Cell-Free Myeloma DNA. Haematologica (2017) 102:1105–11. doi: 10.3324/haematol.2016.161414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swets JA. Measuring the Accuracy of Diagnostic Systems. Science (1988) 240:1285–93. doi: 10.1126/science.3287615 [DOI] [PubMed] [Google Scholar]
- 30. Pugh TJ. Circulating Tumour DNA for Detecting Minimal Residual Disease in Multiple Myeloma. Semin Hematol (2018) 55:38–40. doi: 10.1053/j.seminhematol.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 31. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med (2013) 368:1199–209. doi: 10.1056/NEJMoa1213261 [DOI] [PubMed] [Google Scholar]
- 32. Murtaza M, Dawson SJ, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-Invasive Analysis of Acquired Resistance to Cancer Therapy by Sequencing of Plasma DNA. Nature (2013) 497:108–12. doi: 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- 33. Kwapisz D. The First Liquid Biopsy Test Approved. Is it a New Era of Mutation Testing for Non-Small Cell Lung Cancer? Ann Transl Med (2017) 5:46. doi: 10.21037/atm.2017.01.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR, et al. Plasma Total Cell-Free DNA (cfDNA) is a Surrogate Biomarker for Tumour Burden and a Prognostic Biomarker for Survival in Metastatic Melanoma Patients. Eur J Cancer (2018) 88:1–9. doi: 10.1016/j.ejca.2017.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eskandari M, Manoochehrabadi S, Pashaiefar H, Zaimy MA, Ahmadvand M. Clinical Significance of Cell-Free DNA as a Prognostic Biomarker in Patients With Diffuse Large B-Cell Lymphoma. Blood Res (2019) 54:114–9. doi: 10.5045/br.2019.54.2.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating Liquid Biopsies Into the Management of Cancer. Nat Rev Clin Oncol (2017) 14:531–48. doi: 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 37. Mulligan G, Lichter DI, Di Bacco A, Blakemore SJ, Berger A, Koenig E, et al. Mutation of Nras But Not Kras Significantly Reduces Myeloma Sensitivity to Single-Agent Bortezomib Therapy. Blood (2014) 123:632–39. doi: 10.1182/blood-2013-05-504340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Demuth C, Spindler KG, Johansen JS, Pallisgaard N, Nielsen D, Hogdall E, et al. Measuring Kras Mutations in Circulating Tumor DNA by Droplet Digital Pcr and Next-Generation Sequencing. Transl Oncol (2018) 11:1220–4. doi: 10.1016/j.tranon.2018.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bessi L, Viailly PJ, Bohers E, Ruminy P, Maingonnat C, Bertrand P, et al. Somatic Mutations of Cell-Free Circulating DNA Detected by Targeted Next-Generation Sequencing and Digital Droplet Pcr in Classical Hodgkin Lymphoma. Leuk Lymphoma (2019) 60:498–502. doi: 10.1080/10428194.2018.1492123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.