Abstract
Rubella virus (RV) is the causative agent of the disease known more popularly as German measles. Rubella is predominantly a childhood disease and is endemic throughout the world. Natural infections of rubella occur only in humans and are generally mild. Complications of rubella infection, most commonly polyarthralgia in adult women, do exist; occasionally more serious sequelae occur. However, the primary public health concern of RV infection is its teratogenicity. RV infection of women during the first trimester of pregnancy can induce a spectrum of congenital defects in the newborn, known as congenital rubella syndrome (CRS). The development of vaccines and implementation of vaccination strategies have substantially reduced the incidence of disease and in turn of CRS in developed countries. The pathway whereby RV infection leads to teratogenesis has not been elucidated, but the cytopathology in infected fetal tissues suggests necrosis and/or apoptosis as well as inhibition of cell division of critical precursor cells involved in organogenesis. In cell culture, a number of unusual features of RV replication have been observed, including mitochondrial abnormalities, and disruption of the cytoskeleton; these manifestations are most probably linked and play some role in RV teratogenesis. Further understanding of the mechanism of RV teratogenesis will be brought about by the investigation of RV replication and virus-host interactions.
Rubella virus (RV) is the causative agent of the disease commonly known as German measles. The earliest description of rubella appears to date back to the 1700s, when the clinical manifestations of the disease were described by two German physicians, de Bergan in 1752 and Orlow in 1758 (155). At the time, it was considered to be a derivative of measles, and because of the strong German influence, the illness became popularly known as German measles. There was considerable conjecture about the relationship of rubella to measles and to scarlet fever until in 1814, another German physician, George de Maton, documented the illness as a distinct entity, which had become known as rötheln (155). The disease was renamed rubella (from the Latin for reddish things) in 1866 by Henry Veale, a British Army surgeon, who found the original term “harsh and foreign to our ears” (31).
Several decades passed without significant research into rubella, despite documentation of the viral etiology of the disease by Hiro and Tasaka in 1938 (31). The notion that rubella was only a mild illness of children was dispelled in 1941, when Norman Gregg, an Australian ophthalmic surgeon, reported the devastating teratogenic effects of the virus (47). In the spring and summer of 1940, Australia had experienced an epidemic of rubella, whose spread was probably enhanced by wartime mobilization. Gregg observed an unusually large number of cases of congenital cataracts in newborn children, and the cataracts were often associated with other deformities. He obtained careful histories from the mothers of the affected babies in an attempt to assign a cause to the outbreak. His persistence finally led to the discovery of the link between the congenital abnormalities and rubella infection early in the mother's pregnancy. Gregg had 13 such cases in his own practice, and with colleagues he managed to collate a total of 78 cases of children with cataracts; of these, 68 mothers gave a history of rubella infection early in pregnancy. Thus, the idea that viruses could be teratogenic agents was introduced. Gregg's report initially drew little attention. It was supported by his Australian colleagues, who further reported other defects associated with maternal rubella, but it was not until the comprehensive review by Wesselhoeft published in the New England Journal of Medicine (155) that the true significance of Gregg's earlier observations was accepted.
In 1962, the isolation in cell culture of the etiological agent of rubella was reported by two independent groups (111, 154). Parkman et al. (111) infected African green monkey kidney cells with throat washings from patients with rubella and indirectly demonstrated the presence of RV by resistance to challenge with an echovirus, while Weller and Neva (154) detected cytopathic effects in human amnion cells infected with RV from blood and urine specimens. It has now been shown that RV can be grown in a wide variety of cell culture systems, and this has been the cornerstone of vaccine development.
CLASSIFICATION
RV is classified as the only member of the genus Rubivirus within the family Togaviridae; the name “togavirus” is derived from the Latin “toga,” meaning cloak or shroud, a reference to the virus envelope (99). The genus Alphavirus is the only other genus within this family and comprises at least 26 members, with Sindbis virus, the prototype, and Semliki Forest virus (SFV) being the best-characterized members. While humans are the only known natural hosts for RV, vertebrates and arthropods, such as mosquitoes, are recognized hosts for alphaviruses. RV and the alphaviruses possess similar characteristics in terms of replication strategy and genomic organization. The general characteristics of togaviruses are summarized in Table 1.
TABLE 1.
General properties of togaviruses
| Virus aspect | Description of property |
|---|---|
| Virion | Spherical particle measuring 50–70 nm in diameter |
| Virion envelope composed of host-derived lipid bilayer embedded with spikes made up of the E2, E1, and sometimes E3 glycoproteins | |
| Icosahedral core comprising the viral genome, which is encapsidated with multiple copies of the capsid proteins | |
| Genome | Single-stranded, positive-polarity RNA |
| 3 × 106 to 4 × 106 Da in size | |
| Methyl7guanosine capped at the 5′ terminus and polyadenylated at the 3′ terminus | |
| Two ORFs; the 5′-proximal ORF encodes ns proteins, and the 3′ ORF encodes structural proteins | |
| Replication | The genomic RNA, subgenomic RNA, RI representing partial dsRNA, and RF representing fully dsRNA are produced in RV-infected cells |
| Virus-modified endosomes/lysosomes termed replication complexes are sites of RV replication | |
| The genomic and subgenomic RNA serves as the mRNA for the synthesis of ns and structural proteins, respectively |
CLINICAL FEATURES
Rubella and Its Complications
The virus is transmitted from person to person via respiratory aerosols. In volunteer studies, infection can be induced by aerosol presentation to the nasopharyngeal mucosa. The upper respiratory tract and nasopharyngeal lymphoid tissue appear to be the first sites of virus replication, and the virus then spreads to regional lymph nodes (26, 159). The clinical symptoms of RV infections acquired postnatally are usually mild, and many infections are asymptomatic. The first clinical manifestation of rubella is usually the appearance of a macropapular rash some 16 to 20 days after exposure. The rash first appears on the face and then spreads over the trunk and later over the extremities. Other symptoms typically include low-grade fever, lymphadenopathy, sore throat, and general malaise. Lymphadenopathy can be characteristic, involving the posterior cervical and occipital nodes, which can persist after the rash has resolved.
Rubella can cause complications, with transient joint involvement such as arthritis and arthralgia being the most frequent. Interestingly, these symptoms are more prevalent and severe in RV-infected women than in RV-infected men (142, 159). More serious complications including thrombocytopenic purpura and postinfectious encephalopathy or encephalomyelitis are very occasionally associated with postnatally acquired rubella (34, 159). A rare and usually fatal neurodegenerative disorder termed progressive rubella panencephalitis has also been reported as a late complication of childhood rubella (1, 34, 159).
The major public health concern posed by rubella is its teratogenicity, with maternal infection early in pregnancy leading to the congenital rubella syndrome (CRS) in infants. The time at which infection occurs during gestation can influence the outcome. The earlier in gestation the maternal infection occurs, the more severe is the damage to the fetus. Maternal infection during the first 8 weeks after the last menstrual period results in nearly all fetuses becoming infected and up to 100% of infected fetuses developing congenital defects. The risk of fetal infection and the severity of congenital abnormalities decreases after the first trimester; after 17 weeks gestation, the risk of developing any defects is low (44). The clinical manifestations of CRS are numerous and varied, with deafness being the most common. Other clinical features include cardiac disease, mental retardation, and ocular conditions such as cataracts and glaucoma. Insulin-dependent diabetes mellitus occurs commonly as a late sequela of CRS, and defects such as deafness may not be initially detected (32, 44). Interestingly, cases of CRS have been reported following maternal reinfection, although this does appear to be a rare phenomenon (131). As with primary rubella infection, the gestational age at the time of reinfection influences the chance of fetal abnormalities. No cases of rubella reinfection causing CRS have been reported after 12 weeks gestation.
Pathology of CRS
RV generally establishes a chronic nonlytic infection in the fetus and has the potential to infect any organ (153, 159). Microscopic analyses of aborted infected fetuses revealed cellular damage in multiple sites, with noninflammatory necrosis being common in the structures of the eyes, heart, brain, and ears of aborted RV-infected fetuses (143, 153). Examination of RV-induced cataractous eye lenses from first-trimester fetuses revealed pyknotic nuclei, cytoplasmic vacuoles, and inclusion bodies in primary lens cells; lens development was found to be retarded (143, 153). While the lens is the predominant site of necrosis, other eye structures such as the iris and the retina can be affected. Necrosis is also detected in the endothelial cells within the blood vessels lining the heart and can cause thrombosis of small vessels and necrosis of surrounding tissue; cell destruction of the myocardium is common (143, 153). Vascular necrotic lesions within the walls of the cerebral blood vessels may contribute to ischemic brain damage. As with RV-induced deafness in CRS infants, examination of RV-infected fetuses revealed cellular damage to the epithelium of the cochlear duct and/or stria vascularis (143, 153).
To date there has only been one report of ultrastructural examination of cells from RV-infected fetuses (75). The study reported the presence of nuclear bodies and tubuloreticular complexes in vascular endothelial cells from many organs (75). Loss of cytoplasmic ground substance, swelling of the mitochondria, and dilation of the endoplasmic reticulum (ER) were some of the features observed in cells displaying these tubuloreticular structures. The contribution of RV replication and/or virus-induced host factors to such cellular changes has not been elucidated.
VACCINES
The full impact of the teratogenic properties of RV infection was demonstrated from 1962 to 1965 following a global epidemic of rubella. In the United States, at least 20,000 infants suffered permanent damage as a result of in utero infection. This figure would have been even more overwhelming if not for the large number of therapeutic abortions carried out after maternal rubella infection (25). The consequences of the rubella epidemic added great impetus to the development of a suitable vaccine.
In 1966, Parkman et al. (112) developed the first live attenuated vaccine, HPV-77, by passaging RV 77 times in African green monkey kidney cells. In 1969, following passaging of HPV-77 in duck embryo fibroblasts at Merck, HPV-77 DE-5 became the first rubella vaccine licensed for use in the United States. Soon afterward, other live attenuated RV vaccines became available, and one of these, the RA27/3 strain, became the mainstay of vaccination programs in most developed countries (26, 110, 159).
With the advent of vaccine availability, two different strategies were used to avert further rubella epidemics. In the United States, universal vaccination of all preschool children was used, while in Australia and many European countries, including the United Kingdom, a selective vaccination policy was used concentrating on school-age girls (10 to 15 years old) and susceptible women. Both strategies resulted in a substantial decrease in the number of cases of rubella and CRS. With the latter strategy, rubella infection was still prevalent among males, allowing the virus to circulate to susceptible individuals when opportunity arose. As an indication of the effectiveness of the immunization policies, the United States consistently reported fewer cases of CRS than the United Kingdom did despite having a significantly higher population base (110). As a result, many rubella vaccination policies have been subsequently expanded to be inclusive rather than selective. With the development of the trivalent measles-mumps-rubella vaccine, a combined infant and adolescent immunization strategy has been adopted by many countries. As part of a global effort to eradicate measles, implementation of measles-mumps-rubella vaccine schedules has enhanced rubella immunity (110).
EPIDEMIOLOGY
Rubella has a worldwide distribution. The peak incidence occurs during spring months in countries with a temperate climate, although the disease is present throughout the year (159). In the prevaccine era, rubella epidemics occurred every 6 to 9 years in the United States and at shorter intervals of 3 to 5 years in Europe, including the United Kingdom (110, 159). Since humans are the only known reservoir for RV, maintenance of rubella requires continuous access to a susceptible population. In developed countries before vaccine development, infection was most common in the 5- to 9-year-old group, corresponding to the early school years. With the advent of childhood vaccination in the United States, there was a shift in disease incidence to young adults. In countries practicing the selective vaccination policy, there were much higher notification rates in males, as expected.
From studies with monoclonal antibodies, it was widely agreed that there is essentially only one serotype of RV. With the advent of molecular techniques such as PCR, it is now also possible to look at the genetic makeup of the virus itself and perform molecular epidemiological studies. A number of groups have examined the nucleotide sequence of the RV envelope E1 gene and performed phylogenetic analyses (for a compilation, see reference 35). Geographical isolates were derived from three continents and included wild-type, laboratory, and vaccine strains. The virus strains analyzed were derived from initial rubella isolates collected in the 1960s through to the 1990s, allowing some evolutionary comparisons to be made. Overall, RV could be divided into two genotypes, which differ from each other by 8 to 10% at the nucleotide level. Genotype I was a large intercontinental group containing 60 of the 63 isolates derived from North America, Europe, and Japan, and genotype II represented only 3 Asian isolates from China and India. The diversity seen at the nucleotide level was not evident at the deduced amino acid level, where the two genotypes differed by only 1 to 3%, indicating that they were antigenically very similar. The isolates included strains isolated from patients with CRS; no apparent signature mutations or other changes could be identified to distinguish CRS strains from other strains.
VIRUS STRUCTURE
The mature RV virion is a round or ovoid particle approximately 60 nm in diameter. The virion contains an electron-lucent spherical core composed of multiple copies of the RV capsid protein and a single copy of the viral RNA genome. The RV core is surrounded by a host-derived lipid bilayer containing 5- to 6-nm-long spikes which project from the virion surface; the spikes are composed of the E2 and E1 glycoproteins (34, 98).
Capsid Protein
The capsid protein is a nonglycosylated, phosphorylated, disulfide-linked homodimer with a reported molecular mass of 33 to 38 kDa (18, 92, 108, 146). The capsid protein contains clusters of proline and arginine residues, which have been postulated to be involved in binding to the RV genomic RNA to form the viral nucleocapsids (20, 34). In particular, a 28-amino-acid domain containing a large number of basic residues appears to be directly involved in binding to the RNA genome (88). However, the interaction of the capsid protein with the viral RNA may not be solely dependent on the density of basic residues because other basic regions within the protein were found to bind poorly. It remains to be determined whether other domains of the protein are involved in nucleocapsid formation. On the RV genome, a 29-nucleotide (nt) stretch (nt 347 to 375) interacts with the capsid protein, although it is not clear whether this is sufficient for packaging of the genome (88).
E1 and E2 Glycoproteins
The virion envelope proteins, E1 and E2, are type 1 membrane glycoproteins observed as spikes in the form of E1-E2 heterodimers on the virion surface (18, 66, 108, 144). The E1 and E2 proteins each contain a putative transmembrane (TM) domain, which is 22 and 39 residues in length, respectively (34). For E2, the putative TM domain is followed by a positively charged 7-residue sequence, RRACRRR, and a 20-residue region which acts as a signal sequence for E1; the positively charged 7-residue region is believed to interact with the negatively charged phospholipid head groups of the lipid bilayer. For E1, the TM domain is followed by a 13-residue cytoplasmic domain (58).
The RV E1 glycoprotein migrates as discrete band with a molecular mass of 58 kDa, while the E2 glycoprotein migrates as broad heterogeneous band of 42 to 47 kDa (18, 19, 108). Amino acid sequence analysis of the E1 protein has since revealed that it contains three N-linked glycosylation sites for all strains so far sequenced (127). In contrast, the number of N-linked glycosylation sites of the E2 protein appears to vary depending on the strain. The E2 protein of the M33 and HPV-77 strains possesses four N-linked glycosylation sites, while the E2 protein of the Therien and RA27/3 strains possesses three (127). Studies using RV-infected cells and full-length cDNA clones of E1 and E2 have shown that all the N-linked glycosylation sites are utilized, with N-linked sugars representing approximately 6 kDa and 15 to 20 kDa of the molecular mass of the mature E1 and E2, respectively (60, 127).
The role of N-linked glycosylation on the antigenicity and immunogenicity of E1 has been investigated by several groups. Studies in which recombinant E1 was expressed in Escherichia coli have indicated that glycosylation may be required for correct folding of E1 for the expression of important antigenic and immunogenic epitopes (140). For E2, mutagenesis studies have shown that removal of any of the N-linked sites results in slower glycan processing and lower stability, with the severity of the defect increasing with the number of N-linked glycosylation sites removed (127). In addition to N-linked sugars, the RV E2 protein contains O-linked carbohydrates (89). The presence of these O-linked sugars most probably contributes to the heterogeneous nature of the virion form of E2. Pulse-chase labeling of RV-infected cells has revealed the presence of intracellular forms of E2 (39 kDa), which migrate more rapidly than the virion form of E2 (42 to 47 kDa) (18, 52, 108).
The functions of the RV E1 and E2 glycoproteins have been studied extensively. Using monoclonal antibodies, it has been shown that the E1 protein contains at least six nonoverlapping epitopes, some of which are associated with hemagglutination and neutralization (21, 67, 139, 140, 152, 160, 161). E1 appears to be the main surface protein, with domains involved in the attachment of the virus to the cell. More recent studies have revealed that a 28-residue internal hydrophobic domain of E1 is responsible for the fusogenic activity of RV (164). In addition, this region is involved in the binding to E2 for heterodimer formation (164).
The function of E2 has been more difficult to determine. E2 is disulfide-linked to E1 in the mature virion and is poorly exposed (66, 152). Therefore, the antigenic sites of E2 are less accessible to characterization by monoclonal antibodies (151). However, E2 does contain partial hemagglutination and neutralizing epitopes and may also carry strain-specific epitopes (28, 46).
VIRUS LIFE CYCLE
Attachment and Entry
Molecular characterization of RV has been largely performed in Vero and BHK-21 cells because of their ability to produce high titers of virus, perhaps due to the absence of an interferon system (97). However, RV can establish infections in a variety of cell lines (34), indicating that the host cell receptor is likely to be a ubiquitous molecule. Although the host cell receptor has not been identified, it appears that membrane phospholipids and glycolipids may be involved in viral attachment (94).
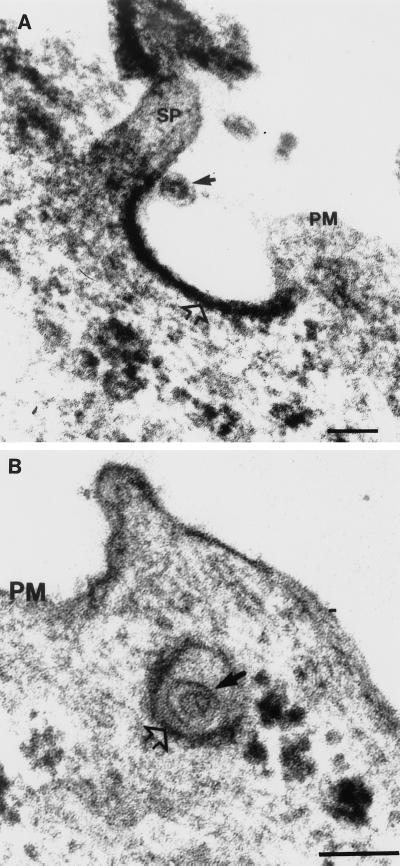
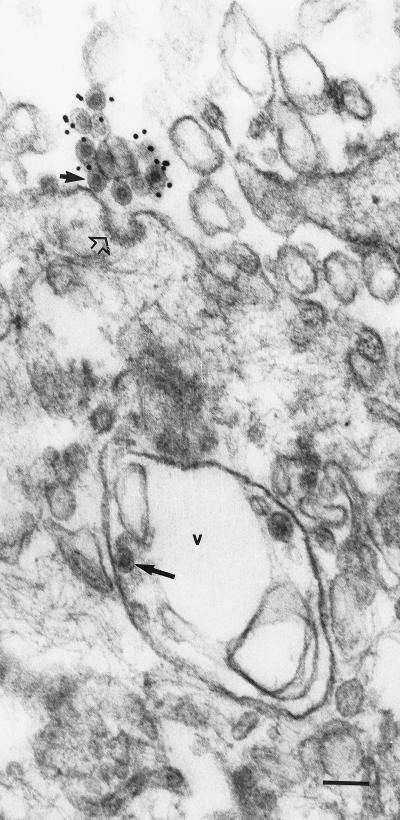
The route of RV entry into the host cell is not well understood. There is some evidence to suggest that RV enters cells via the endocytic pathway, similar to that reported for the alphaviruses (73, 78). Early biochemical studies by Katow and Sugiura (70) showed that exposure of the RV E1 and E2 glycoproteins to pH 6.0 or less induced a conformational change within the glycoproteins that favored the fusion of the viral envelope to the endosomal membrane. This hypothesis was further supported by more recent studies which demonstrated the inhibition of viral replication following the use of lysosomotropic agents (116). Preliminary viral attachment and penetration studies by thin-section electron microscopy (TSEM) indicate that at physiological pH of 7.4, RV enters predominantly via the endocytic route. RV virions could be observed attached to cell surface projections which were either adjacent to or in coated pits (Fig. 1); these events were observed as early as 3 min after the addition of virus to the cell monolayer. Similarly, preembedding immunogold-labeling studies using antibodies to RV revealed that during the viral latent period, gold-labeled RV virions were seen attached to the plasma membrane adjacent to coated pits; unlabeled virions were observed in endosome-like vacuoles (Fig. 2).
FIG. 1.
Attachment and entry of RV into Vero cells. Vero cells were inoculated with RV at a multiplicity of infection of 50 and incubated at 37°C. At 3, 5, 15, 30, and 45 min after the addition of virus, the cell monolayer was harvested and processed for TSEM as described by Lee (81). (A) An RV virion (solid arrow), comprising an electron-lucent core surrounded by a host-derived lipid envelope, can be seen attached to a cell surface projection (SP) adjacent to a coated pit (open arrow). (B) A virion (solid arrow) can be seen located within a coated vesicle (open arrow). Bars, 100 nm. PM, plasma membrane.
FIG. 2.
Detection of an RV virion within an endosome-like vacuole. RV-infected Vero cells were harvested at 4 h p.i. and processed for immunogold-labeling EM using polyclonal antibodies to RV as described previously (84). Gold particles (diameter, 10 nm) were found associated with virions located extracellularly. Note the virion (short solid arrow) attached near a coated pit (open arrow). An unlabeled RV virion (long solid arrow) is seen in an endosome-like vacuole (v). Since the cells were harvested during the viral latent period, it is unlikely that the virion within the endosome-like vacuole represented a newly assembled particle. Bar, 100 nm.
The uncoating event for RV is also not well defined. It has been shown that between pH 5.0 and 5.5, the RV capsid protein undergoes a structural change from having hydrophilic to hydrophobic properties (95). This conformational change in the capsid protein presumably allows uncoating to occur within the endosome, allowing the release of viral genomic RNA into the cytoplasm. Thus, it seems that the low-pH environment of the endosome serves not only to induce virion envelope fusion to the endosomal membrane but also to trigger uncoating of the capsid protein. This is in contrast to the mechanisms of alphavirus uncoating. For SFV, the viral nucleocapsid is uncoated by the binding of the capsid proteins to ribosomes (135).
Replication
RV is characterized by slow replication, which is reflected in the long viral latent period of 8 to 12 h (18, 34). During RV infection, four distinct viral RNA species can be detected. A single-stranded 40S RV genomic RNA (3.8 × 103 kDa) and a 24S subgenomic RNA (1.2 × 103 kDa) that corresponds to the 3′ one-third of the genomic RNA are present in infected cells (68, 109, 134). Both contain a methyl7guanosine cap at the 5′ terminus and a polyadenylate tail at the 3′ terminus. In addition, viral replicative intermediates (RI) of 21S, representing partial double-stranded RNA (dsRNA), and viral replicative forms (RF) of 19 to 20S, representing full dsRNA, have been detected in RV-infected cells (134, 162).
During viral replication, the 40S RV genomic RNA serves as a messenger for the nonstructural (ns) proteins and as a template for the synthesis of a 40S negative-polarity RNA strand. The minus strand in turns acts as a template for the transcription of both the 40S RNA and the 24S RNA (34). Nascent 40S RNA is packaged with the RV capsid protein to form nucleocapsids. In terms of viral kinetics, both the RV 40S RNA and 24S RNA were detected at the end of the viral latent period, with viral structural proteins appearing 4 h later (52). Peak virus production occurs during the period from 36 to 48 h postinfection (p.i.).
One-step multiplication studies have shown that RV is unable to infect every cell at any specific time, irrespective of the titer of the input virus (20, 52, 134, 162). Moreover, the proportion of cells infected by RV at any one time is cell type dependent (52, 134, 162). However, as infection proceeds, the entire culture eventually becomes infected.
Genome Organization
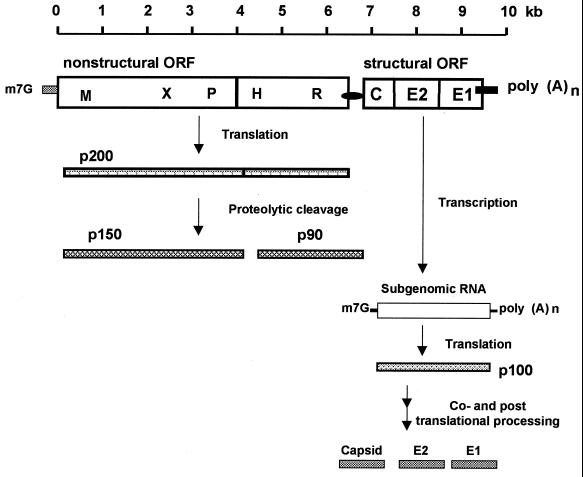
The full-length RV genome measures 9,762 nt and contains two long open reading frames (ORFs) (27) (Fig. 3). The RV 5′-proximal ORF of 6,345 nt encodes the viral ns proteins, p150 and p90, while the 3′ ORF of 3,189 nt encodes the structural proteins, capsid (C), E2, and E1 (27, 166). Thus, the gene order for the RV 40S RNA is 5′-p150-p90-C-E2-E1-3′ (Fig. 3). The complete nucleotide sequence of RV has been determined for three strains, Therien (150), M33 (165), and RA27/3 (121). Nucleotide and deduced amino acid sequencing analysis reveals a high degree of homology between strains, ranging from 97.2 to 99% and 97.6 to 98.9% at the nucleotide and amino acid levels, respectively. The RV genome has an extraordinarily high G+C content of 69.5%, the highest of any known RNA virus to date, and this has undoubtedly contributed to some discrepancies in the reported RV genomic sequences (27, 121, 138, 148).
FIG. 3.
Schematic representation of the translation and processing strategy of the RV ns and structural proteins. The RV genome comprises two long nonoverlapping ORFs, with the 5′ ORF coding for the ns proteins and the 3′ ORF coding for the structural proteins. A polyprotein precursor, p200, is translated from the 5′ ORF of the RV genomic RNA and undergoes cis cleavage to produce two ns proteins, p150 and p90. The locations of the putative amino acid motifs for methyltransferase (M), X motif, papain-like cysteine protease (P), helicase (H), and replicase (R) are indicated on the 5′ ORF. The RV structural proteins are synthesized from a 24S subgenomic RNA transcribed from the 3′ ORF. A polyprotein precursor, p100, is translated from the subgenomic RNA and undergoes several posttranslational modifications to ultimately produce the mature capsid (C), E2, and E1.
Amino acid sequence analysis of the RV 5′ ORF has revealed global amino acid motifs indicative of RNA-dependent RNA polymerase (replicase), helicase, methyltransferase, and proteinase activities (34). In addition, a short region of unknown function, termed the X motif, is found within p150 and is also present in the alphavirus nsP3 protein and the hepatitis E virus 5′ ORF product (27, 34). The amino acid motifs for proteinase and methyltransferase are located in the carboxy terminus of p150, while the motifs for the replicase (GDD) and the helicase (GxGKT and DExx) are located in the carboxy terminus of p90 (27). Thus, the RV gene order for the 5′ ORF is 5′-methyltransferase-X-protease-helicase-replicase-3′ (34). In contrast, the gene order for SIN virus 5′ ORF is 5′-methyltransferase-helicase-protease-X-replicase-3′. This inversion of the motifs relative to each other has led to the hypothesis that genetic rearrangement has occurred during togavirus evolution (34).
Following the complete sequencing of the Therien strain genome, the first cDNA copy of the RV genomic RNA was generated (150). Recently, a full-length infectious clone derived from the M33 strain was also produced (165). The RNA transcripts generated from both the full-length RV clones were found to be infectious, as evidenced by the production of virus that was phenotypically and genetically similar to the parental virus from which the cDNA clone was derived. It is anticipated that the genetic manipulation of such RV infectious clones will contribute further to the understanding of RV replication and pathogenesis.
Structural Proteins: Translation, Processing, and Assembly
The RV structural proteins are translated as a polyprotein precursor, p110, in the order NH2-C-E2-E1-COOH from the 24S subgenomic mRNA (106, 109); two possible AUG initiation codons are present within the RV 24S RNA, and it has been suggest that both AUG codons are used indiscriminately (24, 92). The p110 protein is translocated into the ER by two separate signal peptides, 23 and 20 amino acids in length, located at the amino termini of E2 and E1, respectively. Within the ER, the RV capsid protein is cleaved from E2 and E2 is cleaved from E1 (10, 24, 36, 92, 137). Unlike the alphaviruses, where an autoprotease cleaves the alphavirus capsid protein from the polyprotein precursor, cleavage of RV capsid protein is mediated by a cellular signalase found within the lumen of the ER (24, 36). A unique feature of the RV capsid protein is the retention of the E2 signal peptide on the carboxy terminus of the capsid protein (10, 54, 92, 137). Similarly, the E1 signal peptide of 20 amino acids is retained on the carboxy terminus of the E2 protein after cleavage from E1 by host signalase (36, 54, 59, 107). The respective signal peptides of E2 and E1 direct the insertion of the proteins into the ER (54, 59).
The assembly pathway of RV has not been fully elucidated. Nevertheless, significant progress has been made in understanding RV assembly with the use of cDNA constructs containing the RV structural genes. Of particular significance is the development of cDNA constructs that express RV structural proteins that assemble into rubella virus-like particles (RLP). These RLPs have similar morphology and sites of budding to their wild-type counterpart, and they serve as a convenient tool in dissecting the RV assembly pathway (58, 126). For RV, the assembly of structural proteins begins immediately following translation of these proteins. Following proteolytic cleavage in the ER, the E1 and E2 proteins form disulfide-linked heterodimers while the capsid proteins form disulfide-linked homodimers (11, 152). E1-E2 heterodimer formation is necessary for the transport of E1 from the ER to the Golgi complex and the cell surface; in the absence of E2, E1 is arrested in the post-ER, pre-Golgi complex compartment (57, 61). It has been suggested that E1 and E2 dimerization facilitates the proper folding of E1 (10, 56). An ER retention signal on E1 of 22 amino acids, spanning both the cytoplasmic and transmembrane domains of the protein, functions to retain unassembled E1 subunits and immature E2-E1 dimers in the ER until folding and heterodimer formation are complete (55). In the process, E1 undergoes a conformational change that masks the ER retention signal, thereby allowing the transport of the heterodimers to the Golgi complex. Recently, it has been shown that an internal hydrophobic domain in E1 is involved in E1-E2 interaction, leading to the formation of the heterodimer (164). The E2-E1 heterodimers are retained in the Golgi complex by a retention signal of 18 amino acids located in the TM domain of E2 (62). Recent studies using RLPs have indicated that the TM and cytoplasmic domain of E2 are required for the targeting of the heterodimers to the sites of budding such as the Golgi complex. In contrast, the TM and cytoplasmic domains of E1 were shown not to be required for this process but were necessary for the secretion of RLPs into the medium (41). A better-defined mutagenesis study on the E1 TM and cytoplasmic domain within a RV infectious cDNA has revealed similar findings (165). Collectively, the data suggest that the E1 TM and cytoplasmic domains play a critical role in the very late stages of virus budding.
The role of the capsid protein in RV assembly is less well defined. It has been proposed that maintenance of the E2 signal sequence as part of the capsid protein after cleavage allows the capsid protein to be transported along with the glycoproteins to the Golgi complex (126). Earlier studies postulated that the interaction of the E1 cytoplasmic domain and the RV capsid protein triggered virus budding (58). However, recent studies have revealed that this interaction is not the driving force for virus budding (41). The mechanisms involving the interaction between the nucleocapsid and E2-E1 heterodimers have yet to be elucidated.
While studies with RLPs have provided important insights into RV assembly with respect to virus protein-protein interactions, the precise mechanisms for budding where the virions acquire the host membrane are far from clear. Moreover, recent observations that RV virions and RLPs can be secreted into the apical and basolateral surfaces of polarized cells (40) have raised questions whether the assembly and budding events are similar for both compartments.
Nonstructural Proteins: Translation and Processing
The characterization of RV ns proteins has largely been hampered by the limited production of ns proteins in infected cells. The task was further complicated by the presence of host cell proteins that obscured the detection of the ns proteins because RV does not inhibit host protein synthesis. Early studies reported the detection of several ns proteins, of 200, 150, 87, 75, and 27 kDa in RV-infected cells (18). Pulse-chase studies during hypertonic salt treatment of RV-infected cells demonstrated that the ns200 protein was cleaved to ns150 (18). More recent work employing cDNA constructs containing the RV 5′ ORF of the Therien and M33 strains demonstrated that the 200-kDa protein is an ns polyprotein precursor that is cleaved to produce two products, of 150 and 90 kDa (Fig. 3) (33, 93, 166). Thus, the gene product order for the RV 5′ ORF-encoded protein is NH2-p150-p90-COOH. The 87-kDa protein of Bowden and Westaway (18) in RV-infected cells most probably represents the 90 kDa protein observed in transfection studies (33, 93, 165).
The cleavage of the polyprotein precursor, p200, into two fragments is mediated by a protease residing in p150 with catalytic residues of Cys-1151 and His-1272; the protease cleavage site is found within Gly-1300–Gly-1301 (23). Based on comparative amino acid sequence analysis, the RV protease was proposed to be a Main protease with similarities to cellular proteases (45). Transfection studies have since confirmed that the RV protease is indeed a Main protease because it can function in cis and trans cleavage (87, 165). The p150 protein was recently shown to be localized to RV replication complexes (77). The only other functional activity demonstrated within the RV 5′ ORF is that of the RV helicase (50). NTPase activity has been demonstrated to be associated with the amino acid region encompassing the GxGKT and DExx motifs of the putative helicase (50). The enzymatic functions of the putative replicase have not been shown.
Role of Cellular Proteins in RV Replication
Studies using RV-infected cells showed that the addition of actinomycin D (an inhibitor of DNA-dependent RNA polymerase) in the early stages of RV infection inhibited virus replication, suggesting that cellular or host proteins may also be involved in viral RNA replication (103). Moreover, there are several sequences at the 5′ and 3′ termini of the RV genome that can potentially form stable stem-loop structures representing possible sites for interactions between RV RNA and host proteins (101, 102, 119). A synthetic RNA modeled from the 3′ stem-loop structure has been shown to interact specifically with three phosphorylated cytoplasmic proteins (100, 119). One such protein has been identified as calreticulin, a host protein that is involved in the modulation of genes and that binds specifically to regions located at the 3′ end of the RV RNA (4, 22, 100, 102, 119, 136). Although the 3′ untranslated region contains cis-acting elements necessary for RV replication, the role of calreticulin binding to this region is unclear. It would be of interest to determine whether calreticulin associates with RV replication complexes where active viral RNA synthesis occurs (22).
Other studies have demonstrated that the 5′(+) stem-loop cis elements of RNA could be cross-linked in vitro to the cellular protein, La, implying that it interacts with RV RNA (30, 120). Recently, it was shown that this RNA structure is involved in translation rather that in viral replication (124). The Ro/SS-A antigen has also been suggested to interact with the 5′(+) stem-loop structure, but it is not known whether this interaction is specific (102).
In addition to host proteins interacting with the RV genome, the cellular retinoblastoma (RB) protein interacts with a known RB-binding motif located within the carboxy-terminal half of the RV p90 protein (3). Because the RB protein is involved in the regulation of cell growth, it has been proposed that this interaction may induce RV teratogenesis (3).
Morphogenesis
Numerous investigators have employed TSEM to study RV morphogenesis in a variety of cell lines including RK13, BHK-21, and Vero cells. Early TSEM studies on RV-infected cells reported predominantly on the morphology and maturation of RV. RV maturation is a budding process in which the viral core acquires an envelope membrane after passing through modified host cell membranes (98). The Golgi apparatus, rough ER (RER), cytoplasmic vacuoles, and plasma membrane have been identified as sites of RV maturation (64, 65, 83). This is in contrast to the alphaviruses, where the plasma membrane is the only site of virus budding (159). The maturation of RV at cytoplasmic sites is presumably due to the presence of an ER and a Golgi retention signal on the E1 and E2 glycoproteins, respectively, that facilitate the accumulation and assembly of the proteins in the respective organelles. Mechanisms responsible for triggering virion assembly are poorly defined, and it is not known whether similar mechanisms are involved at the different maturation sites.
Early morphological studies revealed several cellular changes associated with RV infection. However, discrepancies with these findings exist. Cytoplasmic inclusions have been reported in RV-infected RK13 cells (65) but were never detected in a similar cell line studied by other investigators (51). Moreover, the significance of these cytoplasmic inclusions is not known. Annulate lamellae were observed in LLC-MK2 and RK13 cells infected with RV (74, 113), but measurements and descriptions of virus-like particles in these studies did not correlate with the characteristic morphology of RV virions. Thus, it appears unlikely that these structures were associated with RV replication as has been previously reported. In addition, annulate lamellae have been found in other virus-infected cells, indicating that the occurrence of these structures is probably due to a nonspecific cellular response to viral infection (71).
Replication Complexes
Although early TSEM studies have produced several important findings on the morphology and maturation of RV, interest in RV morphogenesis waned with the advent of molecular biology applications. Consequently, in the past two decades, RV research has focused mainly on the characterization of RV replication at the molecular and biochemical level. Nevertheless, interest in RV morphogenesis has continued, leading to the discovery of “replication complexes” in RV-infected cells (81, 83, 84, 85, 91). It is now recognized that the results of these studies complement existing molecular information on RV replication. Furthermore, these morphological studies revealed that RV replication complexes are similar in morphology, function, and biogenesis to alphavirus cytopathic vacuoles type 1 (CPV-1) (2, 37, 38, 48, 49). It must be noted that RV replication complexes were so named in reference to their functional role while their alphavirus counterparts were termed CPV-1 according to their morphology.
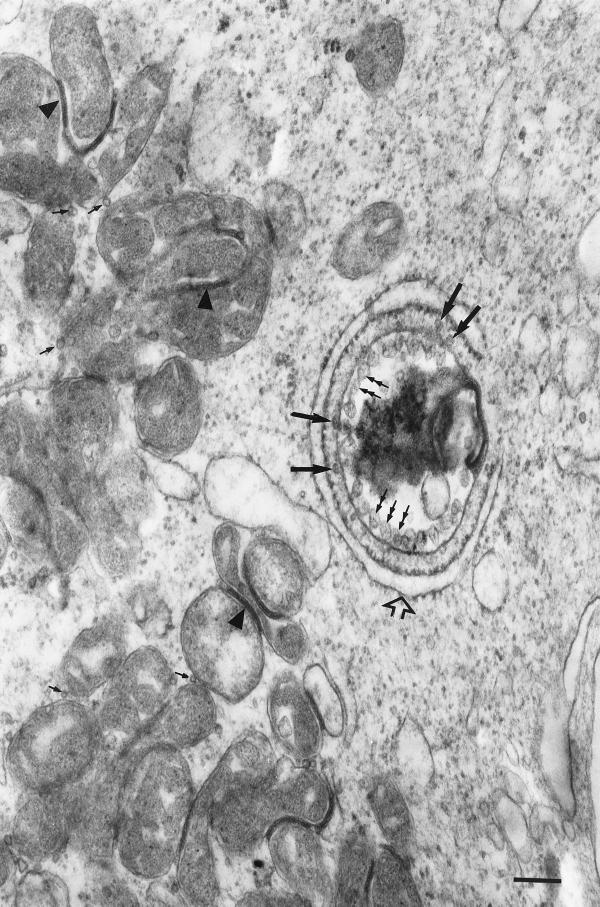
Lee et al. (83) first described RV replication complexes as membrane-bound cytoplasmic vacuoles lined internally with vesicles measuring approximately 60 nm in diameter (Fig. 4). These vesicles contained thread-like inclusions and were found either free in the vacuole or attached to the inner membrane of the vacuole via a membranous neck. Although these vesicles have similar dimensions to those of RV virions, they do not constitute immature or aberrant virion forms. Close examination reveals that the vesicles contain irregular internal structures which are distinct from the spherical electron-lucent cores observed in mature RV virions (83). In addition, aberrant forms of RV virions were reported as multicore structures surrounded by a single membrane (9, 53, 98, 149). Importantly, RV virions have not been detected in the vacuoles of the replication complex where the vesicles were found (83).
FIG. 4.
Cellular changes in RV-infected cells. A typical replication complex is observed with the characteristic vesicles (double-headed arrows) and the close association of the RER (open arrow). RV core particles (long solid arrows) can be seen at the cytoplasmic side of the vesicles of the replication complex. Core particles (small solid arrows) can also be detected in association with the outer membrane of mitochondria. Electron-dense zones (arrowheads) are frequently observed between the outer membranes of adjacent mitochondria. Note the clustering of mitochondria near the replication complex. Bar, 200 nm.
A distinctive feature of RV replication complexes is the close association of the RER with the vacuole complex (Fig. 4). During RV infection, RER was found to associate only with the side of the replication complex vacuole where the vesicles were located, but as the infection progressed, the RER surrounded the whole replication complex (83). RV replication complexes were detected as early as 8 h p.i., coincident with the end of the viral latent period, and peak numbers of these structures were reported at 24 h p.i., coinciding with peak production of the virus (83).
The presence of RV replication complexes in infected cells was first postulated by Bowden et al. (17), who observed discrete cytoplasmic fluorescent foci in RV-infected cells in an immunofluorescence assay using antibodies to dsRNA, a marker for viral RI and RF. When later studies employing immunogold-labeling EM and anti-dsRNA were performed on RV-infected cells, dsRNA was localized within the vacuole of the replication complex, indicating that these virus-induced structures are associated with viral RNA synthesis (84). The precise site of viral replication appears to be the vesicles within the replication complex (84). The localization of RV p150 within these complexes further confirms their role in viral replication (77).
Recently, RV core particles were demonstrated to be associated with replication complexes (Fig. 4) (85). RV replication complex-associated core particles were often detected between the cytoplasmic side of the replication complex and the adjacent RER membrane; these particles were frequently found at the base of the vesicles lining the vacuole complex. Immunogold-labeling studies using monoclonal antibodies to the capsid protein confirmed the identity of the particles (85). Hence, RV replication complexes appear to serve as sites for nucleocapsid assembly. This is not surprising, since the processing of the capsid, E2, and E1 proteins occurs in the RER with the host signal peptidase involved in cleaving E2 from the capsid and E2 from E1. The attachment of the E2 signal peptide at the carboxy terminus of the capsid protein tends to render the protein hydrophobic; thus, the capsid protein is likely to remain attached to the RER membrane (54, 57, 58). Given the close proximity of the RER to the replication complex (Fig. 4), it is conceivable that the assembled capsid protein then associates with newly synthesized genomic RNA as the viral RNA is expelled from the vesicles of the replication complex (Fig. 5) (84). Interestingly, another group has reported the colocalization of RV capsid proteins with p150, thereby indicating the association of capsid proteins with RV replication complexes (77).
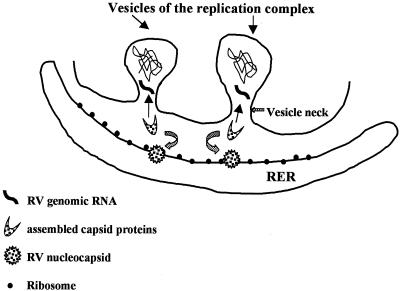
FIG. 5.
Schematic representation of the role of vesicles within the RV replication complex as precise sites of viral RNA synthesis. The vesicles of the replication complex are postulated to provide a protective environment for the synthesis of nascent viral genomic RNA. Newly synthesized viral RNA is then rapidly encapsidated by RV capsid proteins, which are synthesized from the adjacent RER. The mechanisms involved in the translocation of the resulting nucleocapsids for interaction with RV E2-E1 heterodimers have not been defined.
In terms of the biogenesis of RV replication complexes, these structures were initially proposed to be virus-modified lysosomes because degenerating material such as myelin-like membrane whorls were frequently observed within the vacuoles of the complexes (83). More recent studies employing confocal microscopy and immunogold-labeling EM with antibodies specific to lysosome proteins confirmed that the complexes were indeed virus-modified lysosomes similar to their alphavirus counterparts (91). For the alphaviruses, replication complexes were found to be both endosomal and lysosomal in origin. Antibodies to endosomes were not used in these RV studies, but it is most likely that these virus-induced structures are also virus-modified endosomes. Although lysosomes in RV-infected cells have been recruited for virus replication, it is likely that lysosomal functions continue within replication complexes, since acid phosphatase activity was detected in these complexes (91). Furthermore, it appears that these complexes are derived from existing lysosomes during RV infection rather than stimulating the production of new lysosomes for conversion into replication complexes.
While it is well recognized that endosomes and lysosomes play an important role in viral entry via the endocytic pathway, the use of these organelles as sites of viral replication appears to be unique to the togaviruses. A model for the biogenesis of RV replication complexes is proposed based on the collective findings from RV studies (Fig. 6) (81). In this model, the RV virion binds preferentially to a cell surface projection and then translocates into an electron-dense coated pit adjacent to the surface projection. The virion is then transported into the cell when the coated pit invaginates to form a coated vesicle. The virus is subsequently delivered through a series of endosomes with progressively acidic compartments until it reaches an endosome where the environment is sufficiently acidic (pH 5.3) to induce conformational changes within the E1 and capsid protein, resulting in the release of the genomic RNA into the cytoplasm (95). The events from virus uncoating to early formation of the replication complex are unknown. Presumably, the virus ns proteins are synthesized immediately from the newly released viral RNA and remain associated with the endosome where uncoating has occurred. The production of ns proteins, replicating viral RNA, and/or host proteins contributes to the formation of vesicles within the endosome, which provides a protected environment for the transcription of the genomic and possibly subgenomic RNAs. Concomitantly, the RER migrates to the vicinity of the endosome, allowing the translation and processing of the viral structural proteins from the 24S subgenomic RNA to occur. As infection progresses, the vesicles within the endosome increase in number as more ns proteins are synthesized either from newly synthesized RV genomic RNA or from the original viral genomic RNA. Late in infection, the RER is found surrounding the entire vacuole, which has been lined internally with vesicles. While these viral events are occurring, the endosome, which now represents a replication complex, continues in its life cycle and fuses to a lysosome. It is important to bear in mind that the life cycle of both the virus and the endosome/lysosome are intertwined. The decrease in the number of replication complexes late in infection and the detection of vesicles containing thread-like inclusions on the cell surface indicate that the virus-modified lysosome has fused to the plasma membrane, thereby expelling the vesicles and other lysosomal by-products extracellularly.
FIG. 6.
Schematic representation of the biogenesis of RV replication complexes. Step 1, The RV virion attaches to the cell surface and is translocated to the coated pit. Step 2, The coated pit then pinches off to form a coated vesicle that contains the virion. Step 3, The virion passes through a series of endosomes with progressively acidic pH until it arrives at an endosome where the environment is sufficiently acidic to trigger the uncoating process. The E1 and capsid proteins undergo conformational changes that result in the release of the viral genomic RNA into the cytoplasm. Step 4, Release of the viral RNA triggers the transformation of the endosome, and vesicles are induced to form within the endosome. This leads to the formation of the replication complex. Concomitantly, the RER migrates to the vicinity of the virus-modified endosome. At this early stage of the infection, the RER is associated with the side of the vacuole where the vesicles are located. Step 5, As infection progresses, the RER surrounds the entire vacuole, which is lined internally with vesicles. While these events are occurring, the virus-modified endosome fuses to a lysosome as part of its life cycle. Step 6, The replication complex continues in its life cycle as a virus-modified lysosome and eventually expels its lysosomal contents, including the vesicles, after fusion of the lysosomal vacuole membrane to the plasma membrane.
The mechanisms involved in vesicle formation within RV replication complexes are not well understood. However, transfection studies have provided some important clues by showing that SFV genomic RNA, rather than infectious virions, was able to induce the formation of replication complexes (114). Later, confocal microscopy studies by the same group found that the alphavirus nsP1 was localized to endosomes and lysosomes (115). It is not known whether nsP1 alone was sufficient to induce formation of these vesicles, since no EM procedures were performed.
Vesicle formation is not a characteristic confined to togavirus infection. Vesicles play a vital role in the replication of many single-stranded RNA viruses such as the picornaviruses and flaviviruses (14, 15, 90, 104, 105, 156, 157). The vesicles induced by these viruses are postulated to represent replication complexes that are derived from or accumulate in the ER rather than in endosomes and lysosomes. It is unclear, however, why the togaviruses, flaviviruses, and picornaviruses have utilized vesicles for viral replication. It has been postulated that the membranous structure of the replication complex acts as scaffolding, providing an architectural framework for the assembly of all the components of replication, as well as providing a large surface area for viral replication and hence a more rapid synthesis of progeny RNA (156). In addition, the double membrane vesicles may serve to protect the nascent single-stranded viral genomic RNA from degradation by cellular RNases (83).
LINKS TO TERATOGENICITY
For RV, the explicit pathway leading to teratogenicity remains to be elucidated. Many of the steps leading to CRS and the consequences of CRS are well documented, but exactly how the virus causes this dramatic effect has been the subject of much speculation. Cellular damage seen during early gestation of RV-infected fetuses is unlikely to involve the immune system, since no fetal immune response can be detected at this early stage (153). Although the presence of immunoglobulins such as immunoglobulin M (IgM), IgG, and IgA, T cells, natural killer cells, and interferon can be detected by mid-gestation in infected fetuses, the extent to which they limit or contribute to further fetal damage has yet to be determined (153).
A closer examination of some of the unusual features of RV replication and virus-host cell interactions may provide important clues. Previous work has shown two distinctive features of RV replication which may impact on normal host cell function, notably, mitochondrial abnormalities and disruption of the host cell cytoskeleton (17, 82, 85). More recent work has investigated the ability of RV to induce apoptosis, and a mechanism for the teratogenic effect of RV has been proposed (29, 63, 96, 123).
Mitochondrial Changes
A role for mitochondria in the budding and replication of RV has been suggested based on early studies of viral lipid content and metabolic changes in RV-infected cells. Cardiolipin, a phospholipid that is relatively specific to the inner mitochondrial membrane, was reported to be present in RV virions (7). These findings led the investigators to speculate that RV may bud from mitochondria. There were further suggestions of mitochondrial involvement in RV infection when the same group reported a decrease in the level of ATP in RV-infected BHK-21 cells within the first hour of infection coincident with the period of viral adsorption and penetration (6). In addition, there appeared to be an increase in respiration, glycolysis, and alanine synthesis during the same period (8, 145).
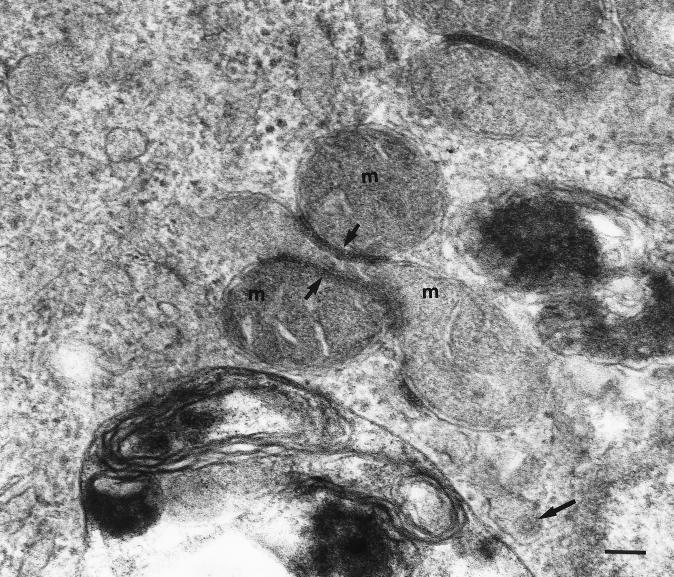
A direct link between mitochondria and RV infection was confirmed more than a decade later, when mitochondrial changes were detected in RV-infected Vero cells by TSEM (Fig. 7) (82). Electron-dense zones associated with mitochondria were identified in three different configurations: between the outer membrane of a mitochondrion and one membrane of the RER, between the outer membranes of two adjacent mitochondria, and between two opposing membranes of the RER. These ultrastructural changes have been designated confronting membranes type 1 (CM-1), confronting membranes type 2 (CM-2), and confronting cisternae (CC), respectively (82). Furthermore, it was noted that during the course of RV infection, the mitochondria appeared to become club shaped (82). It remains to be determined whether mitochondrial function is impaired as a result of such deformities during RV infection. These findings were seen only in RV-infected cells and were not present in the mock-infected preparations or in SFV-infected cells (82). Structures similar to CM-2 have been detected in cells infected with Nodamura virus (42), while CC have been found in a variety of cells including cells infected with herpesviruses (43). However, CM-1 appears unique to RV. The composition of the electron-dense zones within these structures is unknown, although analysis using RV-specific antibodies should show whether viral proteins are localized within these zones. The cellular changes observed in these studies do not correspond to those described by Kistler, who performed an ultrastructural examination of cells from RV-infected human embryos and fetuses (75).
FIG. 7.
Mitochondrial changes in RV-infected cells. Electron-dense zones (short arrows) are frequently observed between the outer membrane of an adjacent mitochondrion (m). The composition of the electron-dense zone is not known. Note an RV virion (long arrow) within a lumen of an ER. Bar, 100 nm.
Another intriguing feature of RV infection is the association of RV core particles with mitochondria (85); no such association has been reported for the alphaviruses. This association is perplexing, since there appears to be no mitochondrial targeting signal on the RV capsid protein. It may be that the capsid protein has a specific affinity for mitochondrial membranes or an affinity for cell membranes in general. The RV capsid protein is a phosphorylated protein with a unique feature in possessing the E2 signal peptide. The precise role of the peptide has not been determined, although in vitro and in vivo studies have revealed that it has an affinity for membranes (54, 56, 57). Further studies are necessary to see whether this signal peptide or phosphorylation play a role in targeting the RV capsid proteins for assembly at mitochondria or whether assembled core particles localize to the mitochondria.
In addition to these observations, pronounced clustering of mitochondria around RV replication complexes was reported; this was similarly observed with the SFV replication complexes (82). In light of these observations, it has been suggested that togavirus replication complexes are sites of a high-energy requirement that induces mitochondria to migrate to their vicinity (82). This phenomenon does not appear to be confined to the togaviruses. Mitochondria in Vero cells infected with African swine fever virus, a dsDNA virus, have also been reported to migrate in large numbers to viral assembly sites (132). The migration was accompanied by a dramatic change in mitochondrial ultrastructure, characteristic of active respiration. In addition, a fourfold increase in the levels of mitochondrial stress proteins and mitochondrial chaperone proteins was observed (132). For RV, cell metabolic and viral kinetic studies need to be performed in parallel with ultrastructural studies to gain a better understanding of RV-induced mitochondrial changes.
Cytoskeletal Changes
The effects of RV infection on the cytoskeletal components of cells have been investigated by immunofluorescence studies using antibodies to actin (17). A significant alteration in the arrangement of actin filaments following RV infection was evident. Instead of the filamentous actin cables observed in uninfected cells, amorphous clumps of fluorescent foci, representing depolymerized actin filaments, were detected in RV-infected cells. Little depolymerization was evident until 16 h after infection, suggesting that the synthesis of some viral product may have been necessary to achieve this effect. As infection progressed, fewer actin filaments were stained, until almost all the actin was found to be disaggregated into large, intensely stained foci; this effect was noted in both RV-infected Vero and BHK-21 cells. Recent studies by Kujala et al. (77) also reported similar findings. Interestingly, no changes were observed to the microtubules, another cytoskeletal component, during RV infection (77).
Apoptosis
Apoptosis is a form of cell death that involves a genetically programmed series of events culminating in the destruction and disposal of unwanted cells. It is now established that apoptosis is characterized by specific morphological and biochemical features that can vary with tissue and cell type. Morphologically, changes within the cell, such as nuclear chromatin condensation, plasma membrane blebbing, and cellular fragmentation into membrane apoptotic bodies, are typical features of apoptosis. Biochemically, degradation of chromatin, as determined by DNA fragmentation assays, is generally characteristic of this form of cell death (133, 158). More specifically, apoptotic events can be characterized by the overexpression of certain regulatory proteins that function to trigger or regulate apoptosis. The proteins within the Bcl-2 family best exemplify the diverse and complex biochemical pathways involved in the regulation of apoptosis. Within the Bcl-2 family are antiapoptotic proteins such as bcl-2 and bcl-xL and proapoptotic proteins such as bax and bcl-xS, which act directly or indirectly to activate a family of proteases called caspases, the major effector proteins for apoptosis (76, 130, 147). The Bcl-2 family of proteins can indirectly promote apoptosis through the regulation of a variety of signal transduction stimuli, e.g., through ceramide, collapse of mitochondrial transmembrane potential, p53 activation, and activation of cytokine receptors such as fas/APO-1/CD 95.
It is becoming increasingly evident that apoptosis plays an important role in the pathogenesis of many viruses (129, 141). Alphaviruses, adenoviruses, Epstein-Barr virus, human papillomavirus, hepatitis B virus, and human immunodeficiency virus are some of the viruses known to induce or interfere with the apoptotic pathway. Some of these viruses possess genes that encode either homologues of the Bcl-2 family of proteins or inhibitors of caspases. Alternatively, some of these viruses have evolved proteins that block apoptosis and, in so doing, prolong the life of the infected cell for virus dissemination (13, 86). The versatility of the virus to control the cellular apoptotic machinery can result in a range of pathogenic outcomes such as persistence, latency, and tissue tropism. The recent large increases in the volume of literature on RV-induced apoptosis have revealed some understanding of the mechanisms of RV persistence and teratogenicity (29, 63, 96, 123).
While RV can establish persistent noncytocidal infection in many cell lines, it can also cause cytopathic effects (CPE) in cell lines such as Vero, BHK-21, and RK13. It is now clear that the RV-induced CPE, seen as cell rounding followed by detachment from the monolayer, is due to apoptotic cell death (29, 63, 96, 123). Characteristic markers of apoptosis such as DNA fragmentation, nuclear chromatin condensation, and annexin V staining have been reported during acute infection with RV. The ability of RV to induce apoptosis varies considerably with cell type and appears to be associated with cells that cause CPE during infection. Active replication is required for RV-induced apoptosis during acute infection, although transfection studies of cells expressing only RV structural proteins indicate that the RV E1, E2, and capsid proteins may not be required for this process (63). The apparent lack of RV structural protein involvement with apoptosis indicates that the trigger for virus-induced apoptosis may be different between RV and the alphaviruses. For Sindbis virus, the prototype alphavirus, the E1 and E2 proteins, and in particular their respective TM domain, can induce apoptosis as shown in cDNA transfection studies (69). However, the mechanisms involved are not known.
Since RV replication requires the formation of replication complexes, it is likely that viral components such as dsRNA (84, 91) and ns proteins (77) found within these structures play a role in the apoptotic process. Recent studies of vaccinia virus infection showed that dsRNA caused apoptosis via the activation of dsRNA-dependent protein kinase, a key player in interferon-mediated host defense against viral infection (5, 39, 72). While this has not been shown for RV, it is possible that the RV RI and RF RNAs may elicit similar responses; interestingly, alpha interferon is present in 90% of RV-infected fetuses and is readily detectable in the sera of fetuses at mid-gestation (21 to 29 weeks) (79). The RV ns proteins may also play a role in RV-induced apoptosis, since determinants of cytopathogenicity have been mapped to these proteins (122). However, the presumed interplay between the ns proteins and cellular proteins involved with the apoptotic pathway remains to be elucidated.
The cellular proteins and biochemical pathways involved in RV-induced apoptosis are also not well characterized. Studies using chemically defined caspase inhibitors have shown indirectly that apoptosis during RV infection is mediated by caspases, and it is likely that the Bcl-2 family of proteins are involved (29, 96). There are conflicting reports on the role of p53 in inducing apoptosis during RV infection. Megyeri et al. (96) demonstrated the involvement of the p53-dependent pathway in RV-induced apoptosis, while Höfmann et al. (63), using a similar cell line, reported that cell death during RV replication is mediated by a p53-independent pathway.
Apoptosis in RV-infected cell cultures occurs asynchronously and is confined mainly to RV-infected cells that have detached from the monolayer as a consequence of CPE (29, 63, 96, 123). Ultrastructural studies of Vero cell monolayers infected with RV confirm these findings, since no evidence of chromatin condensation within the nuclei of cells displaying RV replication complexes and mitochondrial changes were found, even at late stages of infection (J.-Y. Lee, unpublished observations). This asynchronous induction of apoptosis is intriguing, particularly when the majority of the cells are RV-infected. The extent to which viral or host cellular proteins are involved in controlling or limiting apoptosis in these infected cells is not known. The inhibition of apoptosis in the RV-infected monolayer has clear advantages for the virus, since persistence develops as a consequence.
To date, there has been no report on the role of apoptosis in RV teratogenicity. It would be of interest to determine whether the noninflammatory necrosis observed in organs of RV-infected fetuses is caused by RV-induced apoptosis. Further analyses using more sophisticated biochemical and molecular tools in apoptosis are required.
TERATOGENICITY: CONCLUDING REMARKS
The RV teratogenic process most probably begins when placental infection occurs during maternal viremia, leading to dissemination of the virus throughout the fetus. A chronic or persistent infection ensues, which is in general noncytolytic. Paradoxically, although the virus is spread widely in the developing fetus and can be recovered from almost any organ, only small numbers of fetal cells or discrete foci of cells show signs of infection (128, 163). Examination of damaged tissue suggests at least two possible mechanisms that could account for the rubella cytopathology: a direct CPE, which may involve RV-induced apoptosis, and a virus-induced inhibition of cell division. Of interest is that the fetus can still be infected after the first trimester but after 18 to 20 weeks gestation there is unlikely to be any congenital defects. It is believed that by this time the important phase of organogenesis is mostly complete and that as the fetus develops, the immune system may play some role in limiting infection.
Actin is a critical component of the cellular cytoskeleton and plays an important role in cell mitosis. If RV infection directly or indirectly inhibits the assembly of actin, a corresponding inhibition of cell mitosis may result. In the original report by Gregg (47), it was suggested that the congenital eye cataracts were the result of a partial arrest in cell development. In addition, organs of congenitally infected infants are smaller than those of uninfected infants and contain fewer cells (159). A depressed mitotic activity has been shown in congenitally infected embryonic primary cell cultures (128), and a slowing of cell division has also been shown for in vitro RV infections of human fetal cells (117) and BHK-21 cells (145). Furthermore, Plotkin and Vaheri (118) claimed to have extracted a protein from RV-infected human fetal cells that was capable of inhibiting mitosis in uninfected cells. Thus, there is strong evidence that RV infection may be associated with an inhibition of the development of organ precursor cells and that interference of actin assembly may play some role.
Maintenance or regulation of actin assembly is a complex process involving a number of actin-binding proteins and a requirement for ATP. Since RV infection is also associated with mitochondrial changes, at least in vitro (82), it may be that the ability of the mitochondria to carry out normal respiratory function is impaired and one of the consequences is disaggregation of actin. Mitochondria may also be involved in programmed cell death (12, 16). Evidence has been presented that mitochondria contain and release proteins that are an integral part of the apoptotic cascade. However, it remains to be determined if the mitochondrial changes are inducers of apoptosis or are produced as a result of apoptosis.
These features of RV replication are probably interrelated and associated with the teratogenic properties of the virus. A number of animal models have been put forward for investigating RV cytopathology, but none have proved fully suitable. This lack of a small-animal model has hindered research in general and the understanding of the mechanism of teratogenesis in particular. Nevertheless, some of the answers may be found in the genetic makeup of the virus. The complete genome sequence has been determined for a number of the vaccine strains. Since vaccination does not result in congenital abnormalities when inadvertently given in early pregnancy, comparisons of vaccine strains with wild-type strains may reveal important motifs associated with teratogenicity.
The availability of effective vaccines in developed countries and the control of epidemics of rubella in those countries have resulted in reduced research efforts into this fascinating pathogen. Nevertheless, the elucidation of the mechanism of RV teratogenesis will not only provide insights into how the virus manifests its unusual cytopathology but may also provide a paradigm for the teratogenic properties of other viral agents. These aspects alone more than justify further investigations into RV infection.
ACKNOWLEDGMENTS
We acknowledge John Marshall, Victorian Infectious Diseases Reference Laboratory, for the generous provision of electron microscopy facilities.
Jia-Yee Lee is supported by the National Health and Medical Research Council of Australia (project 960389).
REFERENCES
- 1.Abe T, Nakada T, Hatanka H, Tajima M, Hiraiwa M, Ushijima H. Myoclonus in a case of suspected progressive rubella panencephalitis. Arch Neurol. 1983;40:98–100. doi: 10.1001/archneur.1983.04050020060012. [DOI] [PubMed] [Google Scholar]
- 2.Acheson N H, Tamm I. Replication of Semliki Forest virus: an electron microscope study. Virology. 1967;32:128–143. doi: 10.1016/0042-6822(67)90261-9. [DOI] [PubMed] [Google Scholar]
- 3.Atreya C D, Lee N S, Forng R-Y, Höfmann J, Washington G, Marti G, Nakhasi H L. The rubella virus putative replicase interacts with the retinoblastoma tumor suppressor protein. Virus Genes. 1998;16:177–183. doi: 10.1023/a:1007998023047. [DOI] [PubMed] [Google Scholar]
- 4.Atreya C D, Singh N K, Nakhasi H L. The rubella virus binding activity of human calreticulin is localized to the N-terminal domain. J Virol. 1995;69:3848–3851. doi: 10.1128/jvi.69.6.3848-3851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balachandran S, Kim C N, Yeh W C, Mak T W, Bhalla K, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardeletti G. Respiration and ATP levels in BHK-21/13S cells during the earliest stages of rubella virus replication. Intervirology. 1977;8:100–109. doi: 10.1159/000148884. [DOI] [PubMed] [Google Scholar]
- 7.Bardeletti G, Gautheron D C. Phospholipid and cholesterol composition of rubella virus and its host cell BHK21 grown in suspension cultures. Arch Virol. 1976;52:19–27. doi: 10.1007/BF01317861. [DOI] [PubMed] [Google Scholar]
- 8.Bardeletti G, Henry H, Sohier R, Gautheron D. Primary effects of rubella virus on the metabolism of BHK-21 cells grown in suspension culture. Arch Gesamte Virusforsch. 1972;39:26–34. doi: 10.1007/BF01241526. [DOI] [PubMed] [Google Scholar]
- 9.Bardeletti G, Tetkoff J, Gautheron D. Rubella virus maturation and production in two host cell systems. Intervirology. 1979;11:97–103. doi: 10.1159/000149019. [DOI] [PubMed] [Google Scholar]
- 10.Baron M D, Ebel T, Suomalainen M. Intracellular transport of rubella virus structural proteins expressed from cloned cDNA. J Gen Virol. 1992;73:1073–1086. doi: 10.1099/0022-1317-73-5-1073. [DOI] [PubMed] [Google Scholar]
- 11.Baron M D, Forsell K. Oligomerization of the structural proteins of rubella virus. Virology. 1991;185:811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 12.Bernardi P, Scorrano L, Colonna R, Petronilli V, Di Lisa F. Mitochondria and cell death. Mechanistic aspects and methodological issues. Eur J Biochem. 1999;264:687–701. doi: 10.1046/j.1432-1327.1999.00725.x. [DOI] [PubMed] [Google Scholar]
- 13.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G-H, Senkevich T G, Alnemri E S, Moss B, Leonardo M J, Tomaselli K J, Cohen J I. Death effector domain containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bienz K, Egger D, Pfister T. Characteristics of the poliovirus replication complex. Arch Virol Suppl. 1994;S9:147–157. doi: 10.1007/978-3-7091-9326-6_15. [DOI] [PubMed] [Google Scholar]
- 15.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bossy-Wetzel E, Green D R. Apoptosis: checkpoint at the mitochondrial frontier. Mutat Res. 1999;434:243–251. doi: 10.1016/s0921-8777(99)00032-4. [DOI] [PubMed] [Google Scholar]
- 17.Bowden D S, Pedersen J S, Toh B H, Westaway E G. Distribution by immunofluorescence of viral products and actin containing cytoskeletal filaments in rubella virus infected cells. Arch Virol. 1987;92:211–219. doi: 10.1007/BF01317478. [DOI] [PubMed] [Google Scholar]
- 18.Bowden D S, Westaway E G. Rubella virus: structural and non structural proteins. J Gen Virol. 1984;65:933–943. doi: 10.1099/0022-1317-65-5-933. [DOI] [PubMed] [Google Scholar]
- 19.Bowden D S, Westaway E G. Changes in glycosylation of rubella virus envelope proteins during maturation. J Gen Virol. 1985;66:201–206. doi: 10.1099/0022-1317-66-1-201. [DOI] [PubMed] [Google Scholar]
- 20.Bowden D S, Westaway E G. Rubella virus products and their distribution in infected cells. Subcell Biochem. 1989;15:203–231. doi: 10.1007/978-1-4899-1675-4_7. [DOI] [PubMed] [Google Scholar]
- 21.Chaye H, Chong P, Tripet B, Brush B, Gillam S. Localization of virus neutralizing and hemagglutinin epitopes of E1 glycoprotein. Virology. 1992;189:483–492. doi: 10.1016/0042-6822(92)90572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M H, Frey T K. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J Virol. 1999;73:3386–3403. doi: 10.1128/jvi.73.4.3386-3403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J P, Strauss J H, Strauss E G, Frey T K. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol. 1996;70:4707–4713. doi: 10.1128/jvi.70.7.4707-4713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke D M, Loo T W, McDonald H, Gillam S. Expression of rubella virus cDNA coding for the structural proteins. Gene. 1988;65:23–30. doi: 10.1016/0378-1119(88)90413-1. [DOI] [PubMed] [Google Scholar]
- 25.Cochi S L, Edmonds L E, Dyer K, Marks J S, Roveria E Z, Preblud S R, Orenstein W A. Congenital rubella syndrome in the United States, 1970–1985. On the verge of elimination. Am J Epidermiol. 1989;129:349–361. doi: 10.1093/oxfordjournals.aje.a115138. [DOI] [PubMed] [Google Scholar]
- 26.Cooper L Z, Buimovici-Klein E. Rubella. In: Fields B N, Knipe D M, Chanock R M, Melnick J L, Roizman B, Shope R E, editors. Virology. New York, N.Y: Raven Press; 1985. pp. 1005–1020. [Google Scholar]
- 27.Dominguez G, Wang C-Y, Frey T K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology. 1990;177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorsett P H, Miller D C, Green K Y, Byrd F I. Structure and function of rubella virus proteins. Rev Infect Dis. 1985;S7:S150–S156. doi: 10.1093/clinids/7.supplement_1.s150. [DOI] [PubMed] [Google Scholar]
- 29.Duncan R, Muller J, Lee, N. N, Esmaili A, Nakhasi H L. Rubella virus-induced apoptosis varies among cell lines and is modulated by bcl-XL and caspase inhibitors. Virology. 1999;255:117–128. doi: 10.1006/viro.1998.9562. [DOI] [PubMed] [Google Scholar]
- 30.Duncan R, Nakhasi H L. La autoantigen to a 5′ cis-element of rubella virus RNA correlates with element function in vivo. Gene. 1997;201:137–149. doi: 10.1016/s0378-1119(97)00438-1. [DOI] [PubMed] [Google Scholar]
- 31.Forbes J A. Rubella: historical aspects. Am J Dis Child. 1969;118:5–11. doi: 10.1001/archpedi.1969.02100040007002. [DOI] [PubMed] [Google Scholar]
- 32.Forrest J M, Menser M A, Burgess J A. High frequency of diabetes melitis in young adults with congenital rubella. Lancet. 1971;ii:332–334. doi: 10.1016/s0140-6736(71)90057-2. [DOI] [PubMed] [Google Scholar]
- 33.Forng R-Y, Frey T K. Identification of rubella virus nonstructural proteins. Virology. 1995;206:843–853. doi: 10.1006/viro.1995.1007. [DOI] [PubMed] [Google Scholar]
- 34.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frey T K, Abernathy E S, Bosma T J, Starkey W G, Corbett K M, Best J M, Katow S, Weaver S C. Molecular analysis of rubella virus epidemiology across three continents, North America, Europe, and Asia, 1961–1997. J Infect Dis. 1998;178:642–650. doi: 10.1086/515370. [DOI] [PubMed] [Google Scholar]
- 36.Frey T K, Marr L D. Sequence of the region coding for virion proteins C and E2 and the carboxy terminus of the nonstructural proteins of rubella virus: comparison with alphaviruses. Gene. 1988;62:85–99. doi: 10.1016/0378-1119(88)90582-3. [DOI] [PubMed] [Google Scholar]
- 37.Friedman R M, Levin J G, Grimley P M, Berezesky I K. Membrane-associated replication complex in arbovirus infection. J Virol. 1972;10:504–515. doi: 10.1128/jvi.10.3.504-515.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replication occurs on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gale M, Jr, Kwieciszewski B, Dossett M, Nakao H, Katze M. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garbutt M, Chan H, Hobman T C. Secretion of rubella virions and virus-like particles in cultured epithelial cells. Virology. 1999;261:340–346. doi: 10.1006/viro.1999.9857. [DOI] [PubMed] [Google Scholar]
- 41.Garbutt M, Law L M J, Chan H, Hobman T C. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J Virol. 1999;73:3524–3533. doi: 10.1128/jvi.73.5.3524-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garzon S, Strykowski H, Charpentier G. Implications of mitochondria in the replication of Nodurma virus in larvae of Lepidoptera, Galleria mellonella (L.) and in suckling mice. Arch Virol. 1990;113:165–176. doi: 10.1007/BF01316670. [DOI] [PubMed] [Google Scholar]
- 43.Ghadially F N. Ultrastructural pathology of the cell and matrix. London, United Kingdom: Butterworth; 1988. [Google Scholar]
- 44.Gilbert G L. Rubella. In: Gilbert G L, editor. Infectious disease in pregnancy and the newborn infant. 2nd ed. Chur, Switzerland: Harwood Academic Press; 1997. pp. 23–62. [Google Scholar]
- 45.Gorbalenya A E, Koonin E V, Lai M-C. Putative papain-related thiol proteases of positive strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green K Y, Dorsett P H. Rubella virus antigens: localization epitopes involved in hemagglutination and neutralization by using monoclonal antibodies. J Virol. 1986;57:893–898. doi: 10.1128/jvi.57.3.893-898.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregg N M. Congenital cataract following German measles in the mother. Trans Ophthalmol Soc Aust. 1941;3:35–46. [PubMed] [Google Scholar]
- 48.Grimley P M, Berezesky I K, Friedman R M. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J Virol. 1968;2:1326–1338. doi: 10.1128/jvi.2.11.1326-1338.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimley P M, Levin J G, Berezesky I K, Friedman R M. Specific membranous structures associated with the replication of group A arboviruses. J Virol. 1972;10:492–503. doi: 10.1128/jvi.10.3.492-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gros C, Wengler G. Identification of a RNA-stimulated NTPase in the predicted helicase sequence of the rubella virus nonstructural proteins. Virology. 1996;217:367–372. doi: 10.1006/viro.1996.0125. [DOI] [PubMed] [Google Scholar]
- 51.Hamvass J J, Ugovsek S, Iwakata S, Labzoffsky N A. Virus particles in rubella virus infected cell tissue cultures. Arch Gesamte Virusforsch. 1969;26:287–294. doi: 10.1007/BF01242381. [DOI] [PubMed] [Google Scholar]
- 52.Hemphill M L, Forng R-Y, Abernathy E S, Frey T K. Time course of rubella virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology. 1988;162:65–75. doi: 10.1016/0042-6822(88)90395-9. [DOI] [PubMed] [Google Scholar]
- 53.Higashi N. Electron microscopy of viruses in thin sections of cells grown in culture. Prog Med Virol. 1973;15:331–379. [PubMed] [Google Scholar]
- 54.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus glycoprotein E2: the signal peptide is contained in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 55.Hobman T C, Lemon H F, Jewell K. Characterization of an endoplasmic reticulum retention signal in the rubella virus E1 glycoprotein. J Virol. 1997;71:7670–7680. doi: 10.1128/jvi.71.10.7670-7680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hobman T C, Lundström M L, Farquhar M G. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J Cell Biol. 1993;121:269–281. doi: 10.1083/jcb.121.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hobman T C, Lundström M L, Gillam S. Processing and intracellular rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hobman T C, Lundström M L, Mauracher C A, Woodward L, Gillam S, Farquhar M G. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology. 1994;202:574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 59.Hobman T C, Shukin R, Gillam S. Translocation of rubella virus glycoprotein E1 into the endoplasmic reticulum. J Virol. 1988;62:4259–4264. doi: 10.1128/jvi.62.11.4259-4264.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hobman T C, Qiu Z, Chaye H, Gillam S. Analysis of rubella virus E1 glycosylation mutants expressed in COS cells. Virology. 1991;181:768–772. doi: 10.1016/0042-6822(91)90915-x. [DOI] [PubMed] [Google Scholar]
- 61.Hobman T C, Woodward L, Farquhar M C. The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hobman T C, Woodward L, Farquhar M C. Targeting of heterodimeric membrane proteins complex to the Golgi: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol Biol Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Höfmann J, Pletz M W, Liebert U G. Rubella virus-induced cytopathic effect in vitro is caused by apoptosis. J Gen Virol. 1999;80:1657–1664. doi: 10.1099/0022-1317-80-7-1657. [DOI] [PubMed] [Google Scholar]
- 64.Holmes I, Wark M C, Jack I, Grutner J. Identification of two possible virus particles in rubella-infected cell. J Gen Virol. 1968;2:37–42. doi: 10.1099/0022-1317-2-1-37. [DOI] [PubMed] [Google Scholar]
- 65.Holmes I, Wark M C, Warburton M F. Is rubella an arbovirus. II. Ultrastructural morphology and development. Virology. 1969;37:15–25. doi: 10.1016/0042-6822(69)90301-8. [DOI] [PubMed] [Google Scholar]
- 66.Ho-Terry L, Cohen A. Rubella virion polypeptides: characterization by polyacrylamide gel electrophoresis, isoelectric focusing and peptide mapping. Arch Virol. 1982;72:47–54. doi: 10.1007/BF01314449. [DOI] [PubMed] [Google Scholar]
- 67.Ho-Terry L, Cohen A. The role of glycosylation on haemagglutination and immunological reactivity of rubella virus. Arch Virol. 1984;79:139–146. doi: 10.1007/BF01310807. [DOI] [PubMed] [Google Scholar]
- 68.Hovi T, Vaheri A. Rubella virus-specific ribonucleic acids in infected BHK-21 cell. J Gen Virol. 1970;6:77–83. doi: 10.1099/0022-1317-6-1-77. [DOI] [PubMed] [Google Scholar]
- 69.Joe A K, Foo H H, Kleeman L, Levine B. The transmembrane domains of Sindbis virus envelope glycoproteins induce cell death. J Virol. 1998;72:3935–3943. doi: 10.1128/jvi.72.5.3935-3943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katow S, Sugiura A. Low pH-induced conformational change of rubella virus envelope proteins. J Gen Virol. 1988;69:2797–2807. doi: 10.1099/0022-1317-69-11-2797. [DOI] [PubMed] [Google Scholar]
- 71.Kessel R G. Annulate Lamellae: a last frontier in cellular organelles. Int Rev Cytol. 1992;133:43–120. doi: 10.1016/s0074-7696(08)61858-6. [DOI] [PubMed] [Google Scholar]
- 72.Kibler K V, Shors T, Perkins K B, Zeman C C, Banaszak M P, Biesterfeldt J, Langland J O, Jacobs B L. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J Virol. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kielian M, Jungerwirth S. Mechanisms of enveloped virus entry into cells. Mol Biol Med. 1990;7:17–31. [PubMed] [Google Scholar]
- 74.Kim K S W, Boatman E S. Electron microscopy of monkey kidney cell cultures infected with rubella virus. J Virol. 1967;1:205–214. doi: 10.1128/jvi.1.1.205-214.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kistler G S. Cytoplasmic tubuloreticular complexes and nuclear bodies in cells of rubella-infected human embryos and fetuses. Beitr Pathol. 1975;155:101–138. [PubMed] [Google Scholar]
- 76.Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med. 1997;3:614–620. doi: 10.1038/nm0697-614. [DOI] [PubMed] [Google Scholar]
- 77.Kujala P, Ahola T, Ehsani N, Auvinen P, Vihinen H, Kääriäinen L. Intracellular distribution of rubella virus nonstructural proteins P150. J Virol. 1999;73:7805–7811. doi: 10.1128/jvi.73.9.7805-7811.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanzrein M, Schlegel A, Kempf C. Entry and uncoating of enveloped viruses. Biochem J. 1994;302:313–320. doi: 10.1042/bj3020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lebon P, Dafos F, Checoury A, Grangeot-Keros L, Forestier F, Toublanc J E. Presence of an acid-labile alpha interferon in sera from fetuses and children with congenital rubella. J Clin Microbiol. 1985;21:775–778. doi: 10.1128/jcm.21.5.775-778.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reference deleted.
- 81.Lee J-Y. Morphological and functional analysis of rubella virus. Ph.D. thesis. Melbourne, Australia: University of Melbourne; 1993. [Google Scholar]
- 82.Lee J-Y, Bowden D S, Marshall J A. Membrane junctions associated with rubella virus infected cells. J Submicrosc Cytopathol. 1996;28:101–108. [PubMed] [Google Scholar]
- 83.Lee J-Y, Marshall J A, Bowden D S. Replication complexes associated with the morphogenesis of rubella virus. Arch Virol. 1992;122:95–106. doi: 10.1007/BF01321120. [DOI] [PubMed] [Google Scholar]
- 84.Lee J-Y, Marshall J A, Bowden D S. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- 85.Lee J-Y, Marshall J A, Bowden D S. Localization of rubella virus core particles in Vero cells. Virology. 1999;265:110–119. doi: 10.1006/viro.1999.0016. [DOI] [PubMed] [Google Scholar]
- 86.Leopardi R, Roizman B. The herpes simplex virus major regulatory protein ICP4 blocks apoptosis induced by virus or by hyperthermia. Proc Natl Acad Sci USA. 1996;93:9583–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu X, Ropp S L, Jackson R J, Frey T K. The rubella virus nonstructural protease requires divalent cations for activity and functions in trans. J Virol. 1998;72:4463–4466. doi: 10.1128/jvi.72.5.4463-4466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Z, Yang D, Qiu Z, Lim K T, Chong P, Gillam S. Identification of domains in rubella virus genomic RNA and capsid protein necessary for specific interaction. J Virol. 1996;70:2184–2190. doi: 10.1128/jvi.70.4.2184-2190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lundström M L, Mauracher C A, Tingle A J. Characterization of carbohydrates linked to rubella virus glycoproteins E2. J Gen Virol. 1991;72:843–850. doi: 10.1099/0022-1317-72-4-843. [DOI] [PubMed] [Google Scholar]
- 90.Mackenzie J M, Jones M K, Young P. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 91.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J-Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 92.Marr L D, Sanchez A, Frey T K. Efficient in vitro translation and processing of rubella virus structural proteins in the presence of microsomes. Virology. 1991;180:400–405. doi: 10.1016/0042-6822(91)90046-e. [DOI] [PubMed] [Google Scholar]
- 93.Marr L D, Wang C Y, Frey T K. Expression of the rubella virus nonstructural proteins ORF and demonstration of proteolytic processing. Virology. 1994;198:586–592. doi: 10.1006/viro.1994.1070. [DOI] [PubMed] [Google Scholar]
- 94.Mastromarino P, Cioe L, Rieti S, Orsi N. Role of membrane phospholipids and glycolipids in the Vero cells surface receptor for rubella virus. Med Microbiol Immunol. 1990;179:105–114. doi: 10.1007/BF00198531. [DOI] [PubMed] [Google Scholar]
- 95.Mauracher C A, Gillam S, Shukin R, Tingle A J. pH-dependent solubility shift of rubella virus capsid protein. Virology. 1991;181:773–777. doi: 10.1016/0042-6822(91)90916-y. [DOI] [PubMed] [Google Scholar]
- 96.Megyeri K, Berencsi K, Halazonetis T D, Prendergast G C, Gri G, Plotkin S A, Rovera G, Gönczöi E. Involvement of a p53-dependent pathway in rubella virus-induced apoptosis. Virology. 1999;259:74–84. doi: 10.1006/viro.1999.9757. [DOI] [PubMed] [Google Scholar]
- 97.Mifune K, Desmyter J, Williams W E. Effect of exogenous interferon in rubella virus production in carrier cultures of cells defective in interferon production. Infect Immun. 1970;2:132–138. doi: 10.1128/iai.2.2.132-138.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murphy F A. Togavirus morphology and morphogenesis. In: Schlesinger W, editor. The togaviruses. New York, N.Y: Academic Press, Inc.; 1980. pp. 241–326. [Google Scholar]
- 99.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo T M, Summers M D. Virus taxonomy, 6th report of the International Committee on Taxonomy of Viruses (ICTV) Arch Virol. 1995;S10:1–586. [PubMed] [Google Scholar]
- 100.Nakhasi H L, Cao X-Q, Rouault T A, Liu T-Y. Specific binding of host cell proteins to the 3′ terminal stem-loop structure of rubella virus negative-strand RNA. J Virol. 1991;65:5961–5967. doi: 10.1128/jvi.65.11.5961-5967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nakhasi H L, Rouault T A, Haile D J, Liu T-Y, Klausner R D. Specific high affinity binding of host cell proteins to the 3′ regions of rubella virus RNA. New Biol. 1990;2:255–264. [PubMed] [Google Scholar]
- 102.Nakhasi H L, Singh N K, Pogue G P, Cao X-Q, Rouault T A. Identification and characterization of host factors interaction with cis-acting elements of rubella virus. Arch Virol. 1994;9:S255–267. doi: 10.1007/978-3-7091-9326-6_26. [DOI] [PubMed] [Google Scholar]
- 103.Nakhasi H L, Zheng D, Hewlett I K, Liu T-Y. Rubella virus replication: effect on interferon and actinomycin D. Virus Res. 1988;10:1–15. doi: 10.1016/0168-1702(88)90053-6. [DOI] [PubMed] [Google Scholar]
- 104.Ng M L. Ultrastructural studies of Kunjin virus infected Aedes albopictus cells. J Gen Virol. 1987;68:577–582. doi: 10.1099/0022-1317-68-2-577. [DOI] [PubMed] [Google Scholar]
- 105.Ng M L, Pederson J S, Toh B H, Westaway E G. Immunofluorescent sites in Vero cells infected with the flavivirus Kunjin. Arch Virol. 1983;78:177–190. doi: 10.1007/BF01311313. [DOI] [PubMed] [Google Scholar]
- 106.Oker-Blom C. The gene order for rubella virus structural proteins is NH2-C-E2-E1-COOH. J Virol. 1984;51:354–358. doi: 10.1128/jvi.51.2.354-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oker-Blom C, Jarvis D L, Summers M D. Translocation and cleavage of rubella virus envelope glycoproteins: identification and role of the E2 signal sequence. J Gen Virol. 1990;71:3047–3053. doi: 10.1099/0022-1317-71-12-3047. [DOI] [PubMed] [Google Scholar]
- 108.Oker-Blom C, Kalkkinen N, Kääriäinen L, Pettersson R F. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J Virol. 1983;46:964–973. doi: 10.1128/jvi.46.3.964-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oker-Blom C, Ulmanen I, Kääriäinen L, Pettersson R F. Rubella virus 40S genome RNA specifies a 24S subgenomic mRNA that encodes for a precursor to structural proteins. J Virol. 1984;49:403–408. doi: 10.1128/jvi.49.2.403-408.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parkman P D. Making vaccination policy: the experience with rubella. Clin Infect Dis. 1999;28:S140–146. doi: 10.1086/515062. [DOI] [PubMed] [Google Scholar]
- 111.Parkman P D, Buescher E L, Artenstien M S. Recovery of rubella virus from the army recruits. Proc Soc Exp Biol Med. 1962;111:225–230. doi: 10.3181/00379727-111-27750. [DOI] [PubMed] [Google Scholar]
- 112.Parkman P D, Meyer H M, Kirschstein R L, Hoops H E. Attenuated rubella virus. I. Development and laboratory characterization. N Engl J Med. 1966;275:569–574. doi: 10.1056/NEJM196609152751101. [DOI] [PubMed] [Google Scholar]
- 113.Patrizi G, Middlekamp J N. Development and changes of annulate lamellae complexes in rubella virus infected cells. J Ultrastruct Res. 1970;31:405–423. doi: 10.1016/s0022-5320(70)90158-9. [DOI] [PubMed] [Google Scholar]
- 114.Peränen J, Kääriäinen L. Biogenesis of type 1 cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J Virol. 1991;65:1623–1627. doi: 10.1128/jvi.65.3.1623-1627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peränen J, Laakkonen P, Hyvönen M, Kääriäinen L. The alphavirus replicase protein nsP1 is membrane associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 116.Petruzziello R, Orsi N, Macchia S, Rieti S, Frey T K, Mastromarino P. Pathway of rubella virus infectious entry into Vero cells. J Gen Virol. 1996;77:303–308. doi: 10.1099/0022-1317-77-2-303. [DOI] [PubMed] [Google Scholar]
- 117.Plotkin S A, Boue A, Boue J G. The in vitro growth of rubella virus embryo cells. Am J Epidemiol. 1965;81:71–85. doi: 10.1093/oxfordjournals.aje.a120499. [DOI] [PubMed] [Google Scholar]
- 118.Plotkin S A, Vaheri A. Human fibroblasts infected with rubella produce a growth inhibitor. Science. 1967;156:659–661. doi: 10.1126/science.156.3775.659. [DOI] [PubMed] [Google Scholar]
- 119.Pogue G P, Cao X-Q, Singh N K, Nakhasi H L. 5′ sequences of rubella virus RNA stimulate translation of chimeric RNAs and specifically interact with two host-encoded proteins. J Virol. 1993;67:7106–7117. doi: 10.1128/jvi.67.12.7106-7117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pogue G P, Höfmann J, Duncan R, Best J M, Etherington J, Sontheimer R D, Nakhasi H L. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pugachev K V, Abernathy E S, Frey T K. Genomic sequence of the RA27/3 vaccine strain of rubella virus. Arch Virol. 1997;142:1165–1180. doi: 10.1007/s007050050150. [DOI] [PubMed] [Google Scholar]
- 122.Pugachev K V, Abernathy E S, Frey T K. Improvement of specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural. J Virol. 1997;71:562–568. doi: 10.1128/jvi.71.1.562-568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pugachev K V, Frey T K. Rubella virus induces apoptosis in culture cells. Virology. 1998;250:359–370. doi: 10.1006/viro.1998.9395. [DOI] [PubMed] [Google Scholar]
- 124.Pugachev K V, Frey T K. Effects of defined mutations in the 5′ nontranslated region of rubella virus genomic RNA on virus viability and macromolecular synthesis. J Virol. 1998;72:641–650. doi: 10.1128/jvi.72.1.641-650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qiu Z, Hobman T C, McDonalds H L, Seto N O, Gillam S. Role of N-linked oligosaccharides in processing and intracellular transport of E2 glycoprotein of rubella virus. J Virol. 1992;66:3514–3521. doi: 10.1128/jvi.66.6.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qiu Z, Ou D, Wu H, Hobman T C, Gillam S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J Virol. 1994;68:4086–4091. doi: 10.1128/jvi.68.6.4086-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Qiu Z, Tufaro F, Gillam S. The influence of N-linked glycosylation on the antigenicity and immunogenicity of rubella virus E1 glycoprotein. Virology. 1992;190:876–881. doi: 10.1016/0042-6822(92)90929-j. [DOI] [PubMed] [Google Scholar]
- 128.Rawls W E. Congenital rubella: the significance of virus persistence. Prog Med Virol. 1968;10:238–285. [PubMed] [Google Scholar]
- 129.Razvi E S, Welsh R M. Apoptosis in viral infections. Adv Virus Res. 1995;45:1–60. doi: 10.1016/s0065-3527(08)60057-3. [DOI] [PubMed] [Google Scholar]
- 130.Reed J C. Bcl-2 family proteins. Oncogene. 1998;17:3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 131.Robinson J, Lemay M, Vaudry W L. Congenital rubella after anticipated maternal immunity: two cases and a review of the literature. Pediatr Infect Dis J. 1994;13:812–815. doi: 10.1097/00006454-199409000-00013. [DOI] [PubMed] [Google Scholar]
- 132.Rojo G, Chamorro M, Salas M L, Vinuela E, Cuezva J M, Salas J. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol. 1998;72:7583–7588. doi: 10.1128/jvi.72.9.7583-7588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwartzman R A, Cidlowski J A. Apoptosis: the biochemistry and molecular biology of programmed cell death. Endocr Rev. 1993;14:133–151. doi: 10.1210/edrv-14-2-133. [DOI] [PubMed] [Google Scholar]
- 134.Sedwick W D, Sokol F. Nucleic acid of rubella virus and its replication in hamster kidney cells. J Virol. 1970;5:478–489. doi: 10.1128/jvi.5.4.478-489.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Singh I, Helenius A. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J Virol. 1992;66:7049–7058. doi: 10.1128/jvi.66.12.7049-7058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suomalainen M, Garoff H, Baron M D. The E2 signal sequence of rubella virus remains part of the capsid protein and confers membrane association in vitro. J Virol. 1990;64:5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takkinen K, Vidgren G, Ekstrand J, Hellman U, Kalkkinen N, Wernstedt C, Pettersson R F. Nucleotide sequence of the rubella virus capsid protein reveals an unusually high G/C content. J Gen Virol. 1988;69:603–612. doi: 10.1099/0022-1317-69-3-603. [DOI] [PubMed] [Google Scholar]
- 139.Terry G M, Ho-Terry L, Londesborough P, Rees K R. Localization of the rubella E1 epitopes. Arch Virol. 1988;98:189–197. doi: 10.1007/BF01322168. [DOI] [PubMed] [Google Scholar]
- 140.Terry G M, Ho-Terry L, Londesborough P, Rees K R. A bio-engineered rubella E1 antigen. Virology. 1989;104:63–75. doi: 10.1007/BF01313808. [DOI] [PubMed] [Google Scholar]
- 141.Theodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tingle A J, Allen M, Petty R E, Kettyls G D, Chantler J K. Rubella-associated arthritis. Comparative study of joint manifestations associated with natural rubella infection and RA27/3 rubella immunization. Ann Rheum Dis. 1986;45:110–114. doi: 10.1136/ard.45.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Töndury G, Smith D W. Fetal rubella pathology. J Pediatr. 1966;68:867–876. doi: 10.1016/s0022-3476(66)80204-4. [DOI] [PubMed] [Google Scholar]
- 144.Trudel M, Nadon F, Comtois R, Payment P, Bonneau A, Lecomte J. Identification of rubella virus structural proteins by immunoprecipitation. J Virol Methods. 1982;5:191–197. doi: 10.1016/0166-0934(82)90009-x. [DOI] [PubMed] [Google Scholar]
- 145.Vaheri A, Cristofalo V J. Metabolism of rubella virus-infected BHK-21 cells. Enhanced glycolysis and late cellular inhibition. Arch Gesamte Virusforsch. 1967;21:425–436. doi: 10.1007/BF01241741. [DOI] [PubMed] [Google Scholar]
- 146.Vaheri A, Hovi T. Structural and proteins units of rubella virus. J Virol. 1972;9:10–16. doi: 10.1128/jvi.9.1.10-16.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vaux D L, Cory S, Adams J M. Bcl-2 gene promotes haemapoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 148.Vidgren G, Takkinen K, Kalkkinen N, Kääriäinen L, Pettersson R F. Nucleotide sequencing of the genes coding the membrane glycoproteins E1 and E2 of rubella virus. J Gen Virol. 1987;68:2347–2357. doi: 10.1099/0022-1317-68-9-2347. [DOI] [PubMed] [Google Scholar]
- 149.Von Bonsdorff C-H, Vaheri A. Growth of rubella virus in BHK21 cells: electron microscopy of morphogenesis. J Gen Virol. 1969;5:47–51. doi: 10.1099/0022-1317-5-1-47. [DOI] [PubMed] [Google Scholar]
- 150.Wang C-Y, Dominguez G, Frey T K. Construction of rubella virus genome length cDNA clones and synthesis of RNA transcripts. J Virol. 1994;68:3550–3557. doi: 10.1128/jvi.68.6.3550-3557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Waxham M N, Wolinksy J. A model of the structural organization of rubella virions. Rev Infect Dis. 1985;7:S133–S139. doi: 10.1093/clinids/7.supplement_1.s133. [DOI] [PubMed] [Google Scholar]
- 152.Waxham M N, Wolinsky J. Detailed immunologic analysis of the structural polypeptides of rubella virus using monoclonal antibodies. Virology. 1985;143:153–165. doi: 10.1016/0042-6822(85)90104-7. [DOI] [PubMed] [Google Scholar]
- 153.Webster W S. Teratogen update: congenital rubella. Teratology. 1999;58:13–23. doi: 10.1002/(SICI)1096-9926(199807)58:1<13::AID-TERA5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 154.Weller T H, Neva F A. Propagation in tissue culture of cytopathic agents from patients with rubella-like illness. Proc Soc Exp Biol Med. 1962;111:215–225. [Google Scholar]
- 155.Wesselhoeft C. Rubella (German measles) N Engl J Med. 1947;236:943–950. doi: 10.1056/NEJM194706192362506. [DOI] [PubMed] [Google Scholar]
- 156.Westaway E G, Khromykh A A, Mackenzie J M. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- 157.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A. Ultrastructure of Kunjin virus infected cells: colocalization of NS1 and NS3 with double-stranded RNA and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.White E. Life, death and pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 159.Wolinsky J S. Rubella. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Roizman B, editors. Virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lipincott-Raven Publishers; 1996. pp. 899–929. [Google Scholar]
- 160.Wolinsky J S, McCarthy M, Allen-Cannady O, Moore W T, Jin R, Cao S-N, Lovett A, Simmons D. Monoclonal antibody-defined epitope map of expressed rubella virus domains. J Virol. 1991;65:3986–3994. doi: 10.1128/jvi.65.8.3986-3994.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wolinsky J S, Sukholutsky E, Moore W T, Lovett A, McCarthy M, Adame B. An antibody and synthetic peptide-defined rubella virus E1 glycoprotein and neutralization domain. J Virol. 1993;67:961–968. doi: 10.1128/jvi.67.2.961-968.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wong K T, Robinson W S, Merigan T C. Synthesis of viral specific ribonucleic acid in rubella virus-infected cells. J Virol. 1969;4:901–903. doi: 10.1128/jvi.4.6.901-903.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Woods W A, Johnson R T, Hostetler D D, Lepow M L, Robbin F C. Immunofluorescent studies on rubella-infected tissue cultures and human tissues. J Immunol. 1966;96:253–260. [PubMed] [Google Scholar]
- 164.Yang D, Hwang D, Qiu Z, Gillam S. Effects in the mutation in the rubella virus E1 glycoproteins on E1-E2 interaction and membrane fusion activity. J Virol. 1998;72:8747–8755. doi: 10.1128/jvi.72.11.8747-8755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yao J, Gillam S. Mutational analysis, using a full-length rubella virus cDNA clone of rubella virus E1 transmembrane and cytoplasmic domains required for virus release. J Virol. 1999;73:4622–4730. doi: 10.1128/jvi.73.6.4622-4630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yao J, Yang D, Chong P, Hwang D, Liang Y, Gillam S. Proteolytic processing of rubella virus nonstructural proteins. Virology. 1998;246:74–82. doi: 10.1006/viro.1998.9179. [DOI] [PubMed] [Google Scholar]