Abstract
Introduction:
Advances in high-throughput sequencing have greatly advanced our understanding of long non-coding RNA (lncRNAs) in a relatively short period of time. This has expanded our knowledge of cancer, particularly how lncRNAs drive many important cancer phenotypes via their regulation of gene expression.
Areas covered:
Men of African descent are disproportionately affected by PC in terms of incidence, morbidity, and mortality. LncRNAs could serve as biomarkers to differentiate low risk from high-risk disease. Additionally, they may represent therapeutic targets for advanced and castrate-resistant cancer. We review current research surrounding lncRNAs and their association with PC. We discuss how lncRNAs can provide new insights and diagnostic biomarkers in African American men. Finally, we review advances in computational approaches that predict the regulatory effects of lncRNAs in cancer.
Expert opinion:
PC diagnostic biomarkers which offer high specificity and sensitivity are urgently needed. PC specific lncRNAs are compelling as diagnostic biomarkers owing to their high tissue and tumor specificity and presence in bodily fluids Recent studies indicate that PCA3 clinical utility might be restricted to men of European descent. Further work is required to develop lncRNA biomarkers tailored for men of African descent.
Keywords: Prostate cancer, lncRNAs, African American, African Descent, European American, Biomarkers, Diagnostics
1. Introduction
Prostate cancer (PC) is one of the most prevalent cancers worldwide, particularly affecting men living a western lifestyle and of African descent, suggesting risk factors that are genetic, environmental, and socioeconomic in nature. Despite major advances in the diagnosis and treatment of PC, the disease remains a global health problem. PC is the second most diagnosed non-skin cancer in men worldwide [1, 2]. Currently there is considerable interest in lncRNAs and their regulatory roles in developmental processes and disease/cancer states [3, 4]. Prior to high-throughput sequencing technologies becoming common place, lncRNAs were simply considered transcriptional “noise” and largely ignored by the scientific community [5–7]. With increased sequencing sensitivity in combination with innovative bioinformatic tools, in the past decade researchers finally started to uncover the functional roles of lncRNAs. Two emerging discoveries include the realization that they act upon a wide range of cell processes and are differentially expressed in disease states including various cancers [8, 9].
Computational predictions have identified over 10,000 lncRNAs, and researchers have been keen to characterise them functionally, identify their regulatory roles and exploit this knowledge in biomedical research, diagnostics and therapeutic development [10]. In parallel, bioinformatics methods are also being rapidly improved to better predict the regulatory roles of lncRNAs in cancer and determine their full extent as regulatory modulators. This review provides a detailed overview of lncRNAs and particularly in the context of PC. Firstly, we cover lncRNAs, their history, newly discovered functions, and correlation with cancer. We then review PC, racial differences in disease presentation and why lncRNAs are relevant in this context. Lastly, we provide a summary of tools dedicated to deciphering the functional roles of lncRNAs.
2. What is a Long Non-Coding RNA?
lncRNAs are defined as transcripts longer than 200 nucleotides that are putatively not translated into proteins [8]. Both short (<=200 base pairs) and lncRNAs may contain coding regions [11]. Short non-coding transcripts, and particularly microRNAs (miRNAs) have been studied more extensively than lncRNAs and contribute to the development and progression of many cancers [12]. Prior to the genomic era, lncRNAs were considered ‘junk RNA’ and were largely ignored by the scientific community. The first lncRNA to be identified was h19 in 1984. Although at first mistakenly categorised as a messenger RNA (mRNA) [13], h19 has subsequently been recognised as an epigenetic regulator, differentially expressed in several cancers where it acts as a tumor suppressor [3, 14–16]. In 2002, lncRNAs were finally assigned as functional RNA molecules by the Functional Annotation of the Mammalian Genome (FANTOM) project [17]. Like mRNAs, lncRNAs are transcribed by RNA polymerase II, polyadenylated, capped and spliced [18, 19]. Subcellular localisation provides a lens to define lncRNA functional classes [7, 20]. The identification and number of lncRNAs is continually increasing, however, many remain poorly annotated and are not experimentally validated [12, 21].
As of January 2021, LNCBook, a curated lncRNA database contains 268,848 documented lncRNAs (obtained from publicly accessible databases including GENCODE v27, NONCODE v5.0, LNCipedia v4.1 and MiTanscriptome beta). However, there is a large gap between documented lncRNAs and lncRNAs which have a known link to a molecular or biological function. Of the 268,848 recorded lncRNAs on LNCBook, only 1,867 have been categorised as experimentally validated (linked with functional mechanisms/biological processes and identified in relevant publications) [21, 22]. LncRNAs are classified as either 1) intergenic (lincRNAs), transcribed from intergenic regions located between two genes [5, 23, 24] 2) bidirectional (divergent IncRNAs) transcribed on the reverse strand of a coding transcript located no more than 1000 base pairs away [5, 24] 3) intronic lncRNAs, transcribed only from intronic regions of protein coding genes, 4) antisense lncRNAs transcribed from antisense strands of protein coding genes and 5) sense lncRNAs transcribed from sense strands of protein coding genes [6, 25, 26].
Understanding how lncRNAs are regulated and how they in turn regulate their target genes is an important research area [27]. Thus far, several functional roles of lncRNAs have been established. The most common IncRNA functional groups include guides (e.g., HOTAIR) [6, 28], scaffolds (e.g., TERC) [6, 29], signals (e.g., XIST) [6, 7] and decoys (e.g., MALAT1) [6, 30]. There are also a series of documented multifunctional IncRNAs. An example is NEAT1 [31]. NEAT1 was identified as differentially expressed in numerous tumor contexts including lung, colorectal, and prostate cancer [31]. NEAT1 is an important mediator in the regulation of clear cell renal cell carcinoma (ccRCC) progression and predicts poor prognosis and decreased survival in patients with ccRCC [7, 32–34]. Guide IncRNAs can bind and tether (epigenetic complex machinery) to a target gene locus directing gene expression either in cis or in trans. Guide IncRNA can direct chromatin-modifying enzymes (e.g., histone methylases, acetylases, and deacetylases) to a particular gene target locus which can lead to the activation or repression of local genes [35, 36]. Scaffold IncRNAs recruit epigenetic complexes together, thus acting as a molecular scaffold. They recruit RNA-binding factors to form RNA-proteins complexes which promote or inhibit gene transcription [35, 36]. Signal IncRNAs possess signalling properties that promote guidance of epigenetic complexes to specific locations. These IncRNAs possess enhancer functions which alter the structure of chromatin and guide transcriptional complexes to gene targets enabling transcriptional activity [35, 36]. Finally, decoy IncRNAs can lure epigenetic complexes to different genomic locations. These IncRNAs can recruit and decoy a transcriptional complex to a distinct location for suppression of gene expression [35, 36]. Many reviews have documented the molecular functions of lncRNAs (for example, reference 23) however none have considered the computational aspects of predicting lncRNA functions and their targets. Our review addresses this shortcoming.
3. Long Non-Coding RNAs and Prostate Cancer
The number of patients presenting with urological cancers (malignancies of the genital and urinary organs of men and the urinary organs of women) is on the increase [37, 38]. Currently, urological cancers of the prostate, bladder, and renal stand among the top 10 most diagnosed cancers in men [39]. Despite their frequency, biomarkers for urological cancers are limited. Those that are available typically lack sensitivity in low grade disease e.g., urine cytology for bladder cancer and low specificity e.g., the Prostate Specific Antigen (PSA) [39–41]. lncRNAs are now frequently being linked with urological cancers, and many have been designated as playing roles in the development and progression of these malignancies including prostate cancer [42]. Table 1 summarises selected lncRNAs that are differentially expressed in urological cancers, which may have potential as biomarkers or therapeutic targets [6, 7, 31].
Table 1.
Urological cancer associated lncRNAs
| IncRNA | lncRNA functional group * | Urological cancer type | Sample size | Differential Expression | Fold change | Potential Cancer Impact | Hazard ratio | Study reference |
|---|---|---|---|---|---|---|---|---|
| MALAT1 | Scaffold [169] Decoy [170] |
Bladder | 95 | Upregulated | Not described | Poor survival | 1.26 (0.68–2.13) | [171] |
| Prostate | 14 | Upregulated | Not described | Not described | Not described | [80] | ||
| NEAT1 | Scaffold [7] Decoy [7] |
Prostate | 30 | Upregulated | 2.101 | Not described | Not described | [172] |
| LINC00346 | Not described | Bladder | 52 | Upregulated | Not described | promotes bladder cancer migration and invasion | Not described | [173] |
| KCNQ1OT1 | Guide [174] signal [175] | Bladder | 30 | Upregulated | Not described | KCNQ1OT1 facilitates the progress-sion of bladder cancer by targeting MiR-218–5p/HS3ST3B1 | Not described | [176] |
| LOC400891 | Not described | Prostate (All patients underwent radical prostatectomy or laparo-scopic radical prostatectomy) | 81 | Upregulated | Gene chip – 4. 1 QPCR – 2.4 |
Unfavor-able prognosis | Multi-variate Tumor stage - 1.823 Multi-variate LOC400891 expression - 2.116 |
[177] |
| PCA3 | Scaffold [178] | Prostate | 56 | Upregulated | 10–100-fold overexpression | Not described | Not described | [64] |
| PCAT1 | Decoy [179] | High-risk prostate cancer (Gleason ≥ 7) and metastasis prostate cancer | 58 prostate cancer tissues 20 prostate cancer xeno-grafts |
Upregulated | Not described | Not described | Not described | [73] |
| PCAT18 | Not described | Prostate | 160 | Upregulated | 8.8–11.1 fold | potential therapeutic target for metastatic prostate cancer | Not described | [79] |
| PCAT7 | Decoy [84] | Prostate cancer with bone metastasis | 57 | Upregulated | Not described | promotes PCa bone metastasis | 1.85 3.97 |
[84] |
| SChLAP1 | Decoy [180] | Prostate | 208 | Upregulated | Not described | SChLAP1 expression is associated with prostate cancer progression | univariate (hazard ratio = 2.343. multivariate (hazard ratio = 1.99 |
[87] |
| PCGEM1 | Scaffold [181] | Prostate (cells of African American patients) | Not described | Upregulated | Not described | increased expression level with high-risk CaP patients and African-American race is significantly related to PCGEM1 expression in tumor cells | Not described | [91] |
| PCGEM1 | Prostate (African American patients) | 30 patients (15 African-Amer-ican, 15 (Caucasian- Amer-ican) |
Upregulated | Not described | Not described | Not described | [93] | |
| HOTAIR | Scaffold [181] Guide [6] |
Prostate (CRPC) | Not described | Upregulated | Not described | Drives Castration-Resistant Prostate Cancer | Kaplan-Meier analysis of PCa outcome using the GEO: GSE21034 dataset | [145] |
| HORAS5 | Not described | Prostate | Not described | Upregulated | 12.51 | Poorer clinical outcome in human PCa samples | Disease‐free survival based on HORAS5 expression [cBioportal, TCGA PCa database, 456 vs 35 (Elevated HORAS5) patients] |
[138] |
| CTBP1-AS | Decoy [180] | Prostate | 105 prostate cancer samples | Upregulated | Not described | Associated with prostate cancer castration-resistant tumour growth and activates AR signals | Not described | [139, 182] |
| ARLNC1 | Not described | Prostate | 14 prostate cancer cell lines | Upregulated | Not described | Strongly associated with AR signaling in prostate cancer progression | Not described | [147] |
| PCAT29 | Decoy [183] | Prostate | Not described | Downregulated | 0.29 ± 0.15 in tumor tissues as compared to normal tissues | Supp-resses prostate cancer cell proliferation | Not described | [182] |
| GAS5 | Scaffold [184] Decoy [6] |
Prostate | Not described | Downregulated | Not described | Pro-apoptosis | Not described | [185] |
lncRNA functional groups subject to change
Despite major advances in the diagnosis and treatment of PC, the disease remains a global health problem. PC is the second most diagnosed non-skin cancer in men worldwide [43]. Recent estimates indicate that 1 in 7 men will be diagnosed with PC during their lifetime. In 2018 alone, 359,000 recorded deaths from PC occurred globally [44, 45]. Racial differences are common in PC. Men of African descent are adversely affected in terms of both incidence and mortality rates compared to European based counterparts. lncRNAs are now considered to contribute to these racial differences [46, 47].
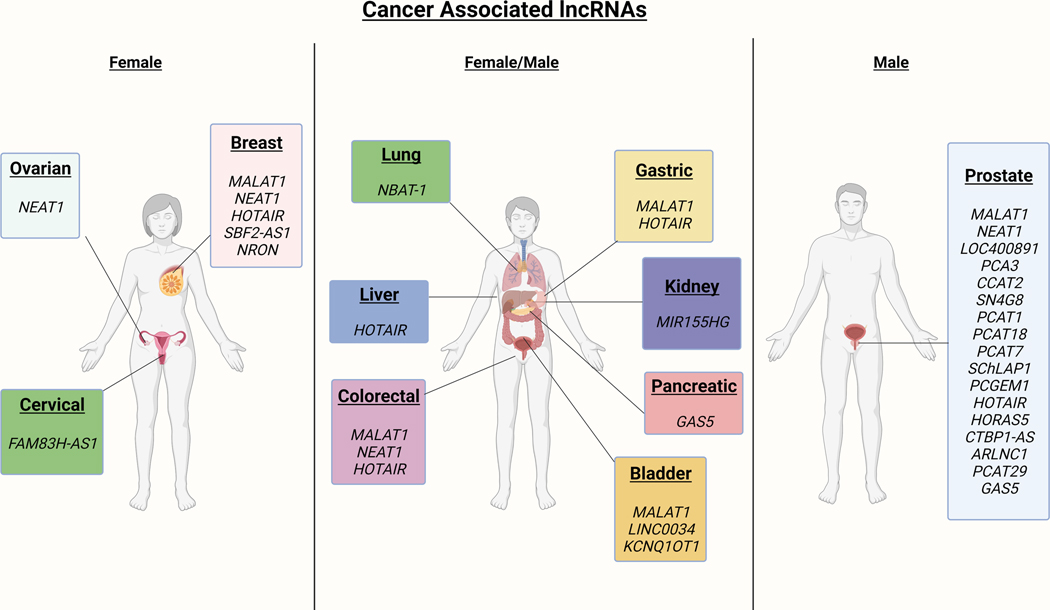
Evidence to date suggests that lncRNAs can alter gene expression at both transcriptional and post-transcriptional levels thus playing roles in important biological processes including carcinogenesis and metastasis [48]. Some lncRNAs function as tumor suppressors, possess the ability to interact with oncogenes, inhibit or accelerate proliferation, and influence apoptosis [30, 49, 50]. Other lncRNAs are commonly aberrantly expressed, deleted, amplified and/or mutated in multiple cancers [9, 51]. lncRNAs have been shown to have roles in numerous forms of cancer [52]. Figure 1 displays lncRNAs correlated with numerous cancer types. The specific cancer types and relevant body tissues are highlighted. Table 2 provides a summary of lncRNAs which have been identified as differentially expressed in various malignancies.
Figure 1. Cancer associated lncRNAs.
Key lncRNAs associated with various cancers are highlighted as to the body tissue and cancer type.
Table 2.
Cancer associated lncRNAs.
| IncRNA | lncRNA class | Cancer Type | Differential Expression | Reference |
|---|---|---|---|---|
| MALAT1 | Antisense | Colorectal | Upregulated | [186] |
| Antisense | Gastric | Upregulated | [187] | |
| Antisense | Breast | Upregulated | [188] | |
| NEAT1 | Intergenic | Ovarian | Upregulated | [189] |
| Colorectal | Upregulated | [190] | ||
| Breast (triple negative breast cancer) | Upregulated | [34] | ||
| HOTAIR | Intergenic | Breast | Upregulated | [191] |
| Gastric | Upregulated | [192] | ||
| Colorectal | Upregulated | [193] | ||
| Liver | Upregulated | [194] | ||
| FAM83H-AS1 | Intergenic | Cervical | Upregulated | [195] |
| SBF2-AS1 | Antisense | Breast | Downregulated | [196] |
| NRON | Antisense | Breast | Downregulated | [197] |
| NBAT-1 | Intergenic | Lung | Downregulated | [198] |
| GAS5 | Antisense | Pancreatic | Downregulated | [199] |
Presently, three areas that merit particular attention; 1) Identification of novel diagnostic and prognostic biomarkers for PC, with an emphasis on diagnostic biomarkers for early stage PC which differentiates low-risk from high-risk tumors [40, 53], 2) An urgent need to better understand biological differences in disease presentation in men of African and Northern European descent [54] and 3) Efficient therapies for advanced and castrate-resistant PC [55]. LncRNAs are emerging as biomarkers or therapeutic targets for these areas, as explained below.
4. lncRNAs as Prostate Cancer Biomarkers
It has been widely accepted that the identification of biomarkers should not exclusively focus upon protein coding genes [56]. A key factor for biomarker utility is whether the proposed biomarker is detectable in bodily fluids. Liquid based biomarkers provide many benefits over traditional tissue-based biomarkers from a ‘no surgery requirement and recovery time’ perspective, painless non-invasive testing, decreased costs, and more rapid diagnostics [57]. lncRNAs are attractive as PC biomarkers owing to their high tissue and tumor specificity [27] and are detectable in bodily fluids, including PCA3 and PCAT18 [57, 58].
Biomarkers with high specificity and sensitivity are particularly needed to help address the over treatment of clinically insignificant PC. The over diagnosis and treatment of low-risk PC remains a common problem, primarily due to use of the Prostate Cancer Specific Antigen (PSA) tests [59] which offer high sensitivity but low specificity [40]. The introduction of the PSA test in the early 1990s was highly criticized for the over-diagnosis and the peak of PC incidence rates [60–62]. Furthermore, PSA tests have poorer diagnostic performance in men of African descent [63]. Therefore, a current need in PC research is the identification of diagnostic biomarkers which can not only offer high specificity and sensitivity but are also universally reliable for all races.
PC specific lncRNAs are compelling as diagnostic biomarkers for clinical use [27]. Possibly the most well-known is PCA3 (Prostate Cancer Specific Antigen 3). PCA3 was first observed as highly expressed in PC tumors in 1999 and has since been characterized as over expressed in 95% of all PC cells [64, 65]. Urine-based measurements of PCA3 provide greater PC specificity than PSA testing [12]. In 2012, the PROGENSA PCA3 assay developed by Gen-Probe Inc was the first lncRNA PC biomarker to obtain Food and Drug Administration (FDA) approval. This lncRNA molecular test is used to help guide urologists in determining the need for repeat biopsies in men aged 50 years or older who had previously presented with a negative biopsy result [66, 67]. However, its practicability as a first use test remains controversial and it cannot predict PC aggressiveness [68, 69]. Several studies have also documented PCA3 expression in PC progression, however further research is required to confirm its feasibility as a PC prognostic biomarker [70]. Additionally, PCA3’s clinical utility might be restricted to men of European descent. O’Malley, et al. examined the clinical significance of the urinary biomarkers (PCA3 & T2ERG) in 718 African American and non-African American men and assessed whether they provided increased utility for the detection of PC. Their results indicated that PCA3 and T2ERG enhanced clinical utility however this result was only observed in non-African American men [71]. A major limitation of this study was the relatively small number of African American samples (72) in comparison to non-African American men. Further research with larger ethnically diverse datasets would facilitate more rigorous assessment.
Other studies have yielded varied outcomes in contrast to the initial findings from the O’Malley study. Feibus, et al. conducted a study using 304 patients, 60% of which were African American and noted that measurements of PCA3 enhanced predictability of high-grade PC in African American men [63] while an earlier study by Adam et al. using South African men challenged these results. Of 105 South African men referred for prostate biopsy, Adam et al, found that PCA3 levels were not significantly increased in black men who had tested positive for the disease [72]. Again, larger cohorts would provide greater statistical power.
Several other lncRNAs have been established as possible biomarkers of PC, including PCAT1 (Prostate Cancer Associated Transcript 1). PCAT1 is a highly PC specific lncRNA [27] and was upregulated in patients presenting with high-risk (Gleason score ≥ 7) PC and metastatic disease [27, 73]. Prensner et al, 2011 identified 121 unannotated PC ncRNAs through the analysis of 102 prostate tumor tissues and cell lines, among these was PCAT1 [74]. PCAT1 was significantly overexpressed in most PC tumors but particularly in tumors that had metastasised. The group remarked that PCAT1 was likely contributing to cell proliferation and potentially could serve as a urine-based biomarker given the prior success with PCA3 [75]. Interestingly, PCAT1 is located at chromosome 8q24, a chromosome location frequently correlated with PC risk due to its common amplification and the presence of PC susceptibility single nucleotide polymorphisms (SNPs) [76, 77]. Yhan et al. and Prensner et al. concluded that PCAT1 supports cell proliferation in PC with Prensner and colleagues noting that it may represent a suitable candidate for biomarker development [73, 78]. PCAT18 is over expressed in PC. Using tissue samples from patients diagnosed at the Stephanshorn clinic in Switzerland and plasma samples collected in Canada, Crea et al. found that PCAT18 was overexpressed in the plasma of patients with localised and advanced forms of the disease compared to healthy individuals. The results from this study displayed a positive correlation between PCAT18 and PC stage, with the highest levels observed in metastatic castrate resistant PC. Thus, PCAT18 could have prognostic value, including to discriminate between localised and metastatic forms of PC, and is a possible therapeutic target for advanced PC [79]. MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1) is a lncRNA originally correlated with lung cancer however it has been found to be over-expressed in several cancers including PC. In 2013, Ren et al. identified MALAT1 as detectable in plasma and serum and suggested it had the potential to be a diagnostic biomarker [80]. Wang et al. identified MALAT1 as over-expressed in urine samples of Chinese PC patients and noted its potential as a urine-based biomarker [81].
lncRNAs correlated with advanced PC include PCAT7 (Prostate Cancer Associated Transcript 7), SChLAP1 (SWI/SNF Complex Antagonist Associated with Prostate Cancer 1) and PCAT18. PCAT7 has previously correlated with disease progression in nasopharyngeal and non-small cell lung cancer [82, 83]. Lang, et al, 2020 noted that PCAT7 has the potential to be a therapeutic target for men presenting with PC bone metastasis via disruption of the constitutive loop between PCA7 and TGF‐β signalling [84]. PCAT7 also has the potential to be a prognostic biomarker. SChLAP1 (SWI/SNF ‘Complex Antagonist Associated With Prostate Cancer’) has been proposed as a biomarker of lethal PC. Cimadamore et al. documented that lncRNA SChLAP1 it is critical in the development and progression of PC [85]. Prensner et al. identified that SChLAP1 is highly expressed in ~25% of PCs analysed (including metastatic PC) and may promote tumor cell invasion (in vitro) as well as metastasis (in vivo) [86–88]. Additionally, Prensner et al. observed SChLAP1 expression in urine and correlated with higher-risk PC patients suggesting its potential as a biomarker to differentiate between low-risk and high-risk tumors [89]. SChLAP1 has also been associated with adverse clinical outcomes in post radical prostatectomy and considered a potential tissue based prognostic biomarker by Mehra, et al [87]. Similarly, Chua et al. suggested SChLAP1 expression was linked with increased biochemical relapse and metastasis PC subsets cribriform architecture and intraductal carcinoma [90].
PCGEM1 is highly PC specific, has been linked to PC progression by androgen receptor transcriptional co-regulators and could also decrease apoptosis in PC cell lines [86, 91, 92]. Petrovics et al. observed that PCGEM1 expression was significantly higher in PC cells of men of African descent compared to men of Northern European descent [91]. An additional study by Lee et al. observed similar results where PCGEM1 was significantly over-expressed in prostate tumors of African-American but not European-American men [93].
Despite a better understanding of the roles of lncRNA and the number identified as potential biomarkers of localized and advanced PC, few lncRNAs have met the success of PCA3. A primary reason for this is that lncRNA research is still in its infancy with thousands of predicted lncRNAs requiring laboratory experimental validation of their utility and confirmation in separate patient cohorts. Moreover, the overwhelming majority of candidate biomarkers fail to achieve clinical utility [58]. The candidate PC biomarkers summarised above require analytical and clinical validation to better assess their utility. To date numerous articles have been published detailing lncRNAs directly related to PC (refer to references 38, 51, 53, 54, 55 and 68), however our review differs in two specific ways, 1) more in-depth detail relating to PC and racial differences and 2) computational methods and software now available for identifying potential lncRNA biomarkers in the disease.
5. Racial Differences in Prostate Cancer
Racial differences are common among cancers, examples include multiple myeloma, colorectal and breast cancer. Multiple myeloma is the most common hematologic cancer in patients of African descent [94]. Although it is a rare disease, diagnoses are twice as high in patients of African descent compared to European descent. Reasons for these differences are currently unknown and the disease remains uncurable [95–97]. In the USA, colorectal cancer patients of African descent experience decreased stage specific survival in comparison to patients of European descent irrespective of socioeconomical background, age, treatment, and stage at diagnosis [98]. Invasive breast cancer mortality rates also differ between patients of African and Northern European descent. In the USA, African American women are more likely to die from the disease than European-American women [99]. African American women also experience worse prognosis and decreased survival times compared to European-American women regardless of their socioeconomical background, age, and stage at diagnosis [100].
Prostate cancer has one of the largest racial disparities of all cancer types. In the U.S.A., African Americans have among the highest mortality and lowest survival rates of any race or ethnic group [101]. PC is also the top cause of cancer mortality among men of African descent living in Caribbean and Sub-Saharan Africa [102]. Similar disparities exist in the UK, where men of African descent are 2–3 times more likely to develop PC than Caucasian men and have a 30% higher mortality rate [103]. Non biological factors contributing to PC racial disparities include diminished societal trust in the medical community, level of education attained, and financial concerns. Access to healthcare plays an important role in PC mortality. However, African American men have higher PSA values and higher tumor grades even at the same cancer stage, compared with non-African American men within the same healthcare system such as the Veterans Administration in the USA [104]. The reasons underlying these racial differences remain an area of active research. Many biological explanations have been presented including dietary factors such as alcohol intake, vitamin D deficiency and lycopene and isoflavone consumption [104–108]. Biological factors including genetic alterations – African Americans have higher rates of variations in tumor suppressor genes such as EPHB2 and are susceptible to higher prevalence of chromosome 8q24 variants [47, 54, 76, 109–111].
An emerging area of research is the role of lncRNAs in cancer development and progression in the context of racial differences. Frequently, lncRNAs are reported as differentially expressed in numerous cancers and are known to be involved in tumorigenesis and metastasis [48]. However, there is a lack of research investigating lncRNA differences between racial groups.
Much remains to be uncovered about the mechanisms by which lncRNAs regulate transcriptional and translational processes that drive oncogenesis. A lncRNA previously found to be significantly correlated with both PC and men of African ancestry is PCGEM1 (Prostate Cancer Gene Expression Marker 1). PCGEM1 is highly PC specific and has been linked to PC progression by androgen receptor transcriptional co-regulators and attenuates apoptotic processes in PC cell lines [86, 91, 92]. Petrovics et al, identified PCGEM1 expression as being significantly higher in PC cells of men of African descent compared to men of Northern European descent [91]. An additional study by Lee et al, 2016 observed similar results where PCGEM1 was significantly over-expressed in prostate tumors of African-American but not in European-American men [93].
Recently, Yuan, et al, 2020 examined genomic and transcriptomic differences among African American and European-American PC patients. The 470 samples which were analysed consisted of RNA-sequencing of 57 African American and 413 European-American patients from The Cancer Genome Atlas project (TCGA) prostate adenocarcinoma (PRAD) portal. The significantly lower amount of African American samples represents a common challenge in health disparities research, with genomic studies typically being constrained to populations of European descent [112].
Yuan, et al, identified 1868 (31%) of the differentially expressed transcripts between African American and European-American patients were lncRNAs. Patient and disease characteristics (age and Gleason score) were accounted for [47]. Of these differentially expressed lncRNAs, all were significantly enriched and regulated at least one target gene identified by the Long Noncoding RNA Heterogeneous Regulatory Network integrator (LongHorn) algorithm. LongHorn predictions are based on reverse-engineered canonical interactions, determined experimentally as part of the Encyclopaedia of DNA Elements (ENCODE) project including the crosslinking and immunoprecipitation assay (eCLIP) and chromatin immunoprecipitation sequencing (ChIP-seq) data [30, 47]. The researchers also showed that nearly 90% of lncRNAs differentially expressed between racial groups remain uncharacterised, underlining the need for continued lncRNA research. Of the 28 characterised lncRNAs differentially expressed between African and European-American men, 13 were PC specific including PCA1 and PCAT10. The researchers noted that the differentially expressed lncRNAs potentially contribute to disease progression and influencing PC racial differences. Further work is required to understand the regulatory functions of uncharacterised lncRNAs differentially expressed between racial groups [47].
Given the evidence highlighting the potential roles of lncRNA in PC tumorigenesis, there is an urgent need to understand the functional consequences of lncRNA differences between African and Northern European PC patients. One of the pressing challenges for integrative computational biology and statistical genetics in racial disparities research is predicting genotype-to-phenotypes. The ability to identify the key drivers among the multitude of interacting molecules is challenging. Additional obstacles include the rapid growth of data, the unavailability of data through issues with incompleteness, inaccuracies, heterogeneity, and data silos [113, 114]. LncRNAs may be prioritised according to their impact upon the gene regulatory network through graph theoretic and systems biology approaches [115, 116]. Understanding the biological underpinnings of racial differences in PC will ultimately provide an opportunity to help eliminate them by identifying possible PC diagnostic and prognostic biomarkers as well as potential therapeutic targets for men of African descent.
6. lncRNAs as Potential Therapeutic Targets for Advanced and Castrate Resistant Prostate Cancer
Advanced PC is traditionally defined as a disease which has spread (metastasized) outside the prostate gland [117]. Cancers generally metastasize preferentially to specific distant organ sites [118]. In most cases of advanced PC, metastasis targets the bones [119]. Androgen Deprivation Therapy (ADT) is clinically used as a palliative treatment for patients diagnosed with advanced PC [120–122]. In healthy men, the androgen testosterone and its derivative dihydrotestosterone are required for normal functioning of the prostate however, researchers have identified it is also vital for PC tumor growth [123]. Despite initial success with ADT, nearly all patients relapse and develop castrate-resistant PC (CRPC) after a mean duration of 2–3 years [124, 125]. CRPC is defined as the progression of PC despite ADT and may present as one or any combination of a steady growth in PSA levels, progression of pre-existing disease, or the emergence of new metastases [126] Currently CRPC remains uncurable and most patients with non-metastatic tumors will develop metastasis within 36 months of diagnosis, 90% of which will experience metastasis to the bone [127, 128]. The survival probability of metastatic PC patients depends on a variety of circumstances from which organ sites the disease has metastasized to, the patient response to treatment and the extent of the metastasis [129]. However, once the disease has spread, survival rates are bleak with 29% five-year survival [130]. Drug resistance in cancer therapies continuously present as a major obstacle [131]. lncRNAs are now considered as possible components to promote drug resistance and enhance drug sensitivity in cancer treatments [131]. Drug resistance refers to intolerance build up against pharmaceutical drugs [132]. Drug sensitivity corresponds to restoring sensitivity to drugs after tolerance has been developed [131].
In PC, there remains an urgent need to identify therapeutic targets for CRPC and advanced forms of the disease. lncRNAs are being proposed as alternative therapeutic targets for slowing the progression of PC, several of which include HOTAIR (Hox Transcript Antisense Intergenic RNA), MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1), and GAS5 (Growth Arrest Specific Transcript 5) [56]. The majority of lncRNAs identified as potential CRPC and metastatic PC therapeutic targets are specifically associated with the androgen receptor (AR) signalling pathway [133, 134]. Notably, the AR is vital in all stages of PC because it stimulates cell growth and survival [135]. Thus, most newly suggested therapeutic targets for PC centre around this [136, 137].
Several lncRNAs proposed as candidate therapeutic targets for CRPC and metastatic PC are primarily associated with the AR, including HORAS5 (LINC00161) [138], CTBP1-AS (C-Terminal Binding Protein 1 Antisense RNA) [139] and PCAT29 (Prostate Cancer Associated Transcript 29) [135]. lncRNAs can be directly influenced by androgen regulation [140] and many have been documented to participate in the advancement of PC through direct contact with the AR [141].
Parolia et al, 2019 found that HORAS5 facilitated the progression of CRPC in an AR dependent manner by regulating AR mRNA stability [138] and found that reduction in HORAS5 expression had a knock-on effect on the expression of the AR and its associated cancerous AR targets such as KIAA0101 [138]. lncRNA CTBP1-AS has been observed to have similar results for both CRPC and hormone dependent PC principally by promoting tumor growth [139]. Yakayama, et al. found that CTBP1-AS can suppress expression levels of CTBP1, a transcriptional corepressor known to regulate expression of tumor suppressors, and several genes involved with cell apoptosis [139, 142]. CTBP1-AS had androgen-dependent functions which could hinder the expression of tumor suppressors in both hormone dependent PC and CRPC suggesting potential as a therapeutic target [139]. PCAT29 is a PC specific lncRNA which has also been linked to the AR [135] Malik. et al, identified PCAT29, as a tumor suppressor of PC which is suppressed by the AR. PCAT29 has PC suppressive effects including hindering cellular proliferation, tumor growth, cellular migration, and metastasis [135]. Lower expression of PCAT29 is correlated with decreased survival in PC patients [135].
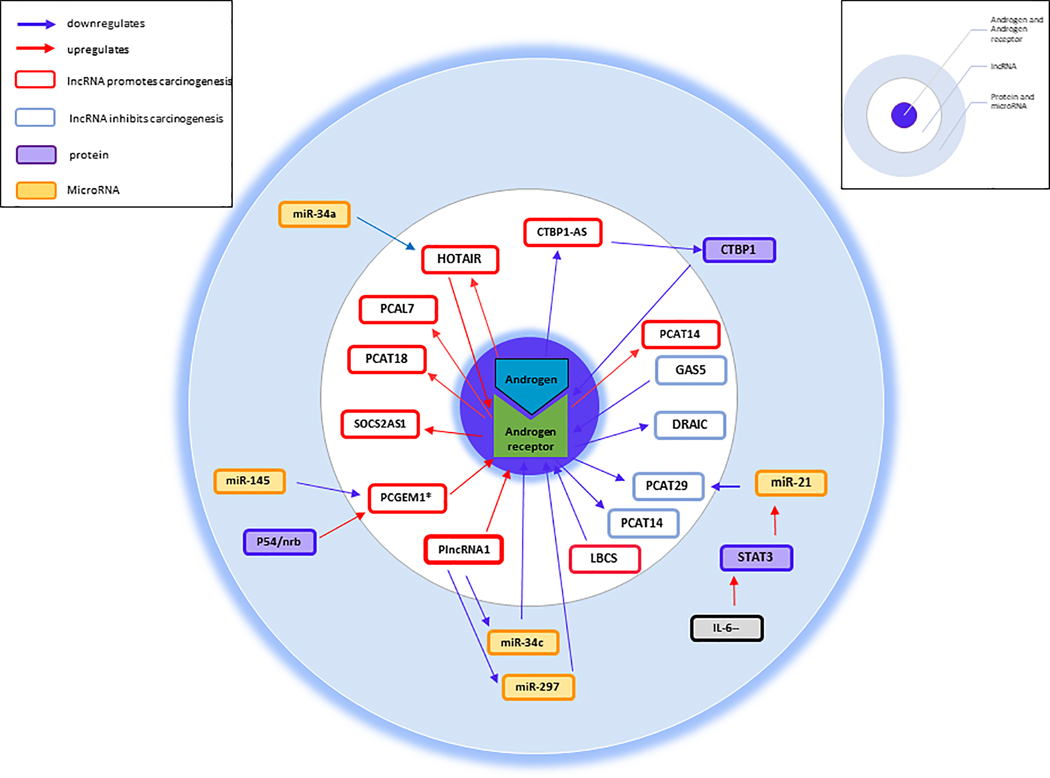
Several other studies have identified lncRNAs linked to PC and the AR. An associated lncRNA is HOTAIR [143]. HOTAIR is categorised as ubiquitously expressed and associates with multiple illnesses such as cardiovascular diseases, and cancers including breast, gastric, colorectal [144]. Zhang, et al, 2015 identified that HOTAIR is repressed by androgens and overexpressed in CRPC [145]. Further research supports the hypothesis that HOTAIR contributes to the progression from hormone dependent to CRPC [146]. Taken together, these results suggest HOTAIR is a key player in numerous cancers including CRPC [145]. Another lncRNA lately associated with the AR is ARLNC1 (AR-regulated long non-coding RNA 1). Zhang, et al, 2018 identified ARLNC1 was strongly correlated with AR signalling in the advancement of PC and deemed it a possible therapeutic target with further detailed study [147]. Several further lncRNAs have been correlated with the androgen receptor (AR) signalling pathway in PC either to inhibit or promote tumor growth. Therefore, lncRNAs can act as key drivers of PC and have potential as therapeutic targets. Figure 2 presents a schematic overview of AR associated lncRNAs that either inhibit or promote PC pathophysiology [133]. This schematic summarizes key lncRNAs and their role in up- and down-regulation of the AR, and the role of these key lncRNAs in promoting and inhibiting carcinogenesis and interplay with miRNAs and proteins.
Figure 2. Androgen receptor (AR) signalling pathway associated PC IncRNAs (Adapted from Aird et al. [118]).
This schematic provides a summary on key lncRNAs and their role in up- and down-regulation of the AR. The role of these key lncRNAs in promoting and inhibiting carcinogenesis and the interplay with miRNAs and proteins is highlighted.
7. LncRNA Interaction Catalogues and Prediction Tools
Exploratory studies suggest that lncRNAs have an extensive interactome; however, current validation data is insufficient; therefore, many lncRNAs lack known interaction partners and their function remains elusive. lncRNA interactions may be acquired from public databases, some of which are considered below. However, existing annotations do not capture all the possible mechanisms and contexts in which lncRNAs may control gene expression. Experimental determination of lncRNA interactions is labour-intensive and costly, not least due to the large space of potential interactions, and so lncRNA function prediction software is required.
7.1. Databases related to lncRNAs
Exploratory studies suggest that lncRNAs have an extensive interactome; however, current validation data is insufficient; therefore, many lncRNAs lack known interaction partners and their function remains elusive. lncRNA interactions may be acquired from public databases, some of which are considered below. However, existing annotations do not capture all the possible mechanisms and contexts in which lncRNAs may control gene expression. Experimental determination of lncRNA interactions is labour-intensive and costly, not least due to the large space of potential interactions, and so lncRNA function prediction software is required.
Presented by Zhou and colleagues, the experimentally validated functional lncRNA database (EVlncRNA) 2.0 is an updated version of the EVlncRNA database which comprises only functional lncRNAs have been experimentally validated though low-throughput techniques [148]. The update, which involved the manual curation of over 19,000 lncRNA-associated publications, includes information on 4010 curated lncRNAs across 124 species, 1082 diseases and 6,244 interactions. Importantly, interaction network integrated into the tool’s online platform (https://www.sdklab-biophysics-dzu.net/EVLncRNAs2/) provides users the ability to easily browse information retained within the database.
Despite the quality of lncRNA annotated within EVlncRNA 2.0, it could be considered much less comprehensive than alternative databases such as RNAInter (RNA Interactome Database) or RISE (RNA Interactome from sequencing experiments) which rely heavily on data from high-throughput studies and thus possess substantially more lncRNA interactions [149–151].
lncTarD database (http://bio-bigdata.hrbmu.edu.cn/LncTarD/) is a manually curated resource retaining information corresponding to 2822 lncRNA-target regulatory interactions that have proven relevance in 177 human diseases. For prostate cancer specifically, the database possesses data on 73 lncRNA-target interactions along with the mechanism of interaction and its functional consequences [74]. LIVE (lncRNA Interaction Validated Encyclopedia) is a database of manually curated and experimentally validated lncRNA interactions [152]. Consequently, LIVE is small with just 572 lncRNA interactions at the time of writing. Importantly, the database contains information relating to lncRNA interactions with multiple cellular components including proteins and transcription factors in addition to disease associations.
lnCaNet (LncRNA-Cancer gene co-expression Network) was first introduced in 2016 and is currently available as a web-based tool (http://lncanet.bioinfo-minzhao.org/)[153]. It operates as a database containing lncRNA-gene co-expression predictions for 11 cancers (colon, lung, breast etc.), with co-expression data sourced from Mitranscriptome, an online data repository of long polyadenylated RNA transcripts identified by RNA-seq [154]. lnCaNet includes lncRNA-gene expression correlations for 2,544 cancer genes and 17,250 lncRNAs
7.2. Prediction Tools
In the absence of experimental evidence, predictive algorithms are used; however, benchmarking has revealed room for improved predictive performance [155, 156]. For RNA-RNA prediction algorithms, most take into consideration the sequence of the RNA molecule when determining interactions. For example, both IntaRNA and LncRRIsearch methods utilise the minimum free energy of interaction, a thermodynamic principle affected by the number, type, and arrangement of nucleotides of an RNA molecule, to predict interactions [157–159]. In addition, the RIScoper tool is capable of extracting RNA-RNA interactions through literature text mining which is highly valuable given that findings would be experimentally validated [160].
lncRNA-DNA interactions mostly occur through RNA-DNA triplex formation, estimations of such structures are useful substitutes in lieu of experimental evidence. For example, the recently developed Triplex Forming Potential Prediction (TriplexFPP) is a machine learning based model which considers triplex formation capacity of a lncRNA based on computational methods in addition to triplex formation sites in DNA based on experimental data in a machine learning model to predict triplex formation [160]. In theory, such a method would be able to outperform more traditional methods such as Triplexator as it able to consider beyond the limits of the computational model through the inclusion of experimental data [160, 161].
Capsule-LPI tool considers sequence, motif, physicochemical and secondary structure features from lncRNAs and proteins within a lncRNA-protein pair to determine lncRNA-protein interactions [162]. Despite its complex mathematical architecture, the authors have compiled Capsule-LPI into a web-based tool (http://csbg-jlu.site/lpc/predict) allowing users to capture lncRNA-protein interactions by providing up to 100 lncRNA and protein sequences in Fasta format. Importantly, under 5-fold cross validation, Capture-LPI was found to possess the highest precision and the second highest sensitivity highlighting the tools utility and its room for improvement.
Interestingly, methods are available to translate interaction information into functional predictions. For example, MechRNA is a pipeline for lncRNA functional prediction based on RNA-RNA and RNA-protein interaction data. This approach utilises the IntaRNA2 algorithm to predict RNA-RNA interactions along with publicly available CLIP-seq (cross-linking immunoprecipitation sequencing) data and the GraphProt computational framework to determine RNA-protein interactions before combining data and predicting function based on several pre-determined possibilities [163, 164].
Possibly the most well-known method for lncRNA regulatory predictions is the LongHorn algorithm, which derives lncRNA interactions based on four predefined models of lncRNA regulation.
These models include activity as decoys, miRNA sponges, co-factors/ guides, and switches [30]. In a seminal publication, Chiu et al. applied the algorithm to TCGA data and predicted lncRNA–gene interactions predictions for individual cancers of interest. The lncRNA-gene predictions are available presently for 13 tumor contexts including prostate adenocarcinoma, lung adenocarcinoma and ovarian serous cystadenocarcinoma. For PC, Longhorn was applied to TCGA data including RNA, miRNA expression and copy number profiles of 371 prostate adenocarcinoma tumors. PLAIDOH (Predicting lncRNA Activity through Integrative Data-driven Omics and Heuristics) was developed at Washington University School of Medicine and applied to identify lncRNAs associated with non-Hodgkin lymphoma [165]. PLAIDOH offers users the opportunity to analyse both publicly and privately owned datasets, analyse statically small datasets and ultimately rank lncRNAs in order of importance based on customizable metrics [165]. One limitation is that it only predicts lncRNA targets that are located within a genomic window limit of 800kb. Using PLAIDOH, researchers have validated predictions through identifying well recognised lncRNA gene regulatory pairs such as HOTAIR matching with HOX genes and familiar lncRNA-protein binding pairs such as NEAT1 and NONO [165], new lncRNA interactions have also been uncovered and subsequently validated, demonstrating the utility of this tool.
lncRNA-screen is interactive platform that operates as a fully functional analysis pipeline which performs all major steps involved with typical RNA-Sequencing processing, its main objective is to facilitate the computational discovery of lncRNA candidates that can be further examined by functional experiments [150]. This pipeline can automatically download and prepare raw FASTQ (sequencing) files from multiple public repositories and performs data quality control, pre-processing, alignment, annotation, differential expression and novel lncRNA identification and classification. After analysis, lncRNA-screen provides users with an interactive HTML summary report that can be searched and filtered to suit users’ individual needs. Information includes locus, number of exons, a genome snapshot and lncRNA–mRNA interactions. lncRNA-screen is limited to lncRNA-gene predictions by physical location (neighbouring genes).
These databases and computational tools may be applied to understand the regulatory impact of lncRNAs upon map oncogenes and tumor suppressors [166, 167]. The resultant predictions are useful for identification of candidate cancer biomarkers and therapeutic targets, in concert with traditional biochemical methods [168].
8. Concluding Remarks
lncRNAs are now established regulators of a vast number of biological processes as well key mediators for the development and progression of various cancers. This is remarkable, considering that lncRNAs were relatively recently branded as ‘junk’ with no regulatory capabilities. Despite significant progress, there is still much to learn about lncRNAs and their potential value as cancer biomarkers or in therapy.
9. Expert Opinion
At present there are three areas of prostate cancer research that require focus: 1) understanding fundamental biological reasons as to why men of African descent are significantly more prone to disease morbidity and mortality, 2) improved therapeutic treatments for advanced and castrate-resistant PC, and 3) the necessity for biomarkers which can differentiate low risk and high-risk disease and assess PC racial disparities. Elucidating the functional roles of lncRNAs will help drive advances in each of these areas and will lead to better diagnosis and treatment of PC.
Despite these advances, opinions in the scientific community as to whether lncRNAs provide realistic therapeutic, prognostic, or diagnostic targets is varied at this time. This will add to the complexity of implementing clinical testing based on lncRNA diagnostics. The Clinical Laboratory Improvement Amendments (CLIA) federal regulatory standards require that clinical laboratories determine and detail their own performance specifications for laboratory-developed tests to guarantee accurate and precise results prior to implementation of the test. Characteristics that must be established typically include accuracy and precision, range for reporting, analytical sensitivity and specificity and reference intervals. Although both a positive and negative attribute, the sheer number of lncRNAs typically predicted by computational tools presents challenges for both clinical and research practice. The hundreds to thousands of lncRNAs which may be identified computationally cannot be feasibly validated in a clinical setting. This elevates the need for more sophisticated computational algorithms based on deep learning approaches which not only predict accurate lncRNAs but also facilitate stringent thresholds and intelligent approaches to triage specific lncRNAs subsets for laboratory-based testing and ultimately clinical implementation.
As precision medicine approaches evolve and become more inclusive the greatest impact that this research will realize is the advancement of knowledge of racial differences among prostate and other cancers. Several studies have determined that lncRNAs may play a role in differences in racial outcomes in cancer. There remain considerable and persistent racial disparities in prostate cancer outcomes. Reduced access to health care services contributes to racial disparities in PC outcomes, but even in equal access health care systems such as the Veterans Administration (VA) in the USA, AA Veterans have higher PSA levels and higher-grade tumors than compared to EA Veterans even when presenting with the same stage of disease. Unfortunately, medical mistrust issues and their associated indicators among AA patients has an adverse effect in recruiting sufficient sample numbers of ethnic minorities and their participation in clinical trials. Distrust in the health care system and fear associated with detection and treatment are responsible for late diagnosis in AA men. There is a paucity of samples from racial minorities in public genomic databases to detect moderately common genomic alterations in AA, which is contributing to the gap in PC disparities. We anticipate that as the lncRNA research field develop and matures greater emphasis will be placed on identifying lncRNAs that play key roles in the development and progression of cancer across all ethnic groups.
This field has already significantly progressed in a relatively short period of time, from the first lncRNA identified (h19) to the realization that lncRNAs do in fact process regulatory capabilities. With novel lncRNAs being identified daily and associated with cancer, pressure to experimentally validate these emerging candidates is increasing. In the future, we expect lncRNA computational tools will become an essential tool for both research and clinical laboratories and for a variety of diseases beyond cancer. We also expect significant growth in curated lncRNA databases with improved functional predictions of lncRNAs. We anticipate increased commercial activities based around lncRNA therapeutic, prognostic and diagnostics.
Article Highlights.
Racial differences continue to persist in prostate cancer
lncRNAs possess regulatory functions and can become diagnostic, prognostic, and therapeutic targets
The vast majority of lncRNAs are yet to be experimentally validated and their molecular function elucidated
lncRNA validation cannot rely solely on experimental validation
There is a clear need for computational software which can predict lncRNA molecular functions
Funding
This paper was funded by support from NIH/NIDA 1U01DA045300-01A1, U54MD010706-CHH and start-up funding from Queens University Belfast to G Hardiman.
Declaration of Interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as:
• Of interest
• • of considerable interest
- 1.Lee DJ, et al. , Recent Changes in Prostate Cancer Screening Practices and Epidemiology. J Urol, 2017. 198(6): p. 1230–1240. [DOI] [PubMed] [Google Scholar]
- 2.Wallace TA, et al. , Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res, 2008. 68(3): p. 927–36. [DOI] [PubMed] [Google Scholar]
- 3.Kung JT, Colognori D, and Lee JT, Long noncoding RNAs: past, present, and future. Genetics, 2013. 193(3): p. 651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, et al. , LncmiRSRN: identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics, 2018. 34(24): p. 4232–4240. [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, and Reik W, Evolution and functions of long noncoding RNAs. Cell, 2009. 136(4): p. 629–41. [DOI] [PubMed] [Google Scholar]
- 6. Salviano-Silva A, et al. , Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis. Noncoding RNA, 2018. 4(1). • •. Detailed review of lncRNAs in different tissue context.
- 7.Dong P, et al. , Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front Genet, 2018. 9: p. 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobuss L, Bar C, and Thum T, Long Non-coding RNAs: At the Heart of Cardiac Dysfunction? Front Physiol, 2019. 10: p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez Calle A, et al. , Emerging roles of long non-coding RNA in cancer. Cancer Sci, 2018. 109(7): p. 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Q, et al. , LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics, 2015. 16 Suppl 3: p. S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi SW, Kim HW, and Nam JW, The small peptide world in long noncoding RNAs. Brief Bioinform, 2019. 20(5): p. 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walsh AL, et al. , Long noncoding RNAs and prostate carcinogenesis: the missing ‘linc’? Trends Mol Med, 2014. 20(8): p. 428–36. • • Extensive review on lncRNAs and their association with prostate cancer.
- 13.Rao MRS, Long Non Coding Biology. 2017: Springer. [Google Scholar]
- 14.Ma L, et al. , The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther, 2016. 17(10): p. 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao Y, et al. , Tumour-suppressor activity of H19 RNA. Nature, 1993. 365(6448): p. 764–7. [DOI] [PubMed] [Google Scholar]
- 16.Brannan CI, et al. , The product of the H19 gene may function as an RNA. Mol Cell Biol, 1990. 10(1): p. 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okazaki Y, et al. , Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature, 2002. 420(6915): p. 563–73. [DOI] [PubMed] [Google Scholar]
- 18.Quinn JJ and Chang HY, Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet, 2016. 17(1): p. 47–62. [DOI] [PubMed] [Google Scholar]
- 19.Lam YWF, Cavallari LH, Pharmacogenomics: Challenges and Opportunities in Therapeutic Implementation. 2013: Academic Press. [Google Scholar]
- 20.Chen LL, Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci, 2016. 41(9): p. 761–772. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, et al. , LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res, 2019. 47(5): p. 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun T, et al. , Emerging players in prostate cancer: long non-coding RNAs. Am J Clin Exp Urol, 2014. 2(4): p. 294–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L, Bajic VB, and Zhang Z, On the classification of long non-coding RNAs. RNA Biol, 2013. 10(6): p. 925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. , The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer, 2013. 12(1): p. 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biosynthesis. What are lncRNAs and lincRNAs. 2013; Available from: https://www.biosyn.com/tew/what-are-lncrnas-and-lincrnas.aspx.
- 26.Wu T and Du Y, LncRNAs: From Basic Research to Medical Application. Int J Biol Sci, 2017. 13(3): p. 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mouraviev V, et al. , Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis, 2016. 19(1): p. 14–20. • • Detailed review of lncRNA functions and their potential as biomarkers and theraputic targets in prostate cancer.
- 28.central, R. Homo sapiens HOX transcript antisense RNA (HOTAIR). Available from: https://rnacentral.org/rna/URS000075C808/9606.
- 29.Wang KC and Chang HY, Molecular mechanisms of long noncoding RNAs. Mol Cell, 2011. 43(6): p. 904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiu HS, et al. , Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep, 2018. 23(1): p. 297–312 e12. • • Provides lncRNA gene target and functional predictions for numerous cancer types including prostate cancer.
- 31.Yu X, et al. , NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif, 2017. 50(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klec C, Prinz F, and Pichler M, Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol, 2019. 13(1): p. 46–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An J, Lv W, and Zhang Y, LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther, 2017. 10: p. 5377–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin VY, et al. , Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis, 2019. 10(4): p. 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhat SA, et al. , Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res, 2016. 1(1): p. 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blewitt M, Noncoding RNAs - long noncoding RNAs introduction, in Epigenetic Control of Gene Expression, Blewitt M, Editor., Coursera: Melbourne, Australia. [Google Scholar]
- 37.Morrison AS, Epidemiology and environmental factors in urologic cancer. Cancer, 1987. 60(3 Suppl): p. 632–4. [DOI] [PubMed] [Google Scholar]
- 38.Fujita K, Urinary biomarkers of urological malignancies. Transl Androl Urol, 2021. 10(4): p. 1827–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu P, Cao Z, and Wu S, New Progress of Epigenetic Biomarkers in Urological Cancer. Dis Markers, 2016. 2016: p. 9864047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filella X, et al. , Emerging biomarkers in the diagnosis of prostate cancer. Pharmgenomics Pers Med, 2018. 11: p. 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HH and Kim SH, Review of non-invasive urinary biomarkers in bladder cancer. Translational Cancer Research, 2020. 9(10): p. 6554–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang G, et al. , Long non-coding RNAs in prostate tumorigenesis and therapy (Review). Mol Clin Oncol, 2020. 13(6): p. 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bray F, et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2018. 68(6): p. 394–424. [DOI] [PubMed] [Google Scholar]
- 44.Schatten H, Cell & Molecular Biology of Prostate Cancer. Vol. 1095. 2018: Springer. [Google Scholar]
- 45.Culp MB, et al. , Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur Urol, 2020. 77(1): p. 38–52. [DOI] [PubMed] [Google Scholar]
- 46.Dovey ZS, et al. , Racial disparity in prostate cancer in the African American population with actionable ideas and novel immunotherapies. Cancer Rep (Hoboken), 2021: p. e1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan J, et al. , Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet, 2020. 16(2): p. e1008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang MC, et al. , Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res, 2019. 9(7): p. 1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 49.Mendell JT, Targeting a Long Noncoding RNA in Breast Cancer. N Engl J Med, 2016. 374(23): p. 2287–9. [DOI] [PubMed] [Google Scholar]
- 50.He Q, et al. , Emerging Roles of lncRNAs in the Formation and Progression of Colorectal Cancer. Front Oncol, 2019. 9: p. 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arun G, Diermeier SD, and Spector DL, Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med, 2018. 24(3): p. 257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlevaro-Fita J, et al. , Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Commun Biol, 2020. 3(1): p. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malik B and Feng FY, Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J Androl, 2016. 18(4): p. 568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rebbeck TR, Prostate Cancer Genetics: Variation by Race, Ethnicity, and Geography. Semin Radiat Oncol, 2017. 27(1): p. 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das R, Feng FY, and Selth LA, Long non-coding RNAs in prostate cancer: Biological and clinical implications. Mol Cell Endocrinol, 2019. 480: p. 142–152. [DOI] [PubMed] [Google Scholar]
- 56.Arriaga-Canon C, et al. , The use of long non-coding RNAs as prognostic biomarkers and therapeutic targets in prostate cancer. Oncotarget, 2018. 9(29): p. 20872–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marrugo-Ramirez J, Mir M, and Samitier J, Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int J Mol Sci, 2018. 19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helsmoortel H, et al. , Detecting long non-coding RNA biomarkers in prostate cancer liquid biopsies: Hype or hope? Noncoding RNA Res, 2018. 3(2): p. 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Etzioni R, et al. , Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst, 2002. 94(13): p. 981–90. [DOI] [PubMed] [Google Scholar]
- 60.Wilt TJ, et al. , Prostate-specific antigen screening in prostate cancer: perspectives on the evidence. J Natl Cancer Inst, 2014. 106(3): p. dju010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Sullivan J, Controversies in PSA screening. Evid Based Med, 2017. 22(6): p. 198. [DOI] [PubMed] [Google Scholar]
- 62.Brawley OW, Trends in prostate cancer in the United States. J Natl Cancer Inst Monogr, 2012. 2012(45): p. 152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feibus AH, et al. , Clinical Use of PCA3 and TMPRSS2:ERG Urinary Biomarkers in African-American Men Undergoing Prostate Biopsy. J Urol, 2016. 196(4): p. 1053–60. [DOI] [PubMed] [Google Scholar]
- 64.Bussemakers MJ, et al. , DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res, 1999. 59(23): p. 5975–9. [PubMed] [Google Scholar]
- 65.Crawford ED, et al. , Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol, 2012. 188(5): p. 1726–31. [DOI] [PubMed] [Google Scholar]
- 66.Yao Y, Ranade K, Jannal B, Genomic Biomarkers for Pharmaceutical Development. 2013: Academic Press. [Google Scholar]
- 67.Gray SG, Epigenetic Cancer Therapy. 2015: Academic Press. [Google Scholar]
- 68.Misawa A, Takayama KI, and Inoue S, Long non-coding RNAs and prostate cancer. Cancer Sci, 2017. 108(11): p. 2107–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dijkstra S, et al. , Personalized management in low-risk prostate cancer: the role of biomarkers. Prostate Cancer, 2012. 2012: p. 327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitobe Y, et al. , Prostate cancer-associated lncRNAs. Cancer Lett, 2018. 418: p. 159–166. [DOI] [PubMed] [Google Scholar]
- 71.O’Malley PG, et al. , Racial Variation in the Utility of Urinary Biomarkers PCA3 and T2ERG in a Large Multicenter Study. J Urol, 2017. 198(1): p. 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adam A, et al. , The role of the PCA3 assay in predicting prostate biopsy outcome in a South African setting. BJU Int, 2011. 108(11): p. 1728–33. [DOI] [PubMed] [Google Scholar]
- 73.Prensner JR, et al. , PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res, 2014. 74(6): p. 1651–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X, et al. , PCAT1 is a poor prognostic factor in endometrial carcinoma and associated with cancer cell proliferation, migration and invasion. Bosn J Basic Med Sci, 2019. 19(3): p. 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prensner JR, et al. , Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol, 2011. 29(8): p. 742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matejcic M, et al. , Germline variation at 8q24 and prostate cancer risk in men of European ancestry. Nat Commun, 2018. 9(1): p. 4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiong T, et al. , PCAT-1: A Novel Oncogenic Long Non-Coding RNA in Human Cancers. Int J Biol Sci, 2019. 15(4): p. 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan Q, et al. , LncRNA PCAT1 and its genetic variant rs1902432 are associated with prostate cancer risk. J Cancer, 2018. 9(8): p. 1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crea F, et al. , Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget, 2014. 5(3): p. 764–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren S, et al. , Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer, 2013. 49(13): p. 2949–59. [DOI] [PubMed] [Google Scholar]
- 81.Wang F, et al. , Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget, 2014. 5(22): p. 11091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y, et al. , Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134–5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun, 2017. 491(2): p. 374–381. [DOI] [PubMed] [Google Scholar]
- 83.Liu Q, et al. , Long Non-Coding RNA Prostate Cancer-Associated Transcript 7 (PCAT7) Induces Poor Prognosis and Promotes Tumorigenesis by Inhibiting mir-134–5p in Non-Small-Cell Lung (NSCLC). Med Sci Monit, 2017. 23: p. 6089–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lang C, et al. , SMAD3/SP1 complex-mediated constitutive active loop between lncRNA PCAT7 and TGF-beta signaling promotes prostate cancer bone metastasis. Mol Oncol, 2020. 14(4): p. 808–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cimadamore A, et al. , Long Non-coding RNAs in Prostate Cancer with Emphasis on Second Chromosome Locus Associated with Prostate-1 Expression. Front Oncol, 2017. 7: p. 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parolia A, et al. , The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Mol Cancer, 2015. 14: p. 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mehra R, et al. , A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia, 2014. 16(12): p. 1121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prensner JR, et al. , The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet, 2013. 45(11): p. 1392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prensner JR, et al. , RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol, 2014. 15(13): p. 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chua MLK, et al. , A Prostate Cancer “Nimbosus”: Genomic Instability and SChLAP1 Dysregulation Underpin Aggression of Intraductal and Cribriform Subpathologies. Eur Urol, 2017. 72(5): p. 665–674. [DOI] [PubMed] [Google Scholar]
- 91.Petrovics G, et al. , Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene, 2004. 23(2): p. 605–11. [DOI] [PubMed] [Google Scholar]
- 92.Fu X, et al. , Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol, 2006. 25(3): p. 135–41. [DOI] [PubMed] [Google Scholar]
- 93.Lee J.e.a., Increased expression of PCGEM1 lncRNA in prostate cancer of African American men. 2016. 6(6). [Google Scholar]
- 94.Waxman AJ, et al. , Racial disparities in incidence and outcome in multiple myeloma: a population-based study. Blood, 2010. 116(25): p. 5501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith CJ, Ambs S, and Landgren O, Biological determinants of health disparities in multiple myeloma. Blood Cancer J, 2018. 8(9): p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cui YS, Song YP, and Fang BJ, The role of long non-coding RNAs in multiple myeloma. Eur J Haematol, 2019. 103(1): p. 3–9. [DOI] [PubMed] [Google Scholar]
- 97.Kazandjian D, Multiple myeloma epidemiology and survival: A unique malignancy. Semin Oncol, 2016. 43(6): p. 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wallace K, et al. , Platelet and hemoglobin count at diagnosis are associated with survival in African American and Caucasian patients with colorectal cancer. Cancer Epidemiol, 2020. 67: p. 101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chatterjee NA, He Y, and Keating NL, Racial differences in breast cancer stage at diagnosis in the mammography era. Am J Public Health, 2013. 103(1): p. 170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gupta V, et al. , Racial disparity in breast cancer: can it be mattered for prognosis and therapy. J Cell Commun Signal, 2018. 12(1): p. 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeSantis CE, et al. , Cancer statistics for African Americans, 2019. CA Cancer J Clin, 2019. 69(3): p. 211–233. [DOI] [PubMed] [Google Scholar]
- 102. Rebbeck TR, et al. , Global patterns of prostate cancer incidence, aggressiveness, and mortality in men of african descent. Prostate Cancer, 2013. 2013: p. 560857. • • Comprehensive review of prosatate cancer racial differences.
- 103.Jones AL and Chinegwundoh F, Update on prostate cancer in black men within the UK. Ecancermedicalscience, 2014. 8: p. 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hardiman G, et al. , Systems analysis of the prostate transcriptome in African-American men compared with European-American men. Pharmacogenomics, 2016. 17(10): p. 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Richards Z, et al. , Prostatic compensation of the vitamin D axis in African American men. JCI Insight, 2017. 2(2): p. e91054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benafif S, et al. , A Review of Prostate Cancer Genome-Wide Association Studies (GWAS). Cancer Epidemiol Biomarkers Prev, 2018. 27(8): p. 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barrington WE, et al. , Difference in Association of Obesity With Prostate Cancer Risk Between US African American and Non-Hispanic White Men in the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA Oncol, 2015. 1(3): p. 342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao J, et al. , Mitochondrial biology and prostate cancer ethnic disparity. Carcinogenesis, 2018. 39(11): p. 1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Freedman ML, et al. , Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A, 2006. 103(38): p. 14068–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Robbins CM, et al. , EphB2 SNPs and sporadic prostate cancer risk in African American men. PLoS One, 2011. 6(5): p. e19494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rawla P, Epidemiology of Prostate Cancer. World J Oncol, 2019. 10(2): p. 63–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bentley AR, Callier SL, and Rotimi CN, Evaluating the promise of inclusion of African ancestry populations in genomics. NPJ Genom Med, 2020. 5: p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hardiman G, An Introduction to Systems Analytics and Integration of Big Omics Data. Genes, 2020. 11(3): p. 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frey LJ, Artificial intelligence and integrated genotype–phenotype identification. Genes, 2019. 10(1): p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koutrouli M, et al. , A Guide to Conquer the Biological Network Era Using Graph Theory. Frontiers in Bioengineering and Biotechnology, 2020. 8(34). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pavlopoulos GA, et al. , Using graph theory to analyze biological networks. BioData Mining, 2011. 4(1): p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moul JW, The evolving definition of advanced prostate cancer. Rev Urol, 2004. 6 Suppl 8: p. S10–7. [PMC free article] [PubMed] [Google Scholar]
- 118.Obenauf AC and Massague J, Surviving at a Distance: Organ-Specific Metastasis. Trends Cancer, 2015. 1(1): p. 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weidle UH, et al. , The Functional Role of Prostate Cancer Metastasis-related Micro-RNAs. Cancer Genomics Proteomics, 2019. 16(1): p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Society AC Hormone Therapy for Prostate Cancer. 2019; Available from: https://www.cancer.org/cancer/prostate-cancer/treating/hormone-therapy.html.
- 121.Service NH Treatment Prostate cancer. 2018; Available from: https://www.nhs.uk/conditions/prostate-cancer/treatment/.
- 122.Wadosky KM and Koochekpour S, Molecular mechanisms underlying resistance to androgen deprivation therapy in prostate cancer. Oncotarget, 2016. 7(39): p. 64447–64470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Crawford ED, et al. , Androgen-targeted therapy in men with prostate cancer: evolving practice and future considerations. Prostate Cancer Prostatic Dis, 2019. 22(1): p. 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dong L, et al. , Metastatic prostate cancer remains incurable, why? Asian J Urol, 2019. 6(1): p. 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karantanos T, Corn PG, and Thompson TC, Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene, 2013. 32(49): p. 5501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hotte SJ and Saad F, Current management of castrate-resistant prostate cancer. Curr Oncol, 2010. 17 Suppl 2: p. S72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moreira DM, et al. , Predictors of Time to Metastasis in Castration-resistant Prostate Cancer. Urology, 2016. 96: p. 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saad F and Hotte SJ, Guidelines for the management of castrate-resistant prostate cancer. Can Urol Assoc J, 2010. 4(6): p. 380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mehtala J, et al. , Overall survival and second primary malignancies in men with metastatic prostate cancer. PLoS One, 2020. 15(2): p. e0227552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Damodaran S, Kyriakopoulos CE, and Jarrard DF, Newly Diagnosed Metastatic Prostate Cancer: Has the Paradigm Changed? Urol Clin North Am, 2017. 44(4): p. 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Barth DA, et al. , lncRNA and Mechanisms of Drug Resistance in Cancers of the Genitourinary System. Cancers (Basel), 2020. 12(8). • Detailed overview of lncRNAs primarily associated with drug resistance in urological cancers including prostate cancer.
- 132.Housman G, et al. , Drug resistance in cancer: an overview. Cancers (Basel), 2014. 6(3): p. 1769–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aird J, et al. , Carcinogenesis in prostate cancer: The role of long non-coding RNAs. Noncoding RNA Res, 2018. 3(1): p. 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shih JW, et al. , Non-Coding RNAs in Castration-Resistant Prostate Cancer: Regulation of Androgen Receptor Signaling and Cancer Metabolism. Int J Mol Sci, 2015. 16(12): p. 28943–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malik R, et al. , The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res, 2014. 12(8): p. 1081–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bajpai P, et al. , Mitochondrial localization, import, and mitochondrial function of the androgen receptor. J Biol Chem, 2019. 294(16): p. 6621–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Altschuler J, Stockert JA, and Kyprianou N, Non-Coding RNAs Set a New Phenotypic Frontier in Prostate Cancer Metastasis and Resistance. Int J Mol Sci, 2021. 22(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Parolia A, et al. , The long noncoding RNA HORAS5 mediates castration-resistant prostate cancer survival by activating the androgen receptor transcriptional program. Mol Oncol, 2019. 13(5): p. 1121–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takayama K, et al. , Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J, 2013. 32(12): p. 1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Scaravilli M, Koivukoski S, and Latonen L, Androgen-Driven Fusion Genes and Chimeric Transcripts in Prostate Cancer. Front Cell Dev Biol, 2021. 9: p. 623809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yang Y, et al. , Androgen Receptor-Related Non-coding RNAs in Prostate Cancer. Front Cell Dev Biol, 2021. 9: p. 660853. • List of lncRNAs regulating the Androgen Receptor in Prostate cancer.
- 142.Wang R, et al. , Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia, 2012. 14(10): p. 905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rinn JL, et al. , Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 2007. 129(7): p. 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carrion K, et al. , The long non-coding HOTAIR is modulated by cyclic stretch and WNT/β-CATENIN in human aortic valve cells and is a novel repressor of calcification genes. PLoS One, 2014. 9(5): p. e96577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang A, et al. , LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep, 2015. 13(1): p. 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mather RL, Wang Y, and Crea F, Is HOTAIR really involved in neuroendocrine prostate cancer differentiation? Epigenomics, 2018. 10(10): p. 1259–1261. [DOI] [PubMed] [Google Scholar]
- 147.Zhang Y, et al. , Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet, 2018. 50(6): p. 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhou B, et al. , EVLncRNAs 2.0: an updated database of manually curated functional long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Research, 2021. 49(D1): p. D86–D91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gong J, et al. , RISE: a database of RNA interactome from sequencing experiments. Nucleic acids research, 2018. 46(D1): p. D194–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gong Y, et al. , lncRNA-screen: an interactive platform for computationally screening long non-coding RNAs in large genomics datasets. BMC Genomics, 2017. 18(1): p. 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lin Y, et al. , RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic acids research, 2020. 48(D1): p. D189–D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.An G, et al. , LIVE: a manually curated encyclopedia of experimentally validated interactions of lncRNAs. Database (Oxford), 2019. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu Y and Zhao M, lnCaNet: pan-cancer co-expression network for human lncRNA and cancer genes. Bioinformatics, 2016. 32(10): p. 1595–7. [DOI] [PubMed] [Google Scholar]
- 154.Iyer MK, et al. , The landscape of long noncoding RNAs in the human transcriptome. Nat Genet, 2015. 47(3): p. 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Umu SU and Gardner PP, A comprehensive benchmark of RNA-RNA interaction prediction tools for all domains of life. Bioinformatics, 2017. 33(7): p. 988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lai D and Meyer IM, A comprehensive comparison of general RNA-RNA interaction prediction methods. Nucleic Acids Res, 2016. 44(7): p. e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pinkney HR, Wright BM, and Diermeier SD, The lncrna toolkit: Databases and in silico tools for lncrna analysis. Non-coding RNA, 2020. 6(4): p. 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fukunaga T, et al. , LncRRIsearch: a web server for lncRNA-RNA interaction prediction integrated with tissue-specific expression and subcellular localization data. Frontiers in genetics, 2019. 10: p. 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mann M, Wright PR, and Backofen R, IntaRNA 2.0: enhanced and customizable prediction of RNA–RNA interactions. Nucleic acids research, 2017. 45(W1): p. W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang Y, Long Y, and Kwoh CK, Deep learning based DNA: RNA triplex forming potential prediction. BMC bioinformatics, 2020. 21(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Buske FA, et al. , Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome research, 2012. 22(7): p. 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li Y, et al. , Capsule-LPI: a LncRNA–protein interaction predicting tool based on a capsule network. BMC bioinformatics, 2021. 22(1): p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maticzka D, et al. , GraphProt: modeling binding preferences of RNA-binding proteins. Genome biology, 2014. 15(1): p. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gawronski AR, et al. , MechRNA: prediction of lncRNA mechanisms from RNA-RNA and RNA-protein interactions. Bioinformatics, 2018. 34(18): p. 3101–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pyfrom SC, Luo H, and Payton JE, PLAIDOH: a novel method for functional prediction of long non-coding RNAs identifies cancer-specific LncRNA activities. BMC Genomics, 2019. 20(1): p. 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ji H, et al. , Identification, functional prediction, and key lncRNA verification of cold stress-related lncRNAs in rats liver. Sci Rep, 2020. 10(1): p. 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.da Silveira WA, et al. , miRmapper: A Tool for Interpretation of miRNA⁻mRNA Interaction Networks. Genes (Basel), 2018. 9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beerenwinkel N, Greenman CD, and Lagergren J, Computational Cancer Biology: An Evolutionary Perspective. PLoS Comput Biol, 2016. 12(2): p. e1004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Amodio N, et al. , MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol, 2018. 11(1): p. 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sun Y and Ma L, New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel), 2019. 11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fan Y, et al. , TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res, 2014. 20(6): p. 1531–41. [DOI] [PubMed] [Google Scholar]
- 172.Nitusca D, et al. , Long Noncoding RNA NEAT1 as a Potential Candidate Biomarker for Prostate Cancer. Life (Basel), 2021. 11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ye T, et al. , Long noncoding RNA linc00346 promotes the malignant phenotypes of bladder cancer. Biochem Biophys Res Commun, 2017. 491(1): p. 79–84. [DOI] [PubMed] [Google Scholar]
- 174.Pandey RR, et al. , Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell, 2008. 32(2): p. 232–46. [DOI] [PubMed] [Google Scholar]
- 175.Mohammad F, et al. , Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development, 2012. 139(15): p. 2792–803. [DOI] [PubMed] [Google Scholar]
- 176.Li Y, et al. , LncRNA KCNQ1OT1 facilitates the progression of bladder cancer by targeting MiR-218–5p/HS3ST3B1. Cancer Gene Ther, 2021. 28(3–4): p. 212–220. [DOI] [PubMed] [Google Scholar]
- 177.Wang J, et al. , Overexpression of long non-coding RNA LOC400891 promotes tumor progression and poor prognosis in prostate cancer. Tumour Biol, 2016. 37(7): p. 9603–13. [DOI] [PubMed] [Google Scholar]
- 178.Bhan A, Soleimani M, and Mandal SS, Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res, 2017. 77(15): p. 3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gao N, et al. , Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front Oncol, 2020. 10: p. 598817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Maass PG, Luft FC, and Bahring S, Long non-coding RNA in health and disease. J Mol Med (Berl), 2014. 92(4): p. 337–46. [DOI] [PubMed] [Google Scholar]
- 181.Xu T, et al. , Pathological bases and clinical impact of long noncoding RNAs in prostate cancer: a new budding star. Mol Cancer, 2018. 17(1): p. 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Al Aameri RFH, et al. , Tonic suppression of PCAT29 by the IL-6 signaling pathway in prostate cancer: Reversal by resveratrol. PLoS One, 2017. 12(5): p. e0177198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sakurai K, et al. , The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res, 2015. 13(5): p. 828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Frank F, et al. , The lncRNA Growth Arrest Specific 5 Regulates Cell Survival via Distinct Structural Modules with Independent Functions. Cell Rep, 2020. 32(3): p. 107933. [DOI] [PubMed] [Google Scholar]
- 185.Pickard MR, Mourtada-Maarabouni M, and Williams GT, Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta, 2013. 1832(10): p. 1613–23. [DOI] [PubMed] [Google Scholar]
- 186.Zheng HT, et al. , High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol, 2014. 7(6): p. 3174–81. [PMC free article] [PubMed] [Google Scholar]
- 187.Zhu K, Ren Q, and Zhao Y, lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol Lett, 2019. 17(6): p. 5335–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kim J, et al. , Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet, 2018. 50(12): p. 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu Y, et al. , Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382–3p/ROCK1 axial. Cancer Sci, 2018. 109(7): p. 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Luo Y, et al. , Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett, 2019. 440–441: p. 11–22. [DOI] [PubMed] [Google Scholar]
- 191.Gupta RA, et al. , Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 2010. 464(7291): p. 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Niinuma T, et al. , Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res, 2012. 72(5): p. 1126–36. [DOI] [PubMed] [Google Scholar]
- 193.Kogo R, et al. , Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res, 2011. 71(20): p. 6320–6. [DOI] [PubMed] [Google Scholar]
- 194.Yang Z, et al. , Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol, 2011. 18(5): p. 1243–50. [DOI] [PubMed] [Google Scholar]
- 195.Barr JA, et al. , Long non-coding RNA FAM83H-AS1 is regulated by human papillomavirus 16 E6 independently of p53 in cervical cancer cells. Sci Rep, 2019. 9(1): p. 3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xia W, et al. , Down-regulated lncRNA SBF2-AS1 inhibits tumorigenesis and progression of breast cancer by sponging microRNA-143 and repressing RRS1. J Exp Clin Cancer Res, 2020. 39(1): p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Niu L, et al. , LncRNA NRON down-regulates lncRNA snaR and inhibits cancer cell proliferation in TNBC. Biosci Rep, 2019. 39(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lei T, et al. , LncRNA NBAT-1 is down-regulated in lung cancer and influences cell proliferation, apoptosis and cell cycle. Eur Rev Med Pharmacol Sci, 2018. 22(7): p. 1958–1962. [DOI] [PubMed] [Google Scholar]
- 199.Lu X, et al. , Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res, 2013. 354(3): p. 891–6. [DOI] [PubMed] [Google Scholar]