Abstract
Runt-related transcription factor 2 (Runx2) has been shown to regulate osteoblast differentiation by directly or indirectly regulating numerous osteoblast-related genes. However, our understanding of the transcriptional mechanisms of RUNX2 is mainly restricted to its transactivation, while the mechanism underlying its inhibitory effect during osteoblast differentiation remains largely unknown. Here, we incorporated the anti-RUNX2 chromatin immunoprecipitation (ChIP) sequencing in MC3T3-E1 cells and RNA-sequencing of parietal bone from Runx2 heterozygous mutant mice, to identify the putative genes negatively regulated by RUNX2. We identified HtrA serine peptidase 1 (Htra1) as a target gene and found ten candidate Htra1 enhancers potentially regulated by RUNX2, among which seven were verified by dual-luciferase assays. Furthermore, we investigated the motifs in the vicinity of RUNX2-binding sites and identified early growth response 1 (EGR1) as a potential partner transcription factor (TF) potentially regulating Htra1 expression, which was subsequently confirmed by Re-ChIP assays. RUNX2 and EGR1 co-repressed Htra1 and increased the expression levels of other osteoblast marker genes, such as osterix, osteocalcin, and osteoprotegerin at the messenger RNA and protein level. Moreover, Alizarin red staining combined with alkaline phosphatase (ALP) staining showed decreased calcified nodules and ALP activity in the siRUNX2+siEGR1 group compared with siRUNX2 group. Our findings revealed the detailed mechanism of the inhibitory function of RUNX2 towards its downstream genes, along with its partner TFs, to promote osteoblast differentiation.
Keywords: Egr1, enhancer, Htra1, osteogenic differentiation, Runx2
1 |. INTRODUCTION
The development of bone tissue is tightly orchestrated by osteogenic cells, which are characterized by successive developmental stages and spatiotemporal gene expression profiles. Various key internal and external factors are related to cell differentiation, proliferation, and apoptosis, including transcription factors, growth factors, and signaling pathways (Vimalraj, Arumugam, Miranda, & Selvamurugan, 2015). Runt-related transcription factor 2 (RUNX2) is an indispensable regulator of osteoblast commitment and differentiation (Komori, 2010; Liu & Lee, 2013). The depletion of Runx2 leads to severe defects in mineralized tissues, such as bone and cartilage (Komori et al., 1997). Previous studies have demonstrated a significant decrease in bone formation in Runx2-null mice, which may even die at birth due to respiratory failure (Komori et al., 1997). Skeletal abnormalities have been observed in Runx2 heterozygous mutant mice, which are characterized by hypoplastic clavicles, open fontanels, and short stature (Otto et al., 1997). RUNX2 is frequently associated with mineralization-related genes, such as those encoding osteocalcin (Ocn), osterix (Osx), and bone sialoprotein (Ibsp/Bsp); it forms a functionally complicated gene-regulatory network during bone development (Nishio et al., 2006). Previous studies have also reported that some generic factors, such as histone deacetylase-3, reciprocally combine with RUNX2 to suppress Ocn expression (Schroeder, Kahler, Li, & Westendorf, 2004). Transducin-like enhancer-1 (TLE-1) modulates ribosomal RNA genes through physical association with RUNX2 during mitosis (Ali et al., 2010). These data indicate that RUNX2 positively or negatively regulates osteoblast gene expression via interactions with a variety of transcription factors, chromatin-modifying enzymes, and cofactors (Nishio et al., 2006; Takahashi et al., 2017; Zhang et al., 2016). With the advent of next-generation sequencing, RUNX2 chromatin immunoprecipitation sequencing (ChIP-seq) has revealed novel downstream genes, signaling pathways, and transcriptional mechanisms on a genome-wide scale (Meyer, Benkusky, & Pike, 2014; Wu et al., 2014). RNA-seq analysis of mouse bone marrow stromal cells (BMSCs) isolated from the parietal bone of Runx2+/− mice (wild-type siblings as controls) identified a large number of upregulated and downregulated genes (Liu et al., 2016). However, the reason why Runx2 haploinsufficiency induces the expression of its downstream genes and the detailed mechanisms involved remains unclear. In the present study, we aimed to explore the regulatory network governed by RUNX2 and determine how RUNX2 interacts with other transcription factors (TFs) during osteogenic differentiation. Thus, we performed a combined analysis of published RUNX2-related RNA-seq and ChIP-seq data to find putative targets directly inhibited by RUNX2 and related cis-regulatory elements, including enhancers. Specifically, we found that RUNX2 repressed HtrA serine peptidase 1 (Htra1) transcription by interacting with early growth response gene-1 (EGR1).
Egr1, an early zinc-finger transcription factor, has been reported to play a significant role in the homeostasis of the skeletal system. Egr1-knockout mice exhibit reduced body size, bone volume and a relatively limited degree of mineralization (Lu et al., 2018). In addition, previous studies have shown that EGR1 regulates the chondrocyte extracellular matrix via PPARγ/RUNX2 signaling pathways (Lu et al., 2018). EGR1 also supports biomineralization in dental stem cells by inducing the expression of DLX3 and BMP2 (Press, Viale-Bouroncle, Felthaus, Gosau, & Morsczeck, 2014). EGR1 modulates epigenetic programming by interacting with polycomb group proteins during chondrogenesis (Spaapen et al., 2013). Functional cooperativity of SMAD3 and EGR1 is involved in luteinizing hormone β subunit promoter activation (Fortin & Bernard, 2010). However, the detailed mechanism whereby EGR1 affects mineralization and extracellular matrix accumulation during skeletal development is still unclear. Therefore, we aimed to unveil the underlying molecular mechanisms involving RUNX2 and EGR1 during the process of osteogenic differentiation.
The target gene in this study, Htra1, belongs to the human HtrA serine protease family. It participates in cell growth, differentiation, apoptosis, osteoarthritis, and bone metabolism (Chien et al., 2004; Grau et al., 2005). Htra1 acts as an essential regulator of matrix mineralization and is secreted by osteoblasts and osteoclasts (Hadfield et al., 2008). Significant growth in bone mass has been observed in Htra1-knockout mice (Graham et al., 2013). Recombinant HTRA1 has been shown to degrade several matrix proteins, including decorin, Type II collagen, fibronectin, fibromodulin, and aggrecan in vitro. Overexpression of Htra1 results in the suppression of extracellular matrix mineralization in 2T3 osteoblasts (Hadfield et al., 2008). Other studies have shown that mineral deposition and osteoblast differentiation are partly suppressed through the Htra1-mediated inhibition of transforming growth factor (TGF)-β signaling (Hadfield et al., 2008; Oka et al., 2004). Taken together, these findings imply that Htra1 is a significant mineralization inhibitor in vivo. However, it remains unknown whether and by what mechanism Htra1 is regulated by RUNX2.
Enhancers are small segments of DNA that serve as operational platforms to recruit TFs (Herz, Hu, & Shilatifard, 2014). Enhancers have been shown to specifically modulate spatio-temporal expression of related genes, resulting in tissue-specific expression patterns of these genes (Visel, Rubin, & Pennacchio, 2009). There are 1 million known enhancers in the human genome, which exceeds the number of protein-coding genes (Rivera & Ren, 2013). Thus, multiple enhancers may cooperatively regulate the expression of a single gene, depending on the circumstances. Active enhancers are characterized by the presence of specific histone modifications, including histone H3 lysine 4 monomethylation (H3K4me1), H3K9 acetylation (H3k9ac), and H3K27 acetylation (Ernst et al., 2011). Enhancers, as regulatory DNA elements, usually contain TF-binding motifs for tissue-specific factors, which are recruited to the enhancer locus and drive the expression of target genes (Visel et al., 2009). Recent studies have confirmed the involvement of active enhancers in the positive and negative regulation of gene expression. For instance, the plasmacytoid dendritic cell-specific RUNX2 super-enhancer activates the myelocytomatosis oncogene in addition to RUNX2 (Kubota et al., 2019) and HOXA9 partly suppresses CEBPA expression through enhancer repression in 32Dcl3 myeloid cells (Peng et al., 2019). In the current study, we confirmed that RUNX2 could inhibit Htra1 expression by directly repressing the activity of seven enhancers. Besides the direct inhibitory effect of RUNX2, we further revealed EGR1 as a partner TF in regulating Htra1 enhancer activity in vitro.
2 |. MATERIALS AND METHODS
2.1 |. Cell isolation and culture
All mouse experiments were approved by the Institutional Animal Care and Use Committee at the School of Stomatology and Hospital of Stomatology of Wuhan University (protocol no. 00265690). Mouse BMSCs were isolated from bone marrow using a previously described protocol (Zhu et al., 2010). Runx2+/− mice were kindly provided by Professor Weiguo Zou (Liu et al., 2016). Eight-week-old Runx2+/− mice and their littermates were sacrificed by cervical dislocation and femurs were obtained. The isolated cells were maintained in α-MEM (Gibco, Carlsbad, CA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 100U/ml penicillin/streptomycin (HyClone). Adherent cells were harvested at passage 0 after reaching 70% confluency.
The mouse pre-osteoblastic cell line, MC3T3-E1 (Sigma-Aldrich, St. Louis, MO) and HEK293FT cells (Invitrogen, Carlsbad, CA) were maintained in α-MEM cell culture medium (Gibco) supplemented with 10% FBS and 100 U/ml penicillin/streptomycin (HyClone) in a humidified 37°C incubator with a 5% CO2 atmosphere. For osteogenic induction, MC3T3-E1 cells were cultured in mineralization medium supplemented with 10mM β-glycerophosphate (Sigma-Aldrich), 10−8 M dexamethasone (Sigma-Aldrich), and 50 mg/ml ascorbic acid (Sigma-Aldrich) (Langenbach, Handschel, & Stark, 2013).
2.2 |. ChIP and Re-ChIP assays coupled with quantitative polymerase chain reaction
MC3T3-E1 cells were exposed to mineralization medium and normal culture medium for 0, 9, and 15 days. A Magna ChIP One-Day Chromatin Immunoprecipitation Kit (#17-100859; Millipore/Upstate, Lake Placid, NY) was used for ChIP assays, following the manufacturer’s instructions. Approximately 1 × 106 MC3T3-E1 cells were subjected to crosslinking with 1% formaldehyde, chromatin preparation, and chromatin immunoprecipitation using an anti-EGRl antibody (#4153; Cell Signaling Technology, Beverly, MA) according to the manufacturer’s instructions. In total, 6 × 106 MC3T3-E1 cells were used for Re-ChIP analyses according to a standard protocol. Briefly, chromatin fragments were precipitated with anti-EGRl or control IgG antibodies in the ChIP assay. For Re-ChIP assays, the first eluted ChIP products were diluted 20 times with a dilution buffer and subjected to the ChIP procedure again. The chromatin-EGRl complex was re-immunoprecipitated using anti-RUNX2 (#12556; Cell Signaling Technology) or rabbit IgG (ABclonal Biotechnology, Woburn, MA) antibodies. Quantitative polymerase chain reaction (qPCR) was performed to analyze the precipitated genomic DNA using primer sets targeting Htra1 enhancers 4, 6, and 7. The sequences of the primers used for ChIP and Re-ChIP assays are shown in Table S1.
2.3 |. RNA extraction and quantitative reverse transcription PCR
Total RNA was isolated from BMSCs or MC3T3-E1 cells using an HP Total RNA Kit (Omega Bio-Tek, Norcross, GA) according to the manufacturer’s instructions. The yield and quality of the RNA samples were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Complementary DNA (cDNA) was synthesized from total RNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative reverse transcription PCR (qRT-PCR) was performed using a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA) using a SYBR Green Kit (Roche Diagnostics GmbH, Mannheim, Germany). Glyceraldehyde 3-phosphate dehydrogenase was used as an internal reference. The 2−ΔΔCt method was used for the relative quantitation of gene expression levels. The sequences of the primers used for qRT-PCR are shown in Table S2.
2.4 |. Vector construction
Genomic DNA was extracted from wild-type mouse livers using a TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). Enhancer fragments, including Htra1-E0 (mm9, chr7: 138079707-138079807), Htra1-E1 (mm9, chr7: 138081131-138081254), Htra1-E2 (mm9, chr7: 138086759-138086863), Htra1-E3 (mm9, chr7: 138089914-138090014), Htra1-E4 (mm9, chr7: 138093068-138093168), Htra1-E5 (mm9, chr7: 138105019-138105146), Htra1-E6 (mm9, chr7: 138108952-138109052), Htra1-E7 (mm9, chr7: 138109623-138109723), Htra1-E8 (mm9, chr7: 138111903-138112003), and Htra1-E9 (mm9, chr7: 138112161-138112270) were cloned from genomic DNA and inserted into pGL3-promoter reporter plasmids (Promega, Madison, WI). The mouse Egr1 sequence was cloned from cDNA templates and inserted into pcDNA3.1(−) (Invitrogen). The primers sequences are shown in Table S1. The deletion mutations of RUNX2-binding sites on Htra1 enhancers were performed using a KOD-Plus Mutagenesis Kit (Toyobo, Osaka, Japan), following the manufacturer’s instructions. Mutant vectors carrying the RUNX2- or EGR1-binding sites were defined as MRUNX2-E0 to MRUNX2-E9 and MEGR1-E0 to MEGR1-E9, respectively. The primers sequences for mutation are shown in Table S3.
2.5 |. Dual luciferase activity assay
Wild-type or mutant pGL3-promoter-containing Htra1-E0 to Htra1-E9 were cotransfected with a RUNX2 overexpression vector (a kind gift from Prof Shuo Chen) (Lin et al., 2019) or an empty vector (pcDNA3.1(−)) (Invitrogen) into HEK293FT/MC3T3-E1 cells, along with a Renilla luciferase reporter plasmid as an internal control. Forty-eight hours after transfection, cells were lysed, and luciferase activity was detected using a Dual-Luciferase Reporter Assay System (Promega). Assays were performed on three independent occasions, according to the manufacturer’s instructions.
2.6 |. Small-interfering RNA and plasmid transfection
Small-interfering RNAs (siRNAs) targeting RUNX2 or EGR1 and negative control siRNAs were purchased from Ambion (Austin, TX). Transfection was performed using Lipofectamine™ 2000 (Invitrogen). siRUNX2 and siEGR1 were transfected into MC3T3-E1 cells at a final concentration of 50 nM.
2.7 |. Co-immunoprecipitation and western blot analysis
Co-immunoprecipitation was performed according to the manufacturer’s instructions. In brief, 4 × 106 MC3T3-E1 cells were collected and lysed with NP-40 lysis buffer (Beyotime, Nantong, China), followed by centrifugation for 10 min at 12,000×g at 4°C. To investigate the interaction between endogenous RUNX2 and EGR1, the clarified supernatants were incubated overnight with protein A/G-agarose and an anti-RUNX2 antibody (Cell Signaling Technology), an anti-EGRl antibody (Cell Signaling Technology), or negative control IgG (ABclonal Biotechnology). The precipitates were then washed five times with NP-40 lysis buffer before analysis by western blotting. Equal amounts of protein were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel. After electrophoresis, the proteins were transferred onto polyvinylidene difluoride membranes and blocked with 5% nonfat milk for 1 hr at room temperature. The membranes were incubated with primary anti-RUNX2 (Cell Signaling Technology) or anti-EGR1 (Cell Signaling Technology) antibodies and then probed with a horseradish peroxidase-conjugated secondary antibody. Western blotting was performed as previously described (Li et al., 2018) and membranes were blotted with antibodies against RUNX2 (Cell Signaling Technology), EGR1 (Cell Signaling Technology), HTRA1 (Santa Cruz Biotechnology, Dallas, TX), OCN (ab93876; Abcam), OSX (sc-22536-R; Santa Cruz Biotechnology), and β-actin (Santa Cruz). Anti-OPN antibody is a kind gift from Prof Shuo Chen. The proteins were detected using enhanced chemiluminescence (ECL) solution (Merck Millipore, Darmstadt, Germany).
2.8 |. Alkaline phosphatase activity detection and alizarin red staining
Alkaline phosphatase (ALP) activity quantitation was measured using an Alkaline Phosphatase Assay Kit (Sigma-Aldrich), according to the manufacturer’s instructions. In brief, assay values were measured at 520 nm using a spectrophotometer and were normalized to total lysate protein concentration, as determined by a BCA Protein Assay Kit (Thermo Fisher Scientific). An ALP staining assay was performed by using an ALP Staining Kit (Sigma-Aldrich). Briefly, cells were washed three times with phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde, which was then replaced with a freshly prepared Tris-buffered staining solution containing BCIP/NBT. Color development was stopped by washing with PBS. For Alizarin Red staining, siRUNX2, siEGR1, and their respective scrambled siRNA controls were transfected individually or together into MC3T3-E1 cells. The normal growth medium was replaced with osteogenic induction medium 72 hr later. After 14 days osteogenic induction, cells were assessed by staining cells with 1% Alizarin Red S, pH 5.5 (Sigma-Aldrich) for 10 min at room temperature and rinsing with distilled water.
2.9 |. Statistical analysis
Each experiment was performed at least three times. Unless otherwise noted, data are presented as mean ± standard deviation. Statistical analysis was performed with a two-tailed Student’s t-test using SPSS statistics 17.0 (IBM, Armonk, NY). A value of p <.05 was considered statistically significant.
3 |. RESULTS
3.1 |. Htra1 is a putative downstream gene negatively regulated by RUNX2 during osteogenic differentiation
To determine whether RUNX2 directly exerts negative effects on gene regulation in osteoblasts, we analyzed anti-RUNX2 ChIP-seq data from MC3T3-E1 cells (Wu et al., 2014) (GEO ID: GSE54013) and RNA-seq data from BMSCs isolated from Runx2+/− parietal bone (wild-type siblings as controls; GEO ID: GSE77007) (Liu et al., 2016) to screen for downstream genes. We used the GREAT tool (McLarren et al., 2000) to identify the nearest genes to the anti-RUNX2 ChIP-seq peaks (distance rule) and compared the list with the genes that were upregulated (by sleuth, q-value <.05, p-value <.05) in Runx2+/− parietal bone. We found 25 genes that were upregulated in Runx2+/− mice compared with wild-type mice (Figure 1a). Of note, RUNX2 ChIP-seq peaks were present near these genes (−5 kb and +1 kb relative to the transcriptional start site). Among the 25 genes that were putatively directly downregulated by RUNX2, Htra1 was selected for further investigation because it is highly expressed in the musculoskeletal system, according to data from the MGI database (Eppig, 2017), and is able to inhibit mineral deposition in osteoblasts (Hadfield et al., 2008). The heatmap of RNA-seq results is shown in Figure S1. To determine the RUNX2-regulated expression pattern of Htra1 during osteoblast differentiation, we harvested BMSCs from Runx2+/− and wild-type mice and observed a 60% decrease in Runx2 messenger RNA (mRNA) levels, but an approximately three-fold increase in Htra1 expression levels in Runx2+/− mice compared with wild-type mice (Figure 1b,c). These results demonstrated that Htra1 was negatively regulated by RUNX2 during the osteogenic differentiation of BMSCs in vitro and in vivo.
FIGURE 1.
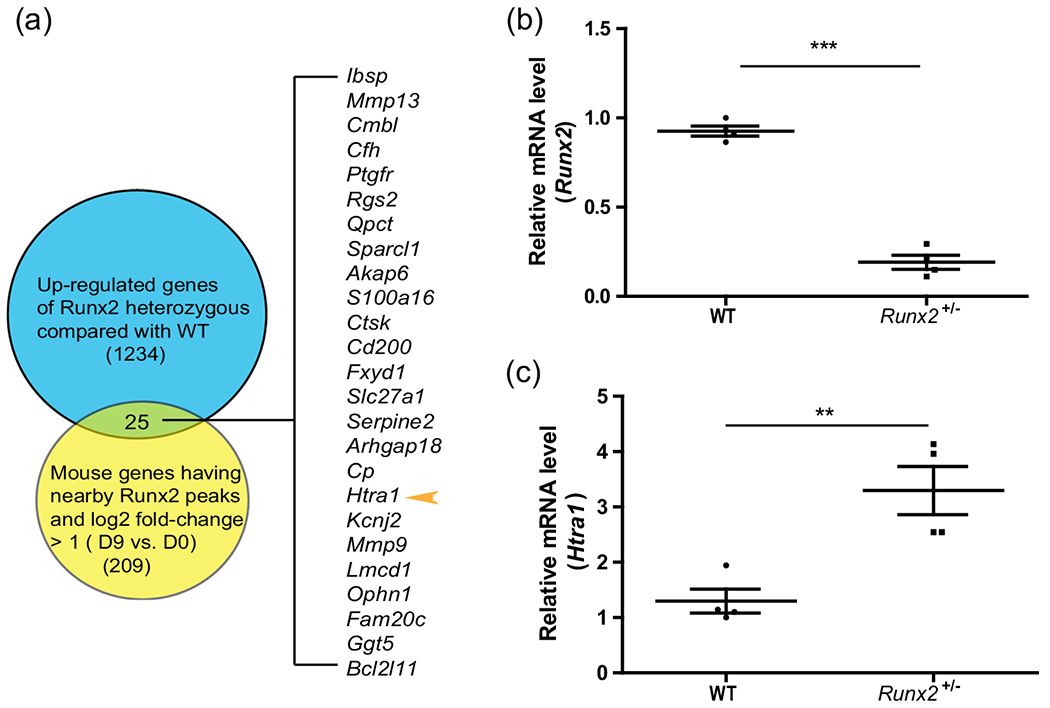
Htra1 was increased in BMSCs isolated from Runx2+/− mutant mice compared with wild-type mice in vivo (a) Venn diagram showing 1,234 upregulated genes from RNA-seq analysis (Runx2+/− mice compared with wild-type mice) (Liu et al., 2016), compared with 209 genes with RUNX2 ChIP-seq peaks nearby (−5 kb and +1 kb relative to the transcriptional start site, log2 fold-change >1, differentiated vs undifferentiated MC3T3-E1 cells) (Wu et al., 2014). (b), (c) qRT-PCR results showing the expression levels of Runx2 and Htra1. All data are presented as means ± SDs. *p < .05, **p < .01, ***p < .0001. qRT-PCR, quantitative reverse transcription polymerase chain reaction; Runx2, Runt-related transcription factor 2; SD, standard deviation
3.2 |. Htra1 enhancer activities are repressed by RUNX2 overexpression
To determine whether RUNX2 directly regulates Htra1 expression through cis-regulatory elements, we integrated the published anti-H3K9ac and anti-H3K4me1 ChIP-seq (GSE ID: GSE41955) (Meyer et al., 2014) data from MC3T3-E1 cells with anti-RUNX2 ChIP-seq (GEO ID: GSE54013) (Wu et al., 2014) data during osteoblastic differentiation from the Gene Expression Omnibus database. We found that the H3K4me1 modification was quite prevalent in Htra1 introns and exons. The previous paper also reported that the majority of RUNX2 binding occurred at intergenic and intronic regions by analyzing anti-RUNX2 ChIP-seq data (Wu et al., 2014), which indicated a critical role of cis-regulatory such as enhancers in RUNX2 regulatory network. Therefore, we hypothesized that RUNX2 directly modulates Htra1 expression through its enhancers/promoters. As shown in Figure 2a, the 10 exonic or intronic enhancers near Htra1 overlapped with RUNX2 ChIP-seq peaks on Day 9 (hereafter referred to as Htra1-E0, which overlapped with the Htra1 promoter, and Htra1-E1 to Htra1-E9) of osteogenic differentiation. Furthermore, the active enhancer signals, H3H9ac and H3K4me1, at D0 and D15, could be detected in these 10 elements. To confirm the enhancer activity, the 10 enhancer candidates were cloned and inserted into the luciferase reporter plasmid, pGL3-promoter. Compared with the empty plasmid, Htra1-E3 to E9 showed a significant increase in luciferase activity. Moreover, overexpression of RUNX2 led to the downregulation of enhancer activity in all 10 enhancer reporter plasmids (Figure 2b), indicating that these 10 enhancers were negative regulatory elements. To determine whether these 10 elements were directly regulated by RUNX2, we used the JASPAR database (Mathelier et al., 2016) to search for RUNX2-binding sites in these 10 enhancers. Only seven of the ten enhancers (Htra1-E0, -E2, -E3, -E4, -E5, -E6, and -E7) contained RUNX2-binding sites, which were mutated in the corresponding reporter plasmids. As for Htra1-E1, E8, and E9, which were also influenced by RUNX2 overexpression, we propose that they were modulated by RUNX2 through an indirect mechanism. We found that at least one mutation in the RUNX2-binding sites led to the recovery of enhancer activity in Htra1-E0, -E2, -E4, -E5, -E6, and -E7 (Figures 2c,d and 2f–i), but not in Htra1-E3 (Figure 2e). These results indicated that RUNX2 directly inhibited Htra1 enhancer activity by binding to Htra1-E0, -E2, -E4, -E5, -E6, and -E7. Sequences of mutated binding sites were listed in Figure S2.
FIGURE 2.
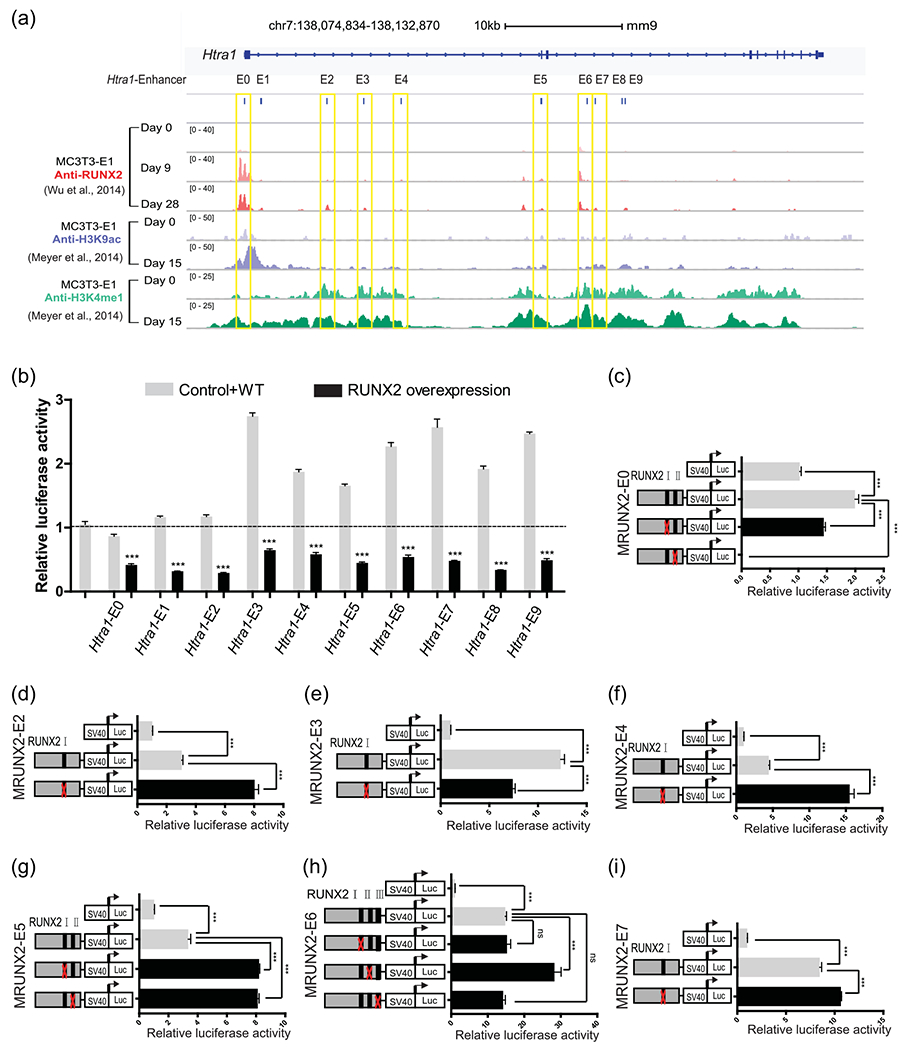
RUNX2 enrichment in 10 enhancers of the Htra1 gene locus (a) Genome browser screenshot of the Htra1 gene locus region, integrating anti-RUNX2 ChIP-Seq at osteogenic differentiation Day 0 (D0), D9, D28 in MC3T3-E1 cells (Wu et al., 2014) and anti-H3K9ac and anti-H3K4me1 at D0 and D15 in MC3T3-E1 cells (Meyer et al., 2014). MC3T3-E1 cells were cultured in osteogenic induction medium or normal growth medium. Ten putative enhancers in this locus were named Htra1-E0 to E9. The seven enhancers containing RUNX2-binding sites are marked by yellow boxes. (b) Dual luciferase assay of the 10 putative enhancers with a RUNX2 overexpression plasmid or an empty vector in HEK293FT cells. (c)–(i) Deletion analysis of RUNX2-binding sites in each putative Htra1 enhancer in MC3T3-E1 cells. All data are presented as means±SDs. *p < .05, **p < 001, ***p < .0001. Runx2, Runt-related transcription factor 2; SD, standard deviation
3.3 |. RUNX2 directly co-operates with EGR1 to regulate osteogenic differentiation by binding to Htra1 enhancers
The enhancer acts as a platform to supply sufficient binding sites for a variety of TFs in a stage-specific manner (Visel et al., 2009). We investigated whether any other TFs could be recruited to Htra1 enhancers and functionally collaborate with RUNX2 to modulate downstream gene expression during osteogenic differentiation. Multiple cooperative TFs are able to bind to closely located motifs (Jolma et al., 2015). Thus, we used the JASPAR database (Mathelier et al., 2016) to search for other TF-binding sites located within the 15-bp region flanking the RUNX2 motifs in seven enhancers and ranked them according to their frequencies (Figure 3a,b). Among the genes that ranked higher than RUNX2, we found that MEIS1 and ZNF354C are not detectable in bone, as reported in the MGI database (Eppig, 2017). Egr1 is highly expressed in bone and Egr1-knockout mice exhibit reduced bone volume (Lu et al., 2018). These data suggested that EGR1 was a potential co-activator of RUNX2. To narrow the range, we also predicted EGR1-binding sites and found that EGR1 only bound to Htra1-E4, -E6, and -E7. To test this hypothesis, we performed anti-EGR1 ChIP-qPCR during osteogenic differentiation. We found that EGR1 was able to bind to Htra1-E4, -E6, and -E7 and that this binding was significantly increased at Day 9 and Day 15 compared with Day 0 (Figure 3c–e). Similar to RUNX2-binding motifs, EGR1 overexpression also repressed the enhancer activities of Htra1-E4, -E6, and -E7. However, at least one mutation of the EGR1-binding motifs in Htra1-E4, -E6, and -E7 increased enhancer activity (Figure 3f–h). Sequences of mutated binding sites were listed in Figure S2.
FIGURE 3.
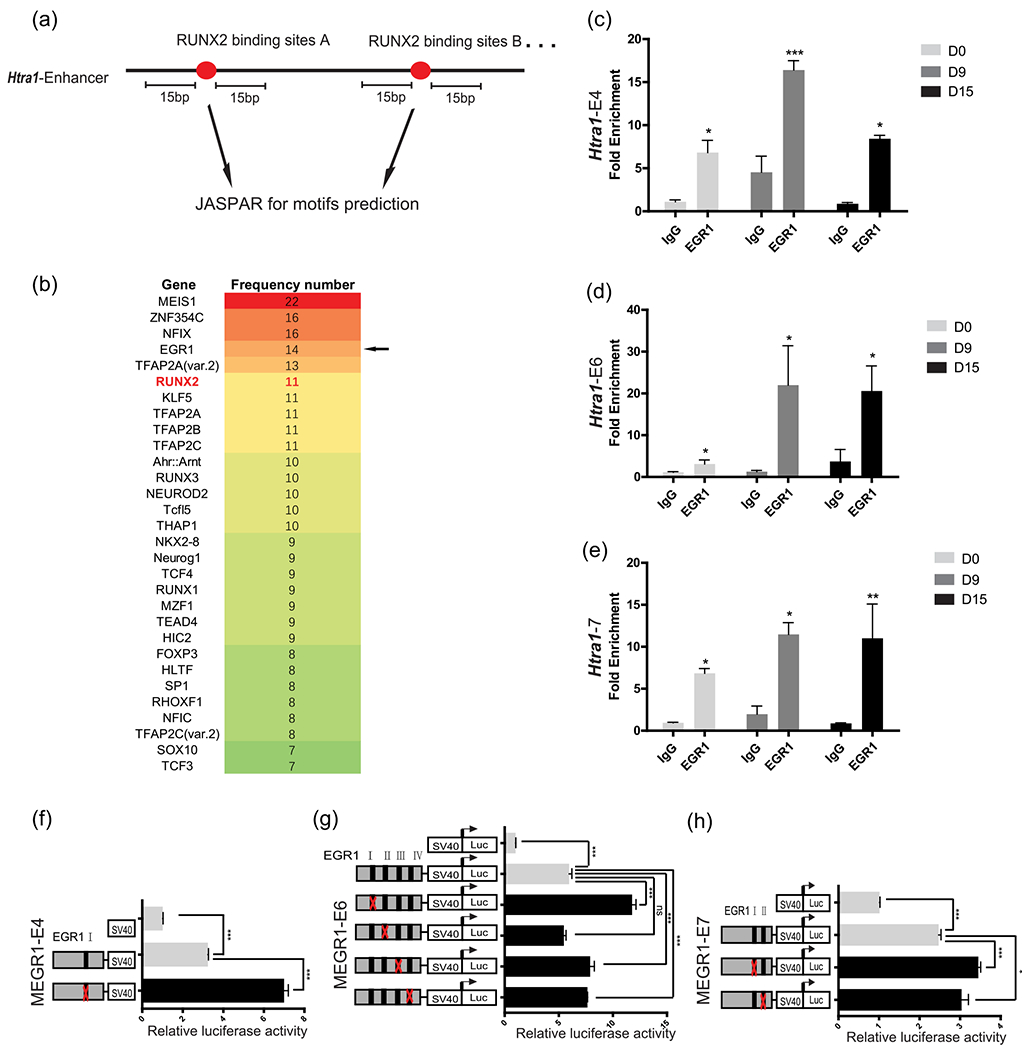
Htra1 enhancers were regulated by EGR1 in vitro (a) Schematic diagram showing the search for putative transcriptional factors within 15 base pairs of RUNX2-binding motifs using the JASPAR database (Mathelier et al., 2016). (b) Predicted transcription factors in Htra1-E0, -E2, -E3, -E4, -E5, -E6, and -E7 were ranked according to their frequencies. (c–e) Anti-EGR1 ChIP-qPCR to detect the enrichment of EGR1 at Htra1-E4, E6, and E7 in MC3T3-E1 cells, which were cultured in osteogenic induction medium or normal growth medium. (f–h) Deletion analysis of EGR1-binding sites at Htra1-E4, E6, and E7 in MC3T3-E1 cells. All data are presented as means ± SDs. *p < .05, **p < .01, ***p < .0001. ChIP-qPCR, chromatin immunoprecipitation- quantitative polymerase chain reaction; EGR1, early growth response 1; Runx2, Runt-related transcription factor 2; SD, standard deviation
3.4 |. EGR1 interacts with RUNX2 to regulate osteogenic differentiation
Given the short distance between the motifs of RUNX2 and EGR1, we also determined whether these two proteins could interact with each other. Considering the physical interaction between RUNX2 and EGR1, we performed co-immunoprecipitation experiments and found that RUNX2 directly bound to EGR1 (Figure 4a). Furthermore, Re-ChIP assays indicated that EGR1 and RUNX2 directly interacted with Htra1-E6, but not Htra1-E4 or Htra1-E7 (Figure 4b). Co-overexpression of RUNX2 and EGR1 further decreased the enhancer activity of Htra1-E6 (Figure 4d). However, no such effect was found for either Htra1-E4 or Htra1-E7 (Figures 4c and 4e). We speculated that Htra1-E4 and Htra1-E7 may be bound by other transcription factors or mediators except for RUNX2, which are also involved in enhancer activation or repression.
FIGURE 4.
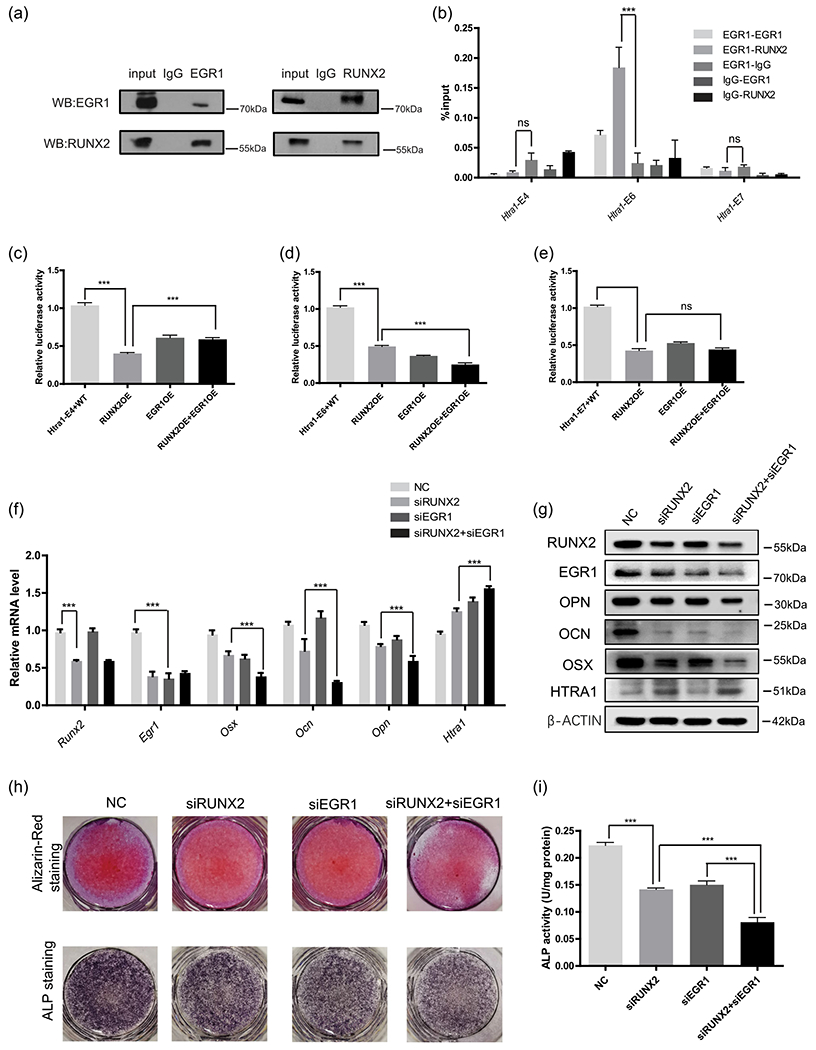
Cooperation of RUNX2 and EGR1 in Htra1 enhancers (a) Co-immunoprecipitation indicated a direct interaction between RUNX2 and EGR1. (b) Re-ChIP experiments were performed to determine whether RUNX2 and EGR1 were co-bound within Htra1-E4, E6, and E7. Relative luciferase activity of Htra1-E4 (c), E6 (d), and E7 (e) was detected after RUNX2 and EGR1 overexpression. RNA (f) and protein levels (g) of RUNX2, EGR1, HTRA1, OSX, OPN, and OCN in MC3T3-E1 cells with the individual or combined knockdown of RUNX2 and EGR1. Alizarin Red staining combined with alkaline phosphatase staining (h) showed calcified nodules and alkaline phosphatase activity (i) in individual or combined knockdown of RUNX2 and EGR1. All data are presented as means±SDs. *p < .05, **p < .01, ***p < .0001. ChIP, chromatin immunoprecipitation; EGR1, early growth response 1; Runx2, Runt-related transcription factor 2; SD, standard deviation
Based on our finding that RUNX2 and EGR1 directly co-regulated Htra1 enhancers, we investigated whether these two proteins could cooperatively regulate Htra1 expression and other osteogenesis-related genes and further induce osteoblast differentiation. We first knocked down RUNX2 and EGR1 expression individually or together via siRNA (Figure 4f,g). We found that the simultaneous knockdown of RUNX2 and EGR1 (siRUNX2+siEGR1) not only led to a further increase in Htra1 mRNA expression levels, but also repressed the expression of mineralization-related genes, such as Osx, Ocn, and Opn at the mRNA (Figure 4f) and protein level (Figure 4g), compared with the siRUNX2 or siEGR1 group. Alizarin Red staining combined with ALP staining showed fewer calcified nodules and decreased ALP activity in the siRUNX2 + siEGR1 group compared with the siRUNX2 group (Figure 4h,i). These findings showed that RUNX2 and EGR1 could co-repress Htra1 expression and increase other osteoblast-related genes. Taken together, our study revealed a detailed trans-inactivation mechanism of RUNX2 and EGR1 on their target gene, Htra1, thus providing more insights into the transcriptional regulatory mechanism of RUNX2 during osteogenic differentiation (Figure 5a).
FIGURE 5.
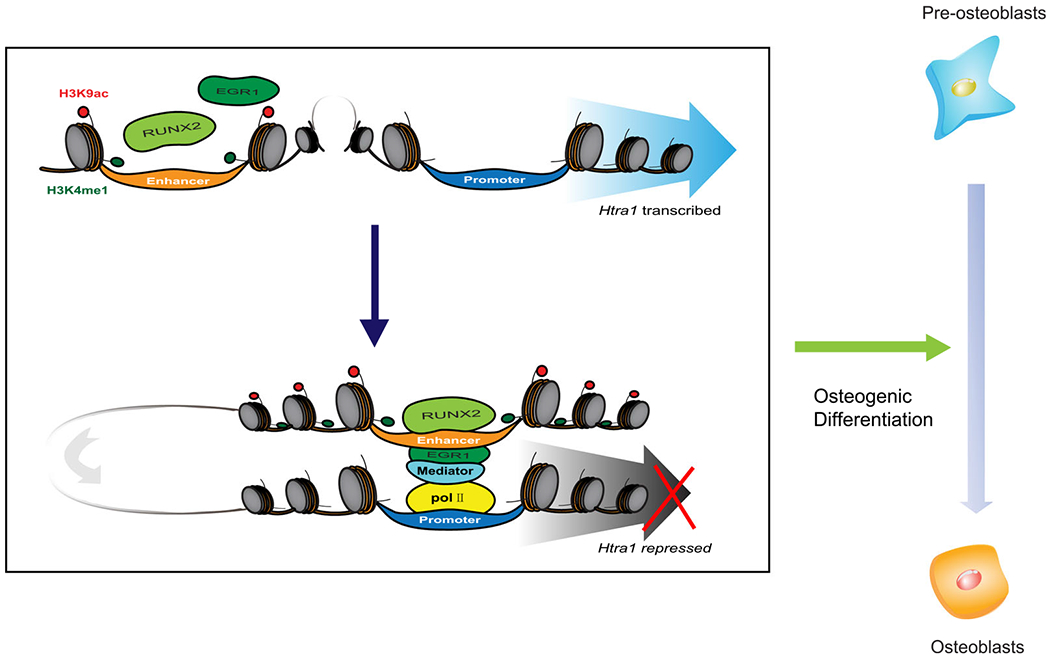
A systematic diagram to show the function of RUNX2 and EGR1 during osteogenic differentiation. In brief, RUNX2 cooperatively combines with EGR1 to repress Htra1 expression and promote osteogenic differentiation. EGR1, early growth response 1; Runx2, Runt-related transcription factor 2
4 |. DISCUSSION
Previous studies have reported that multiple mineralization-related genes, including MMP13, MMP9, Bsp, and Opn (Liu & Lee, 2013) are activated or repressed by RUNX2 during bone formation. Based on these previous findings and bioinformatic analysis in the current study, we identified a novel target gene, Htra1, cooperatively regulated by RUNX2 and EGR1 through an enhancer-repression mechanism, to promote osteogenic differentiation.
The combinatory analysis of RNA-seq in Runx2+/− mice and anti-RUNX2 ChIP-seq revealed 25 candidate genes potentially directly downregulated by RUNX2. We then narrowed the scope based on gene expression patterns. Some of the candidate genes have been reported to be the downstream of RUNX2. For example, RUNX2 overexpression has been shown to enhance MMP13 promoter activity in chondrocytes (Takahashi et al., 2017) and induce IBSP expression in adipose-derived stem cells (Zhang et al., 2016). Cd200 and Ptgfr are expressed at low levels in bone tissue. Ptgfr-knockout mice are unable to deliver normal foetuses at term (Sugimoto et al., 1997) and Cd200-knockout mice have an increased susceptibility to experimentally induced arthritis (Hoek, 2000). As recorded in the MGI database, Htra1 is highly expressed in the musculoskeletal system (Eppig, 2017) and bone volume is increased in Htra1-knockout mice (Graham et al., 2013). However, whether the downregulation of this gene is due to direct modulation by RUNX2 was previously unclear. In this study, we confirmed this regulatory mechanism using reporter assays and ChIP assays. Furthermore, Re-ChIP experiments indicated EGR1 as a partner of RUNX2 during its trans-inactivation.
In the present study, we found that RUNX2 physically interacted with EGR1 through cis-elements, to promote osteogenic differentiation. Previous studies have shown that RUNX2 interacts with different co-activators, co-repressors, and TFs to modulate multiple biological processes. For instance, RUNX2 cooperatively interacts with MED23 in the regulation of osteogenesis (Liu et al., 2016) and combines with histone deacetylase 3 to repress the osteocalcin promoter (Schroeder et al., 2004). Cooperative interactions between ATF4 and RUNX2 stimulate osteoblast-specific Ocn expression (Xiao et al., 2005). Moreover, the interaction of RUNX2 and SMAD6 mediates SMAD ubiquitination regulatory factor 1-induced RUNX2 degradation (Shen et al., 2006). Our study provided additional evidence for the interaction of RUNX2 with its partner TFs during bone development. However, as a master regulator of bone formation, more details of the mechanism of RUNX2 and its partners need to be uncovered in future studies.
Our study revealed a functional interaction between EGR1 and RUNX2 that contributed to osteogenic differentiation. Furthermore, this novel mechanism helps our understanding of the multiplexed transcriptional modulation of RUNX2, which is critical for the correct execution of bone tissue formation. The interaction between RUNX2 and EGR1 as well as their effects on enhancer activity need to be confirmed in vivo in future studies. Although we showed that Htra1-E6 is crucial for RUNX2 and EGR1 regulation in vitro, knockout experiments in transgenic mice may help confirm the biological function of Htra1-E6 during osteogenic differentiation in vivo.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by grants from the National Natural Science Foundation of China (No. 81771066; No. 81420108011) to Z. Chen.
Funding information
National Natural Science Foundation of China, Grant/Award Numbers: 81420108011, 81771066
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
DATA AVAILABILITY STATEMENT
All data included in this study are available upon request by contacting the corresponding authors.
REFERENCES
- Ali SA, Zaidi SK, Dobson JR, Shakoori AR, Lian JB, Stein JL, … Stein GS (2010). Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proceedings of the National Academy of Sciences of the United States of America, 107(9), 4165–4169. 10.1073/pnas.1000620107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J, Staub J, Hu S-I, Erickson-Johnson MR, Couch FJ, Smith DI, … Shridhar V (2004). A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene, 23(8), 1636–1644. 10.1038/sj.onc.1207271 [DOI] [PubMed] [Google Scholar]
- Eppig JT (2017). Mouse Genome Informatics (MGI) Resource: Genetic, genomic, and biological knowledgebase for the laboratory mouse. Institute for Laboratory Animal Research Journal, 58(1), 17–41. 10.1093/ilar/ilx013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen ST, Shoresh N, Ward DL, Epstein BC, … Bernstein EB (2011). Mapping and analysis of chromatin state dynamics in nine human cell type. Nature, 473(7345), 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J, & Bernard DJ (2010). SMAD3 and EGR1 physically and functionally interact in promoter-specific fashion. Cellular Signalling, 22(6), 936–943. 10.1016/j.cellsig.2010.01.019 [DOI] [PubMed] [Google Scholar]
- Graham JR, Chamberland A, Lin Q, Li XJ, Dai D, Zeng W, … Yang Z (2013). Serine protease HTRA1 antagonizes transforming growth factor-beta signaling by cleaving its receptors and loss of HTRA1 in vivo enhances bone formation. PLoS One, 8(9):e74094. 10.1371/journal.pone.0074094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau S, Baldi A, Bussani R, Tian X, Stefanescu R, Przybylski M, … Ehrmann M (2005). Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proceedings of the National Academy of Sciences of the United States of America, 102(17), 6021–6026. 10.1073/pnas.0501823102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KD, Rock CF, Inkson CA, Dallas SL, Sudre L, Wallis GA, … Canfield AE (2008). HtrA1 inhibits mineral deposition by osteoblasts: Requirement for the protease and PDZ domains. Journal of Biological Chemistry 283(9), 5928–5938. 10.1074/jbc.M709299200 [DOI] [PubMed] [Google Scholar]
- Herz HM, Hu D, & Shilatifard A (2014). Enhancer malfunction in cancer. Molecular Cell, 53(6), 859–866. 10.1016/j.molcel.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek RM (2000). Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science, 290(5497), 1768–1771. 10.1126/science.290.5497.1768 [DOI] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, … Taipale J (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature, 527(7578), 384–388. 10.1038/nature15518 [DOI] [PubMed] [Google Scholar]
- Komori T (2010). Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell and Tissue Research, 339(1), 189–195. 10.1007/s00441-009-0832-8 [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, … Kishimoto T (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 89(5), 755–764. 10.1016/s0092-8674(00)80258-5 [DOI] [PubMed] [Google Scholar]
- Kubota S, Tokunaga K, Umezu T, Yokomizo-Nakano T, Sun Y, Oshima M, … Sashida G (2019). Lineage-specific RUNX2 super-enhancer activates MYC and promotes the development of blastic plasmacytoid dendritic cell neoplasm. Nature Communications, 10(1), 1653. 10.1038/s41467-019-09710-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbach F, Handschel J, & Stark A (2013). Effects of dexamethasone, ascorbic acid and beta-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Research & Therapy, 4(5), 117. 10.1186/scrt328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin C, Zhang J, Tao H, Liu H, Yuan G, & Chen Z (2018). Quaking promotes the odontoblastic differentiation of human dental pulp stem cells. Journal of Cellular Physiology, 233(9), 7292–7304. 10.1002/jcp.26561 [DOI] [PubMed] [Google Scholar]
- Lin C, Yu S, Jin R, Xiao Y, Pan M, Pei F, … Chen Z (2019). Circulating miR-338 cluster activities on osteoblast differentiation: Potential diagnostic and therapeutic targets for postmenopausal osteoporosis. Theranostics, 9(13), 3780–3797. 10.7150/thno.34493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, & Lee EH (2013). Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Engineering. Part B, Reviews, 19(3), 254–263. 10.1089/ten.TEB.2012.0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Yao X, Yan G, Xu Y, Yan J, Zou W, & Wang G (2016). Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nature Communications, 7, 11149. 10.1038/ncomms11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Shi TS, Shen SY, Lu WL, Wu J, Zhang KJ, … Xue B (2018). Egr1 deficiency disrupts dynamic equilibrium of. American Journal of Translational Research, 10(6), 1620–1632. [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, … Wasserman WW (2016). JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Research, 44(D1), D110–D115. 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, & Stifani S (2000). The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. Journal of Biological Chemistry, 275(1), 530–538. 10.1074/jbc.275.1.530 [DOI] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, & Pike JW (2014). The RUNX2 cistrome in osteoblasts: Characterization, down-regulation following differentiation, and relationship to gene expression. Journal of Biological Chemistry, 289(23), 16016–16031. 10.1074/jbc.M114.552216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio Y, Dong Y, Paris M, O’Keefe RJ, Schwarz EM, & Drissi H (2006). Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene, 372, 62–70. 10.1016/j.gene.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Oka C, Tsujimoto R, Kajikawa M, Koshiba-Takeuchi K, Ina J, Yano M, … Kawaichi M (2004). HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development, 131(5), 1041–1053. 10.1242/dev.00999 [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell LR, … Owen MJ (1997). Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell, 89(5), 765–771. 10.1016/S0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- Peng L, Guo H, Ma P, Sun Y, Dennison L, Aplan PD, … Friedman AD (2019). HoxA9 binds and represses the Cebpa +8 kb enhancer. PLoS One, 14(5):e0217604. 10.1371/journal.pone.0217604r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press T, Viale-Bouroncle S, Felthaus O, Gosau M, & Morsczeck C (2014). EGR1 supports the osteogenic differentiation of dental stem cells. International Endodontic Journal, 48, 185–192. 10.1111/iej.12299. [DOI] [PubMed] [Google Scholar]
- Rivera CM, & Ren B (2013). Mapping human epigenomes. Cell, 155(1), 39–55. 10.1016/j.cell.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, & Westendorf JJ (2004). Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. Journal of Biological Chemistry, 279(40), 41998–42007. 10.1074/jbc.M403702200 [DOI] [PubMed] [Google Scholar]
- Shen R, Chen M, Wang Y-J, Kaneki H, Xing L, O’Keefe RJ, & Chen D (2006). Smad6 interacts with Runx2 and mediates smad ubiquitin regulatory factor 1-induced Runx2 degradation. Journal of Biological Chemistry, 281(6), 3569–3576. 10.1074/jbc.M506761200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaapen F, van den Akker GG, Caron MM, Prickaerts P, Rofel C, Dahlmans VE, … Voncken JW (2013). The immediate early gene product EGR1 and polycomb group proteins interact in epigenetic programming during chondrogenesis. PLoS One, 8(3):e58083. 10.1371/journal.pone.0058083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Yamasaki A, Segi E, Tsuboi K, Aze Y, Nishimura T, … Narumiya S (1997). Failure of parturition in mice lacking the prostaglandin F receptor. Science, 277(5326), 681–683. 10.1126/science.277.5326.681 [DOI] [PubMed] [Google Scholar]
- Takahashi A, de Andres MC, Hashimoto K, Itoi E, Otero M, Goldring MB, & Oreffo ROC (2017). DNA methylation of the RUNX2 P1 promoter mediates MMP13 transcription in chondrocytes. Scientific Reports, 7(1), 7771. 10.1038/s41598-017-08418-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimalraj S, Arumugam B, Miranda PJ, & Selvamurugan N (2015). Runx2: Structure, function, and phosphorylation in osteoblast differentiation. International Journal of Biological Macromolecules, 78, 202–208. 10.1016/j.ijbiomac.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, & Pennacchio LA (2009). Genomic views of distant-acting enhancers. Nature, 461(7261), 199–205. 10.1038/nature08451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Whitfield WT, Gordon ARJ, Dobson RJ, Tai WLP, Wijnen J. v A., … Lian BJ (2014). Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biology, 15(3), R52. 10.1186/gb-2014-15-3-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, … Franceschi RT (2005). Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. Journal of Biological Chemistry, 280(35), 30689–30696. 10.1074/jbc.M500750200 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ma Y, Fu X, Liu Q, Shao Z, Dai L, … Ao Y (2016). Runx2-modified adipose-derived stem cells promote tendon graft integration in anterior cruciate ligament reconstruction. Scientific Reports, 6, 19073. 10.1038/srep19073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Guo ZK, Jiang XX, Li H, Wang XY, Yao HY, … Mao N (2010). A protocol for isolation and culture of mesenchymal stem cells from mouse compact bone. Nature Protocols, 5(3), 550–560. 10.1038/nprot.2009.238 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


