Abstract
Objective:
DNA methylation is a critical epigenetic modulation in regulating gene expression in cell differentiation process, however, its detailed molecular mechanism during odontoblastic differentiation remains elusive. We aimed to study the global effect of DNA methylation on odontoblastic differentiation and how DNA methylation affects the transactivation of transcription factor (TF) on its target gene.
Methods:
DNA methyltransferase (DNMTs) inhibition assay and following odontoblastic differentiation assay were performed to evaluate the effect of DNA methylation inhibition on odontoblastic differentiation. Promoter DNA methylation microarray and motif enrichment assay were performed to predict the most DNA-methylation-affected TF motifs during odontoblastic differentiation. The enriched target sites and motifs were further analyzed by methylation-specific polymerase chain reaction (MS-PCR) and sequencing. The functional target sites were validated in vitro with Luciferase assay. The regulatory effect of DNA methylation on the enriched target sites in primary human dental pulp cells and motifs were confirmed by in vitro methylation assay.
Results:
Inhibition of DNMTs in preodontoblast cells increased the expression level of Klf4 as well as marker genes of odontoblastic differentiation including Dmp1 and Dspp, and enhanced the efficiency of odontoblastic differentiation. SP1/KLF4 binding motifs were found to be highly enriched in the promoter regions and showed demethylation during odontoblastic differentiation. Mutation of SP1 binding site at −75 within KLF4′s promoter region significantly decreased the luciferase activity. The in vitro methylation of KLF4′s promoter decreased the transactivation of SP1 on KLF4.
Conclusion:
We confirmed that SP1 regulates KLF4 through binding site lying in a CpG island in KLF4′s promoter region which demethylated during odontoblastic differentiation thus enhancing the efficiency of SP1′s binding and transcriptional regulation on KLF4.
Keywords: DNA methylation, human dental pulp cells, Klf4, odontoblastic differentiation, SP1
1 |. INTRODUCTION
Odontoblasts, located at the periphery of the tooth pulp, are derived from neural crest cells,1 form and maintain the dentin which is a major mineralization tissue of teeth. Odontoblastic differentiation is critical for the formation of teeth and tertiary dentin formation when dental caries happen.2 Studies of the odontoblastic differentiation would contribute to a better understanding of tooth development as well as dental pulp regeneration. Such biological process has been extensively studied, and is regulated by several signaling pathways, such as TGF-ß, WNT, and HH pathways3,4 and transcription factors including Sp7, Dlx3, Klf4, etc.2,5,6 Global knockout of Sp7 in mice will lead to severe dentin hypoplasia and obviously abolish the expression of DMP1 and DSPP, which are the classical markers of odontoblasts, in incisors and molars.6 And deficiency of Runx2 in mice can even arrest the development of molars at the late bud stage.7
Transcription factors mainly exert their function through binding to specific DNA motifs in the cis-regulatory elements.8 However, epigenetic modifications of the cis-regulatory elements, such as DNA methylation and histone modification,9 may alter DNA accessibility and thereafter regulating the binding transcription factors and patterns of their downstream gene expression.10–12 ChIP-seq or ChIP-on-chip assay for DNA methylation and histone markers had shown such epigenetic reflection or effect on gene expression thus involved in all kinds of biological processes such as the differentiation of pluripotent stem cells, tumorigenesis, and so on.10,12,13 Previous report on histone modification during human dental pulp cell differentiation revealed that the difference of the dynamic histone enrichment responding to mineralization induction between human dental pulp cell and human dental follicle cell determined the different odontogenic potential of these two cell types.14 In 2013, research based on a large Chinese X-linked hypohidrotic ectodermal dysplasia (XLHED) family found a correlation between hypermethylated EDA promoter and XLHED carriers.15 And following genome-wide scanning study revealed significant differences in the whole genome methylation level of the hypodontia patients comparing with the healthy16 which emphasized the importance of DNA methylation in the incidence of hypodontia in human. Recently, a study of odontoblastic differentiation gave evidence to the potential role of DNA methylation in this process as it reported that knockdown of TET1, a DNA methyl cytosine dioxygenase, could suppress odontoblastic differentiation of human dental pulp cells (hDPCs) through inhibiting hydroxymethylation and subsequent transcription of FAM20C.17 Given all these relative descriptive findings, the more detailed molecular mechanism of how DNA methylation affects odontoblastic differentiation remains elusive.
Here, the promoter DNA methylation microarray was used to evaluate the changes of DNA methylation during mouse odontoblastic differentiation genome-widely. Further analysis revealed SP1/KLF4 binding motif was highly enriched in the demethylated promoter regions after differentiation. We then validated DNA-methylation-dependent activation of KLF4 by SP1 during odontoblastic differentiation of hDPCs. Our findings uncovered the most affected odontoblastic differentiation-related DNA methylation target sites and conserved transcription factor motifs between mouse and human, extending our understanding of the underlying molecular mechanisms during both primary and tertiary dentinogenesis.
2 |. MATERIALS AND METHODS
2.1 |. Cell culture and characterization
Preodontoblast cell (mDPC) were isolated from the first molars of postnatal day 0.5 (PN 0.5) Kunming mice and digested for 1 hour at 37°C in a solution of 3 mg/mL collagenase type I and 4 mg/mL of dispase. mDPC6T is a self-established preodontoblast cell line retaining most of the characteristics of primary cells from mouse dental papilla. hDPCs were isolated as reported before.18 Cells were isolated from six healthy human third molars (from 14 to 20 years old). When reached 90% confluence, the cells from different donors were digested and mixed. HEK293FT cells were purchased from Invitrogen (Invitrogen, Carlsba, CA). All types of the cells used in this study were grown in Dulbecco Modified Eagle Medium (DMEM, Gibco-BRL Life Technologies, Paisley, UK) with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 incubator.
To characterize the stem cell feature of hDPCs, a colony forming assay was carried out by culturing a single cell in a six-well plate and after 6 days a colony-like growth was observed (Supplemental Figure S1 A). Cells were then washed and fixed by 4% paraformaldehyde and stained with 0.1% Crystal Violet (C0775, Sigma-Aldrich, St. Louis, MO). Cluster with more than 50 cells was taken into account as a colony.
Flow cytometry assay was used to further characterize the hDPCs. The experiment were performed according to the protocol described before.18 Results showed that mesenchymal stem cell markers like CD44 (FITC, BioLegend, San Diego, CA) and CD90 (FITC, BioLegend) were dominantly positive in hDPCs population while CD34 (FITC, BioLegend) and CD45 (FITC, BioLegend), which are hematopoietic markers, showed negative signals (Supplemental Figure S1 B–E).
2.2 |. RG108 treatment
RG108, a novel DNA methyltransferase (DNMTs) inhibitor, was purchased from Selleck Chemicals (Houston, TX). The mDPC6T were plated in 6-well plates (Falcon, Franklin Lakes, NJ) at an initial density of 2 × 105 cells/well and started to treat with RG108 when the cells density approaching 50%. To determine the ideal dose of RG108 for following research, different concentrations (45 μM and 90 μM) were chosen, and the treatment lasted for 5 days according to the manufacturer’s protocol. The protein level of DNMT1 was used to evaluate the effect of RG108. After 5 days’ treatment of RG108, this group of mDPC6T represented a DNA methyltransferase inhibited state and were carried into odontoblastic differentiation assay while the other group of untreated cells were used in the same odontoblastic induction as control.
2.3 |. Odontoblastic differentiation assay
For odontoblastic differentiation assay, the treated and untreated mDPC6T or hDPCs were cultured in DMEM supplemented with 10% FBS, 50 mg/ml ascorbic acid (A4544, Sigma-Aldrich), 10 mM sodium β-glycerophosphate (G9422, Sigma-Aldrich), and 10 nM dexamethasone (D4902, Sigma-Aldrich). The cells cultured in DMEM supplemented with only 10% FBS were used as blank control. After 14 days’ induction, cells were prepared for alizarin red (A5533, Sigma-Aldrich) staining to determine the mineralization. RNA and protein were extracted on day 0, 1, 5, 7, 11, and 14 after induction for real-time reverse transcription-polymerase chain reaction (RT-PCR) and Western blot analysis.
2.4 |. Promoter DNA methylation microarray hybridization and data analyses
Primarily cultured mDPCs were maintained in odontoblastic induction medium for 9 days (D9) and were subjected to promoter DNA methylation microarray (MM8 RefSeq Promoter Microarray, Roche NimbleGen, Basel, Switzerland) along with mDPCs not induced (D0). Sample preparation, labeling, hybridization to promoter microarray, array scanning, and raw data cleaning normalization were performed by Aksomics Company (Shanghai, China). Tissue-specific DNA methylated regions were obtained by BEDTools19 if such methylated regions are only positive in D0 or D9 samples. Motif finding was performed by HOMER.20 To display the results in UCSC browsers (http://genome.ucsc.edu/), coordinates for all the DNA methylated regions were converted to MM9 genome build.
The total number of methylated promoters in all mDPCs samples was divided into three bins: low CpG density promoters (LCP), intermediate CpG density (ICP), and high CpG density (HCP) as reported before.21 And the presence of methylation in each promoter region were recorded in Supplementary table.
2.5 |. Bisulfite conversion, methylation-specific PCR (MS-PCR) and sequencing
To check if the demethylation in the promoter of KLF4 also happens during odontoblastic differentiation of hDPCs, the promoter of human KLF4 was chosen for further experiment. For methylation-specific PCR (MS-PCR), methylation-specific primers for KLF4 promoter were designed using MethyPrimer program.22 Total genomic DNA from D0 and D5 hDPCs (before the completion of odontoblastic differentiation) was isolated and treated with EpiTect Bisulfite kit (QIAGEN, Hilden, Germany). MS-PCR for CpG islands was performed with EpiTect® MSP (QIAGEN). And CpG index was calculated as the ratio of methylated DNA to the sum of methylated and unmethylated DNA product. To further validate the CpG site(s) that underwent demethylation during odontoblastic differentiation, the promoter regions of KLF4 was cloned using the converted genomic from D0 and D5, and shuttled into pJet1.2/blunt using CloneJET PCR Cloning Kit (ThermoFisher, Waltham, MA). Three colonies from each group were randomly picked and subjected to Sanger Sequencing (Sangon Biotech, China). Methylated sites were displayed with filled circles and unmethylated sites with hollow circles.
2.6 |. Plasmids construction and transfection
For overexpression of SP1, plasmid coding the complementary DNA (cDNA) of SP1(NM_138473) was obtained from OriGene (OriGene, Rockville, MD). ORF of SP1 was then subcloned into pcDNA3.1(+) (Invitrogen) using Phusion® High-Fidelity DNA Polymerase (NEB, Ipswich, MA), and verified with Sanger sequencing (Sangon Biotech, China). Promoter region of KLF4 was cloned using Phusion® High-Fidelity DNA Polymerase (NEB) into pGL3-Basic (Promega, Fitchburg, WI). Reporter plasmid for KLF4 or empty pGL3-Basic plasmid were maintained in M.SssI (NEB) for in vitro DNA methylation according to manufacturer’s protocol. Each of the putative binding sites of SP1 in the promoter region of KLF4 was deleted using overlapping PCR.
Transfection experiment for overexpression of SP1 or dual-luciferase assays was performed using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were subjected to RNA extraction, protein extraction, or dual-luciferase assay. For luciferase assay, cells were lyzed with lysis buffer provided by Dual-Luciferase® Reporter Assay kit (Promega), and the lysis was subjected to Luciferase Assay System. Triplicate wells were analyzed. And relative luciferase activity was normalized by the ratio of firefly luciferase activity to Renila luciferase activity in the control group (transfected with pcDNA3.1(+), pGL3-Basic, and pRL-TK). To evaluate the effect of DNA methylation in the KLF4 promoter, methylated or unmethylated KLF4 promoter reporter plasmid was co-electroporated into HEK293FT cells with pcDNA3.1(+) or pcDNA3.1-SP1 plasmid and pRL-TK using Amaxa™ Nucleofector™ II (Lonza, Basel, Switzerland) (Cell Line Nucleofector® Solution V, Program Q-001).
2.7 |. Real-time RT-PCR
The total RNA was isolated from cultured cells using the TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed using the RT Premix kit (Takara Bio, Inc., Tokyo, Japan) according to the manufacturer’s instructions. Real-time RT-PCR was performed in an ABI 7900HT Real-time PCR System (Applied Biosystems, Foster City, CA) with SYBR Green Master (Rox) (Roche, Penzberg, Upper Bavaria, Germany). The primers used for Real-time RT-PCR were list as follow: Klf4, 5′-GGGAAGTCGCTTCATGTGAGAG-3′ (forward) and 5′-GCGGGAAGGGAGAAGACACT-3′ (reverse); Dmp1, 5′-CTGTCATTCTCCTTGTGTTCCTTTG-3′ (forward) and 5′-CAAATCAC CCGTCCTCTCTTCA-3′(reverse); Dspp, 5′-ATCATCAGCCAGTCAGAAGCAT-3′ (forward) and 5′-TGCCTTTGT TGGGACCTTCA-3′ (reverse); β-actin 5′-CCTGAGGCTCTTTTCCAGCC-3′ (forward) and 5′-TAGAGGTCTTTACGGATGTCAACGT-3′ (reverse).
Expression of each gene were defined with the threshold cycle (Ct), and relative expression levels were calculated by using the 2−ΔΔCt method after normalization with reference to expression of β-actin. The gene expression ratio was shown as the mean ± standard error of the mean (SEM) from three independent experiments.
2.8 |. Western blot
Cultured cells were collected on specific days and lysed in radioimmunoprecipitation assay (RIPA) buffer. The concentration of total protein in the supernatant was measured by using the bicinchoninic acid (BCA) Protein Assay Kit (Pierce Biotechnology, Rockford, IL). Equal amount protein was loaded and separated by 8% polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Roche). Primary antibodies used were list as below: anti-DMP1 (a kind gift from Professor Chunlin Qin at the Texas A&M Health Science Center),23 anti-SP1 (1:1000, mouse, Santa Cruz, Dallas, TX), anti-DNMT1 (1:1000, rabbit, Santa Cruz), anti-KLF4 (1:2000, rabbit, Proteintech, Rosemont, IL), and anti-DSP (1:1000, rabbit, Santa Cruz).
2.9 |. Chromatin immunoprecipitation quantitative PCR
Chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) was performed as described previously.24 Briefly, equal number of (about 1 × 107) hDPCs on day 0 and day 7 after odontoblastic induction were chemically cross-linked by 2% formaldehyde in room temperature followed by glycine treatment. Cells were resuspended in cell lysis buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, and 0.5% NP-40, 0.5 mM dithiothreitol [DTT]) and diluted with dilution buffer (150 mM NaCl, 16.7 mM Tris, pH 7.5, 3.3 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], and 0.5% Na-Doc) to get about 1 × 106 cells per ChIP. After sheared with sonication, DNA was incubated with Dynabeads (Life Technologies, Carlsba, CA) pretreated by SP1 antibody (Santa Cruz, 5 μg per 100 μ Dynabeads) or IgG Dynabeads (Life Technologies) as control overnight. Beads were then washed sequentially with low- and high-salt wash buffer, LiCl wash buffer, and TE buffer. DNA was eluted using SDS solution and reverse crossed with 5 M NaCl. After purification, DNA was subjected to qPCR using primer as listed below. For validation of SP1 in the 75 bp upstream of KLF4 TSS, we used KLF4–75_F: 5′-GGACCACCACTGACAGGTC-3′, KLF4–75_R: 5′-AGGTCCAGGAGATCGTTGAA-3′; for off-target region where no SP1 motif was spotted, we chose primer set: KLF4-Neg_F: 5′-ACTCGCCTTGCTGATTGTCT-3′ and KLF4-Neg_R: 5′-AATTGGCCGAGATCCTTCTT-3′. ChIP-qPCR was replicated for three times, and fold enrichment for IgG, and Anti-SP1 and off-target control on day 0 and day 9 were normalized to the fold change of input on day 0.
2.10 |. Statistical analysis
All values were represented as the mean ± standard error of the mean (SEM). Statistical significance was determined using the unpaired t-test. P < 0.05 was considered statistically significant.
3 |. RESULTS
3.1 |. Inhibition of DNA methyltransferase enhances the efficiency of odontoblastic differentiation
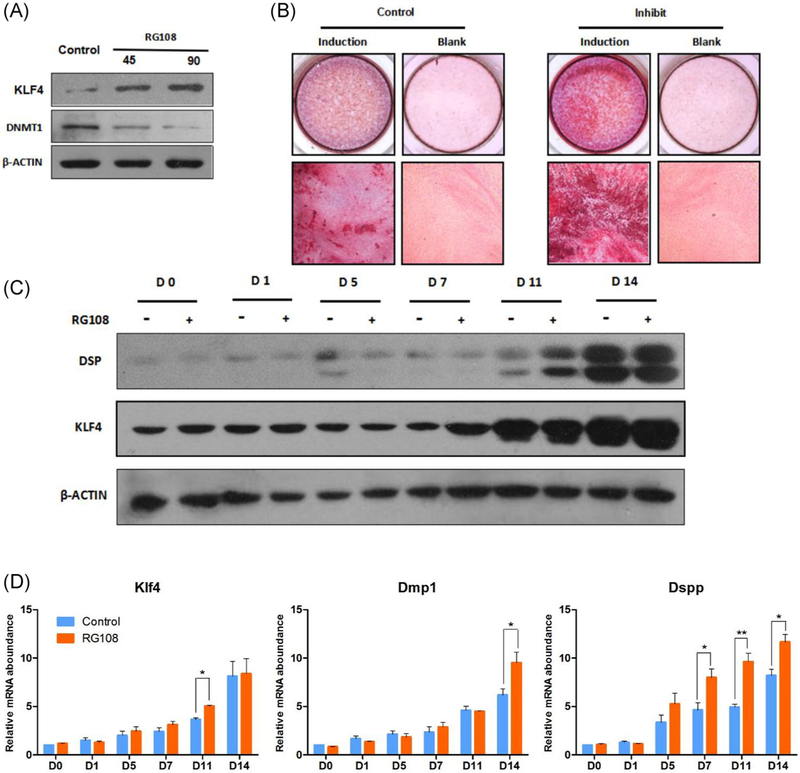
To investigate whether DNA methylation state could affect odontoblastic differentiation, RG108 was used to treat the self-established preodontoblast cell line mDPC6T.25 For the reason that Klf4 is a very important transcription factor and has been proven to promote odontoblastic differentiation in our previous studies, we analyzed the effect of RG108 on Klf4 along with other markers.5,26 RG108 obviously decreased the protein level of DNMT1, which is responsible for maintenance of DNA methylation, and increased the protein level of KLF4 in a dose-dependent manner (Figure 1A). Then the RG108 treated and untreated cells were carried into odontoblastic differentiation assay. After 14 days’ induction, the RG108 treated cell showed enhanced odontogenic potential against untreated cell in Alizarin red staining (Figure 1B). The protein level of KLF4 and DSP during odontoblastic differentiation process was also higher in RG108 treated cells compared with that in untreated cells, especially on day 11 (Figure 1C). On mRNA level, the qRT-PCR assay gave consistent results with the protein level. Along with the odontoblastic differentiation process, Klf4 and marker gene Dspp and Dmp1 showed uptrend in both RG108 treated and untreated cells, and the expression level was obviously higher in RG108 treated group (Figure 1D). All these data above gave a brief evidence to support that inhibition of DNA methyltransferase enhances the efficiency of odontoblastic differentiation of preodontoblast cell.
FIGURE 1.
Inhibition of DNA methyltransferase enhances the efficiency of odontoblastic differentiation. A, After treatment of RG108, protein level of DNMT1 and KLF4 were measured with Western blot. B, RG108 treated and untreated cells were cultured in odontoblastic differentiation medium or normal growth medium for 14 days. Mineralization was determined by alizarin red staining. On induction day of 0, 1, 5, 7, 11, and 14, RNA and protein were extracted from both RG108 treated and untreated cells. The protein level, C, of KLF4, DSP, and mRNA level, D, of Klf4, Dmp1, and Dspp were determined by Western blot and real-time RT-PCR respectively. All data were based on three independent experiments: *P < 0.05. **P < 0.01. RT-PCR, real-time reverse transcription-polymerase chain reaction
3.2 |. SP1/KLF4 binding motif is enriched in the promoter regions and demethylated during odontoblastic differentiation
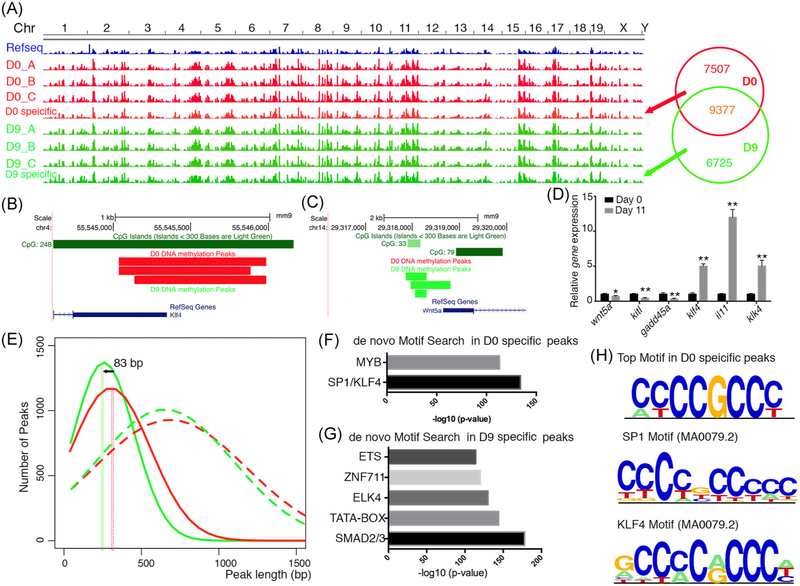
MeDIP-on-chip assay was performed to identify DNA methylation changes around promoter regions during odontoblastic differentiation. All the peaks from each replicate in each group were concatenated and merged to obtain the methylated regions in odontoblast before (day 0, D0) and after (day 9, D9) differentiation.
Subtraction of genomic intervals from the two groups revealed there were 7507 D0-specific DNA methylated peaks which were demethylated in D9, while 6725 DNA regions that were methylated after odontoblastic differentiation (Figure 2A, Supplementary table). Of all the target genes of each peak, transcription level of some genes that were mentioned as tooth/odontoblasts related were analyzed. Wnt5a, Kitl, and Gadd45a were downregulated with their promoter region methylated during odontoblastic differentiation; Klf4, Il11, and Klk4 were upregulated with their promoter regions demethylated during differentiation (Figure 2B–D). Further analysis of the peak lengths in each stage revealed that the methylated region after odontoblastic differentiation were more compacted than those in cells before differentiation (Figure 2E).
FIGURE 2.
SP1/KLF4 binding motif is enriched in the promoter regions and demethylated during odontoblastic differentiation. A, Genome-wide changes of DNA methylation during odontoblastic differentiation. B, UCSC genome browser showing the location of methylated peaks in the promoter of Klf4. C, UCSC genome browser showing the location of methylated peaks in the promoter of Wnt5a. D, Relative expression level for example genes whose promoters were affected by DNA methylation. E, Average diagram of DNA methylation peaks length before and after odontoblastic differentiation. F, Enriched de novo motif in D0-specific DNA methylation peaks. G, Enriched de novo motifs in D9-specific DNA methylation peaks. H, Comparison of top enriched motif in D0 with known SP1 and KLF4 motifs (weighted matrix)
To further characterize these D0-specific or D9-specific regions, de novo motif enrichment analysis was performed in each group. The top motif enriched in D0-specific DNA methylated region (present in ~65% of the D0-specific peaks) corresponded to the SP1/KLF4 motif based on their ChIP-seq data in other cell types (Figure 2F,H). We also noticed the presence of this most enriched motif in the promoter regions of Klf4, Klk4, and Il11 as tested above. However, the top motif enriched in D9-specific DNA methylated regions did not correspond to any specific binding motifs reported in ChIP-seq results or 2018 releases of JASPAR27 (Figure 2G). There were several partially matched possible motifs including SMAD2/3, KLF5, etc. These findings revealed that although demethylation enhanced odontoblastic differentiation, a small fraction of tooth-related genes’ expression patterns correlated with the promoter DNA methylation. Also, a small set of TF motifs were affected by such odontoblastic differentiation-related DNA methylation, such as SP1/KLF4.
3.3 |. SP1 promotes KLF4 expression in human dental pulp cell through binding to a DE-methylated site during odontoblastic differentiation
KLF4 is critical for maintaining the stem cell features, and we previously reported Klf4 played important roles in odontoblastic differentiation.5,26 Driven by the methylation array results of mouse odontoblastic differentiation model, we analyzed the DNA methylation pattern of human KLF4 during odontoblastic differentiation of hDPCs.
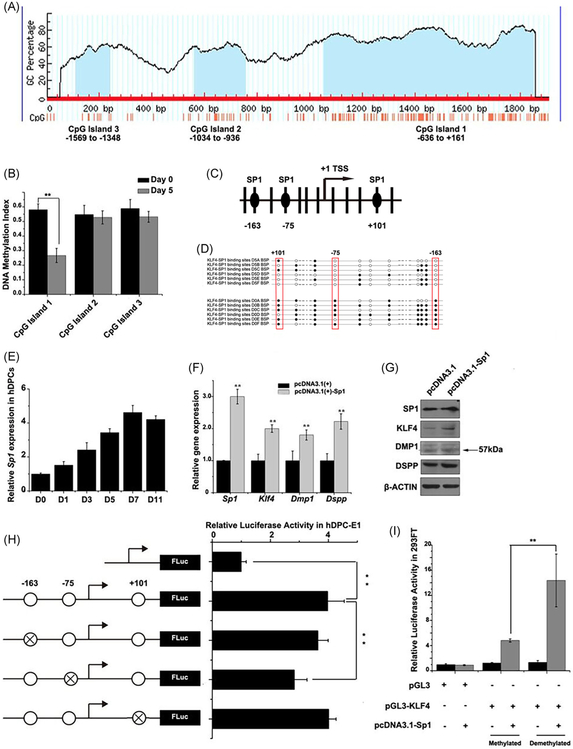
Three CpG islands were found in the promoter region of human KLF4 (RefSeq NC_000009.12 Chromosome 9 Reference GRCh38.p12 Primary Assembly; Figure 3A). MS-PCR revealed the first CpG island exhibited demethylation during odontoblastic differentiation (Figure 3B). We used BSP to footmap which CpG site(s) exhibited demethylation in CpG island 1 of KLF4, and found three CpG sites which were demethylated during odontoblastic differentiation, and interestingly, these three demethylated CpG sites were predicted to be the potential binding site of SP1 (Figure 3C,D).
FIGURE 3.
SP1 promotes KLF4 expression in human dental stem cell through binding to a demethylated site during odontoblastic differentiation. A, There were three CpG islands in the promoter region of human KLF4 which were marked in blue. B, The methylation-specific PCR revealed the first CpG island exhibited demethylation during odontoblastic differentiation while the other two exhibited no change. C, D, To further characterize the pattern of the first CpG island, Bisulfite Genomic Sequence was performed and found three CpG sites within the first CpG island that show demethylated during odontoblastic differentiation and revealed to be binding sites of SP1. E, We analyzed the expression pattern of SP1 during odontoblastic differentiation and found a uptrend along with days. F, Overexpression of SP1 had positive effects on KLF4 and odontoblastic differentiation marker genes including DMP1 and DSPP in both mRNA level, G, and protein level. H, Series luciferase assay were used to validate the three CpG sites, and binding site in − 75 revealed to be the most responsible one as mutation in this site significantly decrease the luciferase activity. I, In vitro methylated KLF4 promoter vector could completely abolish the reporter gene activity and following luciferase assay revealed that SP1 upregulated demethylated KLF4 promoter activity more than the methylated promoter. All data were based on three independent experiments: **P < 0.01. PCR, polymerase chain reaction
Our previous finding in mouse model showed SP1 can promote odontoblastic differentiation of mouse dental papilla cells.28 We validated the expression level of SP1 during odontoblastic differentiation of hDPCs as well and found uptrend of SP1 during the process (Figure 3E). Also, overexpression of human SP1 through transiently transfection in hDPCs, could upregulate KLF4 expression and promote odontoblastic differentiation (Figure 3 F, G).
We further validated the binding sites of SP1 in CpG island 1 using luciferase assay, and found mutation in binding site in − 75 significantly decreased the luciferase activity (Figure 3H). And validated such binding using ChIP-qPCR on day 0 and day 7 of hDPCs after odontoblastic induction. We found significant increase of KLF4 binding enrichment near − 75 on day 7 compared with day 0 (Supplemental Figure S2).
In vitro methylation of KLF4 promoter vector by CpG Methyltransferase (M.SssI) completely abolished the KLF4 promoter reporter gene activity. Luciferase assay following cotransfection of methylated/demethylated KLF4 and SP1 expression plasmid, revealed that the SP1 upregulated demethylated KLF4 promoter activity more than the methylated promoter (Figure 3I). These data consistently proved that demethylation of KLF4 promoter is essential for effective binding of SP1 and subsequent transcriptional regulation on KLF4 during odontoblastic differentiation of hDPCs.
4 |. DISCUSSION
The odontoblasts are a sort of terminally differentiated cells and play critical roles in the dentinogenesis.29 The differentiation process of odontoblasts is also a very important component of tooth development and has been an object of research for many years.2 Lots of studies of odontoblastic differentiation focus on signaling pathways and transcription factors. BMP, FGF, and WNT signaling pathways and many transcription factors such as Sox2, Klf4, and Sp1 have been reported to participate in odontoblastic differentiation process of tooth-related cells in both human and mouse models.5,28,30–33 Besides, many epigenetic factors have been reported to regulate tooth development. Mir-143 and mir-145 have been proven to regulate odontoblastic differentiation in a posttranscriptional manner.26 DNA methylation and histone modification have also been reported to have some effects in tooth-related cells.14–16,34 Recent study indicated a potential role of DNA methylation in odontoblastic differentiation of hDPCs as knockdown of TET1 would suppress odontoblastic differentiation through attenuated hydroxymethylation and transcription of FAM20C.17 In the present study, we explained the effect of DNA methylation on odontoblastic differentiation in a more detailed way, and found that SP1/KLF4 binding motif is responsible for the regulatory effect of SP1 on KLF4 and governed by DNA methylation mechanism during odontoblastic differentiation.
DNMT1 is an enzyme which is responsible for the maintenance of DNA methylation level in the offspring of a given cell.35 We suppressed the expression level of DNMT1 to see whether this action would have some kind of effect. And we found that the suppression of DNMT1 leads to a promotion on odontoblastic differentiation potential of mouse dental papilla cells and upregulation of Klf4, which is a stemness-related transcription factor, as well.36 Given that Klf4 had been proven to promote odontoblastic differentiation in our previous study, we hypothesized that DNA methylation could affect odontoblastic differentiation and participate in the regulation of Klf4 during the process.
Therefore, we tested the DNA methylation changes around promoter regions during odontoblastic differentiation with the help of MeDIP-on-chip assay and found a genome-wide changes from the initial stage to differentiated stage. Among these changes we noticed some genes that were mentioned tooth/odontoblasts related, such as Wnt5a, Kitl, Gadd45a, Klf4, Il11, and Klk4.37–39 By comparing with ChIP-seq data in other cell types, the SP1/KLF4 motif revealed to be the most enriched one in the DNA methylated regions in the initial stage and showed demethylation after odontoblastic differentiation in mouse model. These results were consistent with our previous findings that SP1 could transactivate Klf4 during mouse odontoblastic differentiation and led us to ask whether such DNA-methylation-dependent regulatory relationship was conserved in hDPCs. By analyzing the DNA methylation pattern of KLF4 during odontoblastic differentiation of human dental cells, three CpG islands were spotted in the promoter region of KLF4 in human genome, among which the island 1 showed obvious demethylation during odontoblastic differentiation and happened to have three predicted binding sites of SP1 which were also demethylated during odontoblastic differentiation. The following luciferase assay and in vitro methylation assay validated the necessity of demethylation of CpG island 1 in KLF4 promoter region for the binding and function of SP1.
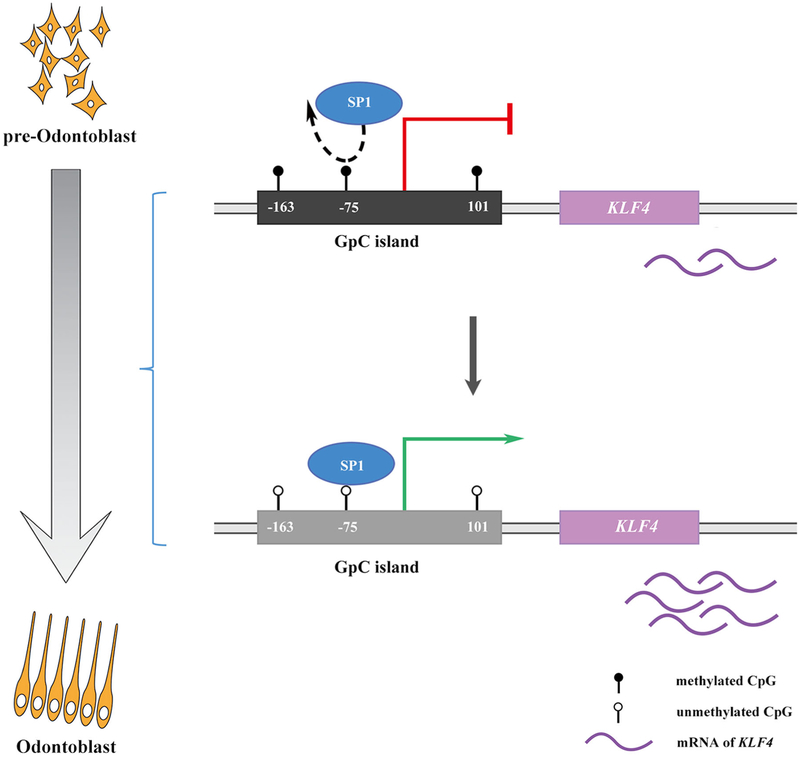
In our previous study, we proved that Sp1 and Klf4 could act as a competing endogenous RNAs pair in odontoblastic differentiation which allowed them to have a similar expression pattern during the process.40 In the current study, we confirmed our hypothesis and proved that SP1 regulated KLF4 through binding site lying in a CpG island in KLF4′s promoter region which demethylated during odontoblastic differentiation thus enhanced the efficiency of SP1′s binding and transcriptional regulation on KLF4 (Figure 4). We regard the present work as a complementary to the regulation networks based on Klf4 during odontoblastic differentiation.
FIGURE 4.
SP1 regulates KLF4 via SP1 binding motif governed by DNA methylation during odontoblastic differentiation of human dental pulp cells. Schematic diagram showing the regulation effect of SP1 on KLF4 based on DNA methylation mechanism
Supplementary Material
ACKNOWLEDGMENT
The study was funded by grants from National Natural Science Foundation of China (No: 81420108011; 81271099) to Prof. Zhi Chen, Young Elite Scientist Sponsorship Program by CAST (No. 2017QNRC001), Natural Science Foundation of Hubei Province (No. 2017CFB515), and National Natural Science Foundation of China (No. 81771057) to Dr. Huan Liu.
Funding information
National Natural Science Foundation of China, Grant/Award Numbers: 81271099, 81420108011, 81771057; Natural Science Foundation of Hubei Province, Grant/Award Number: 2017CFB515; Young Elite Scientist Sponsorship Program by CAST, Grant/Award Number: 2017QNRC001
Footnotes
CONFLICT OF INTERESTS
The authors declare that they have no conflict of interests.
DATA SHARING AND DATA ACCESSIBILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127(8):1671–1679. [DOI] [PubMed] [Google Scholar]
- 2.Balic A, Thesleff I. Tissue interactions regulating tooth development and renewal. Curr Top Dev Biol. 2015;115:157–186. 10.1016/bs.ctdb.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 3.Lim WH, Liu B, Cheng D, et al. Wnt signaling regulates pulp volume and dentin thickness. J Bone Miner Res. 2014;29(4):892–901. 10.1002/jbmr.2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67(2):111–123. [DOI] [PubMed] [Google Scholar]
- 5.Lin H, Liu H, Sun Q, Yuan G, Zhang L, Chen Z. KLF4 promoted odontoblastic differentiation of mouse dental papilla cells via regulation of DMP1. J Cell Physiol. 2013;228(10):2076–2085. 10.1002/jcp.24377 [DOI] [PubMed] [Google Scholar]
- 6.Bae JM, Clarke JC, Rashid H, et al. Specificity protein 7 is required for proliferation and differentiation of ameloblasts and odontoblasts. J Bone Miner Res. 2018;33(6):1126–1140. 10.1002/jbmr.3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoda S, Suda N, Kitahara Y, Komori T, Ohyama K. Delayed tooth eruption and suppressed osteoclast number in the eruption pathway of heterozygous Runx2/Cbfa1 knockout mice. Arch Oral Biol. 2004;49(6):435–442. 10.1016/j.archoralbio.2004.01.010 [DOI] [PubMed] [Google Scholar]
- 8.Latchman DS. Transcription factors: an overview. Int J Exp Pathol. 1993;74(5):417–422. [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A Perceptions of epigenetics. Nature. 2007;447(7143):396–398. 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto T, Furusawa C, Kaneko K. Pluripotency, differentiation, and reprogramming: a gene expression dynamics model with epigenetic feedback regulation. PLoS Comput Biol. 2015;11(8): e1004476. 10.1371/journal.pcbi.1004476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsankov AM, Gu H, Akopian V, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518(7539):344–349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boland MJ, Nazor KL, Loring JF. Epigenetic regulation of pluripotency and differentiation. Circ Res. 2014;115(2):311–324. 10.1161/CIRCRESAHA.115.301517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteller M, Risques RA, Toyota M, et al. Promoter hyper-methylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61(12):4689–4692. [PubMed] [Google Scholar]
- 14.Gopinathan G, Kolokythas A, Luan X, Diekwisch TG. Epigenetic marks define the lineage and differentiation potential of two distinct neural crest-derived intermediate odontogenic progenitor populations. Stem Cells Dev. 2013; 22(12):1763–1778. 10.1089/scd.2012.0711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin W, Ye X, Fan H, Bian Z. Methylation state of the EDA gene promoter in Chinese X-linked hypohidrotic ectodermal dysplasia carriers. PLoS One. 2013;8(4):e62203. 10.1371/journal.pone.0062203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Sun K, Shen Y, et al. DNA methylation is critical for tooth agenesis: implications for sporadic non-syndromic anodontia and hypodontia. Sci Rep. 2016;6:19162. 10.1038/srep19162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Yi B, Feng Z, Meng R, Tian C, Xu Q. FAM20C could be targeted by TET1 to promote odontoblastic differentiation potential of human dental pulp cells. Cell Prolif. 2018;51(2): e12426. 10.1111/cpr.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Lin C, Zhang J, et al. Quaking promotes the odontoblastic differentiation of human dental pulp stem cells. J Cell Physiol. 2018;233(9):7292–7304. 10.1002/jcp.26561 [DOI] [PubMed] [Google Scholar]
- 19.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–766. 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 22.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. [DOI] [PubMed] [Google Scholar]
- 23.Qin C, Brunn JC, Cook RG, et al. Evidence for the proteolytic processing of dentin matrix protein 1. Identification and characterization of processed fragments and cleavage sites. J Biol Chem. 2003;278(36):34700–34708. 10.1074/jbc.M305315200 [DOI] [PubMed] [Google Scholar]
- 24.Hitchler MJ, Rice JC. Genome-wide epigenetic analysis of human pluripotent stem cells by ChIP and ChIP-Seq. Methods Mol Biol. 2011;767:253–267. 10.1007/978-1-61779-201-4_19 [DOI] [PubMed] [Google Scholar]
- 25.Schirrmacher E, Beck C, Brueckner B, et al. Synthesis and in vitro evaluation of biotinylated RG108: a high affinity compound for studying binding interactions with human DNA methyltransferases. Bioconjug Chem. 2006;17(2):261–266. 10.1021/bc050300b [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Lin H, Zhang L, et al. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J Biol Chem. 2013;288(13):9261–9271. 10.1074/jbc.M112.433730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. 10.1093/nar/gkh012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Lin H, Liu H, Zhang L, Yuan G, Chen Z. SP1 promotes the odontoblastic differentiation of dental papilla cells. Dev Growth Differ. 2015;57(5):400–407. 10.1111/dgd.12221 [DOI] [PubMed] [Google Scholar]
- 29.Mitsiadis TA, Rahiotis C. Parallels between tooth development and repair: conserved molecular mechanisms following carious and dental injury. J Dent Res. 2004;83(12):896–902. 10.1177/154405910408301202 [DOI] [PubMed] [Google Scholar]
- 30.Aurrekoetxea M, Irastorza I, García-Gallastegui P, et al. Wnt/beta-catenin regulates the activity of epiprofin/Sp6, SHH, FGF, and BMP to coordinate the stages of odontogenesis. Front Cell Dev Biol. 2016;4:25. 10.3389/fcell.2016.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik Z, Alexiou M, Hallgrimsson B, Economides AN, Luder HU, Graf D. Bone morphogenetic protein 2 coordinates early tooth mineralization. J Dent Res. 2018;97(7):835–843. 10.1177/0022034518758044 [DOI] [PubMed] [Google Scholar]
- 32.Vidovic-Zdrilic I, Vining KH, Vijaykumar A, Kalajzic I, Mooney DJ, Mina M. FGF2 enhances odontoblast differentiation by alphaSMA( + ) progenitors in vivo. J Dent Res. 2018;97:22034518769827–1177. 10.1177/0022034518769827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Zhao Y, Liu X, Chen Y, Liu P, Zhao L. Effect of SOX2 on odontoblast differentiation of dental pulp stem cells. Mol Med Rep. 2017;16(6):9659–9663. 10.3892/mmr.2017.7812 [DOI] [PubMed] [Google Scholar]
- 34.Hoang M, Kim JJ, Kim Y, et al. Alcohol-induced suppression of KDM6B dysregulates the mineralization potential in dental pulp stem cells. Stem Cell Res. 2016;17(1):111–121. 10.1016/j.scr.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28(10):2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 37.Nieminen P, Morgan NV, Fenwick AL, et al. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am J Hum Genet. 2011;89(1):67–81. 10.1016/j.ajhg.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith CEL, Kirkham J, Day PF, et al. A fourth KLK4 mutation is associated with enamel hypomineralisation and structural abnormalities. Front Physiol. 2017;8:333. 10.3389/fphys.2017.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang L, Chen M, He L, et al. Wnt5a regulates dental follicle stem/progenitor cells of the periodontium. Stem Cell Res Ther. 2014;5(6):135. 10.1186/scrt525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Liu H, Lin H, et al. Sp1 is a competitive endogenous RNA of Klf4 during odontoblast differentiation. Int J Biochem Cell Biol. 2017;85:159–165. 10.1016/j.biocel.2017.02.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.