Abstract
KRAS mutations, which are the main cause of the pathogenesis of lethal pancreatic adenocarcinomas, impair the functioning of the GTPase subunit, thus rendering it constitutively active and signaling intracellular pathways that end with cell transformation. In the present study, the AsPC-1 cell line, which has a G12D-mutated KRAS gene sequence, was utilized as a cellular model to test peptide nucleic acid-based antisense technology. The use of peptide nucleic acids (PNAs) that are built to exhibit improved hybridization specificity and have an affinity for complementary RNA and DNA sequences, as well as a simple chemical structure and high biological stability that affords resistance to nucleases and proteases, enabled targeting of the KRAS-mutated gene to inhibit its expression at the translation level. Because PNA-based antisense molecules should be capable of binding to KRAS mRNA sequences, PNAs were utilized to target the mRNA of the mutated KRAS gene, a strategy that could lead to the development of a novel drug for pancreatic cancer. Moreover, it was demonstrated that introducing new PNA to cells inhibited the growth of cancer cells and induced apoptotic death and, notably, that it can inhibit G12D-mutated KRAS gene expression, as demonstrated by RT-PCR and western blotting. Altogether, these data strongly suggest that the use of PNA-based antisense agents is an attractive therapeutic approach to treating KRAS-driven cancers and may lead to the development of novel drugs that target the expression of other mutated genes.
Keywords: pancreatic cancer, KRAS, peptide nucleic acid, peptide nucleic acids, drug candidate
Introduction
Proto-oncogenic RAS mutations are found in the most lethal cancers, as their gain-of-function phenotype underlies the pathogenesis of up to 30% of all human cancers, and these tumors are associated with the worst prognoses (1,2). Such mutations have been implicated in the pathogenesis of numerous cancers, including approximately 97% of pancreatic ductal adenocarcinoma, 52% of colorectal adenocarcinomas, 32% of lung adenocarcinomas (1,3,4), and lower percentages of other cancers (5,6). As an important oncogenic driver in cancer malignancy, KRAS is the most frequently mutated isoform among the three human RAS genes that encode highly homologous RAS proteins-namely, KRAS, NRAS, and HRAS (7). In pancreatic cancer, oncogenic mutations of the KRAS gene are major events that produce a permanent, active KRAS protein which triggers various intracellular pathways involved in malignancy (8). Under normal physiological conditions, the KRAS gene encodes the GTPase transductor protein, which plays a key role in the signal transduction cascades that regulate cell growth, proliferation, migration, differentiation, survival, and apoptosis (7–9). Throughout the regular KRAS GTPase cycle, the KRAS protein switches between its inactive and active forms by binding to guanosine diphosphate (GDP) and guanosine triphosphate (GTP), respectively (10,11). The KRAS protein predominantly exists in a GDP-bound inactive form, but upon stimulation by growth factors it undergoes conformational changes, binds to GTP, and becomes active. Active KRAS then activates a range of molecules that mediate the transmission of signals from the cell surface to the nucleus, leading eventually to cellular processes essential for survival and proliferation (7). Mutations in the KRAS oncogene, which play a pivotal role in driving the progression of pancreatic cancer, occur most frequently at codons 12, 13, and 61. The most common mutation occurs at codon 12 (G12D); it results in the amino acid substitution of aspartate for glycine, interfering with GTP hydrolysis and thereby increasing the proportion of active, GTP-bound KRAS, rendering it constitutively active (8). Despite tremendous effort and decades of intensive studies of KRAS activation, the KRAS mutant has remained difficult to treat with drugs; as a result, a targeted therapy exists currently for only one KRAS mutation (G12C), which is found primarily in non-small-cell lung cancer (12). Much attention has been focused on targeting signaling cascades downstream from KRAS, in particular the PI3K, MAPK, and RAL-GEF pathways (7,13,14). Direct targeting of the GTP-binding pocket of the mutant G12D KRAS protein has proved unsuccessful (7). Moreover, there are no well-defined druggable sites in the surface topology of this protein that are vulnerable to high-affinity, small-sized antagonists, and thus, to date, efforts to inhibit the functioning of mutant G12D KRAS have not translated into clinical benefits (15). Of note, a study that used genetic engineering techniques to shut down KRAS expression in cancer cells showed that PI3K may compensate for KRAS loss and activate MAP-kinase signaling (9,16). Some progress has been made with the treatment of pancreatic cancer using small interfering RNA (siRNA), directed against G12D-mutated KRAS; siRNA has been assessed in vitro and in vivo and shown to induce a significant decrease in KRAS levels, leading to an inhibition of cell proliferation (17). In another study, antisense specific to the KRAS gene sequence has been shown to selectively deplete KRAS mRNA, as well as its protein product, leading eventually to the inhibition of mutant KRAS cell proliferation (18).
In light of the current interest in and success with therapeutic antisense inhibitors of KRAS, we report here a potential candidate treatment for pancreatic cancer that uses PNA-based antisense. In general, PNA applications for developing specific therapeutic agents that can target and inhibit the expression of certain genes (19,20) may prove to be useful. PNAs are synthetic, single-stranded oligonucleotide analogues containing normal nucleobases, covalently attached to backbone of a polyamide structure that consists of repeating N-(2-aminoethyl) glycine units (21–23). They are easily and quickly synthesized and have a simple chemical structure that is resistant to degradation by nucleases and proteases. In addition, they exhibit improved hybridization characteristics, with high specificity and an affinity to complementary sequences of RNA and DNA (24), and they can even differentiate between similar sequences at the level of single-base mismatches (25,26). Moreover, PNA binding to single-stranded RNA or DNA is much stronger than that of complementary DNA sequences (27,28). Due to these advantageous characteristics, for this study, various PNA molecules were hypothesized to be promising therapeutic or diagnostic antisense or antigene molecules and were utilized to regulate gene expression (29–34). When applied as antisense agents, PNAs have been shown to bind to the messenger RNA (mRNA) of the targeted gene and inhibit translation activity (35). PNA-based antisense sequences used as antibacterial agents against key genes (i.e., encoding enzymes involved in the synthesis of DNA, RNA, the cell envelope, fatty acids, and proteins) have also induced inhibition of bacterial growth (22). Other PNA-based antisense sequences have been shown to inhibit the expression of specific genes in primary cells and in the cell lines of mice and humans (36). In contrast to the small interfering RNAs (siRNAs) involved in the endogenous mechanism that regulates gene expression by triggering the degradation of complementary mRNA molecules (37–39), PNA-based antisense sequences can inhibit specific mRNA molecules by means of splicing modulation, blocking, and transcription arrest (36).
Although they may have many advantages in terms of possible applications, the therapeutic use of PNAs remains limited, due to their aggregation, low water solubility, and weak intracellular penetration. To overcome these problems, PNAs can be chemically modified to produce a charged form, or they can be linked to a positively charged peptide such poly-L-lysine. If they are positively charged, however, they are attracted to the negatively charged cell membrane, whereas uncharged PNA molecules might be able to penetrate cells through endocytosis (28,29).
For the current study, we designed and synthetized PNA-based antisense molecules and investigated their ability to pass through the cellular membrane of AsPC-1 cells and their effects on cell growth, proliferation, and cell-cycle checkpoints. The results strongly indicate that the penetration of PNA-based antisense into cells caused apoptosis and arrested the cell cycle, and thus it should be considered for further development as a specific drug candidate for pancreatic cancer treatment.
Materials and methods
RNA preparation, RT-PCR, and the sequencing of the mutated KRAS gene from the AsPC-1 cell line
To ascertain that the G12D-mutated KRAS gene sequence within the AsPC-1 cell line was identical to the known sequence in the GenBank (Accession No. AF493917), we amplified and sequenced a fragment that harbored the G12D mutation. First, forward (KRAS2-F: 5′-TGACTGAATATAAACTTGTGGT-3′) and reverse (KRAS2-R: 5′-CTCATTGCACTGTACTCCTCTTG-3′) primers were designed and utilized to amplify the size of the amplicon (202 bp) that spans the KRAS G12D mutation, using the RT-PCR technique. To this end, total RNA was prepared from 80–90% confluent-trypsinized AsPC-1 cell culture, using GENZOL Tri RNA Pure Kit (Geneaid Biotech Ltd., Taiwan) in keeping with the manufacturer's instructions. The quantity and purity of the total RNA samples were assessed by ultraviolet spectroscopy with a DS-11 spectrophotometer (DeNovix, Inc.). One microgram of total RNA was subjected to a one-step RT-PCR using a Maxime RT-PCR premix kit (INtRON Biotechnology) containing a pellet of 10× RT-PCR buffer, dNTPs, an OptiScript RT-system, hot start i-StarTaq DNA Polymerase, and 0.5 nM KRAS-2 forward and reverse primers. The RT-PCR cycling parameters were as follows: reverse transcription at 45°C for 30 min, denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 1 min, elongation at 72°C for 1 min, and terminal elongation at 72°C for 5 min. The PCR products were electrophoresed in 1% agarose gel, and a DNA band of approximately 200 bp was excised from the gel and sent for sequencing to Hy-Labs Laboratories Ltd. The DNA was sequenced in two orientations. A 202-bp sequence was BLASTed against the NCBI source and found to be identical to the KRAS2 mRNA sequence AF493917.
PNA design, synthesis, and characterization
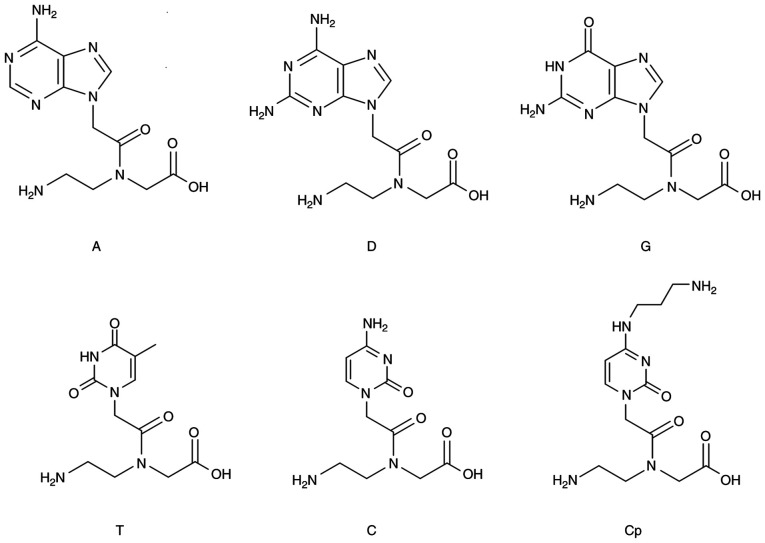
The three PNA-based molecules were designed to target the mutated gene fragment 5′-TGGAGCTGATGGCGTAG-3′. They were synthesized using solid-phase technology, as has been previously described (40–42). The MALDI-TOF MS technique was used to confirm that the synthesis was successful. Reverse-phase HPLC was used for purification and analysis, the wavelength of the UV detector was set to 260 nm, and a linear gradient of 10–25% ACN/water was applied. The chemical structures of the monomers/residues are shown in Fig. 1. The sequences of the three PNA-based molecules are as follows:
Figure 1.
Structures of the monomers used to build the antisense oligonucleotide polymers corresponding to the KRAS gene sequence. Monomer G binds the C nucleotide; monomer D binds the U or T nucleotide; monomer T binds the A nucleotide; monomer C binds the G nucleotide; and monomer Cp is a positively charged monomer that binds the G nucleotide.
PNA1: Composed of PNA-based monomers (A, C, T, G). Three lysine residues are attached at the N-terminal edge. The sequence is CTACGCCATCAGCTCCA-KKK.
PNA3: Monomer D replaces monomer A in targeting the U nucleotide. Three lysine residues are attached at the N-terminal edge. The sequence is CTDCGCCDTCDGCTCCD-KKK.
PNA14: Monomer Cp, a positively charged monomer, replaces monomer C. No lysine residues are linked to this PNA-based molecule. The sequence is CTDCpGCCDTCpDGCTCCpD.
Cell culture and viability tests
AsPC-1 cells were seeded in non-coated tissue culture flasks or well-plates in RPMI1640 medium, supplemented with 10% fetal calf serum and 1% penicillin-streptomycin (Biological Industries), at 37°C in a humidified atmosphere of 5% CO2 in air. The medium was changed three times a week, and the cells were split when they reached 80–90% confluency, using Trypsin EDTA according to the manufacturer's instructions (Biological Industries).
AsPC-1 cell permeability to PNA molecules
To evaluate the permeability of cells to the PNAs, we adapted the protocol recommended by the manufacturer with slight modifications, which are outlined below. The transfection of the PNAs was conducted using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). Initially, AsPC-1 cells were seeded overnight in a 24-well plate at a density of 5×105 per well, until they reached confluency between 90 and 95%. The cells were then treated with 0.5 ml of medium containing 0.5 or 2 µM of fluorescently labeled PNA-1 that was first prepared in 100 µl of Opti-MEM I medium. According to the manufacturer's instructions, each 1 µM of PNA was mixed with 2.5 µl of lipofectamine 2000 (Thermo Fisher Scientific, Inc.) and incubated for 20 min at room temperature before being dripped onto the cells. For the permeability tests, 100 µl of PNA1/lipofectamine mixture were mixed with 400 µl of RPMI1640 medium (without FCS), and the treated cells were incubated for 24 h at 37°C. The cells were washed three times in PBS buffer and fixed with 4% paraformaldehyde solution. They were then stained with DAPI stain for 5 min at 37°C, washed with PBS, and viewed through a Nikon Eclipse Ti2 fluorescent microscope (Nikon Corporation, Tokyo, Japan). The intracellular fluorescence was also measured using a plate reader with an excitation maximum of 493 nm and an emission maximum of 528 nm.
Dissociation curves and melting temperature (Tm) tests
Hybridization reactions were monitored using an Exicycler™ 96 PCR system designed for real-time qPCR (Bioneer, Korea), with qPCRBIO SyGreen Mix (PCR Biosystems Ltd.) and the following hybridization-dissociation protocol: 95°C for 30 sec, cooling to 20°C, incubation at 20°C for 5 min, and gradual reheating to 94°C, then a 1.0°C increase for 30 sec, and finally, incubation for 1 min at 25°C. The SyGreen signal was recorded incrementally (approximately every 20 sec) during the reheating phase, and the average fluorescence measurement during this time was reported. All of the hybridization reactions involved combinations of equal molars of (0.5 µM) PNA, complementary target oligonucleotides, and free PNAs (1.0 µM) in the presence of SyGreen Mix. Dissociation curves and Tm evaluations were obtained using Exicycler™ 96 PCR system analysis software. For controls, we used oligonucleotides without mutations. Three replications of each combination were run in two separate experiments.
Cytotoxicity of the PNA molecules
To evaluate PNA cytotoxicity, AsPC-1 cells were seeded overnight in a 24-well plate at a density of 5×105 per well and treated with 1 to 4 µM of PNAs or 10–20 µM of gemcitabine. The 100 µl PNA/lipofectamine mixture was mixed with 400 µl of RPMI1640 medium (without FCS), and the treated cells were incubated for 48 h at 37°C. The cells were then washed three times in PBS buffer, and viability was determined with a XTT Cell Proliferation Kit (Biological Industries) according to the manufacturer's instructions, using a Thermo Scientific Varioskan LUX Multimode Microplate Reader set at 450 nm and subtracted from the reference absorbance at 620 nm. The experiments were repeated independently three times.
Cell-cycle analysis
AsPC-1 cells were seeded in a 6-well plate at a density of 5×105 cells per well and incubated overnight at 37°C. The medium was changed and the cells were transfected with 2-µM PNAs. At 24 and 48 h after transfection, the cells were trypsinized and collected with the growth media, centrifuged, washed with PBS, and fixed with 70% ethanol for 1 h. Gemcitabine (20 µM) and siRNA (20 µM) served as positive controls; the siRNA was specific to the mRNA sequence of the mutant KRAS gene (5′-GUUGGAGCUGAUGGCGUAGdTdT and 5′-CUACGCCAUCAGCUCCAACdTdT). This was followed by incubation with 0.1% NP-40 for 5 min at 4°C and then with ice and 100 µg/ml of RNase for 30 min. Finally, a 50 µg/ml solution of propidium iodide (PI) was added for 20 min. Cell-cycle phase distributions were determined by flow cytometry, using a Navios flow cytometer (Beckman Coulter, Miami, FL).
Apoptosis assay using Annexin V/PI double staining
Apoptotic cell death was evaluated and quantified by flow cytometry with an Annexin V FITC Detection kit (Mebcyto® Apoptosis kit, MBL) (used according to the manufacturer's recommended procedure) and a PI Double Staining kit. To differentiate between apoptosis and necrosis, the cells were stained with FITC-labeled Annexin V and PI.
The AsPC-1 cells (5×105) were seeded in a six-well plate. The next day, the cells were transfected with PNAs and incubated for 48 h. Adherent and floating cells were both collected in order to detect early and late apoptosis events. Treated and untreated cells were harvested by trypsinization, washed, and suspended in ice-cold PBS. The washed cell pellet was resuspended in ice-cold binding buffer containing FITC-conjugated Annexin V and PI. The samples were then incubated in the dark at room temperature for 15 min before being analyzed with a flow cytometer [Navios flow cytometer (Beckman Coulter)] and Kaluza software.
KRAS expression profile by quantitative real-time PCR
RNA was extracted from the PNA-treated AsPC-1 cells, as well as from non-treated cultured cells, using a GeneJET RNA Purification kit (Thermo Fisher Scientific, Inc.). The RNA was quantified with Nanodrop OneC (Thermo Fisher Scientific, Inc.). First-strand cDNA was synthesized with a qScript cDNA Synthesis kit (Quanta Bio.). The expression level of the mature KRAS was quantified separately, using PerfeCTa SYBR Green FastMix (Quanta Bio.) in accordance with the manufacturer's instructions, and normalized with the use of GAPDH for internal expression control. A real-time PCR was carried out with StepONePlus, and the results were analysed with the comparative delta Ct method, using the StepONePlus analysis software.
Western blot analysis
Following the treatment of the AsPC-1 cells for 48 h, the control and treated cells were collected and lysed with RIPA buffer (Merck) and Complete Protease Inhibitor (Sigma) on ice for 20 min. Protein fractions were separated by centrifugation for 10 min at 13,000 rpm at 4°C. The proteins, in 20-µg aliquots, were separated by SDS-PAGE and then transferred by semi-dry transfer to 0.45-micrometer-pore-size nitrocellulose membranes. The membranes were blocked with 5% milk TBST solution for 1 h at room temperature, followed by overnight incubation with rabbit monoclonal Anti-Ras Antibody (Abcam). They were then washed three times with TBST, each for 10 min, incubated with secondary antibody, peroxidase-conjugated, AffiniPure Goat Anti-Rabbit IgG (Jackson Immune Research Laboratories and Dako) for 1 h at room temperature, and washed three more times. An antigen/antibody complex was detected with an ECL kit Western Bright™ ECL detection reagent (Advansta). The results were analyzed with an Amersham Imager 600 (GE Healthcare).
For reference, and after detection with the specific antibodies, all of the membranes were exposed to mouse Anti-β-actin monoclonal antibody (MP Biomedicals). Donkey polyclonal anti-mouse HRP secondary antibody and an ECL kit were used to detect actin.
Statistics
All analyses were performed using SPSS 19.0 software (IBM, Corp.). All data were expressed as mean value ± Standard Error (SE). Statistical analyses were performed, using one-way analysis of variance (ANOVA) test for comparison among multiple groups followed by Bonferroni test for significancy. The SPSS software served for calculation of differences. P<0.05 was considered to indicate a statistically significant difference.
Results
PNA permeability
Cell membrane and nuclear envelope permeability to PNA were determined by monitoring fluorescently labeled PNA1 (fluorescein isothiocyanate (FITC)-conjugated PNA1) transfected into AsPC-1 cells. The fluorescence intensity was determined using an fmax fluorescence microplate reader (see Fig. 2A). The cells were then stained with DAPI and examined with a fluorescence microscope (Fig. 2B and C).
Figure 2.
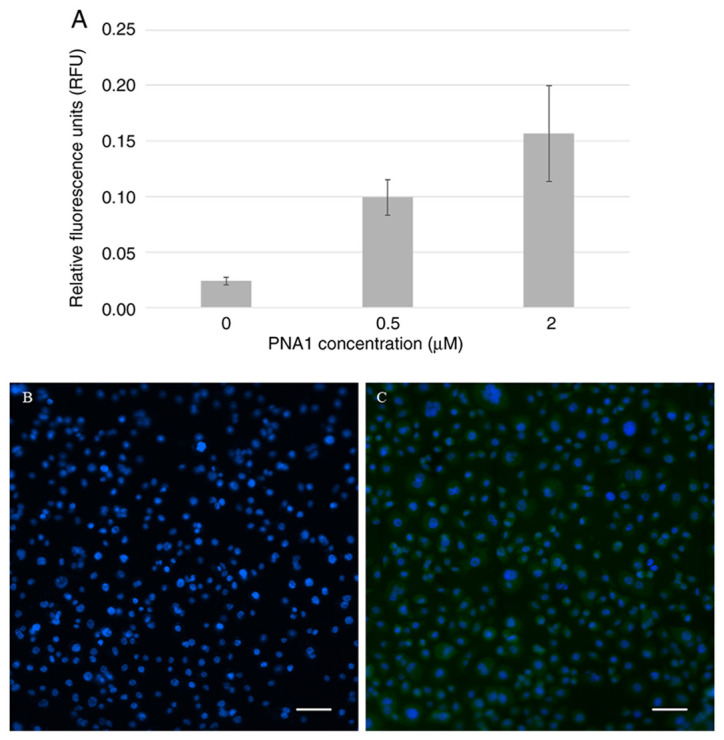
AsPC-1 cell permeability to fluorescently labeled PNA. (A) RFUs of cells incubated with different concentrations of fluorescently labeled PNA1. Cells transfected (B) without or (C) with green fluorescence (FITC)-conjugated PNA1 as observed under a fluorescence microscope. Magnification, ×200. Scale bars, 25 µm. RFU, relative fluorescence unit; PNA, peptide nucleic acid.
The results above demonstrate that the fluorescence signal was positively correlated with the concentration of FITC-PNA1. The fluorescence was observed in the cell cytoplasm.
Effect of PNAs on cell viability
Because the mutated KRAS gene plays a key role in mediating the growth and proliferation of pancreatic cancer cells, the PNAs specific to this gene were expected to affect cell viability and growth. To assess this projected effect, AsPC-1 cells were transfected with different concentrations of PNAs for 48 h, after which the number of viable cells was quantitated using the XTT assay (Fig. 3). Gemcitabine was used as a positive control because it is one of the main chemotherapy drugs used to treat pancreatic cancer. It is not specific to the mutated KRAS gene but acts as a competitive substrate of deoxycytidine triphosphate (dCTP) and is incorporated into DNA during replication, thereby inhibiting DNA chain elongation and cell death by apoptosis.
Figure 3.
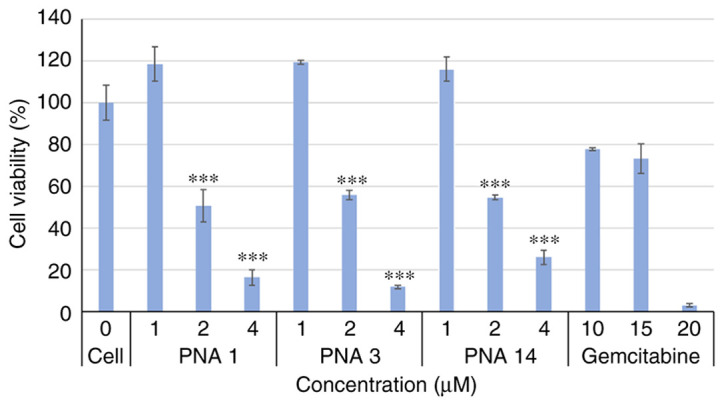
Effects of PNAs on the viability of AsPC-1 cells. AsPC-1 cells were incubated for 48 h with 0, 1, 2 or 4 µM of PNAs and with 10, 15 or 20 µM of gemcitabine (the positive control), and viability was determined using the XTT assay. ***P<0.001 vs. cells in 0 µM. PNA, peptide nucleic acid.
As shown in Fig. 3, exposure of the cells to different PNAs significantly affected cell viability. Increasing PNA concentrations resulted in greater inhibition of cell viability.
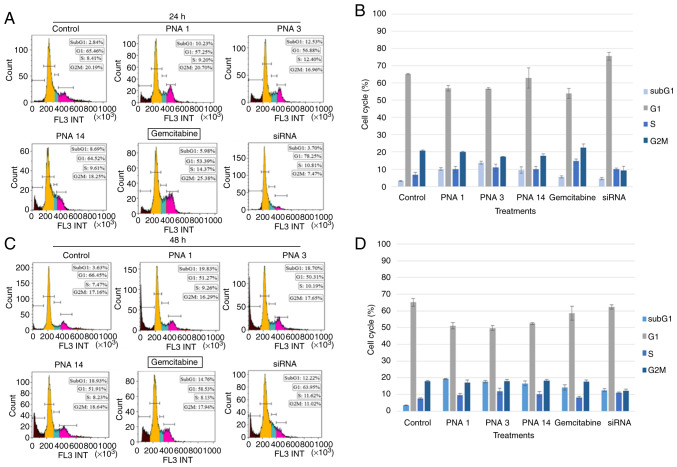
Effects of PNAs on the cell cycle of AsPC-1 cells
PNA binding to the mutated KRAS gene may cause cell-cycle arrest and cell apoptosis (an increase in the number of cells in the sub-G1 phase). To assess this possibility, AsPC-1 cells were treated with 2 µM of PNAs for 24 and 48 h and incubated with propidium iodide (PI) to determine the DNA content, which reflects the cell number. PI signals were then read using a flow cytometer, and the data were analyzed using Kaluza Software. Gemcitabine (20 µM) and siRNA specific to the mRNA sequence of the mutant KRAS gene (20 µM) served as positive controls. The results, which are presented in Fig. 4, show that, compared to the two positive controls, the PNAs induced apoptosis (an increase in the number of cells in the sub-G1 phase), and the number of cells in the G1 phase decreased.
Figure 4.
Effects of PNAs on the cell cycle of AsPC-1 cells. AsPC-1 cells treated with a blank solution (control) or 2 µM of PNAs, and with 20 µM gemcitabine or 20 µM siRNA, served as positive controls. The cells were then washed and stained with PI. The PI-stained cells, indicating the percentage of cells in each phase of the cell cycle, were measured after (A and B) 24 h and (C and D) 48 h treatments with a flow cytometer and analyzed with Kaluza software. PI, propidium iodide; PNA, peptide nucleic acid; siRNA, short interfering RNA.
The percentage of PNA-treated cells (after 48 h treatment) in the G1 phase decreased by approximately 15% relative to the control, and this was mirrored by an approximately 15% increase in the percentage of cells in the sub-G1 phase (see Fig. 4). To verify whether this cell-cycle arrest was due to the induction of apoptosis, the percentage of cells in the sub-G1 phase was quantified, and an apoptosis test was performed (Fig. 5), as discussed below. We also looked at the effect of the PNAs on the percentage of cells in the G2/M phase. At 24 h, the percentage of PNA-treated cells in the G2/M phase decreased by 2–10% relative to the control, and this was mirrored by a 1–10% increase in the percentage of cells in the sub-G1 phase. After the same treatment for 48 h, there was almost a cessation in the G2/M stage of the cell cycle, while the percentage of cells in the sub-G1 phase increased by 9–16% (Figs. 4 and 5). These results strongly indicate that the PNAs induced a halt in cell division and an increase in sub-G1, which triggers cell apoptosis.
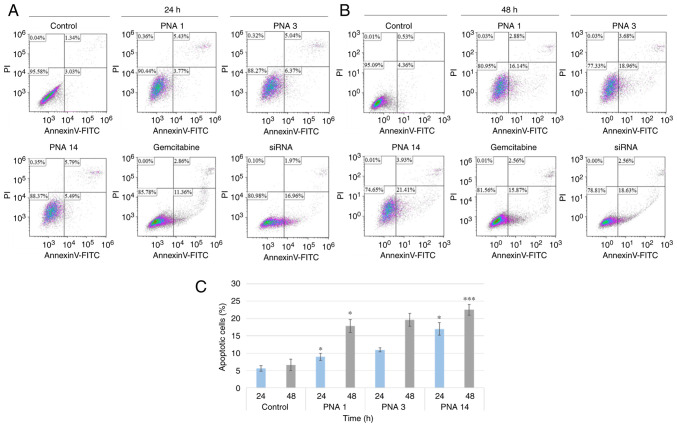
Figure 5.
Effects of PNAs on apoptosis in AsPC-1 cells. AsPC-1 cells were treated with PNAs, gemcitabine, or siRNA for (A) 24 h or (B) 48 h. The control samples were treated with medium. The percentage of cells in the sub-G1 phase was determined using a flow cytometer and analyzed using Kaluza software. (C) Percentages of apoptotic cells, assessed by an Annexin-V FITC Detection Kit. The results are represented in the form of mean ± standard error from three experiments (n=3). *P<0.05 and ***P<0.001 vs. the respective time-point control groups. PNA, peptide nucleic acid; siRNA, short interfering RNA.
Effect of PNAs on apoptosis in AsPC-1 cells
As most cancer cells can escape apoptosis, we set out to determine whether PNAs triggered AsPC-1 apoptosis in vitro. The exposure of cells to 2 µM of PNA1, PNA3, or PNA14 for 48 h resulted in 18, 20, and 20% apoptosis, respectively, whereas in the positive controls treated with 20 µM siRNA or 20 µM gemcitabine, approximately 18 and 22% of the cells were found in the apoptotic state, respectively (Fig. 5). In the untreated control samples, 7% of the cells were apoptotic.
Effect of PNAs on the expression of the KRAS mutant gene in AsPC-1 cells
The PNAs were designed to specifically target the mRNA sequence of the KRAS mutant gene and thus were expected to affect its transcription and translation. To confirm this effect, AsPC-1 cells were transfected with PNAs for 48 h, and the expression of KRAS mRNA and its protein were analyzed. The results are presented in the next two sections.
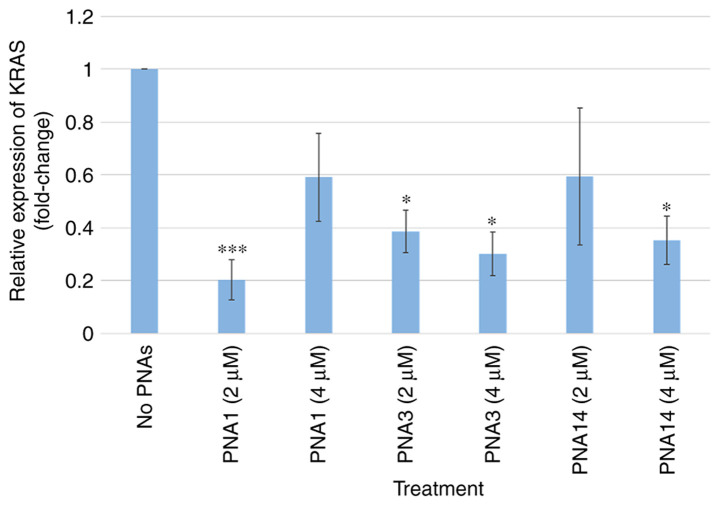
1. Effect of PNAs on KRAS mutant gene transcriptional levels
As shown in Fig. 6, PNA3 and PNA14 are dose-dependent and have a significant effect on the translation of the mutant KRAS gene. The weaker effect of PNA1 at 4 µm compared to PNA1 at 1 µM can be explained by steric hindrance. Collectively, the results clearly show that the PNAs significantly affected the expression of the mutant KRAS gene.
Figure 6.
Reverse transcription-quantitative PCR analysis of mutant KRAS gene expression in PNA-treated AsPC-1 cells. AsPC-1 cells were treated with 2 µM or 4 µM of PNAs for 48 h. The KRAS mRNA was quantitated using forward and reverse primers specific to the KRAS gene. The results are represented in the form of mean ± standard error from three experiments (n=3). *P<0.05, ***P<0.001 vs. no PNAs. PNA, peptide nucleic acid.
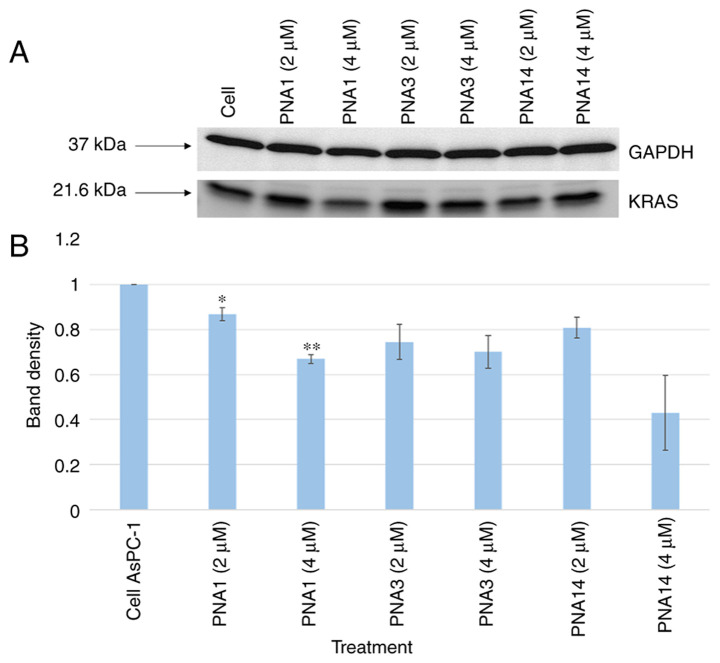
2. Effect of PNAs on KRAS mutant gene translational levels
Total protein lysates were extracted from the PNA-treated cells, resolved with SDS/PAGE, and then transferred to PVDF membranes that had been incubated with mouse anti-human RAS antibodies and mouse anti-human GAPDH antibodies. Treatment with PNA1 resulted in a decrease in total RAS protein levels (Fig. 7).
Figure 7.
Representative western blot of RAS protein expression in PNA-treated AsPC-1 cells. AsPC-1 cells were treated with PNAs for 48 h; then total protein was separated with SDS-PAGE and analyzed using western blot analysis, using specific KRAS and GAPDH antibodies. (A) One representative western blot, while the (B) quantification of band intensity was performed on three western blots by ImageJ software, and the intensities were averaged. *P<0.05, **P<0.001 vs. cell AsPC-1 group. PNA, peptide nucleic acid.
The results of the western blot analysis show an evident effect of PNAs on KRAS protein levels. Overall, it is clear that the three PNAs inhibit gene expression at the level of translation in a dose-dependent manner, as shown in Fig. 7B.
Discussion
PNAs have been proposed as a potential anti-cancer therapy (36). Mutations in the genes of RAS proteins are very common in malignant tumors, especially in gastrointestinal, colorectal, biliary tract, and pancreatic tumors (43). The life expectancy of patients suffering from these tumors is relatively short, as these tumors are generally resistant to current chemical and biological treatments. Even though KRAS is among the most common oncogenes in human cancer, it is a difficult oncogene to target (44,45). Antisense oligonucleotides for the KRAS gene (18), and siRNA products (17) may become therapeutic options for RAS-mutated cancers; however, siRNA technology is restricted by the need for systemic delivery (46).
In this study, the PNAs in question were chemically designed with aqueous solubility and an enhanced ability for cell penetration in mind, owing to the unique distribution of charges over the backbone and the nucleotide-like monomers (rings), and they were attached to a positively charged peptide consisting of three amino acids of lysine (KKK). These structural characteristics not only enhance cell penetration, but also help PNAs to selectively and specifically bind to the KRAS mRNA sequence. Each of the three synthesized PNA has its own chemical unique characteristics, but all three carry a net positive charge that attracts them to the negatively charged cell membrane, hypothetically enabling them to easily penetrate the membrane into the cytoplasm. However, despite these characteristics, repeated experiments have shown that the three PNAs are unable to penetrate cells and thus cannot affect cell functionality. Nevertheless, we were able to introduce them into the cells using lipofectamine, a cationic lipid-based chemical. As demonstrated, lipofectamine effectively and successfully transfected FITC-fluorescently labeled PNA into the cytoplasm of cells. Lipofectamine is a cationic, lipid-based chemical transfectant which forms liposomes that complex with negatively charged nucleic acids, and, through the endocytosis pathway, deliver DNA or RNA into the cell cytosol (47). Despite their positive charge, the PNAs were efficiently delivered into cells with the use of cationic lipofectamine, as demonstrated by florescence measurements and microscopy. It is plausible that lipid subunits of the lipofectamine formed liposomes in the aqueous environment of the growth medium, which transfected the PNAs into the cells. Since lipofectamine cannot be used clinically, in future studies, we ought to investigate the systemic delivery of PNA treatments by other means, possibly using lipid or nanoparticle technology.
Not only did the PNAs examined penetrate cell membranes, they also reached targets and affected important parameters linked to cell viability. Among the measurable parameters were cell membrane integrity and permeability, and the activity of cellular enzymes. The effects of PNAs on cell viability were evaluated using the XTT assay, and a significant, concentration-dependent reduction in cell viability was found. This can be attributed mainly to reduced activity of the mitochondrial dehydrogenase enzyme, and it points to the cytotoxicity of PNAs and their effects on cell membrane leakage. The effects of the three PNAs on cell viability and their maximum inhibition was clearly apparent after two days. Surprisingly, when the cells were exposed to gemcitabine, there was total death with no dose response. Gemcitabine, like other chemotherapies, has a narrow therapeutic window, which is reflected in the results of this experiment.
The affinity of the antisense oligonucleotides for the G12D-mutated KRAS mRNA, and their specificity, were evaluated using melting-temperature (Tm) shift assays; the Tm of the gene fragment increased by 4°C in the presence of PNA (data not shown). In addition, antisense oligonucleotides specific to gene fragments of KRAS inhibited their amplification by a PCR, as could be seen in the agarose gel electrophoresis of the KRAS PCR products (data not shown). To validate their specificity at the cellular level, it is advisable to test the efficacy of PNA-based antisense molecules on cells from a different pancreatic cancer cell line that have a mutation other than the KRAS G12D mutation, such as on HPAF-II cells or on KRAS wild-type pancreatic cancer cells.
KRAS is the predominant mutated RAS gene in cancers and is involved in 84% of all RAS missense mutations; in particular, KRAS mutation is the initiating genetic event for pancreatic ductal adenocarcinoma (PDAC) (43). In general, the proliferation of cells decreases under stressful or damaging conditions, and cell-cycle arrest occurs through the activation of checkpoints in an attempt to repair the damage and ensure proper cell division (48,49). If the damage is too extensive to be repaired, the cells undergo cell death in the form of apoptosis (50–52). Mutated KRAS is persistently GTP-bound and thus remains constitutively active, resulting in an overstimulation of effectors, which causes cells to evade apoptotic signals and to continue proliferating, regardless of extracellular stimuli (48). In this experiment, PNA-based antisense molecules were designed to target the mutation effect by specifically and selectively binding to the G12D region of the KRAS gene sequence. Our results suggest that upon penetrating the cell membrane, PNA-based antisense leads to a significant increase in the percentage of sub-G1 phase (apoptotic) cells in AsPC-1 cells. Targeting the G12D KRAS mutation with PNA-based antisense may cause cells to revert to controlled cell division and make them more responsive to cell-cycle checkpoints, thereby halting uncontrolled division and diverting them to apoptosis in the absence of physiological growth stimuli. As seen in the study, PNA-based antisense strikingly induced apoptosis, compared to the control cells. Moreover, in addition to affecting cell viability, cell-cycle progression, and apoptosis, PNA treatment disrupted the integrity of the typical structure of the AsPC-1 cells, which normally exhibit an epithelial cell-like morphology (data not shown).
Our attempt to design PNA-based antisense that specifically inhibits the expression of the G12D KRAS mutant gene was successful, as evidenced by the inhibition of KRAS mutant gene translation (Fig. 6). We observed a decrease in total RAS protein, probably reflecting the decrease in KRAS gene translation. Since AsPC-1 cells are homozygous for the G12D KRAS mutation (involving two mutant alleles), the PNA-based antisense targets both gene copies; thus, inhibiting the production of the protein product of the KRAS mutant gene would clearly decrease its activity. Like the findings of other studies (53,54), these findings suggest that PNA-based antisense can serve as a powerful tool for sequence-specific inhibition of the oncogenic mutant allele, resulting in the suppression of proliferation pathways and a decrease in cell growth and viability, and this points to the potential usefulness of mutant KRAS as a target for anti-cancer therapy.
PNA-based antisense therapeutics are still limited by lack of an effective method of systemic delivery. More work is needed to develop means of systemic delivery, and possibly administration in conjunction with other drugs, such as gemcitabine. The development of new PNA formulations, for example, utilizing nanoparticle-based delivery methods (55), may offer ways to sustain the delivery of PNA-based therapeutics. Furthermore, in vivo studies will increase our understanding of the delivery and activity of PNAs, as well as of the biological mechanisms involved in the tissue distribution, cellular uptake, and intracellular trafficking of the molecules. Future strategies that refine this approach will be important to maximizing the potential of PNA-based antisense therapeutics to treat a broad spectrum of human diseases.
Furthermore, in order to establish the specificity of PNAs for the G12D KRAS mutated gene sequence in cell-line, other cancer cell lines that do not carry the G12D KRAS mutation must be investigated with PNA-based antisense, since PNAs designed to affect the mutated KRAS gene, which have only a single-point mutation (GGT >GAT, a glycine-to-aspartate substitution in codon 12, G12D), might also affect normal variants of the gene in other cells or other genes that have similar sequences. As well, it is advisable to assess the toxicity of the PNAs by testing their effects on normal pancreatic cells.
Our results suggest that antisense PNAs are capable of inhibiting the expression and downstream function of mutant KRAS in pancreatic cancer cells, exhibiting robust antitumor activity at micromolar doses. PNAs are attractive candidates for the therapeutic targeting of KRAS mutant genes. However, critical issues, such as how to deliver PNAs to target tissues and how to prevent their tendency to aggregate, need to be resolved before they can be tested for pre-clinical and clinical application.
Acknowledgements
Not applicable.
Funding Statement
This research was partially supported by the Israel Innovation Authority (an incubator project that was granted to GeneArrest, Ltd., between the years 2009 and 2015) and The Ministry for Development of the Periphery, Negev and Galilee, 2016 (AS, RA and MF).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AR, AS and MF conceived the study, raised the funds and performed the analysis. EG, RM, HT, MS, MZ and MF carried out the experiments. AR and MF carried out the statistical analysis of the data, interpreted the data and wrote up the conclusions. AS, MS, MZ, AR and MF wrote the first draft of the manuscript. AS, AR and MF edited the manuscript. AR and MF confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
AR and MF are the founders of GeneArrest, Ltd., the IP owner of the novel monomers. We declare that the funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The other authors declare no conflicts of interest.
References
- 1.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: Diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh H, Longo DL, Chabner BA. Improving prospects for targeting RAS. J Clin Oncol. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- 7.Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B. 2019;9:871–879. doi: 10.1016/j.apsb.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 9.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 11.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, Falchook GS, Price TJ, Sacher A, Denlinger CS, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Yang X, Geng M, Huang M. Targeting ERK, an Achilles' Heel of the MAPK pathway, in cancer therapy. Acta Pharm Sin B. 2018;8:552–562. doi: 10.1016/j.apsb.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallejo A, Perurena N, Guruceaga E, Mazur PK, Martinez-Canarias S, Zandueta C, Valencia K, Arricibita A, Gwinn D, Sayles LC, et al. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun. 2017;8:14294. doi: 10.1038/ncomms14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol. 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 16.Takashima A, Faller DV. Targeting the RAS oncogene. Expert Opin Ther Targets. 2013;17:507–531. doi: 10.1517/14728222.2013.764990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zorde Khvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross SJ, Revenko AS, Hanson LL, Ellston R, Staniszewska A, Whalley N, Pandey SK, Revill M, Rooney C, Buckett LK, et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci Transl Med. 2017;9:eaal5253. doi: 10.1126/scitranslmed.aal5253. [DOI] [PubMed] [Google Scholar]
- 19.Marin VL, Roy S, Armitage BA. Recent advances in the development of peptide nucleic acid as a gene-targeted drug. Expert Opin Biol Ther. 2004;4:337–348. doi: 10.1517/14712598.4.3.337. [DOI] [PubMed] [Google Scholar]
- 20.Gambari R. Peptide-nucleic acids (PNAs): A tool for the development of gene expression modifiers. Curr Pharm Des. 2001;7:1839–1862. doi: 10.2174/1381612013397087. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 22.Rajasekaran P, Alexander JC, Seleem MN, Jain N, Sriranganathan N, Wattam AR, Setubal JC, Boyle SM. Peptide nucleic acids inhibit growth of Brucella suis in pure culture and in infected murine macrophages. Int J Antimicrob Agents. 2013;41:358–362. doi: 10.1016/j.ijantimicag.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdel-Aziz M, Yamasaki T, Otsuka M. Synthesis and hybridization property of novel 2′,5′-isoDNA mimic chiral peptide nucleic acids. Bioorg Med Chem Lett. 2003;13:1041–1043. doi: 10.1016/S0960-894X(03)00075-1. [DOI] [PubMed] [Google Scholar]
- 24.Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Adv Drug Deliv Rev. 2003;55:267–280. doi: 10.1016/S0169-409X(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 25.Dean DA. Peptide nucleic acids: Versatile tools for gene therapy strategies. Adv Drug Deliv Rev. 2000;44:81–95. doi: 10.1016/S0169-409X(00)00087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Cola C, Manicardi A, Corradini R, Izzo I, De Riccardis F. Carboxyalkyl peptoid PNAs: Synthesis and hybridization properties. Tetrahedron. 2012;68:499–506. doi: 10.1016/j.tet.2011.11.017. [DOI] [Google Scholar]
- 27.Narenji H, Gholizadeh P, Aghazadeh M, Rezaee MA, Asgharzadeh M, Kafil HS. Peptide nucleic acids (PNAs): Currently potential bactericidal agents. Biomed Pharmacother. 2017;93:580–588. doi: 10.1016/j.biopha.2017.06.092. [DOI] [PubMed] [Google Scholar]
- 28.Wang G, Xu XS. Peptide nucleic acid (PNA) binding-mediated gene regulation. Cell Res. 2004;14:111–116. doi: 10.1038/sj.cr.7290209. [DOI] [PubMed] [Google Scholar]
- 29.Quijano E, Bahal R, Ricciardi A, Saltzman WM, Glazer PM. Therapeutic peptide nucleic acids: Principles, limitations, and opportunities. Yale J Biol Med. 2017;90:583–598. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao XL, Chen BC, Han JC, Wei L, Pan XB. Delivery of cell-penetrating peptide-peptide nucleic acid conjugates by assembly on an oligonucleotide scaffold. Sci Rep. 2015;5:17640. doi: 10.1038/srep17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucarelli F, Marrazza G, Turner AP, Mascini M. Carbon and gold electrodes as electrochemical transducers for DNA hybridisation sensors. Biosens Bioelectron. 2004;19:515–530. doi: 10.1016/S0956-5663(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 32.Barluenga S, Winssinger N. PNA as a biosupramolecular tag for programmable assemblies and reactions. Acc Chem Res. 2015;48:1319–1331. doi: 10.1021/acs.accounts.5b00109. [DOI] [PubMed] [Google Scholar]
- 33.Brandén LJ, Mohamed AJ, Smith CI. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat Biotechnol. 1999;17:784–787. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- 34.Liang KW, Hoffman EP, Huang L. Targeted delivery of plasmid DNA to myogenic cells via transferrin-conjugated peptide nucleic acid. Mol Ther. 2000;1:236–243. doi: 10.1006/mthe.2000.0043. [DOI] [PubMed] [Google Scholar]
- 35.Chiarantini L, Cerasi A, Fraternale A, Millo E, Benatti U, Sparnacci K, Laus M, Ballestri M, Tondelli L. Comparison of novel delivery systems for antisense peptide nucleic acids. J Control Release. 2005;109:24–36. doi: 10.1016/j.jconrel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Montazersaheb S, Hejazi MS, Nozad Charoudeh H. Potential of peptide nucleic acids in future therapeutic applications. Adv Pharm Bull. 2018;8:551–563. doi: 10.15171/apb.2018.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 38.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 39.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/S0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 40.Rayan A, Falah M. Sequence specific double-stranded DNA binding compounds. Patent Application Number WO2009093188. 2009 January 22; Date Filed. [Google Scholar]
- 41.Rayan A, Falah M. Sequence specific double-stranded DNA/RNA binding compounds and uses thereof. Patent Application Number WO2012011114. 2011 July 21; Date Filed. [Google Scholar]
- 42.Komiyama M, Aiba Y, Ishizuka T, Sumaoka J. Solid-phase synthesis of pseudo-complementary peptide nucleic acids. Nat Protoc. 2008;3:646–654. doi: 10.1038/nprot.2008.6. [DOI] [PubMed] [Google Scholar]
- 43.Waters AM, Der CJ. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018;8:a031435. doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl. 2014;53:199–204. doi: 10.1002/anie.201307387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu CF, Wang J. Delivery systems for siRNA drug development in cancer therapy. Asian J Pharm Sci. 2015;10:1–12. doi: 10.1016/j.ajps.2014.08.011. [DOI] [Google Scholar]
- 47.Llovera L, Berthold P, Nielsen PE, Shiraishi T. Cell number and transfection volume dependent peptide nucleic acid antisense activity by cationic delivery methods. Artif DNA PNA XNA. 2012;3:22–27. doi: 10.4161/adna.19906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartwell LH, Weinert TA. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 49.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 50.Chinnasamy S, Zameer F, Muthuchelian K. Molecular and biological mechanisms of apoptosis and its detection techniques. J Oncol Sci. 2020;6:49–64. doi: 10.37047/jos.2020-73477. [DOI] [Google Scholar]
- 51.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 52.Wong RS. Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rachagani S, Senapati S, Chakraborty S, Ponnusamy MP, Kumar S, Smith LM, Jain M, Batra SK. Activated KrasG¹2D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. Br J Cancer. 2011;104:1038–1048. doi: 10.1038/bjc.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smakman N, Veenendaal LM, van Diest P, Bos R, Offringa R, Borel Rinkes IH, Kranenburg O. Dual effect of Kras(D12) knockdown on tumorigenesis: Increased immune-mediated tumor clearance and abrogation of tumor malignancy. Oncogene. 2005;24:8338–8342. doi: 10.1038/sj.onc.1208995. [DOI] [PubMed] [Google Scholar]
- 55.Gupta A, Bahal R, Gupta M, Glazer PM, Saltzman WM. Nanotechnology for delivery of peptide nucleic acids (PNAs) J Control Release. 2016;240:302–311. doi: 10.1016/j.jconrel.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.