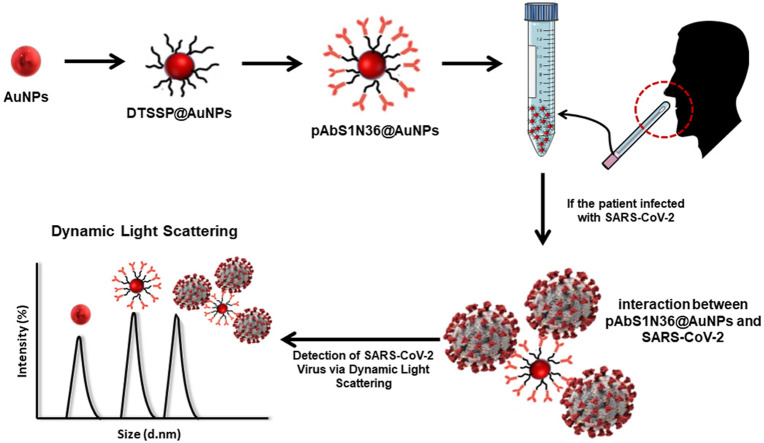
Abstract
Mass testing for the diagnosis of COVID-19 has been hampered in many countries owing to the high cost of genetic material detection. This study reports on a low-cost immunoassay for detecting SARS-CoV-2 within 30 min using dynamic light scattering (DLS). The immunosensor comprises 50-nm gold nanoparticles (AuNPs) functionalized with antibodies against SARS-CoV-2 spike glycoprotein, whose bioconjugation was confirmed using transmission electron microscopy (TEM), UV-Vis spectroscopy, Fourier transform infrared spectroscopy (FTIR), and surface-enhanced Raman scattering spectroscopy (SERS). The specific binding of the bioconjugates to the spike protein led to an increase in bioconjugate size, with a limit of detection (LOD) 5.29 × 103 TCID50/mL (Tissue Culture Infectious Dose). The immunosensor was also proven to be selective upon interaction with influenza viruses once no increase in size was observed after DLS measurement. The strategy proposed here aimed to use antibodies conjugated to AuNPs as a generic platform that can be extended to other detection principles, enabling technologies for low-cost mass testing for COVID-19.
Keywords: SARS-CoV-2, Immunosensors, Bioconjugates, Spectroscopic techniques, Influenza virus
Graphical abstract
1. Introduction
Early diagnosis of COVID-19 caused by SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) [1] is essential to handle the pandemic, which is why the World Health Organization (WHO) [2] has recommended mass testing. Unfortunately, mass testing for COVID-19 has not been possible in many developing countries, including Brazil, because it is mostly performed using expensive methods that require specialized laboratories and personnel. COVID-19 diagnosis has been based on detecting the viral genome, or a specific part of its sequence [3], with reverse transcription-polymerase chain reactions (RT-PCR) as the gold standard test [[4], [5], [6], [7], [8], [9]]. RT-PCR detects the genetic material of the virus (ribonucleic acid, RNA) using a swab, mainly collected in the nose and throat [10]. Less commonly used molecular techniques are reverse transcription loop-mediated isothermal amplification (RT-LAMP) [11,12] and clustered regularly interspaced short palindromic repeats (CRISPR) [13,14], but none of them are amenable to point-of-care diagnostics. Genosensors have been developed to detect the genetic material of SARS-CoV-2 using portable instruments and simple procedures [15], even though they are not commercially available.
Accurate diagnosis can also be achieved by exploiting antibody-antigen interactions in imumunosensors, e.g., used to detect the spike protein [[15], [16], [17], [18], [19]]. The widespread serologic tests to detect IgM and IgG immunoglobulins are not suitable for early diagnosis of COVID-19 because noticeable immune responses take a few days after the infection [[20], [21], [22]]. The presence of IgM antibodies indicates the onset of the immune response whereas the IgG associates to the stage of immunization against SARS-CoV-2 [20]. Detection of virus proteins (S and N, e.g.), on the other hand, yields faster, cheaper tests [23,24], especially if one capitalizes upon extensive knowledge about nanobiosensors [[25], [26], [27]]. These immunosensors may contain gold nanoparticles (AuNPs) whose physicochemical properties are tuned to amplify optical, electrical, and electrochemical signals. For example, the plasmon resonance in AuNPs is varied by changing particle size and shape, and size distribution [28]. AuNPs are suitable for functionalization to anchor active layers as they are biocompatible upon binding of biomolecules [29,30]. Immunosensors based on AuNPs conjugated with antibodies are capable of recognizing antigens, cells, microorganisms, and proteins through varied principles of detection [31]. Bioconjugation affects both the color and size of the AuNPs, thus permitting colorimetric or size assays with dynamic light scattering (DLS) [32,33]. Driskell et al. [34], for instance, have reported the use of DLS to detect the H1N1 virus. To the best of our knowledge, DLS has not been used to detect SARS-CoV-2, using AuNPs functionalized with anti-SARS-CoV-2 spike glycoprotein antibodies, which is the aim of the present study.
2. Experimental details
2.1. Materials and virus samples
Boric acid was purchased from Vetec™ (Duque de Caxias, RJ, Brazil). Gold nanoparticles (50 nm, ∼3.5 × 1010 particles/mL), bovine serum albumin (BSA), and 3,3′-dithiobis (sulfosuccinimidyl propionate) (DTSSP) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Phosphate buffered saline (PBS) solution was purchased from Laborclin (Pinhais, PR, Brazil). Sodium tetraborate was obtained from Dinâmica (Indaiatuba, SP, Brazil). Polyclonal antibody against the N-terminal of the spike protein of SARS-CoV-2 from rabbits was kindly provided by the Virology and Microscopy Laboratory, Universidade de Brasília, Brazil. Samples from influenza and SARS-CoV-2 viruses were kindly donated by Butantan Institute (São Paulo, Brazil). The samples used were A/Brisbane/02/2018 -IVR-190 (H1N1), B/Washington/02/2019 (B), B/Phuket/3073/2013 (B), A/South Australia/34/2019 -IVR-197 (H3N2), and SARS-CoV-2 inactivated by gamma irradiation. The concentrations of the virus samples were determined by TCID50/mL (Tissue Culture Infectious Dose), which defines the dilution of virus required to infect 50% of the cells.
2.2. Antibody-modified AuNPs (pAbS1N@AuNPs)
An aliquot of 1.2 mL (2.6 × 1010 particles/mL) suspension of 50-nm AuNPs had the pH value adjusted to 8.9 with borate buffer (50 mM, pH 8.9). Next, 10 μL of 10 mM of 3,3′-dithiobis (sulfosuccinimidyl propionate) (DTSSP) were added to the AuNPs suspension and incubated at room temperature under light protection for 40 min. This suspension was transferred into a 2 mL Lobind® Eppendorf tube (Eppendorf, Hamburg, Germany) and centrifuged at 7000 g for 5 min at 5 °C. The supernatant containing the excess of DTSSP was removed, and the AuNPs were resuspended in 1100 μL of 2 mM borate buffer pH at 8.9. The centrifugation and resuspension cycle of AuNPs was repeated two more times. Next, the polyclonal antibody (21.6 μg, 32.4 μg, or 43.2 μg) was added to the AuNPs suspension and incubated at room temperature for 90 min. After this 90-min reaction, the suspension was centrifuged at 7,000 g for 5 min at 5 °C. The supernatant containing the excess antibody was removed, and pAbS1N@AuNPs were resuspended in 1100 μL borate buffer (2 mM, pH 8.9) containing 0.25% BSA. For complete removal of the antibody, the centrifugation/resuspension process was repeated twice again. The samples containing 21.6 μg, 32.4 μg, and 43.2 μg antibodies are referred to pAbS1N18@AuNPs, pAbS1N27@AuNPs, and pAbS1N36@AuNPs, respectively.
2.3. Incubation protocol of pAbS1N@AuNPs with viruses
Diluted virus stock was prepared using PBS and 20% sucrose as the diluent for influenza (1 × 1010.3 TCID50/mL for A/Brisbane, 1 × 1010.6 TCID50/mL for B/Washington, 1 × 109 TCID50/mL for B/Phuket, and 1 × 1010.7 TCID50/mL for A/South Australia). Twelve serial dilutions of the virus stock (2.11 × 105 TCID50/mL) were prepared using PBS as the diluent for SARS-CoV-2. First, 90 μL of the respective dilution of virus and 10 μL of pAbS1N18@AuNPs or pAbS1N36@AuNPs were added into a cryotube, and the sample was incubated at room temperature for 30 min on a Kline orbital shaker (400 rpm; Nova Técnica, Piracicaba, SP, Brazil). Then, 50 μL of pAbS1N@AuNPs and the virus mixture were transferred to a small volume cuvette (ZEN0118 model, Malvern Instruments Ltd, Worcestershire, UK) for dynamic light scattering (DLS) measurements.
2.4. Characterization methods
Size and surface morphology for the AuNPs and bioconjugates were characterized by transmission electron microscopy (TEM) using a transmission JEOL 1011, Tokyo, Japan, operated at an accelerating voltage of 100 kV. AuNPs and pAbS1N18@AuNPs were prepared by placing a drop of the sample (5 μL) on a 400-mesh copper grid coated with formvar film. After that, the sample was air-dried, and to enhance the contrast, osmium tetroxide vapor was used. Grids were kept at room temperature until further analysis. Four hundred AuNPs were counted in different images for each sample using ImageJ - Plugin Analyse (set scale) software. The binding affinity of SARS-CoV-2 viruses and pAb1SN36@AuNPs conjugates was confirmed via TEM using a modified negative staining technique [35]. A formvar-carbon coated 400-mesh copper grid was floated on a 40-μL drop of pAb1SN36@AuNPs reagent/sample mixture for 30 min at room temperature. Next, the excess sample was removed with filter paper. The grid was then floated on a 40-μL drop of 0.7% glutaraldehyde for 5 min to fix the sample. After removing excess fixative, the sample was negatively stained by floating the grid on 2% aqueous phosphotungstic acid at pH 7.0 for 60 s. The stain was removed from the grid, and the sample was allowed to dry overnight before obtaining the TEM images. DLS was performed with a Zetasizer Nano ZS90 Model: ZEN3690 (Malvern Instruments Ltd, Worcestershire, UK) equipped with a 4.0 mW 632.8 nm (red) laser and at a scattering detector angle of 90° to measure particle size by using a disposable cuvette (DTS0012 or ZEN0118). Each size measurement consisted of five runs to yield an average and standard deviation. All DLS data were collected and analyzed using Zetasizer software 8.01.4906. The hydrodynamic diameters determined by DLS were obtained by cumulant analysis and referred to as effective diameters.
The electronic spectra for the nanoparticles in suspensions were recorded on a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan) using quartz cells with a 10.00 mm optical length without dilution for AuNPs, pAbS1N18@AuNPs, pAbS1N27@AuNPs, and pAbS1N36@AuNPs samples. The FTIR experiments were performed using a Bruker spectrometer, Vertex 70 model (Billerica, EUA), with an attenuated total reflectance module (ATR). The measurements were averaged over 96 scans, taken at a resolution of 4 cm−1 from 400 to 4000 cm−1. Antibody conjugation was assessed onto the surface of the AuNPs by surface-enhanced Raman scattering (SERS). Silver films were used as a substrate for SERS measurements, and they were prepared using the electrolytic method, with AgNO3 electrolyte solution at 0.1 mg/mL. For SERS studies, a solution of 10 μL BSA, pAbS1N antibody, DTSSP@AuNPs, and pAbS1N36@ AuNPs samples were diluted in 10 μL distilled water, dropped onto the Ag film, then dried with dry nitrogen. Raman and SERS analyses were performed using LabRAM HR Evolution (HORIBA Scientific, Palaiseau, France) equipped with an 1800 lines/mm grating and CCD detector. The measurements were conducted with a 532 nm laser excitation source (10 mW) using a DuoScan Raman imaging system (Horiba Scientific) with a 50X objective. DuoScan was used to distribute the laser beam on the sample surface by obtaining average spectral information of an area of 20 × 20 μm2.
3. Results and discussion
3.1. Conjugation of antibodies onto AuNPs
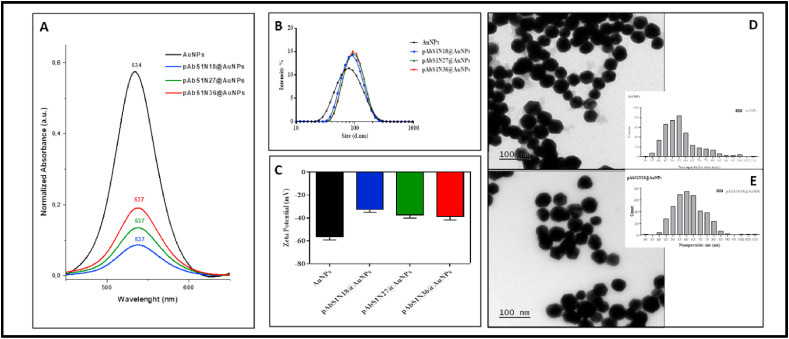
Successful conjugation of polyclonal antibodies onto the AuNPs was experimentally confirmed. The UV–Vis spectra of 50-nm AuNPs (Fig. 1 A) indicate that the conjugation leads to a redshift from 534 nm to 537 nm owing to a change in the refractive index of the nanoparticles. This redshift does not depend upon the specific number of nanoparticles, but its band intensity is proportional to the amount of antibodies used in the bioconjugation process, as expected from the results by Driskel et al. [34]. A second absorbance band is observed within 650–750 nm if larger particles or aggregates are formed [36], turning the dispersion dark blue. Since this effect has not been observed for the bioconjugates (Fig. 1A), one assumes there is no formation of aggregates. DLS data shown in Fig. 1B indicate that the bioconjugates are ca 13% larger than AuNPs. This increased size was also observed in the TEM images of pAbS1N18@AuNPs (Fig. 1D and E), where spherical and hexagonal structures can be noticed with average sizes from 55 nm to 60 nm. These values are smaller than those obtained from DLS (∼80 nm in diameter) because TEM allows obtaining core sizes for the AuNPs; meanwhile, DLS can be related to the hydrodynamic diameter [37,38]. The zeta potentials plotted in Fig. 1C indicate a reduction seen in the number of negative charges after immobilization of pAbS1N antibody from ca ∼ −60 mV to −40 mV. A similar decrease in charge has been reported for protein-coated AuNPs after surface modification with BSA (bovine serum albumin) (or streptavidin) [37]. Our stability studies indicated that AuNPs (before and after bioconjugation) had their sizes kept unchanged for 210 days, with larger hydrodynamic diameter seen for AuNPs containing the antibody pAbS1N (pAbS1N18@AuNPs) as compared to unmodified AuNPs (Figures S-1 and S-2, Supporting Information). The presence of BSA and antibody over the AuNPs surface reduces self-aggregation for AuNPs in aqueous media or buffer solutions [39].
Fig. 1.
(A) UV–Vis spectra (450–650 nm) of 350 μL of AuNPs, pAbS1N18@AuNPs, pAbS1N27@AuNPs, and pAbS1N36@AuNPs. (B) DLS size distribution by intensity; (C) Zeta Potential for AuNPs, pAbS1N18@AuNPs, pAbS1N27@AuNPs, and pAbS1N36@AuNPs. The results correspond to the average of three preparations of binding pAbS1N to AuNPs; TEM micrographs illustrate nanoparticle size and morphology and the corresponding size distribution: (D) AuNPs and (E) pAbS1N18@AuNPs. The bar is 100 nm.
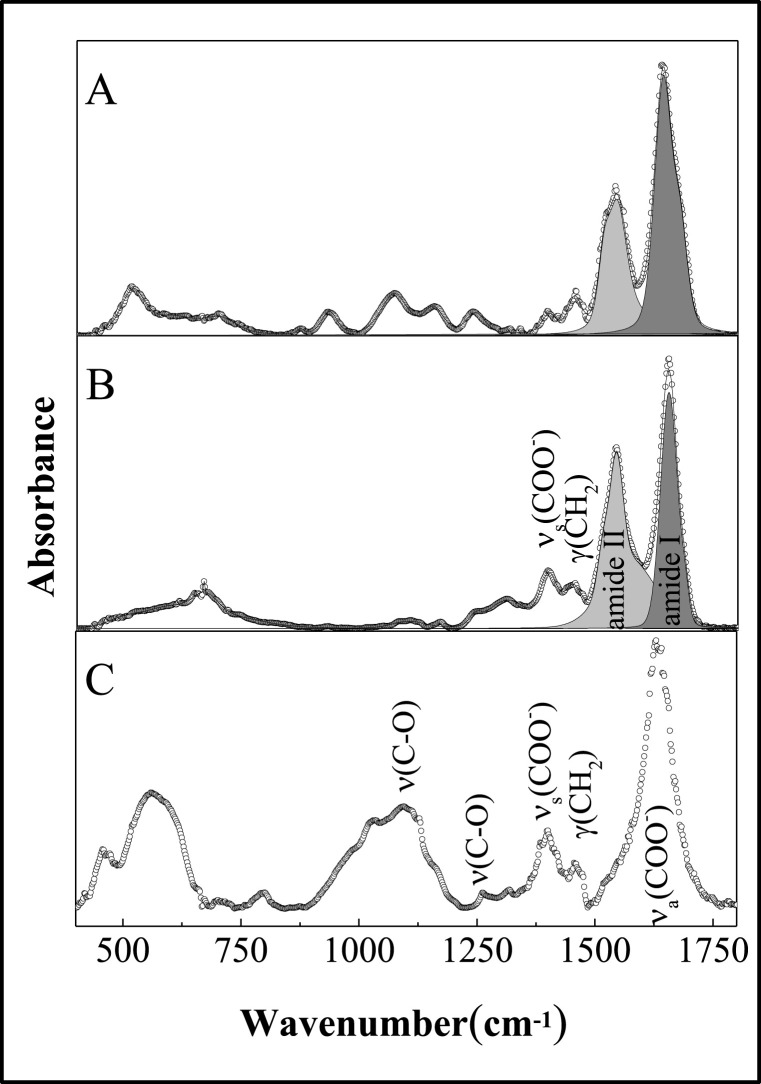
Fig. 2 shows the FTIR spectra (A) for the anti-SARS-CoV-2 spike glycoprotein antibody before (pAbS1N) (B), after coupling to AuNPs (pAbS1N36@AuNPs) and AuNPs (C). The spectrum (Fig. 2C) provides information on AuNPs covered by citrates. The adsorption of citrates on AuNPs is confirmed by the vibration modes of carboxylates, with absorption bands at 1400 cm−1 (νs (COO−)) and 1630 cm−1 (νa (COO−)). The spectrum for the free antibody (pAbS1N) in Fig. 2A displays the typical bands - amide I, amide II, and amide III at 1644, 1540, and 1239 cm−1, respectively. The bands at ca 1400 and 1451 cm−1 can be assigned to C = O symmetric stretching of COO − group and CH 2 scissoring vibration, respectively. The band at 1080 cm−1 can be predominantly attributed to C–O–C and C–O ring vibration. The fingerprint region between 500 and 900 cm−1 can be assigned to vibration in amino acids, e.g., phenylalanine, tyrosine, tryptophan, etc. [40]. Fig. 2B shows the FTIR spectrum of antibody conjugated with AuNPs, with bands at 1654, 1543, and 1243 cm−1 assigned to amide I, amide II and amide II bands, respectively. Note that the bands are shifted to higher wavenumbers compared to the spectrum of AuNPs alone. The intensity ratios of the amide I and amide II bands for pAbS1N and pAbS1N36@AuNPs are ca 1.95 and 1.0, respectively, suggesting that conjugation causes conformational changes in antibodies after Au functionalization. From the FTIR data one infers that the electron-donating effects from free NH2 groups and carboxylate ions are responsible for antibody coupling to terminal succinimidyl ester, thus forming an amide bond by the nucleophilic attack of the amine to the ester carbonyl carbon [41].
Fig. 2.
FTIR spectra for pAbS1N (A), pAbS1N36@AuNPs (B) and AuNPs (C) samples.
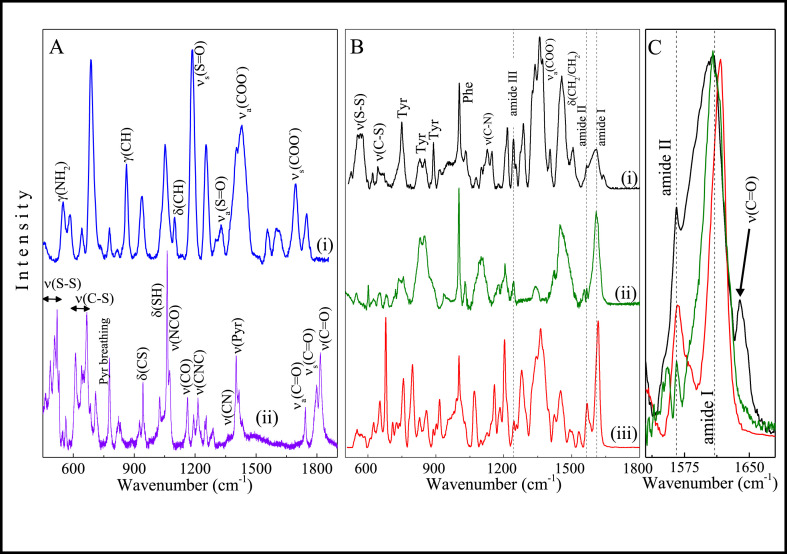
The binding efficiency of antibodies to AuNPs was assessed with Raman spectroscopy and SERS by tracking bioconjugation from the first precoating procedure up to the attachment of the antibody moiety. The Raman spectrum of free DTSSP powder (Fig. 3 A(i)) shows bands primarily due to the presence of 2,5-pyrrolidinedione ring and sulfur-containing groups, as better detailed in Table S-1. The SERS spectrum of DTSSP@AuNPs differs strongly from that of DTSSP powder, with various additional bands appearing due to the SERS effect. These additional bands are also listed in Table S-1. The most striking feature is the redshift of ν(C–S) modes and the quenching of disulfide vibrational modes (490-520 cm−1 region), indicating that disulfide bonds were broken and DTSSP was adsorbed on AuNPs. Fig. 3B shows the SERS spectra of the pAbS1N antibody conjugated with AuNPs (pAbS1N36@AuNPs) compared to those from the pAbS1N antibody and BSA. As expected, no bands from DTSSP are seen in the spectrum of pAbS1N36@AuNPs, which is similar to the spectra of pAbS1N antibody and BSA, with bands arising from amino acids in pAbS1N and BSA, both bioconjugated to AuNPs. The amide bonds in the spectrum of pAbS1N36@AuNPs coincide with those for the pAbS1N antibody (see Fig. 3B(i and ii) and Table S-1). The vibrational energy of these bands is typical of β-sheets which confirm that antibodies are structured [42]. These bands are seen at higher wavenumbers in the spectrum of BSA (see Table S-1), typically from an α-helix-rich structure [43]. One thus infers that the intensity of the dominant Raman signal in the spectrum of pAbS1N36@AuNPs mainly comes from the antibody. Also relevant is the band at 1640 cm−1 (Fig. 3C) for the pAbS1N36@AuNPs sample, absent for both free antibody and BSA. This band can be assigned to the C O stretching of a secondary amide, probably due to the conjugation of AuNPs with antibodies [44].
Fig. 3.
(A) SERS spectrum (i) and Raman spectrum (ii) for DTSSP@AuNPs and DTSSP powder, respectively. (B) SERS spectra of AbS1N36@AuNPs (i), pAbS1N antibody (ii) and BSA (iii). (C) Zoom of SERS spectra in the range 1530–1680 cm−1 for pAbS1N36@AuNPs (black line), pAbS1N antibody (green line), and BSA (red line). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Detection of SARS-CoV-2 virus by using pAbS1N@AuNPs bioconjugates
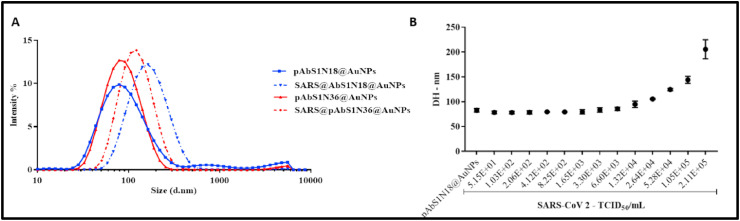
The incubation of pAbS1N18 or pAbS1N36@AuNPs bioconjugates with SARS-CoV-2 at 2.11 × 105 TCID50/mL induced a shift at larger sizes in the DLS data (Fig. 4 A). The shift is larger for pAbS1N18@AuNPs, which was used to estimate the limit of detection (LOD) for the SARS-CoV-2 virus. Fig. 4B indicates that the hydrodynamic diameter increases with increasing the concentration of the virus, SARS-CoV-2, from 1.65 × 103 TCID50/mL to 2.11 × 105 TCID50/mL. The LOD was obtained to be 5.29 × 103 TCID50/mL by using the values of scattered light intensity and standard deviation for the bioconjugate without the virus (σ+ 3s) (Figure S-3, Supporting information) [32].
Fig. 4.
(A) Intensity in DLS measurements versus size for pAbS1N@AuNPs before and after incubation with inactivated SARs-CoV-2 virus at 2.11 × 105 TCID50/mL. (B) Effect of SARS-CoV-2 virus concentration on the hydrodynamic diameter of pAbS1N18@AuNPs after conjugation. In both (A) and (B), the results are given as the mean of two experiments and five DLS measurements.
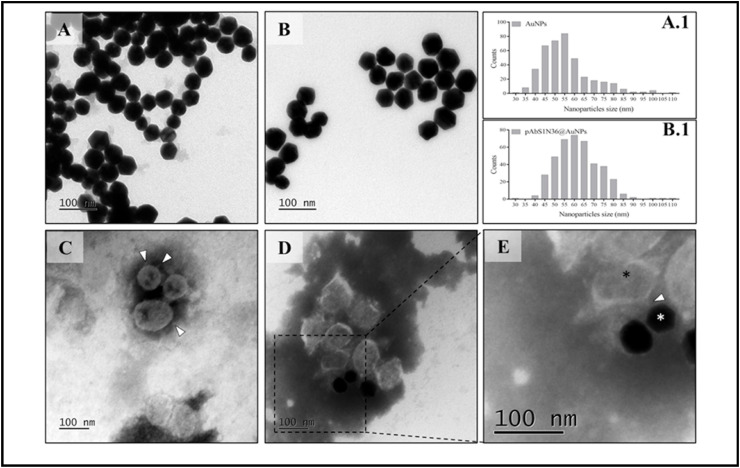
The binding of SARS-CoV-2 to pAbS1N36@AuNPs bioconjugates was also suggested using TEM, with representative images in Fig. 5 A-D. Fig. 5C shows the SARS-CoV-2 viral particles with spike proteins on the virus surface (marked in the image). The binding of pAbS1N36@AuNPs with SARS-CoV-2 can be observed in Fig. 5D, and the interaction of the virus surface spike protein with the bioconjugate is suggested with the magnified image in Fig. 5E.
Fig. 5.
TEM micrographs depicting the interaction of SARS-CoV-2 and the bioconjugate (pAbS1N36@AuNPs). (A) AuNPs; (A.1) histogram of particle size distribution for AuNPs; (B) pAbS1N36@AuNPs; (B.1) histogram of particle size distribution for pAbS1N36@AuNPs; (C) inactivated SARS-CoV-2 virus; (D) reaction between pAbS1N36@AuNPs probes and inactivated SARS-CoV-2 virus at 2.11 × 105 TCID50/mL; and (E) zoomed image from a region in image D, which demonstrates the interaction between pAbS1N36@AuNPs (white asterisk) and SARS-CoV-2 virus (black asterisk). The bar is 100 nm. White arrowheads show the spike protein of the viruses, while the white asterisk shows the nanoparticles.
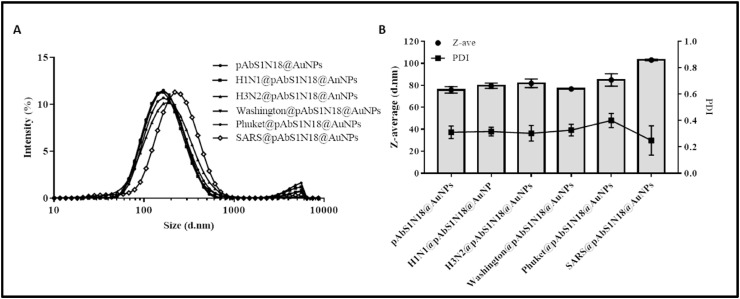
In addition to a suitable sensitivity in detecting SARS-CoV-2, the interaction between antibodies and viruses must be sufficiently specific to avoid giving false-positive results. We have confirmed the selectivity of SARS-CoV-2 detection using DLS by testing pAbS1N18@AuNPs bioconjugate incubated with Influenza Type A Brisbane -H1N1 (H1N1@pAbS1N18@AuNPs), South Australia-H3N2 (H3N2@pAbS1N18@AuNPs), Influenza Type B Washington (Washington@pAbS1N18@AuNPs), and Phuket (Phuket@pAbS1N18@AuNPs) strains. Fig. 6 A shows minor changes in nanoparticle peak size distributions, with the maximum difference being 10 nm for the Phuket strain – compared with almost hundreds of nm after SARS-CoV-2 incubation. The differences in size were confirmed by the hydrodynamic diameter (z-average) values in Fig. 6B, where the SARS@pAbS1N18@AuNPs sample has a larger z-average, indicating an interaction of the antibody with SARS-CoV-2.
Fig. 6.
Selectivity of SARS-CoV-2 virus with the bioconjugate pAbS1N18@AuNPs. (A) Size diameter distribution by intensity for different strains A/Brisbane-H1N1, A/South Australia-H2N3, B/Washington, B/Phuket, and SARS-CoV-2 viruses. (B) Hydrodynamic Diameter (Z-Average) and polydispersity index (PDI) of the samples. All viruses were maintained and analyzed in 20% sucrose PBS solution. The results were presented as the average of one experiment, and five DLS runs measurements.
4. Conclusions
We developed efficient immunosensors based on pAbS1N@AuNPs bioconjugates consisting of AuNPs functionalized with antibodies against SARS-CoV-2 spike protein. The immunosensor was used for rapid (30 min maximum), sensitive detection of SARS-CoV-2 via dynamic light scattering (DLS). The bioconjugates showed to be very stable – at least for 210 days in terms of the hydrodynamic diameter and polydispersity index, which were maintained stabilized due to the presence of amide bonds between antibodies and crosslinker. The specific binding of SARS-CoV-2 spike protein led to a considerable increase in the hydrodynamic diameter for the bioconjugates. Furthermore, selectivity toward SARS-CoV-2 was confirmed in experiments in which no change in the size of the nanoparticles bioconjugates was noted upon interaction with influenza viruses. The main advantages of the method employed here include operational simplicity in the detection mode and offering lower prices for components and each test. The protocols and strategies used for the bioconjugates described here can be extended to colorimetric sensing, and one may envisage its use for low-cost mass testing.
Author contributions
Patricia Bento da Silva: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Jaqueline Rodrigues da Silva: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Mosar Corrêa Rodrigues: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Julia Augusto Vieira: Data curation; Investigation; Visualization. Ikaro Alves de Andrade: Methodology; Validation; Visualization; Roles/Writing - original draft. Tatsuya Nagata: Methodology; Validation; Visualization; Roles/Writing - original draft. Alexandre Silva Santos: Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Sebastião William da Silva: Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Marcia Cristina Oliveira da Rocha: Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Sônia Nair Báo: Methodology; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Pedro M Moraes-Vieira: Methodology; Validation; Visualization. José Proença-Modena: Methodology; Validation; Visualization. Monara K C Angelim: Methodology; Validation; Visualization. Gabriela Fabiano de Souza: Methodology; Validation; Visualization. Stefanie Primon Muraro: Methodology; Validation; Visualization. André Luis Branco de Barros: Conceptualization; Formal analysis; Roles/Writing - original draft. Glêndara Aparecida de Souza Martins: Conceptualization; Project administration; Software; Roles/Writing - original draft. Fátima Ribeiro-Dias: Conceptualization; Software; Roles/Writing - original draft. Giovanna Machado: Conceptualization; Project administration; Software; Roles/Writing - original draft; Writing – review & editing. Melissa Regina Fessel: Methodology; Validation; Visualization. Ana Marisa Chudzinski-Tavassi: Methodology; Validation; Visualization. Célia Machado Ronconi: Conceptualization; Resources; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Debora Gonçalves: Conceptualization; Data curation; Formal analysis; Roles/Writing - original draft; Writing – review & editing. Rui Curi: Conceptualization; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Osvaldo N. Oliveira Jr: Conceptualization; Project administration; Resources; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing. Ricardo Bentes Azevedo: Conceptualization; Funding acquisition; Project administration; Resources; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
CNPq (402816/2020-0), FAPERJ (E26/210.194/2020), CAPES, and FAPESP (2018/22214-6) supported this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123355.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data, (n.d.). https://covid19.who.int/(accessed August 15, 2021).
- 3.Giovannini G., Haick H., Garoli D. Detecting COVID-19 from breath: a game changer for a big challenge. ACS Sens. 2021;6:1408–1417. doi: 10.1021/acssensors.1c00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury R., Luhar S., Khan N., Choudhury S.R., Matin I., Franco O.H. Long-term strategies to control COVID-19 in low and middle-income countries: an options overview of community-based, non-pharmacological interventions. Eur. J. Epidemiol. 2020;35:743–748. doi: 10.1007/s10654-020-00660-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfano V., Ercolano S. The efficacy of lockdown against COVID-19: a cross-country panel analysis. Appl. Health Econ. Health Pol. 2020;18:509–517. doi: 10.1007/s40258-020-00596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burki T. Mass testing for COVID-19. The Lancet Microbe. 2020;1:e317. doi: 10.1016/s2666-5247(20)30205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hosani F., Al-Mazrouei S., Al-Memari S., Al-Yafei Z., Paulo M.S., Koornneef E. A review of COVID-19 mass testing in the United Arab Emirates _ enhanced Reader.pdf. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.661134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., Horiuchi M., Kato K., Imoto Y., Iwata M., Mimura S., Ito T., Tamura K., Kato Y. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), Direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira B.A., de Oliveira L.C., Sabino E.C., Okay T.S. SARS-CoV-2 and the COVID-19 disease: a mini review on diagnostic methods. Rev. Inst. Med. Trop. Sao Paulo. 2020;62:1–8. doi: 10.1590/S1678-9946202062044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M., Li Q., Zhou J., Ai W., Zheng X., Zeng J. Value of swab types and collection time on SARS-COV-2 detection using RT-PCR assay. J. Virol. Methods. 2020;286:113974–113979. doi: 10.1016/j.jviromet.2020.113974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashir J., Yaqinuddin A. Loop mediated isothermal ampli fication (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses. 2020;141:30–34. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR – cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganbaatar U., Liu C. CRISPR-based COVID-19 testing: toward next-generation point-of-care diagnostics. Front. Cell. Infect. Microbiol. 2021;11:1–11. doi: 10.3389/fcimb.2021.663949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soares J.C., Soares A.C., Rodrigues V.C., Oiticica P.R.A., Raymundo-Pereira P.A., Bott-Neto J.L., Buscaglia L.A., De Castro L.D.C, Ribas L.C., Scabini L., Brazaca L.C., Correa D.S., Mattoso L.H.C., De Oliveira M.C.F., De Carvalho A.C.P.L.F., Carrilho E., Bruno O.M., Melendez M.E., Oliveira O.N., Jr. Detection of a SARS-CoV-2 sequence with genosensors using data analysis based on information visualization and machine learning techniques. Mater. Chem. Front. 2021;5(15):5658–5670. doi: 10.1039/d1qm00665g. [DOI] [Google Scholar]
- 16.Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sensor. Actuator. B Chem. 2021;329 doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., Faggioni G., DÁmore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen, Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beduk T., Beduk D., de Oliveira Filho J.I., Zihnioglu F., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Salama K.N., Timur S. Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal. Chem. 2021;93:8585–8594. doi: 10.1021/acs.analchem.1c01444. [DOI] [PubMed] [Google Scholar]
- 20.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., Cipolla M., Viant C., Barnes C.O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K.G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Robbiani D.F., Zhao Z., Gazumyan A., Schwartz R.E., Hatziioannou T., Bjorkman P.J., Mehandru S., Bieniasz P.D., Caskey M., Nussenzweig M.C. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Hu Y., Yu Y., Zhang X., Li B., Wu J., Li J., Wu Y., Xia X., Tang H., Xu J. Positive result of Sars-Cov-2 in faeces and sputum from discharged patients with COVID-19 in Yiwu, China. J. Med. Virol. 2020;92:1938–1947. doi: 10.1002/jmv.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodino K.G., Espy M.J., Buckwalter S.P., Walchak R.C., Germer J.J., Fernholz E., Boerger A., Schuetz A.N., Yao J.D., Binnicker M.J. Evaluation of saline, phosphate-buffered saline, and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdy M.E., Del Carlo M., Hussein H.A., Salah T.A., El-Deeb A.H., Emara M.M., Pezzoni G., Compagnone D. Development of gold nanoparticles biosensor for ultrasensitive diagnosis of foot and mouth disease virus. J. Nanobiotechnol. 2018;16:48–60. doi: 10.1186/s12951-018-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moulahoum H., Ghorbanizamani F., Zihnioglu F., Turhan K., Timur S. How should diagnostic kits development adapt quickly in COVID 19-like pandemic models? Pros and cons of sensory platforms used in COVID-19 sensing. Talanta. 2021;222 doi: 10.1016/j.talanta.2020.121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaculovicova M., Michalek P., Krizkova S., Macka M., Adam V. Nanotechnology-based analytical approaches for detection of viruses. Anal. Methods. 2017:1–56. doi: 10.1039/C7AY00048K. [DOI] [Google Scholar]
- 26.Basso C.R., Tozato C.C., Crulhas B.P., Castro G.R., Junior J.P.A., Pedrosa V.A. An easy way to detect dengue virus using nanoparticle-antibody conjugates. Virology. 2018;513:85–90. doi: 10.1016/j.virol.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Malekzad H., Zangabad P.S., Mirshekari H., Karimi M., Hamblin M.R. Noble metal nanoparticles in biosensors: recent studies and applications. Nanotechnol. Rev. 2017;6:301–329. doi: 10.1515/ntrev-2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu N., Chen C., Yang C., Kao Y., Wu W. Enhanced plasmonic biosensors of hybrid gold nanoparticle-graphene oxide-based label-free immunoassay. Nanoscale Res. Lett. 2018;13:152–153. doi: 10.1186/s11671-018-2565-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aldewachi H., Chalati T., Woodroofe M.N., Bricklebank N., Shadrack B., Gardiner P. Gold nanoparticle-based colorimetric biosensors. Nanoscale. 2017;10:18–33. doi: 10.1039/C7NR06367A. [DOI] [PubMed] [Google Scholar]
- 30.Chang C.-C., Chen C.-P., Wu T.-H., Yang C.-H., Lin C.-W., Chen C.-Y. Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications. Nanomaterials. 2019;9:861–885. doi: 10.3390/nano9060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P., Wang Y., Zhao L., Ji C., Chen D., Nie L. Applications of gold nanoparticles in non-optical biosensors. Nanomaterials. 2018;8:977–1000. doi: 10.3390/nano8120977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jans H., Liu X., Austin L., Maes G., Huo Q. Dynamic light scattering as a powerful tool for gold nanoparticle bioconjugation and biomolecular binding studies. Anal. Chem. 2009;81:9425–9432. doi: 10.1021/ac901822w. [DOI] [PubMed] [Google Scholar]
- 33.Stetefeld J., Mckenna S.A., Patel T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Driskell J.D., Jones C.A., Tompkins S.M., Tripp R.A. One-step assay for detecting influenza virus using dynamic light scattering and gold nanoparticles. Analyst. 2011;136:3083–3090. doi: 10.1039/c1an15303j. [DOI] [PubMed] [Google Scholar]
- 35.Hayat M.A., Miller S.E. McGraw-Hill Pub. Co; 1990. Negative Staining. [Google Scholar]
- 36.Keating C.D., Musick M.D., Keefe M.H., Natan M.J. Kinetics and thermodynamics of Au colloid monolayer undergraduate experiments in surface and nanomaterials chemistry. J. Chem. Educ. 1999;76:949–955. [Google Scholar]
- 37.Wang W., Ding X., Xu Q., Wang J., Wang L., Lou X. Zeta-potential data reliability of gold nanoparticle biomolecular conjugates and its application in sensitive quantification of surface absorbed protein. Colloids Surf. B Biointerfaces. 2016;148:541–548. doi: 10.1016/j.colsurfb.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Zhao W., Chiuman W., Lam J.C.F., McManus S.A., Chen W., Cui Y., Pelton R., Brook M.A., Li Y. DNA aptamer folding on gold nanoparticles: from colloid chemistry to biosensors. J. Am. Chem. Soc. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- 39.Du S., Kendall K., Toloueinia P., Mehrabadi Y., Gupta G., Newton J. Aggregation and adhesion of gold nanoparticles in phosphate buffered saline. J. Nanoparticle Res. 2012;14:758–772. doi: 10.1007/s11051-012-0758-z. [DOI] [Google Scholar]
- 40.Naumann D. Infrared spectroscopy in microbiology. Encycl. Anal. Chem. 2006 doi: 10.1002/9780470027318.A0117. [DOI] [Google Scholar]
- 41.McGee W.M., McLuckey S.A. Efficient and directed peptide bond formation in the gas phase via ion/ion reactions. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:1288–1292. doi: 10.1073/pnas.1317914111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willets K.A. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal. Bioanal. Chem. 2009;394:85–94. doi: 10.1007/s00216-009-2682-3. [DOI] [PubMed] [Google Scholar]
- 43.Chen M.C., Lord R.C. Laser-excited Raman spectroscopy of biomolecules . VIII. Conformational study of bovine serum albumin. Jounal Am. Chem. Soc. 1976;98:990–992. doi: 10.1021/ja00420a021. [DOI] [PubMed] [Google Scholar]
- 44.Da Paz M.C., Santos M.F.M.A., Santos C.M.B., Silva S.W., De Souza L.B., Silva R.C., Lucci C.M., Morais P.C., Azevedo R.B., Lacava Z.G.M. Anti-CEA loaded maghemite nanoparticles as a theragnostic device for colorectal cancer. Int. J. Nanomed. 2012;7:5271–5282. doi: 10.2147/IJN.S32139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.