Abstract
Glutamate is a principal neurotransmitter used extensively by the nervous systems of all vertebrate and invertebrate animals. It is primarily an excitatory neurotransmitter that has been implicated in nervous system development, as well as a myriad of brain functions from the simple transmission of information between neurons to more complex aspects of nervous system function including synaptic plasticity, learning, and memory. Identification of glutamatergic neurons and their sites of glutamate release are thus essential for understanding the mechanisms of neural circuit function and how information is processed to generate behavior. Here, we describe and characterize smFLAG-vGlut, a conditional marker of glutamatergic synaptic vesicles for the Drosophila model system. smFLAG-vGlut is validated for functionality, conditional expression, and specificity for glutamatergic neurons and synaptic vesicles. The utility of smFLAG-vGlut is demonstrated by glutamatergic neurotransmitter phenotyping of 26 different central complex neuron types of which nine were established to be glutamatergic. This illumination of glutamate neurotransmitter usage will enhance the modeling of central complex neural circuitry and thereby our understanding of information processing by this region of the fly brain. The use of smFLAG for glutamatergic neurotransmitter phenotyping and identification of glutamate release sites can be extended to any Drosophila neuron(s) represented by a binary transcription system driver.
Keywords: Drosophila, vGlut, glutamatergic, synaptic vesicle, epitope tag
The development and characterization of a conditional, epitope-tagged glutamatergic synaptic vesicle for the Drosophila model system is reported. The conditional glutamatergic synaptic vesicle marker, smFLAG-vGlut, is demonstrated to be conditionally expressible, specifically expressed in glutamatergic neurons, and subcellularly localized to glutamatergic synaptic vesicles. Additionally, the utility of smFLAG-vGlut is demonstrated by glutamatergic neurotransmitter phenotyping of 26 different neuron types of the central complex of which nine were determined to be glutamatergic.
Introduction
Glutamate is one of the three major neurotransmitters used by organisms throughout the animal kingdom. The ability to identify glutamatergic neurons and define sites of glutamate release are therefore essential to understanding neural circuit function and behavior. Neurons do not have a dedicated neurotransmitter synthesis enzyme for glutamate, as exists for the two other major fast neurotransmitters acetylcholine (cholineacetyltransferase) and GABA (glutamic acid decarboxylase), that can be used as a marker for glutamatergic neurons. The established marker for the identification of glutamatergic neurons is therefore the presence of vesicular glutamate transporters (vGluts) that localize to synaptic vesicles (SVs) and fill them with glutamate (Takamori 2006; Liguz-Lecznar and Skangiel-Kramska 2007). However, it can be challenging to rely on vGlut for glutamatergic neurotransmitter phenotyping. Synaptic release sites are often far from the cell body and intermingled with densely packed synapses from many other neurons, thereby making it difficult to trace vGlut signal present at a synapse back to individual cell bodies.
To overcome this challenge, we have developed a conditional epitope-tagged variant of vGlut via CRISPR/Cas9 genome editing to enable the identification of glutamatergic neurons and distinguish glutamate release sites in the Drosophila model system. Genome editing was performed at the endogenous vGlut locus to ensure the complete regulatory region of vGlut is intact and thus that the epitope-tagged vGlut protein will be expressed in all glutamatergic neurons. Furthermore, expression of epitope-tagged vGlut under its own promoter minimizes the likelihood of subcellular mislocalization due to expression above endogenous levels that typically occurs with ectopic expression from binary transcription systems (GAL4, LexA, Q).
Here, we describe the development and characterization of B2RT-STOP-B2RT-smFLAG-vGlut. The B2 recombinase target site (B2RT) flanked transcription STOP cassette provides conditionality and the spaghetti monster (sm) FLAG tag confers robust signal by virtue of its 10 copies of the FLAG epitope tag (Viswanathan et al. 2015). Immunolocalization, genetic, and electrophysiology data are presented validating smFLAG-vGlut as a conditional marker for glutamatergic SVs. B2RT-STOP-B2RT-smFLAG-vGlut is ideal for use in combination with binary transcription system drivers that express in small neuronal subsets, especially the already extensive and continually expanding collection of split-GAL4 drivers for single neuron types (Aso et al. 2014; Robie et al. 2017; Wolff and Rubin 2018; Dolan et al. 2019; Davis et al. 2020), among others. Last, the utility of B2RT-STOP-B2RT-smFLAG-vGlut for glutamatergic neurotransmitter phenotyping and identification of glutamate release sites is demonstrated using a collection of split-GAL4 drivers representing single neuron types of the central complex.
Materials and methods
Plasmid construction
The pCFD4-vGlut double guide RNA plasmid was generated as previously described (Port et al. 2014) and contains guide RNA sequences ttgatccggaggcagggg and cacgtgtcgcgccccccc. The B2RT-STOP-B2RT-smFLAG-vGlut donor plasmid was assembled in vector pHSG298 (Takara Biosciences) using NEBuilder HiFi (New England Biolabs). The mRuby2_smFP FLAG coding sequence was obtained from plasmid pCAG_mRuby2_smFP FLAG (Addgene plasmid # 59760) as a gift of Loren Looger (Viswanathan et al. 2015). The complete annotated sequence of the donor plasmid is shown in Supplementary Fig. 1.
Genome editing
The pCFD4-vGlut guide RNA plasmid was co-injected with the B2RT-STOP-B2RT-smFLAG-vGlut donor plasmid into embryos of strain nos-Cas9 TH_attP2 (Ren et al. 2013) by Bestgene, Inc. The surviving adults that were injected as embryos were crossed to the balancer stock yw; Sp/CyO. Approximately 100 male CyO progeny were crossed individually to yw; vGlutSS1/CyO and screened for failed complementation on the expectation the desired genome edit would create a lethal vGlut allele. Each male exhibiting failed complementation was subsequently crossed to strain yw; 20XUAS-DSCP-B2; n-syb-GAL4 and third instar larval progeny of the appropriate genotype were assessed for anti-FLAG immunostaining. A stable stock of B2RT-STOP-B2RT-smFLAG-vGlut was established from males showing positive immunostaining in the larval ventral nerve cord (VNC).
Immunostaining
Larval and adult immunostaining were performed as previously described (Certel and Thor 2004; Petersen and Stowers 2011). Primary antibodies and dilution factors: The SYN (3C11) mAb 1:25 developed by E. Buchner (Klagges et al. 1996) was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Mouse anti-vGlut (Banerjee 2021) 1:10; Rabbit anti-Syt (Littleton et al. 1993) 1:1,000; Mouse anti-mCherry (Biorbyt orb256058) 1:200; Rabbit anti-mCherry (Abcam ab213511) 1:500, Rabbit Abfinity anti-GFP (Thermo-Fisher) 1:400, Rabbit anti-HA (Cell Signaling C29F4) 1:500, and Rat anti-FLAG (Novus NBP1-06712) 1:200. Secondary antibodies and dilution factors: Donkey anti-Rat Alexa 488 (Jackson Immunoresearch 712-546-153) 1:400, Donkey anti-Mouse Alexa 488 (Jackson Immunoresearch 715-545-151) 1:400, Goat anti-Rabbit Alexa 488 (Thermo-Fisher A32731) 1:200, Donkey anti-Mouse JF549 (Novus NBP1-75119JF549) 1:200, Goat anti-Rabbit JF549 (Novus NBP1-72732JF549) 1:200, Goat anti-Rabbit JF646 (NBP1-72732JF646). Larval muscles were stained with Phalloidin 405 (Biotium CF405M).
Germline excision
Germline excision of the STOP cassette of B2RT-STOP-B2RT-smFLAG-vGlut was accomplished by crossing to yw; nos-GAL4; 20XUAS-DSCP-B2. Progeny males of the appropriate genotype were crossed to a second chromosome balancer stock to recover potential germline excision chromosomes. Progeny males resulting from the balancer cross were individually crossed to the second chromosome balancer stock again and third instar larva were screened by immunostaining for anti-FLAG immunofluorescence.
Electrophysiology
Wandering third instar larvae were dissected in HL3.1 (0 mM Ca2+) saline as previously described (Feng et al. 2004; Imlach and McCabe 2009). Two-electrode voltage-clamp recordings of larval neuro-muscular glutamatergic synaptic currents were carried out using borosilicate glass electrodes (1B120F-4, World Precision Instruments, FL, USA). Voltage follower electrodes (HS-9Ax0.1) were pulled (Sutter P-1000, CA, USA) to resistances of between 25 and 30 MΩ. Current injection electrodes (HS-9Ax1) were pulled to resistances of 15–20 MΩ and filled with 3 M KCl. Suction electrodes (GC120T-10, Harvard Apparatus, Kent, UK) were fire polished to a diameter of ∼5 µm and filled with recording saline, HL3.1 (1.0 mM Ca2+). mEJC recordings were conducted from muscle 6, segment A3. Muscle fibers were held at −60 mV, recordings were sampled at 20 kHz and lowpass filtered at 0.1 kHz, using pClamp 11 (Molecular Devices, Sunnyvale, CA, USA). mEJC amplitude and frequency were averaged over each 2-min recording. Muscle input resistance was calculated from the muscle voltage response to a −1 nA/500 ms current pulse. Recordings were rejected for analysis if current injection was >10 nA and/or muscle input resistance was <5 MΩ.
Fly strains
Stocks from the Bloomington Drosophila Stock Certer (NIH P40OD018537) were used in this study. Previously described fly strains: 20XUAS-DSCP-B2 and B2RT-STOP-B2RT-GFP-Rab3 (Williams et al. 2019); vGlutSS1 (Sherer et al. 2020), UAS-CD8-mCherry (BDSC # 27392); nos-GAL4 (Tracey et al. 2000) (BDSC # 4442); MBON-6/MB434B (Aso et al. 2014); LH2094 and LH1900 (Dolan et al. 2019); 3XUAS-Syt-smGFP-HA, SS52244, SS02255, SS52245, SS00078, SS52577, SS54295, SS27853, SS02195, SS00090, SS02254, SS52490, SS54549, SS02239, SS52578, SS00117, SS52266, SS52267, SS04778, SS02198, SS46517, SS47398, SS47432, SS47384, SS46525, SS50464, and SS02718 (Wolff and Rubin 2018). The complete genotypes of all experimental fly strains shown in the figures can be found in Supplementary material.
Results
Strategic design of a conditional epitope-tagged glutamatergic SV marker
To develop a conditional glutamatergic SV marker for Drosophila, the endogenous genomic locus of vGlut was chosen for conditional epitope-tagging via CRISPR/Cas9 genome editing. Unlike mammalian species that possess three vGlut homologs expressed in distinct and overlapping neuronal subsets (Liguz-Lecznar and Skangiel-Kramska 2007; Zhang et al. 2018), Drosophila contains only a single vGlut gene (Adams et al. 2000) that is used by all glutamatergic neurons. Thus, conditional tagging of the sole Drosophila vGlut gene is sufficient as a glutamatergic SV marker for all Drosophila glutamate-expressing neurons. In addition, tagging the vGlut genomic locus ensures recapitulation of the complete neuronal expression pattern of vGlut since the entire vGlut regulatory region is present to direct vGlut expression. Finally, expression of tagged-vGlut under its genomic regulatory region should result in endogenous levels of expression and, therefore, an SV subcellular distribution that recapitulates that of native vGlut. However, the possibility exists that the addition of the epitope tag could alter the trafficking, function, or stability of vGlut.
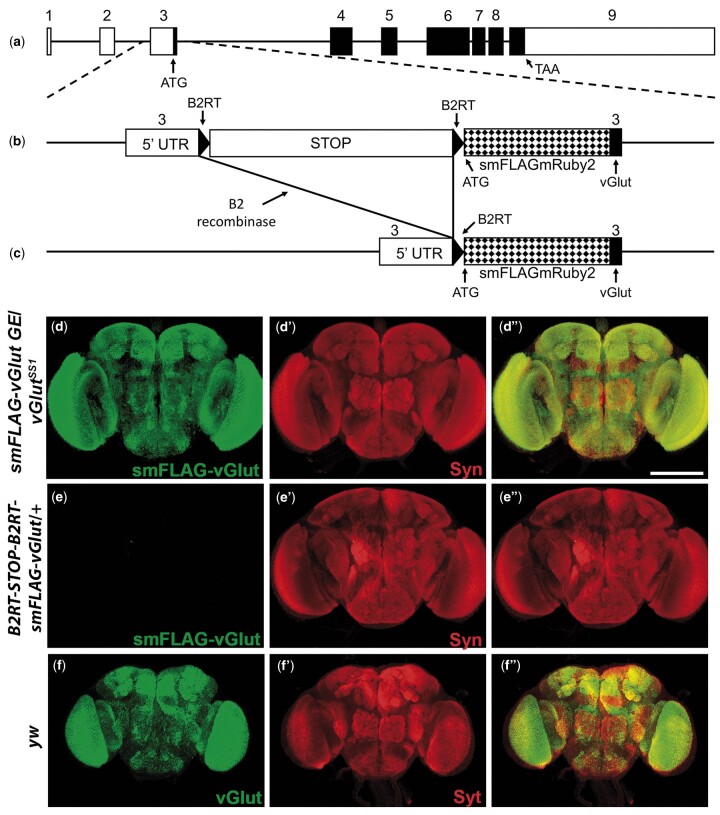
The amino-terminus of vGlut was selected as the location of the epitope tag as vGlut has previously been demonstrated to tolerate amino-terminal fusion of GFP (Daniels et al. 2008). Since tagged vGlut will presumably be expressed at or near relatively low endogenous levels, and thus, detection could be challenging, mRuby2_smFP FLAG was chosen as the tag to enhance sensitivity as it contains 10 copies of the FLAG epitope (Viswanathan et al. 2015). Genome editing was targeted to the third exon of the vGlut gene containing the ATG start codon (Fig. 1a). A transcription STOP cassette flanked by B2 recombinase target sites (B2RTs) (Nern et al. 2011) was inserted in the 5′ UTR just upstream of the translation start site along with mRuby2-_smFP FLAG immediately after the endogenous vGlut ATG translation start codon (Fig. 1b). The STOP cassette prevents transcription of the mRuby2_smFP FLAG-vGlut (hereafter smFLAG-vGlut) prior to excision. However, after excision of the STOP cassette in neurons of interest via expression of the B2 recombinase, smFLAG-vGlut is expressed in neurons that are glutamatergic (Fig. 1c).
Fig. 1.
Strategic design of the conditional glutamatergic synaptic vesicle marker B2RT-STOP-B2RT-smFLAG-vGlut and conditional neuropil-specific expression in adult brain. a) Genomic exon structure of Drosophila vGlut. b) Genome editing at the endogenous vGlut genomic locus included insertion of a transcription STOP cassette flanked by B2 recombinase target sites (B2RTs) in the 5′ UTR upstream of translation start and insertion of the mRuby2_FP FLAG coding sequences in exon 3 at the ATG start codon of vGlut. Prior to excision of the STOP cassette smFLAG-vGlut is not expressed. c) After selective expression of the B2 recombinase in neurons of interest, with a binary transcription system driver and a compatible B2 recombinase responder transgene, the STOP cassette is excised and smFLAG-vGlut is expressed in glutamatergic neurons. d–e) Conditional expression in adult brain. d–d″) smFLAG-vGlut germline excision/vGlutSS1. d) smFLAG-vGlut; d′) Syn; d″) overlay. e–e″) B2RT-STOP-B2RT-smFLAG-vGlut/+. e) smFLAG-vGlut; e′) Syn; e″) overlay. f–f″) yw. f) vGlut; f′) Syt; f″) overlay. smFLAG-vGlut with a germline excision of the STOP cassette exhibits strong neuropil-specific immunostaining in the adult brain only after STOP cassette excision similar to endogenous vGlut. Images in (d) and (e) were collected and processed identically. Syn-Synapsin; Syt-Synaptotagmin. Scale bar: 100 µm.
Conditionality and SV specificity assessment
The primary intent for B2RT-STOP-B2RT-smFLAG-vGlut was for it to be a conditional marker of glutamatergic SVs. To evaluate conditionality, expression of smFLAG-vGlut was assessed with and without germline excision of the STOP cassette in the adult fly brain, the third instar larval VNC, and the third instar larval neuromuscular junction (NMJ). Anti-FLAG immunostaining of smFLAG-vGlut in adult fly brains containing a germline excision of the STOP cassette reveals robust signal broadly distributed throughout the brain (Fig. 1d). In contrast, with the STOP cassette intact no detectable anti-FLAG immunostaining was observed in B2RT-STOP-B2RT-smFLAG-vGlut adult fly brains (Fig. 1e). Similarly, in the third instar larval VNC (Fig. 2a) and at the third larval instar NMJ (Fig. 2d), strong signal was observed in smFLAG-vGlut germline excision larva, but no signal was detected with the STOP cassette present (Fig. 2, b and e, respectively). These results demonstrate the effectiveness of the transcription STOP cassette as there is no detectable constitutive expression, or “leak,” of smFLAG-vGlut in either the larval or adult nervous systems prior to excision.
Fig. 2.
Assessment of the conditionality and synaptic vesicle specificity of B2RT-STOP-B2RT-smFLAG-vGlut in larval VNC and third instar larval NMJ. Third instar larval VNC. a–a″) smFLAG-vGlut germline excision/vGlutSS1. a) smFLAG-vGlut; a′) Syn; a″) overlay. b–b″) B2RT-STOP-B2RT-smFLAG-vGlut/+. b) smFLAG-vGlut; b′) Syn; b″) overlay. c–c″) yw. c) vGlut; c′) Syt; c″) overlay. Third instar larval NMJ. d–d″) B2RT-smFLAG-vGlut germline excision/vGlutSS1. d) smFLAG-vGlut; d′) Syn; d″) overlay. e–e″) B2RT-STOP-B2RT-smFLAG-vGlut/+. e) smFLAG-vGlut; e′) Syn; e″) overlay. f–f″) yw. f) vGlut; f′) Syt; f″) overlay. smFLAG-vGlut with a germline excision of the STOP cassette exhibits strong neuropil-specific immunostaining in the larval VNC and at presynaptic terminals of the third larval instar NMJ similar to endogenous vGlut. No detectable anti-FLAG immunostaining is observed in the larval VNC or third instar larval NMJ prior to excision of the STOP cassette. Images in (a) and (b) and images in (d) and (e) were collected and processed identically. Syn, synapsin; Syt, Synaptotagmin. Scale bars: 100 µm.
To assess the specificity of smFLAG-vGlut for localization to SVs, colabeling with the SV-specific protein Synapsin (SYN) was also included in the above experiments. smFLAG-vGlut expression (Fig. 1d) was restricted to the neuropil regions of the brain as indicated by the localization of SYN (Fig. 1d′). Not unexpectedly, the distribution of SYN is somewhat broader than that of smFLAG-vGlut (Fig. 1d″) because SYN is present not just on glutamatergic SVs but on all SVs independent of neurotransmitter usage. Additional evidence consistent with the SV specificity of smFLAG-vGlut is that its expression closely resembles that of endogenous vGlut (Fig. 1f). Similarly, in the third instar larval VNC (Fig. 2a) and at the third instar larval NMJ (Fig. 2d) smFLAG-vGlut expression overlaps with SYN expression (Fig. 2, a′ and d′, respectively). These patterns of expression of smFLAG-vGlut also closely resemble that of endogenous vGlut in the larval VNC and at the third instar larval NMJ (Fig. 2, c and f, respectively).
An additional experiment was performed evaluating smFLAG-vGlut localization to glutamatergic SVs that involved simultaneous assessment of the distribution of smFLAG-vGlut and native vGlut in the same animal. Immunolabeling was performed on smFLAG-vGlut germline excision/+ adult brains, larval VNCs, and larval NMJs using both anti-FLAG and anti-vGlut antibodies. Although the anti-vGlut antibody recognizes both native vGlut and smFLAG-vGlut, the anti-FLAG antibody will only recognize smFLAG-vGlut. It is thus possible there could be signal from anti-FLAG that does not overlap with signal from anti-vGlut if there is a discrepancy in expression. However, anti-FLAG and anti-vGlut immunolabeling of adult brain (Supplementary Fig. 2,a and a′, respectively), larval VNC (Supplementary Fig. 2,b and b′, respectively), and NMJ (Supplementary Fig. 2,c and c′, respectively) are nearly indistinguishable (Supplementary Fig. 2, a″, b″), and c″. Taken together, these results showing overlapping expression of smFLAG-vGlut and endogenous vGlut in the same animal, combined with results above showing similar distributions of smFLAG-vGlut and endogenous vGlut in different animals, and the overlapping expression of smFLAG-vGlut and SYN, is consistent with the specificity of smFLAG-vGlut for localization to glutamatergic SVs.
The effectiveness of the STOP cassette in suppressing expression of smFLAG-vGlut was also assessed genetically by complementation testing with the vGlut complete loss-of-function allele vGlutSS1. B2RT-STOP-B2RT-smFLAG-vGlut/vGlutSS1 trans-heterozygotes were determined to be noncomplementary for vGlut function with an embryonic lethal phase. As it has previously been determined that vGlut complete loss-of-function mutants exhibit an embryonic lethal phase (Daniels et al. 2006), this result suggests B2RT-STOP-B2RT-smFLAG-vGlut is a complete loss-of-function, or null, allele of vGlut. This genetic result is consistent with the absence of detectable expression of smFLAG-vGlut when the STOP cassette is intact, as shown above.
In contrast, after germline excision of the STOP cassette, smFLAG-vGlut/vGlutSS1 trans-heterozygotes were found to be viable (Fig. 2, a, d, and g) and fertile. The smFLAG-vGlut germline excision chromosome was also determined to be homozygous viable and fertile, and a smFLAG-vGlut germline excision homozygous stock was established that could be propagated over multiple generations. These genetic results imply smFLAG-vGlut is a functional vesicular glutamate transporter.
As an additional test of the functionality of smFLAG-vGlut, electrophysiology was performed at the third instar larval NMJ of smFLAG-vGlut germline excision homozygotes (smFLAG-vGlut GE), smFLAG-vGlut GE in trans to the null allele vGlutSS1, and compared to Canton-S wild-type controls. Mean spontaneous mEJC frequencies were determined to be 1.77 ± 0.31 Hz for smFLAG-vGlut GE homozygotes, 1.98 ± 0.32 Hz for smFLAG-vGlut GE/vGlutSS1, and 1.92 ± 0.35 Hz for Canton-S wildtype controls (Supplementary Fig. 3a). The mean mEJC amplitudes were 0.48 ± 0.01 nA for smFLAG-vGlut GE homozygotes, 0.48 ± 0.02 nA for smFLAG-vGlut GE/vGlutSS1, and 0.47 ± 0.02 nA for Canton-S wildtype controls (Supplementary Fig. 3b). For both measurements, no statistically significant differences were observed between any of the three genotypes for either mEJC frequency or mEJC amplitude. These electophysiology results are consistent with the genetic results above in affirming smFLAG-vGlut as a functional vGlut capable of restoring vGlut activity to mutant alleles.
Assessment of smFLAG-vGlut for SV and neurotransmitter specificity in single neuron types
To determine if smFLAG-vGlut expression is specific for glutamatergic neurons and SVs, the STOP cassette of B2RT-STOP-B2RT-smFLAG-vGlut was excised using a 20XUAS-DSCP-B2 recombinase transgene in combination with split-GAL4 drivers representing single neuron types of known neurotransmitter usage previously determined using independent methods. The split-GAL4 driver for mushroom body output neurons MBON-6 was selected as a positive control for glutamatergic neurons. To visualize the anatomy of each neuron of interest, the plasma membrane marker CD8-mCherry was included in this and subsequent experiments. MBON-6 is a highly polarized neuron with distinct axonal (large arrow), dendritic (small arrow), and cell body (arrowhead) regions (Fig. 3a) (Aso et al. 2014; Tison et al. 2020). smFLAG-vGlut exhibits robust signal in MBON-6 neurons and distributes predominantly to axon terminals (Fig. 3a′), similar to the distribution of the conditional SV marker GFP-Rab3 (Williams et al. 2019) (Fig. 3c′). Virtually no smFLAG-vGlut signal is observed in dendritic regions although it is detectable in the cell body. The cell body expression is presumably nascently translated smFLAG-vGlut protein awaiting transport to presynaptic terminals. High magnification (100X) images of the axon terminals of MBON-6 neurons coexpressing the SV marker Syt-smHA (Fig. 3b) and smFLAG-vGlut (Fig. 3b′) reveals nearly complete colocalization (Fig. 3b″).
Fig. 3.
Assessment of neurotransmitter and synaptic vesicle specificity of B2RT-STOP-B2RT-smFLAG-vGlut in single neuron types. Neuron anatomy is visualized with the plasma membrane marker CD8-mCherry. a–a″) Glutamatergic neuron MBON-6. a) CD8-mCherry; a′) smFLAG-vGlut; a″) overlay. b–b″) High-resolution 100× images of presynaptic terminals of glutamatergic neuron MBON-6. b) Syt-smHA; b′) smFLAG-vGlut; b″) overlay. High-resolution images of MBON-6 presynaptic terminals reveal near precise overlap of smFLAG-vGlut with the synaptic vesicle marker Syt-smHA. c–c″) Glutamatergic neuron MBON-6. c) CD8-mCherry; c′) GFP-Rab3; c″) overlay. GFP-Rab3 is expressed in glutamatergic neuron MBON-6 and distributes predominantly to presynaptic terminals in a pattern highly similar to that of smFLAG-vGlut. d–d″) GABAergic neuron LH1900. d) CD8-mCherry; d′) smFLAG-vGlut; d″) overlay. e–e″) Cholinergic neuron LH2094. e) CD8-mCherry; e′) smFLAG-vGlut; e″) overlay. No expression of smFLAG-vGlut was detected in the GABAergic neuron LH1900 or the cholinergic neuron LH2094. Large arrows—presynaptic terminals; small arrows—dendrites; arrowheads—cell bodies. Scale bars: a, c, d, e)—50 µm; b)—25µm.
Expression of smFLAG-vGlut was also assessed in the known GABAergic and cholinergic neurons LH1900 and LH2094, respectively (Dolan et al. 2019). LH1900 is a GABAergic lateral horn neuron with a distinct cell body (arrowhead) and intermingled axonal and dendritic regions (large arrows) (Tison et al. 2020) (Fig. 3d). LH2094 is a highly polarized cholinergic lateral horn neuron with distinct axonal (large arrow), dendritic (small arrow), and cell body regions (Tison et al. 2020) (Fig. 3e). Expression of smFLAG-vGlut was not detected in LH1900 (Fig. 3d′) or LH2094 (Fig. 3e′) upon excision of the STOP cassette.
Taken together, we have established smFLAG-vGLUT as a conditional marker of glutamatergic SVs by: (1) demonstrating robust smFLAG-vGlut expression in the glutamatergic neuron MBON-6 with a similar subcellular distribution as the SV marker GFP-Rab3; (2) precise spatial colocalization with the SV marker Syt-smHA; and (3) the absence of expression in GABAergic and cholinergic neurons.
Glutamatergic neurotransmitter phenotyping of central complex neurons
With its conditionality and specificity for glutamatergic SVs thus established, the utility of B2RT-STOP-B2RT-smFLAG-vGlut for glutamatergic neurotransmitter phenotyping and identifying sites of glutamate release were demonstrated for 26 distinct Drosophila central complex neuron types of unknown neurotransmitter usage. The central complex is a key region of the insect brain involved in higher-order sensory information processing important for behavioral decision-making (Pfeiffer and Homberg 2014). In flies, a diversity of behaviors are regulated by the central complex including locomotion (Martin et al. 1999; Buchanan et al. 2015), flight (Ilius et al. 1994), courtship (Sakai and Kitamoto 2006), sleep (Donlea et al. 2014), hunger (Park et al. 2016), gravitaxis (Baker et al. 2007), and navigation (Seelig and Jayaraman 2013; Hardcastle et al. 2021). Neurons of the central complex interconnect the central complex neuropils including the protocerebral bridge, noduli, and asymmetric body, as well as surrounding neuropils (Wolff et al. 2015; Wolff and Rubin 2018; Scheffer et al. 2020). Understanding the function of this complex neuropil would be enhanced by mapping neurotransmitter usage of distinct central complex neuron types.
To determine which central complex neurons express glutamate, we took advantage of set of recently described split-GAL4 drivers representing specific neuron types of the central complex (Wolff and Rubin 2018). In total, nine of the 26 neuron types screened were glutamate positive as determined by smFLAG-vGlut expression (Figs. 4 and 5), and 17 were negative (Supplementary Fig. 4). In all images the split-Gal4-expressing neuron is visualized with the plasma membrane marker CD8-mCherry (red, left column) and axonal regions where glutamatergic SVs accumulate are indicated by smFLAG-vGlut (green, indicated with arrows, middle column). Four of the positives are protocerebral bridge neurons including PB.b-LAL.s-PS.s/SS52578 (Fig. 4, a and a″), PBG6-8.sG9.b/SS00117 (Fig. 4, b and b″), PB18.s-GxΔ7 Gy.b/PB18.s-9i1i8c.b/SS52266 (Fig. 4, c and c″), and PBG1/2-9.b-SPS.s/SS52267 (Fig. 4, d and d″). Four additional positives are noduli neurons including LAL.s-GAi.s-NO1i.b/SS46517 (Fig. 5, a and a″), LAL.s-NO2i.b/SS47398 (Fig. 5, b and b″), LAL.s-N03Ai.b/SS47432 (Fig. 5, c and c″), and LAL.s-CREc.s-NO3Pc.b/SS46525 (Fig. 5, d and d″) while the remaining positive neuron was the asymmetric body neuron SLP-AB/SS50464 (Fig. 5, e and e″). In the brains of some neuron types there is additional variable smFLAG-vGlut expression in neurons besides within the one(s) of interest that is discerned by the lack of overlap with the CD8-mCherry plasma membrane marker. This additional expression most likely reflects split-Gal4 driver expression during development that results in the excision of the STOP cassette prior to adulthood. While expression of the split-GAL4 driver in a given glutamatergic neuron during development may be transient and not maintained into the adult stage, any excisions in glutamatergic neurons that occurred during development will be permanent and result in smFLAG-vGlut expression.
Fig. 4.
B2RT-STOP-B2RT-smFLAG-vGlut expression identifies glutamatergic protocerebral bridge neurons. a–a″) PB.b-LAL.s-PS.s/SS52578. b–b″) PBG6–8.sG9.b/SS00117. c–c″) PB18.s-GxΔ7 Gy.b/PB18.s-9i1i8c.b/SS52266. d–d″) PBG1/2-9.b-SPS.s/SS52267. The plasma membrane marker CD8-mCherry (red, left column) allows visualization of the neuroanatomy of each protocerebral neuron. The presence of smFLAG-vGlut expression (arrows, middle column) indicates that each protocerebral neuron is glutamatergic and its subcellular distribution reveals the location of presynaptic terminals where SV fusion and glutamate release occurs. Scale bars: 50 µm.
Fig. 5.
B2RT-STOP-B2RT-smFLAG-vGlut expression identifies glutamatergic noduli and asymmetric body neurons. a–a″) LAL.s-GAi.s-NO1i.b/SS46517. b–b″) LAL.s-NO2i.b/SS47398. c–c″) LAL.s-N03Ai.b/SS47432. d–d″) LAL.s-CREc.s-NO3Pc.b/SS46525. e–e″) SLP-AB/SS50464. The plasma membrane marker CD8-mCherry (red, left column) allows visualization of the neuroanatomy of each neuron. The presence of smFLAG-vGlut expression (arrows, middle column) indicates that each neuron is glutamatergic and its subcellular distribution reveals the location of presynaptic terminals where SV fusion and glutamate release occur. Scale bars: 50 µm.
It should also be noted that while neurons exhibiting smFLAG-vGlut expression can be considered glutamatergic with a high level of confidence, it is possible a neuron that does not express smFLAG-vGlut may still be glutamatergic. This is due to a threshold level of B2 recombinase required for excision of the STOP cassette and the level of B2 recombinase expression being dependent on the strength of the split-GAL4 driver. Thus, there is the possibility that a given neuron that scores negative for smFLAG-vGlut expression is a “false negative” because the split-GAL4 driver is not of sufficient strength to drive expression of the B2 recombinase above the threshold level required for STOP cassette excision. Examples of this are illustrated by noduli neurons LAL.s-GAi.s-NO1i.b/SS46517 (Fig. 5a) and LAL.s-CREc.s-NO3Pc.b/SS46525 (Fig. 5d) where the driver exhibits a significantly higher level of expression in the neuron on the right as compared to the one on the left. In these examples, the threshold level of expression of the B2 recombinase required for excision is only met by the neuron on the right. Consequently, smFLAG-vGlut is only observed in the presynaptic regions of the neurons on the right (arrows, Fig. 5, a′ and d′), but not the neurons on the left (arrowheads, Fig. 5, a′ and d′). Therefore, it is possible that smFLAG-vGlut expression will not be observed in some glutamatergic neurons due to weakness of the split-GAL4 driver.
Discussion
This report describes a conditional epitope-tagged glutamatergic SV marker for Drosophila, B2RT-STOP-B2RT-smFLAG-vGlut, developed via CRISPR/Cas9 genome editing at the endogenous chromosomal locus of the single Drosophila vGlut. Immuno-localization data was presented demonstrating B2RT-STOP-B2RT-smFLAG-vGlut is conditionally expressible and specifically expressed in glutamatergic, but not cholinergic or GABAergic, neurons. Genetic analysis revealed the embryonic lethality of B2RT-STOP-B2RT-smFLAG-vGlut, viability of the smFLAG-vGlut germline excision chromosome in heterozygous combination with a vGlut null allele and also in homozygous condition. Electrophysiology experiments determined that spontaneous mEJP amplitude was statistically indistinguishable between smFLAG-vGliut germline excision homozygotes and wild-type controls. Together, these data strongly support our claim that B2RT-STOP-B2RT-mFLAG-vGlut is a conditional marker of glutamatergic SVs and a functional vesicular transporter of glutamate. smFLAG-vGlut was also shown to have a high sensitivity of detection by virtue of its robust signal even in single neurons. Finally, its utility for glutamatergic neurotransmitter phenotyping and discerning sites of glutamate release were established using split-GAL4 drivers representing 26 neuron types of the central complex, nine of which were shown to be glutamatergic.
B2RT-STOP-B2RT-smFLAG-vGlut provides an alternative and complementary method to single-cell transcriptomics (Henry et al. 2012; Thomas et al. 2012; Yang et al. 2016; Croset et al. 2018; Davis et al. 2020) for identifying glutamate-expressing neurons. These independent methods can potentially be used in parallel to corroborate each other’s results. Confidence in a positive glutamatergic neurotransmitter phenotype indicated by one method would be strengthened upon verification by the other method. B2RT-STOP-B2RT-smFLAG-vGlut does, however, have the additional advantage over single cell transcriptomics in that it provides not just whether a given neuron type is glutamatergic, but also spatial information on the location of glutamate release sites within a glutamatergic neuron. The glutamatergic neurotransmitter phenotyping utilizing B2RT-STOP-B2RT-smFLAG-vGlut presented herein for 26 neuron types of the central complex can be immediately extended to the current existing collection of several hundred Drosophila split-GAL4 drivers specific for single neuron types (Aso et al. 2014; Robie et al. 2017; Wolff and Rubin 2018; Dolan et al. 2019; Davis et al. 2020), among others, and to any additional neuron types for which split-GAL4 drivers are developed in the future.
The glutamatergic phenotype of the four protocerebral bridge neuron types, the four noduli neuron types, and the asymmetric body neuron demonstrated by expression of smFLAG-vGlut in this assessment provides useful information for modeling the neural circuitry of the central complex. Additionally, the recent report that there is no overlap in fast neurotransmitter usage in Drosophila (Deng et al. 2019) suggests these nine glutamatergic neurons are also not cholinergic or GABAergic. This knowledge of glutamate neurotransmitter usage in nine central complex neuron types and likely nonglutamate neurotransmitter usage in 17 other central complex neuron types, will enhance our understanding of sensory information processing and behavioral decision making in Drosophila central complex neurons.
While glutamate has long been known to function as an excitatory neurotransmitter in vertebrates (Curtis et al. 1959) and invertebrates (Usherwood et al. 1968), more recently it has also been shown to be an inhibitory neurotransmitter in Drosophila (Cully et al. 1996; Liu and Wilson 2013). The central complex neurons determined to be glutamatergic by B2RT-STOP-B2RT-smFLAG-vGlut thus almost certainly excite their downstream target neurons that are now comprehensively known owing to the Drosophila connectome (Scheffer et al. 2020), but they also have the potential to be inhibitory dependent on expression of the inhibitory glutamate receptor GluClα. To determine whether a given glutamatergic presynaptic neuron is providing inhibitory input to one of its downstream target neurons, B2RT-STOP-B2RT-smFLAG-vGlut could potentially be used in combination with a recently developed conditionally-expressible tagged variant of GluClα (Fendl et al. 2020). If expression of tagged GluClα is observed on the postsynapse of a downstream target neuron directly across the synaptic cleft of smFLAG-vGlut expression in a presynaptic neuron, this would provide significant evidence for inhibitory glutamatergic input.
B2RT-STOP-B2RT-smFLAG-vGlut is the latest addition to an existing collection of conditional tagged SV markers for Drosophila. This collection includes SV markers for acetylcholine (vAChT) (Pankova and Borst 2017; Tison et al. 2020), monoamines (vMAT) (Sherer et al. 2020), and the neurotransmitter-independent SV marker (Rab3) (Williams et al. 2019). B2RT-STOP-B2RT-smFLAG-vGlut thus fills a missing gap in Drosophila for a conditional glutamatergic SV marker and expands the method of neurotransmitter phenotyping by conditional expression of a tagged neurotransmitter-specific gene to include glutamate. The strategy shown herein to be successful in Drosophila for generating a conditional glutamatergic SV marker is potentially applicable to other species.
Data availability
The complete annotated sequence of the B2RT-STOP-B2RT-smFLAG-vGlut donor construct is shown in Supplemental Information (Supplementary Fig. 1). The B2RT-STOP-B2RT-smFLAG-vGlut donor plasmid and pCFD4-vGlut guide RNA plasmid will be made available upon request. Fly strains original to this publication will be deposited at the Bloomington Drosophila stock center or will be made available upon request.
Supplemental material is available at G3 online.
Conflicts of interest statement
The authors have no conflicts of interest to declare.
Supplementary Material
Acknowledgments
The authors thank the Bloomington Drosophila Stock Center and the Janelia Farm Research Campus for fly strains and the Developmental Studies Hybridoma Bank for antibodies.
Funding
This work was supported by National Institutes of Health (NIH) grant R01GM115510 to SJC and RSS and Swiss National Science Foundation grant 31003A_179587 to BDM.
Literature cited
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo T-TB, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014;3:e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Beckingham KM, Armstrong JD.. Functional dissection of the neural substrates for gravitaxic maze behavior in Drosophila melanogaster. J Comp Neurol. 2007;501(5):756–764. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Vernon S, Jiao W, Choi BJ, Ruchti E, Asadzadeh J, Burri O, Stowers RS, McCabe BD.. Miniature neurotransmission is required to maintain Drosophila synaptic structures during ageing. Nat Commun. 2021;12(1):4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan SM, Kain JS, de Bivort BL.. Neuronal control of locomotor handedness in Drosophila. Proc Natl Acad Sci USA. 2015;112(21):6700–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certel SJ, Thor S.. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development. 2004;131(21):5429–5439. [DOI] [PubMed] [Google Scholar]
- Croset V, Treiber CD, Waddell S.. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife. 2018;7:e34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully DF, Paress PS, Liu KK, Schaeffer JM, Arena JP.. Identification of a Drosophila melanogaster glutamate-gated chloride channel sensitive to the antiparasitic agent avermectin. J Biol Chem. 1996;271(33):20187–20191. [DOI] [PubMed] [Google Scholar]
- Curtis DR, Phillis JW, Watkins JC.. Chemical excitation of spinal neurones. Nature. 1959;183(4661):611–612. [DOI] [PubMed] [Google Scholar]
- Daniels RW, Collins CA, Chen K, Gelfand MV, Featherstone DE, DiAntonio A.. A single vesicular glutamate transporter is sufficient to fill a synaptic vesicle. Neuron. 2006;49(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Gelfand MV, Collins CA, DiAntonio A.. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508(1):131–152. [DOI] [PubMed] [Google Scholar]
- Davis FP, Nern A, Picard S, Reiser MB, Rubin GM, Eddy SR, Henry GL.. A genetic, genomic, and computational resource for exploring neural circuit function. Elife. 2020;9:e50901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, Li Q, Liu X, Cao Y, Li B, Qian Y, Xu R, Mao R, Zhou E, Zhang W, et al. Chemoconnectomics: mapping chemical transmission in Drosophila. Neuron. 2019;101(5):876–893 e874. [DOI] [PubMed] [Google Scholar]
- Dolan M-J, Frechter S, Bates AS, Dan C, Huoviala P, Roberts RJ, Schlegel P, Dhawan S, Tabano R, Dionne H, et al. Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. Elife. 2019;8:e43079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Miesenbock G.. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81(6):1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendl S, Vieira RM, Borst A.. Conditional protein tagging methods reveal highly specific subcellular distribution of ion channels in motion-sensing neurons. Elife. 2020;9:e62953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Ueda A, Wu C-F.. A modified minimal hemolymph-like solution, HL3.1, for physiological recordings at the neuromuscular junctions of normal and mutant Drosophila larvae. J Neurogenet. 2004;18(2):377–402. 10.1080/01677060490894522 15763995 [DOI] [PubMed] [Google Scholar]
- Hardcastle BJ, Omoto JJ, Kandimalla P, Nguyen B-CM, Keleş MF, Boyd NK, Hartenstein V, Frye MA.. A visual pathway for skylight polarization processing in Drosophila. Elife. 2021;10:e63225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry GL, Davis FP, Picard S, Eddy SR.. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 2012;40(19):9691–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilius M, Wolf R, Heisenberg M.. The central complex of Drosophila melanogaster is involved in flight control: studies on mutants and mosaics of the gene ellipsoid body open. J Neurogenet. 1994;9(3):189–206. [DOI] [PubMed] [Google Scholar]
- Imlach W, McCabe BD.. Electrophysiological Methods for Recording Synaptic Potentials from the NMJ of Drosophila Larvae. JoVE. 2009;(24): 10.3791/1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO, Reifegerste R, Reisch D, Schaupp M, Buchner S, Buchner E, et al. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci. 1996;16(10):3154–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguz-Lecznar M, Skangiel-Kramska J.. Vesicular glutamate transporters (VGLUTs): the three musketeers of glutamatergic system. Acta Neurobiol Exp (Wars). 2007;67(3):207–218. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Bellen HJ, Perin MS.. Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development. 1993;118(4):1077–1088. [DOI] [PubMed] [Google Scholar]
- Liu WW, Wilson RI.. Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc Natl Acad Sci USA. 2013;110(25):10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JR, Raabe T, Heisenberg M.. Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J Comp Physiol A. 1999;185(3):277–288. [DOI] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Svoboda K, Rubin GM.. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc Natl Acad Sci USA. 2011;108(34):14198–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankova K, Borst A.. Transgenic line for the identification of cholinergic release sites in Drosophila melanogaster. J Exp Biol. 2017;220(Pt 8):1405–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-Y, Dus M, Kim S, Abu F, Kanai MI, Rudy B, Suh GSB.. Drosophila SLC5A11 mediates hunger by regulating K(+) channel activity. Curr Biol. 2016;26(15):1965–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen LK, Stowers RS.. A Gateway MultiSite recombination cloning toolkit. PLoS One. 2011;6(9):e24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K, Homberg U.. Organization and functional roles of the central complex in the insect brain. Annu Rev Entomol. 2014;59:165–184. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL.. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111(29):E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu L-P, Yang Z, Mao D, Sun L, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA. 2013;110(47):19012–19017., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robie AA, Hirokawa J, Edwards AW, Umayam LA, Lee A, Phillips ML, Card GM, Korff W, Rubin GM, Simpson JH, et al. Mapping the neural substrates of behavior. Cell. 2017;170(2):393–406.e328. [DOI] [PubMed] [Google Scholar]
- Sakai T, Kitamoto T.. Differential roles of two major brain structures, mushroom bodies and central complex, for Drosophila male courtship behavior. J Neurobiol. 2006;66(8):821–834. [DOI] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. A connectome and analysis of the adult Drosophila central brain. Elife. 2020;9:e57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, Jayaraman V.. Feature detection and orientation tuning in the Drosophila central complex. Nature. 2013;503(7475):262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer LM, Catudio Garrett E, Morgan HR, Brewer ED, Sirrs LA, Shearin HK, Williams JL, McCabe BD, Stowers RS, Certel SJ, et al. Octopamine neuron dependent aggression requires dVGLUT from dual-transmitting neurons. PLoS Genet. 2020;16(2):e1008609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S. VGLUTs: ‘exciting’ times for glutamatergic research? Neurosci Res. 2006;55(4):343–351. [DOI] [PubMed] [Google Scholar]
- Thomas A, Lee P-J, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA, et al. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS One. 2012;7(7):e40276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tison KV, McKinney HM, Stowers RS.. Demonstration of a simple epitope tag multimerization strategy for enhancing the sensitivity of protein detection using Drosophila vAChT. G3 (Bethesda). 2020;10(2):495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD Jr., Ning X, Klingler M, Kramer SG, Gergen JP.. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154(1):273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usherwood PN, Machili P, Leaf G.. L-Glutamate at insect excitatory nerve-muscle synapses. Nature. 1968;219(5159):1169–1172. [DOI] [PubMed] [Google Scholar]
- Viswanathan S, Williams ME, Bloss EB, Stasevich TJ, Speer CM, Nern A, Pfeiffer BD, Hooks BM, Li W-P, English BP, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015;12(6):568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Shearin HK, Stowers RS.. Conditional synaptic vesicle markers for Drosophila. G3 (Bethesda). 2019;9(3):737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Iyer NA, Rubin GM.. Neuroarchitecture and neuroanatomy of the Drosophila central complex: a GAL4-based dissection of protocerebral bridge neurons and circuits. J Comp Neurol. 2015;523(7):997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Rubin GM.. Neuroarchitecture of the Drosophila central complex: a catalog of nodulus and asymmetrical body neurons and a revision of the protocerebral bridge catalog. J Comp Neurol. 2018;526(16):2585–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-P, Fu C-C, Sugino K, Liu Z, Ren Q, Liu L-Y, Yao X, Lee LP, Lee T.. Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development. 2016;143(3):411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F-X, Ge S-N, Dong Y-L, Shi J, Feng Y-P, Li Y, Li Y-Q, Li J-L.. Vesicular glutamate transporter isoforms: the essential players in the somatosensory systems. Prog Neurobiol. 2018;171:72–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete annotated sequence of the B2RT-STOP-B2RT-smFLAG-vGlut donor construct is shown in Supplemental Information (Supplementary Fig. 1). The B2RT-STOP-B2RT-smFLAG-vGlut donor plasmid and pCFD4-vGlut guide RNA plasmid will be made available upon request. Fly strains original to this publication will be deposited at the Bloomington Drosophila stock center or will be made available upon request.
Supplemental material is available at G3 online.