Abstract
Gastrodia elata, an obligate mycoheterotrophic orchid, requires complete carbon and mineral nutrient supplementation from mycorrhizal fungi during its entire life cycle. Although full mycoheterotrophy occurs most often in the Orchidaceae family, no chromosome-level reference genome from this group has been assembled to date. Here, we report a high-quality chromosome-level genome assembly of G. elata, using Illumina and PacBio sequencing methods with Hi-C technique. The assembled genome size was found to be 1045 Mb, with an N50 of 50.6 Mb and 488 scaffolds. A total of 935 complete (64.9%) matches to the 1440 embryophyte Benchmarking Universal Single-Copy Orthologs were identified in this genome assembly. Hi-C scaffolding of the assembled genome resulted in 18 pseudochromosomes, 1008 Mb in size and containing 96.5% of the scaffolds. A total of 18,844 protein-coding sequences (CDSs) were predicted in the G. elata genome, of which 15,619 CDSs (82.89%) were functionally annotated. In addition, 74.92% of the assembled genome was found to be composed of transposable elements. Phylogenetic analysis indicated a significant contraction of genes involved in various biosynthetic processes and cellular components and an expansion of genes for novel metabolic processes and mycorrhizal association. This result suggests an evolutionary adaptation of G. elata to a mycoheterotrophic lifestyle. In summary, the genomic resources generated in this study will provide a valuable reference genome for investigating the molecular mechanisms of G. elata biological functions. Furthermore, the complete G. elata genome will greatly improve our understanding of the genetics of Orchidaceae and its mycoheterotrophic evolution.
Keywords: Gastrodia elata, genome assembly, mycoheterotrophic, Orchidaceae, pseudochromosome
Introduction
Mycoheterotrophy represents one extreme end in the mutualism-parasitism continuum of mycorrhizal symbiosis (Leake 1994), upon which the largest number of vascular plant species depend (Leake 2005). In total, more than 450 vascular plant species maintain a fully mycoheterotrophic lifestyle throughout their entire lives without producing green leaves (Merckx and Freudenstein 2010). Full mycoheterotrophy occurs in a wide phylogenetic range of plant species, especially culminating in the Orchidaceae, the most widely distributed plant family on Earth (Leake 1994). This family contains the largest number of fully mycoheterotrophic species (at least 210) (Merckx and Freudenstein 2010).
Gastrodia elata Blume is a fully mycoheterotrophic orchid that is symbiotically associated with at least two fungal partners: a broad range of Mycena spp. are required for seed germination (Xu and Guo 1989; Shunxing and Qiuying 2001; Park and Lee 2013) and Armillaria mellea is essential for plant growth (Zhang and Li 1980). Such mycorrhizal community changes during ontogenetic development have been shown in other species. For example, the fungi that associate with seeds of several Pyrola (Ericaceae family) species differ from those coupled with adult plants (Hashimoto et al. 2012; Hynson et al. 2013; Johansson et al. 2017; Jacquemyn et al. 2018). This indicates that some plants may serially associate with different fungal partners rather than choosing a single best partner. Similarly, the mycorrhizal communities associated with protocorms and adult plants of the orchid Liparis loeselii are also diverse, varying among the different life cycle stages (Waud et al. 2017).
In the last decade, a number of genomic resources have been developed to study the mycoheterotrophic adaptation of G. elata, including transcriptomes (Tsai et al. 2016; Zeng et al. 2017; Wang et al. 2020), proteomic data (Zeng et al. 2018), and a draft genome assembly (Yuan et al. 2018). The previous G. elata genome assembly, determined with the Illumina HiSeq 2500 platform, was highly fragmented (Yuan et al. 2018). In particular, the low contiguity of this genome assembly has limited its application for further research on the genomic evolution of G. elata. Moreover, no chromosome-level genome has ever been assembled for a member of the obligate mycoheterotrophic Orchidaceae family. Therefore, an accurate genome assembly of G. elata is essential for both basic and applied research, which will improve our understanding of genome evolution in the Orchidaceae family and accelerate the genetic improvement for G. elata cultivation in the commercial field for food and medicine.
Here, we present a vastly improved de novo assembly and annotation of the G. elata reference genome using these new sequencing technologies, including single-molecule real-time (SMRT) sequencing from Pacific Biosciences (PacBio) and chromosome conformation capture (Hi-C) (Wingett et al. 2015; Korlach et al. 2017; Pennisi 2017). Notably, this new assembly greatly improves genome completeness and contiguity over the previous version of the reference genome. Last, comparative analysis with other orchid species revealed the emergence of evolutionary novelties and functional diversification of G. elata, leading to the development of the unique mycoheterotrophic lifestyle.
Materials and methods
Sample collection and DNA sequencing
Experimental sample of G. elata was collected from Muju (35˚51’N 127˚39’E; 510-m altitude) in Jeollabuk-do Province, which is located in southern Korea (Figure 1). High-molecular-weight genomic DNA (gDNA) was isolated from a single genotype of G. elata scape, using the modified cetyltrimethylammonium bromide (CTAB) method (Inglis et al. 2018), and the high-quality gDNA was purified using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) after RNase A treatment. The quantity of the extracted DNA was then determined using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Figure 1.

Photograph of Gastrodia elata. The white arrows indicate the mature tuber and scape.
To perform the genomic survey, an Illumina paired-ended DNA library, with an insert size of 550 bp, was prepared according to the Illumina TruSeq DNA PCR-Free Library Prep protocol (Illumina, San Diego, CA, USA). The Agilent 2100 Bioanalyzer High Sensitivity Kit was used to check for quality, and the library was sequenced on the Illumina NovaSeq 6000 platform, using a 150-bp paired-end strategy.
For long-read sequencing, 25 SMRTbell 20 kb DNA libraries were constructed using the following steps, according to the PacBio standard protocol: (1) gDNA shearing using the Covaris g-TUBE (Covaris Inc., Woburn, MA, USA); (2) DNA damage repair; (3) blunt-end ligation with hairpin adapters from the SMRTbell Template Prep Kit 1.0 (PacBio, Menlo Park, CA, USA); (4) 20 kb size-selection using the BluePippin Size Selection System (Sage Science, Beverly, MA, USA); and (5) binding to polymerase using the MagBead Kit (Pacific Biosciences, Menlo Park, CA, USA). Subsequently, SMRT long-read sequencing was performed on a PacBio Sequel platform with the Sequel Sequencing Kit 2.1.
A Dovetail Hi-C library was constructed from a scape tissue according to the manufacturer’s instructions (Dovetail Hi-C Library kit), and sequenced with the Illumina NovaSeq 6000 platform, according to published protocols (Lieberman-Aiden et al. 2009). A scape tissue was cross-linked with PBS/formaldehyde, and then chromatin was prepared with SDS and wash buffer. After normalizing the chromatin plant sample, 800 ng of chromatin was used to make the library. Chromatin was captured by chromatin capture beads and then digested with restriction enzyme. Its end was filled in with biotin and ligated to form Intra-aggregated DNA. After cross-link reversal, 200 ng of DNA was sheared using the Covaris system. Sheared DNA fragments were end-repaired and ligated with Illumina adapter. Ligated DNA was purified using Streptavidin Magnetic Beads. Purified DNA was amplified by PCR to enrich fragments. The quality of the amplified libraries was verified by capillary electrophoresis (Bioanalyzer, Agilent). Sequencing is performed using an Illumina NovaSeq 6000 system following provided protocols for 2 × 150 sequencing. In summary, Hi-C fragment libraries were prepared according to the “Proximo Hi-C protocol” with DpnII digest, and the resulting libraries were sequenced using a 150-bp paired-end strategy.
Genome assembly
Raw Illumina paired-end sequencing reads were filtered using the FASTP v.0.12.6 preprocessor (set to default parameters) to remove low-quality reads, adapters, and reads containing poly-N (Chen et al. 2018). Trimmed Illumina sequencing reads were then used to calculate the percentage of heterozygosity in the genome. For this analysis, Jellyfish v.2.2.10 was first used to compute the histogram of 19 k-mer frequencies (Marcais and Kingsford 2011), and genome heterozygosity was then calculated by the GenomeScope v.2.0 online platform, using the final k-mer count histogram (Vurture et al. 2017).
To perform de novo genome assembly, we used the FALCON-Unzip assembler v0.4 (Chin et al. 2016), with length cutoff parameters (length cutoff = 13 kb, length cutoff pr = 10 kb) and filtered subreads from SMRT Link v.5.0.0 (minimum subread length = 50 bp). To improve accuracy of the assembly, the FALCON-Unzip assembler was polished with the Arrow algorithm, using the PacBio unaligned BAM files as raw data. Then, the error correction was performed with alignment from the short-read using Pilon v.1.23 (Walker et al. 2014).
The falcon-unzip assembly and Dovetail Hi-C reads were then used as input data for HiRise, a software pipeline designed specifically for utilizing proximity ligation data to scaffold genome assemblies (Putnam et al. 2016). Hi-C library sequences were aligned to the draft input assembly using a SNAP read mapper (Zaharia et al. 2011). The separations of Hi-C read pairs mapped within draft scaffolds were analyzed by HiRise to produce a likelihood model for the genomic distance between read pairs. This model was then used to identify and break putative misjoins, score prospective joins, and make joins above a threshold. Finally, organelle genomes were filtered out from public organelle sequences in NCBI using BLAST v.2.4.0 (Altschul et al. 1990), and completeness of the genome assembly was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO) v.3.0.1 with default parameters and the embryophata dataset (Simao et al. 2015).
Transcriptome sequencing
Tissue samples were collected through the 12 development stages of G. elata. The collected samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. Total RNA was extracted from each sample with TRIzol reagent (Invitrogen, Waltham, MA, USA). RNA quality and quantity were checked using the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). The full-length cDNA library was generated using 1 μg of equally mixed RNA from the 12 different tissues and the Clontech SMARTer PCR cDNA Synthesis Kit according to the Isoform Sequencing protocol (PacBio, Menlo Park, CA, USA). PCR optimization was carried out on the full-length cDNA using the PrimeSTAR GXL DNA Polymerase (Clontech, Mountain View, CA, USA) and 12 cycles were sufficient to generate the material required for SMRTbell library preparation. Each cDNA sample was bead cleaned with AMPure PB beads post PCR in preparation for SMRTbell library construction. The sequencing primer from the SMRTbell Template Prep Kit 1.0-SPv3 was annealed to the adapter sequence of the libraries. Each library was bound to the sequencing polymerase with the Sequel Binding Kit v2.1 and the complex formed was then purified using AMPure Purification (Clontech, Mountain View, CA, USA). The libraries were sequenced using 2 SMRTcells v2.0 per library on the Sequel sequencing platform. All libraries had 600-min movies and 240 min of pre-extension time. The full-length isoform sequence was constructed using SMRTLink v.5.1 (Pacific Bioscience, CA, USA) through several steps. First, the qualified sequence was classified based on detection of primers and polyA tail. Then, the isoform sequence was generated with isoform-level clustering that was categorized as high-quality based on over 99% estimated accuracy.
Genome annotation
The G. elata genome was annotated using custom repeat library protocols, ab initio gene prediction, homology search, and full-length transcript evidences. A de novo repeat library was constructed using RepeatModeler v.1.0.3 (Price et al. 2005), including RECON v.1.08 (Bao and Eddy 2002) and RepeatScout v.1.0.5 (Price et al. 2005) with default parameters. Tandem Repeats Finder v.4.09 (Benson 1999) was used to predict consensus sequences, classification information for each repeat, and tandem repeats, including simple repeats, satellites, and low complexity repeats (Benson 1999). To identify highly accurate long terminal repeat retrotransposons (LTR-RTs), we constructed an LTR library with LTR_retriever v.2.9.0 (Ou and Jiang 2018), using combined raw LTR data from LTRharvest v.1.6.1 (Ellinghaus et al. 2008) and LTR_FINDER v.1.0.7 (Xu and Wang 2007). Repetitive elements in the de novo repeat library were identified using RepeatMasker v.4.0.9, and the quality of repetitive elements was assessed using LTR Assembly Index (LAI) program (Ou et al. 2018). Kimura distances (Kimura 1980) for all transposable element (TE) copies from each family found in the library were calculated to estimate the age of TEs.
Genome annotation was performed with MAKER v.2.31.8 (Holt and Yandell 2011), using three rounds of reiterative training (Holt and Yandell 2011). Subsequently, ab initio gene prediction was performed with SNAP v.2006-07-32 (Korf 2004) and Augustus v.3.3.3 (Stanke et al. 2006). MAKER was initially run in est2genome mode based on full-length transcripts from Iso-Seq data. In addition, evidence for protein-coding genes was obtained from the genomes of three orchid plants: Apostasiashenzhenica (GCA_002786265.1) (Zhang et al. 2017), Dendrobiumcatenatum (GCA_001605985.2) (Zhang et al. 2016), and Phalaenopsisequestris (GCA_001263595.1) (Cai et al. 2015). Exonerate v2.4.0 (Slater and Birney 2005), which provides integrated information for the SNAP program, was used to polish MAKER alignments. Other noncoding RNAs were identified using the Barrnap v0.9 (https://vicbioinformatics.com/software.barrnap.shtml). The putative tRNA genes were identified using tRNAscan-SE v2.0.5 (Chan and Lowe 2019). To select the best-supported gene models, we used a quality metric called annotation edit distance (AED), developed by the Sequence Ontology project (Eilbeck et al. 2009). More than 90% of our annotations had an AED score less than 0.5 (Campbell et al. 2014).
For functional annotation, predicted proteins were aligned to the National Center for Biotechnology Information (NCBI) nonredundant protein databases (Marchler-Bauer et al. 2011), using BLAST v.2.4.0 (Altschul et al. 1990) with a maximum e-value cutoff of 1e-5. Protein signatures were annotated using InterProScan v.5.44.79 (Jones et al. 2014) for further BLAST2GO v.5.2.5 (Götz et al. 2008) based gene ontology (GO) analysis (Dimmer et al. 2012). Predicted proteins were also searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to retrieve KEGG-relevant functional annotations.
Gene family identification and phylogenetic analysis
Orthologous gene clusters were classified within the genomes of 15 plant species, including G. elata (Supplementary Table S1), using OrthoMCL v2.0 (OrthoMCL-DB: Ortholog Groups of Protein Sequences) (Li et al. 2003). We then extracted the longest protein sequence isoforms from the gene predictions of each plant species with default parameters to construct a phylogenetic tree, using Orthofinder v2.4.0 (Emms and Kelly 2019) with an e-value cutoff 1e-5 and all-to-all BLASTP analysis of the 15 plant species. MAFFT v.6.861b (Katoh et al. 2009) was used to align each gene family, and the phylogenetic tree was inferred with FastTree v.2.1.10 (Price et al. 2010), with divergence time calibration performed using both PATHd8 (Britton et al. 2007) and TimeTree (Kumar et al. 2017). Last, CAFE v.4.2.1 (Han et al. 2013) was used to predict the likelihood of gene family expansion and contraction with P < 0.01 and automatic searching for the λ value.
Results and discussion
Genome assembly
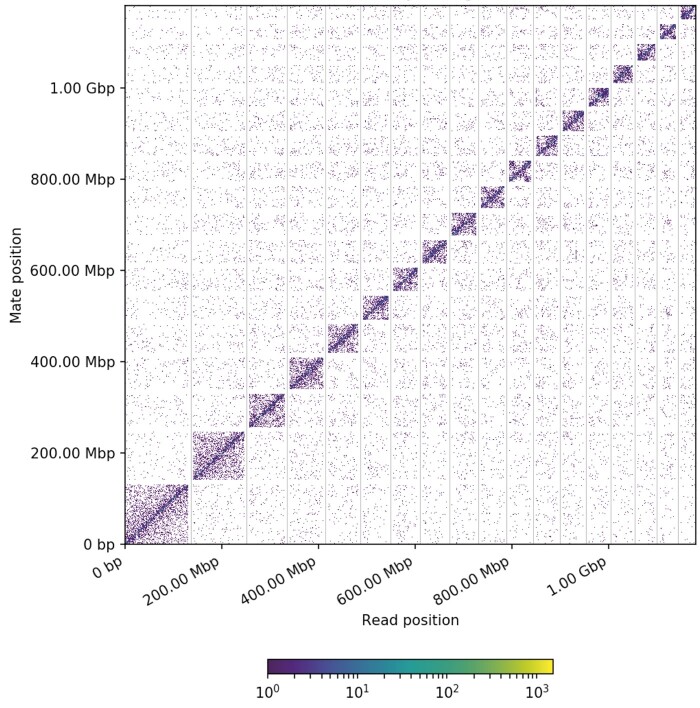
Using Illumina paired-ended sequencing, we first obtained 132.1 Gb of clean data after filtering out adapter sequences and low-quality reads. Prior to genome assembly, size of the G. elata genome was estimated from Illumina sequencing by GenomeScope, which predicted genome size of 1.023 Gbp, with heterozygosity of 0.06% (Supplementary Figure S1). We also performed long-read sequencing of the G. elata genome on the PacBio Sequel platform and obtained 11,449,345 PacBio long reads from 25 SMRT cells, representing a sequencing depth of 84.6X (Supplementary Table S2). FACON-Unzip was used to perform de novo assembly, and after the error correction step, we obtained a de novo assembly of 1.049 Gb, with a contig N50 of 9.18 Mb (Table 1), which is in broad agreement with the estimated genome size (1.023 Gb). Hi-C fragment library sequencing produced 121.6 Gb of clean data after filtering (Table 1). By mapping Hi-C sequencing data onto the genome assembly, we generated 32.33 Gb (52.6X coverage) of high-quality, validated Hi-C data to assemble contigs at the chromosome level (Supplementary Table S3). A total of 488 assembled contigs were anchored onto 18 pseudochromosomes that ranged from 32.1 to 130.6 Mb in length and contained 96.4% of the genome sequences (Figure 2; Supplementary Table S3). This chromosome number agrees with the previous karyotyping result of G. elata (Zhou et al. 2018). The pseudochromosome 10 and 11 showed an off-diagonal pattern, and the Rabl configuration of chromatids might cause it. The Rabl configuration is a description of interphase chromosome arrangement in which telomeres and centromeres are located at opposite sides of the nucleus (Tiang et al. 2012). For validation, the Illumina reads were aligned to the genome, and the percentage of proper pairs aligned was 96.11%.
Table 1.
Assembly statistics of the G. elata genome
| FALCON-Unzip | HiRise | Final | |
|---|---|---|---|
| Number of contigs (scaffolds) | 654 | 514 | 488 |
| Total size of contigs (scaffolds) | 1,048,552,296 | 1,046,143,939 | 1,044,982,141 |
| Longest contig (scaffold) | 25,936,340 | 130,552,502 | 130,552,502 |
| Number of contigs (scaffold) >1M nt | 141 | 18 | 18 |
| Number of contigs (scaffold) >10M nt | 28 | 18 | 18 |
| N50 contig (scaffold) length | 9,175,439 | 50,595,616 | 50,595,616 |
| L50 contig (scaffold) count | 33 | 7 | 7 |
| GC content (%) | 34.27 | 34.27 | 34.27 |
FALCON-Unzip: Assembly result using PacBio data.
HiRise: Scaffolding result using FALCON-Unzip data.
Final: Organelle (plastid) genome removed from HiRise result.
Figure 2.
Genome-wide Hi-C interaction heatmap of G. elata. The 18 assembled scaffolds are ordered by length. The x- and y-axes provide the mapping positions for the first and second reads in each read pair, respectively, grouped into bins. The color of each square indicates the number of read pairs within that bin. Gray vertical and white horizontal lines have been added to indicate the borders between scaffolds. The off-diagonal pattern in the pseudochromosome 10 and 11 may reflect the Rabl configuration of chromatins (Tiang et al. 2012).
Compared with the previous version of draft G. elata genome (Yuan et al. 2018), our genome assembly is greatly improved in terms of the number of scaffolds (488 vs 3779) and the length of scaffold N50 (50.6 vs 4.9 Mb). Among the Orchidaceae, our genome assembly is the first chromosome-level genome assembly of an obligate mycoheterotrophic orchid G. elata, although another chromosome-level reference genome is available for P. aphrodite, which is an epiphytic orchid (Chao et al. 2018). We further used BUSCO to assess the completeness of our genome assembly, based on the embryophyta_odb9 database (Table 2). We found that only 935 (64.9%) of the 1440 highly conserved orthologs are present as complete genes in the G. elata genome, indicating that 451 (31.3%) genes are missing from G. elata, which is consistent with results from the previous genome assembly (Yuan et al. 2018). The gene loss events are frequently observed in plastid genome of mycoheterotrophic orchids (Barrett and Davis 2012; Logacheva et al. 2014; Petersen et al. 2018; Kim et al. 2019), but only a few cases are reported in nuclear genome (Yuan et al. 2018; Jakalski et al. 2020). The extensive gene loss in nuclear genome could also be related to mycoheterotrophic lifestyle and may be associated with the large abundance of repetitive elements in G. elata.
Table 2.
Statistics for genome assessment using BUSCO (embryophyta)
| No. of BUSCOs | Percentage of BUSCOs | |
|---|---|---|
| Complete | 935 | 64.9 |
| Complete and single-copy | 912 | 63.3 |
| Complete and duplicated | 23 | 1.6 |
| Fragmented | 54 | 3.8 |
| Missing | 451 | 31.3 |
Genomic features and repetitive elements
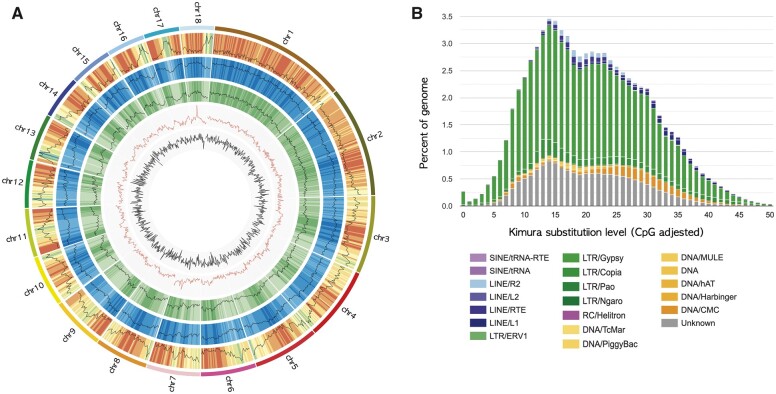
The gene density of orchid genomes, such as P. aphrodite (28.2 genes per Mb) and P. equestris (27.1 genes per Mb), is known to be lower than that of Arabidopsis thaliana (Cai et al. 2015; Chao et al. 2018), which is approximately 204.0 genes per Mb (calculated based on The Arabidopsis Genome Initiative 2000). Here, we found that the average gene density of the G. elata genome is 17.9 genes per Mb, with minimum and maximum densities on the first (Scx7bQ7_8: 10.8 genes per Mb) and 13th (Scx7bQ7_13: 22.5 genes per Mb) chromosomes, respectively (Figure 3A; Table 3). This is even lower than what has been detected in other orchid species. In contrast, the average repeat density was found to be 1406 repeats per Mb, and unlike genes, these are evenly distributed throughout the genome (Figure 3A). Retrotransposable elements, in particular, known to be the dominant form of repeats in angiosperm genomes (Oliver et al. 2013), constitute 74.92% (796.7 Mb) of the G. elata genome. This repetitive element content is higher than what has been found in any other orchid species, such as A. shenzhenica (47%), P. equestris (63.48%), and D. catenatum (64.51%) (Supplementary Figure S2). In addition, Class I (retrotransposons) and Class II (DNA transposons) TEs account for 49.96% and 8.42% of the G. elata genome, respectively (Figure 3B; Table 4). The quality of identified repetitive elements in these orchid species was assessed using LAI value (Supplementary Table S4). Although the LAI value in G. elata is slightly lower than A. shenzhenica and D. catenatum, G. elata shows the highest content of TEs (Supplementary Table S4). The LAI value for P. equestris could not be calculated as the proportion of intact TE was less than 0.05%. Like other sequenced orchid genomes, LTR retrotransposons, mainly Gypsy-type and Copia-type LTRs, are predominant (49.95%), followed in frequency by long interspersed nuclear elements (LINEs), which account for 4.33% of the genome. Of the repetitive elements, 15.32% could not be classified into any known families. In addition, the insertion time of LTR elements was estimated (Supplementary Figure S3), and the most abundant Gypsy-type LTRs were inserted relatively a long time ago and may have become fragmented and thus produce a lower LAI value. In summary, the repeat content of G. elata, especially LTR Gypsy elements, was larger than the other species in Orchidaceae family, which are not mycoheterotrophic.
Figure 3.
(A) Genome overview of the G. elata genome. The pseudochromosomes are in order from longest to shortest in a clockwise manner. The features are arranged in the order of gene density, repeat density, LTR/Gypsy, GC content, and GC skew from outside to inside in 1 Mb intervals across the 18 chromosomes. (B) Kimura distance-based copy divergence analysis of TEs in the G. elata genome. The graph represents the percentage of the genome represented by each repeat type on the y-axis to their corresponding Kimura substitution level (CpG adjusted) illustrated on the x-axis (K-value from 0 to 50). The color chart below the x-axis indicates the repeat types.
Table 3.
Statistics for G. elata genome annotation
| Features | No. of features | Total length of features (bp) |
Average length
of features (bp) |
Density (#/Mb) |
|---|---|---|---|---|
| Gene | 18,698 | 133,969,721 | 7,164.92 | 17.87 |
| CDS | 18,844 | 17,679,423 | 938.20 | 18.01 |
| Exon | 88,096 | 25,125,034 | 285.20 | 84.21 |
| Intron | 69,252 | 109,135,442 | 1,575.92 | 66.20 |
| 3′ UTR | 12,708 | 4,516,303 | 355.39 | 12.15 |
| 5′ UTR | 11,657 | 2,932,215 | 251.54 | 11.14 |
Table 4.
Sequence percentage (%) of annotated TEs proportional to the entire genome of G. elata and three species in the Orchidaceae family
| G. elata | A. shenzhenica | P. equestris | D. catenatum | ||
|---|---|---|---|---|---|
| DNA transposon | DNA | 4.09 | 6.50 | 3.17 | 3.20 |
| LINEa | 4.33 | 4.99 | 4.37 | 7.04 | |
| Retrotransposon | SINEb | 0.01 | 0.08 | 0.04 | 0.07 |
| LTRc | 49.95 | 13.71 | 32.66 | 34.19 | |
| Gypsy | 38.92 | 7.35 | 27.00 | 11.34 | |
| Copia | 4.61 | 3.46 | 4.49 | 19.35 | |
| Other | Unknown | 15.32 | 0.27 | 20.78 | 18.29 |
LINE, long interspersed nuclear element.
SINE, short interspersed nuclear element.
LTR, long terminal repeat.
The LTR elements are known to be the main drivers of gene evolution (Galindo-González et al. 2017), and they could have contributed to the gene loss and formation of unique genes in G. elata.
Gene annotation and comparative analysis
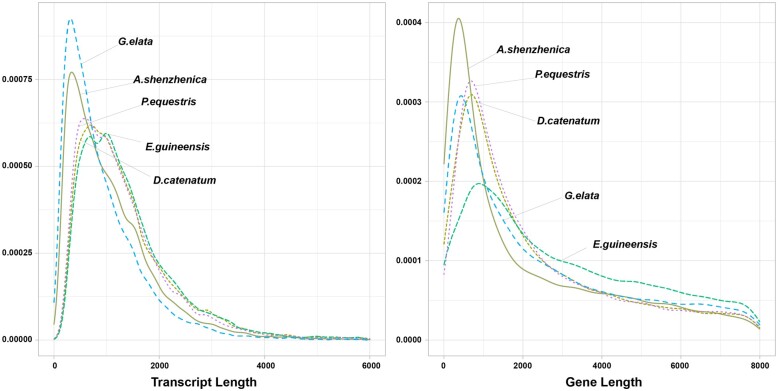
The complete annotated G. elata genome contains a final gene set comprising 18,844 CDS, with an AED less than 0.5 (Table 3). These CDSs total 17.68 Mb, and there is an average of 4.711 exons per gene. Among the final gene set, 15,619 CDSs are annotated in more than one database, including Uniprot, InterPro, Pfam, GO, and KEGG (Supplementary Table S5). The GO term analysis of the predicted proteome identified a number of proteins involved in metabolic and cellular processes, catalytic and binding activity, and cellular anatomical entity (Supplementary Figure S4). To compare gene content in G. elata and related species, we analyzed CDS distribution and gene length in G. elata relative to three other species in the Orchidaceae family (A. shenzhenica, D. catenatum, and P. equestris) and Elaeis guineensis (oil palm), as an outgroup (Figure 4). We found that G. elata shows the highest frequency of shorter length transcripts relative to other species. However, the overall gene-length distribution of G. elata is similar to that of other Orchidaceae species, except A. shenzhenica, which contains a genome that is smaller than the other species in this family (Figure 4; Supplementary Table S6). Last, the number of rRNAs and tRNAs predicted were 439 and 940, respectively.
Figure 4.
The distribution of transcript and gene length between G. elata, the other three species (A. shenzhenica, D. catenatu, and P. equestris) in Orchidaceae, and E. guineensis.
Orthology and gene family contraction and expansion
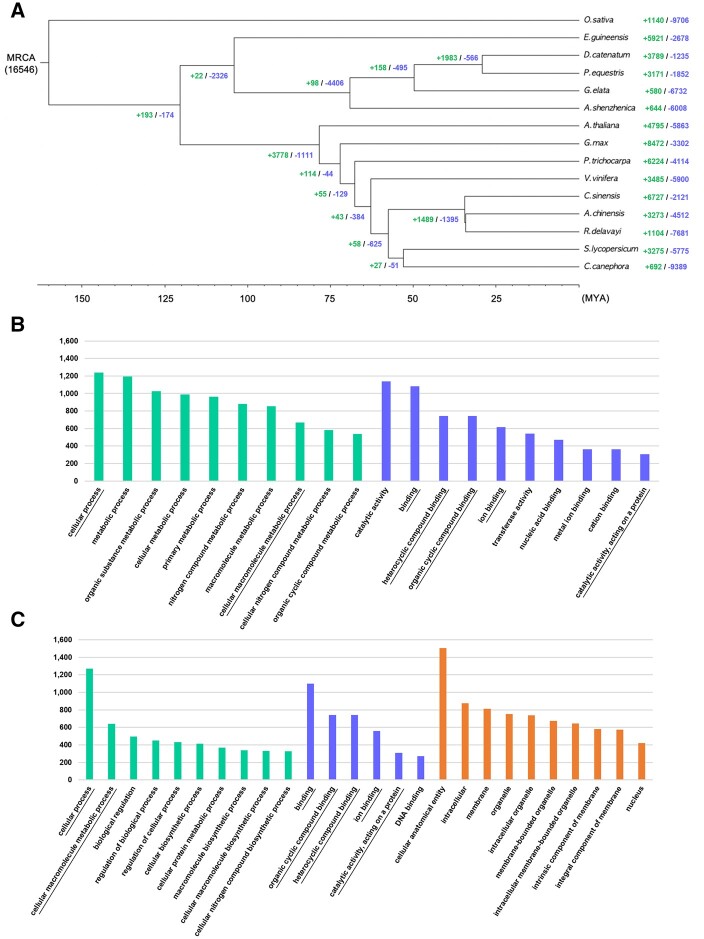
We next constructed a phylogenetic tree with G. elata and 14 other plants (Figure 5A). G.elata was found to cluster with other members of the Orchidaceae family, including A. shenzhenica, D. catenatum, and P. equestris. The tree shows that the Epidendroideae subfamily, which includes G. elata, D. catenatum, and P. equestris diverged from the Apostasioideae subfamily, which includes A. shenzhenica, approximately 65–70 million years ago (Mya). Gene family expansion and contraction analysis showed that substantial contraction occurred throughout divergence within the Orchidaceae family (Figure 5A). Notably, A. shenzhenica experienced gene loss due to a whole-genome duplication event, as previously reported (Zhang et al., 2017). G.elata also experienced extensive gene loss, whereas P. equestris and D. catenatum gained more genes than were lost through evolution. The G. elata genome, specifically, gained 580 gene families but lost 6732 gene families. We then performed GO term analyses of the expanded and contracted gene families in G. elata to assign putative functions (Figure 5, B and C). In the biological process (BP) category, genes related to metabolic process (GO:0008151) and those involved in the metabolism of macromolecules (GO:0043170), such as organic substances (GO:0071704) and nitrogen compounds (GO:0006807), were expanded (Figure 5B; Supplementary Table S7). Although not appearing in the top 50 most frequently identified GO terms (Supplementary Table S7), genes involved in arbuscular mycorrhizal association (GO:00036277) were detected. In the molecular function (MF) category, genes involved in catalytic activity (GO:0003824) and transferase activity (GO:0016740) were expanded (Figure 5B; Supplementary Table S7). Conversely, significantly contracted genes include those related to biosynthetic and metabolic processes in BP, DNA binding (GO:0003677) in MF, and genes in the cellular component (CE) category (Figure 5C; Supplementary Table S8). The loss of genes involved in biosynthetic processes and CE reflects that such features of G. elata may depend on its symbiont partners. In addition, the expansion of genes with novel metabolic processes and binding activities may be rewiring due to the lifestyle transition of G. elata to fully mycoheterotrophic.
Figure 5.
(A) Phylogenetic analysis of G. elata among 15 plants and gene family gain-and-loss analysis including the number of gained gene families (+) and lost gene families (−). (B) The number of genes in the top 10 GO terms of expanded gene families (Supplementary Table S7) and (C) contracted gene families (Supplementary Table S8) in the G. elata genome. The green, blue, and orange colored bars represent the three major GO categories, biological process, MF, and CE, respectively. The overlapping terms in both expanded and contracted gene families are underlined.
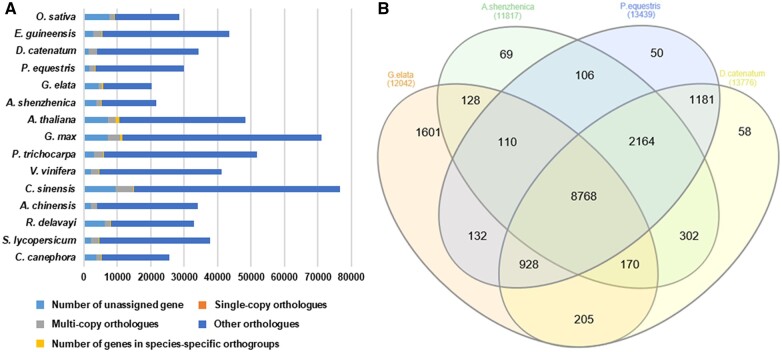
Orthology analysis with the 15 plant species included in our phylogenetic tree identified 16,115 orthologous gene families and 418 species-specific gene families (Supplementary Table S9). G.elata contains the lowest number of protein-coding genes compared to the other plant species and even to the other orchid species (Figure 6A; Supplementary Table S6). Conversely, of all the orchid species, G. elata encodes the highest number of unassigned genes and genes in species-specific orthogroups. We further identified a set of orthologous gene families shared among the orchid species (Figure 6B). This set contains a total of 8768 orthogroups that are conserved across all four orchid genomes, with an additional 928 orthogroups conserved across the three species in the Epidendroideae subfamily (i.e., G. elata, P. equestris, and D. catenatum). G.elata encodes 1601 species-specific orthologs, which is more than the other Epidendroideae species.
Figure 6.
(A) Bar graph of the number of protein-coding genes in the 15 plant species including G. elata. The distribution of number of genes between G. elata and other 14 species by the type of orthogroups. Single-copy orthologs include common orthologs with one copy in all species. Multi-copy orthologs include common orthologs with multiple copy numbers in all species. The number of genes in species-specific orthogroups represents unique genes in specific species. Other orthologs include gene from families shared in 2–14 species. (B) Venn diagram of orthologous gene families between G. elata and other three species (A. shenzhenica, D. catenatu, and P. equestris) in the Orchidaceae family.
We found that the genome of G. elata has an extremely low gene density, proliferation of repeat content, and significant expansion and contraction of genes involved in metabolic processes and biosynthetic processes, respectively. In addition, G. elata has the highest number of unique genes among the compared orchid species. Since nutrient absorption of this obligate mycoheterotrophic plant is entirely dependent on their fungal partners (Merckx et al. 2009), these genomic features may reflect the mycoheterotrophic and symbiotic lifestyle of the G. elata.
Conclusion
Here, we report the first high-quality chromosome-level genome assembly of G. elata. We found an extremely low gene density, proliferation of repeat content, and significant contraction of genes involved in CEs, reflecting on its mycoheterotrophic lifestyle. Consequently, this high-quality reference genome data of G. elata will be important for informing further studies aimed at better understanding genomic interactions and gene expression changes that occur during the development of G. elata with its associated fungi, thereby uncovering the symbiotic mysteries underlying such mycoheterotrophic lifestyles.
Data availability
The G. elata genome project was deposited at NCBI, under BioProject No. PRJNA632604. The raw DNA sequencing reads are available at the Sequence Read Archive (SRA) under Accession Nos. SRR12263394, SRR12263395, and SRR12263396. The raw Iso-Seq data is available under the Accession No. SRR13516450. The genome assembly data have been deposited at GenBank under the Accession No. GCA_016760335.1 (WGS: JACERR000000000.1). The commands and parameters used for the genome assembly and repeat annotation are available in Supplementary Table S10.
Supplementary material is available at G3 online.
Supplementary Material
Acknowledgments
E.-J.P. conceived and designed the study as the lead investigator; E.-K.B., M.-J.K., C.A., and S.-A.L. prepared the materials; K.-T.K. and S.J.L. performed the genome sequencing, assembly, annotation, and further bioinformatics analysis; and E.-J.P., K.-T.K., and E.-K.B. wrote the manuscript. All authors contributed to, reviewed, and approved the final manuscript.
Funding
This study was supported by the RandD Program for Forestry Technology (Grants numbers S111416L0710), and the National Institute of Forest Science project (Grants numbers FG0603-2021-01). This study was supported by (in part) Sunchon National University Research Fund in 2021 (Grant number: 2021-0231).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bao Z, Eddy SR.. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, Davis JI.. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99:1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton T, Anderson CL, Jacquet D, Lundqvist S, Bremer K.. 2007. Estimating divergence times in large phylogenetic trees. Syst Biol. 56:741–752. doi: 10.1080/10635150701613783. [DOI] [PubMed] [Google Scholar]
- Cai J, Liu X, Vanneste K, Proost S, Tsai WC, et al. 2015. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M.. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 48:4.11.11–14.11.39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM.. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y-T, Chen W-C, Chen C-Y, Ho H-Y, Yeh C-H, et al. 2018. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol J. 16:2027–2041. doi: 10.1111/pbi.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O'Donovan C, et al. 2012. The UniProt-GO annotation database in 2011. Nucleic Acids Res. 40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Moore B, Holt C, Yandell M.. 2009. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinform. 10:67. doi: 10.1186/1471-2105-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:1–14. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-González L, Mhiri C, Deyholos MK, Grandbastien M-A.. 2017. LTR-retrotransposons in plants: engines of evolution. Gene. 626:14–25. doi: 10.1016/j.gene.2017.04.051. [DOI] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Fukukawa S, Kunishi A, Suga H, Richard F, et al. 2012. Mycoheterotrophic germination of Pyrola asarifolia dust seeds reveals convergences with germination in orchids. New Phytol. 195:620–630. doi: 10.1111/j.1469-8137.2012.04174.x. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynson NA, Weiß M, Preiss K, Gebauer G, Treseder KK.. 2013. Fungal host specificity is not a bottleneck for the germination of Pyroleae species (Ericaceae) in a Bavarian forest. Mol Ecol. 22:1473–1481. doi: 10.1111/mec.12180. [DOI] [PubMed] [Google Scholar]
- Inglis PW, Pappas MCR, Resende LV, Grattapaglia D.. 2018. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS One. 13:e0206085. doi: 10.1371/journal.pone.0206085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemyn H, Waud M, Brys R.. 2018. Mycorrhizal divergence and selection against immigrant seeds in forest and dune populations of the partially mycoheterotrophic Pyrola rotundifolia. Mol Ecol. 27:5228–5237. doi: 10.1111/mec.14940. [DOI] [PubMed] [Google Scholar]
- Jakalski M, Minasiewicz J, Caius J, May M, Selosse M-A, et al. 2020. The genomic impact of mycoheterotrophy: targeted gene losses but extensive expression reprogramming. bioRxiv. doi: 10.1101/2020.06.26.173617. [DOI] [Google Scholar]
- Johansson VA, Bahram M, Tedersoo L, Koljalg U, Eriksson O.. 2017. Specificity of fungal associations of Pyroleae and Monotropa hypopitys during germination and seedling development. Mol Ecol. 26:2591–2604. doi: 10.1111/mec.14050. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H.. 2009. Multiple alignment of DNA sequences with MAFFT. In: Podada D, editor. Bioinformatics for DNA Sequence Analysis, Methods in Molecular Biology, Vol. 537.Totowa, New Jersey: Humana Press. p. 39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Kim Y-K, Jo S, Cheon S-H, Joo M-J, Hong J-R, et al. 2019. Extensive losses of photosynthesis genes in the plastome of a mycoheterotrophic orchid, Cyrtosia septentrionalis (Vanilloideae: Orchidaceae). Genome Biol Evol. 11:565–571. doi: 10.1093/gbe/evz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics. 5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, Gedman G, Kingan SB, Chin C-S, Howard JT, et al. 2017. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience. 6:1–16. doi: 10.1093/gigascience/gix085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Leake JR. 1994. The biology of mycoheterotrophic (‘saprophytic’) plants. New Phytol. 127:171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x. [DOI] [PubMed] [Google Scholar]
- Leake JR. 2005. Plants parasitic on fungi: unearthing the fungi in myco-heterotrophs and debunking the ‘saprophytic’ plant myth. Mycologist. 19:113–122. doi: 10.1017/S0269-915X(05)00304-6 [DOI] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA.. 2014. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 6:238–246. doi: 10.1093/gbe/evu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27:764–770. doi: 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx V, Bidartondo MI, Hynson NA.. 2009. Myco-heterotrophy: when fungi host plants. Ann Bot. 104:1255–1261. doi: 10.1093/aob/mcp235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx V, Freudenstein JV.. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytol. 185:605–609. doi: 10.1111/j.1469-8137.2009.03155.x. [DOI] [PubMed] [Google Scholar]
- Oliver KR, McComb JA, Greene WK.. 2013. Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol Evol. 5:1886–1901. doi: 10.1093/gbe/evt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Chen J, Jiang N.. 2018. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46:e126. doi: 10.1093/nar/gky730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E-J, Lee WY.. 2013. In vitro symbiotic germination of mycoheterotrophic Gastrodia elata by Mycena species. Plant Biotechnol Rep. 7:185–191. doi: 10.1007/s11816-012-0248-x. [DOI] [Google Scholar]
- Pennisi E. 2017. New technologies boost genome quality. Science. 357:10–11. doi: 10.1126/science.357.6346.10. [DOI] [PubMed] [Google Scholar]
- Petersen G, Zervas A, Pedersen HÈ, Seberg O.. 2018. Genome reports: contracted genes and dwarfed plastome in mycoheterotrophic Sciaphila thaidanica (Triuridaceae, Pandanales). Genome Biol Evol. 10:976–981. doi: 10.1093/gbe/evy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA.. 2005. De novo identification of repeat families in large genomes. Bioinformatics. 21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26:342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shunxing G, Qiuying W.. 2001. Character and action of good strain on stimulating seed germination of Gastrodia elata. Mycosystema. 20:408–412. [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Slater G, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Schoffmann O, Morgenstern B, Waack S.. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Tiang C-L, He Y, Pawlowski WP.. 2012. Chromosome organization and dynamics during interphase, mitosis, and meiosis in plants. Plant Physiol. 158:26–34. doi: 10.1104/pp.111.187161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-C, Wu K-M, Chiang T-Y, Huang C-Y, Chou C-H, et al. 2016. Comparative transcriptome analysis of Gastrodia elata (Orchidaceae) in response to fungus symbiosis to identify gastrodin biosynthesis-related genes. BMC Genomics. 17:212. doi: 10.1186/s12864-016-2508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, et al. 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao Y, Zang P, Xu Y.. 2020. Transcriptome analysis reveals underlying immune response mechanism of fungal (Penicillium oxalicum) disease in Gastrodia elata Bl. f. glauca S. chow (Orchidaceae). BMC Plant Biol. 20:445. doi: 10.1186/s12870-020-02653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waud M, Brys R, Van Landuyt W, Lievens B, Jacquemyn H.. 2017. Mycorrhizal specificity does not limit the distribution of an endangered orchid species. Mol Ecol. 26:1687–1701. doi: 10.1111/mec.14014. [DOI] [PubMed] [Google Scholar]
- Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, et al. 2015. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Guo X.. 1989. Fungus associated with nutrition of seed germination of Gastrodia elata-Mycena osmundicola Lange. Acta Mycol Sin. 8:221–226. [Google Scholar]
- Xu Z, Wang H.. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Jin X, Liu J, Zhao X, Zhou J, et al. 2018. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat Commun. 9:1–11. doi: 10.1038/s41467-018-03423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia M, Bolosky WJ, Curtis K, Fox A, Patterson D, et al. 2011. Faster and more accurate sequence alignment with SNAP. arXiv:1111.5572.
- Zeng X, Li Y, Ling H, Chen J, Guo S.. 2018. Revealing proteins associated with symbiotic germination of Gastrodia elata by proteomic analysis. Bot Stud. 59:8. doi: 10.1186/s40529-018-0224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Li Y, Ling H, Liu S, Liu M, et al. 2017. Transcriptomic analyses reveal clathrin-mediated endocytosis involved in symbiotic seed germination of Gastrodia elata. Bot Stud. 58:31. doi: 10.1186/s40529-017-0185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GQ, Liu KW, Li Z, Lohaus R, Hsiao YY, et al. 2017. The Apostasia genome and the evolution of orchids. Nature. 549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GQ, Xu Q, Bian C, Tsai W-C, Yeh C-M, et al. 2016. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci Rep. 6:19029. doi: 10.1038/srep19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W-J, Li B-F.. 1980. The biological relationship of Gastrodia elata and Armillaria mellea. Acta Bot Sin. 22:57–62. [Google Scholar]
- Zhou HC, Park E-J, Kim HH.. 2018. Analysis of chromosome composition of Gastrodia elata Blume by fluorescent in situ hybridization using rDNA and telomeric repeat probes. Crop Sci. 26:113–118. doi: 10.7783/KJMCS.2018.26.2.113. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The G. elata genome project was deposited at NCBI, under BioProject No. PRJNA632604. The raw DNA sequencing reads are available at the Sequence Read Archive (SRA) under Accession Nos. SRR12263394, SRR12263395, and SRR12263396. The raw Iso-Seq data is available under the Accession No. SRR13516450. The genome assembly data have been deposited at GenBank under the Accession No. GCA_016760335.1 (WGS: JACERR000000000.1). The commands and parameters used for the genome assembly and repeat annotation are available in Supplementary Table S10.
Supplementary material is available at G3 online.