Abstract
Recent developments in a variety of sectors, including health care, research and the direct-to-consumer industry, have led to a dramatic increase in the amount of genomic data that are collected, used and shared. This state of affairs raises new and challenging concerns for personal privacy, both legally and technically. This Review appraises existing and emerging threats to genomic data privacy and discusses how well current legal frameworks and technical safeguards mitigate these concerns. It concludes with a discussion of remaining and emerging challenges and illustrates possible solutions that can balance protecting privacy and realizing the benefits that result from the sharing of genetic information.
Subject terms: Ethics, Genomics
In this Review, the authors describe technical and legal protection mechanisms for mitigating vulnerabilities in genomic data privacy. They also discuss how these protections are dependent on the context of data use such as in research, health care, direct-to-consumer testing or forensic investigations.
Introduction
There are many stories in the media highlighting the multitude of ways by which genomic data are now relied upon, including in basic research, clinical care, discovering relatives and ancestral origins, tracking down criminals, and identification of victims. At the same time, numerous reports from around the world illustrate that some people are concerned about how genomic information that relates to them are used, often stated as challenges to privacy. These apprehensions do have some foundation as people can suffer harm if data about them are used in ways they do not agree with, for example, to examine ancestry1 or to create commercial products2 without the individual’s approval, or if the data are used in a manner that causes an individual to suffer adverse consequences such as stigmatization3, disruption of familial relationships4,5 or loss of employment or insurance. However, the law provides limited, patchy protection6,7.
The concept of privacy and its protection has many facets8. People may wish to control how genomic data about them are used but, in many cases, they only have the choice to opt in (or opt out) based on the terms contained in a consent form or a service agreement9, which frequently goes unread10,11. In other instances, people may not have any choice at all about how genomic data about them are used, such as when data are deemed to be anonymised in accordance with the General Data Protection Regulation (GDPR)12 of the European Union (EU), de-identified in accordance with the Health Insurance Portability and Accountability Act of 1996 (HIPAA)13–15 or considered non-human subject data in accordance with the Common Rule for the Protection of Human Research Participants16 in the United States. Another aspect of privacy is the right to solitude (often voiced as the right to be left alone), a principle first formalized in legal circles in the late 1800s17, which could include the right not to be (re)contacted about ancillary findings generated from genomic testing or discovery-driven investigations into existing genomic data sets18,19 or by previously unknown relatives20,21.
Yet, the right to privacy has never been absolute, in part because many uses of these data, such as clinical care, research, exploring ancestry, finding relatives and identifying criminal suspects and victims of mass casualties, can be valued by users, other stakeholders, or society at large. For example, even though physicians have strong ethical and legal duties of confidentiality that require them not to disclose patients’ information to others, these obligations are not unconditional because the law has created numerous exceptions such as for public health reporting22 or in criminal investigations.
Although the tension between privacy and data utility raises an array of ethical issues23–25 regarding when genomic data can be accessed and used, this Review focuses on the primary tools that are applied to define and protect these boundaries: law (as instantiated in statutes, regulatory regimes and case law), policy and technology. Several reviews on genomic data privacy have been published over the years in response to the evolution of approaches to intrude upon, and protect, privacy. Initially, Malin appraised the robustness of genetic data de-identification26. This study was followed by Erlich and Narayanan who analysed and categorized computational methods for re-identification, in light of new techniques for surname inference, and potential risk mitigation techniques27. Naveed et al. reviewed the privacy and security threats that arise over the course of the genomic data lifecycle, from data generation to its end uses28. Wang et al. studied the technical and ethical aspects of genetic privacy29. Arellano et al. reviewed policies and technologies for protecting the privacy of biomedical data in general30. More recently, Mittos et al. systematically reviewed privacy-enhancing technologies for genomic data and particularly highlighted the challenges associated with using cryptography to maintain privacy over a long period of time31. Grishin et al. reviewed the emerging cryptographic tools for protecting genomic privacy with a focus on blockchains32. Bonomi et al. reviewed privacy challenges as well as technical research opportunities for genomic data applications such as direct-to-consumer genetic testing (DTC-GT) and forensic investigations33. Similarly, numerous articles have addressed the incomplete and inconsistent protection that the law provides from harms to individuals and groups in different settings3,19,34–36.
Our Review diverges from prior work in that we consider it essential to discuss the legal and technological perspectives together. This is because technological interventions can heighten, but also ameliorate, legal risks, whereas some laws provide control or protect people from downstream harm from data use, thereby opening the door to different and perhaps less stringent technological protections. Moreover, recent disruptions associated with mandates for data sharing37,38, the DTC-GT revolution and the coronavirus disease 2019 (COVID-19) pandemic — events that have dramatically accelerated the collection and use of genomic data39–41 — have dramatically changed the social environment in which genomic data are obtained and used. Blending legal and technical protections in a holistic ecosystem of genomic data is challenging because protections are interconnected but vary in the environments in which they were developed, the stakeholders involved and their underlying assumptions. To demystify the connections among and the assumptions behind different legal and technical protections, we partition the ecosystem into four settings: health care, research, DTC and forensic settings.
In this Review, we begin with a brief overview of attacks on privacy in the context of genomic data sharing and subsequently discuss both how to mitigate privacy risks (through technical and legal safeguards) as well as the consequences of failing to do so effectively. Next, we categorize legal protections according to different settings since each setting tends to have unique laws and policies; meanwhile, we identify settings where each technical protection was first introduced and/or has been frequently applied. We consider the particular challenges that can arise in the research setting itself. We then note that genomics researchers also need an appreciation of the larger ecology of the flows of genomic data outside the research and health-care settings in light of their impact on data privacy and public opinion and thus ultimately on public support for genomic research. Thus, we discuss DTC-GT, the obligations that companies that provide these tests owe to users, and the consequences of use by consumers to find relatives and by law enforcement to find criminal suspects. For reference, Fig. 1 illustrates an overview of privacy intrusions and safeguards in the ecology of genomic data flows, and Table 1 summarizes various aspects of the technical literature featured in this Review. In our discussions, a first party refers to the individual to whom the data correspond, whereas a second party refers to the organization (or individual) who collects and/or uses the data for a purpose that the first party is made aware of. By contrast, third parties refer to users (or recipients) of data who have the ability to communicate with the second party only and might include malicious attackers. Examples of third parties include researchers who access data from an existing research study or a pharmaceutical company that partners with a DTC-GT company. We conclude with a discussion of what legal revisions and technical advances may be warranted to balance privacy protection with the benefits to individuals, commercial entities, researchers and society that result from flows of genomic data.
Fig. 1. An overview of privacy intrusions and safeguards in genomic data flows.
The four routes of genomic data flow (as indicated by the arrow colours) represent four settings in which data are used or shared: health care (red), research (gold), direct-to-consumer (DTC; green) and forensic (dark blue). The grey line represents a combination of the first three settings. In the health-care setting, data collected by a health-care entity (for example, Vanderbilt University Medical Center) are protected by the Genetic Information Nondiscrimination Act of 2008 (GINA)128 and the Health Insurance Portability and Accountability Act of 1996 (HIPAA)116,117 for primary uses. In the research setting, data collected by a research entity (for example, 1000 Genomes Project, Electronic Medical Records and Genomics (eMERGE) network or All of Us Research Program) are primarily protected by the Common Rule14,124 for primary uses and protected by the US National Institutes of Health (NIH) data sharing policy37,38 for secondary uses. In the DTC setting, data collected by a DTC entity are protected by the European Union’s General Data Protection Regulation (GDPR)12 and/or the US state privacy laws (for example, California Consumer Privacy Act130, California Privacy Rights Act131 or Virginia Consumer Data Protection Act132) for primary uses and protected by self-regulation (for example, data use agreements36, privacy policies173 or terms of service174) for secondary uses. In the forensic setting, data shared with law enforcement are protected by informed consent192. A first party refers to the individual to whom the data correspond, whereas a second party refers to the organization (or individual) who collects and/or uses the data for a purpose that the first party is made aware of. By contrast, third parties refer to users (or recipients) of data who have the ability to communicate with the second party only and might include malicious attackers. Examples of third parties include researchers who access data from an existing research study or a pharmaceutical company that partners with a DTC genetic testing company. The data flow from a DTC entity to a research entity is represented by the arrow at the bottom. Confidentiality is mostly concerned when data are being used, whereas anonymity and solitude are mostly concerned when data are being shared. Specifically, cryptographic tools31 protect confidentiality against unauthorized access attacks, whereas access control27 and data perturbation approaches83 protect anonymity against privacy intrusions such as re-identification and membership inference attacks. We simplify the figure by omitting the impacts of GDPR and data use agreements in the research setting.
Table 1.
A taxonomy of technical research articles on genomic data privacy featured in this Review
| Attack or protection | Use | Data flow | Data level | Setting | How attacks or protections are achieved | Attributes studied other than genotypes/how data are used | Refs |
|---|---|---|---|---|---|---|---|
| Anonymity | |||||||
| Attack | Secondary | Share | Individual | Health care | Re-ID | Demographics, hospital trail | 42 |
| Research | Re-ID | NA | 84 | ||||
| Pedigree | 51 | ||||||
| Re-ID, genotype imputation | Signal profiles | 90 | |||||
| Re-ID, genotype inference | Diseases | 45 | |||||
| Visual traits/3D facial structures | 46–48 | ||||||
| Re-ID, non-genotypic attribute inference | Demographics, name | 53 | |||||
| Demographics, surname | 54 | ||||||
| Face, traits, demographics | 44,49,50 | ||||||
| Genotype imputation | NA | 85,86 | |||||
| Research, DTC | Genotype imputation | Pedigree | 64 | ||||
| Genotype imputation, genotype inference, genotype reconstruction | Pedigree | 66 | |||||
| Summary | Research | Membership inference | GWAS statistics | 56–58,60,96,97 | |||
| Membership inference, genotype inference | Machine learning model, demographics | 61 | |||||
| GWAS statistics, pedigree | 106 | ||||||
| Membership inference, non-genotypic attribute inference | Disease status | 62 | |||||
| Membership inference, re-ID, genotype imputation | GWAS statistics | 59 | |||||
| Membership inference, re-ID, genotype inference, genotype reconstruction | GWAS statistics, visual traits | 98 | |||||
| Protection | Secondary | Share | Individual | Research | Generalization | RNA sequences | 89 |
| Generalization, suppression, k-anonymity | NA | 88 | |||||
| Masking/hiding, risk assessment | Demographics | 93 | |||||
| Summary | Research | Suppression, risk assessment | NA | 92 | |||
| Beacons | Disease | 95 | |||||
| GWAS statistics, pedigree | 101 | ||||||
| Beacons, differential privacy | GWAS statistics | 99,100 | |||||
| Beacons, risk assessment | GWAS statistics | 102 | |||||
| Differential privacy | GWAS statistics | 103–105,107,108 | |||||
| Generative adversarial network | Disease | 109 | |||||
| Federated learning | GWAS statistics | 149 | |||||
| Risk assessment | NA | 82 | |||||
| Confidentiality | |||||||
| Protection | Primary | Use | Individual | Health care | Homomorphic encryption | Disease susceptibility test | 78 |
| Controlled functional encryption | Relatedness tests | 79 | |||||
| SMC | Disease diagnosis | 147 | |||||
| Research | Homomorphic encryption | GWAS computation | 142,143 | ||||
| Homomorphic encryption, SMC | GWAS computation | 141,150 | |||||
| Homomorphic encryption, TEE | GWAS computation | 154 | |||||
| SMC | GWAS computation | 145,146 | |||||
| TEE | GWAS computation | 153,155 | |||||
| Symmetric encryption, cryptographic hardware | GWAS computation | 151 | |||||
| Research, DTC | Homomorphic encryption | Sequence matching, sequence comparison | 180 | ||||
| SMC | Sequence comparison | 148 | |||||
| Fuzzy encryption | Relative identification | 182 | |||||
| DTC | Private set intersection protocols | Paternity test, genetic compatibility test | 181 | ||||
| Store | Individual | Health care | Honey encryption | NA | 76 | ||
| Secure file format | NA | 77 | |||||
| Secondary | Share | Individual | Research | Blockchain | NA | 158 | |
| Research, DTC | Blockchain | NA | 157 | ||||
| DTC | Blockchain, controlled access, homomorphic encryption, SMC | NA | 161 | ||||
| Summary | Research | Blockchain | Machine learning model | 159 | |||
| Controlled access | GWAS statistics | 81 | |||||
| Solitude | |||||||
| Attack | Secondary | Share | Individual | DTC, forensic | Familial search, genotype imputation, genotype reconstruction | Name, e-mail address | 74,75 |
| Forensic | Familial search, re-ID | Demographics | 72 | ||||
| Familial search, re-ID, genotype imputation | Pedigree | 73 | |||||
| Individual, summary | Research, DTC | Non-genotypic attribute inference, kin genotype reconstruction | Pedigree | 63 | |||
| Attribute inference, kin genotype reconstruction | Pedigree, disease | 65 | |||||
| Protection | Primary | Collect | Individual | Forensic | Controlled access, encryptions | NA | 39 |
| Secondary | Share | Individual | DTC, research | Masking/hiding, risk assessment | Pedigree | 196 | |
DTC, direct-to-consumer; GWAS, genome-wide association study; ID, identification; NA, not applicable; SMC, secure multiparty computation; TEE, trusted execution environment.
Privacy intrusions and protections
Privacy intrusions
Individuals may suffer harm when data about them are used without their permission in ways they do not agree with. In contrast to summary data aggregated across many participants, individual-level data that identify the people to whom they pertain, not surprisingly, pose a greater risk of harm to the person. For example, breaches of identified data might reveal a health condition that the participant had wished not to become public or cause them to suffer adverse consequences such as reputational damage or loss of employment, insurance, or other economic goods3. These disclosures can occur when data holders lose the data, for instance, by misplacing an unencrypted laptop, or when third parties deliberately attack large, identified data collections; therefore, security becomes particularly important when conducting research using identified data.
Much research using genomic data, however, is conducted with additional types of data, such as demographics, social and behavioural determinants of health, and phenotypic information at the molecular and/or clinical level (for example, data derived from electronic health records), from which standard identifying information have been removed. Yet, there has been a vigorous debate about whether genomic data can be de-identified or anonymised on its own or in combination with the accompanying individual information. Over the years, a number of investigators have famously demonstrated their ability to re-identify individuals whose data have been used without common identifiers for genomics research. The following provides a summary of these attacks.
Re-identification
Sharing individual-level genomic data, even without explicit identifiers, creates an opportunity for re-identification42. For example, a data recipient could infer phenotypic information from genomic data that may be leveraged for re-identification purposes27,43. In one study, researchers re-identified individuals in a data set of whole-genome sequences by predicting visual traits, including eye and skin colour44. Similarly, genomic attributes might be inferred from phenotypic traits (for example, physically observable disorders45, visual traits46,47 or 3D facial structures48) for re-identification purposes, although the actual power of these attacks is debatable47,49,50. In addition, known pedigree structures may be leveraged to re-identify genomic records51.
Moreover, potential identifiers may be inferred from the demographic information that is often shared with genomic data, through linkage to other readily accessible data sources. In 2013, participants of the Personal Genome Project52 were re-identified by Sweeney et al. by linking these participants’ data records to publicly available voter registration lists using demographic attributes53. In the same year, Gymrek et al. re-identified certain participants of the 1000 Genomes Project by first inferring surnames from short tandem repeats (STRs) on the Y chromosome, which, in combination with other demographics, were then linked to identified public resources54.
Membership inference
In genome–phenome investigations, such as genome-wide association studies (GWAS), researchers commonly publish only summary statistics that are useful for meta-analyses55. However, in 2008, Homer et al. demonstrated that GWAS summary statistics are vulnerable to membership inference attacks56, whereby it is possible to discover an identified target’s participation in the GWAS as part of a potentially sensitive group. Although the power of this attack was questioned by other researchers57, subsequent studies showed that the inference power can be further improved by leveraging statistics based on allele frequencies58, correlations59 and regression coefficients60. Furthermore, parameters in machine learning (ML) models trained on individual-level genomic data sets have the potential to disclose the genotypes and memberships of the participants61. Identifying an individual’s membership in a GWAS data set could also reveal the participant’s sensitive clinical information such as disease status62.
Reconstruction and familial search
Due to the similarity of relatives’ genomic records, even if someone’s genomic record has never been shared or even generated, their genotypes and predispositions to certain diseases63 can be inferred to a certain degree from their relatives’ shared genotypes64. Recently, more powerful reconstruction attacks have been proposed to infer individuals’ genotypes and phenotypes from their relatives’ genotypes and phenotypes65,66.
In April 2018, the US Federal Bureau of Investigation (FBI) used genomic data from a cold case to arrest a suspected serial murderer known as the Golden State Killer. In this case, law enforcement officers used crime-scene DNA from the then-unidentified suspect and uploaded the sequence data to GEDmatch, a publicly accessible genomic database. Through a process known as long-range familial search, whereby relatives can be identified based on shared blocks of DNA sequence, they found the suspect’s third cousin. From this starting point in the suspect’s wider family, law enforcement officers were then able to make further enquiries, reconstruct a family tree and subsequently trace the suspect. Although this case demonstrated the potential of the forensic use of familial search, now known as forensic or investigative genetic genealogy (FGG/IGG), it sparked privacy concerns67. Acknowledging these concerns, in May 2019, GEDmatch provided users with the opportunity to opt in to allow their data to be used for investigating violent crimes68. By May 2020, most (81%) of GEDmatch’s 1.4 million users still have not opted in69, and users concerned about privacy did delete their data70. Potentially, users who uploaded data to GEDmatch (or a similar database) and their relatives may still be reached out using the long-range familial search technique by anyone (for example, law enforcement officers or hackers) who obtained their genomic data elsewhere71. A study that received a great deal of attention predicted that, in a database of 1 million individuals, 60% of searches using genome data from individuals of European descent as search queries will result in finding a third cousin or closer match to the targeted individuals due to the high number of individuals of European ancestry already in the database72. With the help of correlations between two types of genetic markers (that is, single-nucleotide polymorphism (SNP) and STR markers), the detection of relatives in genomic databases becomes even easier73. Although most records in these databases are not disclosed to end-users, stronger attacks have aimed to reconstruct records in a database by uploading strategically generated artificial records74,75.
Technical protections against intrusions
Security controls
An important element of protecting privacy is preventing access to data by those who are not entitled to them. Some attacks targeting genomic data can be prevented by applying standard security controls (for example, access control27 and cryptographic tools31,76–79) and restricting access to selected trusted recipients27. For example, in response to attacks demonstrated by Gymrek et al.54 and Homer et al.56, the US National Institutes of Health (NIH) and the Wellcome Trust moved certain demographics about the participants80 as well as GWAS summary statistics81 into access-controlled databases. Subsequent studies found the attack to be less powerful under more realistic assumptions82, which contributed to the NIH’s decision in 2018 to derestrict public access to genomic summary statistics33.
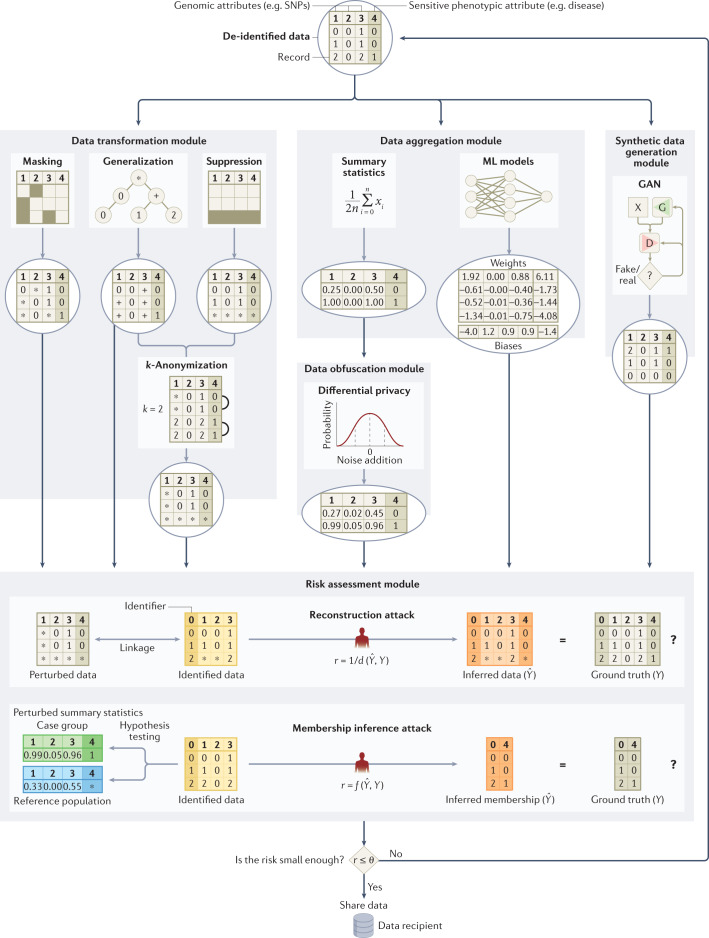
If de-identified data need to be shared with an untrusted third party or the public, the privacy of individuals to whom these data correspond can be protected by perturbing83 (that is, limiting or altering) the data. In the following sections and Fig. 2, we illustrate four approaches for technical protection that perturb data (that is, transformation, aggregation, obfuscation and synthetic data generation) with examples in the context of genomic data sharing.
Fig. 2. Data perturbation approaches for privacy protection in genomic data sharing.
Each module (or submodule) can work independently to protect data as shown by the corresponding data flow. In the transformation module, data can be masked93, generalized88 and/or suppressed according to a privacy protection model (for example, k-anonymity)87. In the aggregation module, data can be aggregated to summary statistics81 or parameters in a machine learning (ML) model61. In the module of synthetic data generation, a synthetic data set can be generated using a generative adversarial network (GAN)110. In the obfuscation module, noise can be added to data using a privacy protection model (for example, differential privacy)103. All contents in each module (or submodule) are examples for illustration purposes only. In the example for the generalization submodule, the plus sign represents a generalization of values one and two for a genomic attribute. In the example for the submodule of summary statistics, the minor allele frequency for each single-nucleotide polymorphism (SNP) marker is computed for each group of individual records. (n represents the number of records in the group; xi represents the value of a genomic attribute for the ith record in a group, which is the number of minor alleles at a SNP position for a record in this example.) In the example for the submodule of ML models, the neural network with three layers has 21 parameters (that is, 16 weights and 5 biases) that need to be learned. In the example for the GAN submodule, X represents the input data set, G represents the generator network and D represents the discriminator network. In the example for the reconstruction attack in the module of risk assessment91, the attacker tries to reconstruct the original data set by linkage and inference66, and the privacy risk is assessed by the data sharer using a distance function. In the example for the membership inference attack in the module of risk assessment92, the attacker tries to infer the membership of each targeted individual by hypothesis testing58, and the privacy risk is assessed by the data sharer using a function that measures the test’s accuracy. The reconstruction attack and the membership inference attack are used here for illustration purposes only and could be replaced with any other attack (for example, a re-identification attack or a familial search attack) or some arbitrary combination of attacks. Data can be sequentially protected by multiple modules and submodules before the privacy risk is mitigated to an acceptable level and finally released. r represents the privacy risk; d represents the distance function; f represents the function measures accuracy; represents the threshold for the privacy risk.
Data transformation
Some have suggested that the number of released genetic variants should be limited because, among millions of SNPs in a person’s genome, less than 100 statistically independent SNPs are required to identify each person uniquely84. However, protecting a genomic data set by hiding a set of genetic variants may not be very effective due to correlations among genetic variants (known as linkage disequilibrium)85 and well-established genotype imputation techniques86.
To thwart re-identification through linkage in general, Sweeney introduced k-anonymity87, a data transformation model, to ensure that each record in a released data set is equivalent to no fewer than (k − 1) other records with the same quasi-identifying values (that is, those which can be relied upon for linkage). Initially developed to address demographics, it was subsequently shown that this model could be applied to genomic data by generalizing nucleotides into broader types based on their biochemical properties to satisfy 2-anonymity88. Another countermeasure based on k-anonymity was proposed89 to thwart recent linkage attacks using signal profiles90 and raw data from functional genomics (for example, RNA sequences)89. Still, given the high dimensionality of genomic data, strategies based on generalization or randomization84 are unlikely to maintain the data at a level of detail that is useful for practical study. Thus, certain legal mechanisms, such as the HIPAA Expert Determination pathway, which we detail later on, tie the notion of de-identification to a re-identification risk assessment based on the capabilities of a reasonable data recipient91. For research, the utility (or usefulness) of genomic data should be maximized when subjecting it to a protection (or transformation) method. As such, Wan et al. demonstrated how to balance the tradeoff between utility and privacy using models based on game theory92,93.
Data aggregation
Although restricting access to data resources, such as the database of genotypes and phenotypes (dbGaP)55, reduces privacy risks, it may also impede research advances. One potential alternative is a semi-trusted registration-based query system94 that processes queries internally and releases only summary results back to the users instead of releasing all individual-level data. For example, Beacon services (for example, the Beacon Network), popularized by the Global Alliance for Genomics and Health (GA4GH), let users query for only one type of information within genomic data sets95, namely the presence of alleles. Although a membership inference attack against Beacon services was demonstrated by Shringarpure and Bustamante96 in 2015 and enhanced later97,98, the effects of this attack can be mitigated by adding noise99,100, imposing query budgets99, adding relatives101 or strategically changing query responses for a subset of genetic variants102.
Data obfuscation
Obfuscating, or adding noise to, summary statistics based on a computational model, such as differential privacy (DP), has been used to counteract membership inference attacks103. However, the role of DP is limited in protecting GWAS and other data sets104,105 because a large amount of noise is required to provide protection27. Even if aggregate statistics are released with significant noise, membership and attribute information can still be inferred106. To preserve privacy, the resulting utility of the DP model is therefore often extremely low61. However, higher data utility could be achieved when assuming a weaker adversarial model107 or combining DP with modern cryptographic frameworks (for example, homomorphic encryption (HE), which we detail later on)108.
Synthetic data generation
Recently, researchers have proposed protecting anonymity by generating synthetic genomic data sets using deep learning models (for example, generative adversarial networks109,110 or restricted Boltzmann machines110). The generated data aim to maintain utility by replicating most of the characteristics of the source data and thus have the potential to become alternatives for many genomic databases that are not publicly available or have accessibility barriers.
Legal implications of data de-identification and use
The question of whether data are considered identifiable or not has important implications for deciding whether the individual to whom they pertain must give consent for their use. It is important to recognize that the laws regarding how genetic and genomic data are handled differ among countries. For illustration, we compare and contrast how regulations in the EU and the United States influence the use of such data.
General Data Protection Regulation
International data privacy legislation is likely to alter the landscape of data privacy protection in genomics research around the world moving forward. The most notable example is the EU’s GDPR, which took effect in 2018 and places restrictions on entities that handle the personal information of citizens of the EU, including genetic information12. The regulations grant data subjects access and deletion rights, impose security and breach notification requirements on entities that handle personal information, and place restrictions on the use and sharing of data without informed consent. Since the GDPR was enacted, there has been heated debate about its impact on the flow of data and hence the conduct of genomics research. Shabani and Marelli, for example, focus on the GDPR’s recognition of the contextual nature of risk, and particularly the risk of re-identification, which they suggest can be ameliorated by compliance with codes of conduct or professional society guidance111. Mitchell et al. suggest that it may be necessary to have more stringent controls as well as to analyse data in place to avoid sharing112. In a subsequent news story, Mitchell also pointed out the complications posed by the emergence of identified ancestry databases113.
The United States has several laws that address the issue of identifiability, some of which have been in place for many years, and which differ in important ways both from each other and from the GDPR113.
United States: HIPAA
One of the most important laws governing patient care and biomedical research is the HIPAA and its Privacy Rule, which is limited in its oversight to data in the possession of three types of covered entities (that is, health-care providers, health plans and health-care clearinghouses) as well as the business associates of such entities114. HIPAA generally requires these entities to obtain patient authorization for uses and disclosures of protected health information outside of treatment, payment, and health-care operations and conveys access rights to individuals115.
However, the protections provided by HIPAA even within ‘covered entities’ contain numerous exceptions116. In particular, HIPAA does not require permission to use or disclose health information, including genomic information, if it has been de-identified through either one of two mechanisms that are colloquially referred to as ‘Safe Harbour’ and ‘Expert Determination’. HIPAA defines de-identified data as follows: “Health information that does not identify an individual and with respect to which there is no reasonable basis to believe that the information can be used to identify an individual is not individually identifiable health information.” The Safe Harbour approach requires the removal of an enumerated list of 18 explicit identifiers (for example, names, social security numbers) and quasi-identifiers (for example, date of birth and 5-digit ZIP code of residence)116 as well as an absence of actual knowledge that the remaining information could be used alone or in combination with other information to identify the individual. By contrast, the alternative Expert Determination pathway requires the application of statistical and/or computational mechanisms to show that the risk of re-identification is very small (a term not explicitly defined by the law)117. Notably, “biometric identifiers, including finger and voice prints”, are listed as one of 18 identifiers, which could lead to the argument that genomic data should be included as well but this issue remains unsettled.
United States: Common Rule
The protections afforded to genomic information shared with researchers depend heavily on the entity carrying out the research and the nature of the information (for example, whether it is shared in identifiable form or is instead converted into de-identified or aggregated data). Human subjects research conducted or funded by agencies within the US Department of Health and Human Services (HHS) and other federal departments is governed by the Federal Policy for the Protection of Human Subjects (that is, the Common Rule), which was initially enacted in 1991 and most recently revised in 2017 (ref.14). Under the Common Rule, such research is subject to oversight by an Institutional Review Board, and investigators must often obtain informed consent before biospecimens and the resulting data can be used for research, thereby enabling the individuals to whom they pertain to have some control over their use. Among many other elements, the regulations require that investigators disclose if they plan to use identifiable information118, to share identifiable data and samples broadly119, to return clinically relevant research results to participants120, or to perform whole-genome sequencing121.
Much research that utilizes genetic data qualifies as minimal risk under the recently revised Common Rule and could therefore be eligible for expedited Institutional Review Board review122 and waiver of consent123. In addition, secondary research involving data that were initially collected for some other clinical or research purpose and has been transformed into a non-identified state (that is, data that have “been stripped of identifiers such that an investigator cannot readily ascertain a human subject’s identity”)124 is currently exempt from Common Rule regulations altogether, especially since a proposal to consider biospecimens and DNA data as identifiable per se was explicitly rejected when the Rule was revised in 2017. Thus, informed consent is not required for such research, a result that is generally much more permissive than the exceptions permitted under HIPAA. However, regulations governing identifiability may change in the future as federal departments and agencies were charged with formally re-examining the definition of ‘identifiable private information’ and ‘identifiable biospecimen’ over time, expecting that emerging technologies, such as whole-genome sequencing, may make genomic data more easily distinguishable.
Other legal issues in the United States
The courts in the United States, especially those at the federal level, have been reluctant to endow individuals with a right to control access to biospecimens or resulting data125–127 or to extend legal protections to discarded DNA22. Moreover, the Genetic Information Nondiscrimination Act of 2008 (GINA)128, which nominally prohibits genetic-based discrimination in the context of health insurance and employment, is limited in its scope, applying only to asymptomatic individuals and offers no protection regarding other types of insurance (for example, life and long-term disability). The Affordable Care Act and the Americans with Disabilities Act fill only some of these gaps6.
State laws
By contrast, over the years, several US state legislatures have enacted laws that convey additional rights or protections to individuals with respect to genetic information about them. For example, some states have deemed genetic information to be the property of the individual being tested and/or require informed consent for genetic testing129. States may also impose security requirements for genetic data or other health records, regulate the retention of biospecimens and data, or convey additional protections to research participants. States, most notably California130,131, Virginia132 and Colorado133, have adopted broad data privacy legislation that provides people with much greater control over some uses of information about them with yet-uncertain implications for genomic information in a variety of settings, including research134. Other states, including Florida135 and New York136, are considering legislation as well. The highly influential Uniform Laws Commission, which proposes statutes for adoption, explicitly defined “genetic sequencing information” as sensitive and thus subject to special protections in its proposed Uniform Personal Data Protect Act approved in July 2021 by the Commission137. These proposed and enacted laws commonly grant more access and correction rights to individuals and impose more restrictions on the use and sharing of personal information without informed consent and thus approach more closely the structure of the GDPR138. Nonetheless, the differences among these statutes themselves and in relation to current federal and international law will doubtless further complicate compliance.
Genomic privacy in context
Context matters
The primary focus of this Review is addressing the complex ethical, legal and technical challenges that arise in protecting privacy in genomic research. Focusing solely on genomic research fails to take into account the potential impact on privacy of the increasing availability of such data in other settings. A wide variety of individuals and entities now collect, use and share genomic data at an unprecedented level. As a result, these data are becoming an increasingly viable resource for parties who might wish to exploit the data, including not only researchers, but also employers, insurers, law enforcement and other individuals33, many of whom have garnered much more media attention than those conducting biomedical investigations. Numerous studies suggest that some people are worried about where genomic data about them go and how they are used, potentially affecting them in ways they neither desire nor expect. In addition to more commonly explored fears of discrimination3, this information can also redefine family relationships, for example, by confirming or disproving paternity, locating previously unknown relatives, or identifying anonymous gamete donors139. These concerns about use and impact, generally couched in terms of desire for genetic privacy, may affect individuals’ willingness to undergo clinical testing or to participate in research3,140. Such reluctance due to privacy concerns, in turn, may exacerbate existing health disparities and stifle scientific progress. Thus, when they design, conduct and discuss their research, investigators need to consider how genomic data are used and how the type of use affects whether or not the data are controlled outside the research setting as well.
Research setting
Technical protections
Researchers often use genomic data accompanied by an array of phenotypic and other information, which they may obtain from individuals directly, through health-care providers or from third parties such as DTC-GT companies. A researcher may also transfer data to third parties for computation or collaboration purposes. Many cryptographic tools can be deployed to protect such use of data from unauthorized access29. Figure 3 illustrates four cryptographic protection approaches with examples in the context of genomic data use cases.
Fig. 3. Cryptographic approaches for privacy protection in the use of genomic data.
a | Homomorphic encryption enables computation by a third party on encrypted data without decrypting any specific record141. In this instance, it is applied to a genome-wide association study142 and a disease susceptibility test78. b | Secure multiparty computation enables multiple parties to jointly compute a function of their inputs without revealing inputs146. Here, three institutions share encrypted data to third parties for summary statistics (for example, minor allele frequency (MAF)) computing145. c | A trusted execution environment, such as Intel Software Guard Extensions (SGX)152, isolates the computation process in an encrypted enclave using central processing unit (CPU) support so that even malicious operating system software cannot see the enclave contents153. Here, an institution computes summary statistics (for example, MAF) in a secure enclave of a third party. d | A blockchain enables encrypted immutable records stored on a decentralized network161. Here, the individual manages the decryption key using a blockchain while sharing encrypted data with researchers32. Avg., average; RAM, random-access memory; SNP, single-nucleotide polymorphism.
Specifically, Fig. 3a illustrates a use case in which an institution that lacks computing capability outsources a computation task (for example, GWAS) to a third party while keeping the data encrypted. Homomorphic encryption (HE) enables computation on encrypted data sets without ever decrypting any specific record and can be utilized when the computation of statistics (for example, counts141, chi-square statistics142 and regression coefficients143) is outsourced to external data centres or public clouds144.
To generate statistically meaningful findings in the research setting, GWAS need many thousands of records that are often distributed among multiple repositories across various institutions, and even across jurisdictions. Secure multiparty computation (SMC), unlike HE, enables multiple parties to jointly compute a function of their inputs without revealing inputs28, as illustrated in Fig. 3b, in which three institutions jointly compute summary statistics (for example, minor allele frequency) over their private data sets. SMC enables the computation of GWAS statistics over distributed encrypted repositories without the local statistics being released145, and it can facilitate quality control and population stratification correction in large-scale GWAS146. SMC can also be applied to sequence matching in other settings147,148. Compared to federated learning, which enables multiple parties to jointly train ML models on genomic data sets over local statistics149, SMC guarantees a much higher security level at the cost of computationally expensive encryption operations. To reduce both the computation overhead and the communication burden, SMC can be combined with HE to support GWAS analyses among a large number (for example, 96) of parties150.
Cryptographic hardware can be leveraged to reduce the burden of computation (for example, secure count queries)151 on encrypted data using HE or SMC152. For example, a trusted execution environment based on Intel Software Guard Extensions (SGX) isolates the computation process in a protected enclave on one’s computer153, as illustrated in Fig. 3c, in which an institution outsources the task of computing summary statistics (for example, minor allele frequency) to a third party. Combining hardware (for example, Intel SGX) and algorithmic tools (for example, HE154 or sketching155 — a data summarization method) can enable users to perform secure GWAS analyses efficiently.
Blockchains can be adopted to incentivize genomic data sharing156 while protecting privacy32,157. For example, researchers have proposed to use blockchains to securely share GWAS data sets158 or parameters of ML models trained on genomic data sets159. Figure 3d illustrates a distributed data sharing system, in which multiple independent parties hold shares of a split decryption key and maintain a blockchain that receives data access requests from researchers and consent from individual participants32. Combined with HE and SMC, blockchains can enable privacy-preserving analysis on genomic data in a personally controlled160 and transparent manner161. However, numerous practical challenges with blockchains remain, including scalability, efficiency and cost157.
Legal protections
Countries around the world have put in place provisions regarding the protection of human research participants, which typically address the need to weigh the risks and benefits to participants, particularly for those who are vulnerable, to assess the scientific merit of protocols, to protect privacy and confidentiality, and to define the role of oversight by research ethics committees and the role of informed consent162. Although the details differ across countries, the most recent version of the Declaration of Helsinki, the foundational document for international research ethics, generally requires consent only for “medical research using identifiable human material or data”163. The Council for International Organizations of Medical Sciences addressed this issue in greater depth in Guideline 11 of its most recent report in 2016 on International Ethical Guidelines for Health-related Research Involving Humans164.
More generally, several international laws influence the ability to access or share genomic data. As noted above, the GDPR provides individuals with substantial control over data about them, typically requiring consent for use and often forbidding the transfer of data to countries whose data protections are not substantially compliant with the GDPR165. Citing several national and individual interests, China heavily regulates when human genomic data can leave the country and requires governmental approval166,167. India168 and many countries in Africa169 have similar practices.
The United States lacks an overarching national data privacy policy and does not typically impose limits on the export of genomic data170. Moreover, the legal protections afforded to genomic information shared with researchers depend heavily on the entity carrying out the research and the nature of the information (for example, whether it is shared in identifiable form or is instead converted into de-identified or aggregated data) as discussed above.
Non-research settings
In recent years, the use of genomic data in non-research settings has garnered an enormous amount of public attention and can have important implications for personal privacy.
Direct-to-consumer setting
Millions of US residents have undergone DTC-GT with companies that purport to provide personal information about a variety of issues, including health, ancestry, family relationships (for example, paternity), and lifestyle and wellness171–173. There are numerous media stories about how consumers use these data to reveal biological relationships, uses that elicit complex responses139, both positive and negative. Some people are pleased to find new relatives or to uncover their biological origins, whereas others are distressed by the results or by unwanted contact. There are, however, virtually no legal constraints on how consumers may use these data, although the legal consequences that may result from their actions could be considerable, including divorce and efforts to avoid support for children19.
The companies offering these services generally fall outside of the purview of the Common Rule and HIPAA (being neither federally funded nor a HIPAA-covered entity, respectively). Instead, the flow of genetic data in the DTC setting is governed largely by self-regulation and notice-and-choice in the form of privacy policies and terms of service172,173. Recent surveys of the privacy policies and terms of service of DTC-GT companies reveal tremendous variability across the industry, with many companies failing to meet best practices and guidelines concerning privacy, secondary uses of genetic information, and sharing of data with third parties172–174.
Although the industry has largely been left to self-regulate, federal agencies have played a limited role in shaping policy with respect to DTC-GT. For example, the US Food and Drug Administration has exercised oversight over a narrow category of DTC health-related tests, although the trend has been to allow these tests to enter the market with little resistance175. The baseline of protection is provided by the Federal Trade Commission, which has the authority to police unfair and deceptive activities across all areas of commerce. Perhaps hindered by its broad mandate and limited resources, the agency to date has only intervened in the DTC-GT space in one case of particularly egregious conduct (that is, unsubstantiated health claims coupled with a lack of security of consumer personal information, including genetic data)176. Instead, the agency has chosen to embrace self-regulation, largely limiting its involvement to the issuance of consumer-facing bulletins177,178 about the implications of genetic testing and broad guidelines for companies offering DTC-GT in the form of a blog post179. For those who are interested, numerous technical strategies exist to permit two users to match genome sequences without disclosing their genomes by using HE180, private set intersection protocols181 or fuzzy encryption182, thereby providing additional privacy protections.
Importantly, millions of people have downloaded their results from DTC-GT and posted them on third-party databases to facilitate finding relatives or to obtain health-related interpretations. These sites are rarely subject to any type of regulation beyond what they specify in their terms of service173. Moreover, these sites reserve the right to change their practices, which may occur as a response to public pressure, but may also be due to changes in business operations. These are the data that facilitate forensic use and are likely to pose the greatest potential for re-identification of genomic data.
Forensic setting
Law enforcement looms large in public opinion about genetic data since it may seek to access genetic information, an issue that has gained intense interest in the wake of high-profile cold cases that were ultimately solved using such information183. Over the years, there has also been an effort to expand government-run forensic databases at the federal, state and local levels184. The FBI currently maintains a nationwide database, the Combined DNA Index System (CODIS), that contains the genetic profiles of over 20 million individuals185 who have been either arrested or convicted of a crime as well as over 1 million forensic profiles derived from crime scenes186.
Law enforcement may also seek to compel the disclosure of genetic information held by an individual or an entity such as a health-care provider, DTC-GT company or researcher. A subpoena is generally all that is required to compel disclosure of genetic information in a patient’s electronic medical record under HIPAA187. Genetic data held by researchers may be shielded by government-issued Certificates of Confidentiality, which purport to assure participants that such data are immune from court orders and outside the reach of law enforcement, but these are issued by default only to research funded by the NIH and other agencies within HHS and may not protect research data that are placed in participants’ electronic health records as well as disclosures required by federal, state and local laws188,189.
Furthermore, law enforcement may also seek to exploit public databases or utilize the services of a DTC-GT company for forensic genealogy purposes in FGG/IGG. To date, law enforcement in the United States has largely focused its efforts on publicly accessible databases (for example, GEDmatch)183 and private databases held by companies that voluntarily cooperate (for example, FamilyTreeDNA)190. For example, law enforcement generated leads in dozens of cold cases by uploading genetic profiles derived from crime scenes to GEDmatch, a public database where individuals can upload their DTC-GT data to learn about where their forebears came from and to locate potential genetic relatives. Similarly, FamilyTreeDNA provides law enforcement access to a version of their Family Finder service, which, like GEDmatch, allows consumers to upload DTC-GT data to locate potential relatives.
In response to public privacy concerns, both GEDmatch and FamilyTreeDNA changed their policies to either require consumers to opt in for their genetic information to be used for law enforcement matching or provide an opportunity to opt out, rather than allowing such searches by default68. This change dramatically reduced the pool of users available to law enforcement, leading them to seek court orders to explore the entire databases of GEDmatch and Ancestry.com, respectively187,191.
In 2019, the US Department of Justice released an interim policy statement designed to signal its intentions regarding privacy and the use of FGG/IGG192. The interim guidelines, which have not been updated since, impose several limitations on federal law enforcement agencies, such as limiting these searches to investigations of serious violent crimes (ill-defined in the guidelines), requirements barring deception on the part of law enforcement when utilizing a DTC service, and requirements that the company seek informed consent from consumers surrounding their cooperation with law enforcement192. At least one local district attorney’s office has developed, and voluntarily adopted, similar guidelines193. Given the recent emergence of these tools, it is perhaps of little surprise that legal regimes are evolving in different ways across the country and around the world194,195.
At the same time, there has been limited research into techniques to mitigate kinship privacy risks196 stemming from the familial genomic searches at the core of FGG/IGG. One general approach is to optimize the choice of SNPs that are masked to minimize the likelihood of successful inference based on relatives’ genomic information196, but little follow-up work has been done on this topic.
Conclusions
As this Review shows, providing appropriate levels of privacy for genomic data will require a combination of technical and societal solutions that consider the context in which the data are applied. Yet, there are challenges to achieving such goals. From a technical perspective, for instance, it is non-trivial to move from privacy-enhancing and security-enhancing technologies that are communicated in a paper or tested in a small pilot study to a full-fledged enterprise-scale solution. This challenge is not unique to genomic data as it is a dilemma for data more generally and for the application domains in which data are applied. In addition, one of the core problems is that it is difficult to build privacy into infrastructure after it has been deployed. Rather, privacy-by-design197, whereby the principles of privacy are articulated at the outset of a project or the point at which data are created and are tailored to the environment to which they are shared, may provide a more systematic and sustainable approach to genomic data protection. However, even if the principles are clearly articulated, there is no guarantee that the technology will support privacy in the long term. For instance, HE, one of the technologies emerging for secure computation over genomic data, is constantly evolving. This may make it difficult to compare genomic data encrypted at one point in time with genomic data created under a more recent version of the technology. Moreover, encryption technologies are not necessarily ideal for long-term management of data198, especially since new computing technologies, such as cheap cloud computing and quantum computing, might make it extremely cheap to crack such encryptions.
Beyond technology, numerous social factors, which inevitably involve tradeoffs between protection and utility, further complicate efforts to protect genomic privacy. Countries, for example, vary dramatically in how much control individuals have over how genomic data about them are used. Some provide individuals granular control while others permit use without consent in many settings, albeit often with stringent security protections. More dramatic is the impact of the growing number of people who post identified genomic data about themselves so that they can find relatives or connect with people who have similar conditions or history. Yet, people who share identified data about themselves increase the potential to re-identify other data about them. In addition, they also reveal information about their relatives, some of whom might have preferred more privacy. These consumer-created databases, unlike medical and research records, frequently have few limitations on use by third parties as has been illustrated by the growth of forensic genealogy. Deciding how to make tradeoffs between protection and use across the entire ecology of genomic data flows requires consideration of both the value of these interests as well as practicable mechanisms of control.
Pressure is growing to protect genomic privacy with security-enhancing technologies and legal regimes for use of genomic data. Nonetheless, it seems clear that simply giving individuals granular control over genomic data that pertain to them, by itself, while attractive to some, risks reifying an unwarranted fear of genomics and is likely to disrupt a wide array of advances in ways that almost surely do not align with the public’s preferences. What may well be needed is a combination of notice and some choice, accountable oversight of uses, and real penalties — both economic and reputational — for inflicting harm on individuals and groups. An additional requirement could be the creation of secure databases for specific purposes (for example, research versus ancestry versus criminal justice) with privacy-protecting tools and individual choice for inclusion that is appropriate for each, which can take the form of law39 as well as private ordering using tools such as data use agreements36. Creating such a complex system will not be elegant and will need to evolve in response to how new laws and privacy-enhancing technologies affect individuals and groups, but simple solutions will not suffice either to protect people and populations from harm or to advance knowledge to improve health.
Acknowledgements
The authors would like to thank their colleagues at the Center for Genetic Privacy and Identity in Community Settings (GetPreCiSe) at Vanderbilt University Medical Center for their constructive feedback. This work was mainly sponsored by GetPreCiSe, a Center for Excellence in Ethical, Legal and Social Implications (ELSI) Research, through a grant from the National Human Genome Research Institute, National Institutes of Health (RM1HG009034). This work was also funded, in part, by the following grants from the National Institutes of Health: R01HG006844 and R01LM009989.
Glossary
- Blockchains
A blockchain is a decentralized digital ledger of records, called blocks, that are linked together using cryptography and are distributed across a peer-to-peer network of computers.
- Short tandem repeats
(STRs). Short tandemly repeated DNA sequences that occur when two or more nucleotides (A, T, C or G) are repeated and the repeated sequences are adjacent to each other.
- Phenome
The complete set of all phenotypes expressed by an organism as a result of genetic variation in populations. A phenotype is an individual’s observable traits such as height, eye colour, blood type, skin colour, hair colour, specific personality characteristics or specific diseases.
- Genome-wide association studies
(GWAS). Observational studies in which genetics research scientists associate specific genetic variations with traits of interest, particularly human diseases. For human disease studies, this method scans the genomes from many different people and looks for genetic markers (for example, single-nucleotide polymorphisms) that occur more frequently in people with a particular disease than in people without the disease.
- Summary statistics
Numbers that give a quick and simple description of a set of records in a data set (for example, mean, median, minimum value, maximum value and standard deviation). A typical example of a summary statistic in a GWAS is a minor allele frequency.
- Forensic or investigative genetic genealogy
(FGG/IGG). A process in which law enforcement seeks to exploit public databases or utilize the services of a direct-to-consumer genetic testing company for forensic purposes.
- Single-nucleotide polymorphism
(SNP). The most common form of DNA variation that occurs when a single nucleotide (A, T, C or G) at a specific position in the genome differs sufficiently (for example, 1% or more) in a species’ population.
- Linkage disequilibrium
Non-random correlations among neighbouring alleles. This occurs due to infrequent recombination events between nearby genomic loci, and hence the alleles are typically co-inherited by the next generation.
- Genotype imputation
A process of estimating missing genotypes from a haplotype or genotype reference panel.
- Differential privacy
(DP). A privacy protection model that publishes summary statistics about a data set while guaranteeing all potential attackers can learn virtually nothing more about an individual than they would learn if that person’s record were absent from the data set.
- Adversarial model
A model that characterizes attackers’ behaviours and incentives with certain assumptions.
- Homomorphic encryption
(HE). A form of encryption that permits computations on encrypted data without revealing the data to any of the parties involved in the cryptographic protocol.
- Secure multiparty computation
(SMC). A form of encryption that enables multiple parties to jointly compute a function of their inputs without revealing inputs.
- Minor allele frequency
The proportion of the second most common of two alleles at a genomic position in a population. An allele corresponds to one of two or more forms of genetic variant at a genetic position. An individual inherits two alleles for each genetic position, one from each parent.
- Population stratification
The systematic difference in allele frequencies between subpopulations of a collection of individuals.
- Private set intersection
A cryptographic technique that allows two parties to compute the intersection of their data without exposing their raw data to the other party.
- Fuzzy encryption
In a fuzzy encryption scheme, the encrypted data can be decrypted by a set of similar keys.
Author contributions
Z.W. and J.W.H. conducted the literature review and drafted the technical and legal parts, respectively. E.W.C. and B.A.M. provided the motivation for this work and designed the organization and structure of the article. E.W.C., Y.V., M.K. and B.A.M. provided detailed edits and critical suggestions on the organization and structure of the article. All authors wrote the manuscript. Z.W. and J.W.H. contributed equally to all aspects of the article. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
1000 Genomes Project: https://www.internationalgenome.org/
All of Us Research Program: https://allofus.nih.gov/
Ancestry.com: https://www.ancestry.com
Beacon Network: https://beacon-network.org/
dbGaP: http://www.ncbi.nlm.nih.gov/gap
eMERGE network: https://emerge-network.org/
FamilyTreeDNA: https://www.familytreedna.com
GEDmatch: http://gedmatch.com
These authors contributed equally: Zhiyu Wan, James W. Hazel.
Change history
3/24/2022
A Correction to this paper has been published: 10.1038/s41576-022-00479-4
References
- 1.Garrison NA. Genomic justice for Native Americans: impact of the Havasupai case on genetic research. Sci. Technol. Hum. Values. 2013;38:201–223. doi: 10.1177/0162243912470009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spector-Bagdady K, et al. “My research is their business, but I’m not their business”: patient and clinician perspectives on commercialization of precision oncology data. Oncologist. 2020;25:620–626. doi: 10.1634/theoncologist.2019-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton EW, Halverson CM, Sathe NA, Malin BA. A systematic literature review of individuals’ perspectives on privacy and genetic information in the United States. PLoS ONE. 2018;13:e0204417. doi: 10.1371/journal.pone.0204417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doe, G. With genetic testing, I gave my parents the gift of divorce. Voxhttps://www.vox.com/2014/9/9/5975653/with-genetic-testing-i-gave-my-parents-the-gift-of-divorce-23andme (2014).
- 5.Copeland, L. The Lost Family: How DNA Testing is Upending Who We Are (Abrams, 2020).
- 6.Clayton EW. Why the Americans With Disabilities Act matters for genetics. JAMA. 2015;313:2225–2226. doi: 10.1001/jama.2015.3419. [DOI] [PubMed] [Google Scholar]
- 7.McKibbin KJ, Malin BA, Clayton EW. Protecting research data of publicly revealing participants. J. Law Biosci. 2021;8:lsab028. doi: 10.1093/jlb/lsab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solove DJ. A taxonomy of privacy. Univ. Pa. Law Rev. 2006;154:477–564. doi: 10.2307/40041279. [DOI] [Google Scholar]
- 9.Niemiec E, Howard HC. Ethical issues in consumer genome sequencing: use of consumers’ samples and data. Appl. Transl. Genom. 2016;8:23–30. doi: 10.1016/j.atg.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obar JA, Oeldorf-Hirsh A. The biggest lie on the Internet: ignoring the privacy policies and terms of service policies of social networking services. Inf. Commun. Soc. 2020;23:128–147. doi: 10.1080/1369118X.2018.1486870. [DOI] [Google Scholar]
- 11.Geier C, Adams RB, Mitchell KM, Holtz B. Informed consent for online research–is anybody reading?: assessing comprehension and individual differences in readings of digital consent forms. J. Empir. Res. Hum. Res. Ethics. 2021;16:154–164. doi: 10.1177/15562646211020160. [DOI] [PubMed] [Google Scholar]
- 12.The European Parliament and The Council Of The European Union. General Data Protection Regulation, Regulation (EU) 2016/679. Official J. Eur. Unionhttps://eur-lex.europa.eu/eli/reg/2016/679/oj (2016).
- 13.Code of Federal Regulations. Title 45, section 164.502: Uses and disclosures of protected health information: general rules (d)(2). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-164/subpart-E/section-164.502#p-164.502(d)(2) (2021).
- 14.Code of Federal Regulations. Title 45, section 164.502: Other requirements relating to uses and disclosures of protected health information (a). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-164/subpart-E/section-164.514#p-164.514(a) (2021).
- 15.Code of Federal Regulations. Title 45, section 164.502: Other requirements relating to uses and disclosures of protected health information (b). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-164/subpart-E/section-164.514#p-164.514(b) (2021).
- 16.Code of Federal Regulations. Title 45, part 46: Protection of human subjects. eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46 (2018). [PubMed]
- 17.Brandeis L, Warren S. The right to privacy. Harv. Law Rev. 1890;4:193–220. doi: 10.2307/1321160. [DOI] [Google Scholar]
- 18.Burke W, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet. Med. 2013;15:854–859. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvik GP, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am. J. Hum. Genet. 2014;94:818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazel JW, et al. Direct-to-consumer genetic testing: prospective users’ attitudes toward information about ancestry and biological relationships. PLoS ONE. 2021;16:e0260340. doi: 10.1371/journal.pone.0260340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garner SA, Kim J. The privacy risks of direct-to-consumer genetic testing: a case study of 23andMe and Ancestry. Wash. Univ. Law Rev. 2019;96:1219. [Google Scholar]
- 22.Clayton EW, Evans BJ, Hazel JW, Rothstein MA. The law of genetic privacy: applications, implications, and limitations. J. Law Biosci. 2019;6:1–36. doi: 10.1093/jlb/lsz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaye J. The tension between data sharing and the protection of privacy in genomics research. Annu. Rev. Genomics Hum. Genet. 2012;13:415–431. doi: 10.1146/annurev-genom-082410-101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoppers BM, Thorogood AM. Ethics and big data in health. Curr. Opin. Syst. Biol. 2017;4:53–57. doi: 10.1016/j.coisb.2017.07.001. [DOI] [Google Scholar]
- 25.Biller-Andorno, N., Capron, A. M. & Elger, B. Ethical Issues in Governing Biobanks: Global Perspectives (Routledge, 2016).
- 26.Malin BA. An evaluation of the current state of genomic data privacy protection technology and a roadmap for the future. J. Am. Med. Inform. Assoc. 2005;12:28–34. doi: 10.1197/jamia.M1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erlich Y, Narayanan A. Routes for breaching and protecting genetic privacy. Nat. Rev. Genet. 2014;15:409–421. doi: 10.1038/nrg3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naveed M, et al. Privacy in the genomic era. ACM Comput. Surv. 2015;48:1–44. doi: 10.1145/2767007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, et al. Genome privacy: challenges, technical approaches to mitigate risk, and ethical considerations in the United States. Ann. NY Acad. Sci. 2017;1387:73–83. doi: 10.1111/nyas.13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arellano AM, Dai W, Wang S, Jiang X, Ohno-Machado L. Privacy policy and technology in biomedical data science. Annu. Rev. Biomed. Data Sci. 2018;1:115–129. doi: 10.1146/annurev-biodatasci-080917-013416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mittos A, Malin B, De Cristofaro E. Systematizing genome privacy research: a privacy-enhancing technologies perspective. Proc. Priv. Enh. Technol. 2019;2019:87–107. [Google Scholar]
- 32.Grishin D, Obbad K, Church GM. Data privacy in the age of personal genomics. Nat. Biotechnol. 2019;37:1115–1117. doi: 10.1038/s41587-019-0271-3. [DOI] [PubMed] [Google Scholar]
- 33.Bonomi L, Huang Y, Ohno-Machado L. Privacy challenges and research opportunities for genomic data sharing. Nat. Genet. 2020;52:646–654. doi: 10.1038/s41588-020-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ram N. Genetic privacy after Carpenter. Va. Law Rev. 2019;105:1357–1425. [Google Scholar]
- 35.Noordyke, M. US state comprehensive privacy law comparison. IAPPhttps://iapp.org/news/a/us-state-comprehensive-privacy-law-comparison/ (2019).
- 36.Hazel JW, Slobogin C. “A world of difference”? Law enforcement, genetic data, and the fourth amendment. Duke Law J. 2020;70:705–774. [Google Scholar]
- 37.Wheeland DG. Final NIH genomic data sharing policy. Fed. Regist. 2014;79:51345–51354. [Google Scholar]
- 38.Rothstein MA. Informed consent for secondary research under the new NIH data sharing policy. J. Law Med. Ethics. 2021;49:489–494. doi: 10.1017/jme.2021.69. [DOI] [PubMed] [Google Scholar]
- 39.Hazel JW, Clayton EW, Malin BA, Slobogin C. Is it time for a universal genetic forensic database? Science. 2018;362:898–900. doi: 10.1126/science.aav5475. [DOI] [PubMed] [Google Scholar]
- 40.Zielinski D, Erlich Y. Genetic privacy in the post-COVID world. Science. 2021;371:566–567. [Google Scholar]
- 41.Shelton JF, et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 42.Malin B, Sweeney L. How (not) to protect genomic data privacy in a distributed network: using trail re-identification to evaluate and design anonymity protection systems. J. Biomed. Inform. 2004;37:179–192. doi: 10.1016/j.jbi.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Kayser M, de Knijff P. Improving human forensics through advances in genetics, genomics and molecular biology. Nat. Rev. Genet. 2011;12:179–192. doi: 10.1038/nrg2952. [DOI] [PubMed] [Google Scholar]
- 44.Lippert C, et al. Identification of individuals by trait prediction using whole-genome sequencing data. Proc. Natl Acad. Sci. USA. 2017;114:10166–10171. doi: 10.1073/pnas.1711125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harmanci A, Gerstein M. Quantification of private information leakage from phenotype-genotype data: linking attacks. Nat. Methods. 2016;13:251–256. doi: 10.1038/nmeth.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Humbert M, Huguenin K, Hugonot J, Ayday E, Hubaux J-P. De-anonymizing genomic databases using phenotypic traits. Proc. Priv. Enh. Technol. 2015;2015:99–114. [Google Scholar]
- 47.Venkatesaramani R, Malin BA, Vorobeychik Y. Re-identification of individuals in genomic datasets using public face images. Sci. Adv. 2021;7:eabg3296. doi: 10.1126/sciadv.abg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sero D, et al. Facial recognition from DNA using face-to-DNA classifiers. Nat. Commun. 2019;10:2557. doi: 10.1038/s41467-019-10617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erlich Y. Major flaws in “Identification of individuals by trait prediction using whole-genome sequencing data”. bioRxiv. 2017 doi: 10.1101/185330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lippert C, et al. No major flaws in “Identification of individuals by trait prediction using whole-genome sequencing data”. bioRxiv. 2017 doi: 10.1101/187542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malin B. Re-identification of familial database records. AMIA Annu. Symp. Proc. 2006;2006:524–528. [PMC free article] [PubMed] [Google Scholar]
- 52.Ball MP, et al. Harvard Personal Genome Project: lessons from participatory public research. Genome Med. 2014;6:10. doi: 10.1186/gm527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sweeney, L., Abu, A. & Winn, J. Identifying participants in the personal genome project by name (a re-identification experiment). Preprint at arXivhttps://arxiv.org/abs/1304.7605 (2013).
- 54.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2013;339:321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 55.Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Homer N, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braun R, Rowe W, Schaefer C, Zhang J, Buetow K. Needles in the haystack: identifying individuals present in pooled genomic data. PLoS Genet. 2009;5:e1000668. doi: 10.1371/journal.pgen.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sankararaman S, Obozinski G, Jordan MI, Halperin E. Genomic privacy and limits of individual detection in a pool. Nat. Genet. 2009;41:965–967. doi: 10.1038/ng.436. [DOI] [PubMed] [Google Scholar]
- 59.Wang R, Li YF, Wang X, Tang H, Zhou X. Learning your identity and disease from research papers: information leaks in genome wide association study. Proc. 16th ACM Conf. Comput. Commun. Secur. 2009;2009:534–544. [Google Scholar]
- 60.Im HK, Gamazon ER, Nicolae DL, Cox NJ. On sharing quantitative trait GWAS results in an era of multiple-omics data and the limits of genomic privacy. Am. J. Hum. Genet. 2012;90:591–598. doi: 10.1016/j.ajhg.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fredrikson M, et al. Privacy in pharmacogenetics: an end-to-end case study of personalized warfarin dosing. Proc. 23rd USENIX Secur. Symp. 2014;2014:17–32. [PMC free article] [PubMed] [Google Scholar]
- 62.Lumley T, Rice K. Potential for revealing individual-level information in genome-wide association studies. JAMA. 2010;303:659–660. doi: 10.1001/jama.2010.120. [DOI] [PubMed] [Google Scholar]
- 63.Humbert M, Ayday E, Hubaux J-P, Telenti A. Addressing the concerns of the Lacks family: quantification of kin genomic privacy. Proc. 2013 ACM Conf. Comput. Commun. Secur. 2013;2013:1141–1152. [Google Scholar]
- 64.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humbert M, Ayday E, Hubaux J-P, Telenti A. Quantifying interdependent risks in genomic privacy. ACM Trans. Priv. Secur. 2017;20:3. doi: 10.1145/3035538. [DOI] [Google Scholar]
- 66.Deznabi I, Mobayen M, Jafari N, Tastan O, Ayday E. An inference attack on genomic data using kinship, complex correlations, and phenotype information. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018;15:1333–1343. doi: 10.1109/TCBB.2017.2709740. [DOI] [PubMed] [Google Scholar]
- 67.Callaway E. Supercharged crime-scene DNA analysis sparks privacy concerns. Nature. 2018;562:315–316. doi: 10.1038/d41586-018-06997-8. [DOI] [PubMed] [Google Scholar]
- 68.Aldhous, P. This genealogy database helped solve dozens of crimes. But its new privacy rules will restrict access by cops. BuzzFeed Newshttps://www.buzzfeednews.com/article/peteraldhous/this-genealogy-database-helped-solve-dozens-of-crimes-but (2019).
- 69.Wood, A. DNA, genealogy led to arrest in series of rapes. Journal Inquirerhttps://web.archive.org/web/20220208235101/https://www.journalinquirer.com/newsletters/dna-genealogy-led-to-arrest-in-series-of-rapes/article_27b25296-ab2d-11ea-8b3e-472861ca42e0.html (2020).
- 70.Zhang, S. How a tiny website became the police’s go-to genealogy database. The Atlantichttps://www.theatlantic.com/science/archive/2018/06/gedmatch-police-genealogy-database/561695/ (2018).
- 71.Murphy, H. Why a data breach at a genealogy site has privacy experts worried. New York Timeshttps://www.nytimes.com/2020/08/01/technology/gedmatch-breach-privacy.html (2020).
- 72.Erlich Y, Shor T, Pe’er I, Carmi S. Identity inference of genomic data using long-range familial searches. Science. 2018;362:690–694. doi: 10.1126/science.aau4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J, Edge MD, Algee-Hewitt BFB, Li JZ, Rosenberg NA. Statistical detection of relatives typed with disjoint forensic and biomedical loci. Cell. 2018;175:848–858. doi: 10.1016/j.cell.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edge MD, Coop G. Attacks on genetic privacy via uploads to genealogical databases. eLife. 2020;9:e51810. doi: 10.7554/eLife.51810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ney P, Ceze L, Kohno T. Genotype extraction and false relative attacks: security risks to third-party genetic genealogy services beyond identity inference. Proc. Netw. Distrib. Syst. Secur. Symp. 2020 doi: 10.14722/ndss.2020.23049. [DOI] [Google Scholar]
- 76.Huang Z, Ayday E, Fellay J, Hubaux J-P, Juels A. Genoguard: protecting genomic data against brute-force attacks. Proc. 2015 IEEE Symp. Secur. Priv. 2015;2015:447–462. doi: 10.1109/SP.2015.34. [DOI] [Google Scholar]
- 77.Huang Z, et al. A privacy-preserving solution for compressed storage and selective retrieval of genomic data. Genome Res. 2016;26:1687–1696. doi: 10.1101/gr.206870.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ayday E, Raisaro JL, Hubaux J-P, Rougemont J. Protecting and evaluating genomic privacy in medical tests and personalized medicine. Proc. 12th ACM Workshop Priv. Electron. Soc. 2013;2013:95–106. doi: 10.1145/2517840.2517843. [DOI] [Google Scholar]
- 79.Naveed M, et al. Controlled functional encryption. Proc. 21st ACM Conf. Comput. Commun. Secur. 2014;2014:1280–1291. [Google Scholar]
- 80.Rodriguez LL, Brooks LD, Greenberg JH, Green ED. The complexities of genomic identifiability. Science. 2013;339:275–276. doi: 10.1126/science.1234593. [DOI] [PubMed] [Google Scholar]
- 81.Zerhouni EA, Nabel EG. Protecting aggregate genomic data. Science. 2008;322:44–44. doi: 10.1126/science.1165490. [DOI] [PubMed] [Google Scholar]
- 82.Craig DW, et al. Assessing and managing risk when sharing aggregate genetic variant data. Nat. Rev. Genet. 2011;12:730–736. doi: 10.1038/nrg3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi X, Wu X. An overview of human genetic privacy. Ann. NY Acad. Sci. 2017;1387:61–72. doi: 10.1111/nyas.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin Z, Owen AB, Altman RB. Genomic research and human subject privacy. Science. 2004;305:183–183. doi: 10.1126/science.1095019. [DOI] [PubMed] [Google Scholar]
- 85.Edge MD, Algee-Hewitt BFB, Pemberton TJ, Li JZ, Rosenberg NA. Linkage disequilibrium matches forensic genetic records to disjoint genomic marker sets. Proc. Natl Acad. Sci. USA. 2017;114:5671–5676. doi: 10.1073/pnas.1619944114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 87.Sweeney L. k-anonymity: a model for protecting privacy. Int. J. Uncertain. Fuzz. 2002;10:557–570. doi: 10.1142/S0218488502001648. [DOI] [Google Scholar]
- 88.Malin BA. Protecting genomic sequence anonymity with generalization lattices. Methods Inf. Med. 2005;44:687–692. doi: 10.1055/s-0038-1634025. [DOI] [PubMed] [Google Scholar]
- 89.Gursoy G, et al. Data sanitization to reduce private information leakage from functional genomics. Cell. 2020;183:905–917. doi: 10.1016/j.cell.2020.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harmanci A, Gerstein M. Analysis of sensitive information leakage in functional genomics signal profiles through genomic deletions. Nat. Commun. 2018;9:2453. doi: 10.1038/s41467-018-04875-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wan Z, et al. A game theoretic framework for analyzing re-identification risk. PLoS ONE. 2015;10:e0120592. doi: 10.1371/journal.pone.0120592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wan Z, et al. Expanding access to large-scale genomic data while promoting privacy: a game theoretic approach. Am. J. Hum. Genet. 2017;100:316–322. doi: 10.1016/j.ajhg.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wan Z, et al. Using game theory to thwart multistage privacy intrusions when sharing data. Sci. Adv. 2021;7:eabe9986. doi: 10.1126/sciadv.abe9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dyke SOM, et al. Registered access: authorizing data access. Eur. J. Hum. Genet. 2018;26:1721–1731. doi: 10.1038/s41431-018-0219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fiume M, et al. Federated discovery and sharing of genomic data using beacons. Nat. Biotechnol. 2019;37:220–224. doi: 10.1038/s41587-019-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shringarpure SS, Bustamante CD. Privacy risks from genomic data-sharing beacons. Am. J. Hum. Genet. 2015;97:631–646. doi: 10.1016/j.ajhg.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.von Thenen N, Ayday E, Cicek AE. Re-identification of individuals in genomic data-sharing beacons via allele inference. Bioinformatics. 2019;35:365–371. doi: 10.1093/bioinformatics/bty643. [DOI] [PubMed] [Google Scholar]
- 98.Ayoz K, Ayday E, Cicek AE. Genome reconstruction attacks against genomic data-sharing beacons. Proc. Priv. Enh. Technol. 2021;2021:28–48. doi: 10.2478/popets-2021-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raisaro JL, et al. Addressing Beacon re-identification attacks: quantification and mitigation of privacy risks. J. Am. Med. Inform. Assoc. 2017;24:799–805. doi: 10.1093/jamia/ocw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cho H, Simmons S, Kim R, Berger B. Privacy-preserving biomedical database queries with optimal privacy-utility trade-offs. Cell Syst. 2020;10:408–416. doi: 10.1016/j.cels.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 101.Ayoz K, Aysen M, Ayday E, Cicek AE. The effect of kinship in re-identification attacks against genomic data sharing beacons. Bioinformatics. 2020;36:i903–i910. doi: 10.1093/bioinformatics/btaa821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wan Z, Vorobeychik Y, Kantarcioglu M, Malin B. Controlling the signal: practical privacy protection of genomic data sharing through Beacon services. BMC Med. Genomics. 2017;10:39. doi: 10.1186/s12920-017-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uhlerop C, Slavkovic A, Fienberg SE. Privacy-preserving data sharing for genome-wide association studies. J. Priv. Confid. 2013;5:137–166. [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson A, Shmatikov V. Privacy-preserving data exploration in genome-wide association studies. Proc. 19th ACM SIGKDD Int. Conf. Knowl. Discov. Data Min. 2013;2013:1079–1087. doi: 10.1145/2487575.2487687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Simmons S, Sahinalp C, Berger B. Enabling privacy-preserving GWASs in heterogeneous human populations. Cell Syst. 2016;3:54–61. doi: 10.1016/j.cels.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Almadhoun N, Ayday E, Ulusoy O. Inference attacks against differentially private query results from genomic datasets including dependent tuples. Bioinformatics. 2020;36:i136–i145. doi: 10.1093/bioinformatics/btaa475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tramèr F, Huang Z, Hubaux J-P, Ayday E. Differential privacy with bounded priors: reconciling utility and privacy in genome-wide association studies. Proc. 22nd ACM Conf. Comput. Commun. Secur. 2015;2015:1286–1297. doi: 10.1145/2810103.2813610. [DOI] [Google Scholar]
- 108.Raisaro JL, et al. Protecting privacy and security of genomic data in i2b2 with homomorphic encryption and differential privacy. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018;15:1413–1426. doi: 10.1109/TCBB.2018.2854782. [DOI] [PubMed] [Google Scholar]
- 109.Bae H, Jung D, Choi H-S, Yoon S. AnomiGAN: generative adversarial networks for anonymizing private medical data. Proc. 25th Pac. Symp. Biocomput. 2019;2020:563–574. [PubMed] [Google Scholar]
- 110.Yelmen B, et al. Creating artificial human genomes using generative neural networks. PLoS Genet. 2021;17:e1009303. doi: 10.1371/journal.pgen.1009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shabani M, Marelli L. Re-identifiability of genomic data and the GDPR: assessing the re-identifiability of genomic data in light of the EU General Data Protection Regulation. EMBO Rep. 2019;20:e48316. doi: 10.15252/embr.201948316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Michell, C., Ordish, J., Johnson, E., Bridgen, T. & Hall, A. The GDPR and Genomic Data–the Impact of the GDPR and DPA 2018 on Genomic Healthcare and Research (PHG Foundation, 2020).
- 113.Petrone, J. Europe’s genomics community wrestling with uncertainty presented by privacy legislation. genomewebhttps://www.genomeweb.com/informatics/europes-genomics-community-wrestling-uncertainty-presented-privacy-legislation (2021).
- 114.Code of Federal Regulations. Title 45, section 160.103: Definitions. eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-160/subpart-A/section-160.103 (2021).
- 115.Code of Federal Regulations. Title 45, section 164.506: Uses and disclosures to carry out treatment, payment, or health care operations. eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-164/subpart-E/section-164.506 (2021).
- 116.Code of Federal Regulations. Title 45, section 164.514: Other requirements relating to uses and disclosures of protected health information (b)(2). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-164/subpart-E/section-164.514#p-164.514(b)(2) (2021).
- 117.Code of Federal Regulations. Title 45, section 164.514: Other requirements relating to uses and disclosures of protected health information (b)(1). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-C/part-164/subpart-E/section-164.514#p-164.514(b)(1) (2021).
- 118.Code of Federal Regulations. Title 45, section 46.116: General requirements for informed consent (b)(9). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46/subpart-A/section-46.116#p-46.116(b)(9) (2021).
- 119.Code of Federal Regulations. Title 45, section 46.116: General requirements for informed consent (d). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46/subpart-A/section-46.116#p-46.116(d) (2021).
- 120.Code of Federal Regulations. Title 45, section 46.116: General requirements for informed consent (c)(8). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46/subpart-A/section-46.116#p-46.116(c)(8) (2021).
- 121.Code of Federal Regulations. Title 45, section 46.116: General requirements for informed consent (c)(9). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46/subpart-A/section-46.116#p-46.116(c)(9) (2021).
- 122.Code of Federal Regulations. Title 45, section 46.110: Expedited review procedures for certain kinds of research involving no more than minimal risk, and for minor changes in approved research (b)(2). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46/subpart-A/section-46.110#p-46.110(b)(2) (2021).
- 123.Code of Federal Regulations. Title 45, section 46.116: General requirements for informed consent (f). eCFRhttps://www.ecfr.gov/current/title-45/subtitle-A/subchapter-A/part-46/subpart-A/section-46.116#p-46.116(f) (2021).
- 124.Guerrini CJ, McGuire AL, Majumder MA. Clearing complexity from the Common Rule NPRM. J. Law Biosci. 2016;3:257–280. doi: 10.1093/jlb/lsw026. [DOI] [Google Scholar]
- 125.California Supreme Court. Moore v. Regents of University of California. Court Listenerhttps://www.courtlistener.com/opinion/2608931/moore-v-regents-of-university-of-california/ (1990).
- 126.US District Court, S.D. Florida, Miami Division. Greenberg v. Miami Children’s Hospital Research Institute. Court Listenerhttps://www.courtlistener.com/opinion/2507167/greenberg-v-miami-childrens-hospital-res-inst-inc/ (2003).
- 127.US District Court, E.D. Missouri, Eastern Division. Washington University v. Catalona. Court Listenerhttps://www.courtlistener.com/opinion/2492481/washington-university-v-catalona/ (2006).
- 128.110th US Congress, The Genetic Information Nondiscrimination Act of 2008, Public Law 110–233. Congress.govhttps://www.congress.gov/bill/110th-congress/house-bill/493/text (2008).
- 129.National Human Genome Research Institute. Genome Statute and Legislation Database. NHGRIhttps://www.genome.gov/about-genomics/policy-issues/Genome-Statute-Legislation-Database (2021).
- 130.California Legislative Information. California Consumer Privacy Act of 2018. California Legislative Informationhttps://leginfo.legislature.ca.gov/faces/billTextClient.xhtml?bill_id=201720180SB1121 (2018).
- 131.Office of the Attorney General. California Privacy Rights Act of 2020, Proposition 24, 1879 (19-0021A1). OAGhttps://oag.ca.gov/system/files/initiatives/pdfs/19-0021A1%20%28Consumer%20Privacy%20-%20Version%203%29_1.pdf (2019).
- 132.Virginia’s Legislative Information System. SB 1392 Consumer Data Protection Act. LIShttps://lis.virginia.gov/cgi-bin/legp604.exe?211+sum+SB1392 (2021).
- 133.Colorado General Assembly. Colorado Privacy Act, SB 21-190, tit. 6, art. 1, part 13. Colorado General Assemblyhttps://leg.colorado.gov/sites/default/files/documents/2021A/bills/2021a_190_rer.pdf (2021).
- 134.Gavejian, J. C., Lazzarotti, J. J. & Atrakchi, M. Virginia passes consumer privacy law; Other states may follow. National Law Reviewhttps://www.natlawreview.com/article/virginia-passes-consumer-privacy-law-other-states-may-follow (2021).
- 135.Brown, G. A. & Lamoureux, C. Florida is the latest state to consider comprehensive data privacy legislation. National Law Reviewhttps://www.natlawreview.com/article/florida-latest-state-to-consider-comprehensive-data-privacy-legislation (2021).
- 136.New York State Senate. New York Privacy Act: Senate Bill S6701. NY Senatehttps://www.nysenate.gov/legislation/bills/2021/S6701 (2021).
- 137.Uniform Law Commission. Uniform Personal Data Protection Act. ULChttps://www.uniformlaws.org/HigherLogic/System/DownloadDocumentFile.ashx?DocumentFileKey=009e3927-eafa-3851-1c02-3a05f5891947 (2021).
- 138.Rippy, S. US state privacy legislation tracker. IAPPhttps://iapp.org/resources/article/us-state-privacy-legislation-tracker (2021).
- 139.Yin Z, Song L, Clayton EW, Malin BA. Health and kinship matter: learning about direct-to-consumer genetic testing user experiences via online discussions. PLoS ONE. 2020;15:e0238644. doi: 10.1371/journal.pone.0238644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Garrison NA, et al. A systematic literature review of individuals’ perspectives on broad consent and data sharing in the United States. Genet. Med. 2016;18:663–671. doi: 10.1038/gim.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kantarcioglu M, Jiang W, Liu Y, Malin B. A cryptographic approach to securely share and query genomic sequences. IEEE Trans. Inf. Technol. Biomed. 2008;12:606–617. doi: 10.1109/TITB.2007.908465. [DOI] [PubMed] [Google Scholar]
- 142.Lauter, K., López-Alt, A. & Naehrig, M. in Progress in Cryptology–LATINCRYPT 2014 (eds Aranha, D. F. & Menezes, A.) 3–27 (Springer, 2015).
- 143.Wang S, et al. HEALER: homomorphic computation of ExAct Logistic rEgRession for secure rare disease variants analysis in GWAS. Bioinformatics. 2016;32:211–218. doi: 10.1093/bioinformatics/btv563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tang H, et al. Protecting genomic data analytics in the cloud: state of the art and opportunities. BMC Med. Genomics. 2016;9:63. doi: 10.1186/s12920-016-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xie W, et al. SecureMA: protecting participant privacy in genetic association meta-analysis. Bioinformatics. 2014;30:3334–3341. doi: 10.1093/bioinformatics/btu561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cho H, Wu DJ, Berger B. Secure genome-wide association analysis using multiparty computation. Nat. Biotechnol. 2018;36:547–551. doi: 10.1038/nbt.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jagadeesh KA, Wu DJ, Birgmeier JA, Boneh D, Bejerano G. Deriving genomic diagnoses without revealing patient genomes. Science. 2017;357:692–695. doi: 10.1126/science.aam9710. [DOI] [PubMed] [Google Scholar]
- 148.Jha S, Kruger L, Shmatikov V. Towards practical privacy for genomic computation. Proc. 2008 IEEE Symp. Secur. Priv. 2008;2008:216–230. doi: 10.1109/SP.2008.34. [DOI] [Google Scholar]
- 149.Wu X, et al. A novel privacy-preserving federated genome-wide association study framework and its application in identifying potential risk variants in ankylosing spondylitis. Brief. Bioinform. 2021;22:bbaa090. doi: 10.1093/bib/bbaa090. [DOI] [PubMed] [Google Scholar]
- 150.Froelicher D, et al. Truly privacy-preserving federated analytics for precision medicine with multiparty homomorphic encryption. Nat. Commun. 2021;12:5910. doi: 10.1038/s41467-021-25972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Canim M, Kantarcioglu M, Malin B. Secure management of biomedical data with cryptographic hardware. IEEE Trans. Inf. Technol. Biomed. 2012;16:166–175. doi: 10.1109/TITB.2011.2171701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Berger B, Cho H. Emerging technologies towards enhancing privacy in genomic data sharing. Genome Biol. 2019;20:128. doi: 10.1186/s13059-019-1741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen F, et al. PRINCESS: Privacy-protecting Rare disease International Network Collaboration via Encryption through Software guard extensionS. Bioinformatics. 2017;33:871–878. doi: 10.1093/bioinformatics/btw758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sadat MN, et al. SAFETY: Secure gwAs in Federated Environment through a hYbrid Solution. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019;16:93–102. doi: 10.1109/TCBB.2018.2829760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kockan C, et al. Sketching algorithms for genomic data analysis and querying in a secure enclave. Nat. Methods. 2020;17:295–301. doi: 10.1038/s41592-020-0761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Roberts JL, Pereira S, McGuire AL. Should you profit from your genome? Nat. Biotechnol. 2017;35:18–20. doi: 10.1038/nbt.3757. [DOI] [PubMed] [Google Scholar]
- 157.Ozercan HI, Ileri AM, Ayday E, Alkan C. Realizing the potential of blockchain technologies in genomics. Genome Res. 2018;28:1255–1263. doi: 10.1101/gr.207464.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y, et al. Enabling privacy-preserving sharing of genomic data for GWASs in decentralized networks. Proc. 12th Int. Conf. Web Search Data Min. 2019;2019:204–212. [Google Scholar]
- 159.Kuo T-T, Gabriel RA, Ohno-Machado L. Fair compute loads enabled by blockchain: sharing models by alternating client and server roles. J. Am. Med. Inform. Assoc. 2019;26:392–403. doi: 10.1093/jamia/ocy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dankar FK. Data privacy through participant empowerment. Nat. Comput. Sci. 2021;1:175–176. doi: 10.1038/s43588-021-00047-6. [DOI] [PubMed] [Google Scholar]
- 161.Grishin D, et al. Citizen-centered, auditable and privacy-preserving population genomics. Nat. Comput. Sci. 2021;1:192–198. doi: 10.1038/s43588-021-00044-9. [DOI] [PubMed] [Google Scholar]
- 162.Office for Human Research Protections, US Department of Health and Human Services. International Compilation of Human Research Standards. HHShttps://www.hhs.gov/ohrp/international/compilation-human-research-standards/index.html (2020).
- 163.World Medical Association. Declaration of Helsinki: medical research involving human subjects. WMAhttps://www.wma.net/what-we-do/medical-ethics/declaration-of-helsinki/ (2013).
- 164.Council for International Organizations of Medical Sciences. International ethical guidelines for health-related research involving humans. CIOMShttps://cioms.ch/publications/product/international-ethical-guidelines-for-health-related-research-involving-humans/ (2016).
- 165.Congressional Research Service. EU data protection rules and US implications. EveryCRSReport.comhttps://www.everycrsreport.com/reports/IF10896.html (2020).
- 166.Ross, L., Zhou, K. & Liu, T. China to strengthen regulation of human genetic resources. WilmerHalehttps://www.wilmerhale.com/en/insights/client-alerts/20200730-china-to-strengthen-regulation-of-human-genetic-resources (2020).
- 167.Chen Y, Song L. China: concurring regulation of cross-border genomic data sharing for statist control and individual protection. Hum. Genet. 2018;137:605–615. doi: 10.1007/s00439-018-1903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Spice Route Legal. Cross-border transfers of genomic data: the Indian framework. Spice Route Legalhttps://spiceroutelegal.com/cross-border-transfers-of-genomic-data-the-indian-framework/ (2020).
- 169.de Vries J, et al. Regulation of genomic and biobanking research in Africa: a content analysis of ethics guidelines, policies and procedures from 22 African countries. BMC Med. Ethics. 2017;18:8. doi: 10.1186/s12910-016-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Needham, K. & Baldwin, C. China’s gene giant harvests data from millions of women. Reutershttps://www.reuters.com/investigates/special-report/health-china-bgi-dna/ (2021).
- 171.Molla, R. Why DNA tests are suddenly unpopular. Voxhttps://www.vox.com/recode/2020/2/13/21129177/consumer-dna-tests-23andme-ancestry-sales-decline (2020).
- 172.Phillips AM. Only a click away — DTC genetics for ancestry, health, love…and more: a view of the business and regulatory landscape. Appl. Transl. Genom. 2016;8:16–22. doi: 10.1016/j.atg.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hazel JW, Slobogin C. Who knows what, and when: a survey of the privacy policies proffered by US direct-to-consumer genetic testing companies. Cornell J. Law Public Policy. 2018;28:35. [PubMed] [Google Scholar]
- 174.Laestadius LI, Rich JR, Auer PL. All your data (effectively) belong to us: data practices among direct-to-consumer genetic testing firms. Genet. Med. 2017;19:513–520. doi: 10.1038/gim.2016.136. [DOI] [PubMed] [Google Scholar]
- 175.US Food and Drug Administration. Direct-to-consumer tests. FDAhttps://www.fda.gov/medical-devices/in-vitro-diagnostics/direct-consumer-tests (2019).
- 176.US Federal Trade Commission. Complaint, In re GeneLink, Inc. & Foru Int’l Corp. FTChttps://www.ftc.gov/system/files/documents/cases/140512genelinkcmpt.pdf (2014).
- 177.Fair, L. DNA test kits: consider the privacy implications. FTChttps://www.consumer.ftc.gov/blog/2017/12/dna-test-kits-consider-privacy-implications (2017).
- 178.US Federal Trade Commission. Direct-to-consumer genetic tests. FTChttps://www.consumer.ftc.gov/articles/0166-direct-consumer-genetic-tests (2018).
- 179.Jillson, E. Selling genetic testing kits? Read on. FTChttps://www.ftc.gov/news-events/blogs/business-blog/2019/03/selling-genetic-testing-kits-read (2019).
- 180.Cheon, J. H., Kim, M. & Lauter, K. in Financial Cryptography and Data Security (eds Böhme, R. & Okamoto, T.) 194–212 (Springer, 2015).
- 181.Baldi P, Baronio R, De Cristofaro E, Gasti P, Tsudik G. Countering gattaca: efficient and secure testing of fully-sequenced human genomes. Proc. 18th ACM Conf. Comput. Commun. Secur. 2011;2011:691–702. [Google Scholar]
- 182.He D, et al. Identifying genetic relatives without compromising privacy. Genome Res. 2014;24:664–672. doi: 10.1101/gr.153346.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Greytak EM, Moore C, Armentrout SL. Genetic genealogy for cold case and active investigations. Forensic Sci. Int. 2019;299:103–113. doi: 10.1016/j.forsciint.2019.03.039. [DOI] [PubMed] [Google Scholar]
- 184.National College of State Legislatures. DNA arrestee laws (NCSL, 2013).
- 185.Federal Bureau of Investigation. The FBI’s Combined DNA Index System (CODIS) hits major milestone. FBIhttps://www.fbi.gov/news/pressrel/press-releases/the-fbis-combined-dna-index-system-codis-hits-major-milestone (2021).
- 186.Federal Bureau of Investigation. CODIS-NDIS statistics. FBIhttps://www.fbi.gov/services/laboratory/biometric-analysis/codis/ndis-statistics (2021).
- 187.Hill, K. & Murphy, H. Your DNA profile is private? A Florida judge just said otherwise. New York Timeshttps://www.nytimes.com/2019/11/05/business/dna-database-search-warrant.html (2019).
- 188.Code of Federal Regulations. Title 42, section 241: Research and investigations generally (d)(1)(E). eCFRhttps://www.govinfo.gov/content/pkg/USCODE-2020-title42/html/USCODE-2020-title42-chap6A-subchapII-partA-sec241.htm (2020).
- 189.Wolf LE, Beskow LM. Certificates of confidentiality: mind the gap. Utah Law Rev. 2021;2021:937–950. doi: 10.26054/0d-80m8-7v82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Haag, M. FamilyTreeDNA admits to sharing genetic data with the FBI. New York Timeshttps://www.nytimes.com/2019/02/04/business/family-tree-dna-fbi.html (2019).
- 191.Aldhous, P. A court tried to force Ancestry.com to open up its DNA database to police. The company said no. BuzzFeed Newshttps://www.buzzfeednews.com/article/peteraldhous/ancestry-dna-database-search-warrant (2020).
- 192.US Department of Justice. Interim policy forensic genetic genealogical DNA analysis and searching. Department of Justicehttps://www.justice.gov/olp/page/file/1204386/download (2019).
- 193.Sacramento County District Attorney’s Office. Memorandum of understanding: investigative genetic genealogy searching. CHIAhttps://chia187.wildapricot.org/page-1841969 (2019).
- 194.Granja R. Long-range familial searches in recreational DNA databases: expansion of affected populations, the participatory turn, and the co-production of biovalue. N. Genet. Soc. 2021;40:331–352. doi: 10.1080/14636778.2020.1853515. [DOI] [Google Scholar]
- 195.Scudder N, Daniel R, Raymond J, Sears A. Operationalising forensic genetic genealogy in an Australian context. Forensic Sci. Int. 2020;316:110543. doi: 10.1016/j.forsciint.2020.110543. [DOI] [PubMed] [Google Scholar]
- 196.Kale G, Ayday E, Tastan O. A utility maximizing and privacy preserving approach for protecting kinship in genomic databases. Bioinformatics. 2018;34:181–189. doi: 10.1093/bioinformatics/btx568. [DOI] [PubMed] [Google Scholar]
- 197.Bednar K, Spiekermann S, Langheinrich M. Engineering privacy by design: are engineers ready to live up to the challenge? Inf. Soc. 2019;35:122–142. doi: 10.1080/01972243.2019.1583296. [DOI] [Google Scholar]
- 198.Oprisanu B, Dessimoz C, De Cristofaro E. How much does GenoGuard really “guard”? An empirical analysis of long-term security for genomic data. Proc. 18th ACM Workshop Priv. Electron. Soc. 2019;2019:93–105. doi: 10.1145/3338498.3358641. [DOI] [Google Scholar]