Abstract
S-Palmitoylation is a reversible post-translational lipid modification that regulates protein trafficking and signaling. The enzymatic depalmitoylation of proteins is inhibited by the beta-lactones Palmostatin M and B, which have been found to target several serine hydrolases. In efforts to better understand the mechanism of action of Palmostatin M, we describe herein the synthesis, chemical proteomic analysis, and functional characterization of analogs of this compound. We identify Palmostatin M analogs that maintain inhibitory activity in N-Ras depalmitoylation assays while displaying complementary reactivity across the serine hydrolase class as measured by activity-based protein profiling. Active Palmostatin M analogs inhibit the recently characterized ABHD17 subfamily of depalmitoylating enzymes, while sparing other candidate depalmitoylases such as LYPLA1 and LYPLA2. These findings improve our understanding of the structure-activity relationship of Palmostatin M and refine the set of serine hydrolase targets relevant to the compound’s effects on N-Ras palmitoylation dynamics.
Keywords: palmitoylation, activity-based protein profiling
The reversibility of the S-palmitoylation of proteins has motivated efforts to discover enzymes such as the serine hydrolases PPT1 (protein palmitoyl thioesterase 1) and LYPLA1 (or APT1, acylprotein thioesterase-1) that display depalmitoylating activity1, 2. As PPT1 is localized to the lysosome3–5, LYPLA1 and its paralog LYPLA26 have been thought to be responsible for much of the protein S-depalmitoylating activity in the cytoplasm of human cells. This hypothesis was initially supported by the discovery of the palmostatins B and M (e.g., Palm B and Palm M). Palm B and Palm M are structurally similar beta-lactones that stabilize the dynamic S-palmitoylation of proteins, such as N-Ras, and also inhibit LYPLA1 and LYPLA2, as well as other serine hydrolases7–10.
As more selective inhibitors were developed for LYPLA1 and LYPLA211, it was found that these compounds do not block N-Ras depalmitoylation in cells10, 12, suggesting that the palmostatins may modulate protein palmitoylation through inhibiting other targets. Consistent with this possibility, activity-based protein profiling (ABPP) experiments revealed that Palm M inhibited several serine hydrolases at pharmacologically relevant concentrations, including a set of closely related and, at that time, uncharacterized enzymes ABHD17A, B, and C (ABHD17s)10. Biochemical and cell biological experiments provided evidence that the ABHD17s exhibit depalmitoylating activity against recombinant N-Ras10, as well as other proteins (e.g., PSD-9513). However, the contribution of ABHD17s to the regulation of endogenous N-Ras palmitoylation and signaling, as well as the broader palmitoylated proteome, remained unclear.
We recently reported the discovery and characterization of ABD957 – a potent, selective, and cell-active covalent inhibitor of the ABHD17s14 – and used this chemical probe to demonstrate that the ABHD17s regulate oncogenic N-Ras palmitoylation and signaling, as well as the growth of NRAS-mutant acute myeloid leukemia (AML) cell lines. However, ABD957 only produced partial stabilization of N-Ras palmitoylation when compared to Palm M. Moreover, Palm M impacted a much larger set of palmitoylated proteins in AML cells than ABD957, reflecting an apparent plasma membrane-delineated activity for ABHD17s. These data, taken together, point to the potential for palmostatins to affect cellular palmitoylation through inhibiting additional enzymes beyond the ABHD17s. Here, we have explored this possibility by synthesizing palmostatin analogs and relating their effects on N-Ras palmitoylation to their serine hydrolase target profiles in AML cells.
We produced a number of palmostatin analogs, preserving the electrophilic beta-lactone while varying the structure of the β-carbon substitution to include modifications of the sulfur oxidation state and replacement of the terminal dimethylamine (Fig. 1a). The compounds were then screened by an established metabolic labeling assay using an alkynylated fatty probe 17-ODYA (17-octadecynoic acid12) for measuring dynamic N-Ras palmitoylation14, as well as broader proteomic palmitoylation. Specifically, we examined a subline of the human NRAS-mutant OCI-AML3 cell line, referred to hereafter as ON cells, where endogenous N-Ras expression was suppressed by RNA interference and a murine green fluorescent protein (GFP)-tagged N-RasG12D fusion protein was expressed14. The rationale underlying this approach was that it allowed us to develop a complementary cell line model (ONK) expressing a different chimeric GFP-N-RasG12D protein in which the C-terminal hypervariable region (HVR) of N-Ras, which contains the palmitoylation site, was replaced by the HVR of K-Ras4b. Such a system enabled assessment of whether the biologic activities of Palm M, ABD957, and other serine hydrolase inhibitors on N-Ras localization signaling were dependent on palmitoylation.
Figure 1. Structures and initial screening of palmostatin analogs.
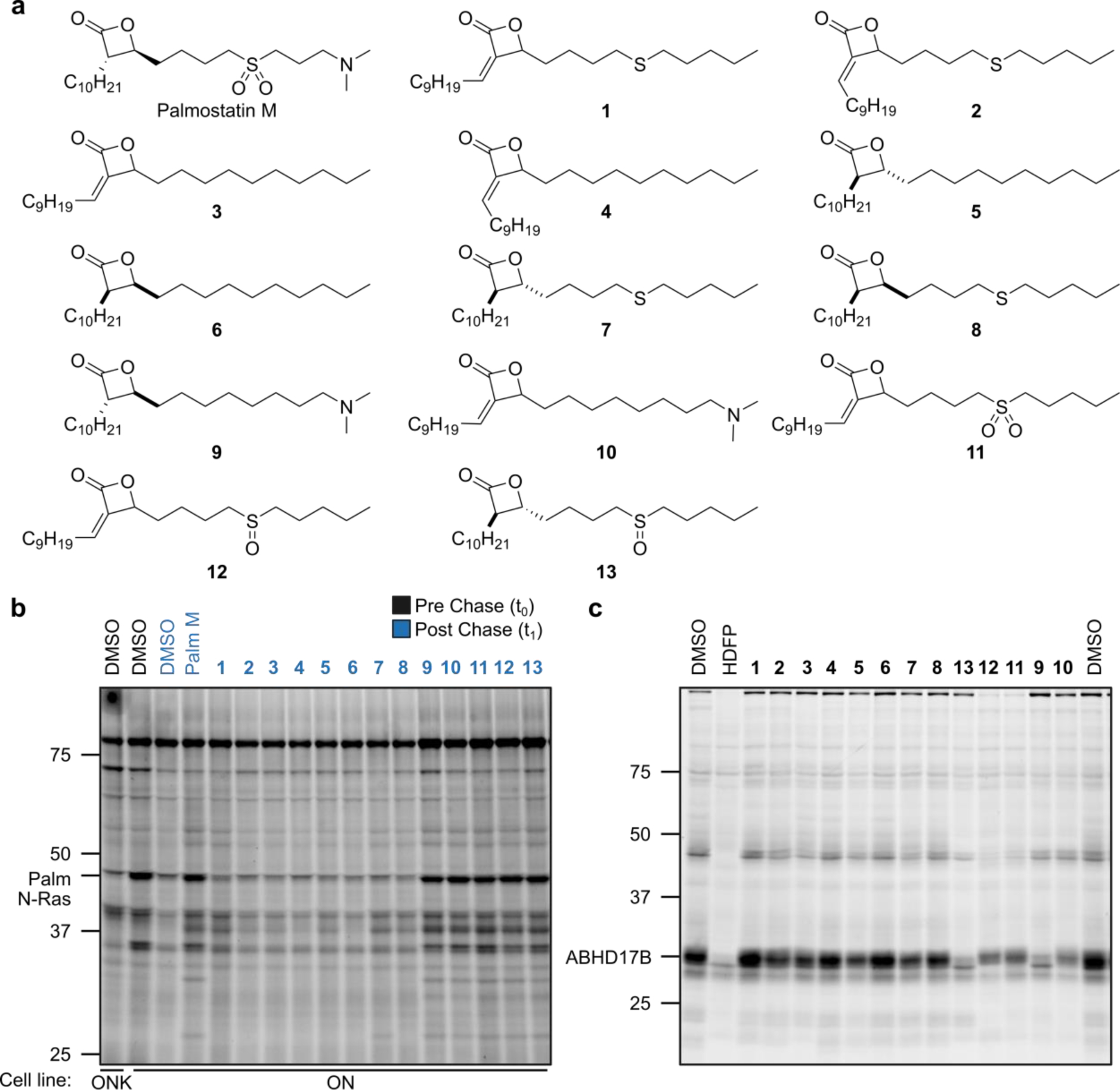
a, Structures of Palmostatin M (Palm M) and analogs. b, Effect of palmostatin analogs on dynamic protein palmitoylation as detected by pulse-chase metabolic labeling of ON and ONK cells with the 17-ODYA probe followed by CuAAC conjugation to a Rh-N3 reporter tag, SDS-PAGE, and in-gel fluorescence scanning. Compounds were screened at 20 μM (Palm M control screened at 10 μM). c, ABHD17 inhibitory activity of palmostatin analogs measured by gel ABPP of HEK293T cells stably expressing ABHD17B. Compounds, including HDFP, were screened at 20 μM and incubated with cells in situ for 4 h prior to lysis, treatment with FP-Rh (1 μM, 1 h), SDS-PAGE, and in-gel fluorescence scanning. The screens in b and c were performed once. Note that the order of compounds 9–13 is differently arranged in b and c.
We first treated ON cells with compounds 1-13 or dimethyl sulfoxide (DMSO) control for 1 h, followed by a 1-h pulse period with 17-ODYA (20 μM) and a 1-h chase period where the cells were re-suspended in growth media lacking 17-ODYA and supplemented with fresh inhibitor or DMSO. Palmitoylated proteins were then conjugated to a rhodamine reporter group (Rh-N3) by copper-catalyzed azide-alkyne cycloaddition (CuAAC)15, 16 and analyzed by SDS-PAGE and in-gel fluorescence scanning (Fig. 1b). A clear SAR emerged, where compounds with β-substitutions that maintained either a terminal dimethylamine (9, 10) or an internal sulfoxide (11) or sulfone (12, 13), blocked the depalmitoylation of N-Ras (~48 kDa protein) and other proteins to similar degree as Palm M (Fig. 1b), while other compounds with purely β-alkyl groups or thioether substitutions exhibited comparatively little or no effect. These findings are consistent with past predictions about the importance of hydrogen bonding and electrostatic interactions emanating from the beta position of palmostatins for productive interactions with enzyme targets9.
In parallel, we screened the compounds for inhibition of ABHD17B using activity-based protein profiling (ABPP)17, 18, where HEK239T cells stably expressing human ABHD17B were treated with compounds (20 μM) or hexadecyl fluorophosphonate (HDFP) as a pan-serine hydrolase inhibitor control, followed by lysis, exposure to an FP-rhodamine probe (FP-Rh19), and analysis by SDS-PAGE and in gel-fluorescence scanning. Compounds 1-8, which were inactive in the N-Ras palmitoylation assay generally showed minimal inhibitory effects against ABHD17B, while compounds 9-13, which blocked N-Ras depalmitoylation, all inhibited ABHD17 to varying extents. These data suggested that ABHD17 inhibition was likely one component required for the effects of palmostatin analogs on dynamic protein palmitoylation. Nonetheless, the similar protein palmitoylation effects of palmostatin analogs that varied in their relative degree of ABHD17B inhibition (e.g., 12 vs. 13) pointed to the potential importance of additional targets of these compounds.
We explored in more detail the activity of 12, alongside an inactive control 1, which only differed from 12 by a single oxygen atom. To visualize the effect of these compounds on N-Ras palmitoylation free of the background of other palmitoylated proteins, we used a previously described14 modification to our pulse-chase assay where an additional anti-GFP immunoprecipitation (IP) step is performed prior to on-bead CuAAC conjugation of Rh-N3 to 17-ODYA-labeled N-Ras. The IP assay results agreed with our global palmitoylation data and showed that 12 completely preserved N-Ras palmitoylation when tested at 20 μM, while 1 was inactive (Fig 2). Neither compound was effective at 2 μM (Fig. 2), and a more complete concentration-dependent analysis readout by either global palmitoylation profiling (Fig. 3a) or GFP-N-Ras immunoprecipitation (Fig. 3b) revealed an IC50 value for blocking N-Ras depalmitoylation of ~3.6 μM for 12 (Fig. 3c).
Figure 2. Palmostatin analog effects on the dynamic palmitoylation of N-Ras.
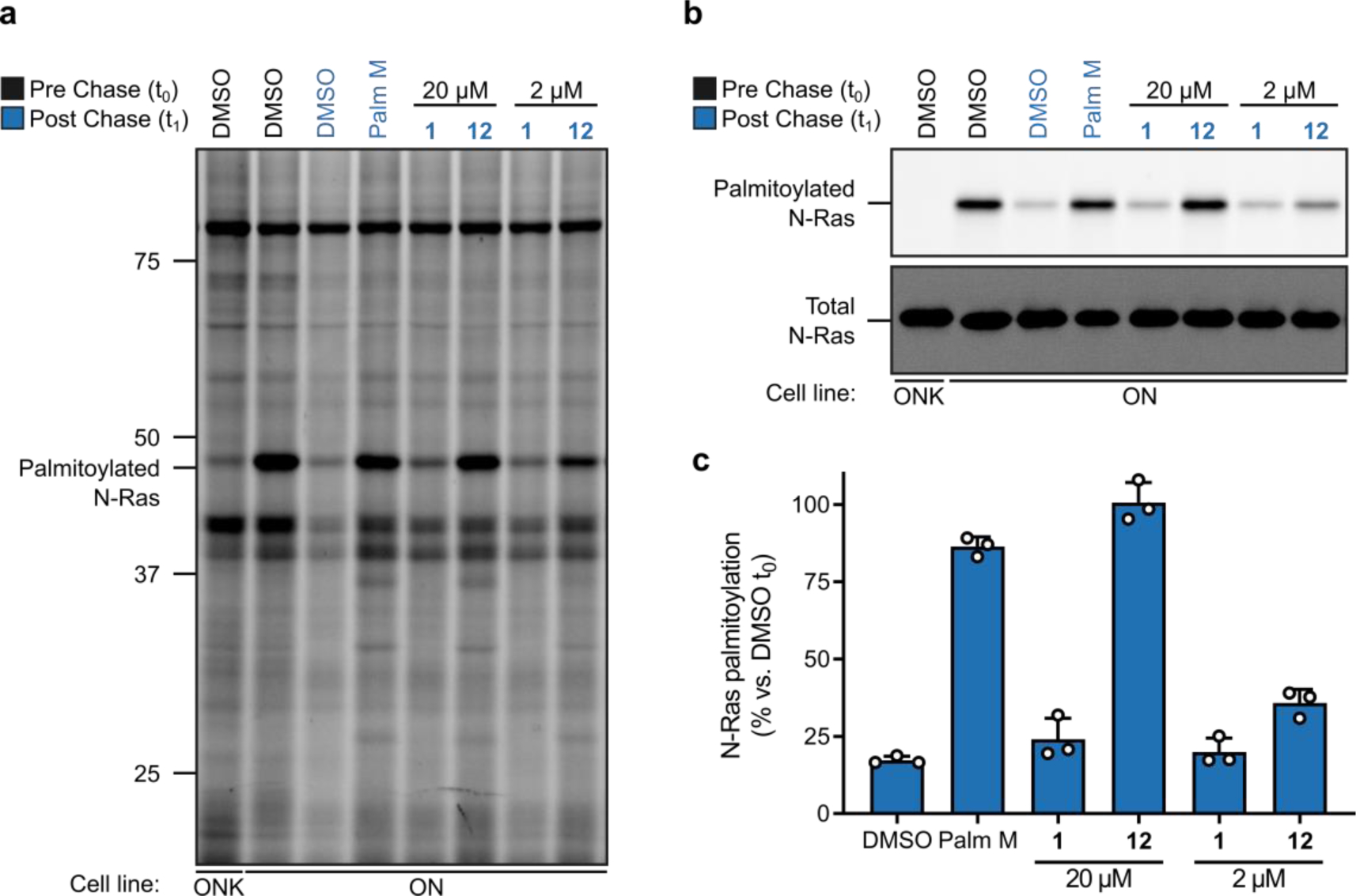
a, b, Effect of active (12) and inactive (1) palmostatin analogs on N-Ras (a, b) and global protein (a) palmitoylation as detected by pulse-chase metabolic labeling of ON cells (or ONK control cells) with the 17-ODYA probe. Data shown are representative of three independent experiments. For b, N-Ras was immunoprecipitated using anti-GFP antibodies prior to visualizing palmitoylation by CuAAC conjugation to a Rh-N3 reporter tag (top panel). Total N-Ras content was measured by Western blotting of GFP enrichments (bottom panel).c, Quantification of palmostatin analog effects on N-Ras palmitoylation; data represent average values ± s.d. from three independent biological replicates, where densitometry was performed on N-Ras signals from anti-GPF immunoprecipitations as shown in b (top panel).
Figure 3. Concentration dependent effects of palmostatin analog 12 on N-Ras and global protein palmitoylation.
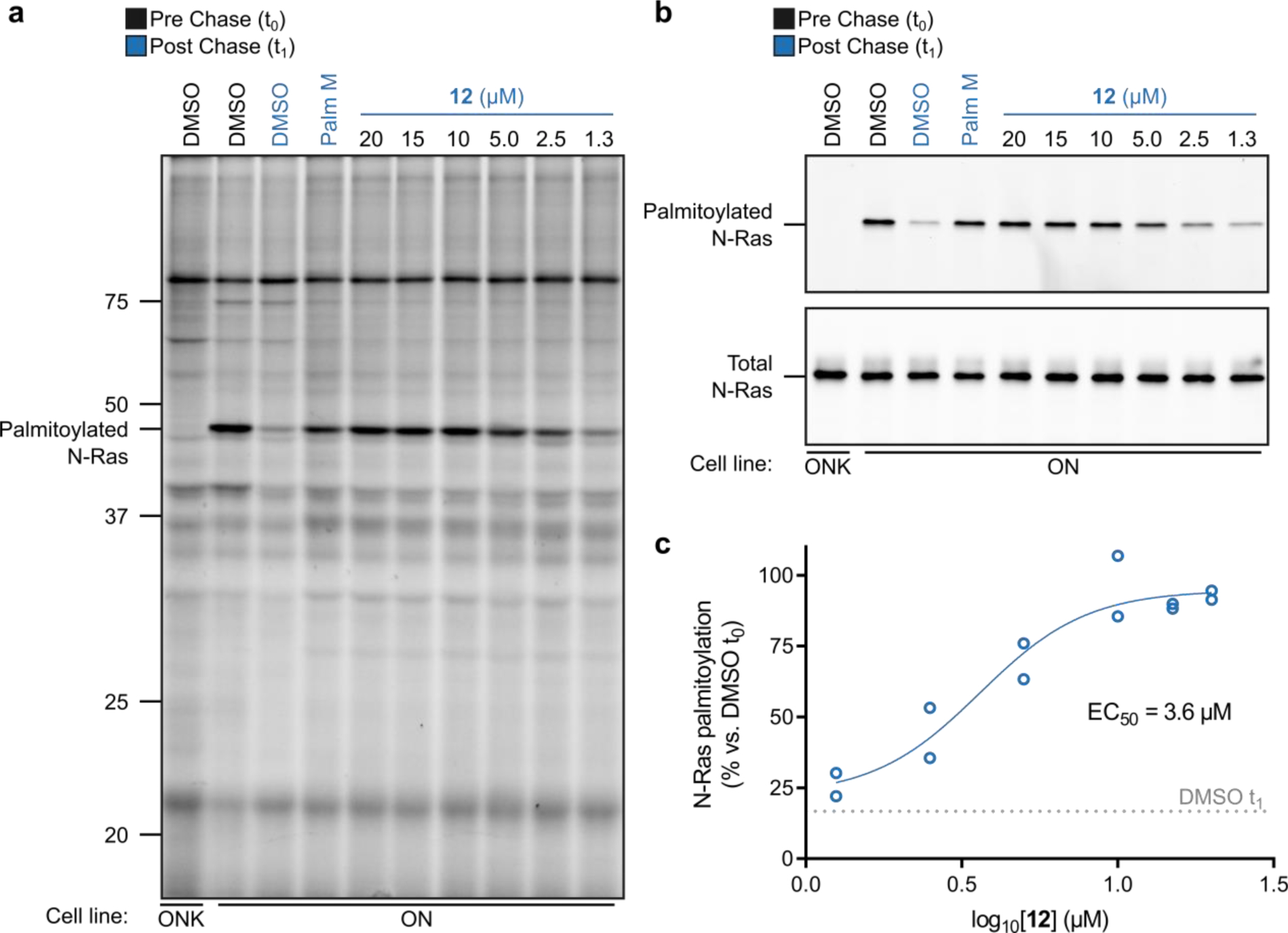
a, Effect of 12 on N-Ras (a, b) and global protein (a) palmitoylation as detected by pulse-chase metabolic labeling of ON cells (or ONK control cells) with the 17-ODYA probe. Data shown are representative of three independent experiments. For b, N-Ras was immunoprecipitated using anti-GFP antibodies prior to visualizing palmitoylation by CuAAC conjugation to a Rh-N3 reporter tag (top panel). Total N-Ras content was measured by Western blotting of GFP enrichments (bottom panel). c, Quantification of concentration-dependent effects of 12 on N-Ras palmitoylation; data represent average values ± s.d. from two independent biological replicates, where densitometry was performed on N-Ras signals from anti-GFP immunoprecipitations as shown in b (top panel).
Considering that the effect of 12 on N-Ras palmitoylation matched that of Palm M and exceeded that of the selective ABHD17 inhibitor ABD95714, we next used a biotinylated FP (FP-biotin17) probe and quantitative mass spectrometry (MS)-ABPP to identify the serine hydrolase targets shared by 12 and Palm M. MS-ABPP of OCI-AML3 cells treated with 12 (20 μM, 2 h, in situ) confirmed substantial engagement (50–75%) of the ABHD17s, along with several other serine hydrolases (> 50% engagement; Fig. 4 and Supp. Table 1). Several of the targets of 12 were also engaged by the inactive control 1 (Fig. 4 and Supp. Table 1). Palm M, which was tested at 10 μM14, substantially engaged most of the targets of 12 or 1 along with several additional enzymes (Fig. 4). Considering the respective effects of each compound in N-Ras depalmitoylation assays, we interpret our ABPP data to designate the subset of serine hydrolases engaged by Palm M and 12, but not 1, as additional candidate depalmitoylating enzymes. Of note, LYPLA1 and LYPLA2 were not among this subset of candidates as they were engaged by inactive compound 1.
Figure 4. Serine hydrolase engagement profiles for palmostatin analogs determined by MS-ABPP using the FP-biotin probe.
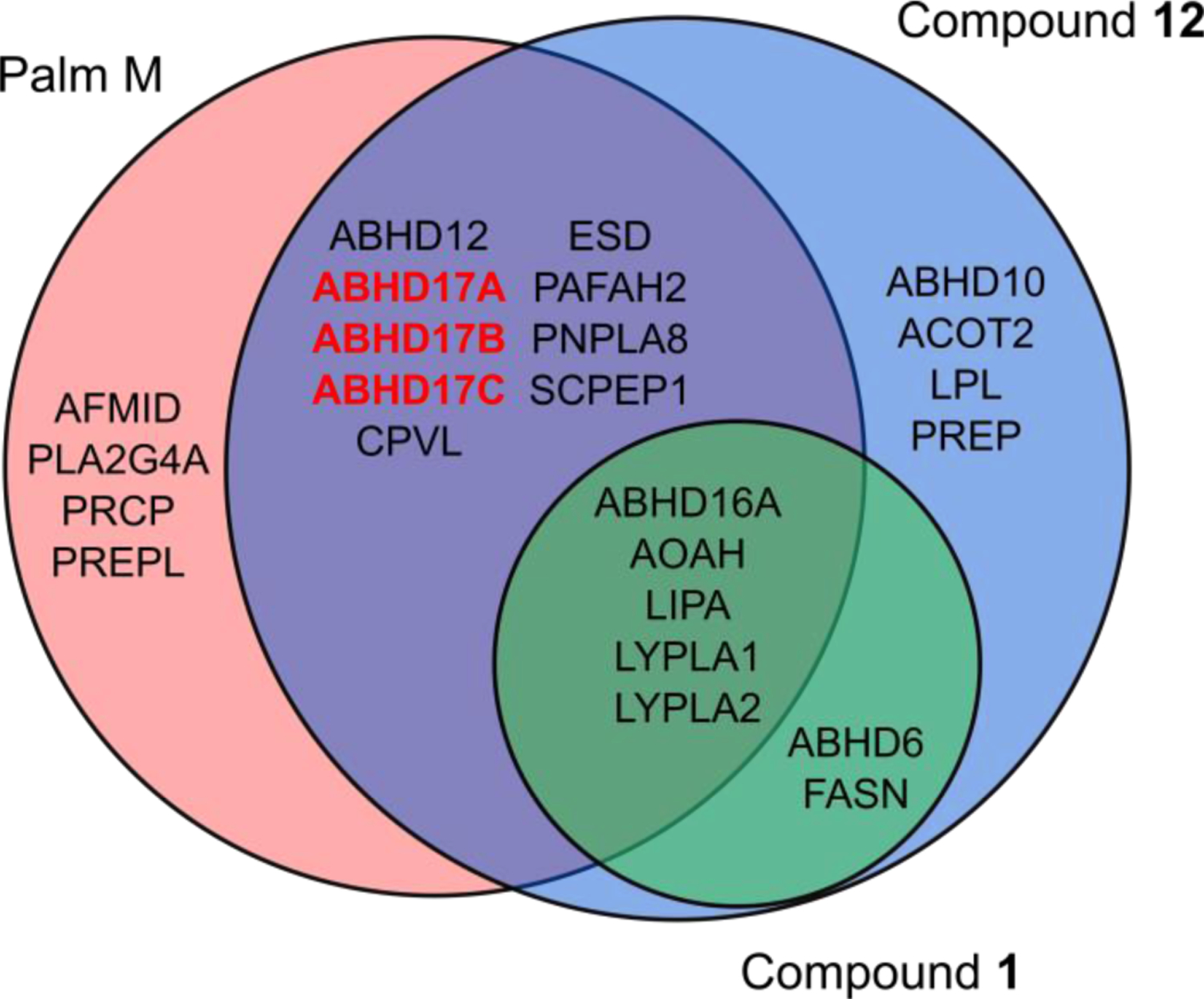
Venn diagram depicting serine hydrolases showing substantial engagement (> 50%) by Palm M (10 μM), 12 (20 μM), and/or 1 (20 μM) in OCI-AML3 cells as determined by MS-ABPP (also see Supp. Table 1). Palm M targets listed from previously published dataset14. MS-ABPP data represent average values from independent experiments (n = 3 for 12, n = 2 for 1).
We finally considered the possibility that enzyme targets outside the serine hydrolase family may contribute to the pharmacological activity of palmostatins. We pursued the discovery of such targets using an alkynylated Palm M probe9 (Palm M-yne, Fig. 5a). We first confirmed that Palm M-yne blocked N-Ras depalmitoylation in ON cells, albeit with slightly lower potency than Palm M (Fig. 5b). We next performed MS-ABPP experiments where we incubated cells with Palm M, HDFP, ABD957, or DMSO, and then exposed lysed cells to Palm M-yne (10 μM, 1 h), followed by CuAAC conjugation with biotin-N3 and processing of samples for quantitative proteomics. In this study, we were principally interested in proteins displaying: i) enrichment by Palm M-yne, and ii) blockade of this enrichment by pre-treatment with Palm M or HDFP, as both compounds exhibit full inhibition of N-Ras depalmitoylation12, 14 (Supp. Table 1). In total, we observed 52 proteins that were enriched by Palm M-yne (> 5-fold compared to DMSO-treated controls) and substantially blocked in their enrichment (> 50%) by pre-treatment with Palm M (Fig. 5c). These proteins included a mix of serine hydrolase- and non-serine hydrolase proteins; however, only serine hydrolase targets of Palm M were also blocked (> 50%) in their enrichment by HDFP (Fig. 5c). Consistent with previous studies14, ABD957 selectively blocked the Palm M-yne enrichment of ABHD17s (Fig. 5c). The chemical proteomics studies with Palm M-yne thus provide further support that the palmostatins likely exert their effects on protein palmitoylation through inhibition of (likely multiple) members of the serine hydrolase class.
Figure 5. Protein engagement profiles for Palmostatin M as determined by MS-ABPP using the Palm M-yne probe.
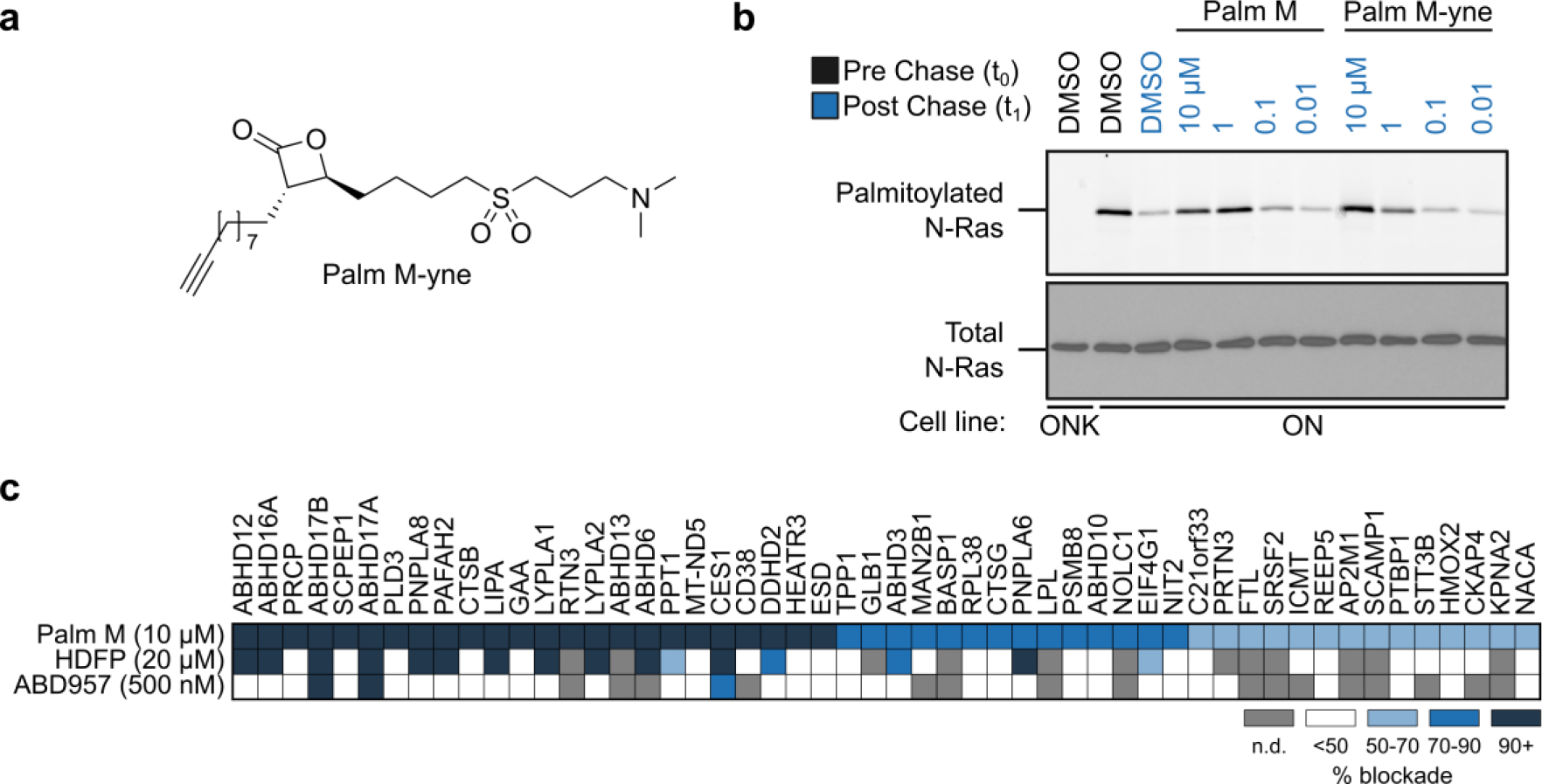
a, Structure of the Palm M-yne probe9. b, Effect of Palm M-yne probe on N-Ras palmitoylation as detected by pulse-chase metabolic labeling of ON cells (or ONK control cells) with the 17-ODYA probe. N-Ras was immunoprecipitated using anti-GFP antibodies prior to visualizing palmitoylation by CuAAC conjugation to a Rh-N3 reporter tag (top panel). Total N-Ras content was measured by Western blotting of GFP enrichments (bottom panel). The concentration-dependent analysis of Palm M and Palm M-yne blockade of N-Ras depalmitoylation was performed once. c, Heatmap summarizing results from competitive MS-ABPP experiments with Palm M (10 μM; n = 3), HDFP (20 μM; n = 2), and ABD957 (500 nM; n = 1) in ON cells treated in situ for 2 h prior to lysis, fractionation, and treatment of the particulate proteome with Palm M-yne (10 μM, 1 h). Values plotted represent percent of blockade of Palm M-yne enrichment by competitor ligands for each protein compared to DMSO control. Heatmap shows only targets that were enriched > 5-fold by the Palm M-yne probe (compared to DMSO control for enrichment; n = 2) and showing > 50% blockade of enrichment by Palm M (compared to DMSO control for competition). For full dataset, see Supp. Table 1. Grey color marks proteins that were not detected or failed to pass quality filters in the indicated experiments. Insufficient numbers of unique peptides were obtained to quantify ABHD17C in this experiment. MS-ABPP data represent average values from independent experiments, as indicated for each condition.
Projecting forward, we could envision multiple ways to test which of the subset of additional serine hydrolase targets of palmostatins might contribute, along with the ABHD17s, to regulation dynamic N-Ras and protein palmitoylation. As selective inhibitors of additional serine hydrolases continue to be developed, these chemical probes could be combined with ABHD17 inhibitors to assess additive effects on the palmitoylated proteome. Alternatively, molecular biology approaches such as genome editing could be used to disrupt candidate palmostatin targets to assess whether loss of individual enzymes amplified the cellular activity of ABHD17 inhibitors. Finally, we cannot exclude the possibility that the palmostatins affect protein palmitoylation through additional mechanisms that do not involve covalent binding to proteins. Regardless, the discovery to date of serine hydrolases that regulate protein palmitoylation in discrete subcellular compartments (e.g., ABHD17s in the plasma membrane10, 14; ABHD10 in the mitochondria20) points to the likely existence of additional members of this enzyme class that contribute in specific ways to controlling dynamic lipid modification states of proteins in human cells.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH (CA193994, CA231991 and CA72614).
References
- 1.Camp LA, Hofmann SL. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J Biol Chem. 1993;268(30): 22566–22574. [PubMed] [Google Scholar]
- 2.Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J Biol Chem. 1998;273(25): 15830–15837. [DOI] [PubMed] [Google Scholar]
- 3.Vesa J, Hellsten E, Verkruyse LA, et al. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 1995;376(6541): 584–587. [DOI] [PubMed] [Google Scholar]
- 4.Verkruyse LA, Hofmann SL. Lysosomal targeting of palmitoyl-protein thioesterase. J Biol Chem. 1996;271(26): 15831–15836. [DOI] [PubMed] [Google Scholar]
- 5.Hellsten E, Vesa J, Olkkonen VM, Jalanko A, Peltonen L. Human palmitoyl protein thioesterase: evidence for lysosomal targeting of the enzyme and disturbed cellular routing in infantile neuronal ceroid lipofuscinosis. EMBO J. 1996;15(19): 5240–5245. [PMC free article] [PubMed] [Google Scholar]
- 6.Toyoda T, Sugimoto H, Yamashita S. Sequence, expression in Escherichia coli, and characterization of lysophospholipase II. Biochim Biophys Acta. 1999;1437(2): 182–193. [DOI] [PubMed] [Google Scholar]
- 7.Dekker FJ, Rocks O, Vartak N, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6(6): 449–456. [DOI] [PubMed] [Google Scholar]
- 8.Hedberg C, Dekker FJ, Rusch M, et al. Development of highly potent inhibitors of the Ras-targeting human acyl protein thioesterases based on substrate similarity design. Angew Chem Int Ed Engl. 2011;50(42): 9832–9837. [DOI] [PubMed] [Google Scholar]
- 9.Rusch M, Zimmermann TJ, Burger M, et al. Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew Chem Int Ed Engl. 2011;50(42): 9838–9842. [DOI] [PubMed] [Google Scholar]
- 10.Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife. 2015;4: e11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adibekian A, Martin BR, Chang JW, et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc. 2012;134(25): 10345–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2011;9(1): 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 Depalmitoylating Enzymes. J Neurosci. 2016;36(24): 6431–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remsberg JR, Suciu RM, Zambetti NA, et al. ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat Chem Biol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41(14): 2596–2599. [DOI] [PubMed] [Google Scholar]
- 16.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67(9): 3057–3064. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96(26): 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77: 383–414. [DOI] [PubMed] [Google Scholar]
- 19.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1(9): 1067–1071. [DOI] [PubMed] [Google Scholar]
- 20.Cao Y, Qiu T, Kathayat RS, et al. ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5. Nat Chem Biol. 2019;15(12): 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


