Abstract
We describe a low-input RNase footprinting approach for the rapid quantification of ribosome-protected fragments with as few as 1000 cultured cells. The assay uses a simplified procedure to selectively capture ribosome footprints based on optimized RNase digestion. It simultaneously maps cytosolic and mitochondrial translation with single-nucleotide resolution. We applied it to reveal selective functions of the elongation factor TUFM in mitochondrial translation, as well as synchronized repression of cytosolic translation after TUFM perturbation. We show the assay is applicable to small amounts of primary tissue samples with low protein synthesis rates, including snap-frozen tissues and immune cells from an individual's blood draw. We showed its feasibility to characterize the personalized immuno-translatome. Our analyses revealed that thousands of genes show lower translation efficiency in monocytes compared with lymphocytes, and identified thousands of translated noncanonical open reading frames (ORFs). Altogether, our RNase footprinting approach opens an avenue to assay transcriptome-wide translation using low-input samples from a wide range of physiological conditions.
RNA translation controls the protein production in a cell and is dynamically regulated during diverse biological processes, such as the stress response, cell differentiation, oncogenesis, and neuronal diseases (Holcik and Sonenberg 2005; Kapur et al. 2017; Costa-Mattioli and Walter 2020; Xu and Ruggero 2020). Quantifying ribosome-protected RNA fragments represents a state-of-art approach to examine the regulation of RNA translation in vivo, and distinguishes protein synthesis from other layers of protein and RNA metabolic processes. Ribosome profiling technology uses RNase to digest RNA fragments not protected by protein complexes, isolates the translating ribosome complexes, and purifies the protected footprints for deep sequencing (Ingolia et al. 2009; Ingolia 2014). This method reveals active translation at single-nucleotide resolution. It has been widely used to study various aspects of protein synthesis, such as the regulation of translation efficiency (Guo et al. 2010; Brar et al. 2012), alternative translation initiation (Ingolia et al. 2011; Lee et al. 2012), ribosome elongation and pausing (Liu et al. 2013; Wu et al. 2019), codon usage (Radhakrishnan et al. 2016), and identification of novel translated open reading frames (ORFs) and micropeptides (fewer than 100 amino acids) encoded in a genome (Ji et al. 2015; Calviello et al. 2016; Chen et al. 2020).
However, current ribosome profiling protocols typically involve complicated experimental procedures to isolate 80S cytosolic ribosome complexes, such as ultracentrifugation through a sucrose cushion/gradient (Ingolia et al. 2009; McGlincy and Ingolia 2017), size-exclusion chromatography (Khajuria et al. 2018; van Heesch et al. 2019), or ribosome immunoprecipitation (Clamer et al. 2018). To study mitochondrial translation, established methods use a sucrose gradient to purify 55S mitoribosomes (Rooijers et al. 2013; Couvillion et al. 2016; Morscher et al. 2018). These complex isolation steps are time-consuming and require many input cells (the protocols typically recommend 1.5 million to 10 million cells) (Ingolia et al. 2012; McGlincy and Ingolia 2017). Additionally, the primary cells from physiological tissues show lower protein synthesis rates than proliferative cultured cells (Liu et al. 2019). These technical barriers have prevented the application of ribosome profiling to many physiological samples with a small number of cells. As a result, most of the current understanding of translational control is based on experiments with cultured cells and is not in the physiological context. To address this technical challenge, here we aimed to develop a low-input RNase footprinting approach to rapidly quantify cytosolic and mitochondrial ribosome footprints simultaneously. And we applied our method to characterize the translation landscape in diverse primary tissue samples.
Results
RNase digestion distinguishes RNA fragments bound by 80S ribosomes versus other RBPs
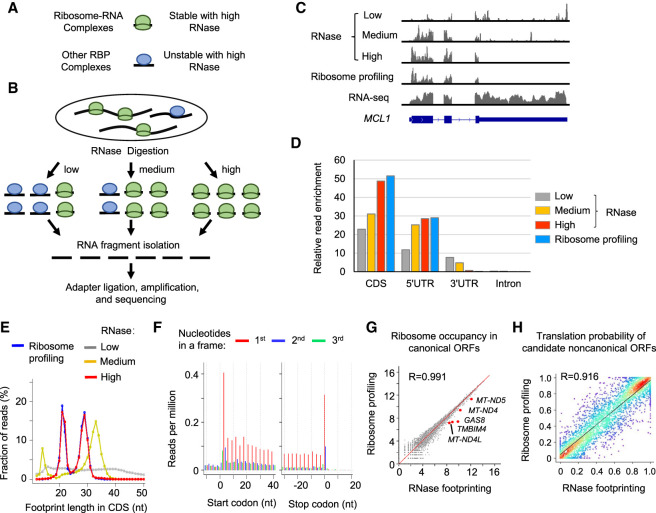
As suggested by previous studies, translating ribosomes (i.e., 80S cytosolic ribosomes and 55S mitoribosomes) have a stronger protection affinity to RNA fragments than other RNA-binding proteins (RBPs) in coding regions. To capture RNA fragments bound by 40S scanning ribosomes or splicing factors, we need to cross-link and stabilize RNA–protein complexes before RNase digestion using formaldehyde (RIP-seq) or UV (CLIP-seq) (Darnell 2010; Archer et al. 2016; Van Nostrand et al. 2017). In contrast, translating ribosomes (i.e., 80S cytosolic ribosomes and mitoribosomes) show stronger protection of RNAs, and the cross-linking step is not needed to capture these footprints (Ingolia et al. 2009; Rooijers et al. 2013; Couvillion et al. 2016). Based on this distinction, we hypothesized that by treating cell lysates with concentrated RNase, we can selectively retain footprints protected by translating ribosomes, while digesting the fragments bound by other RBPs in coding regions (Fig. 1A).
Figure 1.
Different doses of RNase treatment effectively distinguish RNA footprints bound by 80S ribosomes versus other RBPs in coding regions. (A) Schematic illustration of RNA–protein complexes. (B) Experimental steps of isolating RNase footprints for sequencing with three different doses of RNase: low (0.05 U/μg), medium (0.5 U/μg), and high (50 U/μg). (C) Example read distribution across the gene MCL1 with three different doses of RNase. Both RNA-seq and ribosome profiling data are shown for comparison. (D) Relative enrichment of reads in different regions of mRNAs (i.e., coding region [CDS], 5′ UTR, 3′ UTR, and intron). We calculated the fraction of reads in each region and then normalized the value by the relative total length of the region. (E) The distribution of the lengths of RNA fragments in coding regions of mRNAs. (F) RNase footprinting (high-dose) read distribution around the start and stop codons of mRNAs. For footprints showing strong 3-nt periodicity, we adjusted their 5′-end genomic location to the ribosomal A-sites, and plotted the read per million (RPM) values (for details, see Methods). (G) Correlation of read counts in coding regions measured by RNase footprinting (high-dose) and ribosome profiling. The x-axis and y-axis represent log2(read count + 1). We highlighted a few genes showing more than 1.2-fold greater expression levels by RNase footprinting compared with ribosome profiling. (H) The correlation between RibORF-predicted translation probabilities of candidate ORFs using RNase footprinting (high-dose) versus ribosome profiling data in HEK293T cells. We randomly picked 2000 candidate ORFs for the scatter plot colored by density (red color indicates more data points). The Pearson correlation coefficient value is indicated in the plot.
To this end, we treated the cells with three different doses of RNase varying approximately 1000-fold: low (0.05 RNase units/total RNA amount [U/μg]), medium (0.5 U/μg), and high (50 U/μg). Then we isolated the RNA fragments after RNase digestion for sequencing (Fig. 1B,C; Supplemental Fig. S1A). We next compared the read distribution generated by high-dose RNase treatment versus those from lower concentrations as well as published CLIP-seq data for translation factors (Van Nostrand et al. 2020). High-dose RNase treatment resulted in RNA fragments with decreased lengths, stronger enrichment in coding regions of mRNAs, and depletion of those mapped to 3′ UTRs and introns (Fig. 1C–E; Supplemental Fig. S1B). For example, the binding fragments of the cleavage and polyadenylation complexes can be captured with low-dose RNase treatment but were completely digested by concentrated RNase (Supplemental Fig. S1C,D).
With the concentrated RNase treatment, the features of sequencing reads mapped to protein-coding regions of mRNAs reflected those of ribosome footprints, supported by the following evidence. First, the lengths of footprints showed two enriched peaks at 29 nucleotides (nt) and 21 nt, respectively (Fig. 1E). A recent study found that the ∼29-nt footprints represent the ribosome conformational state with occupied A-sites, whereas the ∼21-nt footprints are from the RNase digestion of ribosomes with empty A-sites (Wu et al. 2019). Both states represent actively translating ribosomes. Second, the footprinting reads with the peak sizes (i.e., 20–22 nt and 28–31 nt) showed 3-nt periodicity across coding regions, representing that 80S ribosomes move 3 nt per step to decode RNAs (Fig. 1F; Supplemental Fig. S2A–D; Ingolia et al. 2009). The results are consistent with our hypothesis that 80S ribosome footprints can be selectively retained after concentrated RNase digestion.
RNase footprinting simplifies experimental procedures to profile cytosolic translation and identifies in-frame-translated ORFs
Next, we examined whether RNA fragments from our RNase footprinting can quantitatively measure ribosome occupancy. To this end, we performed detailed comparisons with the ribosome profiling data generated by the conventional protocol, which uses ultracentrifugation through a sucrose cushion to enrich complexes with a higher molecular weight (McGlincy and Ingolia 2017). For each fragment size between 18 nt and 35 nt, we plotted the read distribution around start codons and stop codons of mRNAs (Supplemental Fig. S2D,E). For each fragment size, we observed consistent read distribution patterns between RNase footprinting and ribosome profiling, and the reads are highly enriched in coding regions (Supplemental Fig. S2D–F). Based on these results, we selected sequencing reads with lengths of 18–35 nt in coding regions to quantify ribosome occupancy (McGlincy and Ingolia 2017). The inferred ribosome occupancies using our RNase footprinting data showed a significant correlation with ribosome profiling (R = 0.99) (Fig. 1G; Supplemental Fig. S1E–G), indicating that our RNase footprinting quantitatively measures ribosome binding levels.
A few genes show higher expression (approximately 1.2-fold) from RNase footprinting than from ribosome profiling, such as the mitochondrial transcripts MT-ND5, MT-ND4, and MT-ND4L (Fig. 1G). The data are in line with 55S mitoribosomes having a smaller molecular weight than 80S cytosolic ribosomes. Because RNase footprinting does not perform the centrifugation steps of the conventional protocol, it captures more mitoribosomes. The results are reproducible between biological replicates (R = 0.994) (Supplemental Fig. S1H).
Studies including our work (Michel et al. 2012; Bazzini et al. 2014; Ji et al. 2015; Calviello et al. 2016; Chen et al. 2020) showed ribosome footprints can be used to identify translated noncanonical ORFs based on the continuous 3-nt periodicity read distribution pattern in actively translated regions. Using the ribORF software we developed (Ji 2018b), we selected read fragments showing strong 3-nt periodicity (>50% assigned to first nucleotides of codons in canonical ORFs), adjusted their 5′-end locations to ribosomal A-sites, and used these reads to build a logistic regression classifier to calculate translation probabilities of candidate ORFs. The analyses showed that RNase footprinting reads can accurately distinguish translated canonical ORFs versus internal off-frame ORFs (area under the receiver operating characteristic [ROC] curve = 0.991) (Supplemental Fig. S2G). We trained a model using the RNase footprinting data and another model using ribosome profiling and applied both models to predict the translation probabilities of candidate noncanonical ORFs, including upstream ORFs (uORFs; in 5′ UTRs of mRNAs), downstream ORFs (dORFs; in 3′ UTRs), lncRNA, and pseudogene ORFs. The inferred translation probabilities were well correlated between the two models (R = 0.92) (Fig. 1H). The data further indicated that our RNase footprinting can reliably identify in-frame-translated ORFs.
RNase footprinting identifies nonribosomal footprints in localized noncoding regions
Our above analyses showed consistent footprint distribution in translated regions between RNase footprinting and ribosome profiling. We next compared footprints in other genomic regions and observed major differences in small noncoding RNAs (Supplemental Fig. S3A); 11.5% of our RNase footprinting reads were mapped to small noncoding RNAs, which is 3.7-fold that of ribosome profiling (Supplemental Fig. S3A–C). These reads were found within functional domains of small noncoding RNAs (Supplemental Fig. S3D,E), representing the binding of nonribosomal RNA–protein complexes. The footprints showed highly localized distribution without 3-nt periodicity (Supplemental Fig. S3F,G) and had different lengths compared with ribosome footprints (Supplemental Fig. S3I). For example, the reads in microRNAs were located in mature microRNA regions (Supplemental Fig. S3D) and may represent the binding of RNA-induced silencing complex (RISC). The reads in H/ACA type snoRNAs were mapped to the H-box and ACA-box (Supplemental Fig. S3E). These nonribosomal footprints also exist in conventional ribosome profiling data with a lower amount. The results are consistent with previous reports showing that the ultracentrifugation step during ribosome profiling is not specific to 80S ribosomes but also isolates native stable nonribosomal RNA–protein complexes in abundant structured small noncoding RNAs (Ingolia et al. 2014; Ji et al. 2016). Here, because our RNase footprinting did not perform centrifugation, we retained more of these smaller molecular weight complexes.
Nevertheless, as discussed previously for the ribosome profiling data analyses (Ingolia et al. 2014, 2019; Ji et al. 2015, 2016), the existence of highly localized nonribosomal footprints in noncoding regions does not affect the calculation of ribosome binding in translated ORFs. Based on different distribution patterns of these two types of RNase footprints, we previously developed the percentage of maximum entropy (PME) value to measure the uniformness of read distribution across candidate ORF regions (Ji et al. 2016; Ji 2018a). The PME value effectively distinguishes nonribosomal footprints in small noncoding RNAs versus ribosomal footprints in mRNAs (area under the ROC curve = 0.964) (Supplemental Fig. S3H). The false-positive rate is 0.8% and the true-positive rate is 98.1% when using the cutoff PME < 0.6 to identify nonribosomal footprints.
Low-input RNase footprinting rapidly quantifies RNA translation using 1000–50,000 cultured cells
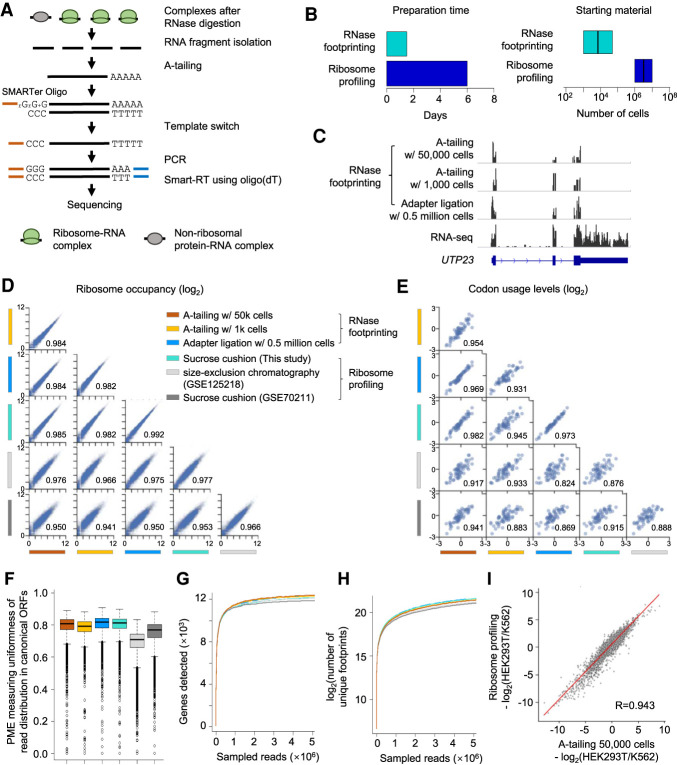
To make the data most comparable, the adapter ligation steps we used during RNase footprinting above were the same as in ribosome profiling (McGlincy and Ingolia 2017). However, these steps tend to lose many input materials. To further reduce the starting material, we made two major modifications to the library generation steps. First, we performed A-tailing of the footprints and reverse-transcription-primed with oligo(dT) in conjunction with template switching using SMARTer oligos (Fig. 2A). A-tailing and the SMARTer oligo-based template switch work well for low-input sequencing library preparation (Picelli et al. 2013; Hornstein et al. 2016). We added four random nucleotides to the 3′-end of SMARTer oligos before the locked nucleic acids GGG, which can increase the sequencing quality. Additionally, we designed 15 random nucleotides in the SMART-RT primer as the unique molecular identifier (UMI) to examine the PCR duplication rate. Second, to enrich ribosome footprints, the conventional protocol performed fragment selection using gel purification after the footprint isolation from the sucrose cushion (McGlincy and Ingolia 2017). In contrast, we performed the size selection of the DNA library after PCR amplification and used computational analyses to select wanted fragment sizes (i.e., 18–35 nt) to calculate ribosome occupancy. Our procedure further reduced required starting materials by skipping intermediate footprint selection steps. Our low-input RNase footprinting method drastically simplified the ribosome profiling procedure and can be performed in ∼1.5 d, the total working time from cell harvesting to final library purification (Fig. 2B; Supplemental Fig. S4).
Figure 2.
Low-input RNase footprinting quantifies ribosome occupancies using 1000 and 50,000 HEK293T cells. (A) The experimental procedure of low-input RNase footprinting. (B) Comparison of preparation time and starting material between RNase footprinting and conventional ribosome profiling. (C) Read distribution representing different RNase footprinting methods across the example gene UTP23. RNA-seq data are shown as the control. (D) Correlation of ribosome occupancies inferred by our RNase footprinting and conventional ribosome profiling data sets. The x-axis and y-axis represent log2(RPM + 1) in coding regions. The Pearson correlation coefficient values are indicated in the plot. (E) Correlation of codon usage levels (log2) at the ribosomal P-sites inferred by our RNase footprinting and conventional ribosome profiling data sets. The Pearson correlation coefficient values are indicated in the plot. (F) The distribution of the percentage of maximum entropy (PME) values, measuring the uniformness of read coverage across codons of canonical ORFs. We included genes with more than 10 reads in the analysis. A PME value closer to one indicates higher uniformness of read distribution. (G,H) We downsampled the sequencing reads and then calculated the number of genes (G) and unique footprints (H) detected by different data sets with a fixed number of reads. (I) The differential ribosome occupancy between HEK293T and K562, comparing our RNase footprinting data to ribosome profiling data. The Pearson correlation coefficient value is indicated in the plot.
Efficient RNase digestion is a key step of our RNase footprinting assay, and we optimized RNase treatment conditions (i.e., RNase concentration and digestion time) for both low and ultralow cell counts (50,000 and 1000 cultured cells, respectively). We determined the RNase amount based on its ratio to total cellular RNA (∼50 U/μg), which is about fivefold higher than used in recent ribosome profiling studies (Wu et al. 2019, 2020). We also found that RNase concentration is critical to achieving efficient digestion, and we added more RNase to adjust the final concentration to 0.5–1 U/μL in the reaction volume. This parameter is especially important for ultra-low-input conditions (e.g., 1000 cultured cells). We optimized other library construction steps including A-tailing, Smart-RT, and PCR conditions (for details, see Methods). We generated RNase footprinting libraries for HEK293T and K562 cells with either 50,000 or 1000 cells (Fig. 2C; Supplemental Fig. S5A). Next, we performed systematic comparisons between our low-input RNase footprinting data versus conventional ribosome profiling generated by this study as well as published work (Iwasaki et al. 2016; Calviello et al. 2020; Martinez et al. 2020).
The following analysis results indicated that our low-input RNase footprinting can accurately examine different aspects of protein synthesis. First, the gene-level ribosome occupancies inferred using 50,000 cells and 1000 cells were significantly correlated with those from the conventional ribosome profiling with millions of input cells (R > 0.97 compared with our data and R > 0.93 compared with published data sets) (Fig. 2D; Supplemental Fig. S5B), although different linker ligation methods used by ribosome profiling data sets can generate variable biases at the 5′- and/or 3′-end of the footprints (Supplemental Fig. S6; Supplemental Table S1; O'Connor et al. 2016). Second, we used the CONCUR software (Frye and Bornelov 2021) to calculate the codon occupancy using the data sets. The overall codon usage levels learned from our low-input RNase footprinting were well correlated with those from the ribosome profiling (R > 0.92 compared with our data and R > 0.82 compared with published data sets) (Fig. 2E; Supplemental Fig. S5C).
Third, based on the combination of the read sequences and UMI, we calculated that the PCR duplication rate in our sequencing library was 5.9% (SD = 2.0) for 50,000 input cells and 9.2% (SD = 1.4) for 1000 cells, which is comparable to high-quality RNA-seq libraries (Fu et al. 2018). Fourth, the reads showed comparable uniformity of distribution across codons (Fig. 2F; Supplemental Fig. S5D), and the downsampling analyses (Hornstein et al. 2016) showed that the same number sequencing reads can detect similar numbers of genes and unique footprints when comparing RNase footprinting with ribosome profiling (Fig. 2G,H; Supplemental Fig. S5E,F). We can quantitatively examine ribosome occupancy for approximately 9000 protein-coding genes (transcript per millions [TPM] value > 3), and approximately 12,000 genes were detectable in total (TPM > 0) (Supplemental Fig. S5G–I). Finally, RNase footprinting reliably inferred differential ribosome binding between HEK293T and K562 cells (R = 0.94 vs. ribosome profiling) (Fig. 2I). Taken together, the data showed that our low-input RNase footprinting is in concordance with established ribosome profiling methods and quantitatively measures cytosolic ribosome binding.
RNase footprinting maps mitochondrial translation
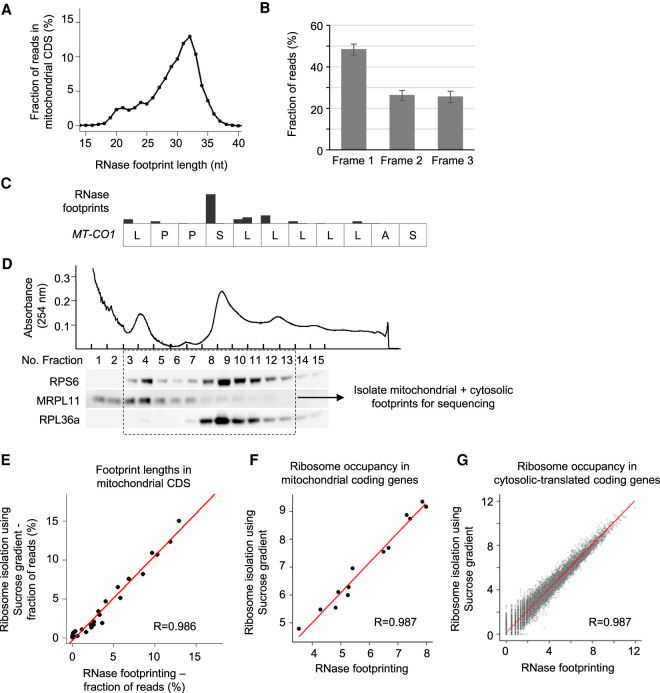
Mitochondria have their own translation machinery to generate the 13 proteins encoded by the mitochondrial genome, which are all subunits of respiratory chain complexes (Pearce et al. 2017). As proteins composing the mitochondrial translation machinery are encoded by the nuclear genome, the cytosolic and mitochondrial translational programs are synchronously regulated during biological processes (Couvillion et al. 2016; Dennerlein et al. 2017; Suhm et al. 2018). Our RNase footprinting reads in mitochondrial protein-coding regions show the features expected of fragments associated with translating ribosomes. The read lengths show a dynamic range between 18 and 35 nt with a peak at 32 nt. Those at the peak size of 32 nt show 3-nt periodicity across the coding regions (Fig. 3A–C). A minor peak at 21 nt suggests that mitoribosome complexes may also have two conformation states, similar to the cytosolic machinery (Fig. 3A).
Figure 3.
RNase footprinting examines mitochondrial translation. (A) The distribution of footprint lengths across mitochondrial protein-coding regions. (B) The fractions of reads in the three codon frames. The 32-nt-long footprints (the peak size) in mitochondrial coding regions were used for the calculation. The SD represents the variation across 13 mitochondria-encoded protein-coding genes. (C) An example transcript region of MT-CO1, showing the 3-nt periodicity of read distribution. (D) We used the sucrose gradient to isolate 55S mitoribosome and 80S ribosome complexes and then extracted associated footprints for sequencing. We performed western blots for ribosomal proteins to show the fractions containing 55S mitoribosomes and 80S ribosomes. (E) The correlation of footprint length distribution in mitochondrial coding regions calculated using our RNase footprinting and sucrose gradient-based ribosome profiling. (F,G) The correlation of ribosome occupancies in mitochondrial-encoded (F) and nuclear-encoded (G) coding genes calculated using the two methods. The RNase footprinting data using 50,000 HEK293T cells were used for the analyses.
To further examine whether our RNase footprinting can quantitatively measure mitoribosome occupancy, we performed a previously described method (Rooijers et al. 2013) that uses a sucrose gradient to isolate 55S mitoribosomes and 80S ribosomes after RNase treatment of 20 million HEK293T cells (Fig. 3D). The resulting footprints in the mitochondrial coding regions showed a consistent length distribution with those from our RNase footprinting (using 50,000 HEK293T cells; R = 0.986) (Fig. 3E). The inferred ribosome occupancies of mitochondrial coding genes (calculated using 18- to 35-nt footprints) showed a significant correlation between the two methods (R = 0.987) (Fig. 3F). A similar correlation was observed for cytosolic coding genes (R = 0.987) (Fig. 3G). These results confirmed that our RNase footprinting provides a simplified method to quantify mitochondrial and cytosolic translation programs simultaneously.
RNase footprinting identifies the selective functions of TUFM in mitochondrial translation
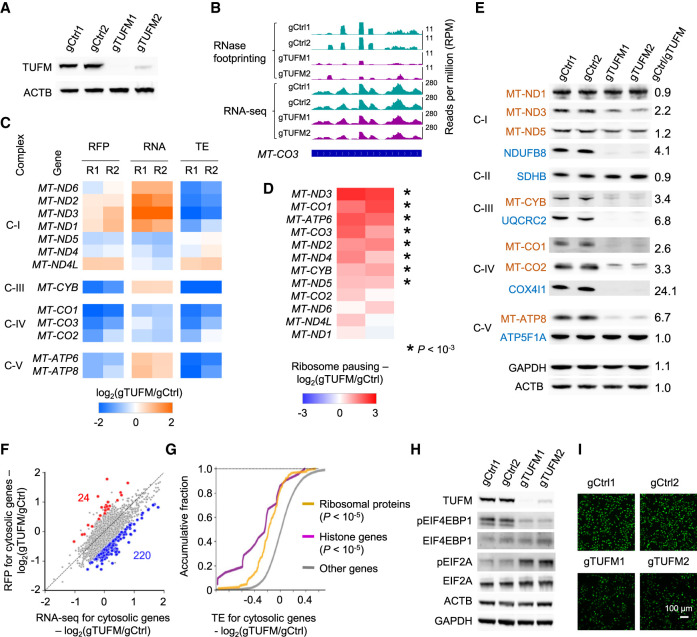
The mitochondrial translation machinery is a macromolecular complex composed of approximately 80 proteins (D'Souza and Minczuk 2018). Currently, the heterogeneity of the complex and specialized functions of individual translation factors remain poorly understood. Using RNase footprinting, we studied the functional roles of the mitochondrial translation elongation factor TUFM in cells. We performed RNase footprinting and RNA-seq in HeLa cells after TUFM knockout (Fig. 4A,B; Supplemental Fig. S7A–C).
Figure 4.
Regulation of mitochondrial and cytosolic RNA translation after TUFM knockout. (A) The western blots showing TUFM knockout. The expression of actin (ACTB) was used as the control. (B) RNase footprinting and RNA-seq read distribution across the MT-CO3 gene in TUFM knockout and control cells. (C) The regulation of ribosome occupancy (ribosome footprints [RFP]), RNA expression (RNA), and translation efficiency (TE) of 13 mitochondria-encoded genes after TUFM knockout. (R1/R2) Replicates 1 and 2. Complexes of the mitochondrial electron transport chain are indicated at left (I, III, IV, and V). (D) The regulation of ribosome pausing of mitochondria-encoded genes after TUFM knockout. The ribosome pausing level was calculated as the ratio of RNase footprinting read density in the first 20% of a transcript versus the remaining. Asterisks indicate significantly increased ribosome pausing after TUFM knockout with a Fisher's exact test (P < 10−3). (E) Western blots showing the expression of mitochondria-encoded (red) and nuclear-encoded (blue) mitochondrial proteins after TUFM knockout. The expression levels of GAPDH and ACTB were used as the control. The ratios of protein expression between the control and TUFM knockout are shown. Complexes I–V of the mitochondrial electron transport chain are indicated at left. (F) Change in mRNA and RFP levels after TUFM knockout. The genes showing differential translation efficiency are highlighted. (Blue) Down-regulated; (red) up-regulated. (G) The cumulative distribution function plot showing the changes in TE of indicated gene sets. The P-values comparing “ribosome proteins” and “histone genes” versus other genes are shown. (H) Western blots showing the expression of translation factors after TUFM knockout. (I) Nascent proteins containing the methionine analog (AHA) were visualized by fluorescence microscopy. An identical setting was used to acquire images, and the percentage of fluorescent cells was 100%. The down-regulation of AHA fluorescence in TUFM knockout cells indicates the inhibition of new protein synthesis.
We calculated the translation efficiency of an mRNA as the ratio of read density in the coding region from RNase footprinting versus that from RNA-seq. The translation efficiencies of the 13 mitochondrial protein-coding genes were repressed at variable levels (from fivefold down-regulation to none) after TUFM knockout (Fig. 4C). Besides occupancy of the transcripts, ribosome elongation and pausing are another important regulatory layer of protein synthesis. Consistent with the fact that TUFM is a mitochondrial translation elongation factor, we found that eight mitochondrial genes showed increased ribosomal pausing (from 1.8-fold to sixfold) at the 5′-end (the first 20%) of transcripts after the gene knockout (Fig. 4D). Ribosome pausing is significantly correlated with the down-regulation of translation efficiency (R = −0.59, P < 0.05) (Supplemental Fig. S7D). Cytosolic-translated genes did not show global increased ribosome pausing, indicating TUFM specifically regulates mitochondrial translation elongation (Supplemental Fig. S7E). The pulse labeling assay with the methionine analog L-azidohomoalanine (AHA) (Dieterich et al. 2007) showed that the nascent protein synthesis of mitochondrial coding genes was generally suppressed at variable levels after TUFM knockout (Supplemental Fig. S7G).
Genes encoding subunits of complexes III, IV, and V of the mitochondrial respiratory chain generally showed decreased translation efficiency and ribosome occupancy, as well as increased ribosome pausing after TUFM knockout (Fig. 4C,D), and their protein expression were drastically down-regulated (Fig. 4E). Heterogeneous regulation was observed for the subunits of complex I. Translation efficiencies of genes such as MT-ND2 and MT-ND6 were decreased after TUFM knockout (Fig. 4C), but ribosome occupancies of the genes were not down-regulated because their RNA expression levels were increased, possibly owing to the compensatory regulation from gene transcription (Fig. 4C). MT-ND1 did not show significant regulation of ribosome occupancy or pausing, and its protein expression was unchanged (Fig. 4C–E). Altogether, these data indicate a selective function of TUFM in regulating mitochondrial translation and show the ability of RNase footprinting to dissect the heterogeneity and redundancy of the translation machinery.
Additionally, we examined the expression of nuclear-encoded respiratory chain subunits, which have been commonly used as markers for complex activity (Baechler et al. 2019; Rudler et al. 2019). For NDUFB8 (complex I), UQCRC2 (complex III), and COX4I1 (complex IV), their protein levels were drastically down-regulated (more than fourfold) (Fig. 4E), but the ribosome occupancies across these transcripts showed modest down-regulation (1.1- to 1.8-fold) (Supplemental Fig. S7F). These data suggest that TUFM knockout translationally suppressed the expression of mitochondria-encoded respiratory chain proteins, causing complex instability and the degradation of nuclear-encoded protein subunits.
RNase footprinting shows the suppression of cytosolic translation upon mitochondrial translation dysfunction
A total of 220 cytosolic mRNAs showed inhibition of translation efficiency greater than 1.5-fold after TUFM knockout (Fig. 4F; Supplemental Fig. S7I,J; Supplemental Table S2). These genes are enriched with ribosomal proteins and histone genes (Fig. 4G; Supplemental Table S3). Only 24 genes showed increased translation efficiency, and nine of them had high ribosome occupancy in their uORFs in 5′ UTRs, such as ATF4 and PPP1R16A (Supplemental Table S2). Gene Ontology analyses showed that the transcriptionally up-regulated genes were enriched in pathways such as “response to extracellular stimulus,” “immune response,” and “regulation of apoptosis” (P < 10−3) (Supplemental Table S3). Western blots showed that the translation initiation factor EIF2A is more phosphorylated and EIF4EBP1 shows decreased phosphorylation after TUFM knockout (Fig. 4H). Using pulse-AHA labeling, we showed that the global nascent protein synthesis rate was decreased by 2.3-fold (Fig. 4I; Supplemental Fig. S7H). Altogether, the data indicate that cytosolic translation activity is inhibited in response to cellular stress induced by mitochondrial dysfunction after TUFM knockout, and the regulated genes and pathways are similar to those mediated by other stress responses (Andreev et al. 2015; Gameiro and Struhl 2018). These results showed that our RNase footprinting data can be used to dissect the synchronized regulation between mitochondrial and cytosolic translation programs.
RNase footprinting reveals base-resolution translation using low amounts of snap-frozen tissues
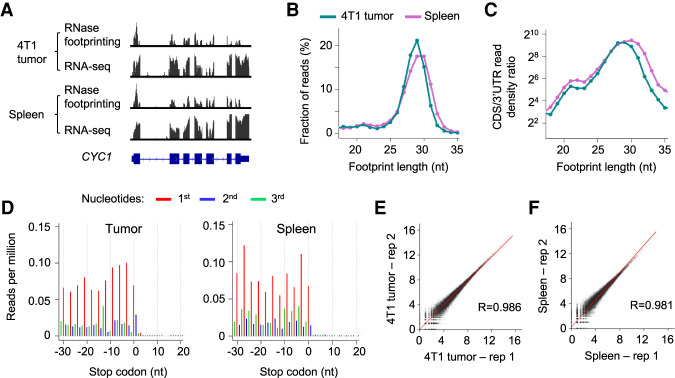
Poorly proliferative primary tissues show lower translational activity than rapidly dividing cultured cells, and this makes it a challenge to quantify the small number of actively translating ribosome complexes in cells. We applied our RNase footprinting approach to assay RNA translation in snap-frozen tumor and spleen samples (with only 1 mg input material) from the mouse 4T1 breast cancer model (Fig. 5A). We calibrated RNase digestion and library preparation conditions based on the cellular total RNA amount (for details, see Methods). The lengths of the ribosome footprints showed enrichment around 29 nt and a minor peak at 21 nt (Fig. 5B). The reads (18–35 nt) were highly enriched in coding regions (Fig. 5C), and those with peak size (∼29 nt) showed continuous 3-nt periodicity across ORFs, indicating active translation (Fig. 5D). The calculated ribosome occupancy levels were highly consistent between replicates (R > 0.98) (Fig. 5E,F). The data indicate that our RNase footprinting can quantify ribosome footprints using snap-frozen tissue samples.
Figure 5.
RNase footprinting maps RNA translation landscape using snap-frozen tumor and spleen tissues. (A) RNase footprinting and RNA-seq read distribution across the example gene CYC1. (B) The distribution of footprint lengths in coding regions of mRNAs. (C) The log2 ratio of read density in coding regions versus 3′ UTRs for each fragment size (18–35 nt). (D) Adjusted read distribution around the stop codons of mRNAs using RNase footprinting data of the 4T1 tumor and spleen tissues. We adjusted 5′-end genomic locations of the 29-nt footprints to the ribosomal A-sites. (E,F) Correlation of ribosome occupancy levels between two replicates of 4T1 tumor (E) and spleen tissues (F). The x-axis and y-axis represent log2(read count + 1) in coding regions.
RNase footprinting reveals the regulation of RNA translation among primary immune cells
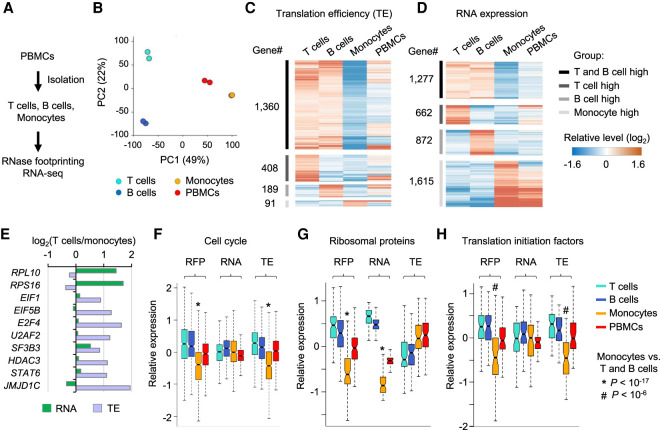
RNA translation is highly regulated during blood development and disorders (Signer et al. 2014; Khajuria et al. 2018). However, it has been a challenge to apply ribosome profiling to clinical samples, because primary peripheral blood mononuclear cells (PBMCs) showed a 30-fold lower protein synthesis rate than did cultured K562 cancer cells (Supplemental Fig. S8A). Using a standard blood draw, we performed RNase footprinting and RNA-seq for mixed PBMCs and purified T cells, B cells, and monocytes (Fig. 6A; Supplemental Fig. S8B–E). Footprinting profiles of these immune cells effectively captured the marker gene expression and classified the cell types (Fig. 6B; Supplemental Fig. S8F–H). Two thousand forty-eight genes showed more than 1.5-fold differential translation efficiency comparing T cells, B cells, and monocytes (Fig. 6C; Supplemental Fig. S9A–C; Supplemental Table S4). These translationally regulated genes showed little overlap with those regulated by gene transcription as identified by RNA-seq (Fig. 6D,E; Supplemental Fig. S9D–G; Supplemental Table S4). The data indicate that RNA translation and gene transcription are two complementary regulatory layers of immune cell activities. Transcriptionally regulated genes are enriched in the genes associated with cell type–specific functions. Genes uniquely activated in T cells and B cells are enriched in the pathways “T-cell activation” and “B-cell activation,” respectively (Supplemental Table S5). Monocyte-specific genes are enriched in the pathway of “inflammatory response” (Supplemental Table S5). Most translationally regulated genes (1360 genes, 66% of total) show consistently higher translation efficiency in both T cells and B cells compared with monocytes (Fig. 6C), representing lymphocyte-specific regulation via translation. These genes are enriched in pathways such as “cell cycle,” “RNA processing,” “regulation of transcription,” and “chromatin organization” (Fig. 6F; Supplemental Fig. S9H–J; Supplemental Table S5).
Figure 6.
Regulation of RNA translation across human primary immune cell types. (A) Experimental steps to isolate T cells, B cells, and monocytes from PBMCs and then perform RNase footprinting and RNA-seq. (B) The principal component analysis (PCA) of ribosome occupancies of T cells, B cells, monocytes, and PBMCs. Genes showing more than twofold regulation across cells were used in the analyses. The percentages of variance explained by PC1 and PC2 are shown in parentheses. (C,D) The heatmap showing genes with regulation of translation efficiency (C) and RNA expression (D) comparing T cells, B cells, and monocytes. (E) Example genes showing differential RNA expression and translation efficiency, comparing T cells versus monocytes. (F–H) Boxplots showing the relative levels of ribosome occupancy (RFP), RNA expression, and translation efficiency (TE) for indicated genesets comparing T cells, B cells, monocytes, and PBMCs: “cell cycle” (F), “ribosomal proteins” (G), and “translation initiation factors” (H). The * or # label indicates the Wilcoxon rank-sum test P-value showing the geneset has significantly lower expression in monocytes, compared with both T cells and B cells.
Ribosomal proteins showed significantly lower RNA expression in monocytes compared to lymphocytes (P < 10−17, Wilcoxon rank-sum test) (Fig. 6G). This is a possible mechanistic explanation for the decreased translational activity observed in monocytes. Translation initiation factors were mildly regulated at the transcriptional level but showed significantly lower translation efficiency in monocytes than lymphocytes (P < 10−8, Wilcoxon rank-sum test) (Fig. 6H). The suppression of initiation factors at the translational level may further decrease the translation activity in monocytes. Altogether, these results show the feasibility and significance of profiling the translatomic landscape of human primary tissue samples, which can lead to novel mechanistic insights into biological systems.
RNase footprinting examines translated noncanonical ORFs in individual's immune cells
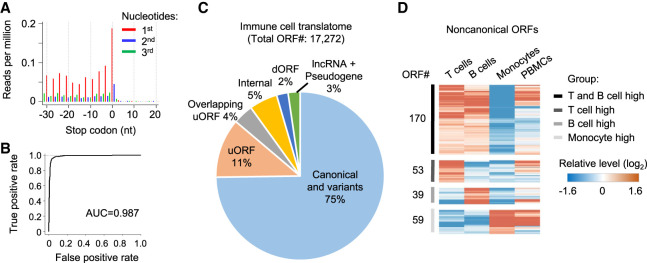
Next, we used the RNase footprinting data to study the immune translatome and examine the regulation of noncanonical ORFs. Using the RibORF software (Ji et al. 2015; Ji 2018b), we selected read fragments that show strong 3-nt periodicity (Fig. 7A) to train a logistic regression model to predict the translation probability of a candidate ORF. Footprinting data from immune cells can accurately distinguish in-frame-translated ORFs versus internal off-frame ones (area under the ROC curve = 0.987) (Fig. 7B). In total, we identified 2639 translated uORFs in 5′ UTRs, 381 dORFs, 265 lncRNA ORFs, and 134 pseudogene ORFs (Fig. 7C; Supplemental Table S6). Three hundred twenty-one of these noncanonical ORFs showed a greater than 1.5-fold differential ribosome occupancy among the individual's primary immune cell types (Fig. 7D; Supplemental Table S7). Our data showed that personalized translatomics data from RNase footprinting can be used to characterize the regulation of noncanonical ORFs.
Figure 7.
Regulation of noncanonical ORFs among primary immune cells. (A) Adjusted RNase footprinting read distribution around the stop codons of mRNAs. For footprints showing strong 3-nt periodicity, we adjusted their 5′-end genomic location to the ribosomal A-sites and plotted the RPM values (for details, see Methods). (B) The ROC curve measuring the RibORF performance in classifying in-frame-translated ORFs versus internal off-frame ORFs using RNase footprinting data in immune cells. The area under the ROC curve (AUC) value is shown. (C) The RibORF-identified genome-wide translated ORFs in immune cells, which were grouped based on their genomic locations. (uORF) ORF in 5′ UTR, (dORF) ORF in 3′ UTR, and (internal) ORF within canonical ORFs. (D) The heatmap showing lncRNA ORFs and uORFs with dynamic regulation of ribosome occupancy comparing T cells, B cells, and monocytes.
Discussion
The original ribosome profiling protocol used the sucrose gradient to isolate 80S ribosome complexes and associated footprints to study RNA translation (Ingolia et al. 2009). Later on, to simplify the experimental procedure, ultracentrifugation through a sucrose cushion or size-exclusion chromatography was used to enrich complexes with higher molecular weights (McGlincy and Ingolia 2017). Here, by leveraging that translating ribosomes (80S ribosomes and 55S mitoribosomes) show stronger protection efficiency than other RBPs, we optimized the digestion conditions using concentrated RNase and effectively retained ribosome-protected fragments while digesting other RNA fragments in coding regions. The simplified procedure to select translating ribosome-protected footprints allowed for rapid ribosome profiling.
In combination with A-tailing and SMARTer oligo-based template switching to add sequencing adapters, our RNase footprinting reveals base-resolution active translation using as few as 1000 cultured cells, which is about 1000-fold fewer starting materials than those of conventional protocols. We showed that our RNase footprinting data can quantitatively measure different aspects of protein synthesis. The calculated gene-level ribosome occupancy, codon usage level, and ORF translation probability are well correlated with conventional ribosome profiling data sets generated by sucrose cushion, sucrose gradient, and size-exclusion chromatography methods. The template switch step (SMART-RT) used by our protocol tends to amplify footprints with the nucleotide G at the 5′-end (Supplemental Fig. S6A; Supplemental Table S1; Wulf et al. 2019). This bias needs to be taken into account when interpreting ribosome occupancy in individual genomic sites. We calibrated our protocol for a variety of primary tissue samples with low protein synthesis rates and showed it worked robustly for snap-frozen tissues and primary immune cells. Our method does not require any specialized equipment. The working hour of our RNase footprinting protocol is about 1.5 workdays (comparable with regular RNA-seq) and is <25% of that needed for conventional ribosome profiling.
We performed quality control of our RNase footprinting data using a set of analyses. The footprint lengths show peaks in ∼29 nt and ∼21 nt (minor in some cells), representing optimized RNase digestion (Martinez et al. 2020). The reads around the peak sizes show 3-nt periodicity across canonical ORFs. Additionally, the reads between 18 and 35 nt in length show high enrichment in coding regions compared with 3′ UTRs. The original ribosome profiling protocol only captured the ∼29-nt footprints and did not examine the ∼21-nt ones. Only recently, several studies highlighted the fraction of ribosome footprints with ∼21 nt (Lareau et al. 2014; Wu et al. 2019). Consistent with the previous study (Wu et al. 2019), the ratio of 29-nt versus 21-nt reads appears to be variable across different cells and tissues. It will be interesting to dissect the biological roles of dynamic regulation of these two ribosome conformational states. During the data analyses, we used 18- to 35-nt reads to calculate ribosome occupancy, but when we calculated codon usage levels and identified actively translated noncanonical ORFs, we only used the reads showing strong 3-nt periodicity (typically those around peak sizes) because these analyses require accurate correction of read location to the ribosomal A-site or P-site.
The RNase footprinting assay was inspired by our previous in-depth analyses of ribosome profiling data (Ji et al. 2015, 2016). We and others previously found that the ultracentrifugation step through a sucrose cushion used by conventional ribosome profiling is not specific for translating 80S ribosomes, and the transcriptomic RNase footprinting nature of the assay also isolates nonribosomal RNA–protein complexes in noncoding RNAs/regions (Ingolia et al. 2014; Ji et al. 2016). Based on the ultracentrifuge parameters, we calculated that RNA fragments in more than 7–10S protein complexes can be isolated for sequencing (Ji et al. 2016). These findings motivated us to skip the ultracentrifuge step. Different from the previously published footprinting method (Silverman et al. 2014), we did not perform the RNA–protein cross-linking steps and treated the cell lysates with highly concentrated RNase. As a result, only the most stable RNA–protein complexes were retained after RNase digestion. The nonribosomal footprints show highly localized distribution without 3-nt periodicity and have different lengths compared with ribosomal ones. Based on these read distribution patterns, we can computationally distinguish the two types of footprints (Ingolia et al. 2014, 2019; Ji et al. 2016). As discussed previously (Ji et al. 2016), the characterization of nonribosomal footprints could provide biological insights into corresponding noncoding regions. Additionally, similar to conventional ribosome profiling, ∼78% of our sequencing reads are fragments from ribosomal RNAs (rRNAs). And ∼10% of reads are mapped to annotated mRNAs and lncRNAs. We recommend a sequencing depth of more than 50 million for each sample. Future study is needed to develop effective methods to deplete unwanted rRNA fragments to increase the fraction of reads usable to study RNA translation. For low-input RNase footprinting conditions, the biotinylated oligonucleotide pulldown approach can decrease RNA amount and cause the PCR overamplification issue. However, the CRISPR-based depletion of unwanted fragments in the DNA library could be a promising approach (Hardigan et al. 2019).
Because of the unbiased assay of transcriptomic RNase footprints, our footprinting data allow the simultaneous quantification of cytosolic and mitochondrial translation. Current assays used to study mitochondrial translation involve complicated procedures using sucrose gradient to purify 55S mitoribosome complexes (Rooijers et al. 2013; Couvillion et al. 2016; Morscher et al. 2018). Our RNase footprinting provides a simplified method to tackle mitochondrial translation and obtains comparable results (i.e., ribosome occupancy and footprint lengths) versus the conventional method. Using the assay, we revealed that TUFM selectively regulates the translation efficiency and ribosome elongation of the 13 mitochondrial-encoded protein-coding genes, and showed that cytosolic translation is synchronously repressed after TUFM knockout. Similar approaches can be used to define the functional roles of other regulators and dissect the cross talk between mitochondrial translation and cytosolic regulation during biological processes.
Our RNase footprinting technique opens up an avenue to examine transcriptomic translation using low-input primary cells from physiological contexts for both basic scientific research and clinical personalized prognosis. For the proof-of-concept experiment, we show that our assay reveals base-resolution active translation in snap-frozen mouse tissues (i.e., tumor and spleen) and primary immune cells (i.e., T cells, B cells, and monocytes) from an individual's standard blood draw. Many genes showed lower translation activity in monocytes than lymphocytes. This is likely owing to the lower RNA expression of ribosomal proteins in monocytes. We analyzed the RNA expression profiles of monocytes and lymphocytes using published RNA-seq data sets and found that this differential regulation appears to be a general phenomenon across human individuals (Supplemental Fig. S9K). Further studies examining RNA translation in more immune cell subtypes and progenitor cells will provide mechanistic insights into translational control during hematopoiesis.
RNA translation is tightly regulated during development, aging, and the progression of human disease (e.g., cancer and neuronal disorders) (Kondrashov et al. 2011; Tahmasebi et al. 2018). Cells in various tissue compartments are under differential stress conditions, potentially impacting RNA translation. In combination with other rare-cell isolation techniques, such as fluorescence-activated cell sorting (FACS) or laser-capture microdissection, our RNase footprinting method can be used to examine RNA translation of individual cell types from heterogeneous tissue samples. The results will refine our understanding of the translational control underlying physiological and pathophysiological conditions. We envision that our RNase footprinting method can work for single-cell ribosome profiling with the optimization of RNase digestion and library preparation procedures. Recently, two single-cell protocols were developed to examine RNA translation. Ribo-STAMP uses the fusion protein of translation factor-APOBEC1 (a cytidine deaminase enzyme) to identify RNA targets bound by ribosomes at the single-cell level (Brannan et al. 2021). Although this method can examine the expression of actively translated RNAs, it cannot reveal which ORFs are translated, and the transfection of the fusion protein is hardly applicable to primary tissue samples. Separately, a single-cell method based on MNase digestion was developed to obtain ∼35-nt ribosome footprints for sequencing without the ultracentrifuge step to isolate the ribosome complex (Reid et al. 2015; VanInsberghe et al. 2021). However, because RNase digestion has been most commonly used for ribosome profiling and the biological regulation of RNase footprints has been well characterized, the further development of our RNase footprinting method toward the single-cell level would be valuable to understand the heterogeneous translational response across cell types.
Recent studies showed that micropeptides encoded by noncanonical ORFs can serve as novel cancer biomarkers/targets (Prensner et al. 2021) and can be used to design novel neoantigens for immunotherapy (Laumont et al. 2018; Chong et al. 2020; Ruiz Cuevas et al. 2021; Ouspenskaia et al. 2022). A potentially exciting application of RNase footprinting is to define the comprehensive translatome and noncanonical ORFs using patient samples, which can lead to novel clinical disease diagnosis and personalized treatment strategies.
Methods
Low-input RNase footprinting
The protocol has two major steps.
Step 1. Cell harvesting, RNase treatment, and footprint purification
For cultured adherent cells, cells were seeded in one well of a multiwell plate the day before harvest and were grown to 70%–80% confluency. Cells were supplemented with 100 μg/mL cycloheximide and incubated for 3 min at 37°C. To harvest cells, we quickly washed the cells twice with cold PBS containing 100 μg/mL cycloheximide. Lysis buffer was immediately added to each well (20 mM Tris at pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/mL cycloheximide, 1% v/v Triton X-100). The cells were left to lyse for 10 min on ice. Cell lysates were collected in a 1.5-mL microcentrifuge tube, followed by 10 times trituration through a 26-gauge needle. For suspension-cultured cells and primary immune cells, cells were pelleted in a 1.5-mL microcentrifuge tube and were washed twice with cold PBS. Cells were immediately lysed in the lysis buffer for 10 min on ice, followed by 10 times trituration through a 26-gauge needle. For primary tissue samples, powdered tissue was collected in a 1.5-mL microcentrifuge tube and was lysed in the lysis buffer. Tissue lysate was incubated for 10 min on ice, followed by 10 times trituration through a 26-gauge needle. After centrifugation at 1000g for 5 min, the supernatant was collected in a new 1.5-mL microcentrifuge tube.
The RNase treatment condition was calibrated based on the total RNA amount in the cell lysate. RNA concentration in the lysate was measured by Qubit (Invitrogen). RNase digestion conditions were then determined by the following two steps. First, we calculated the required RNase amount with the following equation: the amount of RNase (U) = 50 × total RNA amount (μg). Second, we found that the RNase concentration in the reaction volume is also critical to achieving efficient digestion, and we added more RNase to adjust the final RNase concentration to 0.5–1 U/μL. The adjustment is especially important for ultralow cell counts (e.g., 1000 cultured cells). RNase with an even higher concentration can cause overdigestion of ribosome footprints.
Fifty thousand cultured cells (we generally obtained ∼1 μg of total RNA from cultured HEK293T, K562, and HeLa cells) and 1 mg primary tissues (∼1 μg of total RNA) were lysed in 90 μL buffer and digested with 50 U RNase 1. One thousand cultured cells (10–20 ng of total RNA) and immune cells (50–100 ng of total RNA was obtained from our PBMC, T cells, B cells, and monocytes) were lysed in 60 μL buffer and digested with 60 U RNase I. Our RNase condition was optimized based on the Lucigen unit definition (10 U/μL, catalog no. N6901K). The cell lysate was digested for 1.5 h at room temperature (25°C). We added 400 μL of TRIzol (Ambion) and 100 μL of chloroform (Sigma-Aldrich) to extract RNase footprints, and the footprints were precipitated overnight with isopropanol.
Step 2. Sequencing library construction using A-tailing and SMARTer oligo-based template switching
The following are the experimental conditions used for 50,000 cultured cells (HEK293T, K562, or HeLa cells) or 1 mg primary tissues. Precipitated RNase footprints were suspended in 5 μL of TNK reaction mixture (0.5 μL of 10 × T4 polynucleotide kinase [PNK] buffer, 1 μL of PNK [New England Biolabs], 0.5 μL of SUPERase•In RNase inhibitor [Invitrogen], and 3 μL of RNase-free water). The footprints were end-repaired for 90 min at 37°C followed by incubation for 5 min at 65°C to inactivate PNK. The end-repaired RNase footprints were 3′ polyadenylated with 10 U of Escherichia coli poly(A) polymerase (New England Biolabs) by incubating for 2 h at 37°C (supplemented with 2 μL of first-strand buffer [Invitrogen, 250 mM Tris-HCl at pH 8.3, 375 mM KCl, 15 mM MgCl2], 2 μL of E. coli poly(A) polymerase [New England Biolabs], 0.25 μL of 10 mM ATP, 0.5 μL of SUPERase•In RNase inhibitor, and 0.25 μL of RNase-free water). The resulting polyadenylated RNA was reverse-transcribed by a modified SMART-RT reaction. The RNA was first mixed with 2.5 μL of 10 μM RT primer and annealed by heating to 72°C for 5 min and cooled on ice immediately. The reaction mixture was then supplemented with SMART-RT mixture (2.5 μL of 10 mM dNTP, 2 μL of SuperScript II reverse transcriptase [Invitrogen], 0.5 μL of SUPERase•In RNase inhibitor, 4 μL of SuperScript II first-strand buffer, 1.5 μL of 100 mM DTT, 6 μL of 5 M betaine, 0.12 μL of 1 M MgCl2, 0.4 μL of 100 μM template-switching oligos, and 0.5 μL of RNase-free water) and incubated for 1 h at 42°C, followed by 10 cycles of heating for 2 min at 50°C and then cooling for 2 min to 42°C. The enzyme was inactivated by heating for 10 min at 70°C. The RNA fragments hybridized to DNA were then removed by incubating the reaction mixture for 15 min at 37°C with 5 U of RNase H (New England Biolabs).
The resultant 30 μL of cDNA was combined with 50 μL of 2× Ultra II Q5 master mix (New England Biolabs), 0.5 μM first-primers, and water to obtain a final reaction volume of 100 μL. This first round of PCR was performed with an initial 3-min denaturation at 98°C, followed by one cycle of 20-sec denaturation at 98°C, 30-sec annealing at 65°C, and 90-sec extension at 72°C, and then six cycles of 20-sec denaturation at 98°C, 20-sec annealing at 67°C, and 60-sec extension at 72°C. PCR round 1 was finished with a 3-min extension at 72°C. The PCR products were purified using a DNA Clean & Concentrator column (Zymo Research) and eluted in 20 μL of water. This library was amplified by a second round of PCR using a set of outer primers. Purified PCR1 products were combined with 25 μL of 2× Ultra II Q5 master mix (New England Biolabs) and 0.5 μM second-primers. We performed an initial 3-min denaturation at 98°C, followed by four cycles of 20-sec denaturation at 98°C, 20-sec annealing at 67°C, and 30-sec extension at 72°C, and finished with a 3-min extension at 72°C. The final DNA library was separated and visualized in a 4% agarose gel. DNA fragments with 15- to 35-bp insert bands were excised and recovered using the DNA gel recover kit (Zymo Research). We designed two DNA sequences with 15-nt- and 35-nt-length inserts as markers to guide the library size selection. The primers used for the library preparation are shown in Supplemental Table S8.
For the library construction of ultra-low-input conditions (i.e., 1000 cultured cells and immune cells), we increased the polyadenylation tailing incubation time to 4 h to achieve >80% polyadenylation of footprints. We also increased the PCR cycles (six cycles for the first round and five cycles for the second) to generate the sequencing library.
RNase footprinting data analyses
We performed paired-end sequencing (2 × 151 nt) of the footprinting libraries. Because the RNase footprint lengths are short (<35 nt for ribosome footprints), we used the first ends of reads to perform mapping and gene expression calculation. We first trimmed the 3′ sequencing adapters from the reads (AAAAAAAAAA for the A-tailing methods, and AGATCGGAAG for the data sets generated by linker ligation methods). For the libraries using SMARTer oligo-based template switching, we trimmed the first 7 nt, including the random 4 nt and three locked Gs in the 5′ sequencing adapters. The trimmed reads were first mapped to the rRNA sequences (5S, 5.8S, 18S, and 28S) using Bowtie 2 (version 2.2.6) (Langmead and Salzberg 2012). Approximately 78% of reads were mapped to rRNAs. The unmappable reads were then mapped to a reference genome (hg38 for human or mm10 for mouse) and transcriptome using TopHat (version 2.1.0) (Kim et al. 2013). The transcriptome annotation was based on GENCODE (release 28 for human and release 20 for mouse) (Frankish et al. 2019), and we modified the annotation by incorporating the mitochondrial transcripts as annotated by the UCSC Genome Browser.
We used the uniquely mappable reads with lengths of 18–35 nt for the calculations of ribosome occupancy because actively translating ribosomes show footprints of these defined lengths. To obtain read counts in RefSeq-defined protein-coding regions, we excluded the regions overlapping with uORFs (Ji et al. 2015) and used HTSeq (version 0.9.1) (Anders et al. 2015) to generate gene-level read counts. The ribosome occupancy levels were then measured as TPM values. To examine the read distribution across codons, we used the RibORF software (Ji et al. 2015; Ji 2018b) to plot the read distribution around the start and stop codons of mRNAs, adjust the reads’ genomic locations based on the offset distance between the 5′-end of fragments and ribosomal A-site, and examine 3-nt periodicity. We selected the reads showing strong 3-nt periodicity to identify translated noncanonical ORFs using RibORF. For more details, see Supplemental Methods.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under SuperSeries accession number (GSE151989, GSE151986, GSE151987, GSE151988, and GSE153411). The source codes for the data analyses are provided as a Supplemental Code file as well as in GitHub (https://github.com/zhejilab/RNaseFootprinting).
Supplementary Material
Acknowledgments
We thank Marcus Peter, Alfred George, Derek Walsh, Feng Yue, and members of Zhe Ji's laboratory for helpful discussions. This work was supported by grants to Z.J.: the National Institutes of Health (R35GM138192, R01HL161389, and R00CA207865), Lynn Sage Breast Cancer Research Foundation, Lynn Sage Scholar fund, and the Searle Leadership Fund in the Life Sciences from Northwestern University.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276139.121.
Competing interest statement
Northwestern University has filed patent applications related to the work described here. The title of the patent application is “Methods for mapping personalized translatome.” The U.S. Provisional Application was filed on December 4, 2020, application number 63/121,680.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 4: e03971. 10.7554/eLife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T. 2016. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535: 570–574. 10.1038/nature18647 [DOI] [PubMed] [Google Scholar]
- Baechler SA, Factor VM, Dalla Rosa I, Ravji A, Becker D, Khiati S, Miller Jenkins LM, Lang M, Sourbier C, Michaels SA, et al. 2019. The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis. Nat Commun 10: 83. 10.1038/s41467-018-07922-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33: 981–993. 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan KW, Chaim IA, Marina RJ, Yee BA, Kofman ER, Lorenz DA, Jagannatha P, Dong KD, Madrigal AA, Underwood JG, et al. 2021. Robust single-cell discovery of RNA targets of RNA-binding proteins and ribosomes. Nat Methods 18: 507–519. 10.1038/s41592-021-01128-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. 10.1126/science.1215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, Ohler U. 2016. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 13: 165–170. 10.1038/nmeth.3688 [DOI] [PubMed] [Google Scholar]
- Calviello L, Hirsekorn A, Ohler U. 2020. Quantification of translation uncovers the functions of the alternative transcriptome. Nat Struct Mol Biol 27: 717–725. 10.1038/s41594-020-0450-4 [DOI] [PubMed] [Google Scholar]
- Chen J, Brunner AD, Cogan JZ, Nuñez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD, et al. 2020. Pervasive functional translation of noncanonical human open reading frames. Science 367: 1140–1146. 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong C, Müller M, Pak H, Harnett D, Huber F, Grun D, Leleu M, Auger A, Arnaud M, Stevenson BJ, et al. 2020. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun 11: 1293. 10.1038/s41467-020-14968-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamer M, Tebaldi T, Lauria F, Bernabò P, Gómez-Biagi RF, Marchioretto M, Kandala DT, Minati L, Perenthaler E, Gubert D, et al. 2018. Active ribosome profiling with RiboLace. Cell Rep 25: 1097–1108.e5. 10.1016/j.celrep.2018.09.084 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P. 2020. The integrated stress response: from mechanism to disease. Science 368: eaat5314. 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. 2016. Synchronized mitochondrial and cytosolic translation programs. Nature 533: 499–503. 10.1038/nature18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. 2010. HITS-CLIP: panoramic views of protein–RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–286. 10.1002/wrna.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennerlein S, Wang C, Rehling P. 2017. Plasticity of mitochondrial translation. Trends Cell Biol 27: 712–721. 10.1016/j.tcb.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. 2007. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2: 532–540. 10.1038/nprot.2007.52 [DOI] [PubMed] [Google Scholar]
- D'Souza AR, Minczuk M. 2018. Mitochondrial transcription and translation: overview. Essays Biochem 62: 309–320. 10.1042/EBC20170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Bornelov S. 2021. CONCUR: quick and robust calculation of codon usage from ribosome profiling data. Bioinformatics 37: 717–719. 10.1093/bioinformatics/btaa733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu PH, Beane T, Zamore PD, Weng Z. 2018. Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics 19: 531. 10.1186/s12864-018-4933-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Struhl K. 2018. Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep 24: 1415–1424. 10.1016/j.celrep.2018.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan AA, Roberts BS, Moore DE, Ramaker RC, Jones AL, Myers RM. 2019. CRISPR/Cas9-targeted removal of unwanted sequences from small-RNA sequencing libraries. Nucleic Acids Res 47: e84. 10.1093/nar/gkz425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327. 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- Hornstein N, Torres D, Das Sharma S, Tang G, Canoll P, Sims PA. 2016. Ligation-free ribosome profiling of cell type-specific translation in the brain. Genome Biol 17: 149. 10.1186/s13059-016-1005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2014. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet 15: 205–213. 10.1038/nrg3645 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. 10.1016/j.celrep.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Hussmann JA, Weissman JS. 2019. Ribosome profiling: global views of translation. Cold Spring Harb Perspect Biol 11: a032698. 10.1101/cshperspect.a032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. 2016. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534: 558–561. 10.1038/nature17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z. 2018a. Rfoot: transcriptome-scale identification of RNA-protein complexes from ribosome profiling data. Curr Protoc Mol Biol 124: e66. 10.1002/cpmb.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z. 2018b. RibORF: identifying genome-wide translated open reading frames using ribosome profiling. Curr Protoc Mol Biol 124: e67. 10.1002/cpmb.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, Struhl K. 2015. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4: e08890. 10.7554/eLife.08890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Huang H, Regev A, Struhl K. 2016. Transcriptome-scale RNase-footprinting of RNA-protein complexes. Nat Biotechnol 34: 410–413. 10.1038/nbt.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE, Ackerman SL. 2017. Regulation of mRNA translation in neurons: a matter of life and death. Neuron 96: 616–637. 10.1016/j.neuron.2017.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, et al. 2018. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173: 90–103.e19. 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Ishijima J, Shiroishi T, Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3: e01257. 10.7554/eLife.01257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumont CM, Vincent K, Hesnard L, Audemard E, Bonneil E, Laverdure JP, Gendron P, Courcelles M, Hardy MP, Côté C, et al. 2018. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med 10: eaau5516. 10.1126/scitranslmed.aau5516 [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. 2013. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell 49: 453–463. 10.1016/j.molcel.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Molinaro G, Shu H, Stackpole EE, Huber KM, Richter JD. 2019. Optimization of ribosome profiling using low-input brain tissue from fragile X syndrome model mice. Nucleic Acids Res 47: e25. 10.1093/nar/gky1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez TF, Chu Q, Donaldson C, Tan D, Shokhirev MN, Saghatelian A. 2020. Accurate annotation of human protein-coding small open reading frames. Nat Chem Biol 16: 458–468. 10.1038/s41589-019-0425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Ingolia NT. 2017. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126: 112–129. 10.1016/j.ymeth.2017.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. 2012. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res 22: 2219–2229. 10.1101/gr.133249.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, Rabinowitz JD. 2018. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554: 128–132. 10.1038/nature25460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor PB, Andreev DE, Baranov PV. 2016. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat Commun 7: 12915. 10.1038/ncomms12915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenskaia T, Law T, Clauser KR, Klaeger S, Sarkizova S, Aguet F, Li B, Christian E, Knisbacher BA, Le PM, et al. 2022. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat Biotechnol 40: 209–217. 10.1038/s41587-021-01021-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SF, Rebelo-Guiomar P, D'Souza AR, Powell CA, Van Haute L, Minczuk M. 2017. Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem Sci 42: 625–639. 10.1016/j.tibs.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10: 1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Prensner JR, Enache OM, Luria V, Krug K, Clauser KR, Dempster JM, Karger A, Wang L, Stumbraite K, Wang VM, et al. 2021. Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat Biotechnol 39: 697–704. 10.1038/s41587-020-00806-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen YH, Martin S, Alhusaini N, Green R, Coller J. 2016. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167: 122–132.e9. 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Shenolikar S, Nicchitta CV. 2015. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods 91: 69–74. 10.1016/j.ymeth.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijers K, Loayza-Puch F, Nijtmans LG, Agami R. 2013. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat Commun 4: 2886. 10.1038/ncomms3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudler DL, Hughes LA, Perks KL, Richman TR, Kuznetsova I, Ermer JA, Abudulai LN, Shearwood AJ, Viola HM, Hool LC, et al. 2019. Fidelity of translation initiation is required for coordinated respiratory complex assembly. Sci Adv 5: eaay2118. 10.1126/sciadv.aay2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Cuevas MV, Hardy MP, Holly J, Bonneil E, Durette C, Courcelles M, Lanoix J, Côté C, Staudt LM, Lemieux S, et al. 2021. Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep 34: 108815. 10.1016/j.celrep.2021.108815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman IM, Li F, Alexander A, Goff L, Trapnell C, Rinn JL, Gregory BD. 2014. RNase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biol 15: R3. 10.1186/gb-2014-15-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhm T, Kaimal JM, Dawitz H, Peselj C, Masser AE, Hanzén S, Ambrožič M, Smialowska A, Björck ML, Brzezinski P, et al. 2018. Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metab 27: 1309–1322.e6. 10.1016/j.cmet.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Tahmasebi S, Khoutorsky A, Mathews MB, Sonenberg N. 2018. Translation deregulation in human disease. Nat Rev Mol Cell Biol 19: 791–807. 10.1038/s41580-018-0034-x [DOI] [PubMed] [Google Scholar]
- van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, et al. 2019. The translational landscape of the human heart. Cell 178: 242–260.e29. 10.1016/j.cell.2019.05.010 [DOI] [PubMed] [Google Scholar]
- VanInsberghe M, van den Berg J, Andersson-Rolf A, Clevers H, van Oudenaarden A. 2021. Single-cell Ribo-seq reveals cell cycle-dependent translational pausing. Nature 597: 561–565. 10.1038/s41586-021-03887-4 [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Nguyen TB, Gelboin-Burkhart C, Wang R, Blue SM, Pratt GA, Louie AL, Yeo GW. 2017. Robust, cost-effective profiling of RNA binding protein targets with single-end enhanced crosslinking and immunoprecipitation (seCLIP). Methods Mol Biol 1648: 177–200. 10.1007/978-1-4939-7204-3_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen JY, Cody NAL, Dominguez D, et al. 2020. A large-scale binding and functional map of human RNA-binding proteins. Nature 583: 711–719. 10.1038/s41586-020-2077-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Zinshteyn B, Wehner KA, Green R. 2019. High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol Cell 73: 959–970.e5. 10.1016/j.molcel.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Peterson A, Zinshteyn B, Regot S, Green R. 2020. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182: 404–416.e14. 10.1016/j.cell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf MG, Maguire S, Humbert P, Dai N, Bei Y, Nichols NM, Corrêa IR Jr, Guan S. 2019. Non-templated addition and template switching by Moloney murine leukemia virus (MMLV)-based reverse transcriptases co-occur and compete with each other. J Biol Chem 294: 18220–18231. 10.1074/jbc.RA119.010676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ruggero D. 2020. The role of translation control in tumorigenesis and its therapeutic implications. Annu Rev Cancer Biol 4: 437–457. 10.1146/annurev-cancerbio-030419-033420 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.