Abstract
Assembly of KU and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) at DNA double strand breaks (DSBs) forms DNA-PK holoenzyme as a critical initiating step for non-homologous end joining (NHEJ) repair of DSBs produced by radiation and chemotherapies. Advanced cryo-electron microscopy (cryo-EM) imaging together with breakthrough macromolecular X-ray crystal (MX) structures of KU and DNA-PKcs recently enabled visualization of the ~600kDa DNA-PK assembly at near atomic resolution. These important static structures provide the foundation for definition and interpretation of functional movements crucial to mechanistic understanding that can be tested through solution state structure analysis. We herein therefore leverage Cryo-EM and MX structures for the interpretation of synchrotron small-angle X-ray scattering (SAXS) data on DNA-PK conformations in solution to inform the structural mechanism for NHEJ initiation. SAXS, which measures thermodynamic solution-state conformational states and assemblies outside of cryo- and solid-state conditions, unveils the inherent flexibility of KU, DNA-PKcs and DNA-PK. The combined structural measurements reveal mobility of KU80 C-terminal region (KU80CTR), motion/plasticity of HEAT (DNA-PKcs Huntingtin, Elongation Factor 3, PP2 A, and TOR1) regions, allosteric switching upon DNA-PKcs autophosphorylation, and dimeric arrangements of DNA-PK assembly. Importantly, the results uncover displacement of the N-terminal HEAT domain during autophosphorylation as suitable for a regulated release mechanism of DNA-PKcs from DNA-PK to control unproductive access to toxic and mutagenic DNA repair intermediates. These integrated analyses show that the marriage of SAXS with cryo-EM leverages the strengths of both techniques to enable assessment of functional conformations and flexibility defining atomic-resolution molecular mechanisms for DSB repair.
Keywords: Structural dynamics, X-ray scattering, Cryo-Electron microscopy, Genome stability, DNA break repair
1. Introduction
A major enabling step in understanding non-homologous end joining (NHEJ) DNA repair has been realized through breakthrough comprehensive and high-resolution visualizations of DNA-PKcs by combining cryo-EM [1, 2] with X-ray crystallographic [3] results. Defining the structural mechanisms for NHEJ are important for cancer etiology and therapeutic strategies since this is the primary DSB repair pathway in humans. Alternative DSB repair pathways either involve homology-directed repair or the more error prone alternative end joining that depends upon XRCC1 complexes [4–7]. To obtain structural insights before the current cryo-EM structures, it was necessary to build up models of the NHEJ complexes one component at a time [8]. Importantly, such insights pertain to genome instability seen in some cancers associated with translocations that depend upon the actions of NHEJ proteins and that are particularly associated with non-B DNA-forming sequences [9, 10]. Moreover, as DNA-PK has functional interactions with poly(ADP-ribose) polymerase (PARP1) [11] and both PARP1 and poly(ADP-ribose) glycohydrolase (PARG) inhibitors are actively being pursued for targeting the DNA damage response to kill cancer cells, these DNA-PK complexes are certainly relevant to ongoing cancer research [12, 13].
Currently, in cases where cryo-EM provided only near atomic resolution, integration of high-resolution crystal structures of the assembly and components [3, 14] into the cryo-EM maps enabled the reconstruction of atomistic models for the larger KU-DNA-DNA-PKcs (DNA-PK) assembly [15]. Yet, these are flexible complexes. In order to create a tractable sample for cryo-EM analysis, crosslinking agents were required to stabilize the complex. Such crosslinking limits assessment of flexibility but implies the complex is functionally dynamic. Indeed, the complex is expected to undergo allosteric transitions for function. We have therefore been developing and applying solution state small angle X-ray scattering (SAXS) techniques to complement cryo-EM and X-ray crystallographic structures, bridge size restrictions on techniques and provide a perspective on functionally-relevant solution behavior.
Previously we employed SAXS data to evolve understanding of static structures into dynamic multistate functional conformations and to visualize flexible or unfolded regions [16–19]. Due to rotational averaging, the information content of SAXS is dramatically reduced compared to a density maps generated from cryo-EM or crystallography. Two advantages of SAXS, however, are that it provides objective data on flexibility [20] and that SAXS profiles can be efficiently calculated from atomistic models and directly matched to experimental data [21]. As a result, multistate data-based models that mimic dynamic rearrangements, such as domain motions, transient complexation, and the presence of unfolded regions, can be robustly determined by SAXS-based atomistic modeling [22, 23]. Moreover, high-throughput SAXS data collection techniques, including size-exclusion, chromatography-coupled SAXS (SEC-SAXS)[24], enable the collection of many samples at multiple different conditions, facilitating the integration of SAXS with cryo-EM and crystallography [25–27]. E.g. such SEC-SAXS unveiled the basis for the regulation of the cancer-important P53 protein [28]. Indeed, first defining and then controlling functional conformations by small molecules can control activities and even repair pathway choice as seen for the MRE11 complex [6, 29].
Here, we build upon published SAXS data and studies that characterized KU, KU-DNA, DNA-PKcs [27, 30], and the DNA-PK assembly [30] and incorporate newly available atomic structures. Integrating atomic structures/models with new approaches in SAXS-based atomistic modeling [21, 23, 31] allowed us to derive dynamic models of the DNA-PK assembly and its components. From these analyses, we characterized a displacement of the KU80 C-Terminal Region (KU80CTR) from the KU core, show an inherent swing-like motion of the DNA-PKcs M and N- HEAT regions and identify a significant contraction of this N-HEAT region upon DNA-PKcs autophosphorylation. Interestingly, similar functional contractions and swing-like motions have been seen in the MRE11-RAD5O-NBS1 complex that acts in the homology-directed DSB repair as the major alternative pathway to NHEJ[7, 32]. We moreover define and validate the solution state of the DNA-PK assembly that suggests stabilization of the DNA-PKcs HEAT regions upon recruitment to the KU-DNA complex. Intriguingly, we show that DNA-PKcs and DNA-PK can form a head to head interaction that appears to be a likely suitable precursor of the pre-synaptic NHEJ complex.
2. Materials and methods
2.1. Purification of proteins
DNA-PKcs and KU were purified from HeLa cells as described [33, 34]. KUΔCTR (KU70/KU80 1–569) were expressed and purified from baculovirus-infected insect cells as described [35]. Large-scale preparation of phosphorylated DNA-PKcs for structural analysis were prepared as described by [27]. 20-bp double-stranded DNA containing a short DNA stem-loop on one end for the preparation of KU-DNA complex was annealed and purified according [27]. 20-bp double-stranded DNA containing a short DNA stem-loop on one end and a 5’-nucleotide (nt) overhang on the other for the preparation of the DNA-PK complex was annealed and purified according [30].
2.2. SAXS experiment
For small-angle X-ray scattering coupled with multi-angle light scattering in line with size-exclusion chromatography (SEC-SAXS-MALS) experiments, 60 μL samples containing either 5 mg/mL of KUΔCTR and 5 mg/mL KU were prepared in 50 mM Hepes 7.5, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT. SEC-SAXS-MALS were collected at the ALS beamline 12.3.1 LBNL Berkeley, California [36]. X-ray wavelength was set at λ=1.127 Å and the sample-to-detector distance was 2100 mm resulting in scattering vectors, q, ranging from 0.01 Å-1 to 0.4 Å-1. The scattering vector is defined as q = 4πsinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20°C [37] and data was processed as described [24]. Briefly, a SAXS flow cell was directly coupled with an online Agilent 1260 Infinity HPLC system using a Shodex KW803 column. The column was equilibrated with running buffer with a flow rate of 0.5 mL/min. 55 μL of each sample was run through the SEC and 3 second X-ray exposures were collected continuously during a 30min elution. The SAXS frames recorded prior to the protein elution peak were used to subtract all other frames. The subtracted frames were investigated by the radius of gyration Rg derived by the Guinier approximation I(q) = I(0) exp(−qRg)2/3 with the limits qRg<1.5. The elution peak was mapped by comparing integral of ratios to background and Rg relative to the recorded frame using the program SCÅTTER (Fig. S1). Eluent was subsequently split 3 to 1 between SAXS line and a series of UV @ 280 and 260 nm, multi-angle light scattering (MALS), quasi-elastic light scattering (QELS), and refractometer detector. MALS experiments were performed using an 18-angle DAWN HELEOS II light scattering detector connected in tandem to an Optilab refractive index concentration detector (Wyatt Technology). System normalization and calibration was performed with bovine serum albumin using a 45 μL sample at 10 mg/mL in the same SEC running buffer and a dn/dc value of 0.19. The light scattering experiments were used to perform analytical scale chromatographic separations for MW determination of the principle peaks in the SEC analysis. UV, MALS, and differential refractive index data was analyzed using Wyatt Astra 7 software to monitor the homogeneity of the sample across the elution peak complimentary to the above-mentioned SEC-SAXS signal validation (see Fig. S1).
The DNA-PKcs and KU-DNA-DNA-PKcs (DNA-PK) complex was prepared and measured by SEC-SAXS-MALS experiment (see Fig. 3A) as described by [30]. Additionally, SAXS data of KU, KU-DNA, DNA-PKcs, phosphorylated DNA-PKcs were measured by high-throughput SAXS experiment (HT-SAXS) as described by [27]. To improve the signal to noise ratio at higher q range SAXS curved derived from SEC-SAXS and HT-SAXS experiments were merged for KU and DNA-PKcs samples (see Fig. 1A and 2A, Table 1). DNA-PKcs in complex with 40-bp duplex with a Y-shaped structure at one end (DNA-PKcs-40bp Y-DNA) and 40bp DNA with the hairpin (40bp H-DNA) [27] and 40bp DNA with two blunt ends were measured by high-throughput SAXS experiment (HT-SAXS) as described by [27].
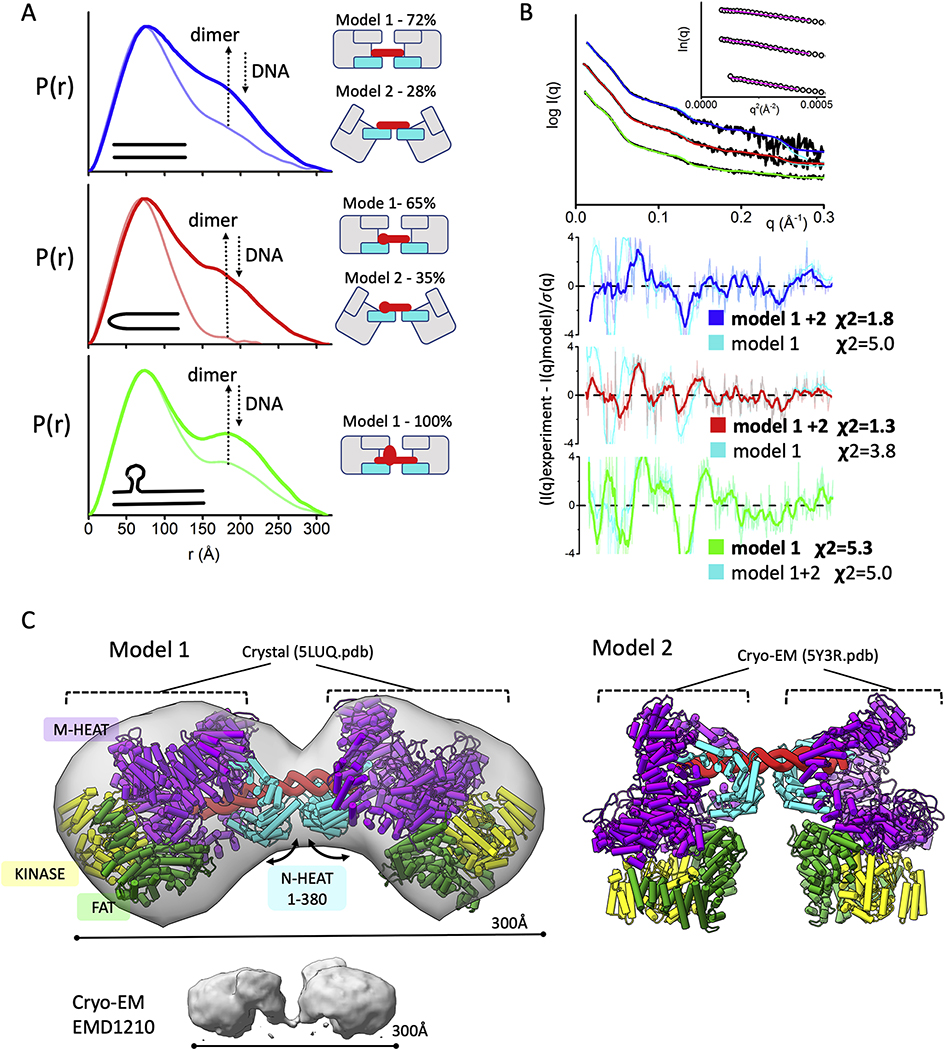
Fig. 3.
Formation of dumbbell DNA-PKcs-DNA dimers
A) P(r) functions for DNA-PKcs - 40bp DNA (blue), 40bp H-DNA (red) and 40bp Y-DNA (green) with the ratio 2:1 (DNA-PKcs :DNA), calculated from the experimental SAXS shown in the panel B. The light-colored P(r) functions are shown for the samples with equimolar DNA-PKcs:DNA molar ratio. The left panel shows cartoon representations of the atomistic models and its weights for each DNA-PKcs-DNA complex that were used to match the SAXS data shown in the panel B.
B) Experimental (black) and theoretical SAXS profiles for the single (cyan) and multistate-model of DNA-PKcs in the complex with 40bp DNA (blue), 40bp H-DNA (red) and 40bp Y-DNA (green) with the DNA-PKcs:DNA ratio 2:1. SAXS fits are shown together with the fit residuals and goodness of fit values (χ2). Guinier plots for experimental SAXS curves are shown in the inset.
C) Two atomistic models of DNA-PKcs dimer (model 1 and model 2) bridged by 40bpDNA(red). Model 2 was built based on the DNA-PK cryo-EM structure [15] by replacing KU with the DNA-PKcs. Model 1 was built by replacing both DNA-PKcs with DNA-PKcs-crystal structure [3]. Conformational variability in the N-HEAT 1–380 region as seen between DNA-PKcs from DNA-PK (PDBID 5Y3R) and DNA-PKcs crystal structure (PDBID: 5LUQ) results in altering of DNA-PKcs tilt. The N-HEAT 1–380, M-HEAT, FAT and kinase regions are colored as indicated. cryo-EM map for putative DNA-PKcs-DNA-KU complex [57] is shown in the bottom panel.
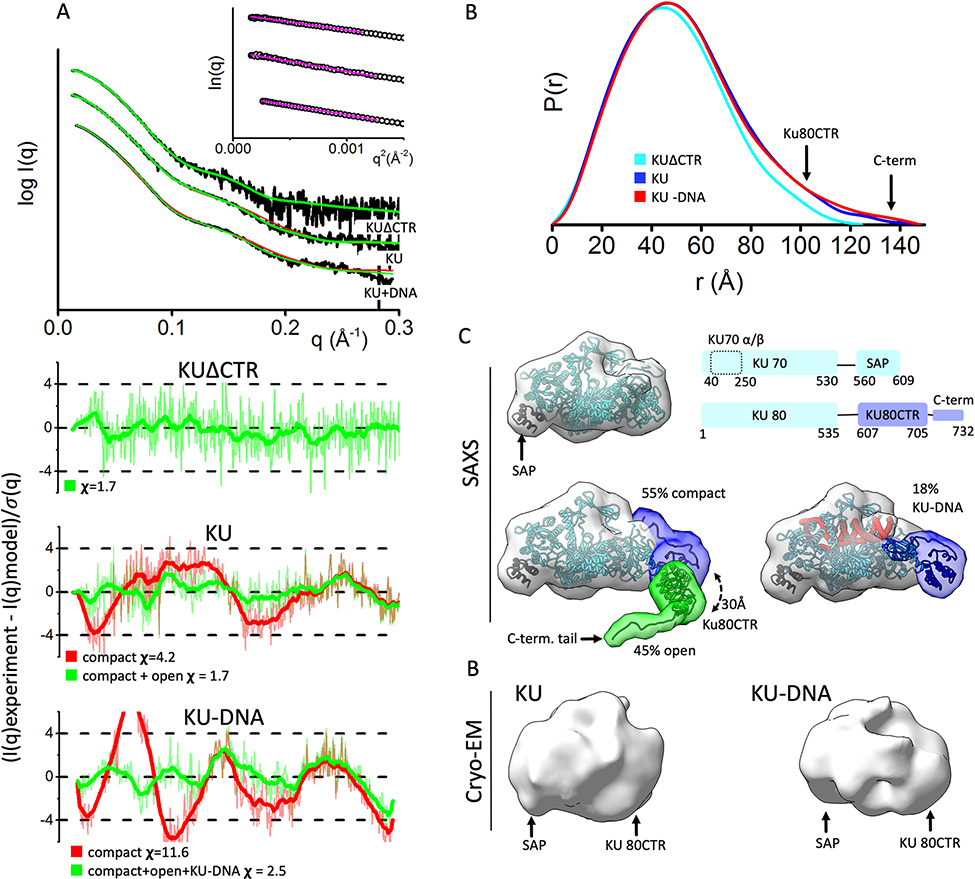
Fig. 1.
KU80CTR is located in close proximity to the KU70 α/β region in DNA free and DNA bound states.
A) Experimental (black) and theoretical (colored as indicated) SAXS profiles for the solution state models of KUΔCTR, KU and KU-DNA. SAXS fits are shown together with the fit residuals and goodness of fit values (χ2). Guinier plots for experimental SAXS curves are shown in inset.
B) Normalized pair distribution P(r) functions for experimental SAXS curves of KUΔCTR (cyan), KU (blue) and KU-DNA (red).
C) top panel: Crystal structure of KU [14]. A schematic representation highlighting the domains of KU: two KU70 regions composed of the KU core region and the SAP domain; and three KU80 regions composed of the KU80 core region, the KU80CTR domain and the KU80 C-terminal helix. bottom panel: conformers in selected multistate model of KU and KU-DNA used to calculate theoretical SAXS. The weight of each model is indicated. The two conformers (compact and open) used to fit experimental SAXS curves of KU and KU-DNA. The KU-DNA conformer with 18% weight is shown independently. Atomistic models are displayed in ribbon style together with molecular envelop calculated at the 25Å resolution to match the resolution of cryo-EM maps shown at the bottom panel.
D) cryo-EM maps of KU (EMD#:1270) and KU-DNA complex (EMD#1271) [46]
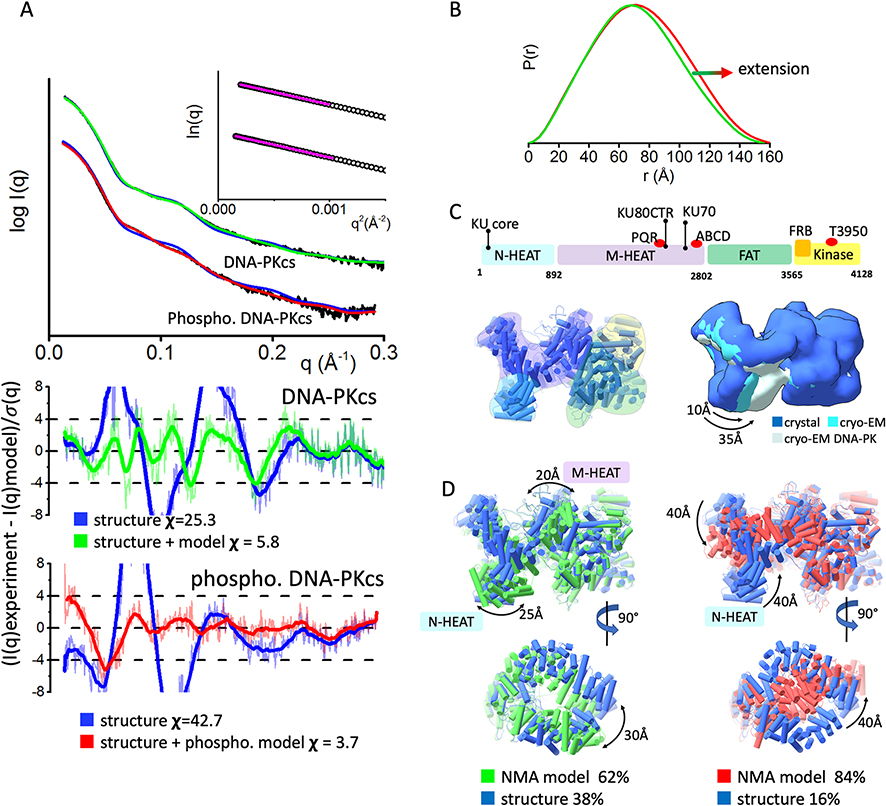
Fig. 2.
Inherent dynamicity of DNA-PKcs HEAT region and its rearrangement during the autophosphorylation.
A) Experimental (black) and theoretical (colored as indicated) SAXS profiles for the solution state models of DNA-PKcs and autophosphorylated DNA-PKcs. SAXS fits are shown together with the fit residuals and goodness of fit values (χ2). Guinier plots for experimental SAXS curves are shown in the inset.
B) Pair distribution P(r) functions, normalized at the maxima, for experimental SAXS curves of DNA-PKcs and autophosphorylated DNA-PKcs (taken from [27]).
C) Top panel: A schematic representation highlighting the four super secondary structural components of DNA-PKcs: the two HEAT region composed of the N-terminal domain (N-HEAT); the M-HEAT region and the Head regions, which contains the FAT and kinase regions. The KU binding area, FRB domain, autophosphorylation clusters PQR and ABCD, and highly conserved T3950 autophosphorylation site are shown above the schematic.
Left panel: Crystal structure of DNA-PKcs with highlighted N-HEAT, M-HEAT, FAT and kinase regions. middle panel: Comparison of the crystal structure and cryo-EM structure from [1, 3], and cryo-EM structure of DNA-PKcs taken from the DNA-PK complex [15]. For better visualization of conformational variability in the HEAT region, atomic models are displayed as a molecular envelop at the 20Å resolution.
D)Two orthogonal views of multi-state model used to match experimental SAXS curves of DNA-PKcs and autophosphorylated DNA-PKcs. The models were superimposed on each other at the FAT region. Weight for each model is indicated.
Table 1.
Structural parameters from SAXS and MALS data
| SAXS sample SASBDB# | Dmax (Å) | Rg (Å) from Guinier plot | Rg (Å) from P(r) | MW Seq. monomer (kDA) | MW SAXS (kDa) | MW MALS (kDa) | Model fit χ2 | data source |
|---|---|---|---|---|---|---|---|---|
| KUΔCTR SASDJV4 | ~ 125 | 37.9±0.6 | 37.9 | 134 | ~ 140 | 145 | 1.7 | SEC-SAXS (This study) |
| KU SASDJU4 | ~ 155 | 41.5±1.4 | 42.7 | 153 | ~ 160 | 160 | 1.7 | SEC-SAXS (This study) merged with SAXS from [27] |
| KU-DNA SASDJW4 | ~ 155 | 40.7±0.3 | 41.9 | 173 | ~ 170 | 190 | 2.6 | Data from [27] |
| DNA-PKcs SASDJX4 | ~ 155 | 57.1±1.7 | 54.4 | 469 | ~ 480 | 480 | 5.9 | SEC-SAXS from [30] merged with SAXS from [27] |
| Autophospho. DNA-PKcs SASDJY4 | ~ 160 | 57.2±1.7 | 56.0 | 469 | ~ 480 | ND | 3.7 | SAXS from [27] |
| DNA-PK monomer SASDJZ4 | ~ 230 | 65.1±0.6 | 66.8 | 640 | ~ 590 | 640 | 3.7 | SEC-SAXS from [30] |
| DNA-PK monomer/dimer SASDJ25 | ~ 300 | 75.4±1.5 | 83.5 | 640 | ~ 720 | 680–700 | 1.0 | SEC-SAXS from [30] |
| DNA-PKcs – 40bp DNA | ~ 315 | 80.9 ± 4.2 | 92.9 | 499 | ~860 | ND | 1.8 | SAXS from [27] |
| DNA-PKcs – 40bp H-DNA | ~ 315 | 87.7 ± 6.3 | 96.4 | 497 | ~900 | ND | 1.3 | SAXS from [27] |
| DNA-PKcs – 40bp Y-DNA | ~ 315 | 90.2 ± 4.1 | 101.2 | 501 | ~880 | ND | 5.0 | SAXS from [27] |
2.3. SAXS data evaluation.
Final merged SAXS profiles where used for further analysis including Guinier plot which determined an aggregation free state (see Fig. 1A, 2A, 3B). The program SCÅTTER was used to compute the pair distribution function P(r). The distance r where P(r) approach zero intensity identifies the maximal dimension of the macromolecule (Dmax). P(r) functions of KUΔCTR, KU, KU-DNA (Fig. 1B) and DNA-PK (Fig. 3C) were normalized based on the molecular weight of the assemblies as determined from SAXS curves by SCÅTTER using volume of correlation Vc (see Table 1) [38, 39]. To better visualized broadening of phosphorylated DNA-PKcs, the P(r) functions for DNA-PKcs and phosphorylated DNA-PKcs were normalized at their maxima (see Fig. 2B).
2.4. Solution Structure Modeling
The pool of KU conformers from our previous study [27] was used to fit the experimental SAXS curve of KU using FOXS [23, 40, 41] and a multistate state model was selected using MultiFOXS [23] (see Fig. 1C). The same pool of conformers plus the pool of KU-DNA models from [27] was used to fit experimental SAXS of KU-DNA. A three-state model was selected by MultiFOXS [23] (see Fig. 1C).
To fit SAXS of DNA-PKcs and phosphorylated DNA-PKcs we initially built missing loops, including the ~2576–2776 region that contains the ABCDE phosphorylation sites [42, 43] in to the X-ray crystal [3] and Cryo-EM structures [1] using MODELLER [44]. First we applied conformational sampling BILBOMD [22] to mimic the plasticity of the added loops. Next we optimized SAXS fit by large movement of DNA-PKcs domains using normal mode analysis (NMA) program SREFLEX [31]. The SREFLEX program uses NMA in Cartesian space to estimate the flexibility of atomistic models to improve their agreement with experimental SAXS curve. The DNA-PKcs crystal structure [3] with added missing loops was divided into 4 regions that include residues 1–370 (N-terminal N-HEAT region), 371–1800 (N-HEAT and M-HEAT), 1801–2800 (second region of M-HEAT) and 2801–4119 (FAT and Kinase region). SREFLEX optimized the position of each domain relative to the others including normal mode elastic movement in the region. The generated 8 NMA models plus the crystal [3] and Cryo-EM [1] structures (with added missing loops) is a pool of the models that were fitted to the experimental SAXS by FoXS [23, 40, 41] followed by selection of two-state model by MultiFOXS [23].
To fit the SAXS data of DNA-PKcs- 40-bp duplex with a Y-shaped structure at one end (40bp Y-DNA), 40bp DNA with the hairpin (40bp H-DNA) and 40bp DNA with two blunt ends (40bpDNA) we built atomistic model of DNA-PKcs dumbbell dimer by replacing KU in the DNA-PK structure [15] with DNA-PKcs (model 2). Alternative model with the different conformation of N-HEAT 1–380 region were built by replacing DNA-PKcs with the conformers as seen in the crystal structure (model 1) (Fig. 3C). Due to conformational flexibility of N-HEAT 1–380 region the DNA-PKcs may adopts different tilt relative to the linear DNA (Fig. 3C). Thus alternative models with various tilts of DNA-Pcs were built (Model 3–9). We used the pool of the models that include dimer-models and DNA-PKcs monomer to fit the SAXS curve for all three DNA complexes using program FoXS [23, 40, 41], followed by selection of two-state model by MultiFOXS [23].
To fit the SAXS curve of the monomeric DNA-PK assembly we initially built missing loops in the Cryo-EM structure [15] using MODELLER [44]. The missing KU80CTR domain was added in the close proximity of previously identify KU80CTR binding site at the M-HEAT region [3]. DNA-PK model was fitted to the experimental SAXS data by FoXS [23, 40, 41].
To fit experimental SAXS curves of the DNA-PK monomer/ dimer mixture we initially modeled a DNA-PKcs dimer by molecular docking of two DNA-PKcs monomers using a rigid docking, geometric shape-matching algorithm PatchDock [45]. The docking of DNA-PKcs (taken from DNA-PK structure PDBID: 5Y3R [15]) was performed without symmetry operator. The docking of the DNA-PKcs crystal structure [3] (PDBID: 5ULQ) and SAXS-based atomistic model was performed with symmetry operator. The top scoring model derived without symmetry operator resemble top scoring models with symmetry operator, and was further used to build the DNA-PK dimer model (Supplementary Fig. S2). DNA-PK dimers were constructed by aligning two DNA-PK monomers with the top scoring docking model of DNA-PKcs dimer. Experimental SAXS curve of DNA-PK monomer/ dimer mixture was fitted by both monomer and dimer model using FoXS [23, 40, 41] followed by selection of two-state model by MultiFOXS [23].
Data and the related models were deposited in the SASBDB data base (https://www.sasbdb.org/). The SASBDB data base accession codes and experimental SAXS parameters are reported in Table 1.
3. Results
3.1. KU80 C-terminal region (CTR) dynamicity in DNA-free and DNA-bound states
Enhanced SAXS experiments and analysis identify and verify a preferentially close interaction between the flexibly linked KU80CTR domain and the main KU70 α/β domain. Analysis of SAXS data collected on statics samples assuming homogeneous monomers in solution had suggested the complete disassociation of KU80CTR relative to the KU core in both DNA-free and DNA-bound states [27]. Our new measurements, obtained with size exclusion chromatography coupled with in-line SAXS and multi-angle light scattering (SEC-SAXS-MALS, Fig. 1A, Supplementary Fig. S1), separated small amounts of transiently self-associating KU dimers, which had not been considered previously. Analyzing the SEC-SAXS from the monomeric peak demonstrated smaller radius of gyration (Rg) values and less elongated P(r) functions (Fig. 1B, Supplementary Fig. S1 and Table 1) than those observed in the static experiment [27]. Further modelling through conformational sampling of the KU80CTR and KU80 C-terminal regions was applied [22] and, based on the goodness-of-fit (χ2) between theoretical and experimental SAXS curves [41], the best single or multi-state models [23], were selected. The single state model showed a compact arrangement of KU80CTR neighboring the KU70 α/β domain (Fig. 1C). Significant improvement in the SAXS fit was achieved by selecting the multistate model (χ2 single state 4.2 vs. χ2 two-state 2.2, Fig. 1A). The multistate model included 55% of the conformers with the KU80CTR domain in close proximity to the KU70 α/β domain and 45% of the conformers with detached KU80CTR, ~30Å from the KU core (Fig. 1C). The larger weighting of compact structures in solution validates and extends the models observed in cryo-EM studies showing relatively compact low resolution molecular envelopes, which suggested close contacts between KU80CTR and KU70 α/β domains [46] (Fig. 1D). However, a significant population of detached KU80CTR may remain of mechanistic importance in recruiting of DNA-PKcs.
Next, SAXS analysis of the KU-DNA complex was prepared using 16-bp double-stranded DNA (dsDNA) with a short stem loop at one end as template [27]. Similar to prior SAXS analysis of the KU-DNA complex [27], only small differences between KU and KU-DNA measurements were observed. Particularly, narrowing of the P(r) functions (Fig. 1B) indicates a less hollow structure that can be explained by insertion of DNA between the bridge and β barrel KU-regions [14]. Again, conformational sampling was performed on KU80CTR to determine the movement of KU80CTR relative to the KU-DNA core. In the search for representative multistate solution models, the KU-DNA experimental SAXS data was fit against a pool of ~10000 conformers including KU and KU-DNA models with various conformations of KU80CTR. Surprisingly, the best fit multistate model showed only 18% of the KU-DNA complex with the KU80CTR in close proximity to the KU core (Fig. 1C). This partial occupancy of the KU-DNA complex suggests dissociation of KU from unprotected DNA. These results are in agreement with multiple assays under non-reducing condition [47].
3.2. Conformational plasticity in the DNA-PKcs HEAT region
DNA-PKcs (4128 amino acids, ~469 kDa) [48] is composed of FAT, Kinase, FAT-C domains that form the “head” or crown and a large, flexible HEAT region, also called the palm region, that can be divided into the M-HEAT [49–51] and the recently visualized N-terminal HEAT region (N-HEAT)[1, 3]. DNA-PKcs structures, together with previously reported cryo-EM low-resolution molecular envelopes [49, 50] suggest that the DNA-PKcs HEAT region is flexibly attached to the “head” region allowing it to move during autophosphorylation [27] and rearrange upon interaction with the KU-DNA complex [15]. Our previous solution scattering studies have shown that DNA-PKcs undergoes self-association at higher protein concentrations [27]. Dimer-free DNA-PKcs SAXS data revealed a typical globular particle with a Dmax of 155 Å (Fig. 2B and Table 1).
To test how the existing atomic resolution structures match the solution state, we compared existing DNA-PKcs structures to experimental SAXS curves. The available atomic models are missing loop regions corresponding to amino acids 2576–2776 which contains the ABCDE cluster of phosphorylation sites [42, 43] located between the M-HEAT and FAT domain, and a conserved DNA-PK signature motif [52]. Without these domains, fits of the X-ray crystal [3] and Cryo-EM [1, 15] structures were poor (χ2crystal =128 crystal and χ2cryo-EM =160) (Fig. 2A). To further test whether this discrepancy was due to the flexibility of the missing regions, these sections were built using MODELER[44] and conformational sampling was applied using BILBOMD [22] to mimic the plasticity of the added regions. The goodness of SAXS fit improved by including flexibility in the ~2576–2776 region (χ2 single state =25.3); however, the remaining discrepancy between theoretical and experimental SAXS curves in the low-resolution range (q 0.05–0.15Å−1, see Fig. 2A), suggested larger conformational rearrangements of the DNA-PKcs domains in solution.
To visualize these allosteric changes, the conformational sampling protocol was applied using normal mode analysis (NMA) [31]. DNA-PKcs was divided into 4 regions (see Materials and methods) and constrained NMA sampling was applied to optimize the position of each region relative to the each other including normal mode movement inside each region. The best SAXS fit model was found by searching the two-state models [23] from the pool of existing structures and 10 models derived from NMA conformational sampling (NMA-model). The best two-state model included 38% of the original structure and 62% of the NMA-model, and was in excellent agreement with the experimental SAXS profile (χ2 two-state = 5.8, Fig. 2A). The NMA-model showed a ~25Å displacement of the N-HEAT domain (Fig. 2D) and agreed with the conformational variability of this region, as shown by comparison of the cryo-EM and crystal structures (Fig. 2C). Additionally, the NMA-model showed a smaller ~10Å movement of the M-HEAT region. In sum, a multistate model of DNA-PKcs was determined experimentally that shows the flexibility of the HEAT region in solution.
3.3. Displacement of the N-terminal HEAT domain during autophosphorylation suggests the release mechanism of DNA-PKcs from DNA-PK.
Autophosphorylation of DNA-PKcs prompts its release from KU-DNA double strand break (DSB) complexes in vitro and in vivo [27, 53–56]. In prior studies, the in vitro autophosphorylated form of purified DNA-PKcs was isolated and analyzed using SAXS [27]. Comparison of the SAXS data of DNA-PKcs with and without autophosphorylation revealed different scattering profiles over the entire observed scattering range (Fig. 2A). These changes were visualized in the P(r) function as broadening (Fig. 2B) and indicated a large conformational change involving the relocation of DNA-PKcs domains rather than extension of a single domain or a local change at the phosphorylation site. Calculated SAXS profiles from atomic resolution DNA-PKcs structures disagree with experimental SAXS profiles (χ2cryo-EM = 42.7). The large discrepancies seen between the theoretical and experimental profiles at low-resolution (q range 0.05–0.15Å-1, see Fig. 2A), further suggest a large conformational rearrangement.
Using a similar process to our analysis of non-phosphorylated DNA-PKcs, constrained NMA conformational sampling was performed to modify the position of the N-HEAT, M-HEAT, FAT, and Kinase regions (see Materials and methods). The best fit model was found by searching the two-state models from the pool of existing structures and multiple NMA derived models. The best two-state model, included 16% of conformations near the crystal structure [3] and 84% of the NMA-model. The two-state model fit was a great improvement over the single model fit and gave an excellent match to the experimental SAXS curve (χ2 two-state = 5.8, Fig. 2A). The NMA-model showed large (~40Å) displacements of both the N- and M-HEAT regions leading to closure of the aperture between these domains (Fig. 2D). Together with the extension of the M-HEAT region, the NMA-model explains the observed broadening of the P(r) function and the increasing Rg values relative to the non-phosphorylated DNA-PKcs. This atomistic model of autophosphorylated DNA-PKcs uncovers rearrangement of the entire HEAT region suggesting inaccessibility of the KU/N-HEAT binding site [15]. These results show that DNA-PKcs autophosphorylation conceals the interface between DNA-PKcs and KU. We hypothesize that by making the N-HEAT/KU binding site inaccessible, KU is forced to the detach from DNA-PKcs which subsequently allows other NHEJ processing enzymes, like DNA ligase Ligase IV and polynucleotide kinase phosphatase (PNKP), to bind the DSB.
3.4. Two DNA-PKcs bridged by 40bp DNA form an “dumbbell” arrangement.
Previously we showed similarity between the low resolution (33Å) DNA-PK cryo-EM map (EMD1210),[57] and SAXS envelopes of the dumbbell arrangement of two DNA-PKcs molecules bridged by a 40-bp duplex with a Y-shaped structure at one end (40bp Y-DNA) [27] (Fig. 3). We also found that a more compact dumbbell dimer is formed in the presence of 40bp DNA with the hairpin (40bp H-DNA) and 40bp DNA with two blunt ends (40bpDNA) [27]. Distinct dumbbell arrangements in the presence of 40bpY-DNA were interpreted as a bridging of two DNA-PKcs with the bulky head regions pointing outward, whereas the more compact DNA-PKcs - 40bp H-DNA and DNA-PKcs - 40bp-DNA dimers were explained by a head to head arrangement.
Here we used SAXS with atomistic models to test formation of the dumbbell arrangement and consider the capability of DNA-PKcs to bind various DNA ends in the absence of KU. The DNA-binding site within the N-HEAT and M-HEAT aperture is rich in positively-charged residues mostly located on the N-HEAT 1–380 region [15]. The lack of specific protein-DNA contacts and the opened DNA-binding site may allow DNA-PKcs to accommodate a wide spectrum of DNA ends [15]. Disappearance of the dimeric state in the excess of DNA (Fig. 3A) [27] further supports the notion that the DNA-PKcs-DNA interaction is not stably maintained without KU [15, 58]. By preserving the DNA binding site and mimicking conformational variability of the N-HEAT 1–380 region, DNA-PKcs can adopt different tilts relative to the linear DNA (Fig. 3C). To fit the SAXS data, we used a pool of dimer models build based on DNA-PK structure [15] (see Methods). The best fit for the DNA-PKcs-40bp DNA dimer was obtained by a two-state model that includes 56% of the dimer using DNA-PKcs crystal structure (model 1) [3] and 44% of the dimer using DNA-PKcs taken from the DNA-PK structure (model 2) [15]. This two-state model gives a significant improvement in the SAXS fit over the single model (χ2 one-state = 5.0 vs. χ2 two-state = 1.8, Fig. 3B). Model 1 and model 2 (65% and 35%) were also selected to give best SAXS fit of DNA-PKcs - 40bp H-DNA data with the significant improvement over the single model fit (χ2 one-state = 3.8 vs. χ2 two-state = 1.3, Fig. 3B). On the other hand, the extended dimer of DNA-PKcs – 40bp Y-DNA was well matched with extended single model 1, whereas only a non-significant improvement in the SAXS fit was obtained by two-state model (χ2 one-state = 5.0 vs. χ2 two-state = 5.3, Fig. 3B). These observations suggest a dominant presence of the extended dumbbell assembly: they further agree with well separated peaks in P(r) function (Fig. 3A) and distinct volumes in the SAXS envelope (Fig. 3C). Yet, this ability of DNA-PKcs to assemble on both ends of the DNA structure as visualized here and in previous studies [57] may not be physiologically relevant, as in vivo DNA damage-induced DSBs would each have only one exposed DNA end. In contrast, the self-association in the DNA-PKcs dimer, provides an appropriate protein arrangement for the initial NHEJ step and is further described in the next section.
3.5. DNA-PK dimer reconstruction
In prior work [30], the overall DNA-PK complex architecture was characterized in solution with KU bound to a short dsDNA oligomer mimicking a DSB. For the current experiments, DNA-PKcs was mixed with KU that had been pre-incubated with 20 bp DNA duplex containing a short DNA stem-loop on one end and a 5’-nucleotide (nt) overhang on the other (20 bpDNA). SEC-SAXS-MALS measurements, complimented by SDS-PAGE analysis of the SEC fractions, confirmed the DNA-PK assembly. DNA-PK eluted as an asymmetric peak (Fig. 3A), whereas MALS-analysis showed a decrease in the molecular weight (MW) of DNA-PK from 750 kDa at the beginning of elution peak to ~650 kDa at the tail (Fig. 4A). Thus, under these experimental conditions, we found that DNA-PK was primarily forming a 1:1:1 assembly with a theoretical MW of 642 kDa. Analysis of SAXS frames across the primary elution peak also showed a decrease in Rg from ~75 Å at the peak to ~65 Å at the tail (Fig. 4A, Table 1). These SEC-SAXS-MALS results suggest formation of a transient DNA-PK dimer that slowly interconverts over the elution time into a DNA-PK 1:1:1 assembly (DNA-PKcs/KU-DNA). The formation of a DNA-PK dimer agrees with the tendency of DNA-PKcs to self-associate [27, 59] (Fig. 4C inset).
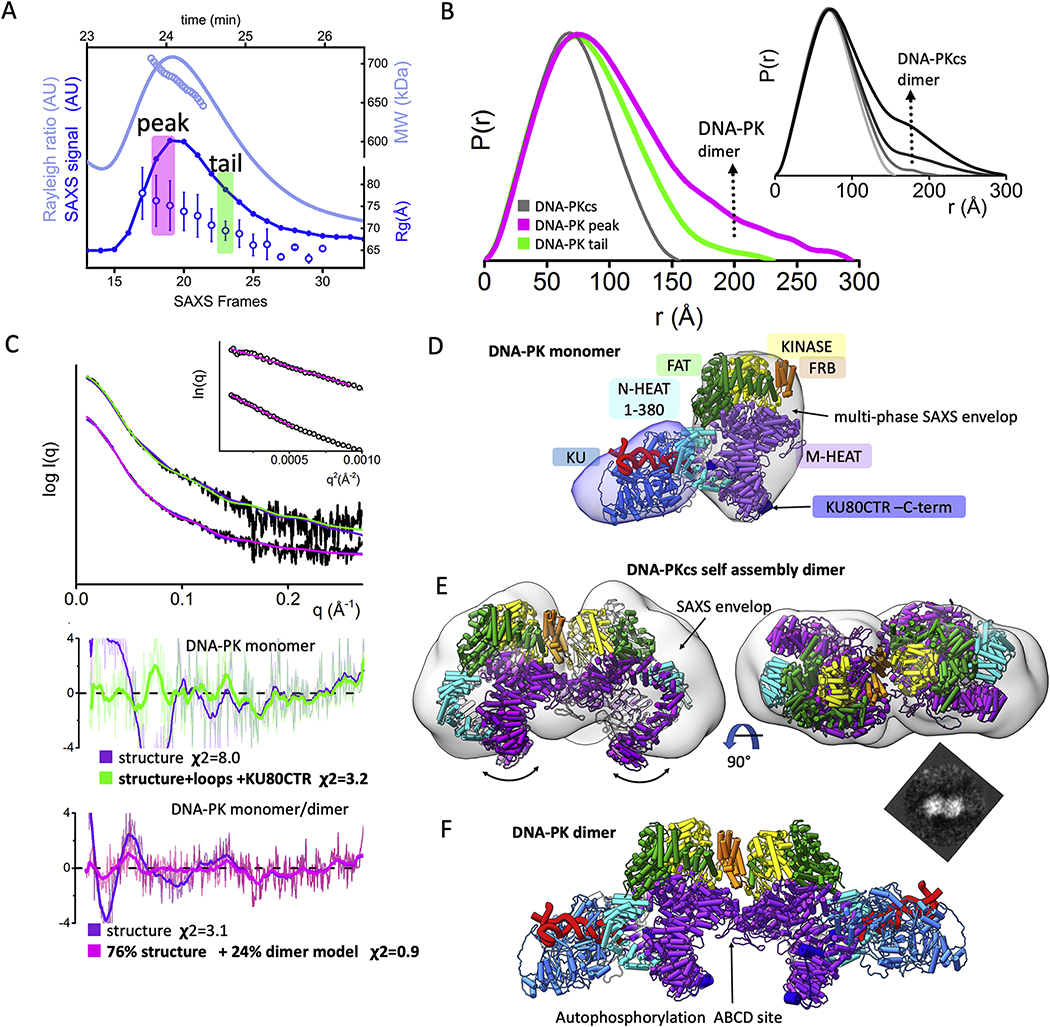
Fig. 4.
Solution state of DNA-PK and its dimerization through FRB domain.
A) SEC-MALS-SAXS chromatograms for DNA-PK assembly. Solid lines represent the MALS signal shown as Rayleigh signal (light blue) or integrated SAXS signal (dark blue) in arbitrary units, while symbols represent molecular mass (light blue) and Rg values for each collected SAXS frame (dark blue) versus elution time (taken from [30].
B) Experimental (black) and theoretical (colored as indicated) SAXS profiles for the solution state of DNA-PK in monomeric and dimeric state. SAXS fits are shown together with the fit residuals and goodness of fit values (χ2). Guinier plots for experimental SAXS curves are shown in inset.
C) Normalized pair distribution P(r) functions for experimental SAXS curves of DNA-PK assemblies measured at the peak and tail (green) of the elution peak (magenta) in comparison to monomeric DNA-PKcs taken from [27]. Inset: Normalized P(r) functions calculate for the experimental SAXS curves of DNA-PKcs collected at protein concentrations 1.5, 3, 7 and 15 mg/ml (from light gray to black) indicates self-association of DNA-PKcs at higher concentrations (taken from [27]).
D) The cryo-EM structure of DNA-PK [15] is superimposed on to the multi-phase SAXS envelop of DNA-PK taken from [30].
E) Two orthogonal views of DNA-PKcs docking model representing the self-association dimer. The DNA-PKcs self-association dimer superimposed on to the SAXS envelop reconstruction for the SAXS data collected at the 15 mg/ml protein concentration (taken from [27]. Additional top scoring models are shown in the supplemental Fig. 2. The 2D EM projection of DNA-PKcs self-association dimer (taken from [59]).
F) Proposed atomic model of DNA-PK – dimer that was used in 24% weight to match the SAXS curve measured at the SEC elution peak (see panel A).
To further analyze both monomeric and dimeric solution states of DNA-PK, the primary SEC elution peak was deconvoluted into two SAXS profiles (peak and tail, Fig. 4B). The P(r) function calculated for the SAXS curve at the elution peak showed an elongated assembly with a Dmax of ~300 Å, while the P(r) from the tail was narrower with a Dmax of ~220Å (Fig. 4C). Due to the expected transient dimerization of DNA-PK, the SAXS curves collected at the peak and tail of the SEC were fit separately.
The monomeric DNA-PK model built based on a cryo-EM structure [15] matched well to the SAXS profile from the elution tail (Fig. 4B). However, adding the missing parts of DNA-PKcs and KU, including the KU80CTR domain to the model improved the fit (χ2structure = 8.0 vs. χ2full model = 3.6) (Fig. 4B). Further refinement of KU80CTR location relative to the KU core was not possible due to low signal-to-noise in the SAXS data. Nevertheless, the overall arrangement of DNA-PK supported and extended reported multiphase SAXS envelops [30] and further confirmed the overall DNA-PK architecture in solution (Fig. 4D).
To build the DNA-PK dimer model, we initially modelled the DNA-PKcs dimer through the molecular docking of two DNA-PKcs monomers using a rigid docking, geometric shape-matching algorithm [45]. The best scoring model showed two DNA-PKcs monomers [3] in a mirror symmetry with a head-to-head arrangement (Fig. 4E). Overall, the observed arrangement of self-associated DNA-PKcs resembled the previously reported V-like SAXS envelop [27] and low resolution 2D EM projection [59] (Fig. 4E–inset). Furthermore, the docking model suggested contact between FKBP12-rapamycin-binding domains (FRB) which are conserved within the PI3K kinase family members (Fig. 4E). While the role of the FRB domain is unclear, there is evidence from mTOR studies that it might act as a gatekeeper that restricts access to the catalytic site [60]. The stand-alone FRB domain appears to function as gate to the buried and conserved T3950 autophosphorylation site [3] that deactivates the kinase [53]. The dimerization interface of DNA-PKcs modeled in this study may alter FRB positioning and control access to the T3950 autophosphorylation site.
Building upon these results, atomic models of the DNA-PK dimer were constructed by aligning two DNA-PK monomers with the DNA-PKcs dimer. Exchange between the two states occurred over the course of purification, so that a pure dimeric state could not be isolated from the monomeric one. Thus, the SAXS curve generated from the SEC peak was fit against a library of atomic models including DNA-PKcs structures, DNA-PKcs models, DNA-PK monomer, and DNA-PK dimer. The best fit two-state model was obtained by including 24% of DNA-PK dimer and 76% of DNA-PK monomer conferring an excellent match to the experimental SAXS profile (χ2 = 0.9, Fig. 4BE). The SAXS modeling together with the determined MW demonstrated the tendency of DNA-PK to form a head-to-head dimer with a V-shape arrangement. In the model, KU-DNA was positioned at the extremity of the dimer with the DNA entering an aperture in DNA-PKcs between the N- and M-HEAT regions.
4. Discussion
The promise of macromolecular structural biology is to visualize structures that unveil critical functional mechanisms. This promise is too often not fully realized due to missing or incomplete knowledge of functional conformations assessed outside of cryo- and solid-state conditions [17]. On the other hand, SAXS, which can be high-throughput and directly measures thermodynamic solution-state conformational states and assemblies, is resolution limited. Yet, SAXS can be combined with other measurements to interrogate atomic level information [61]. Here we assess practical and robust methods to join SAXS with cryo-EM and X-ray crystallographic data as applied to the dynamic structure of DNA-PKcs and its interactions with KU and DNA in solution. The results reported provide insights into the dynamic architectural changes whereby these complexes orchestrate non-sequential NHEJ repair [62–64]. Previously our SAXS results predicted XRCC4 and XLF protein filaments for DNA end protection and alignment in DSBR[65, 66], and these filaments were subsequently identified in single-molecule experiments as well as in cells [67, 68]. Importantly, the new results presented here uncover displacement of the N-terminal HEAT domain during autophosphorylation as suitable for a regulated release mechanism of DNA-PKcs from DNA-PK to control unproductive access to toxic and mutagenic DNA repair intermediates. Such release of unproductive complexes has been seen before for FEN1 and XPG nuclease functions in base and nucleotide excision repair [69–71]. Overall, we present new SAXS data, revisit solution structure modeling of the KU heterodimer, and combine SAXS measurements with atomic resolution static structures.
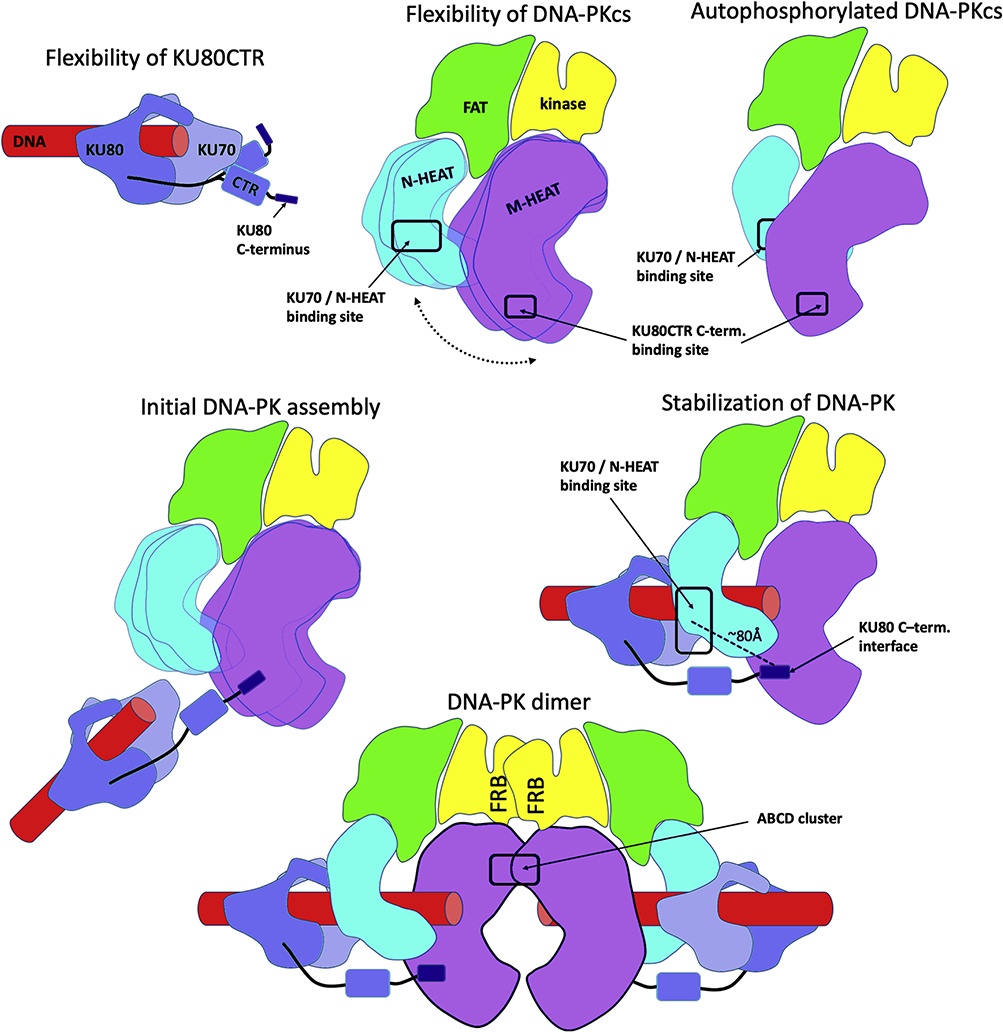
Our improved experiments on KU reveal that a major population of KU80CTR is mostly located in close proximity to the KU core. However, a nearly equal part (45%) of the solution state involves an extension of KU80CTR domain through the flexible linker (Fig. 1C) that is in apparent disagreement with the static picture of KU visualized by cryo-EM [46]. Yet, our results support and extend earlier studies showing proteolytic sensitivity of the KU80CTR [72, 73] and subsequent structural studies revealing a disordered linker region [74]. We reason that the detachment of KU80CTR from KU core remains functionally important for its role in the recruiting of DNA-PKcs. By superimposing DNA-PK and DNA-PKcs-KU80CTR structures [3, 15], we can determine the distance between the KU core and KU80CTR (Fig. 4D). When KU is bound to DNA-PKcs, the KU80CTR region needs to be far more extended from the KU core (~60Å) than in the free state (Fig. 5). Additionally, the C-terminal interaction motif of KU80CTR [75], which is predicted to have helical propensity, is even more distant (~80Å) from the KU core [3]. Thus, the KU80CTR domain including the KU80CTR C-terminus needs to undergo a large displacement during KU interaction with DNA-PKcs. Such a dramatic rearrangement is enabled by the length of the KU80CTR linker (~60 residues). To initiate complexation, the flexibly tethered KU80CTR C-terminus helix needs to find the binding site near the “PQR” autophosphorylation cluster [3, 76]. This initial KU-tethering is followed by recruitment of the KU core to the N-HEAT binding site, allowing insertion of the DNA end into the M- / N-HEAT aperture (Fig. 5). The relatively compact arrangement of KU80CTR in the absence and presence of DNA suggests that this KU80CTR rearrangement is initiated by interaction between the KU80CTR C-terminus and the M-HEAT domain [3] rather than by DNA binding (Fig. 5).
Fig. 5.
DNA-PK functional flexibility as cartoons. i) Minimal flexibility of KU80CTR and extension of its C-terminus. ii) Flexibility of HEAT domain relatively to the DNA-PKcs head region. iii) N and M-HEAT rearrangement upon the DNA-PKcs autophosphorylation. iv) Extension of the KU80 “arm” during initial DNA-PK assembly. v) Stabilization of the DNA-PK assembly by multiple protein-protein and protein-DNA interactions. vi) Proposed DNA-PK dimer arrangement.
Alone, DNA-PKcs interacts with DNA with low affinities [58]. However, its ability to assemble on various ends of the DNA structure can lead to formation of dumbbell arrangement with two DNA-PKcs binding DNA ends [27]. This DNA-PKcs dimer arrangement is evidently not physiologically relevant, because in vivo DNA damage induced DSBs would each have only one exposed DNA end. A tighter interaction between DNA-PKcs and DNA is supported by KU (Fig. 5) [15, 27, 56]. DNA-PKcs and KU together form a DNA-binding tunnel where DNA duplex fills the aperture between the N-HEAT and the M-HEAT region of DNA-PKcs [15]. We find that in solution the N-HEAT region undergoes large motions in free DNA-PKcs (Fig. 5), in agreement with the conformational variability between reported DNA-PKcs structures [1–3] (Fig. 2C), as well between two DNA-PKcs chains from the crystal structure [3]. Larger movement of N-HEAT and M-HEAT regions in solution (Fig. 2D) correlate with much smaller changes in conformation and position of the HEAT regions observed in the crystal and cryo-EM structure. In the DNA-PK structure [15] the N-HEAT domain is displaced by ~35Å inwards towards the FAT domain upon binding with KU-DNA (Fig. 2C). Our solution-based modeling shows similar movement of the N-HEAT domain in the absence of KU-DNA (Fig. 2D), which supports the importance of inherent flexibility in this domain upon DNA-PK activation. Conformational adaptability of the N-HEAT region, bending of KU70 [15], and bridging of the KU80CTR C-terminus with the M-HEAT domain all contribute to a lock-in mechanism that stabilizes the formation of the DNA-PK assembly (Fig. 5). Such a mechanism would ensure that assembly of DNA-PK controls a molecular switch needed to transfer the activation signal from the KU80CTR binding site to the kinase located in the head region ~100Å away (Fig. 5).
Formation of a small but significant population of dimeric DNA-PK in solution (Fig. 4B) suggests that self-association of DNA-PKcs [27] (Fig. 4E) may support the dimerization of the DNA-PK complex. Here modeled head-to-head arrangement of the DNA-PKcs dimer resembles the arrangement of the closely related ATM kinase dimer [77]. This arrangement raises the question of whether the FRB dimerization controls the autophosphorylation, which activates the release of DNA-PKcs from the NHEJ presynaptic complex [43, 54, 78].
The main rearrangement of DNA-PKcs during autophosphorylation shows widening of the M-HEAT region (Fig. 2D) that allows the N-terminal domain (1–370) of the N-HEAT to slide in to the cradle formed by wider M-HEAT region (Fig. 5). We hypothesize that this mechanism may conceal the binding site for KU70 (Fig. 5) and trigger release of DNA-PKcs from the DNA ends.
Autophosphorylation-induced domain rearrangement furthermore has the potential to affect the interaction of DNA-PKcs with accessory proteins, which may further regulate NHEJ in vivo. These collective results and ideas support and extend our previous structural understanding of DNA-PK activation and flexibility. Specifically, integrating SAXS measurements with the atomistic modeling utilizing crystal and cryo-EM structures provides data-based dynamic models that suggest how KU and DNA-PKcs combine and assemble enzymatically to promote structural and catalytic activities for NHEJ initiation and the choreography of DSB repair. Going forward will be interesting to see how RNA may regulate or active these DNA-PK complexes [79]. Furthermore, in this and other efforts to combine advanced cryo-EM with SAXS, the use of gold-label SAXS may provide a means to reduce SAXS samples needs substantially [80].
5. Synopsis and perspective
NHEJ mechanisms must be fast, flexible and accurate while accommodating alignment components and active ligase conformations along with two DNA ends. These features cannot be conveyed by a single structure. However single structures do provide a starting point for analyzing solution based information that typically contains averaged or single distance information like FRET or SAXS. Here we describe our analysis of SAXS data on the central DNA-PK assembly and its components DNA-PKcs, and KU in the context of new cryo-EM and crystallographic structures. While the static cryo-EM or crystallographic structures alone cannot decipher functional dynamicity at the observed atomic resolution, by adding a solution-based atomistic modeling we are able to visualize conformational switches likely to be required for assembly and disassembly of DNA-PK complex. In general, these combined SAXS and atomic structural analyses on this biologically and medically important and exemplary DNA repair complex provide and test an enabling approach for robust data-based models of functional mechanisms in dynamic macromolecular complexes with relevance to cancer biology and therapeutics.
Supplementary Material
Acknowledgements
This work was supported by NIH grants P01 CA092584 and R35 CA220430. J.A.T is further supported by the Cancer Prevention and Research Institute of Texas and a Robert A. Welch Chemistry Chair. SAXS data collection at SIBYLS is funded through DOE BER Integrated Diffraction Analysis Technologies (IDAT) program and NIGMS grant P30 GM124169-01, ALS-ENABLE.
Abbreviations
- ATM
ataxia telangiectasia mutated
- cryo-EM
cryo-electron microscopy
- ds
double-stranded
- DSB
DNA double strand break
- DSBR
DNA double strand break repair
- DNA-PK
DNA-activated/dependent protein kinase
- DNA-PKcs
DNA-PK catalytic subunit
- FAT
FRAP, ATM and TRRAP
- FRB
FKBP12-rapamycin-binding domains
- HEAT
Huntingtin, Elongation factor, PP2A-A subunit, TOR
- KU
KU70/KU80 heterodimer
- KU80CTR
KU80 C-terminal region
- MX
macromolecular X-ray crystal
- NHEJ
non-homologous end joining
- N-HEAT
N-terminal HEAT domain
- M-HEAT
central HEAT domain
- NMA
normal mode analysis
- MALS
multi-angle light scattering
- PI3K
phosphatidyl inositol 3 kinase
- SAP
SAF-A/B, Acinus and PIAS
- SAXS
small-angle X-ray scattering
- SEC
size exclusion chromatography
- SEC-SAXS-MALS
size exclusion chromatography coupled with in-line SAXS and multi-angle light scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sharif H, Li Y, Dong Y, Dong L, Wang WL, Mao Y, et al. Cryo-EM structure of the DNA-PK holoenzyme. Proc Natl Acad Sci U S A. 2017;114:7367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baretic D, Maia de Oliveira T, Niess M, Wan P, Pollard H, Johnson CM, et al. Structural insights into the critical DNA damage sensors DNA-PKcs, ATM and ATR. Progress in biophysics and molecular biology. 2019;147:4–16. [DOI] [PubMed] [Google Scholar]
- [3].Sibanda BL, Chirgadze DY, Ascher DB, Blundell TL. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science. 2017;355:520–4. [DOI] [PubMed] [Google Scholar]
- [4].Eckelmann BJ, Bacolla A, Wang H, Ye Z, Guerrero EN, Jiang W, et al. XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cells. NAR Cancer. 2020;2:zcaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dutta A, Eckelmann B, Adhikari S, Ahmed KM, Sengupta S, Pandey A, et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic acids research. 2017;45:2585–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Progress in biophysics and molecular biology. 2015;117:182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Syed A, Tainer JA. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu Rev Biochem. 2018;87:263–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst). 2014;17:110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Molecular cell. 2014;55:829–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bacolla A, Tainer JA, Vasquez KM, Cooper DN. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic acids research. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Spagnolo L, Barbeau J, Curtin NJ, Morris EP, Pearl LH. Visualization of a DNA-PK/PARP1 complex. Nucleic acids research. 2012;40:4168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. [DOI] [PubMed] [Google Scholar]
- [13].Houl JH, Ye Z, Brosey CA, Balapiti-Modarage LPF, Namjoshi S, Bacolla A, et al. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nature communications. 2019;10:5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. [DOI] [PubMed] [Google Scholar]
- [15].Yin X, Liu M, Tian Y, Wang J, Xu Y. Cryo-EM structure of human DNA-PK holoenzyme. Cell research. 2017;27:1341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hammel M Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS). Eur Biophys J. 2012;41:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quarterly reviews of biophysics. 2007;40:191–285. [DOI] [PubMed] [Google Scholar]
- [18].Brosey CA, Tainer JA. Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Current opinion in structural biology. 2019;58:197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, et al. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nature methods. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schneidman-Duhovny D, Hammel M. Modeling Structure and Dynamics of Protein Complexes with SAXS Profiles. Methods Mol Biol. 2018;1764:449–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic acids research. 2016;44:W424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL 2nd, Tsutakawa SE, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nature methods. 2009;6:606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Knott GJ, Cress BF, Liu JJ, Thornton BW, Lew RJ, Al-Shayeb B, et al. Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, et al. Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou Y, Millott R, Kim HJ, Peng S, Edwards RA, Skene-Arnold T, et al. Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis. Structure. 2019;27:1485–96 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular cell. 2014;53:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hammel M, Yu Y, Radhakrishnan SK, Chokshi C, Tsai MS, Matsumoto Y, et al. An Intrinsically Disordered APLF Links Ku, DNA-PKcs and XRCC4-DNA Ligase IV in an Extended Flexible Non-Homologous End Joining Complex. J Biol Chem. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Panjkovich A, Svergun DI. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys Chem Chem Phys. 2016;18:5707–19. [DOI] [PubMed] [Google Scholar]
- [32].Kashammer L, Saathoff JH, Lammens K, Gut F, Bartho J, Alt A, et al. Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Molecular cell. 2019;76:382–94 e6. [DOI] [PubMed] [Google Scholar]
- [33].Goodarzi AA, Lees-Miller SP. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair (Amst). 2004;3:753–67. [DOI] [PubMed] [Google Scholar]
- [34].Chan DW, Mody CH, Ting NS, Lees-Miller SP. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem Cell Biol. 1996;74:67–73. [DOI] [PubMed] [Google Scholar]
- [35].Radhakrishnan SK, Lees-Miller SP. DNA requirements for interaction of the C-terminal region of Ku80 with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). DNA repair. 2017;57:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J Appl Crystallogr. 2013;46:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dyer KN, Hammel M, Rambo RP, Tsutakawa SE, Rodic I, Classen S, et al. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol Biol. 2014;1091:245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu Rev Biophys. 2013;42:415–41. [DOI] [PubMed] [Google Scholar]
- [40].Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic acids research. 2010;38:W540–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J. 2013;105:962–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, et al. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. The Biochemical journal. 2002;368:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase Is required for efficient end processing during DNA double-strand break repair. Molecular and cellular biology. 2003;23:5836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. [DOI] [PubMed] [Google Scholar]
- [45].Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic acids research. 2005;33:W363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO reports. 2007;8:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Andrews BJ, Lehman JA, Turchi JJ. Kinetic analysis of the Ku-DNA binding activity reveals a redox-dependent alteration in protein structure that stimulates dissociation of the Ku-DNA complex. J Biol Chem. 2006;281:13596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–56. [DOI] [PubMed] [Google Scholar]
- [49].Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM Structure of the DNA-Dependent Protein Kinase Catalytic Subunit at Subnanometer Resolution Reveals alpha Helices and Insight into DNA Binding. Structure. 2008;16:468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure. 2005;13:243–55. [DOI] [PubMed] [Google Scholar]
- [51].Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lees-Miller JP, Cobban A, Katsonis P, Bacolla A, Tsutakawa SE, Hammel M, et al. Uncovering DNA-PKcs ancient phylogeny, unique sequence motifs and insights for human disease. Progress in biophysics and molecular biology. 2020;this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, et al. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Molecular and cellular biology. 2007;27:1581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, et al. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic acids research. 2004;32:4351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. The Journal of cell biology. 2007;177:219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Progress in biophysics and molecular biology. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Molecular cell. 2006;22:511–9. [DOI] [PubMed] [Google Scholar]
- [58].Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic acids research. 1999;27:4679–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Boskovic J, Rivera-Calzada A, Maman JD, Chacon P, Willison KR, Pearl LH, et al. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. The EMBO journal. 2003;22:5875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rambo RP, Tainer JA. Super-Resolution in Solution X-Ray Scattering and Its Applications to Structural Systems Biology. Annu Rev Biophys. 2013. [DOI] [PubMed] [Google Scholar]
- [62].Yano K, Chen DJ. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell cycle (Georgetown, Tex. 2008;7:1321–5. [DOI] [PubMed] [Google Scholar]
- [63].Cottarel J, Frit P, Bombarde O, Salles B, Negrel A, Bernard S, et al. A noncatalytic function of the ligation complex during nonhomologous end joining. The Journal of cell biology. 2013;200:173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang C, Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: new developments in nonhomologous end joining. International journal of radiation oncology, biology, physics. 2013;86:440–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, et al. XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair. J Biol Chem. 2011;286:32638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF regulates filament architecture of the XRCC4.ligase IV complex. Structure. 2010;18:1431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wang JL, Duboc C, Wu Q, Ochi T, Liang S, Tsutakawa SE, et al. Dissection of DNA double-strand-break repair using novel single-molecule forceps. Nat Struct Mol Biol. 2018;25:482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc Natl Acad Sci U S A. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tsutakawa SE, Sarker AH, Ng C, Arvai AS, Shin DS, Shih B, et al. Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations. Proc Natl Acad Sci U S A. 2020;117:14127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tsutakawa SE, Thompson MJ, Arvai AS, Neil AJ, Shaw SJ, Algasaier SI, et al. Phosphate steering by Flap Endonuclease 1 promotes 5’-flap specificity and incision to prevent genome instability. Nature communications. 2017;8:15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rashid F, Harris PD, Zaher MS, Sobhy MA, Joudeh LI, Yan C, et al. Single-molecule FRET unveils induced-fit mechanism for substrate selectivity in flap endonuclease 1. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Paillard S, Strauss F. Site-specific proteolytic cleavage of Ku protein bound to DNA. Proteins. 1993;15:330–7. [DOI] [PubMed] [Google Scholar]
- [73].Mo X, Dynan WS. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Molecular and cellular biology. 2002;22:8088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Harris R, Esposito D, Sankar A, Maman JD, Hinks JA, Pearl LH, et al. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR). Journal of molecular biology. 2004;335:573–82. [DOI] [PubMed] [Google Scholar]
- [75].Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic acids research. 1999;27:3494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–15. [DOI] [PubMed] [Google Scholar]
- [77].Baretic D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, et al. Structures of closed and open conformations of dimeric human ATM. Sci Adv. 2017;3:e1700933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Molecular and cellular biology. 2005;25:10842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lees-Miller SP, Beattie TL, Tainer JA. Noncoding RNA joins Ku and DNA-PKcs for DNA-break resistance in breast cancer. Nat Struct Mol Biol. 2016;23:509–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hura GL, Tsai CL, Claridge SA, Mendillo ML, Smith JM, Williams GJ, et al. DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci U S A. 2013;110:17308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.