Abstract
Motivation
In proteomics, database search programs are routinely used for peptide identification from tandem mass spectrometry data. However, many low-quality spectra cannot be interpreted by any programs. Meanwhile, certain high-quality spectra may not be identified due to incompleteness of the database or failure of the software. Thus, spectrum quality (SPEQ) assessment tools are helpful programs that can eliminate poor-quality spectra before the database search and highlight the high-quality spectra that are not identified in the initial search. These spectra may be valuable candidates for further analyses.
Results
We propose SPEQ: a spectrum quality assessment tool that uses a deep neural network to classify spectra into high-quality, which are worthy candidates for interpretation, and low-quality, which lack sufficient information for identification. SPEQ was compared with a few other prediction models and demonstrated improved prediction accuracy.
Availability and implementation
Source code and scripts are freely available at github.com/sor8sh/SPEQ, implemented in Python.
1 Introduction
Tandem mass spectrometry (MS/MS) is the main approach used for protein identification in proteomics studies (Hernandez et al., 2006). In this technique, high-performance liquid chromatography is coupled with tandem MS to identify peptides from complex mixtures of proteins (Aebersold and Mann, 2003). First, proteins are digested into smaller peptides using enzymes, such as Trypsin. The resulting peptides are then subject to liquid chromatography tandem mass spectrometry ( LC–MS/MS) analysis. Each of these experiments may produce thousands of MS/MS spectra, each supposedly corresponding to a peptide. A database search engine, such as MS-GF+ (Kim and Pevzner, 2014), Comet (Eng et al., 2013), MaxQuant (Cox and Mann, 2008), PEAKS DB (Zhang et al., 2012) or Mascot (Perkins et al., 1999) is usually used to identify the peptide for each spectrum. Here, experimental spectra produced using the mass spectrometer are compared with theoretical spectra predicted from peptides in a protein database. Other approaches can be used to interpret the experimentally produced spectra, such as de novo sequencing, where identification is made without a database and instead by interpreting the peaks of an experimental spectrum. Several programs, such as PEAKS (Ma et al., 2003), Novor (Ma, 2015), PepNovo (Frank and Pevzner, 2005) and pNovo 3 (Yang et al., 2019), have been developed for de novo sequencing.
The aforementioned database search analysis inevitably causes false-positive and false-negative identifications. With much research, the false positives can now be reliably controlled by the false-discovery rate (FDR). Usually, a target-decoy method (Elias and Gygi, 2007; Moosa et al., 2020) is used to establish a score threshold. Only the peptide-spectrum matches (PSMs) with scores above the threshold are reported by the analysis. An FDR of 1% is often used to ensure that there are (on average) at most 1% false identifications in the reported PSMs.
However, the false negatives of the database search are rarely studied. Typically, 50% or more of the MS/MS spectra cannot be confidently identified by database search. Many of these unidentified spectra are due to poor spectrum quality, e.g. the spectrum does not contain enough information for any meaningful interpretation. But an unknown portion of these unidentified spectra are actually high-quality spectra. Their missing is solely because of the limitation of the data analysis, such as inadequate software, sub-optimal search parameters, unspecified post-translational modifications (PTMs) and incomplete sequence databases (Nesvizhskii et al., 2006). These spectra can be regarded as the false negatives of the data analysis. A SPEQ score that labels the high-quality spectra would be useful in dealing with these false-negative spectra, as elaborated in the following.
First, the amount of the unidentified high-quality spectra may potentially provide a proxy to estimate the level of false negatives. If proved to be true by future research, this approach can be attractive as there is no established method in proteomics to estimate the false-negative rate of a data analysis. Certain statistical approaches such as PeptideProphet (Keller et al., 2002) may be used to estimate the false negatives caused by the correctly assigned but low-scoring PSMs. However, the ones caused by the unassigned or wrongly assigned spectra remain unknown. Second, a small portion (such as 1%) of the uninterpreted spectra with the highest-quality score can be analyzed by a more time-consuming method or by a human expert manually to troubleshoot for the reasons for their missing. The identified reasons can be used by a data analyst to adjust the search strategy, by a tool developer to improve the software or by a lab scientist to improve the MS experiment. A similar idea called Preview was described earlier by Kil et al. (2011), where a subset of spectra are searched to determine the best search parameters before the full search. The SPEQ score would be helpful here to select the subset. Third, when the quality score function is sufficiently developed to have a nearly perfect accuracy, one can safely discard the low-quality spectra at the very beginning of the data analysis. This should improve both the speed of the downstream data analysis, and reduce the false positives caused by the low-quality spectra.
In recent years, several related quality assessment tools have been developed. Bern et al. (2004) used a set of handcrafted features and a support vector machine to conduct spectral quality assessments. Salmi et al. (2006) combined previous work with more handcrafted features and used a decision tree and random forest to conduct classification. Meanwhile, Flikka et al. (2006) utilized multiple machine learning classifiers to classify spectra based on 17 manually extracted features. Similarly, Nesvizhskii et al. (2006) used a linear discriminant function to combine 15 scoring features selected by a human expert into one discriminant score. Na and Paek (2006) proposed a new score function based on Cumulative Intensity Normalization to filter spectra based on their score. To describe the quality of tandem mass spectra, Wu et al. (2008) first proposed a method that mapped each tandem spectrum into a feature vector before using fisher linear discriminant analysis to construct the classifier. More recently, Ma et al. (2011) evaluated spectral quality via sequence tagging. Handcrafted features played a primary role in these studies, which subsequently combined them with other classification methods. Thus, the challenge was to find the most optimal set of spectral features that could separate as many spectra containing useful information from noisy spectra as possible. Moreover, none of these software tools have been actively maintained, causing them either not anymore available or fail to work on today’s computer systems or for the data produced with today’s mass spectrometers.
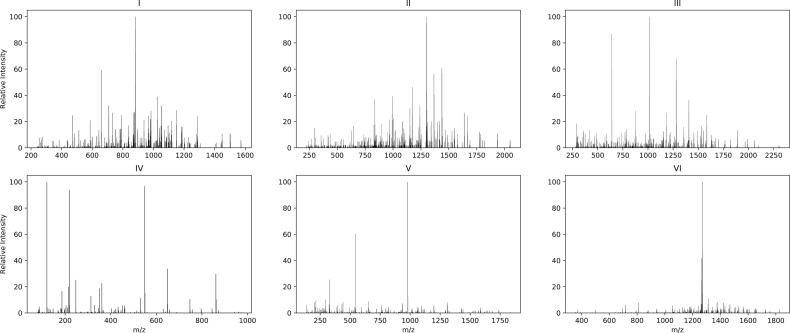
Figure 1 illustrates how the spectra of different qualities can be classified. These scans are manually selected as examples to highlight some of the differences between low- and high-quality spectra but may not represent all the differences. In these examples, if a significant peak was defined as a peak with a relative intensity of around 5% or higher, as seen in Figure 1, the high-quality spectra consisted of many peaks, and the number of significant peaks in that spectrum was relatively high compared with the low-quality spectrum. Moreover, the m/z differences between significant peaks in a high-quality spectrum encode meaningful information in terms of the mass of different amino acid residues. In contrast, the low-quality spectrum had fewer peaks, and the significant peaks were sparse. These observable differences can be combined with other proteomic features to build a program for spectrum qualification. However, handcrafting all these features is tedious, particularly because different types of mass spectrometers may require different features.
Fig. 1.
The first row (I, II and III) shows three scans that were assigned confidently by MS-GF+ (1% FDR). In the second row (IV, V and VI), three scans that were not assigned confidently by MS-GF+ are shown. Scans in the first row are considered high-quality samples, while the bottom row contains low-quality samples
In this manuscript, we present SPEQ, a new method to predict the SPEQ by using a deep neural network (DNN) model. In comparison to earlier machine learning-based methods, DNN does not require the features to be handcrafted. Instead, the input of the new model is just the whole spectrum, and the model’s training will automatically extract beneficial features from the training data. Therefore, in contrast to previous related works, the extracted features differ for each type of dataset and are related to specific characteristics, such as the instrument or experimental methods used to generate the dataset. In addition, as one of the main characteristics of the deep learning method, SPEQ benefits from the availability of large proteome training datasets and offers better performance with relatively less development time compared with other methods. SPEQ’s performance and usefulness were tested in different scenarios to demonstrate that:
SPEQ has a better prediction accuracy than the other models tested.
Most of the high-quality but unidentified spectra have a confident de novo sequence tag, suggesting they are indeed spectra of peptides.
A small portion of the unidentified spectra with the highest-quality score can be examined to troubleshoot a data analysis, leading to the identification of more spectra.
2 Materials and methods
In this section, we describe the datasets that are used in our experiments, including the data preprocessing steps and the testing procedure. Details about the implemented neural network used in SPEQ are also presented.
2.1 Testing data
We used four datasets generated using different high-resolution instruments in the present study.
2.1.1 Quadrupole time-of-flight dataset
This dataset was previously used by Flikka et al. (2006) for the development of their SpectrumQuality tool, and was downloaded from http://services.cbu.uib.no/software/spectrumquality. This dataset (quadrupole time-of-flight (Q-TOF) N-terminal in the original paper) contains 10 055 MS/MS spectra from extreme amino-terminal peptides of proteins, measured with a Q-TOF mass spectrometer. Mascot was used to search the human proteins IPI database and IPI-derived N-terminally truncated databases for the peptide identification. Spectra with a score equal to or above the Mascot identity threshold, when the confidence level was set to 95%, were considered as ‘good’, while all other spectra were labeled ‘bad’. The same SPEQ labels made by Flikka et al. (2006) were kept and used in the present article. This gives 1683 positive and 8372 negative spectra in this dataset.
2.1.2 Orbitrap human dataset
This proteome dataset was first provided by Bruderer et al. (2017) and is available with identifier PXD005573 (Fig4_HeLa-1m_DDA_R01_T0.raw) at the ProteomeXchange repository. It is a data-dependent acquisition dataset obtained on a Q Exactive HF instrument using a HeLa lysate. ProteoWizard (Chambers et al., 2012) was used to centroid each profile spectrum before converting the dataset from a raw mass spectrometer output file to an XML file.
Two different database search programs, MS-GF+ and Comet, were used to search the UniProt Homo sapiens proteome (UP000005640) for the peptide identification. The search parameters are the following: precursor tolerance = 20 ppm; fixed PTM = Carbamidomethyl on C; variable PTM = Oxidation on M, Acetylation at protein N-term and Deamidation on N and Q; enzyme = Semi-tryptic. A decoy database generated with the de-Bruijn method (Moosa et al., 2020) was also searched together to determine the FDR. A spectrum is considered high quality if a database peptide can be identified with <1% FDR by either MS-GF+ or Comet. This gives 95 646 positive and 123 702 negative spectra in this dataset.
2.1.3 NIST dataset
This dataset consists of a Homo sapiens hair peptide spectral library and was downloaded from https://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:cdownload. It is created from data generated by an Orbitrap Fusion Lumos instrument. A total of 6280 spectra with 2240 unique peptide sequences are available in this dataset. Using MS-GF+, these spectra were then searched against the FASTA file provided along with the spectral library (with 20 183 proteins in it). The search parameters are the same as used in the Orbitrap human dataset. Spectra with a peptide identification with <1% FDR were labeled as high quality. This gives 3932 positive and 2348 negative spectra in this dataset.
2.1.4 Orbitrap mouse dataset
The last dataset used in this study was first published by McDonagh et al. (2014) and is available with identifier PXD001054 (BMD_2013_07_31_Gastro_Aged_2.mgf) at the ProteomeXchange repository. Similar to the second dataset, this dataset is also obtained on a Q Exactive instrument. However, the proteins in this dataset are from the Mus musculus (Mouse) species. The dataset has 22 686 spectra, which were searched against the UniProt Mus musculus proteome (UP000000589) with MS-GF+. The search parameters are the same as used in the Orbitrap human data, except that the precursor tolerance is set to 10 ppm to match the original publication (McDonagh et al., 2014). Spectra with a peptide identification with <1% FDR were labeled as high quality. This gives 3648 positive and 19 038 negative spectra in this dataset.
2.2 SPEQ method
2.2.1 Vector representation
As an input, SPEQ takes a spectrum in Mascot Generic Format (MGF). In an MGF file, for each spectrum, the peak list is provided as tuples, where a peak is an (m/z value, raw intensity) tuple. The intensity is first converted to a value between 0 and 100 by normalizing against the intensity of the most abundant peak in the spectrum. Then we transform each peak list into a vector by binning the m/z range using a 1.000507 m/z as the bin width and a 0.4 m/z as the offset. Each dimension of the vector corresponds to the maximum relative peak intensity of all the peaks in the same m/z bin. If a bin has no peak in it, then the dimension of the vector is set to 0. A similar procedure was used in Comet (Eng et al., 2015) to convert the spectrum to a vector representation. By choosing this data representation, we could induce the m/z distance between peaks of a spectrum to the model, which is related to the quality of the spectrum. In addition, we also include the precursor’s charge state and m/z as input of the model.
2.2.2 Neural network
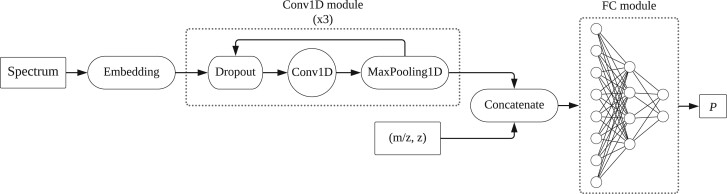
A DNN was used to predict the quality of a spectrum from its vector representation. The implementation was conducted in Python 3.7 (https://www.python.org). We used the Pyteomics toolkit (Goloborodko et al., 2013) to read the MGF files and TensorFlow (Abadi et al., 2015) to build the SPEQ model. As shown in Figure 2, the model used in SPEQ consisted of a one-dimensional convolutional module for automatic feature extraction. This was connected to a fully connected neural network that was used to conduct the classification.
Fig. 2.
The architecture of the DNN model. A feature extraction part using three Conv1D modules is connected to a three-layer FC module. A Conv1D is a 1D convolutional layer, and an FC is a fully connected network. The inputs of the model are the vector representation of the MS/MS spectrum, and the mass to charge ratio (m/z) and charge state (z) of the precursor ion
The first part contained an embedding layer on top of three convolutional blocks, where each block had a dropout layer connected to a 1D convolutional layer and a 1D MaxPooling layer at the end. This part was designed to process the vector representation and extract features from the peaks of the input spectrum. The parameters of each convolutional layer (first layer: kernel size 11 with strides 5, second layer: kernel size 51 with strides 10, third layer: kernel size 3 with strides 1) were selected so that the important information within a spectrum was best induced to the model. In the next part, the output of the first part was concatenated to the two additional features: the precursor’s charge state and m/z. The obtained vector was fed to a three-layer fully connected neural network. After applying a sigmoid activation function on the last layer, we obtained P, which is the probability of a spectrum being high quality; it can also be interpreted as the quality score for the input spectrum. After applying a threshold on this probability, the output was assigned a label of ‘good’ or ‘bad’ for the input spectrum. The model was compiled with the Adam optimizer (Kingma and Ba, 2014), and was trained using a binary cross-entropy loss function.
2.3 Other methods tested
Three other methods, a baseline model, SpectrumQuality and Bern’s model were tested together with the SPEQ method. The baseline model simply uses the number of peaks in the spectrum as the quality score. SpectrumQuality is the method published by Flikka et al. (2006) and the software was downloaded from http://services.cbu.uib.no/software/spectrumquality.
The Bern’s model was previously published by Bern et al. (2004). In this model, each input is a histogram of m/z differences from a spectrum. The histogram is a 187-length vector, where an element i is the bin of m/z difference of [] (187 is the maximum mass of an amino acid residue). As mentioned by Bern et al. (2004), due to the time complexity of the algorithm used in the proposed data representation, this method requires significant training and testing time. The software that uses the Bern’s model has not been published by the author and is not available for use. Therefore, the results presented here are derived from our implementation based on the model described by Bern et al. (2004). To obtain a score for each sample, we used the Epsilon-Support Vector Regression (SVR) package available in scikit-learn (Pedregosa et al., 2011), with radial basis functions as the kernel. In the SVR model, the width parameter, γ, must be set. This value was set to 500 in the study by Bern et al. (2004). However, this led to very low classification performance in our experiments (area under the receiver operating characteristic (ROC) curve around 50%). Instead, γ was set to ‘auto’ first. A linear search was also performed to find the best γ. These two approaches gave a very similar performance. The better performance of the two was used in each comparison.
2.4 Testing procedure
SPEQ’s performance was evaluated in three different aspects. First, the prediction accuracy of SPEQ and other models was compared. A 5-fold cross-validation was used to measure the prediction accuracy. More specifically, the dataset was first divided into five parts . Following this, each part (Di) was used once as the test set, while the four other partitions () were combined and used as the training set. This process was conducted five times, and the final reported performance of the model was the average of these five processes. The dividing procedure ensured that spectra with the same m/z and z were in the same part. This way, repeated scans of the same precursor were not simultaneously present in the training and testing set. Moreover, a cross-species validation was also performed, where the models were trained on the Orbitrap human dataset and tested on the independent Orbitrap mouse dataset. The results of this accuracy test are provided in Section 3.1.
Second, the unidentified spectra from a database search analysis were subject to additional analyses to check whether they can be interpreted by other analytical methods. In this test, the Orbitrap human dataset was first searched with MS-GF+ with the parameters provided in Section 2.1.2. Using the labels generated from the MS-GF+ search, we trained the SPEQ model and used the model to assign a quality score (SPEQ score) to all the spectra. The unidentified spectra were further searched with the Comet software with the same parameters. The spectra unidentified by the first two analyses were de novo sequenced with the Novor software (Ma, 2015). A de novo sequence that contains at least five amino acids with a high confidence score (>70) is regarded as a confident de novo tag. It is expected that a higher SPEQ score in the spectra unidentified by the first search is associated with a higher percentage of spectra assigned by either Comet or de novo sequencing. The results of this test are provided in Section 3.2.
The third test is to demonstrate the usefulness of the SPEQ score in ‘troubleshooting’ a data analysis. In this test, the Orbitrap human dataset was first searched with MS-GF+ with the parameters provided in Section 2.1.2. It was suspected that many of the unidentified spectra were because they contain a PTM unspecified in the search parameter. However, searching with too many variable PTMs on the whole dataset is prohibitively slow. To troubleshoot, SPEQ was used to score all the unidentified spectra. The top 1% of these unidentified spectra according to the SPEQ score were selected to conduct a search with many additional variable PTMs. From the identified peptides, a few most common PTMs were selected. Then a third-round search was conducted to identify more peptides using all of the unidentified spectra and these few additionally selected variable PTMs. The results of this ‘troubleshooting’ test are provided in Section 3.3.
3 Results
3.1 Prediction accuracy
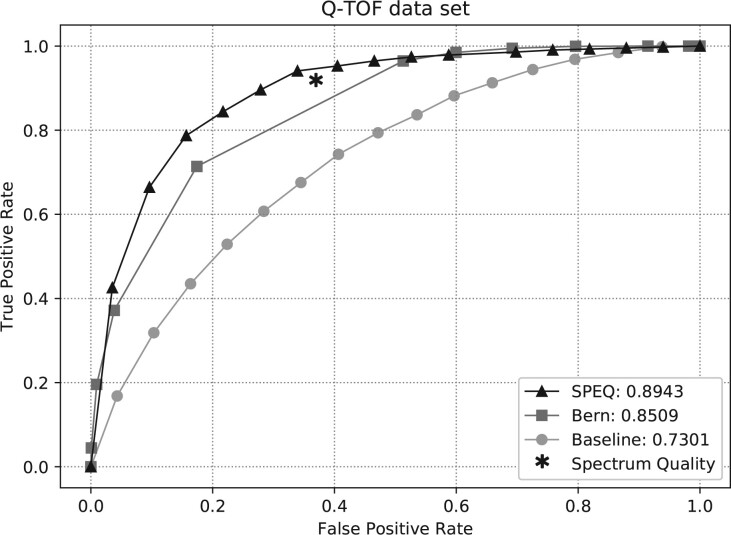
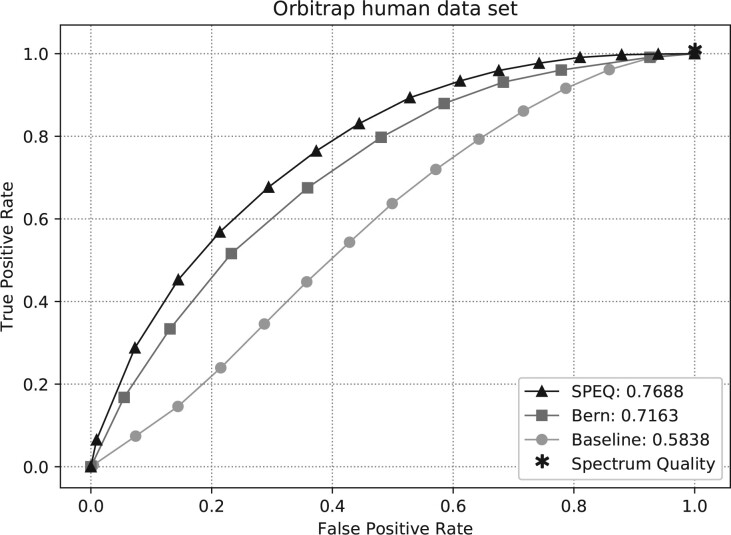
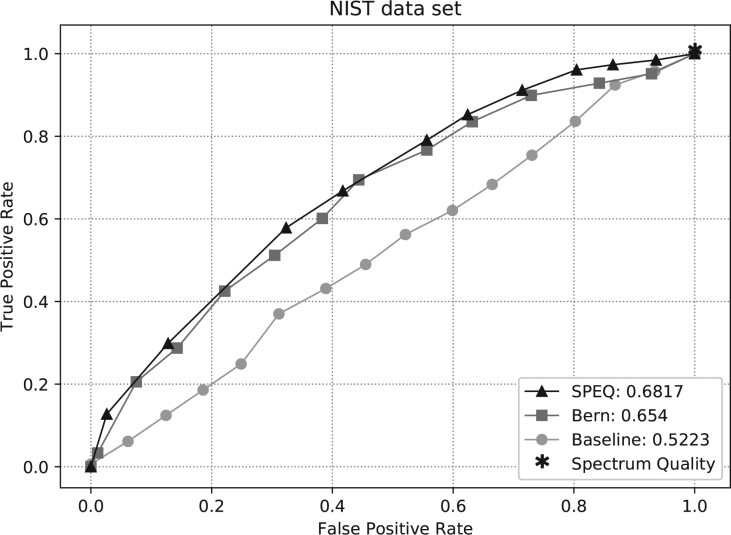
Figures 3–5 show the ROC curves of the predictions made by each model on the Q-TOF, Orbitrap human and NIST datasets, respectively. The area under curve (AUC) of the ROC curve for each method is also provided in the figures. The figures clearly show that SPEQ’s prediction accuracy outperforms all other tools in all datasets. Note that, SpectrumQuality’s curve has only one data point. This is because it does not output a quality score, but only classifies the spectrum into two classes. Also, SpectrumQuality failed to make any valid prediction on the Orbitrap human dataset (Fig. 4) and the NIST dataset (Fig. 5). Instead, it assigned high quality to every spectrum. This is likely because both datasets were obtained from Orbitrap instruments, while the model was developed based on data from the Q-TOF instruments.
Fig. 3.
ROC curves and AUC of different tools on the Q-TOf dataset
Fig. 4.
ROC curves and AUC of different tools on the Orbitrap human dataset
Fig. 5.
ROC curves and AUC of different tools on the NIST dataset
All models’ performances were low for the NIST dataset. This is likely because the spectra in the NIST dataset have already been selected when they were collected in the spectrum library, and lack extremely low-quality spectra. It is a harder task to distinguish between the high and medium-quality spectra than between the high and low-quality spectra.
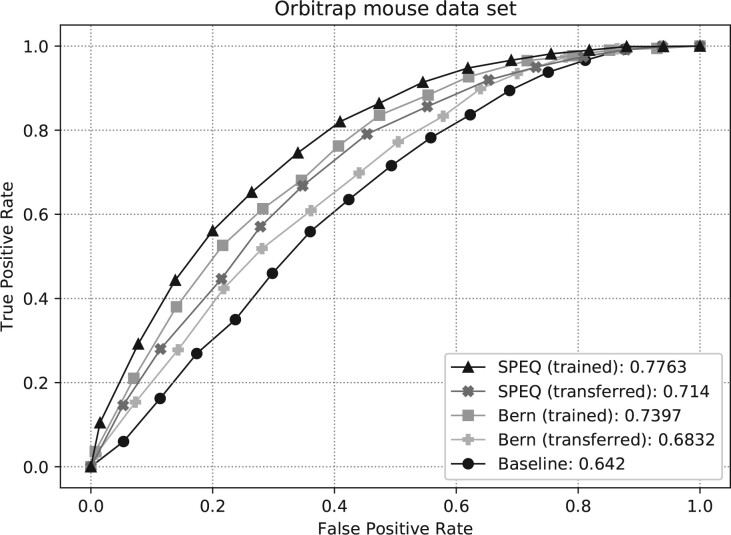
Figure 6 shows the ROC curves and their AUC of different methods on the Orbitrap mouse dataset. The SpectrumQuality tool failed to make any valid prediction here and is not included in the figure. To produce the curves, the SPEQ and Bern’s methods were either trained on the same dataset and tested with a 5-fold cross-validation, or trained on the Orbitrap human dataset. Not surprisingly, for both models, the cross-validation test on the same dataset produced better accuracy than training on a different dataset. Nevertheless, SPEQ achieved a decent prediction accuracy when training and testing were performed on different datasets. Also, SPEQ’s performance is better than both the Bern’s method and the baseline method in both testing scenarios.
Fig. 6.
ROC curves and AUC of different methods on the Orbitrap mouse dataset. The SPEQ (trained) and Bern (trained) curves were obtained when the models were trained with the same Orbitrap mouse dataset and tested with 5-fold cross-validation. The SPEC (transferred) and Bern (transferred) curves were obtained when the models were trained with the Orbitrap human dataset and tested with the Orbitrap mouse dataset
3.2 Unidentified high-quality spectra
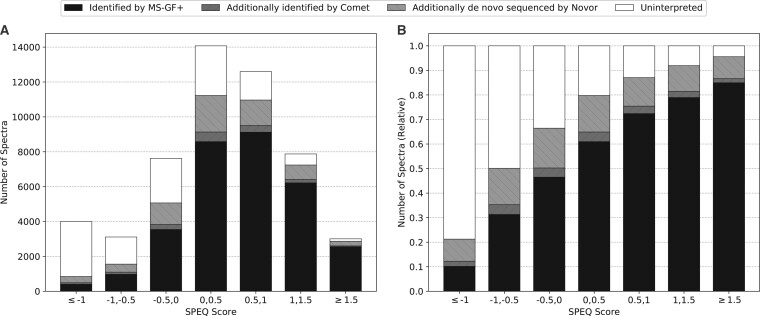
Figure 7 shows the results of the test for the unidentified spectra (as described in Section 2.4). After searching the 52 285 spectra in the Orbitrap human dataset using MS-GF+, 20 885 spectra failed to be confidently identified. These unidentified spectra were subject to a second database search with Comet and de novo sequencing with Novor. The histogram of Figure 7A shows the number of spectra in each SPEQ score interval, and the distribution of the four categories of spectra:
Fig. 7.
(A) The number of identified and unidentified spectra in different intervals of the quality score assigned by SPEQ. (B) The relative ratio of identified and unidentified spectra in different intervals of the SPEQ score. The SPEQ score used in this figure is the logit of the probability predicted by SPEQ. The black, dark gray and light gray bars represent the spectra identified by MS-GF+, Comet but not MS-GF+, and Novor but not the other two tools, respectively. The white bars represent the spectra unidentified by any of the three tools
identified in the first search by MS-GF+,
not identified in the first search but identified by Comet in the second search,
not identified by the first two searches but containing a confident de novo sequence tag, and
not interpreted by any of these analyses.
Figure 7B is the same as Figure 7A except that the y-axis becomes the percentage in each SPEQ score interval. To plot these figures, the logit of the probability predicted by SPEQ was used as the SPEQ score for a more proper division of the score intervals.
As can be seen, the percentage of spectra confidently identified by at least one engine grew as the quality score provided by SPEQ increased. This suggests that high-scoring spectra not identified by the first search engine can often be further interpreted in other analyses, while low-score spectra are usually not interpretable. It is more likely for a spectrum containing valuable information to obtain a higher score compared with an uninterpretable scan.
Moreover, when the score is high enough (e.g. ≥1.5), the majority of the spectra not identified by any of two database search tools contain confident de novo sequencing tags. This strongly suggests that these spectra are indeed produced by peptides, but unidentified because of the inadequate data analysis.
3.3 SPEQ-directed troubleshooting
For the troubleshooting test (as described in Section 2.4), using the Orbitrap human dataset, the first search with MS-GF+ used the following variable PTMs.
Oxidation on M,
Acetyl at protein N-term and
Deamidated on N and Q.
Note that, the original publication (Bruderer et al., 2017) of the dataset used only the first two variable PTMs in this list. But adding the third PTM resulted in more identifications in the first search, which identified 31 400 of the 52 285 spectra. Among the 20 885 unidentified spectra, the top 1% spectra (according to their SPEQ score) were searched again with the following variable PTMs:
Oxidation on M,
Deamidated on N and Q,
Carbamidomethyl at peptide N-term,
Pyro-Glu at peptide N-term on Q and E,
Acetyl at protein N-term,
Acetyl on K,
Methyl on K and
Phospho on S, T and Y.
The search took 3.75 min and identified 24 confident peptides with PTMs listed above. Three of the most common PTMs in these peptides were:
Pyro-Glu at peptide N-term on Q,
Carbamidomethyl at peptide N-term and
Deamidated on N and Q.
Because deamidation was used in the first search already, for the 24 peptides identified here, deamidation always appeared together with another PTM in the same peptide.
A third search was conducted to search with these three most common PTMs and the 20 885 unidentified spectra. The search finished in 12.06 min and confidently identified 829 spectra that contain at least one of these three PTMs. To compare, searching all the 20 885 unidentified spectra with the longer list of PTMs took 78.44 min and identified 863 spectra.
The results here demonstrate that the SPEQ score can indeed be used to select a small portion (1%) of the unidentified spectra for troubleshooting, and the factors identified by the troubleshooting can be used to adjust the search strategy in additional searches to identify more peptides.
4 Discussion
In this work, we present SPEQ, a software tool that uses deep learning to predict the quality of an MS/MS spectrum. The prediction accuracy of SPEQ was evaluated by the ROC curves on several different datasets. SPEQ performed better (with higher AUC) than the other tools compared (Figs 3–5). This is still the case when the testing and training data are from independent experiments of two different species (Fig. 6).
The use of deep learning may be an important reason for the improvement of SPEQ. With sufficiently large training data, deep learning can automatically discover the features important for the prediction. This is in contrast to the traditional machine learning that requires the tool developers to handcraft features. Handing off the feature extraction to the learning algorithm not only saves the developers’ time, but also allows the learning algorithm to discover new features that the tool developers may not be able to. This is particularly interesting in a cross-disciplinary area such as bioinformatics, where sometimes the tool developer may not be the most knowledgeable domain expert to handcraft the features. Also, this makes the model more adaptive to different types of MS instruments and experimental methods.
Experiments were also carried out to demonstrate the potential usefulness of SPEQ in proteomics data analysis. In general, a quality assessment tool helps the proteomics data analysis in two ways. First, it provides some hope in dealing with the false negatives. The proteomics research community has established a standard way (the FDR) to control false positives of a data analysis. However, there is no established way to know the level of false negatives in a data analysis. The results of Figure 7 show a strong correlation between the SPEQ score and the percentage of the false-negative spectra (i.e. the spectra that were not identified by the initial database search but identifiable with additional efforts). This suggests that the quality score assigned by SPEQ (or by another tool) can potentially be used to estimate the level of false negatives. However, more research is needed to prove this can indeed provide an accurate estimation.
Second, the quality score can be used to direct the allocation of resources (either computing power or human experts’ time) to focus on the high-quality spectra, which have the best chance to be interpreted by the data analysis effort. This is demonstrated in the article with the ‘troubleshooting’ experiment, where the top 1% of the unidentified spectra were analyzed with a much more extensive and costly search by selecting a long list of variable PTMs. This revealed that some peptides contain PTMs that had not been specified in the original database search. By adding back the most frequent PTMs found by the troubleshooting, more spectra were identified. This troubleshooting practice is in line with the Preview idea proposed by Kil et al. (2011), where only a subset of spectra was searched first to determine the best search parameters for the full search. The quality score can be also used here to select the best subset of spectra for the pre-search.
While SPEQ improved the prediction accuracy relative to other tools, its accuracy is still not ideal. For example, there is still a big gap between 1 and the AUC of SPEQ (0.7688) on the Orbitrap human dataset. Since the spectra were labeled according to whether they are confidently identified by database search, the false negatives of the database search would have created mislabels in the training and testing data. The mislabels in the testing data could have an adversary effect on the AUC. In another word, the actual performance of SPEQ (and other tools) may be better than indicated here. Meanwhile, if the number of mislabeled spectra in the training data can be reduced, the machine learning algorithm will learn a better model too. Other possible ways to improve the model include different neural network structures, larger training data and other learning strategies such as transferred learning. In fact, if the scoring function can be sufficiently developed in the future, the low-quality spectra can be excluded from the analysis from the very beginning. This will not only improve the data analysis speed, but also reduce the false positives created by these low-quality spectra.
Overall, we have developed the SPEQ tool for spectrum quality assessment based on deep learning, and demonstrated its usefulness. Further improvement of the quality score by the bioinformatics community is needed, and will greatly enhance the usefulness of the quality assessment. The availability of SPEQ may help other proteomics data analyses and support other bioinformatics researchers to further improve the accuracy of SPEQ assessment. SPEQ is written in Python and the source code is freely available at github.com/sor8sh/SPEQ.
Acknowledgement
The authors thank Dr Shengheng Guan for his valuable discussion and assistance during this research.
Funding
This work was supported by a Natural Sciences and Engineering Research Council discovery grant [RGPIN-2016-03998] and by Genome Canada and Ontario Genomics Institute through a Bioinformatics and Computational Biology program [OGI-166].
Conflict of Interest: none declared.
Contributor Information
Soroosh Gholamizoj, David R. Cheriton School of Computer Science, University of Waterloo, Waterloo, ON N2L 3G1, Canada.
Bin Ma, David R. Cheriton School of Computer Science, University of Waterloo, Waterloo, ON N2L 3G1, Canada.
References
- Abadi M. et al. (2015) TensorFlow: large-scale machine learning on heterogeneous systems. www.tensorflow.org (13 November 2020, date last accessed).
- Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature, 422, 198–207. [DOI] [PubMed] [Google Scholar]
- Bern M. et al. (2004) Automatic quality assessment of peptide tandem mass spectra. Bioinformatics (Oxford, England), 20, i49–54. [DOI] [PubMed] [Google Scholar]
- Bruderer R. et al. (2017) Optimization of experimental parameters in data-independent mass spectrometry significantly increases depth and reproducibility of results. Mol. Cell. Proteomics, 16, 2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M.C. et al. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol., 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008) Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol., 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Elias J.E., Gygi S.P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods, 4, 207–214. [DOI] [PubMed] [Google Scholar]
- Eng J.K. et al. (2013) Comet: an open-source ms/ms sequence database search tool. Proteomics, 13, 22–24. [DOI] [PubMed] [Google Scholar]
- Eng J.K. et al. (2015) A deeper look into comet—implementation and features. J. Am. Soc. Mass Spectrometry, 26, 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flikka K. et al. (2006) Improving the reliability and throughput of mass spectrometry-based proteomics by spectrum quality filtering. Proteomics, 6, 2086–2094. [DOI] [PubMed] [Google Scholar]
- Frank A., Pevzner P. (2005) Pepnovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem., 77, 964–973. [DOI] [PubMed] [Google Scholar]
- Goloborodko A.A. et al. (2013) Pyteomics—a python framework for exploratory data analysis and rapid software prototyping in proteomics. J. Am. Soc. Mass Spectrometry, 24, 301–304. [DOI] [PubMed] [Google Scholar]
- Hernandez P. et al. (2006) Automated protein identification by tandem mass spectrometry: issues and strategies. Mass Spectrometry Rev., 25, 235–254. [DOI] [PubMed] [Google Scholar]
- Keller A. et al. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem., 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kil Y. et al. (2011) Preview: a program for surveying shotgun proteomics tandem mass spectrometry data. Anal. Chem., 83, 5259–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Pevzner P.A. (2014) Ms-gf+ makes progress towards a universal database search tool for proteomics. Nat. Commun., 5, 5277–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D., Ba J. (2014) Adam: a method for stochastic optimization. In: 3rd International Conference on Learning Representations, {ICLR} 2015, San Diego, CA, USA, May 7-9, 2015.
- Ma B. (2015) Novor: real-time peptide de novo sequencing software. J. Am. Soc. Mass Spectrometry, 26, 1885–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B. et al. (2003) Peaks: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrometry, 17, 2337–2342. [DOI] [PubMed] [Google Scholar]
- Ma Z.-Q. et al. (2011) Scanranker: quality assessment of tandem mass spectra via sequence tagging. J. Proteome Res., 10, 2896–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh B. et al. (2014) Differential cysteine labeling and global label-free proteomics reveals an altered metabolic state in skeletal muscle aging. J. Proteome Res., 13, 5008–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa J.M. et al. (2020) Repeat-preserving decoy database for false discovery rate estimation in peptide identification. J. Proteome Res., 19, 1029–1036. [DOI] [PubMed] [Google Scholar]
- Na S., Paek E. (2006) Quality assessment of tandem mass spectra based on cumulative intensity normalization. J. Proteome Res., 5, 3241–3248. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I. et al. (2006) Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell. Proteomics, 5, 652–670. [DOI] [PubMed] [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Perkins D.N. et al. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Salmi J. et al. (2006) Quality classification of tandem mass spectrometry data. Bioinformatics, 22, 400–406. [DOI] [PubMed] [Google Scholar]
- Wu F.-X. et al. (2008) Quality assessment of peptide tandem mass spectra. BMC Bioinformatics, 9, S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. (2019) PNOVO 3: precise de novo peptide sequencing using a learning-to-rank framework. Bioinformatics (Oxford, England), 35, i183–i190. 31510687[pmid], PMC6612832[pmcid], 5529238[PII]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. (2012) Peaks db: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics, 11, M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]