Abstract
Proline (Pro) catabolism and reactive oxygen species production have been linked in mammals and Caenorhabditis elegans, while increases in leaf respiration rate follow Pro exposure in plants. Here, we investigated how alternative oxidases (AOXs) of the mitochondrial electron transport chain accommodate the large, atypical flux resulting from Pro catabolism and limit oxidative stress during Pro breakdown in mature Arabidopsis (Arabidopsis thaliana) leaves. Following Pro treatment, AOX1a and AOX1d accumulate at transcript and protein levels, with AOX1d approaching the level of the typically dominant AOX1a isoform. We therefore sought to determine the function of both AOX isoforms under Pro respiring conditions. Oxygen consumption rate measurements in aox1a and aox1d leaves suggested these AOXs can functionally compensate for each other to establish enhanced AOX catalytic capacity in response to Pro. Generation of aox1a.aox1d lines showed complete loss of AOX proteins and activity upon Pro treatment, yet full respiratory induction in response to Pro remained possible via the cytochrome pathway. However, aox1a.aox1d leaves displayed symptoms of elevated oxidative stress and suffered increased oxidative damage during Pro metabolism compared to the wild-type (WT) or the single mutants. During recovery from salt stress, when relatively high rates of Pro catabolism occur naturally, photosynthetic rates in aox1a.aox1d recovered slower than in the WT or the single aox lines, showing that both AOX1a and AOX1d are beneficial for cellular metabolism during Pro drawdown following osmotic stress. This work provides physiological evidence of a beneficial role for AOX1a but also the less studied AOX1d isoform in allowing safe catabolism of alternative respiratory substrates like Pro.
The alternative oxidase of plant mitochondria contributes to proline catabolism by preventing oxidative stress in plant tissues and this aids recovery of photosynthetic rates following salinity stress.
Introduction
Plant respiration is a tightly regulated metabolic network that is capable of adapting to changing cellular needs and growth conditions by altering its metabolic behavior (Plaxton and Podestá, 2006; O'Leary et al., 2019). Through the complete or partial oxidation of substrates, plant respiration serves three main purposes: ATP synthesis, biosynthetic precursor synthesis, and cellular redox balancing (O'Leary et al., 2019). The principal respiratory substrates in nonphotosynthesizing plant tissues are carbohydrates, metabolized sequentially via glycolysis and the tricarboxylic acid (TCA) cycle, yielding reducing power to feed the mitochondrial electron transport chain (mETC; O'Leary and Plaxton, 2016). However, under certain conditions, often associated with starvation and stress, plants rely on alternative respiratory substrates such as amino acids or derivatives of fatty acids, which are metabolized through various additional respiratory pathways leading to the mETC (Millar et al., 2011; Rasmusson and Møller, 2011; Cabassa-Hourton et al., 2016; Launay et al., 2019; O'Leary et al., 2020). In photosynthesizing cells, mitochondrial respiration also has a clear function to enhance photosynthesis by participating in photorespiration (glycine oxidation) and oxidizing excess reductant exported by the plastid (Raghavendra and Padmasree, 2003; Tcherkez et al., 2017; Del-Saz et al., 2018). Because of its size and functional complexity, our understanding of the control and regulation of plant respiration as a network is limited (Sweetlove et al., 2010). In turn, our ability to understand how variability in respiratory metabolism contributes to plant performance in nature (O'Leary et al., 2017) or in the field (Amthor et al., 2019; Reynolds et al., 2021) is constrained.
Like other aspects of the respiratory network, the plant mETC is highly flexible from a metabolic standpoint (Rasmusson et al., 2008; Millar et al., 2011). First, multiple nonproton pumping alternative enzymes exist which bypass components of the standard mETC (i.e. complexes I–IV; Millar et al., 2011). In particular, plant mitochondria contain various isoforms of alternative oxidase (AOX), an enzyme that catalyzes the transfer of electrons directly from ubiquinol to O2, thereby bypassing complexes III and IV of the standard mETC and partly uncoupling respiration from ATP generation (Millar et al., 2011; Vanlerberghe, 2013). Because AOX activity appears to constitute an energetically wasteful process, researchers have long sought to understand the countervailing benefits of AOX activity in nonthermogenic tissues. Electron transport through the mETC contributes to reactive oxygen species (ROS) production, especially when electron flux is high or the activity of mETC components is disrupted (Møller, 2001; Huang et al., 2016). Under these scenarios, AOX activity could function to reduce the redox poise of the ubiquinone pool and other mETC components to thereby to mitigate ROS formation (Millar et al., 2001). Previous reports have shown that the overexpression of AOX in transgenic Nicotiana tabacum cells (Maxwell et al., 1999) and drought-treated N. tabacum leaves (Dahal and Vanlerberghe, 2017), activation of AOX in isolated durum wheat (Triticum durum) mitochondria (Pastore et al., 2001) and upregulation of AOX in salt-stressed Arabidopsis seedlings (Gong et al., 2020) all resulted in less ROS abundance and oxidative damage. A second potential benefit of AOX activity is that it allows the mETC to run faster and be more responsive toward redox balancing, albeit with lower rates of ATP production. AOX also supports photosynthetic activity, and it would appear to do this by allowing the mETC to rapidly dissipate excess reducing power, thus contributing to the overall cellular redox balance (Vishwakarma et al., 2015; Jiang et al., 2019). Despite some remaining uncertainty about the precise benefits of AOX expression and function, available evidence suggests its presence is favorable for plant growth (Vanlerberghe, 2013; Del-Saz et al., 2018; Selinski et al., 2018b).
In Arabidopsis, the AOX gene family consists of five genes: AOX1a–AOX1d and AOX2. Analysis of the distribution and conservation of AOX family genes throughout the plant kingdom is complicated, but AOX1d genes, AOX1a–c genes and AOX2 genes represent three separate clades that are present in many plant families (Costa et al., 2014). Transcriptomic and proteomic analyses have consistently indicated that AOX1a is the dominant AOX isoform in Arabidopsis in all vegetative tissues (Clifton et al., 2006). AOX1b is expressed during early flowering stages while AOX1c and AOX2 are expressed at low levels regardless of developmental stages (Clifton et al., 2006). Both AOX1a and AOX1d demonstrate a clear pattern of elevated expression under a range of stress conditions such as salinity (Van Aken et al., 2009; Feng et al., 2013) and low nitrogen stress (Watanabe et al., 2010) and also in response to chemical inhibition of mETC complexes, such as with antimycin A (Strodtkotter et al., 2009). Manipulation of the AOX1a expression in Arabidopsis under diverse environmental treatments such as cold (Fiorani et al., 2005), drought (Giraud et al., 2008), high CO2 concentrations (Gandin et al., 2012), high arsenic (Demircan et al., 2020), and ecologically relevant doses of ultraviolet-B (UV-B) (Garmash et al., 2020) have highlighted the potential of AOX1a to enable acclimation to unfavorable growth conditions. AOX1a expression is particularly strongly upregulated by chemical disruption of complexes I, III, IV, and V, and has been linked to mitochondrial retrograde signaling (Sweetlove et al., 2002; Clifton et al., 2005; Schwarzländer et al., 2011; Umbach et al., 2012) and the alleviation of excess reducing power arising from impediments to normal mETC activity (Jiang et al., 2019). Comparable genetic studies with altered expression of AOX2 have been performed on soybean (Glycine max; Chai et al., 2012) and Chinese dwarf cherry (Cerasus humilis; Sun et al., 2021). Selinski et al. (2018a) showed that Arabidopsis AOX isoforms are differentially activated by various TCA cycle intermediates, suggesting sub-functionalization of the AOX gene family. Still there is limited information on the physiological function of AOX genes other than AOX1a in plants (Clifton et al., 2006; Strodtkotter et al., 2009; Konert et al., 2015).
Changes in cellular metabolite levels strongly influence respiratory rates in leaves (O'Leary et al., 2017; O'Leary et al., 2020). In particular, external proline (Pro) exposure is followed by cellular Pro accumulation, which induces a strong stimulation of Pro respiratory catabolism in Arabidopsis leaves, leading to a doubling of the respiration rate over 14 h (O'Leary et al., 2020). Pro is an atypical respiratory substrate in plants whose accumulation and metabolism under osmotic stress and recovery is well-established (Yoshiba et al., 1995; Hare et al., 1998; Verbruggen and Hermans, 2008; Verslues and Sharma, 2010). Here, we suspected that Pro’s ability to potently stimulate respiration presented an opportunity to study metabolic flexibility at the level of the mETC. The two-step pathway of Pro oxidative catabolism to Glu is understood. The mitochondrial enzyme Pro dehydrogenase (ProDH) oxidizes Pro to pyrroline-5-carboxylate (P5C) and likely supplies electrons directly to ubiquinol via its FADH cofactor (Szabados and Savouré, 2010; Servet et al., 2012; Cabassa-Hourton et al., 2016; Launay et al., 2019). In the second step, P5C spontaneously transforms into glutamate-5-semialdehyde, which is oxidized to Glu by mitochondrial P5C dehydrogenase (P5CDH) with the concomitant reduction of NAD+ to NADH (Forlani et al., 1997; Verslues and Sharma, 2010; Trovato et al., 2019). Pro catabolism increases ROS production via ProDH activity in mammalian cells (Donald et al., 2001) and Caenorhabditis elegans (Zarse et al., 2012), while in plants, the absence of P5CDH led to increased mitochondrial ROS following Pro exposure (Miller et al., 2009). What is currently unclear is how mETC components adjust to accommodate the rapid and nonstandard electron flux change resulting from scenarios requiring large scale Pro catabolism. In this study, we provide evidence that Pro exposure specifically upregulates AOX1d and AOX1a expression, greatly increasing the AOX capacity within the mETC. Our results indicate that AOX1a and AOX1d both act to limit oxidative stress during Pro catabolism and thereby facilitate plant recovery from osmotic stress.
Results
Leaf exposure to Pro leads to an increase in AOX1a and AOX1d transcript and protein abundances
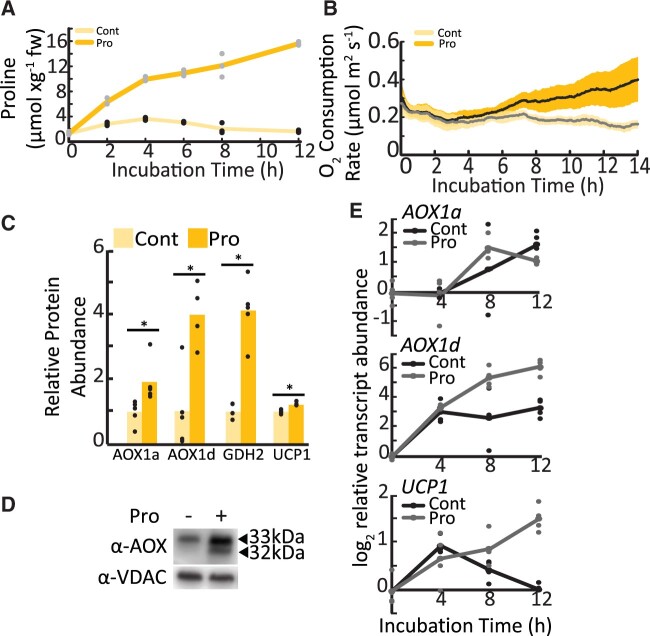
Exogenous exposure of leaf discs to 10 mM Pro in the dark causes time-dependent increases of Pro content in leaf tissues. Pro levels measured in our system were similar in magnitude to Pro accumulation in leaves of drought-treated Arabidopsis plants (Figure 1A;Miller et al., 2009). This Pro treatment also leads to a pronounced PDH-dependent stimulation of leaf disc oxygen consumption rates (OCR; Figure 1B;O'Leary et al., 2020), a situation that may evoke strong deviations from typical energy and redox balances. We hypothesized that increased leaf OCR following Pro exposure requires stimulation of mechanisms that uncouple the mETC from ATP production and retain redox balance (e.g. AOX or uncoupling protein [UCP] expression).
Figure 1.
Transcript and protein abundance of AOX1a and AOX1d after Pro exposure in leaf tissues. A, Pro amount in a leaf disc during incubation in the presence or absence of 10 mM Pro. Lines represent mean values; replicate data points are shown (n = 4). Adapted from O'Leary et al. (2020). B, Time-dependent OCR stimulation in single leaf discs floated on buffer solution with or without Pro. Shaded area indicates the 95% confidence intervals (n = 6). C, Mitochondrial proteins that significantly increased in abundance following 10-mM Pro treatment of leaf tissue, shown relative to nontreated samples. Asterisks represent significant differences between treatments (t test, n ≥ 4; P < 0.05). D, Immunoblot analysis of enriched mitochondrial fractions from WT leaf tissue incubated in the presence or absence of Pro. Bands corresponding to AOX isoforms are indicated. E, Transcript analysis of AOX1a, AOX1d, and UCP1 in control and Pro-treated leaf discs. Lines represent mean values; replicate data points are shown (n ≥ 3).
Multiple reaction monitoring (MRM) was used to quantify a variety of mitochondrial protein abundances in the wild-type (WT) control and 14 h Pro-exposed leaf samples, including 24 components of the classical electron transport chain, and 43 components of the TCA cycle and enzymes of organic and amino acid metabolism (Supplemental Table S1). Only eight proteins showed significant abundance differences in response to Pro, with four being upregulated and four being downregulated. AOX1d and glutamate dehydrogenase 2 (GDH2) increased four-fold, AOX1a increased two-fold, and uncoupling protein 1 (UCP1) displayed a minor induction (Figure 1C). On the other hand, isocitrate dehydrogenase subunit 1, pyruvate dehydrogenase complex subunit 1-2 and E1 α-subunit, and succinyl CoA ligase β-subunit showed modest but significant decreases after Pro treatment (Supplemental Table S1). Notably, GDH can function downstream of Pro catabolism and catalyzes glutamate deamination to 2-OG; thus GDH2 likely functions to connect Pro catabolism to the TCA cycle. Analysis of the abundance of 24 components distributed across the complexes I–V of the classical ETC showed no Pro-dependent changes in abundance (Supplemental Table S1). Immunoblots against the constitutively expressed mitochondrial membrane protein, voltage-dependent anion channel (VDAC), confirmed that leaf disc Pro treatment does not lead to a general increase in mitochondrial protein content (Supplemental Figure S1A and Supplemental Table S1).
The large upregulation of multiple AOX isoforms was confirmed by immunoblotting of enriched mitochondrial samples from 14 h Pro-treated leaf samples (Figure 1D). Transcript analysis of leaf discs also confirmed the time-dependent upregulation of AOX1a, AOX1d, and UCP1 in response to 12 h Pro treatment, suggesting the enhanced expression of these uncoupling proteins was controlled at least partly at the level of transcription (Figure 1E). In contrast, AOX1b, AOX1c, and AOX2 transcripts did not increase in response to Pro (Supplemental Figure S1B). Together these data indicated that alterations to AOX expression represented the largest change to mETC functionality we could find upon Pro exposure.
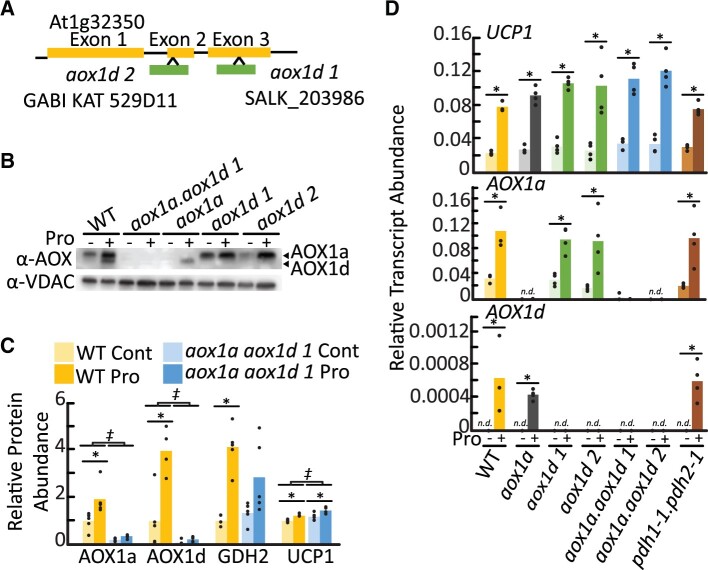
We distinguished two AOX immunoreactive bands in WT leaf tissues that increased in abundance following Pro exposure (Figure 1D). Similar doublet bands have been previously observed on AOX immunoblots of isolated Arabidopsis mitochondria following various treatments, and Konert et al. (2015) identified an upper band as AOX1a and a lower band as AOX1d using tandem mass-spectrometry. To establish the molecular nature of the two AOX bands in our treatments, we probed enriched mitochondrial samples isolated from an established aox1a line (SAIL_303_D08) and two aox1d lines, aox1d 1 (SALK_203986) and aox1d 2 (GABI_529D11) that we characterized here (Figure 2A;Supplemental Figure S2A). The immunoblot banding pattern observed in the aox1a and aox1d samples confirmed the association of the upper and lower immunoreactive bands with AOX1a (33 kDa) and AOX1d (32 kDa), respectively (Figure 2B). Both aox1d lines lacked any detectable AOX1d transcript and protein, and AOX1d expression was only detectable upon Pro treatment in WT and aox1a plants (Figure 2, B–D). Although AOX1d transcript levels remained much lower than those of AOX1a, the AOX1d protein accumulated to appreciable amounts under Pro treatment. Therefore, the aox1a line was not an effective AOX knockout in this instance. To achieve an effective AOX knockout, two independent aox1a.aox1d lines were constructed. AOX1a and AOX1d transcripts were absent (Figure 2D;Supplemental Figure S2A) and no AOX protein could be detected by immunoblotting of enriched mitochondrial samples following leaf tissue Pro treatment in both aox1a.aox1d lines (Figure 2B;Supplemental Figure S2B). There was also no observable growth phenotype between WT and any of the aox lines under our standard growth conditions (Supplemental Figure S2C). We repeated the mitochondrial MRM analysis on samples from aox1a.aox1d 1 leaf tissue and confirmed the loss of AOX1a and AOX1d abundance (Figure 2C;Supplemental Tables S1 and S2). aox1a.aox1d plants displayed a minor induction of UCP1 transcript in both control and Pro-exposed leaf tissues indicating a potential compensation for the lack of AOX (Supplemental Tables S1 and S2).
Figure 2.
Characterization of aox1a, aox1d, and aox1a.aox1d lines. A, Gene model of AOX1d (At1g32350) indicating T-DNA insertion locations corresponding to SALK_203986 and GABI KAT_529D11. Genotyping data are shown in Supplemental Figure 2. B, Immunoblot analysis of enriched mitochondrial fractions from leaf tissue pre-incubated in the presence or absence of Pro. Each lane contains 20 µg of protein. Bands corresponding to AOX1a and AOX1d are indicated. WT lanes presented here are the same as Figure 1D. C, Multiple reaction monitoring protein abundance measurements. Relative abundance is calculated in comparison to WT nontreated samples. Asterisks represent significant treatment effects, double daggers represent significant genotypic effects (two-way ANOVA; n ≥ 4; P < 0.05). D, Transcript analysis of AOX1a, AOX1d, and UCP1 following 14-h Pro treatment of leaf discs. Bars represent average transcript abundance; replicate data points are shown. Asterisks represent significant differences between treatments (t test, n ≥ 3; P < 0.05).
We investigated whether the presence of Pro itself or the metabolism of Pro via the mETC acted as the signal to stimulate AOX expression. The accumulation of AOX1a, AOX1d, and UCP1 transcripts following Pro treatment was unaffected in pdh1-1.pdh2-1 leaf tissue that lack expression of ProDH (Figure 2D). Previous work using these mutants has demonstrated that ProDH is needed for Pro catabolism via the mETC (Cabassa-Hourton et al., 2016; O'Leary et al., 2020). This result suggests that elevated Pro levels even in the absence of high Pro respiratory flux is sufficient to signal AOX expression.
Pro-dependent respiratory flux is maintained in the absence of AOX
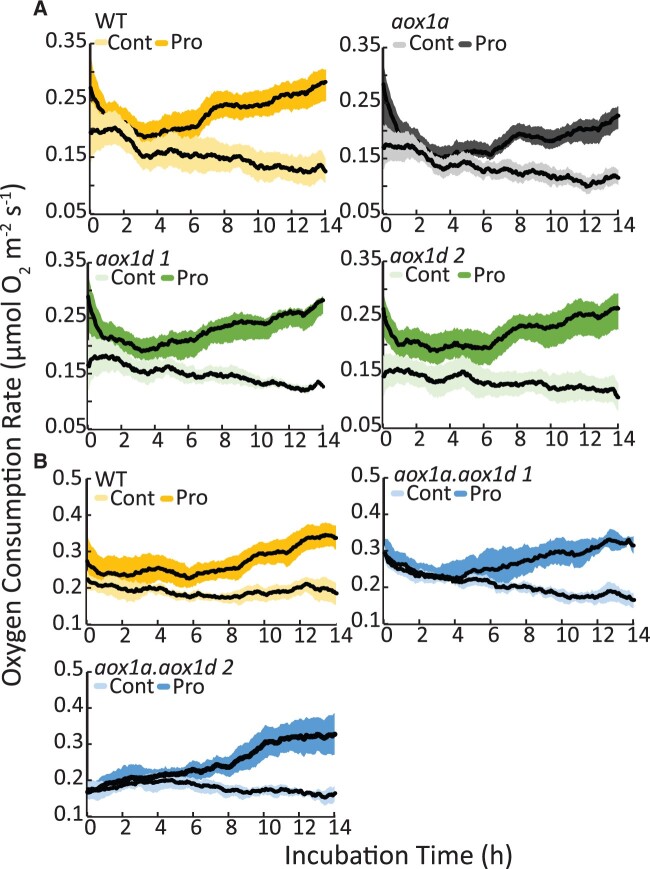
To understand if either AOX1a or AOX1d had a role in Pro-dependent respiration, we measured OCR in WT, aox1a, aox1d, and aox1a.aox1d leaf discs exposed to Pro. All genotypes showed similar Pro-inductions of OCR (Figure 3). This indicates that AOX is not required, and that the cytochrome oxidase pathway (COX) capacity is sufficient to carry electron transport chain flux resulting from high rates of Pro catabolism.
Figure 3.
The effect of the loss of AOX1a or AOX1d on Pro stimulated RN in AOX knockout lines. Time-dependent stimulation in respiration after Pro exposure. Single leaf discs of (A) WT, aox1a, aox1d 1, aox1d 2 and (B) WT, aox1a.aox1d 1, and aox1a.aox1d 2 were floated on 10-mM Pro respiration buffer. Control-treated leaf discs were floated on respiration buffer lacking Pro. Traces show moving average respiration rates (n = 6). The shaded area shows the 95% confidence intervals.
We note that additional leaf disc experiments conducted with commonly used AOX inhibitors, salicylhydroxamic acid (SHAM) and n-propyl gallate (nPG), produced contradictory results. While the presence of SHAM did not affect Pro uptake (Supplemental Figure S3A), SHAM completely inhibited Pro-dependent respiratory stimulation in leaf discs of all genotypes including both aox1a.aox1d lines (Supplemental Figure S3B). This effect of SHAM was therefore independent of AOX expression level in leaf tissue. Furthermore, OCR of enriched mitochondrial fractions derived from control and Pro-treated leaf tissue was not substantially inhibited by nPG when either Pro or NADH and succinate were available as respiratory substrates, confirming that AOX is not essential for Pro-dependent respiration (Supplemental Figure S3C). While both SHAM and nPG are AOX inhibitors, SHAM is widely used for leaf tissues (Bartoli et al., 2005; Yoshida et al., 2006) and nPG is commonly used for inhibition of AOX in isolated and crude mitochondrial extracts (Cabassa-Hourton et al., 2016; Launay et al., 2019). As SHAM can affect multiple oxidases beyond AOX (Ordog et al., 2002), we hypothesize that additional SHAM sensitive oxidases are involved in Pro signaling in leaf tissue. Therefore, the use of genetics rather than AOX inhibitors is preferred here to allow for a more direct interpretation of Pro-dependent respiratory fluxes over longer time periods.
AOX1d contributes to increased alternative pathway capacity upon Pro treatment
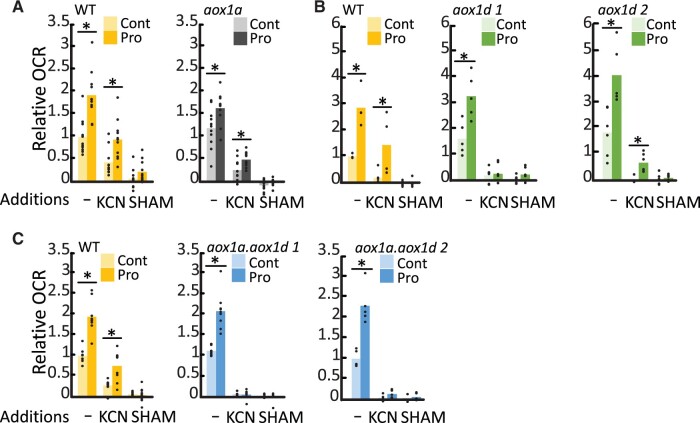
We wanted to verify whether AOX isoforms and AOX1d in particular, could substantially contribute to mETC capacity in leaves following Pro exposure. OCR measurements of Pro pre-treated leaf discs were performed using Clark-type oxygen electrodes to observe rapid OCR changes, wherein KCN additions were used to measure AOX capacity. In agreement with the above Q2 measurement, in the absence of inhibitors there was an approximately two-fold stimulation of OCR resulting from Pro pre-treatment and Pro stimulation of OCR was not significantly different in the absence of AOX1a and/or AOX1d (Figure 4). The addition of potassium cyanide (KCN) revealed a substantial induction of AOX capacity by Pro-pre-treatment in WT leaf discs. In comparison, AOX capacity (following KCN addition) was significantly less in aox1a (Figure 4A) and aox1d (Figure 4B), and absent in both aox1a.aox1d lines (Figure 4C). In all instances, KCN-resistant respiration was rapidly inhibited by SHAM. These results indicated that AOX capacity is upregulated in response to Pro due to contributions from both AOX1a and AOX1d.
Figure 4.
Both AOX1a and AOX1d contribute to the increase in AOX capacity after Pro exposure. Electrode-based OCR measurements of control or Pro-treated leaf discs were made before (−) and after the sequential addition of KCN and SHAM. Separate experiments compared (A) aox1a, (B) aox1d lines, and (C) aox1a.aox1d lines to the WT. Data are expressed relative to uninhibited OCR in corresponding WT control samples. Replicate data points are shown. Asterisks represent significant differences between treatments (t test; n ≥ 4; P < 0.05).
AOX1a and AOX1d are required to limit Pro-induced oxidative stress
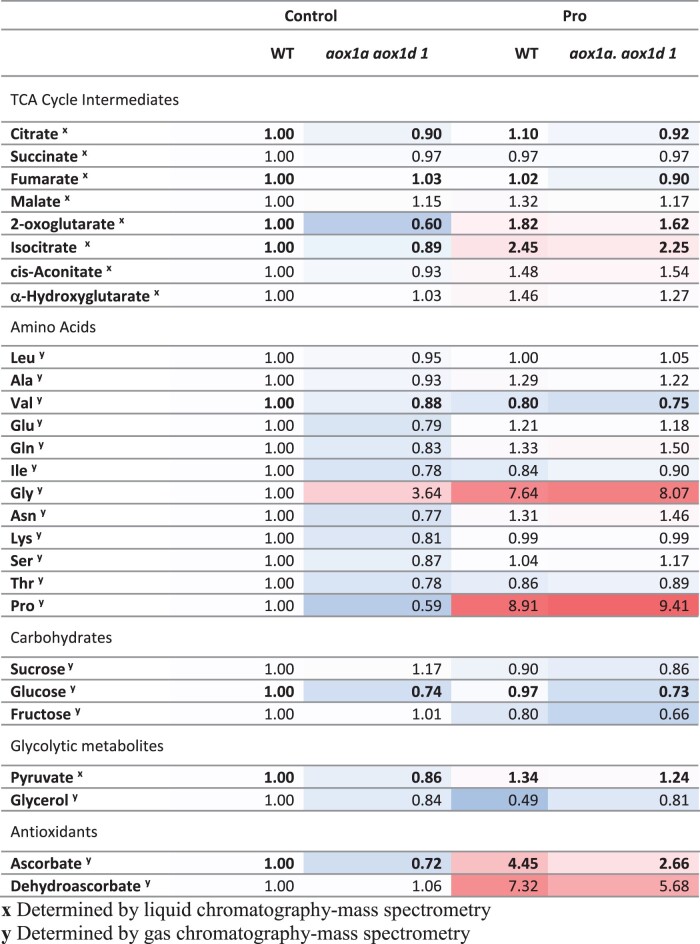
To understand the potential benefit of enhanced AOX capacity on respiratory metabolism following Pro exposure, we performed a time course metabolomics analyses in aox1a.aox1d and WT leaf discs following Pro treatments for 0, 4, or 8 h to identify Pro- and AOX-dependent alterations to metabolite levels (Figure 5;Supplemental Table S3). Statistical analysis revealed strong genotypic and treatment effects on metabolite levels, but significant genotype by treatment effects were not observed. Pro treatment caused a strong accumulation of both ascorbate (Asc; reduced) and dehydroascorbate (DHA; oxidized ascorbate), indicating an activation of antioxidant mechanisms. Notably, aox1a.aox1d samples displayed significantly less Asc across all treatments indicating possible differences in oxidative stress (Figure 5;Supplemental Table S3). Further genotypic effects were observed, wherein the absence of AOX led to minor significant decreases in citrate, fumarate, 2-oxoglutarate, isocitrate, valine, glucose, and pyruvate across all treatment types (Figure 5;Supplemental Table S3).
Figure 5.
Quantification of AOX- and Pro-dependent changes in cellular metabolites. Measurement of cellular metabolites in WT and aox1a.aox1d 1 leaf discs following 4h Pro treatment. Average metabolite abundances were calculated relative to WT control samples. Red and blue coloring indicate higher and lower relative metabolite abundances. Boldface indicates significant genotypic differences between WT and aox1a.aox1d samples (three-way ANOVA; P < 0.05). See Supplemental Table 1 for full data set.
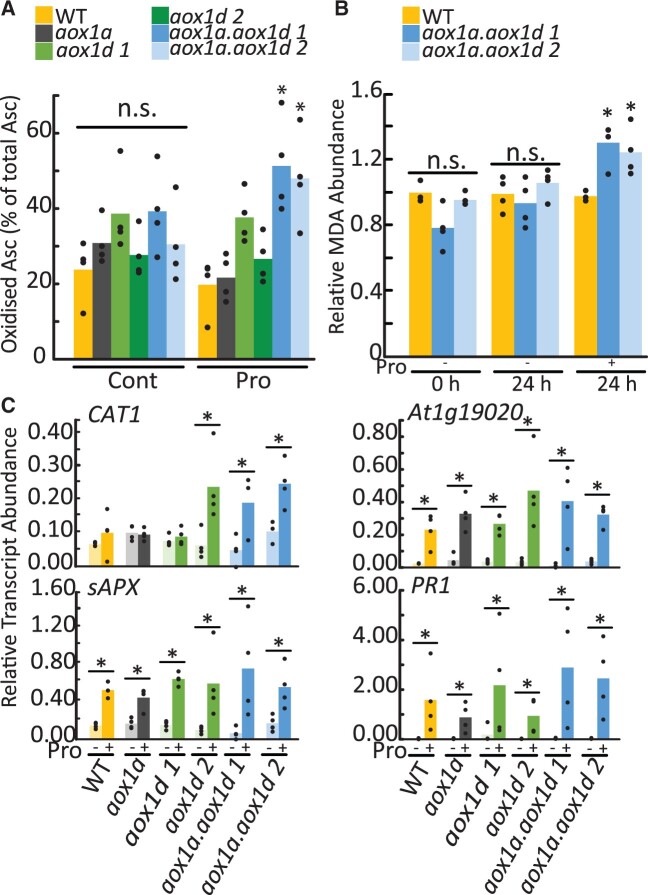
Quantification of actual Asc:DHA ratios using gas chromatography–mass spectrometry is not possible as the oxidation state of Asc is not strictly preserved. Therefore, we conducted additional measures to verify the redox status of Asc pools and oxidative damage in Pro-treated leaf discs. Following 4 h of Pro exposure, the Asc redox status in aox1a.aox1d lines, but not single aox1a or aox1d knockouts, was more oxidized in comparison to the WT (Figure 6A;Supplemental Table S4). Our observation is similar to previous observations of altered Asc:DHA ratios in salinity-treated Arabidopsis seedlings (Huang et al., 2005) and N. tabacum seedlings (Yamamoto et al., 2005). The impact of cellular oxidative stress following Pro treatment was then assessed using an assay to quantify malondialdehyde (MDA) levels. MDA is a byproduct of lipid peroxidation that reflects accumulated cellular oxidative damage. A significant increase in MDA abundance in aox1a.aox1d but not WT leaf discs occurred after 24 h of Pro exposure (Figure 6B;Supplemental Table S4), but not at earlier time points (Supplemental Figure S4). These results suggest a potential role for both AOX1a and AOX1d to maintain redox balance and limit oxidative stress during periods of rapid Pro catabolism. Lastly, we analyzed transcript abundance of genes defined as hallmarks of oxidative stress. At1g19020 and PATHOGENESIS-RELATED 1 (PR1) expression is upregulated by ROS (Green and Fluhr, 1995; Gadjev et al., 2006); whereas CATALASE 1 (CAT1) and ASCORBATE PEROXIDASE (sAPX) represent the induction of antioxidant systems, (Vishwakarma et al., 2015). At1g19020, PR1, and sAPX were significantly upregulated following 14h Pro exposure in all genotypes (Figure 6C;Supplemental Table S4). Pro-dependent increases in CAT1 were observed in aox1d 2 and aox1a.aox1d lines but not in the other genotypes (Figure 6C;Supplemental Table S4).
Figure 6.
Quantification of oxidative stress markers following Pro exposure. A, Redox status of the Asc pool in leaf discs in the presence and absence of Pro treatment. Replicate data points are shown. Asterisks indicate significant treatment specific difference compared to the WT (two-way ANOVA; n = 4; P < 0.05). B, MDA abundance following exposure to Pro. Bars represent average MDA abundance relative to WT 0 h. Replicate data points are shown. Asterisks indicate significant treatment-specific difference compared to the WT (one-way ANOVA; n = 4; P < 0.05). C, Transcript analysis of CAT1, sAPX, At1g19020, and PR1 following 14-h Pro treatment of leaf discs. Bars represent relative average transcript abundance; replicate data points are shown. Asterisks represent significant differences between treatments (two-way ANOVA; n ≥ 3; P < 0.05).
AOX1a and AOX1d aid in post-salinity recovery
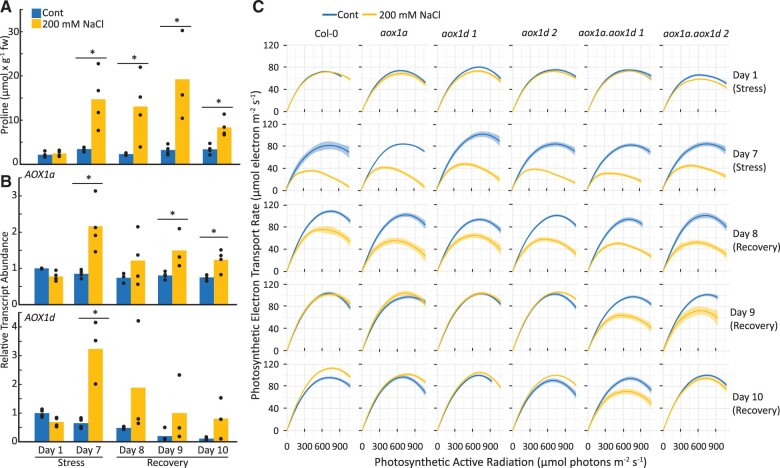
To test the impact of loss of AOX1a and AOX1d at a physiological level, we assessed the ability of plants to recover from acute salt stress. Pro accumulates greatly in plant tissues during salinity stress and is quickly catabolized during subsequent recovery (Verslues and Sharma, 2010). Here, we show that Pro levels rose and fell in WT, aox1a, aox1d, and aox1a.aox1d leaf tissue following 7 d of salinity stress treatment and 3 d of recovery (Figure 7A;Supplemental Figure S5). The accumulation of Pro content during this salinity stress was comparable to the exogenous Pro treatment (Figure 1A). The salinity stress treatment induced AOX1a and AOX1d expression in the WT, similar to results from earlier studies (Smith et al., 2009; Van Aken et al., 2009; Figure 7B). Much evidence suggests that AOX activity plays a role in maintaining photosynthetic rates through cellular redox balancing (Vishwakarma et al., 2014, 2015) and the photosynthetic electron transport rate (ETR) is known to be sensitive to saline stress (Stepien and Johnson, 2008). Therefore, we measured the photosynthetic ETR during and after salinity stress. Following 7 d of salinity stress, all genotypes showed similar reductions in their ETR response curves (Figure 7C). However, following salt removal and re-watering of the plants, both aox1a.aox1d lines displayed slower rates of recovery in their ETR response curves in comparison to the WT or the aox single knockout lines (Figure 7C). There were no observable differences in growth rate between aox1a.aox1d lines and the WT during the first 3 d of recovery (Supplemental Figure S5B). Other photosynthetic parameters, including nonphotochemical chlorophyll fluorescence quenching, qL, and Fv/Fm, were also measured and were mostly unaffected during salinity and recovery (Supplemental Figure S6). While it is not clear that the interaction between Pro catabolism and mETC activity is the direct cause of the photosynthetic phenotype that we observed here, these results lead us to conclude that AOX1a and AOX1d expression are responsive to conditions of elevated Pro in vivo and appear to be involved in optimizing photosynthetic capacity during Pro drawdown and recovery from salt stress.
Figure 7.
The effects of salinity and post-salinity recovery on Pro content, AOX1a and AOX1d expression levels, and photosynthetic capacity. A, Pro content from WT leaf discs obtained during salinity stress and post-salinity recovery. Bars represent average Pro content; replicate data points are shown. Asterisks represent significant differences between treatments (t test; n ≥ 3; P < 0.05). B, Transcript analysis of AOX1a and AOX1d from WT leaf discs obtained during salinity stress and post-salinity recovery. Bars represent average relative transcript abundance; replicate data points are shown. Asterisks represent significant differences between treatments (t test; n ≥ 3; P < 0.05). C, Photosynthetic ETR response curves of WT and aox lines during salinity stress and post-salinity recovery. Each curve represents the average photosynthetic ETR response curve from four biological replicates with each biological replicate measured in over 6–12 areas of interest on leaves. The shaded area shows the 95% confidence intervals.
Discussion
AOX upregulation alleviates oxidative stress caused by Pro catabolism
This study explored the relationship between Pro catabolism and AOX activity in plants. It is well known that Pro accumulation in plant cells can lead to the upregulation of Pro catabolic enzymes (ProDH and P5CDH), whose activities supply reductant directly or indirectly to the plant mETC (Cabassa-Hourton et al., 2016; Launay et al., 2019; O'Leary et al., 2020). However, results from this and previous studies suggest that Pro exposure influences the expression of other mETC components as well, in particular AOX (Figure 1, C–E; Supplemental Table S1). Previously, mitochondria isolated from Pro-treated Arabidopsis seedlings displayed increased AOX capacity, which was particularly evident when Pro was used as the respiratory substrate (Cabassa-Hourton et al., 2016). Similarly, mitochondria isolated from senescing Arabidopsis leaves, which have high levels of ProDH expression, show a pronounced upregulation of Pro respiratory capacity via both COX and AOX pathways (Launay et al., 2019). Third, we previously observed strong, Pro-dependent, respiratory stimulation of mature Arabidopsis leaf discs OCR that was inhibited by the absence of both PDH1 and PDH2 (O'Leary et al., 2020). Our results here confirm that these high rates of Pro catabolism also coincide with induced AOX capacity, thereby linking Pro exposure with AOX expression across several tissue types in Arabidopsis.
We initially hypothesized that enhanced AOX capacity may function to catalyze the increased electron flux emanating from Pro catabolism (Supplemental Figure S3B). We reasoned that although enhanced Pro catabolism could drive an approximate doubling of the respiration rate, there was no obvious need for a doubling of cellular ATP production; uncoupling the mETC could alleviate this imbalance. Previously, mitochondria isolated from Pro-treated seedlings displayed a significant decrease in their ADP/O ratio, indicative of uncoupling (Cabassa-Hourton et al., 2016). However, here, leaves lacking detectable AOX expression still displayed full respiratory induction by Pro (Figures 3, B and 4, C). Therefore, while COX pathway capacity can be sufficient to catalyze high rates of Pro catabolism in leaf discs (Figure 4C), AOX induction in the WT likely contributes to Pro oxidation in vivo, for reasons beyond simply supplying additional respiratory capacity.
The expression of AOX, specifically AOX1a, inversely correlates with both ROS abundance and oxidative damage in plant cells under different stress conditions (Maxwell et al., 1999; Pastore et al., 2001; Giraud et al., 2008; Vishwakarma et al., 2015; Gong et al., 2020). Since Pro catabolism via ProDH stimulates ROS production across in mammalian cells (Donald et al., 2001) and C. elegans (Zarse et al., 2012), we tested whether the loss of AOX led to enhanced oxidative stress during Pro catabolism. Total Asc levels and Asc peroxidase transcript were strongly upregulated following Pro treatment in WT and AOX-deficient leaf discs, suggesting the induction of Asc-dependent antioxidant defense mechanisms (Figures 5 and 6;Supplemental Table S3; Panchuk et al., 2002, Sofo et al., 2015). The redox poise of the Asc:DHA pairing in aox1a.aox1d lines was significantly more oxidized, suggesting that AOX-depleted leaf tissues were more likely to be experiencing oxidative stress (Figure 6A). Indeed, more lipid-based oxidative damage (Figure 6B) and increased expression of catalase transcripts (Figure 6C) were detected following prolonged Pro exposure in aox1a.aox1d lines compared to the WT. Therefore, enhanced AOX expression following Pro exposure is beneficial because it lowers oxidative damage associated with Pro catabolism.
Both AOX1a and AOX1d are responsible for AOX capacity in mETC upon Pro exposure
It is largely unknown if AOX isoforms other than AOX1a have specific purposes to fulfill in the respiratory pathways of plants. Do these AOX isoforms share the same functionality as AOX1a under certain perturbed metabolic conditions or do they act independently? While the upregulation of AOX1d transcripts during senescence (Clifton et al., 2006) and various stressful treatments (Strodtkotter et al., 2009; Feng et al., 2013; Demircan et al., 2020) is known, the perceived low abundance of AOX1d, compared to AOX1a, has limited further investigation (Figure 1C). In this study, we provided genetic and biochemical evidence of AOX1d’s role in Pro catabolism and its apparent functional redundancy to AOX1a in this role. Upon Pro exposure the fold increase of AOX1d was substantially greater than that of AOX1a (Figure 1, C and D), and Pro treatment of aox1a leaf discs still caused a significant increase in AOX capacity (Figure 4A). In contrast, aox1a.aox1d leaf tissue displayed no detectable AOX protein or activity (Figure 2, B–D and Figure 4B) but rather displayed signs of enhanced redox imbalance compared to single aox1a or aox1d lines following Pro exposure (Figure 6A). Therefore, AOX1d functions in support of Pro catabolism, but judging from AOX1d’s expression profile from transcriptomic analyses (Clifton et al., 2006; Winter et al., 2007; Van Aken et al., 2009) its role may be more general in nature, such as supporting mETC redox balancing under stress and senescence.
A lack of characterized knockout lines has hindered progress in understanding the AOX1d isoform function in plants. The data we present here coupled to the complex evolutionary relationship of the AOX gene family in plants, which indicate that AOX1a–c exist in a separate phylogenetic clade from AOX1d (Costa et al., 2014), suggest more attention is due to this isoform and its orthologs. Whether a Pro catabolizing function is conserved across AOX1d orthologs remains to be tested and the role of AOX1d in different metabolic stress scenarios also needs to be assessed. Results from this study also warn about the over interpretation of the effects of nonspecific oxidase inhibitors such as SHAM and nPG on whole cells or intact tissues with regard to AOX function, again focusing on the need to use genetic resources to explore the roles of different AOX isoforms.
AOX1a and AOX1d aid in post-salinity recovery
The inhibition of AOX (via chemical or genetic means) leads to reduced photosynthetic capacity under stressful growth conditions (Giraud et al., 2008; Strodtkotter et al., 2009; Vishwakarma et al., 2014, 2015; Dinakar et al., 2016; Watanabe et al., 2016). Previous studies demonstrating the link between AOX expression and photosynthetic performance have only worked with aox1a plants (Strodtkotter et al., 2009; Vishwakarma et al., 2014, 2015; Dinakar et al., 2016; Watanabe et al., 2016). Here, we show that aox1a.aox1d leaves displayed a greater reduction in photosynthetic capacity during recovery from salinity stress compared to WT, aox1a, or aox1d plants (Figure 7C). This pattern is consistent with our other measures of oxidative stress (Figure 6) and indicates a level of isoform redundancy between AOX1a and AOX1d with regard to enhancing photosynthetic capacity.
Interestingly, the negative impact of severe salinity stress on photosynthetic capacity did not differ between any of the genotypes while the stress was ongoing, but only during recovery (Figure 7C). During recovery from salinity, Pro can be returned back to control levels in a matter of days (Verslues and Sharma, 2010; Figure 7A). The benefit of increased AOX capacity towards photosynthesis therefore coincides with this acute period of enhanced Pro metabolism. In actively photosynthesizing cells, AOX is thought to optimize photosynthetic rates by better allowing the mETC to safely oxidize excess reductant exported by the plastid (Dahal et al., 2015; Del-Saz et al., 2018; Alber and Vanlerberghe, 2021). Vishwakarma et al. (2014) first observed higher NADH/NAD+ and NADPH/NADP+ ratios in high light-treated aox1a Arabidopsis plants. Later, it was reported that antimycin A-treated aox1a plants displayed higher NADH/NAD+ ratios (Vishwakarma et al., 2015). Both observations suggest that the presence of AOX in WT plants allows for faster re-oxidation of NADH and NADPH to ensure optimal photosynthetic capacity. During salinity recovery, mitochondria may be required to balance the excess reducing power generated concurrently from photosynthesis and from Pro catabolism. As observed in our study, the importance of mETC activity and AOX capacity toward cellular redox balancing may be heightened under such conditions. However, it should be noted that our observations of Pro-induced respiratory regulation and oxidative stress were made in darkness, so these observations may need to be independently confirmed in the light.
Advantage of AOX activity in different metabolic scenarios
While the earlier view that AOX activity is, an energetically wasteful process has largely been dispelled, replacing this view with evidence for the specific metabolic benefits of AOX action has been slow. Our results demonstrate two specific outcomes of AOX expression that are beneficial in metabolism from an energetic standpoint. First, AOX activity can limit oxidative damage to cellular macromolecules under demanding respiratory conditions imposed by Pro catabolism (Figure 6B). The ATP savings from a reduced rate of macromolecule turnover due to oxidative damage need to be weighed against the loss of ATP production because of AOX activity. Secondly, AOX action supports recovery of the photosynthetic rate and therefore directly increases the overall energy/carbon budget available to the cell under conditions of recovery from salinity (Figure 7C). These expanded energetic accounting considerations are consistent with the phenotypical observation that AOX expression is generally positive for plant performance (Vanlerberghe, 2013; Dahal et al., 2015; Del-Saz et al., 2018; Selinski et al., 2018b).
Materials and methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana) accession Col-0 (N6000) was used as the WT. The following mutant lines were used: aox1a (At3g22370; SAIL_030_D08; Giraud et al., 2008), aox1d 1 (At1g32350; SALK_203986), aox1d 2 (GABI_529_D11), and pdh1-1.pdh2-1 (At3g30775 and At5g38710; Funck et al., 2010, O'Leary et al., 2020). Two independent aox1a aox1d lines were constructed by crossing aox1a with the two aox1d lines (aox1a.aox1d 1 and aox1a.aox1d 2; Supplemental Figure S2A). Primers used for genotyping of aox1d 1 and aox1d 2 can be found in Supplemental Table S5.
Seeds were sown on 3:1:1 potting soil: perlite: vermiculite supplemented with slow releasing fertilizer and covered with a transparent cover until seedlings were established. Seedlings were then transplanted into individual pots with soil kept moist with a regular watering schedule. Plants were grown in a controlled growth chamber maintaining a short-day photoperiod of 8 h light (21°C): 16 h dark (18°C; 2300-h to 0700-h light) with a photon flux of 100 µmol m−2s−1 and a relative humidity of 65%. For respiratory measurements leaf disc samples (7 mm in diameter) were taken from mature leaves of 8- to 10-week-old plants, which were in the vegetative stage. For treatments prior to crude mitochondrial extracts, 8- to 10-week-old leaves were cut into strips (approximately four to five strips per leaf) and floated adaxial side up on respiration buffer in the presence or absence of Pro.
For salt stress treatments, eight biological replicates (four control, four salt-treated) of all six genotypes (i.e. WT, aox1a, aox1d 1, aox1d 2, aox1a.aox1d 1, and aox1a.aox1d 2) were placed in two separate square trays. Each individual pot contained 50 g of soil. To induce salinity conditions, salinity-treated plants were watered with 1 L of 200 mM NaCl solution and control-treated plants were watered with 1 L of water. The solution in both trays was replaced every day during the treatment period. For post-salinity recovery, both trays were subjected to multiple cycles of washing which included placing pots on paper towels for an hour and transferring them to 1 L of water for another hour. Arabidopsis plants were subjected to salinity stress (for 7 d) and re-watered (for 3 d).
Respiratory measurements
Leaf disc respiratory measurements were performed using the Q2 oxygen sensor (Astec Global) in sealed 850 µL capacity tubes at 21°C by the method of O'Leary et al. (2020, 2022). Leaf discs were sampled 4 h into their night period and floated on the adaxial side up on 600 µL of leaf respiration buffer (50 mM HEPES, pH 6.6, 10 mM MES, 2 mM CaCl2) with additional metabolites or chemicals as described. All external metabolite concentrations were 10 mM unless otherwise indicated. All Q2 experiments sampled a minimum of six leaf discs from different pots. To obtain RN respiratory traces, 2 h moving slopes of O2 depletion over time were calculated.
Leaf disc respiratory measurements using the Clark-type oxygen electrode (Hansatech) were conducted following 14 h of Pro exposure, with eight leaf discs in 1.5 mL of respiration buffer (50 mM HEPES, 10 mM MES, 2 mM CaCl2, pH 6.6). AOX capacity was measured as the difference in OCR between the addition of 2 mM KCN and 4 mM SHAM.
Mitochondrial respiration rate measurements were performed using the Clark-type oxygen electrode (Hansatech) with 120–240 µg of enriched mitochondria in 1 mL of mitochondrial respiration buffer (0.3 M sucrose, 5 mM KH2PO4, 10 mM TES, 10 mM NaCl, 2 mM MgSO4, 0.1% (w/v) BSA in MilliQ water, pH 7.2 at 25 °C; Jacoby et al., 2015). To quantify Pro-dependent respiration, 250 µM ADP and 6 mM Pro were used to activate the mETC. To quantify complexes I and II dependent respiration, 5 mM succinate, 1 mM NADH, and 250 µM ADP were used to activate the mETC. Electron capacity via the COX pathway was measured as oxygen consumption rates sensitive to KCN treatment. KCN (2 mM) was used to inhibit the COX pathway and determine AOX capacity. nPG (0.1 mM) was used to inhibit the AOX pathway and determine background respiratory activity.
Transcript analysis
Leaf discs were subjected to metabolite incubations as described above. Following 14 h of metabolite exposure, leaf samples were collected and snap frozen in liquid nitrogen. For transcript analysis of WT plants during salinity stress, leaf samples were harvested after obtaining photosynthetic ETR curves at the respective time points. In all cases, two leaf discs were combined to form one biological replicate and powdered using a mixer mill. RNA extraction, cDNA synthesis and RT-qPCR were performed in accordance with O'Leary et al. (2020). Analysis of amplification curves was done using LinregPCR (Ruijter et al., 2009). Absolute N0 values of three technical replicates were averaged for each of the biological replicates (n = 4). Relative transcript abundance was obtained by normalizing to two housekeeping genes (AtACTIN2 and AtUBIQUITIN10). The two normalized values were then combined using geometric mean calculation. Primers used in RT-qPCR can be found in Supplemental Table S5.
Quantification of Pro content
Two leaf discs were harvested from two leaves from each pot and combined to form one biological replicate. Leaf samples were powdered using a mixer mill and measurement of Pro content was performed as detailed previously in O'Leary et al. (2020). Samples were extracted once in 200 µL of 80% (v/v) ethanol at 80 °C for 20 min and spun down at full speed for 1 min. Resulting supernatant (100 µL) was added to 200 µL of reaction mix (1% [w/v] ninhydrin, 60% [v/v] acetic acid, and 20% [v/v] ethanol). The samples were incubated at 95°C for 20 min, cooled on ice and transferred to a microtiter plate for measurement of A520. Pro concentrations were interpolated using a standard curve.
Extraction of crude mitochondria
Enriched mitochondria extracts were isolated at 4°C from 12 g of mature leaves (8- to 10-week old) following a 14 h incubation in the presence or the absence of Pro. Extraction of crude mitochondria was performed as detailed in Millar et al. (2007). Harvested leaves were ground using a pre-chilled mortar and pestle and 120 mL of extraction buffer (10 mM TES, 0.3 M sucrose, 25 mM tetrasodiumpyrophosphate, 2 mM EDTA, 10 mM KH2PO4, 1% [w/v] polyvinylpyrrolidone 40, 1% [w/v] BSA, adjusted to pH 7.5 with 85% [v/v] phosphoric acid; 20mM ascorbic acid and 5 mM cysteine was added before use). Ground tissue was filtered through two layers of Miracloth and centrifuged for 5 min at 2,500g at 4°C. The supernatant was transferred and centrifuged for 20 min at 18,000g at 4°C. The resulting pellet was washed twice with sucrose wash buffer (0.3 M sucrose, 10 mM TES, 7.4 mM KH2PO4, adjusted to pH 7.5 with NaOH). Washing of the pellet was carried out twice by subjecting samples to the same low and high-speed centrifugation mentioned above. The enriched mitochondria pellet was resuspended in 250 µL of sucrose wash buffer. Enriched mitochondrial protein was quantified using Coomassie Plus (Bradford) Assay Kit (Thermo Fisher Scientific) with BSA as a standard.
Immunoblotting
Enriched mitochondria pellets were extracted in SDS sample buffer (4% [w/v] SDS, 20% [v/v] glycerol, pH 6.8, 125 mM Tris–HCl, 1 µL of bromophenol blue, and 50 mM DTT) containing 2 mM PMSF and cOmplete protease inhibitor cocktail (Roche) according to the manufacturer’s instructions. Samples were run on a 12% precast SDS–PAGE gel (Bio-rad, catalog no. 456046), transferred onto a PVDF membrane and blocked in 2% (w/v) skim milk. The blot was probed at 4°C with orbital shaking overnight with anti-AOX antibody (Agrisera, catalog no. AS04054) at 1:2,000 dilution or anti-VDAC antibody (Agrisera, catalog no. AS07212) at 1:5,000 dilution. Detection of the mitochondrial VDAC protein was used as a loading control. After secondary antibody (Anti-Rabbit, Sigma Aldrich, catalog no. A0545) incubation, the blot was exposed to ECL Western Blotting Substrate (Thermo Fisher Scientific, catalog no. 32106) and imaged using an Amersham 680 Imager CCD camera (GE Life Sciences).
Oxidative stress measurements
Lipid peroxidation was quantified using the thiobarbituric acid reactive substances assay as detailed previously in Hodges et al. (1999) and Calzadilla et al. (2016) with modifications. Following 24 h of Pro exposure, leaf disc samples were collected and snap frozen in liquid nitrogen. Four biological replicates were collected; each biological replicate had eight leaf discs combined and powdered using a mixer mill. 5% (w/v) meta-phosphoric acid (750 µL) and of 2% (w/v) butyl hydroxytoluene (15 µL) were added to the powdered samples, vortexed, and centrifuged for 20 min at 15,000g at 4°C. The resulting supernatant (600 µL) was added to a mixture of 2% (w/v) butyl hydroxytoluene (40 µL), 1% (w/v) thiobarbituric acid (200 µL), and 25% (v/v) HCl (200 µL) and heated at 95°C for 30 min with shaking at 450 rpm. 1-Butanol (700 µL) was added, and the resulting mixture was centrifuged for 2 min at 15,000g. The resulting supernatant (200 µL) was used for quantifying A532 and A600 using a spectrophotometer. The absorbance of MDA was calculated as the difference between A532 and A600.
Asc measurements were performed based on the methods of Foyer et al. (1983), Queval and Noctor (2007) with modifications. Following 4 h of Pro exposure, leaf disc samples were collected and snap frozen in liquid nitrogen. Four biological replicates were collected; each biological replicate had 10 leaf discs combined and powdered using a mixer mill. Ice cold 1 M HClO4 (700 µL) was added to each sample and subjected to centrifugation for 10 min at 16,000g at 4°C. The resulting supernatant (500 µL) was transferred into a new tube containing 50 µL of 0.2-M NaH2PO4 (pH 5.6). The extract was then neutralized with 5 M K2CO3 to pH 5–6 and subjected to centrifugation for 5 min at 16,000g at 4°C. To assay reduced Asc, triplicate aliquots of 40 µL of neutralized supernatant were added to plate wells containing 55 µL of water and 100 µL 0.2 M NaH2PO4 (pH 5.6) and A265 was recorded using a spectrophotometer. To assay total Asc, 150 µL of neutralized supernatant was added to a mixture of 210-µL 0.12 M NaH2PO4 (pH 7.5) and 20 µL of 25 mM DTT and incubated at room temperature for 30 min in the dark. Total Asc was then measured in the same manner as reduced Asc. Oxidized Asc is the difference between total and reduced Asc.
Protein and metabolite mass spectrometry
Targeted mass spectrometry via MRM was employed to quantify mETC protein abundance. Pre-chilled 100% acetone (400 µL) was added to 100 µg of enriched mitochondria and incubated overnight at –20°C. The resulting pellets were washed with acetone thrice. Samples were alkylated, trypsin digested, desalted, and cleaned as previously described by Petereit et al. (2020). Samples were loaded onto an AdvanceBio Peptide Map column (2.1 × 250 mm, 2.7-μm particle size; part number 651750-902, Agilent), using an Agilent 1290 Infinity II LC System coupled to an Agilent 6495 Triple Quadrupole MS as described previously (James et al., 2019).The list of peptide transitions used for MRM is provided in Supplemental Table S6. Peptide abundances from each sample were normalized against VDAC.
Metabolite abundance was measured using gas-chromatography and liquid chromatography mass spectrometry as previously detailed in O’Leary et al. (2017) and Le et al. (2021), respectively.
Photosynthetic ETR measurement
Chloroplastic ETR measurements were performed using an IMAGING-PAM M-Series (MAXI version) (Walz; Effeltrich, Germany). ETR response curves were obtained using an inbuilt script (settings: 2, corresponding to the following PAR: 0, 1, 11, 21, 36, 56, 81, 111, 146, 186, 231, 281, 336, 396, 461, 531, 611, 701, 801, 926, 1,076-µmol photons m−2 s−1, 20 s interval between each measurement). The ETR of technical replicates (6–12 areas of interest) was combined to get the leaf ETR, and the ETR of different leaves for the same treatment were plotted to get the treatment ETR trend. A Loess regression was adjusted to each curve with a 95% confidence interval using the R package ggplot2.
Statistical analysis
All statistical analyses were performed using XLSTAT 2021.3.1. Statistical tests and replicate numbers are as indicated in figure legends. Biological replicates indicate samples that were collected from different plants grown at the same time. Post hoc analyses following ANOVA were performed using the Tukey’s (honestly significant difference) method. All experimental results were repeated at least once with separate batches of plants.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers that can be found in Supplemental Table 1.
Data availability
All data are provided in supplemental files.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript and mitochondrial protein abundance in Pro-treated leaf tissues.
Supplemental Figure S2. Characterization of aox1d and aox1a.aox1d 2 lines.
Supplemental Figure S3. The effect of SHAM and nPG on Pro-stimulated respiration.
Supplemental Figure S4. Time-dependent accumulation of MDA levels in WT and aox1a.aox1d Pro-treated leaf discs.
Supplemental Figure S5. The effect of salinity and post-salinity recovery on Pro accumulation.
Supplemental Figure S6. The effect of salinity and post-salinity recovery on nonphotochemical chlorophyll fluorescence quenching, qL, and Fv/Fm.
Supplemental Table S1. Relative abundance of mitochondrial proteins measured using targeted MRM.
Supplemental Table S2. Two-way ANOVA for proteins that show significant differences following Pro exposure.
Supplemental Table S3. Quantification of cellular metabolites in leaf discs following Pro treatment.
Supplemental Table S4. ANOVAfor Asc, MDA content, and transcript (CAT1, sAPX, At1g19020, and PR1) abundance quantified in Pro-exposed leaf discs.
Supplemental Table S5. Primers used for transcript analysis as outlined in the methods section.
Supplemental Table S6. The MRM transitions used to measure relative abundance of mitochondrial proteins
Supplementary Material
Acknowledgments
We thank Ricarda Fenske for optimizing the method and running the targeted MRM for the mitochondrial protein abundance measurements. We thank Chun-Pong Lee for a critical reading of the manuscript.
Funding
G.G.K.O. was supported by Research Training Program Fee Offset–International Student and UWA Safety-Net Top-Up Scholarships. This work was supported by Australian Research Council funding to A.H.M (CE140100008).
Conflict of interest statement. There are no conflicts of interest.
G.G.K.O., B.M.O., and A.H.M. conceived and designed the project. G.G.K.O. performed the experiments and data analysis. B.M.O. and A.H.M. supervised the experiments. G.G.K.O. and S.S. performed the antioxidant analysis. S.S. optimized experimental conditions for salinity treatments and ETR measurements. G.G.K.O. and B.M.O. wrote the manuscript, S.S. and A.H.M. edited the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: A. Harvey Millar (harvey.millar@uwa.edu.au).
References
- Alber NA, Vanlerberghe GC (2021) The flexibility of metabolic interactions between chloroplasts and mitochondria in Nicotiana tabacum leaf. Plant J 106: 1625–1646 [DOI] [PubMed] [Google Scholar]
- Amthor JS, Bar-Even A, Hanson AD, Millar AH, Stitt M, Sweetlove LJ, Tyerman SD (2019) Engineering strategies to boost crop productivity by cutting respiratory carbon loss. Plant Cell 31:297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Gomez F, Gergoff G, Guiamét JJ, Puntarulo S (2005) Up-regulation of the mitochondrial alternative oxidase pathway enhances photosynthetic electron transport under drought conditions. J Exp Bot 56: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Cabassa-Hourton C, Schertl P, Bordenave-Jacquemin M, Saadallah K, Guivarc'h A, Lebreton S, Planchais S, Klodmann J, Eubel H, Crilat E, et al. (2016) Proteomic and functional analysis of proline dehydrogenase 1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. Biochem J 473: 2623–2634 [DOI] [PubMed] [Google Scholar]
- Calzadilla PI, Signorelli S, Escaray FJ, Menéndez AB, Monza J, Ruiz OA, Maiale SJ (2016) Photosynthetic responses mediate the adaptation of two Lotus japonicus ecotypes to low temperature. Plant Sci 250: 59–68 [DOI] [PubMed] [Google Scholar]
- Chai TT, Simmonds D, Day DA, Colmer TD, Finnegan PM (2012) A GmAOX2b antisense gene compromises vegetative growth and seed production in soybean. Planta 236: 199–207 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta Bioenerg 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Costa JH, McDonald AE, Arnholdt-Schmitt B, Fernandes de Melo D (2014) A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion 19 (Pt B) :172–183 [DOI] [PubMed] [Google Scholar]
- Dahal K, Martyn GD, Vanlerberghe GC (2015) Improved photosynthetic performance during severe drought in Nicotiana tabacum overexpressing a nonenergy conserving respiratory electron sink. New Phytol 208: 382–395 [DOI] [PubMed] [Google Scholar]
- Dahal K, Vanlerberghe GC (2017) Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol 213: 560–571 [DOI] [PubMed] [Google Scholar]
- Del-Saz NF, Ribas-Carbo M, McDonald AE, Lambers H, Fernie AR, Florez-Sarasa I (2018) An in vivo perspective of the role(s) of the alternative oxidase pathway. Trends Plant Sci 23: 206–219 [DOI] [PubMed] [Google Scholar]
- Demircan N, Cucun G, Uzilday B (2020) Mitochondrial alternative oxidase (AOX1a) is required for the mitigation of arsenic-induced oxidative stress in Arabidopsis thaliana. Plant Biotechnol Rep 14: 235–245 [Google Scholar]
- Dinakar C, Vishwakarma A, Raghavendra AS, Padmasree K (2016) Alternative oxidase pathway optimizes photosynthesis during osmotic and temperature stress by regulating cellular ROS, malate valve and antioxidative systems. Front Plant Sci 7: 68–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald SP, Sun X-Y, Hu C-AA, Yu J, Mei JM, Valle D, Phang JM (2001) Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res 61: 1810–1815 [PubMed] [Google Scholar]
- Feng H, Guan D, Sun K, Wang Y, Zhang T, Wang R (2013) Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochim Biophys Sin 45: 985–994 [DOI] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139: 1795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani G, Scainelli D, Nielsen E (1997) [delta]1-Pyrroline-5-carboxylate dehydrogenase from cultured cells of potato (purification and properties). Plant Physiol 113: 1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C, Rowell J, Walker D (1983) Measurement of the ascorbate content of spinach leaf protoplasts and chloroplasts during illumination. Planta 157: 239–244 [DOI] [PubMed] [Google Scholar]
- Funck D, Eckard S, Muller G (2010) Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin A, Duffes C, Day DA, Cousins AB (2012) The absence of alternative oxidase AOX1A results in altered response of photosynthetic carbon assimilation to increasing CO(2) in Arabidopsis thaliana. Plant Cell Physiol 53: 1627–1637 [DOI] [PubMed] [Google Scholar]
- Garmash EV, Velegzhaninov IO, Ermolina KV, Rybak AV, Malyshev RV (2020) Altered levels of AOX1a expression result in changes in metabolic pathways in Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation. Plant Sci 291: 110332. [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LHM, Clifton R, Carroll A, Estavillo G, Tan Y-F, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Li S, Zheng Y, Duan H, Xiao F, Zhuang Y, He J, Wu G, Zhao S, Zhou H, et al. (2020) SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J 102: 1157–1171 [DOI] [PubMed] [Google Scholar]
- Green R, Fluhr R (1995) UV-B-Induced PR-1 accumulation is mediated by active oxygen species. Plant Cell 7: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA, Van Staden J (1998) Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ 21: 535–553 [Google Scholar]
- Hodges DM, Delong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611 [DOI] [PubMed] [Google Scholar]
- Huang C, He W, Guo J, Chang X, Su P, Zhang L (2005) Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J Exp Bot 56: 3041–3049 [DOI] [PubMed] [Google Scholar]
- Huang S, Van Aken O, Schwarzländer M, Belt K, Millar AH (2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants Plant Physiol 171: 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby RP, Millar AH, Taylor NL (2015) Assessment of respiration in isolated plant mitochondria using Clark-type electrodes. Methods Mol Biol 1305: 165–185 [DOI] [PubMed] [Google Scholar]
- James AM, Haywood J, Leroux J, Ignasiak K, Elliott AG, Schmidberger JW, Fisher MF, Nonis SG, Fenske R, Bond CS, et al. (2019) The macrocyclizing protease butelase 1 remains autocatalytic and reveals the structural basis for ligase activity. Plant J 98: 988–999 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Watanabe CKA, Miyagi A, Kawai-Yamada M, Terashima I, Noguchi K (2019) Mitochondrial AOX supports redox balance of photosynthetic electron transport, primary metabolite balance, and growth in Arabidopsis thaliana under high light. Int J Mol Sci 20:3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konert G, Trotta A, Kouvonen P, Rahikainen M, Durian G, Blokhina O, Fagerstedt K, Muth D, Corthals GL, Kangasjärvi S (2015) Protein phosphatase 2A (PP2A) regulatory subunit B′γ interacts with cytoplasmic ACONITASE 3 and modulates the abundance of AOX1A and AOX1D in Arabidopsis thaliana. New Phytol 205: 1250–1263 [DOI] [PubMed] [Google Scholar]
- Launay A, Cabassa-Hourton C, Eubel H, Maldiney R, Guivarc'h A, Crilat E, Planchais S, Lacoste J, Bordenave-Jacquemin M, Clement G, et al. (2019) Proline oxidation fuels mitochondrial respiration during dark-induced leaf senescence in Arabidopsis thaliana. J Exp Bot 70: 6203–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le XH, Lee C-P, Millar AH (2021) The mitochondrial pyruvate carrier (MPC) complex mediates one of three pyruvate-supplying pathways that sustain Arabidopsis respiratory metabolism. Plant Cell 33: 2776–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Liddell A, Leaver CJ (2007) Isolation and subfractionation of mitochondria from plants. In LA Pon and EA Schon, eds, Methods in Cell Biology, Vol. 80. Academic Press, Cambridge, MA, pp 65–90: [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA (2011) Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Millar H, Considine MJ, Day DA, Whelan J (2001) Unraveling the role of mitochondria during oxidative stress in plants. IUBMB Life 51: 201–205 [DOI] [PubMed] [Google Scholar]
- Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A (2009) Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem 284: 26482–26492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- O'Leary BM, Asao S, Millar AH, Atkin OK (2019) Core principles which explain variation in respiration across biological scales. New Phytol 222: 670–686 [DOI] [PubMed] [Google Scholar]
- O'Leary BM, Lee CP, Atkin OK, Cheng R, Brown TB, Millar AH (2017) Variation in leaf respiration rates at night correlates with carbohydrate and amino acid supply. Plant Physiol 174: 2261–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary BM, Oh GGK, Lee CP, Millar AH (2020) Metabolite regulatory interactions control plant respiratory metabolism via target of rapamycin (TOR) kinase activation. Plant Cell 32: 666–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary BM, Oh GGK, Millar AH (2022) High-throughput oxygen consumption measurements in leaf tissue using oxygen sensitive fluorophores. Methods Mol Biol 2363: 63–75 [DOI] [PubMed] [Google Scholar]
- O'Leary BM, Plaxton WC (2016) Plant respiration. IneLS, John Wiley & Sons, Ltd, Chichester, pp 1–11 [Google Scholar]
- Ordog SH, Higgins VJ, Vanlerberghe GC (2002) Mitochondrial alternative oxidase is not a critical component of plant viral resistance but may play a role in the hypersensitive response. Plant Physiol 129: 1858–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore D, Trono D, Laus MN, Di Fonzo N, Passarella S (2001) Alternative oxidase in durum wheat mitochondria. activation by pyruvate, hydroxypyruvate and glyoxylate and physiological role. Plant Cell Physiol 42: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Petereit J, Duncan O, Murcha MW, Fenske R, Cincu E, Cahn J, Pružinská A, Ivanova A, Kollipara L, Wortelkamp S, et al. (2020) Mitochondrial CLPP2 assists coordination and homeostasis of respiratory complexes. Plant Physiol 184: 148–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Podestá FE (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198 [Google Scholar]
- Queval G, Noctor G (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (. 2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Geisler DA, Moller IM (2008) The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8: 47–60 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Møller IM (2011) Mitochondrial electron transport and plant stress. InKempken F, ed., Plant Mitochondria. Springer, New York, NY, pp 357–381 [Google Scholar]
- Reynolds M, Atkin OK, Bennett M, Cooper M, Dodd IC, Foulkes MJ, Frohberg C, Hammer G, Henderson IR, Huang B, et al. (2021) Addressing research bottlenecks to crop productivity. Trends Plant Sci 26: 607–630 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, Van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, König A-C, Sweetlove LJ, Finkemeier I (2011) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Selinski J, Hartmann A, Deckers-Hebestreit G, Day DA, Whelan J, Scheibe RCP (2018a) Alternative oxidase isoforms are differentially activated by tricarboxylic acid cycle intermediates. Plant Physiol 176: 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J, Scheibe R, Day DA, Whelan J (2018b) Alternative oxidase is positive for plant performance. Trends Plant Sci 23: 588–597 [DOI] [PubMed] [Google Scholar]
- Servet C, Ghelis T, Richard L, Zilberstein A, Savoure A (2012) Proline dehydrogenase: a key enzyme in controlling cellular homeostasis. Front Biosci (Landmark Ed) 17: 607–620 [DOI] [PubMed] [Google Scholar]
- Smith CA, Melino VJ, Sweetman C, Soole KL (2009) Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol Plant 137: 459–472 [DOI] [PubMed] [Google Scholar]
- Sofo A, Scopa A, Nuzzaci M, Vitti A (2015) Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int J Mol Sci 16: 13561–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P, Johnson GN (2008) Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol 149: 1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strodtkotter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, et al. (2009) Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297 [DOI] [PubMed] [Google Scholar]
- Sun LJ, Zhao XY, Ren J, Yan SP, Zhao XY, Song XS (2021) Overexpression of Cerasus humilis ChAOX2 improves the tolerance of Arabidopsis to salt stress. 3 Biotech 11: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97 [DOI] [PubMed] [Google Scholar]
- Tcherkez G, Gauthier P, Buckley TN, Busch FA, Barbour MM, Bruhn D, Heskel MA, Gong XY, Crous KY, Griffin K, et al. (2017) Leaf day respiration: low CO2 flux but high significance for metabolism and carbon balance. New Phytol 216: 986–1001 [DOI] [PubMed] [Google Scholar]
- Trovato M, Forlani G, Signorelli S, Funck D (2019) Proline metabolism and its functions in development and stress tolerance. InHossain MA, Kumar V, Burritt DJ, Fujita M, Mäkelä PSA, eds, Osmoprotectant-Mediated Abiotic Stress Tolerance in Plants: Recent Advances and Future Perspectives. Springer International Publishing, Cham, pp 41–72 [Google Scholar]
- Umbach AL, Zarkovic J, Yu J, Ruckle ME, McIntosh L, Hock JJ, Bingham S, White SJ, George RM, Subbaiah CC, et al. (2012) Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS One 7: e44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C (2008) Proline accumulation in plants: a review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Sharma S (2010) Proline Metabolism and Its Implications for Plant-Environment Interaction. SPIE, Bellingham, WA; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma A, Bashyam L, Senthilkumaran B, Scheibe R, Padmasree K (2014) Physiological role of AOX1a in photosynthesis and maintenance of cellular redox homeostasis under high light in Arabidopsis thaliana. Plant Physiol Biochem 81: 44–53 [DOI] [PubMed] [Google Scholar]
- Vishwakarma A, Tetali SD, Selinski J, Scheibe R, Padmasree K (2015) Importance of the alternative oxidase (AOX) pathway in regulating cellular redox and ROS homeostasis to optimize photosynthesis during restriction of the cytochrome oxidase pathway in Arabidopsis thaliana. Ann Bot 116: 555–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe CK, Hachiya T, Takahara K, Kawai-Yamada M, Uchimiya H, Uesono Y, Terashima I, Noguchi K (2010) Effects of AOX1a deficiency on plant growth, gene expression of respiratory components and metabolic profile under low-nitrogen stress in Arabidopsis thaliana. Plant Cell Physiol 51: 810–822 [DOI] [PubMed] [Google Scholar]
- Watanabe CKA, Yamori W, Takahashi S, Terashima I, Noguchi K (2016) Mitochondrial alternative pathway-associated photoprotection of photosystem II is related to the photorespiratory pathway. Plant Cell Physiol 57: 1426–1431 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Bhuiyan MNH, Waditee R, Tanaka Y, Esaka M, Oba K, Jagendorf AT, Takabe T (2005) Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J Exp Bot 56: 1785–1796 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K (1995) Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7: 751–760 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K (2006) Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol 47: 22–31 [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M (2012) Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab 15: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in supplemental files.