Abstract
Glandular trichomes (GTs) are epidermal structures that provide the first line of chemical defense against arthropod herbivores and other biotic threats. The most conspicuous structure on leaves of cultivated tomato (Solanum lycopersicum) is the type-VI GT (tVI-GT), which accumulates both flavonoids and volatile terpenoids. Although these classes of specialized metabolites are derived from distinct metabolic pathways, previous studies with a chalcone isomerase 1 (CHI1)-deficient mutant called anthocyanin free (af) showed that flavonoids are required for terpenoid accumulation in tVI-GTs. Here, we combined global transcriptomic and proteomic analyses of isolated trichomes as a starting point to show that the lack of CHI1 is associated with reduced levels of terpenoid biosynthetic transcripts and enzymes. The flavonoid deficiency in af trichomes also resulted in the upregulation of abiotic stress-responsive genes associated with DNA damage and repair. Several lines of biochemical and genetic evidence indicate that the terpenoid defect in af mutants is specific for the tVI-GT and is associated with the absence of bulk flavonoids rather than loss of CHI1 per se. A newly developed genome-scale model of metabolism in tomato tVI-GTs helped identify metabolic imbalances caused by the loss of flavonoid production. We provide evidence that flavonoid deficiency in this cell type leads to increased production of reactive oxygen species (ROS), which may impair terpenoid biosynthesis. Collectively, our findings support a role for flavonoids as ROS-scavenging antioxidants in GTs.
Flavonoid deficiency in glandular trichomes of tomato leads to a cell type-specific increase in the accumulation of reactive oxygen species and downregulation of terpenoid biosynthesis.
Introduction
Many plants have anatomical structures and specialized cell types that produce defense-related compounds. Prominent examples include resin ducts, laticifers, and glandular trichomes (GTs). Confining the high-level production and storage of specialized metabolites to these structures is thought to minimize adverse effects of autotoxicity or interference with primary metabolism needed for normal plant growth and development (Kliebenstein, 2013; Lange and Turner, 2013; Lange, 2015; Guo et al., 2018). Owing to their abundance and ease of isolation, GTs have received attention as a source of plant compounds that have practical uses as medicines, flavors, and food additives (Duke et al., 2000; Gibon et al., 2009; Schilmiller et al., 2010; Tissier, 2018). A better understanding of factors affecting GT metabolic pathways is therefore of both fundamental and applied interest.
In providing a physicochemical interface between the plant and its environment, GTs constitute an important line of chemical defense against various plant consumers. Studies of mutants that are impaired in the development or biosynthetic capacity of GTs have helped to discern the role of these structures in plant anti-insect defense. Defects in GT development in the odorless-2 (od-2) mutant of cultivated tomato (Solanum lycopersicum), for example, are associated with increased susceptibility to attack by larvae of several beetle species (Kang et al., 2010a). Loss of resistance to herbivores was also observed in the hairless (hl) and anthocyanin-free (af) mutants of tomato, which are defective in the production of specialized metabolites in the abundant type VI GTs (tVI-GTs) (Kang et al., 2010b, 2014, 2016). Transgenic tobacco (Nicotiana tabacum) plants that overexpress a B-type cyclin gene were shown to accumulate reduced levels of terpenoids in GTs, which corresponded with a reduced resistance to insect feeding (Gao et al., 2017). Conversely, metabolic engineering of S. lycopersicum GTs to produce defensive metabolites found in wild tomato species resulted in enhanced herbivore resistance (Bleeker et al., 2012).
The metabolic activity of tVI-GTs in tomato is dominated by the synthesis and accumulation of flavonoids and terpenoids (Kang et al., 2010b; McDowell et al., 2011), which constitute two diverse classes of specialized metabolites in plants. Available evidence indicates that trichome-borne terpenoids serve an important function in resistance of tomato to insect herbivores (Kennedy, 2003; Li et al., 2004; Bleeker et al., 2009, 2012). Although the role of GT-localized flavonoids in protection against biotic stress is less clear, the flavonol quercetin and its glycosides (e.g. rutin), which are highly abundant in tVI-GTs, exert growth-retarding effects on insects when added to artificial diets (Isman and Duffey, 1982, 1983). An increasing body of literature further indicates that flavonols are critical for detoxification of reactive oxygen species (ROS) in various tissues and cell types of tomato, including root hairs, guard cells, and pollen (Maloney et al., 2014; Watkins et al., 2017; Muhlemann et al., 2018). In addition to providing a chemical screen for ultraviolet (UV) radiation, it has been proposed that the antioxidant properties of some GT-borne flavonoids may reduce oxidative damage caused by short-wavelength solar radiation (Tattini et al., 2000; Agati and Tattini, 2010). Whether flavonoid production in GTs has a role in mitigating the effects of stress-associated ROS production has yet to be addressed through the use of flavonoid-deficient mutants.
The flavonoid and terpenoid biosynthetic pathways are metabolically distinct and generally assumed to operate independently. We recently observed, however, that the anthocyanin free (af) mutant of tomato, which fails to produce a specific isoform, chalcone isomerase 1 (CHI1), of the flavonoid biosynthetic enzyme CHI, not only lacks anthocyanins and flavonoids in all tissues but is also severely impaired in volatile terpenoid production in tVI-GTs (Kang et al., 2014). The underlying mechanism by which CHI1 promotes trichome-borne production of volatile terpenoids remained to be determined. This study was designed to address this question, using tVI-GT-specific transcriptomic and proteomic datasets as a starting point. We show that the loss of CHI1 results in the downregulation of terpenoid biosynthetic genes and enzymes in tVI-GTs of the af mutant, concomitant with upregulation of genes involved in DNA repair and response to radiation. A series of follow-up experiments provided evidence that these phenotypes are linked to elevated ROS accumulation under flavonoid-deficient conditions in af trichomes. These findings are consistent with a role for flavonoids as ROS-scavenging antioxidants in a specialized cell type that is adapted to function under high levels of solar radiation.
Results
Loss of CHI1 results in global changes in metabolic and stress response pathways in tVI-GTs
To assess the global contribution of CHI1 to metabolic and cellular processes in tVI-GTs, we obtained transcriptome datasets with tVI gland cells isolated from the af mutant and appropriate isogenic control lines. Because the af mutant was generated by X-ray mutagenesis (Von Wettstein-Knowles, 1968), this line could potentially harbor other mutations that confound the interpretation of gene expression profiles. Therefore, as an isogenic “wild-type” control, we used two independent transgenic lines (designated CHI4 and CHI9) in which the wild-type CHI1 gene was expressed in the af mutant background under control of the native CHI1 promoter, fully complementing the af phenotypes (Kang et al., 2014). Transcriptome data sets were generated by mRNA sequencing (RNA-seq) of RNA isolated from af and the two complemented lines (CHI4 and CHI9), using four biological replicates per genotype (12 independent samples) and two technical replicates per sample (Supplemental Figure S1; Supplemental Dataset S1). The following work flow was used to identify a robust set of genes that are differentially expressed in tVI-GTs as a result of CHI1 deficiency (Supplemental Table S1). First, the RNA-seq data were processed with three independent analysis programs (EdgeR;Robinson et al. (2010), TCC; Sun et al. (2013), and DESeq2; Love et al. (2014)) to identify differentially expressed genes (DEGs) in the af versus CHI4 and af versus CHI9 comparisons (threshold of log2 > 1.0 or less than −1.0 and adjusted P-value of permutation test <0.05). Second, we compared the DEGs obtained for each of the three data analysis tools to arrive at a list of common DEGs within each of the two genotypic comparisons. Finally, we compared the list of common DEGs between the two genotypic comparisons to arrive at a set of consensus DEGs. Using this filtering approach, the transcript abundance of 516 and 642 genes was determined to be lower in the af mutant compared to CHI4 and CHI9, respectively. Among these genes, all three analysis tools identified an overlapping, consensus set of 384 transcripts whose abundance was lower in af than in both CHI4 and CHI9 (Supplemental Table S1). Likewise, the transcript abundance of 1,602 and 1,533 genes was determined to be higher in the af mutant compared to CHI4 and CHI9, respectively, with a consensus set of 930 genes expressed to higher levels in af relative to both CHI4 and CHI9 (Supplemental Table S1).
We used the consensus list of 384 downregulated and 930 upregulated genes in af for gene ontology (GO) enrichment analysis to gain insights into processes within tVI-GT cells that are affected by the loss of CHI1. Genes that were downregulated in the af mutant showed strong enrichment of GO categories related to metabolic pathways operating mainly in the chloroplast (Table 1;Supplemental Table S2). Most prominent among these pathways were isoprenoid/terpenoid metabolism, lipid metabolism, amino acid metabolism, and photosynthesis-related processes. GO analysis of the 930 genes that are upregulated in tVI gland cells of the af mutant revealed enrichment of functional categories related to abiotic stress responses. Categories related to DNA damage and repair were particularly overrepresented among upregulated genes in af trichomes, as were responses to heat, high light, and ROS (Table 1;Supplemental Table S2). These data indicate that the loss of CHI1 in af GTs both impairs the expression of genes involved in chloroplast metabolism and also enhances transcriptional responses related to high light and temperature stress.
Table 1.
Gene ontologies of differentially regulated transcripts in tVI-GTs of the tomato af mutant
| GO ID | GO description of genes downregulated in af mutanta | FDR | P-value |
|---|---|---|---|
| 0055114 | Oxidation–reduction process | 1.14E-11 | 2.77E-15 |
| 0044711 | Single-organism biosynthetic process | 4.87E-10 | 6.70E-13 |
| 1901565 | Organonitrogen compound catabolic process | 4.87E-10 | 7.05E-13 |
| 0006629 | Lipid metabolic process | 1.47E-09 | 2.48E-12 |
| 0008299 | Isoprenoid biosynthetic process | 1.64E-09 | 2.96E-12 |
| 0015979 | Photosynthesis | 1.99E-08 | 4.79E-11 |
| 1901606 | Amino acid catabolic process | 6.82E-07 | 1.97E-09 |
| 0016114 | Terpenoid biosynthetic process | 1.80E-06 | 6.30E-09 |
| 0042440 | Pigment metabolic process | 2.65E-06 | 9.91E-09 |
|
| |||
| GO ID | GO description of genes upregulated in af mutanta | FDR | P-value |
|
| |||
| 0006310 | DNA recombination | 3.97E-03 | 2.37E-06 |
| 0009408 | Response to heat | 4.08E-03 | 3.59E-06 |
| 0006298 | Mismatch repair | 4.91E-03 | 5.33E-06 |
| 0051716 | Cellular response to stimulus | 8.00E-03 | 1.54E-05 |
| 0009314 | Response to radiation | 1.04E-02 | 2.22E-05 |
| 0007165 | Signal transduction | 1.19E-02 | 2.88E-05 |
| 0009628 | Response to abiotic stimulus | 1.38E-02 | 3.48E-05 |
| 1901700 | Response to oxygen-containing compound | 1.71E-02 | 5.19E-05 |
Differential gene expression was determined by an RNA-seq experiment in which transcript abundance in isolated tVI trichomes from the CHI1-deficienct af mutant was compared to transcript levels in two CHI1-complemented control lines.
To further investigate the effects of CHI1-deficiency on cellular processes in tVI gland cells, we performed a quantitative proteomics analysis of extracts obtained from isolated tVI-GTs. Because the CHI4 and CHI9 lines showed very similar patterns of global gene expression (Supplemental Figure S1), we used only the CHI9 isogenic line as a control for proteomic analysis of three independent trichome gland preparations for each genotype (af and CHI9). Differential protein abundance in tVI-GTs of CHI9 versus af was assessed by isobaric mass-tag labeling (see “Materials and methods”) and was calculated on the basis of adjusted spectral counts (filter of log2 > 0.3 or less than −0.3 and adjusted P-value of permutation test <0.05). Comparison of the aggregate spectral-count data in all six samples showed that the protein content of GTs from the CHI9 and af lines was distinct (Supplemental Figure S1). Of 4,992 unique proteins represented by more than one peptide in our dataset, the abundance of 133 proteins was determined to be higher in tVI-GTs of the af mutant compared to CHI9, whereas the abundance of 52 proteins was lower in the same comparison (Supplemental Dataset S2). These results indicate that the overall effects of CHI1 deficiency were much less pronounced at the protein abundance level than they were at the transcript level.
We found that proteins with functions in isoprenoid biosynthesis, volatile oxylipin biosynthesis, and photosynthesis were less abundant in GTs of the af mutant relative to the CHI9 control (Supplemental Dataset S2). In agreement with the terpenoid deficiency of af plants (Kang et al., 2014), mutant GTs contained reduced levels of several terpenoid biosynthetic enzymes, including 1-deoxy-D-xylulose 5-phosphate synthase (Solyc11g010850), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (Solyc03g114340), undecaprenyl pyrophosphate synthase (Solyc08g005680), and various terpene synthases (Solyc08g005640, Solyc08g005670, and Solyc06g059930). Two additional chloroplast-localized proteins, lipoxygenase C and fatty acid hydroperoxide lyase (encoded by Solyc01g006540 and Solyc07g049690, respectively) involved in the formation of green leaf volatiles were also downregulated in the af trichomes. Further validation of the proteomics approach came from the finding that CHI1 (encoded by Solyc05g010320) protein levels were lower in af compared to CHI9, which accords with the fact that CHI1 does not accumulate in the af mutant (Kang et al., 2014).
Among the 133 proteins that overaccumulated in af GTs were numerous stress-associated enzymes, including glutathione S-transferases, heat shock proteins, and molecular chaperones (Supplemental Dataset S2). We also found that af trichomes had increased levels of the UV-B receptor (UVR8; encoded by Solyc5g018620) and two UVR8-like proteins as well (encoded by Solyc02g085780 and Solyc04g081200). UVR8 mediates responses of tomato to UV-B stress by activating the expression of CHALCONE SYNTHASE (CHS) and other flavonoid biosynthetic genes (Li et al., 2018). Consistent with this, our RNA-seq data showed that CHS and most other flavonoid biosynthetic genes were upregulated in the af mutant (Figure 1; Supplemental Dataset S1). Two exceptions to this trend were the gene encoding flavonol synthase (Solyc10g083440), which had low expression levels across all samples, and the CHI1 gene that is mutated in af (Figure 1).
Figure 1.
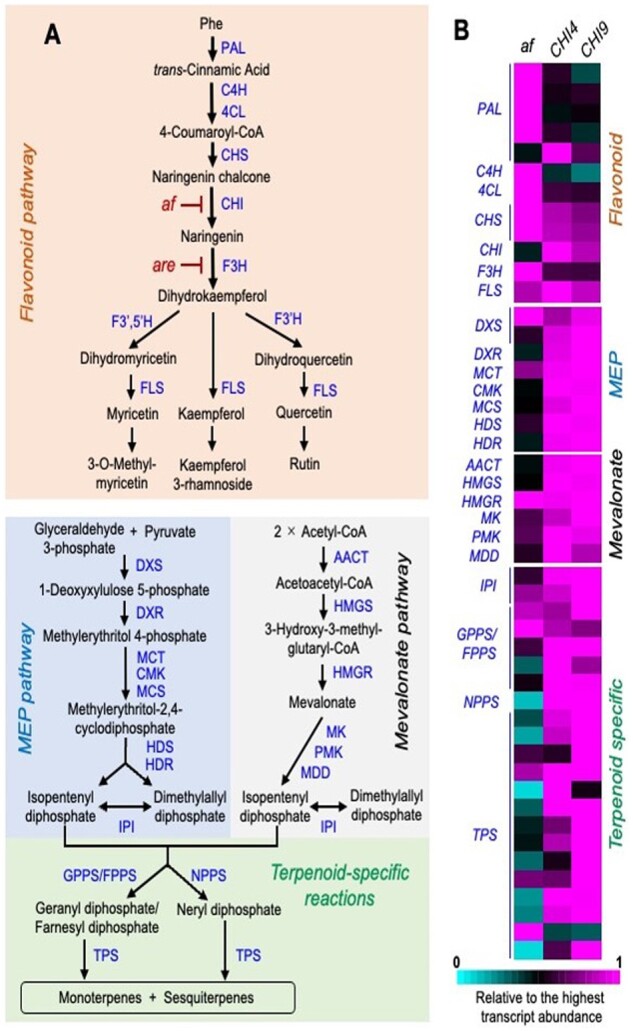
Loss of CHI1 in the af mutant results in higher expression of tVI-GT-expressed genes involved in flavonoid biosynthesis and lower expression of genes involved in terpenoid biosynthesis. A, Metabolic scheme of the flavonoid pathway (orange), MEP pathway (blue), mevalonate pathway (gray), and terpenoid-specific reactions (green). B, Heatmap representation of the relative transcript abundance of genes involved in flavonoid, MEP, mevalonate, and terpenoid-specific pathways. The color code depicts the relative expression level of a given gene in af, CHI4, and CHI9 GTs normalized to the highest expression value among genotypes, with the cut-off for expression level set at 50 transcripts per kilobase million. The complete list of gene identifiers and expression levels is provided in Supplemental Dataset S1. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-hydroxy-cinnamoyl CoA ligase; F3H, flavanone-3-hydroxylase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid-3′5′-hydroxylase; FLS, flavonol synthase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT, 2C-methyl-D-erythritol 4-phosphate cytidyltransferase; CMK, 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase; MCS, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, 1-hydroxy-2-methyl-2-butenyl 4-phosphate synthase; HDR, 1-hydroxy-2-methyl-2-butenyl 4-phosphate reductase; AACT, acetoacetyl-CoA thiolase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MDD, mevalonate 5-diphosphate decarboxylase; IPI, isopentenyl diphosphate isomerase; GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; NPPS, neryl diphosphate synthase; TPS, terpene synthase. The flavonoid biosynthetic reaction that is impaired in the af and are mutants are indicated in red.
Terpenoid deficiency in the af mutant is specific to glandular trichomes
We next investigated the extent to which the flavonoid deficiency in the af mutant disrupts various isoprenoid-related processes in tVI-GTs and whole leaves. In-depth analysis of the RNA-seq data revealed that the af mutation broadly downregulates the expression of genes involved in the biosynthesis of monoterpenes and sesquiterpenes in tVI-GTs (Figure 1). Among these genes were those encoding enzymes in the methylerythritol 4-phosphate (MEP) pathway, which provides precursors for monoterpene biosynthesis, as well as the mevalonic acid (MVA) pathway, which is largely responsible for precursor supply into the sesquiterpene biosynthetic pathway (Hemmerlin et al., 2012). Reverse transcription-quantitative PCR (RT-qPCR) assays confirmed the attenuated expression in af trichomes of 11 terpenoid biosynthetic genes encoding enzymes that span the MEP and MVA pathways (Supplemental Figure S2). Analysis of volatile metabolites by gas chromatography–mass spectroscopy (GC–MS) confirmed that af trichomes accumulate very low levels of 10 individual terpenoids, including monoterpenes (e.g. β-phellandrene) and sesquiterpenes (e.g. β-caryophyllene) that are abundant in wild-type tVI-GTs (Supplemental Figure S3). We found that the total level of terpene volatiles (mono and sesquiterpenes) in tVI-GTs of the af mutant was <10% of that in GTs from the isogenic control lines (Figure 2A). These data demonstrate that the absence of CHI1 in tVI-GTs abrogates the expression of terpenoid biosynthetic genes and enzymes, as well as the production of volatile terpenoids in this cell type.
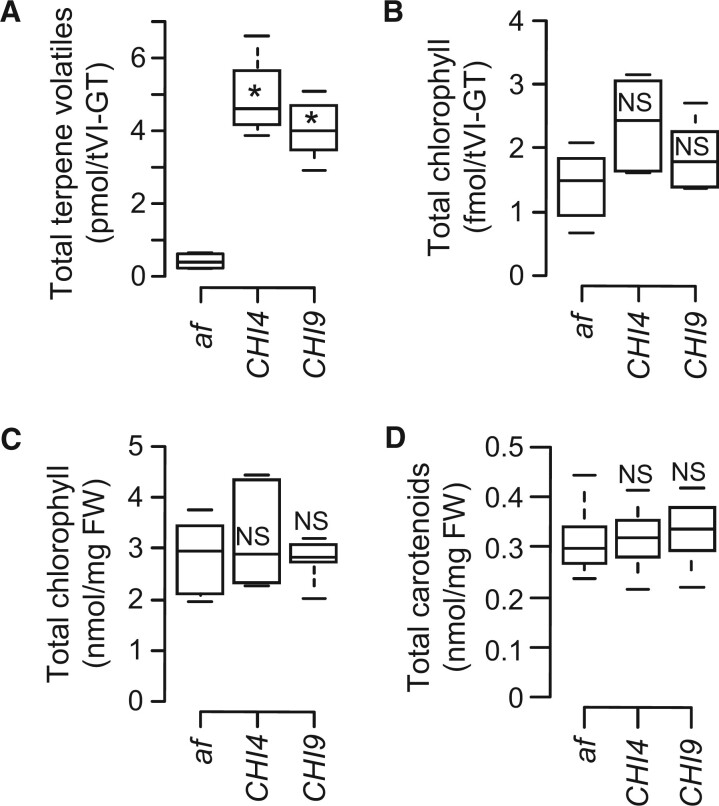
Figure 2.
Downregulation of terpenoid biosynthesis in the af mutant is restricted to tVI-GTs. A, tVI-GTs of the af mutant accumulate low levels of volatile terpenoids (combined mono- and sesquiterpenes) relative to two CHI1-complemented lines (CHI4 and CHI9) (n = 6 independent collections of hand-picked trichomes per genotype). Quantitative data for individual terpenoids are shown in Supplemental Figure S3. B and C, Total chlorophyll (chl a and chl b) levels in isolated tVI-GTs (B, n = 5 independent collections of hand-picked trichomes per genotype) and in whole leaves (C, n = 12 plants per genotype from four independent experiments). D, Bulk carotenoid content in leaves of the indicated genotypes (n = 12 plants per genotype from four independent experiments). Center lines, box range, whiskers, and points indicate the median, lower to upper quartile, 1.5× interquartile, and outliers, respectively. Statistical significance was analyzed by Dunnett’s multiple comparison method and is indicated with an asterisk for P ≤ 0.01. NS, no statistical difference in comparison of the af mutant to isogenic control lines.
Compared to the large negative effect of CHI1 deficiency on terpenoid accumulation in tVI-GTs, the levels of chlorophylls, which contain a C20 phytol side chain derived from the general terpenoid pathway, were not significantly different in tVI-GTs of the af mutant compared to the two control lines (Figure 2B). Consistent with this finding, RNA-seq data showed that the expression level of chlorophyll metabolic genes in af GTs was comparable to that in trichomes of the isogenic control lines (Supplemental Dataset S1). Analysis of whole leaf samples showed that the levels of bulk chlorophylls and carotenoids (C40 tetraterpenes), derived mainly from mesophyll cells, were also similar between af and the two complemented lines (Figure 2, C and D). These data indicate that the loss of CHI1 has a strong negative effect on the accumulation of terpene volatiles in tVI-GTs but most likely does not impact other terpenoid pathway-derived products (e.g. chlorophylls) in tVI-GTs and other cell types of the leaf.
Nuclear localization of CHI1 is not required for terpenoid accumulation in tVI-GTs
Previous studies have shown that canonical CHI enzymes and their corresponding green fluorescent protein (GFP) fusion proteins accumulate not only in the cytosol, which is the site of flavonoid biosynthesis, but also in the nucleus (Saslowsky et al., 2005; Dastmalchi and Dhaubhadel, 2015). This observation led to the suggestion that CHI may perform a “moonlighting” function in regulating gene expression (Saslowsky et al., 2005). To investigate the subcellular localization of CHI1 in gland cells of the tVI-GTs, we generated a transgenic line (pCHI1::CHI1–GFP) of tomato that stably expresses CHI1–GFP in the af mutant background and under the control of the native CHI1 promoter. Confocal microscopy of intact tVI-GTs from pCHI1::CHI1–GFP plants showed that the CHI1–GFP fusion protein was located in both the cytosol and the nucleus of gland cells (Figure 3A), thus confirming the dual localization of CHI1 in this cell type. The reticulate pattern of CHI1–GFP signal within the cytosol is consistent with the action of CHI on the cytosolic face of the ER (Saslowsky et al., 2005).
Figure 3.
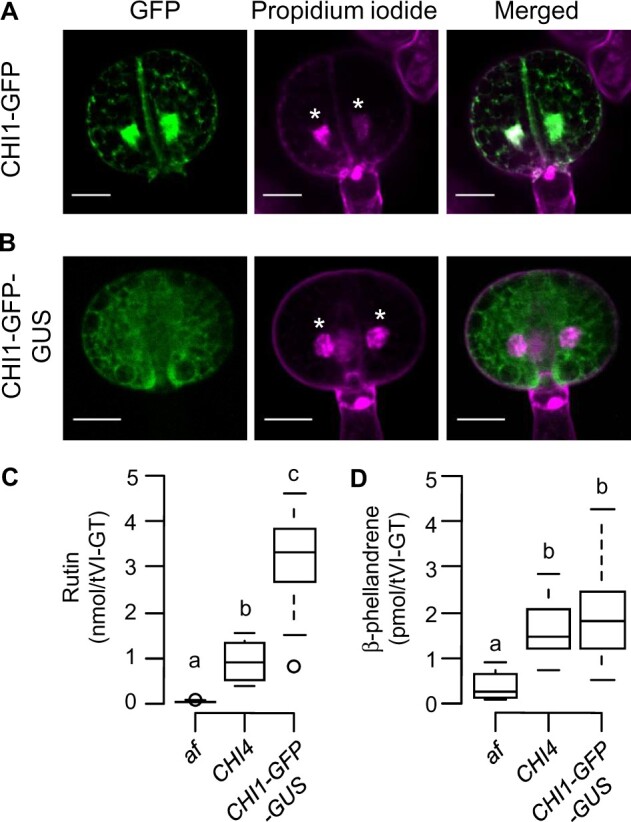
Exclusion of CHI1 from nuclei does not impair terpenoid accumulation in tVI-GTs. A and B, CHI1–GFP (A) or CHI1–GFP–GUS line (B) fusion proteins were expressed in the af mutant background under the control of the native CHI1 promoter. Additional complementation phenotypes in the line expressing CHI1–GFP–GUS (line #7) are provided in Supplemental Figure S4. Confocal microscopy was used to localize the GFP fusion proteins within the four-celled glandular head of tVI-GTs in the corresponding transgenic line. Left, GFP fluorescence; middle, propidium iodide (PI) fluorescence (nuclear stain); right, merged GFP and PI images. Asterisks denote nuclei and scale bars correspond to 20 µM. C and D, Quantification of rutin (C) and β-phellandrene (D) levels in tVI-GTs of the indicated genotypes (n = 8–15 collections of hand-picked trichomes per genotype for (C), and 13 collections of hand-picked trichomes per genotype for (D)). Center lines, box range, whiskers, and points indicate the median, lower to upper quartile, 1.5× interquartile, and outliers, respectively. Different lowercase letters indicate a statistical difference among genotypes by Tukey’s multiple comparisons (P < 0.05).
The presence of CHI1–GFP in nuclei of tVI gland cells raised the possibility that CHI1 may act in the nucleus to positively regulate terpenoid production through the control of terpene biosynthetic gene expression. To test this hypothesis, we transformed the af mutant with a transgene (pCHI1::CHI1–GFP–GUS) that expresses a CHI1–GFP–GUS fusion protein under the control of the native CHI1 promoter. We reasoned that the high molecular mass (predicted 115 kDa) of CHI1–GFP–GUS should exclude its diffusion through the nuclear pore complex (Grossman et al., 2012), thereby attenuating any function of CHI1 in the nucleus. Confocal microscopy of tVI glands from pCHI1::CHI1–GFP–GUS plants showed that CHI1–GFP–GUS maintained the cytosolic, reticulated pattern of CHI1–GFP but, unlike CHI1–GFP, was not detectable in the nucleus (Figure 3B). We next assessed pCHI1::CHI1–GFP–GUS plants and appropriate control lines (af and CHI4) for the accumulation of the dominant flavonoid (rutin) and terpenoid (β-phellandrene) components of tVI-GTs. Consistent with the ability of pCHI1::CHI1–GFP–GUS to complement the anthocyanin deficiency in the af mutant (Supplemental Figure S4), CHI1–GFP–GUS fully restored the accumulation of rutin in tVI-GTs relative to the af mutant and CHI4 controls (Figure 3C). Importantly, we also found that CHI1–GFP–GUS fully complemented the β-phellandrene deficiency of af tVI-GTs (Figure 3D). These data indicate that the role of CHI1 in promoting terpenoid accumulation in tVI-GTs does not likely require a nuclear pool of CHI1 but rather depends on the canonical function of the enzyme in flavonoid biosynthesis.
Genome-scale modeling predicts metabolic perturbations in glandular trichomes of the af mutant
We have previously employed mathematical modeling as a tool for formulating hypotheses concerning GT metabolism in silico, thereby reducing the number of follow-up experiments required for testing the strongest hypotheses (Rios-Estepa et al., 2008, 2010; Lange and Rios-Estepa, 2014; Johnson et al., 2017; Zager and Lange, 2018; Turner et al., 2019). To apply this methodology to this study, we developed a genome-scale model (termed Solyc_tVI-GT_2021v1) of metabolism in tVI-GTs of tomato, which incorporates publicly available information and several datasets acquired as part of this study (details describing the model development process are provided in Supplemental Methods and Data File S1). The first-generation model featured 782 unique, stoichiometrically balanced reactions (Figure 4A;Supplemental Table S3). Using a custom script, each reaction in the model was associated with the appropriate tomato gene(s) and enzyme(s), which allowed us to integrate our transcriptomic and proteomic datasets (Supplemental Table S3). Due to the low abundance of many peptides in the proteomics dataset, simulations focused on constraining fluxes with transcriptome data.
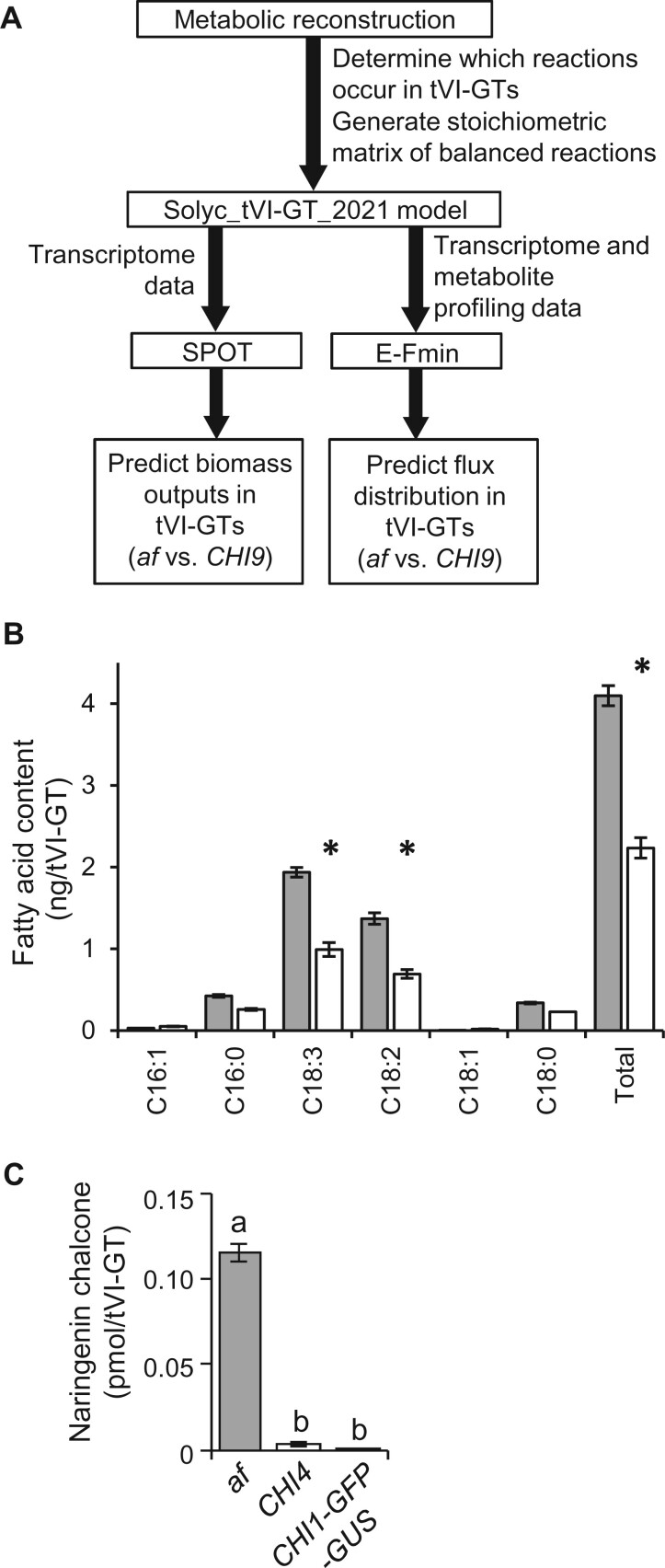
Figure 4.
Experimental testing of predictions of a mathematical model of metabolism in tomato tVI-GTs. A, Overview of workflow for generating the Solyc_tVI-GT_2021 model. B, Quantitative fatty acid analysis of isolated tVI-GTs from the af mutant (gray bars) and CHI9 isogenic control (open bars) to test the Solyc_tVI-GT_2021 prediction that flux through fatty acid biosynthesis is higher in the af mutant compared to CHI9. Asterisks denote P < 0.01 in comparisons between af and CHI9 (n = 5–6). C, Comparison of the amount of naringenin chalcone in tVI-GTs of the indicated genotypes (n = 8–15 collections of hand-picked trichomes per genotype) to test the Solyc_tVI-GT_2021 prediction that only the flavonoid pathway reactions upstream of CHI1, but not those downstream, carry significant flux in the af mutant. Bars represent mean ± standard error. Statistical difference across genotypes, based on Tukey’s multiple comparison test (P < 0.05), is indicated by lowercase letters.
Transcriptome data obtained with isolated tVI-GTs were incorporated using a modified SPOT algorithm (Kim et al., 2016). As an initial test for the validity of the reaction network architecture, we asked whether Solyc_tVI-GT_2021v1 could be employed to predict biomass outputs in the form of metabolites accumulated in tVI-GTs. Consistent with recent metabolic studies of tomato tVI-GTs (Balcke et al., 2017), inputs of the model included sucrose as carbon source, ammonia, oxygen, hydrogen sulfide, inorganic phosphate, water, and carbon dioxide. Metabolic outputs of the model included flavonoids, monoterpenes, sesquiterpenes, acyl sugars, sterols, cell wall carbohydrates, and amino acid pools for protein synthesis. Notably, Solyc_tVI-GT_2021v1 accurately predicted the lack of flavonoid and monoterpene end products in the af mutant, whereas these classes of metabolites were projected to accumulate to appreciable levels in trichomes of the CHI9 isogenic control line (Supplemental Table S3).
As the next step in our in silico analysis, experimentally determined concentrations of more than 50 metabolic end products, including flavonoids, monoterpenes, sesquiterpenes, acyl sugars, sterols, and cell wall carbohydrates, were integrated into Solyc_tVI-GT_2021v1 as part of a biomass equation. We also included the quantity of polyphenol oxidase, which accounts for ∼70% of the total protein in tVI-GTs (Yu et al., 1992) and therefore represents a major sink of amino acids in this cell type (Supplemental Methods and Data File S1). The E-Fmin algorithm (Song et al., 2014) was then employed to obtain a prediction for flux minimization, as a function of gene expression, through the metabolic network by employing the biomass equation as the constraint. Expectedly, flux was predicted to be particularly low toward flavonoid and monoterpene end products in the af mutant, whereas trichomes of the CHI1-complemented line (CHI9) were predicted to have a higher flux through the reactions that generate these compounds. Flux through many other pathways was also predicted to be lower in the af mutant relative to CHI9, which is likely explained by the fact that there is a lower demand for carbon flux through the entire network when flavonoid and terpenoid end product concentrations are substantially reduced or entirely eliminated from consideration.
A notable exception to the general pattern of decreased flux through metabolic pathways in af GTs were the reactions of fatty acid biosynthesis, which were predicted by Solyc_tVI-GT_2021v1 to carry more flux (increased by 15%) in the af mutant relative to CHI9 (Supplemental Table S3). These results could not have been expected because fatty acid quantities had not been determined and integrated into the biomass equation. To further test the utility of our model, we quantified total fatty acids in isolated tVI-GTs. Indeed, the total fatty acid content in tVI-GTs was determined to be 1.8-fold higher in the af mutant compared to CHI9 (Figure 4B). This difference was primarily the result of increased accumulation of polyunsaturated fatty acids. For example, the levels of linoleic acid (18:2) and linolenic acid (18:3) were both increased approximately two-fold in af compared to CHI9 tVI-GTs (Figure 4B). Our RNA-seq data set showed that seven tomato homologs of functionally characterized fatty acid desaturases (FADs) in Arabidopsis thaliana (Li-Beisson et al., 2013) were expressed to varying levels in tVI-GTs. Among these expressed SlFAD genes, the abundance of transcripts derived from Solyc01g006430 was ∼10-fold greater than transcripts from all other SlFAD genes combined (Supplemental Dataset S1). The protein encoded by Solyc01g006430 (also known as SlFAD2-1) is an ortholog of the endoplasmic reticulum-localized FAD2 enzyme in Arabidopsis, which catalyzes the conversion of 18:1 to 18:2 (Okuley et al., 1994; Lee et al., 2020). Consistent with the higher level of 18:2 in trichomes of the af mutant, SlFAD2-1 transcripts accumulated to modestly but significantly higher levels in the mutant compared to CHI4 and CHI9 control lines (Supplemental Figure S5; Supplemental Dataset S1). This insight into the effect of CHI1 deficiency on fatty acid content led us to update Solyc_tVI-GT_2021 (second-generation model, version 2) to incorporate the reactions relevant for synthesizing unsaturated fatty acids (the first-generation model contained only reactions to produce a generic saturated fatty acid), which enabled flux predictions at higher resolution and accuracy for this pathway (Supplemental Table S3).
Following these successful tests, we asked whether Solyc_tVI-GT_2021v2 could provide further insights into how the flavonoid and terpenoid pathways are coordinated in tVI-GTs. As mentioned above, the SPOT algorithm predicted an overall decreased flux through the flavonoid pathway in the af mutant relative to isogenic controls. We then asked how this might affect metabolite profiles. Our simulations indicated that all reactions of the flavonoid pathway in tVI-GTs of control lines carry flux (Supplemental Table S4) and, as previously established experimentally (Kang et al., 2014), rutin, kaempferol rhamnoside, quercetin trisaccharide, and 3-O-methylmyricetin would thus be expected to accumulate. The model also predicted that only the flavonoid pathway reactions upstream of CHI1, but not those downstream (see Figure 1), should carry flux in the af mutant (Supplemental Table S4). The loss of CHI1 activity in af therefore suggested that naringenin chalcone (an CHI1 substrate) would accumulate in tVI-GTs of the mutant. To test these predictions of the model, flavonoids were extracted from af mutant and CHI9 control plants and quantified by high performance liquid chromatography (HPLC)-MS. Notably, decreases of rutin (signature flavonoid of CHI9) and β-phellandrene (signature terpenoid of CHI9) in the af mutant (Figure 3, C and D) were accompanied by a large increase in the concentration of naringenin chalcone (Figure 4C), which agrees with model predictions.
Correlation of flavonoid and terpenoid accumulation across genotypes
To further investigate the relationship between flavonoid and terpenoid production, we employed anthocyanin-deficient mutants to test the hypothesis that bulk flavonoid deficiency correlates with low levels of terpenoids. Comparison of the af mutant and its isogenic control line (CHI9) revealed a strong positive correlation (Pearson correlation coefficient R = 0.992) between total flavonoid and terpenoid content based on leaf dip measurements (Figure 5A). Similar results (R = 0.994) were obtained for the anthocyanin reduced (are) mutant, which is impaired in the gene encoding flavonoid 3-hydroxylase (Figure 1; Maloney et al., 2014), and its wild-type parent VF36 (Figure 5B). Unlike the reduced density of tVI-GTs on af leaves (Rick et al., 1976; Kang et al., 2014), the flavonoid deficiency in leaves of the are mutant was not associated with altered trichome density (Supplemental Figure S6). Thus, the reduced terpenoid content in leaves of flavonoid-deficient mutants is likely attributed to factors other than trichome abundance.
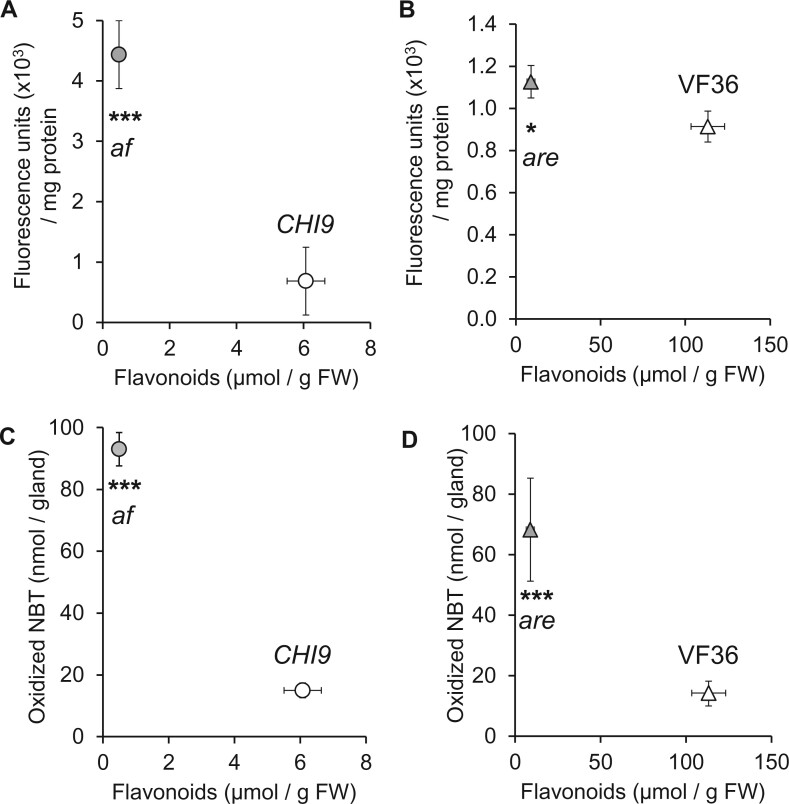
Figure 5.
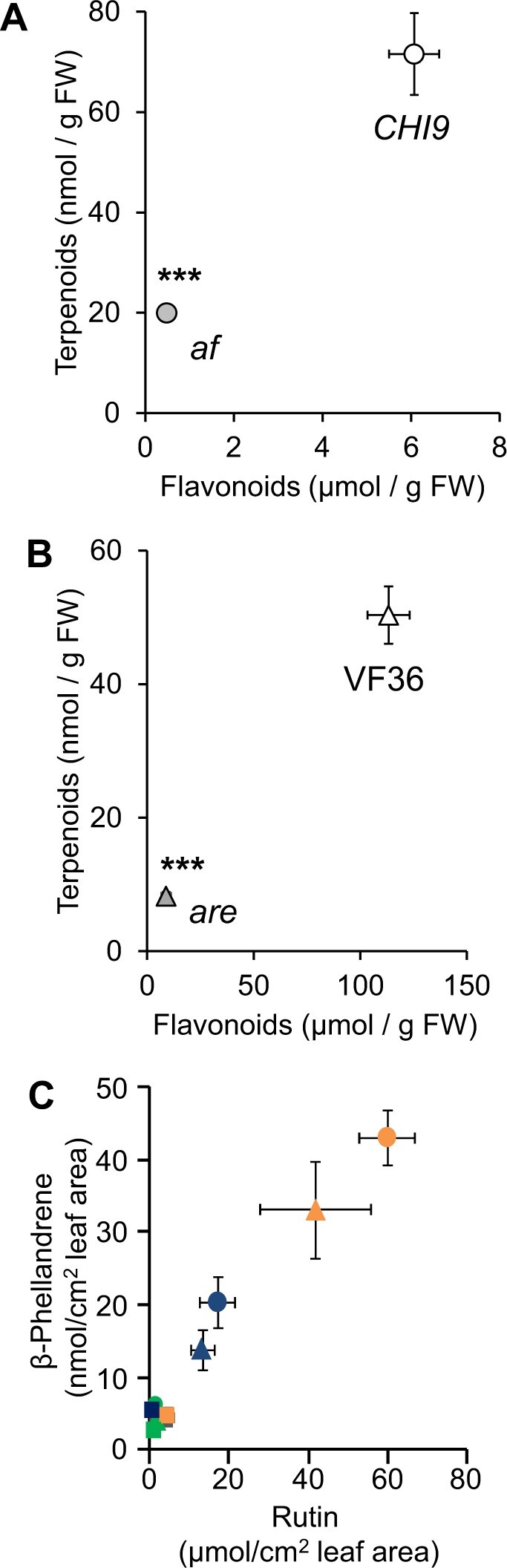
Loss of flavonoids in tomato af and are mutants correlates with decreased terpenoid accumulation. A, Flavonoid-to-terpenoid correlation in the af mutant (gray circle) and CHI9 control (open circle). B, Flavonoid-to-terpenoid association in the are mutant (gray triangle) and VF36 parent (open triangle). C, Flavonoid-to-terpenoid correlation under various light conditions. Data were obtained for rutin (most abundant flavonoid in tVI-GTs) and β-phellandrene (most abundant terpenoid in tVI-GTs) from CHI4 (circle), CHI9 (triangle), and af (square) under three light conditions (100 μE m−2 s−1, green symbols; 200 μE m−2 s−1, blue symbols; 500 μE m−2 s−1, orange symbols). For all graphs, data points represent the average of concentration measurements from leaf dips, with bars corresponding to standard error (if invisible, error bars are smaller than symbol) (n = 5). Where appropriate, statistical significance of pairwise comparisons based on an unpaired Student’s t test is indicated by asterisks (*P ≤ 0.01; **P ≤ 0.001; ***P ≤ 0.001).
The reactivity and broad biological activities of chalcones (Sahu et al., 2012) raised the possibility that elevated levels of naringenin chalcone in the af mutant (Figure 4C) is a causal factor in the downregulation of the terpenoid pathway. To test this hypothesis, we analyzed the flavonoid composition in the af and are mutants. The af mutant had 12-fold lower total flavonoid levels in tVI-GTs compared to the CHI9 controls, with a concomitant relative increase in naringenin chalcone (70% of flavonoids in af, 0.7% in the CHI9 control) (Figure 5A;Supplemental Figure S7). The flavonoid content in tVI-GTs of the are mutant was also about 12-fold lower than in the wild-type control (VF36), but none of the early pathway intermediates accumulated to high levels (Figure 5B;Supplemental Figure S7). These results suggest that the correlation between total flavonoid and total terpenoid amounts across genotypes is not dependent on the accumulation of a particular intermediate (e.g. naringenin chalcone) in flavonoid biosynthetic mutants but rather results from a deficiency in bulk flavonoid levels. We further tested this hypothesis by investigating the flavonoid-to-terpenoid correlation in leaves from af, CHI4, and CHI9 plants grown at various light intensities, which is known to affect the levels of these two classes of compounds (Jaakola and Hohtola, 2010; Banerjee and Sharkey, 2014). We observed that the content of rutin and β-phellandrene, which are the most abundant flavonoid and terpenoid derivatives, respectively, in tVI-GTs, correlated (R = 0.93) under various light intensities and across genotypes (Figure 5C).
Decreased flavonoid levels in mutants correlate with increased superoxide levels in tVI-GTs across genotypes
Based on the observation that loss of CHI1 has pleiotropic effects on metabolism in tVI-GTs, we further explored the potential global effects of flavonoid deficiency. Plant flavonoids are often implicated in protection against UV-B light and ROS, as well as sunlight irradiance outside the UV spectrum (Pietta, 2000; Jenkins, 2009; Agati and Tattini, 2010). Consistent with a role for flavonoids in protection against photooxidative stress in tVI-GTs, GO categories related to DNA repair and response to radiation were among the biological processes most significantly overrepresented in the set of genes expressed to higher levels in the af mutant relative to CHI1-complemented controls (Table 1;Supplemental Table S2). We therefore evaluated the hypothesis that the observed loss of flavonoids in the af mutant might compromise processes involved in cellular protection of tVI-GTs. First, we compiled a list of previously reported UV-B-responsive genes (Brown et al., 2005; Oravecz et al., 2006; Brown and Jenkins, 2008) and identified the putative tomato orthologs using the BLASTX search algorithm. Of 132 genes in this list, 36 (27%) were more than two-fold upregulated in tVI-GTs of the af mutant compared to CHI9 (Supplemental Table S5). The same approach was employed to generate a consensus list of ROS-responsive genes in tomato (Lai et al., 2012). Of 134 genes in this list, 47 transcripts (37%) were more than two-fold upregulated in tVI-GTs of the af mutant (Supplemental Table S6).
We next employed a fluorescence assay using the 2',7'-dichlorodihydrofluorescein diacetate indicator (Egan et al., 2007) to directly test the hypothesis that flavonoid deficiency in GTs is associated with increased production of ROS. The fluorescence signal from this reporter of active-oxygen species was 6.5-fold higher in isolated tVI-GTs of the af mutant compared to that observed in the CHI9 controls (P-value of 0.005; Supplemental Figure S8). ROS accumulation was 26% higher in tVI-GTs of the are mutant when compared to VF36 controls (P-value of 0.02). We also observed a negative correlation between flavonoid concentrations and ROS accumulation for both the af/CHI9 (Pearson correlation coefficient R = −0.66) and the are/VF36 comparisons (R = −0.49) (Figure 6, A and B). As an independent method for ROS evaluation, we used a staining assay that is based on the reaction of the superoxide anion radical () with nitroblue tetrazolium (NBT; Liu et al., 2009). The results of these experiments showed that tVI-GTs of the af mutant, when compared to those of CHI9 controls, contained 6.2-fold higher concentrations of as an indicator for ROS-related stress. We also found that concentrations in tVI-GTs of the are mutant were 4.8-fold higher compared to those in VF36 controls (Supplemental Figure S8). A strong negative correlation was observed for flavonoid concentrations and NBT-detectable ROS for both the af/CHI9 (R = −0.97) and the are/VF36 comparisons (R = −0.77) (Figure 6, C and D). Importantly, there was no significant difference in the NBT staining of whole leaves of these mutants and the corresponding wild-type controls (Supplemental Figure S8), suggesting that increased ROS stress is specific for tVI-GTs of tomato mutants impaired in flavonoid biosynthesis. We also considered the possibility that increased accumulation in flavonoid-deficient tVI-GTs reflects decreased expression of ROS-quenching enzymes such as superoxide dismutase (SOD) and catalase (Sies, 2017). Although genes encoding multiple SOD and catalase isoforms were expressed in tVI-GTs, we found no evidence for differential expression of these genes in comparisons between af and CHI-complemented controls (Supplemental Table S7). These data indicate that the flavonoid deficiency in tVI-GTs strongly correlates both with elevated abundance of a cytotoxic ROS species (i.e. ) and increased expression of stress-responsive genes.
Figure 6.
Loss of flavonoids in tomato af and are mutants correlates with increased levels of ROS. A, Flavonoid-to-ROS correlation (measured as fluorescent signal generated by oxidation of the H2DCFDA reagent) in the af mutant (gray circle) and CHI9 control (open circle). B, Flavonoid-to-ROS correlation in the are mutant (gray triangle) and VF36 parent (open triangle). C, Flavonoid-to- correlation (measured as oxidized NBT) in the af mutant (gray circle) and CHI9 control (open circle). D, Flavonoid-to-ROS correlation in the are mutant (gray triangle) and VF36 parent (open triangle). Data points represent the average, with bars corresponding to standard error (if invisible, error bars are smaller than symbol) (n = 5). Statistical significance of pairwise comparisons based on a Student’s t tests is indicated by asterisks (*P ≤ 0.01; **P ≤ 0.001; ***P ≤ 0.001).
Discussion
CHI1 is required for robust expression of terpenoid biosynthetic genes in tVI-GTs
In addition to lacking anthocyanin pigments, the af mutant of tomato has a lower density of tVI-GTs, reduced emission of leaf volatiles, and increased susceptibility to coleopteran herbivores (Rick et al., 1976). The pleiotropic nature of this mutant led to the suggestion that af may define a regulatory factor rather than a flavonoid biosynthetic component (De Jong et al., 2004). Subsequent map-based cloning studies, however, demonstrated that Af encodes the flavonoid pathway enzyme CHI1, and confirmed that loss of CHI1 is responsible not only for the anthocyanin defect but also the deficiency in GT-borne terpenoid volatiles (Kang et al., 2014). In this study, we tested various hypotheses to explain the unexpected dependence of terpenoid production in GTs on CHI1. Transcriptomic and proteomic datasets acquired with isolated tVI-GTs of the af mutant and appropriate isogenic control lines showed that genes involved in terpenoid biosynthesis were generally downregulated in the af mutant, as was the abundance of several of the corresponding pathway enzymes. This finding demonstrates that the absence of CHI1 in tVI-GTs has a major negative effect on the expression of terpenoid biosynthetic components and thus provides an explanation for the low levels of volatile terpenoids observed in the mutant (Rick et al., 1976; Kang et al., 2014). Our data further suggest that the terpenoid deficiency in af plants may be specific for tVI-GTs and does not extend to other isoprenoid derivatives such as chlorophyll. Our conclusion that CHI1 is required for full expression of terpenoid-related genes in tVI-GTs is consistent with the emerging view that plant flavonoids modulate a wide variety of cellular processes, including gene expression (Grotewold, 2006; Pourcel et al., 2013).
There is an increasing number of examples in which a protein serving a canonical function also performs a second, so-called moonlighting role (Jeffery, 2009). The finding that CHI1 is located both in the cytosol and in the nucleus of GT cells raised the possibility that the protein may have a moonlighting function related to the expression of flavonoid-related genes. However, our analysis of stable transgenic lines that express a functional CHI1 fusion protein (CHI1–GFP–GUS) that is excluded from the nucleus ruled out the hypothesis that a nuclear role of CHI1 is required for the accumulation of terpenoid volatiles in tVI-GTs. This finding supports the notion that terpenoid production depends on the canonical function of CHI1 as a cytosolic enzyme in the flavonoid pathway. Although nuclear localization of CHI has been reported for several plant species (Saslowsky et al., 2005; Dastmalchi and Dhaubhadel, 2015; this study), a potential role for CHI in this subcellular compartment, if any, remains unknown.
We found that the total number of genes upregulated in af GTs was more than twice that of downregulated genes. A similar proportion of differentially upregulated versus downregulated genes was reported in CHI-deficient seedlings of Arabidopsis (Pourcel et al., 2013). Among the transcripts that overaccumulated in tVI-GTs of the af mutant were those encoding the flavonoid biosynthetic enzymes (Figure 1). This finding suggests that compensatory processes are activated in af plants in response to the lack of flavonoids in tVI-GTs. Similar observations were reported for mutants of Arabidopsis, where the impaired expression of a flavonoid biosynthetic gene resulted in a higher abundance of other transcripts or proteins related to flavonoid biosynthesis (Pelletier et al., 1999; Pourcel et al., 2013). The coordinated upregulation of flavonoid-associated genes in af GTs may reflect the increased activity of stress-responsive factors that modulate transcription. An attractive candidate for such a factor is the UV-B receptor UVR8, which exhibited increased mRNA and protein levels in af GTs.
Model-based identification of altered flux distribution in the af mutant
To inform strategies for experimental testing of hypotheses, we developed a genome-scale metabolic reconstruction of tomato tVI-GTs (Solyc_tVI-GT_2021) based on published bioanalytical information about this cell type. Incorporation of the gene expression and protein abundance data sets generated is this study allowed us to assess flux distribution through pathways of primary and specialized metabolism in tVI-GTs. Using the SPOT algorithm (Kim et al., 2016), our model expectedly predicted, for both transcriptomics and proteomics data, particularly high fluxes through the TCA cycle, sugar breakdown, and processing, the Calvin–Benson cycle, amino acid metabolism, and the pentose phosphate pathway. However, while simulations incorporating transcriptome data predicted high fluxes through flavonoid and terpenoid biosynthesis, these fluxes were returned as zero when incorporating proteomic (rather than transcriptomic) datasets. This discrepancy appears to reflect the generally low abundance of proteins involved in specialized metabolism. Thus, successful integration of proteomics data into modeling efforts will be aided by deep protein coverage across all relevant pathways.
The development of testable hypotheses is an important benefit of using models for simulating flux distribution. In this study, we focused mainly on evaluating differences in flux distribution between tVI-GTs of the af mutant and those of isogenic controls. Our model correctly predicted reduced concentrations of total flavonoids and terpenoids in the af mutant, and also predicted increased accumulation of naringenin chalcone. Following up on this prediction, we expanded our analytical platform beyond the major flavonoid pathway end products and demonstrated that the naringenin chalcone intermediate indeed accumulated to high levels in tVI-GTs of the af mutant. This observation raised the possibility that naringenin chalcone might constitute a flavonoid signal with potential roles in crosstalk with the terpenoid pathway. However, a deficiency in volatile terpenoids was also observed in tVI-GTs of the are mutant that does not overaccumulate naringenin chalcone, which argues against this hypothesis (see below). Another example of an unexpected prediction of our model was that flux through fatty acid biosynthesis is elevated in the af mutant; subsequent bioanalytical assays confirmed that tVI-GTs of the af mutant contained higher concentrations of polyunsaturated fatty acids in comparison to trichomes isolated from control lines. The deep coverage of lipid-associated genes and proteins represented in our omics datasets provides an attractive starting point for further analysis of biochemical mechanisms that contribute to altered lipid homeostasis in tVI-GTs of flavonoid-deficient mutants. A high content in unsaturated fatty acids has been discussed as potentially playing a role in quenching ROS in GTs (Balcke et al., 2017). In accordance with this notion, we found that the elevated content of unsaturated fatty acids correlated with increased ROS stress in the af mutant.
These examples from our study illustrate the utility of mathematical modeling for hypothesis generation. Once such hypotheses have been tested experimentally, the model can be updated to more accurately reflect measured values, which in turn allows new predictions. This iterative process has the potential to generate nontrivial predictions that provide novel insights into how metabolism is regulated in GTs. In the future, it will be important to expand the scope of our GT-specific models by assessing the tissue-level context of specialized metabolism, including processes and cell types involved in CO2 fixation and the transport of nitrogen and carbon (e.g. sucrose) sources into tVI-GTs. Recent studies demonstrating the prevalence of sugar import over photosynthetic CO2 fixation in tomato tVI-GTs provide an important advance toward addressing this challenge (Balcke et al., 2017). On the computational side, approaches for multi-scale modeling of plant metabolism have been described (de Oliveira Dal’Molin et al., 2010; Grafahrend-Belau et al., 2013; Bogart and Myers, 2016). It will also be desirable to further study temporal changes during the development of tVI-GTs or under different growing conditions. The first kinetic metabolic models for GTs were developed to quantitatively capture the dynamics of terpenoid biosynthesis in mint (Mentha) GTs (Rios-Estepa et al., 2008, 2010). A model of flavonoid biosynthesis has also been generated for flavonoid biosynthesis in tomato tissues (Groenenboom et al., 2013), which could be adjusted to the specifics of tVI-GT metabolism. More broadly, these concepts may guide future efforts to assess how the interaction of different cell types and tissues affects the production of GT-borne compounds for anti-herbivore defense and other roles in plant resilience to environmental stress.
Flavonoid deficiency in tVI-GTs correlates with increased ROS levels
Our finding that attenuated production of volatile terpenoids in tVI-GTs of the af mutant extends to the tomato are mutant provides insight into how the flavonoid and terpenoid pathways may be coordinated in this unique cell type. First, unlike the accumulation of naringenin chalcone in the af mutant, the exceedingly low concentrations of this intermediate in are tVI-GTs argue against a role for this compound as a terpenoid-modulating signal. Second, the coordinate reduction in flavonoids and terpenoids in the are mutant appears to exclude a direct role for CHI1 in promoting terpenoid production. Rather, these findings support an alternative hypothesis in which terpenoid production in tVI-GTs depends on accumulation of flavonoid pathway end products. How might the flavonoid content of GTs promote terpenoid production in these cells? It is well established that various flavonoids, including flavonols such as quercetin and kaempferol that are deficient in the af and are mutants, have potent antioxidant and ROS-scavenging properties (Heim et al., 2002). Of particular relevance to our findings, flavonols are known to modulate cellular redox pathways in various tissues of tomato, including root hairs, stomata, and pollen tubes (Muhlemann et al., 2018; Gayomba et al., 2017). We suggest that increased ROS production arising from a genetic defect in flavonoid biosynthesis indirectly impairs the biosynthesis of terpenoids. The downregulation of multiple terpenoid biosynthetic genes in af trichomes suggests the involvement of stress-responsive changes in transcription factor activity, but additional studies are needed to test this hypothesis. In addition to transcriptional control, it is also possible that the iron–sulfur clusters of the MEP pathway enzymes 1-hydroxy-2-methyl-2-butenyl 4-phosphate synthase (HDS) and 1-hydroxy-2-methyl-2-butenyl 4-phosphate reductase (HDR; see Figure 1) predispose these enzymes to inactivation by oxidative stress (Banerjee and Sharkey, 2014). Given the many ways in which ROS modulate cellular processes in plant cells (Mittler, 2017), additional studies are needed to test the hypothesis that elevated ROS production is linked to reduced expression of terpenoid biosynthetic genes.
Our analysis of the af and are mutants showed that decreased terpenoid production is a major metabolic consequence of flavonoid deficiency in tVI-GTs. However, the unbiased transcriptome and proteome analysis performed with af GTs suggest that the metabolic and cellular effects of flavonoid deficiency are more pleiotropic. A particularly striking effect was the upregulation in af tVI-GTs of genes involved in DNA damage repair and responses to UV stress. This observation is consistent with a role for flavonoids in ROS detoxification and protection against photooxidative damage in a cell type that is exposed to high levels of solar irradiance. The detrimental effects of short-wavelength light on plant genome integrity (Hu et al., 2016) suggests that disruption of ROS homeostasis in af GTs results in DNA damage. A previous transcriptomic survey provided evidence that genes involved in several ROS-response pathways are expressed at high levels in tomato tVI-GTs (Balcke et al., 2017). Our own cell type-specific transcriptome data established that ROS-responsive genes were expressed at higher levels in tVI-GTs of the af mutant compared to control lines, which is consistent with the increased occurrence of superoxide anions in tVI-GTs of the af and are mutants. A negative correlation between flavonoid content and ROS production has also been described for tomato guard cells (Watkins et al., 2014, 2017).
Although our data support a model in which flavonoid deficiency in tVI-GTs attenuates terpenoid biosynthesis by a mechanism involving increased ROS accumulation, we cannot exclude the possibility that other cellular processes in tVI-GTs depend on flavonoid production. For example, a recent study showed that kaempferol, which is produced in tVI-GTs (Kang et al., 2010b), contributes to the biosynthesis of the essential respiratory cofactor ubiquinone (Soubeyrand et al., 2018). Evidence for the importance of mitochondrial respiration as an energy source to drive metabolism in tVI-GTs (Balcke et al., 2017; Johnson et al., 2017; this study) suggests that reduced pools of kaempferol may compromise specialized metabolism, which should be addressed in future studies. This and other examples of the diverse functions for flavonoids (Taylor and Grotewold, 2005; Gayomba et al., 2017) highlight the general importance of this class of specialized metabolites in governing cellular functions under changing environmental conditions. The experimentally amenable tVI-GT of tomato provides an attractive single cell-type system in which to study the role of flavonoids in photoprotection and ROS scavenging, and how these fundamental cellular processes govern specialized metabolism and plant resilience to environmental stress.
Materials and methods
Plant material and growth conditions
Seed for the tomato (S. lycopersicum) af mutant (LA1049) was obtained from the C.M. Rick Tomato Genetics Resource Center. Seed for the are mutant (LA1526) and its wild-type control (VF36) was generously provided by Dr. Gloria Muday. For comparisons to af, the previously described CHI4 and CHI9 complemented lines, which express the wild-type CHI1 gene under control of its native promoter (Kang et al., 2014), were used as isogenic wild-type controls. Plants were grown in Jiffy-7 peat pots (Hummert International, Earth City, MO, USA) and maintained in growth chambers under a 16-h light/8-h dark photoperiod, at 28°C, 60% humidity, and with a light intensity of 250 μE m−2 s−1. To analyze the effect of variable light intensity on metabolism in tVI-GTs, plants grown in the same growth chamber were placed at three discrete distances from the light source located at the top of the chamber: proximal to the light source, 500 μE m−2 s−1; distal to the light source, 100 μE m−2 s−1; and an intermediate distance to the light source, 200 μE m−2 s−1.
Stable expression of CHI1 fusion proteins
The effect of CHI1 localization on flavonoid and terpenoid biosynthesis in tVI-GTs was analyzed in stable transgenic lines of tomato expressing a CHI1–GFP or CHI1–GFP–GUS fusion protein. A PCR product containing the CHI1 cDNA under the control of its native promoter (pCHI1::CHI1) was amplified (Supplemental Table S8) from a complementation vector (Kang et al., 2014) and transferred to pK7FWG2 and pKGWFS7 binary vectors (Karimi et al., 2002). The resulting pCHI1::CHI1–GFP and pCHI1::CHI1–GFP–GUS constructs were introduced into Agrobacterium tumefaciens strain AGL-0 and used to transform af cotyledon explants as described previously (Li and Howe, 2001; Li et al., 2004; Gonzales-Vigil et al., 2011). Leaves from stably transformed plants grown in soil were harvested for mRNA, protein, and metabolite profiling (see below).
The localization of fusion proteins was assessed using an Olympus FluoView FV1000 confocal laser scanning microscope configured on an Olympus IX81 inverted microscope and a 20x UPLSAPO objective (NA 0.75) (Olympus Corporation, Tokyo, Japan). Fluorescence for eGPF was excited using a 488 nm Argon gas laser (10% of 10 mW laser) and detected through a 500–545 nm band pass emission filter using a photomultiplier tube for detection (high voltage = 820). Fluorescence for propidium iodide (PI) was excited using a 559 nm solid state laser (10% of 10 mW) and detected through a 570–620 nm band pass filter using a photomultiplier tube for detection (high voltage = 572). Fluorescence emissions for the eGFP and PI signals were collected sequentially to prevent detection of any emission crossover between the two detection channels.
Quantitation of anthocyanins, flavonoids, and terpenoid volatiles
Leaf anthocyanin levels were determined as previously described (Campos et al., 2016). Flavonoid quantitation and profiling were performed using the leaf-dip method (Kang et al., 2010b). Extracts were analyzed on a Xevo G2-XS QTOF-MS system (Waters, Broadway, NY, USA) following separation on an Ascentis Express C18 HPLC column (10 cm × 2.1 mm, 2.0 μm; Sigma-Aldrich, St Louis, MO, USA) as described by Kang et al. (2010b). Flavonoids were monitored at m/z 271.069 (naringenin chalcone), 609.153 (rutin), 463.153 (quercetin hexoside), 447.101 (quercetin rhamnoside), and 741.196 (quercetin dihexosyl rhamnoside). Peak areas were normalized to either the number of collected tVI-GTs or to tissue weight, and the intensity of the signal for the internal standard (propyl-4-hydroxybenzoate, monitored at m/z = 179.079). Analytes were quantified based on calibration curves obtained with authentic standards, with the exception of quercetin dihexosyl rhamnoside, for which a normalized peak area was calculated.
Terpenoids were extracted from the cell surface using a leaf-dip method (Kang et al., 2010b) and the resulting extracts were analyzed on a 5975B GC–MS system (Agilent Technologies, Santa Clara, CA, USA). Alternatively, metabolites were extracted from tVI-GTs that were manually hand-picked using a glass micropipette (Kang et al., 2010b). Separation of analytes was achieved by injection of the extract onto a VF-5MS column (50 m × 0.25 mm × 0.25 μm; Agilent Technologies, Santa Clara, CA, USA) with the following GC–MS settings: injector at 280°C, transfer line at 280°C, and helium carrier gas at 1 mL/min. The mass spectrometer operated in scan mode with 70 eV electron ionization. The initial oven temperature of 40°C was held for 2 min and then ramped in four steps: 40–90°C at 40°C/min, 90–110°C at 15°C/min, 110–250°C at 25°C/min, and 250–320°C at 40°C/min. The final temperature of 320°C was held for 2 min before returning to starting conditions. Peak areas were normalized to an internal standard (tetradecane) and either the number of collected tVI-GTs or the weight of the leaf disk. Analytes were quantified based on calibration curves obtained with authentic monoterpene and sesquiterpene standards. For commercially unavailable β-phellandrene and δ-elemene, α-phellandrene, and β-caryophyllene were used as surrogate standards, respectively.
Quantitation of chlorophylls and carotenoids
Two different extractions methods were performed using leaves of 4-week-old plants: (1) 10,000 tVI-GTs, hand-picked from leaves using a glass micropipette (Kang et al., 2010b) under a dissection microscope, were extracted with 200 μL MeOH and (2) leaf disks (5 mm diameter) obtained with a hole puncher were immediately submerged in 1 mL MeOH. Extracts were analyzed on a Xevo TQD QQQ-MS (Waters, Broadway, NY, USA) following separation on an Ascentis Express C18 HPLC column (5 cm × 2.1 mm, 2.7 μm; Sigma-Aldrich, St Louis, MO, USA). Analytes were separated at a flow rate of 0.3 mL min−1 using a linear gradient from 65% solvent A (acetonitrile:water = 3:2, v:v) and 35% solvent B (acetonitrile:isopropanol = 1:9, v:v) to 100% solvent B at 4 min, followed by 100% B for another 1 min. Chlorophyll a and b were quantified by monitoring the m/z 893→ 555 and m/z 907 → 569 transitions, respectively. Bulk chlorophyll and carotenoid levels in leaf samples were determined spectrophotometrically as previously described (Lichtenthaler and Wellburn, 1983).
Quantitation of sterols and fatty acids
Isolated tVI-GTs, obtained as described by McCaskill et al. (1992), were flash frozen in liquid nitrogen and then homogenized with mortar and pestle. The homogenate was transferred to a screw-cap glass vial containing 4 mL CHCl3/MeOH (2:1; v:v), with 100 mg/mL epi-cholesterol as internal standard, and analyte extraction was allowed to proceed at 75°C for 1 h. The supernatant was removed and evaporated to dryness (Genevac EZ-Bio, Ipswich, UK). The residue was resuspended in 2-mL potassium hydroxide in MeOH (6%; w:v) and sterol esters were saponified at 90°C for 1 h. After the solvent had cooled to 25°C, 2 mL hexane and 2 mL H2O were added and the mixture vortexed at the highest speed setting for 30 s (Vortex-Genie 2, Scientific Instruments, West Palm Beach, FL, USA). Extracts were centrifuged at 1,800g for 2 min to allow for phase separation. The upper organic phase was transferred to a 2 mL glass vial and evaporated to dryness. Immediately before analysis by GC–MS, N-methyl-N-(trimethylsilyl) trifluoroacetamide (50 mL) was added, and the tube vortexed as above for 20 s. The solution was transferred to a 100 mL glass insert and the mixture was incubated for 5 min at 25°C. The GC–MS analysis was performed according to Lange et al. (2015). Analyte quantitation was achieved based on calibration curves obtained with authentic, derivatized sterol standards.
Fatty acids were analyzed as described previously (Browse et al., 1986). Briefly, tVI-GT homogenate prepared as described above was suspended in 1.5 mL of 2.5% (v:v) H2SO4 in MeOH, containing 200 mg/mL heptadecanoic acid as internal standard. Following incubation at 80°C for 1 h, 1.5 mL water and 0.4 mL n-hexanes were added, the mixtures was vortexed at the highest speed setting for 30 s (Vortex-Genie 2, Scientific Instruments, West Palm Beach, FL, USA), and then centrifuged (2,500g, 5 min, 25°C). The upper organic phase was transferred to a 2 mL glass vial and a 1-µL aliquot analyzed by GC–MS. Calibration curves obtained with authentic methyl ester standards were used to quantify derivatized fatty acids.
Determination of nonextractable residues
tVI-GT homogenate prepared as described above (McCaskill et al., 1992) was lyophilized for 12 h (Lyp-Lock 12, Labconco). Following the addition of 90% aqueous MeOH (1.5 mL), the mixture was sonicated in a water bath at 25°C for 30 min and then centrifuged for 2 min at 2,500g. The solvent was decanted and soluble constituents were removed from the residue by successive extractions with 1.5 mL MeOH (twice), 1 M sodium chloride, 1% (w/v) SDS, water (twice), EtOH, CHCl3:MeOH (1:1; v/v), and t-butyl methyl ether. The remaining pellet containing nonextractable residue was allowed to dry in a fume hood for 2 h, before recording the weight. This value was used to calculate the molar concentration of cellulose as dominant carbohydrate sink.
Quantitation of polyphenol oxidase as amino acid sink
Homogenates from isolated tVI-GTs (McCaskill et al., 1992) were prepared as described above with the exception that a proteinase inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA) was included in the isolation buffer. The homogenate was extracted with a buffer containing 100 mM Tris–HCl (pH 6.8), 150 mM NaCl, 10% (v:v) glycerol, 4% (v:v) sodium dodecyl sulphate (SDS), 200-mM dithiothreitol (DTT), mixed briefly by vortexing, and sonicated in a water bath for three 30-s intervals (with 30 s cooling on ice between intervals). Following centrifugation at 14,000g for 20 min at 4°C, the supernatant was transferred to 2-mL Eppendorf tubes containing 300-mL water, 400-mL MeOH, and 100-mL CHCl3, and the mixture was vortexed for 5 min at high speed on a multi-tube vortexer (VWR Scientific, Radnor, PA, USA). Samples were then centrifuged at 14,000g for 5 min at 4°C. The lower organic phase was removed and discarded, and 400-mL MeOH was added to the remaining protein extract. Following mixing as above, samples were centrifuged at 14,000g for 5 min at 4°C. The resulting pellet was dried and protein content determined using the Bradford assay. Equal quantities of protein were then processed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE). The band for polyphenol oxidase at ∼70 kDa constituted, based on the relative intensity (MultiImage II, AlphaInnotech, San Leandro, CA, USA), 60% of the total protein in tVI-GTs, which allowed us to calculate its concentration in the extract. The known amino acid composition of polyphenol oxidase (Thipyapong and Steffens, 1997) was then used to calculate the quantity of each amino acid.
Global transcript profiling of type VI trichomes
tVI-GTs were isolated from leaves of 5-week-old plants as described previously (Schilmiller et al., 2010), using four independent sets of plants per genotype as experimental replicates. Total RNA was extracted from frozen trichomes using RNeasy Plant and RNase-free DNase kits (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. RNA quality was monitored with a BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Sequencing libraries were prepared from total RNA with the TruSeq RNA Library Prep Kit (Illumina, San Diego, CA, USA) and RNA-seq was performed using a HiSeq 2500 instrument (Illumina, San Diego, CA, USA) for paired-end 100-bp reads. Sequence reads were trimmed using the Trimmomatic software (Bolger et al., 2014) to eliminate adaptor sequences and low-quality reads. The remaining reads were mapped to the S. lycopersicum L. reference genome sequence (ITAG 2.5 genome) using Tophat2 software (Kim et al., 2013). This pipeline applied gene annotations based on ITAG 2.4 gene models provided from Sol Genomics Network (https://solgenomics.net). The mapped reads were then aligned using SAMtools (Li et al., 2009) and further analyzed by HTSeq (Anders et al., 2015). Genes expressed differentially between the af mutant and isogenic control lines were identified using three independent programs (EdgeR, DESeq2, and TCC). The threshold for differential expression was defined as log2FC > 1 or less than −1, with an adjusted P < 0.05. GO categories were assessed as described by Martel et al. (2015). Enrichment analysis of GO terms employed Fisher’s exact test (adjusted for false discovery rate) and was implemented using the Blast2GO program (Conesa and Götz, 2008).
RT-qPCR
Total RNA (100 ng; obtained from isolated tVI-GTs as by Schilmiller et al., 2010) was converted to first-strand cDNA using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Foster City, CA, USA) and diluted to 0.1 ng RNA equivalents per microliter. Amplification was monitored using a 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with the Power SYBR Green PCR master mix and gene-specific primers (listed in Supplemental Table S8). Quantitation was based on calibration curves constructed with a nucleic acid sample of known concentration. Expression level of the specific genes was calculated relative to an Actin gene (Solyc10g080500). Standard curves were constructed with serial dilutions of RNA from a CHI4 sample.
Quantitative proteomic analysis
tVI-GTs were isolated from leaves of 5-week-old af and CHI9 plants as described previously (Schilmiller et al., 2010). The trichome isolation was repeated three times using independent sets of plants as a source of biological replicates. Soluble proteins were extracted from frozen GTs using Buffer 1, which contained 100-mM Tris–HCl (pH 6.8), 150-mM NaCl, 10% (v:v) glycerol, 4% (v:v) SDS, 200-mM DTT, and the Complete Mini EDTA-free proteinase inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA). Proteins were then precipitated by adding four volumes of MeOH and CHCl3 as previously described (Schilmiller et al., 2010). Precipitated protein was resuspended in Buffer 2, which was identical to Buffer 1 with the exception that it contained 1% (w:v) SDS. Total protein was quantified using the Protein Assay kit (Bio-Rad, Hercules, CA, USA) based on calibration curves obtained with bovine serum albumin.
Protein samples were digested with trypsin using the Filter-Aided Sample Preparation protocol (Wiśniewski et al., 2009) and spin ultrafiltration units with a nominal molecular weight cutoff of 30,000 Da. The resulting peptides were de-salted using custom-made reversed-phase tips (Stop and Go Extraction) and dried by vacuum centrifugation. Samples were then processed with the TMTsixplex isobaric label reagent set (ThermoFisher Scientific, Rockford, IL, USA) according to manufacturer’s instructions. After labeling, all six samples were combined and dried by vacuum centrifugation. The sample was re-suspended in 0.3% formic acid, de-salted as above and dried again by vacuum centrifugation. The combined peptide samples were separated over a pH gradient (pH 3–10) into 12 fractions using an Agilent OffGel 3100 fractionator (Agilent Technologies, St Clara, CA, USA) according to manufacturer’s instructions. Each fraction was de-salted as above, dried by vacuum centrifugation, and frozen at −20°C until use.
Dried fractions were reconstituted in 50-µL 2% acetonitrile/0.1% trifluoroacetic acid, injected onto an Acclaim PepMap 100 C18 trapping column (0.1 mm × 20 mm, 5 µm, 100 Å; ThermoFisher Scientific), which was washed with Buffer A (0.1% formic acid) for 5 min on an EASY-nLC 1000 HPLC system (ThermoFisher Scientific). Bound peptides were then eluted onto an Acclaim PepMap RSLC C18 resolving column (0.075 mm × 150 mm, 2 µm, 100 Å; ThermoFisher Scientific) and separated with a gradient from 95% buffer A/5% buffer B (99.9% acetonitrile, 0.1% formic acid) to 70% Buffer A/30% Buffer B in 84 min, a short ramp to 100% B at 85 min, and a final hold at 100% B for 10 min.
Eluted peptides were sprayed into the ion source of a Q-Exactive Hybrid Quadrupole-Orbitrap MS (ThermoFisher Scientific, Waltham, MA, USA) and spectra acquired with 70,000 resolution at m/z 200. The top ten ions in each survey scan were subjected to automatic higher energy collision-induced dissociation with fragment spectra acquired at 35,000 resolution. Conversion of MS/MS (tandem MS) spectra to peak lists and quantitation of tandem mass tag (TMT) reporter ions was done using the Proteome Discoverer software (version 1.4.1.14). Peptide-to-spectrum matching was performed using the Sequest HT, Mascot and X!Tandem search algorithms against the ITAG 2.4 tomato protein sequence database appended with common laboratory contaminants (downloaded from www.solgenomics.org and www.thegpm.org, respectively). The output from all three search algorithms was then combined and analyzed using the Scaffold Q + S software (version 4.4.5) (Proteome Software, Portland, OR, USA) to probabilistically validate protein identifications and quantification. Assignments validated using the Scaffold 1% false discovery rate confidence filter were considered true.
Quantitation of ROS
Quantitation of total ROS levels in tomato tVI-GTs was performed according to Egan et al. (2007). Briefly, 100 mg isolated tVI-GTs, obtained as described by McCaskill et al. (1992), were homogenized in 1.0 mL of a 10 mM Tris–HCl buffer (adjusted to pH 7.2). The homogenate was then centrifuged at 12,000g for 20 min at 4°C. A 100-µL aliquot of the clear supernatant was incubated with 10 µL of 1 mM H2DCFDA reagent (Life Technologies, Oregon, USA) in the dark for 10 min. Fluorescence was then measured (excitation at 492, emission detected at 525 nm) in a spectrophotometer (BioTek Instruments Synergy H1, Vermont, USA). Protein concentration of the cleared supernatant was determined using a Bradford assay.
For the quantitation of ROS caused by accumulation of , leaf tissue (20 g fresh weight per plant) was removed and vacuum-infiltrated with 100 mM potassium phosphate buffer (pH 7.3) containing 0.3 mM NBT at 1.1 bar for 20 min. Leaflets were then imbibed in this buffer for an additional four h in the dark at 25°C. Control plants received the same treatment but with buffer lacking NBT. tVI-GTs were then isolated and homogenized as described by McCaskill et al. (1992). With minor modifications, a previously described protocol (Liu et al., 2009) was used to quantify NBT as an indicator of superoxide. GT homogenate was suspended in 0.5 mL 2 M KOH and sonicated in a water bath for 1 h at 25°C. CHCl3 (0.5 mL) was added, the mixture shaken at 4°C for 10 min, and phases were separated by centrifugation at 2,500g for 5 min. The organic layer was transferred to a new glass vial and extracted twice with CHCl3. The solvent was removed in vacuum (Genevac EZ-Bio, Ipswich, UK) from the combined organic phases. The dried residue was suspended in 0.8-mL dimethylsulfoxide and the absorbance measured at 680 nm and 730 nm for quantification of monoformazan and diformazan, respectively.
Metabolic reconstruction and flux balance analysis
The development of a genome-scale model of metabolism in tVI-GTs, and the subsequent modeling efforts to better understand flux distribution in these specialized anatomical structures, are described in detail in Supplemental Methods and Data File S1.
Statistical analysis
Transcriptomic and proteomic data sets were analyzed as described above. Metabolite correlation analyses were performed using the Dunnett’s and Tukey–Kramer methods with R software (version 3.3.0). Principal component and Pearson’s correlation coefficient analyses for all datasets were performed with the “prcomp” and “cor” programs within the R software environment.
Phylogenetic analysis
Amino acid sequences of Arabidopsis and tomato FADs were obtained from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and the Sol Genomics Network (SGN, https://solgenomics.net/) genomic databases. Phylogenetic trees were constructed using the Neighbor-Joining method with 1,000 replicates for bootstrap evaluation.
Accession numbers
RNA-seq data for analysis of gene expression in tVI-GTs of the af mutant and CHI1-complemented CHI4 and CHI19 lines were deposited in the NCBI Gene Expression Omnibus (GEO) as accession GSE118065.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Hierarchical clustering of samples from independent transcriptomic and proteomic experiments.
Supplemental Figure S2 . RT-q PCR analysis of expression levels for selected terpenoid biosynthetic genes in the MEP and MVA pathways.
Supplemental Figure S3 . GC–MS measurement of specific terpenoids in extracts obtained from hand-collected tVI-GTs of the af mutant and two isogenic control lines.
Supplemental Figure S4 . Phenotypic characterization of transgenic af plants expressing a CHI1–GFP–GUS fusion protein under the control of the native CHI1 promoter.
Supplemental Figure S5 . Transcript abundance of FAD genes in tVI-GTs.
Supplemental Figure S6 . Type VI trichome density on leaves of the tomato are mutant.
Supplemental Figure S7 . Flavonoid composition in tVI-GTs of various tomato genotypes.
Supplemental Figure S8 . Accumulation of ROS in various tomato genotypes.
Supplemental Table S1 . Number of DEGs in tVI-GTs of the af mutant.
Supplemental Table S2 . GO enrichment analysis of genes that are differentially regulated in tVI-GTs of the af mutant.
Supplemental Table S3 . Stoichiometric matrix for modeling metabolism in tVI-GTs and flux predictions using the EF-min and SPOT algorithms.
Supplemental Table S4 . SPOT flux predictions through reactions of flavonoid biosynthesis in the tomato af mutant and CHI9 control plants.
Supplemental Table S5 . Genes reported in the literature to respond to UV-B treatment and expression levels of putative tomato orthologs in tVI-GTs.
Supplemental Table S6 . Genes reported in the literature to respond to ROS and expression levels of putative tomato orthologs in tVI-GTs.
Supplemental Table S7 . Expression of SOD and catalase genes in tVI-GTs of the af mutant and control lines.
Supplemental Table S8 . Oligonucleotide primers used in this study.
Supplemental Methods and Data File S1. Detailed description of development of models used for metabolic reconstruction and flux balance analysis.
Supplemental Dataset S1 . Transcript abundance based on RNA-seq analysis of tVI-GTs of af mutant and CHI1-complemented control lines.
Supplemental Dataset S2 . Proteomics data acquired from tVI-GTs of af mutant and CHI9 control line.
Supplementary Material
Acknowledgments
We acknowledge Katerina Lay, Michael Das, Onyinye Nnamdi-Nwosu, and George Kapali for technical assistance during the course of this study. We thank Dr Gloria Muday (Wake Forest University) for providing seeds of the tomato are mutant, and the C.M. Rick Tomato Genetics Resource Center at the University of California, Davis, for providing other tomato seed stocks. We thank Jim Klug and Cody Keilen (MSU) as well as Julie Thayer, Sue Vogtman, and Devon Thrasher (WSU) for operating and maintaining plant growth facilities. We thank Curtis Wilkerson and Douglas Whitten in the MSU Research Technology Support Facility (RTSF) for valuable advice with proteomic analyses, as well as Dan Jones and Tony Schilmiller in the RTSF Mass Spectrometry and Metabolomics Core Facilities for their support with metabolite analyses. We are grateful to Melinda Frame for assistance with confocal microscopy and Guo-Qing Song in the MSU Plant Biotechnology Resource and Outreach Center for performing tomato transformations. We are grateful for access to equipment at the M.J. Murdock Metabolomics Laboratory and the Kamiak High-Performance Computing Cluster at Washington State University (WSU).
B.M.L. and G.A.H. conceived the project and supervised the research; K.S., J.J.Z., B.M.L., B.S.A., and G.A.H. designed the experiments; K.S. J.J.Z., and B.S.A. performed experiments; K.S., J.J.Z., B.S.A., B.M.L., and G.A.H. analyzed the data. K.S., J.J.Z., B.M.L., and G.A.H. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Gregg A. Howe (howeg@msu.edu).
Funding
This study was supported by the National Science Foundation grants to G.A.H. [award 1456864] and B.M.L. [award 1457127], and a Japan Society for the Promotion of Science Fellowship for Young Scientists to K.S. [award 24·841]. G.A.H. also acknowledges support from the Michigan AgBioResearch Project MICL02278 from Michigan State University. This publication was made possible in part by a predoctoral training award to B.S.A. from Grant T32-GM110523 from the National Institute of General Medical Sciences of the National Institutes of Health.
Conflict of interest statement. J.J.Z. and B.M.L. have financial interests in Dewey Scientific LLC. This company did not sponsor the current study and was not involved in the generation or interpretation of the data presented here.
References
- Agati G, Tattini M (2010) Multiple functional roles of flavonoids in photoprotection. New Phytol 186:786–793 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcke GU, Bennewitz S, Bergau N, Athmer B, Henning A, Majovsky P, Jiménez-Gómez JM, Hoehenwarter W, Tissier A (2017) Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 29:960–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Sharkey TD (2014) Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat Prod Rep 31:1043–1055 [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Diergaarde PJ, Ament K, Guerra J, Weidner M, Schütz S, de Both MTJ, Haring MA, Schuurink RC (2009) The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol 151:925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Mirabella R, Diergaarde PJ, VanDoorn A, Tissier A, Kant MR, Prins M, de Vos M, Haring MA, Schuurink RC (2012) Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proc Natl Acad Sci USA 109: 20124–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogart E, Myers CR (2016) Multiscale metabolic modeling of C4 plants: connecting nonlinear genome-scale models to leaf-scale metabolism in developing maize leaves. PLoS One 11:e0151722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102:18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Jenkins GI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146:576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the “16:3” plant Arabidopsis thaliana. Biochem J 235:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7:12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastmalchi M, Dhaubhadel S (2015) Soybean chalcone isomerase: evolution of the fold, and the differential expression and localization of the gene family. Planta 241:507–523 [DOI] [PubMed] [Google Scholar]
- De Jong WS, Eannetta NT, De Jong DM, Bodis M (2004) Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet 108:423–432 [DOI] [PubMed] [Google Scholar]
- Duke SO, Canel C, Rimando AM, Telle MR, Duke MV, Paul RN (2000) Current and potential exploitation of plant glandular trichome productivity. Advances in Botanical Research. Academic Press, Cambridge, MA, pp 121–151 [Google Scholar]
- Egan MJ, Wang ZY, Jones MA, Smirnoff N, Talbot NJ (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci USA 104:11772–11777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Gao Y, Xiong C, Yu G, Chang J, Yang Q, Yang C, Ye Z (2017) The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci 262:103–114 [DOI] [PubMed] [Google Scholar]
- Gayomba SR, Watkins JM, Muday GK (2017) Flavonols regulate plant growth and development through regulation of auxin transport and cellular redox status. In Yoshida K, ed, Recent Advances in Polyphenol Research, 1st edn, John Wiley & Sons, Ltd., Hoboken, NJ, pp 143–170 [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Hoehne M, Guenther M, Stitt M (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32:859–874 [DOI] [PubMed] [Google Scholar]
- Gonzales-Vigil E, Bianchetti CM, Phillips GN, Howe GA (2011) Adaptive evolution of threonine deaminase in plant defense against insect herbivores. Proc Natl Acad Sci USA 108:5897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafahrend-Belau E, Junker A, Eschenröder A, Müller J, Schreiber F, Junker BH (2013) Multiscale metabolic modeling: dynamic flux balance analysis on a whole-plant scale. Plant Physiol 163:637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenenboom M, Gomez-Roldan V, Stigter H, Astola L, van Daelen R, Beekwilder J, Bovy A, Hall R, Molenaar J (2013) The flavonoid pathway in tomato seedlings: transcript abundance and the modeling of metabolite dynamics. PLoS One 8:e68960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M (2012) Functional architecture of the nuclear pore complex. Annu Rev Biophys 41:557–584 [DOI] [PubMed] [Google Scholar]
- Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780 [DOI] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA (2018) Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44:72–81 [DOI] [PubMed] [Google Scholar]
- Heim KE, Tagliaferro AR, Bobilya DJ (2002) Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J Nutr Biochem 13:572–584 [DOI] [PubMed] [Google Scholar]
- Hemmerlin A, Harwood JL, Bach TJ (2012) A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog Lipid Res 51:95–148 [DOI] [PubMed] [Google Scholar]
- Hu Z, Cools T, De Veylder L (2016) Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol 67:439–462 [DOI] [PubMed] [Google Scholar]
- Isman MB, Duffey SS (1982) Toxicity of tomato phenolic compounds to the fruitworm, Heliothis zea. Entomol Exp Appl 31:370–376 [Google Scholar]
- Isman MB, Duffey SS (1983) Pharmacokinetics of chlorogenic acid and rutin in larvae of Heliothis zea. J Insect Physiol 29:295–300 [Google Scholar]
- Jaakola L, Hohtola A (2010) Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ 33:1239–1247 [DOI] [PubMed] [Google Scholar]
- Jeffery CJ (2009) Moonlighting proteins—an update. Mol Biosyst 5:345–350 [DOI] [PubMed] [Google Scholar]
- Jenkins GI (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Johnson SR, Lange I, Srividya N, Lange BM (2017) Bioenergetics of monoterpenoid essential oil biosynthesis in nonphotosynthetic glandular trichomes. Plant Physiol 175:681–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Campos ML, Zemelis-Durfee S, Al-Haddad JM, Jones AD, Telewski FW, Brandizzi F, Howe GA (2016) Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. J Exp Bot 67:5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Liu G, Shi F, Jones AD, Beaudry RM, Howe GA (2010a) The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol 154:262–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, McRoberts J, Shi F, Moreno JE, Jones AD, Howe GA (2014) The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol 164:1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Shi F, Jones AD, Marks MD, Howe GA (2010b) Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot 61:1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195 [DOI] [PubMed] [Google Scholar]
- Kennedy GG (2003) Tomato, pests, parasitoids, and predators: tritrophic interactions involving the genus Lycopersicon. Annu Rev Entomol 48:51–72 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Lane A, Kelley JJ, Lun DS (2016) E-Flux2 and SPOT: validated methods for inferring intracellular metabolic flux distributions from transcriptomic data. PLoS One 11:e0157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ (2013) Making new molecules—evolution of structures for novel metabolites in plants. Curr Opin Plant Biol 16:112–117 [DOI] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA 109: 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM (2015) The evolution of plant secretory structures and emergence of terpenoid chemical diversity. Annu Rev Plant Biol 66:139–159 [DOI] [PubMed] [Google Scholar]
- Lange BM, Rios-Estepa R (2014) Kinetic modeling of plant metabolism and its predictive power: peppermint essential oil biosynthesis as an example. Methods Mol Biol 1083:287–311 [DOI] [PubMed] [Google Scholar]
- Lange BM, Turner GW (2013) Terpenoid biosynthesis in trichomes—current status and future opportunities. Plant Biotechnol J 11:2–22 [DOI] [PubMed] [Google Scholar]
- Lange I, Poirier BC, Herron BK, Lange BM (2015) Comprehensive assessment of transcriptional regulation facilitates metabolic engineering of isoprenoid accumulation in Arabidopsis. Plant Physiol 169:1595–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Padilla CS, Gupta C, Galla A, Pereira A, Li J, Goggin FL (2020) The FATTY ACID DESATURASE2 family in tomato contributes to primary metabolism and stress responses. Plant Phyiol 182:1083–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. Arabidopsis Book 11:e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11:591–592 [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. ; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li Y, Deng H, Sun X, Wang A, Tang X, Gao Y, Zhang N, Wang L, Yang S, Liu Y, Wang S (2018) Tomato UV-B receptor SlUVR8 mediates plant acclimation to UV-B radiation and enhances fruit chloroplast development via regulating SlGLK2. Sci Rep 8: 6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Howe GA (2001) Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol Biol 46:409–419 [DOI] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16:126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Weisman D, Ye YB, Cui B, Huang YH, Colón-Carmona A, Wang ZH (2009) An oxidative stress response to polycyclic aromatic hydrocarbon exposure is rapid and complex in Arabidopsis thaliana. Plant Sci 176:375–382 [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney GS, DiNapoli KT, Muday GK (2014) The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol 166:614–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Zhurov V, Navarro M, Martinez M, Cazaux M, Auger P, Migeon A, Santamaria ME, Wybouw N, Diaz I, et al. (2015) Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Mol Plant Microbe Interact 28:343–361 [DOI] [PubMed] [Google Scholar]
- McCaskill D, Gershenzon J, Croteau R (1992) Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 187:445–454 [DOI] [PubMed] [Google Scholar]
- McDowell ET, Kapteyn J, Schmidt A, Li C, Kang JH, Descour A, Shi F, Larson M, Schilmiller A, An L, et al. (2011) Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol 155: 524–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2017) ROS are good. Trends Plant Sci 22:11–19 [DOI] [PubMed] [Google Scholar]
- Muhlemann JK, Younts TLB, Muday GK (2018) Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc Natl Acad Sci USA 115:E11188–E11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6:147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin CG, Quek L-E, Palfreyman RW, Brumbley SM, Nielsen LK (2010) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Ádám É, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18:1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier MK, Burbulis IE, Winkel-Shirley B (1999) Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol Biol 40:45–54 [DOI] [PubMed] [Google Scholar]
- Pietta PG (2000) Flavonoids as antioxidants. J Nat Prod 63:1035–1042 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Irani NG, Koo AJK, Bohorquez-Restrepo A, Howe GA, Grotewold E (2013) A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J 74:383–397 [DOI] [PubMed] [Google Scholar]
- Rick CM, Quiros CF, Harry Lange W, Allen Stevens M (1976) Monogenic control of resistance in the tomato to the tobacco flea beetle: probable repellance by foliage volatiles. Euphytica 25: 521–530 [Google Scholar]
- Rios-Estepa R, Lange I, Lee JM, Lange BM (2010) Mathematical modeling-guided evaluation of biochemical, developmental, environmental, and genotypic determinants of essential oil composition and yield in peppermint leaves. Plant Physiol 152:2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Estepa R, Turner GW, Lee JM, Croteau RB, Lange BM (2008) A systems biology approach identifies the biochemical mechanisms regulating monoterpenoid essential oil composition in peppermint. Proc Natl Acad Sci USA 105:2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu NK, Balbhadra SS, Choudhary J, Kohli DV (2012) Exploring pharmacological significance of chalcone scaffold: a review. Curr Med Chem 19:209–225 [DOI] [PubMed] [Google Scholar]
- Saslowsky DE, Warek U, Winkel BSJ (2005) Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 280:23735–23740 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL (2010) Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol 153:1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H (2017) Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol 11:613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HS, Reifman J, Wallqvist A (2014) Prediction of metabolic flux distribution from gene expression data based on the flux minimization principle. PLoS One 9:e112524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyrand E, Johnson TS, Latimer S, Block A, Kim J, Colquhoun TA, Butelli E, Martin C, Wilson MA, Basset GJ (2018) The peroxidative cleavage of kaempferol contributes to the biosynthesis of the benzenoid moiety of ubiquinone in plants. Plant Cell 30:2910–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Nishiyama T, Shimizu K, Kadota K (2013) TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattini M, Gravano E, Pinelli P, Mulinacci N, Romani A (2000) Flavonoids accumulate in leaves and glandular trichomes of Phillyrea latifolia exposed to excess solar radiation. New Phytol 148:69–77 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8:317–323 [DOI] [PubMed] [Google Scholar]
- Thipyapong P, Steffens JC (1997) Tomato polyphenol oxidase: differential response of the polyphenol oxidase F promoter to injuries and wound signals. Plant Physiol 115:409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A (2018) Plant secretory structures: more than just reaction bags. Curr Opin Biotechnol 49: 73–79 [DOI] [PubMed] [Google Scholar]
- Turner GW, Parrish AN, Zager JJ, Fischedick JT, Lange BM (2019) Assessment of flux through oleoresin biosynthesis in epithelial cells of loblolly pine resin ducts. J Exp Bot 70:217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wettstein-Knowles P (1968) Mutations affecting anthocyanin synthesis in the tomato: I. Genetics, histology, biochemistry. Hereditas 60:317–346 [Google Scholar]
- Watkins JM, Chapman JM, Muday GK (2017) Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol 175:1807–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JM, Hechler PJ, Muday GK (2014) Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol 164:1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362 [DOI] [PubMed] [Google Scholar]
- Yu H, Kowalski SP, Steffens JC (1992) Comparison of polyphenol oxidase expression in glandular trichomes of Solanum and Lycopersicon species. Plant Physiol 100:1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zager JJ, Lange BM (2018) Assessing flux distribution associated with metabolic specialization of glandular trichomes. Trends Plant Sci 23:638–647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.